Abstract
Background and Purpose:
Candidemia is one of the most important fungal infections caused by Candida species. Infections and mortality caused by Candida species have been on a growing trend during the past two decades. The resistance of yeasts to antifungal drugs and their epidemiological issues have highlighted the importance of accurately distinguishing the yeasts at the species level. The technique applied for yeast identification should be fast enough to facilitate the imminent initiation of the appropriate therapy. Candidemia has not been studied comprehensively in Iran yet. Regarding this, the current study aimed to assess the epidemiology of candidemia at Tehran hospitals and compare the results with the previous findings.
Materials and Methods:
This study was conducted on 204 positive blood cultures obtained from 125 patients hospitalized in several hospitals located in Tehran, Iran, within a period of 13 months. The yeast isolation and species identification were accomplished using several phenotypic methods (i.e., production of germ tube in human serum, culture on CHROMagar Candida, and Corn meal agar containing Tween 80) and molecular methods, such as polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). In addition, unknown cases were subjected to PCR sequencing. These methods were then compared in terms of accuracy, sensitivity, and speed of identification.
Results:
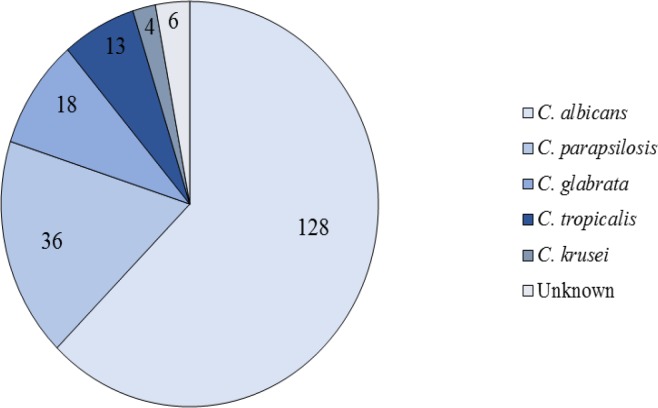
According to the results, C. albicans (62.4%) was the most common isolate, followed by C. parapsilosis (n=36, 17.5%), C. glabrata (n=18, 8.8%), C. tropicalis (n=13, 6.3%), Trichosporon asahii (n=3, 1.5%), C. kefyr (n=2, 1.0%), C. lusitaniae (n=2, 1.0%), C. intermedia (n=1, 0.5%), C. guilliermondii (n=1, 0.5%), and C. krusei (n=1, 0.5%), respectively.
Conclusion:
As the findings indicated, the most common species causing candidemia were C. albicans, C. parapsilosis, and C. glabrata, respectively. Children less than one year old and people with cancer were at higher risk for candidemia, compared to other groups. Moreover, phenotypic and molecular methods resulted in the identification of 65.2% and 96.6% of the isolates, respectively. Consequently, PCR-RFLP could be concluded as a more favorable technique for species identification.
Key Words: Candidemia, Blood culture, Epidemiology, PCR-RFLP
Introduction
Candidemia is one of the most important fungal infections caused by Candida species [1, 2]. Deaths due to Candida infections have been on a growing trend during the past two decades [1]. According to the literature, the patients admitted to the Intensive Care Units have a higher rate of mortality caused by candidemia, compared to others (from 60% to 80%) [1-6]. Given the resistance of the yeast to antifungal drugs and their epidemiological issues, the species-level identify-cation of the yeast should be performed with a high accuracy [1, 2, 7-10]. In addition, the detection methods should be fast enough in order to facilitate the eminent initiation of the appropriate therapy. Therefore, there is an increasing demand to improve the detection ability and speed of the methods employed for the isolation and detection of the yeasts [1, 4, 5, 7, 10, 15].
In our previous study, which was conducted in one of the greatest hospitals of Tehran, Iran, during Aug. 2008 to Nov. 2009, C. parapsilosis was identified as the most common isolate obtained from candidemia patients [7]. Therefore, we decided to expand our study by investigating different large hospitals (i.e., perform a multicenter study) to find more accurate results, especially from the epidemiologic aspect.
With this background in mind, the present study was conducted to determine the common Candida species in Tehran hospitals. To this end, we managed to isolate yeasts from the blood cultures of the patients suffering from candidemia in several hospitals of Tehran. Several traditional techniques and some new molecular methods (e.g., polymerase chain reaction-restriction fragment length polymer-phism [PCR-RFLP]) were performed to identify Candida species. Moreover, as a secondary goal of this research, we compared several Candida identification methods based on different metrics. In particular, we compared the experimental results in terms of the accuracy, sensitivity, and speed of each method, and provided a detailed discussion regarding the more effective identification method by considering all aspects.
Materials and Methods
A total of 40,412 blood samples obtained from patients admitted to nine hospitals of Tehran (i.e., Imam Khomeini, Children's Medical Center, Baqiyatallah, Ali Asghar, Shariati, Tehran Heart Center, Shohadaye Tajrish, Rasoul Akram, and Firoozgar) were cultured within February 2014 to March 2015. The study was approved by the Medical Research Ethics Committee of Yazd University of Medical Sciences, Yazd, Iran (IR.IAU.YAZD.REC.1397,23). In order to isolate yeast colonies from blood cultures, the blood cultures were subcultured on Sabouraud dextrose agar (Merck, Germany) and incubated at 25-30°C for 48 h. Subsequently, the colonies were transferred into tubes containing 35% glycerol in distilled water and kept at 20°C for further study [7]. In this study, six standard strains, including C. albicans (ATCC 10231), C. parapsilosis (ATCC 90018), C. glabrata (ATCC 90030), C. krusei (ATCC 6258), C. tropicalis (ATCC 0750), and C. lusitaniae (JCM 1619), were employed as quality controls.
The yeasts were first identified according to morphological characteristics and physiological properties using several phenotypic techniques, such as the production of germ tube in human serum [1, 7, 11], culture on CHROMagar Candida (BioMerieux, France) [1, 7, 11, 12], and Corn meal agar (Difco, USA), containing Tween 80 [7, 11, 12]. After the initial morphological identification, the extraction of yeast DNA was carried out using the direct PCR and boiling method [1, 16, 17]. We performed PCR-RFLP based on a standard method described by Mirhendi et al. [13, 14, 18]. In the mentioned study, universal primers, namely ITS1 (5-TCCGTAGGTGAACCTGCGG-3) (CinnaGen, Iran) and ITS4 (5-TCCTCCGCTTATTGATATGC-3) (CinnaGen, Iran), were used to facilitate the amplification of the ITS1-5.8S rRNA ITS2 regions.
The PCR amplification was performed in a final volume of 50 µL. Each reaction contained 1 µL template DNA, 0.5 µM of the primers, 1.5 mM MgCl2 (Sinaclon, Iran), 400 µM of deoxynucleoside triphosphate (dNTP) (Sinaclon, Iran), 5 µL of 10x PCR buffer (Sinaclon, Iran), and 1.25 U of Taq DNA polymerase (Fermentas, Lithuania). The PCR process included an initial denaturation step at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 56°C for 45 sec with an extension of 72°C for 1 min, and a final extension step at 72°C for 7 min.
The presence of specific 400-900 base-pair PCR products was examined through staining with ethidium bromide (Sinaclon, Iran) after electro-phoresis on 1.5% agarose gel (Promega, USA). The amplified PCR products were digested with the restriction enzyme MspI (Thermo Fisher Scientific, USA) in order to distinguish the common yeast species, including C. albicans, C. tropicalis, C. glabrata, C. parapsilosis, and C. krusei [1, 18]. The remaining unknown yeasts (n= 6, 3.4%) were sent for DNA sequencing.
Results
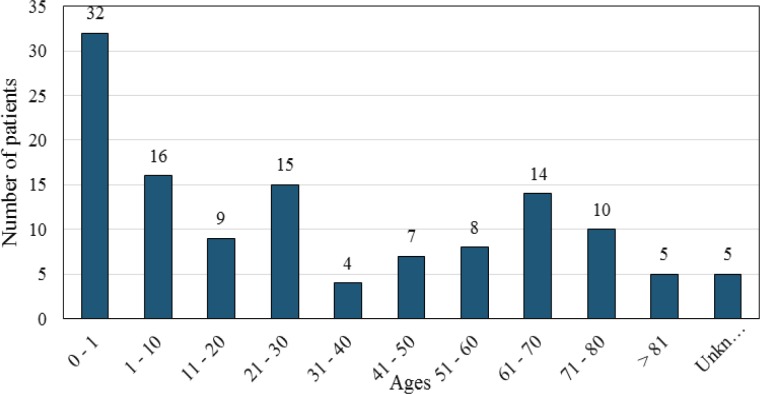
Out of the 40,412 tested samples, 204 cases were positive for yeasts. The positive blood samples belonged to 125 patients, including 72 females (57.6%) and 53 males (42.4%) with the mean ages of 36.1 and 20.7 years, respectively. The age range of the patients was 0-90 years. Figure 1 depicts the effect of patients' age on the frequency of Candida species occurrence in blood cultures. Our comprehensive study of the medical records of patients showed that all patients with candidemia had an underlying disease (see Table 1).
Figure 1.
Number of patients with candidemia based on different age groups
Table 1.
Frequency of different underlying diseases in patients with candidemia
| Underlying Diseases | Frequency (%) |
|---|---|
| Infections caused by iatrogenic factors | 1.8 |
| Impaired immune system (e.g., neutropenia, lupus, and Parkinson) | 1.8 |
| Liver diseases | 2.7 |
| Cardiac diseases | 3.5 |
| Bacterial and viral infections | 4.4 |
| Digestive disorders | 5.3 |
| Diabetes | 5.8 |
| Renal failure | 7.1 |
| Pulmonary disorders | 8.0 |
| Metabolic and genetic disorders | 9.7 |
| Brain and neurologic | 11.1 |
| Sepsis | 11.9 |
| Cancer: Hematologic malignancy | 2.7 |
| Cancer: Other tumors | 9.7 |
| Other | 14.6 |
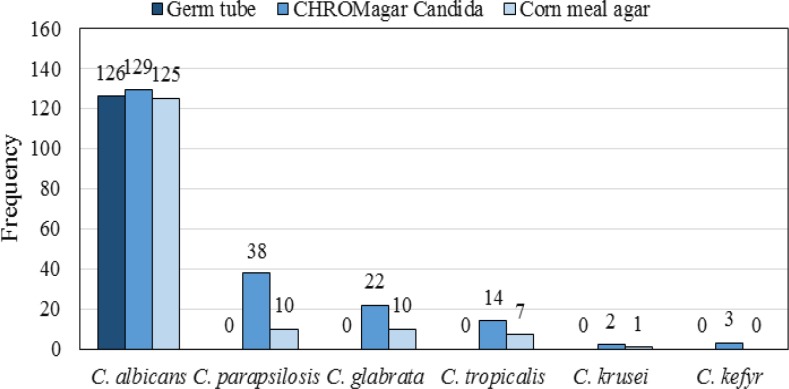
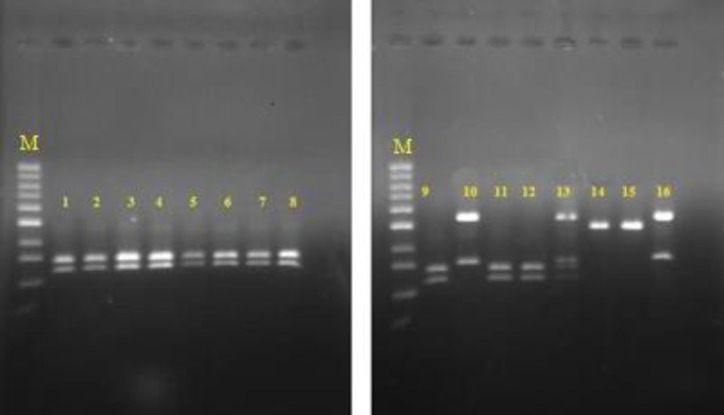
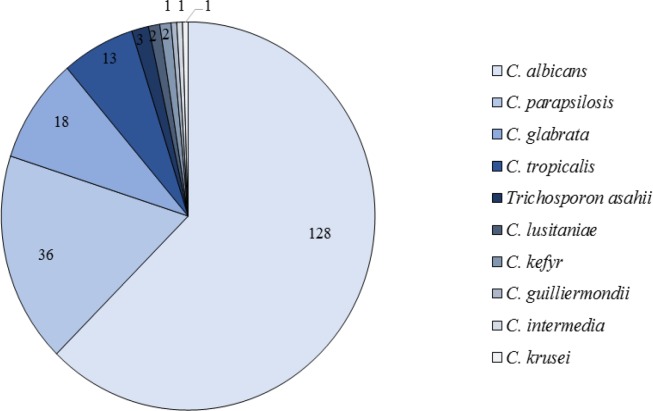
Other predisposing factors for candidemia in our experiments were the use of antibiotics, antifungal drugs, and corticosteroid, as well as surgery. Table 2 presents the different predisposing factors and their frequency. According to the available information, 45 patients (36%) passed away in these hospitals. Figure 2 illustrates the results of different phenotypic methods used for the identification of Candida species in positive blood cultures. The electrophoresis of PCR products is displayed in Figure 3. Figure 4 demonstrates the RLFP-PCR profiles of the samples using MspI restriction enzyme. Figure 5 reports the frequency of the species obtained by using PCR-RFLP. As can be seen in Figure 5, six cases remained still unknown after the implementation of PCR-RFLP. As a result, these unknown cases, along with three uncertain cases as control specimens, were subjected to DNA sequencing. Figure 6 illustrates the contribution of each Candida species in the positive blood cultures.
Table 2.
Predisposing factors for candidemia
| Candidemia Predisposing Factors | Frequency (%) |
|---|---|
| Prematurity | 0.9 |
| Surgery | 16.0 |
| Organ transplants | 2.1 |
| Antibiotics | 33.1 |
| Antifungal drugs | 18.7 |
| Corticosteroid | 12.7 |
| Receiving immunosuppressive therapy | 1.5 |
| Chemotherapy | 3.9 |
| Iatrogenic illness | 11.1 |
Figure 2.
Comparison of phenotypic tests with respect to the number of isolated species
Figure 3.
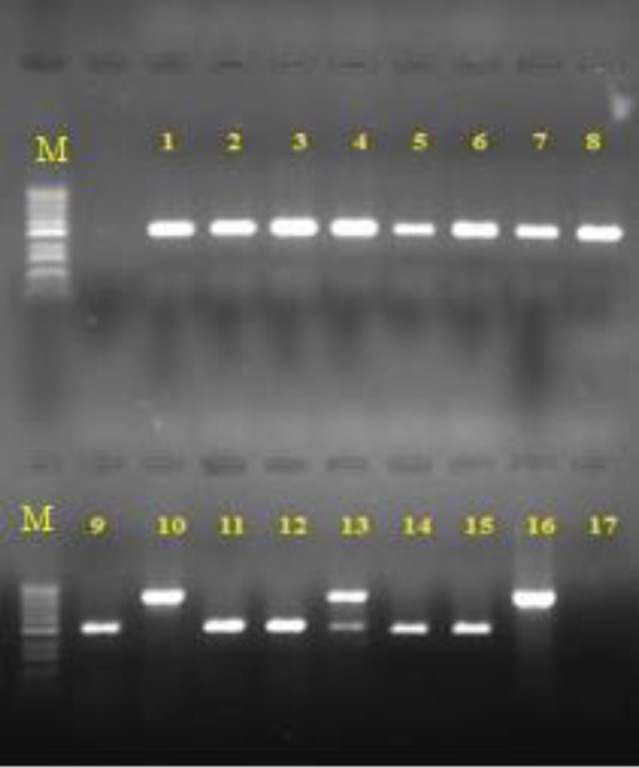
Electrophoresis of polymerase chain reaction products; lanes 1-9, 11, 12) C. albicans (535 bp), lanes 14 and 15) C. parapsilpsis (520 bp), lanes 10 and 16) C. glabrata (871 bp), lane 13) a combination of C. albicans and C. glabrata, lane 17) negative control, and lane M) 100 bp DNA size marker
Figure 4.
Polymerase chain reaction-restriction fragment length polymorphism profile of samples; lanes 1-9, 11, 12) C. albicans (297, 238 bp), lanes 14, 15) C. parapsilpsis (520 bp), lanes 10, 16) C. glabrata (557, 314 bp), lane 13) a combination of C. albicans and C. glabrata, and lane M) 100 bp DNA size marker
Figure 5.
Frequency of the species isolated from the blood cultures of patients with candidemia using polymerase chain reaction-restriction fragment length polymorphism
Figure 6.
Overall frequency of the species isolated from the blood cultures of patients with candidemia
Discussion
Several methods have been proposed to detect and distinguish Candida species in a positive blood culture. It is important to employ an accurate and fast technique for the identification of Candida species in order to select a proper therapy imminently [1, 2, 10]. Therefore, in this research, we used three well-known phenotypic methods and two genotypic methods, namely PCR-RFLP and DNA sequencing, to detect Candida species in 204 positive blood cultures. This section includes some discussions about our experiments and comparisons of the applied techniques. In the current study, C. albicans was recognized as the dominant causative agent of candidemia; in this regard, this species was observed in 62.4% of the tested positive blood cultures.
Moreover, C. parapsilosis (17.5%) and C. glabrata (8.8%) were the second and third common species detected in our evaluated positive blood cultures, respectively. In a study performed by Ghahri et al. (2012) [7], C. parapsilosis was reported as the most common species identified in candidemia patients admitted to Baqiyatallah Hospital, Tehran, Iran. Therefore, the current research was initiated to holistically study the different etiologic agents of candidemia in patients hospitalized in several important hospitals of Tehran.
This is in line with the results reported from different regions of the world, such as those obtained by Nieto-Rodriguez et al. [19] in the USA, Yildiz et al. [6] in Turkey, Wille et al. [2] in Brazil, and Razzaghi et al. [20] in Kashan, Iran. In the present study, C. parapsilosis and C. glabrata had the second and third ranks of occurrence, respectively. On the other hand, in the study of Razzaghi et al. [20], C. glabrata and C. parapsilosis had the second and third ranks, respectively. This difference is mainly due to the investigation of different hospitals and the number of positive blood cultures in these two studies. Therefore, the results achieved in the previous studies conducted in Tehran are limited to that period and specific location.
In the current study, out of the 40,412 blood cultures sent to the laboratory, 204 cases (0.51%) were positive. This ratio was also obtained in our previous study performed on 5,141 blood cultures. Regarding this, it could be concluded that the rate of candidemia occurrence in patients admitted to Tehran hospitals has remained almost unchanged from 2011 to 2017. In the current study, PCR-RFLP method failed to identify six isolates (3.4%); therefore, they were subjected to DNA sequencing.
As indicated in Figure 2, CHROMagar Candida showed a better function in species identification than other phenotypic methods used in this study. This method facilitated the detection of Candida species in 84.8% of the positive blood cultures. Accordingly, it resulted in the detection of C. albicans (63.2%), C. parapsilosis (18.6%), C. glabrata (10.8%), C. tropicalis (6.9%), and C. krusei (0.9%) in the positive blood cultures. Moreover, in the current study, there were some positive blood cultures having more than one Candida species. CHROMagar Candida is the only phenotypic method that can detect the mixed cultures of Candida species in the positive blood cultures.
As an example, we found combinations of C. albicans and C. glabrata, as well as C. glabrata and C. parapsilosis three and four times, respectively. The aforementioned discussion showed that CHROMagar Candida outperformed other phenotypic methods in terms of the ability of Candida species detection. In the present study, phenotypic methods resulted in the detection of Candida species in 65.2% of the isolates. However, only 3.4% of the isolates remained unknown when using the PCR-RFLP. The results of PCR-RFLP were consistent with those of the phenotypic methods in 97.5% of the cases. This clearly confirmed the efficiency of the phenotypic methods. In terms of the speed of detection, PCR-RFLP outperformed other phenotypic methods as it could be performed in about 5 h, while other phenotypic methods needed about 48-72 h.
Considering the epidemiologic aspects, most of the patients with candidemia were female (57.6%) with a mean age of 36.1 years. The analysis of the age of the patients with candidemia revealed that children had the highest risk factor for this disease in comparison with other age groups. The Candida species detected in children were mostly C. albicans (n=49) and C. parapsilosis (n=19), respectively. Regarding the adults (i.e., less than 60 years old), C. albicans, C. parapsilosis, and C. tropicalis were identified in 47, 9, and 9 cases, respectively. Furthermore, considering the elderly group (i.e., more than 60 years old), composing 23.2% of the patients with candidemia, similar to other age categories, C. albicans was more common than other Candida species. The second common Candida species in the elderly group was C. glabrata, which rarely happened in other age groups.
In the current study, the most common predisposing factors for candidemia were antibiotic therapy, antifungal drugs, and surgery (Table 1). Moreover, cancer and sepsis were among the high-risk factors for candidemia. This study was performed for the partial fulfillment of the requirements for the degree of Master of Science; consequently, the study had limited time period. Moreover, the current study received no funding support to cover the expenses of performing phenotypic and genotypic methods.
Conclusion
As the findings of the present study indicated, Candida albicans was the most common Candida species of candidemia in patients admitted to the hospitals of Tehran, followed by C. parapsilosis and C. glabrata, respectively. In addition, PCR-RFLP outperformed other phenotypic methods in terms of accuracy and detection speed.
Acknowledgments
Authors would like to appreciate Dr. Seyed Ahmad Hosseini and Mr. Reza Davarzani in Resalat Laboratory for providing the facilities of this research. We had no funding support for this study.
Author’s contribution
M. Gh. and S. Z. S. managed the project, performed the isolation and identification of the blood cultures, analyzed the data, and wrote the first draft of the manuscript. S. B. contributed in performing the molecular methods (e.g., PCR-RFLP). A. A. I. F., S. S., and Z. B. contributed in providing the blood cultures and data from the hospitals of Tehran.
Conflicts of interest
The authors declare no conflicts of interest regarding the publication of this study.
Financial disclosure
The authors declare no financial interests related to the materials of the study.
References
- 1.Ghahri M, Mirhendi H, Zomorodian K, Kondori N. Identification and antifungal susceptibility patterns of Candida strains isolated from blood specimens in Iran. Arch Clin Infect Dis. 2013;8(3):e14529. [Google Scholar]
- 2.Wille MP, Guimaraes T, Furtado GH, Colombo AL. Historical trends in the epidemiology of candidaemia: analysis of an 11-year period in a tertiary care hospital in Brazil. Mem Inst Oswaldo Cruz. 2013;108(3):288–92. doi: 10.1590/S0074-02762013000300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blot S, Dimopoulos G, Rello J, Vogelaers D. Is Candida really a threat in the ICU? Curr Opin Crit Care. 2008;14(5):600–4. doi: 10.1097/MCC.0b013e32830f1dff. [DOI] [PubMed] [Google Scholar]
- 4.Gudlaugsson O, Gillespie S, Lee K, Berg JV, Hu J, Messer S, et al. Attributable mortality of nosocomial candidemia. Clin Infect Dis. 2003;37(9):1172–7. doi: 10.1086/378745. [DOI] [PubMed] [Google Scholar]
- 5.Quiles-Melero I, Garcia-Rodriguez J, Romero-Gomez M, Gomez-Sanchez P, Mingorance J. Rapid identification of yeasts from positive blood culture bottles by pyrosequencing. Eur J Clin Microbiol Infect Dis. 2011;30(1):21–4. doi: 10.1007/s10096-010-1045-5. [DOI] [PubMed] [Google Scholar]
- 6.Yıldız Hİ, Berktas M, Yaman G, Guducuoglu H, Cıkman A. Yogun bakım unitesinden gelen hasta orneklerinden izole edilen kandida turleri ve antifungal duyarlılıkları. VAN Med J. 2016;23(2):143–7. [Google Scholar]
- 7.Ghahri M, Mirhendi H, Fooladi AI, Beyraghi S. Species identification of Candida strains isolated from patients with candidemia, hospitalized in Tehran, by enzymatic digestion of ITS–rDNA. J Isfahan Med Sch. 2012;29:167. [Google Scholar]
- 8.Zirkel J, Klinker H, Kuhn A, Abele-Horn M, Tappe D, Turnwald D, et al. Epidemiology of Candida blood stream infections in patients with hematological malignancies or solid tumors. Med Mycol. 2012;50(1):50–5. doi: 10.3109/13693786.2011.587211. [DOI] [PubMed] [Google Scholar]
- 9.Chen LY, Liao SY, Kuo SC, Chen SJ, Chen YY, Wang FD, et al. Changes in the incidence of candidaemia during 2000–2008 in a tertiary medical Centre in northern Taiwan. J Hosp Infect. 2011;78(1):50–3. doi: 10.1016/j.jhin.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Shrivastava G, Bajpai T, Bhatambare GS, Chitnis V, Deshmukh AB. Neonatal candidemia: clinical importance of species identification. Sifa Med J. 2015;2(2):37. [Google Scholar]
- 11.Farasat A, Ghahri M, Mirhendi H, Beiraghi S. Identification of Candida species screened from catheter using patients with PCR-RFLP method. Eur J Exp Biol. 2012;2(3):651–6. [Google Scholar]
- 12.Atalay MA, Koc AN, Demir G, Sav H. Investigation of possible virulence factors in Candida strains isolated from blood cultures. Niger J Clin Prac. 2015;18(1):52–5. doi: 10.4103/1119-3077.146979. [DOI] [PubMed] [Google Scholar]
- 13.Mirhendi H, Makimura K, Khoramizadeh M, Yamaguchi H. A one-enzyme PCR-RFLP assay for identification of six medically important Candida species. Nihon Ishinkin Gakkai Zasshi. 2006;47(3):225–9. doi: 10.3314/jjmm.47.225. [DOI] [PubMed] [Google Scholar]
- 14.Mirhendi S, Makimura K. PCR-detection of CandidaAlbicans in blood using a new primer pair to diagnosis of systemic candidiasis. Iran J Public Health. 2003;32(1):1–5. [Google Scholar]
- 15.Makimura K, Murayama SY, Yamaguchi H. Detection of a wide range of medically important fungi by the polymerase chain reaction. J Med Microbiol. 1994;40(5):358–64. doi: 10.1099/00222615-40-5-358. [DOI] [PubMed] [Google Scholar]
- 16.Williams DW, Wilson MJ, Lewis M, Potts A. Identification of Candida species by PCR and restriction fragment length polymorphism analysis of intergenic spacer regions of ribosomal DNA. J Clin Microbial. 1995;33(9):2476–9. doi: 10.1128/jcm.33.9.2476-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghahri M. Genotyping of CandidaAlbicans strains isolated from different clinical candidiasis using rapid-PCR in Tehran. [PhD Thesis] Tehran: Tarbiat Modares University; 2010. (Persian) [Google Scholar]
- 18.Mirhendi H, Bruun B, Schonheyder HC, Christensen J, Fuursted K, Gahrn-Hansen B, et al. Differentiation of CandidaGlabrata C Nivariensis and C Bracarensis based on fragment length polymorphism of ITS1 and ITS2 and restriction fragment length polymorphism of its and d1/d2 regions in rDNA. Eur J Clin Microbiol Infect Dis. 2011;30(11):1409–16. doi: 10.1007/s10096-011-1235-9. [DOI] [PubMed] [Google Scholar]
- 19.Nieto-Rodriguez JA, Kusne S, Manez R, Irish W, Linden P, Magnone M, et al. Factors associated with the development of candidemia and candidemia-related death among liver transplant recipients. Ann Surg. 1996;223(1):70–6. doi: 10.1097/00000658-199601000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Razzaghi R, Momen-Heravi M, Erami M, Nazeri M. Candidemia in patients with prolonged fever in Kashan, Iran. Curr Med Mycol. 2016;2(3):20–6. doi: 10.18869/acadpub.cmm.2.3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]