Abstract
Background:
Cisplatin-based chemotherapy is currently part of the standard of care for bladder cancer (BC). Unfortunately, some patients respond poorly to chemotherapy and have acquired or developed resistance. The molecular mechanisms underlying this resistance remain unclear. Here, we introduce a multidimensional proteomic analysis of a cisplatin-resistant BC model that provides different levels of protein information, including that of the global proteome and phosphoproteome.
Methods:
To characterize the global proteome and phosphoproteome in cisplatin-resistant BC cells, liquid chromatography-mass spectrometry/mass spectrometry experiments combined with comprehensive bioinformatics analysis were performed. Perturbed expression and phosphorylation levels of key kinases associated with cisplatin resistance were further studied using various cell biology assays, including western blot analysis.
Results:
Analyses of protein expression and phosphorylation identified significantly altered proteins, which were also EGF-dependent and independent. This suggests that protein phosphorylation plays a significant role in cisplatin-resistant BC. Additional network analysis of significantly altered proteins revealed CDK2, CHEK1, and ERBB2 as central regulators mediating cisplatin resistance. In addition to this, we identified the CDK2 network, which consists of CDK2 and its 5 substrates, as being significantly associated with poor survival after cisplatin chemotherapy.
Conclusions:
Collectively, these findings potentially provide a novel way of classifying higher-risk patients and may guide future research in developing therapeutic targets.
Keywords: global proteome, phosphoproteomics, cisplatin resistance, bladder cancer, biological network
Introduction
Bladder cancer (BC) is a common malignancy of the urinary tract. Resection of the tumor (when possible) and cisplatin-based chemotherapy is the present standard of care. However, resistance to chemotherapy has been a clinical challenge for long-term remission [11]. Chemo-resistance can rapidly develop and a majority of patients ultimately experience disease progression and a poor prognosis [4]. The failure of conventional treatment following remission typically results in a < 15% chance of 5-year survival [10, 18, 39]. Therefore, therapeutic strategies that can re-sensitize tumors to chemotherapy would significantly benefit BC patient care. Additionally, a molecular signature panel capable of predicting cisplatin resistance in BC would greatly improve prognostic and clinical outcomes for high-risk patients. Unfortunately, despite multiple independent studies aimed at identifying the subset of patients who will develop chemo-resistance, research on predictive biomarker(s) is still in its infancy [3, 36, 38].
Overexpression, amplification, or mutations of EGFR family members are often observed in BC. This suggests that the proteins involved in the EGFR signaling pathways may play important roles in the carcinogenesis and maintenance of BC, making them relevant therapeutic targets. Our group had previously established cisplatin-sensitive (T24S) and resistant (T24R) T24 human BC cell lines [21]. The viability of the resistant cells was reduced by inhibition of EGFR kinase activity [21]. Either expression or phosphorylation of the proteins involved in signaling pathways can contribute to alterations of pathway activations. Thus, analyses of the global proteome and phosphoproteome would allow for direct interrogation of the altered activation of signaling pathways and provide comprehensive profiles regarding protein quantity and phosphorylation [6, 8, 41]. A network model describing such protein alterations in cancer cells could provide novel insights into the molecular mechanisms of resistance and contribute to identifying new biomarkers or therapeutic targets.
To obtain such a systems-level insight into cisplatin-resistance, we used high-resolution mass spectrometry and investigated the temporal changes in protein abundance and phosphorylation in T24S and T24R cells after EGF stimulation. Our comprehensive proteomic analysis identified proteins involved in EGFR signaling that were perturbed in cisplatin-resistant cells. This included substrates of EGF receptors and downstream kinases. Further network analysis of our proteomic and phosphoproteomic data identified downstream kinases of EGFR signaling pathways.
MATERIALS AND METHODS
Reagents.
Antibodies against CDK2 (1:1000, Novus Biologicals; Littleton, CO), and β-actin (1:5000, Sigma) were used for this study. The following antibodies: P-CDK2 (Thr160) (2561, 1:1000), P-ERBB2 (Tyr1196) (6942, 1:1500), P-CHEK1 (Ser345) (2348, 1:750), phosphom-TOR (Ser2448) (5536; 1:1000), p-Rb (Ser807/811) (8516, 1:1000), p-tyrosine (P-Tyr100) (9411, 1:2000), and HRP-conjugated secondary antibodies (7074, 1:1000; 7076, 1:1000) were obtained from Cell Signaling Technologies.
Cell culture.
T24, J82, and RT4 BC cells were obtained and cultured, according to instructions provided by the American Type Culture Collection (ATCC) (Manassas, VA). Media was supplemented with 10% fetal bovine serum, 2% glutamine, and 1% antibiotics (Invitrogen, Carlsbad, CA). Cells were maintained in a humidified incubator with 5% CO2 at 37 °C [21].
Protein extraction and quantification.
The cell pellets were kept at −80 °C for 30 mins. Afterwards, the cells were lysed with RIPA buffer [50 mM Tris-HCl, 150 mM NaCl, 0.1% (w/v) sodium dodecyl sulfate (SDS), 1% (v/v) NP-40, 1 mM PMSF, 1X protease inhibitor cocktail, 1X phosphatase inhibitor cocktail, PhoSTOP] and left on ice for 15 mins while undergoing sonication. The cellular lysates were centrifuged at 14,000 g for 15 mins at 4 °C. The supernatant was collected and 500 μg of protein for each cell line was digested using the filter aided sample preparation (FASP) method [43].
Filter aided sample preparation (FASP).
Proteins were reduced in a sodium dodecyl sulfate (SDS)-lysis buffer (4% (w/v) SDS; 0.1 M Tris/HCl pH 7.6; 0.1 M DTT) at 37 °C for 45 mins. The proteins were then boiled at 95 °C for 10 mins and then sonicated for 10 mins. The solution was transferred to a 30 k membrane filter (Microcon devices, YM-30, Millipore, MA) and centrifuged at 14,000 g and 20 °C for 60 mins. The concentrates were diluted in their filters with 0.2 ml of UA solution (8 M urea in 0.1 M Tris/HCl, pH 8.5) and centrifuged at 14,000 g for 30 mins to remove the remaining SDS (x3). After that, the concentrates were mixed with 0.1 ml of 50 mM indole-3-acetic acid (IAA) in UA solution and incubated in darkness at room temperature for 25 mins. Following centrifugation, the resulting product was diluted with 0.2 ml of UA solution and concentrated again. This step was repeated 3 times. Next, the concentrates were washed with 0.2 ml of 100 mM triethyl ammonium bicarbonate (TEAB) and centrifuged at 14,000 g for 30 mins (x3). Subsequently, 10 μg of trypsin prepared in 0.1 ml of 100 mM TEAB (with the enzyme to protein ratio of 1:50) was added to the filter and the samples were incubated at 37 °C overnight. The second digestion was done using trypsin with an enzyme to protein ratio of 1:100 for 6 hrs. Peptides were collected by centrifuging the filter units at 14,000 g for 30 mins. TEAB (75 μL, 100 mM) was added to the filter and centrifuged at 14,000 g for 20 mins (x3). Finally, the peptide solutions were dried in a speed vacuum concentrator and stored at −80 °C.
TMT labeling of peptide.
Peptides were labeled with TMT™ Isobaric Mass Tagging Reagent, according to the manufacturer’s instructions (ThermoScientific, Foster City, CA). For each sample of 500 μg peptides, 250 μl of 200 mM TEAB were added. TMT reagents were resuspended in anhydrous acetonitrile and then added to each sample (126: Cisplatin sensitive/EGF 0 min, 127: Cisplatin sensitive/EGF 10 mins, 128: Cisplatin sensitive/EGF 30 mins, 129: Cisplatin resistant/EGF 0 min, 130: Cisplatin resistant/EGF 10 mins, 131: Cisplatin resistant/EGF 30 mins). After one hour, the reaction was quenched with 5% hydroxylamine. The chemically tagged samples were pooled into one tube and concentrated via speed vacuum centrifugation. The labeled samples were then mid-pH fractionated.
Mid-pH reversed phase liquid chromatography (RPLC) fractionation.
Mid-pH RPLC fractionation was performed to separate peptides based on their hydrophobicity. An Accucore™ 150 C18 LC column (150 × 2.1 mm, 4 μm) was used for fractionation with mid-pH buffers A and B; (A) the mobile phase was 10 mM TEAB in water (pH 7.5) and (B) the mobile phase was 10 mM TEAB in 90% CAN (pH 7.5). The samples were then divided into 15 smaller samples using the Agilent 1260 Series HPLC System (Agilent Technologies, Santa Clara, CA). The resultant gradient was 0–10 mins, 5% B; 10–70 mins, 5–35% B; 70–80 mins, 70% B; 80–105 mins, 5% B. The separated peptides were collected and dried in a speed vacuum. Each fraction was vacuum dried and stored at −80 °C until liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) experiments.
Phosphopeptide enrichment using immobilized metal affinity chromatography (IMAC).
IMAC beads were prepared from Ni-NTA magnetic agarose beads. Ni-NTA beads (500 ml) were washed (3×) with deionized water (DIW). The beads were then reacted with 100 mM of EDTA (pH 8.0) for 30 mins with end-over-end rotation to remove nickel ions. The reacted EDTA solution was eliminated and the beads were washed 3× with DIW. The NTA beads were treated with 10 mM of aqueous FeCl3 solution for 30 mins with end-over-end rotation. Iron-chelated IMAC beads were washed (x3) with DIW. IMAC beads were aliquoted into 10 microcentrifuge tubes and each bead was washed with 400 μl of 80% acetonitrile (CAN)/0.1% trifluoracetic acid (TFA). In total, 10 fractions, which were obtained by combining some fractions from 15 (#1 and #11, #2 and #12, #3 and #13, #4 and #14, #5 and #15, #6, #7, #8, #9, #10), were used to enrich for phosphopeptides. The 10 fractionated peptide samples were suspended in 500 μl of 80% ACN/0.1% TFA and were reacted with IMAC beads again for 30 mins with end-over-end rotation. Phosphopeptides were eluted using 125 μl of 1:1 ACN/2.5% ammonia in 2 mM phosphate buffer (pH 10) after incubating for 1.5 mins. All phosphopeptides were acidified immediately with 10% TFA. The collected phosphopeptides were dried in a speed vacuum and purified in C18 spin columns for LC-MS/MS analysis.
LC-MS/MS analysis.
All peptide samples were separated on an ultra performance liquid chromatography (UPLC) system that was equipped with analytical columns (75 μm x 50 cm, C18, 3 μm, 100 Å) and trap columns (75 μm x 2 cm, C18, 3 μm, 100 Å). The temperature of the analytical columns was set to 60 °C. The solvents A and B were 0.1% formic acid in water and 0.1% formic acid in acetonitrile, respectively. A gradient made after 180 mins (1–40% solvent B for 160 mins, 40–80% solvent B for 5 mins, 80% solvent B for 10 mins, and 1% solvent B for 5 mins) was used for global proteome profiling analysis. A gradient made after 134 mins (5–40% solvent B for 120 mins, 40–80% solvent B for 2 mins, 80% solvent B for 10mins, and 1% solvent B for 2 mins) was used for phosphoproteome analysis. The flow rate of all experiments was set to 250 nl/min. The eluted peptides from LC were mass-analyzed on a Q Exactive Orbitrap Mass Spectrometer (ThermoScientific, Bremen, Germany). The electric potential of electrospray ionization was kept at 2.0 kV and the temperature of desolvation capillary was set to 250 °C. Full MS scans were acquired for a mass range of 400–2000 Th at a resolution of 70,000. The 10 most abundant ions, with charges of 2, 3, 4, and 5, were dynamically selected with an isolation width of 2.0 Th and fragmented with an exclusion duration of 30 secs with a normalized collision energy (NCE) of 30 for higher energy collisional dissociation (HCD). The LC-MS/MS scans were acquired at a resolution of 17,500 with a fixed initial m/z of 100 Th. Maximum ion injection times were 120 ms for both the full MS and LC-MS/MS scans. The automated gain control (AGC) target value was set to 1.0 × 106 for both the MS and LC-MS/MS scans.
Identification of peptides and proteins.
MS and MS/MS spectra were converted from Thermo. RAW files into the mzML format using Msconvert (ProteoWizard) and searched with MSGF+ against the Universal Protein Resource Human Database (Uniprot released 2016_02, 20,198 entries, http://www.uniprot.org/). Search parameters were as follows: 20 ppm tolerance for precursor ion mass error and fixed modification for carbamidomethylation (+57.0214 Da) to cysteine, variable modification for methionine oxidation (+15.994915 Da), phosphorylation (+79.966330 Da) to serine, threonine, tyrosine, TMT (+229.162932) to N-termini and lysine and acetylation (+42.010564) to N-terminal of protein. The peptide identification stringency was set to a maximum of 1% peptide-to-spectrum matches (PSMs) FDR in the target-decoy way. The intensities of all TMT-6 plex reporter ions were extracted using the MAGIC software [27]. When multiple phosphopeptides were assigned to a phosphoprotein, the phosphopeptide that had the largest comparative fold change [44] was used as the representative fold change of the phosphoprotein. The mass spectrometry proteomics data has been deposited to the PRIDE [17] repository with the dataset identifier, PXD005308.
Identification of proteins with altered expression or phosphorylation.
TMT intensities of the peptides were normalized using the quantile normalization method [5]. Protein abundance was estimated by summing all TMT reporter ion intensities of different fully tryptic peptides belonging to the same protein groups. Of these qualified proteins, those with more than 2 non-redundant peptides were chosen for the next analyses. We then identified differentially expressed proteins (DEPs) using an integrative statistical method, as reported previously [14]. Briefly, for each protein, the t-test and log2-median-ratio test were applied to its abundances for all 3 replicates. An empirical null distribution was estimated by applying the Gaussian Kernel Density Estimation method to log2 fold-changes that were obtained after performing all possible permutations on all the TMT labels [1]. Using the distributions for each protein, we computed the adjusted p-values for the two tests. We then combined the p-values to compute the overall p-value using Stouffer’s method. Finally, we computed the false discovery rate (FDR) for the overall p-values using Storey’s method. We selected DEPs that had a combined FDR < 0.05, fold change ≥ 1.5, and were detected as proteins in at least 2 of the 3 replicates.
For the phosphoproteome, we applied differential phosphorylation analysis at the peptide level to identify significantly altered phosphopeptides between cell types or time points. For this, the same normalization and statistical testing methods described above were used. Phosphopeptides were selected and considered as significantly altered if they had a FDR < 0.05 and fold-change ≥ 1.5. Finally, differentially phosphorylated proteins (DPPs) were identified as proteins that contained significantly altered phosphopeptides that were uniquely assigned to the protein. The Phospho-UMC filter was used to localize the site of phosphorylation. This simple method uses unique mass class (UMC) information to differentiate phosphopeptides with different phosphorylation sites [28]. Briefly, MS features within 10 ppm, but emerging over a period of time during their LC elution, were identified and grouped into an UMC. Then, the links between the LC-MS/MS data and the UMCs were made. The resultant LC-MS/MS data were subjected to a database search using the MSGF+ engine and peptide identifications within 1% FDR were used.
To explore the cellular processes and subcellular localizations represented by the DEPs and DPPs, functional enrichment analyses of gene ontology biological processes (GOBPs), gene ontology cellular components (GOCCs), and Kyoto encyclopedia of genes and genomes (KEGG) pathways [19] were performed using the DAVID software [13]. The GOBPs, GOCCs, and KEGG pathways represented by DEPs and DPPs were identified as those with p < 0.05.
Reconstruction of cellular network models and identification of key kinases.
To identify key kinases regulating DPPs, data for more than 12,000 kinase-substrate interactions (KSI) was collected from the SIGNOR database [34]. For each kinase, the number of targets in the DEPs was calculated using the KSI data. The p-value for the number of substrates was then computed based on a hypergeometric distribution from Fisher’s exact test. Of the kinases with p < 0.05, those belonging to DEPs or DPPs were selected as key kinases. We estimated an empirical distribution of network density by randomly sampling the same number of proteins in each kinase group with KSIs 100,000 times. Significance levels of the kinases were computed using the right-sided test.
Western blot analysis.
After EGF treatment (10 ng/ml), cells were quickly harvested, flash frozen in liquid nitrogen, and stored at −80 °C. Total protein was extracted using a lysis buffer [1% Nonidet P-40, 50 mM Tris (pH 7.4), 10 mM NaCl, 1 mM NaF, 5 mM MgCl2, 0.1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride] with a complete protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany). The solution was centrifuged at 12,500 ×g for 15 mins. After measuring for protein concentration, 25 μg of protein per line were subjected to SDS-PAGE gel running and then transferred onto nitrocellulose membranes for western blot analysis. After blocking for 1 hr with 10% bovine serum albumin (BSA) in phosphate buffered saline with Tween 20 (PBST), the membranes were incubated with specific antibodies in PBST, overnight. β-actin was used as the loading control. The blots were visualized using enhanced chemiluminescence.
Gene silencing and cell viability assay.
For transient cell transfections of siRNA oligonucleotides, T24R cells were grown to 80% confluence in 6-well culture plates and transfected with siRNAs targeting CDK2, CHEK1, ERBB2, and control (ThermoScientific) using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. Cisplatin-resistant BC cells (T24R, J82R, and RT4R) were incubated with cisplatin and/or CDK2 inhibitor for 72 hr. Post-treatment cell survival was determined by measuring cell viability using MTS reagents (Promega Corporation, Madison, WI), according to the provided protocols.
Tissue micro-array (TMA) construction.
Patient consent and IRB approval was given prior to tissue collection, and TMA slides were constructed from a cohort of 42 BC patients at CedarsSinai Medical Center (IRB #: Pro00044997). Detailed information was not available except that a majority of the patients were male and over 60 years old. Although tumors were pathologically graded prior to tissue collection, the annotations and image analysis were blindly conducted by two trained scientists, including one certified pathologist. Tumor and normal bladder tissue samples were received from all patients. Circular TMA cores 1 mm in diameter were constructed for all tissue samples onto glass slides. In total, there were 82 TMA cores per slide.
Immunohistochemistry (IHC) analysis.
Antibodies against CDK2 (Novus Biologicals; Littleton, CO) were incubated on their respective slides for 32 mins. Following antibody incubation, antigen retrieval was done at a high pH for 64 mins. Ultraview DAB Detection (Ventana Medical Systems; Tucson, AZ) was used for nuclear and cytoplasmic counterstaining. These slides were scanned and uploaded onto the Leica Biosystems cloud drive.
Digitalized image analysis.
Following staining with both the antibodies of interest and pan-CK, the slides were scanned using the Aperio Turbo Scanscope AT (Leica Biosystems; Buffalo Grove, IL). High resolution images of each slide were uploaded onto the Leica Biosystems cloud drive for annotations and analysis.
Annotation and analysis of slides were done using the Leica Tissue Image Analysis (IA) 2.0 software program (Copyright 2012 by SlidePath Ltd.). Stromal and structural tissues were not included in the annotations. Although these structures may provide additional insight, the focus of the experiment was based on tumor epithelium only. Each annotated core had a minimum threshold of 100,000 cells to be analyzed. Following annotations, the “Measure Stained Cells Algorithm” option in the Leica Tissue IA software was used. For each antibody slide, color definition preferences were defined and algorithm input parameters were optimized based on several cores. These individualized parameters were used to analyze the annotated regions on each slide.
For the color definition, haematoxylin was set as the nuclear counter stain, and DAB was set as the nuclear, cytoplasmic, and membrane marker. Parameters in the program were based on a numbered greyscale between 0–255, with 0 being the minimum intensity (black) and 255 being the maximum (white). The max threshold in our analysis was set to 220, with 180 being the threshold for positive or negative staining. The max nuclear window size radius was set to 37 (default) and the nuclear area threshold was set to be 0–1,500 mm2; any nuclei or cells out of these parameters were eliminated from analysis. The minimum percent of stained area in the nucleus to be considered as positive was set to 20%. The threshold for positive identification of cytoplasmic staining was set at a higher threshold for some of the antibodies due to differences in background and staining. The maximum threshold range was set for cytoplasmic staining detection between 220–240, with a minimum percent of stained area set to 70%.
After analysis, data regarding the nuclear h-score, % of positive nuclei, % of positive nuclear area in tissue, cytoplasmic h-score, and % positive cytoplasm of cells was collected and used for comparative graphing. These scores were chosen to represent the level and intensity of nuclear and cytoplasmic staining in the epithelial cells. Test runs of the analysis were conducted with a pathologist to assure that the program correctly identified and graded the nuclear and cytoplasmic components of epithelial cells.
Comparative graphing and statistical analysis.
R Studio was used for analyzing and graphing the data. Using the ggplot2 package, violin plots were constructed to demonstrate the differences in nuclear h-score, % of positive nuclei, % of positive nuclear area in tissue, cytoplasmic h-score, and % of cells with positive cytoplasmic staining between tumor and normal adjacent to tumor (NAT) cores. Statistical analysis was done via a two-sided Wilcoxon rank-sum test. Statistical significance was defined as p < 0.05.
RESULTS
Global proteome and phosphoproteome in cisplatin-resistant BC cells
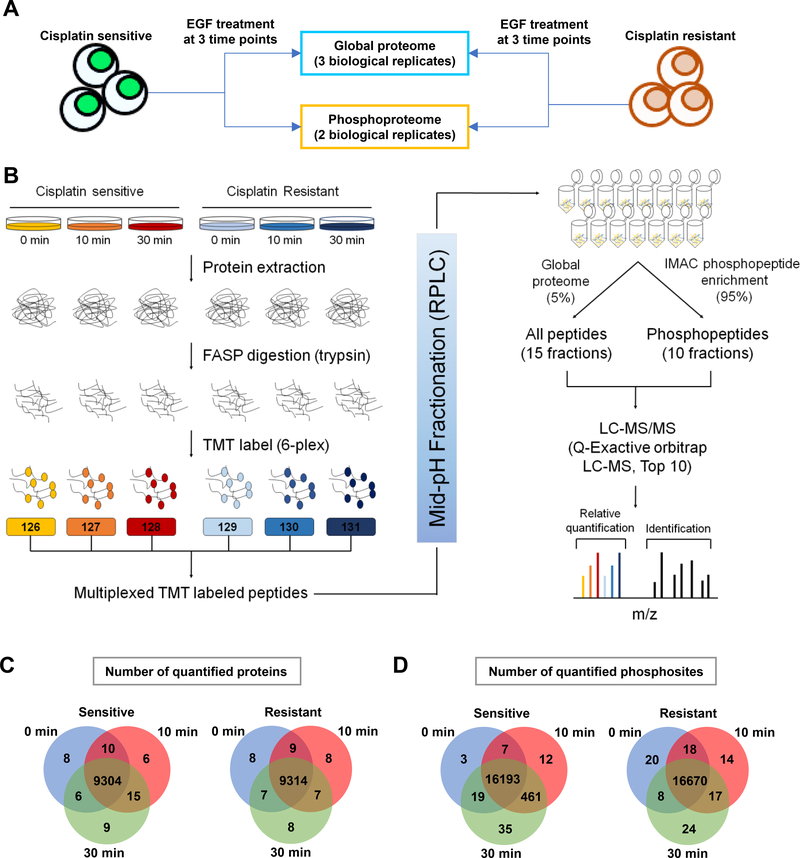
We sought to understand the molecular mechanisms of cisplatin resistance using a paired cell culture system consisting of T24S and T24R cells. Alterations of EGFR signaling pathways have been frequently observed and reported in various cancer types. It has also been considered a critical module associated with resistance mechanisms against several kinase inhibitors and other drugs, including cisplatin [9, 12, 15, 29]. Based on this knowledge, we applied a multiplexed TMT and LC/LC-MS/MS approach to samples and replicates harvested after 0 min, 10 mins, and 30 mins of EGF stimulation (10 ng/ml) (Fig. 1A). After a FASP peptide digestion, each conditioned peptide was respectively labeled with 6-plex TMT reagents. For mid-pH fractionation, we used TEAB as a buffer solvent. Several benefits of using TEAB include compatibility with TMT labeling, high volatility (allowing us to skip the desalting step), and low harmful impact on separation columns. After fractionation, 5% of the peptides from each of the 15 fractions were used for global proteome analysis. The remaining 95% of the peptides were subjected to an IMAC enrichment experiment and analyzed using an orbitrap-based high resolution/accurate-mass mass spectrometer (Fig. 1B). This serial enrichment strategy was done to simultaneously observe changes in either global or phosphopeptides from the same peptide pool [32]. We then searched for acquired mass spectra using MS/MS automated selected ion chromatography (MASIC) in the Uniprot Human Protein Database (2016) with MSGF+ TMT reporter ion extraction [20] (see methods). From the 3 biological replicates of the global proteome, we identified an average of 48,044 non-redundant peptides (9,405 protein groups) (Supplementary Table 1 and Fig. 1C). We also identified an average of 13,980 phosphopeptides and 16,700 phosphosites (Supplementary Table 2) belonging to 3,939 protein groups (Supplementary Table 3 and Fig. 1D).
Figure 1. Quantitative measuring of the proteome and phosphoproteome using TMT labeling.
A. Experimental design to study the effects of EGF stimulation across 3 time points. A total of 3 and 2 biological replicates from T24S and T24R cells were analyzed for their global proteome and phosphoproteome, respectively. B. Overall experimental workflow for multiplex TMT labeling and comprehensive profiling of the global proteome and phosphoproteome. Both T24S and T24R cells were treated with EGF (0, 10, or 30 mins). Proteins were extracted using RIPA buffer and peptides were FASP digested with trypsin. Peptide samples were derived at 6 different time points, labeled using TMT reagents, mixed, and separated using mid-pH reverse phase chromatography. Fractions were combined in a non-contiguous way into 15 fractions for proteome analysis (5% of total proteins) and 10 fractions for phosphoproteome analysis (95% of total proteins). All peptides and phosphopeptides were analyzed on a Q Exactive Mass Spectrometer. Protein identification and. quantification was achieved using the MSGF+ search engine and MASIC (reporter ion intensity extractor). C and D. Numbers of identified (C) proteins or (D) phosphopeptides from T24S and T24R cells. Venn diagrams depict the number of common and uniquely identified proteins or phosphopeptides in T24S and T24R cells after 0, 10 and 30 mins of EGF stimulation
Altered proteins in cisplatin-resistant BC cells
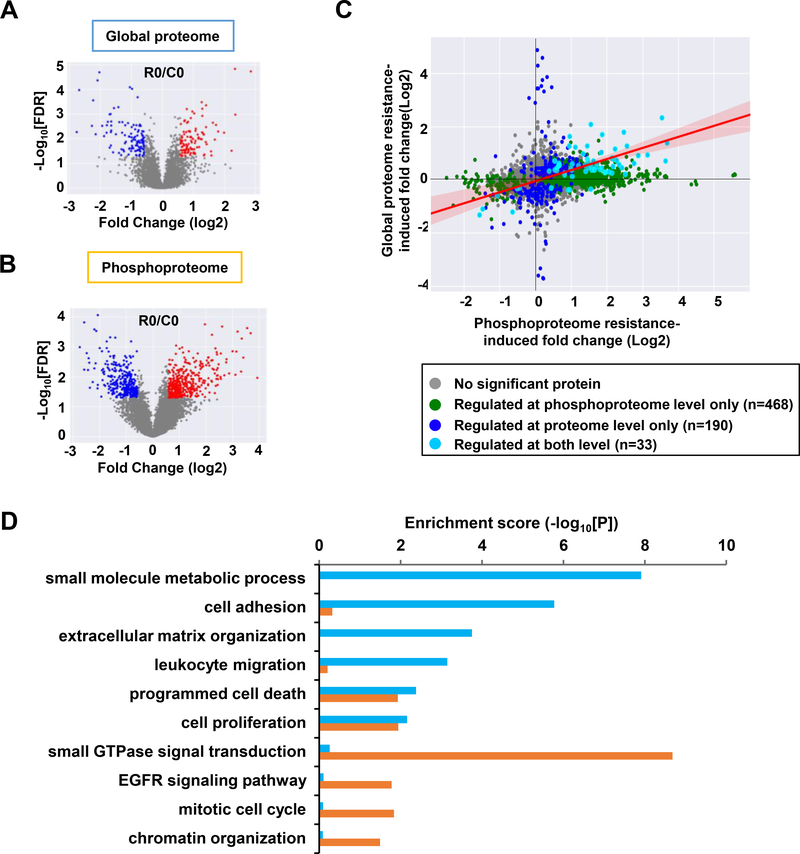
To identify proteins with altered expression and phosphorylation between T24R and T24S cells before any EGF stimulation, we performed separate statistical hypothesis testing for the global proteome and phosphoproteome. From the global proteome, we identified 223 DEPs between T24R and T24S cells (Fig. 2A). When analyzing the phosphoproteome, we identified 899 differentially phosphorylated peptides belonging to 501 DPPs (Fig. 2B). Comparing the list of DEPs and DPPs (Supplementary Table 4), we observed that 33 proteins exhibited a positive correlation (Spearman’s rho = 0.50; P <0.001), with significant changes at both the phosphorylation and expression levels (Fig. 2C). Functional enrichment analysis, using the DAVID [13], showed that the DEPs and DPPs were enriched for common and distinct cellular processes (Fig. 2D). DEPs exhibited predominant enrichment for cell adhesion and migrationrelated processes, including extracellular matrix organization and leukocyte migration. DPPs, on the other hand, displayed direct association with the EGFR signaling pathway and downstream processes, such as small GTPase signal transduction, mitotic cell cycle, and chromatin organization. Both DEPs and DPPs were enriched for cell survival and proliferation-related processes. These results suggest that DPPs in cisplatin-resistant BC cells represent the activation of EGFR signaling pathways or EGF-driven resistance mechanisms.
Figure 2. Altered proteins in the global proteome and phosphoproteome between T24R and T24S cells.
A and B. Volcano plots display differentially expressed (A) proteins or (B) differentially phosphorylated peptides. Red and blue dots represent upregulated and downregulated proteins or peptides, respectively. C. Protein expression and phosphorylation levels were compared and represented in the scatterplot. Proteins and phosphorylation sites were considered to be perturbed by cisplatin resistance based on the integrative hypothesis testing method (FDR < 0.05). Comparison between biological replicates of MS-based quantitative proteomic and phosphoproteomic experiments of T24R and T24S cells. Each dot represents one protein. D. Enrichment analysis using DAVID software showed that differentially expressed proteins (cyan) and differentially phosphorylated peptides (purple) were enriched for various cellular functions.
Altered proteins belonging to the EGFR signaling pathway are involved in cisplatin resistance
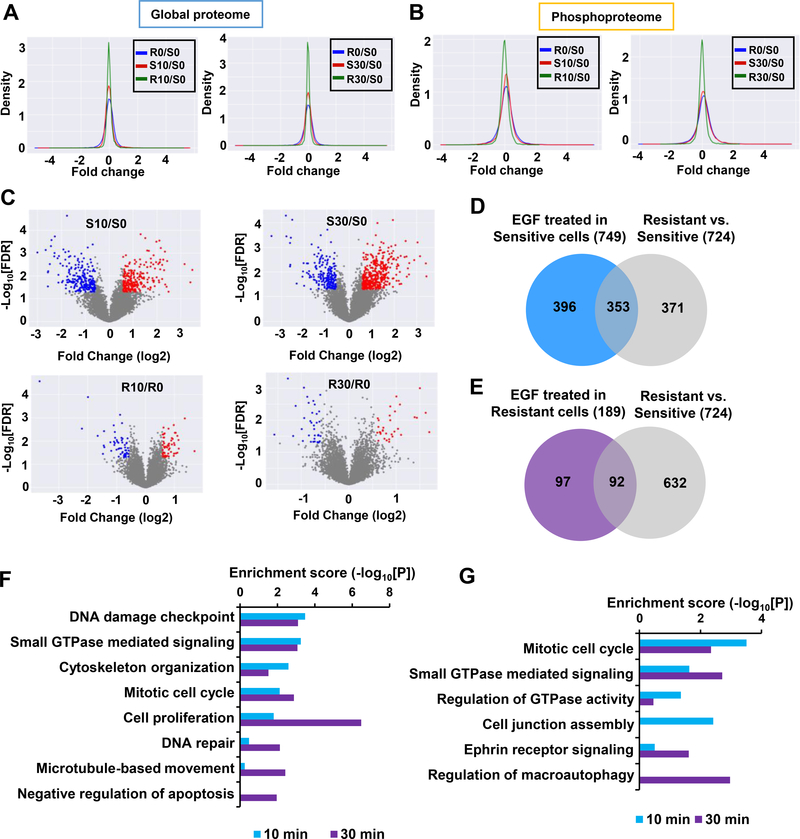
Using our global proteome and phosphoproteome data, we next performed differential analyses of EGF stimulation in T24R and T24S cells. For the global proteome in both T24R and T24S cells, only the tails of the density distribution in the log2 fold change increased with EGF stimulation (Fig. 3A and Supplementary Fig. 1A). In contrast, the global median of the phosphoproteome in T24S cells increased in abundance when stimulated with EGF (Fig. 3B). The largest changes in EGF-induced phosphorylation were observed in T24S cells, which exhibited phosphosite changes of 4% (656) and 5.3% (887) after 10 mins and 30 mins of EGF stimulation, respectively. T24R cells were altered less, with the percentage of altered phosphorylation sites being 1.1% and 0.7% after 10 mins and 30 mins, respectively (Supplementary Fig. 1B). This suggested that the effects of EGF stimulation impacts T24S cells more. Volcano plots for differentially phosphorylated proteins were consistent with the density plots shown in Fig. 3B (Fig. 3C).
Figure 3. Distinct protein alteration patterns in T24R and T24S cells after EGF stimulation.
A and B. Density plots are shown for the log2 fold changes of the (A) global proteome and (B) phosphoproteome for T24R and T24S cells after 10 mins and 30 mins of EGF treatment. Only phosphopeptides or proteins quantified in at least two samples were plotted. Log2 fold changes in T24S cells at 10 mins (S10) vs. 0 min (S0) of EGF treatment, T24S cells at 30 mins (S30) vs. 0 min (S0) of EGF treatment, T24R cells at 10 mins (R10) vs. 0 min (R0) of EGF treatment, T24R cells at 30 mins (R30) vs. 0 min (R0) of EGF treatment, and R0 vs. S0 are displayed. C. Volcano plots display differentially expressed phosphopeptides in T24R and T24S cells after 10 mins and 30 mins of EGF treatment. Red and blue dots indicate up- and downregulated phosphopeptides, respectively. Only those quantified in at least two samples were plotted. D and E. Number of phosphoproteins associated with cisplatin resistance mechanisms that are involved in EGFR signaling in (D) T24S and (E) T24R cells. Venn diagram depicts the overlapping altered phosphoproteins in (D) T24S or (E) T24R cells with or without EGF treatment. F and G. Enriched cellular functions of the overlapping proteins after 10 mins and 30 mins of EGF treatment in (F) T24S and (G) T24R cells.
We next sought to select altered proteins (APs) that were associated with cisplatin resistance and were part of the EGFR signaling pathway. Both DEPs and DPPs were considered as APs because either protein expression or phosphorylation can reflect alterations in signaling pathways. A total of 749 APs (454 APs at 10 mins and 547 APs at 30 mins) were identified in T24S cells (Supplementary Table 5). Of these 749 APs, 353 overlapped with the 724 APs identified from T24S vs. T24R cells with no EGF treatment. Interestingly, 49% (353 out of 724) of the APs responsible for cisplatin resistance were regulated by EGF stimulation (Fig. 3D). This suggested that a large proportion of APs are involved in downstream EGFR signaling and are likely to also be associated with cisplatin resistance. In contrast, T24R cells exhibited a relatively smaller number of APs (n=92) that overlapped with those found between T24R and T24S cells (Fig. 3E). Functional enrichment analysis of the 353 APs (Supplementary Table 6) in T24S cells revealed known associations of cisplatin resistance with DNA damage, repair, and cell cycle regulation (Fig. 3F). This was replicated with the 92 APs (Supplementary Table 7) found in T24R cells (Fig. 3G). The results were consistent with previous reports demonstrating inherent and persistent downstream EGFR signaling in resistant cells [21].
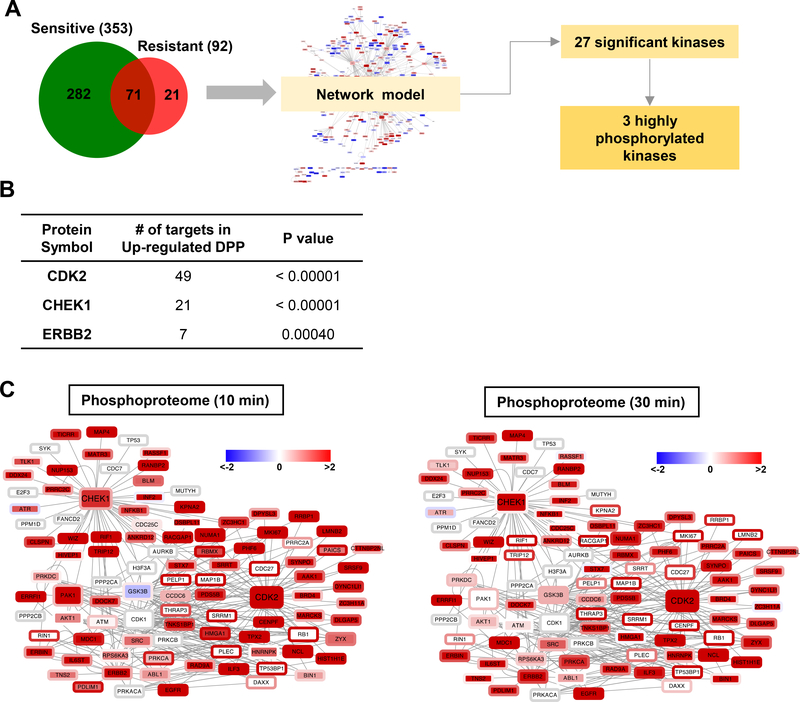
Several key kinases involved in cisplatin resistance were identified
Given this total list of 374 APs, 353 from T24S and 92 from T24R (with overlap), we next aimed to identify the key regulators of the observed phosphorylation profiles linked to cisplatin resistance. To this end, we focused on the kinases that can regulate the phosphorylation levels of these differential sites by performing a kinase enrichment analysis. Using kinase-substrate interactions obtained from the SIGNOR database [34], we constructed a network model consisting of 121 kinases and 329 substrates with 893 interactions (Supplementary Fig. 2). Kinase enrichment analysis of the network model identified 3 out of the 27 kinases (Supplementary Table 8), checkpoint kinase 1 (CHEK1), cyclin-dependent kinase 2 (CDK2), and Erb-B2 receptor tyrosine kinase 2 (ERBB2), as having a significant number of interactions with APs when stimulated by EGF (Fig. 4A, B). Based on these 3 key kinases and their substrates, we reconstructed a subnetwork consisting of 101 nodes connected by 265 edges (Fig. 4A). The subnetwork was visualized using Cytoscape [37]. Notably, most of the substrates were highly phosphorylated in T24R, rather than T24S cells (Fig. 4C).
Figure 4. Identification of key kinases linked to cisplatin resistance through the activation of EGFR signaling.
A. Schematic diagram describing the process of reconstructing the networks of proteins significantly altered by EGF stimulation in T24R and T24S cells. Venn diagram depicts number of altered proteins involved in cisplatin resistance in downstream EGFR signaling. Using kinase-substrate interaction information of the significantly altered proteins and their upstream kinases, 27 kinases were initially selected. Of those, 3 kinases were found to be significant altered in either protein expression or phosphorylation after EGF treatment. B. List of the 3 key kinases with the number of significantly altered substrates and number of interactions. C. Network model describing the interactions of the 3 kinases and their substrates. Node and border color represent proteins phosphorylation ratios in T24R vs. T24S cells after 10 mins or 30 mins of EGF treatment, respectively. Red and green indicate up- and downregulated phosphorylation following EGF treatment, respectively.
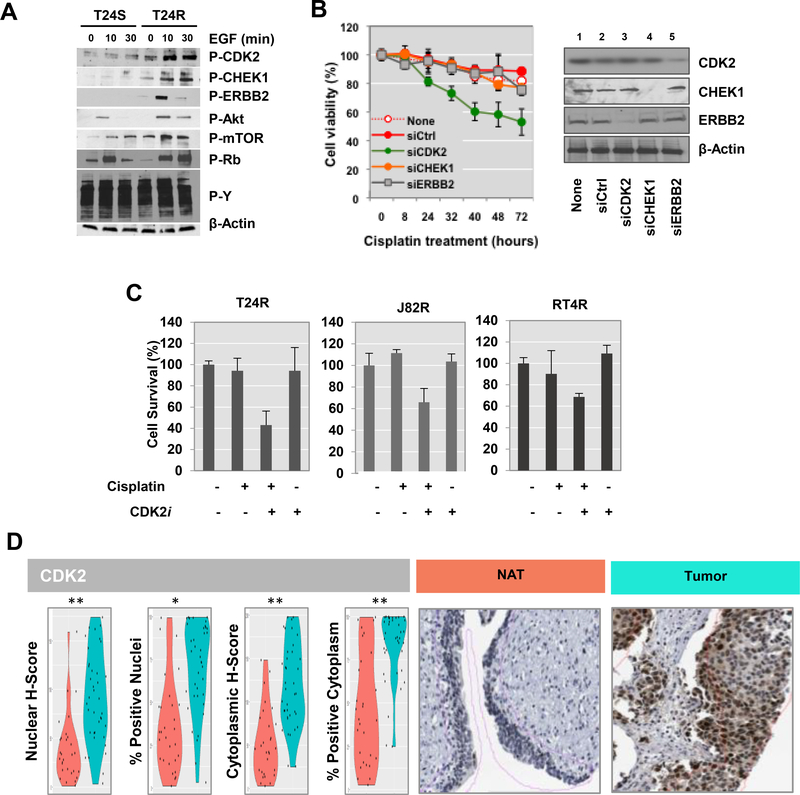
Validation with western blot analysis confirmed that phosphorylation of CDK2, CHEK1, ERBB2, mTOR, AKT, and RB significantly increased in T24R cells after treatment with 10 ng/ml of EGF (Fig. 5A). These results provide potential molecular mechanisms through which these 3 major kinases mediate cisplatin resistance. To further identify genes that have a critical effect on cisplatin resistance, we knocked-down the 3 kinases, CDK2, CHEK1, and ERBB2, in T24R cells using siRNAs. Western blot analysis was conducted to confirm knockdown of these kinases (Fig. 5B, right). T24R cells were significantly sensitized to cisplatin when CDK2 was knocked-down, suggesting that CDK2 may be important in maintaining resistance (Fig. 5B). We next tested whether a potent and selective inhibitor of CDK2, dinaciclib (SCH 727965) [33], could overcome resistance in three independent cisplatin-resistant BC cell lines, T24R, J82R, and RT4R (Fig. 5C). Consistent to the data shown in Fig. 5B, inhibition of CDK2 sensitized resistant cells when they were incubated in combination with cisplatin (Fig. 5C).
Figure 5. Functional role of CDK2 associated with cisplatin resistance.
A. Western blot analysis of the key protein kinases. After stimulation with 10 ng/ml of EGF treatment for 0, 10, or 30 mins, cells were harvested for protein extraction and western blot analysis. Representative western blot images were selected after experiments were repeated at least 3 times. B. Gene silencing of CDK2 enhanced cisplatin sensitivity in T24R cells. T24R cells transiently transfected with siRNAs targeting CDK2, CHEK1, or ERBB2 were incubated in culture medium with 10 μM cisplatin. T24R cells transfected with control siRNAs (siCtrl) were used as controls. The cell viability rate was measured at various time points (0, 8, 24, 32, 40, 48, and 72 hrs). Experiments were done in triplicate. *p<0.05 (Student’s t-test). Representative western blot analysis data demonstrated that the levels expression of CDK2, CHEK1, or ERBB2 in these experiments were downregulated by targeting siRNAs. C. Cisplatin resistant BC cell lines (R24R, J82R, or RT4R) were treated with cisplatin alone, CDK2 inhibitor (CDKi), or a combination of both for 72 hr. Cell survival rates were quantified as describe in Methods. C. Increased expression levels of CDK2 in BC. IHC analysis was performed to measure the protein expression of CDK2 in bladder tissues from BC patients. Violin plots showing expression of CDK2 in NAT and tumor tissue. Representative images of NAT and tumor cores are shown.
To further investigate the correlation of high CDK2 expression in BC, IHC analysis was done on TMAs. There were statistically significant differences in comparative scores for CDK2 expression between tumor and normal adjacent to tumor (NAT) cores (Fig. 5D). We applied a digital annotation algorithm to the core images in an effort to quantitatively assess this observation. Our image algorithm was set so that nuclear and cytoplasmic staining could be detected as positive at a color spectrum darker than that for typical DAB staining. Using the scores from our digital image analysis, we observed significantly upregulated expression of CDK2 in tumor cores. Comparative images of the tumor to NAT tissue show increased nuclear and cytoplasmic staining in the tumor epithelium (Fig. 5D). Collectively, these results demonstrate that our proteome-driven signature may be a useful predictor of survival for patients who have already undergone cisplatin chemotherapy.
Clinical implications of the key kinases and their substrates
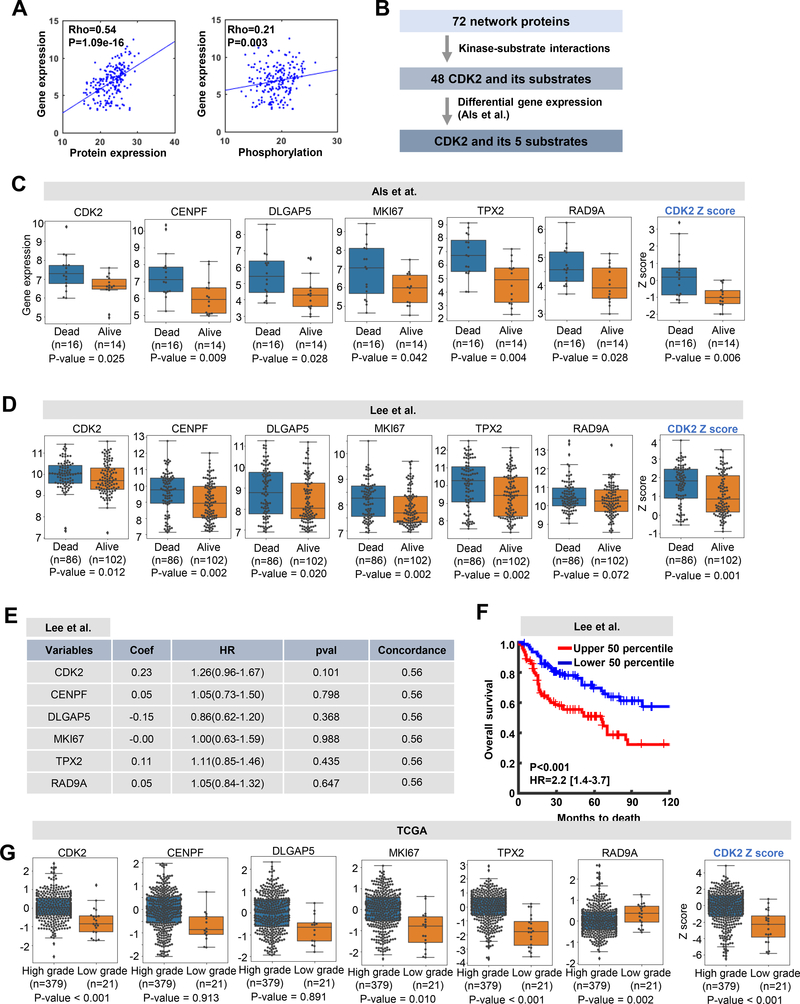
Based on the significance of CDK2 and its substrates to cisplatin resistance, we examined the association of gene expression of CDK2 and its substrates to clinical outcomes. For this, we utilized the transcriptome dataset (GSE5287) from Als et al. [2], which contains the gene expression profiles of 30 BC patients along with their survival information after cisplatin chemotherapy. We first assessed the correlation of APs in the global network model in terms of gene expression versus protein expression or phosphorylation levels (Fig. 6A). Significant positive correlations between gene expression versus protein expression (P=1.09e-16) and gene expression versus protein phosphorylation (P=0.003) were observed (Fig. 6A). This suggests that gene expression profiling can reflect protein expression and phosphorylation, supporting the possibility that the gene expression profiles of APs in BC patients can be indicators of dysregulated networks. We then searched for CDK2 substrates from the network model, resulting that 48 proteins were selected based on the kinase-substrate interaction information from the SIGNOR database (Fig. 6B). For further selection, we examined differential gene expression of the 48 proteins, including CDK2 and its substrates, using the BC dataset from Als et al. (GSE5287) [2]. For this we stratified the samples into those who survived 5 or more years (alive) and those who deceased within 5 years after chemotherapy (dead). This allowed us to identify genes in the CDK2 network that were significantly associated with poor survival after cisplatin chemotherapy (Fig. 6C). As a result, 6 genes out of 72 were found to have higher expression in the dead group. These were CDK2, CENPF, DLGAP5, MKI67, TPX2, and RAD9A.
Figure 6. CDK2 network may have clinical implications in BC.
A. Correlation of 72 network proteins in levels of gene expression vs. protein expression (left) or phosphorylation (right). Dots on the scatter plot represent proteins and lines represent regression lines. Correlation was measured using the Spearman’s rank correlation method. B. Selection criteria used to identify the 6 proteins from the 72 network proteins. C and D. Box plots display differential gene expression of the selected 6 proteins from the network model using the gene expression datasets from (C) Als et al. and (D)Lee et al. Box plots at the far right display differential representation of CDK2 network scores computed by the Z-score method. Significance levels of differential gene expression between BC patients who were alive and deceased after cisplatin treatment were computed via the Wilcoxon rank-sum test. E. Regression coefficient and significance of the 6 genes based on multivariate Cox regression analysis using Lee at al. dataset. F. Kaplan-Meier curves for overall survival based on the expression of the CDK2 network genes are shown for the high (upper 50th percentile; n=97) and low (lower 50th percentile; n=91) z-score groups from the Lee et al. patient dataset. P-values and hazard ratios (HRs) were taken from a Cox regression analysis. G. Box plots display differential expression of the 6 genes in the TCGA BC cohort. Wilcoxon rank-sum test was performed to compute significance of differential expression between high and low grade samples.
We further investigated the clinical association of the 6 genes to the CDK2 network using two gene expression datasets from Als et al. (GSE5287) [2] and Lee et al. (GSE13507) [24]. The Als et al. dataset showed significantly higher expression of the 6 genes in the CDK2 network and its summarized score (Z-score) in the dead group (Fig. 6C). The same was true in Lee et al. dataset (Fig. 6D). We then examined the association of the 6 genes in CDK2 network with recurrence-free survival (RFS) by performing multivariate Cox regression analysis using Lee et al. dataset (Fig. 6E). Although individual genes showed no significant correlation with RFS, the summarized score of the 6 genes presented significant association with RFS (Fig. 6F). We further confirmed that expression of the 6 gene and its summarized score in the CKD2 network were significantly segregated high vs low grade BC samples from The Cancer Genome Atlas (TCGA) [35] (Fig. 6G). Collectively, these results suggest that our AP-derived CDK2 network genes may be helpful in predicting prognosis and treatment options for BC patients.
DISCUSSION
Our present work characterized the enriched global proteome and phosphoproteome in cisplatin-resistant BC. By characterizing the signaling network alterations associated with cisplatin resistance, we also demonstrated the utility of quantitative phosphoproteomics in highlighting perturbed networks between cisplatin-resistant and sensitive BC cells. This approach revealed a vastly different cellular context between T24R and T24S cells, suggesting that there are potential key regulators modulating the perturbation of signaling networks. Measuring the abundance of proteins and mapping the phosphorylated kinase-substrate interactions facilitated a comprehensive biological interpretation of the complex network context behind cisplatin resistance. Expanded analysis of transcriptome data from BC patients further provided strong evidence that concurrent molecular alterations in gene expression to protein modification can be the core basis of disease.
Our experimental results illustrated that the global phosphoproteomics profile in cisplatin-resistant cells is significantly altered and that several key kinases may be mediating this. These kinases could potentially be used as therapeutic targets for blocking chemoresilience. We also observed an increase in the phosphorylation of CDK2, CHEK1, and ERBB2 in cisplatin-resistant cells. Our comprehensive proteomics approach revealed many novel phosphorylation sites of CDK2, CHEK1, ERBB2, and their substrates, suggesting new intervention points in cisplatin resistance-specific signaling networks. The experimental results from this study implicate the active involvement of CDK2, CHEK1, and ERBB2 in contributing to resistance against cisplatin-based chemotherapies.
CHEK1 is a serine/threonine kinase that is involved in the checkpoint-mediated control of the cell cycle and the activation of DNA repair in response to DNA damage or the presence of unreplicated DNA. The ATR-CHEK2 signaling pathway was recently proposed to play an important role in regulating the response to cisplatin in BC [16]. Inhibition of the ATR-CHEK2 pathway with selective inhibitors can sensitize cells to cisplatin [7, 26]. ERBB2 (HER2), a member of the EGF receptor family of receptor tyrosine kinases, has been known to be associated with aggressive variants of BC. Mutated ERBB2 is well documented as a cancer driver gene and is primarily found in high-risk tumors. Mutations in ERBB2 have also been found to be independent risk factors associated with death in BC patients [22]. Thus, ERBB2targeting treatment has been an attractive option in improving clinical outcomes for BC patients.
Our kinase activity-based network modeling suggested that one of the most significantly increased sites of phosphorylation in resistant cells is CDK2. The CDK2 network consists of several putative substrates, including CDC27, DAXX, and CENPF (Fig. 4A). CDK2, a Ser/Thr protein kinase, phosphorylates substrates at the S/TPx(x)R/K consensus motif. CDK2 is originally known to function in cell cycle regulation; increased CDK2 kinase activity is essential for the G1 to S stage transition as well as the phosphorylation of the Rb protein. During the S phase, active CDK2 and cyclin A complexes predominate and phosphorylate E2F [30]. CDK2 inhibitors (e.g., ribociclib, dinociclib, seliciclib et al.), which deregulate E2F, have been suggested as promising pharmacological strategies against various cancer types. However, a comprehensive overview of CDK2 and its substrates has not yet been established and the biological effects or regulatory mechanisms of CDK2 remain elusive.
Our findings suggest that inhibition of CDK2, a key player of cisplatin resistance, could induce cisplatin sensitivity. Therefore, CDK2 may be a potential target in the modulation of cisplatin resistance modulating human BC. This is consistent with previous reports. Alterations of CDK2 that are associated with cisplatin resistance in vitro have been reported in other cancer types, such as ovarian [31, 40], oral [25], testicular [23] and cervical [42]. While our results are promising, we are aware of the limitations in our study, mainly owing to a lack in quantity of clinical specimens for phosphoproteomic analysis. Our study was also restricted to cell linederived signatures; although, it was further validated in the clinical setting. Since this finding was based on a retrospective study from a single institution, the efficacy of the signature could be impaired. Future prospective validation studies using high-throughput assays should be followed up in multiple centers to consolidate our signature’s analytical validity and reproducibility in the routine clinical practice setting.
Supplementary Material
Supplementary Figure S1. Altered proteins and phosphoproteins in cisplatin-resistant and sensitive cells. A. Venn diagram depicts number of DPPs in sensitive cells after 10 mins (S10) vs. 0 min (S0) of EGF treatment, sensitive cells after 30 mins (S30) vs. 0 min (S0) of EGF treatment, resistant cells after 10 mins (R10) vs. 0 min (R0) of EGF treatment, resistant cells after 30 mins (R30) vs. 0 min (R0) of EGF treatment, and resistant (R0) vs. sensitive (S0). B. Venn diagram depicts number of differentially phosphorylated sites in sensitive cells after 10 mins (S10) vs. 0 min (S0) of EGF treatment, sensitive cells after 30 mins (S30) vs. 0 min (S0) of EGF treatment, resistant cells after 10 mins (R10) vs. 0 min (R0) of EGF treatment, resistant cells after 30 mins (R30) vs. 0 min (R0) of EGF treatment, and resistant (R0) vs. sensitive (S0).
Supplementary Figure S2. Network model of altered proteins associated with cisplatin resistance. Node size represents the number of kinase-substrate interactions; larger node indicates many interactions with other proteins in the network. Node color represents differential expression between cisplatin-resistant and sensitive cells.
Supplementary Table S1. List of identified peptides and their estimated quantity in the global proteome.
Supplementary Table S2. Phosphopeptides with modification sites.
Supplementary Table S3. List of identified peptides and their estimated quantity from phosphoproteome.
Supplementary Table S4. List of DEPs and DPPs between cisplatin-resistant and sensitive cells.
Supplementary Table S5. Lists of 454 and 547 altered proteins after 10 mins and 30 mins of EGF treatment.
Supplementary Table S6. List of 353 shared DEPs and/or DPPs in control cells.
Supplementary Table S8. List of 27 kinases significantly enriched by altered phosphoproteins in cisplatin-resistant cells.
Supplementary Table S7. List of 92 shared DEPs and/or DPPs in resistant cells.
Highlights:
A molecular signature panel predicting cisplatin resistance in bladder cancer
Altered protein abundance and phosphorylation in cisplatin resistance
EGFR signaling pathways perturbed in cisplatin-resistant cells
CDK2 may be a central regulator of cisplatin resistance
ACKNOWLEDGEMENTS
The authors acknowledge support from the Bio and Medical Technology Development Program (project no. 2012M3A9B6055305) though the National Research Foundation of Korea funded by the Korean Ministry of Education, Science and Technology. We also appreciate support from the Multiomics Research Program (NRF-2012M3AB9036675), which is funded by the Korean Ministry of Science, ICT, and Future Planning. This work was also supported by the National Institutes of Health grants (1U01DK103260, 1R01DK100974, U24 DK097154, NIH NCATS UCLA CTSI UL1TR000124), Department of Defense (W81XWH-15–1-0415), Centers for Disease Controls and Prevention (1U01DP006079), IMAGINE NO IC Research Grant, Steven Spielberg Discovery Fund in Prostate Cancer Research Career Development Awards, and the U.S.-Egypt Science and Technology Joint Fund (to J.K.). J.K. is a former recipient of the Interstitial Cystitis Association Pilot Grant, Fishbein Family IC Research Grant, New York Academy of Medicine, and Boston Children’s Hospital Faculty Development. Any opinions, findings, conclusions, or recommendations expressed in this article are those of the authors alone and do not necessarily reflect the views of any of the previously mentioned sponsors. In addition, this article is derived from the Subject Data funded in whole or part by National Academies of Sciences, Engineering, and Medicine (NAS) and The United States Agency for International Development (USAID). Any opinions, findings, conclusions, or recommendations expressed in this article are those of the authors alone, and do not necessarily reflect the views of USAID or NAS.
Footnotes
Competing interests
The authors declare that there are no conflicts of interest that could be perceived as prejudicing the impartiality of this review.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].B.A.W.A. A., Applied smoothing techniques for data analysis: the kernel approach with SPlus illustrations., Clarendon Press;Oxford University Press, Oxford New York, 1997. [Google Scholar]
- [2].Als AB, Dyrskjot L, von der Maase H, Koed K, Mansilla F, Toldbod HE, Jensen JL, Ulhoi BP, Sengelov L, Jensen KM, Orntoft TF, Emmprin and survivin predict response and survival following cisplatin-containing chemotherapy in patients with advanced bladder cancer, Clin Cancer Res, 13 (2007) 4407–4414. [DOI] [PubMed] [Google Scholar]
- [3].Apolo AB, Grossman HB, Bajorin D, Steinberg G, Kamat AM, Practical use of perioperative chemotherapy for muscle-invasive bladder cancer: summary of session at the Society of Urologic Oncology annual meeting, Urol Oncol, 30 (2012) 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Black PC, Dinney CP, Growth factors and receptors as prognostic markers in urothelial carcinoma, Curr Urol Rep, 9 (2008) 55–61. [DOI] [PubMed] [Google Scholar]
- [5].Bolstad BM, Irizarry RA, Astrand M, Speed TP, A comparison of normalization methods for high density oligonucleotide array data based on variance and bias, Bioinformatics, 19 (2003) 185–193. [DOI] [PubMed] [Google Scholar]
- [6].Casado P, Hijazi M, Britton D, Cutillas PR, Impact of phosphoproteomics in the translation of kinase-targeted therapies, Proteomics, 17 (2017). [DOI] [PubMed] [Google Scholar]
- [7].Chen S, Chen X, Xie G, He Y, Yan D, Zheng D, Li S, Fu X, Li Y, Pang X, Hu Z, Li H, Tan W, Li J, Cdc6 contributes to cisplatin-resistance by activation of ATR-Chk1 pathway in bladder cancer cells, Oncotarget, 7 (2016) 40362–40376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cutillas PR, Role of phosphoproteomics in the development of personalized cancer therapies, Proteomics Clin Appl, 9 (2015) 383–395. [DOI] [PubMed] [Google Scholar]
- [9].Eckstein N, Servan K, Girard L, Cai D, von Jonquieres G, Jaehde U, Kassack MU, Gazdar AF, Minna JD, Royer HD, Epidermal growth factor receptor pathway analysis identifies amphiregulin as a key factor for cisplatin resistance of human breast cancer cells, J Biol Chem, 283 (2008) 739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gakis G, Efstathiou J, Lerner SP, Cookson MS, Keegan KA, Guru KA, Shipley WU, Heidenreich A, Schoenberg MP, Sagaloswky AI, Soloway MS, Stenzl A, ICUD-EAU International Consultation on Bladder Cancer 2012: Radical cystectomy and bladder preservation for muscle-invasive urothelial carcinoma of the bladder, Eur Urol, 63 (2013) 45–57. [DOI] [PubMed] [Google Scholar]
- [11].Gofrit ON, Re: Long-Term Outcomes in Patients with Muscle-Invasive Bladder Cancer After Selective Bladder-Preserving Combined-Modality Therapy: A Pooled Analysis of Radiation Therapy Oncology Group Protocols 8802, 8903, 9506, 9706, 9906, and 0233, Eur Urol, 68 (2015) 165–166. [DOI] [PubMed] [Google Scholar]
- [12].Granados ML, Hudson LG, Samudio-Ruiz SL, Contributions of the Epidermal Growth Factor Receptor to Acquisition of Platinum Resistance in Ovarian Cancer Cells, PLoS One, 10 (2015) e0136893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Huang da W, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nat Protoc, 4 (2009) 44–57. [DOI] [PubMed] [Google Scholar]
- [14].Hwang D, Rust AG, Ramsey S, Smith JJ, Leslie DM, Weston AD, de Atauri P, Aitchison JD, Hood L, Siegel AF, Bolouri H, A data integration methodology for systems biology, Proc Natl Acad Sci U S A, 102 (2005) 17296–17301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Inoue M, Koga F, Yoshida S, Tamura T, Fujii Y, Ito E, Kihara K, Significance of ERBB2 overexpression in therapeutic resistance and cancer-specific survival in muscle-invasive bladder cancer patients treated with chemoradiation-based selective bladder-sparing approach, Int J Radiat Oncol Biol Phys, 90 (2014) 303–311. [DOI] [PubMed] [Google Scholar]
- [16].Isono M, Hoffmann MJ, Pinkerneil M, Sato A, Michaelis M, Cinatl J Jr., Niegisch G, Schulz WA, Checkpoint kinase inhibitor AZD7762 strongly sensitises urothelial carcinoma cells to gemcitabine, Journal of experimental & clinical cancer research : CR, 36 (2017) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jones P, Cote RG, Martens L, Quinn AF, Taylor CF, Derache W, Hermjakob H, Apweiler R, PRIDE: a public repository of protein and peptide identifications for the proteomics community, Nucleic Acids Res, 34 (2006) D659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kamat AM, Hegarty PK, Gee JR, Clark PE, Svatek RS, Hegarty N, Shariat SF, Xylinas E, Schmitz-Drager BJ, Lotan Y, Jenkins LC, Droller M, van Rhijn BW, Karakiewicz PI, ICUD-EAU International Consultation on Bladder Cancer 2012: Screening, diagnosis, and molecular markers, Eur Urol, 63 (2013) 4–15. [DOI] [PubMed] [Google Scholar]
- [19].Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M, KEGG for integration and interpretation of large-scale molecular data sets, Nucleic Acids Res, 40 (2012) D109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kim S, Pevzner PA, MS-GF+ makes progress towards a universal database search tool for proteomics, Nat Commun, 5 (2014) 5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim WT, Kim J, Yan C, Jeong P, Choi SY, Lee OJ, Chae YB, Yun SJ, Lee SC, Kim WJ, S100A9 and EGFR gene signatures predict disease progression in muscle invasive bladder cancer patients after chemotherapy, Ann Oncol, 25 (2014) 974–979. [DOI] [PubMed] [Google Scholar]
- [22].Koga F, Yoshida S, Tatokoro M, Kawakami S, Fujii Y, Kumagai J, Neckers L, Kihara K, ErbB2 and NFkappaB overexpression as predictors of chemoradiation resistance and putative targets to overcome resistance in muscle-invasive bladder cancer, PLoS One, 6 (2011) e27616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Koster R, di Pietro A, Timmer-Bosscha H, Gibcus JH, van den Berg A, Suurmeijer AJ, Bischoff R, Gietema JA, de Jong S, Cytoplasmic p21 expression levels determine cisplatin resistance in human testicular cancer, The Journal of clinical investigation, 120 (2010) 35943605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lee JS, Leem SH, Lee SY, Kim SC, Park ES, Kim SB, Kim SK, Kim YJ, Kim WJ, Chu IS, Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors, J Clin Oncol, 28 (2010) 2660–2667. [DOI] [PubMed] [Google Scholar]
- [25].Lee MR, Lin C, Lu CC, Kuo SC, Tsao JW, Juan YN, Chiu HY, Lee FY, Yang JS, Tsai FJ, YC-1 induces G0/G1 phase arrest and mitochondria-dependent apoptosis in cisplatinresistant human oral cancer CAR cells, BioMedicine, 7 (2017) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li CC, Yang JC, Lu MC, Lee CL, Peng CY, Hsu WY, Dai YH, Chang FR, Zhang DY, Wu WJ, Wu YC, ATR-Chk1 signaling inhibition as a therapeutic strategy to enhance cisplatin chemosensitivity in urothelial bladder cancer, Oncotarget, 7 (2016) 1947–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lynn KS, Chen CC, Lih TM, Cheng CW, Su WC, Chang CH, Cheng CY, Hsu WL, Chen YJ, Sung TY, MAGIC: an automated N-linked glycoprotein identification tool using a Y1-ion pattern matching algorithm and in silico MS(2) approach, Anal Chem, 87 (2015) 24662473. [DOI] [PubMed] [Google Scholar]
- [28].Madar, Reduction of Ambiguity in Phosphorylation-site Localization in Large-scale Phosphopeptide Profiling by Data Filter using Unique Mass Class Information, Bull. Korean Chem. Soc, 35 (2014) 845. [Google Scholar]
- [29].Mandic R, Rodgarkia-Dara CJ, Krohn V, Wiegand S, Grenman R, Werner JA, Cisplatin resistance of the HNSCC cell line UT-SCC-26A can be overcome by stimulation of the EGFreceptor, Anticancer Res, 29 (2009) 1181–1187. [PubMed] [Google Scholar]
- [30].Morgan DO, Cyclin-dependent kinases: engines, clocks, and microprocessors, Annu Rev Cell Dev Biol, 13 (1997) 261–291. [DOI] [PubMed] [Google Scholar]
- [31].Pan H, Wang F, Rankin GO, Rojanasakul Y, Tu Y, Chen YC, Inhibitory effect of black tea pigments, theaflavin3/3’-gallate against cisplatin-resistant ovarian cancer cells by inducing apoptosis and G1 cell cycle arrest, International journal of oncology, 51 (2017) 1508–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Park JM, Park JH, Mun DG, Bae J, Jung JH, Back S, Lee H, Kim H, Jung HJ, Kim HK, Lee H, Kim KP, Hwang D, Lee SW, Integrated analysis of global proteome, phosphoproteome, and glycoproteome enables complementary interpretation of disease-related protein networks, Sci Rep, 5 (2015) 18189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Paruch K, Dwyer MP, Alvarez C, Brown C, Chan TY, Doll RJ, Keertikar K, Knutson C, McKittrick B, Rivera J, Rossman R, Tucker G, Fischmann T, Hruza A, Madison V, Nomeir AA, Wang Y, Kirschmeier P, Lees E, Parry D, Sgambellone N, Seghezzi W, Schultz L, Shanahan F, Wiswell D, Xu X, Zhou Q, James RA, Paradkar VM, Park H, Rokosz LR, Stauffer TM, Guzi TJ, Discovery of Dinaciclib (SCH 727965): A Potent and Selective Inhibitor of Cyclin-Dependent Kinases, ACS medicinal chemistry letters, 1 (2010) 204–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Perfetto L, Briganti L, Calderone A, Cerquone Perpetuini A, Iannuccelli M, Langone F, Licata L, Marinkovic M, Mattioni A, Pavlidou T, Peluso D, Petrilli LL, Pirro S, Posca D, Santonico E, Silvestri A, Spada F, Castagnoli L, Cesareni G, SIGNOR: a database of causal relationships between biological entities, Nucleic Acids Res, 44 (2016) D548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, Hinoue T, Laird PW, Hoadley KA, Akbani R, Castro MAA, Gibb EA, Kanchi RS, Gordenin DA, Shukla SA, Sanchez-Vega F, Hansel DE, Czerniak BA, Reuter VE, Su X, de Sa Carvalho B, Chagas VS, Mungall KL, Sadeghi S, Pedamallu CS, Lu Y, Klimczak LJ, Zhang J, Choo C, Ojesina AI, Bullman S, Leraas KM, Lichtenberg TM, Wu CJ, Schultz N, Getz G, Meyerson M, Mills GB, McConkey DJ, Network TR, Weinstein JN, Kwiatkowski DJ, Lerner SP, Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer, Cell, 171 (2017) 540–556 e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Schwentner C, Stenzl A, Gakis G, Monitoring high-risk bladder cancer, Curr Opin Urol, 22 (2012) 421–426. [DOI] [PubMed] [Google Scholar]
- [37].Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome Res, 13 (2003) 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sonpavde G, Sternberg CN, Neoadjuvant chemotherapy for invasive bladder cancer, Curr Urol Rep, 13 (2012) 136–146. [DOI] [PubMed] [Google Scholar]
- [39].Sternberg CN, Bellmunt J, Sonpavde G, Siefker-Radtke AO, Stadler WM, Bajorin DF, Dreicer R, George DJ, Milowsky MI, Theodorescu D, Vaughn DJ, Galsky MD, Soloway MS, Quinn DI, ICUD-EAU International Consultation on Bladder Cancer 2012: Chemotherapy for urothelial carcinoma-neoadjuvant and adjuvant settings, Eur Urol, 63 (2013) 58–66. [DOI] [PubMed] [Google Scholar]
- [40].Sun Y, Jin L, Liu JH, Sui YX, Han LL, Shen XL, Interfering EZH2 Expression Reverses the Cisplatin Resistance in Human Ovarian Cancer by Inhibiting Autophagy, Cancer biotherapy & radiopharmaceuticals, 31 (2016) 246–252. [DOI] [PubMed] [Google Scholar]
- [41].Tan VM, Cheng LC, Drake JM, Complementing genomics and transcriptomics: Phosphoproteomics illuminates systems biology in prostate cancer, Mol Cell Oncol, 3 (2016) e1246075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang B, Huang Z, Gao R, Zeng Z, Yang W, Sun Y, Wei W, Wu Z, Yu L, Li Q, Zhang S, Li F, Liu G, Liu B, Leng L, Zhan W, Yu Y, Yang G, Zhou S, Expression of Long Noncoding RNA Urothelial Cancer Associated 1 Promotes Cisplatin Resistance in Cervical Cancer, Cancer biotherapy & radiopharmaceuticals, 32 (2017) 101–110. [DOI] [PubMed] [Google Scholar]
- [43].Wisniewski JR, Zougman A, Nagaraj N, Mann M, Universal sample preparation method for proteome analysis, Nat Methods, 6 (2009) 359–362. [DOI] [PubMed] [Google Scholar]
- [44].Zhang B, Chambers MC, Tabb DL, Proteomic parsimony through bipartite graph analysis improves accuracy and transparency, Journal of proteome research, 6 (2007) 35493557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Altered proteins and phosphoproteins in cisplatin-resistant and sensitive cells. A. Venn diagram depicts number of DPPs in sensitive cells after 10 mins (S10) vs. 0 min (S0) of EGF treatment, sensitive cells after 30 mins (S30) vs. 0 min (S0) of EGF treatment, resistant cells after 10 mins (R10) vs. 0 min (R0) of EGF treatment, resistant cells after 30 mins (R30) vs. 0 min (R0) of EGF treatment, and resistant (R0) vs. sensitive (S0). B. Venn diagram depicts number of differentially phosphorylated sites in sensitive cells after 10 mins (S10) vs. 0 min (S0) of EGF treatment, sensitive cells after 30 mins (S30) vs. 0 min (S0) of EGF treatment, resistant cells after 10 mins (R10) vs. 0 min (R0) of EGF treatment, resistant cells after 30 mins (R30) vs. 0 min (R0) of EGF treatment, and resistant (R0) vs. sensitive (S0).
Supplementary Figure S2. Network model of altered proteins associated with cisplatin resistance. Node size represents the number of kinase-substrate interactions; larger node indicates many interactions with other proteins in the network. Node color represents differential expression between cisplatin-resistant and sensitive cells.
Supplementary Table S1. List of identified peptides and their estimated quantity in the global proteome.
Supplementary Table S2. Phosphopeptides with modification sites.
Supplementary Table S3. List of identified peptides and their estimated quantity from phosphoproteome.
Supplementary Table S4. List of DEPs and DPPs between cisplatin-resistant and sensitive cells.
Supplementary Table S5. Lists of 454 and 547 altered proteins after 10 mins and 30 mins of EGF treatment.
Supplementary Table S6. List of 353 shared DEPs and/or DPPs in control cells.
Supplementary Table S8. List of 27 kinases significantly enriched by altered phosphoproteins in cisplatin-resistant cells.
Supplementary Table S7. List of 92 shared DEPs and/or DPPs in resistant cells.