An ethylene responsive C2H2-type zinc finger transcription factor, AdDof3, regulates starch degradation in kiwifruit via trans-activation of the AdBAM3L promoter.
Abstract
Ripening, including softening, is a critical factor in determining the postharvest shelf-life of fruit and is controlled by enzymes involved in cell wall metabolism, starch degradation, and hormone metabolism. Here, we used a transcriptomics-based approach to identify transcriptional regulatory components associated with texture, ethylene, and starch degradation in ripening kiwifruit (Actinidia deliciosa). Twelve differentially expressed structural genes, including seven involved in cell wall metabolism, four in ethylene biosynthesis, and one in starch degradation, and 14 transcription factors (TFs) induced by exogenous ethylene treatment and inhibited by the ethylene signaling inhibitor 1-methylcyclopropene were identified as changing in transcript levels during ripening. Moreover, analysis of the regulatory effects of differentially expressed genes identified a zinc finger TF, DNA BINDING WITH ONE FINGER (AdDof3), which showed significant transactivation on the AdBAM3L (β-amylase) promoter. AdDof3 interacted physically with the AdBAM3L promoter, and stable overexpression of AdBAM3L resulted in lower starch content in transgenic kiwifruit leaves, suggesting that AdBAM3L is a key gene for starch degradation. Moreover, transient overexpression analysis showed that AdDof3 up-regulated AdBAM3L expression in kiwifruit. Thus, transcriptomics analysis not only allowed the prediction of some ripening-regulating genes but also facilitated the characterization of a TF, AdDof3, and a key structural gene, AdBAM3L, in starch degradation.
Ripening is a programmed process involving substantial changes in fruit quality properties, such as color, aroma, flavor, and texture (Prasanna et al., 2007; Klee and Giovannoni, 2011). The overall process of fruit ripening, however, is that of senescence, accompanied by fruit quality deterioration and postharvest loss (Seymour et al., 2013). Thus, the postharvest control of ripening is critical for the fruit industry. In tomato (Solanum lycopersicum), multiple regulators of fruit ripening have been identified, such as Ripening-inhibitor (RIN; Vrebalov et al., 2002), Colorless nonripening (CNR; Manning et al., 2006), Never-ripe (Nr; Wilkinson et al., 1995), APETALA2a (AP2a; Karlova et al., 2011), etc. Transgenic studies of such genes in tomato fruit, or the use of mutants, has shown the impacts of these genes on fruit ripening. However, such studies have been less frequent in other fruit crops, especially perennial fruit, where transgenic studies and the availability of mutants are limited. Moreover, the function of these ripening regulators may differ in various crops, as the tomato rin mutant (Vrebalov et al., 2002) and FaMADS9 (MCM1/AGAMOUS/DEFICIENS/SRF [MADS] box) antisense transgenic strawberry (Fragaria × ananassa) fruit (Seymour et al., 2011) inhibit or retard ripening, while MdMADS8/9-suppressed apple (Malus × domestica) has a phenotype of small fruit with reduced flesh (Ireland et al., 2013). These findings highlight the need for investigations of ripening regulation in different fruit types and species.
Kiwifruit (Actinidia deliciosa), one of the most recently domesticated fruit crops, now has global distribution (Huang and Ferguson, 2001). Its rise as an important economic crop has led to extensive research on the regulation of kiwifruit ripening. It is a typical climacteric fruit exhibiting an ethylene burst and also is particularly sensitive to ethylene (McDonald and Harman, 1982). Postharvest ripening indices for kiwifruit are similar to those in other fruit, including starch degradation/soluble solids accumulation, ethylene biosynthesis, cell wall metabolism, and volatile emission (Atkinson et al., 2011). A number of structural genes related to these physiological changes have been characterized, but most are associated with multiple gene families involved with the regulation of fruit softening (CkPGA/B/C, polygalacturonase [Wang et al., 2000]; AdXTH4/5/6/7/8/10/13, xyloglucan endotransglucosylase/hydrolase [Atkinson et al., 2009]; AdBAM3L/3.1/9, β-amylase; AdAMY1, α-amylase; and AdAGL3, α-glucosidase [Hu et al., 2016]) and prove difficult to manipulate in relation to genetic improvement. Other genes have been identified with multiple quality traits; for example, knockdown of AdACO1 (1-aminocyclopropane-1-carboxylic acid oxidase) resulted in less ethylene production and firm fruit but lower levels of the quality attributes of aroma and flavor (Atkinson et al., 2011).
In terms of regulating fruit ripening, targeting transcription factors (TFs) is an alternative option. In tomato, the above-mentioned CNR and RIN genes encode SQUAMOSA promoter-binding protein and MADS TFs (Dong et al., 2013). Other TFs also have been shown to be involved in fruit ripening, mostly characterized in tomato, including AP2/Ethylene Response Factor (e.g. SlERF6 [Lee et al., 2012] and SlERF.B3-SRDX [Liu et al., 2014]), NAC (e.g. SlNAC1/4; Ma et al., 2014; Zhu et al., 2014; Meng et al., 2016), and Homeobox (e.g. LeHB1; Lin et al., 2008). In other fruit species, potential roles of TFs in fruit ripening have been identified (Xie et al., 2016), but few have full or partial functional characterization. Citrus spp. CitERF13 is a regulator of ethylene-driven fruit postharvest degreening via binding and regulation of the CitPPH (pheophorbide hydrolase) promoter (Yin et al., 2016); a jasmonate-responsive MdMYC2 TF mediates the jasmonate regulation of ethylene biosynthesis and apple fruit ripening (Li et al., 2017).
In this study, we analyzed three major ripening traits in kiwifruit: texture (firmness and cell wall components), ethylene production, and starch degradation (as well as total soluble solids [TSS]). We performed transcriptomic analysis on ethylene-treated and 1-methylcyclopropene (1-MCP)-treated fruit with the aim of identifying the most responsive TFs and structural genes during fruit ripening. Using the dual-luciferase assay, multiple TFs showed regulatory effects on different promoters, including transactivation by DNA BINDING WITH ONE FINGER (AdDof3) on a starch degradation gene (AdBAM3L). Furthermore, electrophoretic mobility shift assays (EMSAs) indicated specific cis-elements of the AdBAM3L promoter for AdDof3 binding. We investigated the functions of AdDof3 and AdBAM3L, as well as their in vivo regulation in fruit, with stable transformation (analysis using leaves) or transient overexpression in kiwifruit (analysis with the core tissues at two sites within a single fruit). This strategy provided insights into the molecular basis of starch degradation during kiwifruit ripening.
RESULTS
Analyses of Kiwifruit Ripening
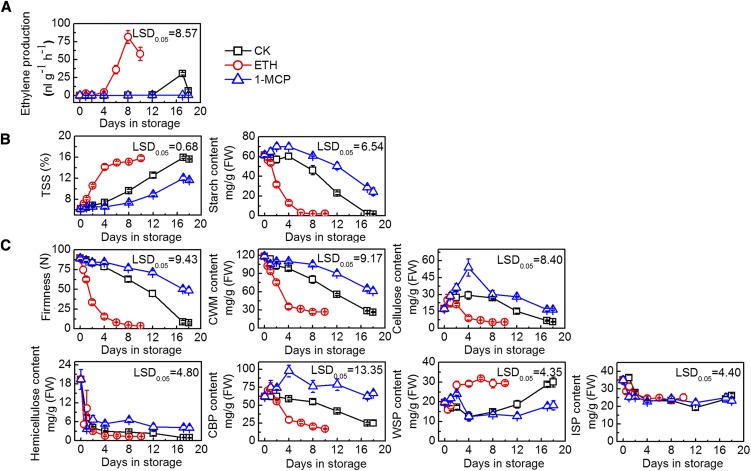
Ethylene treatment significantly accelerated fruit ripening, with treated fruit reaching the ethylene climacteric peak of 81.4 nL g−1 h−1 at 8 d in storage (DIS), which was higher and occurred earlier than that of control fruit, which peaked at 17 d with 30.2 nL g−1 h−1 production. Ethylene production was inhibited in 1-MCP-treated fruit and, therefore, was undetectable over the whole experimental period (Fig. 1A).
Figure 1.
Effects of ethylene and 1-MCP treatment on kiwifruit ripening and softening. Fruit were treated with 100 μL L−1 ethylene (ETH), 1 μL L−1 1-MCP, or air (control [CK]) for 24 h at 20°C. A, Ethylene production of cv Hayward kiwifruit during storage. Error bars represent se from three replicates. B, TSS and starch content in cv Hayward fruit. For TSS and starch content, error bars represent se from 10 and three replicates, respectively. C, Firmness, cell wall material (CWM) content, cellulose content, hemicellulose content, and pectin content (covalent binding pectin [CBP], water soluble pectin [WSP], and ionic soluble pectin [ISP]) of fruit in storage. Error bars for firmness represent se based on 12 replicates; all others were from three replicates. FW, Fresh weight. lsd values represent lsd at P = 0.05.
Starch degradation is considered the first sign of kiwifruit postharvest ripening and also contributes to the TSS. Starch content decreased from 61.5 mg g−1 at 0 DIS to 1.5 mg g−1 at 18 DIS in control fruit. In ethylene-treated fruit, starch content decreased rapidly to 1.9 mg g−1 by 8 DIS. 1-MCP-treated fruit exhibited a significantly slower rate of decrease, with 24.3 mg g−1 starch remaining at 18 DIS (Fig. 1B). TSS showed the opposite trends to starch content, increasing from 6.2% at 0 DIS to 15.6% at 18 DIS for the control fruit. In ethylene- and 1-MCP-treated fruit, TSS reached 15.2% at 8 DIS and 11.7% at 18 DIS, respectively (Fig. 1B).
Changes in fruit firmness were similar to those of starch content. Ethylene-treated fruit reached an eating-ripe stage (firmness of 7.9 n) at 6 DIS, whereas control fruit reached a similar firmness (7.4 n) at 18 DIS (Fig. 1C). In parallel with fruit softening, extractable CWM also decreased, with the decrease accelerated by ethylene and inhibited by 1-MCP (Fig. 1C). With regard to main cell wall components, in general, only cellulose and CBP showed similar patterns to that of fruit firmness. Both cellulose and CBP contents were relatively lower in ethylene-treated and higher in 1-MCP-treated fruit. WSP content showed an increasing trend during ripening (Fig. 1C). Both hemicellulose and ISP decreased during fruit ripening, but their decreasing rate and contents were similar among the three treatments (Fig. 1C).
Expression of TF and Structural Genes during Ripening
RNA sequencing (RNA-seq) provided an overview of genes differentially expressed during ripening and in response to exogenous ethylene and 1-MCP treatments. The transcript abundances of genes were estimated by fragments per kilobase of exon per million fragments mapped (FPKM). The boxplot distribution of the log10FPKM values in Supplemental Figure S1A showed that the median and quartile values of expression across the libraries compared for differential expression were comparable. De novo assembly also predicted a total of 4,542 genes that have not appeared in the Hong Yang genome database annotated by COG, GO, KEGG, KOG, Pfam, eggnog, Swiss-Prot, and nr databases (Supplemental Fig. S1B). At 1 and 4 DIS, 6,326 and 3,994 differentially expressed genes (DEGs) were found between the control and ethylene treatments, while only 25 and 34 DEGs were found between the control and 1-MCP treatments (Supplemental Fig. S1, C and D). KEGG pathway analysis revealed that the DEGs between control and ethylene treatments were enriched mainly in carbon metabolism, biosynthesis of amino acids, and starch and Suc metabolism at both 1 and 4 DIS (Supplemental Fig. S1, E and F).
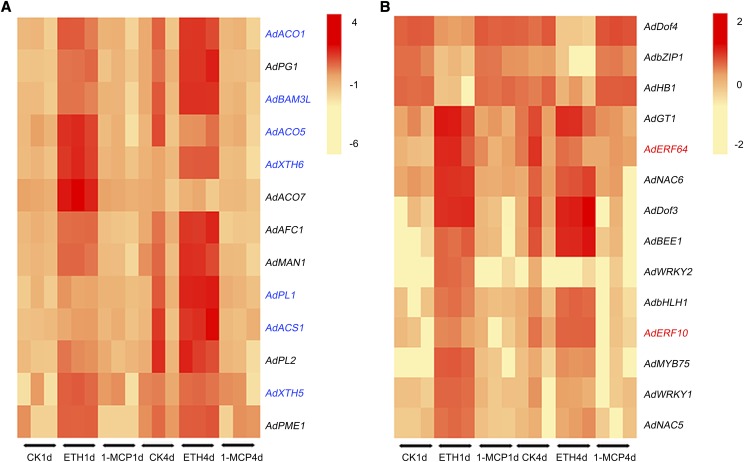
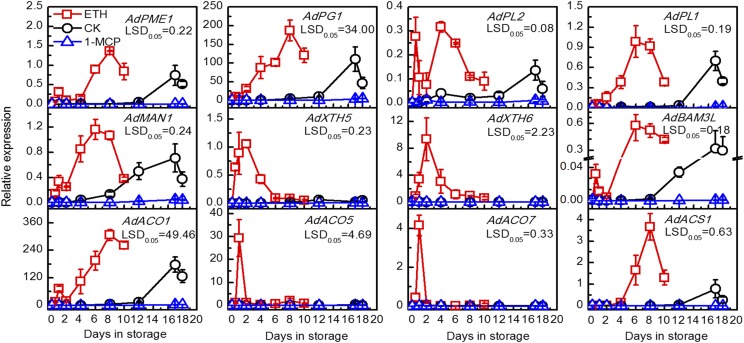
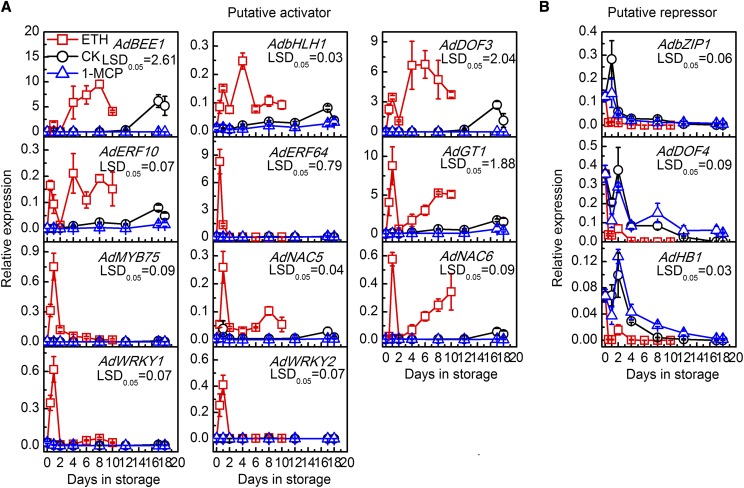
The selective thresholds were set using a false discovery rate ≤ 0.01, estimated absolute log2fold change > 2, FPKM ratio of control/ethylene > 40 or ethylene/1-MCP > 100, and highest FPKM values > 9. Thirteen target genes, including four ethylene biosynthesis genes, eight cell wall-related genes, and one starch degradation gene (Fig. 2A), and 14 TFs (Fig. 2B) emerged from all DEGs (Supplemental Table S1). Expression of the 13 structural genes was positively associated with kiwifruit postharvest ripening and softening, as all genes were induced by ethylene treatment and suppressed by 1-MCP (Fig. 2). This was verified by reverse transcription quantitative PCR (RT-qPCR) for all genes (Fig. 3) except AdAFC1 (acid β-fructofuranosidase; Supplemental Fig. S2). However, these structural genes also showed different responses to exogenous and internal ethylene. AdXTH5, AdXTH6, AdACO5, and AdACO7 were responsive to ethylene treatment and peaked at 1 or 2 DIS, then declined to similar basal levels of the control and 1-MCP-treated fruit. AdPME1 (pectin methyl esterase), AdPL2 (pectin lyase), AdMAN1 (endo-β-mannanase), AdBAM3L, and AdACO1 were up-regulated rapidly by ethylene treatment and then showed a significantly higher expression peak at the ethylene climacteric peak (17 DIS for control and 4–8 DIS for ethylene-treated fruit). The expression of AdPG1, AdPL1, and AdACS1 (1-aminocyclopropane-1-carboxylate synthase) followed the pattern of the ethylene climacteric peak (Fig. 3). In contrast to the expression patterns of structural genes, 11 TFs were putative activators and were up-regulated by ethylene (Fig. 4A), while AdbZIP1 (basic leucine zipper protein), AdDof3, and AdHB1 were putative repressors down-regulated by ethylene treatment (Fig. 4B). In general, all of these structural genes and TFs could be potential candidates involved in programming kiwifruit ripening. It should be noted that we only looked at three key ripening processes in this study, and other structural genes involved in fruit ripening also could be targets for these TFs.
Figure 2.
Comparison of DEGs between control, ethylene-treated, and 1-MCP-treated kiwifruit. Fruit were treated with 100 μL L−1 ethylene (ETH), 1 μL L−1 1-MCP, or air (control [CK]) for 24 h at 20°C, and comparisons were made at 1 and 4 d. A, DEGs of 13 structural genes with putative function in kiwifruit ethylene biosynthesis, cell wall modification, and starch degradation. B, DEGs of 14 transcriptional factors. There were three replicates at each point. Transcript abundance is indicated by color. The names in black represent new genes, which were not included in the cv Hong Yang genome database; those in blue and red are published structural and TF genes, respectively.
Figure 3.
Expression of structural genes in response to ethylene or 1-MCP treatment during kiwifruit ripening. Fruit were treated with 100 μL L−1 ethylene (ETH), 1 μL L−1 1-MCP, or air (control [CK]) for 24 h at 20°C. Gene expression was analyzed by RT-qPCR. Error bars represent se based on three replications. lsd values represent lsd at P = 0.05.
Figure 4.
Expression of TFs in response to ethylene or 1-MCP treatment during kiwifruit ripening. Fruit were treated with 100 μL L−1 ethylene (ETH), 1 μL L−1 1-MCP, or air (control [CK]) for 24 h at 20°C. Gene expression was analyzed by RT-qPCR. A, Expression of putative activators. B, Expression of putative repressors. BEE, Brassinosteroid enhanced expression; GT, Trihelix TF. Error bars represent se based on three replications. lsd values represent lsd at P = 0.05.
In Vivo Regulation of Ripening-Associated TFs and Structural Genes
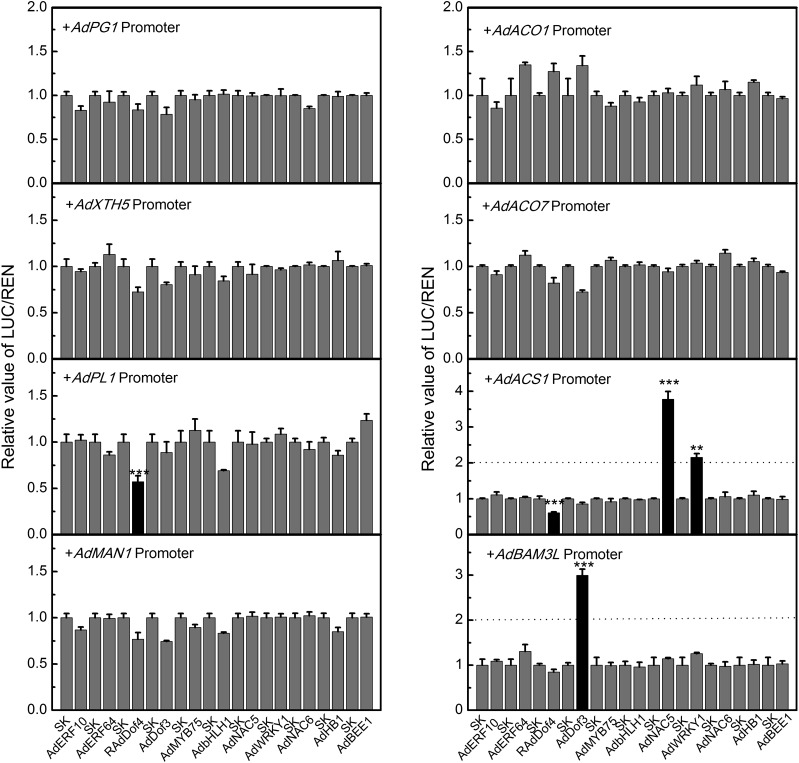
The regulatory effects of ripening-associated TFs on structural genes were tested using the dual-luciferase assay. Although these TFs and structural genes had very obvious associations at the transcript level, limited in vivo interactions were found between them (Fig. 5). AdDof4 acted as a repressor on promoters of AdPL1 and AdACS1, where luciferase activity (the ratio of firefly luciferase and Renilla luciferase) was reduced to approximately 0.5. AdNAC5 and AdWRKY1 both targeted the AdACS1 promoter, with 3.7- and 2.3-fold induction, respectively. The AdBAM3L promoter was only up-regulated by AdDof3, with an approximately 3-fold induction. Except for these regulatory effects, the relations of the other ripening-associated TFs and structural genes remained unclear.
Figure 5.
Regulatory effects of TFs on promoters of ethylene biosynthesis, cell wall-modifying, and starch degradation genes as determined by dual-luciferase assays. The ratio of firefly luciferase and Renilla luciferase (LUC/REN) of the empty vector plus promoter was set as 1. SK represents the empty pGreen II 0029 62-SK vector. Error bars indicate se from three replicates (**, P < 0.01 and ***, P < 0.001).
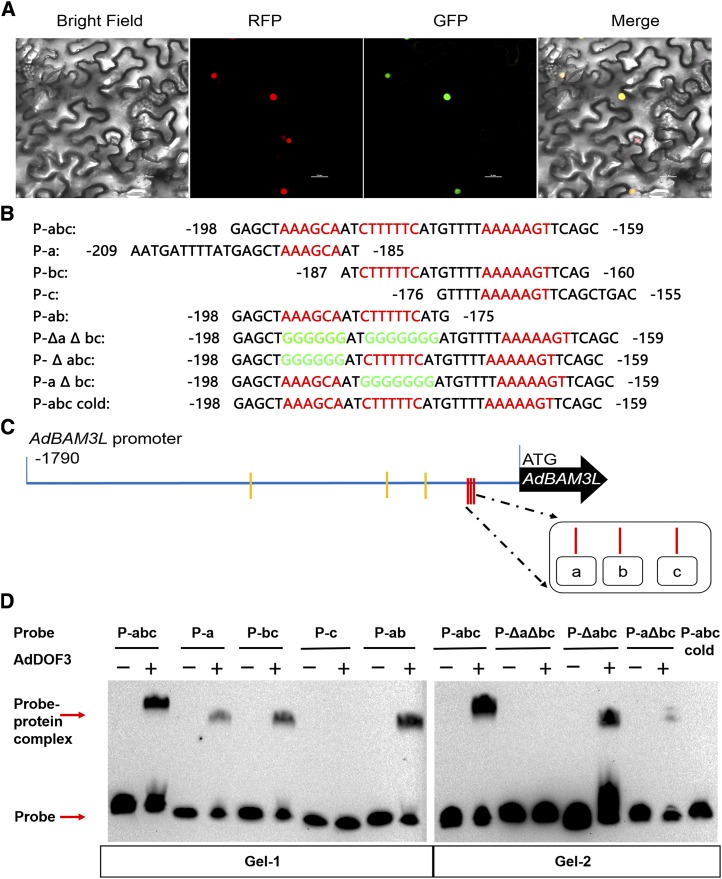
Investigation of AdDof3 Binding Elements on the AdBAM3L Promoter
Based on the dual-luciferase assays (Fig. 5) and RNA-seq results (Supplemental Fig. S1), the mechanism of AdDof3 regulation on the AdBAM3L promoter was selected for further investigation. First, subcellular localization results indicated that AdDof3 was located at the nucleus (Fig. 6A), which is similar to most TFs. The core binding sequence for the Dof family is AAAG/CTTT (Yanagisawa and Schmidt, 1999). In the region of the AdBAM3L promoter (−1,790 to −1 bp), six motifs were found (Fig. 6C). EMSA results indicated that the region (−160 to −200 bp) that contains the other three motifs showed the binding band in the presence of AdDof3 (Fig. 6, B and D).
Figure 6.
Subcellular localization of AdDof3 and EMSA. A, Subcellular localization of AdDof3-GFP in transgenic Nicotiana benthamiana leaves (expressed with nucleus-located mCherry). AdDof3 was inserted into the pCAMBIA1300-sGFP vector. The GFP fluorescence of AdDof3-GFP is indicated. Bars = 25 μm. B, Oligonucleotides used for the EMSA with the Dof core sequences are in red. The mutated bases are indicated in green. C, Core sequences (AAAG/CTTT) of Dof protein-binding sites in the AdBAM3L promoter. D, EMSA of 3′ biotin-labeled dsDNA probes with the AdDof3 DNA-binding domain proteins. Recombinant AdDof3 was purified from E. coli cells and used for DNA-binding assays with P-abc, P-a, P-bc, P-c, P-ab, and mutated P-ΔaΔbc, P-Δabc, and P-aΔbc together with cold unlabeled competitor as the probes. Water was added in place of AdDof3 protein as a control.
In order to determine the exact binding site between −160 and −200 bp, eight different probes were designed with deletion or mutagenesis (Fig. 6B). EMSA showed that AdDof3 could bind to the probes of P-abc (probe with three motifs), P-a, P-bc, and P-ab but not the probe P-c (Fig. 6D). These results suggested that AdDof3 binds physically to the AdBAM3L promoter (sites a and b), which was further confirmed by mutated probes. In the presence of AdDof3, P-Δabc (probe with b and c motifs and mutated a motif) and P-aΔbc showed binding signals with different intensities, while P-ΔaΔbc failed to generate any visible binding signal (Fig. 6D). Moreover, the shifted band disappeared with the addition of an unlabeled competitor with the same sequence P-abc.
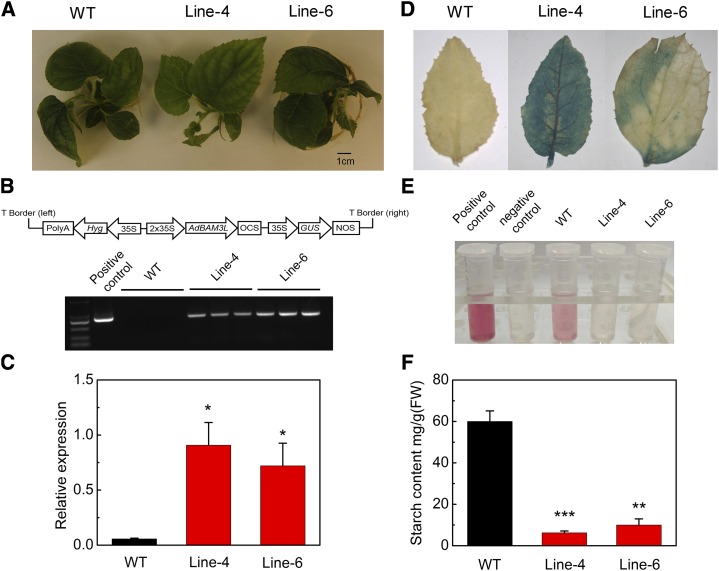
Stable Transformation Suggests That AdBAM3L Is an Important Gene for Starch Degradation in Kiwifruit
For most perennial fruits, it is difficult to perform stable transformation due to low efficiency and the long periods required for growth and development. Here, AdBAM3L was overexpressed in cv Qinmei tissue cultured plantlets. Although the aim of our research is fruit ripening, only leaves from 5-month plantlets on Murashige and Skoog (MS) medium were used for transgenic verification and phenotype analyses (Fig. 7A), due to the very slow growth rate of kiwifruit plantlets (Supplemental Fig. S3). First, the integration of AdBAM3L into the genome was confirmed by conventional PCR analyses (Fig. 7B). Further analysis by RT-qPCR (Fig. 7C) and GUS staining (Fig. 7D) confirmed the overexpression of AdBAM3L in the two transgenic lines. The expression of AdBAM3L in transgenic plants (lines 4 and 6) was more than 10-fold higher than that in the wild type (Fig. 7C). Starch analyses indicated that both transgenic lines had lower starch contents in leaves than the wild-type plants (Fig. 7, E and F). At the same period (about 5 months), leaves of wild-type plants contained 60 mg g−1 fresh weight starch, while the transgenic lines (lines 4 and 6) contained only 6.2 and 9.98 mg g−1 fresh weight starch, respectively (Fig. 7F).
Figure 7.
Overexpression of AdBAM3L in kiwifruit plants. A, Five-month-old plants on MS medium. B, Schematic map of the AdBAM3L-pCAMBIA1301 construct and PCR analysis of the wild type (WT) and two independently regenerated transgenic lines. The positive control used a plasmid containing the AdBAM3L-pCAMBIA1301 construct as a template. C, Expression of AdBAM3L in the wild type and transgenic lines. D, GUS staining of wild-type and AdBAM3L transgenic plants. E, Starch content reflected by quinoneimine dye. The color intensity represents starch concentration. Positive control, d-Glc standard (Megazyme International Ireland); negative control, water. F, Starch content in wild-type and transgenic plant leaves. FW, Fresh weight. Error bars in C and F indicate se from three replicates (*, P < 0.05; **, P < 0.01; and ***, P < 0.001).
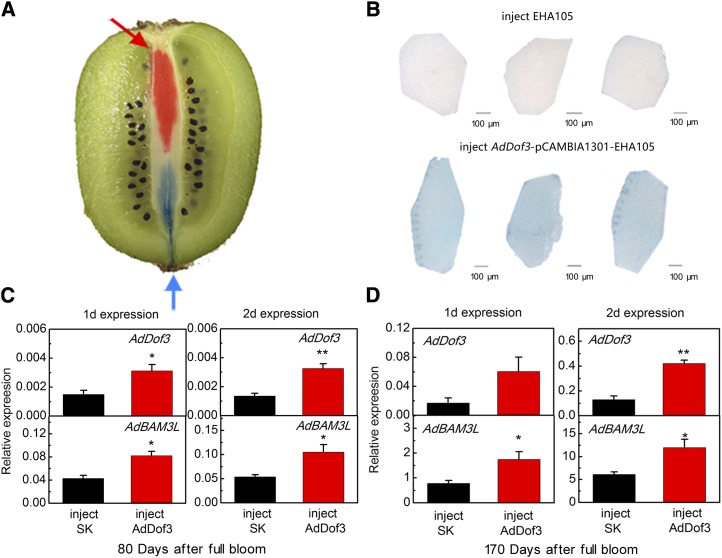
Transient Overexpression Analyses Suggest the Regulation of AdBAM3L by AdDof3 during Kiwifruit Ripening
The results from the dual-luciferase assay, EMSA, and stable transformation analyses suggest a regulatory pathway between AdDof3 and AdBAM3L. AdDof3 could contribute to kiwifruit ripening via physical binding and activation on the promoter of AdBAM3L, a key gene of starch degradation. Further experiments were designed to test the proposed regulatory model in kiwifruit. Transient overexpression experiments were carried out in cv Hayward fruit core tissue because this tissue has high permeability and provides the ideal single fruit control with two ends (Fig. 8A). RT-qPCR analyses indicated that the expression of AdDof3 and AdBAM3L was similar in the two different fruit ends (Supplemental Fig. S4). Thus, one end of the core tissue was infiltrated with Agrobacterium tumefaciens strain EHA105 as a control and the other was injected with AdDof3-pCAMBIA1301-EHA105 (Fig. 8B). GUS staining showed that the strain with the vector carrying AdDof3 not only infiltrated into the kiwifruit core tissue but also was translated and functioned normally, while the control tissue had no visible GUS staining (Fig. 8B).
Figure 8.
Transient overexpression of AdDof3 and its up-regulation of AdBAM3L in the core tissue of cv Hayward fruit. A, Schematic diagram for injection with differential color inks. The arrows show injection sites. B, GUS staining of kiwifruit core tissue segments injected with AdDof3-pCAMBIA1301-EHA105 or EHA105 at 1 d after injection. The segments were photographed separately. Bars = 100 μm. C, Gene expression of endogenous AdDof3 and AdBAM3L in immature kiwifruit at 80 DAFB. Injection of A. tumefaciens strain (GV3101) with the empty SK vector was the control, and the AdDof3 recombined SK vector was the treatment. D, Gene expression of endogenous AdDof3 and AdBAM3L in mature kiwifruit harvested at 170 DAFB. Error bars in C and D indicate se from three replicates (*, P < 0.05 and **, P < 0.01).
Fruit at two different stages of maturity were selected for transient expression experiments, including immature fruit (80 d after full bloom [DAFB]) and commercially mature fruit (170 DAFB). Both AdDof3 and AdBAM3L expression levels were much higher in the mature fruit core tissue than in immature fruit (Fig. 8, C and D). The abundances of AdDof3 and AdBAM3L transcripts increased at 1 and 2 d after AdDof3 infiltration in both stages. Injecting with AdDof3 resulted in 16.1% (1 d after injection) and 25.2% (2 d after injection) reductions in 80-DAFB samples and 12.5% (1 d after injection) and 9% (2 d after injection) reductions in 170-DAFB samples (Supplemental Fig. S5). However, these reductions were not significant at P < 0.05.
DISCUSSION
Characterization of Genes Associated with Postharvest Ripening of Kiwifruit by Transcriptomics Analysis
Kiwifruit is an ideal species for studying fruit ripening and softening and contains four distinct softening phases (Atkinson et al., 2011). Here, the measurement of three characteristic indices (ethylene production, firmness, and starch content) indicated that ripening and softening of cv Hayward kiwifruit was, as expected, accelerated by ethylene and retarded by 1-MCP, as reported previously in kiwifruit (Koukounaras and Sfakiotakis, 2007; Atkinson et al., 2011; Mworia et al., 2012).
Transcriptomic analysis indicated that there were more than 5,000 DEGs between ethylene-treated and control fruit at 1 d, which is consistent with general ideas on the multigenic traits of fruit ripening. Our selective analysis identified 12 structural genes related to ethylene biosynthesis, cell wall, and starch degradation. As only three characteristic traits were selected in this research, it is likely that additional DEGs will exist for the other ripening-related traits, such as aroma, sugar, and acid degradation (Nieuwenhuizen et al., 2015; Tang et al., 2016). In addition, 14 TFs reached the selective threshold set for DEGs, with 11 putative activators and three putative repressors, based on their sequence similarity with previously published sequences. Unlike the structural genes, these 14 TFs could be potential regulators of any aspect of kiwifruit ripening, including other ripening traits in addition to softening, starch, and ethylene. Most of these structural genes have been reported previously from gene expression or activity analyses (AdXTH5/6 [Atkinson et al., 2009], AdBAM3L [Hu et al., 2016], and AdACO1 [Xu et al., 1998]) or functional analysis (AdACS1; Atkinson et al., 2011). However, some new genes, such as a PG gene (AdPG1) and two PLs (AdPL1/2), were identified in this work and may contribute to kiwifruit pectin solubilization and softening. For hemicellulose degradation, the isolation of another new gene, AdMAN1, suggests a potential metabolic pathway for hemicellulose degradation in addition to the role of the XTHs (Cutillasiturralde et al., 1994; Atkinson et al., 2009). It is particularly interesting that AdPG1 could not be found in the current version of the kiwifruit genome database. Of the TFs identified, only AdERF10 and AdERF64 have been reported previously from gene expression analysis (Yin et al., 2010; Zhang et al., 2016), and all others were newly identified regulator genes potentially implicated in controlling kiwifruit ripening and quality.
Characterization of Multiple Links between TFs and Ripening-Related Genes
Ripening-associated TFs have been reported widely in various fruit; however, most (especially from perennial fruit) have only been characterized through correlations between gene transcripts and fruit ripening. Few TFs have reported regulatory effects on target genes: a number of tomato fruit ripening regulators (RIN [Vrebalov et al., 2002], CNR [Manning et al., 2006], Nr [Wilkinson et al., 1995], and AP2a [Karlova et al., 2011]), a few banana (Musa acuminata) ripening-related genes (MabHLH6, basic helix-loop-helix [Xiao et al., 2018]; MaDREB1–MaDREB4, dehydration-responsive element-binding protein [Kuang et al., 2017]; MaERF9; and MaDof23 [Feng et al., 2016]), apple ERF genes (MdCBF [Tacken et al., 2010] and MdERF2 [Li et al., 2016]), and papaya (Carica papaya) CpERF9 (Fu et al., 2016). Here, among 14 TFs, at least four (AdDof3, AdDof4, AdNAC5, and AdWRKY1) showed significant regulatory activity on different target kiwifruit ripening genes, suggesting potential roles in ripening and softening. AdDof3 was an activator on the AdBAM3L promoter, AdNAC5 and AdWRKY1 were activators for the AdACS1 promoter, and AdDof4 was a repressor on both AdPL1 and AdACS1 promoters. In kiwifruit, AdERF9 has been characterized as a regulator of the AdXTH5 promoter (designated AdXET5; Yin et al., 2010). Thus, the roles of these TFs on fruit ripening-related genes may provide new information on kiwifruit ripening regulation.
Compared with the model fruit tomato and perennial fruit banana and apple, our results appear to show previously uncharacterized links between TFs and ripening-related genes. In tomato, PL was characterized recently as an important regulator for fruit softening (Yang et al., 2017) and long shelf life (Uluisik et al., 2016), but its transcriptional regulation remains unclear. In tomato, high-resolution mapping showed that a quantitative trait locus contained an ERF and three PME genes (Chapman et al., 2012). In apple, an ERF gene (MdCBF) also was characterized as an activator of the apple MdPG (Tacken et al., 2010). In kiwifruit, the physiology analysis also confirmed that pectin degradation is important for postharvest softening. Thus, the regulation of AdDof4 on AdPL1 suggests a regulatory link between TFs and pectin modification.
In general, the interactions between the four TFs (AdDof3, AdDof4, AdNAC5, and AdWRKY1) and ripening-related genes were not only beneficial to understand kiwifruit ripening but also provided new examples for other fruit species. The differences between our results in kiwifruit and previous findings from other fruit, while probably reflecting species differences, also could be due to the selective threshold setting. Thus, homologs of the ripening-related TFs from other fruit also may exist in kiwifruit. Moreover, the structural genes were only limited to ethylene response and cell wall and starch degradation; thus, the 10 other TFs still retain the potential for fruit ripening regulation via other ripening-related genes.
Functional Verification of the Role of AdBAM3L as a Key Gene in Starch Degradation
The two AdBAM3L overexpressed transgenic lines of kiwifruit showed obvious phenotypes. The transgenic lines had low gene expression levels and lower starch contents. In addition, Suc content was higher in transgenic lines, but Fru and Glc contents were similar (Supplemental Fig. S6). Thus, the transformation and expression analysis (both postharvest ripening and on-tree development) suggests that AdBAM3L is a key gene for kiwifruit starch degradation (Fig. 3; Supplemental Fig. S7). For perennial fruit, stable transformation is extremely difficult and has only been used in a few species (e.g. papaya and apple). Thus, most fruit-related genes (except for tomato) have been characterized with in vitro molecular biology platforms. Among the three structural genes (AdPL1, AdBAM3L, and AdACS1) that could be regulated by differentially expressed TFs, AdBAM3L was one of the predicted starch degradation-related genes from our previous research, using various postharvest treatments and RT-qPCR (Hu et al., 2016). Thus, the functional verification of AdBAM3L with overexpression analysis in kiwifruit is an extension of previous results, with the approach providing a shortcut to overcoming long-term transformation in kiwifruit (Supplemental Fig. S3) and also the basis for selections of AdDof3-AdBAM3L interactions for further analysis. The associations between the expression of BAM, as well as some other structural genes, and ripening have been reported previously in fruit, such as kiwifruit (Hu et al., 2016; Tang et al., 2016), banana (Xiao et al., 2018), and Poncirus trifoliate (Peng et al., 2014). However, none of these genes had been characterized functionally with stable transformation.
In Vitro and in Vivo Regulation of AdBAM3L by AdDof3
The dual-luciferase assay showed that AdDof3 transactivated the promoter of AdBAM3L, which is further supported by the associations between AdDof3 and AdBAM3L expression in both developing kiwifruit (Supplemental Fig. S7) and fruit undergoing postharvest ripening (Fig. 2). EMSA analyses further indicated that AdDof3 bound physically to AAAG/CTTT elements within the AdBAM3L promoter, which is similar to Dof homologs from other plants. For instance, a maize (Zea mays) Dof class protein, PBF (prolamin-box binding factor), can transactivate the γ-zein gene promoter by binding to the AAAG motif (Marzábal et al., 2008). Moreover, ZmDof3 recognized and bound to the AAAG core sequence in promoters of the starch biosynthesis genes Du1 and Su2 in maize and functioned as a positive regulator (Qi et al., 2017). Similar to these plants, AdDof3 is a direct activator on the AdBAM3L promoter and may contribute to starch degradation in kiwifruit. Banana MabHLH6 also is an activator of starch degradation genes via physical binding to target promoters (Xiao et al., 2018). Here, AdDof3 provided another type of regulator on fruit starch degradation.
Moreover, while MabHLH6 showed regulatory function on starch degradation genes based on EMSA and dual-luciferase assays, its regulatory function in banana fruit remains unclear. Transient expression experiments have been used widely for gene function analysis in various fruit species (Akagi et al., 2009; Li et al., 2017), but the precision of such analyses is largely influenced by bias that is generated between different fruit tissues (discs or slices) and also different fruits. Our results indicate that kiwifruit core tissue had very high water permeability and that the two ends of the fruit had similar gene expression levels (Fig. 8), thus providing an ideal system for transient overexpression analysis. With the benefit of this system, as well as GUS staining (Fig. 8), AdDof3 was shown to regulate AdBAM3L in vivo in cv Hayward kiwifruit in both immature (80 DAFB) and mature (170 DAFB) fruit. However, analysis of the starch contents in transient overexpressed tissues indicated that AdDof3 could reduce starch contents compared with the empty control (SK), which is consistent with its regulation of starch degradation genes. However, the reduction was not statistically significant (Supplemental Fig. S5). To find out whether such an effect is significant requires more extended transient overexpression experiments or stable transformation.
CONCLUSION
Transcriptomic analysis predicted 12 structural genes (for ethylene biosynthesis, cell wall degradation, and starch degradation) and 14 TFs that showed high potential for fruit ripening regulation. Moreover, analysis of the relationships between these TFs and structural genes indicated previously uncharacterized novel potential transcriptional regulation links, including four TFs (AdDof3, AdDof4, AdNAC5, and AdWRKY1) and three structural genes (AdBAM3L, AdACS1, and AdPL1). Most significantly, using stable transformation, EMSA, and transient analysis, AdBAM3L was confirmed as a key regulator of kiwifruit starch degradation, which could be transactivated by AdDof3 via binding on AAAG/CTTT elements. Thus, these findings advance our understanding of the regulation of fruit starch degradation, a critical step for both fruit initial ripening and the ultimate fruit flavor contributed by soluble sugars.
MATERIALS AND METHODS
Plant Material and Treatments
Mature kiwifruit (Actinidia deliciosa var deliciosa ‘Hayward’) was harvested from a commercial orchard in Shanxi, China, in 2015, with mean TSS of 6.19%. Fruit of uniform size free from visible defects were selected and divided into three batches for three treatments. Each treatment contained three biological replicates of approximately 200 fruit. The fruit were treated with ethylene (100 μL L−1, 24 h, 20°C), 1-MCP (1 μL L−1, 24 h, 20°C), or air as the control (24 h, 20°C) in 20-L air-tight containers. After treatment, the fruit were transferred to normal air at 20°C. At each sampling point, three replicates of four fruit were collected from each batch. The outer pericarp (without skin or seeds) of the fruit was cut into small pieces and rapidly frozen in liquid nitrogen and then stored at −80°C for further experiments.
Fruit Physiological Properties
A number of kiwifruit postharvest quality properties were measured, including ethylene production, TSS and starch content, firmness, and CWM. Ethylene was measured by gas chromatography (Agilent Technologies 7890A GC System) and firmness using a TA-XT2i texture analyzer (Stable Micro Systems), with the same parameters as described in our previous report (Zhang et al., 2016). TSS were measured using an Atago digital hand-held refractometer. Two ends of each fruit were sliced and then three drops of juice were squeezed from each slice onto the refractometer. Ethylene was measured with three replicates (four fruit in each replicate) for each treatment. Firmness and TSS were measured with 10 single fruit replicates.
Total starch was measured on three replicates of 0.1-g frozen samples using a total starch assay kit (Megazyme International Ireland), following a method described previously (Stevenson et al., 2006; Hu et al., 2016). Starch contents were measured from frozen fruit flesh with three biological replicates.
CWM extraction and isolation was performed as described previously with slight modifications (Vicente et al., 2013; Minas et al., 2014). The CWM extractions were performed on three biological replicates. For each extraction, approximately 3 g of frozen fruit flesh was placed in 20 mL of 80% (v/v) ethanol and boiled for 20 min. After centrifuging at room temperature, the low-Mr solutes and insoluble materials were separated. The supernatant was discarded, and sediments were washed sequentially with 80% (v/v) ethanol, chloroform:methanol (1:1), and acetone and dried at 40°C for 24 h. The dried residue was collected and weighed. Approximately 50 mg of CWM from each sample was used to determine the contents of different cell wall components. First, they were suspended in 6 mL of 50 mm acetic acid-sodium acetic buffer (pH 6.5) and stirred at 20°C for 6 h, then they were centrifuged and vacuum filtered. The filtrate were designated as WSP. Second, the residue was suspended in 6 mL of 50 mm acetic acid-sodium acetic buffer (pH 6.5) with 50 mm EDTA for 6 h of continuous shaking. After centrifugation, the supernatant was filtered and measured as ISP. Third, the EDTA-insoluble pellet was extracted with 6 mL of 50 mm Na2CO3 at 4°C for 18 h, then turned to 20°C shaking for 2 h. The slurry was centrifuged, and the supernatant was filtered as CBP. Subsequently, the residue was extracted with 3 mL of 4 m KOH at 20°C for 5 h of continuous shaking, and the supernatant was designated as hemicellulose content. Finally, the insoluble residue was washed with 3 mL of 0.3 m acetic acid and 6 mL of 80% (v/v) ethanol and then centrifuged. The pellet was dried at 40°C and weighted as α-cellulose. Pectin and hemicellulose were measured according to reported protocols (Yemm and Willis, 1954; Blumenkrantz and Asboe-Hansen, 1973). For pectin, 3 mL of H2SO4 was added to 0.5 mL of extracting solution and boiled for 20 min. The mixture was cooled down to room temperature, and 0.2 mL of carbazole-anhydrous ethanol (1.5 g L−1) was added. After 30 min of standing, absorbance was measured at 530 nm. Hemicellulose content was determined by the anthrone-sulfuric acid method. The hemicellulose extracting solution comprised 5 mL of anthrone reagent (2 g of anthrone dissolved in 80% [v/v] H2SO4 and diluted with 80% H2SO4 to 1,000 mL). The mixture was heated in a 100°C water bath for 10 min. After cooling to room temperature, absorbance measurements were made at 625 nm. The results were expressed as mg GalA (GalUA) g−1 fresh weight and mg Glu (Glc) g−1 fresh weight, respectively.
RNA Extraction and RNA-seq
Total RNA was extracted from frozen kiwifruit flesh following our previous protocol (Yin et al., 2008). For RNA-seq, at least 1 µg of RNA from each sampling point (1 and 4 d) and each treatment (control, 1-MCP, and ethylene) was sent for sequencing. Three replicates were used for RNA-seq.
RNA-seq and bioinformatics analyses were conducted by Biomarker. Library construction was carried out following the manufacturer’s instructions for the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs; E7530) and NEBNext Multiplex Oligos for Illumina (New England Biolabs; E7500), and libraries were sequenced with the Illumina HiSeq 4000 sequencing platform. Transcriptome analysis used reference genome-based reads mapping. The clean reads filtered from raw data were mapped to the cv Hong Yang (Actinidia chinensis) genome database using Tophat2 software (Kim et al., 2013). Low-quality reads were removed by perl script (unknown nucleotides > 5% or low Q value ≤ 20%). FPKM were used to estimate gene expression levels by the Cufflinks software (Trapnell et al., 2010). The false discovery rate was used to identify the P value threshold in multiple texts. Sequences were compared against various protein databases, including NCBI (the National Center for Biotechnology Information), Nr (nonredundant protein), and Swiss-Prot (a manually annotated and reviewed protein sequence database) by a cutoff E value of 10−5. Gene function was annotated with GO (Gene Ontology), KO (KEGG ortholog database), Swiss-Prot, and Nr annotation. GO enrichment analysis of the DEGs was carried out using Wallenius noncentral hypergeometric distribution based on GOseq R packages (Young et al., 2010), which can adjust for gene length bias in DEGs. KEGG is a database resource for understanding high-level functions and utilities of the biological system (Kanehisa et al., 2008). KOBAS software was used to test the statistical enrichment of DEGs in KEGG pathways (Mao et al., 2005).
cDNA Synthesis and RT-qPCR
For cDNA synthesis, TURBO DNase (Ambion) was used to remove contaminating genomic DNA. The Reverse Transcription System (Promega) was used for cDNA synthesis. Three biological replicates with three independent RNA extractions and cDNA synthesis were performed for each sampling point and each treatment.
RT-qPCR was carried out using a LightCycler 480 instrument (Roche) with LightCycler 480 SYBR Green I Master (Roche). The specificity of primers was double checked by melting curves and product resequencing (Yin et al., 2010). Primers for RT-qPCR analysis are listed in Supplemental Table S2. Kiwifruit Actin (GenBank no. EF063572) was employed as the housekeeping gene (Zhang et al., 2006).
Genomic DNA Extraction
Kiwifruit genomic DNA was extracted from leaves. Approximately 0.1 g of tissue was put into 900 μL of TPS buffer (100 mm Tris-HCl, 10 mm EDTA, and 1 m KCl) for 1 h in a water bath at 75°C, then centrifuged at 12,000 rpm for 10 min and added with an equal volume of isopropyl alcohol to the supernatant to subside genomic DNA. After 5 min, the tube was centrifuged again and the supernatant was discarded. The pellet (genomic DNA) was dissolved in sterile water. Three individual plants (5 months old) from each line were used as three replicates. The PCR template using a plasmid containing the target sequence acted as the positive control, and water was the negative control.
Gene Isolation, Promoter Cloning, and Promoter Motif Analysis
Based on the RNA-seq results, differentially expressed sequences (DES) associated with ethylene biosynthesis, starch degradation, and cell wall metabolism were selected. Genes induced by ethylene and repressed by 1-MCP relative to the control were assigned as candidates, and the threshold of DES was set as 50-fold based on FPKM values between control, ethylene, and 1-MCP treatments on days 1 and 4 separately. The full-length coding sequences for these DES were obtained from the kiwifruit genome database (Huang et al., 2013), and the sequences were verified by PCR using gene-specific primers (Supplemental Table S3) with cDNA from cv Hayward.
Promoters of postharvest ripening-related genes (the structural genes) were isolated according to the genome database, except for AdPG1 (not present in the genome database). With the promoter sequences from the genome database, the forward primers (FP) were located in promoter regions and reverse primers (RP) were from coding sequences. For AdPG1, genome walking was carried out to obtain its promoter, with the GenomeWalker kit (Clontech), using primary RP (5′-TGCATGGCCCGCTAAACATAGTC-3′) and secondary RP (5′-GCCGCAAGCTGAATCCCATGCG-3′). The analysis of cis-elements within promoter regions was conducted using the online Web site http://jaspardev.genereg.net. All promoter sequences used in the following experiments are listed in Supplemental Table S4.
Dual-Luciferase Assays
The dual-luciferase assay was used to investigate the regulatory roles of different TFs on target promoters, according to our previous protocols (Min et al., 2012). Full-length sequences of 11 TFs were inserted into pGreen II 0029 62-SK vector (SK), while the promoters of eight softening-related genes were recombined to the pGreen II 0800-LUC vector. Primers used for vector constructions are listed in Supplemental Table S5. All the constructs were transferred into Agrobacterium tumefaciens (GV3101), and the cultures were adjusted to an OD600 of 0.75 with infiltration buffer (150 mm acetosyringone, 10 mm MES, and 10 mm MgCl2, pH 5.6). The ratio of A. tumefaciens mixtures of TFs and promoters was 10:1, which were then infiltrated into the leaves of Nicotiana benthamiana. The N. benthamiana plants were cultivated in a glasshouse for 3 d. Firefly luciferase and Renilla luciferase were assayed with the Dual-Luciferase Reporter Assay System (Promega). For each TF-promoter interaction, triplicate transient assay measurements were performed. Those with significant regulatory effects were confirmed by at least three independent experiments.
Recombinant Protein and EMSA Analysis
According to the results of the dual luciferase assay, the regulatory action of AdDof3 on the AdBAM3L promoter was analyzed further by an EMSA.
In order to obtain the recombinant protein, the full-length AdDof3 was amplified with FP (5′-GCTGATATCGGATCCGAATTCATGCCTCCGGAAACTTCCG-3′) and RP (5′-GCAAGCTTGTCGACGGAGCTCGACTTGAGACCTTTGCCTG-3′) and inserted into the pET-32a (Novagen) vector with double digestions of EcoRI and SacI. The construct was purified and transformed into Escherichia coli strain BL21 (DE3). The recombinant protein was induced by 0.5 mm isopropyl β-d-1-thiogalactopyranoside at 20°C for 18 h and purified as follows. The E. coli cells were lysed with the buffer (20 mm Tris-HCl, pH 8, 0.5 m NaCl, 10 mm β-mercaptoethanol, and 10% glycerol) and then subjected to sonication on ice at 200 W with a 3-s/2-s on/off cycle for 30 min and centrifuged at 9,000g for 20 min at 4°C. Following this, the Ni-NTA resin (TRAN) was added to the supernatant to combine His-tagged proteins, and the His-tagged proteins were eluted with gradient imidazole-containing buffers (75, 100, 125, 150, 175, 250, and 500 mm). The portions eluted by 500 mm imidazole were used further in the following EMSA experiment (Supplemental Fig. S8).
The probes were 3′ biotin end labeled by HuaGene and converted to dsDNA probes by annealing complementary oligonucleotides. EMSA was performed using the LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific; 20148). The binding specificity was examined by mutant probes and competition probe (1,000-fold unlabeled oligonucleotides).
Stable Transformation and Analysis in Kiwifruit
To obtain transgenic kiwifruit, the 1,641-bp AdBAM3L coding sequence was amplified with primers (FP, 5′-AGAGAACACGCCCGGGGATCCATGGCTTTAACATTACATTG-3′; RP, 5′-CTTGCATGCCTGCAGGTCGACTTACACAAAAGCAGCCTCCT-3′) and inserted downstream of the cauliflower mosaic virus 35S promoter into the modified pCAMBIA1301 vector via the BamHI/SalI restriction sites. The pCAMBIA1301 vector also contained a GUS reporter gene following one cauliflower mosaic virus 35S promoter (Fig. 7B). The construct was then introduced into A. tumefaciens strain EHA105.
Leaves of plantlet cv Qinmei in tissue culture were used for transformation. First, the leaves were cut into pieces (1 cm × 1 cm) and cultured in coculture medium for 24 h in dark. They were then coincubated for 10 min with A. tumefaciens cultures containing 1 mL L−1 acetosyringone solution (100 mm) and put back into coculture medium with sterilized filter paper for 48 h in the dark. Then, the explants were transferred and screened on shoot induction medium under long-day conditions (16 h of light/8 h of dark) until the regenerated shoots reached ∼2 cm and then were transferred to rooting medium. All the processes were performed at 25°C, and the representative status of transformed plants at different stages are shown in Supplemental Figure S3. The coculture medium components were MS + 1 mL L−1 Nitsch & Nitsch vitamin solution (NV) + 4 mg L−1 6-benzylaminopurine + 1 mg L−1 naphthaleneacetic acid solution; the shoot induction medium was MS + 1 mL L−1 NV + 4 mg L−1 6-benzylaminopurine + 1 mg L−1 naphthaleneacetic acid solution + 5 mg L−1 hygromycin + 50 mg L−1 meropenem; the rooting medium contained MS + 1 mL L−1 NV + 5 mg L−1 hygromycin + 50 mg L−1 meropenem.
Transgenic plants overexpressing AdBAM3L were identified by DNA detection, RT-qPCR, GUS staining, and starch content analysis. Verification of DNA level used genomic DNA from the transgene plants as a template with primers across the 35S promoter region (−424 bp) and coding sequence region (Supplemental Table S6). The methods of RT-qPCR and starch content have been described above, with reduced amounts of materials (0.25 g for RNA and 0.05 for starch). Three biological replicates were carried out for each line.
GUS Staining
Histochemical staining was conducted to confirm the expression of the GUS reporter cotransformed with AdBAM3L. The staining buffer was 0.1 m sodium phosphate buffer (pH 7), 10 mm EDTA, 1 mm ferricyanide, 1 mm ferrocyanide, 0.5% Triton X-100, and 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide. The leaves of transgenic kiwifruit plantlets were immersed in the staining buffer under vacuum for 30 min and then incubated overnight at 37°C. Ethanol at 75% was used for degradation of the chlorophyll of stained leaves.
Subcellular Localization of AdDof3
The AdDof3 full-length coding sequence without the stop codon was amplified using specific primers (FP, 5′-GGACGAGCTCGGTACCATGCCTCCGGAAACTTCCG-3′; RP, 5′-TGCTCACCATGTCGACCTTGAGACCTTTGCCTGGAG-3′) and then was fused to the pCAMBIA1300-sGFP vector (KpnI/SalI). The construct (35S-AdDof3-GFP) was transformed into A. tumefaciens strain GV3101 and then expressed transiently in transgenic N. benthamiana (expressed with nucleus-located mCherry) leaves. The GFP and RFP fluorescence was imaged with a fluorescence microscope (Zeiss).
Transient Overexpression in cv Hayward Fruit
The in vivo regulation of AdDof3 on AdBAM3L was investigated by transient overexpression in cv Hayward fruit with core tissue. In order to eliminate the variation across different fruit, the controls (empty SK vector) and AdDof3 (same as the dual-luciferase assays) were infiltrated separately into two different ends of core tissue within intact fruit (Fig. 8A). The infiltrated materials analyzed by GUS staining were injected into strain EHA105 as a control and strain EHA105 with the pCAMBIA1301 vector with AdDof3 and the GUS reporter gene (Fig. 8B). The primers for AdDof3 for GUS staining were FP (5′-AGAGAACACGCCCGGGGATCCATGCCTCCGGAAACTTCCG-3′) and RP (5′-ACGACGGCCAGTGCCAAGCTTCTACTTGAGACCTTTGCCTG-3′).
The fruit at 80 and 170 DAFB were harvested from Shaanxi in 2017. The buffers together with A. tumefaciens carrying constructs were the same as for the dual-luciferase assay. Either 0.2 mL of AdDof3 or empty vector was infiltrated into the core from the two ends, and then fruit were stored in an incubator at 25°C for 2 d. The material was collected at 1 and 2 d with three biological replicates.
Statistical Analysis
lsd analysis and Student’s t test were conducted by DPS7.05 (Zhejiang University). Figures were drawn with Origin 8.0 (Microcal Software). The heat maps were conducted with the log10FPKM values using online software (https://console.biocloud.net/static/index.html#/drawtools/intoDrawTool).
Accession Numbers
All sequence reads are available at GenBank SRR6885590 to SRR6885601.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RNA-seq analysis of DEGs between control, ethylene-treated, and 1-MCP-treated kiwifruit.
Supplemental Figure S2. AdAFC1 gene expression.
Supplemental Figure S3. Transgenic plants overexpressing AdBAM3L regenerated and cultured on MS medium for up to 22 weeks.
Supplemental Figure S4. RT-qPCR analyses of AdDof3 and AdBAM3L expression at apical and basal ends of kiwifruit core tissue.
Supplemental Figure S5. Starch contents in transient overexpressed core tissue with AdDof3 or empty vector.
Supplemental Figure S6. Sugar content in transgenic AdBAM3L kiwifruit.
Supplemental Figure S7. Expression of AdDof3 and AdBAM3L during the development of cv Hayward kiwifruit.
Supplemental Figure S8. AdDof3 protein purification.
Supplemental Table S1. Ethylene-responsive structural genes and TFs.
Supplemental Table S2. Primers for RT-qPCR.
Supplemental Table S3. Primers for full-length amplification.
Supplemental Table S4. Sequences (5′–3′) for promoter isolation.
Supplemental Table S5. Primers for vector construction in dual-luciferase assays.
Supplemental Table S6. PCR primers for DNA detection in transgenic kiwifruit plants.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Zhenghe Li (Zhejiang University) for providing the transgenic N. benthamiana that expressed with nucleus-located mCherry.
Footnotes
This research was supported by the National Key Research and Development Program (2016YFD0400102), the National Natural Science Foundation of China (31722042), the Natural Science Foundation of Zhejiang Province, China (LR16C150001), the Fok Ying Tung Education Foundation, China (161028, 20170101210004), and the 111 Project (B17039).
References
- Akagi T, Ikegami A, Tsujimoto T, Kobayashi S, Sato A, Kono A, Yonemori K (2009) DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol 151: 2028–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson RG, Johnston SL, Yauk YK, Sharma NN, Schröder R (2009) Analysis of xyloglucan endotransglucosylase/hydrolase (XTH) gene families in kiwifruit and apple. Postharvest Biol Technol 51: 149–157 [Google Scholar]
- Atkinson RG, Gunaseelan K, Wang MY, Luo L, Wang T, Norling CL, Johnston SL, Maddumage R, Schröder R, Schaffer RJ (2011) Dissecting the role of climacteric ethylene in kiwifruit (Actinidia chinensis) ripening using a 1-aminocyclopropane-1-carboxylic acid oxidase knockdown line. J Exp Bot 62: 3821–3835 [DOI] [PubMed] [Google Scholar]
- Blumenkrantz N, Asboe-Hansen G (1973) New method for quantitative determination of uronic acids. Anal Biochem 54: 484–489 [DOI] [PubMed] [Google Scholar]
- Chapman NH, Bonnet J, Grivet L, Lynn J, Graham N, Smith R, Sun G, Walley PG, Poole M, Causse M, et al. (2012) High-resolution mapping of a fruit firmness-related quantitative trait locus in tomato reveals epistatic interactions associated with a complex combinatorial locus. Plant Physiol 159: 1644–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutillasiturralde A, Zarra I, Fry SC, Lorences EP (1994) Implication of persimmon fruit hemicellulose metabolism in the softening process: importance of xyloglucan endotransglycosylase. Physiol Plant 91: 169–176 [Google Scholar]
- Dong T, Hu Z, Deng L, Wang Y, Zhu M, Zhang J, Chen G (2013) A tomato MADS-box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiol 163: 1026–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng BH, Han YC, Xiao YY, Kuang JF, Fan ZQ, Chen JY, Lu WJ (2016) The banana fruit Dof transcription factor MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes. J Exp Bot 67: 2263–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu CC, Han YC, Qi XY, Shan W, Chen JY, Lu WJ, Kuang JF (2016) Papaya CpERF9 acts as a transcriptional repressor of cell-wall-modifying genes CpPME1/2 and CpPG5 involved in fruit ripening. Plant Cell Rep 35: 2341–2352 [DOI] [PubMed] [Google Scholar]
- Hu X, Kuang S, Zhang AD, Zhang WS, Chen MJ, Yin XR, Chen KS (2016) Characterization of starch degradation related genes in postharvest kiwifruit. Int J Mol Sci 17: 2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Ferguson AR (2001) Kiwifruit in China. New Zeal J Crop Hort 29: 1–14 [Google Scholar]
- Huang S, Ding J, Deng D, Tang W, Sun H, Liu D, Zhang L, Niu X, Zhang X, Meng M, et al. (2013) Draft genome of the kiwifruit Actinidia chinensis. Nat Commun 4: 2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland HS, Yao JL, Tomes S, Sutherland PW, Nieuwenhuizen N, Gunaseelan K, Winz RA, David KM, Schaffer RJ (2013) Apple SEPALLATA1/2-like genes control fruit flesh development and ripening. Plant J 73: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA (2011) Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23: 923–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Koukounaras A, Sfakiotakis E (2007) Effect of 1-MCP prestorage treatment on ethylene and CO2 production and quality of ‘Hayward’ kiwifruit during shelf-life after short, medium and long term cold storage. Postharvest Biol Technol 46: 174–180 [Google Scholar]
- Kuang JF, Chen JY, Liu XC, Han YC, Xiao YY, Shan W, Tang Y, Wu KQ, He JX, Lu WJ (2017) The transcriptional regulatory network mediated by banana (Musa acuminata) dehydration-responsive element binding (MaDREB) transcription factors in fruit ripening. New Phytol 214: 762–781 [DOI] [PubMed] [Google Scholar]
- Lee JM, Joung JG, McQuinn R, Chung MY, Fei Z, Tieman D, Klee H, Giovannoni J (2012) Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J 70: 191–204 [DOI] [PubMed] [Google Scholar]
- Li T, Jiang Z, Zhang L, Tan D, Wei Y, Yuan H, Li T, Wang A (2016) Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J 88: 735–748 [DOI] [PubMed] [Google Scholar]
- Li T, Xu Y, Zhang L, Ji Y, Tan D, Yuan H, Wang A (2017) The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 29: 1316–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D (2008) A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J 55: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Diretto G, Pirrello J, Roustan JP, Li Z, Giuliano G, Regad F, Bouzayen M (2014) The chimeric repressor version of an Ethylene Response Factor (ERF) family member, Sl-ERF.B3, shows contrasting effects on tomato fruit ripening. New Phytol 203: 206–218 [DOI] [PubMed] [Google Scholar]
- Ma N, Feng H, Meng X, Li D, Yang D, Wu C, Meng Q (2014) Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. BMC Plant Biol 14: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21: 3787–3793 [DOI] [PubMed] [Google Scholar]
- Marzábal P, Gas E, Fontanet P, Vicente-Carbajosa J, Torrent M, Ludevid MD (2008) The maize Dof protein PBF activates transcription of γ-zein during maize seed development. Plant Mol Biol 67: 441–454 [DOI] [PubMed] [Google Scholar]
- McDonald B, Harman JE (1982) Controlled-atmosphere storage of kiwifruit. 1. Effect on fruit firmness and storage life. Sci Hortic (Amsterdam) 17: 113–123 [Google Scholar]
- Meng C, Yang D, Ma X, Zhao W, Liang X, Ma N, Meng Q (2016) Suppression of tomato SlNAC1 transcription factor delays fruit ripening. J Plant Physiol 193: 88–96 [DOI] [PubMed] [Google Scholar]
- Min T, Yin XR, Shi YN, Luo ZR, Yao YC, Grierson D, Ferguson IB, Chen KS (2012) Ethylene-responsive transcription factors interact with promoters of ADH and PDC involved in persimmon (Diospyros kaki) fruit de-astringency. J Exp Bot 63: 6393–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minas IS, Vicente AR, Dhanapal AP, Manganaris GA, Goulas V, Vasilakakis M, Crisosto CH, Molassiotis A (2014) Ozone-induced kiwifruit ripening delay is mediated by ethylene biosynthesis inhibition and cell wall dismantling regulation. Plant Sci 229: 76–85 [DOI] [PubMed] [Google Scholar]
- Mworia EG, Yoshikawa T, Salikon N, Oda C, Asiche WO, Yokotani N, Abe D, Ushijima K, Nakano R, Kubo Y (2012) Low-temperature-modulated fruit ripening is independent of ethylene in ‘Sanuki Gold’ kiwifruit. J Exp Bot 63: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuizen NJ, Chen X, Wang MY, Matich AJ, Perez RL, Allan AC, Green SA, Atkinson RG (2015) Natural variation in monoterpene synthesis in kiwifruit: transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-like transcription factors. Plant Physiol 167: 1243–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, Zhu X, Duan N, Liu JH (2014) PtrBAM1, a β-amylase-coding gene of Poncirus trifoliata, is a CBF regulon member with function in cold tolerance by modulating soluble sugar levels. Plant Cell Environ 37: 2754–2767 [DOI] [PubMed] [Google Scholar]
- Prasanna V, Prabha TN, Tharanathan RN (2007) Fruit ripening phenomena: an overview. Crit Rev Food Sci Nutr 47: 1–19 [DOI] [PubMed] [Google Scholar]
- Qi X, Li S, Zhu Y, Zhao Q, Zhu D, Yu J (2017) ZmDof3, a maize endosperm-specific Dof protein gene, regulates starch accumulation and aleurone development in maize endosperm. Plant Mol Biol 93: 7–20 [DOI] [PubMed] [Google Scholar]
- Seymour GB, Ryder CD, Cevik V, Hammond JP, Popovich A, King GJ, Vrebalov J, Giovannoni JJ, Manning K (2011) A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria × ananassa Duch.) fruit, a non-climacteric tissue. J Exp Bot 62: 1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour GB, Chapman NH, Chew BL, Rose JKC (2013) Regulation of ripening and opportunities for control in tomato and other fruits. Plant Biotechnol J 11: 269–278 [DOI] [PubMed] [Google Scholar]
- Stevenson DG, Johnson SR, Jane J, Inglett GE (2006) Chemical and physical properties of kiwifruit (Actinidia deliciosa) starch. Starch 58: 323–329 [Google Scholar]
- Tacken E, Ireland H, Gunaseelan K, Karunairetnam S, Wang D, Schultz K, Bowen J, Atkinson RG, Johnston JW, Putterill J, et al. (2010) The role of ethylene and cold temperature in the regulation of the apple POLYGALACTURONASE1 gene and fruit softening. Plant Physiol 153: 294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Zheng Y, Dong J, Yu J, Yue J, Liu F, Guo X, Huang S, Wisniewski M, Sun J, et al. (2016) Comprehensive transcriptome profiling reveals long noncoding RNA expression and alternative splicing regulation during fruit development and ripening in kiwifruit (Actinidia chinensis). Front Plant Sci 7: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uluisik S, Chapman NH, Smith R, Poole M, Adams G, Gillis RB, Besong TMD, Sheldon J, Stiegelmeyer S, Perez L, et al. (2016) Genetic improvement of tomato by targeted control of fruit softening. Nat Biotechnol 34: 950–952 [DOI] [PubMed] [Google Scholar]
- Vicente AR, Manganaris GA, Minas IS, Goulas V, Lafuente MT (2013) Cell wall modifications and ethylene-induced tolerance to non-chilling peel pitting in citrus fruit. Plant Sci 210: 46–52 [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wang ZY, MacRae EA, Wright MA, Bolitho KM, Ross GS, Atkinson RG (2000) Polygalacturonase gene expression in kiwifruit: relationship to fruit softening and ethylene production. Plant Mol Biol 42: 317–328 [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ (1995) An ethylene-inducible component of signal transduction encoded by never-ripe. Science 270: 1807–1809 [DOI] [PubMed] [Google Scholar]
- Xiao YY, Kuang JF, Qi XN, Ye YJ, Wu ZX, Chen JY, Lu WJ (2018) A comprehensive investigation of starch degradation process and identification of a transcriptional activator MabHLH6 during banana fruit ripening. Plant Biotechnol J 16: 151–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XL, Yin XR, Chen KS (2016) Roles of APETALA2/Ethylene Responsive Factors in regulation of fruit quality. Crit Rev Plant Sci 35: 120–130 [Google Scholar]
- Xu ZC, Hyodo H, Ikoma Y, Yano M, Ogawa K (1998) Biochemical characterization and expression of recombinant ACC oxidase in Escherichia coli, and endogenous ACC oxidase from kiwifruit. Postharvest Biol Technol 14: 41–50 [Google Scholar]
- Yanagisawa S, Schmidt RJ (1999) Diversity and similarity among recognition sequences of Dof transcription factors. Plant J 17: 209–214 [DOI] [PubMed] [Google Scholar]
- Yang L, Huang W, Xiong F, Xian Z, Su D, Ren M, Li Z (2017) Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol J 15: 1544–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yemm EW, Willis AJ (1954) The estimation of carbohydrates in plant extracts by anthrone. Biochem J 57: 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XR, Chen KS, Allan AC, Wu RM, Zhang B, Lallu N, Ferguson IB (2008) Ethylene-induced modulation of genes associated with the ethylene signalling pathway in ripening kiwifruit. J Exp Bot 59: 2097–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XR, Allan AC, Chen KS, Ferguson IB (2010) Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol 153: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XR, Xie XL, Xia XJ, Yu JQ, Ferguson IB, Giovannoni JJ, Chen KS (2016) Involvement of an ethylene response factor in chlorophyll degradation during citrus fruit degreening. Plant J 86: 403–412 [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene Ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang AD, Hu X, Kuang S, Ge H, Yin XR, Chen KS (2016) Isolation, classification and transcription profiles of the Ethylene Response Factors (ERFs) in ripening kiwifruit. Sci Hortic (Amsterdam) 199: 209–215 [Google Scholar]
- Zhang B, Chen K, Bowen J, Allan A, Espley R, Karunairetnam S, Ferguson I (2006) Differential expression within the LOX gene family in ripening kiwifruit. J Exp Bot 57: 3825–3836 [DOI] [PubMed] [Google Scholar]
- Zhu M, Chen G, Zhou S, Tu Y, Wang Y, Dong T, Hu Z (2014) A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol 55: 119–135 [DOI] [PubMed] [Google Scholar]