GhCPK33 negatively regulates defense against fungal infection by promoting the phosphorylation and degradation of a protein required for jasmonic acid biosynthesis.
Abstract
Verticillium wilt, caused by the soil-borne fungus Verticillium dahliae, is a destructive vascular disease in plants. Approximately 200 dicotyledonous plant species in temperate and subtropical regions are susceptible to this notorious pathogen. Previous studies showed that jasmonic acid (JA) plays a crucial role in plant-V. dahliae interactions. V. dahliae infection generally induces significant JA accumulation in local and distal tissues of the plant. Although JA biosynthesis and the associated enzymes have been studied intensively, the precise mechanism regulating JA biosynthesis upon V. dahliae infection remains unknown. Here, we identified the calcium-dependent protein kinase GhCPK33 from upland cotton (Gossypium hirsutum) as a negative regulator of resistance to V. dahliae that directly manipulates JA biosynthesis. Knockdown of GhCPK33 by Agrobacterium tumefaciens-mediated virus-induced gene silencing constitutively activated JA biosynthesis and JA-mediated defense responses and enhanced resistance to V. dahliae. Further analysis revealed that GhCPK33 interacts with 12-oxophytodienoate reductase3 (GhOPR3) in peroxisomes. GhCPK33 phosphorylates GhOPR3 at threonine-246, leading to decreased stability of GhOPR3, which consequently limits JA biosynthesis. We propose that GhCPK33 is a potential molecular target for improving resistance to Verticillium wilt disease in cotton.
Plants are constantly threatened by the aggression of a wide spectrum of pathogens in the terrestrial phyllosphere (Li et al., 2016a; Xin et al., 2016; Hillmer et al., 2017; Motte and Beeckman, 2017). Plants have evolved a multilayered surveillance system to launch prompt and effective defense responses to limit pathogen invasion (Akira et al., 2006). Plants have evolved two strategies to perceive pathogens. One involves the interaction between plasma membrane-localized recognition receptors and pathogen-associated molecular patterns (PAMPs), damage-associated molecular patterns (DAMPs), or extracellular effectors. The second class of perception is the interaction between intracellular recognition receptors and corresponding pathogen-derived intracellular effectors (Boller and Felix, 2009; Tsuda et al., 2009; Stotz et al., 2014; Wu et al., 2014; Li et al., 2017; Xu et al., 2017). One of the earliest physiological events during plant immune responses is a significant increase in cytosolic Ca2+ (Ma et al., 2008; Boller and Felix, 2009; Boudsocq et al., 2010; Gao et al., 2013b). Cytosolic Ca2+ serves as a vital second messenger in plant cells during pathogen infection (Martí et al., 2013; Webb, 2013; Brandt et al., 2015). Plants possess four major classes of Ca2+ sensors: calmodulins, calmodulin-like proteins, calcineurin B-like proteins, and Ca2+-dependent protein kinases (CPKs; Bender and Snedden, 2013). CPK proteins are thought to play a key role in Ca2+ signaling transduction and can be vastly and temporally activated by PAMP-triggered immunity (PTI) and effector-triggered immunity elicitation. They show multiple subcellular localizations to optimally and rapidly respond to diverse Ca2+ distribution and substrate preferences (Dammann et al., 2003).
Jasmonic acid (JA) plays an important role in defense against pathogens, and both PTI and effector-triggered immunity modulate JA synthesis following the recognition of microbial pathogens (Tsuda et al., 2009; Zhang et al., 2017). After wounding or pathogen infection, elevation of JA content is usually seen within a few minutes or a few hours (Hettenhausen et al., 2013b). JA biosynthesis begins with the cleavage of α-linolenic acid from plastidial membrane lipids by lipases and its oxygenation by 13-lipoxygenases to generate 13(S)-hydroperoxyoctadecatrienoic acid, which is further converted to (9S,13S)-12-oxophytodienoic acid (OPDA) by allene oxide synthase (AOS) and allene oxide cyclase (AOC). OPDA is transported into the peroxisome and reduced by OPDA reductase3 (OPR3) to generate 8-(3-oxo-2-(pent-2-enyl)cyclopentyl)octanoic acid (OPC-8), and OPC-8 is subjected to β-oxidation, which is finally converted to JA (Hettenhausen et al., 2013a; Fragoso et al., 2014; Chini et al., 2018). Mutants that are impaired in the JA biosynthesis or signaling pathways show a great reduction in antifungal secondary metabolite biosynthesis and become susceptible to pathogen infection (Fradin et al., 2011; De Geyter et al., 2012; Campos et al., 2014; Jiang and Yu, 2016). Several CPK proteins have been reported to participate in regulating JA biosynthesis. For instance, simultaneously silencing CDPK4 and CDPK5 in Nicotiana attenuata results in significant JA accumulation upon wounding and herbivory, which suggests that NaCDPK4 and NaCDPK5 are negative regulators of JA synthesis (Yang et al., 2012; Hettenhausen et al., 2013b). A CPK28 loss-of-function mutant in Arabidopsis (Arabidopsis thaliana) shows phase-specific and spatially restricted alterations in JA-mediated gene expression and JA content (Matschi et al., 2015). Although several studies have reported that CPKs are involved in JA synthesis, the mechanism by which the CPKs suppress or activate JA biosynthesis remains unknown.
Verticillium wilt is a type of vascular disease caused by the soil-borne fungus Verticillium dahliae, and it is the most destructive disease of cotton (Gossypium hirsutum) production (Gao et al., 2013a; Yang et al., 2015). Unfortunately, there is no Verticillium wilt immune germplasm in the widely planted upland cotton (Yang et al., 2015). Previous studies in Arabidopsis show that the unobstructed JA signaling pathway plays an important role in plant resistance to V. dahliae (Fradin et al., 2011); in cotton, the plants hyperaccumulate JA and JA-Ile within a few hours of V. dahliae infection, and the plants with constitutively activated JA signaling show enhanced resistance to V. dahliae (Hu et al., 2018). Repressing the transcriptional level of GhSILENCE-INDUCED STEM NECROSIS by RNA interference enhances cotton resistance to V. dahliae by enabling the hyperaccumulation of JA and JA-Ile (Sun et al., 2014). A WRKY transcription factor from Gossypium barbadense, GbWRKY1, negatively regulates cotton defense against V. dahliae by promoting JASMONATE ZIM-DOMAIN1 (JAZ1) expression, which blocks the JA signaling pathway (Li et al., 2014).
In this study, we identified from an RNA sequencing database a CPK gene (GhCPK33) that is induced by V. dahliae V991 (Zhang et al., 2018). Further experiments demonstrated that GhCPK33 is localized to the peroxisome and functions as a negative regulator of cotton defense against V. dahliae by manipulating JA synthesis via the phosphorylation of 12-oxophytodienoate reductase3 (GhOPR3), which decreases its stability. Our study provides novel insight into the phosphorylation network between GhCPK33 and GhOPR3, which directly regulates JA biosynthesis and the JA-dependent V. dahliae response in cotton. This study also confirms that, in plants, CPK participates directly in regulating stress-induced JA biosynthesis. GhCPK33 could be a potential molecular target to improve Verticillium wilt disease resistance in plants.
RESULTS
Identification and Expression Profile of the GhCPK33 Gene
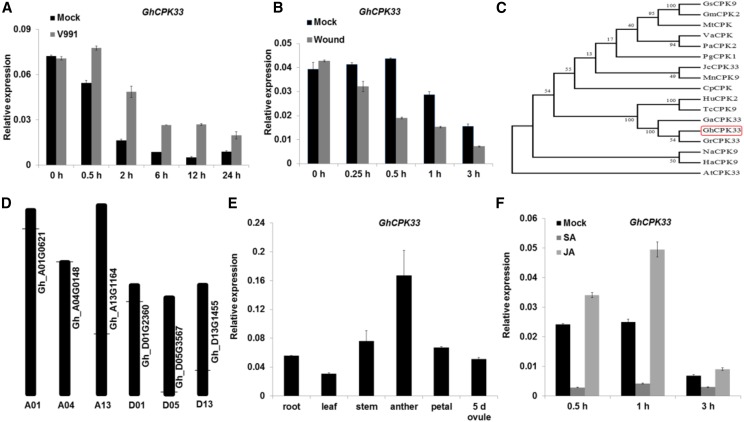
Because CPK proteins are important in plant immunity, we analyzed all CPK genes in the RNA sequencing data derived from cotton cv YZ1 roots infected with the V. dahliae isolate V991 (Zhang et al., 2018). Among the CPK genes, a gene named GhCPK33 was induced in the cotton root by V. dahliae strain V991, but it was suppressed by mechanical wounding (Fig. 1, A and B). We cloned the full-length cDNA of GhCPK33 from cotton cv YZ1, which consists of 1614 nucleotides and encodes a protein of 537 amino acids with an N-terminal myristoylation sequence and palmitoylation site, and a predicted peroxisomal targeting signal 2 according to Flynn et al. (1998, Supplementary Figure S1). Phylogenetic analysis performed with the protein sequences showed that the closest orthologs of GhCPK33 are GaCPK33 (Gossypium arboreum), GrCPK33 (Gossypium raimondii), TcCPK9 (Theobroma cacao), and HuCPK2 (Herrania umbratica; Fig. 1C). There are six copies of GhCPK33 (Gh_D01G2360, Gh_A01G0621, Gh_D13G1455, Gh_A13G1164, Gh_D05G3567, and Gh_A04G0148) in the cotton genome (Fig. 1D). RT-qPCR analysis confirmed that GhCPK33 was expressed in several organs (Fig. 1E) and was induced strongly in the roots by treatment with JA but was suppressed by salicylic acid (SA; Fig. 1F). This finding is consistent with the possibility that GhCPK33 may be involved in the cotton defense response to V. dahliae infection and participate in the JA-mediated signaling pathway.
Figure 1.
Identification and expression profile of GhCPK33. A, Reverse transcription quantitative PCR (RT-qPCR) analysis of GhCPK33 expression at different time points following inoculation with V. dahliae. Total RNA was extracted from the roots of mock and V. dahliae-inoculated plants. The values are means ± sd, n = 3, and normalized to those of GhUB7. B, RT-qPCR analysis of GhCPK33 expression at different time points following mechanical wounding. The values are means ± sd, n = 3, and normalized to those of GhUB7. Total RNA was extracted from the roots of mock and mechanically wounded plants. C, Phylogenetic analysis of GhCPK33 and CPKs from other plants: Glycine soja (Gs), Glycine max (Gm), Medicago truncatula (Mt), Vitis amurensis (Va), Prunus avium (Pa), Panax ginseng (Pg), Jatropha curcas (Jc), Morus notabilis (Mn), Carica papaya (Cp), Herrania umbratica (Hu), Theobroma cacao (Tc), Gossypium arboreum (Ga), Gossypium raimondii (Gr), Gossypium hirsutum (Gh), Nicotiana attenuata (Na), Helianthus annuus (Ha), and Arabidopsis thaliana (At). The neighbor-joining tree was constructed using the MEGA5 program. D, Distribution of GhCPK33 genes in the whole genome. There are six copies of GhCPK33 in the cotton genome, and the horizontal lines indicate the relative position of each copy on the chromosomes. E, RT-qPCR analysis of GhCPK33 in different tissues. Total RNA was isolated from roots, leaves, stems, anthers, petals, and 5-d ovules of the wild-type cotton cv YZ1. The values are means ± sd, n = 3, and normalized to those of GhUB7. F, RT-qPCR analysis of GhCPK33 expression in JA- and SA-treated cotton. Total RNAs were extracted from the roots of mock and JA or SA-treated plants. The values are means ± sd, n = 3, and normalized to those of GhUB7.
GhCPK33 Negatively Regulates Cotton Resistance to V. dahliae by Manipulating JA Synthesis
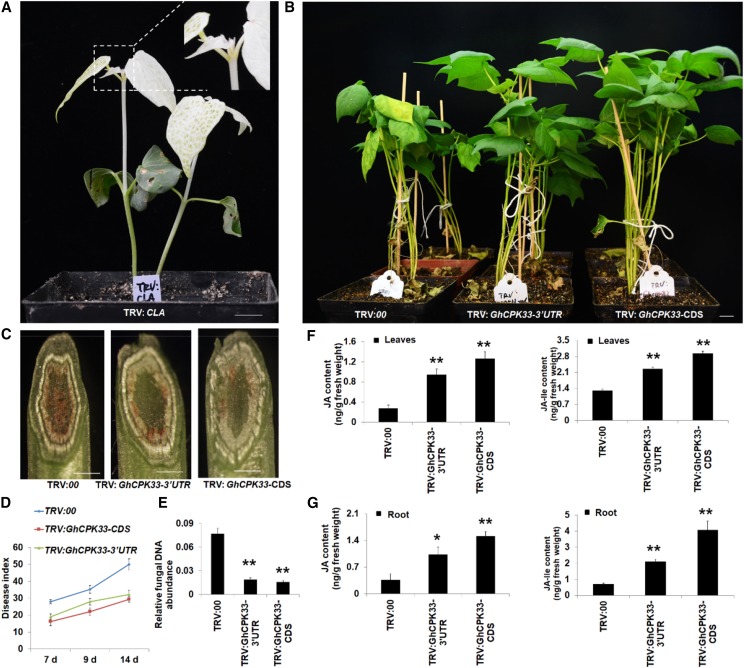
To investigate the putative biofunction of GhCPK33 in the interaction between cotton and V. dahliae, the tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) strategy was used to knock down the expression of GhCPK33 in cotton. Given that there are six copies of GhCPK33 in the upland cotton genome, we constructed two different pTRV2 vectors that contain either a fragment of the GhCPK33 3′ untranslated region (TRV:GhCPK33-3′UTR, Gh_D01G2360 and Gh_A01G0621 have the same 3′ UTR sequence) or the coding sequence (TRV:GhCPK33-CDS; Supplemental Fig. S2, A and B) to suppress all six genes. The pTRV2 vector without DNA insertion (TRV:00) or a fragment of GhCLA (TRV:CLA [cloroplastos alterados1]) was used as a control or a VIGS efficiency indicator, respectively (Supplemental Fig. S2C). After the true leaves of the plants that were inoculated with TRV:CLA-expressing agrobacteria showed a photobleaching phenotype (approximately 16 d; Fig. 2A), the young leaves and roots from TRV:00, TRV:GhCPK33-3′UTR, and TRV:GhCPK33-CDS plants were harvested to determine the expression level of GhCPK33 (Supplemental Fig. S3, A and B). When reaching the three-leaf stage, the seedlings were inoculated with V991. The results showed that silencing of GhCPK33 enhanced the resistance to V991 compared with the control plants (Fig. 2B). The typical disease symptoms were more severe in the TRV:00 plants than in either the TRV:GhCPK33-3′UTR or the TRV:GhCPK33-CDS plant (Fig. 2C). The disease index analysis supported these results (Fig. 2D), and fungal DNA abundance detection also revealed that less fungal DNA was detected in GhCPK33-silenced plants than in control plants (Fig. 2E). Moreover, the TRV:GhCPK33-CDS plants showed better resistance capability than the TRV:GhCPK33-3′UTR plants according to the disease index and fungal biomass (Fig. 2, D and E), which suggest that the dosage of GhCPK33 was important for the cotton-V. dahliae interaction.
Figure 2.
GhCPK33 negatively regulates cotton defense against V. dahliae via manipulating JA biosynthesis. A, Photobleaching phenotype of the plants inoculated with TRV:CLA for 16 d. Bar = 1 cm. B, Disease symptoms of TRV:00, TRV:GhCPK33-3′UTR, and TRV:GhCPK33-CDS plants following inoculation with V. dahliae strain V991 and photographed 14 d after inoculation. Bar = 1 cm. C, V991 hyphal growth in TRV:00, TRV:GhCPK33-3′UTR, and TRV:GhCPK33-CDS plants after inoculation for 14 d. Bars = 1 mm. D, Disease indices of TRV:00, TRV:GhCPK33-3′UTR, and TRV:GhCPK33-CDS plants were determined after inoculation with V991 for 7, 9, and 14 d. The values are means ± sd, n = 3. E, Relative fungal DNA abundance detection. Total DNA of stems from each line (inoculated with V991 for 14 d) was extracted as a template for fungal biomass detection by RT-qPCR. GhUB7 was used as the control. The values are means ± sd, n = 3. F and G, JA and JA-Ile contents in leaves (F) and roots (G) from TRV:00, TRV:GhCPK33-3′UTR, and TRV:GhCPK33-CDS plants. Values are means ± sd, n = 6. Statistical analyses were performed using Student’s t test: *, P < 0.05 and **, P < 0.01.
GhCPK33 Is a Negative Regulator of JA Synthesis
We further examined the possible reason(s) for the enhanced disease resistance in the VIGS-silenced GhCPK33 plants. Several studies in Arabidopsis and N. attenuata have shown that the CPK protein may participate in regulating JA synthesis (Szczegielniak et al., 2012; Yang et al., 2012; Hettenhausen et al., 2013b; Matschi et al., 2015). Therefore, we determined the JA and JA-Ile contents in the leaves and roots from TRV:00, TRV:GhCPK33-CDS, and TRV:GhCPK33-3′UTR plants. The results show that silencing GhCPK33 caused significant JA and JA-Ile accumulation (Fig. 2, F and G) and activation of the JA signaling pathway (Supplemental Figs. S4 and S5). Genes involved in JA synthesis, such as GhLOX2, GhLOX3, GhLOX4, GhLOX6, GhAOS, GhAOC, and GhOPR3, and genes involved in the JA signaling pathway, such as GhMYC2, GhMYC3, GhERF1, and GhPR4, were up-regulated in the GhCPK33 knockdown plants (Supplemental Figs. S4 and S5), which suggests that GhCPK33 negatively regulates cotton resistance to V. dahliae via manipulating JA biosynthesis.
GhCPK33 Interacts with GhOPR3, and the Complex Localizes in Peroxisomes
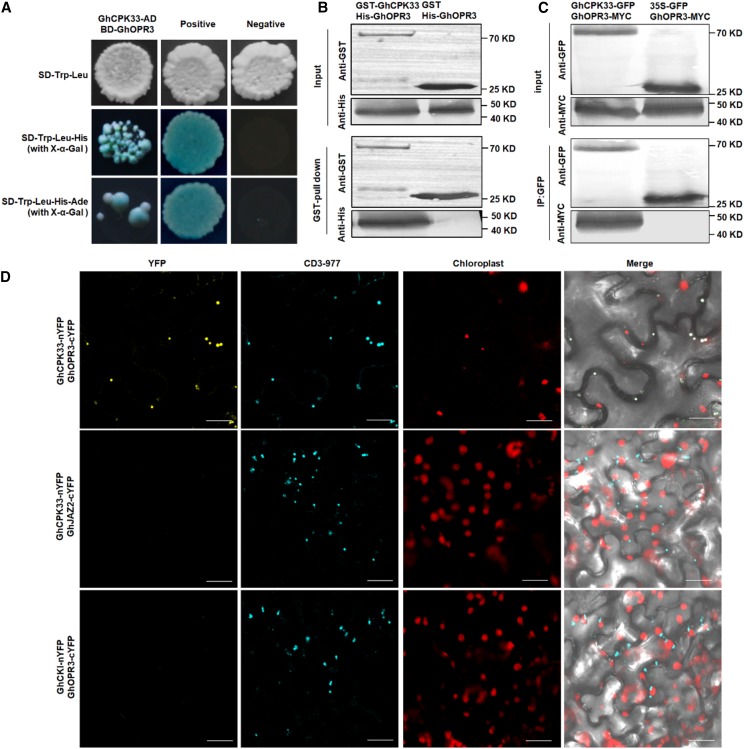
To elucidate the regulatory function of GhCPK33 in JA biosynthesis, full-length GhCPK33 was used to screen a yeast two-hybrid (Y2H) library of cotton to identify potential interacting proteins. Several candidate GhCPK33-interacting proteins were identified, among which we focused on GhOPR3, which showed 76% identity with the amino acid sequence of AtOPR3 (Supplemental Fig. S6). AtOPR3 was verified to be involved in JA synthesis (Sanders et al., 2000; Stintzi and Browse, 2000). To confirm this interaction, 1:1 Y2H was performed between GhCPK33 fused with the Gal4 activation domain (GhCPK33-AD) and GhOPR3 fused to the Gal4 binding domain (GhOPR3-BD). It was observed that GhCPK33 interacts with GhOPR3 in yeast (Fig. 3A). GST pull-down, coimmunoprecipitation (Co-IP), and biomolecular fluorescence complementation (BiFC) assays were performed between GhCPK33 and GhOPR3. We observed that GST-GhCPK33, but not GST alone, precipitated His-GhOPR3 in vitro (Fig. 3B; Supplemental Fig. S7A) and that GhCPK33-GFP, but not GFP alone, precipitated GhOPR3-MYC in Nicotiana benthamiana leaves (Fig. 3C). The BiFC assay revealed that the interaction between GhCPK33 and GhOPR3 occurred in the peroxisomes (Fig. 3D). Additionally, GhOPR3-GFP and GhCPK33-GFP fusion proteins colocalized with the peroxisome marker CD3-977 in N. benthamiana leaves (Supplemental Fig. S7B).
Figure 3.
GhCPK33 interacts with GhOPR3 in vitro and in vivo. A, Y2H assays showing the interaction between GhCPK33 and GhOPR3. Transformed yeast cells were grown on synthetic dextrose (SD) media, and the blue colonies on SD-Trp-Leu-His (with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid [X-α-Gal]) and SD-Trp-Leu-His-Ade (with X-α-Gal) media indicate positive interactions. B, GST pull-down assay showing direct interactions between His-OPR3 and GST-GhCPK33 fusion proteins. His-GhOPR3 protein was incubated with immobilized GST or GST-GhCPK33 proteins, and immunoprecipitated fractions were detected by an anti-His antibody or an anti-GST antibody. C, Co-IP assay showing direct interaction between GhCPK33 and GhOPR3 in vivo. The clones carrying GhCPK33-GFP and GhOPR3-MYC, and those carrying 35S:GFP and GhOPR3-MYC, were coinfiltrated into N. benthamiana leaves by Agrobacterium tumefaciens infiltration. Total proteins were immunoprecipitated with GFP-coupled beads, and the immunoblot was probed with anti-GFP and anti-Myc antibodies, respectively. D, BiFC assay showing that the interaction between GhCPK33-nYFP and GhOPR3-cYFP formed a functional YFP in the peroxisomes. CD3-977 served as a CFP peroxisome marker. The interactions between GhCPK33-nYFP and GhJAZ2-cYFP, and between GhCKI-nYFP and GhOPR3-cYFP, were used as negative controls for the BiFC assay. Merge = merging of YFP, CFP, and chloroplast autofluorescence. All the experiments were repeated at least three times with similar results. Bars = 20 μm.
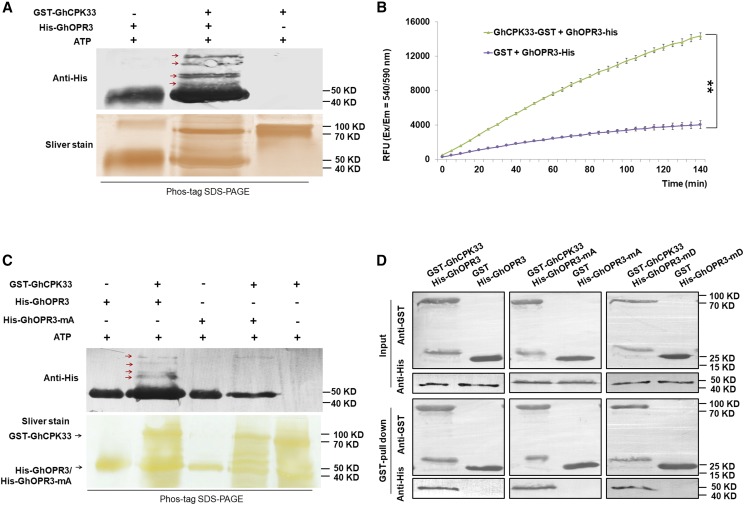
GhCPK33 Phosphorylates GhOPR3 at Site Thr-246
Bioinformatic analysis indicated that GhOPR3 contains several consensus CAMK (Ca2+/calmodulin-dependent protein kinase) phosphorylation sites (Supplemental Fig. S8A). To determine whether GhCPK33 phosphorylates GhOPR3, GST-GhCPK33 and His-GhOPR3 were subjected to in vitro phosphorylation analysis by an in gel assay, and the results demonstrated that GhCPK33 specifically phosphorylates GhOPR3 (Fig. 4A). A universal fluorometric kinase assay (Universal Fluorometric Kinase Assay Kit; Sigma-Aldrich) further confirmed that GhCPK33 phosphorylates GhOPR3 (Fig. 4B). To identify the GhOPR3 site(s) phosphorylated by GhCPK33, we collected His-GhOPR3 (reaction without GhCPK33, used as a control) and the identified phosphorylated His-GhOPR3 by Phos-tag SDS-PAGE and analyzed the products by liquid chromatography-tandem mass spectrometry. The most abundant phosphopeptide contained Thr-246 as the phosphorylation site (Supplemental Fig. S8B). To further verify the phosphorylation site of GhOPR3, the Thr-246 site was mutated to Ala (GhOPR3-mA). His-tagged GhOPR3-mA recombinant protein was generated, and its phosphorylation by GhCPK33 was examined. The results demonstrated that the T246A substitution greatly reduced the phosphorylation of GhOPR3 (Fig. 4C). Furthermore, to investigate whether Thr-246 impacts the interaction between GhCPK33 and GhOPR3, GST pull-down assays were performed between the His-tagged phosphodeficient GhOPR3-mA protein and GST-GhCPK33 and between the His-tagged phosphomimic GhOPR3-mD protein (Thr-246 mutated to Asp) and GST-GhCPK33. The results showed that His-GhOPR3-mA and His-GhOPR3-mD had normal physical interactions with GhCPK33 (Fig. 4D).
Figure 4.
GhCPK33 phosphorylates GhOPR3. A, In vitro phosphorylation assays between GhOPR3 and GhCPK33 using Phos-tag SDS-PAGE. GST-GhCPK33 was used to phosphorylate purified His-tagged GhOPR3 protein. The reaction mixtures without kinase or without substrate were used as controls. The reaction mixtures were subjected to SDS-PAGE with Phos-tag, and the phosphorylated protein was immunoblotted with anti-His antibody. The red arrows indicate phosphorylated GhOPR3. B, Measurement of GhCPK33 kinase activity using GhOPR3 as a substrate. GST indicates the purified mixture from pGEX-4T-1 without the GhCPK33 DNA insertion and served as the negative control for GST-GhCPK33. The values are means ± sd, n = 3. Statistical analyses were performed using Student’s t test: **, P < 0.01. All the experiments were repeated at least three times with similar results. C, In vitro phosphorylation assays of recombinant GhOPR3 or GhOPR3-mA (the Thr-246 site was mutated to Ala) by GhCPK33 using Phos-tag SDS-PAGE. The red arrows indicate phosphorylated GhOPR3. D, GST pull-down assay showing direct interactions between GST-GhCPK33 and His-OPR3, His-GhOPR3-mA, or His-GhOPR3-mD (the Thr-246 site was mutated to Asp) fusion proteins. His-tagged protein was incubated with immobilized GST or GST-GhCPK33 proteins, and the immunoprecipitated fractions were detected by an anti-His antibody or anti-GST antibody. All the experiments were repeated at least three times with similar results.
Phosphorylation by GhCPK33 Decreases the Stability of GhOPR3
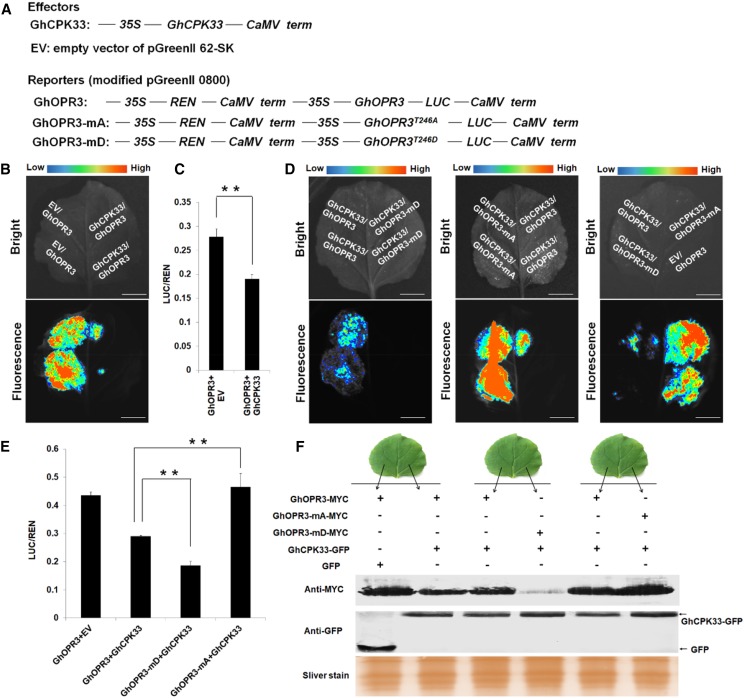
To further determine the biofunction of GhCPK33-mediated GhOPR3 phosphorylation in vivo, the dual-luciferase expression vectors with native GhOPR3, phosphodeficient GhOPR3-mA, phosphomimic GhOPR3-mD, and GhCPK33 were generated under the control of the 35S promoter, as indicated in Figure 5A. Because several reports show that the phosphorylation status of a protein may affect its stability (Monaghan et al., 2014; Li et al., 2016b), we speculated that GhCPK33 may decrease the stability of GhOPR3 via phosphorylation. As shown in Figure 5B, equal concentrations and volumes of the indicated constructs were coinfiltrated into N. benthamiana leaves by A. tumefaciens infiltration. The LUC (firefly luciferase) luminescence intensity, which indicates the protein level of recombinant GhOPR3-LUC, decreased in the presence of GhCPK33. The result suggests that GhCPK33 decreases the stability of GhOPR3. The dual-luciferase reporter assay also was performed in cotton protoplasts to confirm this conclusion. The ratio of LUC activity to the control REN (Renilla luciferase) activity was calculated to indicate the protein level of GhOPR3, and the result showed that LUC/REN activity decreased significantly in the presence of GhCPK33 (Fig. 5C). To investigate whether the effect of GhCPK33 on GhOPR3 stability depends on phosphorylation, the indicated constructs were similarly coinfiltrated into N. benthamiana leaves or cotton protoplasts. The results showed that, in the presence of GhCPK33, compared with the native form GhOPR3, GhOPR3-mA showed increased stability and GhOPR3-mD showed significantly decreased stability (Fig. 5, D and E). Furthermore, constructs with C-terminal MYC-tagged GhOPR3 (GhOPR3-MYC, GhOPR3-mA-MYC, and GhOPR3-mD-MYC) and C-terminal GFP-tagged GhCPK33 (GhCPK33-GFP) were generated, and equal concentrations and volumes of the indicated constructs were infiltrated into N. benthamiana leaves, as shown in Figure 5F. Total protein was used to detect the GhOPR3 protein level. Again, we observed that GhCPK33-mediated phosphorylation decreased the stability of GhOPR3 (Fig. 5F). These observations indicate that GhCPK33 negatively manipulates JA synthesis via phosphorylating GhOPR3 and decreases the stability of GhOPR3; thus, the GhCPK33 knockdown plants demonstrated a significant accumulation of JA and JA-Ile. Moreover, even in the absence of GhCPK33, GhOPR3-mD accumulated to a lower level than GhOPR3 and GhOPR3-mA, which suggests that the Thr-246-phosphorylated GhOPR3 might be less stable in vivo (Supplemental Fig. S9, A–C). In addition, when we treated N. benthamiana leaves (which were coinfiltrated with GhOPR3-mD and GhCPK33 constructs, as indicated in Supplemental Fig. S9, D and E) with the proteasome inhibitor MG132, the levels of GhOPR3-mD increased greatly, as indicated by the enhanced GhOPR3-mD-LUC fluorescence intensity and the distinct GhOPR3-mD-MYC protein band (Supplemental Fig. S9, D and E). These findings indicated that GhOPR3-mD is degraded by the 26S proteasome.
Figure 5.
GhCPK33-phosphorylated GhOPR3 shows decreased protein stability. A, Diagrams of the modified dual-luciferase reporter system. 35S, Promoter of cauliflower mosaic virus 35S RNA gene; REN, Renilla luciferase; CaMV term, cauliflower mosaic virus terminator; LUC, firefly luciferase. B, LUC activity indicates the accumulation level of GhOPR3 with or without GhCPK33 in N. benthamiana leaves. C, GhOPR3-LUC protein levels in cotton protoplasts following treatment with empty vector (EV) or GhCPK33. The LUC/REN activity ratios are means ± sd, n = 3. D, LUC activity indicates the level of GhOPR3, GhOPR3-mA, or GhOPR3-mD in the presence of GhCPK33 in N. benthamiana leaves. E, GhOPR3-LUC, GhOPR3-mA-LUC, or GhOPR3-mD-LUC protein accumulation levels of the reporter in the presence of GhCPK33 in cotton protoplasts. The LUC/REN activity ratios are means ± sd, n = 3. F, Western blotting showing the accumulation of GhOPR3-MYC, GhOPR3-mA-MYC, or GhOPR3-mD-MYC when GhCPK33 also is expressed. All the experiments were repeated at least three times with similar results. Bars in B and D = 1 cm.
DISCUSSION
The plant small-molecule hormone JA plays a vital role in regulating plant defense against pathogens and herbivores (Nickstadt et al., 2004; Howe and Jander, 2008; Hettenhausen et al., 2013; Campos et al., 2014; Sun et al., 2014; Zhang et al., 2016, 2017). The transient accumulation of JA is one of the early responses in plants to infection by pathogens and attack by herbivores. In addition, wounding and the concomitant production of pathogen-specific DAMPs, PAMPs, HAMPs (herbivore-associated molecular patterns), and effectors allow the plant to recognize the invader and employ an invader-specific response (Campos et al., 2014). Although JA biosynthesis and signaling pathways have been studied extensively, and nearly all enzymes involved in JA synthesis have been cloned, the mechanism by which plants precisely regulate JA accumulation (biosynthesis and degradation) after the perception of invaders is not well understood.
Pleiotropic Functions of CPKs in Plant Defense against Pathogens
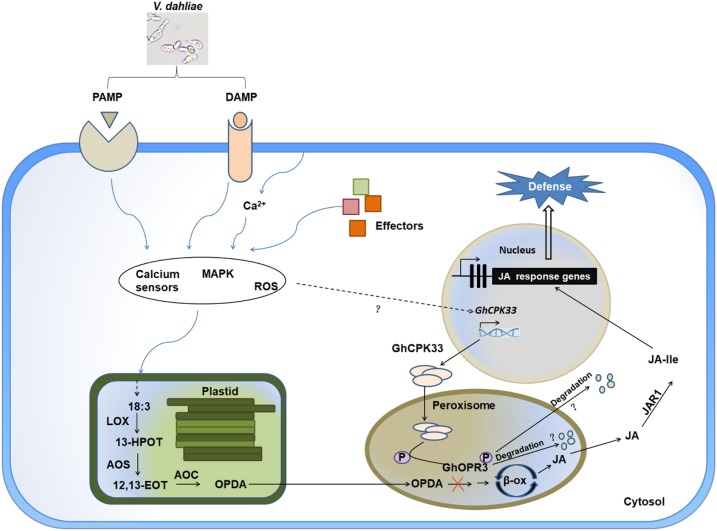
Positively charged Ca2+ and negatively charged phosphate ions are two major primary signaling elements in most organisms (Clapham, 2007). The Ca2+ concentration, oscillations, amplitude, and duration time affect almost all aspects of cell survival and development (Ali et al., 2007; Ma et al., 2008; Ranf et al., 2011; Webb, 2013; Zhou et al., 2015). Phosphate ions are involved in protein phosphorylation and setting up an omnipresent and unique signal for protein interactions and cell communication (Ubersax and Ferrell, 2007; Bah et al., 2015; Wang et al., 2018b). The addition of a phosphate group to a protein may affect its biofunction in vivo, including its activity, localization, interaction, and stability (Ubersax and Ferrell, 2007; Bah et al., 2015; Wang et al., 2018b). The CPK proteins are the core regulators that link Ca2+ signaling to the phosphorylation of a specific substrate. The Arabidopsis CPK5 positively regulates plant innate immunity via phosphorylating the respiratory burst oxidase homolog D protein to generate reactive oxygen species at local and distal sites (Dubiella et al., 2013). AtCPK28 functions as a negative regulator of plant immune signaling by phosphorylating BIK1 to control its turnover and to negatively regulate plant resistance to bacterial pathogens. Phosphorylation of BIK1 by AtCPK28 leads to its degradation by the 26S proteasome and results in the attenuation of PTI signaling (Monaghan et al., 2014, 2015; Wang et al., 2018a). Additionally, several reports in rice (Oryza sativa), maize (Zea mays), wheat (Triticum aestivum), potato (Solanum tuberosum), and pepper (Capsicum annuum) confirm that CPK proteins participate in the plant defense response via regulating reactive oxygen species accumulation, biosynthesis of defense-associated phytohormones, and other aspects of the plant immune system by phosphorylating specific substrate proteins (Geng et al., 2013; Shen et al., 2016; Wei et al., 2016; Bundó and Coca, 2017; Fantino et al., 2017; Wang et al., 2018b). In our study, we demonstrate that the cotton calcium-dependent protein kinase CPK33 phosphorylates the key JA biosynthesis-related protein OPR3 to regulate plant immunity through modulating stress-induced JA accumulation (Fig. 6). Our results uncover a multilayered CPK protein-mediated plant defense system, furthering our understanding of the complex interaction between plants and pathogens.
Figure 6.
Schematic model illustrating the proposed role of GhCPK33 in plant defense against V. dahliae. GhCPK33 negatively regulates plant defense against V. dahliae via phosphorylating GhOPR3 to block JA biosynthesis. Knocking down the expression level of GhCPK33 results in the accumulation of JA and JA-Ile and constitutively activates the JA-mediated defense response, enhancing plant resistance to V. dahliae. +P, Phosphorylation.
V. dahliae May Target GhCPK33 to Suppress Plant Immunity
In our research, the VIGS-silenced GhCPK33 plants showed enhanced resistance to V. dahliae with a significant accumulation of JA and JA-Ile and activation of the JA-mediated defense signaling pathway (Fig. 2; Supplemental Figs. S4 and S5). This demonstrates that GhCPK33 is a negative regulator of JA biosynthesis and negatively regulates cotton defense against V. dahliae. Subsequent experiments showed that GhCPK33 can phosphorylate GhOPR3, thereby decreasing the stability of GhOPR3 and inhibiting JA biosynthesis (Figs. 4 and 5). Compared with the mock treatment, the induced expression pattern shows that the expression level of GhCPK33 is up-regulated by V. dahliae infection; however, the overall expression level decreases as the infection progresses (Fig. 1A). By contrast, the expression of GhCPK33 is down-regulated by mechanical wounding (Fig. 1B). Therefore, we speculate that V. dahliae may induce the expression of GhCPK33 to facilitate its infection by suppressing JA accumulation and JA signaling; somehow, the host is able to detect V. dahliae infection-associated mechanical wounding and suppress the expression of GhCPK33 to maintain JA biosynthesis. Moreover, chemically pure oligogalacturonide (OGA), which is recognized as a DAMP, can suppress the expression of GhCPK33 in cotton suspension cultured cells (Supplemental Fig. S9F). This result shows that plants have evolved other mechanisms to sense pathogen infection-associated cell wall damage and, in response, modulate the expression of GhCPK33 to regulate JA biosynthesis. As shown in Figure 6, further research will be directed to the mechanism by which V. dahliae regulates the expression of GhCPK33 and by which the host limits the expression of GhCPK33 after the perception of V. dahliae.
Calcium, GhCPK33, and GhOPR3 Function Interdependently to Manipulate V. dahliae-Induced JA synthesis
Cytosolic Ca2+ concentration is regarded as an essential and important second messenger in numerous plant processes (Clapham, 2007; Zhu et al., 2007; Brandt et al., 2015), including the perception of and defense against pathogens (Lecourieux et al., 2006; Ali et al., 2007; Ma et al., 2008; Gao et al., 2013a). Upon V. dahliae infection, there is a clear Ca2+ accumulation in the cytosol of cotton root cells, but it is unknown how V. dahliae induces Ca2+ accumulation in host cells (Cheng et al., 2016). The alteration of the Ca2+ concentration could be sensed by a Ca2+-dependent signaling network involving protein interactions and phosphorylation cascades to trigger subsequent changes in protein activity or defense-associated hormone accumulation (Bender and Snedden, 2013; Monaghan et al., 2014, 2015; Scholz et al., 2014; Matschi et al., 2015). However, it is unknown how calcium accumulation and/or calcium sensors modulate JA synthesis. In our research, we found that GhCPK33, a peroxisome-localized Ca2+-dependent protein kinase (Supplemental Fig. S7B), interacts with and phosphorylates GhOPR3 at Thr-246 (Figs. 3 and 4). The phosphorylated GhOPR3 proteins show reduced protein stability, resulting in an interrupted JA biosynthesis pathway (Fig. 5). A recent study in Arabidopsis uncovered an OPDA-OPR3-independent JA biosynthesis pathway, which uses 4,5-didehydrojasmonate as substrate for JA synthesis via the reducing action of OPR2 (Chini et al., 2018). In that research, JA content was no different between the opr2-1 mutant and the wild type under standard conditions, but it was decreased dramatically in the opr3-3 mutant, and only the opr2-1/opr3-3 double mutant accumulated less JA than the opr3-3 mutant upon mechanical wounding. These results suggest that the biofunction of the OPR2-dependent JA biosynthesis pathway is amplified in the absence of OPR3 and that it is only a subsidiary pathway for JA biosynthesis (Chini et al., 2018). Upon wounding or pathogen infection, the rapid and dramatic accumulation of JA in plant cells is dependent mainly on the OPDA-OPR3-mediated pathway. Our work provides solid evidence of the direct interaction between calcium sensors and JA biosynthesis and uncovers a novel strategy to regulate JA biosynthesis via manipulating the OPR3 protein levels in cotton. However, there are still several outstanding questions that need further research, such as the quantification of Ca2+ concentration in peroxisomes and whether the activation of GhCPK33 depends directly on the peroxisome Ca2+ concentration or on other molecules.
The amino acid sequence alignment of the OPR family (OPR1, OPR2, and OPR3) from cotton and Arabidopsis showed that Thr-246 is not conserved between OPR3 and other subfamilies either in cotton or in Arabidopsis (Supplemental Fig. S10). We also identified the OPR3 homologs from other plant species, including rice, maize, oilseed rape (Brassica napus), and N. attenuata, and the amino acid sequence alignments demonstrated that half of the investigated plant species have the Thr-246 site (Supplemental Fig. S11). This implies that the model in Figure 6 may operate only in the plant species in which Thr-246 is conserved. In those plant species without the conserved Thr-246 site, OPR3 may contain other phosphorylation site(s) targeted by other protein kinases or they may employ other strategies to regulate OPR3 protein levels.
MATERIALS AND METHODS
Plant Materials and Culture Conditions
Seeds of upland cotton (Gossypium hirsutum ‘YZ1’) and Nicotiana benthamiana were sown in soil, and the seedlings were grown in a greenhouse maintained at a constant temperature of 25°C under long-day conditions with an 8-h/16-h dark/light photoperiod and a relative humidity of 60%.
Phylogenetic Analysis
The neighbor-joining method was used to produce the phylogenetic tree of GhCPK33 and CPKs in the collected plant species using MEGA5 software (Tamura et al., 2011).
Treatment with Verticillium dahliae, Methyl Jasmonate, SA, and Mechanical Wounding
Treatments with V. dahliae, methyl jasmonate, and SA were performed on three-leaf-stage seedlings according to a procedure described previously (Hu et al., 2018). For the mechanical wounding treatment, four cv YZ1 seedlings were cultivated in a pot with Hoagland solution (Arnon and Hoagland, 1940) in a growth chamber until the three-leaf stage was reached, and the roots were cut to lengths of approximately 1 cm using scissors. The roots of the four individual seedlings were collected from each treatment at each time point.
Gene Cloning and VIGS
The fragments containing part of the 3′ UTR and the coding sequence of GhCPK33 were amplified from the cDNA of cv YZ1 using the corresponding primer pairs, GhCPK33-VIGS-3′-F/R and GhCPK33-CDS-VIGS-F/R, and then were cloned into the TRV:00 plasmid at the BamHI-KpnI sites using the In-Fusion HD Cloning Kit (Clontech) according to the protocol provided by the manufacturer. The recombinant plasmids were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. TRV vectors were agroinfiltrated into two fully expanded cotyledons of cv YZ1 cotton plants, as described previously (Gao et al., 2011, 2013a; Yang et al., 2015), in a 25°C controlled growth chamber with a 16-h/8-h light/dark photoperiod. At least 20 plants were inoculated with each construct. The TRV:CLA (Gao et al., 2011) construct served as a positive marker for evaluating VIGS efficiency. At 16 d after inoculation, the expression levels of the target gene were examined, and the successfully silenced plants were subjected to infection by V. dahliae V991. The primers used in our study are listed in Supplemental Table S1.
RT-PCR and RT-qPCR
Total RNA was extracted from cotton tissues using a modified guanidine thiocyanate method (Deng et al., 2012). First-strand cDNA was generated from 3 μg of total RNA using SuperScript III reverse transcriptase (Invitrogen). To determine gene expression levels, RT-PCR and RT-qPCR analyses were performed according to a procedure described previously (Hu et al., 2018). The primers used in our study are listed in Supplemental Table S1.
Pathogen Preparation, Infection, and Disease Assay
Pathogen cultivation was performed as we described previously (Xu et al., 2011). In brief, a highly aggressive defoliating V. dahliae isolate, V991, was used and was inoculated on a potato dextrose agar (PDA)plate for 1 week at 25°C. Next, the fungal colonies were transformed into Czapek medium (3% [w/v] saccharose, 0.3% [w/v] NaNO3, 0.1% [w/v] K2HPO4, 0.1% [w/v] KCl, 0.1% [w/v] MgSO4, and 0.002% [w/v] FeSO4) on a shaker at 120 rpm at 25°C for 4 d until the spore concentration reached ∼108 spores mL−1. To determine the plant performance in response to the fungal pathogens, the spores of V991 were adjusted to a concentration of 105 spores mL−1 with sterile distilled water, and the infected VIGS cotton plants were inoculated with V991 by root dip in the spore suspension for 2 min according to Xu et al. (2011). The disease index was scored using at least 20 plants per treatment and repeated at least three times. The severity of the disease symptoms on each cotton seedling was scored using a 0 to 4 rating scale according to Xu et al. (2012) with some modifications, where 0 means no visible wilting or yellowing symptoms; 1 means 0% to 25% (inclusive) true leaves wilted or dropped off; 2 means 25% to 50% (inclusive) true leaves wilted or dropped off; 3 means 50% to 75% (inclusive) true leaves wilted or dropped off; and 4 means 75% to 100% (inclusive) true leaves wilted or dropped off. The plant disease index was calculated according to the following formula:
where n denotes the disease rating from 0 to 4.
Measurement of Fungal DNA Abundance
To measure the fungal biomass in the cotton seedlings, stems were collected from each line at the same position (1 cm around the cotyledonary node). Total DNA was extracted and used as a template for RT-qPCR to detect the amount of fungal DNA. The cotton endogenous gene GhUB7 was used as a control, and the internal transcribed spacer region of the ribosomal DNA was targeted using the fungus-specific ITS1-F primer in combination with the V. dahliae-specific reverse primer ST-VE1-R, generating a 200-bp amplicon for fungal DNA detection. The primers used in our study are listed in Supplemental Table S1.
Measurement of Phytohormones
The measurement of the endogenous concentrations of JA and JA-Ile in leaf or root samples (∼100 mg) was performed using an internal standard method. In brief, 100-mg samples were immediately ground in liquid nitrogen and extracted with 300 µL of cold extraction buffer (80% [v/v] methanol with 15 ng mL−1 [+/−]9-,10-dihydro-JA) overnight at 4°C. After centrifugation at 12,000 rpm and 4°C for 20 min, the supernatant was collected, and the residual pellet was reextracted with 200 µL of cold extraction buffer (80% [v/v] methanol with 15 ng mL−1 [+/−]9-,10-dihydro-JA) for 1 h, and after centrifugation at 12,000 rpm and 4°C for 20 min, the supernatants were combined. (+/−)9-,10-Dihydro-JA (Olchemim) was used as an internal standard, JA and JA-Ile (Sigma-Aldrich) were used as external standards and also prepared with the extraction buffer (80% [v/v] methanol with 15 ng mL−1 [+/−]9-,10-dihydro-JA). Further quantitative analyses of JA and JA-Ile were performed as described previously (Liu et al., 2012).
Y2H Assay
A yeast (Saccharomyces cerevisiae) two-hybrid assay was performed using the Match-maker Gold Yeast Two-Hybrid System (Clontech) according to the manufacturer’s instructions. To generate the bait construct BD-CPK33, full-length GhCPK33 cDNA was amplified using the primer pair GhCPK33-cotton Y2H-F/R. PCR-amplified fragments were cloned in frame with GAL4BD in the vector pGBKT7 using the In-Fusion HD Cloning Kit (Clontech). BD-CPK33 was introduced into the yeast strain Y2H. The library that we used for this experiment was a cotton cDNA library prepared from cotton leaves and roots under several stress conditions using Match-maker Library Construction and Screening Kits (Clontech). To confirm the interaction between GhCPK33 and GhOPR3, the full-length coding sequence of GhCPK33 was amplified using the primer pair GhCPK33-AD-F/R and cloned into pGADT7 to generate GhCPK33-AD and then introduced into yeast strain Y187. The GhOPR3-BD construct was amplified using the corresponding primer pairs and introduced into yeast strain Y2H, as described above. Mating between yeast strains containing different constructs was performed according to the experimental objectives and selected on SD-Leu-Trp, SD-Leu-Trp-His (with X-α-Gal), and SD-Leu-Trp-His-Ade (with X-α-Gal) media. The primers used in this study are listed in Supplemental Table S1.
Recombinant Protein Expression and Pull-Down Assays
The full-length sequence of GhOPR3 was cloned into the pET-28-a vector (Novagen) by Gateway cloning technology to produce His-GhOPR3 proteins. The full lengths of GhOPR3-mA and GhOPR3-mD were amplified with primer pairs GhOPR3-mA-F/R and GhOPR3-mD-F/R by overlap extension PCR and cloned into the pET-28-a vector (Novagen) by Gateway cloning technology to produce His-tagged GhOPR3-mA and GhOPR3-mD proteins. The full-length coding sequence of GhCPK33 was cloned into pGEX-4T-1 (Pharmacia) by Gateway cloning technology to generate the GST-GhCPK33 protein. The primers used for these constructs are listed in Supplemental Table S1. GST and His fusion recombinant protein production was induced by isopropyl β-d-1-thiogalactopyranoside in the Escherichia coli strain BL21. The soluble GST and His fusion proteins were purified using the MagneGST Protein Purification System (Promega) or the MagneHis Protein Purification System (Promega) following the manufacturer’s instructions. The purified mixtures from pET-28-a and pGEX-4T-1 without the transgene insertions served as negative controls for the His-tagged fusion protein and GST-tagged fusion protein, respectively. Pull-down assays between GST-GhCPK33 and His-GhOPR3 were performed as reported previously (Deng et al., 2017). The primers used in this study are listed in Supplemental Table S1.
Co-IP Assay
The Co-IP assay was performed according to a previous report with minor modifications (Deng et al., 2017). In brief, the full lengths of GhCPK33 and GhOPR3 were amplified using the primer pairs GhCPK33-GFP-F/R and GhOPR3-MYC-F/R and inserted into the vectors pGWB451 and pGWB417 by Gateway cloning technology to generate the constructs GhCPK33-GFP and GhOPR3-MYC containing C-terminally tagged GFP protein and MYC protein, respectively. The plasmid 35S:GFP was used as a control. All plasmids were transformed individually into the A. tumefaciens strain GV3101. Clones carrying GhCPK33-GFP and GhOPR3-MYC, and those carrying 35S:GFP (pGWB452) and GhOPR3-MYC, were coinfiltrated into N. benthamiana leaves by A. tumefaciens infiltration (Chen et al., 2008). The infiltrated plants were grown at 25°C for 48 to 72 h, and the infiltrated leaf tissues were collected and frozen in liquid nitrogen. About 5 g of infiltrated leaf tissues was ground into powder in liquid nitrogen and homogenized in 5 mL of extraction buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% [v/v] Triton X-100, 5% [v/v] glycerol, 1 mm DTT, 1% [w/v] protease inhibitor cocktail [Roche], and 1% [w/v] PhosSTOP [Roche]). Immunoblotting and Co-IP experiments were performed following the instructions for the Dynabeads Co-Immunoprecipitation Kit (Invitrogen) using the Enhanced HRP-DAB Chromogenic Substrate Kit (Tiangen) to visualize the results. The primers used in this study are listed in Supplemental Table S1.
BiFC Assays and Subcellular Localization
To generate the BiFC constructs, the full lengths of GhCPK33 and GhOPR3 were amplified with primer pairs GhCPK33-1301-F/R and GhOPR3-1301-F/R and inserted into linearized pS1301nYFP or pS1301cYFP vectors (Yuan et al., 2010), which were digested with BamHI and SalI using an in-fusion enzyme to obtain GhCPK33-nYFP and GhOPR3-cYFP. The full lengths of GhJAZ2 (Hu et al., 2016) and GhCKI (Min et al., 2015) were amplified with primer pairs GhJAZ2-1301-F/R and GhCKI-1301-F/R and inserted into linearized pS1301nYFP or pS1301cYFP vectors to obtain GhJAZ2-cYFP and GhCKI-nYFP and used as negative controls for the BiFC assays. To determine the subcellular localization of GhCPK33 and GhOPR3, the full lengths of those genes were amplified with the corresponding primer pairs and cloned into vector pMDC83 by BP and LR recombination reactions to generate the C-terminally fused GFP constructs. CD3-977 was used as a peroxisome marker (Nelson et al., 2007). All vectors were transformed into N. benthamiana plants via the A. tumefaciens strain GV3101 (Chen et al., 2008). Fluorescence signals in leaf epidermal cells were observed using a confocal microscope (Olympus FV1200). The primers used in this study are listed in Supplemental Table S1.
In Vitro Phosphorylation Assay
Using GST-GhCPK33 (E. coli induced and expressed as described above) as the kinase and His-GhOPR3, His-GhOPR3-mA, or His- GhOPR3-mD (E. coli induced and expressed as described above) as the substrate, the kinase and substrate were mixed in a ratio of 2:1 with kinase assay buffer (25 mm Tris-HCl, pH 8.8, 0.5 mm DTT, 10 mm MgCl2, 0.1 mm CaCl2, and 0.1 mm ATP) in a total volume of 50 μL. The reactions were incubated at 30°C for 1 h and then stopped by heating at 65°C for 5 min. The results of the phosphorylation assay were detected by Phos-tag SDS-PAGE (12% [w/v] acrylamide gels, 0.375 m Tris-HCl, 0.1% [w/v] SDS, 0.1 mm MnCl2, and 0.05 mm Phos-tag AAL-107), following which the gels were visualized by silver staining or blotting with His antibody.
Kinase Activity Assay
The kinase activity of GhCPK33 (E. coli induced and expressed as described above) toward His-tagged GhOPR3 was measured using the Universal Fluorometric Kinase Assay Kit (Sigma-Aldrich) following the manufacturer’s instructions. The fluorescence intensities (λex = 540 nm/λem = 590 nm) were detected using a Multimode Plate Reader (Perkin-Elmer).
Dual-Luciferase Reporter Assays in N. benthamiana and Cotton Protoplasts
The full lengths of native GhOPR3, phosphodeficient GhOPR3-mA, and phosphomimic GhOPR3-mD were amplified with primer pair GhOPR3-LUC-F/R using the GhOPR3-pET-28a, GhOPR3-mA-pET-28a, and GhOPR3-mD-pET-28a plasmids as templates, respectively. The PCR-amplified fragments were inserted into linearized pGreenII 0800 vector digested with NotI and NcoI using an in-fusion enzyme to obtain the GhOPR3-pGreenII 0800, GhOPR3-mA-pGreenII 0800, and GhOPR3-mD-pGreenII 0800 vectors. The full length of the 35S promoter was amplified with primer pair 35S-0800-F/R from pGWB418 and inserted into linearized modified pGreenII 0800 vector (GhOPR3-pGreenII 0800, GhOPR3-mA-pGreenII 0800, or GhOPR3-mD-pGreenII 0800) digested with PstI and BamHI using an in-fusion enzyme to obtain the dual-luciferase reporter system. The full length of GhCPK33 was amplified with primer pair GhCPK33-62-SK-F/R and inserted into the linearized pGreenII 62-SK vector digested with PstI and BamHI using an in-fusion enzyme; the pGreenII 62-SK vector without any DNA insertion was used as an empty vector control. All of the plasmids were transformed individually into A. tumefaciens strain GV3101 for transient expression in N. benthamiana. The indicated constructs were adjusted to the same concentration (GhOPR3-pGreenII 0800, GhOPR3-mA-pGreenII 0800, and GhOPR3-mD-pGreenII 0800 were adjusted to OD600 = 0.2, empty vector and GhCPK33-pGreenII 62-SK were adjusted to OD600 = 0.6), and the pairs of the coinfiltrated constructs were mixed in equal volumes. The prepared constructs were coinfiltrated into N. benthamiana leaves in equal volumes to ensure comparability. The infiltrated plants were grown at 25°C for 72 to 96 h, and infiltrated leaves were then sprayed with 1 mm luciferin (Promega) and put in the dark for 5 min. The LUC luminescence was visualized using a cryogenically cooled CCD camera (Lumazome PyLoN 2048B), and the exposure time was set to 15 min. For MG132 treatment, N. benthamiana leaves were treated with 100 µm MG132 or DMSO (control) for 12 h in the dark. The primers used in this study are listed in Supplemental Table S1.
The dual-luciferase reporter assays in cotton protoplasts were performed as described previously with modifications. Specifically, the effector and reporter (6 μg each) were cotransformed into protoplasts using polyethylene glycol 4000. The transformed protoplasts were cultured at 25°C in the dark for 36 h; LUC and REN activity assays were performed according to the manufacturer’s instructions for the Dual Luciferase Reporter Assay System (Promega) and detected using the Multimode Plate Reader (Perkin-Elmer).
Transient Expression in N. benthamiana to Determine GhOPR3 Protein Stability
The full lengths of the phosphodeficient form GhOPR3-mA and the phosphomimic form GhOPR3-mD were amplified with primer pair GhOPR3-MYC-F/R using the GhOPR3-mA-pET-28a and GhOPR3-mD-pET-28a plasmids as templates, respectively. The PCR fragments were inserted into vector pGWB417 by Gateway cloning to generate the constructs GhOPR3-mA-MYC and GhOPR3-mD-MYC. All plasmids were transformed individually into the A. tumefaciens strain GV3101. The GhOPR3-MYC, 35S:GFP, and GhCPK33-GFP constructs described above (see “Co-IP Assay”) also were used in this experiment. The constructs were adjusted to the same concentration (GhOPR3-MYC, GhOPR3-mA-MYC, and GhOPR3-mD-MYC were adjusted to OD600 = 0.2, 35S:GFP and GhCPK33-GFP were adjusted to OD600 = 0.6), and the pairs of coinfiltrated constructs were mixed in equal volumes. Equal volumes of the prepared constructs were coinfiltrated into N. benthamiana leaves to ensure comparability. The infiltrated plants were grown at 25°C for 72 to 96 h. For the MG132 treatment, N. benthamiana leaves were treated with 100 µm MG132 or dimethyl sulfoxide (control) for 12 h in the dark. About 100 mg of infiltrated leaf tissues was ground into powder in liquid nitrogen and homogenized in 200 μL of extraction buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% [v/v] Triton X-100, 5% glycerol, 1 mm DTT, 1% protease inhibitor cocktail [Roche], and 1% PhosSTOP [Roche]), with a ratio of sample weight to extraction buffer volume of 1:2 (w/v). Next, 20 μL of the total protein samples was subjected to SDS-PAGE and immunoblotted with anti-MYC or anti-GFP antibody to determine protein levels. The primers used in this study are listed in Supplemental Table S1.
Elicitation of Cotton Suspension Cultured Cells
The treatments with OGAs and water were performed according to a procedure described by Hu et al. (2018). Five-milliliter cell suspension cultures were treated with 5 mL of distilled water or OGAs (final concentration, 0.5 mg mL−1). The cell suspension cultures were harvested at 10 h postelicitation and frozen at 80°C until further analysis.
Accession Numbers
Sequence data from this article can be found in the CottonFGD database (https://cottonfgd.org) or GenBank databases under the following accession numbers: GhCPK33 (six copies in the whole genome), Gh_D01G2360, Gh_A01G0621, Gh_D13G1455, Gh_A13G1164, Gh_D05G3567, and Gh_A04G0148; GhOPR3, Gh_D05G0339.1; and GhUB7, DQ116441.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of GhCPK33.
Supplemental Figure S2. Schematic diagrams of the VIGS constructs.
Supplemental Figure S3. Expression of GhCPK33 in control and GhCPK33-silenced plant leaves.
Supplemental Figure S4. Expression of genes involved in JA biosynthesis and the JA signaling pathway in leaves from control and GhCPK33-silenced plants.
Supplemental Figure S5. Expression of genes involved in JA biosynthesis and the JA signaling pathway in roots from control and GhCPK33-silenced plants.
Supplemental Figure S6. Amino acid sequence alignment of GhOPR3 and AtOPR3.
Supplemental Figure S7. Purification of GhOPR3 and GhCPK33 proteins and subcellular localization of GhCPK33:GFP and GhOPR3:GFP in vivo.
Supplemental Figure S8. Predicted GhOPR3 CAMK phosphorylation sites and identified GhCPK33-phosphorylated site.
Supplemental Figure S9. Dual-luciferase reporter system to determine GhOPR3-mD stability in vivo.
Supplemental Figure S10. Amino acid alignment of the OPR families (OPR1, OPR2, and OPR3) from cotton and Arabidopsis.
Supplemental Figure S11. Amino acid alignment of GhOPR3 and OPR3 proteins from other plant species.
Supplemental Table S1. Oligonucleotides used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We are indebted to Dongqin Li and Hongbo Liu (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University) for phytohormone qualification and to Huazhi Song and De Zhu for assistance with the laser scanning confocal microscope.
Footnotes
This work was supported by the National Natural Science Foundation of China (U1703231 and 31471541) and the International Science and Technology Cooperation Program of China (Grant 2015DFA30860).
Articles can be viewed without a subscription.
References
- Akira S, Uematsu S, Takeuchi O (2006) Pathogen recognition and innate immunity. Cell 124: 783–801 [DOI] [PubMed] [Google Scholar]
- Ali R, Ma W, Lemtiri-Chlieh F, Tsaltas D, Leng Q, von Bodman S, Berkowitz GA (2007) Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI, Hoagland DR (1940) Crop production in artificial culture solutions and in soils with special reference to factors influencing yields and absorption of inorganic nutrients. Soil Sci 50: 463–485 [Google Scholar]
- Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, Sonenberg N, Kay LE, Forman-Kay JD (2015) Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 519: 106–109 [DOI] [PubMed] [Google Scholar]
- Bender KW, Snedden WA (2013) Calmodulin-related proteins step out from the shadow of their namesake. Plant Physiol 163: 486–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J (2010) Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B, Munemasa S, Wang C, Nguyen D, Yong TY, Yang PG, Poretsky E, Belknap T, Waadt R, Alemán F, et al. (2015) Calcium specificity signaling mechanisms in abscisic acid signal transduction in Arabidopsis guard cells. eLife 4: 03599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundó M, Coca M (2017) Calcium-dependent protein kinase OsCPK10 mediates both drought tolerance and blast disease resistance in rice plants. J Exp Bot 68: 2963–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos ML, Kang JH, Howe GA (2014) Jasmonate-triggered plant immunity. J Chem Ecol 40: 657–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HQ, Han LB, Yang CL, Wu XM, Zhong NQ, Wu JH, Wang FX, Wang HY, Xia GX (2016) The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. J Exp Bot 67: 1935–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Monte I, Zamarreño AM, Hamberg M, Lassueur S, Reymond P, Weiss S, Stintzi A, Schaller A, Porzel A, et al. (2018) An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat Chem Biol 14: 171–178 [DOI] [PubMed] [Google Scholar]
- Clapham DE. (2007) Calcium signaling. Cell 131: 1047–1058 [DOI] [PubMed] [Google Scholar]
- Dammann C, Ichida A, Hong B, Romanowsky SM, Hrabak EM, Harmon AC, Pickard BG, Harper JF (2003) Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol 132: 1840–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Geyter N, Gholami A, Goormachtig S, Goossens A (2012) Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci 17: 349–359 [DOI] [PubMed] [Google Scholar]
- Deng F, Tu L, Tan J, Li Y, Nie Y, Zhang X (2012) GbPDF1 is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol 158: 890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Zhai K, Xie Z, Yang D, Zhu X, Liu J, Wang X, Qin P, Yang Y, Zhang G, Li Q, Zhang J, et al. (2017) Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355: 962–965 [DOI] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T (2013) Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA 110: 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantino E, Segretin ME, Santin F, Mirkin FG, Ulloa RM (2017) Analysis of the potato calcium-dependent protein kinase family and characterization of StCDPK7, a member induced upon infection with Phytophthora infestans. Plant Cell Rep 36: 1137–1157 [DOI] [PubMed] [Google Scholar]
- Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GC, Joosten MH, Thomma BP (2011) Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol 156: 2255–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso V, Rothe E, Baldwin IT, Kim SG (2014) Root jasmonic acid synthesis and perception regulate folivore-induced shoot metabolites and increase Nicotiana attenuata resistance. New Phytol 202: 1335–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Long L, Xu L, Gao WH, Sun LQ, Liu LL (2013a) Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol Cell Proteomics 12: 3690–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wheeler T, Li Z, Kenerley CM, He P, Shan L (2011) Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J 66: 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J, et al. (2013b) Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng S, Li A, Tang L, Yin L, Wu L, Lei C, Guo X, Zhang X, Jiang G, Zhai W, et al. (2013) TaCPK2-A, a calcium-dependent protein kinase gene that is required for wheat powdery mildew resistance enhances bacterial blight resistance in transgenic rice. J Exp Bot 64: 3125–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettenhausen C, Baldwin IT, Wu J (2013a) Nicotiana attenuata MPK4 suppresses a novel jasmonic acid (JA) signaling-independent defense pathway against the specialist insect Manduca sexta, but is not required for the resistance to the generalist Spodoptera littoralis. New Phytol 199: 787–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettenhausen C, Yang DH, Baldwin IT, Wu J (2013b) Calcium-dependent protein kinases, CDPK4 and CDPK5, affect early steps of jasmonic acid biosynthesis in Nicotiana attenuata. Plant Signal Behav 8: e22784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer RA, Tsuda K, Rallapalli G, Asai S, Truman W, Papke MD, Sakakibara H, Jones JDG, Myers CL, Katagiri F (2017) The highly buffered Arabidopsis immune signaling network conceals the functions of its components. PLoS Genet 13: e1006639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Jander G (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Hu H, He X, Tu L, Zhu L, Zhu S, Ge Z, Zhang X (2016) GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor GhMYB25-like. Plant J 88: 921–935 [DOI] [PubMed] [Google Scholar]
- Hu Q, Min L, Yang X, Jin S, Zhang L, Li Y, Ma Y, Qi X, Li D, Liu H, et al. (2018) Laccase GhLac1 modulates broad-spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and jasmonic acid synthesis. Plant Physiol 176: 1808–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Yu D (2016) The WRKY57 transcription factor affects the expression of Jasmonate ZIM-Domain genes transcriptionally to compromise Botrytis cinerea resistance. Plant Physiol 171: 2771–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourieux D, Ranjeva R, Pugin A (2006) Calcium in plant defence-signalling pathways. New Phytol 171: 249–269 [DOI] [PubMed] [Google Scholar]
- Li B, Meng X, Shan L, He P (2016a) Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 19: 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, He X, Luo X, Xu L, Liu L, Min L, Jin L, Zhu L, Zhang X (2014) Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol 166: 2179–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang W, Gao J, Yin K, Wang R, Wang C, Petersen M, Mundy J, Qiu JL (2016b) MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell 28: 2866–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhu Z, Chern M, Yin J, Yang C, Ran L, Cheng M, He M, Wang K, Wang J, et al. (2017) A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170: 114–126.e15 [DOI] [PubMed] [Google Scholar]
- Liu H, Li X, Xiao J, Wang S (2012) A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice-bacterium interaction. Plant Methods 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Smigel A, Tsai YC, Braam J, Berkowitz GA (2008) Innate immunity signaling: cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol 148: 818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí MC, Stancombe MA, Webb AA (2013) Cell- and stimulus type-specific intracellular free Ca2+ signals in Arabidopsis. Plant Physiol 163: 625–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschi S, Hake K, Herde M, Hause B, Romeis T (2015) The calcium-dependent protein kinase CPK28 regulates development by inducing growth phase-specific, spatially restricted alterations in jasmonic acid levels independent of defense responses in Arabidopsis. Plant Cell 27: 591–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L, Hu Q, Li Y, Xu J, Ma Y, Zhu L, Yang X, Zhang X (2015) LEAFY COTYLEDON1-CASEIN KINASE I-TCP15-PHYTOCHROME INTERACTING FACTOR4 network regulates somatic embryogenesis by regulating auxin homeostasis. Plant Physiol 169: 2805–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J, Matschi S, Shorinola O, Rovenich H, Matei A, Segonzac C, Malinovsky FG, Rathjen JP, MacLean D, Romeis T, et al. (2014) The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16: 605–615 [DOI] [PubMed] [Google Scholar]
- Monaghan J, Matschi S, Romeis T, Zipfel C (2015) The calcium-dependent protein kinase CPK28 negatively regulates the BIK1-mediated PAMP-induced calcium burst. Plant Signal Behav 10: e1018497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motte H, Beeckman T (2017) PHR1 balances between nutrition and immunity in plants. Dev Cell 41: 5–7 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nickstadt A, Thomma BP, Feussner I, Kangasjärvi J, Zeier J, Loeffler C, Scheel D, Berger S (2004) The jasmonate-insensitive mutant jin1 shows increased resistance to biotrophic as well as necrotrophic pathogens. Mol Plant Pathol 5: 425–434 [DOI] [PubMed] [Google Scholar]
- Ranf S, Eschen-Lippold L, Pecher P, Lee J, Scheel D (2011) Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J 68: 100–113 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz SS, Vadassery J, Heyer M, Reichelt M, Bender KW, Snedden WA, Boland W, Mithöfer A (2014) Mutation of the Arabidopsis calmodulin-like protein CML37 deregulates the jasmonate pathway and enhances susceptibility to herbivory. Mol Plant 7: 1712–1726 [DOI] [PubMed] [Google Scholar]
- Shen L, Yang S, Yang T, Liang J, Cheng W, Wen J, Liu Y, Li J, Shi L, Tang Q, et al. (2016) CaCDPK15 positively regulates pepper responses to Ralstonia solanacearum inoculation and forms a positive-feedback loop with CaWRKY40 to amplify defense signaling. Sci Rep 6: 22439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz HU, Mitrousia GK, de Wit PJ, Fitt BD (2014) Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci 19: 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhu L, Xu L, Yuan D, Min L, Zhang X (2014) Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat Commun 5: 5372–5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczegielniak J, Borkiewicz L, Szurmak B, Lewandowska-Gnatowska E, Statkiewicz M, Klimecka M, Cieśla J, Muszyńska G (2012) Maize calcium-dependent protein kinase (ZmCPK11): local and systemic response to wounding, regulation by touch and components of jasmonate signaling. Physiol Plant 146: 1–14 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax JA, Ferrell JE Jr (2007) Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol 8: 530–541 [DOI] [PubMed] [Google Scholar]
- Wang J, Grubb LE, Wang J, Liang X, Li L, Gao C, Ma M, Feng F, Li M, Li L, et al. (2018a) A regulatory module controlling homeostasis of a plant immune kinase. Mol Cell 69: 493–504.e6 [DOI] [PubMed] [Google Scholar]
- Wang J, Wang S, Hu K, Yang J, Xin X, Zhou W, Fan J, Cui F, Mou B, Zhang S, et al. (2018b) The kinase OsCPK4 regulates a buffering mechanism that fine-tunes innate immunity. Plant Physiol 176: 1835–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AA. (2013) Focus issue: calcium signaling. Plant Physiol 163: 457–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Shen F, Hong Y, Rong W, Du L, Liu X, Xu H, Ma L, Zhang Z (2016) The wheat calcium-dependent protein kinase TaCPK7-D positively regulates host resistance to sharp eyespot disease. Mol Plant Pathol 17: 1252–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Shan L, He P (2014) Microbial signature-triggered plant defense responses and early signaling mechanisms. Plant Sci 228: 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XF, Nomura K, Aung K, Velásquez AC, Yao J, Boutrot F, Chang JH, Zipfel C, He SY (2016) Bacteria establish an aqueous living space in plants crucial for virulence. Nature 539: 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Yang L, Zhang J, Guo X, Zhang X, Li G (2012) Prevalence of the defoliating pathotype of Verticillium dahliae on cotton in central China and virulence on selected cotton cultivars. J Phytopathol 160: 369–376 [Google Scholar]
- Xu G, Yuan M, Ai C, Liu L, Zhuang E, Karapetyan S, Wang S, Dong X (2017) uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545: 491–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhu L, Tu L, Liu L, Yuan D, Jin L, Long L, Zhang X (2011) Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J Exp Bot 62: 5607–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CL, Liang S, Wang HY, Han LB, Wang FX, Cheng HQ, Wu XM, Qu ZL, Wu JH, Xia GX (2015) Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae. Mol Plant 8: 399–411 [DOI] [PubMed] [Google Scholar]
- Yang DH, Hettenhausen C, Baldwin IT, Wu J (2012) Silencing Nicotiana attenuata calcium-dependent protein kinases, CDPK4 and CDPK5, strongly up-regulates wound- and herbivory-induced jasmonic acid accumulations. Plant Physiol 159: 1591–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Chu Z, Li X, Xu C, Wang S (2010) The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell 22: 3164–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Ding Z, Wu K, Yang L, Li Y, Yang Z, Shi S, Liu X, Zhao S, Yang Z, et al. (2016) Suppression of jasmonic acid-mediated defense by viral-inducible microRNA319 facilitates virus infection in rice. Mol Plant 9: 1302–1314 [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang M, Li N, Wang H, Qiu P, Pei L, Xu Z, Wang T, Gao E, Liu J, et al. (2018) Long noncoding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant Biotechnol J 16: 1172–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Corwin JA, Copeland D, Feusier J, Eshbaugh R, Chen F, Atwell S, Kliebenstein DJ (2017) Plastic transcriptomes stabilize immunity to pathogen diversity: the jasmonic acid and salicylic acid networks within the Arabidopsis/Botrytis pathosystem. Plant Cell 29: 2727–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Lan W, Chen B, Fang W, Luan S (2015) A calcium sensor-regulated protein kinase, CALCINEURIN B-LIKE PROTEIN-INTERACTING PROTEIN KINASE19, is required for pollen tube growth and polarity. Plant Physiol 167: 1351–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ, et al. (2007) Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]