Abstract
Objective
Effect of a probiotic on the gut microbiome and peripheral immune function in healthy controls and relapsing-remitting multiple sclerosis (RRMS) patients.
Methods
MS patients (N=9) and controls (N=13) were orally administered a probiotic containing Lactobacillus, Bifidobacterium and Streptococcus twice daily for two months. Blood and stool specimens were collected at baseline, after completion of the 2-month treatment, and 3 months after discontinuation of therapy. Frozen peripheral blood mononuclear cells (PBMCs) were used for immune cell profiling. Stool samples were used for 16S rRNA profiling and metabolomics.
Results
Probiotic administration increased the abundance of several taxa known to be depleted in MS such as Lactobacillus. We found that probiotic use decreased the abundance of taxa previously associated with dysbiosis in MS including Akkermansia and Blautia.
Predictive metagenomic analysis revealed a decrease in the abundance of several KEGG (Kyoto Encyclopaedia of Genes and Genomes) pathways associated with altered gut microbiota function in MS patients such as methane metabolism following probiotic supplementation.
At the immune level, probiotic administration induced an anti-inflammatory peripheral immune response characterized by decreased frequency of inflammatory monocytes, decreased mean fluorescence intensity (MFI) of CD80 on classical monocytes as well as decreased HLA-DR MFI on dendritic cells. Probiotic administration was also associated with decreased expression of MS risk allele HLA-DQA1 in controls. Probiotic induced increased in the abundance of Lactobacillus and Bifidobacterium were associated with decreased expression of MS risk allele HLA.DPB1 in controls.
Interpretation
Our results suggest that probiotic could have a synergistic effect with current MS therapies.
Introduction
The gut microbiome has been implicated in several autoimmune disorders including inflammatory bowel disease, rheumatoid arthritis and MS1–4. Studies in experimental autoimmune encephalomyelitis (EAE), the mouse model of MS, have shown that perturbation of the gut microbiota composition affects susceptibility to EAE5,6. Others have shown that colonization of mice with Bacteroides fragilis ameliorates EAE7 and that colonization of mice with segmented filamentous bacteria (SFB), a Th17 inducer, exacerbates EAE8. Tryptophan-derived AHR ligands produced by intestinal commensals can reach the CNS to modulate astrocyte function and suppress inflammation and neurodegeneration9. A recent study showed that the human gut derived commensal Prevotella suppresses EAE in an HLA class II transgenic mouse model10.
Alterations in the gut microbiome of MS patients has been reported by a number of investigators. We have found increased Methanobrevibacter and decreased Butyricimonas in the gut microbiome of MS patients1. Others have reported dysbiosis in the gut microbiome of MS subjects3,4. Recently, it was shown that high frequency of intestinal Th17 cells correlates with alterations in gut microbiota composition as well as increased disease activity in MS11. A twin study performed in discordant MS twins reported that the transfer of feces from the affected twin to germ-free mice induce more severe EAE symptoms than feces from the healthy twin12. Consistent with these fecal transfer experiments findings, another group observed that germ-free mice colonized with MS derived feces develop more severe EAE symptoms than mice colonized with feces from healthy subjects13. These findings suggest that manipulation of the gut microbiome could potentially be beneficial to MS patients.
The multistrain probiotic (probiotic LBS) contains 8 bacteria and has been administered to animals and humans with good safety profile. Probiotic LBS has been shown to induce IL-10+ and IL-10-dependent TGF-β-bearing regulatory cells in the gut of a mouse model of colitis14. Furthermore, LBS has been shown to promote an anti-inflammatory response in the gut of a mouse model of peanut allergy and diabetes15,16. A recent study conducted in ART-treated HIV-1 positive patients reports an association between LBS administration and decreased T cell activation in the peripheral blood and in gut associated lymphoid tissue17. LBS immunomodulatory properties extend to the central nervous system. For example, LBS has been shown to promote neuroprotection in a mouse model of traumatic spinal cord injury18. Another study reports that LBS administration was associated with decreased microglial cell activation as well as decreased CNS monocytes infiltration leading to improved sickness behavior in a mouse model of liver inflammation19. In humans, LBS has been shown to be beneficial to patients with pouchitis, ulcerative colitis and diabetes20–22. Little is known about the effect of LBS on peripheral immune function in humans. The goal of our study is to investigate the effect of LBS on the gut microbiome and peripheral immune function in healthy controls and MS patients.
Methods
Study population
RRMS patients and healthy controls were recruited from the Partners MS Center at Brigham and Women’s Hospital. RRMS subjects treated with glatiramer acetate (N=7); untreated (N=2) and healthy controls (N=13) were orally administered LBS twice daily (total 3,600 billion CFU/day) for two months. Subjects characteristics are shown in Table 1. All cases had relapsing-remitting MS (McDonald criteria) and none of them had an active relapse at the time of study enrollment. Exclusion criteria for both MS patients and controls were as follows: antibiotic or probiotic use within the past 3 months; corticosteroids use within the past month; interferon treatment within 30 days prior to initial visit; teriflunomide, fingolimod or dimethyl fumarate use within the past 3 months; natalizumab use within the past six months; treatment with rituximab, cyclophosphamide or other immunosuppressive drugs within the past year; history of gastroenteritis within the past month; travel to Latin America, Mexico, Africa or India in the past month; history of bowel surgery, inflammatory bowel disease, rheumatoid arthritis, systemic lupus erythematosus, type 1 diabetes or other autoimmune diseases; chronic medical disease other than MS compromising organ function; and pregnancy. A dietary survey was administered to all subjects before initiation of therapy and at the 2-month visit.
Table 1.
Demographics of Study Population
| Healthy | Multiple sclerosis | P-value (t-test) | |
|---|---|---|---|
| N=13 | N=9 | ||
| Age | 35±14 | 50±10 | 0.01 |
| Male (%) | 38.5 | 44.4 | |
| Female (%) | 61.5 | 55.6 | |
| Body mass index | 25.8±4.1 | 31.1±5.6 | 0.03 |
| Caucasian | 7 | 8 | |
| Black | 1 | 0 | |
| Others | 5 | 1 | |
| Disease Duration (years) | NA | 8.8±6.5 | |
| EDSS Score | NA | 1.4±0.9 | |
| Untreated | NA | 2 | |
| Glatiramer acetate | NA | 7 |
The high concentration probiotic formulation (900 billion Colony Forming Units (CFU) per sachet) we used contains four strains of Lactobacillus (L. paracasei DSM 24734, L. plantarum DSM 24730, L. acidophilus DSM 24735, L. delbruckei subspecies bulgaricus DSM 24734), three strains of Bifidobacterium (B. longum DSM 24736, B. infantis DSM 24737, B. breve DSM 24732) and one strain of Streptococcus (Streptococcus thermophilus DSM 24731). This probiotic was originally sold under the brand name VSL3 and since January 2016, it is sold in the USA under the brand name Visbiome, in Europe under the brand name Vivomixx and in Korea as DeSimone Formulation.
This study was approved by the Partners Human Research Committee and all participants signed an informed consent before any data collection or study procedure.
Sample collection
Blood and stool specimens were collected prior to, at discontinuation of therapy and 3 months thereafter (Fig 1). Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque density gradient centrifugation (Pharmacia LKB Biotechnology, Piscataway, NJ) and stored in liquid nitrogen until use.
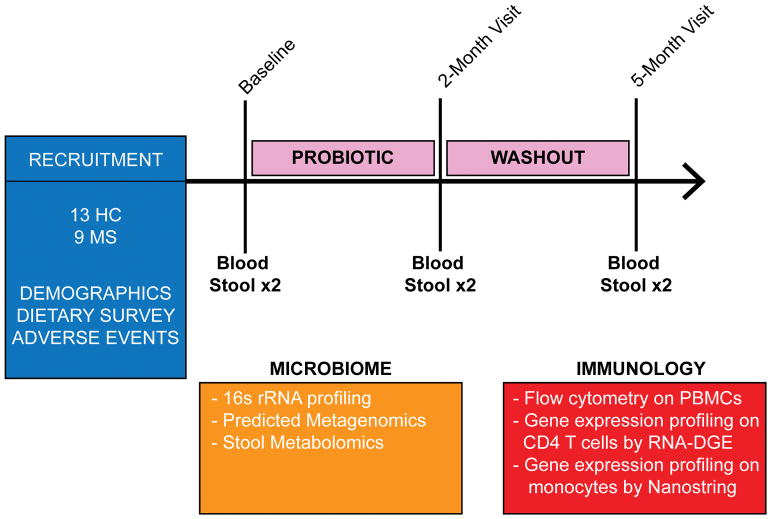
Figure 1. Study design flowchart.
Blood and fecal samples were collected from healthy subjects (n=13) and multiple sclerosis (MS) patients (n=9) prior to (baseline), at discontinuation of probiotic LBS (2-month visit) and 3 months thereafter (5-month visit). Bacteria DNA was extracted from feces samples obtained at all 3 time points and 16S rRNA sequencing was performed using Illumina MiSeq.16S data was used to obtain predicted metagenomics by PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States). Feces samples were also used for stool metabolomics profiling at all 3 time points. Immune cells profiling was performed in healthy subjects and MS patients at all 3 time points by flow cytometry. Gene expression profiling was performed on peripheral monocytes and CD4 T cells from healthy controls and MS patients using a Nanostring immunology panel array and high-throughput eukaryotic digital gene expression RNA-Seq (RNA-DGE) respectively.
Stool samples were obtained from patients by providing them with stool containers. Subjects collected two samples at each time point produced at any time of day with no specific dietary restrictions. Collection containers were then placed in boxes with provided ice packs for immediate shipment to our laboratory via overnight delivery at a maintained temperature of 0°C. On receipt of samples, they were frozen at −80°C until DNA extraction.
16S rRNA Microbial Community Profiling
Fecal DNA was isolated (MoBio PowerLyzer PowerSoil Kit) and the V4 region of the bacterial 16S rRNA gene was amplified using barcoded forward primers (515f, 926R) from the Earth Microbiome Project23 and http://www.earthmicrobiome.org/protocols-and-standards/16s/. Samples were sequenced by paired end 250 base pair reads at the Harvard Medical School Biopolymer Facility using the MiSeq platform (Illumina). Sequence quality was evaluated with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The median Phred quality scores were above Q30 for the first 230 basepairs, and above Q20 for the entire sequence for forward read, however, the median Phred Q scores for read 2 dropped below Q20 within the first 50 bases of the reverse read. Thus, we utilized only the forward read of 250 bases, corresponding to the V4 region of the microbial 16S rRNA. Downstream analysis was performed in QIIME (Qualitative Insights into Microbial Ecoloy)24. Sequences were de-multiplexed and quality filtered in which reads are truncated if two consecutive bases fall below a quality score of Q20 (1% error), and reads that are <75% of full length are discarded23. OTUs were picked using the open reference method sumaclust (http://metabarcoding.org/sumatra) and sortmeRNA25. Taxonomy is picked against the Greengenes 13.8 release database (http://greengenes.secondgenome.com) using a 97% similarity threshold. Alpha diversity was calculated with the phylogentic diversity_whole_tree, Shannon diversity, and richness metrics, and beta-diversity was calculated using weighted and unweighted UniFrac Distances.
Prediction Metagenomic
Metagenomic predictions were made using PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States)26 and summarized as KEGG (Kyoto Encyclopaedia of Genes and Genomes) pathways.
Stool metabolomics
Fecal samples were prepared for NMR analysis by vortex mixing for 5 min 80 mg of stool with 1 ml of deionized water. The obtained mix was then centrifuged for 15 min at 15K rpm and 4°C. 700μl of supernatant was added to 100 μl of a D2O solution of 3-(trimethylsilyl)-propionic-2,2,3,3-d4 acid sodium salt (TSP) 10 mM, used as NMR chemical-shift reference, buffered at pH 7.00 by means of 1M phosphate buffer. Finally, the so obtained sample was centrifuged again at the above conditions. 1H-NMR spectra were recorded at 298 K with an AVANCE III spectrometer (Bruker, Milan, Italy) operating at a frequency of 600.13 MHz. Following Ventrella et al., the signals from broad resonances originating from large molecules were suppressed by a CPMG-filter composed by 400 echoes with a τ of 400 μs and a 180° pulse of 24 μs, for a total filter of 330 ms27. The HOD residual signal was suppressed by means of presaturation. This was done by employing the cpmgpr1d sequence, part of the standard pulse sequence library. Each spectrum was acquired by summing up 256 transients using 32 K data points over a 7184 Hz spectral window, with an acquisition time of 2.28s. In order to apply NMR as a quantitative technique28, the recycle delay was set to 5s, keeping into consideration the relaxation time of the protons under investigation. The signals were assigned by comparing their chemical shift and multiplicity with the Human Metabolome Database29 and Chenomx software data bank (Chenomx Inc., Canada, ver 8.1). 1H-NMR spectra baseline-adjusted by means of the simultaneous peak detection and baseline correction algorithm (SPDBC) implemented in the baseline R package30. Briefly, the algorithm tries to iteratively locate peaks as consistent deviations from noise and interpolate over the spectra after peak removal. To each spectrum a linear correction was then applied to make the points pertaining to the baseline randomly spread around zero. No manual alignment of the signals was necessary, different to previous investigations31. Differences in water content among samples were taken into consideration by probabilistic quotient normalization32, applied to the entire spectra array.
FACS analysis
Frozen PBMCs from all time points of one individual were processed and analyzed together to minimize batch-to-batch variability and to allow for comparisons of MFIs. Surface staining of PBMCs was performed as previously described33. Non-purified CD4 T cells were activated with plate bound anti CD3 (5μg/mL, BD Biosciences), soluble anti human CD28 (1μg/mL, BD Biosciences) and IL-2 (20ng/ml, R&D Systems) for 48 hours. CD4 T cells were stimulated with PMA (50ng/mL, Sigma), ionomycin (250ng/mL, Sigma) and Golgistop for 4 hours. CD4 T cells were stained with anti-human CD4 PE-CY7 and violet fluorescent reactive dye (VVD, Life Technologies) and then cells were fixed and permeabilized with BD fixation and permeabilization buffer and stained for IL17 PE, IL-10 APC, IFN-γ FITC and GMCSF APC. Antibodies used for surface and intracellular staining of PBMCs are listed in the table below. Efluor 506 was obtained from BD Biosciences. All antibodies were titrated to obtain optimal signal-to-noise ratio. Samples were acquired using a BD LSR II flow cytometer (BD Bioscience) and analyzed using Flowjo software.
| Antibody | Company | Catalog # | Clone |
|---|---|---|---|
| CD4-PE-Cy7 | BD Biosciences | 560649 | RPA-T4 |
| CD4-FITC | BD Biosciences | 555346 | RPA-T4 |
| CD4-PE | Biolegend | 300508 | RPA-T4 |
| IL-10-APC | Biolegend | 506806 | JES3-19F1 |
| IL-17A-PE | Biolegend | 512306 | BL168 |
| IFNγ-FITC | BD Biosciences | 340449 | 25723.11 |
| GMCSF-APC | Biolegend | 502310 | BVD2-21C11 |
| CD3-Amcyan | BD Biosciences | 339186 | SK7 |
| CD25-PE | eBioscience | 12-0259-42 | BC96 |
| CD127-FITC | eBioscience | 11-1278-41 | EBioRDR5 |
| CD39-PECy7 | Biolegend | 328212 | A1 |
| LAP-PE | Biolegend | 349604 | TW4-2F8 |
| CD8-FITC | BD Biosciences | 555366 | RPA-T8 |
| CCR7-PE | BD Biosciences | 560765 | 150503 |
| CD45RA-APC | BD Biosciences | 550855 | HI100 |
| CD14-AF700 | BD Biosciences | 557923 | M5E2 |
| CD16-PE | BD Biosciences | 555407 | 3G8 |
| CD45-PE-Cy5 | BD Biosciences | 555484 | HI30 |
| LIN-FITC | BD Bioscience | 340546 | 3G8 |
| CD11c-APC | BD Biosciences | 559877 | B-ly6 |
| CD80-FITC | BD Biosciences | 557226 | L307.4 |
| HLA-DR-PE | BD Biosciences | 555561 | TU36 |
| CD303-PE-Cy7 | Biolegend | 354214 | 201A |
| CD3 | BD Biosciences | 555329 | UCHT1 |
| CD28 | BD Biosciences | 555725 | CD28.2 |
Gene expression studies
Monocytes and CD4 T cells were sorted from PBMCs of healthy controls (N=9 and N=7, respectively) and MS patients (N=7 and N=6, respectively) using Miltenyi Biotec positive selection kit Purified monocytes were treated with TNFα for 4 hours after which they were lysed and the expression of immune genes were analyzed using a Nanostring array (nCounter, Gene expression code set, Human immunology kit) as previously described33. Purified CD4 T cells were activated with plate bound anti CD3 (5μg/mL, BD Pharmingen), soluble anti human CD28 (1μg/mL, BD Pharmingen) and IL-2 (20ng/ml, R&D Systems). After 48 hours, CD4 T cells were stained with 7-aminoactinomycin D (7-AAD) viability dye to sort for viable CD4 T cells. Total RNA was extracted from viable T cells using the micro RNAeasy kit (Qiagen) and the RNA samples were submitted to the Broad Technology Labs for high-throughput eukaryotic digital gene expression RNA-Seq.
Statistical Analyses
Statistical differences for alpha and beta diversities were calculated with PRISM 7.0 software (GraphPad) using paired one-way ANOVA with the Tukey post-test to assess significance at the 0.05 level and to correct for multiple comparisons. Differences in beta-diversity visualized on the PCoA plots were calculated with the ADONIS test at 999 permutations34 using the R vegan package35. For Nanostring, FACS, stool metabolomics, compositional differences in taxonomic abundance and predicted metagenome studies, we assessed the change between time points in the healthy controls and MS patients using the Wilcoxon signed rank test. For this analysis, each pair of time points was compared, and the two groups were treated separately. To assess if the change in one of the outcome measures was associated with the change in another outcome measure, we calculated the relevant change for both measures and compared them using Spearman’s correlation coefficient. False discovery rate (FDR) was calculated using R software and the p. adjust function from the stats package.
Results
Product tolerability and safety
All study participants were greater than 90% compliant with probiotic supplementation. Patients were given the exact number of LBS sachets needed for the duration of the study and compliance was monitored by asking patients to return back unused sachets. No serious adverse reactions were reported by the patients or identified by the clinicians. None of the patients experienced a relapse during or after probiotic supplementation.
Administration of probiotic LBS is associated with changes in the structure and composition of the gut microbiome in healthy controls and multiple sclerosis subjects
To assess for temporal fluctuations in the gut microbiota composition, each subject provided two stool samples about 2–7 days apart at baseline, ON LBS and OFF LBS (6 stool samples in total per subject). We compared the 16S profile of the paired stool samples for each time point using the ADONIS test of clustering and found no difference in their community structure in both controls and MS patients (p>0.05 for all paired stool samples in both groups). Given that the temporal fluctuations in the gut microbiota composition were negligible, from here on, we merged the gut microbiome findings from the paired stool samples at each time point.
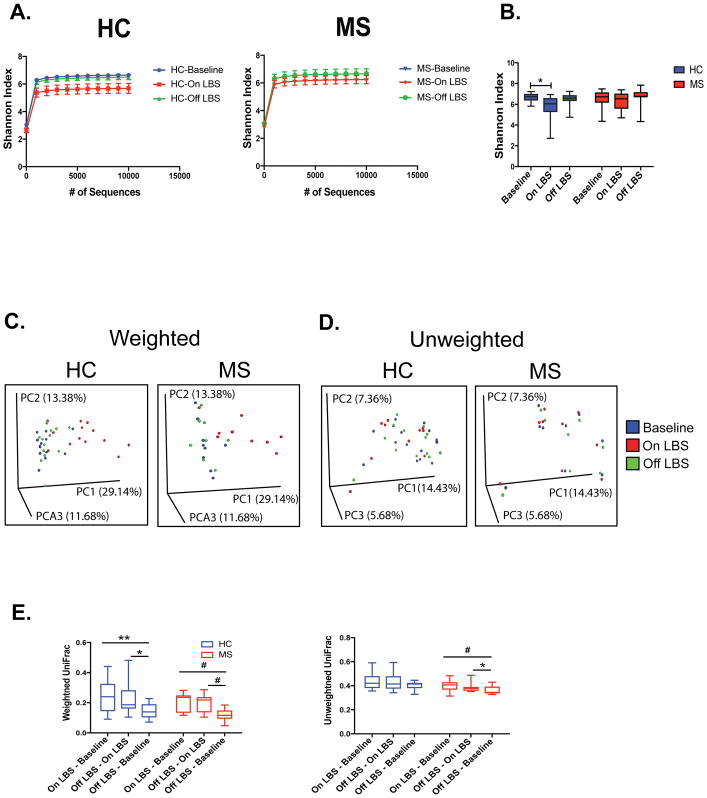
To assess for overall differences in microbial community structure in MS patients and controls, we calculated measures of alpha- and beta-diversity. Alpha diversity is a measurement of the ecological diversity within a given sample (i.e. types of microbial species in a microbiota sample), whereas beta-diversity compares the bacterial composition between samples (i.e. whether the microbial species are similar between microbiota samples). Shannon Index, an alpha diversity measurement of both richness (number of species present) and evenness (whether there are predominant and rare species or equal representation of all species), was measured at multiple sequencing depths using rarefaction curves (Fig 2A). We found that administration of LBS was associated with decreased alpha diversity in healthy controls (Fig 2B). No change in alpha diversity was observed in MS subjects following LBS administration (Fig 2B).
Figure 2. LBS effect on alpha and beta diversity of the gut microbiome.
Rarefaction curves were calculated at multiple sequence depths on MiSeq platform for Shannon entropy to compare differences in alpha-diversity at the indicated time points in A) healthy controls (n=13) and multiple sclerosis (MS) patients (n=9). Shannon index was also measured at a depth of 10000 reads in B) healthy controls (HC) and MS patients at the indicated time points determined by paired one-way ANOVA with Tukey’s post-test, *p<0.05. Principal coordinate analysis of weighted and unweighted UniFrac distances colored according to time points in C–D) healthy controls and MS patients. E) Average individual UniFrac distances calculated per subject assessed by paired one-way ANOVA with Tukey’s post-test, *p<0.05; **p<0.01; #p<0.1.
To determine whether overall microbial community structure was different among the 3 time points, we calculated differences in beta diversity using the unweighted and weighted UniFrac metric and visualized the communities by principal coordinate analysis (PCoA). LBS administration significantly changed the overall microbial community structure in controls, as assessed by the ADONIS test for clustering of weighted UniFrac distances (p=0.048, Fig 2C). We observed a similar trend in MS patients (p=0.10, Fig 2C). The overall microbial community structure shifted back to baseline level following discontinuation of LBS in controls and MS patients (p=0.58, p=0.54 respectively; Fig 2C). No shift was observed with unweighted UniFrac distances, which measures the presence versus absence of the microbiota, likely due to the fact that taxa closely related to the strains within LBS are known to inhabit the gut, albeit at lower levels (Fig 2D). We next computed UniFrac distances for each subject, comparing baseline, ON LBS, and OFF LBS microbiota samples (Fig 2E). Baseline compared to ON LBS, and OFF LBS compared to ON LBS had higher weighted UniFrac distances than comparing baseline to OFF LBS (paired one-way ANOVA with Tukey post-test), further suggesting that the individual microbiota compositions resembled their starting microbiota before treatment.
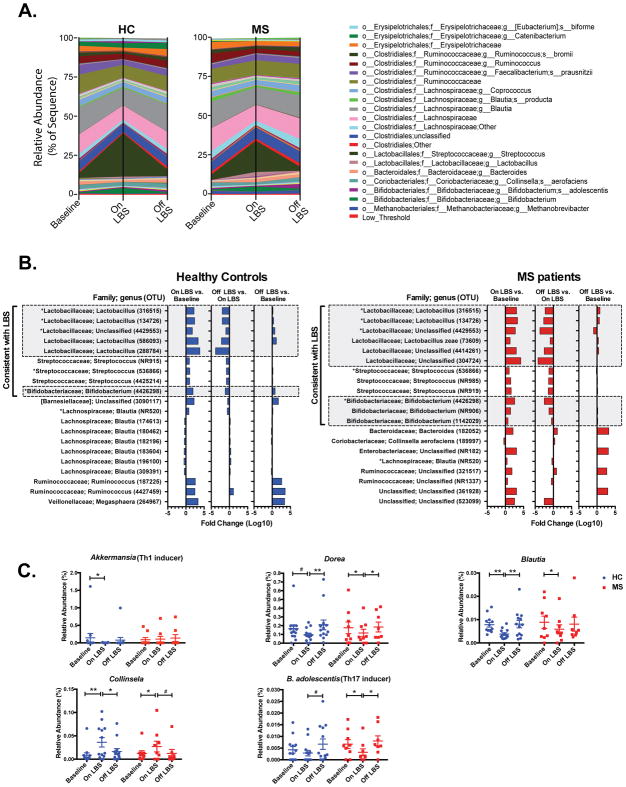
Consistent with these findings, we observed a change in the relative abundance of several genera following LBS supplementation in both controls and MS patients (Fig 3A). We noted an increase in the relative abundance of several operational taxonomic units (OTUs) following LBS supplementation with enriched taxa predominated by Lactobacillus, Streptococcus and Bifidobacterium (p<0.05, Fig 3B). We performed open reference OTU picking, thus reference based OTUs number and new reference (NR) OTUs unique to this study are listed in parentheses in Figure 3B.
Figure 3. LBS effect on gut microbiota composition.
Compositional differences in fecal microbiota before and after LBS administration in A) healthy controls (HC) and multiple sclerosis (MS) patients. B) Log10 fold change in abundance of top 20 most affected operational taxonomic units (OTUs) following LBS administration and discontinuation in controls and MS patients determined by Wilcoxon signed rank test at p<0.05. C) Effect of LBS on relative abundance of the indicated taxa in HC and MS patients determined by Wilcoxon signed rank test, *p<0.05; **p<0.01; #p<0.1. OTU = Operational taxonomic unit; NR = New Reference.
Of note, previous studies have reported decreased abundance of Lactobacillus genera in MS patients4. Furthermore, a recent study reported that supplementation of MS patients with a cocktail of three Lactobacillus species and one Bifidobacterium species was associated with improved EDSS score as well as decreased depression and stress36. Others studies conducted in the EAE mouse model have demonstrated that administration of a cocktail of Lactobacillus species and/or Bifidobacterium species either before or after EAE induction ameliorate EAE37,38. Hence, these studies suggest that increased in the relative abundance of Lactobacillus and Bifidobacterium taxa following LBS supplementation could be beneficial to MS patients.
Following LBS administration, we observed increase in the relative abundance of family Veillonellaceae in controls (Fig 3B) and genus Collinsela (Fig 3C), two taxa that are depleted in the gut of MS patients. LBS supplementation was also associated with decreased in the abundance of taxa that have been reported to be elevated in the gut of MS patients including genera Akkermansia (Th1 inducer), Blautia and Dorea (Fig 3B–C). We also observed decreased in the relative abundance of B. adolescentis (Th17 inducer), following LBS supplementation in controls and MS patients (Fig 3C). The relative abundance of most of these OTUs returned to baseline levels following discontinuation of LBS (Fig 3B–C). Thus, LBS supplementation can reverse MS induced alterations in the gut microbiota composition as well as inhibits the growth of pro-inflammatory bacteria.
Functional analysis of gut microbiota
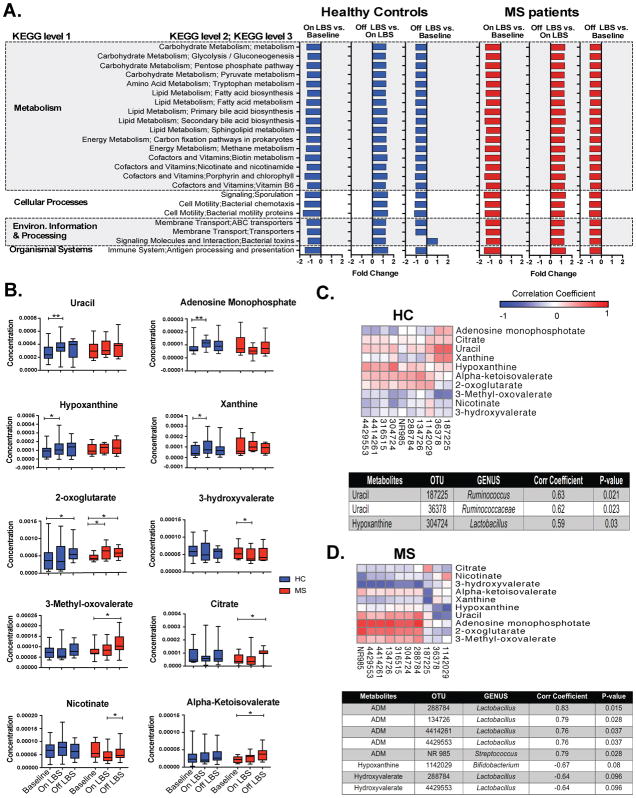
We used PICRUSt to assess the functional content of the microbiota based on the 16S data. The abundance of several KEGG pathways was decreased in both controls and MS patients following LBS administration including pathways related to Metabolism, Cellular Processes, Environmental Information and Processing and Organismal Systems (P<0.05, Fig 4A). ABC transporters, Carbon fixation pathways in prokaryotes, Porphyrin and chlorophyll metabolism and Sporulation are four KEGG pathways that have been reported to be increased in MS patients4. The abundance of these four pathways was reduced following LBS supplementation (p<0.05, Fig 4A). Methane metabolism is another pathway whose abundance was reduced following administration of LBS (p<0.05, Fig 4A). Methane is metabolized by methanogens and we have previously reported increased abundance of a methanogen, Methanobrevibacter smithii, in MS patients. Hence these findings suggest that LBS can reverse MS-related alterations in gut microbiota function.
Figure 4. LBS effect on gut microbiota function.
PICRUSt was used to infer the functional content of the gut microbiota based on 16S data. A) Fold change of the abundance of KEGG (Kyoto Encyclopaedia of Genes and Genomes) pathways following administration and discontinuation of LBS in healthy controls and multiple sclerosis (MS) patient assessed by Wilcoxon signed rank test with FDR (false discovery rate) <0.05. B) Effect of LBS on stool metabolites concentration in controls and MS patients determined by Wilcoxon signed rank test, *p<0.05; **p<0.01. Spearman’s correlations between microbiota abundances and stool metabolites concentration following LBS supplementation in C–D) healthy controls (HC) and MS patients. Corr Coefficient = Spearman’s correlation coefficient; OTU = Operational taxonomic unit; ADM = Adenosine monophosphate; NR = New Reference.
Next, we performed stool metabolomics profiling in controls and MS patients at all 3 time points. We noted increased concentration of uracil (p=0.006), adenosine monophosphate (p=0.027), hypoxanthine (p=0.040) and xanthine (p=0.017) following LBS supplementation in controls (fig 4B). We observed increased 2-oxoglutarate level (p=0.040) following discontinuation of LBS in controls (Fig 4B). In MS patients, we noted increased level of 2-oxoglutarate (p=0.039) following LBS administration (Fig 4B). We also found decreased 3-hydroxyvalerate level (p=0.027) following LBS administration in MS patients (Fig 4B). We observed increased level of 3-Methyl-oxovalerate (p=0.008), citrate (p=0.023), nicotinate (p=0.039) and alpha-ketoisovalerate (p=0.016) following LBS discontinuation in MS patients (Fig 4B). We then investigated potential associations between gut microbiota and metabolites changes. We correlated microbial abundance with a set of metabolites whose concentration were significantly changed following LBS administration. In controls, we found a positive correlation between two Ruminococcaceae OTUs (187225 and 36378) and uracil levels following LBS administration (Fig 4C). We also observed a positive correlation between Lactobacillus OTU 304724 and hypoxanthine concentration in controls (Fig 4C). In MS patients, we found an association between increased in the relative abundance of the following 4 Lactobacillus OTUs: 288784, 134726, 4414261 and 4429553) and adenosine monophosphate (Fig 4D). We also observed a positive correlation between Streptococcus NROTU 985 and adenosine monophosphate in MS patients (Fig 4D). We observed a trend toward a negative association between level of Bifidobacterium OTU 1142029 and hypoxanthine concentration in MS patients (Fig 4D). We also found a trend toward a negative association between two Lactobacillus OTUs: 288784 and 4429553 and hydroxyvalerate following LBS administration in MS (Fig 4D). Thus, changes in the gut microbiota composition following LBS administration were associated with changes in stool metabolomics profile in controls and MS.
Changes in T cells frequencies in peripheral blood
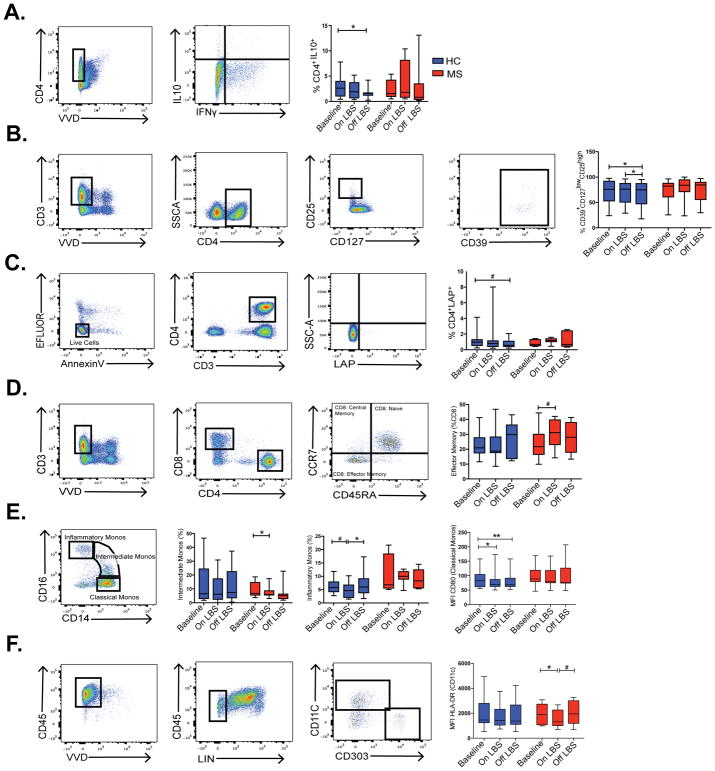
PBMCs were isolated from blood samples at all 3 time points and subsequently used for immune cell profiling by flow cytometry. Given prior reports of increased latency-associated peptide (LAP) and IL10+ T regulatory cells (T regs) in the gut following LBS administration, we conducted experiments to investigate the effect of probiotic LBS on peripheral T regulatory cells. No significant change in the relative frequencies of CD4+CD127lowCD25high T regs (data not shown) and CD4+ IL-10+ T regs were observed following administration of LBS (Fig 5A). Discontinuation of LBS was associated with decreased relative frequency of CD4+IL-10+ T regs (P=0.027, Fig 5A) as well as decreased relative frequency of CD39+CD127lowCD25high T regs (p=0.011 and p=0.048, Fig 5B) in controls. We also observed a trend toward decreased frequency of LAP+ T regs in controls (p=0.06, Fig 5C). Subsequently, we evaluated the frequencies of CD4+ and CD8+ T-cells. We found a trend toward increased in frequency of effector memory CD8 T cells following probiotic supplementation in MS patients (p=0.062, Fig 5D). The relative frequencies of Th1 and Th17 cells were trending down following LBS administration in both controls and MS patients (data not shown). We did not observe a change in the frequencies of naïve CD4 or naïve CD8 T cells, central memory CD4 T cells, effector memory CD4 T cells or central memory CD8 T cells following LBS supplementation in our cohort.
Figure 5. LBS effect on immune cells profile.
FACS analysis was used to compare the frequencies of A–D) CD4+ IL-10+ T cells, CD39+ CD127low CD25high T cells, CD4+ LAP+ T cells and effector memory CD8 T cells in healthy controls (HC) and multiple sclerosis patients (MS) at the indicated time points determined by Wilcoxon signed rank test. *p<0.05; #p<0.1. FACS analysis was used to compare E–F) the frequencies of intermediate monocytes, inflammatory monocytes as well as CD80 mean fluorescence intensity (MFI) on classical monocytes and HLA-DR MFI on dendritic cells in HC and MS patients at the indicated time points determined by Wilcoxon signed rank test, *p<0.05; **p<0.01; #p<0.1. The following markers were used for the lineage pool: CD3, CD14, CD16, CD19, CD20, CD56.
Next, we performed high-throughput eukaryotic digital gene expression RNA-Seq on CD4 T cells isolated from PBMCs of controls and MS patients. We did not observe a significant change in the expression of pro-inflammatory and anti-inflammatory cytokines in CD4 T cells following LBS administration in our cohort.
Changes in monocytes frequencies and activation in peripheral blood
Next, we performed immune cell profiling of monocytes from healthy control and MS patients at the 3 time points and found that administration of LBS was associated with decreased frequency of intermediate monocytes (CD14highCD16low) in MS patients (p=0.039, Fig 5E). We observed a trend toward decreased frequency of inflammatory monocytes after LBS supplementation in controls (p=0.068, Fig 5E). Discontinuation of LBS was associated with increased frequency of inflammatory monocytes in healthy controls (p=0.033, Fig 5E). In healthy controls, we also observed decreased MFI of costimulatory marker CD80 on classical monocytes (p=0.035, Fig 5E) following administration of LBS. In MS patients, we observed decreased MFI of HLA-DR on myeloid derived dendritic cells (CD45+LIN-CD11C+) following administration of LBS (p=0.0156, Fig 5F). Of note, we did not observe a change in the frequencies of B cells, NK cells, myeloid or plasmacytoid dendritic cells following administration of LBS in our cohort.
Gene expression profiling of peripheral monocytes
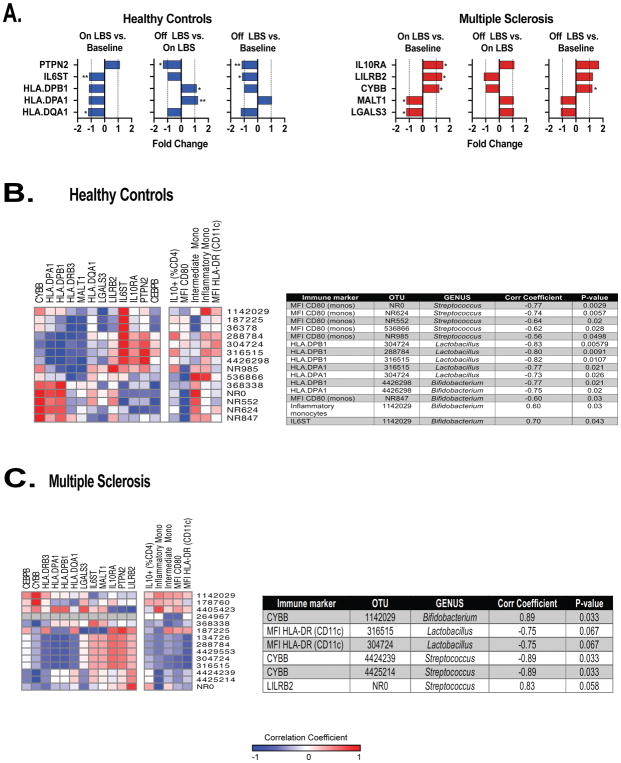
Next, we performed gene expression profiling in monocytes isolated from PBMCs of controls and MS patients using Nanostring immunology panel to measure the expression of 568 immune-related genes. We found decreased expression of MS risk allele HLA.DQA1 (P=0.011) as well as decreased expression of pro-inflammatory genes HLA.DPA1 (p=0.055) and IL6ST (p=0.0039) in monocytes following LBS administration in controls (Fig 6A). In controls, we found increased expression of HLA.DPA1 as well as MS risk allele HLA.DPB1 following discontinuation of LBS (p=0.0078 and p=0.039, respectively; Fig 6A). We found decreased expression of anti-inflammatory gene PTNP2 following discontinuation of LBS in controls (p=0.0078, Fig 6A). Of note, we continued to observe decreased expression of IL6ST following discontinuation of LBS (p=0.039, Fig 6A). We also continued to observe a trend toward decreased expression of HLA.DQA1 following LBS cessation (p=0.097, Fig 6A).
Figure 6. LBS effect on gene expression profile in peripheral monocytes.
A) Gene expression was measured in circulating monocytes using the Nanostring Immunology panel array at the indicated time points in healthy controls (n=9) and MS patients (n=7) assessed by Wilcoxon signed rank test, *p<0.05; **p<0.01. Spearman’s correlation between immune markers and operational taxonomic units (OTUs) relative abundance following LBS supplementation in B–C) healthy controls and MS patients. OTU = Operational taxonomic unit; NR = New Reference; Mono/monos = Monocytes; MFI = Mean fluorescence intensity; Corr coefficient = Correlation coefficient.
We found increased expression of anti-inflammatory genes IL-10RA (p=0.031), LILRB2 (p=0.031) and CYBB (p=0.031) as well as decreased expression of pro-inflammatory genes MALT1 (p=0.031) and LGALS3 (p=0.031) in monocytes following administration of LBS in MS patients (Fig 6A). The expression of CYBB gene remained decreased upon cessation of LBS (p=0.031, Fig 6A). We also observed a trend toward decreased expression of LILRB2 following LBS discontinuation (p=0.094, Fig 6A).
Thus, consistent with the findings from the FACS analysis showing that LBS administration was associated with decreased activation of monocytes as well decreased frequency of pro-inflammatory monocytes in controls and MS patients, at the transcriptomic level, we found decreased expression of pro-inflammatory genes in monocytes following LBS administration.
Correlations between gut microbiota and immune markers in peripheral blood
Next, we investigated potential associations between gut microbiota and immune changes. We correlated microbial abundance with a set of immune markers that were significantly changed following LBS administration. Given that most immune markers and microbial abundance changes were observed following LBS administration, we focused on that time point for our correlation studies. We found a negative correlation between CD80 MFI on classical monocytes and relative abundance of the following five Streptococcus OTUs: NR0 (p=0.0029), NR624 (P=0.0057), NR552 (p=0.02), 536866 (p=0.028), and NR985 (p=0.0498) in controls (Fig 6B). Decreased level of expression of HLA.DPA1 and HLA.DPB1 in monocytes was associated with increased relative abundance of the following three Lactobacillus OTUs: 304724 (p=0.026 and p=0.00579 respectively), 288784 (p=0.0091 for HLA.DPB1) and 316515 (p=0.021, p=0.0107 respectively) in controls (Fig 6B). We observed a negative correlation between relative abundance of Bifidobacterium OTU 4426298 and level of expression of HLA.DPA1 and HLA.DPB1 in monocytes from controls (p=0.02, p=0.021 respectively; Fig 6B). Bifidobacterium NR847 had a negative correlation with MFI CD80 on monocytes in controls (Fig 6B). In controls, Bifidobacterium OTU 1142029 had a positive correlation with MFI CD80 and IL6ST (p=0.03, p=0.043 respectively; Fig 6B). In MS, OTU 1142029 had a positive correlation with anti-inflammatory gene CYBB (p=0.033). Thus, OTU 1142029 was associated with pro-inflammatory immune markers in controls and with an anti-inflammatory marker in MS patients.
In MS, we found a trend toward a negative correlation between the following two Lactobacillus OTUs: 316515 and 304724 (p=0.066, p=0.066, respectively) and MFI of HLA-DR on dendritic cells (Fig 6C). CYBB had a negative correlation with the following two Streptococcus OTUs: 4424239 (p=0.033) and 4425214 (p=0.033) in MS patients (Fig 6C). We found a trend toward a positive correlation between Streptococcus NROTU0 and LILRB2 in MS patients (p=0.058, Fig 6C). Hence, Lactobacillus, Streptococcus and Bifidobacterium genera that were enriched in the gut of controls and MS patients following LBS supplementation had a negative correlation with pro-inflammatory and a positive correlation with anti-inflammatory immune markers.
Correlations between stool metabolites and immune markers in peripheral blood
We correlated stool metabolites concentration with a set of immune markers that were significantly changed following LBS administration. In controls, we observed that hypoxanthine had a negative correlation with HLA.DPA1, HLA.DPB1 and a positive correlation with PTPN2 (p=0.0020, p=0.031, p=0.037 respectively; Table 2). Of note, we mentioned above that hypoxanthine had a positive correlation with Lactobacillus OTU 304724. Hence, increased in the relative abundance of OTU 304724 is associated with elevated level of stool hypoxanthine which is associated with decreased expression of HLA.DPA1 and HLA.DPB1 in monocytes. We did not find significant correlations between stool metabolites and immune markers in MS patients following LBS supplementation.
Table 2.
Correlation Between Stool Metabolites and Immune Markers
| Immune marker | Metabolite | Corr Coefficient | P-value |
|---|---|---|---|
| HLA.DPA1 | Hypoxanthine | −0.90 | 0.0020 |
| HLA.DPB1 | Hypoxanthine | −0.73 | 0.031 |
| PTPN2 | Hypoxanthine | 0.72 | 0.037 |
| HLA.DQA1 | Hydroxyvalerate | −0.77 | 0.021 |
| HLA.DQA1 | Nicotinate | −0.60 | 0.096 |
Spearman’s correlation between immune markers and stool metabolites in healthy controls. Corr Coefficient = Spearman’s correlation coefficient.
Discussion
Our studies showed that LBS supplementation was associated with an enrichment of taxa depleted in MS including genus Lactobacillus in both controls and MS patients. We also observed a depletion of taxa associated with dysbiosis in MS such as genera Blautia and Dorea. In addition, LBS administration was also associated with a decreased in the abundance of KEGG pathways implicated in MS including methane metabolism, ABC transporters, Carbon fixation pathways in prokaryotes as well as Porphyrin and chlorophyll metabolism.
We found changes in stool metabolomics profile following LBS administration. Notably, increased levels of Lactobacillus OTU 304724 was associated with increased level of stool hypoxanthine level which was associated with decreased expression of MS risk allele HLA.DPB1 as well as HLA.DPA1.
At the immune level, LBS effect was predominantly seen on monocytes and dendritic cells. LBS administration induced an anti-inflammatory peripheral immune response characterized by decreased frequency of intermediate monocytes in MS patients as well as decreased MFI of costimulatory marker CD80 on monocytes and a trend toward decreased frequency of inflammatory monocytes in controls. Administration of LBS was also associated with decreased MFI of HLA-DR on dendritic cells in MS subjects. In controls, gene expression studies in monocytes revealed decreased expression of pro-inflammatory genes including MS risk alleles HLA.DQA1 and HLA.DBP1 following LBS supplementation. Similarly, in MS patients, we observed decreased expression of pro-inflammatory genes MALT1 and LGALS3 in monocytes following LBS administration. On the other hand, discontinuation of LBS had the opposite effect on peripheral immune function. Lactobacillus, Streptococcus and Bifidobacterium genera had a negative correlation with the following pro-inflammatory immune markers: CD80 MFI on classical monocytes, MFI HLA-DR on dendritic cells and expression of HLA.DPA1 and HLA.DPB1 on monocytes.
While our study provides initial insights into understanding the potential role for probiotics in the management of MS, our study has certains limitations. The small sample size may have precluded the detection of other important differences.
Given our limited sample size, the minimum attainable p-value for the Wilcoxon signed rank test for the comparison of baseline to on probiotics was 0.0039, and this p-value was attained for several of the OTUs. When we corrected for multiple comparisons using the false discovery rate (FDR), none of the OTUs were significant when controlling the FDR at 0.05, but this was due to the limit on the minimum attainable p-value from our sample size. For this reason, we did not calculate FDR for stool metabolomics, FACS, gene expression studies and correlations studies. Also, patients were stable and on treatment which may have made it difficult for us to observe the full anti-inflammatory potential of LBS.
However, the study has several strengths including the relative homogeneity of MS cases, the exclusion of conditions that can affect the gut microbiome and the fact that by study design, each individual served as their own control. Of note, the probiotic effect was similar in both groups despite the age and BMI differences.
Despite the study’s small size, our results have important implications. First that high dose probiotic supplementation is safe and well tolerated by MS patients taking glatiramer acetate. Second, that LBS supplementation can reverse MS induced alterations in the gut microbiota composition and function. Third, that probiotic LBS induced changes in the gut microbiome are associated with an anti-inflammatory peripheral immune response in controls and MS subjects. Hence, these findings suggest that LBS could have a synergistic effect with current MS disease modifying therapies. Of note, most of the LBS induced changes in the gut microbiome and peripheral immune system did not persist following discontinuation of LBS thereby implying that probiotic supplementation will need to be done continuously. This study is the first report on the effect of LBS on the gut microbiome and peripheral immune function in the context of MS. This study is a proof of principle study to investigate whether it is possible to modulate the gut microbiome of MS patients by using probiotics. Given that all the gut microbiome studies so far were conducted in RRMS patients, we focused on that patient population. We also opted for a simple study design in which all patients were on a single disease modifying drug (DMD) and we chose glatiramer acetate. It must be emphasized that we have no information on the long-term safety of LBS in the MS population and do not know if the beneficial changes in the gut and immune system will be maintained in MS patients who are on LBS for a long period of time. Additional studies are required to evaluate the long-term safety and immunomodulatory properties of LBS in both relapsing-remitting and progressive MS patients as well as in patients on different types of DMDs. Such studies will help determine whether there are synergistic effects of LBS with certain DMDs and not with others. In addition, it will also be important to determine if LBS induced anti-inflammatory peripheral immune responses are associated with improved clinical outcome in MS patients.
Acknowledgments
This study was sponsored by Teva Neuroscience and the Ann Romney Center for Neurologic Diseases.
Footnotes
Author contributions
Conception and design of study: HLW, JS, BH, RG, SC, BG, PK, SKT
Acquisition and analysis of data: SKT, KR, LL, BH, ET, IP, HL and LMC
Drafting of text or figures preparation: SKT, ET, LMC
Potential Conflicts of Interest:
This study was supported in part by Teva Neuroscience (Teva manufactures the drug glatiramer acetate used in this study).
References
- 1.Jangi S, Gandhi R, Cox LM, et al. Alterations of the human gut microbiome in multiple sclerosis. Nature Communications. 2016;7:1–11. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang D, Jia H, Feng Q, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015:1–13. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 3.Miyake S, Kim S, Suda W, et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS ONE. 2015;10(9):e0137429–16. doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, Chia N, Kalari KR, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Nature Publishing Group. 2016:1–10. doi: 10.1038/srep28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, et al. Role of Gut Commensal Microflora in the Development of Experimental Autoimmune Encephalomyelitis. The Journal of Immunology. 2009;183(10):6041–6050. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- 6.Berer K, Mues M, Koutrolos M, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479(7374):538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 7.Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, et al. Central Nervous System Demyelinating Disease Protection by the Human Commensal Bacteroides fragilis Depends on Polysaccharide A Expression. The Journal of Immunology. 2010;185(7):4101–4108. doi: 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
- 8.Lee YK, Menezes JS, Umesaki Y, et al. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108:4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothhammer V, Mascanfroni ID, Bunse L, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med. 2016;22(6):586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangalam A, Shahi SK, Luckey D, et al. Human Gut-Derived Commensal Bacteria Suppress CNS Inflammatory and Demyelinating Disease. CellReports. 2017;20(6):1269–1277. doi: 10.1016/j.celrep.2017.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cosorich L, Dalla-Costa G, Sorini C, et al. High Frequency of intestinal TH17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Sci Adv. 2017;3:1–9. doi: 10.1126/sciadv.1700492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berer K, Gerdes LA, Cekanaviciute E, et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci USA. 2017;114(40):10719–10724. doi: 10.1073/pnas.1711233114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cekanaviciute E, Yoo BB, Runia TF, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci USA. 2017;114(40):10713–10718. doi: 10.1073/pnas.1711235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Giacinto C, Marinaro M, Sanchez M, et al. Probiotics Ameliorate Recurrent Th1-Mediated Murine Colitis by Inducing IL-10 and IL-10-Dependent TGF-β-Bearing Regulatory Cells. The Journal of Immunology. 2005;174(6):3237–3246. doi: 10.4049/jimmunol.174.6.3237. [DOI] [PubMed] [Google Scholar]
- 15.Barletta B, Rossi G, Schiavi E, et al. Probiotic VSL#3-induced TGF-β ameliorates food allergy inflammation in a mouse model of peanut sensitization through the induction of regulatory T cells in the gut mucosa. Mol Nutr Food Res. 2013;57(12):2233–2244. doi: 10.1002/mnfr.201300028. [DOI] [PubMed] [Google Scholar]
- 16.Dolpady J, Sorini C, Di Pietro C, et al. Research Article Oral Probiotic VSL#3 Prevents Autoimmune Diabetes by Modulating Microbiota and Promoting Indoleamine 2,3-Dioxygenase-Enriched Tolerogenic Intestinal Environment. Journal of Diabetes Research. 2015:1–12. doi: 10.1155/2016/7569431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.d’Ettorre G, Rossi G, Scagnolari C, et al. Probiotic supplementation promotes a reduction in T-cell activation, an increase in Th17 frequencies, and a recovery of intestinal epithelium integrity and mitochondrial morphology in ART-treated HIV-1-positive patients. Immunity, Inflammation and Disease. 2017;26:2–17. doi: 10.1002/iid3.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kigerl KA, Hall JCE, Wang L, et al. Gut dysbiosis impairs recovery after spinal cord injury. J Exp Med. 2016;213(12):2603–2620. doi: 10.1084/jem.20151345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Mello C, Ronaghan N, Zaheer R, et al. Probiotics Improve Inflammation-Associated Sickness Behavior by Altering Communication between the Peripheral Immune System and the Brain. Journal of Neuroscience. 2015;35(30):10821–10830. doi: 10.1523/JNEUROSCI.0575-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bibiloni R, Fedorak RN, Tannock GW, et al. VSL#3 Probiotic-Mixture Induces Remission in Patients with Active Ulcerative Colitis. Am J Gastroenterology. 2005;100(7):1539–1546. doi: 10.1111/j.1572-0241.2005.41794.x. [DOI] [PubMed] [Google Scholar]
- 21.Gionchetti P, Rizzello F, Helwig U, et al. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology. 2003;124(5):1202–1209. doi: 10.1016/s0016-5085(03)00171-9. [DOI] [PubMed] [Google Scholar]
- 22.Jafarnejad S, Saremi S, Jafarnejad F, Arab A. Clinical Study Effects of a Multispecies Probiotic Mixture on Glycemic Control and Inflammatory Status in Women with Gestational Diabetes: A Randomized Controlled Clinical Trial. Journal of Nutrition and Metabolism. 2016:1–8. doi: 10.1155/2016/5190846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28(24):3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 26.Langille MGI, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ventrella D, Laghi L, Barone F, et al. Age-Related 1H NMR Characterization of Cerebrospinal Fluid in Newborn and Young Healthy Piglets. PLoS ONE. 2016;11(7):e0157623–13. doi: 10.1371/journal.pone.0157623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simmler C, Napolitano JG, McAlpine JB, et al. Universal quantitative NMR analysis of complex natural samples. Current Opinion in Biotechnology. 2014;25:51–59. doi: 10.1016/j.copbio.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wishart DS, Tzur D, Knox C, et al. HMDB: The Human Metabolome Database. Nucleic Acids Research. 2007;35(Database):D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liland KH, Almøy T, Mevik BH. Optimal choice of baseline correction for multivariate calibration of spectra. Applied spectroscopy. 2010;64(9):1007–1016. doi: 10.1366/000370210792434350. [DOI] [PubMed] [Google Scholar]
- 31.Barbara G, Scaioli E, Barbaro MR, et al. Gut microbiota, metabolome and immune signatures in patients with uncomplicated diverticular disease. Gut. 2017;66(7):1252–1261. doi: 10.1136/gutjnl-2016-312377. [DOI] [PubMed] [Google Scholar]
- 32.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic Quotient Normalization as Robust Method to Account for Dilution of Complex Biological Mixtures. Application in 1H NMR Metabonomics. Anal Chem. 2006;78(13):4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 33.Mazzola MA, Raheja R, Murugaiyan G, et al. Identification of a novel mechanism of action of fingolimod (FTY720) on human effector T cell function through TCF-1 upregulation. Journal of Neuroinflammation. 2015:1–12. doi: 10.1186/s12974-015-0460-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. [Google Scholar]
- 35.Oksanen J, Blanchet FG, Kindt R, et al. R package version 20–9. Vegan: community ecology package. [Google Scholar]
- 36.Kouchaki E, Tamtaji OR, Salami M, et al. Clinical and metabolic response to probiotic supplementation in patients with multiple sclerosis: A randomized, double-blind, placebo-controlled trial. Clinical Nutrition. 2017;36(5):1245–1249. doi: 10.1016/j.clnu.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 37.Salehipour Z, Haghmorad D, Sankian M, et al. Bifidobacterium animalis in combination with human origin of Lactobacillus plantarum ameliorate neuroinflammation in experimental model of multiple sclerosis by altering CD4+ T cell subset balance. Biomedicine & Pharmacotherapy. 2017;95:1535–1548. doi: 10.1016/j.biopha.2017.08.117. [DOI] [PubMed] [Google Scholar]
- 38.Lavasani S, Dzhambazov B, Nouri M, et al. A Novel Probiotic Mixture Exerts a Therapeutic Effect on Experimental Autoimmune Encephalomyelitis Mediated by IL-10 Producing Regulatory T Cells. PLoS ONE. 2010;5(2):e9009–11. doi: 10.1371/journal.pone.0009009. [DOI] [PMC free article] [PubMed] [Google Scholar]