An EAR-motif-containing AP2/ERF transcription factor IDS1 recruits a transcriptional repression complex to negatively regulate rice salt tolerance.
Abstract
Perception and transduction of salt stress signals are critical for plant survival, growth, and propagation. Thus, identification of components of the salt stress-signaling pathway is important for rice (Oryza sativa) molecular breeding of salt stress resistance. Here, we report the identification of an apetala2/ethylene response factor transcription factor INDETERMINATE SPIKELET1 (IDS1) and its roles in the regulation of rice salt tolerance. By genetic screening and phenotype analysis, we demonstrated that IDS1 conferred transcriptional repression activity and acted as a negative regulator of salt tolerance in rice. To identify potential downstream target genes regulated by IDS1, we conducted chromatin immunoprecipitation (ChIP) sequencing and ChIP-quantitative PCR assays and found that IDS1 may directly associate with the GCC-box-containing motifs in the promoter regions of abiotic stress-responsive genes, including LEA1 (LATE EMBRYOGENESIS ABUNDANT PROTEIN1) and SOS1 (SALT OVERLY SENSITIVE1), which are key genes regulating rice salt tolerance. IDS1 physically interacted with the transcriptional corepressor topless-related 1 and the histone deacetylase HDA1, contributing to the repression of LEA1 and SOS1 expression. Analyses of histone H3 acetylation status and RNA polymerase II occupation on the promoters of LEA1 and SOS1 further defined the molecular foundation of the transcriptional repression activity of IDS1. Our findings illustrate an epigenetic mechanism by which IDS1 modulates salt stress signaling as well as salt tolerance in rice.
Abiotic stresses such as soil salinity, drought, cold, and heat can influence plant growth and development. Plants have evolved several strategies to cope with salt stress, including the salt overly sensitive (SOS) pathway, transcriptional cascade, and responsive gene expression (Ji et al., 2013; Zhu, 2016; Yang and Guo, 2018). Rice (Oryza sativa) is the dominant food crop in Asia. A common factor affecting rice grain yield is soil salinity. Many genes conferring salt stress tolerance in rice have been isolated, such as genes involved in signal transduction and transcription regulation (Dubouzet et al., 2003; Hu et al., 2006; Huang et al., 2009), ion transporters (Ren et al., 2005; Senadheera et al., 2009), and metabolic pathways (Garg et al., 2002). Among these genes, APETALA37 (AP37), ZINC FINGER PROTEIN179 (ZFP179), and LATE EMBRYOGENESIS ABUNDANT PROTEIN1 (LEA1) are key regulators that positively modulate rice salt tolerance (Oh et al., 2009; Sun et al., 2010; Duan and Cai, 2012).
Transcriptional factors (TFs), which regulate gene expression, rely on a DNA-binding domain (DBD) that recognizes a specific DNA sequence (Mitsuda and Ohme-Takagi, 2009). In plants, several TFs regulating salinity tolerance in rice have been identified, such as APETALA2/ethylene response factors (AP2/ERFs), NAMATAF1/2 and CUC TFs (NACs), and zinc finger proteins (ZFPs; Hu et al., 2006; Liu et al., 2012; Schmidt et al., 2013; Shen et al., 2017). AP2/ERF TFs have a conserved function domain (AP2 domain) that recognizes and binds cis-elements, i.e. the GCC-box (AGCCGCC) and dehydration responsive element/C-repeat, RCCGCC element, in the promoter regions of target genes throughout the genome. This superfamily, containing more than 100 members in rice, consists of AP2/ERF proteins and regulates plant development, propagation, and stress response. For example, a rice AP2/ERF type TF, SERF1 (SALT-RESPONSIVE ERF1), responds to salt stress and participates in regulation of grain filling and gibberellin-meditated seedling establishment (Schmidt et al., 2014). Some AP2/ERF proteins, like GmRAV-03, JcERF011, and ERF109 are involved in the regulation of salt tolerance in plants (Bahieldin et al., 2016; Tang et al., 2016b; Zhao et al., 2017). Rice INDETERMINATE SPIKELET1 (IDS1), a typical AP2 type TF, has been reported to play roles in inflorescence architecture and the establishment of floral meristems (Lee and An, 2012).
TFs may act as transcriptional activators or repressors. Transcriptional repressors can further be classified as either active or passive repressors. Passive repressors are proteins that indirectly affect transcription, for example, by steric interference of the transcriptional activator. Active repressors, which normally possess a functional repression domain, confer repressive activity through recruiting the transcriptional repression components (Licausi et al., 2013). For repression domains, a typical motif calling the ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motif, which bears the consensus sequence pattern of either LxLxLx or DLNxxP, is one of the most predominant repressive motifs identified in plants (Kagale and Rozwadowski, 2011). In Arabidopsis (Arabidopsis thaliana), the EAR motifs recruit some transcriptional repression complex members, such as the corepressors TOPLESS/TOPLESS-LIKE PROTEIN (TPL/TPR) or the SIN3A ASSOCIATED PROTEIN18 (AtSAP18). TPL/TPR proteins are widely involved in the regulation of hormone signaling, such as auxin, jasmonic acid, abscisic acid (ABA), ethylene, and strigolactones, and affect plant embryogenesis, leaf development, morphogenesis, drought response, and ripening (Szemenyei et al., 2008; Pauwels et al., 2010; Jiang et al., 2013; Tao et al., 2013; Han et al., 2016; Tang et al., 2016a; Liu et al., 2017).
In this study, we focused on the identification of functional EAR-motif-containing TFs that may participate in the modulation of rice salt tolerance. We identified one AP2 type transcription factor, IDS1, as a key negative regulator of rice salt tolerance. According to our results, IDS1 bound to the promoter regions of LEA1 and SOS1, which are positive modulators of rice salt tolerance and repressed their expression. More importantly, we showed that IDS1 could physically interact with the corepressor TPR1 and the histone deacetylase HDA1 through its EAR motif and form an IDS1-TPR1-HDA1 transcriptional repression complex, contributing to the transcriptional repression activity of IDS1. These results revealed an epigenetic mechanism for TPL- and HDAC-dependent regulation of salt-responsive genes as well as rice salt tolerance under stress conditions.
RESULTS
Identification of IDS1 from Rice EAR-Containing TFs by Salt Tolerance Analysis
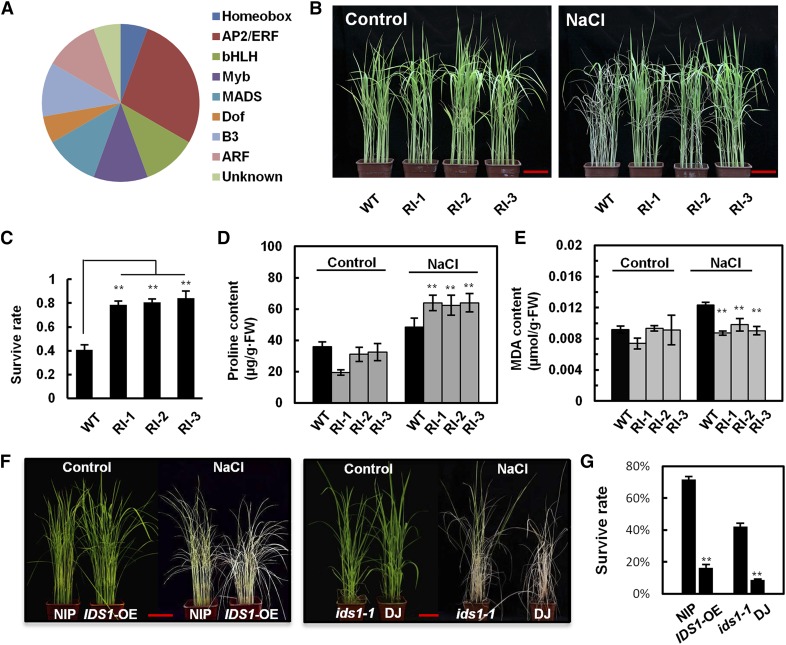
Many studies have confirmed the important role of the EAR motif in transcriptional regulation. Thus, in this study, we searched for EAR-motif-containing (DLNxxP or LxLxLx) TFs in rice to identify new TFs involved in salt tolerance regulation. Based on PatMatch1.2 (www.arabidopsis.org), 374 EAR motif TFs were identified from the database (Plant Transcription Factor Database v2.0; http://planttfdb.cbi.pku.edu.cn). These TFs belong to different TF families, including AP2/ERF, ARF, B3, BES1, C2H2, Dof, EIL, HD-ZIP, MYB, NAC, bHLH, and WRKY. Next, we conducted gene expression analysis using the rice database (http://www.ricearray.org/). Results showed that 57 TFs were either up- or down-regulated after salt treatment, indicating that these 57 TFs might be relevant to rice salt stress signaling. To experimentally confirm the potential function of these TFs in the regulation of salt resistance, we constructed artificial microRNA (amiRNA) interfering transgenic seedlings using rice ssp. japonica cv Nipponbare (NIP) as the background, in which the 57 TFs were separately knocked down at the transcriptional level. We then evaluated their survival rates and yield traits in high saline and alkali soil. In total, 20 of the TF RNAi seedlings showed substantially increased or decreased salt tolerance compared with the wild-type background material (Fig. 1A). Among these seedlings, IDS1 RNAi lines (RI-1, RI-2, RI-3) showed greatly enhanced salt tolerance (Fig. 1, B and C; Supplemental Figs. S1 and S2). Under salt stress, plants rely on ion transport and compartmentation, and synthesis and accumulation of osmotic agents for survival (Fedina et al., 2002). We measured the amounts of Pro and malondialdehyde (MDA) in IDS1 RNAi lines and found that after salt treatment, Pro accumulated while MDA decreased in these lines when compared with wild type. These results indicated better osmotic tolerance and membrane integrity after the knockdown of IDS1, supporting the hypothesis that IDS1 may weaken the rice tolerance to salt stress (Fig. 1, D and E).
Figure 1.
Identification of an EAR-motif-containing transcription factor IDS1 in rice using a salt tolerance assay. A, Pie chart showing different types of EAR-motif-containing TFs with increased or decreased salt tolerance in their amiRNA interference seedlings. B, Salt tolerance phenotype of IDS1-knockdown rice transgenic plants. The seedlings of wild-type (WT) and amiRNA-mediated IDS1-knockdown lines RI-1, RI-2, and RI-3 were separately treated with or without (control) 120 mm NaCl for 10 d (n ≥ 30). C, The statistical quantification of survival rates of wild-type NIP and the three independent IDS1 knockdown transgenic lines (n ≥ 30). D and E, Determination of the Pro and MDA contents in wild-type NIP and IDS1-knockdown transgenic lines with or without NaCl treatment. Error bars represent sds (n ≥ 30). F, Salt tolerance phenotypes of IDS1 overexpression (IDS1-OE), and ids1-1 mutant rice plants. G, Survival rates of wild-type NIP, IDS1-OE, and ids1-1 mutant seedlings as well as wild-type DJ seedlings treated with or without (control) 120 mm NaCl for 14 d (n ≥ 30). Error bars represent sds among three independent replicates. **P < 0.01 (Student’s t test). Scale bar, 5 cm.
IDS1 Negatively Regulates Salt Resistance
To further confirm our observations that IDS1 knockdown led to compromised salt tolerance, we used an IDS1 overexpression (IDS1-OE) transgenic rice line and a T-DNA insertion mutant line ids1-1 to test salt tolerance phenotypes. According to immunoblot and reverse transcription quantitative PCR (RT-qPCR) assays, IDS1 was highly accumulated in IDS1-OE plants (Supplemental Fig. S3), and the expression of IDS1 was blocked in the ids1-1 mutant (Supplemental Fig. S4). Under salt stress treatment, the survival rate of IDS1-OE was significantly lower than that in wild-type. In contrast, the ids1-1 mutant had a higher survival rate when compared to wild-type plants (Fig. 1, F and G). Meanwhile, the expression levels of abiotic stress-responsive genes were significantly increased in ids1-1 seedlings compared to wild-type (Supplemental Fig. S5). These results indicate that IDS1 plays a negative role in rice salt tolerance.
To examine the effects of abiotic stress on the transcriptional regulation of IDS1 expression, we first examined the expression level of IDS1 in rice seedlings after 120 mm NaCl treatment within 24 h using RT-qPCR. IDS1 expression was induced by NaCl 4 h posttreatment (Supplemental Fig. S6A). To analyze the spatiotemporal expression pattern of IDS1, we generated ProIDS1:GUS transgenic rice line. GUS staining was stronger in salt-treated seedlings compared to the control group (Supplemental Fig. S6B). These results suggested that IDS1 was transcriptionally induced by salt stress.
IDS1 Confers Transcriptional Repression Activity
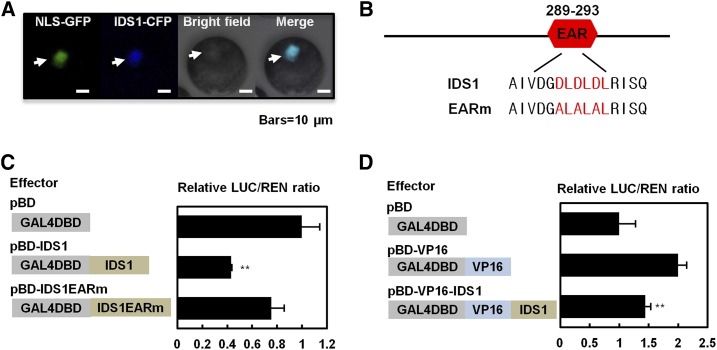
IDS1 is predicted to be an AP2/ERF type TF. Subcellular localization of IDS1 was detected using the IDS1-GFP-fused protein, which was transiently expressed in rice protoplasts. The fluorescent signal of IDS1-GFP colocalized with nuclear localization signal NLS-CFP, indicating that IDS1 is a nuclear protein (Fig. 2A). Since a canonical EAR motif (DLDLDL) is located in the C-terminal region of IDS1 (Fig. 2B), we predicted that IDS1 may possess a transcriptional repression activity. To confirm this prediction, we performed a transcriptional activity assay using an effector-reporter system by comparing luciferase (LUC) expression levels after the accumulation of GAL4 DNA binding domain (BD), BD-IDS1 fusion protein and BD-IDS1 with a mutated EAR motif (BD-IDS1EARm, ALALAL) (Fig. 2, B and C). In the assay, Renilla luciferase (REN) driven by the 35S promoter was used as an internal control. As shown in Figure 2C, luciferase activity was significantly reduced in BD-IDS1 compared with that in BD, suggesting that IDS1 may confer transcriptional repression activity. Nevertheless, the LUC activity in BD-IDS1m showed no obvious difference with that in the BD control (Fig. 2C), indicating that the transcriptional repression activity of IDS1 may largely depend on its functional EAR motif. To further confirm the repression activity of IDS1, the IDS1 protein was fused with the activator VP16 to generate VP16-IDS1. According to our results, IDS1 significantly suppressed the activation activity of VP16 since the VP16-IDS1-accumulated sample exhibited relatively lower luciferase activity when compared to the VP16-expressing positive control (Fig. 2D). These results suggested that IDS1 may function as a transcriptional repressor.
Figure 2.
IDS1 confers transcriptional repression activity. A, Subcellular localization of IDS1. Arrows mark the nuclei. NLS, nuclear localization sequence. Scale bars, 10 μm. B, Wild-type and loss-of-function mutated amino acid sequence of IDS1 EAR motif. C and D, Transient dual-luciferase expression assays illustrating the transcriptional repression activity of IDS1. The reporters luciferase (LUC) and renilla luciferase (REN) and the effectors (pBD, pBD-IDS1, pBD-VP16, and pBD-IDS1-VP16) were separately constructed as shown in the left columns of C and D. The activities of LUC and REN were determined 16 h after transformation, and the relative LUC/REN ratios represent the transcriptional activation activities. Error bars represent sds among three independent replicates. **P < 0.01 (Student’s t test).
IDS1 Binding Profiles in Rice Seedlings
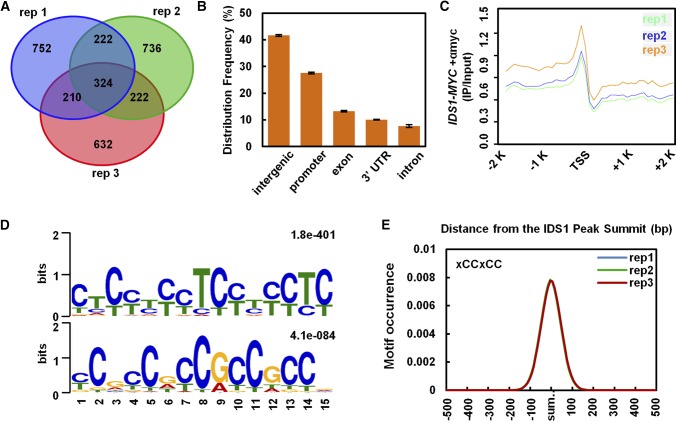
To further understand how IDS1 regulates plant salt tolerance, we comprehensively identified IDS1 target genes by investigating global binding profiles of IDS1. Chromatin immunoprecipitation sequencing (ChIP-seq) was performed using IDS1-OE transgenic seedlings with three technical replicates (rep1, rep2, rep3; Supplemental Fig. S7). Each two replicates shared a large number of target genes that covered more than 50% of one set (Fig. 3A; Supplemental Data Set 1), indicating high sequencing quality. Further genome distribution analysis revealed that the IDS1 binding sites were highly enriched in the promoter regions, where they accounted for 30% of all the peaks. This was much higher than in the gene body regions (Fig. 3B). To investigate the detailed IDS1 binding profile in the entire genome, the distance between each peak and its nearest gene was calculated. The line chart of these distances in the ±2-kb region around the transcription start site (TSS) confirmed that the IDS1 binding sites were strongly enriched in the promoter regions of target genes and reached one peak at about 250 bp upstream of TSSs (Fig. 3C). These distribution patterns indicated that IDS1 acted as a TF.
Figure 3.
IDS1 binding site analysis in the rice genome. A, Venn diagram depicting genes reproducibly associated with IDS1 binding peaks in the ChIP-seq analysis of three replicates. B, Relative distribution of IDS1 binding peaks in the rice genome. Error bars represent sds among three independent replicates. C, IDS1 binding peaks highly enriched in −2,000 to 0 bp of promoter regions in seedlings. D, MEME motif search identifying two dominant IDS1-binding motifs, defined as GCC-box or GCC-box-like (xCCxCC) motifs. E, Distribution of IDS1 binding motifs in the 200-bp regions surrounding the IDS1 binding peak summits.
To identify the IDS1 binding motifs, the ±100-bp flanking sequences around peak summits were submitted to Multiple EM for Motif Elicitation (MEME) to calculate the statistically predominant motifs de novo (Egea et al., 2018). Two similar motifs were found with conserved sequences TCCTCC and GCCGCC. Thus, they could be combined as one motif, xCCxCC, which is known as GCC- or GCC-like-box (Fig. 3D). Therefore, we further focused on the GCC- or GCC-like-box motifs for further analysis. Motif alignment with peaks database has been conducted with MAST (Dong et al., 2015). By calculating the occurrence plots of the GCC- and GCC-like-box within all peaks, the motifs were observed to be enriched precisely in the peak summits and to decrease to the background level in the flanking ±500-bp sites (Fig. 3E).
Genes Associated with IDS1 Binding Sites Are Enriched in the Abiotic Stress-Responsive Pathway
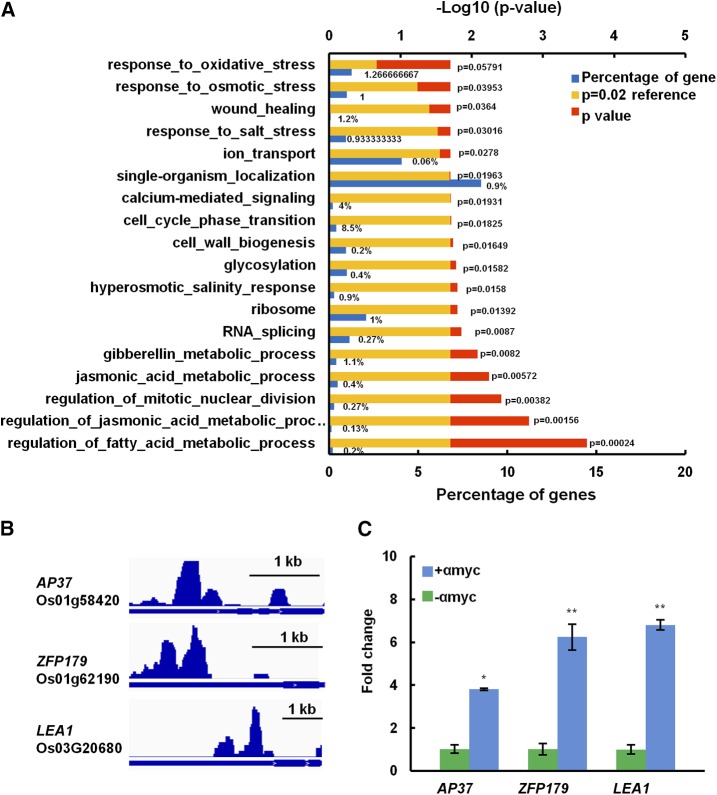
We then performed a gene ontology enrichment analysis to investigate in which biological processes IDS1-targeted downstream genes may be involved. IDS1 participated in diversified pathways that affect not only plant growth and development but also stress responses (Fig. 4A). Several important plant stress tolerance-related genes were identified (Supplemental Data Set 2). Among these genes, AP37, ZFP179, and LEA1 positively regulate rice salt tolerance (Oh et al., 2009; Sun et al., 2010; Duan and Cai, 2012). According to the ChIP-seq assay, the peak summits for IDS1 binding sites were located within 2 kb upstream of the AP37, ZFP179, and IDS1 TSS (Fig. 4B). ChIP-qPCR further confirmed that IDS1 could indeed associate with the promoter regions of these genes in vivo (Fig. 4C).
Figure 4.
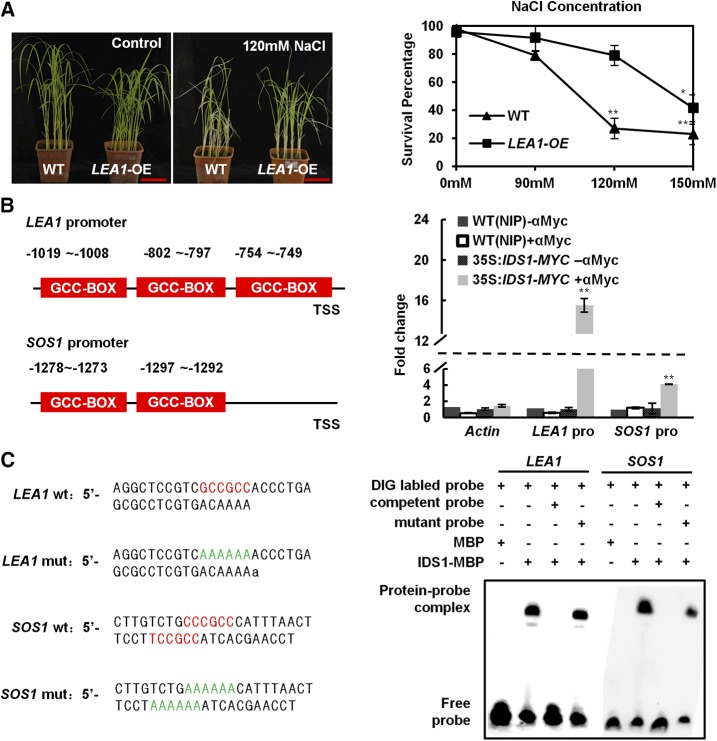
Genes associated with IDS1 binding sites. A, Enriched pathways of genes associated with IDS1 binding sites. B, IDS1 binding profiles in the promoter regions of abiotic stress-responsive genes. C, Validation of IDS1 binding sites in the promoter regions of genes in B by ChIP-qPCR analysis. The fold enrichment was normalized against the promoter of Actin. αmyc, antibodies against myc tag. Error bars represent sds for three independent replicates. *P < 0.05. **P < 0.01 (Student’s t test).
IDS1 Binds to LEA1 and SOS1 Promoters in Vivo and in Vitro
LEA1 and SOS1 play important roles in regulating rice and Arabidopsis salt tolerance, respectively (Duan and Cai, 2012). In this study, we also found that the LEA1-OE line exhibited a more energetic status and a significantly higher survival rate than the background material NIP after NaCl treatment, supporting the previously reported conclusion (Fig. 5A). Considering that the sequential GCC-boxes exist in the LEA1 and SOS1 promoters (approximately 800 bp and 1200 bp upstream from the LEA1 or SOS1 TSS, respectively), we hypothesized LEA1 and SOS1 as two potential downstream target genes of IDS1. First, we performed ChIP-qPCR to test our prediction. According to the ChIP-qPCR assay, IDS1 was indeed enriched on the promoter regions of LEA1 and SOS1 in vivo (Fig. 5B). Next, we conducted electrophoretic mobility shift assays (EMSAs) to test the direct association of IDS1 with LEA1 and SOS1 promoters in vitro. Results show that IDS1 could combine exclusively with the wild-type probes of LEA1 and SOS1 promoters (LEA1 wt and SOS1 wt), which contain the intact GCC-box, but not the probes with a mutated GCC-box (LEA1 mut and SOS1 mut, in which the GCC-boxes were replaced by AAAAAA), further proving that IDS1 could directly associate with LEA1 and SOS1 promoters (Fig. 5C). These results together confirmed that LEA1 and SOS1 were two downstream target genes that are directly regulated by IDS1.
Figure 5.
IDS1 directly binds to the LEA1 promoter. A, Improvement of salt tolerance in LEA1-OE compared to wild-type (WT) seedlings under NaCl stress. Left, salt treatment phenotype; right, the statistics of survival percentage. The seedlings of wild-type and LEA1-OE lines were separately treated with or without (control) NaCl for 10 d (n ≥ 30). Scale bar, 5 cm. Error bars represent sds among three independent replicates. *P < 0.05 and **P < 0.01 (Student's t test). B, ChIP-qPCR assay detected IDS1 enrichment level on the LEA1 and SOS1 promoters. Schemes illustrate the 2-kb promoters of LEA1 and SOS1 (left). The red boxes show the positions of GCC-box motifs. Validation of IDS1 direct binding sites in the LEA1 and SOS1 promoters by ChIP-qPCR analysis (right). Three biological replicates were performed. Error bars represent sds among three independent replicates. **P < 0.01 (Student’s t test). C Direct binding of IDS1 to the LEA1 and SOS1 promoters in EMSAs. The wild-type probes containing GCC-box motifs (red color) were derived from LEA1 and SOS1 promoters, while LEA1 mut and SOS1 mut show probes with mutated GCC-box motifs (AAAAAA, green color). Competition was performed with 125-fold wild-type or mutated cold probes. MBP protein was used as a negative control.
IDS1 Represses Expression of Target Genes at the Transcriptional Level
Since IDS1 has transcriptional repression activity, we hypothesized that the increased resistance to salt tolerance in the ids1-1 mutant may be due to the elevated expression levels of the IDS1-targeted salt-responsive genes. As shown in Figure 6A and Supplemental Figure S8, the expression levels of LEA1, SOS1, AP37, and ZFP179 were all significantly increased in the ids1-1 mutant compared to in the wild type. Consistently, the expression levels of LEA1 and SOS1 were downregulated in IDS1-OE plants (Fig. 6A). These results were further supported by the transcriptional activity assays in Nicotiana benthamiana. Our results showed that IDS1-GFP specifically repressed the expression of the LUC gene driven by native promoters of LEA1 and SOS1, but not by those with mutated GCC-boxes (Fig. 6, B and C). In summary, our data supported the conclusion that IDS1, as a transcriptional repressor, may negatively regulate the expression of its downstream target genes, such as LEA1 and SOS1.
Figure 6.
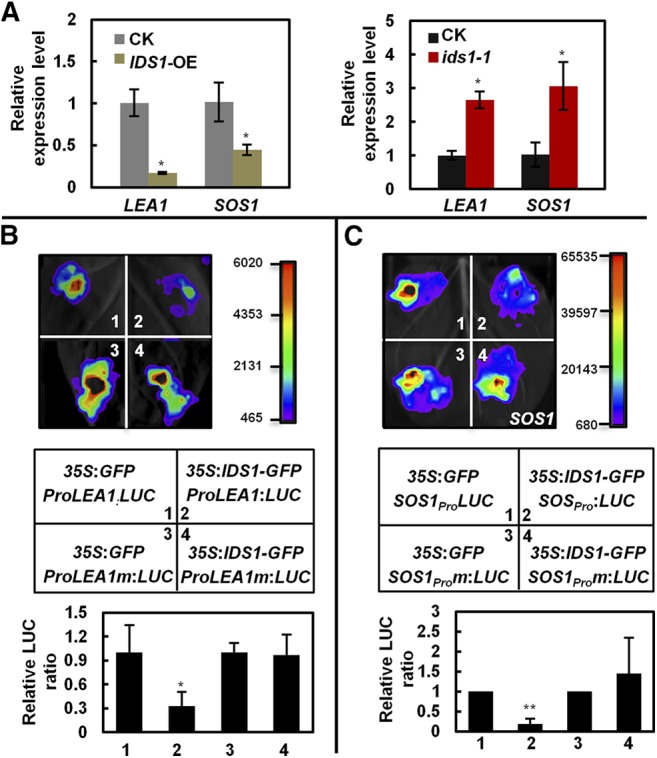
IDS1 negatively regulates the expression of LEA1 and SOS1 in rice. A, Transcription levels of LEA1 and SOS1 in IDS1-OE (left) and ids1-1 (right) rice materials. ACTIN was the internal control. Error bars represent sds among three independent replicates. B and C, Transient expression assays illustrating the transcriptional repression of LEA1 (B) and SOS1 (C) by IDS1. The 2-kb promoters of LEA1 and SOS1 containing the wild-type or mutated GCC-box motifs were separately used to drive the LUC gene expression and were coexpressed with 35S:GFP or 35S:IDS1-GFP in N. benthamiana leaves. LUC activities were determined 48 h postinfiltration. Quantification of the relative luminescence intensities (n = 4) is shown in the bottom of B and C. The mean values of each combination were normalized against controls. Error bars represent sds among three independent replicates. *P < 0.05 and **P < 0.01 (Student’s t test).
IDS1 Interacts with Transcriptional Corepressor TPR1
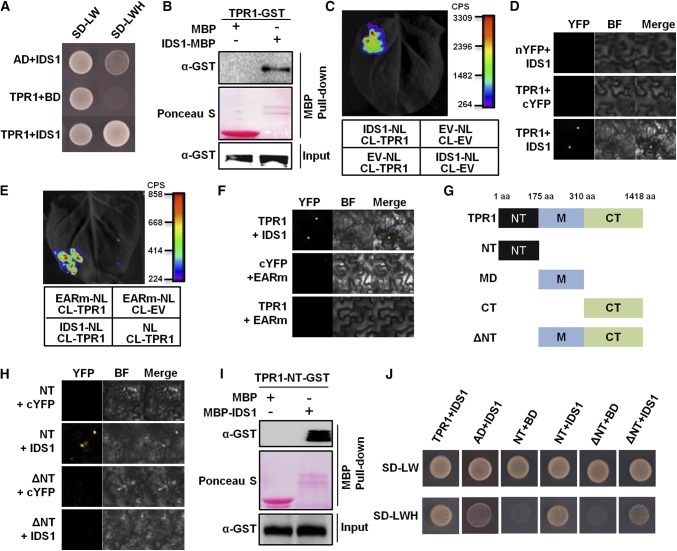
The EAR motif can recruit transcriptional corepressors such as TPL family proteins or AtSAP18 in Arabidopsis to form a transcriptional repression complex. To determine if IDS1 may interact with rice corepressor homologs, interactions between TPR1 and IDS1 and SAP18 and IDS1 were detected using several biological approaches. In yeast two-hybrid (Y2H), pull–down, and firefly LUC complementation imaging (LCI) assays, IDS1 notably interacted with TPR1 in vitro and in vivo (Fig. 7, A–C), but not with SAP18 (Supplemental Fig. S9). The bimolecular fluorescence complementation (BiFC) assay further showed that the interaction between IDS1 and TPR1 occurred specifically in the nucleus (Fig. 7D). In addition, according to the LCI and BiFC assays, mutation in the IDS1 EAR motif eliminated the interaction between IDS1 and TPR1, indicating that the IDS1-TPR1 association may depend on the IDS1 EAR motif (Fig. 7, E and F). To map the interacting domain of TPR1 with IDS1, we divided the TPR1 into four truncated parts, i.e. NT (N terminus, including the LiSH and C-terminal to lissencephaly homology domains), MD (middle domain), CT (C terminus, containing repeated WD40 domains), and the ΔNT (NT region deletion) (Fig. 7G). BiFC, pull-down, and Y2H assays all showed that NT of TPR1 mediated the physical interaction with IDS1 (Fig. 7, H–J). To experimentally confirm the corepressor function of TRP1,we coexpressed 35S:IDS1 and 35S:TPR1 in N. benthamiana with a LUC gene reporter driven by promoters of IDS1 targeting genes. As shown in Supplemental Figure S10, TPR1 enhanced the transcriptional repression activity of IDS1.
Figure 7.
IDS1 directly interacts with the transcriptional corepressor TPR1. A, Y2H assay showing the physical interaction between IDS1 and TPR1. SD-LW, synthetic dextrose medium lacking Leu and Trp; SD-LWH, SD medium lacking Leu, Trp, and His. B, Pull-down assay illustrating that IDS1 directly interacts with TPR1. C, Firefly LCI assay detecting the interaction between IDS1 and TPR1. Left, a representative leaf image; right, the colored scale bar indicates the luminescence intensity (CPS). NL, N terminus of LUC; CL, C terminus of LUC; EV, empty vector. D, BiFC assay revealing the interaction of IDS1 with TPR1. BF, bright field. E and F, LCI and BiFC assays showing that the EARm version of IDS1 failed to interact with TPR1. G, Schemes showing the full-length as well as truncated versions of TPR1 protein. NT, N-terminal domain; MD, middle domain; CT, C-terminal domain; ΔNT, deletion of the NT; aa, amino acids. H, BiFC assay showing that the NT of TPR1 is sufficient for the interaction with IDS1. I, Pull-down assay showing the interaction between TPR1-NT and IDS1. J, Y2H assay confirming the interaction between TPR1-NT and IDS1.
IDS1 Directly Interacts with Rice Histone Deacetylase HDA1
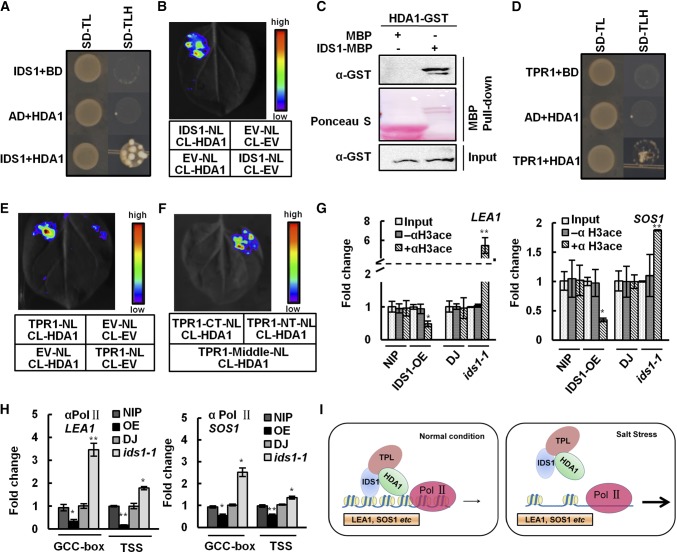
To identify new IDS1-interacting proteins besides TPR1 in rice, we screened a rice Y2H library using IDS1 as bait. HDA1, a histone deacetylase, was finally identified as a candidate. As shown in Figure 8A, AD-IDS1 and BD-HDA1 were coexpressed in yeast cells, and the transformed cells could grow on medium lacking Leu, His, and Trp, indicating the interaction between IDS1 and HDA1. Moreover, the LCI assay also showed that IDS1 could interact with HDA1 in planta (Fig. 8B). Similarly, the in vitro pull-down assay showed HDA-GST could exclusively interact with IDS1-maltose binding protein (MBP), but not MBP, further confirming the direct interaction between IDS1 and HDA (Fig. 8C). These results demonstrated that IDS1 could directly interact with HDA1 in vitro and in vivo.
Figure 8.
IDS1, TPR1, and histone deacetylase HDA1 physically interact with each other and lead chromatin remodeling in LEA1 and SOS1 promoter regions. A, Y2H assay showing the interaction of IDS1 and HDA1. B, LCI showing the interaction of IDS1 and HDA1. C, Pull-down assay confirming the interaction between IDS1 and HDA1. D, Y2H assay revealing the interaction between TPR1 and HDA1. E, LCI showing the interaction of TPR1 and HDA1. F, LCI showing that the N-terminal domain of TPR1 mediates its interaction with HDA1. TPR1-NT, TPR1-MD, and TPR1-CT represent truncated versions of TPR1 as shown above. ChIP assays by using the anti-acetyl-histone H3 (G) or Pol II antibodies (H) were performed to measure the histone acetylation levels (G) and Pol II assembling (H) at the promoter regions of LEA1 and SOS1 in the IDS1-OE, ids1-1 mutant, and wild-type (NIP and DJ) seedlings. The specific amplicons were described as above. I, A model for the IDS1 function in rice salt tolerance. Error bars (in G and H) represent sds among three independent replicates. *P < 0.05 and **P < 0.01 (Student’s t test).
TPR1 Associates with HDA1 to Form a Transcription Repression Complex
Several studies demonstrate that TPL/TPR proteins may interact with histone deacetylase family members and regulate embryogenesis, circadian transcription, and stress responses (Long et al., 2006; Szemenyei et al., 2008; Wang et al., 2013; Tang et al., 2016a). Thus, we detected the physical interaction between TPR1 and HDA1. As expected, the Y2H assay proved that TPR1 and HDA1 could indeed interact with each other in yeast cells (Fig. 8D), and this interaction was further confirmed by an LCI assay in N. benthamiana (Fig. 8E). To define the interaction domains of TPR1 and HDA1, the NT, MD, and CT of TPR1 were fused with the N-terminal part of LUC and separately coexpressed with HDA1, which was fused with the C-terminal part of LUC. The LCI results showed that the strongest interaction signal occurred between the TPR1-NT and CT-HDA1, demonstrating that the N-terminal part of TPR1 is not only responsible for the interaction with IDS1 but also mediated the association with HDA1 (Fig. 8F). Besides the physical interactions among IDS1, TPR1, and HDA1, the IDS1, TPR1, and HDA1 genes also showed similar expression patterns in rice roots, stems, and leaf tissues (Supplemental Fig. S10). These results indicate that the three members may function together and may form a transcriptional repression complex in vivo (Supplemental Fig. S11). Interestingly, the EARm version of IDS1 could still interact with HDA1, implying that the EAR motif is dispensable in the interaction between IDS1 and HDA1 (Supplemental Fig. S12).
IDS1 Mediates Transcriptional Repression by Chromatin Remodeling
Histone acetylation is a marker of gene transcription activation status. HDA1 plays an important role in controlling gene expression by changing histone acetylation levels. We tried to clarify whether this mechanism is involved in the transcriptional repression of IDS1-regulated target genes in vivo. To this end, histone acetylation levels in the IDS1-OE, ids1-1 mutant, and the wild type were checked using ChIP-qPCR with an antibody against acetylated histone H3. H3 acetylation levels on the promoter regions of LEA1 and SOS1 were increased in ids1-1 mutants and accordingly decreased in IDS1-OE seedlings compared to wild type (Fig. 8G). These results were consistent with the changes of expression levels of IDS1-targeting genes in the ids1-1 mutant and IDS1-OE. In NIP protoplasts, the transcription levels of LEA1 and SOS1 were increased upon exposure to the HDAC inhibitor trichostatin A (TSA) compared to the sample without TSA treatment, confirming that HDACs may potentially participate in the transcriptional repression of LEA1 and SOS1 (Supplemental Fig. S13). Previous studies have shown that nucleosomes with H3me and H3Ac modifications together with RNA polymerase II (Pol II) occupy the promoters of most protein-coding genes and represent a hallmark of transcription initiation and elongation (Guenther et al., 2007). Transcription initiation requires the assembly of the preinitiation complex, the mediator complex, and Pol II with an unphosphorylated CTD (C-terminal domain). However, active transcription requires highly phosphorylated CTD with Ser-2 and Ser-5 positions of conserved YSPTSPS heptapeptide repeats. The Ser-5 phosphorylation peaks emerge around the TSS (Baugh et al., 2009). Thus, a ChIP-qPCR assay, using anti-Pol II CTD YSPTSPS S5P antibody, was performed using ids1-1 mutants, IDS1-OE, and wild-type plants to detect the enrichment levels of active Pol II on the TSS of promoter regions of IDS1 target genes. The results showed that Pol II proteins significantly concentrate on the TSS of LEA1 and SOS1 in ids1-1 mutants (Fig. 8H). Together, the above results demonstrated that IDS1 may antagonize the histone acetylation and active Pol II occupation levels on the promoters of IDS1-targeting genes.
DISCUSSION
IDS1 Acts as a Negative Regulator of Rice Salt Tolerance
Understanding the salt response mechanism of rice is important for molecular improvement of rice salt tolerance. Many rice salt-responsive genes have been cloned and characterized (Hu et al., 2006; Liu et al., 2012; Jin et al., 2013; Schmidt et al., 2013; Toda et al., 2013; Shen et al., 2017). Several genes have been used as molecular markers in salt tolerance breeding, which can shorten breeding and selection. SKC1, a major QTL gene for salt tolerance, which is an HKT-type sodium transporter, was cloned by QTL mapping with a nearly isogenic line of salt-tolerant rice and a salt-susceptible rice variety (Ren et al., 2005). Hitomebore salt tolerant1), a B-type response regulator named OsRR22, was isolated by screening EMS mutagenesis lines of a local rice cultivar. The hst1 mutant was used to breed a salt-tolerant variety (Takagi et al., 2015). Our previous studies and these data show that ERF transcription factors play a role in rice abiotic stress responses. This suggests that there may be many ERF TFs participating in salt tolerance (Wang et al., 2012; Zhang et al., 2012, 2013; Xiao et al., 2016). We have used bioinformatics analysis combined with amiRNA library screening to identify several ERF TFs involved in rice salt tolerance. Among these, IDS1 was proven to be a negative regulator of the salt response.
The IDS1-TPR1-HDA1 Module Acts as Transcriptional Repressor Complex
Epigenetic mechanisms, including DNA/RNA methylation, histone modification, and noncoding RNA-mediated gene expression adjustment, are important mechanisms used by plants to cope with abiotic environmental stress (Kinoshita and Seki, 2014). Most of the methylated cytosine sites can persist in subsequent generations when plants are exposed to salt stress (Jiang et al., 2014). Changes of chromatin structure by histone modification can change target gene expression and help plants to adapt to and survive harsh environments. In Arabidopsis, HD2C and HDA6 regulate ABA and salt response by increasing the expression of ABA response genes ABI1 and ABI2 via histone deacetylation (Luo et al., 2012). In crops, the histone acetylation level of promoter regions of some cell cycle genes in maize (Zea mays) and peroxidize genes in beet (Beta vulgaris) can change under salt stress (Zhao et al., 2014; Yolcu et al., 2016). Detailed molecular mechanisms of histone modification linked with salinity response in rice require additional clarification. Our data showed that the IDS1-TPR1-HDA1 module regulated expression levels of the salt-responsive genes LEA1 and SOS1. The IDS1-mediated salt response recruited TPR1-HDA1 to inhibit target gene expression, and this can be released in rice under salt stress (Fig. 8I). Our results established a model in which IDS1 recruits TPR1 and HDA1 to target stress-specific genes participating in osmotic stress, ion transport, transcriptional cascade, and metabolic processes.
The transcription repression activity of IDS1 was endowed by its EAR motif. Much evidence indicates that the EAR motif is responsible for repression of target gene expression. In addition, EAR motif-containing TFs can activate the expression of other TFs. The function of EAR-containing TFs on target gene regulation is still unclear (Ohta et al., 2001; Kagale and Rozwadowski, 2011; Barah et al., 2016). For stress-related EAR-containing TFs, there are at least two additional functions in plant cells: suppression of the stress response genes under stress-free environments and activation of other target genes in biological processes such as the cell cycle, plant cell differentiation, and meristem development. The EAR domain of IDS1 in salt response interacts with the N terminus of TPR1 to recruit the histone deacetylation enzyme. The conserved amino acid residues, DLDLDL, are the interacting sites indicating that the epigenetic mechanism of transcription repression is conserved in plants.
The NT, containing the C terminus to lissencephaly homology domain, was considered to be a binding partner of TPL/TPRs (Szemenyei et al., 2008). Our results demonstrated that the NT of TRP1 interacts with the IDS1. The EAR motif usually mediates the interaction with corepressors like the TPL/TPR protein or the SAP18 in Arabidopsis. We showed that the EAR motif in IDS1 is indispensable for the interaction with TPR1, but not SAP18. In a transient expression system, TPR1 could facilitate IDS1 to repress the expression of LEA1 and SOS1.
IDS1 Maintains the Balance of Yield and Survival of Rice
IDS1 was initially identified as an AP2 type TF in maize, which determines spikelet meristem fate and controls the number of floral meristems (Chuck et al., 1998). In rice, IDS1 is involved in inflorescence architecture, affecting yield. The ids1 and snb ids1 (supernumerary bract) mutants show no obvious differences compared with the wild type in vegetative growth, flowering time, tiller number, and plant height. However, the two mutants have severe reproductive defects with more bracts, including rudimentary glumes, lemma, and palea (Lee and An, 2012). These data show that IDS1 controls yield and salt response and indicates that IDS1 may have multiple roles in balancing the energy consumption of rice to develop the best strategies in response to environmental changes.
A single gene can have several different pathways to control the development of different tissues. For example, WOX11, wuschel-related homeobox 11, regulates cell proliferation of the crown root meristem by recruiting the ADA2-GCN5 histone acetyltransferase module and also controls shoot development by recruiting histone H3K27me3 demethylase (Zhou et al., 2017; Cheng et al., 2018). While IDS1 suppresses the salt response genes by recruiting TPR1 and HDA1, IDS1 activates many B, C, and E function genes such as OsMADS2, OsMADS4, OsMADS16, OsMADS3, OsMADS1, OsMADS5, OsMADS7, and OsMADS8, in panicle formation. These data indicate that IDS1 plays several important roles, involving distinct molecular mechanisms, in response to environmental stimuli and developmental signals.
CONCLUSION
This study focused on functions of IDS1 in the regulation of rice salt tolerance. IDS1 bound to the GCC-box motif of the rice genome at a global level, including GCC-boxes in the promoter regions of LEA1, AP37, ZFP179, and SOS1. IDS1 repressed expression of target genes at the transcriptional level. We demonstrated that IDS1 interacted with the transcriptional repression complex in vitro as well as in vivo and caused chromatin remodeling in the promoter regions of target genes. We propose that IDS1 modulates target gene expression at the transcriptional level to regulate rice plant salt tolerance. Additional research will be needed to determine the molecular factors that regulate IDS1 expression under salt stress.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
To construct the IDS1 amiRNA interference vector, 21-nt cDNA of IDS1 was fused with the pNW55 stem-loop structure and cloned into binary pCAMBIA5300. The construct was introduced into rice (Oryza sativa ssp. japonica cv Nipponbare [NIP]) by Agrobacterium tumefaciens-mediated transformation. For the overexpression construct, the full-length cDNA of IDS1 was cloned into the pENTR/D-TOPO vector (Invitrogen) and subcloned into the expression vector with a myc tag. The construct was transformed into NIP. The loss-of-function mutant ids1-1 was ordered from the Rice T-DNA Insertion Sequence Database rice mutant library (http://cbi.khu.ac.kr/RISD_DB.html).
The rice plants were grown in the greenhouse with a 12-h-light/12-h-dark cycle at 25°C to 30°C. Nicotiana benthamiana plants were grown in a greenhouse at 24°C with a 16-h-light/8-h-dark cycle.
Salt Treatment
The salt tolerance of the amiRNA interference plants, overexpression plants, and T-DNA insertion mutants was compared to that of the wild type. Plants of different genotypes were germinated in petri dishes and transplanted in soil. At the four-leaf stage, salt stress treatments were conducted using different concentrations of NaCl for several days until leaf curvature was observed. Plants were allowed to recover for 10 d and then the survival rates were calculated. Plants with green leaves and regenerating shoots were considered survivors. Three replicates were performed for each experiment.
RNA Extraction and Gene Expression Analyses
Total RNA was extracted from the rice materials using Trizol (Invitrogen) reagent. About 2 μg of total RNA was used for further reverse transcription using M-MLV reverse transcriptase (Promega). For determination of gene expression, SYBR Premix Ex Taq (Perfect Real Time; TaKaRa) was used for the RT-qPCR assays. Expression levels of target genes were normalized to the internal control gene Actin1 or Ubiquitin. Each assay was repeated three times independently, and the statistical significance was evaluated by Student’s t test (significance, P < 0.05). Nucleotide sequences of the primers used in RT-qPCR assays are listed in Supplemental Data Set 3. The RT-qPCR primers for abiotic stress responsive genes followed the information in Schmidt et al. (2013).
ChIP Assays
NIP and its transgenic lines generated from the transformation of ProUbi:IDS1-myc, wild-type DJ (Dongjin), and the ids1-1 mutant were used for the ChIP assays (Lu et al., 2013). A total of 4 g of 4-week-old seedlings were harvested. Plant materials were cross linked with 1% (v/v) formaldehyde under vacuum for 10 min. The samples were subjected to nucleus isolation and sonication and then incubated with different antibodies, including anti-c-myc antibodies (Roche; 11667149001), anti-acetyl-histone H3 (Millipore; 06-599) and anti-RNA polymerase II CTD repeat YSPTSPS (phospho S5) (Abcam; ab5408), with protein G magnetic beads (Invitrogen; 10003D) together. The DNA was then precipitated and stored at −80°C.
For ChIP-seq, isolated DNA samples were fragmented and amplified for 14 cycles. Then DNA fragments of 100 to 500 bp were purified for construction of a sequencing library.
Analysis of ChIP-Seq Data
After quality control analysis, the clean reads of ChIP-seq data were mapped to Oryza_sativa.IRGSP-1.0 with Bowtie2 (Langmead and Salzberg, 2012). Then the uniquely mapped reads were used for peak identification, and the peaks were searched with MACS (Zhang et al., 2008).
Motif Search and Classification
For all peaks present in three replicates, 200 bp around the peak summits (upstream 100 bp and downstream 100 bp) were subjected to MEME (http://meme-siute.org/) to calculate motifs.
ChIP-qPCR
The prepared DNA was applied for qPCR using low rox EVER Green PCR Master Mix (Abm) with ABI QuantStudio 7 real-time PCR detection system. The enrichment levels were normalized to the input sample and fold enrichment was calculated against the Actin promoter. The negative control had no antibodies added. Primers are listed in Supplemental Data Set 3.
Luciferase Transient Expression Assays
To test the transcriptional repression activity of IDS1 in N. benthamiana protoplasts, a dual-luciferase reporter assay was conducted. For the effectors generation, IDS1 was fused with the GAL4 DBD to generate pBD-IDS1. IDS1 containing a transactivating domain VP16 in its N terminus was also fused with the GAL4 DBD to generate a pBD-VP16-IDS1 effector. The GAL4 DBD with or without VP16 (pBD and pBD-VP16) was used as the positive and the negative controls, respectively. The activities of LUC and REN were determined using the Dual-Luciferase Reporter Assay System (Promega, E1910), and the relative LUC/REN ratio was calculated. For each replication, three independent transformations were conducted.
To evaluate the transcriptional repression activity of IDS1, the 2-kb LEA1 and SOS1 promoters were first fused with LUC to generate the reporter constructs ProLEA1:LUC or ProSOS1:LUC. For generation of the effectors, the IDS1-GFP coding sequence was cloned into the expression vector. The coding sequence of GFP was also cloned into the expression vector as the negative control. All the reporter and effector constructs were separately introduced into A. tumefaciens strain GV3101. A. tumefaciens harboring the indicated reporter or effector constructs were coinfiltrated into N. benthamiana leaves, and the LUC signals were detected and quantified 48 h postinfiltration using Night-SHADE LB 985 (Berthold).
EMSAs
The MBP tagged IDS1 protein was expressed in Escherichia coli strain Transetta-DE3 (Transgen Biotech, CD801) and purified using the amylose resin (New England Biolabs; E8021V) following manufacturer’s instructions. The LEA1 promoter probes containing the GCC-box cis-elements were synthesized and labeled with digoxigenin-11-ddUTP at the 3′ end by using the DIG gel shift kit (Roche; 03353591910). EMSAs were performed as described by Liu et al. (2017). The MBP protein alone was used as the negative control. Each experiment was independently replicated three times.
Pull-Down Assays
The indicated proteins were fused with MBP or GST tags and expressed in the E. coli strain Transetta-DE3 (Transgen Biotech; CD801). The supernatant containing MBP- and GST-tagged soluble proteins was mixed and incubated with 20 μL amylose resin beads (New England Biolabs; E8021V) for 2 h. The beads were then harvested and washed four times with Tris buffer. The eluted samples were analyzed by immunoblots using the indicated antibodies.
LCI and BiFC Assays
The LCI and BiFC assays for protein interaction detection were performed in N. benthamiana leaves using A. tumefaciens-mediated transient expression. For the LCI assay, the indicated genes were separately fused with the N- or C-terminal parts of the reporter gene LUC and then coinfiltrated into N. benthamiana leaves. LUC activities were analyzed 48 h postinfiltration using Night-SHADE LB 985 (Berthold). For the BiFC assay, the indicated genes were fused with the N- or C-terminal parts of the yellow fluorescent protein and coexpressed in N. benthamiana leaves. After 48 h postinfiltration, the yellow fluorescent protein fluorescence signals were imaged with a confocal microscope (Carl Zeiss; LSM880).
Y2H Assays
The coding sequences of indicated genes were first cloned into the pGADT7 (AD) and pGBKT7 (BD) plasmids to generate the GAL4-AD and GAL4-BD derivatives. Next, the GAL4-AD and GAL4-BD derivatives were cotransformed into the yeast (Saccharomyces cerevisiae) strain AH109 and grown on synthetic dextrose medium lacking Leu and Trp (SD-l/W). Then, the yeast cells were screened on synthetic dextrose selection medium lacking Leu, Trp, and His (SD-l/W/H). A total of three independent replications were performed.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: IDS1, LOC_Os03g60430; TPR1, LOC_Os03g14980; HDA1, LOC_Os06g38470; LEA1, LOC_Os06g06760; SOS1, LOC_Os12g44360; SAP18, LOC_Os02g02960; ACTIN, LOC_Os03g50890; DREB2A, LOC_Os01g07120; LEA, LOC_Os01g50910; ABI5, LOC_Os01g64000; SNAC2, LOC_Os01g66120; bZIP23, LOC_Os02g52780; SNAC1, LOC_Os03g60080; LEA3, LOC_Os05g46480; SALT-RESPONSIVE ERF1, LOC_Os05g34730; ZFP182, LOC_Os03g60560; NAC5, LOC_Os11g08210; AP37, LOC_Os01g58420; dehydrin, LOC_Os11g26760; ZFP179, LOC_ Os01g62190; ZFP252, LOC_Os12g39400. All the ChIP-seq data sets have been deposited in ArrayExpress under accession number E-MTAB-6504. The ChIP-seq raw data files were deposited in the SRA under accession number SRP130178.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence analysis of IDS1.
Supplemental Figure S2. Determination of IDS1 expression levels in IDS1-knockdown transgenic lines.
Supplemental Figure S3. Overexpression of IDS1.
Supplemental Figure S4. T-DNA insertion mutant of IDS1.
Supplemental Figure S5. Expression levels analysis of representative abiotic stress responsive genes in ids1-1 transgenic plants by RT-qPCR.
Supplemental Figure S6. IDS1 expression responses to salt treatment in rice.
Supplemental Figure S7. Overview of ChIP assays and ChIP-seq data.
Supplemental Figure S8. Expression level analysis of IDS1-targeted genes in ids1-1 with RT-qPCR.
Supplemental Figure S9. IDS1 does not interact with SAP18.
Supplemental Figure S10. IDS1, TPR1, and HDA1 show similar expression patterns.
Supplemental Figure S11. EARm version of IDS1 could interact with HDA1.
Supplemental Figure S12. TPR1 could facilitate IDS1 to repress the expression of LEA1 and SOS1 in the N. benthamiana transient expression system.
Supplemental Figure S13. Determination of the transcription levels of LEA1 and SOS1 in NIP protoplasts with or without TSA treatment.
Supplemental Table S1. Overview of ChIP-seq results.
Supplemental Data Set 1. ChIP-seq results of NIP-IDS1 in seedlings.
Supplemental Data Set 2. Representative salt stress response genes associated with IDS1 binding sites.
Supplemental Data Set 3. Primers used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Prof. Lizhong Xiong (Huazhong Agricultural University) for the gift of the HDA1 entry vector, Prof. Shouyi Chen (Institute of Genetics and Developmental Biology, CAS) for providing vectors for the transcriptional regulation activity assay in protoplasts, and Prof. Renee Sung (University of California, Berkeley) for critical reading of the manuscript.
Footnotes
This work was supported by National Key Special Programs for Breeding from the Ministry of Science and Technology of China (2016YFD0100403), the National Science Foundation of China (31671610), the China Postdoctoral Science Foundation (2017M611064), a Basic Research Grant from the Chinese Academy of Agricultural Sciences (Y2018LM03), and the Key Research and Development Program of Xinjiang Uygur Autonomous Region of China (2018B01006-2).
Articles can be viewed without a subscription.
References
- Bahieldin A, Atef A, Edris S, Gadalla NO, Ali HM, Hassan SM, Al-Kordy MA, Ramadan AM, Makki RM, Al-Hajar AS, et al. (2016) Ethylene responsive transcription factor ERF109 retards PCD and improves salt tolerance in plant. BMC Plant Biol 16: 216–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barah P, B N MN, Jayavelu ND, Sowdhamini R, Shameer K, Bones AM (2016) Transcriptional regulatory networks in Arabidopsis thaliana during single and combined stresses. Nucleic Acids Res 44: 3147–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Demodena J, Sternberg PW (2009) RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science 324: 92–94 [DOI] [PubMed] [Google Scholar]
- Cheng S, Tan F, Lu Y, Liu X, Li T, Yuan W, Zhao Y, Zhou DX (2018) WOX11 recruits a histone H3K27me3 demethylase to promote gene expression during shoot development in rice. Nucleic Acids Res 46: 2356–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley RB, Hake S (1998) The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev 12: 1145–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Wang Q, Manik SM, Song Y, Shi S, Su Y, Liu G, Liu H (2015) Nicotiana sylvestris calcineurin B-like protein NsylCBL10 enhances salt tolerance in transgenic Arabidopsis. Plant Cell Rep 34: 2053–2063 [DOI] [PubMed] [Google Scholar]
- Duan J, Cai W (2012) OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS One 7: e45117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33: 751–763 [DOI] [PubMed] [Google Scholar]
- Egea I, Pineda B, Ortíz-Atienza A, Plasencia FA, Drevensek S, García-Sogo B, Yuste-Lisbona FJ, Barrero-Gil J, Atarés A, Flores FB, et al. (2018) The SlCBL10 calcineurin B-like protein ensures plant growth under salt stress by regulating Na+ and Ca2+ homeostasis. Plant Physiol 176: 1676–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedina IS, Georgieva K, Grigorova I (2002) Light-dark changes in proline content of barley leaves under salt stress. Biol Plant 45: 59–63 [Google Scholar]
- Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99: 15898–15903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YC, Kuang JF, Chen JY, Liu XC, Xiao YY, Fu CC, Wang JN, Wu KQ, Lu WJ (2016) Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and expansins during fruit ripening. Plant Physiol 171: 1070–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103: 12987–12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Pardo JM, Batelli G, Van Oosten MJ, Bressan RA, Li X (2013) The salt overly sensitive (SOS) pathway: established and emerging roles. Mol Plant 6: 275–286 [DOI] [PubMed] [Google Scholar]
- Jiang C, Mithani A, Belfield EJ, Mott R, Hurst LD, Harberd NP (2014) Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res 24: 1821–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, et al. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Xue Y, Wang R, Xu R, Bian L, Zhu B, Han H, Peng R, Yao Q (2013) Transcription factor OsAP21 gene increases salt/drought tolerance in transgenic Arabidopsis thaliana. Mol Biol Rep 40: 1743–1752 [DOI] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K (2011) EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6: 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Seki M (2014) Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol 55: 1859–1863 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, An G (2012) Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 69: 445–461 [DOI] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199: 639–649 [DOI] [PubMed] [Google Scholar]
- Liu D, Chen X, Liu J, Ye J, Guo Z (2012) The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J Exp Bot 63: 3899–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cheng X, Liu P, Sun J (2017) miR156-targeted SBP-box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol 174: 1931–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523 [DOI] [PubMed] [Google Scholar]
- Lu Z, Yu H, Xiong G, Wang J, Jiao Y, Liu G, Jing Y, Meng X, Hu X, Qian Q, et al. (2013) Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25: 3743–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Wang YY, Liu X, Yang S, Lu Q, Cui Y, Wu K (2012) HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot 63: 3297–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Ohme-Takagi M (2009) Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol 50: 1232–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Kim YS, Kwon CW, Park HK, Jeong JS, Kim JK (2009) Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol 150: 1368–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E, et al. (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Schmidt R, Mieulet D, Hubberten HM, Obata T, Hoefgen R, Fernie AR, Fisahn J, San Segundo B, Guiderdoni E, Schippers JH, et al. (2013) Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25: 2115–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Schippers JH, Mieulet D, Watanabe M, Hoefgen R, Guiderdoni E, Mueller-Roeber B (2014) SALT-RESPONSIVE ERF1 is a negative regulator of grain filling and gibberellin-mediated seedling establishment in rice. Mol Plant 7: 404–421 [DOI] [PubMed] [Google Scholar]
- Senadheera P, Singh RK, Maathuis FJ (2009) Differentially expressed membrane transporters in rice roots may contribute to cultivar dependent salt tolerance. J Exp Bot 60: 2553–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Lv B, Luo L, He J, Mao C, Xi D, Ming F (2017) The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci Rep 7: 40641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SJ, Guo SQ, Yang X, Bao YM, Tang HJ, Sun H, Huang J, Zhang HS (2010) Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J Exp Bot 61: 2807–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Takagi H, Tamiru M, Abe A, Yoshida K, Uemura A, Yaegashi H, Obara T, Oikawa K, Utsushi H, Kanzaki E, et al. (2015) MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat Biotechnol 33: 445–449 [DOI] [PubMed] [Google Scholar]
- Tang N, Ma S, Zong W, Yang N, Lv Y, Yan C, Guo Z, Li J, Li X, Xiang Y, et al. (2016a) MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell 28: 2161–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Qin S, Guo Y, Chen Y, Wu P, Chen Y, Li M, Jiang H, Wu G (2016b) Genome-wide analysis of the AP2/ERF gene family in physic nut and overexpression of the JcERF011 gene in rice increased its sensitivity to salinity stress. PLoS One 11: e0150879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q, Guo D, Wei B, Zhang F, Pang C, Jiang H, Zhang J, Wei T, Gu H, Qu LJ, et al. (2013) The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda Y, Tanaka M, Ogawa D, Kurata K, Kurotani K, Habu Y, Ando T, Sugimoto K, Mitsuda N, Katoh E, et al. (2013) RICE SALT SENSITIVE3 forms a ternary complex with JAZ and class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell 25: 1709–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kim J, Somers DE (2013) Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci USA 110: 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wan L, Zhang L, Zhang Z, Zhang H, Quan R, Zhou S, Huang R (2012) An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol Biol 78: 275–288 [DOI] [PubMed] [Google Scholar]
- Xiao G, Qin H, Zhou J, Quan R, Lu X, Huang R, Zhang H (2016) OsERF2 controls rice root growth and hormone responses through tuning expression of key genes involved in hormone signaling and sucrose metabolism. Plant Mol Biol 90: 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Guo Y (2018) Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol 217: 523–539 [DOI] [PubMed] [Google Scholar]
- Yolcu S, Ozdemir F, Güler A, Bor M (2016) Histone acetylation influences the transcriptional activation of POX in Beta vulgaris L. and Beta maritima L. under salt stress. Plant Physiol Biochem 100: 37–46 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang J, Quan R, Pan X, Wan L, Huang R (2013) EAR motif mutation of rice OsERF3 alters the regulation of ethylene biosynthesis and drought tolerance. Planta 237: 1443–1451 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Zhang R, Huang R (2012) The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J 71: 273–287 [DOI] [PubMed] [Google Scholar]
- Zhao L, Wang P, Hou H, Zhang H, Wang Y, Yan S, Huang Y, Li H, Tan J, Hu A, et al. (2014) Transcriptional regulation of cell cycle genes in response to abiotic stresses correlates with dynamic changes in histone modifications in maize. PLoS One 9: e106070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao SP, Xu ZS, Zheng WJ, Zhao W, Wang YX, Yu TF, Chen M, Zhou YB, Min DH, Ma YZ, et al. (2017) Genome-wide analysis of the RAV family in soybean and functional identification of GmRAV-03 involvement in salt and drought stresses and exogenous ABA treatment. Front Plant Sci 8: 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Jiang W, Long F, Cheng S, Yang W, Zhao Y, Zhou DX (2017) Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem. Plant Cell 29: 1088–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2016) Abiotic stress signaling and responses in plants. Cell 167: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]