Abstract
Glioblastoma is characterized by widespread genetic and transcriptional heterogeneity, yet little is known about the role of the epigenome in glioblastoma disease progression. Here, we present genome-scale maps of DNA methylation in matched primary and recurring glioblastoma tumors, using data from a highly annotated clinical cohort that was selected through a national patient registry.. We demonstrate the feasibility of DNA methylation mapping in a large set of routinely collected formalin-fixed paraffin-embedded (FFPE) samples, and we validate bisulfite sequencing as a multi-purpose assay that allowed us to infer a range of different genetic, epigenetic, and transcriptional characteristics of the profiled tumor samples. Based on these data, we identified subtle differences between primary and recurring tumors, links between DNA methylation and the tumor microenvironment, and an association of epigenetic tumor heterogeneity with patient survival. In summary, this study establishes an open resource for dissecting DNA methylation heterogeneity in a genetically diverse and heterogeneous cancer, and it demonstrates the feasibility of integrating epigenomics, radiology, and digital pathology for a national cohort, thereby leveraging existing samples and data collected as part of routine clinical practice.
Keywords: Glioblastoma, epigenetic heterogeneity, DNA methylation, disease progression, tumor microenvironment, transcriptional subtypes, bioinformatics, integrative data analysis, medical epigenomics
Introduction
Glioblastoma is a brain cancer with devastating prognosis. Even under the best available care, median survival is little more than one year, and very few patients live for more than three years1,2. Despite intense efforts, there has not been much therapeutic progress over the last decade, and a series of phase III clinical trials with targeted agents have failed to improve overall survival3–5.
Glioblastoma shows extensive temporal and spatial heterogeneity, which appears to contribute to therapeutic resistance and inevitable relapse. Prior research on tumor heterogeneity in glioblastoma has focused mainly on its genomic and transcriptomic dimensions6–15. The role of the epigenome in glioblastoma disease progression is much less understood, although recent studies using DNA methylation microarrays have identified characteristic differences in the DNA methylation profiles between subgroups of glioblastoma patients16–19.
Research in other cancers has demonstrated the power of DNA methylation sequencing for analyzing epigenetic heterogeneity. For example, DNA methylation heterogeneity has been linked to clonal progression in prostate cancer20, low-grade glioma21, esophageal squamous cell carcinoma22, and hepatocellular carcinoma23; and new measures of DNA methylation heterogeneity such as epi-allele burden, proportion of discordantly methylated reads (PDR), and DNA methylation inferred regulatory activity (MIRA) have been linked to clinical variables in acute myeloid leukemia24, chronic lymphocytic leukemia25, and Ewing sarcoma26.
To investigate the contribution of epigenetics to temporal and spatial heterogeneity in glioblastoma, we profiled DNA methylation in a glioblastoma progression cohort of isocitrate dehydrogenase (IDH) wildtype patients (primary glioblastoma) with matched samples from primary and recurring tumors (2-4 successive tumor resections per patient). This cohort was selected through the population-based Austrian Brain Tumor Registry27 and reflects routine clinical practice for glioblastoma at eight contributing medical centers across Austria.
DNA methylation profiling was performed on archival formalin-fixed paraffin-embedded (FFPE) samples using reduced representation bisulfite sequencing (RRBS)28–30. Compared to Infinium microarrays, our optimized RRBS protocol yields higher genomic coverage31 and works well on degraded DNA from FFPE samples of variable quality32. Moreover, it provides single-CpG and single-allele resolution: In RRBS, each sequencing read captures the DNA methylation status of individual CpGs in one single allele from one single cell, which enables high-resolution analysis of epigenetic tumor heterogeneity24–26.
Our full dataset comprises 499 RRBS-based DNA methylation profiles, of which 426 were derived from FFPE samples. These data not only provide a map of epigenetic heterogeneity in glioblastoma, but also allowed us to infer transcriptional subtypes, copy number aberration, MGMT promoter methylation, and glioma CpG island methylator phenotype (G-CIMP). Our study thus validates the feasibility and relevance of DNA methylation sequencing in large cohorts of routinely collected clinical FFPE tumor samples.
To put the DNA methylation data into a broader disease context, we obtained comprehensive patient and tumor data that allowed us to identify DNA methylation patterns predictive of immune cell infiltration and tumor microenvironment, progression-linked loss of DNA methylation at the promoters of Wnt signaling genes, and stronger survival associations for recurring tumors than for primary tumors.
In summary, our study provides a detailed picture of temporal and spatial heterogeneity in glioblastoma, a widely usable resource of DNA methylation profiles and associated annotation data, proof-of-concept for DNA methylation sequencing in large FFPE sample sets collected as part of routine diagnostics, and an integrative analysis of DNA methylation with various types of clinical, histopathological, and radiological information.
Results
DNA methylation sequencing in a cohort of matched primary and recurring glioblastoma tumor samples
To investigate the DNA methylation dynamics associated with glioblastoma disease progression, we established a richly annotated dataset of primary glioblastoma (wildtype IDH status) patients who underwent surgery at least twice, with tumor samples routinely collected during the initial tumor resection and at least once upon recurrence (Figure 1a). In total, 112 such patients were selected through the population-based Austrian Brain Tumor Registry27 (Fig. 1b and Supplementary Table 1) and included in our progression cohort.
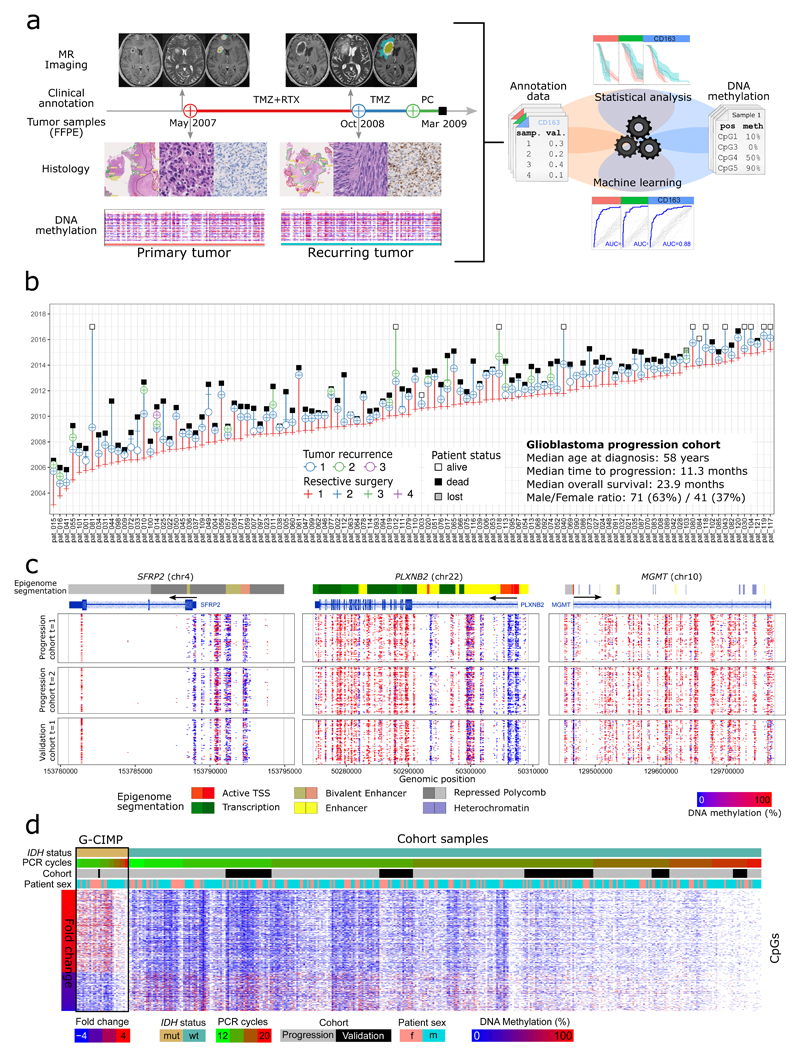
Figure 1. DNA methylation landscape of glioblastoma disease progression.
(a) Integrative analysis of longitudinal DNA methylation data (RRBS) with matched magnetic resonance (MR) imaging data (morphology, segmentation), clinical annotation data (treatment, progression, IDH mutation status), and histopathological data (segmentation, morphology, immunohistochemistry) using statistical methods and machine learning. TMZ: Temozolomide; RTX: Radiation therapy; PC: Palliative care.
(b) Overview of the progression cohort, summarizing the disease courses of 112 primary glioblastoma patients with IDH-wildtype status and at least two tumor resections (ordered by time of first surgery).
(c) DNA methylation profiles for primary (t=1) and recurring (t=2) tumors at three glioblastoma-linked gene loci (SFRP2, PLXNB2, MGMT). Gene annotations and epigenome segmentations for astrocytes were obtained from the WashU EpiGenome Browser (session URL: http://epigenome-segmentation.computational-epigenetics.org).
(d) DNA methylation levels at individual CpGs indicative of the glioma CpG island methylator phenotype (G-CIMP), shown for IDH-mutated reference samples (which are G-CIMP positive) and for the IDH-wildtype primary glioblastoma samples from the study cohort (which are G-CIMP-negative). The DNA methylation fold change between IDH-mutated and IDH-wildtype samples is based on published data for G-CIMP marker CpGs38.
Because a second surgery at relapse is rarely performed in patients who are frail or have many comorbidities, the patients in the progression cohort were on average younger (median age at diagnosis: 58 years) and lived longer (median overall survival: 23.9 months) than the average registry patient (median age at diagnosis: 63 years, median overall survival: 8 months). To control for this selection bias, we included a validation cohort of 105 primary glioblastoma patients (wildtype IDH status) who underwent resective surgery only once. These patients (median age at diagnosis: 65 years; median overall survival: 8.8 months) were much more representative of the unselected Austrian patient population (Supplementary Fig. 1a,b).
For each of these tumor samples, we established genome-scale DNA methylation profiles using RRBS, and we collected detailed time/sample-matched patient and tumor data including magnetic resonance (MR) imaging, quantitative pathology capturing tumor morphology, proliferative activity, and microenvironment of the tumors, and clinical variables including patient survival. In the presented analyses, we systematically integrated these datasets using statistical methods and machine learning (Figure 1a). All data are publicly available through the Supplementary Website (http://glioblastoma-progression.computational-epigenetics.org/).
RRBS profiling was successful for all tested samples, and 96% of the resulting DNA methylation profiles included more than 500,000 unique CpGs (Supplementary Table 2). The median number of unique CpGs for FFPE samples (1,839,096) was lower than for fresh-frozen samples (3,846,772) but higher than for an ethanol-based fixation method33 (1,005,828) tested by one of the centers that contributed tumor samples (Supplementary Fig. 1c and Supplementary Table 2). Bisulfite conversion rates, which assess the selective chemical conversion of unmethylated cytosines, were indicative of high-quality data: ~99% of genomic cytosines outside of CpGs were read as thymines, the mean underconversion rate on unmethylated spike-in controls was ~1%, and the mean overconversion rate on methylated spike-in controls was ~2% (Supplementary Fig. 1d and Supplementary Table 2).
The DNA methylation profiles showed the typical distribution of DNA methylation across promoters, enhancers, and genome-wide tiling regions (Supplementary Fig. 1e), with a tendency toward lower DNA methylation levels in lower quality samples (Supplementary Fig. 1f). To check for batch effects in the DNA methylation data34, we applied multidimensional scaling to the entire dataset and found the expected clustering of the samples by material type (FFPE vs. fresh-frozen) and disease diagnosis, while none of the investigated sources of batch effects (center, date, etc.) had a readily discernable effect on our dataset (Supplementary Fig. 1g).
Comparing locus-specific DNA methylation between matched primary and recurring tumors (Supplementary Fig. 1h), we observed high correlations across the genome (r > 0.94 for 5-kilobase tiling regions). Accordingly, zooming into selected genomic loci revealed inter-patient heterogeneity but no major differences between primary and recurring tumors (Figure 1c and Supplementary Fig. 2a, b).
We specifically investigated the promoter of the MGMT gene (Figure 1c), whose DNA methylation status correlates with sensitivity to alkylating chemotherapy35. The MGMT promoter was unmethylated in the majority of samples based on our RRBS data (Supplementary Fig. 2c), with the caveat that a robust and accurate assessment of DNA methylation at the MGMT promoter is technically challenging35–37. Patients with a methylated MGMT promoter in their recurring tumors showed significantly better progression-free survival (PFS) and overall survival (OS) compared to patients with unmethylated MGMT promoters (Supplementary Fig. 2d and Supplementary Table 3), and a similar association with OS was observed for the primary tumors when combining the progression and validation cohort (Supplementary Fig. 2d).
To provide further context for our IDH-wildtype primary glioblastoma cohort, we performed RRBS on primary and recurring tumors of 14 IDH-mutated patients who were diagnosed with oligodendroglioma, astrocytoma, or secondary glioblastoma from the same population. The DNA methylation profiles of these tumors showed G-CIMP characteristics as expected based on previous research38 (Figure 1d), thus providing additional validation for our genome-scale DNA methylation sequencing of FFPE material.
We also evaluated bioinformatic inference as a way of obtaining copy number aberration (CNA) profiles directly from RRBS data. We indeed detected various CNAs previously described in glioblastoma (Supplementary Fig 3a). Of particular interest, chromosome 10q deletions in recurring tumors (which affect MGMT and correlate with increased sensitivity to alkylating chemotherapy39) were associated with longer survival (Supplementary Fig. 3b). Based on the inferred CNA data, we also verified that none of our primary glioblastoma samples harbored the 1p19q co-deletion, thereby excluding the presence of any misclassified cases of anaplastic oligodendroglioma (Supplementary Fig. 3c). Finally, we validated the accuracy of our RRBS-based CNA analysis in 43 glioblastoma samples for which we generated low-coverage whole genome sequencing data, and we observed high concordance between both methods (Supplementary Fig. 4a,b).
Prediction of transcriptional subtypes in glioblastoma based on DNA methylation
Large-scale genome/transcriptome profiling efforts have defined three transcriptional subtypes of glioblastoma with distinct molecular and clinical characteristics: classical, mesenchymal, and proneural16,40. (A fourth “neural” subtype has also been described but appears to be an artifact of contaminating non-tumor tissue and has been disregarded in recent research15,41.) Because transcriptome profiling is challenging to perform on routinely collected FFPE samples, we tested whether these transcriptional subtypes can be inferred from RRBS data – which would remove the dependence on high-quality RNA for transcriptional subtyping (Figure 2a).
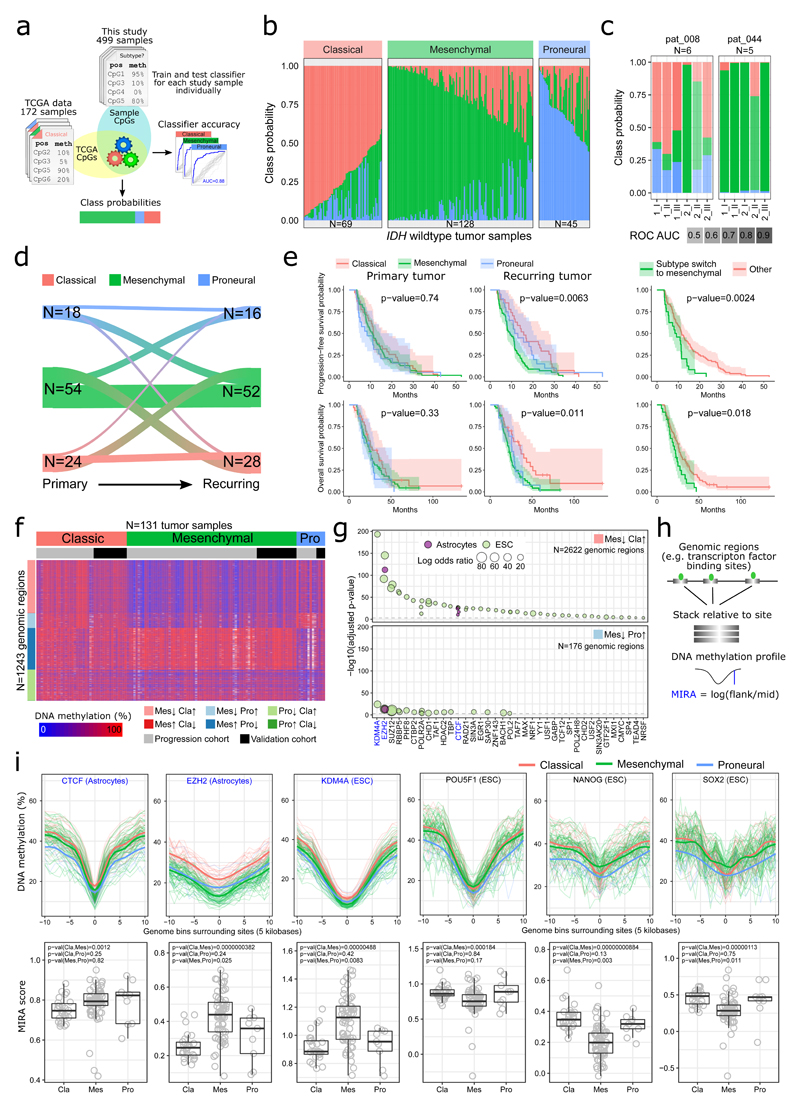
Figure 2. Glioblastoma transcriptional subtypes inferred from DNA methylation profiles.
(a) Overview of the machine learning approach for predicting transcriptional subtypes from DNA methylation.
(b) Transcriptional subtype heterogeneity in the progression cohort, as indicated by class probabilities of the RRBS-based subtype classifier. Samples are grouped and ordered by their dominant subtype.
(c) Distribution of class probabilities across different regions of the same tumor (indicated by Roman numbers) and across different surgeries (indicated by Arabic numbers) for two patients with multisector samples. N denotes the total number of tumor samples profiled for each patient.
(d) Riverplot depicting transitions in the predicted transcriptional subtype between primary and recurring tumors. The number of tumor samples (N) in each state is indicated. Only patients for which the sample-specific subtype classifier achieved high accuracy (ROC AUC > 0.8) were included in the analyses for panel d to f.
(e) Kaplan-Meier plots showing progression-free and overall survival for patients of the progression cohort stratified by predicted transcriptional subtypes (left) and subtype switching (right). The number of patients (N) is provided in Supplementary Table 3.
(f) Heatmap displaying the DNA methylation levels of the most differential CpGs between the three transcriptional subtypes, excluding CpGs with more than 100 missing values. Only tumor samples that were classified with high class probabilities (>0.8) for their dominant subtype were included in this analysis.
(g) LOLA region set enrichment analysis for differentially methylated CpGs between transcriptional subtypes (binned into 1-kilobase tiling regions). Region sets from astrocytes or embryonic stem cells with an adjusted p-value below 0.001 are shown.
(h) Schematic depicting the calculation of ‘DNA methylation inferred regulatory activity’ (MIRA) scores. High MIRA scores reflect local demethylation at the binding sites of a specific transcription factor across the genome, which indicates high regulatory activity of that factor.
(i) DNA methylation profiles (top row) and corresponding MIRA scores (bottom row) for three sets of transcription factor binding sites that were hypomethylated in the mesenchymal subtype (CTCF, EZH2, KDM4A) and for the binding sites of three key regulators of pluripotency (POU5F1/OCT4, NANOG, SOX2). The number of tumor samples (N) in each group is provided in Supplementary Table 4.
Using machine learning (L2-regularized logistic regression) and training data from The Cancer Genome Atlas (http://cancergenome.nih.gov/), we validated RRBS-based prediction of transcriptional subtypes in 37 fresh-frozen glioblastoma samples for which we generated matched RRBS and RNA sequencing data. Most samples were concordantly assigned by the DNA methylation-based and RNA-based classifiers (Supplementary Fig. 5a,b); and the receiver operating characteristic (ROC) area under curve (AUC) values exceeded 0.8 for all three transcriptional subtypes (Supplementary Fig. 5c). Among the (few) discordant cases, we observed reduced confidence in the RNA-based predictions, suggesting that the true accuracy of DNA methylation based subtype classification may be roughly on par with RNA-based classification (Supplementary Fig. 5d).
Training and applying individual classifiers for RRBS-based subtype prediction, we obtained subtype prediction probabilities with high ROC AUC values (>80%) for most tumor sample in our dataset (Supplementary Fig. 6a). According to these predictions, all three transcriptional subtypes were well represented among the IDH-wildtype primary glioblastoma samples of the progression and validation cohort (Fig. 2b and Supplementary Fig. 6b,c). In contrast, the IDH-mutant oligodendroglioma/astrocytoma/glioblastoma tumor samples were almost invariably assigned to the proneural subtype (Supplementary Fig. 6d).
Although the subtype prediction probabilities primarily reflect the confidence with which a sample was assigned to each of the transcriptional subtypes, we used them also as an indicator of the relative contribution of each subtype to individual tumor samples, thus providing an initial assessment of intra-tumor heterogeneity. Most tumor samples showed signatures of more than one transcriptional subtype (Figure 2b and Supplementary Fig. 6b), consistent with recent single-cell RNA-seq data that identified similar heterogeneity within individual samples11. Moreover, five out of six patients with multi-sector samples displayed at least two different predominant transcriptional subtypes (Figure 2c and Supplementary Fig. 6e-g), and about half of the patients changed their predominant transcriptional subtype between the primary and recurring tumor (Figure 2d).
Predicted transcriptional subtypes in the recurring tumor (but not in the primary tumor) were associated with patient survival (Figure 2e and Supplementary Fig. 6h) – the mesenchymal subtype being associated with the worst prognosis and the classical subtype with the best prognosis. Moreover, patients whose tumors switched to the mesenchymal subtype had particularly poor PFS and OS (Figure 2e).
To investigate epigenetic differences between transcriptional subtypes, we compared the DNA methylation profiles between tumors that were confidently assigned to one specific subtype (Figure 2f). Performing genomic region enrichment analysis for the differentially methylated CpGs using LOLA42, we identified strong enrichment of binding sites for the chromatin proteins EZH2, KDM4A, RBBP5, and SUZ12 among regions hypomethylated in the mesenchymal subtype (Figure 2g).
We also calculated ‘DNA methylation inferred regulatory activity’ (MIRA) scores26 for each tumor (Figure 2h). MIRA scores measure the local depletion of DNA methylation across all binding sites of a specific chromatin protein, and high MIRA scores indicate a strong dip in DNA methylation levels indicative of high regulatory activity of the chromatin protein. We observed significantly increased MIRA scores for CTCF, EZH2, and KDM4A in the mesenchymal subtype, while MIRA scores for key regulators of pluripotency (NANOG, SOX2, POU5F1) were decreased (Figure 2i; Supplementary Fig. 7a,b; Supplementary Table 4).
To investigate the role of EZH2 in more detail, we measured the fraction of EZH2 positive cells in the tumors by immunohistochemistry but did not observe an association with MIRA scores for EZH2 (Supplementary Fig. 7c) nor with transcriptional subtypes (Supplementary Fig. 7d). These results indicate that the increased regulatory activity of EZH2 in the mesenchymal subtype (predicted by DNA methylation depletion at EZH2 binding sites) is not the result of an increased fraction of EZH2 expressing cells but may rather reflect cell-intrinsic epigenome regulation.
Finally, to connect the differences observed between transcriptional subtypes to the wider landscape of epigenome regulation, we obtained cell type specific enhancer annotations from the Roadmap Epigenomics Project43, including embryonic stem cells (as an undifferentiated cell state) and astrocytes (used as reference due to the presumed similarity between astrocytes and the cell-of-origin from which glioblastoma develops). Applying MIRA analysis to these two sets of enhancer regions, we indeed observed characteristic, subtype-specific patterns of regulatory activity (Supplementary Fig. 7e-f).
Linking DNA methylation differences to changes in the tumor microenvironment
To test whether the RRBS profiles capture relevant aspects of the tumor microenvironment, we analyzed them together with matched histopathology and MR imaging data (Figure 3, Supplementary Figs. 8a-f and 9a-g). Specifically, the tumor microenvironment was assessed by immunohistochemistry for markers of immune cell types (CD3, T cells; CD8, cytotoxic T cells; CD68, macrophages), subpopulations (CD163, infiltrating macrophages; FOXP3, anti-inflammatory T cells; CD45ro, memory T cells), and functional characteristics (CD80, co-stimulatory signal to T-cell activation; Ki-67 / MIB-1: cellular proliferation status).
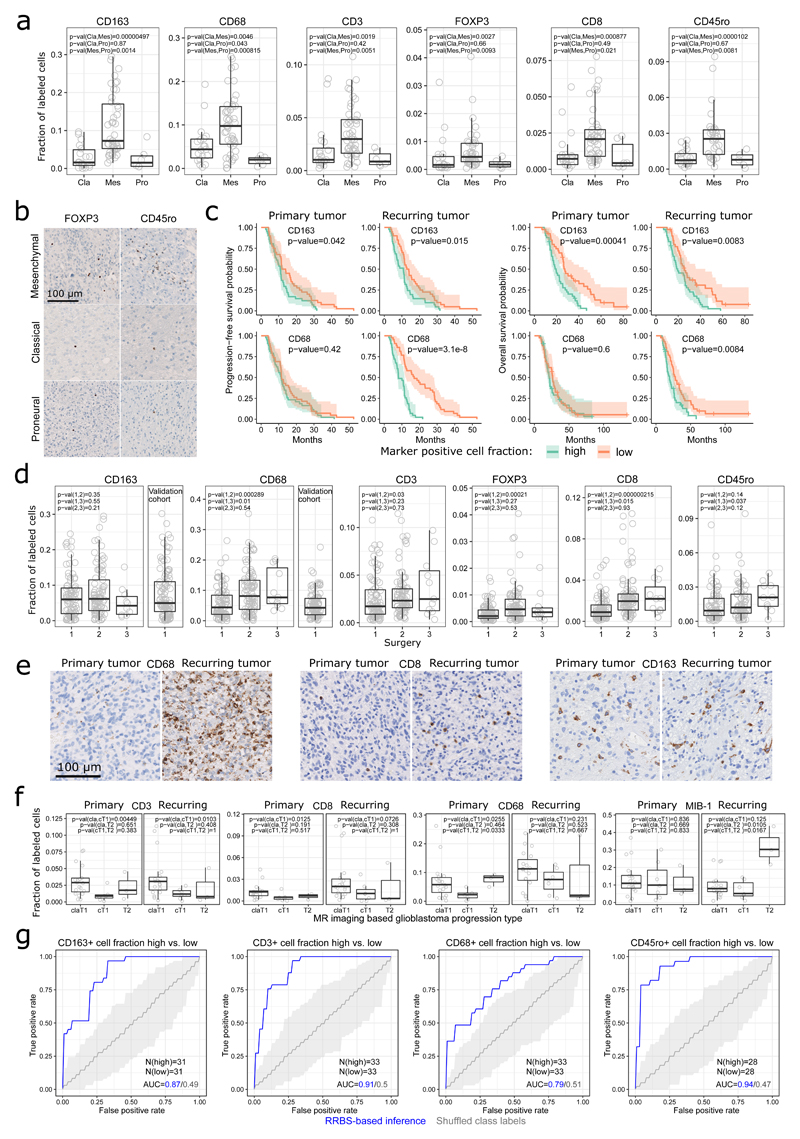
Figure 3. DNA methylation and the tumor microenvironment.
(a) Level of tumor-infiltrating immune cells across transcriptional subtypes (Cla: classical, Mes: mesenchymal, Pro: proneural), as measured by quantitative immunohistochemistry for the indicated marker proteins.
(b) Illustrative immunohistochemical stainings for FOXP3 and CD45ro in three individual patients (one for each subtype).
(c) Kaplan-Meier plots showing progression-free and overall survival for patients in the progression cohort stratified by the level of CD163 positive and CD68 positive immune cell infiltration. The number of patients (N) is provided in Supplementary Table 3.
(d) Level of tumor-infiltrating immune cell for samples originating from the patients’ first surgery (primary tumor), second surgery (recurring tumor), or third surgery (where available) in the progression cohort, and for the first (and only) surgery in the validation cohort.
(e) Illustrative immunohistochemical stainings for three marker proteins (CD68, CD8, CD163) in three individual patients undergoing with changing levels of tumor-infiltrating immune cells between primary and recurring tumors.
(f) Level of tumor-infiltrating immune cells (CD3, CD8, CD68) and proliferating cells (MIB-1 positive) compared between patients that were assigned to different progression types based on magnetic resonance (MR) imaging: Classic T1 (claT1), cT1 relapse / flare-up (cT1), and T2 diffuse (T2).
(g) ROC curves for DNA methylation based prediction of immune cell infiltration levels. N denotes the number of tumor samples in each group.
The number of tumor samples (N) for the association tests (panels a, d, f) is provided in Supplementary Table 4.
We observed significant differences in the immune cell infiltration between the three transcriptional subtypes (Figure 3a-b, Supplementary Fig. 8a). The highest number of immune cells was found in tumors of the mesenchymal subtype (in concordance with previous work44). This subtype also showed lower cell density and large necrotic areas in histopathology (Supplementary Fig. 8b-d), as well as a tendency toward increased tumor size, fewer vital tumor areas, and increased edema in the MR imaging data (Supplementary Fig. 9a-c).
Combining data across subtypes, high levels of CD68 positive cells (macrophages of all types) were associated with poor prognosis in recurring tumors; and high levels of CD163 positive cells (tumor infiltrating macrophages) were associated with poor prognosis in both primary and recurring tumors specifically in the progression cohort (Figure 3c, Supplementary Fig. 8f). Our results thus confirm the described prognostic value of CD68 and CD163 stainings45,46.
Comparing matched primary and recurring tumors, we observed significantly higher immune cell infiltration upon recurrence (Figure 3d,e); lower cell density and fewer necrotic areas in histopathology (Supplementary Fig. 8b,c,e); and decreased necrotic volume, decreased contrast-enhancing (active) tumor mass, increased edema, and stable tumor size in MR imaging (Supplementary Fig 9a,b,d). On average, patients with less necrotic or contrast-enhancing (active) tumor mass upon recurrence had longer progression-free survival (Supplementary Fig. 9e).
When we stratified patients according to prognostically relevant progression types (classic T1, cT1 relapse / flare-up, and T2 diffuse)47 based on MR imaging (Supplementary Fig. 9f,g), we found that primary and recurring tumors from patients displaying the cT1 relapse / flare-up subtype had lower infiltration of pro-inflammatory immune cell types (CD3, CD8, CD68) and a lower fraction of MIB-1-positive, proliferating cells (Figure 3f).
Using machine learning, we were able to predict various patient and tumor properties from the RRBS profiles (Figure 3g and Supplementary Fig. 10). For example, DNA methylation distinguished well between samples with high vs. low percentage of necrosis in histopathology (progression cohort ROC AUC = 0.87, validation cohort ROC AUC = 0.95). The same was true for high vs. low levels of specific immune cell infiltrates including CD163 positive cells (progression cohort 0.87, validation cohort 0.84), CD68 positive cells (progression cohort 0.79, validation cohort 0.75), CD45ro positive cells (progression cohort 0.94), CD3 positive cells (progression cohort 0.91), and CD8 positive cells (progression cohort 0.83). These results suggest that DNA methylation data can be used to infer immune cell infiltration in a similar way as it has been shown for RNA expression profiles15,48, with the advantage of full compatibility with routinely collected FFPE material.
Linking DNA methylation differences to tumor cell-intrinsic properties
Histopathological analysis assesses not only the tumor microenvironment but also tumor cell-intrinsic properties such as cell proliferation and subcellular morphology, which constitute an additional layer of information that we integrated with the DNA methylation data.
In our dataset, the percentage of proliferating cells (based on immunohistochemistry for MIB-1) was significantly lower in tumors of the mesenchymal subtype (Figure 4a) but showed no consistent changes between primary and recurring tumors (Supplementary Fig. 11a). Nevertheless, high proliferation in the recurring tumors (but not in the primary tumors) was associated with longer PFS (Figure 4b and Supplementary Fig. 11b). This perhaps counter-intuitive observation may be explained by the fact that early relapses tend to occur while the patient receives chemotherapy (which impedes cell proliferation); moreover, high proliferation may render tumors more sensitive to cytostatic chemotherapy.
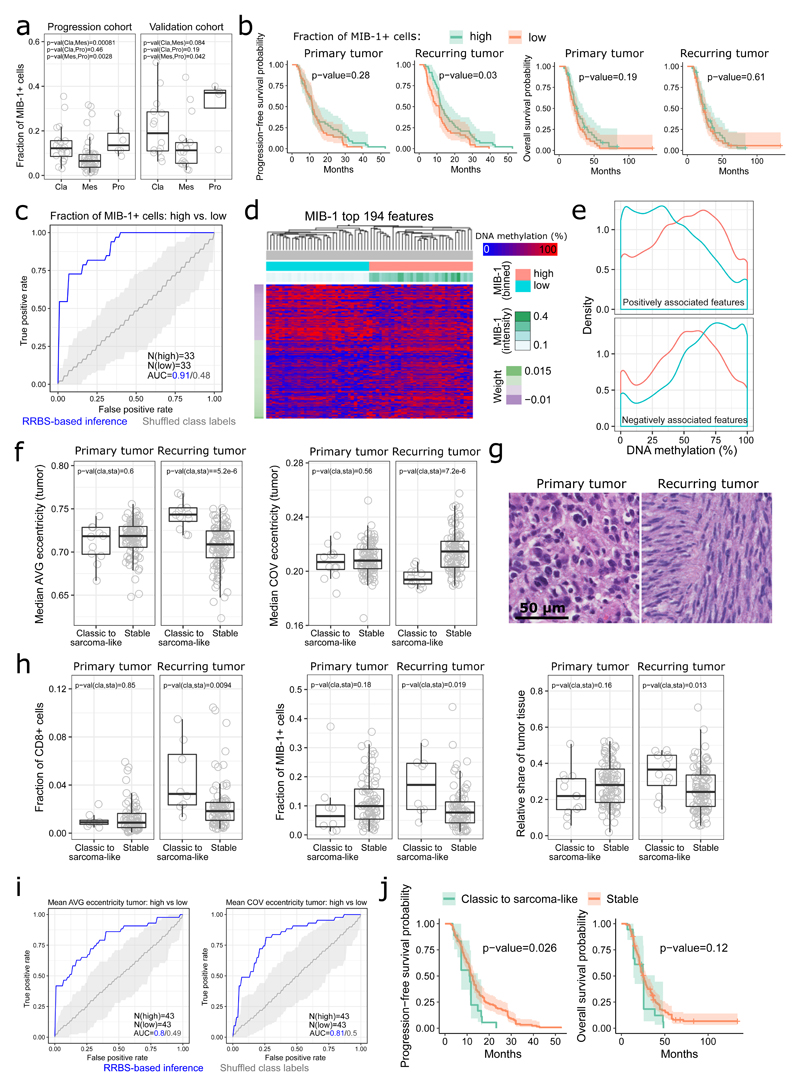
Figure 4. DNA methylation and histopathological tumor characteristics.
(a) Fraction of proliferating (MIB-1 positive) cells across transcriptional subtypes (Cla: classical, Mes: mesenchymal, Pro: proneural).
(b) Kaplan-Meier plots showing progression-free and overall survival stratified by the fraction of proliferating (MIB-1 positive) cells.
(c) ROC curves for DNA methylation based prediction of the fraction of proliferating (MIB-1 positive) cells. N denotes the number of tumor samples in each group.
(d) Clustered heatmap for the column-scaled DNA methylation levels of the most predictive genomic regions (5-kilobase tiling regions) from the classifier predicting the fraction of proliferating (MIB-1 positive) cells.
(e) Histogram showing the distribution of DNA methylation levels across the most predictive genomic regions from the classifier predicting the fraction of proliferating (MIB positive) cells.
(f) Average nuclear eccentricity (AVG) and its coefficient of variation (COV) for tumors that shift to a sarcoma-like phenotype during disease progression and those that retain a stable histological phenotype.
(g) Illustrative H&E stains of matched primary and recurring tumors from one patient that shifted to a sarcoma-like phenotype.
(h) Comparison of additional tumor properties (as indicated by the y-axis labels) between tumors that shifted to a sarcoma-like phenotype during disease progression and those that retained a stable histological phenotype.
(i) ROC curves for DNA methylation based prediction of average nuclear eccentricity (AVG) and its coefficient of variation (COV). N denotes the number of tumor samples in each group.
(j) Kaplan-Meier plots showing progression-free and overall survival stratified by whether or not their tumors shifted to a sarcoma-like phenotype.
The number of tumor samples (N) for the association tests (panels a, f, h) is provided in Supplementary Table 4.
The number of patients (N) in the Kaplan-Meier analysis (panels b, j) is provided in Supplementary Table 3.
DNA methylation patterns discriminated with high accuracy between tumors characterized by high vs. low proliferation rates (progression cohort ROC AUC = 0.91, validation cohort ROC AUC = 0.84) (Figure 4c and Supplementary Fig. 11c). This was not due to differences in mean DNA methylation levels indicative of global hypomethylation or hypermethylation (Figure 4d-e and Supplementary Fig. 11d). Rather, highly proliferating tumors showed intermediate DNA methylation levels at discriminatory regions, while tumors with low proliferation rates showed more extreme DNA methylation patterns (Figure 4e and Supplementary Fig. 11e).
Glioblastoma is a histologically polymorphic cancer, a property that we quantified by determining nuclear eccentricity (a measure of elongated shape) and the variability of this measure. Neither value was significantly different between primary and recurring tumors (Supplementary Fig. 11f). However, tumors that shifted to a sarcoma-like phenotype (i.e., secondary gliosarcoma)49 upon recurrence showed an increase in nuclear eccentricity and a decrease in its variability (Figure 4f-g), accompanied by an increase in CD8 immune cell infiltration, cell proliferation (MIB-1 positive cells), and relative tumor mass in the recurring tumor (Figure 4h).
DNA methylation patterns predicted nuclear eccentricity (ROC AUC = 0.8) and its variability (0.81) in the progression cohort (Figure 4i) but not in the validation cohort (where ROC AUC values were below 0.7 for both measures), which might be due to decreased variability of these features in the validation cohort (Supplementary Fig. 11f). Patients with shape-shifting tumors (i.e., classic to sarcoma-like) displayed significantly shorter PFS and a trend towards reduced OS (Figure 4j).
DNA methylation heterogeneity and temporal dynamics between primary and recurring tumors
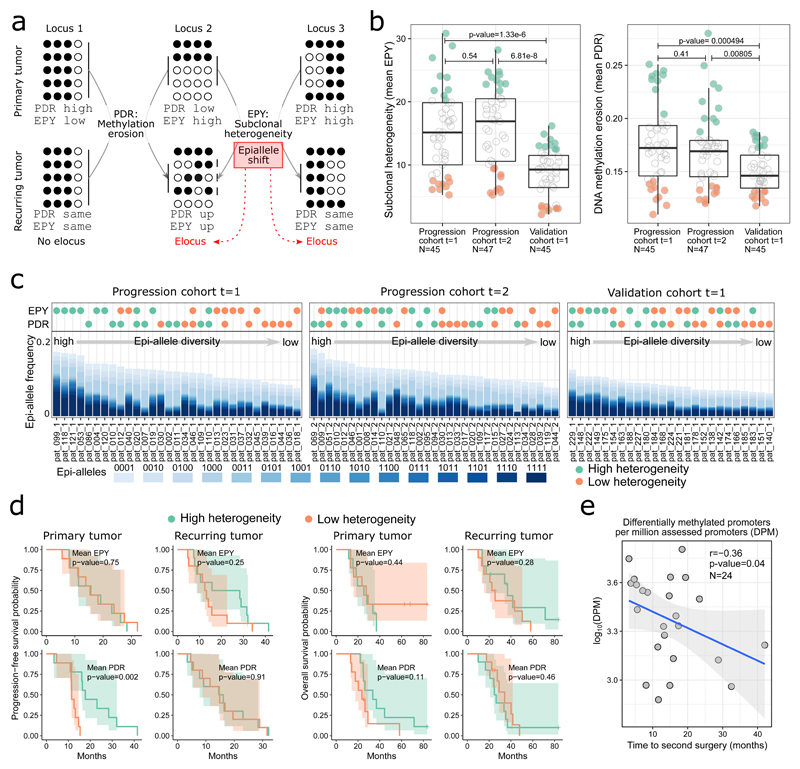
Each RRBS read captures the DNA methylation pattern of an individual cell, allowing us to investigate intra-tumor heterogeneity without the need for single-cell sequencing. We used two complementary approaches to investigate epigenetic heterogeneity at gene promoters (Figure 5a). First, erosion of DNA methylation patterns was measured by the proportion of discordant reads (PDR)25. This score is high for regions with many reads that contain both methylated and unmethylated CpGs. Second, subclonal heterogeneity was measured by epi-allele entropy (EPY)50. This score is high for regions with many different allelic DNA methylation patterns.
Figure 5. DNA methylation heterogeneity in glioblastoma disease progression.
(a) Illustration of ‘proportion of discordant reads’ (PDR) and ‘epi-allele entropy’ (EPY) as complementary measures of epigenetic tumor heterogeneity. Genomic regions can have high values for one but not the other measure (Locus 1 and 2, top row) or for both measures simultaneously (Locus 3, top row). Regions that undergo extensive changes in their epi-allele composition (i.e., in the distribution of subclonal DNA methylation patterns) are referred to as eloci50 (Locus 2 and 3, bottom row).
(b) Sample-wise PDR and EPY scores averaged across promoter regions for first surgery (t=1, primary tumor) and second surgery (t=2, recurring tumor) of the progression cohort and first (and only) surgery of the validation cohort. The samples with the 20% highest and lowest scores are shown in green and orange, respectively. N denotes the number of tumor samples.
(c) Relative epi-allele frequencies across promoter regions for all samples highlighted in green and/or orange in panel b. Samples are ordered by epi-allele diversity, and samples with high epi-allele diversity (left) tend to show high PDR and/or EPY (green dots). For clearer visualization, the “0000” majority epi-allele with a frequency of ~70% to 90% is not displayed. 0: unmethylated, 1: methylated
(d) Kaplan-Meier plots showing progression-free and overall survival stratified by PDR and EPY values, respectively. The number of patients (N) is provided in Supplementary Table 3.
(e) Scatterplot depicting the (negative) association of the number of differentially methylated promoters between primary and recurring tumor samples (normalized to the number of assessed promoters) and the time between first and second surgery. r: Pearson correlation. The p-value was calculated with a one-sided Pearson’s test. N denotes the number of tumor samples.
In the progression cohort, both approaches identified substantial intra-tumor heterogeneity and patient-to-patient variability, but no clear trend toward higher or lower heterogeneity in primary or recurring tumors (Figure 5b and Supplementary Fig. 12a). In the validation cohort (which comprises older patients with shorter survival), both heterogeneity measures had significantly lower values than in the progression cohort (Figure 5b).
To investigate the epi-allele composition from which the PDR and EPY scores are calculated (i.e., the distribution of observed DNA methylation patterns at four subsequent CpGs covered by the same RRBS read), we focused on the samples with the highest (green) and lowest (orange) mean PDR or EPY scores (Figure 5b). As expected for gene promoters, the most frequently observed epi-allele was consistently that of an unmethylated read (DNA methylation pattern: 0000). In contrast, there was large sample-to-sample variation in the ratio of epi-alleles that were fully methylated (1111) or heterogeneously methylated (e.g., 1001 or 0111) (Figure 5c, bottom panel). Samples with high mean PDR and/or EPY scores generally showed greater diversity in their epi-allele composition than those with low heterogeneity scores (Figure 5c, upper panel).
Heterogeneity scores (PDR and EPY) were weakly correlated with tumor size measured by MR imaging (Supplementary Fig. 12b), and we observed a significant positive association between increased DNA methylation erosion (mean PDR) and progression-free survival specifically in the primary tumors of the progression cohort (Figure 5d). In contrast, no such association was found for epi-allele entropy nor in the validation cohort (Figure 5d and Supplementary Fig. 12c).
We next investigated temporal heterogeneity by analyzing promoter-associated changes in DNA methylation between primary and recurring tumors of the same patient. Larger numbers of DNA methylation differences were observed for patients with a shorter time between first and second surgery (Figure 5e), suggesting that aggressive tumors may be characterized by high epigenetic plasticity.
The number of genomic regions that displayed changes in their epi-allele composition (so-called eloci50, illustrated in Fig. 5a) varied considerably between patients (Supplementary Fig. 12d); but these patient-to-patient differences were not associated with clinical outcome (Supplementary Fig. 12e) nor with the time span between first and second surgery (Supplementary Fig. 12f). The regions identified as eloci also did not show any evidence of selection based on their position relative to the nearest gene (Supplementary Fig. 12g). However, we did observe an enrichment of EZH2 binding sites among these regions (Supplementary Fig. 12h), providing further support for connections between EZH2 and epigenome deregulation in glioblastoma.
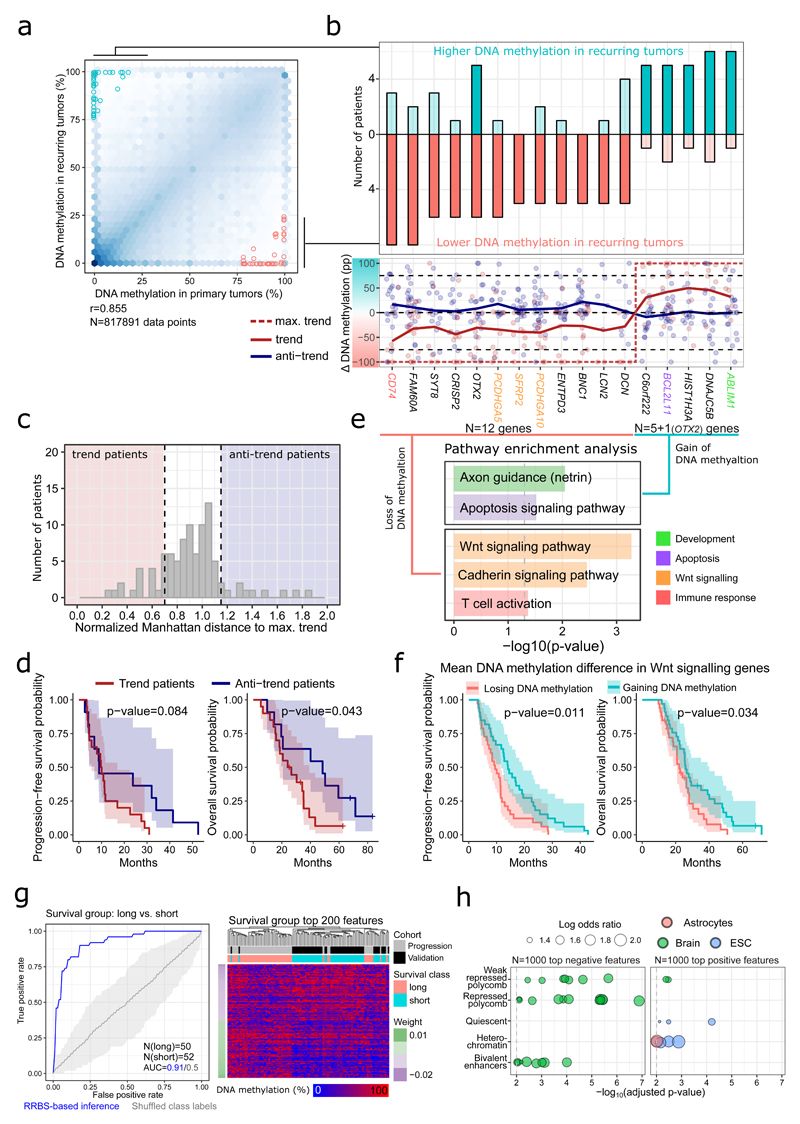
In search for regulatory mechanisms that may be involved in glioblastoma disease progression, we focused on the small number of promoters with strong progression-associated change in DNA methylation across multiple patients (Figure 6a). Most of these promoters showed consistent trends toward either gain or loss of DNA methylation upon recurrence (Figure 6b, upper panel). When we classified the individual patients into those that followed the cohort-level trend in differential DNA methylation (trend patients) and those that did not (anti-trend patients) (Figure 6b, lower panel, and Figure 6c), the trend patients showed worse prognosis (Figure 6d), suggesting that some of the observed differences may indeed contribute to aggressive recurrent tumors.
Figure 6. DNA methylation differences between primary and recurring tumors.
(a) Scatterplot depicting the association of DNA methylation at promoter regions between primary and recurring tumors. Promoters that were differentially methylated in at least five patients are highlighted. r: Pearson correlation. N denotes the number of data points (samples × promoters).
(b) Barplots (top) depicting the number of patients that show significant gain or loss of DNA methylation at the differentially methylated promoters highlighted in panel a; dot and line plots (bottom) showing the change in DNA methylation associated with disease progression (percentage points, pp) for patients following (red) or not following (blue) the cohort trend. Trend lines were calculated using the loess() function in R.
(c) Histogram showing the number of “trend” and “anti-trend” patients based on the Manhattan distance between the maximal trend at differentially methylated promoters (DNA methylation values of 0% or 100%, respectively) and the observed difference in DNA methylation for each patient.
(d) Kaplan-Meier plots showing progression-free and overall survival for trend and anti-trend patients according to panel c.
(e) Gene set enrichment analysis of those genes that recurrently gain (top) or lose (bottom) promoter DNA methylation during disease progression. N denotes the number of genes.
(f) Kaplan-Meier plots showing progression-free and overall survival stratified by progression-linked changes in DNA methylation at the promoters of Wnt signaling genes (top and bottom 30% of patients included).
(g) ROC curve and clustered heatmap for DNA methylation based prediction of overall survival, comparing patients with survival above the median in the progression cohort (“long”) to patients with survival below the median in the validation cohort (“short”). N denotes the number of tumor samples in each group.
(h) LOLA region set enrichment analysis for the top-2,000 tiling regions (5 kilobases) most predictive of overall survival. Region sets from astrocytes, brain tissue, or embryonic stem cells with an adjusted p-value below 0.001 are shown.
The number of patients (N) in the Kaplan-Meier analysis (panels d, f) is provided in Supplementary Table 3.
Biological pathway analysis identified an enrichment of annotations referring to neural development and apoptosis signaling among genes whose promoters gained DNA methylation during disease progression; in contrast, genes whose promoters lost DNA methylation were enriched in the Wnt signaling pathway and T cell activation (Figure 6e). Corroborating this observation, when we classified patients according to whether they gained or lost DNA methylation across the promoters of Wnt signaling genes, we observed a significant association between loss of DNA methylation and reduced survival (Figure 6f).
Finally, to test the global association between DNA methylation and glioblastoma patient survival, we trained and evaluated a machine learning classifier that distinguishes between the patient group with shortest survival (patients from the validation cohort whose survival was below the cohort median) and the patient group with longest survival (patients from the progression cohort whose survival was above the cohort median). Cross-validation confirmed the predictive power of DNA methylation (ROC AUC: 0.91, Figure 6g), and the predictive regions were enriched for poised enhancers and genomic regions that are polycomb-repressed in brain tissue or inactive (quiescent, heterochromatin) in astrocytes and embryonic stem cells (Figure 6h).
Discussion
Longitudinal analysis of matched tumor samples has great potential for charting cancer progression and therapy resistance, yet such cohorts are rare and difficult to obtain51. By leveraging a national patient registry and routinely collected FFPE tumor blocks, we established a progression cohort comprising 112 primary glioblastoma patients that had undergone at least two tumor resections. To account for bias toward younger and healthier patients in the progression cohort, we also assembled a validation cohort (n = 105) that is broadly representative of primary glioblastoma patients in the Austrian population. We established genome-scale DNA methylation maps for all tumors and collected matched clinical, histopathological, and MR imaging data, thereby enabling a comprehensive analysis of glioblastoma disease progression and epigenetic tumor heterogeneity.
From a technical perspective, our study was facilitated by an optimized RRBS protocol that provided single-CpG and single-allele DNA methylation data for a median of ~2 million and up to ~4 million unique CpGs based on small quantities of FFPE material. The RRBS data allowed us to infer a broad range of tumor properties, including known biomarkers such as G-CIMP status, MGMT promoter methylation, and chromosome 1p19q co-deletion. We also established the utility of RRBS data for predicting glioblastoma transcriptional subtypes16,40, thus extending this candidate biomarker to routinely collected FFPE samples (which are challenging to profile using RNA-seq). Finally, the single-allele resolution of RRBS provided initial insights into epigenetic tumor heterogeneity in glioblastoma, and it distinguishes our approach from recent studies that used the Infinium microarray platform for diagnostic classification of brain tumors17,52.
Connecting the DNA methylation data to a detailed histopathological characterization of the tumors, we found DNA methylation to be predictive of immune cell infiltration, extent of necrosis, and shape of tumor cell nuclei. Moreover, DNA methylation depletion at regulatory elements identified footprints of increased EZH2 binding activity in glioblastoma tumors of the mesenchymal subtype, which were characterized by the worst survival. We also observed associations between immune cell infiltration and MR imaging based subtypes of glioblastoma disease progression, and we identified DNA methylation patterns that predicted the percentage of proliferating cells inside a tumor, which was positively associated with progression-free survival.
Between primary and recurring tumors, patient-specific differences in the DNA methylation profiles were largely retained, but we found several recurrent progression-associated changes. For example, we observed characteristic demethylation of Wnt signaling gene promoters in a subset of patients, which was associated with worse prognosis. Aberrant activation of the Wnt signaling pathway is seen in various cancers including glioblastoma, where it has been linked to stemness, invasiveness, angiogenesis, and therapeutic resistance53.
DNA methylation erosion (measured by the PDR score) and subclonal heterogeneity (EPY score) were variable across patients but similar between primary and recurring tumors – arguing against a therapy-induced increase in epigenetic heterogeneity, but also against a decrease in epigenetic heterogeneity due to strong selective sweeps driven by therapy-resistant subclones. Nevertheless, we found an association between high levels of DNA methylation erosion in the primary tumors and longer progression-free survival, suggesting that epigenetic heterogeneity may not always be a driver of progression (as it seems to be the case in leukemia24,25) but may under certain circumstances become a liability for a fast-growing solid tumor such as glioblastoma.
We observed characteristic similarities and differences between the progression cohort with its bias toward younger and healthier patients, and the population-representative validation cohort. While the frequency and heterogeneity of transcriptional subtypes were similar in both cohorts, the measures of epigenetic heterogeneity were significantly lower in the validation cohort. Finally, DNA methylation patterns accurately distinguished between the longest surviving and the shortest surviving patients in the two cohorts.
In summary, our study establishes a rich and openly available resource characterizing the DNA methylation dynamics of glioblastoma progression in a highly annotated clinical cohort with matched MR imaging and detailed histopathological data. This work highlights the feasibility of establishing large, well-annotated patient cohorts out of routine clinical practice across multiple centers, utilizing a national patient registry and a DNA methylation assay that is compatible with routinely collected FFPE samples. Given that robust protocols are available for measuring DNA methylation in routine clinical diagnostics54, epigenetic biomarkers are likely to contribute to improved diagnosis, prognosis, and personalized therapy in glioblastoma and other cancers.
Online Methods
Sample acquisition
All primary glioblastoma patients (IDH wildtype status) were selected from the Austrian Brain Tumor Registry27 (Supplementary Table 1). For the progression cohort, only patients with a first surgery at diagnosis and at least one additional surgery upon recurrence were included; for the validation cohort, only patients with one surgery at diagnosis and no further resective surgeries were included. Tumor samples and clinical data were provided by the following institutions: Medical University of Vienna (including Hospital Rudolfstiftung Vienna), Kepler University Hospital Linz, Paracelsus Private Medical University Salzburg, Medical University of Innsbruck, University Hospital of St. Poelten, State Hospital Klagenfurt, General Hospital Wiener Neustadt, and Medical University of Graz. All samples were deposited in the neurobiobank of the Medical University of Vienna (ethics vote EK078-2004). The progression cohort comprised 159 patients with matched FFPE samples for the primary tumor and at least one recurring tumor. After screening for sufficient tumor purity (>50%), 47 patients were excluded and 112 patients were retained, each with at least two and up to four time points (283 tumor samples in total, including 6 patients with multi-sector sampling for spatial heterogeneity). The validation cohort comprised 105 patients with FFPE samples for the primary tumor. In all cases, the diagnosis of primary glioblastoma (IDH wildtype status) was confirmed by central pathology review according to the 2016 update of the WHO classification49 including targeted assessment of the IDH R132H mutational status. In addition to the primary glioblastoma patients, 14 patients (33 tumor samples) with IDH-mutated oligodendroglioma/astrocytoma/glioblastoma and 5 patients (5 brain samples) who underwent temporal lobe surgery due to epilepsy (Medical University of Vienna) were included as controls. Informed consent was obtained according to the Declaration of Helsinki, and the study was approved and overseen by the ethics committee of the Medical University of Vienna (ethics votes EK550/2005, EK1412/2014, EK 27-147/2015).
DNA and RNA isolation
Hematoxylin-eosin (H&E) stained slides of all available FFPE blocks for a given patient and surgery were assessed for tumor cell content and tumor purity by an expert neuropathologist, and the most suitable FFPE block was selected for DNA extraction (for the 6 patients with multi-sector sampling, several distinct regions from one FFPE block were processed as separate samples). Where the tumor purity in the selected FFPE block was lower than 50%, vital tumor without artificial damage was enriched by macro-dissection of the FFPE block (with secondary transferal of the region-of-interest to a new FFPE block) or by macro-dissection of tissue shavings. Five tissue shavings per FFPE block cut at a thickness of 5 μm each were used for DNA extraction with the QIAamp DNA FFPE Tissue Kit (Qiagen) following manufacturer’s instructions. In addition to the analysis of FFPE samples, matched DNA and RNA samples for validation of the subtype predictions were isolated from fresh-frozen tumor samples with the AllPrep DNA/RNA Mini Kit (Qiagen) following manufacturer’s instructions.
DNA methylation profiling by RRBS
RRBS was performed as described previously29 using 100 ng of genomic DNA for most samples, while occasionally going lower (down to 2 ng) or higher (up to 200 ng) when DNA quantity/quality was limiting (Supplementary Table 2). To assess bisulfite conversion efficiency independent of CpG context, methylated and unmethylated spike-in controls were added at a concentration of 0.1%. DNA was digested using the restriction enzymes MspI and TaqI in combination (as opposed to only MspI in the original protocol) to increase genome-wide coverage. Restriction enzyme digestion was followed by fragment end repair, A-tailing, and adapter ligation. The amount of effective library was determined by qPCR, and samples were multiplexed in pools of 10 with similar qPCR threshold cycle (Ct) values. The pools were then subjected to bisulfite conversion followed by library enrichment by PCR. Enrichment cycles were determined using qPCR and ranged from 12 to 21 (median: 16). After confirming adequate fragment size distributions on Bioanalyzer High Sensitivity DNA chips (Agilent), libraries were sequenced on Illumina HiSeq 3000/4000 machines using the 50 or 60 basepair single-read setup.
Low-coverage whole genome sequencing
Concentration and fragmentation of DNA extracted from fresh-frozen tumor tissue were assessed with the Qubit 2.0 fluorometric quantitation system (Life Technologies) and agarose gel electrophoresis, respectively. One sample with an insufficiently low DNA concentration was excluded. Libraries were prepared from 1 μg input material using the TruSeq DNA PCR-Free HT Library Prep Kit (Illumina) with IDT for Illumina TruSeq UD Indexes (Integrated DNA Technologies). Briefly, genomic DNA was sheared using a Covaris S220 focused-ultrasonicator instrument, DNA fragments were cleaned, end-repaired, and 3’ A-tailed, followed by ligation of the sequencing adapters. After quality control, individual libraries were diluted, equimolarly pooled, and sequenced on Illumina HiSeq 3000/4000 machines using the 50 basepair single-read setup.
RNA sequencing
Concentration of total RNA extracted from fresh-frozen tissue material was measured using the Qubit 2.0 fluorometric quantitation system (Life Technologies), and the quality was assessed by determining the RNA integrity number (RIN) with an Experion automated electrophoresis system (Bio-Rad). Samples with a RIN score below 5 were excluded (7 samples). Libraries for transcriptome profiling were prepared from a target amount of 1 μg input RNA with the TruSeq Stranded mRNA LT sample preparation kit (Illumina) using Sciclone and Zephyr liquid handling workstations (PerkinElmer). Final library concentrations were quantified with the Qubit 2.0 fluorometric quantitation system, and the fragment size distribution was assessed using the Experion automated electrophoresis System. Individual libraries were diluted and pooled equimolarly, followed by sequencing on Illumina HiSeq 3000/4000 machines using the 50 basepair single-read setup.
DNA methylation data processing
RRBS data were processed using a custom pipeline based on Pypiper (v0.2) (http://pypiper.readthedocs.io/) and Looper (v0.3) (http://looper.readthedocs.io/). Adapter sequences were trimmed, and 60 basepair reads were cropped to 50 basepairs using Trimmomatic (v0.32)55. Trimmed reads were aligned to the human reference genome (GRCh38) using BSMAP (v2.90) in RRBS mode56,57, and DNA methylation calling was performed with a Python script (biseqMethCalling.py) published previously29. To assess bisulfite conversion efficiency, unmapped reads were aligned to the spike-in reference sequences using Bismark (v0.12.2)58, and DNA methylation calls for methylated and unmethylated controls were extracted from the alignment file. CpGs in repetitive regions according to the UCSC RepeatMasker track were excluded from further analysis. DNA methylation was analyzed at single-CpGs resolution and in binned format with mean DNA methylation values calculated across 5-kilobase regions, CpG islands (as defined in the UCSC Genome Browser), enhancers in astrocytes (as defined by the Roadmap Epigenomics Project) or GENCODE promoter regions (1 kilobase upstream to 500 bases downstream of the annotated transcription start site).
DNA copy number analysis
Low-coverage whole genome sequencing reads were aligned to human reference genome (GRCh38) using BWA’s (v0.7.12) aln and samse commands59. Samtools (v1.4) was used for sorting and indexing the aligned short reads60. Copy number aberrations were detected with CNVkit (v0.9.1)61. This analysis relied on generating expected read counts in each genomic window (~50 kilobases) from a set of normal samples and calculating the log2 ratios of the read counts from the tumor samples relative to the normal samples. We used 20 whole genome samples from the Genom Austria project (http://www.personalgenomes.org/at) to generate expected normal/germline read counts for each genomic window. In order to make the high-coverage Genom Austria data comparable with the low-coverage tumor data, we down-sampled them to 36 million reads with 50 basepair read length. The batch pipeline recommended in the CNVkit manual (http://cnvkit.readthedocs.io/en/stable/pipeline.html#batch) was applied for generating reference values, calculating log2 ratios in the tumor samples, segmentation of the log2 ratios, and finally estimating the integer copy number values.
Copy number aberrations were also inferred from the RRBS data, using the R/Bioconductor package ‘CopywriteR’62 applied to the BSMAP-aligned BAM files (bin size: 100 kilobases). Data from five normal brain controls were merged at the level of aligned BAM files to serve as the shared control for all analyses. Each individual sample was normalized either against the merged control or against the cohort median, whichever showed the less extreme (i.e., more conservative) value for a given bin. Genomic segments identified by CopywriteR were classified as amplified or deleted if their normalized absolute copy number value deviated more than one cohort standard deviation (mean standard deviation across all bins in a given segment) from zero. Amplified or deleted segments for each sample were plotted in an overview graph sorted by segment length.
RNA-seq data processing
RNA-seq data were processed as described previously63: Adapter sequences were removed using trimmomatic (v0.32)55, only retaining reads with a minimum length of 25 basepairs after adapter trimming. Reads were aligned to the human transcriptome (GRCh38 including ncRNAs, Ensembl release 83) using Bowtie1 (v1.1)64, mapping up to 100 different positions for each read. Normalized transcript-wise expression estimates (RPKM values) were calculated based on the transcriptome alignments using the R package ‘BitSeq’ (v1.14.0)65, running the getExpression() function with the “uniform” option set to “FALSE”.
Gene annotation and enrichment analysis
Glioblastoma associated genes from a recent publication66 were annotated as oncogene, tumor suppressor gene, or drug resistance gene based on a published classification of cancer genes67. Genes not contained in this classification were manually annotated according to their known or suspected molecular functions as described in GeneCards (http://www.genecards.org). Enrichment analysis for gene sets and pathways was performed with Enrichr68,69, using an R interface (https://github.com/definitelysean/enrichR) to query the Panther_2016 database (http://www.pantherdb.org/) for enrichments with an adjusted p-value below 0.05.
Patient survival analysis
Kaplan-Meier survival analysis was performed using the functions survfit() and survdiff() of the R package ‘survival’. For continuous variables, patients with values above the median were classified as “high” and compared to those with values below the median (classified as “low”) unless indicated otherwise. Survival curves were plotted with ggsurvplot() from the R package ‘survminer’. The significance of differences in overall or progression-free survival was calculated using the log-rank test. Cox proportional hazards regression was performed using the function coxph() from the R package ‘survival’.
Inference of transcriptional subtypes from RRBS data
Glioblastoma transcriptional subtypes were predicted from DNA methylation data at the level of single CpGs using L2-regularized logistic regression as implemented in the R package ‘LiblineaR’. Classifiers were trained and evaluated on Infinium 27k DNA methylation data for glioblastoma tumors16 obtained from the TCGA data portal (https://portal.gdc.cancer.gov/). TCGA data were restricted to IDH-wildtype, non-G-CIMP tumors, resulting in a total of 172 samples included in the analysis. Furthermore, neural subtype samples were excluded because this subtype of glioblastoma had previously been associated with tumor margin and contamination with non-tumor brain tissue15,41. For each tumor sample in our cohort, a classifier was trained and evaluated on the TCGA data, using those CpGs that were covered also in the sample of which the transcriptional subtype was to be predicted (1,417 CpGs on average). This approach was successful for all but two samples in which too few shared CpGs were covered for reliable prediction. After performance evaluation by 10-fold cross-validation and calculation of the cross-validated receiver operating characteristic (ROC) area under curve (AUC) values, a final classifier was built using all selected TCGA samples. This classifier was used for RRBS-based prediction of the transcriptional subtype (including class probabilities) on the respective tumor sample.
Inference of transcriptional subtypes from RNA-seq data
To infer transcriptional subtypes from the RNA-seq data, we used the published set of subtype discriminatory genes, expression values, and subtype calls for the core TCGA samples from the study that first defined these transcriptional subtypes40. The corresponding data were obtained from the TCGA website (https://tcga-data.nci.nih.gov/docs/publications/gbm_exp/). We used these data to create reference expressions profiles for each of the three subtypes (classical: 38 samples, mesenchymal: 56 samples, proneural: 53 samples) by calculating subtype-wise average expression values for each of the 513 genes that had been assigned to one subtype. To simulate heterogeneity in terms of transcriptional subtype composition, we created all possible “mix profiles” (N = 5,151) in steps of 1% from the three “pure” reference profiles by calculating a weighted mean expression value for each of the genes. Finally, we calculated the Pearson correlation between the expression profile of the to-be-classified tumor samples and all “mix profiles”. For visualization, all correlations for a given sample were color-coded and plotted in a triangular coordinate system using the R package ‘ggtern’. The “mix profile” with the highest correlation score for a given sample was used to determine the majority transcriptional subtype, and the highest correlation score itself was used as a measure of confidence in the RNA-based transcriptional subtype inference.
Differential DNA methylation analysis
Differentially methylated CpGs between groups of tumor samples (e.g. transcriptional subtypes) were identified with a custom R script that uses a two-sided Wilcoxon rank-sum test. Groups containing less than five samples were excluded from the analysis, and only CpGs covered by at least five reads per sample in at least 30% of samples were included. CpGs in repetitive regions (“RepeatMasker”, “Simple Repeats”, and “WM + SDust” tracks from the UCSC Genome Browser, downloaded 6 September 2016) were also excluded. For the retained CpGs, differential DNA methylation between groups of samples was assessed using the wilcox.test() function in R, and p-values were adjusted for multiple testing using the Benjamini-Hochberg method (p.adjust() in R). CpGs with multiple-testing adjusted p-values smaller than 0.05 and with a median difference of beta values larger than 0.1 were considered significant.
Differentially methylated CpGs between sample pairs (i.e., primary tumor versus matched recurring tumor) were identified with a custom R script that uses a two-sided Fisher’s exact test. This test was applied to the methylated and unmethylated read counts derived from the BSMAP-aligned reads by the biseqMethCalling.py script. P-values were adjusted for multiple testing using the Benjamini-Hochberg method. To identify differentially methylated gene promoters, p-values were combined using a generalization of Fischer’s method70 as implemented in RnBeads71. Promoter-specific DNA methylation levels for each sample were calculated as the mean of all CpGs in the promoter region. Promoter regions were defined as the genomic region 1 kilobase upstream to 500 basepairs downstream of a given transcription start site as annotated by GENCODE72. High-confidence differentially methylated promoters (Figure 6) were defined by a DNA methylation difference greater than 75 percentage points, an adjusted p-value below 0.001, and an average RRBS read coverage greater than 20 reads. A cohort-level trend (“max. trend” in Figure 6b) of DNA methylation changes during progression was defined based on recurrently differentially methylated promoters (Figure 6b). “Trend” patients were defined as those whose DNA methylation profiles are most similar to the “max. trend” (low normalized Manhattan distance); “Anti-trend” patients were defined as those whose methylation profiles are most different from the “max. trend” (high normalized Manhattan distance) (Figure 6c).
Region set enrichment analysis using LOLA
To identify shared biologically patterns among the differentially methylated regions, the LOLA software was used to perform region set enrichment analysis against a compendium of publicly available region sets42. To reduce potential biases from co-located CpGs, CpGs were merged into 1-kilobase tiling regions across the genome prior to LOLA analysis. The hypermethylated or hypomethylated regions were used as the LOLA query set, and the set of all differentially methylated tiling regions were used as background (“universe”). For a focused analysis, only region sets from astrocytes (as a differentiated cell type related to glioblastoma) and embryonic stem cells (as a highly undifferentiated cell type) in the LOLA Core database were included. P-values were corrected for multiple testing using the Benjamini-Yekutieli (BY) method (p.adjust() in R), and all enrichments with an adjusted p-value below 0.001 were considered significant. In a control experiment, to assess potential effects of imbalance between hypermethylated and hypomethylated region sets, the analysis was repeated using the same number (defined by the minimum number of differentially methylated regions in either set) of highest-ranking regions from both sets. LOLA analysis was also applied to the regions identified as eloci in any patient. Here, all assessed loci were used as background (“universe”), all cell types in the LOLA Core database were included, and enrichments with a BY corrected p-value of below 0.001 were considered significant. Finally, for the analysis of regulatory regions predictive of survival, chromatin segmentations were obtained from the Roadmap Epigenomics Project43, transferred from genome assembly hg19 to hg38 using the liftOver tool73, and used in the region set enrichment analysis in replacement of the LOLA Core database. All regions considered by the machine learning classifier were used as LOLA background (“universe”), only region sets from astrocytes, brain tissue, and embryonic stem cells were included, and enrichments with a BY corrected p-value below 0.001 were considered significant.
DNA methylation inferred regulatory activity (MIRA)
MIRA scores for selected sets of transcription factor binding sites from the LOLA Core database42 or epigenome regulatory region sets from the Roadmap Epigenomics Project43 (transferred from genome assembly hg19 to hg38 using the liftOver tool73) were calculated as described previously26,74. Briefly, aggregated DNA methylation profiles around the center of the designated sites (2.5 kilobases upstream and downstream, split into 21 bins) were created for each sample and for each set of transcription factor binding sites or genomic segmentation regions. MIRA scores were calculated as the log ratio between aggregated DNA methylation values for the center bin (bin 0, reflecting the center of the designated site) and the average of two flanking bins (bins -5 and +5).
Immunohistochemistry
FFPE blocks were cut at a thickness of 3 μm, and sections were stained on a Dako autostainer system using the following antibodies: CD3 (1:200 Thermo Scientific #RM-9107-S1), CD8 (1:100 Dako Cytomation #M7103), CD45Ro (1:500 Dako Cytomation #M0742), CD80 (1:100 Abcam #ab1341120), CD163 (1:1000 Novocastra #NCL-L-CD163), CD68 (1:5000 Dako Cytomation #M0814), HLA-DR (1:400 Dako Cytomation #M0775), Ki-67 (MIB-1) (1:200 Dako Cytomation #M7240), CD34 (1:100 Novocastra #NCL-l-END). Antigens were retrieved by heating the sections in 10 mM sodium citrate (pH 6.0) at 95°C for 20 min, followed by antibody incubation for 30 min at room temperature. The Dako FLEX+ Mouse detection system was used according to manufacturer’s recommendations. For antibodies against IDH1 (1:60 Dianova #DIA-H09) and FoxP3 (1:25 BioLegend #320116), a Ventana BenchMark automated staining system was used, followed by visualization using the Ultra View detection kit. All sections were counterstained with hematoxylin. EZH2 (1:50 Cell Signaling Technology #5246) stainings were performed manually starting with antigen retrieval by heating the sections in 10 mM sodium citrate (pH 6.0) at 95°C for 30 min, followed by antibody incubation for 12 hours at 4°C and visualization with the Dako EnVision detection system. For each antibody, positive and negative controls were included in every 30-slide-batch. For negative controls, the primary antibody was omitted and the Universal Negative Control rabbit (Dako) for polyclonal rabbit antibodies or purified mouse myeloma IgG1 (Zymed Laboratories, San Francisco, CA) for monoclonal mouse antibodies was used.
Histopathological analysis of whole slide scans
Slides were scanned using a Hamamatsu NanoZoomer 2.0 HT slide scanner. The Hamamatsu NDP.view2 software was used to manually annotate relevant regions in the H&E slide scans (necrosis, hemorrhage, preexisting brain parenchyma, fibrotic scar, and artificially damaged tissue) by an expert neuropathologist. The remaining tissue area was assigned to the tumor. The H&E slide scans were downsampled to 10× magnification and exported using the NDPITools plugin in Fiji75–77 along with XML files featuring the annotations. To obtain a cell nuclei mask, the Color Deconvolution method of Fiji was used to obtain an 8-bit greyscale image of the hematoxylin stain78. Automated local Phansalkar thresholding was used to segment cell nuclei (parameters k = 0.2, r = 0.5, radius = 8)79, followed by the binary Close and Open operations. The Watershed method was used to separate clustered nuclei80.
MATLAB R2014b (MathWorks) was used for further image analysis. The binary nucleus mask was analyzed in blocks of 160 × 160 pixels (~146 μm × 146 μm). This block size was empirically found to provide a good compromise between spatial resolution and nuclear content required for statistical analysis. The centroids of the nuclei were localized and the nuclear density as well as morphological measures (nucleus area and eccentricity) were assessed in each block (using MATLAB’s regionprops() function). In each block, the mean, standard deviation, coefficient of variation, median, and mode of the aforementioned characteristics were calculated. Using the binary annotation masks, each block was assigned to its corresponding region if >90% of the pixels in that block were uniformly annotated. Subsequently, for each annotated region, the mean, standard deviation, coefficient of variation, median, and mode of the previously obtained block-specific statistical parameters were calculated.
For automated analysis of scanned immunohistochemical slides, whole slide scans were downsampled to 5× magnification. First, the Color Deconvolution method of Fiji was used to separate the hematoxylin from the DAB stain. The 8-bit greyscale hematoxylin- and DAB- images were then each thresholded using Phansalkar thresholding, and stained cells/nuclei were counted with the Analyze Particles algorithm. For each slide, DAB+ cells were normalized to the total cell count.
Radiological evaluation of glioblastoma patients
MR images of sufficient quality for first diagnosis as well as recurrence were available for 54 of the glioblastoma patients included in this study, which were contributed by six radiology departments. The dataset included T1-weighted images with contrast enhancement (CE) and fluid-attenuated inversion recovery (FLAIR)/T2-weighted axial images, which were reviewed for topographic tumor location to assess solitary vs. multicentric tumors and local vs. distant recurrences. Multicentric glioblastomas were defined as at least two spatially distinct lesions that are not contiguous and whose surrounding abnormal FLAIR/T2 signals do not overlap81. Tumor segmentation was performed with BraTumIA82, which uses multi-modal MRI sequences for fully automated volumetric tumor segmentation. T1, T1 contrast enhanced, T2, and FLAIR sequences were used to segment four tumor tissue types: necrotic, cystic, edema/non-enhancing, and enhancing tumor. Due to differences in MRI protocols across the study sites, the multi-modal sequences were affine registered to the T1 sequence with SPM122 and resampled to 1 mm × 1 mm × 3 mm voxel size prior to segmentation. The BraTumIA-derived segmentations were reviewed by an expert radiologist, and errors in the automatic segmentation were manually corrected.
Evaluation of MR imaging-based progression phenotypes
MR imaging-based tumor progression was assessed according to the Response Assessment in Neuro-Oncology (RANO) standard83. Serial T1-weighted images with CE and FLAIR/T2-weighted images were available for 43 patients. Progression subtypes were classified as described previously47,84: (i) Classic T1 (incomplete disappearance of T1-CE during therapy followed by T1-CE increase at progression), (ii) cT1 relapse / flare-up (complete disappearance of T1-CE during therapy followed by T1-CE reoccurrence at progression), (iii) primary non-responder (increase and/or additional T1-CE lesions at first MR imaging follow-up after start of therapy), (iv) T2-circumscribed (bulky and inhomogeneous T2/FLAIR progression, no or single faintly speckled T1-CE lesions at progression), and (v) T2-diffuse (complete decrease in T1-CE during therapy but exclusive homogeneous T2/FLAIR signal increase with mass effect at progression).
DNA methylation based prediction of tumor properties
Tumor properties such as immune cell infiltration and tumor cell morphology were predicted from DNA methylation data using a machine learning approach based on the R package ‘LiblineaR’. The DNA methylation data were prepared by calculating for each sample the mean DNA methylation levels in 5-kilobase tiling regions across the genome. Tiling regions covered in less than 90% of the samples and samples covering less than 80% of the selected tiling regions were excluded from the analysis, and the filtered data matrix (samples × tiling regions) was subjected to imputation using the function impute.knn() from the R-package ‘impute’, with the parameter k (i.e., the number of nearest neighbors considered) set to 5. Tumor properties represented by continuous response variables were converted into categorical variables by setting the 20% highest values to ‘high’, the 20% lowest values to ‘low’, and the remaining samples to ‘NA’ (missing value). Imputed beta values were used to train and evaluate the classifiers using LiblineaR(). In the confirmatory hierarchical clustering based on the most predictive features identified by the classifiers, the beta values were scaled across samples prior to clustering, but displayed in the heatmap as unscaled beta values for better visualization and comparability. LiblineaR() was set to use support vector classification by Crammer and Singer85 as model type, and the appropriate cost parameter was estimated from the imputed data matrix using the function heuristicC() from the same package. For each tumor property, the performance of the classifiers was determined through 10-fold cross-validation, and 100 control runs with randomly shuffled labels were included to detect potential overfitting. ROC curves and ROC AUC values were determined using the functions prediction() and performance() of the R-package ‘ROCR’. Finally, we trained a classifier on the entire dataset using the selected model and cost parameter, which was then used for further analysis including the extraction of the most predictive features, hierarchical clustering, and the prediction of additional samples (for the transcriptional subtypes and for model testing in the validation cohort).
Estimation of DNA methylation heterogeneity
The proportion of discordant reads (PDR), a measure of DNA methylation erosion and disorder, was calculated as described in the original publication25. Briefly, the number of concordantly or discordantly methylated reads with at least four valid CpG measurements was determined for each CpG using a custom Python script. The PDR at each CpG was then calculated as the ratio of discordant reads compared to all valid reads covering that CpG. CpGs at the end of a read were disregarded to remove potential biases due to the end-repair step of RRBS library preparation. Because the PDR and epi-allele entropy (EPY) calculation is highly sensitive to differences in the read composition of the underlying RRBS library, we focused this analysis on RRBS libraries with a similar number of PCR cycles (13-15) to ensure high consistency between samples (Supplementary Fig. 12a).
The epi-allele entropy (EPY), a measure of subclonality within a tumor, was calculated using a slightly modified version of methclone (v0.1)50. We calculated epi-allele entropies separately for each of the samples and, independently, for each matched pair of primary and recurring tumors. Input files to methclone were created by aligning the trimmed RRBS reads to the human reference genome (GRCh38) using Bismark58. To find suitable thresholds for read coverage and entropy change, we performed a series of analyses with read thresholds set to 20, 40, and 60 reads and entropy change thresholds set to -40,-50,-60,-70,-80, and -90. We did not observe significant differences in the results except for the expected effects at the extremes of the spectrum. Therefore, we chose a moderate read threshold of 40 reads for methclone to consider a locus, and loci with a combinatorial entropy change below -80 were classified as epigenetic shift loci (eloci) between primary and recurring tumors or normal brain control samples (as described in the original publication)50. For each pair, we then calculated epi-allele shifts per million loci (EPM), dividing the number of eloci by the total number of assessed loci normalized to 1 million loci50.
Sample-wise PDR and epi-allele entropy values were calculated by averaging across all promoters that were covered in more than 75% of the samples. Promoter regions were defined as the genomic region 1 kilobase upstream to 500 basepairs downstream of a given transcription start site as annotated by GENCODE72.
Statistics and reproducibility
All boxplots were created using the geom_boxplot() function of the R package ‘ggplot2’ with standard settings. The upper and lower hinges represent the 75th and 25th percentiles, respectively. The center line represents the median. The whiskers extend to the largest and smallest values within 1.5 times the interquartile range.
All violin plots were created using the geom_violin() function of the R package ‘ggplot2’ with standard settings, representing data point density along the y-axis.
P-values in the enrichment analyses (LOLA: region sets, Enrichr: gene sets) were calculated using a two-sided Fisher’s exact test. Adjustment for multiple testing was performed using the Benjamini & Yekutieli (BY) method (LOLA) and the Benjamini-Hochberg (BH) method (Enrichr).
P-values accompanying Kaplan-Meier analysis were calculated using a two-sided log-rank test. No adjustment for multiple testing was performed.
P-values indicating the statistical significance of differences between groups of samples in continuous histopathological, MRI, and molecular variables were calculated using a two-sided Wilcoxon rank sum test. No adjustment for multiple testing was performed.
P-values indicating the statistical significance of correlations were calculated using a two-sided Pearson’s test.
Shaded regions around central estimates in ROC curves, Kaplan-Meier plots, and scatterplots denote 95% confidence intervals.
Histopathology and MR imaging pictures represent individual patients and are provided for illustration.
Multidimensional scaling analysis was performed by calculating the Euclidean distance between samples using the R dist() function, followed by multidimensional scaling using the R isoMDS() function.
Data and code availability
All data are available through the Supplementary Website (http://glioblastoma-progression.computational-epigenetics.org/). Genome browser tracks support locus-specific inspection of the DNA methylation data, and a graphical data explorer enables interactive analysis of associations in the dataset (Supplementary Fig. 13). The Supplementary Website also provides lists of regions predictive of immune cell infiltration; pre-computed R objects with DNA methylation profiles and clinical annotation data for follow-up analysis; raw and segmented histopathological image data; and raw as well as segmented MR imaging data. The processed DNA methylation data are also openly available from the NCBI GEO repository (accession number: GSE100351), and the raw sequencing data are available from EBI EGA (accession number: EGAS00001002538) as controlled access. To protect patient privacy, interested researchers need to apply via a data access committee, which will grant all reasonable requests by bona fide researchers. Finally, in the spirit of reproducible research86, the Supplementary Website makes the source code underlying the presented analyses publicly available.
Supplementary Material
Acknowledgements
We thank all patients who have donated their samples for this study. We also thank Gloria Wilk, Martina Muck, Susanne Schmid, and Ulrike Andel for technical assistance with immunohistochemical stainings, macrodissection, and tumor tissue shavings; Simon Mages for contributing to the interactive data visualization; the Biomedical Sequencing Facility at CeMM for assistance with next generation sequencing; and all members of the Bock lab for their help and advice.
The study was funded in part by an Austrian Science Fund grant (FWF KLI394) to A.W., a Marie Curie Career Integration Grant (European Union’s Seventh Framework Programme grant agreement no. PCIG12-GA-2012-333595) to CB, an ERA-NET project (EpiMark FWF I 1575-B19) to C.B., an Austrian Science Fund grant (FWF I2714-B31) to G.L. and K.-H. N., and an ERC Starting Grant (European Unions’s Horizon 2020 research and innovation program, grant agreement no. 640396) to B.B. Moreover, C.B. is supported by a New Frontiers Group award of the Austrian Academy of Sciences and by an ERC Starting Grant (European Union’s Horizon 2020 research and innovation programme, grant agreement no. 679146). Activities of the Austrian Brain Tumor Registry are supported by unrestricted research grants of Roche Austria to JAH and the Austrian Society of Neurology to SO. Part of the samples used for this research project were kindly provided by Biobank Graz.
Footnotes
Author contributions
J. Klughammer, A.W., and C.B. designed the study. B.K., T.R., K.-H.N., J.F., N.P., M.N., M.A., M.M., T.S., G.L., B.B., J.A.H., and A.W. established and annotated the cohort. A.K. and P.D. performed DNA methylation profiling. D.A. performed low-coverage whole genome sequencing. M.S. performed RNA-seq. J. Klughammer performed the data analysis. N.F., N.C.S, and B.E. contributed to data analysis. P.M., C.F.F., J. Kerschbaumer, C.T., A.E.G., G.S., M.K., S.O., F.M., S.W., J.T., J.B., J. Pichler, J.H., S.K., K.M.A., G.v.C., F.P., C.S., J. Preiser, T.H., P.A.W., W.K., F.W., T.B.-K., M.S., S.S., K.D., M.P., E.K., G.W., and C.M. contributed tumor samples and clinical data. J. Klughammer, A.W., and C.B. wrote the manuscript with contributions from all authors.
Conflict of interest
The optimized RRBS protocol that was used in this study has been licensed to Diagenode s.a. (Liège, Belgium) and commercialized as a kit and service.
References
- 1.Ferlay J, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. International Agency for Research on Cancer; 2013. [accessed on 30/06/2018]. [Internet]. Available from: http://globocan.iarc.fr. [Google Scholar]
- 2.Woehrer A, Bauchet L, Barnholtz-Sloan JS. Glioblastoma survival: has it improved? Evidence from population-based studies. Current opinion in neurology. 2014;27:666–674. doi: 10.1097/WCO.0000000000000144. [DOI] [PubMed] [Google Scholar]
- 3.Chinot OL, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. The New England journal of medicine. 2014;370:709–722. doi: 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert MR, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. The New England journal of medicine. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stupp R, et al. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): a multicentre, randomised, open-label, phase 3 trial. The Lancet. Oncology. 2014;15:1100–1108. doi: 10.1016/S1470-2045(14)70379-1. [DOI] [PubMed] [Google Scholar]
- 6.Kim H, et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome research. 2015;25:316–327. doi: 10.1101/gr.180612.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J, et al. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell. 2015;28:318–328. doi: 10.1016/j.ccell.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 8.Kumar A, et al. Deep sequencing of multiple regions of glial tumors reveals spatial heterogeneity for mutations in clinically relevant genes. Genome biology. 2014;15:530. doi: 10.1186/s13059-014-0530-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JK, et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet. 2017 doi: 10.1038/ng.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer M, et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:851–856. doi: 10.1073/pnas.1320611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snuderl M, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48:768–776. doi: 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q, et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell. 2018;33:152. doi: 10.1016/j.ccell.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capper D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ceccarelli M, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturm D, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 20.Brocks D, et al. Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell reports. 2014;8:798–806. doi: 10.1016/j.celrep.2014.06.053. [DOI] [PubMed] [Google Scholar]
- 21.Mazor T, et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell. 2015;28:307–317. doi: 10.1016/j.ccell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao JJ, et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. 2016;48:1500–1507. doi: 10.1038/ng.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin DC, et al. Genomic and Epigenomic Heterogeneity of Hepatocellular Carcinoma. Cancer Res. 2017;77:2255–2265. doi: 10.1158/0008-5472.CAN-16-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med. 2016;22:792–799. doi: 10.1038/nm.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landau DA, et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26:813–825. doi: 10.1016/j.ccell.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheffield NC, et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat Med. 2017;23:386–395. doi: 10.1038/nm.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wohrer A, et al. The Austrian Brain Tumour Registry: a cooperative way to establish a population-based brain tumour registry. J Neurooncol. 2009;95:401–411. doi: 10.1007/s11060-009-9938-9. [DOI] [PubMed] [Google Scholar]
- 28.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klughammer J, et al. Differential DNA Methylation Analysis without a Reference Genome. Cell Rep. 2015;13:2621–2633. doi: 10.1016/j.celrep.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veillard AC, Datlinger P, Laczik M, Squazzo S, Bock C. Diagenode premium RRBS technology: cost-effective DNA methylation mapping with superior coverage. Nature Methods (Application Note) 2016;13 [Google Scholar]
- 31.Bock C, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu H, et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. 2010;7:133–136. doi: 10.1038/nmeth.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stefanits H, et al. KINFix--A formalin-free non-commercial fixative optimized for histological, immunohistochemical and molecular analyses of neurosurgical tissue specimens. Clinical neuropathology. 2016;35:3–12. doi: 10.5414/NP300907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 2012;13:705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 35.Weller M, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2010;6:39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- 36.Bienkowski M, et al. Clinical Neuropathology practice guide 5-2015: MGMT methylation pyrosequencing in glioblastoma: unresolved issues and open questions. Clin Neuropathol. 2015;34:250–257. doi: 10.5414/NP300904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikeska T, et al. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagn. 2007;9:368–381. doi: 10.2353/jmoldx.2007.060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wemmert S, et al. Patients with high-grade gliomas harboring deletions of chromosomes 9p and 10q benefit from temozolomide treatment. Neoplasia. 2005;7:883–893. doi: 10.1593/neo.05307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowman RL, Wang Q, Carro A, Verhaak RG, Squatrito M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro-oncology. 2017;19:139–141. doi: 10.1093/neuonc/now247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheffield NC, Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2016;32:587–589. doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beier CP, et al. The cancer stem cell subtype determines immune infiltration of glioblastoma. Stem Cells Dev. 2012;21:2753–2761. doi: 10.1089/scd.2011.0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strojnik T, et al. Prognostic impact of CD68 and kallikrein 6 in human glioma. Anticancer Res. 2009;29:3269–3279. [PubMed] [Google Scholar]
- 46.Prosniak M, et al. Glioma grade is associated with the accumulation and activity of cells bearing M2 monocyte markers. Clin Cancer Res. 2013;19:3776–3786. doi: 10.1158/1078-0432.CCR-12-1940. [DOI] [PubMed] [Google Scholar]
- 47.Nowosielski M, et al. Progression types after antiangiogenic therapy are related to outcome in recurrent glioblastoma. Neurology. 2014;82:1684–1692. doi: 10.1212/WNL.0000000000000402. [DOI] [PubMed] [Google Scholar]
- 48.Gentles AJ, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015 doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Louis DN, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 50.Li S, et al. Dynamic evolution of clonal epialleles revealed by methclone. Genome Biol. 2014;15:472. doi: 10.1186/s13059-014-0472-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aldape K, et al. Glioma Through the Looking GLASS: Molecular Evolution of Diffuse Gliomas and the Glioma Longitudinal AnalySiS Consortium. Neuro Oncol. 2018 doi: 10.1093/neuonc/noy020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sahm F, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18:682–694. doi: 10.1016/S1470-2045(17)30155-9. [DOI] [PubMed] [Google Scholar]
- 53.McCord M, Mukouyama YS, Gilbert MR, Jackson S. Targeting WNT Signaling for Multifaceted Glioblastoma Therapy. Front Cell Neurosci. 2017;11:318. doi: 10.3389/fncel.2017.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bock C, et al. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol. 2016;34:726–737. doi: 10.1038/nbt.3605. [DOI] [PubMed] [Google Scholar]
- 55.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xi Y, et al. RRBSMAP: a fast, accurate and user-friendly alignment tool for reduced representation bisulfite sequencing. Bioinformatics. 2012;28:430–432. doi: 10.1093/bioinformatics/btr668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol. 2016;12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuilman T, et al. CopywriteR: DNA copy number detection from off-target sequence data. Genome Biol. 2015;16:49. doi: 10.1186/s13059-015-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J, et al. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 2016;17:178–187. doi: 10.15252/embr.201540946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Glaus P, Honkela A, Rattray M. Identifying differentially expressed transcripts from RNA-seq data with biological variation. Bioinformatics. 2012;28:1721–1728. doi: 10.1093/bioinformatics/bts260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sahm F, et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta neuropathologica. 2016;131:903–910. doi: 10.1007/s00401-015-1519-8. [DOI] [PubMed] [Google Scholar]
- 67.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–9. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Makambi K. Weighted inverse chi-square method for correlated significance tests. Journal of Applied Statistics. 2003;30:225–234. [Google Scholar]
- 71.Assenov Y, et al. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods. 2014;11:1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hinrichs AS, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lawson JT, Tomazou EM, Bock C, Sheffield NC. MIRA: An R package for DNA methylation-based inference of regulatory activity. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deroulers C, et al. Analyzing huge pathology images with open source software. Diagnostic pathology. 2013;8:92. doi: 10.1186/1746-1596-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: An open platform for biomedical image analysis. Molecular reproduction and development. 2015;82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Analytical and quantitative cytology and histology. 2001;23:291–299. [PubMed] [Google Scholar]
- 79.Phansalkar N, More S, Sabale A, Joshi M. Adaptive local thresholding for detection of nuclei in diversity stained cytology images; Communications and Signal Processing (ICCSP), 2011 International Conference on; IEEE; 2011. pp. 218–220. [Google Scholar]
- 80.Vincent L, Soille P. Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE transactions on pattern analysis and machine intelligence. 1991;13:583–598. [Google Scholar]
- 81.Liu Q, et al. Genetic, epigenetic, and molecular landscapes of multifocal and multicentric glioblastoma. Acta neuropathologica. 2015;130:587–597. doi: 10.1007/s00401-015-1470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Porz N, et al. Multi-modal glioblastoma segmentation: man versus machine. PloS one. 2014;9:e96873. doi: 10.1371/journal.pone.0096873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wen PY, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 84.Nowosielski M, et al. Radiologic progression types are treatment specific: An exploratory analysis of a phase 3 study of bevacizumab plus radiotherapy plus temozolomide for patients with newly diagnosed glioblastoma (AVAglio) Journal of Clinical Oncology. 2016;34:2048–2048. [Google Scholar]
- 85.Crammer K, Singer Y. On the algorithmic implementation of multiclass kernel-based vector machines. J Mach Learn Res. 2002;2:265–292. [Google Scholar]
- 86.Gentleman R, Temple Lang D. Statistical analyses and reproducible research. Bioconductor project working papers. Working paper 2. 2004 http://biostats.bepress.com/bioconductor/paper2.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available through the Supplementary Website (http://glioblastoma-progression.computational-epigenetics.org/). Genome browser tracks support locus-specific inspection of the DNA methylation data, and a graphical data explorer enables interactive analysis of associations in the dataset (Supplementary Fig. 13). The Supplementary Website also provides lists of regions predictive of immune cell infiltration; pre-computed R objects with DNA methylation profiles and clinical annotation data for follow-up analysis; raw and segmented histopathological image data; and raw as well as segmented MR imaging data. The processed DNA methylation data are also openly available from the NCBI GEO repository (accession number: GSE100351), and the raw sequencing data are available from EBI EGA (accession number: EGAS00001002538) as controlled access. To protect patient privacy, interested researchers need to apply via a data access committee, which will grant all reasonable requests by bona fide researchers. Finally, in the spirit of reproducible research86, the Supplementary Website makes the source code underlying the presented analyses publicly available.