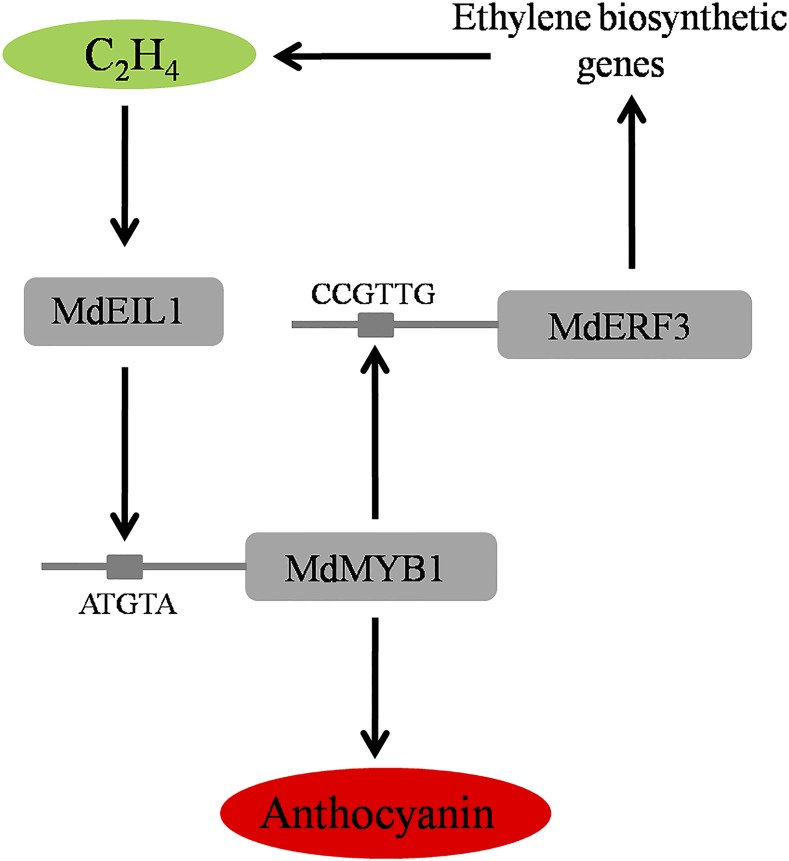
MdEIL1 directly activates MdMYB1 expression to promote anthocyanin accumulation, and MdMYB1 directly activates MdERF3 to promote ethylene production.
Abstract
Ethylene regulates climacteric fruit ripening, and EIN3-LIKE1 (EIL1) plays an important role in this process. In apple (Malus domestica), fruit coloration is accompanied by ethylene release during fruit ripening, but the molecular mechanism that underlies these two physiological processes is unknown. In this study, we found that ethylene treatment markedly induced fruit coloration as well as the expression of MdMYB1, a positive regulator of anthocyanin biosynthesis and fruit coloration. In addition, we found that MdEIL1 directly bound to the promoter of MdMYB1 and transcriptionally activated its expression, which resulted in anthocyanin biosynthesis and fruit coloration. Furthermore, MdMYB1 interacted with the promoter of ETHYLENE RESPONSE FACTOR3, a key regulator of ethylene biosynthesis, thereby providing a positive feedback for ethylene biosynthesis regulation. Overall, our findings provide insight into a mechanism involving the synergistic interaction of the ethylene signal with the MdMYB1 transcription factor to regulate ethylene biosynthesis and fruit coloration in apple.
For many fleshy fruits, the fruit-ripening process is accompanied by dramatic changes in fruit characteristics, including color changes (degradation of chlorophylls and accumulation of pigments), degradation of starches, accumulation of soluble sugars and volatile compounds, as well as the release of ethylene (Klee and Giovannoni, 2011). Fruit ripening is crucial for cultivated fruits and determines the quality and storage ability of fruits. Therefore, it is important to elucidate the regulatory mechanisms of fruit ripening, which will provide information for improving fruit quality.
Ethylene is essential for the ripening of climacteric fruits, and a rapid burst of ethylene production and a rise in respiration occur at the transition point of fruit ripening (Theologis, 1992; Giovannoni, 2004). Ethylene biosynthesis and the signal transduction pathway have been studied intensively in many species, including tomato (Solanum lycopersicum) and apple (Malus domestica; Alexander and Grierson, 2002; Alonso and Stepanova, 2004; Lin et al., 2009; Klee and Giovannoni, 2011; Gapper et al., 2013; Grierson, 2013; Ireland et al., 2013; Li et al., 2016, 2017).
In plants, S-adenosyl Met is a precursor in ethylene biosynthesis and is first catalyzed to 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthase (ACS). Then, ACC is oxidized and converted to active ethylene by ACC oxidase (ACO; Yang and Hoffman, 1984; Wang et al., 2002). During the signal transduction process, ethylene activates the downstream signaling pathway through several components, including CONSTITUTIVE TRIPLE RESPONSE1 and ETHYLENE INSENSITIVE2 (EIN2). Subsequently, the primary responsive transcription factors (TFs), EIN3/EIN3-LIKEs (EILs), induce the secondary responsive TFs, ETHYLENE RESPONSE FACTORs (ERFs), which, in turn, activate the expression of downstream ethylene-responsive related genes, ultimately inducing the ethylene response (Alonso and Stepanova, 2004; Lin et al., 2009; Klee and Giovannoni, 2011; Gapper et al., 2013).
ACS and ACO proteins are essential for ethylene biosynthesis as well as fruit ripening and have been characterized extensively in tomato (Barry et al., 2000; Lin et al., 2009; Klee and Giovannoni, 2011; Gapper et al., 2013), apple (Sunako et al., 1999; Harada et al., 2000; Dandekari et al., 2004; Oraguzie et al., 2004; Dougherty et al., 2016), and other species. To date, a series of proteins that regulate ethylene biosynthesis and fruit ripening through ACSs and ACOs have been identified. In tomato, the MADS box protein RIPENING INHIBITOR promotes fruit ripening through transcriptionally activating the expression of LeACS2 (Ito et al., 2008; Fujisawa et al., 2013). In banana (Musa nana), MaERF11 recruits HISTONE DEACETYLASE1 to suppress the expression of MaACO1 (Han et al., 2016). In apple, MdMADS8 and MdMADS9 interact with the promoters of MdACS1 and MdACO1 and activate their expression, promoting ethylene production (Ireland et al., 2013). In apple, MdERF3 promotes, whereas MdERF2 represses, the expression of MdACS1 (Li et al., 2016, 2017). Aside from transcriptional regulation, posttranslational modification also is important in the regulation of ethylene biosynthesis. The Bric-à-brac, Tramtrack, and Broadcomplex ubiquitin ligase ETHYLENE OVERPRODUCER1 targets several ACSs for degradation to negatively regulate ethylene biosynthesis (Wang et al., 2004). In addition, the phosphorylation of ACS6 by MPK6 represses 26S proteasome pathway-mediated degradation (Joo et al., 2008), whereas this phosphorylation is suppressed by PROTEIN PHOSPHATASE2A (Skottke et al., 2011). These findings suggest that both the transcriptional and posttranslational regulations are important cues that modulate ethylene synthesis.
EIN3 and its EIL homologs are key TFs that initiate the ethylene-mediated downstream transcriptional cascade (Chao et al., 1997; Solano et al., 1998). In Arabidopsis (Arabidopsis thaliana), the MAP kinase (MAPK/MPK) cascade signaling and Glc signaling pathways interact with EIN3 to regulate its phosphorylation and protein stability (Yanagisawa et al., 2003; Yoo et al., 2008). In addition, EIN3/EIL stability is tightly regulated by the EIN3-BINDING F-BOX PROTEIN1/2-mediated ubiquitin-proteasome degradation pathway (Guo and Ecker, 2003; Potuschak et al., 2003). As a central TF, EIN3 activates the expression of a wide range of downstream genes, including ERF1, HOOKLESS1, C-REPEAT BINDING FACTOR, EPITHELIUM-SPECIFIC ETS1, and others, to regulate hypocotyl elongation, apical hook formation, abiotic stress tolerance, and other ethylene response-related processes (Zhang et al., 2011; Shi et al., 2012; Shen et al., 2016). In banana, MaEIL5 interacts physically with NAM, ATAF1/2, CUC22, which may be involved in ethylene-mediated fruit ripening (Shan et al., 2012). In kiwifruit (Actinidia chinensis), AdEIL2 and AdEIL3 regulate the fruit-ripening processes by activating the transcription of AdACO1 and stimulating ethylene production (Yin et al., 2010). Taken together, EIN3/EILs may emerge as key regulators in ethylene signaling transduction.
Anthocyanins are secondary metabolites that are widely distributed in flowers and fruits and attract animals for pollination and seed dispersal (Cipollini and Levey, 1997; Schaefer et al., 2004). They are synthesized via the phenylpropanoid pathway, and many synthesis-related catalytic enzymes have been identified in a variety of plant species, including PHENYLALANINE AMMONIA LYASE (PAL), CHALCONE SYNTHASE (CHS), CHALCONE ISOMERASE (CHI), FLAVANONE 3-HYDROXYLASE (F3H), DIHYDROFLAVONOL 4-REDUCTASE (DFR), ANTHOCYANIDIN SYNTHASE, and UDP-GLUCOSE/FLAVONOID 3-O-GLUCOSYLTRANSFERASE (UFGT; Koes et al., 2005; Tanaka et al., 2008). The MYB/bHLH/WD40 complex has been recognized as central to the regulation of anthocyanin accumulation (Ramsay and Glover, 2005; Gonzalez et al., 2008; Hichri et al., 2011). MYB TF-regulated anthocyanin accumulation has been well studied in Arabidopsis (Borevitz et al., 2000), apple (Ban et al., 2007; Espley et al., 2007; Allan et al., 2008), pear (Pyrus bretschneideri; Yao et al., 2017), peach (Prunus persica; Zhou et al., 2016), and other species (Jaakola, 2013; Xu et al., 2015). In apple, the homologs of Arabidopsis PRODUCTION OF ANTHOCYANIN PIGMENT1/2 (PAP1/2), MdMYB1/10/A, are responsible for anthocyanin biosynthesis. They are known as positive regulators for anthocyanin biosynthesis and fruit coloration by directly targeting the downstream anthocyanin-associated genes (Ban et al., 2007; Espley et al., 2007; Allan et al., 2008).
In plants, a series of developmental and environmental factors, including internal hormones and external environments, influence anthocyanin biosynthesis (Lancaster and Dougall, 1992; Jaakola, 2013). Recent studies have shown that MYB TFs play important roles in the internal and external cue-modulated anthocyanin accumulation by both transcriptional and posttranscriptional regulation (Jaakola, 2013; Xu et al., 2015). For example, light regulates anthocyanin accumulation by modulating the expression levels and protein stabilities of the MYB TFs (Takos et al., 2006; Li et al., 2012; An et al., 2017a). Nitrate acts as a negative factor in regulating anthocyanin accumulation mainly by decreasing the expression of PAP1/2 in Arabidopsis (Rubin et al., 2009). It is also well known that the expression of MdMYB1 and its alleles is up-regulated in fruit ripening, accompanied by fruit coloration and ethylene release (Faragher and Brohier, 1984; Whale and Singh, 2007; Whale et al., 2008). However, further research is required to clarify how these anthocyanin-related MYB TFs are regulated by ethylene and other stimuli.
Apple is a climacteric fruit that exhibits a burst of respiration and ethylene production during fruit ripening after harvest (Faragher and Brohier, 1984; Song and Bangerth, 1996). Multiple ethylene-regulated proteins that are involved in fruit ripening have been identified by a proteomic approach (Zheng et al., 2013). In apple, it is crucial to clarify the regulatory mechanism of ethylene release and fruit coloration during fruit maturation, which determines the storage life and quality of the fruit. Previous studies have found that ethylene release is increased and fruit pigment is accumulated during fruit ripening (Faragher and Brohier, 1984; Whale and Singh, 2007; Whale et al., 2008), but less is known about whether cross talk exists between these two physiological processes. In this study, we found that ethylene treatment markedly induced fruit coloration as well as the expression of MdEIL1 and MdMYB1. MdEIL1 bound directly to the promoter of MdMYB1 to increase the ethylene-mediated anthocyanin accumulation and fruit coloration. In addition, MdMYB1 also induced ethylene release by transcriptionally activating the expression of MdERF3, a positive regulator of ethylene biosynthesis, which accounts for more ethylene release in red-flesh apple fruits. Overall, these results provide new insights into the regulatory mechanism of the synergistic interaction of the ethylene signal with the MdMYB1 TF to regulate anthocyanin accumulation and fruit coloration.
RESULTS
Ethephon Treatment Promotes Anthocyanin Biosynthesis in Apple Fruits
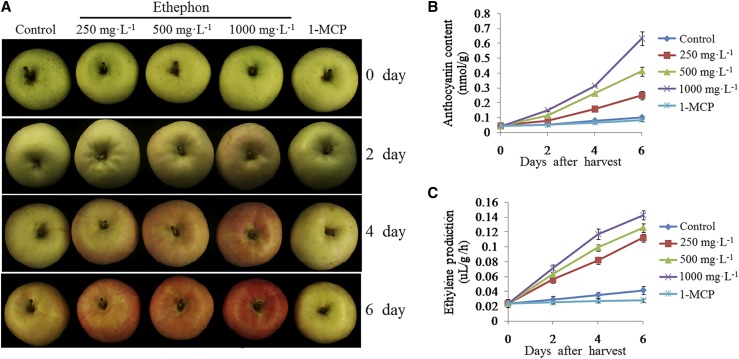
A previous study showed that the onset of anthocyanin accumulation coincided with the start of rapid ethylene release during apple ripening (Faragher and Brohier, 1984). For an in-depth investigation of the role of ethylene in the coloration of apples, cv Red Delicious apples harvested 120 d after full bloom (DAFB) were treated with different concentrations of ethephon solutions (0, 250, 500, and 1,000 mg L−1) or 1-methylcyclopropene (1-MCP, an ethylene inhibitor; 1 µL L−1) and stored in a phytotron at 24°C with constant light (70 μmol m−2 s−1) for 6 d. The anthocyanin contents and ethylene production of cv Red Delicious apples were then measured. Anthocyanin accumulation and ethylene production increased significantly with the ethephon treatment but were suppressed by the 1-MCP treatment (Fig. 1). These results indicated a positive association between ethylene production and anthocyanin synthesis and that the ethephon treatment contributed to anthocyanin accumulation in the apples.
Figure 1.
Ethephon induces anthocyanin accumulation in cv Red Delicious apples. A, The cv Red Delicious apples were grown in bags beginning at 30 DAFB. They were debagged at 120 DAFB and treated with the indicated concentrations of ethephon solutions (250, 500, and 1,000 mg L−1) or 1-MCP (1 µL L−1) and then were stored in a phytotron at 24°C with constant high light (70 μmol m−2 s−1) for 6 d. Untreated fruits were used as the control. Representative images were taken. The assay was performed in three replicates. B and C, Measurements of anthocyanin content (B) and ethylene production (C) in ethephon- or 1-MCP-treated and control apples. The assay was performed in three replicates. The results represent means of these three replicates. Error bars indicate sd.
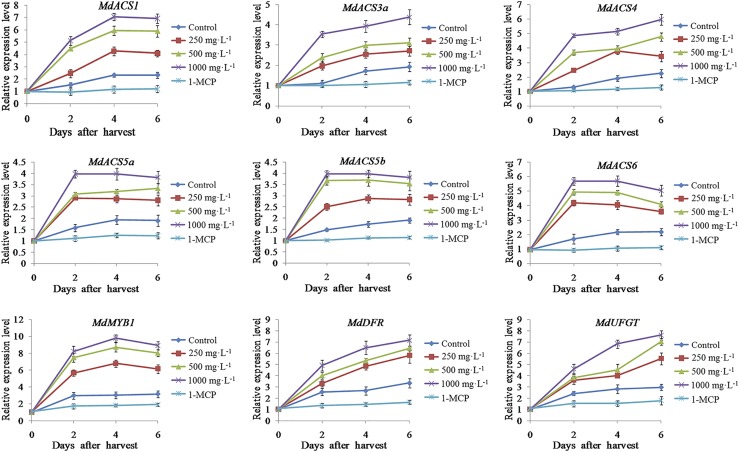
In addition, the expression levels of the ethylene- and anthocyanin-associated genes were analyzed. The expression of ethylene synthetic genes (MdACS1, MdACS3a, MdACS4, MdACS5a, MdACS5b, and MdACS6), anthocyanin synthetic genes (MdDFR and MdUFGT), and the regulatory gene MdMYB1 was induced by the ethephon treatment but was suppressed by the 1-MCP treatment (Fig. 2). These results suggested that ethylene promoted apple anthocyanin synthesis and fruit coloration through regulating the transcription of MdMYB1 and anthocyanin synthetic genes.
Figure 2.
Expression analysis of ethylene and anthocyanin synthetic genes in ethephon- or 1-MCP-treated and control cv Red Delicious apples by reverse transcription-quantitative PCR (RT-qPCR). Ethylene biosynthetic genes are MdACS1, MdACS3a, MdACS4, MdACS5a, MdACS5b, and MdACS6, and anthocyanin biosynthetic genes are MdMYB1, MdDFR, and MdUFGT. The value for day 0 was set to 1. RT-qPCR was performed with three technical replicates and three biological replicates. The results represent means of these three replicates. Error bars indicate sd.
MdMYB1 Is a Direct Target of MdEIL1
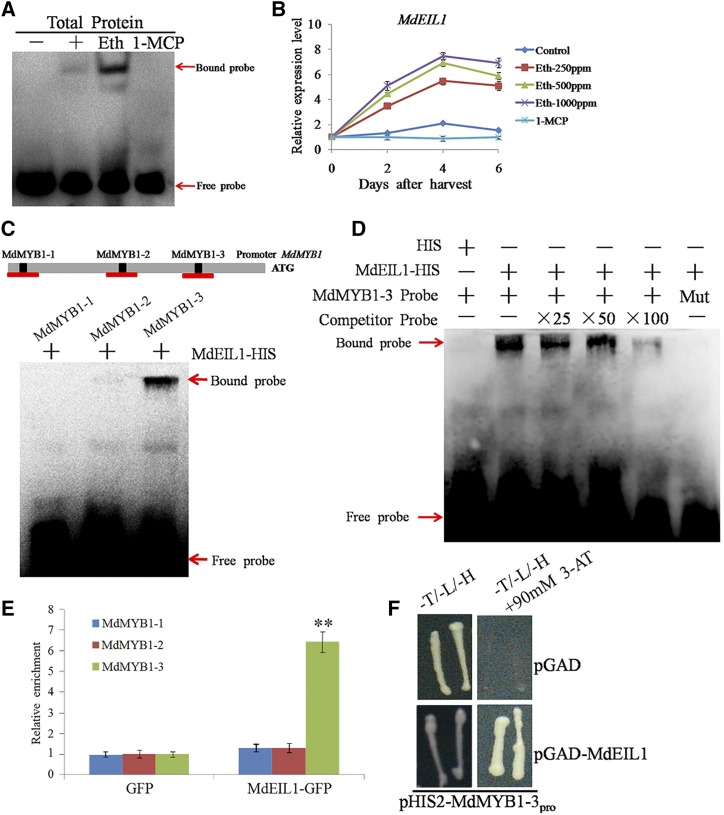
Considering that the transcription of MdMYB1 was induced by ethylene (Fig. 2) and that MdMYB1 has an essential role in regulating anthocyanin synthesis and fruit coloration in apple (Takos et al., 2006; Espley et al., 2007), we speculated that ethylene affects anthocyanin biosynthesis by regulating the transcription of MdMYB1. To verify this hypothesis, we analyzed the potential binding sequences of ethylene-related genes, such as the dehydration-responsive element, the GCC box, and the ATGTA motif in the MdMYB1 promoter region (Fujimoto et al., 2000; Liu et al., 2006; Zhang et al., 2011). Three ATGTA motifs (sites MdMYB1-1, MdMYB1-2, and MdMYB1-3) were found (Supplemental Fig. S1; Supplemental Table S1). To screen the potential ethylene-responsive element-binding factors, mixed biotin-labeled probes containing the three different ATGTA sequences were prepared for a DNA-affinity trapping assay. As shown in Figure 3A, when the total protein extracts were incubated with the mixed biotin-labeled probes, a blurry DNA-protein complex was observed. The band became clearer in the sample treated with ethephon, whereas it disappeared when treated with 1-MCP. Subsequently, the binding proteins were analyzed using mass spectrometry, and the candidate protein (GenBank accession no. MDP0000423881) bound to the ATGTA sequence was named MdEIL1 through multiple sequence alignment and phylogeny evolution analysis (Supplemental Fig. S2). MdEIL1 had a similar expression pattern to MdMYB1 in response to the ethephon and 1-MCP treatments (Fig. 3B), indicating an association between these two genes.
Figure 3.
Binding of MdEIL1 to the MdMYB1 promoter. A, Identification of the MdEIL1 protein that binds the cis-element of the MdMYB1 promoter with an electrophoretic mobility shift assay (EMSA). The total proteins were extracted from ethephon- or 1-MCP-treated and untreated apple plants. – and + represented samples without or with the addition of total protein extracted from the untreated apple plants, respectively. Eth and 1-MCP represent samples with the addition of total protein extracted from the ethephon- or 1-MCP-treated apple plants, respectively. B, Expression analysis of MdEIL1 in ethephon- or 1-MCP-treated and control apples by RT-qPCR. The value for day 0 was set to 1. RT-qPCR was performed with three technical replicates and three biological replicates. The results represent means of these three replicates. Error bars indicate sd. C, Top, schematic diagram of the MdMYB1 promoter showing the potential MdEIL1-binding sites (MdMYB1-1, MdMYB1-2, and MdMYB1-3). The predicted ATGTA sequences are indicated by black boxes. Red lines represent the fragments amplified in the ChIP-PCR assay. Bottom, EMSA showing the MdEIL1-HIS fusion protein bound to the MdMYB1-3 site of the MdMYB1 promoter and the free and bound DNAs separated on an acrylamide gel. D, EMSA showing that the MdEIL1-HIS fusion protein bound directly to the MdMYB1-3 site of the MdMYB1 promoter. Unlabeled probes were used as competitors. Mut represents a mutated probe in which the ATGTA motif was replaced by CTTGC. E, ChIP-PCR assay of MdEIL1 binding to the promoter of the MdMYB1 gene. Chromatin from the empty vector control (GFP) and 35S:MdEIL1-GFP apple calli (MdEIL1-GFP) were immunoprecipitated with anti-GFP antibodies or without antibodies. Three regions (MdMYB1-1, MdMYB1-2, and MdMYB1-3) were examined by RT-qPCR. The enrichment of the wild type was set to 1. RT-qPCR was performed with three technical replicates and three biological replicates to examine the enrichment of MdMYB1 fragments. The results represent means of these three replicates. Error bars indicate sd. Asterisks denote Student’s t test significance: **, P < 0.01. F, Y1H assay showing MdEIL1 interaction with the promoter of MdMYB1. The promoter fragment of MdMYB1 (MdMYB1-3) was fused to the pHIS2 vector, and the MdEIL1 gene was fused to the pGAD vector. The columns represent the addition of the pHIS2-MdMYB1-3pro vector. The rows represent the addition of the pGAD and pGAD-MdEIL1 vectors.
To verify whether MdEIL1 bound to the MdMYB1 promoter, EMSAs were performed using an MdEIL1-HIS fusion protein. As shown in Figure 3C, MdEIL1 specifically bound to the MdMYB1-3 site. When unlabeled probes were added as competitors, the binding was reduced, whereas the band disappeared when the mutated probes were added (Fig. 3D). These results indicated that MdEIL1 bound to the ATGTA motif of the MdMYB1 promoter in vitro.
To further confirm the binding of MdEIL1 to the MdMYB1 promoter in vivo, a chromatin immunoprecipitation (ChIP)-PCR assay was conducted using 35S:MdEIL1-GFP transgenic apple calli and the empty vector GFP transgenic apple calli as a control. The fragment containing MdMYB1-1 had higher enrichment in MdEIL1 transgenic apple calli, suggesting that MdEIL1 bound to the promoter of MdMYB1 in vivo (Fig. 3E).
In addition, a yeast one-hybrid (Y1H) assay was performed. The MdMYB1-3 sequence was cloned to a pHIS2 vector, and the coding sequence (CDS) of MdEIL1 was inserted into the pGADT7 vector. The optimal concentration (90 mm) of 3-amino-1,2,4-triazole (3-AT; Supplemental Fig. S3) was used for the experiments. The results showed that the coexpressed yeast strains of pGAD-MdEIL1 and pHIS2-MdMYB1-3 were able to grow on a SD/-Trp/-Leu/-His/90 mm 3-AT plate, but no growth was observed on the control plate (Fig. 3F). These results provide evidence that MdEIL1 binds to the MdMYB1 promoter.
MdEIL1 Positively Regulates the Expression of MdMYB1
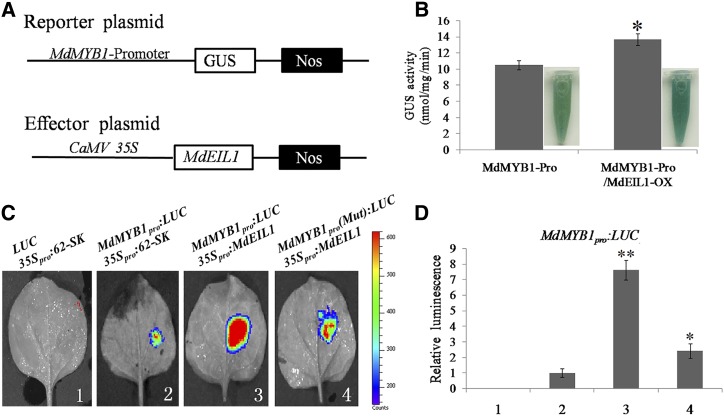
A GUS transactivation assay was performed in apple calli to determine how MdEIL1 regulates the expression of MdMYB1. The 2,000-bp sequence of the MdMYB1 promoter was cloned into the pBI101-GUS vector as an effector plasmid (MdMYB1-Pro), and the effector plasmid was then transformed into wild-type apple calli. The 35S:MdEIL1 construct was cotransformed into the MdMYB1-Pro transgenic apple calli (MdMYB1-Pro/MdEIL1-OX; Fig. 4A). GUS activity analysis demonstrated that the MdMYB1-Pro/MdEIL1-OX transgenic apple calli had higher GUS activity than MdMYB1-Pro alone (Fig. 4B), suggesting that MdEIL1 activated the transcription of MdMYB1.
Figure 4.
Regulation of the MdMYB1 promoter by MdEIL1. A, Schematic representation of the GUS reporter vector containing the MdMYB1 promoter and the effector vector containing MdEIL1. B, GUS staining analysis and GUS activity detection of pMdMYB1-GUS and pMdMYB1-GUS/MdEIL1-OX transgenic apple calli. MdMYB1-Pro, MdMYB1-Promoter-GUS transgenic apple calli; MdMYB1-Pro/MdEIL1-OX, MdMYB1-Promoter-GUS and 35S:MdEIL1-GFP cotransformed apple calli. The GUS activity of MdMYB1-Pro was used as the reference. Transgenic apple calli were stained using GUS buffer. GUS staining analysis and GUS activity detection were performed in three replicates. The results represent means of these three replicates. Error bars indicate sd. The asterisk denotes Student’s t test significance: *, P < 0.05. C, Transient expression assays showing MdEIL1 promotion of MdMYB1 expression. The promoter fragment of MdMYB1 was cloned into the pGreenII 0800-LUC vector to generate the reporter construct. The effector (35Spro:MdEIL1) was generated by recombining the MdEIL1 gene into the pGreenII 62-SK vector. In MdMYB1pro(Mut), the ATGTA motif was replaced by CTTGC. D, Quantitative analysis of luminescence intensity. The value for column 2 (MdMYB1pro:LUC-35Spro:62-SK) was set to 1. The transient expression assay was performed in three replicates. The results represent means of these three replicates. Error bars indicate sd. Asterisks denote Student’s t test significance: *, P < 0.05 and **, P < 0.01.
To verify the transcriptional activation results, a transient transactivation assay was conducted in tobacco (Nicotiana tabacum) leaves. The CDS of MdEIL1 was fused to the pGreenII 62-SK vector as an effector, and the promoter sequences of MdMYB1 and the mutated MdMYB1 promoter fragments were inserted into the pGreenII 0800-LUC vectors to generate reporters [MdMYB1pro and MdMYB1(Mut)pro, respectively; Supplemental Fig. S4]. Agrobacterium tumefaciens containing recombinant plasmids were mixed and coinjected into the tobacco leaves. The luminescence detection indicated that the coexpression of 35Spro:MdEIL1 with MdMYB1Pro:LUC resulted in much stronger luminescence compared with the controls, whereas 35Spro:MdEIL1 was unable to induce the expression of MdMYB1Pro(Mut):LUC (Fig. 4, C and D). These results indicated that MdEIL1 directly activated the expression of MdMYB1.
MdEIL1 Promotes Anthocyanin Biosynthesis and Fruit Coloration
Based on the results that MdEIL1 interacted with the MdMYB1 promoter and activated its transcription (Figs. 3 and 4), we speculated that MdEIL1 may play a crucial role in regulating anthocyanin biosynthesis. Hence, the overexpression vector 35S:MdEIL1-GFP was constructed and transformed into wild-type apple calli (Supplemental Fig. S5A), and two transgenic lines (MdEIL1#1 and MdEIL1#2) were selected for the following assays. The coloration assays showed that the overexpression of MdEIL1 significantly increased the anthocyanin accumulation compared with the wild type (Supplemental Fig. S5, B and C). In addition, RT-qPCR was performed to test the expression levels of anthocyanin synthetic genes in wild-type and MdEIL1 transgenic apple calli. Overexpression of MdEIL1 up-regulated the expression of anthocyanin synthetic genes (MdDFR, MdUFGT, MdF3H, MdCHI, and MdCHS) and the regulatory gene MdMYB1 (Supplemental Fig. S5D). To further characterize its function, 35S:MdEIL1-GFP was transformed into wild-type Arabidopsis, and three lines (MdEIL1-L1, MdEIL1-L2, and MdEIL1-L4) were selected. Coloration assays and RT-qPCR analysis demonstrated that MdEIL1 positively regulated anthocyanin synthesis in Arabidopsis by up-regulating anthocyanin synthesis-related genes, including AtPAP1, AtPAP2, AtPAL1, AtDFR, AtUFGT, AtCHI, and AtCHS (Supplemental Fig. S5, E–G).
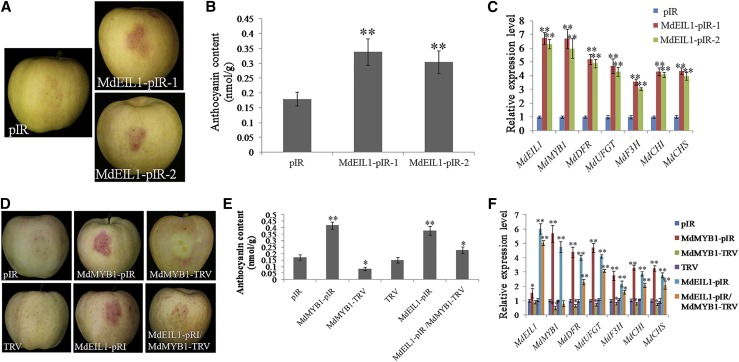
Moreover, viral vector-mediated overexpression was conducted using 120-DAFB apples. MdEIL1-pIR vectors (IL60-1+MdEIL1-IL60-2) were generated, and pIR vectors (IL60-1+IL60-2) were used as a control. Compared with the empty vector control (pIR), overexpression of MdEIL1 promoted anthocyanin accumulation in the apple skin around the injection sites (Fig. 5, A and B). The expression levels of MdMYB1, MdDFR, MdUFGT, MdF3H, MdCHI, and MdCHS were elevated in the MdEIL1-pIR injection areas compared with the controls (Fig. 5C). Therefore, these results demonstrated that MdEIL1 played a positive role in anthocyanin synthesis and fruit coloration.
Figure 5.
MdEIL1 promotes anthocyanin biosynthesis in an MdMYB1-dependent manner. Overexpression of MdEIL1 promotes anthocyanin biosynthesis. A, Apple peel injection assays. At 120 DAFB, cv Red Delicious apples were debagged, injected with the mixed vectors, and stored in a phytotron at 15°C with constant high light (70 μmol m−2 s−1) for 4 d. pIR, IL60-1+IL60-2; MdEIL1-pIR, IL60-1+MdEIL1-IL60-2. B and C, Detection of anthocyanin contents (B) and expression analysis of anthocyanin biosynthetic genes (MdMYB1, MdDFR, MdUFGT, MdF3H, MdCHI, and MdCHS; C) in fruit peels around the injection sites. The anthocyanin content of pIR was set as a control. The value for pIR was set to 1. The apple peel injection assay was performed in three replicates. RT-qPCR was performed with three technical replicates and three biological replicates. The results represent means of these three replicates. Error bars indicate sd. Asterisks denote Student’s t test significance: **, P < 0.01. Overexpression of MdEIL1 promotes anthocyanin biosynthesis in an MdMYB1-dependent manner. D, Apple peel injection assays. At 120 DAFB, cv Red Delicious apples were debagged, injected with the mixed vectors or A. tumefaciens solutions, and stored in a phytotron at 15°C with constant high light (70 μmol m−2 s−1) for 4 d. pIR, IL60-1+IL60-2; MdMYB1-pIR, IL60-1+MdMYB1-IL60-2; MdMYB1-TRV, TRV1+MdMYB1-TRV2; MdEIL1-pIR, IL60-1+MdEIL1-IL60-2; MdEIL1-pRI/MdMYB1-TRV, IL60-1+MdMYB1-IL60-2/TRV1+MdMYB1-TRV2. E and F, Detection of anthocyanin contents (E) and expression analysis of anthocyanin biosynthetic genes (MdMYB1, MdDFR, MdUFGT, MdF3H, MdCHI, and MdCHS; F) in fruit peels around the injection sites. The anthocyanin content of pIR was set as a control. The value for pIR was set to 1. The apple peel injection assay was performed in three replicates. RT-qPCR was performed with three technical replicates and three biological replicates. The results represent means of these three replicates. Error bars indicate sd. Asterisks denote Student’s t test significance: *, P < 0.05 and **, P < 0.01.
MdEIL1 Promotes Anthocyanin Accumulation in an MdMYB1-Dependent Pathway
To examine whether MdEIL1 regulated MdMYB1-modulated anthocyanin synthesis and fruit coloration, wild-type, transgenic (MdMYB1-OX, MdMYB1-Anti, and MdEIL1#1), and cotransformed (MdEIL1#1/MdMYB1-Anti) apple calli were used for coloration assays. MdMYB1 or MdEIL1 overexpression promoted, while MdMYB1 suppression inhibited, the synthesis of anthocyanin (Supplemental Fig. S6, A and B). Moreover, MdEIL1-promoted anthocyanin accumulation was inhibited significantly in the MdEIL1#1/MdMYB1-Anti apple calli (Supplemental Fig. S6, A and B). MdEIL1-induced up-regulation of anthocyanin synthetic genes was inhibited in the MdEIL1#1/MdMYB1-Anti apple calli (Supplemental Fig. S6C). These results indicated that MdEIL1 promoted the anthocyanin accumulation in an MdMYB1-dependent pathway.
To provide further evidence for MdEIL1-modulated fruit coloration by regulating MdMYB1, MdMYB1-TRV suppression vectors (TRV1+MdMYB1-TRV2) were obtained and TRV vectors (TRV1+TRV2) were used as a control. As shown in Figure 5, D and E, overexpression of MdMYB1 or MdEIL1 promoted anthocyanin accumulation in apple skin around the injection sites, whereas MdMYB1 suppression inhibited anthocyanin accumulation. MdEIL1-promoted fruit coloration was inhibited significantly in the MdEIL1-pIR/MdMYB1-TRV coinjected sites, which was further confirmed by RT-qPCR analysis (Fig. 5F). These results implied that MdEIL1 might promote anthocyanin accumulation and fruit coloration in an MdMYB1-dependent pathway.
Red-Flesh Apples Promote Ethylene Production
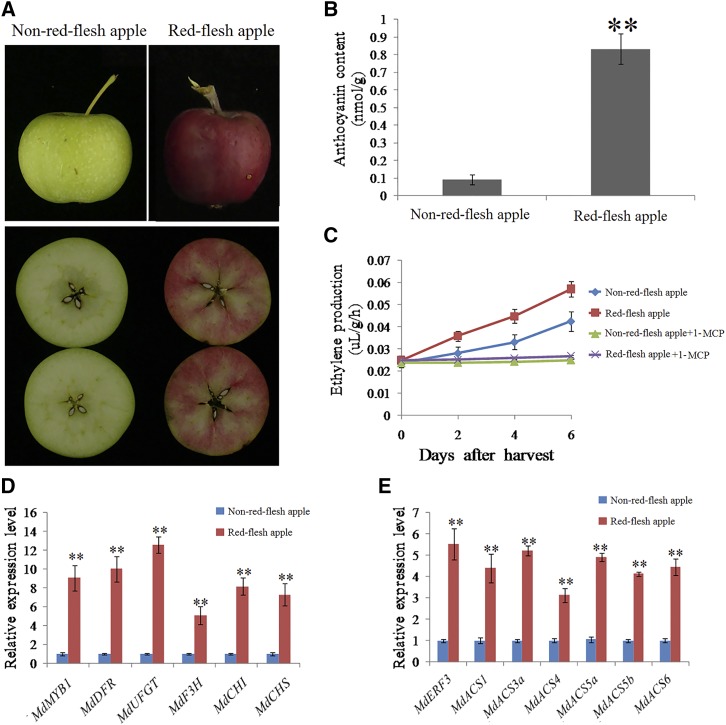
In apples, red flesh occurs due to the constitutive expression of MdMYB10, an allele of MdMYB1 (Ban et al., 2007; Espley et al., 2007). MdMYB10 transgenic apples emit a more volatile aroma (Espley et al., 2007), which is similar to the effects of ethylene (Song and Bangerth, 1996), indicating that MdMYB1 also may regulate ethylene synthesis and signal transduction in addition to being regulated by ethylene. To investigate this hypothesis, 140-DAFB red-flesh apples and non-red-flesh apples were used to detect anthocyanin contents and ethylene production. The results showed that the red-flesh apples had higher anthocyanin contents and ethylene production rates (Fig. 6, A–C). Ethylene production was inhibited significantly when red-flesh and non-red-flesh apples were treated with 1-MCP (Fig. 6C). Compared with the non-red-flesh apples, expression levels of anthocyanin synthetic and regulatory genes (MdMYB1, MdDFR, MdUFGT, MdF3H, MdCHI, and MdCHS) and ethylene synthetic and regulatory genes (MdERF3, MdACS1, MdACS3a, MdACS4, MdACS5a, MdACS5b, and MdACS6) were up-regulated extensively in red-flesh apples (Fig. 6, D and E). These results indicated that ethylene production rates were induced and determined by the MdMYB1 locus in red-flesh apples.
Figure 6.
Analysis of anthocyanin content and ethylene production in red-flesh and non-red-flesh apples. A to C, Representative red-flesh and non-red-flesh apples (A) and measurements of anthocyanin content (B) and ethylene production (C). Apple cross-breeding groups (red-flesh and non-red-flesh apples) were harvested at 140 DAFB for anthocyanin measurements. They were treated or untreated with 1-MCP (1 µL L−1) and stored at room temperature for 6 d for ethylene measurements. The anthocyanin content of non-red-flesh apples was used as the control. The assay was performed in three replicates. The results represent means of these three replicates. Error bars indicate sd. Asterisks denote Student’s t test significance: **, P < 0.01. D and E, Expression of anthocyanin biosynthetic genes (MdMYB1, MdDFR, MdUFGT, MdF3H, MdCHI, and MdCHS; D) and ethylene biosynthetic genes (MdERF3, MdACS1, MdACS3a, MdACS4, MdACS5a, MdACS5b, and MdACS6; E) in untreated red-flesh and non-red-flesh apples. The value for non-red-flesh apple was set to 1. RT-qPCR was performed with three technical replicates and three biological replicates. The results represent means of these three replicates. Error bars indicate sd. Asterisks denote Student’s t test significance: **, P < 0.01.
MdMYB1 Activates the Transcription of MdERF3
Elevated ethylene production rates in red-flesh apples led us to consider whether MdMYB1 was responsible for the production of ethylene. We first analyzed ethylene production and the expression levels of ethylene synthetic genes using the MdMYB1 overexpression (MdMYB1-OX) and suppression (MdMYB1-Anti) apple calli. Overexpression of MdMYB1 increased ethylene production and up-regulated the expression of ethylene synthetic genes (MdERF3, MdACS1, MdACS3a, MdACS4, MdACS5a, MdACS5b, and MdACS6) in MdMYB1-OX. In contrast, MdMYB1 suppression decreased ethylene production and repressed the expression of these genes in the MdMYB1-Anti transgenic calli (Supplemental Fig. S7, A–C).
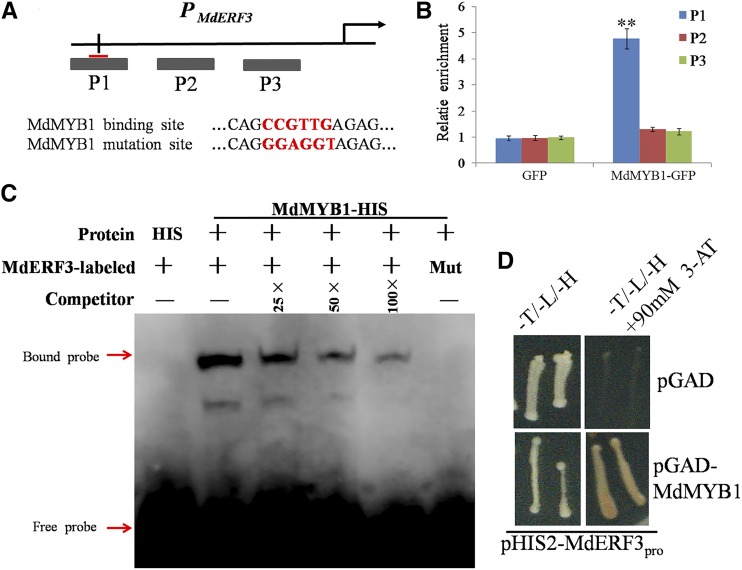
As an important MYB TF, MdMYB1 regulates target genes by binding the CAACGG/CCGTTG sequence in apples (Hu et al., 2016). Interestingly, a potential binding site was found in the MdERF3 promoter (Fig. 7A; Supplemental Table S2). To evaluate whether MdMYB1 regulated ethylene production by binding to the promoter of MdERF3, a ChIP-PCR assay was conducted using 35S:MdMYB1-GFP transgenic apple calli. The results demonstrated that MdMYB1 overexpression enhanced the enrichment of the promoter of MdERF3 (Fig. 7B), indicating that MdMYB1 actually bound to the MdERF3 promoter in vivo.
Figure 7.
Binding of MdMYB1 to the MdERF3 promoter. A, Schematic diagram of the MdERF3 promoter showing the potential MdMYB1 binding sites. The predicted CCGTTG sequences are indicated by the black line. In the mutated probe (Mut), the CCGTTG motif was replaced by GGAGGT. B, ChIP-PCR assay of MdMYB1 binding to the promoter of the MdERF3 gene. Chromatin from the empty vector control (GFP) and 35S:MdMYB1-GFP apple calli (MdMYB1-GFP) were immunoprecipitated with and without anti-GFP antibodies. Three regions (P1, P2, and P3) were examined by RT-qPCR. The enrichment of GFP was set to 1. RT-qPCR was performed with three technical replicates and three biological replicates to examine the enrichment of MdERF3 fragments. The results represent means of these three replicates. Error bars indicate sd. Asterisks denote Student’s t test significance: **, P < 0.01. C, EMSA results showing that the MdMYB1-HIS fusion protein bound directly to the MdMYB1 promoter. Unlabeled probes were used as competitors. In the mutated probe (Mut), the CCGTTG motif was replaced by GGAGGT. D, Y1H assay showing MdMYB1 interaction with the MdERF3 promoter. The promoter of MdERF3 was fused to the pHIS2 vector, and the MdMYB1 gene was fused to the pGAD vector. The columns represent the addition of the pHIS2-MdERF3pro vector. The rows represent the addition of the pGAD and pGAD-MdMYB1 vectors.
To further verify the binding of MdMYB1 to the promoter of MdERF3, EMSA and Y1H experiments demonstrated that MdMYB1 bound directly to the MdERF3 promoter in vitro (Fig. 7, C and D).
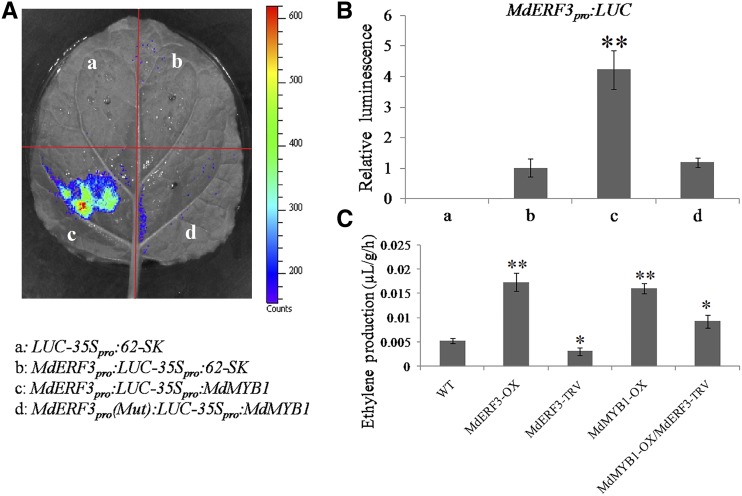
Subsequently, the transcriptional regulation of MdERF3 by MdMYB1 was investigated using a transient transactivation assay in tobacco leaves. The results showed that coexpressed MdMYB1 plus the promoter of MdERF3 increased luminescence, whereas MdMYB1 failed to activate MdERF3 when replacing the CCGTTG motif with GGAGGT (Fig. 8, A and B). These results indicated that MdMYB1 activates the transcription of MdERF3 by binding its promoter. In addition, the ethylene release by apple calli was tested. As shown in Figure 8C, overexpression of MdMYB1 increased ethylene release, whereas the suppression of MdERF3 in MdMYB1-OX calli significantly inhibited MdMYB1-modulated ethylene release, indicating that MdMYB1 promoted ethylene biosynthesis in an MdERF3-dependent pathway.
Figure 8.
MdMYB1 positively regulates the expression of MdERF3. A, Transient expression assays showing that MdMYB1 promoted the expression of MdERF3. The promoter fragment of MdERF3 was cloned into the pGreenII 0800-LUC vector to generate the reporter construct. The effector (35Spro:MdMYB1) was generated by recombining MdMYB1 into the pGreenII 62-SK vector. In MdERF3pro(Mut), the CCGTTG motif was replaced by GGAGGT. B, Quantitative analysis of luminescence intensity. The value for column b (MdERF3pro:LUC-35Spro:62-SK) was set to 1. The transient expression assay was performed in three replicates. The results represent means of these three replicates. Error bars indicate sd. Asterisks denote Student’s t test significance: **, P < 0.01. C, Ethylene production of apple calli. Wild-type apple calli (WT), MdERF3 overexpression calli (MdERF3-OX), MdMYB1 overexpression calli (MdERF3-OX), MdERF3-TRV transient single-transgenic calli (MdERF3-TRV), and MdERF3-TRV transient calli in the background of MdMYB1-OX (MdMYB1-OX/MdERF3-TRV) were used for measurements of ethylene production. The ethylene production of the wild type was used as the reference. The assay was performed in three replicates. The results represent means of these three replicates. Error bars indicate sd. Asterisks denote Student’s t test significance: *, P < 0.05 and **, P < 0.01.
DISCUSSION
Ethylene is an important hormone in regulating fruit ripening, especially in climacteric fruit (Adams-Phillips et al., 2004; Oraguzie et al., 2004; Gapper et al., 2013; Li et al., 2017). Recently, a series of genes involved in fruit ripening were identified, most of which are ethylene-related genes, which are responsible for fruit firmness (Goulao et al., 2008), pigmentation (Kondo et al., 2002), and other biological processes. In apples, ethylene treatment markedly increases the enzymatic activity of anthocyanin synthetic structural genes and anthocyanin accumulation during fruit ripening (Faragher and Brohier, 1984). However, the molecular mechanism of how ethylene promotes anthocyanin accumulation and fruit coloration during fruit ripening is unclear. In this study, we found that ethephon treatment markedly enhanced anthocyanin accumulation and fruit coloration. In addition, expression levels of MdEIL1, which encodes an important TF in the ethylene signaling pathway, and MdMYB1, a critical regulator in anthocyanin synthesis, were apparently induced by ethephon. Moreover, it was suggested that MdEIL1 plays a role in ethylene biosynthesis, anthocyanin accumulation, and fruit coloration mediated by MdMYB1.
MdEIL1 Interacts with the MdMYB1 Promoter to Regulate Ethylene-Modulated Anthocyanin Accumulation and Fruit Coloration
A previous study demonstrated that anthocyanin accumulation and fruit coloration are accompanied by rapid ethylene production during fruit ripening (Faragher and Brohier, 1984). However, less is known about how anthocyanin biosynthesis and fruit ripening are regulated during fruit maturation. In grape (Vitis vinifera), ethylene triggers the expression of gene related to anthocyanin biosynthesis and enhances the color of grape skins (El-Kereamy et al., 2003). Our findings showed that ethylene treatment apparently induced anthocyanin biosynthesis and fruit coloration in apples and induced the transcription of MdMYB1 and anthocyanin biosynthetic genes (Figs. 1A and 2), indicating that MdMYB1 may play a role in ethylene-modulated anthocyanin biosynthesis and fruit coloration.
As primary response regulators, EIN3/EIL proteins play important roles in ethylene-mediated plant growth and development (Chao et al., 1997; Solano et al., 1998; Zhang et al., 2011; Shen et al., 2016). Considering that the expression of MdEIL1 was induced by ethylene, it is reasonable to speculate that MdEIL1 may participate in ethylene-modulated anthocyanin biosynthesis and fruit coloration, which was proved through transgenic apple calli and Arabidopsis (Fig. 5, A–C; Supplemental Fig. S5). ATGTA is the core ethylene-responsive element that is essential for binding by EIN3 (Zhang et al., 2011), which also was present in the promoter sequence of MdMYB1 (Fig. 3; Supplemental Fig. S1). Subsequently, a series of experiments demonstrated that MdEIL1 interacted directly with the MdMYB1 promoter and transcriptionally activated its expression rather than a protein interaction between MdEIL1 and MdMYB1 or other anthocyanin biosynthesis regulators (Figs. 3 and 4; Supplemental Figs. S3 and S8); this ultimately promoted anthocyanin accumulation and fruit coloration through an MdMYB1-dependent pathway (Fig. 5, D–F; Supplemental Fig. S6). These findings describe a molecular mechanism for the regulation of anthocyanin biosynthesis and fruit coloration by ethylene as mediated by the MdEIL1-MdMYB1 signal pathway.
A Positive Feedback Regulatory Loop Mediated by MdMYB1 in Ethylene Biosynthesis
In apples, MdMYB1 and its alleles have been identified as key regulators in anthocyanin biosynthesis and fruit coloration (Takos et al., 2006; Ban et al., 2007; Espley et al., 2007). Our previous research demonstrated that MdMYB1 also regulates malate accumulation by directly facilitating the gene expression of a related tonoplast transporter (Hu et al., 2016). In addition to apples, the homologs of MdMYB1 have been well studied in Arabidopsis (Borevitz et al., 2000) and other fruit tree species (Allan et al., 2008; Jaakola, 2013), especially in grapes (Kobayashi et al., 2004), peaches (Zhou et al., 2016), and pears (Yao et al., 2017), and were shown to act as positive regulators in anthocyanin accumulation and fruit coloration.
Recently, PyMYB114, a homolog of MdMYB1, was identified as a candidate responsible for the red skin color of Chinese pears. PyMYB114 interacted with the ERF/APETALA2 TF PyERF3 to coregulate anthocyanin biosynthesis and fruit coloration. The transcript abundance of PyERF3 was correlated significantly with that of PyMYB114 (Yao et al., 2017), indicating that, in addition to protein interactions, PyMYB114 transcriptionally regulated the expression of PyERF3. Interestingly, the previous study also found that, compared with the wild type, MdMYB10 transgenic apple fruits exhibited a phenotype with a smaller fruit size and tendencies of cracking, which may be attributable to the hastening of the ripening; the fruits also emitted a more volatile aroma (Espley et al., 2007). These physiological phenotypes in MdMYB10 transgenic apples are similar to the effects of ethylene during fruit ripening (Song and Bangerth, 1996), indicating a possible correlation between MdMYB10 and ethylene.
In this study, elevated ethylene production was found in the red-flesh apples (Fig. 6). A series of in vivo and in vitro experiments showed that MdMYB1 actually bound to the promoter of MdERF3 and activated its expression, then positively regulated ethylene synthesis (Figs. 7 and 8). These data demonstrate the regulatory mechanism of the elevated ethylene production in the red-flesh apples and indicate a positive feedback regulatory loop mediated by MdMYB1 in ethylene synthesis. This positive feedback regulatory loop may bring about a rapid and remarkable up-regulation of ethylene production, leading to the subsequent increase in ethylene- and signaling-modulated fruit ripening mediated by MdMYB1.
Role of MdEIL1-MdMYB1 Signaling in Plant Genetic Evolution and Biological Adaptation Modulated by Ethylene
To adapt to the environment, fruit trees have evolved a complex developmental process. During the initial stages of fruit development, the synthesis of ethylene is suppressed. A low level of ethylene inhibits the softening of fruits and the degradation of chlorophyll. Ethylene synthesis is activated when the seeds mature, promoting fruit softening, soluble accumulation, and fruit coloration (Liu et al., 2015). The fully ripe fruits are edible and brightly colored to attract animals to spread more offspring (Shang et al., 2011), which is a perfect evolutionary strategy for fruit trees.
In this study, we found that, during fruit maturation, the ethylene-MdEIL1 signal promotes MdMYB1-mediated anthocyanin synthesis and fruit coloration and ethylene synthesis through the MdMYB1-MdERF3 pathway; this is followed by strengthening of the ethylene-MdEIL1 signal-mediated fruit ripening, which promotes seed dissemination and species reproduction. This signal cascade partially explains the regulation network for anthocyanin accumulation and fruit ripening mediated by ethylene. However, less is known about the activation of the ethylene signal during fruit ripening.
The Differential Expression of MdMYB1 May Affect the MdEIL1-MdMYB1-MdERF3 Transcriptional Cascade
Apple is a typical climacteric fruit, showing a burst of respiration and increased ethylene production during fruit ripening. Our data showed that ethylene production promoted the transcription of MdMYB1 and the accumulation of anthocyanin in the red-skinned apple, which partially requires the function of MdEIL1. However, ethylene production could not promote the accumulation of anthocyanin in the non-red-skinned apple, the underlying mechanism of which remains unknown.
Previous studies showed that the expression of MdMYB1 in red-skinned apple is much higher than that in non-red-skinned apple (Takos et al., 2006). Different apple species have sequence polymorphisms in the MdMYB1 promoter region, which are used to design molecular markers to differentiate between red-skinned and non-red-skinned apples (Takos et al., 2006; Yuan et al., 2014). We speculated that the polymorphism in the MdMYB1 promoter region might affect MdEIL1 binding to the MdMYB1 promoter, which leads to the differential expression of MdMYB1 in the red-skinned and non-red-skinned apples. Recent studies reported that DNA methylation is involved in the regulation of the transcription of MYB1 in apple and pear (Wang et al., 2013; Tian et al., 2017). Different methylation levels in the MdMYB1 promoter region of striped apples result in different anthocyanin contents in red and green stripes, and green stripes have higher DNA methylation levels than red stripes (Telias et al., 2011). Interestingly, cv Granny Smith apple, a green-skinned apple cultivar, turns red quickly when removing bagging treatments during fruit ripening, which also is associated with the DNA methylation level in the MdMYB1 promoter region (Zhang et al., 2013). Based on these studies, we speculate that the sequence polymorphism and DNA methylation level in the MdMYB1 promoter region may affect the ethylene-regulated MdMYB1 expression and anthocyanin accumulation, which contributes to the formation of red-skinned and non-red-skinned apples. However, this hypothesis needs further investigation.
The Interplay between the Ethylene Signal and MdMYB1 in the Regulation of Ethylene Biosynthesis, Anthocyanin Accumulation, and Fruit Coloration
Taking into account the observations in this study and the results of existing research on the regulation of the ripening process (Giovannoni, 2004; Lin et al., 2009; Klee and Giovannoni, 2011; Gapper et al., 2013), we propose the underlying mechanism that regulates the ethylene release and the fruit coloration during fruit ripening (Fig. 9). In this model, at the onset of fruit ripening, the ethylene signal is activated, which then induces the expression and protein stability of MdEIL1; MdEIL1 then interacts with the promoter of MdMYB1 and transcriptionally activates its expression. The activated MdMYB1 regulates anthocyanin accumulation and fruit coloration by regulating the expression of genes in the anthocyanin biosynthetic pathway. In addition to pigment accumulation, MdMYB1 regulates ethylene release by inducing the transcription of MdERF3, a critical regulator in ethylene biosynthesis, thus regulating ethylene synthesis and further strengthening the ethylene-mediated anthocyanin accumulation and fruit coloration.
Figure 9.
Proposed model of the mechanism regulating anthocyanin accumulation and ethylene release during fruit ripening. MdEIL1 binds directly to the promoter of MdMYB1 and enhances its action, leading to increased anthocyanin accumulation. In addition, MdMYB1 binds to the promoter of MdERF3 and activates its expression, resulting in enhanced ethylene production. C2H4, Ethylene; ATGTA, MdEIL1-binding sequence; CCGTTG, MdMYB1-binding sequence. Solid arrows show positive regulation.
MdMYB1 is a TF that has multiple internal and external responses in addition to ethylene (Takos et al., 2006; Li et al., 2012, 2017; An et al., 2017a; Zhou et al., 2017). Thus, an elaborate regulatory network between ethylene signaling and other cues mediated by MdMYB1 is central to fruit ripening. It is also important to identify other factors that participate in MdEIL1-MdMYB1-mediated ethylene biosynthesis and fruit coloration to unveil the regulatory mechanism between ethylene and MdMYB1 during fruit ripening. Overall, our findings provide insight into the transcriptional regulatory mechanism of the synergistic interaction of the ethylene signal with the MdMYB1 TF to regulate ethylene synthesis and fruit coloration. These results provide important information for fruit breeding with regard to improved coloration, softening, and storage ability.
MATERIALS AND METHODS
Fruit Materials and Treatments
‘Red Delicious’ apples (Malus domestica) were used for the ethephon and 1-MCP treatments, as well as the injection assays. For the ethephon treatment of the fruits, 120-DAFB apples were dipped into different concentrations (0, 250, 500, and 1,000 mg L−1) of the ethephon solutions for 1 min. After drying naturally, they were kept in individual and closed containers. The containers were stored in a phytotron at 24°C with constant light (70 μmol m−2 s−1) for 6 d. For the 1-MCP treatment, the apples were exposed to 1 µL L−1 1-MCP for 12 h at room temperature in an air-tight container. After the 1-MCP treatment, the air-tight container was transferred to a phytotron at 24°C with constant light (70 μmol m−2 s−1) for 6 d. During the treatments, anthocyanin contents and ethylene production were measured at certain times. The two groups of fruits were treated simultaneously, and the tests were conducted three times.
Measurements of Anthocyanin Contents and Ethylene Production
The apples were peeled and collected after treatments. The isolated apple peels were dipped into anthocyanin extraction solution (95% [v/v] anhydrous ethanol:1.5 mol L−1 HCl, 17:3) for 24 h in darkness. A spectrophotometer was used for the absorbance measurements at 530, 620, and 650 nm. The results were determined based on the following equation: optical density (OD) = (A530 – A620) – [0.1 × (A650 – A620)] (An et al., 2017a).
The cv Orin apple calli for coloration assays and injection areas for the fruit injection assays were collected for measurements of anthocyanin. The ethylene production of the apples and apple calli was determined as described by Tan et al. (2013) and Li et al. (2016), respectively. The apple fruits or apple calli were placed in individual containers and stored in a phytotron (24°C). One-milliliter syringes were used for the collection of gas samples. The antisense viral vector MdERF3-TRV2 was obtained by cloning the CDS of MdERF3 into the TRV2 vector. The TRV1 vector was used as an auxiliary plasmid. The TRV vectors were introduced into Agrobacterium tumefaciens LBA4404. The mixed vectors and A. tumefaciens solutions were transformed into the apple calli by a vacuum air pump. The MdERF3-TRV transient transformation of the apple calli was used for the measurements of ethylene production. All experiments were repeated at least three times.
RT-qPCR Analysis
Apple peels and apple calli were used for RNA extraction using the RNA Plant Plus reagent (Tiangen; An et al., 2017a). The ethephon-treated apple peels and the apple peels around the injection area were collected using a peeler. The expression levels of the anthocyanin synthetic genes and ethylene synthetic genes were tested using specific primers that are shown in Supplemental Table S3.
Generation of Transgenic Apple Calli and Transgenic Arabidopsis
The overexpression vectors MdEIL1-GFP and MdMYB1-GFP were generated by cloning the CDSs of MdEIL1 and MdMYB1 into the transformed vector pCAMBIA1300-GFP. The suppression vector MdMYB1-Anti was generated by cloning the DNA fragment of the MdMYB1 reverse complement into pCAMBIA1300. The individual pCAMBIA1300-GFP was used as a control.
The transgenic apple calli (MdEIL1-OX, 35S:MdEIL1-GFP; MdMYB1-OX, 35S:MdMYB1-GFP; MdMYB1-Anti, 35S:MdMYB1-Anti; and GFP, the empty vector control) were obtained by A. tumefaciens-mediated genetic transformation. The wild-type apple calli and A. tumefaciens-inserted recombined vectors were incubated for 30 min at 24°C. Subsequently, the transformed apple calli were screened in a selective medium containing antibiotics (An et al., 2017a).
The transgenic Arabidopsis (Arabidopsis thaliana) plants were obtained by the floral dip transformation method (Clough and Bent, 1998).
EMSA
The fusion proteins of MdEIL1-HIS and MdMYB1-HIS were generated through prokaryotic expression in vitro. The CDSs of MdEIL1 and MdMYB1 were cloned into the PET32a vector containing a His (HIS) target to generate recombined vectors. Then, these recombined vectors were transformed into Escherichia coli BL21 (DE3). Three millimolar isopropyl β-d-1-thiogalactopyranoside was used to induce protein production. The fusion proteins were purified using the ProFound PolyHIS Protein Kit (Thermo).
The 3′ biotin-labeled probes and the LightShift Chemiluminescent EMSA Kit (Thermo) were prepared for the subsequent EMSA. Briefly, the fusion proteins and biotin-labeled probes were mixed in a binding buffer for 20 min at 24°C. The HIS protein was used as a negative control, and unlabeled probes were used for probe competition (An et al., 2017a).
ChIP-PCR Assays
ChIP-PCR assays were performed as described by An et al. (2017b). The transgenic apple calli MdEIL1-OX and MdMYB1-OX containing the GFP targets were applied to the ChIP-PCR assays using the EpiTect ChIP OneDay Kit (Qiagen). The empty vector pCAMBIA1300-GFP-overexpressing apple calli (GFP) was used as a control. A GFP-specific antibody was used in this study. The enriched DNA fragments were examined by qPCR using the primers shown in Supplemental Table S3.
Y1H Assays
The CDSs of MdEIL1 and MdMYB1 were inserted into the pGADT7 vector to generate the recombined constructs pGAD-MdEIL1 and pGAD-MdMYB1, and the promoter fragments of MdMYB1 (MdMYB1-3) and MdERF3 were cloned into the pHIS2 vector. The cotransformed Y2H Gold yeast strains were plated on -Trp/-Leu/-His medium, supplementing an appropriate concentration of 3-AT.
GUS Activity Detection
The reporter plasmid was obtained (MdMYB1-Pro) by cloning the promoter fragment of MdMYB1 into pBI101-GUS. MdEIL1-pCAMBIA1300-GFP was used as an effector plasmid. The reporter plasmid and effector plasmid were introduced into the wild-type apple calli separately or together.
GUS staining was conducted using a GUS staining buffer. The apple calli were incubated with the GUS staining buffer (1 mm 5-bromo-4-chloro-3-indolyl-β-GlcA, 0.1 mm EDTA, 0.5 mm ferroyanide, and 0.1% [v/v] Triton X-100, pH 7) at 37°C in the dark. For the quantitative analysis of GUS activity, approximately 0.5 g of the apple calli was extracted with 1 mL of extraction buffer (50 mm NaHPO4, 10 mm β-mercaptoethanol, 10 mm Na2EDTA, and 0.1% [v/v] Triton X-100, pH 7). The total protein concentration was determined using the RC DC Protein Assay Kit (Bio-Rad). The extract was added to the GUS reaction buffer and was incubated at room temperature. After the reaction had proceeded for 0, 5, 10, 15, 30, and 60 min, the reaction mixture was added to the stop solution (1 m sodium carbonate). Fluorescence was measured using a spectrofluorometer at 365 and 450 nm.
Transient Expression Assays
The transient expression assays were performed using tobacco (Nicotiana tabacum) leaves (An et al., 2017a). The promoter fragments of MdMYB1 and MdERF3 were cloned into the pGreenII 0800-LUC vectors to generate the reporter constructs. The effectors (35Spro:MdEIL1 and 35Spro:MdMYB1) were generated by recombining the MdEIL1 and MdMYB1 genes into the pGreenII 62-SK vector. The recombinant plasmids were transformed into A. tumefaciens LBA4404. The bacteria were mixed and coinjected into the tobacco leaves. A living imaging apparatus was used for luminescence detection.
Coloration Assays of Apple Calli
Wild-type and transgenic apple calli were subcultured in a medium and stored in a phytotron at 24°C with constant light (70 μmol m−2 s−1). The treated apple calli were collected for measurements of anthocyanin contents and the detection of gene expression.
Apple Injection Assays
Fruit injection assays were carried out as described previously (Li et al., 2012). The overexpression viral vectors MdEIL1-IL60-2 and MdMYB1-IL60-2 were generated by inserting the CDSs of MdEIL1 and MdMYB1 into the IL60-2 vector. The IL60-1 vector was used as an auxiliary plasmid. The antisense viral vector MdMYB1-TRV2 was obtained by cloning the CDS of MdMYB1 into the TRV2 vector. The TRV1 vector was used as an auxiliary plasmid. The TRV vectors were introduced into A. tumefaciens LBA4404. The mixed vectors and the A. tumefaciens solutions were injected into the fruit peels. Then, the fruits were stored in a phytotron at 24°C with constant light (70 μmol m−2 s−1) for coloration.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: MdEIL1 (MDP0000423881), MdERF3 (MDP0000787281), MdACS1 (MDP0000370791), MdACS3a (MDP0000145123), MdACS4 (MDP0000262872), MdACS5a (MDP0000923426), MdACS5b (MDP0000435100), MdACS6 (MDP0000133334), MdMYB1 (MDP0000259614), MdDFR (MDP0000494976), MdUFGT (MDP0000405936), MdF3H (MDP0000323864), MdCHI (MDP0000759336), MdCHS (MDP0000686666), AtPAP1 (AT1G56650.1), AtPAP2 (AT1G66390.1), AtPAL (AT2G37040.1), AtDFR (AT5G42800.1), AtUFGT (AT5G54060.1), AtCHI (AT3G55120.1), and AtCHS (AT5G13930.1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Schematic diagram of the MdMYB1 promoter showing the potential MdEIL1-binding sites (MdMYB1-1, MdMYB1-2, and MdMYB1-3).
Supplemental Figure S2. Phylogenetic analysis and protein sequence alignment of MdEIL1 with Arabidopsis EIN3/EILs.
Supplemental Figure S3. pHIS2-MdMYB1-3pro and pHIS2-MdERF3pro transformed into Y187 yeast strains to screen for optimal 3-AT concentration for reporter gene inhibition.
Supplemental Figure S4. Schematic representation of the LUC reporter vector containing the indicated fragment of MdMYB1 or MdERF3 promoters and the effector vector containing MdEIL1 or MdMYB1 genes.
Supplemental Figure S5. Overexpression of MdEIL1 in apple calli and Arabidopsis increases anthocyanin content.
Supplemental Figure S6. MdEIL1 promotes anthocyanin synthesis in an MdMYB1-dependent manner.
Supplemental Figure S7. MdMYB1 regulates ethylene response.
Supplemental Figure S8. MdEIL1 does not interact with MdMYB1, MdbHLH3, and MdbHLH33 in yeast two-hybrid assays.
Supplemental Table S1. MdEIL1 binding motif in the MdMYB1 promoter.
Supplemental Table S2. MdMYB1 binding motif in the MdERF3 promoter.
Supplemental Table S3. Primers used for gene expression analysis and vector construction in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Zhi-Long Bao of Shandong Agricultural University for language editing.
Footnotes
This work was supported by grants from the Natural Science Foundation of China (31772288 and 31601742), the Ministry of Education of China (IRT15R42), the Shandong Province Government (SDAIT-06-03), and the Ministry of Agriculture of China (CARS-27, 2016-X11).
Articles can be viewed without a subscription.
References
- Adams-Phillips L, Barry C, Giovannoni J (2004) Signal transduction systems regulating fruit ripening. Trends Plant Sci 9: 331–338 [DOI] [PubMed] [Google Scholar]
- Alexander L, Grierson D (2002) Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot 53: 2039–2055 [DOI] [PubMed] [Google Scholar]
- Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13: 99–102 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN (2004) The ethylene signaling pathway. Science 306: 1513–1515 [DOI] [PubMed] [Google Scholar]
- An JP, Qu FJ, Yao JF, Wang XN, You CX, Wang XF, Hao YJ (2017a) The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic Res 4: 17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JP, Wang XN, Yao JF, Ren YR, You CX, Wang XF, Hao YJ (2017b) Apple MdMYC2 reduces aluminum stress tolerance by directly regulating MdERF3 gene. Plant Soil 418: 255–266 [Google Scholar]
- Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48: 958–970 [DOI] [PubMed] [Google Scholar]
- Barry CS, Llop-Tous MI, Grierson D (2000) The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol 123: 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Cipollini ML, Levey DJ (1997) Secondary metabolites of fleshy vertebrate-dispersed fruits: adaptive hypotheses and implications for seed dispersal. Am Nat 150: 346–372 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dandekari AM, Teo G, Defilippi BG, Uratsu SL, Passey AJ, Kader AA, Stow JR, Colgan RJ, James DJ (2004) Effect of down-regulation of ethylene biosynthesis on fruit flavor complex in apple fruit. Transgenic Res 13: 373–384 [DOI] [PubMed] [Google Scholar]
- Dougherty L, Zhu Y, Xu K (2016) Assessing the allelotypic effect of two aminocyclopropane carboxylic acid synthase-encoding genes MdACS1 and MdACS3a on fruit ethylene production and softening in Malus. Hortic Res 3: 16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kereamy A, Chervin C, Roustan JP, Cheynier V, Souquet JM, Moutounet M, Raynal J, Ford C, Latché A, Pech JC, et al. (2003) Exogenous ethylene stimulates the long-term expression of genes related to anthocyanin biosynthesis in grape berries. Physiol Plant 119: 175–182 [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faragher JD, Brohier RL (1984) Anthocyanin accumulation in apple skin during ripening: regulation by ethylene and phenylalanine ammonia-lyase. Sci Hortic (Amsterdam) 22: 89–96 [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Shima Y, Ito Y (2013) A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25: 371–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper NE, McQuinn RP, Giovannoni JJ (2013) Molecular and genetic regulation of fruit ripening. Plant Mol Biol 82: 575–591 [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ. (2004) Genetic regulation of fruit development and ripening. Plant Cell (Suppl) 16: S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53: 814–827 [DOI] [PubMed] [Google Scholar]
- Goulao LF, Cosgrove DJ, Oliveira CM (2008) Cloning, characterization and expression analyses of cDNA clones encoding cell wall-modifying enzymes isolated from ripe apples. Postharvest Biol Technol 48: 37–51 [Google Scholar]
- Grierson D. (2013) Ethylene and the control of fruit ripening. In Seymour GB, Poole M, Giovannoni JJ, Tucker GA, eds, The Molecular Biology and Biochemistry of Fruit Ripening. John Wiley & Sons, New York, pp 43–73 [Google Scholar]
- Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Han YC, Kuang JF, Chen JY, Liu XC, Xiao YY, Fu CC, Wang JN, Wu KQ, Lu WJ (2016) Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and expansins during fruit ripening. Plant Physiol 171: 1070–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Sunako T, Wakasa Y, Soejima J, Satoh T, Niizeki M (2000) An allele of the 1-aminocyclopropane-1-carboxylate synthase gene (Md-ACS1) accounts for the low level of ethylene production in climacteric fruits of some apple cultivars. Theor Appl Genet. 101: 742–746 [Google Scholar]
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62: 2465–2483 [DOI] [PubMed] [Google Scholar]
- Hu DG, Sun CH, Ma QJ, You CX, Cheng L, Hao YJ (2016) MdMYB1 regulates anthocyanin and malate accumulation by directly facilitating their transport into vacuoles in apples. Plant Physiol 170: 1315–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland HS, Yao JL, Tomes S, Sutherland PW, Nieuwenhuizen N, Gunaseelan K, Winz RA, David KM, Schaffer RJ (2013) Apple SEPALLATA1/2-like genes control fruit flesh development and ripening. Plant J 73: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T (2008) DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J 55: 212–223 [DOI] [PubMed] [Google Scholar]
- Jaakola L. (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci 18: 477–483 [DOI] [PubMed] [Google Scholar]
- Joo S, Liu Y, Lueth A, Zhang S (2008) MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J 54: 129–140 [DOI] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H (2004) Retrotransposon-induced mutations in grape skin color. Science 304: 982. [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Kondo S, Hiraoka K, Kobayashi S, Honda C, Terahara N (2002) Changes in the expression of anthocyanin biosynthetic genes during apple development. J Am Soc Hortic Sci 127: 971–976 [Google Scholar]
- Lancaster JE, Dougall DK (1992) Regulation of skin color in apples. Crit Rev Plant Sci 10: 487–502 [Google Scholar]
- Li T, Jiang Z, Zhang L, Tan D, Wei Y, Yuan H, Li T, Wang A (2016) Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J 88: 735–748 [DOI] [PubMed] [Google Scholar]
- Li T, Xu Y, Zhang L, Ji Y, Tan D, Yuan H, Wang A (2017) The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 29: 1316–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Mao K, Zhao C, Zhao XY, Zhang HL, Shu HR, Hao YJ (2012) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol 160: 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Zhong S, Grierson D (2009) Recent advances in ethylene research. J Exp Bot 60: 3311–3336 [DOI] [PubMed] [Google Scholar]
- Liu M, Pirrello J, Chervin C, Roustan JP, Bouzayen M (2015) Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiol 169: 2380–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhao TJ, Liu JM, Liu WQ, Liu Q, Yan YB, Zhou HM (2006) The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box. FEBS Lett 580: 1303–1308 [DOI] [PubMed] [Google Scholar]
- Oraguzie NC, Iwanami H, Soejima J, Harada T, Hall A (2004) Inheritance of the Md-ACS1 gene and its relationship to fruit softening in apple (Malus × domestica Borkh.). Theor Appl Genet 108: 1526–1533 [DOI] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10: 63–70 [DOI] [PubMed] [Google Scholar]
- Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21: 3567–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer HM, Schaefer V, Levey DJ (2004) How plant-animal interactions signal new insights in communication. Trends Ecol Evol 19: 577–584 [Google Scholar]
- Shan W, Kuang JF, Chen L, Xie H, Peng HH, Xiao YY, Li XP, Chen WX, He QG, Chen JY, et al. (2012) Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J Exp Bot 63: 5171–5187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Venail J, Mackay S, Bailey PC, Schwinn KE, Jameson PE, Martin CR, Davies KM (2011) The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol 189: 602–615 10.1111/j.1469-8137.2010.03498.x [DOI] [PubMed] [Google Scholar]
- Shen X, Li Y, Pan Y, Zhong S (2016) Activation of HLS1 by mechanical stress via ethylene-stabilized EIN3 is crucial for seedling soil emergence. Front Plant Sci 7: 1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Tian S, Hou L, Huang X, Zhang X, Guo H, Yang S (2012) Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24: 2578–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skottke KR, Yoon GM, Kieber JJ, DeLong A (2011) Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet 7: e1001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Bangerth F (1996) The effect of harvest date on aroma compound production from “Golden Delicious” apple fruit and relationship to respiration and ethylene production. Postharvest Biol Technol 8: 259–269 [Google Scholar]
- Sunako T, Sakuraba W, Senda M, Akada S, Ishikawa R, Niizeki M, Harada T (1999) An allele of the ripening-specific 1-aminocyclopropane-1-carboxylic acid synthase gene (ACS1) in apple fruit with a long storage life. Plant Physiol 119: 1297–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142: 1216–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Li T, Wang A (2013) Apple 1-aminocyclopropane-1-carboxylic acid synthase genes, MdACS1 and MdACS3a, are expressed in different systems of ethylene biosynthesis. Plant Mol Biol Report 31: 204–209 10.1007/s11105-012-0490-y [DOI] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54: 733–749 [DOI] [PubMed] [Google Scholar]
- Telias A, Lin-Wang K, Stevenson DE, Cooney JM, Hellens RP, Allan AC, Hoover EE, Bradeen JM (2011) Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol 11: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis A. (1992) One rotten apple spoils the whole bushel: the role of ethylene in fruit ripening. Cell 70: 181–184 [DOI] [PubMed] [Google Scholar]
- Tian J, Li KT, Zhang SY, Zhang J, Song TT, Zhu YJ, Yao YC (2017) The structure and methylation level of the McMYB10 promoter determine the leaf color of Malus crabapple. HortScience 52: 520–526 [Google Scholar]
- Wang KL, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. Plant Cell (Suppl) 14: S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KLC, Yoshida H, Lurin C, Ecker JR (2004) Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950 [DOI] [PubMed] [Google Scholar]
- Wang Z, Meng D, Wang A, Li T, Jiang S, Cong P, Li T (2013) The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear. Plant Physiol 162: 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whale SK, Singh Z (2007) Endogenous ethylene and color development in the skin of ‘Pink Lady’ apple. J Am Soc Hortic Sci 132: 20–28 [Google Scholar]
- Whale SK, Singh Z, Behboudian MH, Janes J, Dhaliwal SS (2008) Fruit quality in ‘Cripp’s Pink’ apple, especially colour, as affected by preharvest sprays of aminoethoxyvinylglycine and ethephon. Sci Hortic (Amsterdam) 115: 342–351 [Google Scholar]
- Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20: 176–185 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525 [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35: 155–189 [Google Scholar]
- Yao G, Ming M, Allan AC, Gu C, Li L, Wu X, Wang R, Chang Y, Qi K, Zhang S, et al. (2017) Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J 92: 437–451 [DOI] [PubMed] [Google Scholar]
- Yin XR, Allan AC, Chen KS, Ferguson IB (2010) Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol 153: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K, Wang C, Wang J, Xin L, Zhou G, Li L, Shen G (2014) Analysis of the MdMYB1 gene sequence and development of new molecular markers related to apple skin color and fruit-bearing traits. Mol Genet Genomics 289: 1257–1265 [DOI] [PubMed] [Google Scholar]
- Zhang L, Li Z, Quan R, Li G, Wang R, Huang R (2011) An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol 157: 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XJ, Wang LX, Liu YL, Chen XX, Yang YZ, Zhao ZY (2013) Differential gene expression analysis of ‘Granny Smith’ apple (Malus domestica Borkh.) during fruit skin coloration. S Afr J Bot 88: 125–131 [Google Scholar]
- Zheng Q, Song J, Campbell-Palmer L, Thompson K, Li L, Walker B, Cui Y, Li X (2013) A proteomic investigation of apple fruit during ripening and in response to ethylene treatment. J Proteomics 93: 276–294 [DOI] [PubMed] [Google Scholar]
- Zhou H, Peng Q, Zhao J, Owiti A, Ren F, Liao L, Wang L, Deng X, Jiang Q, Han Y (2016) Multiple R2R3-MYB transcription factors involved in the regulation of anthocyanin accumulation in peach flower. Front Plant Sci 7: 1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LJ, Li YY, Zhang RF, Zhang CL, Xie XB, Zhao C, Hao YJ (2017) The small ubiquitin-like modifier E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low-temperature conditions in apple. Plant Cell Environ 40: 2068–2080 [DOI] [PubMed] [Google Scholar]