Multi-biparental populations generate a large set of metabolite QTL (mQTL), and consolidation of a metabolic pathway may confer plant disease resistance activity.
Abstract
Metabolomic analysis coupled with advanced genetic populations represents a powerful tool with which to investigate the plant metabolome. However, genetic analyses of the rice (Oryza sativa) metabolome have been conducted mainly using natural accessions or a single biparental population. Here, the flag leaves from three interconnected chromosome segment substitution line populations with a common recurrent genetic background were used to dissect rice metabolic diversity. We effectively used multiple interconnected biparental populations, constructed by introducing genomic segments into Zhenshan 97 from ACC10 (A/Z), Minghui 63 (M/Z), and Nipponbare (N/Z), to map metabolic quantitative trait loci (mQTL). A total of 1,587 mQTL were generated, of which 684, 479, and 722 were obtained from the A/Z, M/Z, and N/Z chromosome segment substitution line populations, respectively, and we designated 99 candidate genes for 367 mQTL. In addition, 1,001 mQTL were generated specifically from joint linkage analysis with 25 candidate genes assigned. Several of these candidates were validated, such as LOC_Os07g01020 for the in vivo content of pyridoxine and its derivative and LOC_Os04g25980 for cis-zeatin glucosyltransferase activity. We propose a novel biosynthetic pathway for O-methylapigenin C-pentoside and demonstrated that LOC_Os04g11970 encodes a component of this pathway through fine-mapping. We postulate that the methylated apigenin may confer plant disease resistance. This study demonstrates the power of using multiple interconnected populations to generate a large number of veritable mQTL. The combined results are discussed in the context of functional metabolomics and the possible features of assigned candidates underlying respective metabolites.
A vast number of metabolites are produced by plants, many of which are essential for plants to interact with the environment (Schwab, 2003; Saito and Matsuda, 2010) and represent important constituents of the human diet and development (Keurentjes, 2009; Saito and Matsuda, 2010; De Luca et al., 2012). For example, the water-soluble B6 group of metabolites not only function in cellular defense against oxidative stress in plants (Herrero et al., 2011) but also may reduce the incidence of important human diseases, such as hypertension and diabetes (Hellmann and Mooney, 2010; Fitzpatrick et al., 2012). Similarly, it has been reported that specialized metabolites such as flavonoids are involved in biotic and abiotic stress tolerance in plants (Luo et al., 2009; Kaur et al., 2010; Saito et al., 2013) and confer health-promoting effects against chronic diseases and certain cancers in humans (Niggeweg et al., 2004; Butelli et al., 2008). Therefore, in view of the importance of metabolites, it is necessary both to further study their in planta functions and explore their value for humans. However, this is a somewhat daunting task, since it was estimated that more than 200,000 metabolites are produced in the plant kingdom (Dixon and Strack, 2003), and these metabolites exhibit severe qualitative and quantitative variation in abundance between species (Keurentjes et al., 2006; Morohashi et al., 2012). Therefore, it is necessary to unveil the plant metabolome in a more efficient and systemic way.
Metabolomics represents one of the major tools of experimental systems biology and, as such, has driven the understanding of the complex genetic traits both in isolation (Ritchie et al., 2015; Fernie and Tohge, 2017) and in combination with other omics tools (Keurentjes et al., 2008; Tohge et al., 2016; Wen et al., 2016; Westhues et al., 2017; Zhu et al., 2018). In recent years, tremendous progress has been made in utilizing metabolomics as a tool to explore metabolic diversity in various species and to dissect the underlying biosynthetic and regulatory pathways by studying germplasm collections (Riedelsheimer et al., 2012; Angelovici et al., 2013; Chen et al., 2014; Shang et al., 2014). Although recently becoming more popular given the constraint of hidden population structure and limited collective resources for association mapping (Flint-Garcia et al., 2003), metabolic quantitative trait loci (mQTL) analyses of natural variation thus far have been carried out largely in single biparental segregating populations of multiple species, including Arabidopsis (Arabidopsis thaliana; Knoch et al., 2017), tomato (Solanum lycopersicum; Liu et al., 2016; Rambla et al., 2017), rice (Oryza sativa; Xu et al., 2016), wheat (Triticum aestivum; Hill et al., 2015), and maize (Zea mays; Wen et al., 2015). Although simple to develop and efficient for QTL mapping (Xu et al., 2017), the combination of only two genomes in biparental populations may result in inadequate allelic diversity for QTL mapping (Jannink, 2007). Consequently, this may limit the discovery of mQTL to the diversity exhibited by the two parents. To circumvent this problem, nested association mapping populations of maize (Yu et al., 2008) and multiple advanced genetic intercross populations from multiple crops (Kover et al., 2009; Huang et al., 2012a; Bandillo et al., 2013; Pascual et al., 2015; Sannemann et al., 2015) were proposed as multiparental population solutions. In the meantime, it has been demonstrated that utilizing multi-biparental populations also can improve the efficiency of mapping (Bardol et al., 2013; Yao et al., 2016).
Rice is one of the most important crops worldwide and provides starch and many other metabolites that serve as nutrients to a considerable proportion of global human consumers. mQTL mapping or genome-wide association study coupled with metabolomics analyses (mGWAS) have been performed previously in rice to dissect the genetic basis of metabolite content regulation (Gong et al., 2013; Chen et al., 2014). The mGWAS (Chen et al., 2014) suggested the importance of investigating the biochemical diversity of the two major subspecies (indica and japonica) that have evolved with high genetic diversity from the wild progenitors (Huang et al., 2011; Chen et al., 2017). In view of the connection between metabolites and crop yield or nutrition (Fridman et al., 2004; Li et al., 2013; Tieman et al., 2017), it is envisaged that such mGWAS and mQTL studies will provide important insights in understanding the genetic basis for the biochemical diversity of rice cultivars and facilitating the use of functional metabolomics approaches (Fernie and Tohge, 2017) in breeding elite varieties with both increased resistance to stress and enhanced nutritional properties (Okazaki and Saito, 2016). To elucidate the genetic basis of these important metabolic traits, we performed metabolite profiling of 281 metabolites in three independent chromosome segment substitution line (CSSL) populations, which were generated from four parents, including two typical indica cultivars (Minghui 63 [MH63] and Zhenshan 97 [ZS97]), one japonica cultivar (Nipponbare [Nip]), and one wild accession (Oryza rufipogon ACC10), using ZS97 as a common recurrent background and the other three genotypes as independent donors. We demonstrate that utilizing three connected CSSL populations results in greater mQTL identification and more facile pathway elucidation. Furthermore, joint linkage analysis was proven to be even more powerful in mapping some of the metabolites. These combined observations suggest that populations from diverse parents may accelerate gene identification and pathway elucidation for metabolites. We subsequently identified and validated candidate genes for pyridoxine (one of the vitamers of vitamin B6) and cis-zeatin O-glucoside. Moreover, the biosynthetic pathway underlying the production of methylated apigenin was postulated and tested. The combined results, therefore, demonstrate the enhanced power of using such a genetic design strategy in the identification and validation of metabolites as well as in identifying the genes involved in their respective biosynthetic pathways. Considering the potential disease resistance activities of the candidate assigned metabolites, such discovery (i.e. those of biologically functional metabolites and their underlying biosynthetic pathways) achieved by metabolic analysis will provide key leads for further studies of specific metabolites. Such studies ultimately may be essential in attempts to metabolically fortify plants in order to secure future rice yields.
RESULTS
Metabolic Profiling of Rice Flag Leaves
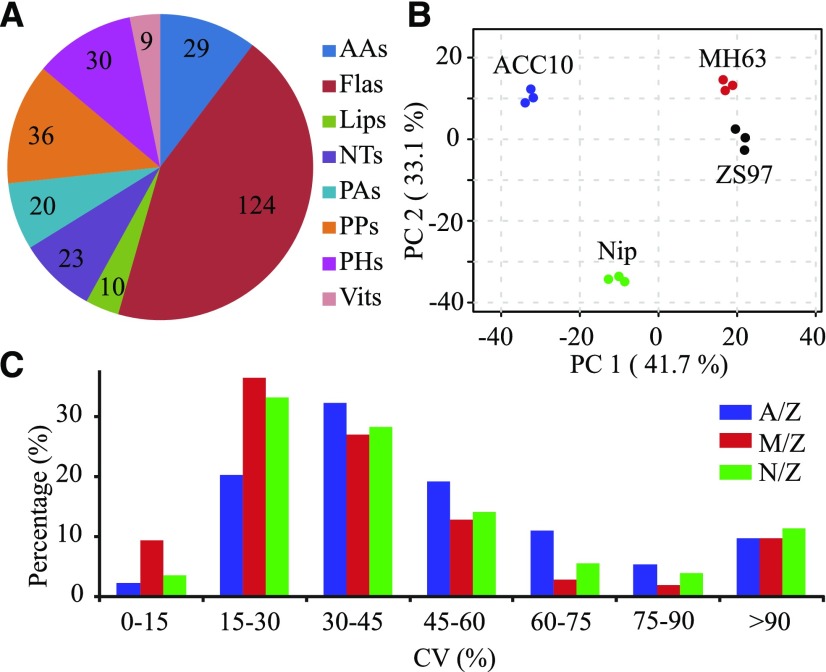
Flag leaves at heading stage were collected from the four parental lines, namely ACC10 (a wild accession of O. rufipogon), MH63 (an indica cultivar), Nip (a japonica cultivar), and ZS97 (an indica cultivar), and individuals from the three CSSL populations created using these parental lines (the A/Z, M/Z, and N/Z populations), to perform metabolic profiling analysis. By utilizing a previously established widely targeted metabolomics method (Chen et al., 2013), we detected a total of 281 metabolites (82 of them were confirmed by standards) in rice flag leaves, comprising 29 amino acids and their derivatives (AAs), 124 flavonoids (Flas), 10 lipids (Lips), 23 nucleotides, 20 phenolamides (PAs), 30 phytohormones and their derivatives (PHs), 36 polyphenols (PPs), and nine vitamins (Vits; Fig. 1A; Supplemental Table S1). A principal component analysis of the metabolic diversity indicated that the four parental lines exhibited major differences in their metabolomes (Fig. 1B). The first and second principal components (PC1 and PC2) explained 41.7% and 33.1% of total variance, respectively, and these components were contributed mainly by different metabolite classes (Supplemental Table S2): when assessing the 100 most divergent metabolite abundances among the three parents (ACC10, MH63, and Nip) compared with that in ZS97 (Supplemental Table S3), Flas and AAs were the most divergent in Nip and ACC10, respectively, whereas PPs were the most divergent in MH63 (Supplemental Fig. S1A). We next evaluated the coefficient of variation (CV) on the 281 metabolites in the three independent CSSL populations (Fig. 1C), which revealed that a lower proportion of relatively high variances (CV of 30%–90%) was apparent in the M/Z lines compared with the A/Z and N/Z populations, suggesting that these metabolites were less variable among the M/Z lines. Moreover, the M/Z lines have relatively higher heritability of metabolites when compared with that in the remaining two populations (Supplemental Fig. S1B). These observations on CV and heritability among the three CSSL populations are consistent with the closer relationship between MH63 and ZS97 (Fig. 1B; Supplemental Fig. S2, A and B) and the lower metabolite diversity in the M/Z population than in the other two CSSL populations (Supplemental Fig. S2C).
Figure 1.
Schematic representation of metabolic profiles of different lines. A, A total of 281 metabolites, comprising AAs (29), Flas (124), Lips (10), nucleotides (NTs; 23), PAs (20), PPs (36), PHs (30), and Vits (nine), were detected in this study. B, Principal component analysis of the 281 metabolites in the four parental lines ACC10, MH63, Nip, and ZS97. PC1 and PC2 refer to the first and second principal components, respectively. C, Distribution of CV among the three independent CSSL populations. A/Z, M/Z, and N/Z represent the ACC10/ZS97, MH63/ZS97, and Nip/ZS97 CSSL populations, respectively.
Bin Mapping of mQTL in Different CSSL Populations
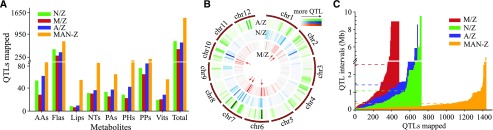
Following the metabolite profiling of the different lines, we next performed mQTL analyses based on the high-density maps of the CSSL populations (Supplemental Table S4). Totals of 684, 479, and 722 mQTL were mapped for 239, 216, and 232 metabolites in the A/Z, M/Z, and N/Z populations, respectively, and they constituted 1,587 nonduplicated loci when considering mQTL for the same metabolites from the three respective CSSL populations (A/Z, M/Z, and N/Z) that were within 1 Mb of one another to be redundant (Fig. 2A; Supplemental Tables S5 and S6). We observed the clustering of multiple hotspots, some of which were colocalized in different populations (indicated by red arrows in Fig. 2B). For example, 43, 42, and 54 mQTL were mapped within the 8- to 12-Mb block on chromosome 6, which respectively comprised 60.6%, 45.7%, and 61.4% of the total mQTL mapped on chromosome 6 from the A/Z, M/Z, and N/Z populations (Fig. 2B; Supplemental Table S7). Additional hotspots were mapped mainly on chromosomes 1, 7, and 10 (Fig. 2B; red-marked cells in Supplemental Table S7).
Figure 2.
Statistical results of mQTL. A, Number of mQTL mapped for each class of metabolites (AAs, Flas, Lips, nucleotides [NTs], PAs, PHs, PPs, and Vits; Total represents the sum of all classes) in the four populations. B, Heat map of hotspots mapped on each chromosome from the three independent CSSL populations (A/Z, M/Z, and N/Z). mQTL were counted for each of the continuous chromosomal regions spanning 2-Mb intervals. Red arrows indicate the comapped hotspots from at least two populations. C, Intervals of mQTL mapped from the four CSSL populations. The dashed lines indicate the average intervals for each population of the corresponding color.
Given that the variation among the four parental lines was contributed by different classes of metabolites (Supplemental Figs. S1 and S2; Supplemental Tables S2 and S3), it was perhaps unsurprising that the mapping outcome of the respective CSSL populations also was quite different. More mQTL were mapped in the A/Z population for most metabolic classes (except Flas, which were highest in the N/Z lines; Fig. 2A), and a better mapping outcome could be expected for AAs, PAs, PHs, and Vits in the A/Z population and Flas in the N/Z lines when considering the average number of mQTL mapped per metabolite (Supplemental Fig. S1C; Supplemental Tables S5 and S6). In addition, more candidate genes for Flas were uniquely assigned from the N/Z lines, whereas more candidates were obtained for AAs, PAs, and Vits in the A/Z population (Supplemental Tables S8 and S9). The preferential mapping of AAs in the A/Z CSSL populations and Flas in the N/Z population coincided with increased variation in the levels of the corresponding metabolites in the respective populations (Supplemental Tables S10 and S11). The M/Z population exhibited less total mQTL as well as less mQTL per each metabolite class and broader mapping interval mapped in each metabolite class (Fig. 2; Supplemental Fig. S1C).
Aside from utilizing the three mapping populations (M/Z, A/Z, and N/Z) individually, a joint linkage analysis was conducted on the integrated population MAN-Z (indicating that the chromosome segments of ZS97 were substituted by that from MH63, ACC10, and Nip) in order to enhance the mapping output. Following this approach, more mQTL were obtained for each metabolite class (Fig. 2A; Supplemental Tables S5–S7) with increased resolution (Fig. 2C) when compared with the results obtained from the individual CSSL populations. This trend also is reflected in the number of mQTL identified per metabolite, especially for Lips and PHs (Supplemental Fig. S1C). For instance, 12 mQTL were mapped with the highest log of the odds (LOD) value of 9 for Lips from the three CSSL populations, whereas 54 mQTL were obtained with the highest LOD value of 19.5 following analysis of the joint population (Supplemental Table S12). Similarly, 70 and 140 mQTL were mapped for PHs from the three CSSL populations and the joint population, respectively (Supplemental Table S13). The specifically mapped loci that were obtained only by this approach and were absent from any of the three CSSL populations (Supplemental Table S14) further exemplified the enhanced mapping performance of the integrated population.
Identification of Candidate Genes Underlying Metabolic Traits
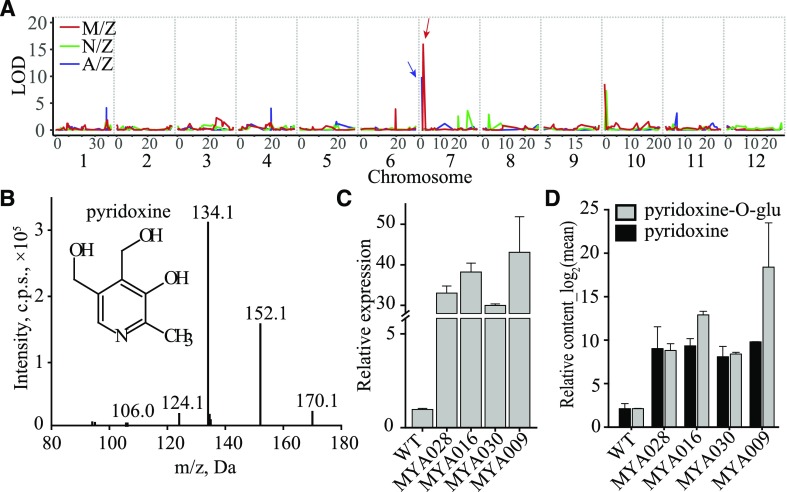
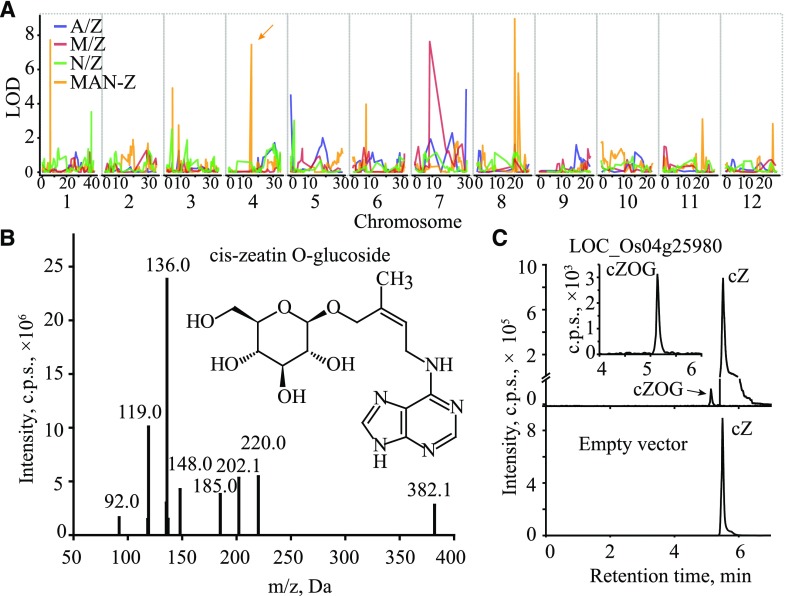
We then went on to assign candidate genes underlying the mQTL. Generally, annotations of genes within or around the mapped intervals (as well as the chemical characters of corresponding metabolites) were evaluated for candidate gene selection (Chen et al., 2014). For example, several AAs (Asp, m0018; His, m0036; and serotonin, m0051) were mapped in the 37.99- to 38.47-Mb and 38.1- to 38.47-Mb intervals on chromosome 1 from the A/Z and N/Z populations, respectively (Supplemental Table S8). LOC_Os01g65660/LOC_Os01g65670, which encodes a putative amino acid transporter and lies within the mapping interval, was assigned as a candidate due to its high sequence identity to Arabidopsis Amino Acid Permease5 (AAP5; At1g44100), which is essential for the uptake and transportation of amino acids in Arabidopsis (Svennerstam et al., 2008). Similarly, multiple loci were mapped for pyridoxine, one of the vitamers of vitamin B6, including a commonly mapped region in the block 9.8 to 823.7 kb of chromosome 7 exhibiting high LOD values (Fig. 3A; Supplemental Table S15). After manually screening the annotations of genes within this block, LOC_Os07g01020 was selected as a candidate gene given its high shared sequence identity to At5g01410 and At2g38230 (85% and 81% at the amino acid level, respectively). These genes are reported to catalyze the biosynthesis of vitamin B6 in Arabidopsis (Leuendorf et al., 2010). Our candidate gene was confirmed in vivo by the increased accumulation of pyridoxine and pyridoxine O-glucoside in lines of the rice cv Zhonghua 11 overexpressing LOC_Os07g01020 (Fig. 3, B–D). A total of 99 candidate genes responsible for 367 mQTL mapped from the three independent CSSL populations were assigned via this strategy (Table 1; Supplemental Tables S8 and S9).
Figure 3.
Linkage of LOC_Os07g01020 to in vivo contents of pyridoxine and its derivative. A, mQTL mapped for pyridoxine from the three CSSL populations. Vertical gray lines indicate the separation of chromosomes, and the comapped QTL on chromosome 7 from the A/Z and M/Z populations are pointed to by arrows with corresponding colors. B, Structure and liquid chromatography-tandem mass spectrometry (LC-MS/MS) fragmentation of pyridoxine. C, Expression levels of LOC_Os07g01020 in transgenic lines. WT, Transgenic background variety ZH11. Data are means ± se (n = 3). D, Contents of pyridoxine and pyridoxine O-glucoside in corresponding transgenic positive individuals. Data are means ± se (n = 3).
Table 1. Summary of novel candidate genes that were newly disclosed by examining the mQTL data from various populations.
Oapi C-pen, O-Methylapigenin C-pentoside; Tri O-rham-O-malhex, tricine O-rhamnosyl-O-malonylhexoside. More information is given in Supplemental Table S8.
| Metabolite | Population | Chromosome | Interval | LOD | PVE | Candidate Gene | Description |
|---|---|---|---|---|---|---|---|
| Mb | % | ||||||
| Asp | A/Z | 1 | 3.04–3.87 | 13.6 | 35.55 | LOC_Os01g08270 | Aminotransferase |
| Peonidin O-hexoside | A/Z | 1 | 22.10–22.24 | 14.6 | 12.07 | LOC_Os01g39480 | Anthocyanin regulatory Lc protein |
| N-Feruloyl agmatine | A/Z | 7 | 9.00–14.82 | 11.1 | 12.60 | LOC_Os07g23960 | Transferase |
| Pyridoxine | A/Z | 7 | 0.00–0.82 | 9.9 | 27.93 | LOC_Os07g01020 | SOR/SNZ family protein |
| Pro | A/Z | 10 | 15.07–15.77 | 16.5 | 44.02 | LOC_Os10g30090 | Amino acid permease |
| Met | A/Z | 11 | 19.50–20.48 | 25.1 | 84.94 | LOC_Os11g35040 | Aminotransferase |
| Ser | M/Z | 6 | 8.09–9.66 | 9.5 | 21.80 | LOC_Os06g16420 | Amino acid transporter |
| Cyanidin 3-O-rutinoside | M/Z | 8 | 25.49–26.10 | 47.0 | 87.87 | LOC_Os08g40440 | Dihydroflavonol-4-reductase |
| Oapi C-pen | N/Z | 4 | 5.99–15.49 | 69.8 | 11.82 | LOC_Os04g11970 | O-Methyltransferase |
| Apigenin 8-C-glucoside | N/Z | 6 | 12.05–14.41 | 25.7 | 28.18 | LOC_Os06g23560 | Glucosyltransferase |
| Val | N/Z | 7 | 5.90–6.90 | 10.5 | 22.21 | LOC_Os07g12330 | Trp/Tyr permease |
| Indole-3-carboxaldehyde | N/Z | 7 | 7.65–8.40 | 10.8 | 22.45 | LOC_Os07g14610 | IAA-amino acid hydrolase |
| Tri O-rham-O-malhex | N/Z | 7 | 5.90–6.90 | 29.3 | 17.98 | LOC_Os07g11440 | Chalcone synthase |
| Apigenin 5-O-glucoside | MAN-Z | 2 | 7.78–8.00 | 19.8 | 2.28 | LOC_Os02g14630 | Glucosyltransferase |
| LPC (1-acyl 18:3) | MAN-Z | 4 | 18.53–18.84 | 7.2 | 6.37 | LOC_Os04g31070 | Acyl-desaturase |
| Cis-zeatin O-glucoside | MAN-Z | 4 | 15.49–15.53 | 8.1 | 5.25 | LOC_Os04g25980 | Cytokinin-O-glucosyltransferase |
| LOC_Os04g25970 | |||||||
| Linoleic acid | MAN-Z | 8 | 21.75–21.83 | 11.5 | 8.02 | LOC_Os08g34220 | ω-6 fatty acid desaturase |
| Ala | MAN-Z | 12 | 26.20–26.67 | 14.5 | 13.90 | LOC_Os12g42850 | Amino acid permease |
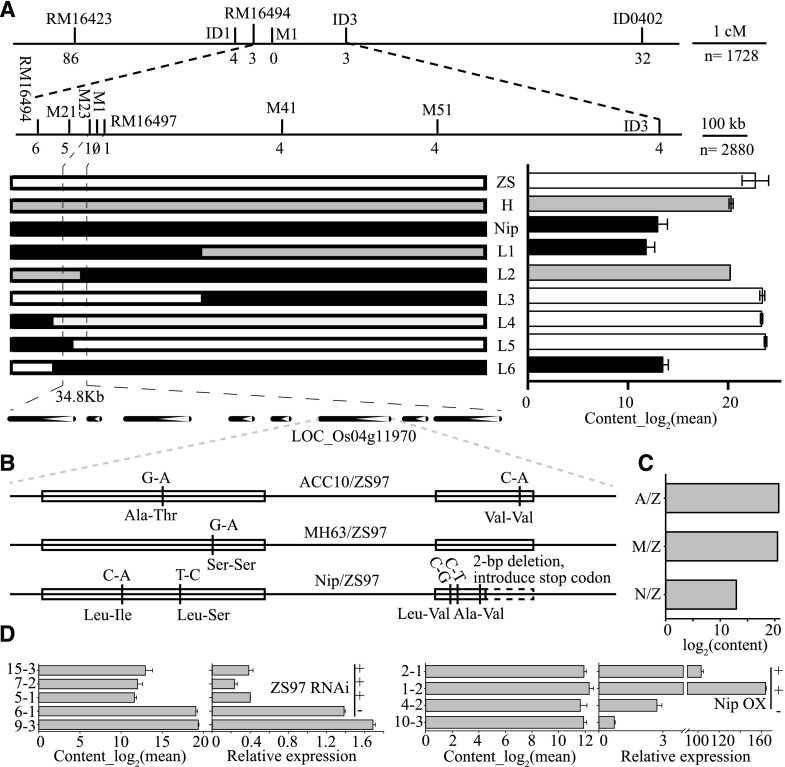
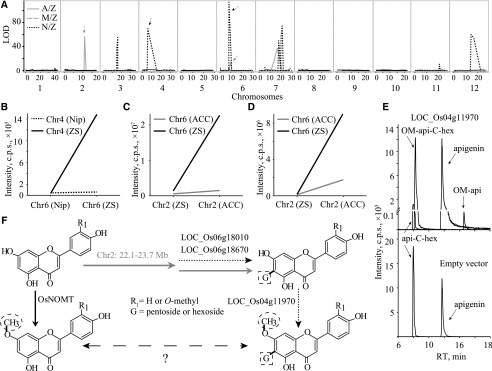
The above strategy in candidate gene assignment is workable for loci with relatively narrow intervals but difficult for an mQTL that is across a wide range of chromosomal regions. In this study, several flavonoids, specifically m0424, O-methylapigenin C-pentoside; m0467, O-methylapigenin 6-C-hexoside; m0506, O-methylluteolin C-hexoside; and m0545, O-methylchrysoeriol C-hexoside, were mapped to the block 5.99 to 15.49 Mb on chromosome 4 with LOD values of 69.8, 116.6, 82.1, and 71.8, respectively, from the N/Z population (Supplemental Table S16). Map-based cloning was conducted on the F2 progeny of IL4 and ZS97, generating 1,728 individuals, and the QTL was mapped to an approximately 1.4-Mb region between the two molecular markers RM16494 and ID3 on chromosome 4. Further analysis on an additional 2,880 plants from the BC1F2 population further narrowed the interval to a 34.8-kb segment between the markers M23 and RM16497, which contains two transposons, two conserved hypothetical proteins, three expressed proteins, and one putative O-methyltransferase. The O-methyltransferase coding gene, LOC_Os04g11970, thus was considered the most likely candidate gene (Fig. 4A). Sequence variation among the three parents compared with ZS97 indicated a 2-bp deletion near the 3′ end of the gene in Nip that introduces a stop codon (Fig. 4B), which we believed to be the causal polymorphism for decreased content of O-methylapigenin C-pentoside (m0424) in the N/Z CSSL population (Fig. 4C). The correlation between the polymorphism and metabolite content (m0424 as an example) was confirmed via further study of the rice germplasms (Supplemental Fig. S3A). The function of the candidate gene also was confirmed in vivo, with the contents of the corresponding flavonoids being significantly lower in the RNA interference (RNAi) plants of the ZS97 background (Fig. 4D; Supplemental Fig. S3B).
Figure 4.
Fine-mapping, sequence variation, and validation of LOC_Os04g11970. A, Fine-mapping narrowed the interval to a 34.8-kb segment in which LOC_Os04g11970 was selected as the candidate gene. ZS, ZS97 homozygous (white bar); H, heterozygote (gray bar); Nip, Nip homozygous (black bar); L1 to L6 represent recombinant plants. The metabolic data of m0545 (putative O-methylchrysoeriol C-hexoside) were log2 transformed, and data are means ± se (n = 3). B, Sequence variation of LOC_Os04g11970 among the three parents compared with that in ZS97, which are indicated by vertical lines. White boxes represent exons for the gene. C, Contents of m0424 (putative O-methylapigenin C-pentoside) in CSSL individuals with substituted segments harboring LOC_Os04g11970 from the three donor parents. D, Expression levels of LOC_Os04g11970 and contents of O-methylapigenin C-pentoside in RNAi individuals of ZS97 background (left) and overexpression (OX) lines of Nip background (right). The transgenic lines harboring the designated genes (LOC_Os04g11970) and empty vectors are labeled with + and −, respectively. Data are means ± se (n = 3).
For mQTL specifically identified from the joint linkage analysis (MAN-Z population), candidate genes also were assigned and validated. Specifically, the 18.53- to 18.84-Mb interval on chromosome 4 was mapped (Supplemental Table S12), corresponding to the abundance of four 1-acyl lysophosphatidylcholines [LPC (1-acyl 16:0), LPC (1-acyl 18:1), LPC (1-acyl 18:3), and LPC (1-acyl 18:2)]. A putative acyl desaturase-encoding gene, LOC_Os04g31070, which showed 81% identity at the amino acid level to At2g43710 that encodes a desaturase involved in fatty acid desaturation (Yang et al., 2016), was assigned as the candidate gene for this mQTL. Further phylogenetic analysis on additional acyl desaturase-coding genes (Supplemental Fig. S4) indicated that LOC_Os04g31070 shares highest sequence similarity with At2g43710 among others, thus strengthening our claim of its involvement in fatty acid metabolism. For PHs such as salicylic acid (SA), which serves as an important plant defense hormone (Herrera-Vásquez et al., 2015), the 33.3- to 34.3-Mb interval on chromosome 1 was acquired for its derivative methyl SA as a specific mQTL from the MAN-Z population (Supplemental Table S17). We tentatively assigned LOC_Os01g57770 as the candidate gene for its high sequence similarity to tobacco (Nicotiana tabacum) SABP2, encoding a protein reported to display high affinity to bind SA (Kumar and Klessig, 2003) and catalyze the conversion of methyl SA to SA (Forouhar et al., 2005). Similarly, the 15.49- to 15.53-Mb block on chromosome 4 was mapped specifically from the MAN-Z population (Fig. 5A; Supplemental Table S13), and the candidate genes LOC_Os04g25970/LOC_Os04g25980 (annotated as encoding cytokinin-O-glucosyltransferase) were assigned for cis-zeatin O-glucoside (mr2132) abundance, and an in vitro assay using recombinant enzyme confirmed that LOC_Os04g25980 does indeed have cis-zeatin glucosyltransferase activity (Fig. 5, B and C). An additional 25 candidate genes were assigned to mQTL generated specifically from the joint analysis (Table 1; Supplemental Table S8).
Figure 5.
In vitro validation of a candidate gene that mapped specifically from the MAN-Z population. A, Mapping result of cis-zeatin O-glucoside (mr2132), in which the 15.49- to 15.53-Mb interval (indicated by the arrow) was mapped specifically from the MAN-Z population. Vertical gray lines indicate the separation of chromosomes. B, Structure and LC-MS/MS fragmentation of cis-zeatin O-glucoside. C, LC-MS chromatograms of in vitro enzyme assays showing the enzyme activity of recombinant LOC_Os04g25980. Protein extract from Escherichia coli containing the pGEX-6p-1 empty vector was used as a negative control. cZ, Cis-zeatin; cZOG, cis-zeatin O-glucoside.
Dissection of Genetic Models and Elucidation of the Metabolic Pathway
Having assigned and validated candidate genes for the above metabolites, we next studied the mapping information for one of the metabolites in more detail. First, we evaluated the interaction between significant loci and elucidated the biosynthetic pathway from apigenin (m0164) to O-methylapigenin C-pentoside (m0424) by utilizing a previously described genetic logistic approach (Kliebenstein et al., 2001, 2002). Three loci were mapped for O-methylapigenin C-pentoside on chromosomes 2 (specifically mapped from the A/Z population), 4 (specifically mapped from the N/Z population), and 6 (mapped from the A/Z and N/Z populations; Fig. 6A), whereas only two of them (loci from chromosomes 2 and 6) were obtained for apigenin 6-C-pentoside. This enabled us to place the locus on chromosome 4 downstream of that on chromosome 6, which was confirmed by the epistasis analysis of these loci (Fig. 6B). We furthermore placed the locus on chromosome 2 upstream of the locus on chromosome 6 based on the epistasis analyses of the two loci for O-methylapigenin C-pentoside (Fig. 6C) and apigenin 6-C-pentoside (Fig. 6D). Taken together, these analyses suggest that the loci on chromosomes 2 (22.11- to 23.7-Mb interval mapped from the A/Z population), 6 (8.23- to 9.78-Mb and 8.37- to 10.61-Mb intervals mapped from the A/Z and N/Z populations, respectively), and 4 (5.99- to 15.49-Mb interval mapped from the N/Z population; Supplemental Table S5) acted sequentially in the accumulation of O-methylapigenin C-pentoside (m0424) from apigenin (m0164). LOC_Os06g18010 and LOC_Os06g18670 were assigned as candidates for mQTL on chromosome 6 based on a previous report (Brazier-Hicks et al., 2009; Chen et al., 2016), and LOC_Os04g11970 was assigned as the underlying locus on chromosome 4 via fine-mapping (Fig. 4), whereas the candidate gene underlying the locus on chromosome 2 remains unclear.
Figure 6.
Pathway dissection for the biosynthesis of O-methylapigenin C-pentoside. A, Mapping result for metabolite m0424 (putative O-methylapigenin C-pentoside) in the three CSSL populations. Vertical gray lines indicate the separation of chromosomes. B, Epistasis analysis of loci on chromosomes 6 and 4 for O-methylapigenin C-pentoside (m0424). C, Epistasis analysis of loci on chromosomes 2 and 6 for metabolite O-methylapigenin C-pentoside (m0424). D, Epistasis analysis of loci on chromosomes 2 and 6 for apigenin 6-C-pentoside (m0400). E, LC-MS chromatograms of in vitro enzyme assays showing the enzyme activity of recombinant LOC_Os04g11970. Protein extract from E. coli containing the pGEX-6p-1 empty vector was used as a negative control. OM-api, O-Methylapigenin; api-C-hex, apigenin C-hexoside; OM-api-C-hex, O-methylapigenin C-hexoside; RT, retention time. F, Predicted pathway from m0164 (standard apigenin) to m0424 (putative O-methylapigenin C-pentoside) based on the epistasis analyses and a previous report (Shimizu et al., 2012). The CSSL populations in which the respective QTLs mapped are indicated with different arrows consistent with those in A: gray solid arrows for A/Z and black dashed arrows for N/Z populations.
Combining the epistasis analyses and the annotations of the assigned candidates, we assumed that apigenin was first glycosylated (catalyzed by LOC_Os06g18010/LOC_Os06g18670) and then O-methylated (by LOC_Os04g11970) to form O-methylapigenin C-pentoside. To confirm this notion, we performed in vitro enzyme assays using apigenin and apigenin C-hexoside (at the same molar concentration) as substrates. The extremely low (∼1%) relative catalytic activity of LOC_Os04g11970 with apigenin compared with that with apigenin C-hexoside (Fig. 6E) indicated that LOC_Os04g11970 likely is an apigenin hexoside methyltransferase in vivo. Consistent with this notion, knockdown of LOC_Os04g11970 in the ZS97 background (functional for LOC_Os04g11970 and LOC_Os06g18010/LOC_Os06g18670; Fig. 6B) resulted in decreased levels of O-methylapigenin C-pentoside (m0424) and other Flas (Fig. 4D; Supplemental Fig. S3B), whereas no significant differences in the contents of these Flas were observed between the wild type and LOC_Os04g11970-overexpressing lines in the Nip background (nonfunctional for LOC_Os04g11970 and LOC_Os06g18010/LOC_Os06g18670; Fig. 6B; Supplemental Fig. S3B). Moreover, in assuming that methylated apigenin may confer disease resistance activities, a more severe disease index appeared for individuals with lowered LOC_Os04g11970 expression levels (LOC_Os04g11970-RNAi lines; Supplemental Fig. S5), further supporting the notion that LOC_Os04g11970 functions as a methyltransferase. On the basis of all these data, the pathway for the biosynthesis of O-methylapigenin C-glycoside from apigenin is proposed (Fig. 6F).
DISCUSSION
Metabolomics studies coupled with genetic approaches have been proven to be efficient and powerful in mapping mQTL underlying phenotypic diversity (Suhre et al., 2011; Li et al., 2013), in which two mapping strategies were applied (Chapman et al., 2012; Angelovici et al., 2013; Gong et al., 2013; Wen et al., 2014). Within the two strategies, the association mapping is constrained by multiple drawbacks, such as hidden population structures (Flint-Garcia et al., 2003) and hard-to-detect rare variants (Breseghello and Sorrells, 2006). Alternatively, numerous populations are available for linkage mapping, and the biparental populations are most widely used (Bernardo, 2008), for they are simple to develop and efficient for detecting QTLs (Xu et al., 2017).
However, single biparental populations were mainly employed in previous mQTL studies (Matsuda et al., 2012; Alseekh et al., 2015; Knoch et al., 2017), which may have caused many overlooked mQTL, since the recombination of only two genomes may result in inadequate allelic diversity for QTL mapping (Jannink, 2007). Thus, we applied a design using three connected biparental populations, which provides richer allelic diversity without obvious additional background noise by introducing segments from various rice subspecies, specifically indica MH63, japonica Nip, and wild accession ACC10, to the same genetic background ZS97. The observed metabolic variation among the CSSL individuals (Supplemental Fig. S2C) implies the causation of genetic difference (introduced segments in this study); thus, numerous mQTL were obtained.
It has long been assumed that the significant variation between the two rice subspecies indica and japonica was driven by divergent natural selection (Kovach et al., 2007). Meanwhile, selective breeding of cultivated rice has caused significant changes in agricultural traits (Huang et al., 2012b). As one of the phenotypic traits subject to change, metabolite contents, although perhaps only selected indirectly, experience progressive changes corresponding to genetic reshaping during selection (Shang et al., 2014; Zhu et al., 2018). Thus, it is pivotal to understand the connection between genetic divergence (genotype) and metabolic difference (phenotype) in a more efficient way if we are to fully exploit the functional metabolomics tools coupled with other omics tools in facilitating a more comprehensive and more precise manner to conduct future rice breeding programs.
Here, we demonstrated that the three donor parents ACC10, MH63, and Nip varied in metabolic diversity compared with ZS97 for different metabolite classes (Supplemental Fig. S2). By introducing segments from the respective parents, new allelic variations (single-nucleotide polymorphisms [SNPs]/insertions/deletions [InDels]) affecting metabolite contents were harbored among the CSSL individuals, and these metabolites could be mapped preferentially from designated biparental populations. Indeed, AAs were mapped preferentially in the A/Z population (61 mQTL mapped, whereas 28 and 53 mQTL were mapped from the M/Z and N/Z populations, respectively; Fig. 2A; Supplemental Table S6). This coincided with the highly varied AA content diversity from the respective donor parents compared with ZS97, in that 16 of 29 amino acids displayed a variation above 2.5-fold between ACC10 and ZS97, but only 10 displayed variation above this threshold in the comparison between Nip and ZS97 and zero reached this threshold in the comparison between MH63 and ZS97 (Supplemental Table S10).
That said, the ability to predict the best populations for mQTL breeding had not been tested empirically when this suggestion was made, and the results presented here comparing three different germplasm sources suggest that predictions can be made faithfully and, therefore, that such predictions may be useful in breeding more nutritious rice (with higher amino acids contents). This is not only the case for the AA contents in this population, as similar observations were made for flavonoids that were mapped preferentially in the N/Z population (Supplemental Table S6), which was consistent with the highest content diversity between its parental lines (Supplemental Table S11; Supplemental Fig. S2A). By contrast, the M/Z population was less preferred in almost all statistical features (Fig. 2), presumably because of the lower metabolic diversity between its parental lines (MH63 and ZS97; Supplemental Fig. S2). However, it is important to note that this additionally could be the consequence of the small population size and the lower marker density in the M/Z population (Supplemental Table S4).
Mapping and identifying loci or genes underlying different metabolite contents is an essential initial step in rendering functional metabolomics approaches (Fernie and Tohge, 2017) as an important additional tool in existing genomics-assisted strategies for crop improvement (Fernie and Schauer, 2009). Many of these metabolites play important roles in the interaction between plants and their environment (Luo et al., 2009; Kaur et al., 2010; Herrero et al., 2011; Saito et al., 2013; Chen et al., 2018), and candidates assigned here may reinforce the findings of previous reports or provide new resources for further functional validation. For instance, grain protein content is an important nutritional index for rice. Although amino acid permease/transporter genes responsible for it were reported at the molecular level (Zhao et al., 2012; Taylor et al., 2015), few of them have been genetically mapped. Here, we tentatively assigned several amino acid permease/transporter coding genes responsible for amino acid contents (Supplemental Table S8), and one of them was designated LOC_Os01g65670, underlying the mQTL responsible for the contents of multiple amino acids in rice (Supplemental Fig. S6). This gene has been reported previously as OsAAP6, which affects rice nutritional quality by enhancing grain protein content (Peng et al., 2014). Moreover, numerous mQTL for metabolites that may confer disease resistance or plant defense activities, such as flavonoids (Treutter, 2006; Luo et al., 2009; Saito et al., 2013), Ala (Park et al., 2009), His (Seo et al., 2016), serotonin (Jin et al., 2015), vitamin B6 (Zhang et al., 2014, 2015), methyl salicylate (Park et al., 2007), and zeatin (Choi et al., 2011; Grosskinsky et al., 2013), were mapped with possible candidate genes assigned or validated (Supplemental Table S8).
Furthermore, we suggest that flavonoid-methylated apigenins may possess disease resistance activities, which supports our notion that such metabolic studies are informative for future crop improvement. The basis of this suggestion is, first, that 7-O-methylation of the two structurally similar substrates, apigenin and naringenin, is catalyzed by the same enzyme, OsNOMT (Shimizu et al., 2012), whereas the product corresponding to naringenin O-methylation, sakuranetin, was reported to play roles in multiple biotic and abiotic stress processes (Kodama et al., 1992). Second, the association of O-methylation of a metabolite with increased resistance against biotic stresses has been demonstrated previously for the conversion of 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside (DIMBOA-Glc) to 2-hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one glucoside (Dafoe et al., 2011; Glauser et al., 2011). Third, lowering the contents of methylated apigenins (O-methylapigenin C-pentoside as an example; Fig. 4D) resulted in a more severe disease index against rice bacterial blight appearing in RNAi ZS97 individuals (Supplemental Fig. S5). Therefore, the above resource of metabolites associated with mQTL loci and candidate genes will be highly informative in breeding rice with improved disease resistance after intensive studies. Nonetheless, to conduct such studies more efficiently, some elusive features of the pathway underlying the methylated apigenin proposed in this study (Fig. 6F) should be addressed faithfully. We postulate that an alternative route exists within the raised pathway (Fig. 6F) when it is considered that the methylation of both apigenin and naringenin can be catalyzed by the same enzyme, OsNOMT (Shimizu et al., 2012), and the fact that DIMBOA-Glc is stored as an inactive form in the vacuole, being transformed to active DIMBOA by the action of glucosidases during tissue maceration (Meihls et al., 2013). For this purpose, future experiments that are required include probing the flux between O-methylapigenin and O-methylapigenin C-glycoside under different circumstances to confirm the alternative pathway indicated by the dashed line with arrowheads at both sides in Figure 6F. They also should compare the similarities and differences in the biological function(s) of methylated apigenins with sakuranetin to provide insights on the potential of utilizing methylated apigenins as biologically active compounds in plant disease defense. Taken together, the reconstructed pathway (Supplemental Fig. S7) including metabolites that may be involved in plant defense processes (and the corresponding candidate genes involved in their synthesis and/or degradation) provides fruitful candidates for future studies in disease resistance signaling pathways. We intend to update these lists with every future study of the genotypes used here.
CONCLUSION
In summary, in this study, we demonstrated the power of using interconnected biparental populations not only in providing higher resolution mQTL but also in improving the analysis of epistatic interactions in a manner that allows reconstruction of the biosynthetic pathway for O-methylapigenin C-pentoside, a metabolite that we postulate to possess disease resistance activity. As such, this study uses genetics to deliver on the promise, suggesting on theoretical grounds that metabolomics can be used as an approach to elucidate metabolic pathways (Weckwerth and Fiehn, 2002). Since the flux of primary metabolites can be traced by isotopes, which is not as facile for secondary metabolism, the approach described here and in the studies of Kliebenstein et al. (2001, 2002 likely will prove highly useful for fully defining the currently fragmentary pathway structures of plant secondary metabolism, thus improving our fundamental understanding of plant metabolism. Ultimately, more comprehensive pathway networks will facilitate better decisions in crop improvement endeavors.
MATERIALS AND METHODS
CSSL Population Construction
Three CSSL populations were used to dissect the genetic architecture of the rice (Oryza sativa) metabolome. The three CSSL populations, named N/Z, M/Z, and A/Z, were developed previously using a marker-assisted selection backcross scheme in which the recurrent parent was indica variety ZS97 and the donor parents were the japonica variety Nip, the indica variety MH63, and the Oryza rufipogon accession IRGC105491 (ACC10), respectively. There were 146, 132, and 155 lines in the N/Z, M/Z, and A/Z CSSL populations, respectively (Chen et al., 2007; Zhang et al., 2010; Sun et al., 2015). Each line contained a substituted donor segment of a particular chromosomal region within the common genetic background of ZS97, and all the substitution segments together covered the entire donor genome in each CSSL population.
All the CSSLs were genotyped using an Infinium RICE6K array (Illumina) containing 5,102 SNP/InDel markers distributed evenly on the 12 rice chromosomes with an average density of 12 SNPs/InDels per Mb. The released assembly version 6.1 of genomic pseudomolecules of japonica Nip, consistent with that described when this chip was developed (Yu et al., 2014), was used as the reference genome to indicate the physical positions of markers and annotated genes. The chip hybridization, polymorphism calling, genotyping, and map construction for the CSSLs were conducted as described previously (Yu et al., 2014). Bin genotyping of a single CSSL population was constructed using polymorphic genotyping as described previously (Paran and Zamir, 2003). In order to increase the mapping resolution, joint analysis (Li et al., 2015; Hoyos-Villegas et al., 2016) was conducted on the MAN-Z population, which was integrated from the three CSSL populations. The bin genotyping of the MAN-Z population was defined according to that of a single population.
Metabolic Profiling
All the CSSL lines were grown in a two-row plot for each line with 10 individuals per row in the experimental field of Huazhong Agricultural University at Wuhan (30.4_N, 114.2_E) under normal field management conditions. Since there was about a 4-week difference in maturity for each of the three CSSL populations and among parental lines (roughly, July 20 for ACC10, August 1 for ZH97, August 20 for Nip and MH63; July 25 to August 25 for A/Z and M/Z and August 1 to September 10 for N/Z), samples were collected at the day of 50% panicle emergence for each line. All samples were collected within a narrow time window (from 8 to 9:30 am) to minimize the variation caused by sampling time. Three biological replicates of flag leaves were collected from independent plants at the heading stage and instantly snap frozen by liquid nitrogen prior to being freeze dried for metabolite extraction. The extracts were absorbed (CNWBOND Carbon-GCB SPE Cartridge, 250 mg, 3 mL; Shanghai ANPEL Scientific Instrument) and analyzed using an LC-electrospray ionization-MS/MS system as described previously (Chen et al., 2013). The quantification of metabolites was conducted using a scheduled multiple reaction monitoring (MRM) method (Dresen et al., 2010), with an MRM detection window of 80 s and a target scan time of 1.5 s.
Statistical Analysis
Metabolite data were first log2 transformed to improve the normality of distribution, and the biological replicates were then averaged for downstream analysis (Gong et al., 2013). The principal component analyses of the data were conducted by an online tool (http://www.metaboanalyst.ca/; Xia et al., 2015). The values of the CV were calculated for each metabolite: σ/μ, where σ and μ are the sd and mean of each metabolite in the CSSL populations, respectively, and the heritability of the detected metabolites was calculated as described (Chen et al., 2014): H2 = VarG/(VarG + VarE), in which VarG and VarE are for variations of genotype and environment.
QTL Mapping
QTL analyses of the log2-transformed metabolite data with the bins as markers were performed using a likelihood ratio test based on a stepwise regression (RSTEP-LRT) method in CSL mode in the software IciMapping version 4.0 (http://www.isbreeding.net/software/; Wang et al., 2006). The threshold value of the condition number for reducing multicollinearity among marker variables and the probability in stepwise regression were set as 1,000 and 0.001 by default, respectively. The LOD threshold was set as 2.5 to declare the presence of a putative QTL in a given bin. Additional study was performed based on the mQTL mapped in at least two of three replicates.
Fine-Mapping of LOC_Os04g11970
Several flavonoids (m0424, O-methylapigenin C-pentoside; m0467, O-methylapigenin C-hexoside; m0506, O-methylluteolin C-hexoside; and m0545, O-methylchrysoeriol C-hexoside) were mapped to the wide range (5.99–15.49 Mb) of chromosome 4 from the N/Z population. One of the mapped metabolites (m0545), owing to its extremely high content in ZS97 compared with that in Nip, was selected as an indicator to narrow down the mapping interval. Subsequently, IL4, one of the N/Z CSSLs (Supplemental Table S18), was selected to construct an F2 population by crossing with ZS97 because its metabolite content (i.e. content of m0545) was comparable to that of Nip. By analyzing the metabolite contents and marker profiles among the generated 1,728 individuals, the mapping interval was narrowed down to an approximately 1.4-Mb segment between molecular markers RM16494 and ID3. Afterward, a BC1F2 population consisting of 2,880 lines was generated with ZS97 as a recurrent parent, and the interval was further located to the 34.8-kb segment between molecular markers M23 and RM16497, in which LOC_Os04g11970 was selected as the candidate gene.
Plasmid Construction and Rice Transformation
Primers Designed
Primers used in this study are listed in Supplemental Table S19.
RNAi Vector Construction and Transformation
A 310-bp cDNA fragment amplified from ZS97 by primers DS970F and DS970R (Supplemental Table S19) was inserted into the vector PDS1301 (a modified vector of PCAMBIA 1301; Chu et al., 2006) and transformed into ZS97.
Overexpression Vector Construction and Transformation
Overexpression vectors (pJC034 for LOC_Os07g01020 and pUC1301 for LOC_Os04g11970; Sun et al., 2015) for rice were constructed with the 35S promoter replaced by the maize (Zea mays) ubiquitin promoter. The overexpression constructs were generated by directionally inserting the full cDNAs from ZS97 first into the entry vector pDONR207 and then into the destination vectors using the Gateway recombination reaction (Invitrogen). The constructs were introduced into Agrobacterium tumefaciens strain EHA105 and then transferred into japonica ZH11 (LOC_Os07g01020) or Nip (LOC_Os04g11970) as described previously (Hiei et al., 1994). Targeted metabolite analysis for each construct was performed on at least three independent T2 progeny using the MRM method as described above.
Expression Analyses
Total RNA was isolated from rice flag leaves using an RNA extraction kit (TRIzol reagent; Invitrogen). First-strand cDNA then was synthesized using 3 μg of RNA with 200 units of M-MLV reverse transcriptase (Invitrogen). Quantitative reverse transcription-PCR was performed on an optical 96-well plate in an AB StepOnePlus PCR system (Applied Biosystems) using SYBR Premix reagent F-415 (Thermo Scientific). All the experiments were performed following the manufacturers’ protocols. The relative expression quantities were calculated using the relative quantification method (Livak and Schmittgen, 2001) using Ubiquitin as an endogenous control.
In Vitro Validation of Candidate Genes
First, full-length cDNAs (LOC_Os04g25980 from MH63 and LOC_Os04g11970 from ZS97) were amplified using the respective primers listed in Supplemental Table S19. Clones were digested by BamHI/EcoRI and directionally ligated to pGEX-6p-1. Error-free recombinant vectors were transformed into BL21 (DE3) pLysS cells (Novagene), which was cultured subsequently on a Luria-Bertani plate. A single colony was grown in Luria-Bertani medium overnight, and recombinant proteins were expressed after induction by the addition of 0.4 mm isopropyl β-d-1-thiogalactopyranoside and growing continually for 12 h at 16°C. Cells were harvested and suspended in 50 mm sodium phosphate buffer (pH 7.8) and lysed by sonication. The crude extract was collected and clarified by centrifugation at 12,000g for 1 h at 4°C, and supernatant of the crude enzyme was stored at −80°C.
Standard in vitro enzyme assays for the role of enzymes encoded by LOC_Os04g25980 (cis-zeatin as substrate) and LOC_Os04g11970 (apigenin and apigenin C-hexoside as substrate) were performed in a total volume of 20 μL containing 3 μL of purified protein (i.e. the purified expression product of LOC_Os04g25980 or LOC_Os04g11970), 100 μm substrate, 100 μm SAM (in validating LOC_Os04g11970) or 750 μm UDP-Glc (in validating LOC_Os04g25980), and 5 mm MgCl2 in 100 mm Tris-HCl buffer (pH 7.4). After incubating at 37°C for 30 min, the reaction was stopped by adding 60 μL of methanol. The reaction mixture then was filtered through a 0.2-μm filter (Millipore) before being used for LC-MS analysis. Each enzyme assay, as well as those that utilize expressed protein from pGEX-6p-1 empty vector, was repeated three times to eliminate possible false results.
Pathogen Inoculation and Disease Scoring
Bacterial inoculum was prepared as described previously (Lin et al., 1996). The leaf-clipping method (Sun et al., 2004) was conducted to inoculate Xanthomonas oryzae pv oryzae strain PXO61 on RNAi and normal ZS97 individuals at the booting stage in the field. Lesion lengths were scored (in cm) at 3 weeks after inoculation, and Student’s t test (P < 0.05) was performed to analyze the data.
Accession Numbers
Sequence data from this article can be found on the Web sites http://rice.plantbiology.msu.edu/ and http://www.arabidopsis.org/ for rice and Arabidopsis genes, respectively, with respective locus identifiers provided in this study.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Statistical information regarding mQTL analysis among the CSSL populations.
Supplemental Figure S2. Metabolic profiles of multiple lines.
Supplemental Figure S3. Metabolite contents of rice germplasms and transgenic plants of LOC_Os04g11970.
Supplemental Figure S4. Phylogenetic analysis of acyl desaturase-coding genes.
Supplemental Figure S5. Potential role of methylated apigenin in disease resistance against rice bacterial blight.
Supplemental Figure S6. An mQTL responsible for multiple amino acid contents.
Supplemental Figure S7. Pathway for metabolites that may possess disease resistance activities by consolidating newly assigned candidate genes.
Supplemental Table S1. Metabolites detected in the screening.
Supplemental Table S2. Contribution of each metabolite in the principal components.
Supplemental Table S3. Main principal components of the three parental lines compared with ZS97.
Supplemental Table S4. Bin information of different populations.
Supplemental Table S5. Mapping results of the CSSL populations.
Supplemental Table S6. Numbers of mQTL mapped of the 281 metabolites in different populations.
Supplemental Table S7. Numbers of mapped mQTL within chromosomal blocks from the four populations.
Supplemental Table S8. List of candidate genes corresponding to respective mQTL.
Supplemental Table S9. Numbers of candidate genes assigned to metabolite classes.
Supplemental Table S10. Ratios of AA metabolites in the three donor parents compared with ZS97.
Supplemental Table S11. Ratios of Fla metabolites in the three donor parents compared with ZS97.
Supplemental Table S12. Mapping results for lipids in the four populations.
Supplemental Table S13. Mapping results for PH metabolites in the four populations.
Supplemental Table S14. QTLs specifically mapped from the MAN-Z population.
Supplemental Table S15. Mapping results of pyridoxine in the three CSSL populations.
Supplemental Table S16. Candidate gene selection for mQTL with a wide interval.
Supplemental Table S17. Mapping results of the two phytohormones and their main derivatives.
Supplemental Table S18. RICE6K SNP array of IL4.
Supplemental Table S19. Primers used in this study.
Supplemental Table S20. Orthologs of candidates raised in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Footnotes
This work was supported by the National Science Fund for Distinguished Young Scholars (No. 31625021), by The Ministry of Science and Technology of China (2016YFD0100500) to J.L., by the National Natural Science Foundation of China (No. 31671656) and the National High Technology Research and Development of China (No. 2014AA10A604) to S.Y., and by the National Natural Science Foundation of China (No. 31770328) to W.C.
References
- Alseekh S, Tohge T, Wendenberg R, Scossa F, Omranian N, Li J, Kleessen S, Giavalisco P, Pleban T, Mueller-Roeber B, et al. (2015) Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell 27: 485–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Lipka AE, Deason N, Gonzalez-Jorge S, Lin H, Cepela J, Buell R, Gore MA, Dellapenna D (2013) Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. Plant Cell 25: 4827–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandillo N, Raghavan C, Muyco PA, Sevilla MAL, Lobina IT, Dilla-Ermita CJ, Tung CW, McCouch S, Thomson M, Mauleon R, et al. (2013) Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice (N Y) 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardol N, Ventelon M, Mangin B, Jasson S, Loywick V, Couton F, Derue C, Blanchard P, Charcosset A, Moreau L (2013) Combined linkage and linkage disequilibrium QTL mapping in multiple families of maize (Zea mays L.) line crosses highlights complementarities between models based on parental haplotype and single locus polymorphism. Theor Appl Genet 126: 2717–2736 [DOI] [PubMed] [Google Scholar]
- Bernardo R. (2008) Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci 48: 1649–1664 [Google Scholar]
- Brazier-Hicks M, Evans KM, Gershater MC, Puschmann H, Steel PG, Edwards R (2009) The C-glycosylation of flavonoids in cereals. J Biol Chem 284: 17926–17934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breseghello F, Sorrells ME (2006) Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172: 1165–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, et al. (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Chapman NH, Bonnet J, Grivet L, Lynn J, Graham N, Smith R, Sun G, Walley PG, Poole M, Causse M, et al. (2012) High-resolution mapping of a fruit firmness-related quantitative trait locus in tomato reveals epistatic interactions associated with a complex combinatorial locus. Plant Physiol 159: 1644–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Jing X, Liao B, Zhu Y, Xu J, Liu R, Zhao Y, Li X (2017) BioNano genome map resource for Oryza sativa ssp. japonica and indica and its application in rice genome sequence correction and gap filling. Mol Plant 10: 895–898 [DOI] [PubMed] [Google Scholar]
- Chen Q, Mu J, Zhou H, Yu S (2007) Genetic effect of japonica alleles detected in indica candidate introgression lines. Zhongguo Nong Ye Ke Xue 40: 2387–2394 [Google Scholar]
- Chen W, Gong L, Guo Z, Wang W, Zhang H, Liu X, Yu S, Xiong L, Luo J (2013) A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol Plant 6: 1769–1780 [DOI] [PubMed] [Google Scholar]
- Chen W, Gao Y, Xie W, Gong L, Lu K, Wang W, Li Y, Liu X, Zhang H, Dong H, et al. (2014) Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat Genet 46: 714–721 [DOI] [PubMed] [Google Scholar]
- Chen W, Wang W, Peng M, Gong L, Gao Y, Wan J, Wang S, Shi L, Zhou B, Li Z, et al. (2016) Comparative and parallel genome-wide association studies for metabolic and agronomic traits in cereals. Nat Commun 7: 12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Holmes EC, Rajniak J, Kim JG, Tang S, Fischer CR, Mudgett MB, Sattely ES (2018) N-Hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis. Proc Natl Acad Sci USA 115: E4920–E4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Choi D, Lee S, Ryu CM, Hwang I (2011) Cytokinins and plant immunity: old foes or new friends? Trends Plant Sci 16: 388–394 [DOI] [PubMed] [Google Scholar]
- Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL, Zhang Q, Wang S (2006) Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev 20: 1250–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafoe NJ, Huffaker A, Vaughan MM, Duehl AJ, Teal PE, Schmelz EA (2011) Rapidly induced chemical defenses in maize stems and their effects on short-term growth of Ostrinia nubilalis. J Chem Ecol 37: 984–991 [DOI] [PubMed] [Google Scholar]
- De Luca V, Salim V, Atsumi SM, Yu F (2012) Mining the biodiversity of plants: a revolution in the making. Science 336: 1658–1661 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Strack D (2003) Phytochemistry meets genome analysis, and beyond. Phytochemistry 62: 815–816 [DOI] [PubMed] [Google Scholar]
- Dresen S, Ferreirós N, Gnann H, Zimmermann R, Weinmann W (2010) Detection and identification of 700 drugs by multi-target screening with a 3200 Q TRAP LC-MS/MS system and library searching. Anal Bioanal Chem 396: 2425–2434 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Schauer N (2009) Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet 25: 39–48 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Tohge T (2017) The genetics of plant metabolism. Annu Rev Genet 51: 287–310 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick TB, Basset GJ, Borel P, Carrari F, DellaPenna D, Fraser PD, Hellmann H, Osorio S, Rothan C, Valpuesta V, et al. (2012) Vitamin deficiencies in humans: can plant science help? Plant Cell 24: 395–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint-Garcia SA, Thornsberry JM, Buckler ES IV (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54: 357–374 [DOI] [PubMed] [Google Scholar]
- Forouhar F, Yang Y, Kumar D, Chen Y, Fridman E, Park SW, Chiang Y, Acton TB, Montelione GT, Pichersky E, et al. (2005) Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc Natl Acad Sci USA 102: 1773–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D (2004) Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Glauser G, Marti G, Villard N, Doyen GA, Wolfender JL, Turlings TCJ, Erb M (2011) Induction and detoxification of maize 1,4-benzoxazin-3-ones by insect herbivores. Plant J 68: 901–911 [DOI] [PubMed] [Google Scholar]
- Gong L, Chen W, Gao Y, Liu X, Zhang H, Xu C, Yu S, Zhang Q, Luo J (2013) Genetic analysis of the metabolome exemplified using a rice population. Proc Natl Acad Sci USA 110: 20320–20325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskinsky DK, Edelsbrunner K, Pfeifhofer H, van der Graaff E, Roitsch T (2013) Cis- and trans-zeatin differentially modulate plant immunity. Plant Signal Behav 8: e24798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H, Mooney S (2010) Vitamin B6: a molecule for human health? Molecules 15: 442–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Vásquez A, Salinas P, Holuigue L (2015) Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front Plant Sci 6: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero S, González E, Gillikin JW, Vélëz H, Daub ME (2011) Identification and characterization of a pyridoxal reductase involved in the vitamin B6 salvage pathway in Arabidopsis. Plant Mol Biol 76: 157–169 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hill CB, Taylor JD, Edwards J, Mather D, Langridge P, Bacic A, Roessner U (2015) Detection of QTL for metabolic and agronomic traits in wheat with adjustments for variation at genetic loci that affect plant phenology. Plant Sci 233: 143–154 [DOI] [PubMed] [Google Scholar]
- Hoyos-Villegas V, Song QJ, Wright EM, Beebe SE, Kelly JD (2016) Joint linkage QTL mapping for yield and agronomic traits in a composite map of three common bean RIL populations. Crop Sci 56: 2546–2563 [Google Scholar]
- Huang BE, George AW, Forrest KL, Kilian A, Hayden MJ, Morell MK, Cavanagh CR (2012a) A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol J 10: 826–839 [DOI] [PubMed] [Google Scholar]
- Huang X, Zhao Y, Wei X, Li C, Wang A, Zhao Q, Li W, Guo Y, Deng L, Zhu C, et al. (2011) Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet 44: 32–39 [DOI] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, Wang ZX, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, et al. (2012b) A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannink JL. (2007) Identifying quantitative trait locus by genetic background interactions in association studies. Genetics 176: 553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B, Zhou X, Jiang B, Gu Z, Zhang P, Qian Q, Chen X, Ma B (2015) Transcriptome profiling of the spl5 mutant reveals that SPL5 has a negative role in the biosynthesis of serotonin for rice disease resistance. Rice (N Y) 8: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Heinzel N, Schöttner M, Baldwin IT, Gális I (2010) R2R3-NaMYB8 regulates the accumulation of phenylpropanoid-polyamine conjugates, which are essential for local and systemic defense against insect herbivores in Nicotiana attenuata. Plant Physiol 152: 1731–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJ. (2009) Genetical metabolomics: closing in on phenotypes. Curr Opin Plant Biol 12: 223–230 [DOI] [PubMed] [Google Scholar]
- Keurentjes JJ, Fu J, de Vos CH, Lommen A, Hall RD, Bino RJ, van der Plas LH, Jansen RC, Vreugdenhil D, Koornneef M (2006) The genetics of plant metabolism. Nat Genet 38: 842–849 [DOI] [PubMed] [Google Scholar]
- Keurentjes JJ, Sulpice R, Gibon Y, Steinhauser MC, Fu J, Koornneef M, Stitt M, Vreugdenhil D (2008) Integrative analyses of genetic variation in enzyme activities of primary carbohydrate metabolism reveal distinct modes of regulation in Arabidopsis thaliana. Genome Biol 9: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Gershenzon J, Mitchell-Olds T (2001) Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics 159: 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D, Pedersen D, Barker B, Mitchell-Olds T (2002) Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics 161: 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoch D, Riewe D, Meyer RC, Boudichevskaia A, Schmidt R, Altmann T (2017) Genetic dissection of metabolite variation in Arabidopsis seeds: evidence for mQTL hotspots and a master regulatory locus of seed metabolism. J Exp Bot 68: 1655–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama O, Miyakawa J, Akatsuka T, Kiyosawa S (1992) Sakuranetin, a flavanone phytoalexin from ultraviolet-irradiated rice leaves. Phytochemistry 31: 3807–3809 [Google Scholar]
- Kovach MJ, Sweeney MT, McCouch SR (2007) New insights into the history of rice domestication. Trends Genet 23: 578–587 [DOI] [PubMed] [Google Scholar]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, Durrant C, Mott R (2009) A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Klessig DF (2003) High-affinity salicylic acid-binding protein 2 is required for plant innate immunity and has salicylic acid-stimulated lipase activity. Proc Natl Acad Sci USA 100: 16101–16106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuendorf JE, Osorio S, Szewczyk A, Fernie AR, Hellmann H (2010) Complex assembly and metabolic profiling of Arabidopsis thaliana plants overexpressing vitamin B6 biosynthesis proteins. Mol Plant 3: 890–903 [DOI] [PubMed] [Google Scholar]
- Li C, Li Y, Bradbury PJ, Wu X, Shi Y, Song Y, Zhang D, Rodgers-Melnick E, Buckler ES, Zhang Z, et al. (2015) Construction of high-quality recombination maps with low-coverage genomic sequencing for joint linkage analysis in maize. BMC Biol 13: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Peng Z, Yang X, Wang W, Fu J, Wang J, Han Y, Chai Y, Guo T, Yang N, et al. (2013) Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet 45: 43–50 [DOI] [PubMed] [Google Scholar]
- Lin XH, Zhang DP, Xie YF, Gao HP, Zhang Q (1996) Identifying and mapping a new gene for bacterial blight resistance in rice based on RFLP markers. Phytopathology 86: 1156–1159 [Google Scholar]
- Liu Z, Alseekh S, Brotman Y, Zheng Y, Fei Z, Tieman DM, Giovannoni JJ, Fernie AR, Klee HJ (2016) Identification of a Solanum pennellii chromosome 4 fruit flavor and nutritional quality-associated metabolite QTL. Front Plant Sci 7: 1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luo J, Fuell C, Parr A, Hill L, Bailey P, Elliott K, Fairhurst SA, Martin C, Michael AJ (2009) A novel polyamine acyltransferase responsible for the accumulation of spermidine conjugates in Arabidopsis seed. Plant Cell 21: 318–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Okazaki Y, Oikawa A, Kusano M, Nakabayashi R, Kikuchi J, Yonemaru J, Ebana K, Yano M, Saito K (2012) Dissection of genotype-phenotype associations in rice grains using metabolome quantitative trait loci analysis. Plant J 70: 624–636 [DOI] [PubMed] [Google Scholar]
- Meihls LN, Handrick V, Glauser G, Barbier H, Kaur H, Haribal MM, Lipka AE, Gershenzon J, Buckler ES, Erb M, et al. (2013) Natural variation in maize aphid resistance is associated with 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell 25: 2341–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Casas MI, Falcone Ferreyra ML, Falcone Ferreyra L, Mejía-Guerra MK, Pourcel L, Yilmaz A, Feller A, Carvalho B, Emiliani J, et al. (2012) A genome-wide regulatory framework identifies maize pericarp color1 controlled genes. Plant Cell 24: 2745–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggeweg R, Michael AJ, Martin C (2004) Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat Biotechnol 22: 746–754 [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Saito K (2016) Integrated metabolomics and phytochemical genomics approaches for studies on rice. Gigascience 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paran I, Zamir D (2003) Quantitative traits in plants: beyond the QTL. Trends Genet 19: 303–306 [DOI] [PubMed] [Google Scholar]
- Park KS, Paul D, Kim JS, Park JW (2009) L-Alanine augments rhizobacteria-induced systemic resistance in cucumber. Folia Microbiol (Praha) 54: 322–326 [DOI] [PubMed] [Google Scholar]
- Park SW, Kaimoyo E, Kumar D, Mosher S, Klessig DF (2007) Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318: 113–116 [DOI] [PubMed] [Google Scholar]
- Pascual L, Desplat N, Huang BE, Desgroux A, Bruguier L, Bouchet JP, Le QH, Chauchard B, Verschave P, Causse M (2015) Potential of a tomato MAGIC population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnol J 13: 565–577 [DOI] [PubMed] [Google Scholar]
- Peng B, Kong H, Li Y, Wang L, Zhong M, Sun L, Gao G, Zhang Q, Luo L, Wang G, et al. (2014) OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat Commun 5: 4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambla JL, Medina A, Fernández-Del-Carmen A, Barrantes W, Grandillo S, Cammareri M, López-Casado G, Rodrigo G, Alonso A, García-Martínez S, et al. (2017) Identification, introgression, and validation of fruit volatile QTLs from a red-fruited wild tomato species. J Exp Bot 68: 429–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedelsheimer C, Czedik-Eysenberg A, Grieder C, Lisec J, Technow F, Sulpice R, Altmann T, Stitt M, Willmitzer L, Melchinger AE (2012) Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat Genet 44: 217–220 [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D (2015) Methods of integrating data to uncover genotype-phenotype interactions. Nat Rev Genet 16: 85–97 [DOI] [PubMed] [Google Scholar]
- Saito K, Matsuda F (2010) Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol 61: 463–489 [DOI] [PubMed] [Google Scholar]
- Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR (2013) The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol Biochem 72: 21–34 [DOI] [PubMed] [Google Scholar]
- Sannemann W, Huang BE, Mathew B, Leon J (2015) Multi-parent advanced generation inter-cross in barley: high-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol Breed 35: 86 [Google Scholar]
- Schwab W. (2003) Metabolome diversity: too few genes, too many metabolites? Phytochemistry 62: 837–849 [DOI] [PubMed] [Google Scholar]
- Seo S, Nakaho K, Hong SW, Takahashi H, Shigemori H, Mitsuhara I (2016) L-Histidine induces resistance in plants to the bacterial pathogen Ralstonia solanacearum partially through the activation of ethylene signaling. Plant Cell Physiol 57: 1932–1942 [DOI] [PubMed] [Google Scholar]
- Shang Y, Ma Y, Zhou Y, Zhang H, Duan L, Chen H, Zeng J, Zhou Q, Wang S, Gu W, et al. (2014) Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 346: 1084–1088 [DOI] [PubMed] [Google Scholar]
- Shimizu T, Lin F, Hasegawa M, Okada K, Nojiri H, Yamane H (2012) Purification and identification of naringenin 7-O-methyltransferase, a key enzyme in biosynthesis of flavonoid phytoalexin sakuranetin in rice. J Biol Chem 287: 19315–19325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wägele B, Altmaier E, Deloukas P, Erdmann J, Grundberg E, et al. (2011) Human metabolic individuality in biomedical and pharmaceutical research. Nature 477: 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Zhou Q, Yao Y, Qiu X, Xie K, Yu S (2015) Identification of genomic regions and the isoamylase gene for reduced grain chalkiness in rice. PLoS ONE 10: e0122013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q (2004) Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J 37: 517–527 [DOI] [PubMed] [Google Scholar]
- Svennerstam H, Ganeteg U, Näsholm T (2008) Root uptake of cationic amino acids by Arabidopsis depends on functional expression of amino acid permease 5. New Phytol 180: 620–630 [DOI] [PubMed] [Google Scholar]
- Taylor MR, Reinders A, Ward JM (2015) Transport function of rice amino acid permeases (AAPs). Plant Cell Physiol 56: 1355–1363 [DOI] [PubMed] [Google Scholar]
- Tieman D, Zhu G, Resende MFR Jr, Lin T, Nguyen C, Bies D, Rambla JL, Beltran KS, Taylor M, Zhang B, et al. (2017) A chemical genetic roadmap to improved tomato flavor. Science 355: 391–394 [DOI] [PubMed] [Google Scholar]
- Tohge T, Wendenburg R, Ishihara H, Nakabayashi R, Watanabe M, Sulpice R, Hoefgen R, Takayama H, Saito K, Stitt M, et al. (2016) Characterization of a recently evolved flavonol-phenylacyltransferase gene provides signatures of natural light selection in Brassicaceae. Nat Commun 7: 12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutter D. (2006) Significance of flavonoids in plant resistance: a review. Environ Chem Lett 4: 147–157 [Google Scholar]
- Wang J, Wan X, Crossa J, Crouch J, Weng J, Zhai H, Wan J (2006) QTL mapping of grain length in rice (Oryza sativa L.) using chromosome segment substitution lines. Genet Res 88: 93–104 [DOI] [PubMed] [Google Scholar]
- Weckwerth W, Fiehn O (2002) Can we discover novel pathways using metabolomic analysis? Curr Opin Biotechnol 13: 156–160 [DOI] [PubMed] [Google Scholar]
- Wen W, Li D, Li X, Gao Y, Li W, Li H, Liu J, Liu H, Chen W, Luo J, et al. (2014) Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun 5: 3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Li K, Alseekh S, Omranian N, Zhao L, Zhou Y, Xiao Y, Jin M, Yang N, Liu H, et al. (2015) Genetic determinants of the network of primary metabolism and their relationships to plant performance in a maize recombinant inbred line population. Plant Cell 27: 1839–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Liu H, Zhou Y, Jin M, Yang N, Li D, Luo J, Xiao Y, Pan Q, Tohge T, et al. (2016) Combining quantitative genetics approaches with regulatory network analysis to dissect the complex metabolism of the maize kernel. Plant Physiol 170: 136–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhues M, Schrag TA, Heuer C, Thaller G, Utz HF, Schipprack W, Thiemann A, Seifert F, Ehret A, Schlereth A, et al. (2017) Omics-based hybrid prediction in maize. Theor Appl Genet 130: 1927–1939 [DOI] [PubMed] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, Wishart DS (2015) MetaboAnalyst 3.0: making metabolomics more meaningful. Nucleic Acids Res 43: W251–W257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Xu Y, Gong L, Zhang Q (2016) Metabolomic prediction of yield in hybrid rice. Plant J 88: 219–227 [DOI] [PubMed] [Google Scholar]
- Xu Y, Li P, Yang Z, Xu C (2017) Genetic mapping of quantitative trait loci in crops. Crop J 5: 175–184 [Google Scholar]
- Yang W, Dong R, Liu L, Hu Z, Li J, Wang Y, Ding X, Chu Z (2016) A novel mutant allele of SSI2 confers a better balance between disease resistance and plant growth inhibition on Arabidopsis thaliana. BMC Plant Biol 16: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N, Lee CR, Semagn K, Sow M, Nwilene F, Kolade O, Bocco R, Oyetunji O, Mitchell-Olds T, Ndjiondjop MN (2016) QTL mapping in three rice populations uncovers major genomic regions associated with African rice gall midge resistance. PLoS ONE 11: e0160749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Xie W, Li J, Zhou F, Zhang Q (2014) A whole-genome SNP array (RICE6K) for genomic breeding in rice. Plant Biotechnol J 12: 28–37 [DOI] [PubMed] [Google Scholar]
- Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Qiu X, Dong H, Yu S (2010) Development and characterization of chromosome segment substitution lines using O. rufipogon as donor. Mol Plant Breed 8: 1113–1119 [Google Scholar]
- Zhang Y, Liu B, Li X, Ouyang Z, Huang L, Hong Y, Zhang H, Li D, Song F (2014) The de novo biosynthesis of vitamin B6 is required for disease resistance against Botrytis cinerea in tomato. Mol Plant Microbe Interact 27: 688–699 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jin X, Ouyang Z, Li X, Liu B, Huang L, Hong Y, Zhang H, Song F, Li D (2015) Vitamin B6 contributes to disease resistance against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea in Arabidopsis thaliana. J Plant Physiol 175: 21–25 [DOI] [PubMed] [Google Scholar]
- Zhao H, Ma H, Yu L, Wang X, Zhao J (2012) Genome-wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa L.). PLoS ONE 7: e49210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Wang S, Huang Z, Zhang S, Liao Q, Zhang C, Lin T, Qin M, Peng M, Yang C, et al. (2018) Rewiring of the fruit metabolome in tomato breeding. Cell 172: 249–261.e12 [DOI] [PubMed] [Google Scholar]