Three Auxin Response Factors control hypocotyl elongation in Arabidopsis under environmental conditions that require rapid growth.
Abstract
The hormone auxin regulates growth largely by affecting gene expression. By studying Arabidopsis (Arabidopsis thaliana) mutants deficient in AUXIN RESPONSE FACTORS (ARFs), we have identified three ARF proteins that are required for auxin-responsive hypocotyl elongation. Plants deficient in these factors have reduced responses to environmental conditions that increase auxin levels, including far-red-enriched light and high temperature. Despite having decreased auxin responses, the ARF-deficient plants responded to brassinosteroid and gibberellin, indicating that different hormones can act partially independently. Aux/IAA proteins, encoded by IAA genes, interact with ARF proteins to repress auxin response. Silencing expression of multiple IAA genes increased hypocotyl elongation, suggesting that Aux/IAA proteins modulate ARF activity in hypocotyls in a potential negative feedback loop.
Plant organs grow to their final size by cell expansion. Several plant hormones, including auxin, brassinosteroids, ethylene, and GAs, regulate expansion growth, and environmental signals such as temperature and light can affect abundance of or responsiveness to these hormones (Wolters and Jürgens, 2009; Depuydt and Hardtke, 2011; Leivar and Monte, 2014). Integration of these signals with metabolic signals determines the degree of cell expansion and, hence, the final size of stems, leaves, and other organs.
Hypocotyl elongation after seed germination is sensitive to multiple hormonal and environmental signals, and occurs largely by the expansion of cells produced in the embryo. In Arabidopsis (Arabidopsis thaliana), mutations in genes regulating light perception, hormone response, the circadian rhythm, or transcription can each affect hypocotyl elongation. Many such mutations also affect growth in other organs, indicating that common mechanisms regulate expansion growth in multiple tissues. Thus, the hypocotyl is a useful model tissue in which to explore general control mechanisms of cell expansion.
High temperature and far-red-enriched light stimulate hypocotyl elongation, and the circadian rhythm gates hypocotyl elongation, in part by regulating auxin response. Integrated signals from these and other inputs affect the levels and activity of PHYTOCHROME INTERACTING FACTOR (PIF) transcription factors (Gray et al., 1998; Covington and Harmer, 2007; de Lucas et al., 2008; Feng et al., 2008; Leivar et al., 2008; Lorrain et al., 2008; Koini et al., 2009; Shin et al., 2009; Stavang et al., 2009; Nusinow et al., 2011; Li et al., 2012; Sairanen et al., 2012; Box et al., 2015; Nieto et al., 2015; Soy et al., 2016). PIFs can increase auxin biosynthesis and/or signaling (Franklin et al., 2011; Nozue et al., 2011; Hornitschek et al., 2012; Li et al., 2012; Sun et al., 2012; Goyal et al., 2016), and they can interact with BRASSINAZOLE RESISTANT/BRI1 EMS SUPPRESSOR (BZR/BES) and AUXIN RESPONSE FACTOR (ARF) transcription factors, which regulate responses to brassinosteroid and auxin (Oh et al., 2014). GAs also can modulate PIF activity and stability (de Lucas et al., 2008; Feng et al., 2008; Li et al., 2016). PIF, BZR/BES, and ARF transcription factors share many target genes that promote cell expansion, and may activate some of these genes cooperatively (Oh et al., 2014).
ARFs bind to conserved DNA sequence elements in auxin-responsive promoters (Ulmasov et al., 1997, 1999a, 1999b; Boer et al., 2014). Class II ARFs have Q-rich middle domains that can activate gene expression. Conversely, the same ARF proteins can repress gene expression by interacting with Aux/IAA proteins that recruit histone deacetylase complexes (Tiwari et al., 2004; Szemenyei et al., 2008; Vernoux et al., 2011; Piya et al., 2014; Guilfoyle, 2015). Auxin-stimulated turnover of Aux/IAA proteins switches gene-repressing ARF-Aux/IAA complexes to gene-activating ARF complexes (Salehin et al., 2015). In addition, class I ARF proteins lacking the Q-rich middle domain can repress gene expression, possibly independently of recruiting Aux/IAA proteins (Ulmasov et al., 1999a; Tiwari et al., 2003; Ellis et al., 2005; Vernoux et al., 2011; Piya et al., 2014).
Loss-of-function mutations in several Arabidopsis arf genes decrease auxin-induced gene expression and cause growth defects associated with decreased auxin response. For example, arf6 and arf8 mutations decrease inflorescence stem and flower organ growth (Nagpal et al., 2005; Tabata et al., 2010; Reeves et al., 2012), and nph4/arf7 and arf19 mutations decrease leaf cell expansion and lateral root formation (Okushima et al., 2005; Wilmoth et al., 2005). nph4/arf7 arf19 double mutants also have decreased sensitivity to root growth inhibition by auxin. Similarly, gain-of-function iaa mutations that reduce auxin-induced turnover of Aux/IAA proteins also inhibit auxin-responsive gene induction (Timpte et al., 1994; Nagpal et al., 2000; Gray et al., 2001; Tian et al., 2002; Overvoorde et al., 2005). Many of these mutations cause phenotypes similar to those of loss-of-function arf mutations.
Several gain-of-function iaa mutations affect hypocotyl elongation. shy2/iaa3-2, shy2/iaa3-3, and axr2/iaa7-1 gain-of-function mutations cause short hypocotyls (Timpte et al., 1992; Reed et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000; Chapman et al., 2012), suggesting that interacting ARF proteins normally promote hypocotyl elongation. However, whereas an arf6 arf8 double mutant had somewhat shorter hypocotyls than did wild-type plants in darkness (Nagpal et al., 2005), arf8 single mutant light-grown seedlings had longer hypocotyls than did wild-type seedlings (Tian et al., 2004). Similarly, the axr3-1 mutation stabilized AXR3/IAA17 protein and caused accelerated hypocotyl elongation at early time points, although the final hypocotyl length was less than that of wild-type seedlings (Leyser et al., 1996; Nagpal et al., 2000) and mutations that stabilize IAA18 increased hypocotyl elongation (Uehara et al., 2008; Ploense et al., 2009). These contrasting phenotypes suggest that quantitative or tissue-specific aspects of ARF activity may influence the kinetics and extent of hypocotyl growth, and that additional redundancy may exist among the ARF genes that promote elongation.
To explore how ARF proteins regulate hypocotyl elongation, we characterized hypocotyl growth in higher order mutants deficient in multiple class II ARF genes known to affect cell expansion and in plants deficient in multiple IAA genes. We found that three ARF proteins together have a major role in controlling hypocotyl elongation downstream of auxin, high temperature, far-red enrichment, and circadian regulation; that other hormone response pathways act partially independently of these ARF proteins; and that Aux/IAA proteins repress hypocotyl elongation.
RESULTS
ARF6, NPH4/ARF7, and ARF8 Promote Hypocotyl Elongation
The class II ARF proteins ARF6 (At1g30330), NPH4/ARF7 (At5g20730), ARF8 (At5g37020), and ARF19 (At1g19220) promote cell expansion in leaf, inflorescence, and/or flower organs (Nagpal et al., 2005; Wilmoth et al., 2005). These four ARF proteins form a clade together with MP/ARF5 (At1g19850; Remington et al., 2004), can each mediate auxin-induced gene expression (Ulmasov et al., 1999a; Tiwari et al., 2003; Wilmoth et al., 2005), and interact strongly with Aux/IAA proteins in yeast two-hybrid assays (Vernoux et al., 2011; Piya et al., 2014). ARF6, NPH4/ARF7, and ARF8 were each expressed in dissected hypocotyls (Chapman et al., 2012; Kohnen et al., 2016).
We assessed hypocotyl growth in seedlings with combinations of arf6, nph4/arf7, arf8, and arf19 null mutations. We did not include mp/arf5 mutants in these experiments because these have defective vasculature and lack a hypocotyl entirely (Berleth and Jürgens, 1993). We also silenced ARF6 and ARF8 by driving the expression of the microRNA precursor MIR167a with the broadly expressed PRPS5a:GAL4-VP16 transactivation line (Weijers et al., 2005). The mature microRNA miR167 targets ARF6 and ARF8 transcripts for turnover, and overexpressing MIR167a behind the P35S viral promoter confers phenotypes very similar to those of arf6-2 arf8-3 mutant plants (Wu et al., 2006). In most experiments, transactivated PRPS5a>MIR167a F1 seedlings had hypocotyl lengths and adult phenotypes indistinguishable from those of arf6-2 arf8-3 plants, and nph4 PRPS5a>MIR167a and arf6-2 nph4 arf8-3 genotypes had phenotypes indistinguishable from each other (Supplemental Figs. S1, A–D, and S2, B–D and G). PRPS5a>MIR167a and nph4 PRPS5a>MIR167a F1 plants were used for many experiments below because they did not require genotyping of individual seedlings in populations segregating for the sterile arf6-2 arf8-3 mutant combination.
Dark-grown seedlings lacking combinations of ARF6, NPH4/ARF7, and ARF8 had shorter hypocotyls than did wild-type seedlings (Fig. 1, A and B, control data; Supplemental Fig. S1A). Hypocotyl lengths of single mutants were 80% to 90% of wild-type hypocotyl lengths, those of plants lacking two of these ARFs were 50% to 80% of wild-type hypocotyl lengths, and those of plants deficient in all three ARFs were 20% to 30% of wild-type hypocotyl lengths. As found previously, dark-grown nph4-1 seedlings had partially open apical hooks (Stowe-Evans et al., 1998). The arf19-4 mutation did not affect elongation in the nph4-1 or arf6-2 nph4-1 arf8-3 background (Supplemental Fig. S1, A and C). Thus, arf6-2 nph4-1 arf8-3 triple and arf6-2 nph4-1 arf8-3 arf19-4 quadruple mutants had equally short hypocotyls (of 2–3 mm [i.e. 1–2 mm growth from the mature embryo]; Supplemental Fig. S1C). These results indicate that, together, ARF6, NPH4/ARF7, and ARF8 control hypocotyl elongation in darkness. Therefore, we focused further experiments on plants deficient in these three ARFs.
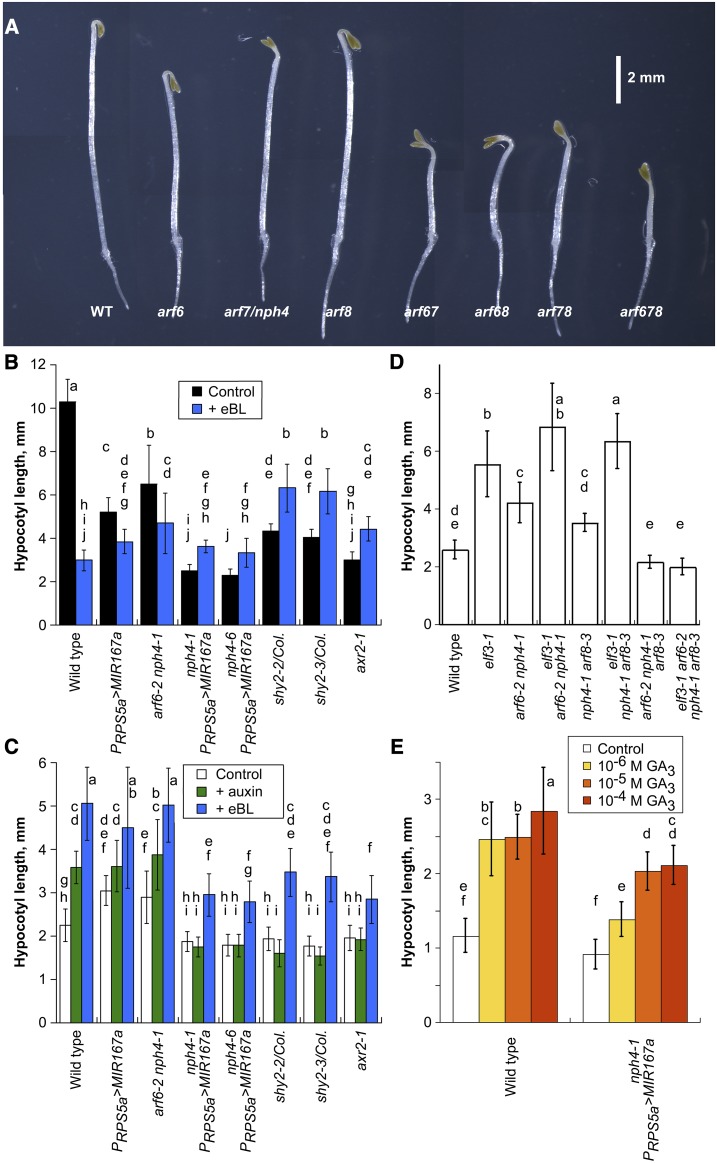
Figure 1.
Hypocotyl lengths of seedlings deficient in ARFs. A, Three-day-old dark-grown seedlings of the indicated genotypes. Transactivation of MIR167a was used for the arf6 arf8 and arf6 arf7 arf8 combinations. WT, Wild type. Bar = 2 mm. B, Hypocotyl lengths of 3-d-old dark-grown seedlings of the indicated genotypes in the absence or presence of 1 µm epibrassinolide (eBL). n = 16 to 27. Supplemental Figure S1 shows data for additional genotypes without brassinolide. C, Hypocotyl lengths of 4-d-old short-day-grown seedlings of the indicated genotypes grown in the absence or presence of the synthetic auxin 30 nm 533 or 1 µm eBL. n = 17 to 26. Supplemental Figure S1 shows data for additional genotypes without added hormones. D, Effects of elf3-1 on hypocotyl lengths of 4-d-old short-day-grown seedlings. n = 15 to 103. E, Response to GA3. The GA biosynthesis inhibitor paclobutrazol was included on the plates at 1 µm to decrease endogenous GA production. Seeds were subjected to 6 d of cold treatment before plating to ensure germination even in the absence of new GA synthesis or exogenous GA. n = 12 to 31. In B to E, data are means ± sd, and lowercase letters indicate statistically distinguishable values as assessed by Tukey’s honestly significant difference test.
Dark-grown shy2-2, shy2-3, and axr2-1 seedlings, with mutations that stabilize the Aux/IAA proteins SHY2/IAA3 or AXR2/IAA7 (Timpte et al., 1992; Reed et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000), had hypocotyls either the same length as or slightly longer than those of nph4 PRPS5a>MIR167a seedlings (Fig. 1B). These results suggest that the stabilized Aux/IAA proteins in these mutants inhibit gene activation by ARF6, NPH4/ARF7, and ARF8 in hypocotyls, albeit incompletely in some cases.
Partial ARF Deficiency Increases Hypocotyl Elongation in Light-Grown Seedlings
In diurnal conditions, wild-type hypocotyls elongate much less than in darkness. When grown in short days at 22°C, arf6 nph4 arf8 triple mutant, arf6 nph4 arf8 arf19 quadruple mutant, nph4 PRPS5a>MIR167a, shy2-2, shy2-3, and axr2-1 seedlings had hypocotyls only slightly shorter than, and often not statistically distinguishable from, those of wild-type seedlings (typically about 2 mm after 4 d; Fig. 1C, control data; Supplemental Fig. S1, B and D). However, even in these diurnal cycles, these mutants had shorter hypocotyls than did wild-type seedlings when subjected to some hormone treatments or environmental conditions that increase wild-type hypocotyl growth (see below).
Whereas seedlings grown in darkness had hypocotyl lengths corresponding to their ARF6/7/8 gene dosage, short-day-grown seedlings deficient in one or two of these ARFs often had slightly longer hypocotyls than did wild-type seedlings (Fig. 1C; Supplemental Fig. S1B). These results indicate that, although ARF6, NPH4/ARF7, and ARF8 together promote hypocotyl elongation, under day/night cycles, greater elongation occurred when ARF activity was partially compromised by knocking out just one or two of these genes. Thus, the same ARF proteins also can inhibit growth under these conditions.
We used time-lapse imaging to determine the diurnal stage at which ARF proteins may inhibit growth. In short days, hypocotyls of wild-type plants grew in the latter part of the night and early in the day, with the growth rate peaking at about dawn 2, 3, and 4 d after transfer from cold to growth conditions (Supplemental Fig. S3A; Nozue et al., 2007; Michael et al., 2008; Stewart et al., 2011). nph4-1 arf8-3 seedlings also grew most quickly at these stages, but they had a higher maximum hypocotyl growth rate and longer growth window than wild-type seedlings during the second period of rapid growth 3 d after transfer to growth conditions (Supplemental Fig. S3A).
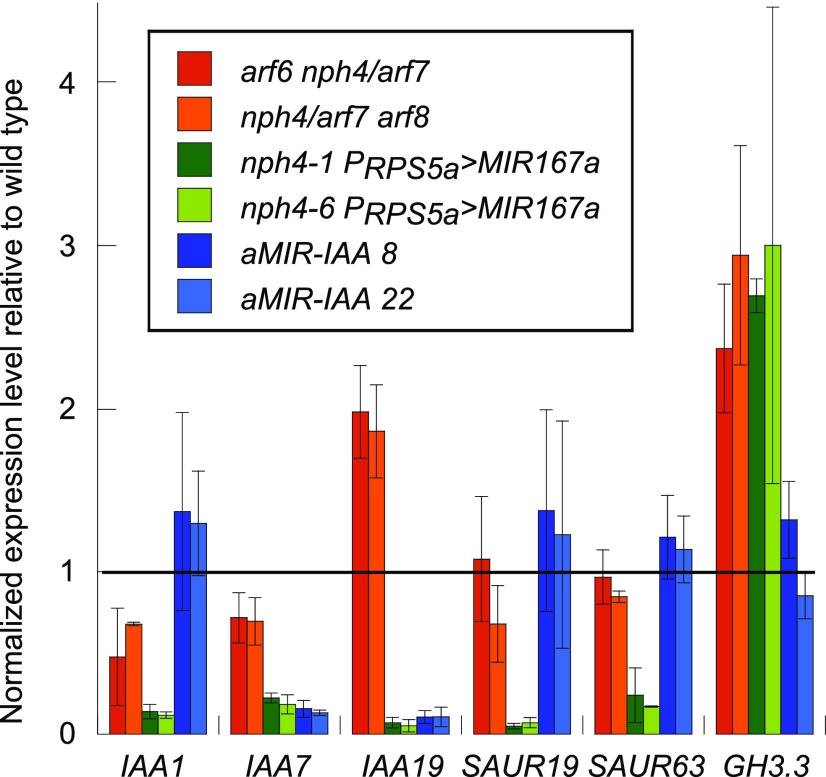
To explore the regulatory basis for these phenotypes, we assayed the expression levels of several auxin-responsive genes in ARF-deficient seedlings grown in short days. nph4 PRPS5a>MIR167a seedlings had decreased expression of the auxin-induced genes IAA1, AXR2/IAA7, IAA19, SAUR19 (SMALL AUXIN UP-RNA19), and SAUR63 (Fig. 2), consistent with an overall decrease in auxin response. SAUR19 and SAUR63 promote hypocotyl elongation (Chae et al., 2012; Spartz et al., 2012), whereas the IAA genes encode Aux/IAA proteins that can inhibit ARF action. These results indicate that these three ARF proteins are required for expression both of growth outputs mediated by SAUR proteins and of Aux/IAA-mediated negative feedback. The At2g23170/GH3.3 gene, which also is auxin-inducible and encodes an auxin-amino acid-conjugating enzyme, was overexpressed in nph4 PRPS5a>MIR167a seedlings, and may have been activated independently of auxin response. In arf6-2 nph4-1 and nph4-1 arf8-3 seedlings with partial ARF deficiency, expression of IAA1 and AXR2/IAA7 was reduced slightly, although to a much lesser degree than in nph4 PRPS5a>MIR167a seedlings. In contrast, MSG2/IAA19 was expressed at about twice the level as in wild-type seedlings (Fig. 2). SAUR19 and SAUR63 were expressed at similar levels in wild-type, arf6-2 nph4-1, and nph4-1 arf8-3 seedlings.
Figure 2.
Gene expression in mutant seedlings. Real-time quantitative PCR (RT-qPCR) of the indicated genes is shown normalized to the UBQ transcript level. Tissue was harvested in the morning, 1 to 2 h after dawn. The horizontal line indicates normalized wild-type expression level of 1 for each gene. n = 2 to 3 per genotype. Error bars indicate sd.
ARF Proteins Are Required for Auxin-Induced Hypocotyl Elongation
We explored seedling hypocotyl elongation responses to the synthetic auxin 533, which promoted growth more strongly than did the natural auxin IAA (Savaldi-Goldstein et al., 2008). At the most effective dose (30 nm), short-day-grown wild-type seedling hypocotyl growth was stimulated by about 75% (Fig. 1C; Supplemental Fig. S4A). In contrast, auxin did not increase hypocotyl lengths of nph4 PRPS5a>MIR167a, axr2-1, or shy2 seedlings (Fig. 1C; Supplemental Fig. S4A).
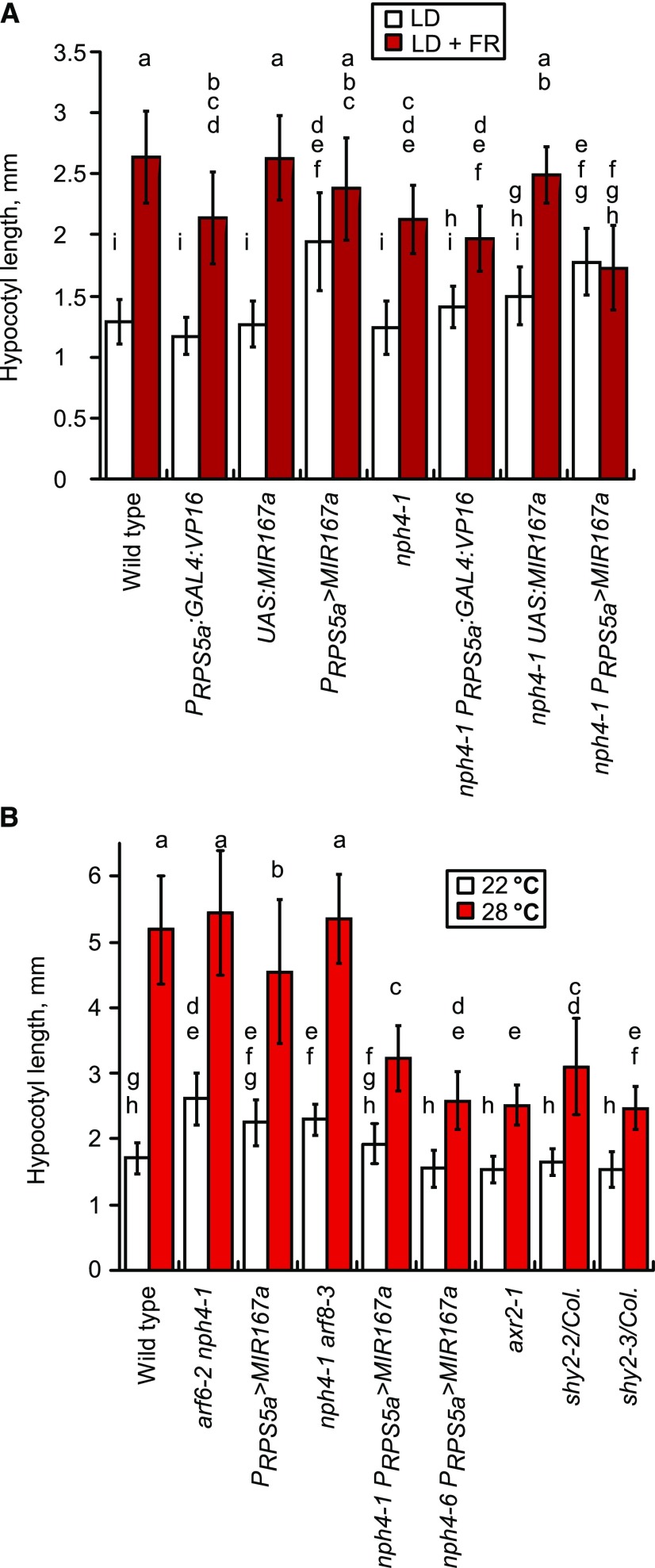
Both shade light simulated by far-red enrichment, which creates a low red:far red (R:FR) ratio, and high growth temperature increase hypocotyl elongation, in part by inducing changes in auxin levels (Gray et al., 1998; Tao et al., 2008; Keuskamp et al., 2010; Franklin et al., 2011). Indeed, nph4 PRPS5a>MIR167a and arf6-2 nph4-1 arf8-3 seedlings lacked a hypocotyl elongation response to low R:FR light (Fig. 3A; Supplemental Fig. S4B). When grown at 28°C, nph4 PRPS5a>MIR167a, shy2, and axr2-1 seedlings increased hypocotyl elongation less than half as much as did wild-type seedlings compared with growth at 22°C, although they did retain some high-temperature response (Fig. 3B).
Figure 3.
Hypocotyl elongation responses to treatments that affect auxin levels. A, Hypocotyl growth response to low R:FR light. LD, long days; LD + FR, long days with supplemental far-red light. n = 15 to 16. B, Hypocotyl growth after 4 short days at 22°C or 28°C. n = 17 to 29. Data are means ± sd, and lowercase letters indicate statistically distinguishable values as assessed by Tukey’s honestly significant difference test.
Defects in the evening protein complex cause increased hypocotyl elongation in short days because of altered circadian rhythm outputs and are thought to arise from increased PIF expression and resultant auxin biosynthesis (Zagotta et al., 1996; Nozue et al., 2011; Nusinow et al., 2011). The elf3-1 mutant is deficient in a component of the evening complex and also may affect PIF4 protein activity directly (Nieto et al., 2015). Consistent with the model that auxin response is required for the elf3-1 effect on hypocotyl elongation, hypocotyls of an elf3-1 nph4-1 arf6-2 arf8-3 quadruple mutant were as short as those of the nph4-1 arf6-2 arf8-3 triple mutant (Fig. 1D). However, the elf3-1 arf6-2 nph4-1 arf8-3 quadruple mutant flowered earlier and had smaller leaves than the arf6-2 nph4-1 arf8-3 triple mutant (Supplemental Fig. S2, E and F), indicating that ELF3 can act independently of these ARF proteins at later stages of development.
In these assays, lower order arf mutants retained some response. Thus, exogenous auxin and low R:FR light each stimulated hypocotyl elongation of PRPS5a>MIR167a, nph4 arf6, and nph4 arf8 seedlings, in most cases to an extent similar to that seen in the wild type (Figs. 1C and 3A; Supplemental Fig. S4, A and B). Similarly, high temperature stimulated hypocotyl elongation in arf double mutants to a greater extent than in the triple mutant (Fig. 3B), and elf3-1 enhanced the long-hypocotyl phenotype of arf6-2 nph4-1 and nph4-1 arf8-3 double mutant combinations (Fig. 1D).
In a recent transcriptomic study, elf3-1 seedlings overexpressed a number of auxin-regulated genes, including SAUR63 (Ezer et al., 2017). elf3-1 SAUR63:GUS seedlings, with a gain-of-function form of the growth-promoting protein SAUR63 driven by its native auxin-inducible promoter (Chae et al., 2012), had hypocotyls longer than either elf3-1 or SAUR63:GUS seedlings, about 3 times the length of wild-type hypocotyls (Supplemental Fig. S1E).
GA and Brassinosteroid Can Act Independently of ARF6, NPH4/ARF7, and ARF8
Although hypocotyls of seedlings deficient in ARF6, NPH4/ARF7, and ARF8 failed to elongate in response to exogenous auxin, they did elongate relative to their length in the embryo, and they elongated more in darkness than in short days, indicating that additional growth-promoting pathways were still active. The hormones brassinolide and GAs each can promote hypocotyl elongation. Exogenous eBL and GA3 each increased elongation of short-day-grown nph4-1 PRPS5a>MIR167a seedling hypocotyls, although to a lesser extent than in the wild type (Fig. 1, C and E). eBL also increased hypocotyl elongation of shy2-2, shy2-3, and axr2-1 seedlings. Conversely, the brassinosteroid biosynthesis inhibitor brassinazole (Asami et al., 2000) and the GA biosynthesis inhibitor paclobutrazol (Rademacher, 2000) each decreased hypocotyl elongation of these seedlings in short days (Fig. 1E, note the short hypocotyl lengths in the absence of GA3; Supplemental Fig. S4D). While these inhibitors may have delayed germination, which could have decreased hypocotyl elongation at the single time points measured in these assays, taken together these results suggest that both brassinosteroid and GA response pathways are active in the ARF-deficient hypocotyls.
Whereas the concentration of exogenous eBL we used promotes hypocotyl elongation in light-grown seedlings, it inhibits elongation in wild-type dark-grown seedlings. eBL also inhibited hypocotyl elongation in dark-grown PRPS5a>MIR167a and nph4-1 arf6-2 seedlings deficient in two ARF genes. However, eBL stimulated elongation in dark-grown nph4 PRPS5a>MIR167a, shy2, and axr2-1 seedlings (Fig. 1B). Conversely, dark-grown nph4 PRPS5a>MIR167a had shorter hypocotyls when grown in the presence of the inhibitor brassinazole than in its absence (Supplemental Fig. S4C).
ARF6, NPH4/ARF7, and ARF8 Affect Leaf and Inflorescence Growth
arf6 nph4 arf8 triple mutant, nph4 PRPS5a>MIR167a, and arf6 nph4 arf8 arf19 quadruple mutant plants also had more severe defects in rosette leaf growth and inflorescence stem elongation than did the lower order mutants (Supplemental Figs. S2, B–D, and S5). In these respects, the triple and quadruple mutants resembled gain-of-function axr2-1 or shy2 plants having increased AXR2/IAA7 or SHY2/IAA3 protein level (Wilson et al., 1990; Tian and Reed, 1999). However, arf6 nph4 arf8 and nph4 PRPS5a>MIR167a inflorescence stems grew upright, unlike the agravitropic inflorescences of axr2-1 plants. arf6 nph4 arf8 and nph4 PRPS5a>MIR167a plants had closed flower buds resembling those of arf6 arf8 or PRPS5a>MIR167a plants, and with similar final flower organ lengths (Nagpal et al., 2005; Supplemental Fig. S2, B–D and G). Thus, ARF6, NPH4/ARF7, ARF8, and ARF19 each promotes growth in leaves and inflorescences, whereas ARF6 and ARF8 are the main drivers of rapid organ growth in opening flowers.
Wild-Type Aux/IAA Proteins Inhibit Hypocotyl Elongation
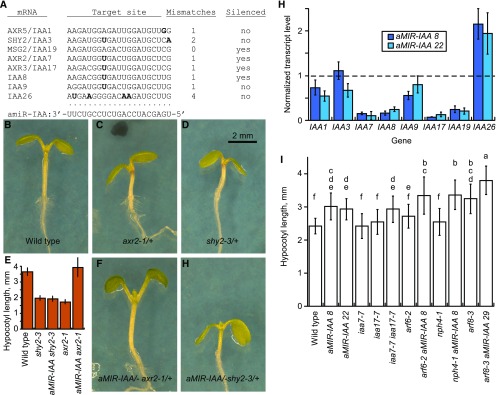
ARF6, NPH4/ARF7, and ARF8 have gene activation activity but can be converted to repressors by multimerization with Aux/IAA proteins (Ulmasov et al., 1999a; Tiwari et al., 2003; Han et al., 2014; Korasick et al., 2014; Nanao et al., 2014; Dinesh et al., 2015). Auxin relieves this repression by stimulating turnover of Aux/IAA proteins. To assess how IAA genes regulate hypocotyl elongation, we generated plants overexpressing an artificial microRNA designed to target multiple IAA genes (Fig. 4A). We tested expression of eight representative IAA genes by RT-qPCR in 4-d-old seedlings of two stable single-locus P35S:aMIR-IAA lines. AXR2/IAA7, IAA8, AXR3/IAA17, and MSG2/IAA19 transcript levels were each lower in P35S:aMIR-IAA than in wild-type seedlings (Figs. 2 and 4H). In contrast, AXR5/IAA1, SHY2/IAA3, IAA9, and IAA26 transcript levels were similar between wild-type and P35S:aMIR-IAA seedlings. The apparent increase in IAA26 expression could potentially reflect a compensatory negative feedback leading to increased expression of some IAA genes. Consistent with these expression data, P35S:aMIR-IAA suppressed the morphological phenotypes of the axr2-1 gain-of function mutation in AXR2/IAA7 but did not appreciably suppress the phenotypes of the shy2-3 mutation in SHY2/IAA3 (Fig. 4, C–G). The growth-promoting genes SAUR19 and SAUR63 had normal expression levels in aMIR-IAA seedlings (Fig. 2).
Figure 4.
aMIR-IAA plants. A, Predicted amiR-IAA sequence base pairing to selected IAA genes. G:U base pairs are allowed. IAA gene mRNA sequences are shown 5′ to 3′. Bases not expected to pair with aMIR-IAA are indicated in boldface. B to D, F, and G Photographs of 7-d-old seedlings of the indicated genotypes. E, Mean hypocotyl lengths of the seedlings shown in B to D, F, and G, showing suppression of axr2-1 but not shy2-3. n = 9 to 13. Error bars indicate sd. H, RT-qPCR of IAA genes in two different P35S:aMIR-IAA lines. n = 3. Error bars indicate sd. The horizontal dashed line indicates a normalized wild-type expression level of 1 for each gene. I, Hypocotyl lengths in short days. Error bars indicate sd. n = 13 to 46.
In the wild-type, arf6-2, nph4-1, and arf8-3 backgrounds, P35S:aMIR-IAA increased hypocotyl elongation in short days, indicating that Aux/IAA proteins were inhibiting elongation (Fig. 4I). Time-lapse measurements of growth indicated that the increase in growth rate occurred primarily surrounding dawn 3 d after transfer from cold to growth conditions, as was observed for nph4/arf7-1 arf8-3 (Supplemental Fig. S3B). P35S:aMIR-IAA seedlings did elongate in response to exogenous auxin (Supplemental Fig. S4A), suggesting that additional Aux/IAA proteins are present and could regulate ARFs in these plants.
We found previously that the axr2/iaa7-5 loss-of-function allele in ecotype Wassilewskija had a slightly longer hypocotyl than did wild-type plants (Nagpal et al., 2000). Unlike the Wassilewskija mutant, the Columbia mutant axr2/iaa7-7 had a similar hypocotyl length to wild-type seedlings when grown in short days. However, hypocotyls of axr2/iaa7-7 axr3/iaa17-7 double mutant seedlings were longer than those of wild-type seedlings and as long as those of aMIR-IAA seedlings (Fig. 4I). Thus, decreased AXR2/IAA7 and AXR3/IAA17 activity can account for the increased hypocotyl growth of aMIR-IAA seedlings, although decreased levels of other Aux/IAA proteins also may contribute.
DISCUSSION
Our results show that ARF6, NPH4/ARF7, and ARF8 together account for the hypocotyl elongation response to auxin. They likely activate genes in hypocotyl epidermal cells, including SAUR genes, cell wall-loosening functions, and genes acting in GA and brassinosteroid biosynthesis or signaling pathways (Chapman et al., 2012; Procko et al., 2016).
ARF-regulated hypocotyl elongation appears to be most important under specific environmental conditions. For example, ARF-deficient seedlings grew very little in darkness, lacked the shade-avoidance response to far-red enrichment, and had a decreased response to high temperature. Consistent with these results, both far-red-enriched light and high temperature can increase endogenous auxin production in cotyledons and increase hypocotyl elongation (Gray et al., 1998; Tao et al., 2008; Koini et al., 2009; Franklin et al., 2011; Li et al., 2012; Sun et al., 2012; Zheng et al., 2016). In contrast, ARF-deficient seedlings had almost normal hypocotyl growth in diurnal cycles at 22°C under fluorescent light. Thus, for hypocotyl elongation, the auxin response might act mainly under conditions that cause particularly rapid growth. Similarly, ARF6 and ARF8 promote petal and stamen filament growth just before flowers open, when these organs elongate especially quickly (Nagpal et al., 2005; Tabata et al., 2010; Reeves et al., 2012).
As the arf mutants could respond to both brassinosteroid and GA, it is likely that the responses to these two hormones are adequate for baseline hypocotyl growth under diurnal conditions. ARF6 can interact with PIF4 and BZR1 proteins, suggesting that, at many promoters, complexes of BES/BZR, ARF, and PIF proteins together activate genes required for growth (Oh et al., 2014). Indeed, ARF-deficient seedlings responded less to eBL or GA3 than did wild-type seedlings, suggesting that full BZR/BES and PIF effects may require auxin response (Nemhauser et al., 2004; Zhou et al., 2013). Conversely, ARFs are probably not sufficient on their own to activate maximal hypocotyl elongation in the absence of PIF and BES/BZR factors, because higher order bes/bzr and pif mutants also have short hypocotyls in darkness and in diurnal conditions (Yin et al., 2005; Leivar et al., 2008, 2012; Shin et al., 2009), and inhibition of brassinosteroid or GA biosynthesis also reduces hypocotyl elongation.
Brassinolide inhibited hypocotyl elongation of wild-type seedlings in darkness but promoted elongation in light. In contrast, it promoted elongation of hypocotyls of ARF-deficient seedlings grown under either condition. Exogenous brassinolide or increased brassinolide signaling also promoted hypocotyl elongation in GA-deficient seedlings (Bai et al., 2012; Gallego-Bartolomé et al., 2012). In each of these cases, when the hypocotyls were short, brassinolide promoted elongation. These results raise the possibility that the brassinolide response may depend on the growth state of the seedling rather than on the light environment per se.
ARFs also were required for the long hypocotyl growth in elf3-1 seedlings deficient in the evening complex. Epistasis of the nph4-1 arf6-2 arf8-3 triple mutation combination to the elf3-1 mutation for hypocotyl elongation is consistent with the model that elf3-1 acts by increasing auxin response in hypocotyls (Nozue et al., 2011; Nusinow et al., 2011).
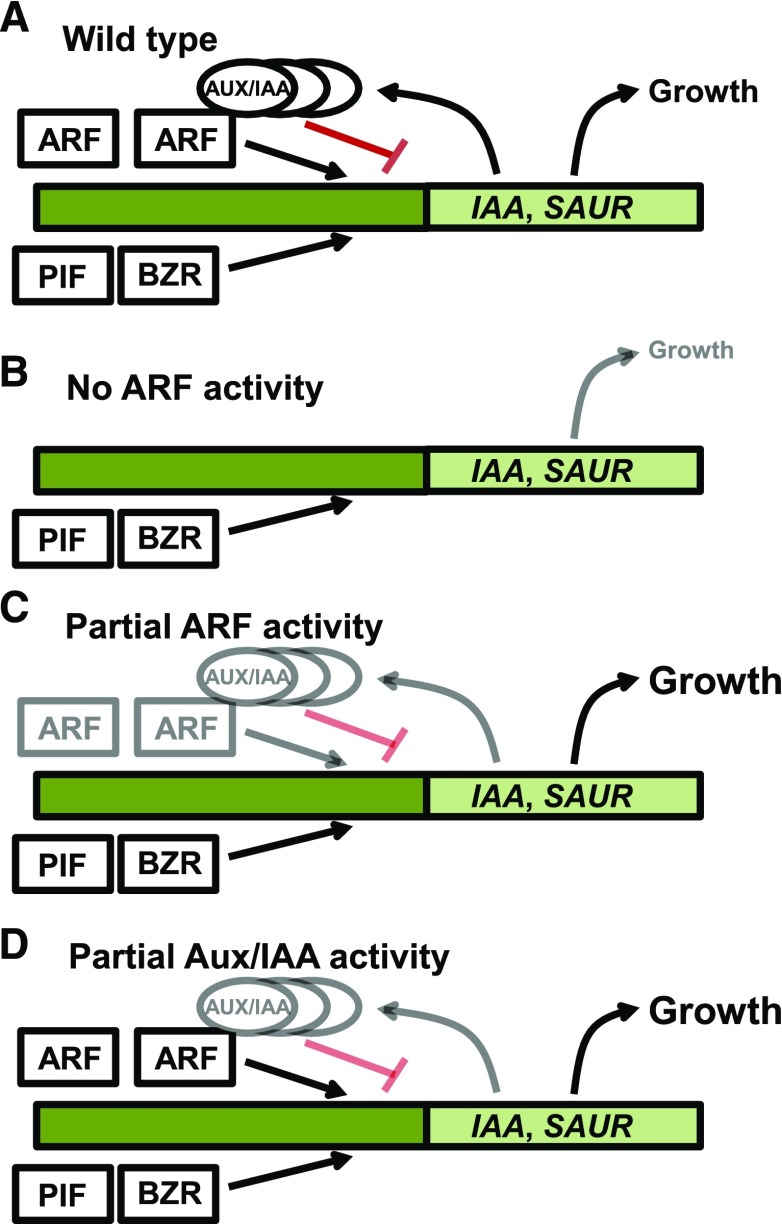
Seedlings lacking combinations of just two of the three ARFs and seedlings deficient in multiple IAA genes each had longer hypocotyls than did wild-type seedlings in short days. These results support the model that ARF-Aux/IAA protein complexes may normally limit expression of growth-promoting genes under diurnal conditions (Fig. 5). The C-terminal domains of ARF6, NPH4/ARF7, and ARF8 each can interact strongly with Aux/IAA proteins (Vernoux et al., 2011; Piya et al., 2014). Under conditions of increased auxin level, turnover of Aux/IAA proteins would normally convert these repressing ARF-Aux/IAA complexes to activating ARF complexes. Partial ARF deficiency may increase expression of growth-promoting target genes through a combination of decreased ARF-Aux/IAA complex repression and increased recruitment or activity of PIF, BZR/BES, or other activating factors that the missing ARF-Aux/IAA complexes would normally counteract (Fig. 5C). arf double mutants with slightly elongated hypocotyls had slightly increased or decreased levels of different IAA gene transcripts, suggesting that feedback-regulated IAA expression levels may buffer changes in ARF levels. Such feedback also may rewire auxin responsiveness under persistent shade conditions, leading to altered steady-state levels of IAA gene transcription (Pucciariello et al., 2018). The mechanisms may also involve genes not assayed here, or more subtle changes in the kinetics or tissue-specific expression of relevant target genes. Partial ARF deficiency can cause increased growth in both hypocotyls and fruits (Tian et al., 2004; Goetz et al., 2006; de Jong et al., 2009).
Figure 5.
Conceptual model for ARF-Aux/IAA control of hypocotyl growth. A, In wild-type seedlings, ARF, PIF, and BZR/BES transcription factors regulate expression of growth-promoting genes such as SAUR genes and of IAA genes encoding Aux/IAA repressors of transcription. When the auxin level is high, Aux/IAA proteins are turned over and ARF transcription factors can activate gene expression. When auxin levels are low, ARF-Aux/IAA protein complexes are more persistent and repress gene expression. B, In the absence of ARF6, NPH4/ARF7, and ARF8, elongation depends primarily on PIF and BZR/BES factors and is reduced in darkness and other conditions that require an auxin response. C, An intermediate ARF level, as in arf double mutants, may decrease both gene activation by ARF transcription factors and gene repression by ARF-Aux/IAA complexes. Levels of IAA gene transcription may be increased or decreased depending on the gene. These combined effects produce an altered regulatory balance. In darkness, this decreases growth, whereas in diurnal conditions, the balance of gene expression favors slightly increased growth. Additional feedbacks and non-cell-autonomous effects such as PIF regulation of auxin biosynthesis genes likely also contribute to the final output. D, A reduced Aux/IAA level, as in aMIR-IAA plants, decreases repression by ARF-Aux/IAA complexes, leading to increased ARF activation of SAUR genes and slightly increased growth.
The elongated hypocotyls of axr2/iaa7 axr3/iaa17 double loss-of-function mutants are consistent with AXR2/IAA7 and AXR3/IAA17 each regulating ARF activity in hypocotyls (Fig. 5D). However, these seedlings still responded to auxin, so other IAA genes may also contribute to hypocotyl growth control. New genome-engineering technologies may make it feasible to knock out more IAA genes at once to reveal more fully their roles in growth control and the integration of environmental and stress stimuli (Shani et al., 2017). Mutants of the moss Physcomitrella patens lacking multiple IAA genes have a constitutively active auxin response (Lavy et al., 2016).
Collectively, ARF6, NPH4/ARF7, ARF8, and ARF19 also regulate growth of inflorescence stems, leaves, and flower organs. Moreover, the phenotypes of higher order arf mutants resemble many of those seen for mutants with gain-of-function mutations in IAA genes that decrease the turnover of the corresponding Aux/IAA protein (Reed, 2001). Our results here thus validate the notions that those iaa mutant phenotypes arose primarily from inhibition of ARF-mediated gene activation, and that use of such mutant IAA genes as tools in ectopic expression studies reveals functions of ARFs in affected tissues (Procko et al., 2016). The MIR167a transactivation system described here will also be a useful tool for further studies of auxin responses and of brassinolide or GA responses in the absence of confounding auxin responses.
MATERIALS AND METHODS
Plant Genotypes
All Arabidopsis (Arabidopsis thaliana) genotypes were in the Columbia background. Mutations arf6-2, arf8-3, arf19-4, axr2-1, elf3-1, nph4-1, and nph4-6 have been described previously (Wilson et al., 1990; Zagotta et al., 1996; Harper et al., 2000; Nagpal et al., 2000, 2005; Wilmoth et al., 2005). shy2-2 and shy2-3 mutations were originally identified in Landsberg erecta (Tian and Reed, 1999) and were introgressed for eight and nine generations, respectively, into Columbia for experiments in this work. axr2/iaa7-7 (SALK_089809) and axr3/iaa17-7 (SALK_011820; Alonso et al., 2003; http://signal.salk.edu/cgi-bin/tdnaexpress) were obtained from the Arabidopsis Biological Resource Center at Ohio State University. Plants expressing the GAL4:VP16 fusion gene encoding a strong transcriptional activator behind the PRPS5a promoter were described previously (Weijers et al., 2003). PRPS5a:GAL4:VP16 driver line 5 conferred stronger phenotypes than line 10 when crossed with UAS:MIR167a responder lines. UAS:MIR167a was constructed by inserting the MIR167a sequence (Wu et al., 2006) into pSDM7006 (Weijers et al., 2003), and the resulting construct was transformed into Columbia plants by the floral dip method (Clough, 2005). We characterized two single-locus UAS:MIR167a insertion lines and found that line 9 gave stronger phenotypes than line 2 when crossed with PRPS5a:GAL4:VP16 driver lines. F1 progeny of crosses between the stronger driver and responder lines were used for the experiments presented. We refer to these F1 plants carrying both transgenes as PRPS5a>MIR167a. These transgenes were crossed into nph4 mutant backgrounds, and nph4 PRPS5a:GAL4:VP16 driver and nph4 UAS:MIR167a responder lines were then crossed to each other to obtain nph4 PRPS5a>MIR167a seedlings.
The artificial microRNA sequence against IAA genes was engineered in the MIR167a precursor backbone (Wu et al., 2006) by site-directed mutagenesis using primers listed in Supplemental Table S1 to replace the miR167 and miR167* sequences with amiR-IAA and amiR-IAA* sequences. aMIR-IAA then was cloned into pB2GW7 behind the P35S promoter (Karimi et al., 2002), and transgenic lines were generated by floral dip.
Phenotypic Analyses
For hypocotyl length measurements, Arabidopsis seedlings were grown on petri plates containing 0.5× Murashige and Skoog salts (Murashige and Skoog, 1962) and 0.6% (w/v) phytoagar (Research Products International) without Suc. Hormones or inhibitors were diluted from stocks dissolved in DMSO (533), ethanol (GA3 or brassinazole), or methanol (paclobutrazol) and added to plates when they were poured. Surface-sterilized seeds were given 1 to 2 d of cold treatment before incubation under growth conditions. Plates were oriented vertically in a growth chamber at 22°C or 28°C. For dark growth experiments, seedlings were first exposed to light for 6 to 18 h to induce germination, and hypocotyls were measured after 3 d of growth in darkness at 22°C. For short-day conditions, plates were incubated under an 8-h-light/16-h-dark photoperiod (100–120 µmol m−2 s−2), and hypocotyl lengths were measured after 4 d unless indicated otherwise. Under diurnal conditions, most growth was achieved after 4 d (Supplemental Fig. S3). Images of seedlings were scanned, and hypocotyl lengths were then measured using ImageJ (Abramoff et al., 2004). For shade-avoidance assays, seeds were sown directly onto soil (Primasta potting substrate) and stratified for 4 d at 4°C in darkness. Seedlings were then grown for 4 d in a 16/8-h (light/dark) photoperiod at 130 µmol m−2 s−1 white light (R:FR ratio = 1.8; Philips HPI 400 W) before transfer to low R:FR light (R:FR ratio = 0.15; Philips HPI 400 W supplemented with Philips Green Power FR LED research modules) or control white light conditions and measured after a further 3 d. In cases where segregating populations were assayed, seedling genotypes were assessed subsequently either by diagnostic PCR assays or by the characteristic closed-flower-bud phenotype of arf6 arf8 mutant plants (Nagpal et al., 2005).
For time-lapse imaging of hypocotyl elongation, seedlings were grown on vertically oriented 1× Murashige and Skoog plates and imaged over time using near-infrared image detection following Brooks et al. (2010) with slight modifications. In particular, images were acquired using an AVT Guppy F-146 Mono CCD Camera (Edmund Optics) and flexible arm-mounted LED lights (LED890-66-60, 890-nm peak wavelength IR LEDs from Roithner Lasertechnick, with heatsinks [47 × 20 TO-66]) instead of back lighting. Hypocotyl lengths were measured for images taken every 36 min using ImageJ, starting with the earliest time point for which hypocotyl length could be measured. Hypocotyl lengths at each time point for six seedlings per genotype (or fewer for the very first time points) were averaged and then smoothed by averaging over a window of five successive time points centered at each time point. Data were converted to rates by subtracting successive smoothed values, normalized to length and time scales, and graphed.
Flower organs and rosettes were measured as described previously (Nagpal et al., 2005; Wilmoth et al., 2005; Reeves et al., 2012). RT-qPCR was performed as described previously (Reeves et al., 2012) using the primers listed in Supplemental Table S1.
Accession Numbers
The Arabidopsis Genome Initiative numbers of featured genes are as follows: ARF6 (At1g30330), NPH4/ARF7 (At5g20730), ARF8 (At5g37020), ARF19 (At1g19220), AXR5/IAA1 (At4g14560), SHY2/IAA3 (At1g04240), AXR2/IAA7 (At3g23050), IAA8 (At2g22670), IAA9 (At5g65670), AXR3/IAA17 (At1g04250), MSG2/IAA19 (At3g15540), IAA26 (At3g16500), ELF3 (At2g25930), SAUR19 (At5g18010), SAUR63 (At1g29440), and GH3.3 (At2g23170).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Hypocotyl lengths of seedlings with altered auxin response.
Supplemental Figure S2. Adult phenotypes of PRPS5a>MIR167a transactivated plants and arf mutants.
Supplemental Figure S3. Time-lapse growth rates of seedlings grown in short days.
Supplemental Figure S4. Hormone responses of arf mutant seedlings.
Supplemental Figure S5. Rosettes of plants with decreased auxin response.
Supplemental Table S1. Primers used in this work.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Ty Hedrick for help setting up the time-lapse imaging apparatus.
Footnotes
This work was supported by U.S. National Science Foundation Grant IOS-1147045 to J.W.R. and Punita Nagpal.
Articles can be viewed without a subscription.
References
- Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophoton Int 11: 36–42 [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth T, Jürgens G (1993) The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118: 575–587 [Google Scholar]
- Boer DR, Freire-Rios A, van den Berg WA, Saaki T, Manfield IW, Kepinski S, López-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, et al. (2014) Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156: 577–589 [DOI] [PubMed] [Google Scholar]
- Box MS, Huang BE, Domijan M, Jaeger KE, Khattak AK, Yoo SJ, Sedivy EL, Jones DM, Hearn TJ, Webb AAR, et al. (2015) ELF3 controls thermoresponsive growth in Arabidopsis. Curr Biol 25: 194–199 [DOI] [PubMed] [Google Scholar]
- Brooks TL, Miller ND, Spalding EP (2010) Plasticity of Arabidopsis root gravitropism throughout a multidimensional condition space quantified by automated image analysis. Plant Physiol 152: 206–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, Reed JW (2012) Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J 71: 684–697 [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Greenham K, Castillejo C, Sartor R, Bialy A, Sun TP, Estelle M (2012) Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS ONE 7: e36210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ. (2005) Floral dip: Agrobacterium-mediated germ line transformation. Methods Mol Biol 286: 91–102 [DOI] [PubMed] [Google Scholar]
- Covington MF, Harmer SL (2007) The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH (2009) The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J 57: 160–170 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Depuydt S, Hardtke CS (2011) Hormone signalling crosstalk in plant growth regulation. Curr Biol 21: R365–R373 [DOI] [PubMed] [Google Scholar]
- Dinesh DC, Kovermann M, Gopalswamy M, Hellmuth A, Calderón Villalobos LI, Lilie H, Balbach J, Abel S (2015) Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response. Proc Natl Acad Sci USA 112: 6230–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW (2005) AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132: 4563–4574 [DOI] [PubMed] [Google Scholar]
- Ezer D, Jung JH, Lan H, Biswas S, Gregoire L, Box MS, Charoensawan V, Cortijo S, Lai X, Stöckle D, et al. (2017) The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat Plants 3: 17087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, Alabadí D, Blázquez MA (2012) Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 109: 13446–13451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM (2006) AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18: 1873–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal A, Karayekov E, Galvão VC, Ren H, Casal JJ, Fankhauser C (2016) Shade promotes phototropism through phytochrome B-controlled auxin production. Curr Biol 26: 3280–3287 [DOI] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ. (2015) The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Park Y, Kim I, Kim EH, Yu TK, Rhee S, Suh JY (2014) Structural basis for the auxin-induced transcriptional regulation by Aux/IAA17. Proc Natl Acad Sci USA 111: 18613–18618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E (2000) The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12: 757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LA, Peeters AJ, Pierik R (2010) Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci USA 107: 22740–22744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnen MV, Schmid-Siegert E, Trevisan M, Petrolati LA, Sénéchal F, Müller-Moulé P, Maloof J, Xenarios I, Fankhauser C (2016) Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 28: 2889–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC (2014) Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci USA 111: 5427–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M, Prigge MJ, Tao S, Shain S, Kuo A, Kirchsteiger K, Estelle M (2016) Constitutive auxin response in Physcomitrella reveals complex interactions between Aux/IAA and ARF proteins. eLife 5: e13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, Erickson E, Quail PH (2012) Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24: 1398–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser HMO, Pickett FB, Dharmasiri S, Estelle M (1996) Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J 10: 403–413 [DOI] [PubMed] [Google Scholar]
- Li K, Yu R, Fan LM, Wei N, Chen H, Deng XW (2016) DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat Commun 7: 11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. (2012) Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, Chory J (2008) A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol 6: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, et al. (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118 [DOI] [PubMed] [Google Scholar]
- Nanao MH, Vinos-Poyo T, Brunoud G, Thévenon E, Mazzoleni M, Mast D, Lainé S, Wang S, Hagen G, Li H, et al. (2014) Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun 5: 3617. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C, López-Salmerón V, Davière JM, Prat S (2015) ELF3-PIF4 interaction regulates plant growth independently of the Evening Complex. Curr Biol 25: 187–193 [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Nozue K, Harmer SL, Maloof JN (2011) Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol 156: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY (2014) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, et al. (2005) Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17: 3282–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piya S, Shrestha SK, Binder B, Stewart CN Jr, Hewezi T (2014) Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front Plant Sci 5: 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploense SE, Wu MF, Nagpal P, Reed JW (2009) A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 136: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Burko Y, Jaillais Y, Ljung K, Long JA, Chory J (2016) The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev 30: 1529–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucciariello O, Legris M, Costigliolo Rojas C, Iglesias MJ, Hernando CE, Dezar C, Vazquez M, Yanovsky MJ, Finlayson SA, Prat S, et al. (2018) Rewiring of auxin signaling under persistent shade. Proc Natl Acad Sci USA 115: 5612–5617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher W. (2000) Growth retardants: effects on gibberellin biosynthesis and other metabolic pathways. Annu Rev Plant Physiol Plant Mol Biol 51: 501–531 [DOI] [PubMed] [Google Scholar]
- Reed JW. (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6: 420–425 [DOI] [PubMed] [Google Scholar]
- Reed JW, Elumalai RP, Chory J (1998) Suppressors of an Arabidopsis thaliana phyB mutation identify genes that control light signaling and hypocotyl elongation. Genetics 148: 1295–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PH, Ellis CM, Ploense SE, Wu MF, Yadav V, Tholl D, Chételat A, Haupt I, Kennerley BJ, Hodgens C, et al. (2012) A regulatory network for coordinated flower maturation. PLoS Genet 8: e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW (2004) Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135: 1738–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairanen I, Novák O, Pěnčík A, Ikeda Y, Jones B, Sandberg G, Ljung K (2012) Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell 24: 4907–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M (2015) SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27: 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Baiga TJ, Pojer F, Dabi T, Butterfield C, Parry G, Santner A, Dharmasiri N, Tao Y, Estelle M, et al. (2008) New auxin analogs with growth-promoting effects in intact plants reveal a chemical strategy to improve hormone delivery. Proc Natl Acad Sci USA 105: 15190–15195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E, Salehin M, Zhang Y, Sanchez SE, Doherty C, Wang R, Mangado CC, Song L, Tal I, Pisanty O, et al. (2017) Plant stress tolerance requires auxin-sensitive Aux/IAA transcriptional repressors. Curr Biol 27: 437–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J, Leivar P, González-Schain N, Martín G, Diaz C, Sentandreu M, Al-Sady B, Quail PH, Monte E (2016) Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters. Proc Natl Acad Sci USA 113: 4870–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang JA, Gallego-Bartolomé J, Gómez MD, Yoshida S, Asami T, Olsen JE, García-Martínez JL, Alabadí D, Blázquez MA (2009) Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J 60: 589–601 [DOI] [PubMed] [Google Scholar]
- Stewart JL, Maloof JN, Nemhauser JL (2011) PIF genes mediate the effect of sucrose on seedling growth dynamics. PLoS ONE 6: e19894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe-Evans EL, Harper RM, Motchoulski AV, Liscum E (1998) NPH4, a conditional modulator of auxin-dependent differential growth responses in Arabidopsis. Plant Physiol 118: 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Tabata R, Ikezaki M, Fujibe T, Aida M, Tian CE, Ueno Y, Yamamoto KT, Machida Y, Nakamura K, Ishiguro S (2010) Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol 51: 164–175 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Reed JW (1999) Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126: 711–721 [DOI] [PubMed] [Google Scholar]
- Tian CE, Muto H, Higuchi K, Matamura T, Tatematsu K, Koshiba T, Yamamoto KT (2004) Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J 40: 333–343 [DOI] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW (2002) Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14: 301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpte CS, Wilson AK, Estelle M (1992) Effects of the axr2 mutation of Arabidopsis on cell shape in hypocotyl and inflorescence. Planta 188: 271–278 [DOI] [PubMed] [Google Scholar]
- Timpte C, Wilson AK, Estelle M (1994) The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 138: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H (2008) Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol 49: 1025–1038 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1997) ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1999a) Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA 96: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1999b) Dimerization and DNA binding of auxin response factors. Plant J 19: 309–319 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. (2011) The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Van Hamburg JP, Van Rijn E, Hooykaas PJ, Offringa R (2003) Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol 133: 1882–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Sauer M, Meurette O, Friml J, Ljung K, Sandberg G, Hooykaas P, Offringa R (2005) Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell 17: 2517–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW (2005) NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J 43: 118–130 [DOI] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M (1990) A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol Gen Genet 222: 377–383 [DOI] [PubMed] [Google Scholar]
- Wolters H, Jürgens G (2009) Survival of the flexible: hormonal growth control and adaptation in plant development. Nat Rev Genet 10: 305–317 [DOI] [PubMed] [Google Scholar]
- Wu MF, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133: 4211–4218 [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR (1996) The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J 10: 691–702 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Guo Y, Novák O, Chen W, Ljung K, Noel JP, Chory J (2016) Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat Plants 2: 16025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XY, Song L, Xue HW (2013) Brassinosteroids regulate the differential growth of Arabidopsis hypocotyls through auxin signaling components IAA19 and ARF7. Mol Plant 6: 887–904 [DOI] [PubMed] [Google Scholar]