Summary
Microrchidia (MORC) proteins comprise a family of proteins that have been identified in prokaryotes and eukaryotes. They are defined by two hallmark domains: a GHKL‐type ATPase and an S5‐fold. In plants, MORC proteins were first discovered in a genetic screen for Arabidopsis thaliana mutants compromised for resistance to a viral pathogen. Subsequent studies expanded their role in plant immunity and revealed their involvement in gene silencing and genome stabilization. Little is known about the role of MORC proteins of cereals, especially because knockout (KO) mutants were not available and assessment of loss of function relied only on RNAi strategies, which were arguable, given that MORC proteins in itself are influencing gene silencing. Here, we used a Streptococcus pyogenes Cas9 (SpCas9)‐mediated KO strategy to functionally study HvMORC1, one of the current seven MORC members of barley. Using a novel barley RNA Pol III‐dependent U3 small nuclear RNA (snRNA) promoter to drive expression of the synthetic single guide RNA (sgRNA), we achieved a very high mutation frequency in HvMORC1. High frequencies of mutations were detectable by target sequencing in the callus, the T0 generation (77%) and T1 generation (70%–100%), which constitutes an important improvement of the gene‐editing technology in cereals. Corroborating and extending earlier findings, SpCas9‐edited hvmorc1‐ KO barley, in clear contrast to Arabidopsis atmorc1 mutants, had a distinct phenotype of increased disease resistance to fungal pathogens, while morc1 mutants of either plant showed de‐repressed expression of transposable elements (TEs), substantiating that plant MORC proteins contribute to genome stabilization in monocotyledonous and dicotyledonous plants.
Keywords: CRISPR/Cas9, gene‐editing, barley, wheat, rice, MORC1, Blumeria, Fusarium
Introduction
Gene‐editing methods have arisen as an efficient tool for rapid analysis of gene function. From the agricultural perspective, these new methods can be harnessed to create crop plants with desired traits for agronomic purposes with significantly less undesirable side effects on the plant genome. While traditional plant breeding methods involve chemical and radiation mutagenesis that often create random deleterious and chimeric mutations across genomes, modern gene‐editing tools allow precise modification of the genome at a desired position (Lowder et al., 2015; Malzahn et al., 2017; Xiong et al., 2015). Genome modification requires an engineered nuclease to create double‐strand breaks (DSBs) at defined targets, which then triggers cellular DNA repair mechanism, depending on the DNA repair pathway and presence of a repair template. There are two known DSB repair pathways, nonhomologous end joining (NHEJ) and homologous recombination (HR). NHEJ in most instances leads to random insertions or deletions (indels) of nucleotides at the repair site. In case DSB generates overhangs, NHEJ can also introduce gene insertions or precise gene modifications with a double‐stranded DNA fragment with compatible overhangs (Cristea et al., 2013; Maresca et al., 2013). In the presence of a DNA template with homology to the separated chromosome ends, DSBs can be repaired by HR, although this mechanism is rather exceptional at least in somatic cells. Nevertheless, this process can be used to insert DNA fragments and precisely modify genes (Bortesi and Fischer, 2015).
Zinc finger nucleases (ZFNs) and transcription activator‐like effector nucleases (TALENs) have shown promising results in achieving site‐directed DNA breaks. Both enzymes use a dimeric Fok1 nuclease for creating DNA breaks (Christian et al., 2010; Smith et al., 2000). In 2013, the type II clustered regularly interspaced short palindromic repeat (CRISPR)‐associated Cas9 system was discovered in Streptococcus pyogenes (Sp), which emerged as a powerful tool to induce precise mutations in the human genome (Cong et al., 2013; Mali et al., 2013). It promises high on‐target activity and low off‐target effects compared to RNAi (Smith et al., 2017). Subsequent implementation of SpCas9 as RNA‐guided, sequence‐specific nuclease (SSN) for genome editing in plants led to comparably fast and reliable results (Li et al., 2013). SpCas9‐mediated DNA editing involves introduction of two components, the Cas9 protein and a synthetic single guide RNA (sgRNA), into the target cell (genome) to be mutated. The sgRNA (~80 nucleotide [nt] total length) consists of a ~20 nt sequence with sequence similarity to the target gene and a synthetic RNA sequence that adopts functions of CRISPR RNA (crRNA) and transactivating crRNA (tracrRNA) of the original bacterial system (Deltcheva et al., 2011; Jinek et al., 2012; Sorek et al., 2013). SpCas9 induces DSBs by recruiting the sgRNA. An important requirement for DNA cleavage is the presence of a conserved protospacer adjacent motif (PAM), usually carrying the sequence 5′‐NGG‐3′ (for SpCas9) downstream of the target DNA (Gasiunas et al., 2012; Jinek et al., 2012). Since 2013, the SpCas9 system has successfully been applied for gene‐editing in plants such as Arabidopsis thaliana, tobacco and tomato (Brooks et al., 2014; Li et al., 2013; Nekrasov et al., 2013), as well as cereals such as rice, wheat, barley and sorghum (Jiang et al., 2013; Miao et al., 2013; Zhang et al., 2014; reviewed in Ma et al., 2016; Malzahn et al., 2017). Engineering disease resistance in major crops is especially promising because many resistant traits are recessively inherited (Hückelhoven et al., 2003; van Schie and Takken, 2014). A prominent example is powdery mildew resistance in cereals, which is conferred by recessive alleles of the locus mildew‐o (mlo; Acevedo‐Garcia et al., 2014). Significantly, SpCas9‐mediated simultaneously editing each of the three Mlo homeologs in allohexaploid bread wheat (Triticum aestivum) resulted in mlo‐based disease resistance against the wheat powdery mildew fungus Blumeria graminis f. sp. tritici (Wang et al., 2014). A limitation of the technology was the low mutation frequencies shown in the above study for wheat (5.6% in the T0 generation). Plant RNA Pol III‐dependent promoters from small nuclear RNA (snRNA)‐encoding genes (e.g. U3 snRNA and U6 snRNA) have been used to express sgRNA that guides the Cas9 protein to its target in the genome (Brooks et al., 2014; Jiang et al., 2013; Li et al., 2013; Miao et al., 2013; Nekrasov et al., 2013; Zhang et al., 2014). Lawrenson et al. (2015) exploited the wheat promoter of the TaU6 snRNA gene for SpCas9‐mediated gene‐editing of barley HvPM19, which encodes an ABA‐inducible plasma membrane protein. Holme et al. (2017) edited HvPAPhy, a barley phytase gene using a similar construct. Mutation frequencies of 10%–44% were observed in the T0 generation, and induced mutations were transmitted to T1 plants independently of the T‐DNA construct. Kapusi et al. (2017) used the SpCas9 system to disrupt a barley Endo‐N‐acetyl‐β‐D‐glucosaminidase (ENGase) gene by employing the rice OsU6 promoter to drive the sgRNA, reaching a SpCas9‐induced mutation frequency of 78%. However, these studies on cereals had some limitations concerning the mutation and/or transformation efficiency, thereby either accessing a mutation enrichment method using restriction enzymes to identify mutated plants in T0 generation (Holme et al., 2017; Lawrenson et al., 2015) or studying a large number of explants (Kapusi et al., 2017) to identify SpCas9‐positive plants (~10%), which reduces the overall efficiency of the SpCas9 gene‐editing system. These results indicate a need to improve the efficiency of SpCas9‐mediated gene‐editing in cereals.
In this study, we exemplarily used HvMORC1 (GenBank: HG316119.1), one of the seven members of the barley microrchidia (MORC) GHKL (gyrase, Hsp90, histidine kinase, MutL) ATPase subfamily (Koch et al., 2017), to further improve application of the SpCas9‐mediated gene‐editing system in the cereal model barley. Plant MORC genes were first discovered in a genetic screen for Arabidopsis knockout (KO) mutants with compromised resistance against the turnip crinkle virus (TCV), suggesting that they play a role in plant immunity (Kang et al., 2008, 2010, 2012). Subsequent studies in Arabidopsis revealed their involvement in gene silencing and transposable element repression (Lorković et al., 2012; Moissiard et al., 2012, 2014). Unlike Arabidopsis atmorc mutants, barley became more resistant to fungal pathogens, such as powdery mildew fungus Blumeria graminis f. sp. hordei (Bgh), when HvMORC2, a paralog of HvMORC1, was partially silenced by expressing MORC2‐targeting silencing constructs with inverted promoters in transgenic plants (Langen et al., 2014). Consistent with this, transient overexpression of either of the five at that time‐known HvMORC paralogs compromised resistance to Bgh. Yet, functional analysis of cereal MORC proteins has been hampered by the unavailability of respective KO mutants. Hence, we anticipated that the MORC gene family is an excellent model for SpCas9‐mediated gene‐editing applications in barley.
Using a novel barley U3 snRNA promoter to drive the sgRNA, we achieved an unprecedentedly high mutation frequency. Distinct hvmorc1‐KO mutations were detectable by target sequencing in the transgenic calli, the T0 generation (77%) and T1 (70%–100%) generation, which represents an important improvement of the technology. Extending earlier findings that were based on hvmorc2‐KD mutants generated by RNAi‐mediated knockdown (KD) strategies, SpCas9‐edited hvmorc1‐KO barley showed increased disease resistance to biotrophic Bgh and necrotrophic Fusarium graminearum. However, in contrast to barley hvmorc1‐KD mutants, hvmorc1‐KO barley, alike atmorc1 mutants, showed de‐repressed expression of transposable elements (TEs), suggesting that barley MORCs also are involved in genome stabilization.
Results
Identification and characterization of a barley RNA Pol III‐dependent snRNA promoter
As RNAi‐mediated KD may result in low efficiency and thus substantial residual amounts of transcript and protein, we further analysed the function of MORCs using stable KO mutant barley lines generated by SpCas9‐based nuclease. To ensure efficient transcription of sgRNAs in barley cells, we first set out to identify suitable regulatory elements by focusing on cereal U3 snRNA promoters. A U3 snRNA promoter from rice has been previously characterized (OsU3; Qu et al., 1996). This promoter has been used for SpCas9‐mediated gene‐editing in rice (e.g. Zhang et al., 2014) and maize (Xing et al., 2014). A suitable HvU3 regulatory element (GenBank: CAJX011995286.1) was identified by similarity to the wheat TaU3 promoter (GenBank: X63065.1; Marshallsay et al., 1992) from the database of barley cultivar ‘Bowman’ (http://webblast.ipk-gatersleben.de/barley/). U3 snRNA promoter sequences from barley, rice and wheat were compared for the presence of features characteristic for Pol III‐dependent promoters in monocotyledonous plants: TATA box, an upstream sequence element (USE), and monocot‐specific promoter (MSP) elements (Figure 1a–c). MSPs are conserved G + C‐rich sequences with a consensus of RGCCCR in either direction, usually present −30 to −130 bp upstream of the USE (Connelly et al., 1994). Consistent with this, the barley HvU3 promoter (pHvU3) contains a TATA box, located 23 base pairs (bp) upstream of the transcription initiation site, and the USE with the consensus sequence 5′‐TCCCACCTCG 25 bp upstream of TATA box, along with five MSPs, thus matching with the characteristics of RNA Pol III‐dependent promoters from monocotyledons (Waibel and Filipowicz, 1990).
Figure 1.

U3 promoter sequences from rice (a); wheat (b) and barley (c). Sequence motifs are underlined: TATA box, upstream sequence element (USE) and monocot‐specific promoter (MSP) element.
Assessment of barley and rice U3 snRNA promoter activities
We first studied the activity of barley and rice U3 snRNA promoter fragments in the tissue of immature barley embryos; 638 and 380 bp upstream of the predicted transcription start sites of HvU3 and OsU3, respectively, were cloned into pGY1‐35s:GFP (Figure S1). The U3 regulatory elements replaced the 35S promoter in pGY1 to drive expression of GFP. Resulting constructs were delivered to tissues from excised immature embryos of spring barley cultivar (cv.) Golden Promise by particle bombardment. Foci of GFP expression were detected 48 h after bombardment in embryonic cells transformed with either construct (Figure 2a,b). Although foci occurred at rather low frequencies in comparison with our routine observations, when bombarding constructs for Pol‐II promoter‐driven expression, the results demonstrate activity of both the HvU3 and OsU3 promoter fragments in barley. Notably, this also suggests that U3‐driven transcripts can, at least to some extent, engage the translational machinery in the cytoplasm.
Figure 2.
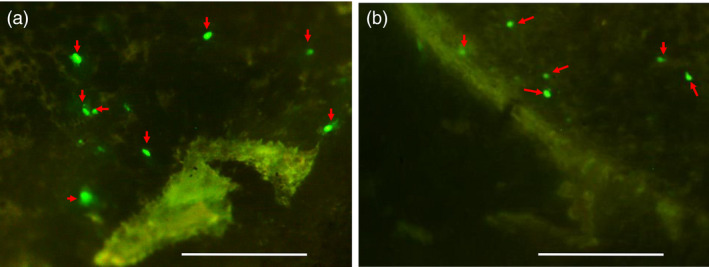
GFP expression in immature embryos of barley cv. Golden Promise 48 h after biolistic transformation with (a) pGY1‐pH vU3: GFP , containing 638 bp of the promoter sequence upstream of the coding region of the U3 snRNA promoter from barley, and (b) pGY1‐pO sU3: GFP , containing 380 bp of the promoter sequence upstream of the coding region of the U3 snRNA from rice, to drive GFP expression. Arrows mark GFP fluorescence. Bar scale 0.5 mm.
SpCas9‐induced mutation of HvMORC1
The barley genome contains seven MORC genes, all of which are assumed to act as negative regulators of immunity as deduced from overexpression and RNAi‐mediated KD studies (Koch et al., 2017; Langen et al., 2014). To further address MORCs’ function in barley, HvMORC1 was targeted by SpCas9 to generate loss‐of‐function alleles. A target site in the 5′ part of the HvMORC1 gene upstream of the ATPase domain with no potential off‐targets in any of the seven barley MORCs (Figure S2) or the barley genome (see Experimental procedures) was chosen, and a respective sgRNA was designed (Figure 3a). Two constructs HvU3:sgRNA and OsU3:sgRNA, containing either pHvU3 or pOsU3 driving sgRNA expression and SpCas9 under control of the maize ubiquitin promoter (ZmUbi:Cas9), were transformed into immature embryos by agro‐transformation (Figure 3b,c; see also Figure S3). Genome editing activity from transformation of the construct with HvU3‐driven sgRNA was analysed in calli 6 weeks after transformation. Genomic DNA was extracted from randomly chosen embryonic calli grown on hygromycin selective media. The target region was amplified by PCR, and amplicons were analysed by direct sequencing of both strands. From two calli, the wild‐type (wt) sequence was obtained (Figure 4a; calli 1 and 6), and chromatograms were not indicative of any nuclease activity at the target site. In contrast, a homozygous deletion of 2 bp was obtained for one callus and also a bi‐allelic lesion (Figure 4a, calli 2 and 5). For the remaining five calli, varying degrees of nuclease activity were detected in chromatograms, and precise alleles could not be deduced as multiple sequences were detected (Figure 4a,b), suggesting that samples used for PCR originated from a mixed population of cells containing different molecular lesions at the target site. Thus, nuclease activity was detected in almost 77% (7/9) of analysed calli by direct sequencing, and bi‐allelic disruptive mutations affecting both alleles were also readily detected, suggesting occurrence of loss‐of‐function lines already in the T0 generation. We used TIDE (Tracking of Indels by Decomposition; Brinkman et al., 2014) to further access the spectrum and frequency of SpCas9‐induced mutations in calli. Overall, mainly indels of −2, −1 and +1 nt were detected by decomposition of chromatograms, and frequencies were comparable. Deletions of up to 5 nt were also detected, but frequencies were low. Among all calli (including those without detectable mutations), the wt sequence represented in average only 40% (±34%) of all sequence information, suggesting highly efficient genome editing when expressing sgRNAs under control of the HvU3 promoter.
Figure 3.

MORC domain structure and constructs used for targeted KO of HvMORC1 by SpCas9‐mediated gene‐editing. (a) Targeted area (20 nt) of HvMORC1 domain with the PAM sequence in grey shade. The hallmark domains of HvMORC1: a GHKL‐type ATPase, an S5‐fold and a CC‐domain, are highlighted. (b, c) Schematic representation of the T‐DNA regions containing all components for Agrobacterium‐mediated, SpCas9‐based HvMORC1 gene‐editing. Construct with barley U3 promoter (b) and construct with rice U3 promoter (c). Hygromycin, hygromycin phosphotransferase gene [hpt]; pZmUbi, ubiquitin promoter of Zea mays; t35s, CaMV 35S terminator; LB, RB, left and right border sequences of the T‐DNA; sgRNA, synthetic single guide RNA.
Figure 4.

Mutations in the 20 bp target region of HvMORC1 in 6‐week‐old T0 calli expressing the construct HvU3:sgRNA _ZmUbi:Cas9. (a) Overview of sequences obtained from calli. The PAM (CGG) sequence is highlighted in grey. (b) Example of a typical chromatogram, which could not be resolved into two or less distinct alleles. Note peaks with multiple overlaying signals indicating the presence of at least three different alleles within the sample.
Selection of homozygous hvmorc1‐KO barley in the T1 generation
Genome editing activities were further analysed in T1. T0 plantlets regenerating on hygromycin selective medium were randomly selected and propagated in soil to obtain seeds for T1 generation. Notably, the parental T0 lines were not checked for the presence of either T‐DNA construct or mutations in HvMORC1. In T1 offspring, target sites were PCR‐amplified and amplicons were analysed by sequencing. For construct HvU3:sgRNA, 71 2‐week‐old plants from 12 different T1 lines (five to six plants per line) were analysed, and mutations could be detected in all T1 populations (100% efficiency). Similarly, 60 plants from 10 independent T1 lines (six plants per line) carrying OsU3:sgRNA were analysed. Mutations were detected in seven of these T1 populations (70% efficiency). For both transformation events, homozygous indel mutations (Figure 5a,b) were identified within the 20 bp target sequence, at a frequency of 38% (HvU3) or 42% (OsU3). Heterozygous mutations showed the presence of double peaks in the sequencing chromatogram (Figure 5c,d). While homozygous mutations are bi‐allelic, heterozygous mutations theoretically could be either mono‐allelic or bi‐allelic (with different mutations on both chromosomes). Further assessment of T1 populations with bi‐allelic mutations showed that in each population plants homozygous for each allele could be discovered (Figure 5e). These results confirm that SpCas9 can induce different mutations on different chromosomal strands of the same T0 plants, resulting in homozygous plants with two different mutation patterns in the T1 generation. Notably, we also identified mutated T1 plants that did not contain a T‐DNA construct (nontransgenic): 11 of 73 tested plants (15%) were devoid of the construct, indicating segregation of T‐DNA construct and the lesions within HvMORC1.
Figure 5.

SpCas9‐induced mutations in independent barley T1 lines. (a–b) Homozygous mutations in T1 plants containing the HvU3:sgRNA _ZmUbi:Cas9 and the OsU3:sgRNA _ZmUbi:Cas9 construct, respectively. (c) Heterozygous mutants have characteristic double peaks in the chromatogram, for example starting 4 bp upstream of the PAM sequence (grey). (d) Mutation patterns of heterozygous mutants determined after sequencing using specific primers (Table S1) from both directions. (e) Examples for mutations in homozygous bi‐allelic T1 lines. The PAM (CGG) sequence is highlighted in grey, the 20 bp target region in HvMORC1 is underlined, and insertions are marked in bold.
Enhanced sgRNA accumulation by HvU3 promoter‐driven expression in barley
To further corroborate the suitability of the HvU3 promoter for genome editing approaches in barley, expression of sgRNA under control of either the HvU3 or OsU3 promoter was quantified in T‐DNA‐positive lines using quantitative RT‐PCR (RT‐qPCR). To normalize for potential copy number variations and/or transgene insertions at different genome locations, sgRNA expression was normalized to the expression of T‐DNA‐encoded SpCas9 (Figure 6) or hygromycin phosphotransferase (Figure S4). We observed clearly higher expression of pHvU3‐driven sgRNA transcripts compared to pOsU3 under both instances.
Figure 6.

Relative expression of sgRNA under control of barley and rice U3 promoters (pH vU3 and pO sU3) in leaves of hvmorc1‐KO T2 homozygous mutants measured by RT‐PCR and normalized against SpCas9. Error bars indicate standard deviation of three repetitions. Asterisks indicate statistical significant difference (Student's t‐test: ***P < 0.001).
hvmorc1‐KO mutants show increased resistance to fungal pathogens
Arabidopsis lines deficient in MORC1 and MORC2 are severely impaired in resistance to viral, bacterial, oomycete and fungal pathogens (Kang et al., 2008, 2012), while, in contrast, RNAi‐mediated reduced transcript levels of HvMORC2 in barley enhanced resistance (Langen et al., 2014). To substantiate the opposing function of MORCs in barley vs. Arabidopsis, mutated hvmorc1‐KO T1 plants (consisting of both homozygous and heterozygous bi‐allelic mutations) from pHvU3:sgRNA_pZmUbi:Cas9 construct (hvmorc1‐L3, hvmorc1‐L13 and hvmorc1‐L16; see Figure 5e) were tested for powdery mildew resistance. Detached leaves were inoculated with conidia of BghA6 (virulent on cv. Golden Promise). Mutant lines developed less fungal colonies 6 days postinoculation (dpi) compared to wt plants (hvmorc1‐L3: 71.5%; ‐L13: 71.8%; hvmorc1‐L16: 76%; Figure 7a). These results were consistent with our expectation that barley MORC paralogs respond similar to Bgh (Langen et al., 2014). T1 plants from hvmorc1‐L10 and hvmorc1‐L13 that were homozygous for frameshift mutations (hvmorc1‐1 and hvmorc1‐4; Figure S5) in the 5′ region of HvMORC1 were propagated for analyses of T2 plants. We further studied hvmorc1‐1 and hvmorc1‐4 T2 homozygous plants for their response to the mycotoxin‐producing fungus Fusarium graminearum. Detached leaves of hvmorc1‐KO and wt plants were inoculated with macrospores of F. graminearum, and fungal DNA was quantified by quantitative PCR at five dpi. Fungal biomass was significantly reduced in hvmorc1 KO mutant tissues (Figure 7b).
Figure 7.

SpCas9‐mediated KO of HvMORC1 results in enhanced resistance against fungal pathogens. (a) hvmorc1‐KO T1 barley cv. Golden Promise lines (hvmorc1‐L3, hvmorc1‐L13, hvmorc1‐L16) display enhanced resistance against powdery mildew. Detached second leaves of 14‐day‐old plants were inoculated with 3–5 conidia per mm2. Bgh colonies were counted at six dpi. Shown is the average number of Bgh colonies on 1.5 cm2 leaf area (n = 14). The experiment was repeated twice with similar results. Error bars indicate standard error. Asterisks indicate statistical significant difference (Student's t‐test *P < 0.05, **P < 0.01). (b) hvmorc1‐KO T2 homozygous mutants show enhanced resistance against Fusarium graminearum (Fg). For inoculation, 20 ul of Fg conidia (5 × 104 conidia mL−1) was drop‐inoculated on detached third leaves of 21‐day‐old plants. Quantification of Fg on leaves was performed five dpi by quantitative RT‐PCR based on the ratio of fungal tubulin (FgTub) to plant ubiquitin (HvUbi). Significant changes are marked: ***P < 0.001 (Student's t‐test). Presented are mean of 10 leaves. Bars represent standard deviation of three repetitions.
hvmorc1‐KO mutants show enhanced expression of PR genes
We investigated whether enhanced resistance of hvmorc1‐KO lines are associated with constitutive activation of defence responses. To this end, we measured expression of defence‐related genes. Expression of HvPR1b (GenBank: X74940.1), HvPR2 (GenBank: AF479647.2) and HvPR5 (GenBank: AM403331.1) in hvmorc1‐1 and hvmorc1‐4 T2 homozygous plants was determined by RT‐qPCR at 0, 24, 48 and 72 hpi with Bgh. Without pathogen stimulus (0 hpi), all of these genes were expressed to higher levels in hvmorc1‐KO mutants compared to wt (Figure 8a–c). Upon Bgh inoculation, differences in PR expression of hvmorc1‐KO vs. wt were even more pronounced, noticeably at an early infection stage (24 hpi). Most strikingly, expression of PR1b was strongly induced in the hvmorc1‐KO mutant. We concluded that compromised HvMORC1 functions de‐repress at least parts of the plant defence system.
Figure 8.

Relative PR gene expression in leaves of SpCas9‐generated hvmorc1‐KO T2 homozygous mutants vs. wt measured by RT‐qPCR and normalized to plant ubiquitin. Expression of SA pathway marker genes HvPR1b (a), HvPR2 (b) and HvPR5 (c). Detached second leaves of 14‐day‐old plants were inoculated with 10 to 15 conidia per mm2 (n = 5). Error bars indicate standard deviation of three repetitions. Asterisks indicate statistical significant difference (Student's t‐test: *P < 0.05, **P < 0.01, ***P < 0.001).
hvmorc1‐KO mutants show de‐repressed transposable elements
In Arabidopsis atmorc1 and atmorc6 mutants, expression of transposable elements (TEs) located around the pericentromeric region is strongly increased (Moissiard et al., 2012), while transposon de‐repression has not been observed in barley hvmorc1‐KD mutants that were only partially silenced for HvMORC1 (Langen et al., 2014). We refined the analysis of TE expression using the hvmorc1‐KO lines. To this end, expression of long terminal repeat (LTR) and non‐LTR retrotransposons (Long INterspersed Elements; LINE) with sequence similarity to those de‐repressed in Arabidopsis atmorc mutants (Langen et al., 2014; Moissiard et al., 2012) was measured by RT‐qPCR in hvmorc1‐1 and hvmorc1‐4 T2 homozygous mutated plants. In contrast to partially silenced hvmorc1‐KD mutants, SpCas9‐generated hvmorc1‐KO lines showed significant transposon de‐repression as compared to wt (Figure 9), although the degree of TE de‐repression was lower than previously reported for Arabidopsis atmorc mutants. This suggests that HvMORC1, such as AtMORC1, is involved in genome stabilization.
Figure 9.

Expression of transposons (TEs) in second leaves of 14‐day‐old SpCas9‐generated hvmorc1‐KO T2 homozygous mutants (hvmorc1‐1 and hvmorc1‐4) vs. wt assayed by RT‐qPCR and normalized to plant ubiquitin (n = 5). Error bars indicate standard deviation of three repetitions. Asterisks indicate statistical significant difference (Student's t‐test: *P < 0.05, **P < 0.01, ***P < 0.001).
hvmorc1‐KO mutants show increased expression of HvMORC2 in immature embryos
In Arabidopsis, AtMORC1 and AtMORC2 interact with AtMORC6 to form distinct heteromers to achieve gene silencing. Additionally, the function of AtMORC6 is epistatic to both AtMORC1 and AtMORC2 (Moissiard et al., 2014). We assessed the effect of knocking out HvMORC1 on the expression of other barley MORC homologs. Expression of HvMORC2 (GenBank: HG316120) and HvMORC6a (GenBank: HG316122) was measured in immature embryos of T1 hvmorc1‐KO homozygous plants hvmorc1‐1 and hvmorc1‐4 and leaves of their T2 progenies. An increased expression of HvMORC2 was observed in embryos of hvmorc1‐1 and hvmorc1‐4 compared to wt, while expression of HvMORC6a was similar in all genotypes (Figure 10a). This raises the possibility that, in the absence of HvMORC1, there is an increased expression of HvMORC2 to maintain the cellular concentration of heteromeric complexes involving HvMORC6 for transcriptional repression of TEs, especially in immature embryonic tissue. Notably, HvMORC2 showed no significant increase in expression in leaves of hvmorc1‐KO mutants (Figure 10b). Both immature embryos and leaves of hvmorc1‐KO show reduced transcript level of HvMORC1 (Figure 10a,b), which could be a result of mRNA degradation by non‐sense‐mediated mRNA decay pathway that identifies and removes mRNA with premature STOP codons (Reviewed in Baker and Parker, 2004).
Figure 10.

Relative expression of barley MORC genes in immature embryos and leaves of hvmorc1‐ KO mutants and wt assayed by RT‐qPCR and normalized to plant ubiquitin. (a) Expression of HvMORC1, HvMORC2 and HvMORC6a in immature embryos of T1 homozygous mutants (hvmorc1‐1 and hvmorc1‐4). (b) Expression of HvMORC1, HvMORC2 and HvMORC6a in leaves of T2 homozygous mutants (hvmorc1‐1 and hvmorc1‐4). mRNA was extracted from second leaves of 14‐day‐old plants and immature embryos (n = 5). Error bars indicate standard deviation of three repetitions. Asterisks indicate statistical significant difference (Student's t‐test: *P < 0.05, ***P < 0.001).
Discussion
Identification and transient expression of the barley U3 snRNA promoter
Monocot and dicot RNA Pol III promoters from snRNA genes have been used to express sgRNA for genome editing. Diverse Arabidopsis promoters such as AtU3b, AtU3d, AtU6, AtU6‐1, AtU6‐26 and AtU6‐29 have been shown to be functional in dicotyledons (Brooks et al., 2014; Fauser et al., 2014; Feng et al., 2014; Gao et al., 2015; Ma et al., 2015; Mao et al., 2013; Nekrasov et al., 2013; Xing et al., 2014). For the expression of sgRNA in cereals, promoter variants OsU3 and OsU6 for rice (Ma et al., 2015; Shan et al., 2013), TaU3 and ZmU6 for maize (Svitashev et al., 2015; Xing et al., 2014), and TaU6 for wheat and barley (Lawrenson et al., 2015; Wang et al., 2014) have been used. In the newly isolated barley U3 promoter, the USE element lies 25 bp upstream of TATA box, which conforms to the consensus sequence of RNA Pol III‐dependent promoters (Figure 1c). In maize, deletion or substitution of MSPs decreases the transcription efficiency by 30%–60% (Connelly et al., 1994; Qu et al., 1996). Thus, it was crucial to check the functionality of promoters to be used in our SpCas9 system. RNA Pol III is able to produce functional mRNA with a low efficiency in human cells (Gunnery and Mathews, 1995), but similar studies have not been carried out in plants. In our study, upon transient transformation both barley and rice (RNA Pol III‐dependent) U3 promoters coupled with GFP were expressed in barley immature embryos (Figure 2a,b), confirming their functionality. This finding also suggests that in plants some protein‐coding genes might be RNA Pol III‐dependent.
Highly efficient genome editing in barley by HvU3‐driven sgRNA expression
Several studies have been published that reported the use of SpCas9 system in barley (Holme et al., 2017; Kapusi et al., 2017; Lawrenson et al., 2015). We used a single T‐DNA vector similar to the study by Lawrenson et al. (2015), main difference to this previous work being the use of the U3 promoters of either barley or rice for sgRNA expression (Figures 1 and 3). Using these promoters, we achieved both stringent selection of transgenic plants on hygromycin and highly efficient genome editing with bi‐allelic mutations occurring already in T0 (Figure 4a). Although we cannot exclude an extraordinary high efficiency of the sgRNA used in our study, we assume that U3 promoters are highly suitable for sgRNA expression in barley genome editing applications. Notably, HvU3‐driven sgRNA showed highest transcript accumulation as compared to OsU3 (Figures 6 and S4). We provide these regulatory elements, and also TaU6 and OsU6 promoter fragments used in previous experiments, as part of a convenient toolkit to the plant research community. Our toolkit, which is similar to a previously reported toolkit for genome editing in dicot plants (Ordon et al., 2017), provides simple and rapid (Golden Gate‐based) cloning procedures and high multiplexing capacity for expression of four or up to eight sgRNAs. A description of the toolkit with cloning manual (Appendix S1) and vector maps (Appendix S2) is provided.
SpCas9 nuclease‐induced mutations in barley
The aim of our study was to further increase the mutation frequency and select mutated plants growing on hygromycin selective medium using simple PCR and Sanger sequencing. Using HvU3, we obtained 77.7% mutation frequency in 6‐week‐old T0 callus (Figure 4a), which is seemingly high for barley. In separate experiments, T0 plants growing on hygromycin selective medium were selected for seed propagation. Later in T1 offspring of those plants, we obtained mutation frequencies of 100% and 77% in HvMORC1 using HvU3 and OsU3 promoters, respectively. We obtained 38%–41% bi‐allelic homozygous plants and 15% plants were T‐DNA‐free T1 generation. The T‐DNA‐free homozygous plants do not contain any inserted DNA fragment/gene and carry the same mutation on both chromosomes, thus being ideal for gene function studies. Hence, we show here that it is possible to get a high frequency of mutation in barley using the SpCas9 technique. No doubt in future this technology would be the first choice of gene modification for plant pathologists, breeders and biochemists.
Higher HvMORC2 expression compensates for KO of HvMORC1 in hvmorc1‐KO embryos
Previous work suggested that AtMORC1 and AtMORC2 do not interact with each other but both interact with AtMORC6, leading to the proposal that AtMORC6 mediates gene silencing by forming mutually exclusive heterodimers with either AtMORC1 or AtMORC2, or as a homodimer (Liu et al., 2014; Moissiard et al., 2014), and the function of AtMORC6 is epistatic to both AtMORC1 and AtMORC2 (Moissiard et al., 2014). Supportive of the former reports, we found an increased expression of HvMORC2 in immature embryos of hvmorc1 compared to wt, while expression of HvMORC6a was not changed (Figure 10a). These data suggest that in the absence of HvMORC1, there is an increased expression of HvMORC2 to maintain the cellular concentration of heteromeric complexes involving HvMORC6 for transcriptional repression of TEs, although it is not resolved whether the targets of MORC1‐MORC6a and MORC2‐MORC6a complexes are identical, different or overlapping. Notably, HvMORC2 showed no significant increase in expression in leaves of hvmorc1‐KO (Figure 10b), suggesting that the cell machinery is epigenetically programmed to identify and compensate for defects in DNA methylation during reproductive stage. Finally, constitutive HvMORC6a expression is higher compared to HvMORC1 and HvMORC2 (Figure 10a,b), further arguing for a prominent cellular requirement of HvMORC6a.
Barley MORC1 modulates plant immunity and regulates TE expression
While in Arabidopsis and potato, MORCs positively regulate resistance to microbial pathogens, they are negative regulators in tobacco and tomato (Kang et al., 2008, 2010, 2012; Manosalva et al., 2015). In barley, RNAi‐mediated KD of HvMORC2 also resulted in higher resistance, resembling the situation in tobacco and tomato (Langen et al., 2014). Given MORC proteins are also influencing gene silencing, there remains a technical uncertainty to assess the loss of function using RNAi. In the present study, a complete KO of HvMORC1 also enhances plant immunity against fungal pathogens Bgh and F. graminearum (Figure 7a,b), confirming similar immune functions of barley paralogs MORC1 and MORC2. Enhanced resistance to fungal pathogens correlated with elevated transcript level of PR genes in hvmorc1‐KO mutants (Figure 8a–c). PR expression was further enhanced in response to Bgh, particularly during initial phase of fungal colonization, providing hvmorc1‐KO mutants an early advantage over wt plants. It appears HvMORC1 controls at least part of the plant's immune system, possibly thereby avoiding autoimmune reactions of an overshooting defence system.
Several lines of evidence suggest that MORC proteins also have nuclear targets. For example, in vitro assays demonstrated that AtMORC1 and HvMORC1 bind DNA/RNA, display endonuclease activity and are transferred from cytoplasmic locations to the nucleus in response to PAMP signals such as flagellin (Kang et al., 2012; Langen et al., 2014). Furthermore, MORC proteins from a range of prokaryotes and eukaryotes have been shown to play roles in chromatin modification and/or DNA recombination and repair (Iyer et al., 2008; Pastor et al., 2014; Perry and Zhao, 2003). The identification of AtMORC1 and/or AtMORC6 in three independent forward genetic screens of Arabidopsis mutants defective for transcriptional gene silencing (TGS) provided the first insight into nuclear MORC protein function (Brabbs et al., 2013; Lorković et al., 2012; Moissiard et al., 2012). In plants, TGS plays an important role in repressing TEs, intergenic regions, DNA repeats and some genes; it is mediated by the RNA‐directed DNA methylation (RdDM) pathway (Law and Jacobsen, 2010; Matzke et al., 2009, 2015). RdDM utilizes small RNAs to recruit the DNA methylation machinery to targeted sequences. DNA methylation in turn leads to recruitment of histone‐modifying enzymes, and the combined effect of these repressive epigenetic marks establishes chromatin in a silenced state. De‐repression of silenced reporter genes as well as TEs was observed in most atmorc mutants, suggesting that these proteins play a role in epigenetic gene silencing (Bordiya et al., 2016; Brabbs et al., 2013; Harris et al., 2016; Lorković et al., 2012; Moissiard et al., 2012, 2014). In the present study, we found an increased expression of barley TEs in homozygous hvmorc1‐KO mutants (Figure 9). A huge part (84%) of the barley genome consists of mobile and repeat structures, 76% of which are retrotransposons. Some 99.6% of retrotransposons are long terminal repeat (LTR) transposons, while 0.31% are non‐LTR retrotransposons (International Barley Genome Sequencing Consortium (IBSC), 2012). Notably, RNAi‐mediated KD of HvMORC1 did not result in detectable de‐repression of barley TEs (Langen et al., 2014), suggesting that the remaining MORC protein activity (degree of gene KD was approx. 50%) was sufficient to repress TEs, which can explain the different phenotypes of RNAi‐generated hvmorc1‐KD vs. SpCas9‐mediated hvmorc1‐KO plants. Yet, when comparing morc1 mutants from barley and Arabidopsis, the different degrees of transposon de‐repression are conspicuous (barley up to 14‐fold [this study] vs. Arabidopsis up to 500‐fold [Moissiard et al., 2014;] as compared to the respective wt plants). However, in two subsequent studies, lower expression of transposons (AtCopia28/RomaniaT5) was observed in atmorc1 (Moissiard et al., 2014; Zhang, 2016). Moreover, a previous report showed that barley retrotransposons are responsive to various biotic and abiotic environmental cues (Alzohairy et al., 2012). Consistent with our study, the barley LTRs did not show high transcript level in response to such triggers.
While the link between MORC proteins role in immunity and TGS is currently unknown, the discovery that Pseudomonas syringae pv. tomato (Pst) infection alters AtMORC1 binding at genomic regions preferentially associated with TEs provides an important clue (Bordiya et al., 2016). A growing number of studies suggest that TEs are key regulatory elements that control stress‐associated gene expression (Dowen et al., 2012). Thus, the finding that Pst infection reduces AtMORC1 binding at loci associated with heterochromatic TEs led Bordiya et al. (2016) to propose that loss of AtMORC1 binding at these sites disrupts a silencing complex and thus up‐regulates heterochromatic TE expression. The de‐repressed TEs could serve as enhancers of proximal gene expression in barley. It is tempting to speculate that elevated resistance of hvmorc1‐KO mutants results from barley MORC role in genome stabilization, which is attenuated in the mutants resulting in higher expression of TEs and concomitantly PR gene expression.
Experimental procedures
Plant material and fungal inoculation
Seeds of barley (Hordeum vulgare) cv. ‘Golden Promise’ were germinated for 3 days on filter paper. Seedlings were transferred to soil (Fruhstorfer Erde Typ T) and cultivated in a growth chamber at 22 °C/18 °C (day/night cycle) with 60% relative humidity and a photoperiod of 16 h (240 μmol/m2/s photon flux density). After complete emergence (12–14 day), the second leaves were detached, laid on 0.5% (w/v) water agar and inoculated with BghA6 (Langen et al., 2014) at a density of 2 to 5 conidia mm−2. For expression analysis, a high density of 10 to 15 conidia mm−2 was used. F. graminearum (strain 1003; Jansen et al., 2005) was regularly cultured on SNA (synthetic nutrient‐poor agar) plates containing 0.1% KH2PO4, 0.1% KNO3, 0.1% MgSO4·7H2O, 0.05% KCL, 0.02% glucose, 0.02% sucrose and 1.4% agar. Plates were incubated at room temperature under constant illumination from one near‐UV tube (Phillips TLD 36 W/08, http://www.philips.de) and one white light tube (Phillips TLD 36 W/830HF, http://www.philips.de). Sterile 0.02% Tween water (v/v) was poured on 2‐week‐old plates, and conidial suspension was scrubbed using a glass rod and filtered through a miracloth (Calbiochem, http://www.merck-chemicals.de). Conidia concentration was adjusted to 5 × 10−4 spore mL−1; 20 μL of spore suspension was drop‐inoculated on detached barley leaves kept on 0.5% water agar plates. Square Petri plates with detached leaves were kept at room temperature under one white tube (Phillips TLD 36 W/830HF, http://www.philips.de). Progression of infection was routinely monitored. For quantification of fungal invasion, leaf samples were harvested at 5 dpi and DNA was extracted (Doyle and Doyle, 1987), which was later used to determine the amount of fungal DNA by quantitative RT‐PCR.
Generation of vectors to study barley and rice U3 promoter activity using GFP reporter gene in transient assay system
A 638 bp upstream of barley U3 coding sequence (GenBank: CAJX011995286.1) and a 380 bp sequence upstream of rice U3 coding sequence (Miao et al., 2013) were amplified with primers containing restriction sites XhoI and NcoI (Table S1). Both barley and rice U3 promoters were coupled with the reporter gene for the green fluorescent protein (GFP) by replacing the CMV35s promoter in pGY1‐35s:GFP (Schweizer et al., 1999) using restriction enzyme XhoI and NcoI to generate plasmid constructs—pGY1pHvU3:GFP and pGY1pOsU3:GFP.
Generation of CRISPR/Cas9 constructs
Twenty bp target sequences with NGG (PAM) at 3′ end were selected using CRISPR sgRNA design online tool (https://atum.bio/eCommerce/cas9/input) for HvMORC1 (GenBank: HG316119.1). The designed 20 bp target sequences was blasted (BlastN) against nucleotide collection of Hordeum vulgare (taxid: 4513) at NCBI for putative off‐targets, and ACTTCGGGTGCACCCGCGCG was selected. Cloning overhangs (HvU3: agca/aaac; OsU3: ggca/aaac) were added and guide sequences cloned as hybridized oligonucleotides. To adapt OsU3, HvU3, OsU6 and TaU6 elements for the multiplexing system, existing BsaI and BpiI sites were removed, and promoter fragments were cloned together with a ccdB cassette and the sgRNA backbone into a pUC57 derivative as previously described (Ordon et al., 2017). Recipient vectors were assembled by modular cloning as previously described (Engler et al., 2014; Ordon et al., 2017). Details on cloning procedures and primer sequences are available upon request.
Plant transformation
Plasmids were electroporated (Gene Pulser, Biometra) into Agrobacterium tumefaciens strain AGL1 (Lazo et al., 1991), and the resulting Agrobacterium was used to transform spring barley ‘Golden Promise’ as described (Imani et al., 2011; Tingay et al., 1997). Transient barley transformation was performed as described (Schweizer et al., 1999). Immature barley embryos were shot using a particle inflow gun (PDS‐1000/He, BIO‐RAD) with DNA‐coated on 1‐μm gold particles. One microgram of plasmid per shot was used with a rupture disc of 650 psi.
DNA isolation and quantitative PCR analysis
DNA/RNA extraction and quantitative RT‐PCR were performed as described (Doyle and Doyle, 1987; Imani et al., 2011). Primer pairs used for expression analysis are listed in Table S1.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Figure S1 Maps of constructed plasmid.
Figure S2 Target site conservation in MORC genes.
Figure S3 638 bp sequence of barley RNA Pol III promoter (TATA box is underlined) (A); 380 bp sequence of rice RNA Pol III promoter (TATA box is underlined) (B); Sequence of sgRNA is underlined with terminator (C).
Figure S4 Relative expression of sgRNA under control of barley and rice U3 promoter (pHvU3 and pOsU3) in leaves of hvmorc1‐KO T2 homozygous mutants measured by RT‐PCR and normalized against Hygromycin gene.
Figure S5 SpCas9‐induced frame‐shift mutations in HvMORC1 leads to premature STOP codons Predicted HvMORC1 open reading frames (in red) with premature stop codons after Cas9 induced mutation (b‐d) compared to wt (A) using online tool (http://web.expasy.org/translate/).
Table S1 Oligonucleotide primers used in this study (restriction sites are underlined).
Appendix S1 Cloning manual for pMGE genome editing vectors.
Appendix S2 Annotated sequence files (Genbank) for pMGE vectors.
Supplementary File
Acknowledgements
We thank E. Swidtschenko and C. Dechert for excellent technical assistance. This work was supported by the German Research Council (Deutsche Forschungsgemeinschaft, DFG; Ko 1208/23‐1) to K.H.K. and the Deutscher Akademischer Austauschdienst (DAAD) to N.K. JO was supported by grant STU642‐1 / 1 from the German Research Council (DFG) to JS, and seed funding from the Collaborative Research Center 648 (Collaborative Research Center 648) to JS. JS and JO acknowledge Ulla Bonas for continous support.
Contributor Information
Karl‐Heinz Kogel, Email: Karl-Heinz.Kogel@agrar.uni-giessen.de.
Jafargholi Imani, Email: Jafargholi.Imani@agrar.uni-giessen.de.
References
- Acevedo‐Garcia, J. , Kusch, S. and Panstruga, R. (2014) Magical mystery tour: MLO proteins in plant immunity and beyond. 2014. New Phytol. 204, 273–281. [DOI] [PubMed] [Google Scholar]
- Alzohairy, A.M. , Yousef, M.A. , Edris, S. , Kerti, B. , Gyulai, G. and Bahieldin, A. (2012) Detection of LTR retrotransposons reactivation induced by in vitro environmental stresses in barley (Hordeum vulgare) via RT‐qPCR. Life Sci. J. 9, 1–8. [Google Scholar]
- Baker, K.E. and Parker, R. (2004) Nonsense‐mediated mRNA decay: terminating erroneous gene expression. Curr. Opin. Cell Biol. 16, 293–299. [DOI] [PubMed] [Google Scholar]
- Bordiya, Y. , Zheng, Y. , Nam, J.C. , Bonnard, A.C. , Choi, H.W. , Lee, B.K. , Kim, J. et al. (2016) Pathogen infection and MORC proteins affect chromatin accessibility of transposable elements and expression of their proximal genes in Arabidopsis. Mol. Plant Microbe Interact. 29, 674–687. [DOI] [PubMed] [Google Scholar]
- Bortesi, L. and Fischer, R. (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 41–52. [DOI] [PubMed] [Google Scholar]
- Brabbs, T.R. , He, Z. , Hogg, K. , Kamenski, A. , Li, Y. , Paszkiewicz, K.H. , Moore, K.A. et al. (2013) The stochastic silencing phenotype of Arabidopsis morc6 mutants reveals a role in efficient RNA‐directed DNA methylation. Plant J. 75, 836–846. [DOI] [PubMed] [Google Scholar]
- Brinkman, E.K. , Chen, T. , Amendola, M. and van Steensel, B. (2014) Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, C. , Nekrasov, V. , Lippman, Z.B. and Eck, J.V. (2014) Efficient gene‐editing in tomato in the first generation using the CRISPR/Cas9 system. Plant Physiol. 3, 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian, M. , Cermak, T. , Doyle, E.L. , Schmidt, C. , Zhang, F. , Hummel, A. , Bogdanove, A.J. et al. (2010) Targeting DNA double‐strand breaks with TAL effector nucleases. Genetics, 186, 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, L. , Ran, F.A. , Cox, D. , Lin, S. , Barretto, R. , Habib, N. , Hsu, P.D. et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly, S. , Marshallsay, C. , Leader, D. , Brown, J.W.S. and Filipowicz, W. (1994) Small nuclear RNA genes transcribed by either RNA polymerase II or RNA polymerase III in monocot plants share three promoter elements and use a strategy to regulate gene expression different from that used by their dicot plant counterparts. Mol. Cell. Biol. 14, 5910–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea, S. , Freyvert, Y. , Santiago, Y. , Holmes, M.C. , Urnov, F.D. , Gregory, P.D. and Cost, G.J. (2013) In vivo cleavage of transgene donors promotes nuclease‐mediated targeted integration. Biotechnol. Bioeng. 110, 871–880. [DOI] [PubMed] [Google Scholar]
- Deltcheva, E. , Chylinski, K. , Sharma, C.M. , Gonzales, K. , Chao, Y. , Pirzada, Z.A. , Eckert, M.R. et al. (2011) CRISPR RNA maturation by trans‐encoded small RNA and host factor RNase III. Nature, 471, 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen, R.H. , Pelizzola, M. , Schmitz, R.J. , Lister, R. , Dowen, J.M. , Nery, J.R. , Dixon, J.E. et al. (2012) Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl Acad. Sci. USA, 109, E2183–E2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, J.J. and Doyle, J.L. (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15. [Google Scholar]
- Engler, C. , Youles, M. , Gruetzner, R. , Ehnert, T.M. , Werner, S. , Jones, J.D. , Patron, N.J. et al. (2014) A golden gate modular cloning toolbox for plants. ACS Synth. Boil. 3, 839–843. [DOI] [PubMed] [Google Scholar]
- Fauser, F. , Schiml, S. and Puchta, H. (2014) Both CRISPR/Cas‐based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana . Plant J. 79, 348–359. [DOI] [PubMed] [Google Scholar]
- Feng, Z. , Mao, Y. , Xu, N. , Zhang, B. , Wei, P. , Yang, D.L. , Wang, Z. et al. (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas‐induced gene modifications in Arabidopsis. Proc. Natl Acad. Sci. USA, 111, 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y. , Zhang, Y. , Zhang, D. , Dai, X. , Estelle, M. and Zhao, Y. (2015) Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc. Natl Acad. Sci. USA, 112, 2275–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas, G. , Barrangou, R. , Horvath, P. and Siksnys, V. (2012) Cas9‐crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA, 109, E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnery, S. and Mathews, M.B. (1995) Functional mRNA can be generated by RNA polymerase III. Mol. Cell. Biol. 15, 3597–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, C.J. , Husmann, D. , Liu, W. , El Kasmi, F. , Wang, H. , Papikian, A. , Pastor, W.A. et al. (2016) Arabidopsis AtMORC4 and AtMORC7 form nuclear bodies and repress a large number of protein‐coding genes. PLoS Genet. 12, e1005998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holme, I.B. , Wendt, T. , Gil‐Humanes, J. , Deleuran, L.C. , Starker, C.G. , Voytas, D.F. and Brinch‐Pedersen, H. (2017) Evaluation of the mature grain phytase candidate HvPAPhy_a gene in barley (Hordeum vulgare L.) using CRISPR/Cas9 and TALENs. Plant Mol. Biol. 95, 111–121. [DOI] [PubMed] [Google Scholar]
- Hückelhoven, R. , Dechert, C. and Kogel, K.‐H. (2003) Over‐expression of barley BAX Inhibitor‐1 induces enhanced accessibility to Blumeria graminis and breakdown of mlo‐mediated penetration resistance in barley. Proc. Nat. Acad. Sci. USA, 100, 5555–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imani, J. , Li, L. , Schäfer, P. and Kogel, K.‐H. (2011) STARTS—a stable root transformation system for rapid functional analyses of proteins of the monocot model plant barley. Plant J. 67, 726–735. [DOI] [PubMed] [Google Scholar]
- International Barley Genome Sequencing Consortium (IBSC) . (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature, 491, 711–716. [DOI] [PubMed] [Google Scholar]
- Iyer, L.M. , Abhiman, S. and Aravind, L. (2008) MutL homologs in restriction modification systems and the origin of eukaryotic MORC ATPases. Biol. Direct. 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, C. , von Wettstein, D. , Schäfer, W. , Kogel, K.‐H. , Felk, A. and Maier, F.J. (2005) Infection patterns in barley and wheat spikes inoculated with wild‐type and trichodiene synthase gene disrupted Fusarium graminearum . Proc. Natl Acad. Sci. USA, 102, 16892–16897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. , Zhou, H. , Bi, H. , Fromm, M. , Yang, B. and Weeks, D.P. (2013) Demonstration of CRISPR/Cas9/sgRNA‐mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41, e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M. , Chylinski, K. , Fonfara, I. , Hauer, M. , Doudna, J.A. and Charpentier, E. (2012) A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H.‐G. , Kuhl, J.C. , Kachroo, P. and Klessig, D.F. (2008) CRT1, an Arabidopsis ATPase that interacts with diverse resistance proteins and modulates disease resistance to turnip crinkle virus. Cell Host Microbe, 3, 48–57. [DOI] [PubMed] [Google Scholar]
- Kang, H.‐G. , Oh, C.S. , Sato, M. , Katagiri, F. , Glazebrook, J. , Takahashi, H. , Kachroo, P. et al. (2010) Endosome associated CRT1 functions early in resistance gene‐mediated defense signaling in Arabidopsis and tobacco. Plant Cell, 22, 918–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H.‐G. , Hyong, W.C. , von Einem, S. , Manosalva, P. , Ehlers, K. , Liu, P.P. , Buxa, S.V. et al. (2012) CRT1 is a nuclear‐translocated MORC endonuclease that participates in multiple levels of plant immunity. Nat. Commun. 3, 1297. [DOI] [PubMed] [Google Scholar]
- Kapusi, E. , Corcuera‐Gómez, M. , Melnik, S. and Stoger, E. (2017) Heritable genomic fragment deletions and small indels in the putative ENGase gene induced by CRISPR/Cas9 in barley. Front Plant Sci. 8, 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, A. , Kang, H.‐G. , Steinbrenner, J. , Dempsey, D.A. , Klessig, D.F. and Kogel, K.‐H. (2017) MORC proteins: novel players in plant and animal health. Front Plant Sci. 8, 1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen, G. , von Einem, S. , Koch, A. , Imani, J. , Pai, S.B. , Manohar, M. , Ehlers, K. et al. (2014) The compromised recognition of turnip crinkle virus1 subfamily of microrchidia ATPases regulates disease resistance in barley to biotrophic and necrotrophic pathogens. Plant Physiol. 164, 866–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, J.A. and Jacobsen, S.E. (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenson, T. , Shorinola, O. , Stacey, N. , Li, C. , Ostergaard, L. , Patron, N. , Uauy, C. et al. (2015) Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA‐guided Cas9 nuclease. Genome Biol. 16, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo, G.R. , Stein, P.A. and Ludwig, R.A. (1991) A DNA transformation‐competent Arabidopsis genomic library in Agrobacterium. Nat. Biotechnol. 9, 963–967. [DOI] [PubMed] [Google Scholar]
- Li, J.F. , Norville, J.E. , Aach, J. , McCormack, M. , Zhang, D. , Bush, J. , Church, G.M. et al. (2013) Multiplex and homologous recombination‐mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z.W. , Shao, C.R. , Zhang, C.J. , Zhou, J.X. , Zhang, S.W. , Li, L. , Chen, S. et al. (2014) The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA‐directed DNA methylation loci. PLoS Genet. 10, e1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković, Z.J. , Naumann, U. , Matzke, A.J.M. and Matzke, M. (2012) Involvement of a GHKL ATPase in RNA‐directed DNA Methylation in Arabidopsis thaliana . Curr. Biol. 22, 933–938. [DOI] [PubMed] [Google Scholar]
- Lowder, L.G. , Zhang, D. , Baltes, N.J. , Paul, J.W. III , Tang, X. , Zheng, X. , Voytas, D.F. et al. (2015) A CRISPR/Cas9 toolbox for multiplexed plant gnome editing and transcriptional regulation. Plant Physiol. 169, 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Zhang, Q. , Zhu, Q. , Liu, W. , Chen, Y. , Qiu, R. , Wang, B. et al. (2015) A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant, 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Zhu, Q. , Chen, Y. and Liu, Y.G. (2016) CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol. Plant, 9, 961–974. [DOI] [PubMed] [Google Scholar]
- Mali, P. , Yang, L. , Esvelt, K.M. , Aach, J. , Guell, M. , DiCarlo, J.E. , Norville, J.E. et al. (2013) RNA‐guided human genome engineering via Cas9. Science, 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzahn, A. , Lowder, L. and Qi, Y. (2017) Plant genome editing with TALEN and CRISPR. Cell Biosci. 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosalva, P. , Manohar, M. , Kogel, K.‐H. , Kang, H.‐G. and Klessig, F. (2015) The GHKL ATPase MORC1 modulates species‐specific plant immunity in Solanaceae. Mol. Plant Microbe Interact. 28, 927–942. [DOI] [PubMed] [Google Scholar]
- Mao, Y. , Zhang, H. , Xu, N. , Zhang, B. , Gou, F. and Zhu, J.K. (2013) Application of the CRISPR‐Cas system for efficient genome engineering in plants. Mol. Plant, 6, 2008–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca, M. , Lin, V.G. , Guo, N. and Yang, Y. (2013) Obligate ligation‐gated recombination (ObLiGaRe): custom‐designed nuclease‐mediated targeted integration through nonhomologous end joining. Genome Res. 23, 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshallsay, C. , Connelly, S. and Filipowicz, W. (1992) Characterization of the U3 and U6 snRNA genes from wheat: U3 snRNA genes in monocot plants are transcribed by RNA polymerase III. Plant Mol. Biol. 19, 973–983. [DOI] [PubMed] [Google Scholar]
- Matzke, M. , Kanno, T. , Daxinger, L. , Huettel, B. and Matzke, A.J. (2009) RNA‐mediated chromatin‐based silencing in plants. Curr. Opin. Cell Biol. 21, 367–376. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A. , Kanno, T. and Matzke, A.J. (2015) RNA‐directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 66, 243–267. [DOI] [PubMed] [Google Scholar]
- Miao, J. , Guo, D. , Zhang, J. , Huang, Q. , Qin, G. , Zhang, X. , Wan, J. et al. (2013) Targeted mutagenesis in rice using CRISPR‐Cas system. Cell Res. 23, 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard, G. , Cokus, S.J. , Cary, J. , Feng, S.H. , Billi, A.C. , Stroud, H. , Husmann, D. et al. (2012) MORC family ATPases required for heterochromatin condensation and gene silencing. Science, 336, 1448–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard, G. , Bischof, S. , Husmann, D. , Pastor, W.A. , Hale, C.J. , Yen, L. , Stroud, H. et al. (2014) Transcriptional gene silencing by Arabidopsis microrchidia homologues involves the formation of heteromers. Proc. Natl Acad. Sci. USA, 111, 7474–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov, V. , Staskawicz, B. , Weigel, D. , Jones, J.D. and Kamoun, S. (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA‐guided endonuclease. Nat. Biotechnol. 31, 691–693. [DOI] [PubMed] [Google Scholar]
- Ordon, J. , Gantner, J. , Kemna, J. , Schwalgun, L. , Reschke, M. , Streubel, J. , Boch, J. et al. (2017) Generation of chromosomal deletions in dicotyledonous plants employing a user‐friendly genome editing toolkit. Plant J. 89, 155–168. [DOI] [PubMed] [Google Scholar]
- Pastor, W.A. , Stroud, H. , Nee, K. , Liu, W. , Pezic, D. , Manakov, S. , Lee, S.A. et al. (2014) MORC1 represses transposable elements in the mouse male germline. Nat. Commun. 5, 5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, J. and Zhao, Y. (2003) The CW domain, a structural module shared amongst vertebrates, vertebrate‐infecting parasites and higher plants. Trends Biochem. Sci. 28, 576–580. [DOI] [PubMed] [Google Scholar]
- Qu, F. , Zhai, W. , Chen, H. , Zhu, L.H. and Morris, T.J. (1996) Cloning, characterization and transient expression of the gene encoding a rice U3 small nuclear RNA. Gene, 172, 217–220. [DOI] [PubMed] [Google Scholar]
- van Schie, C.C.N. and Takken, F.L.W. (2014) Susceptibility genes 101: how to be a good host. Ann. Rev. Phytopath. 52, 551–581. [DOI] [PubMed] [Google Scholar]
- Schweizer, P. , Pokorny, J. , Abderhalden, O. and Dudler, R. (1999) A transient assay system for the functional assessment of defense‐related genes in wheat. Mol. Plant Microbe Interact. 12, 647–654. [Google Scholar]
- Shan, Q.W. , Wang, Y.P. , Li, J. , Zhang, Y. , Chen, K.L. , Liang, Z. , Zhang, K. et al. (2013) Targeted genome modification of crop plants using a CRISPR‐Cas system. Nat. Biotechnol. 31, 686–688. [DOI] [PubMed] [Google Scholar]
- Smith, J. , Bibikova, M. , Whitby, F.G. , Reddy, A.R. , Chandrasegaran, S. and Carroll, D. (2000) Requirements for double‐strand cleavage by chimeric restriction enzymes with zinc finger DNA‐recognition domains. Nucleic Acids Res. 28, 3361–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, I. , Greenside, P.G. , Natoli, T. , Lahr, D.L. , Wadden, D. , Tirosh, I. , Narayan, R. et al. (2017) Evaluation of RNAi and CRISPR technologies by large‐scale gene expression profiling in the connectivity map. PLoS Biol. 15, e2003213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R. , Lawrence, C.M. and Wiedenheft, B. (2013) CRISPR‐mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 82, 237–266. [DOI] [PubMed] [Google Scholar]
- Svitashev, S. , Young, J.K. , Schwartz, C. , Gao, H.R. , Falco, S.C. and Cigan, A.M. (2015) Targeted mutagenesis, precise gene‐editing, and site‐specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169, 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingay, S. , McElroy, D. , Kalla, R. , Fieg, S. , Wang, M. , Thornton, S. and Brettell, R. (1997) Agrobacterium tumefaciens‐mediated barley transformation. Plant J. 11, 1369–1376. [Google Scholar]
- Waibel, F. and Filipowicz, W. (1990) RNA‐polymerase specificity of transcription of Arabidopsis U snRNA genes determined by promoter element spacing. Nature, 346, 199–202. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Cheng, X. , Shan, Q. , Zhang, Y. , Liu, J. , Gao, C. and Qiu, J.L. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Xing, H.L. , Dong, L. , Wang, Z.P. , Zhang, H.Y. , Han, C.Y. , Liu, B. , Wang, X.C. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, J.S. , Ding, J. and Li, Y. (2015) Genome‐editing technologies and their potential application in horticultural crop breeding. Hortic. Res. 2, 15019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. (2016) Functional analysis of Arabidopsis thaliana matrix metalloproteinases and MORC in plant immunity (Doctoral dissertation). Retrieved from Giessener Elektronische Bibliothek (http://geb.uni-giessen.de/geb/volltexte/2016/12043/pdf/ZhangFei_2016_04_14.pdf)
- Zhang, H. , Zhang, J. , Wei, P. , Zhang, B. , Gou, F. , Feng, Z. , Mao, Y. et al. (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene‐editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Maps of constructed plasmid.
Figure S2 Target site conservation in MORC genes.
Figure S3 638 bp sequence of barley RNA Pol III promoter (TATA box is underlined) (A); 380 bp sequence of rice RNA Pol III promoter (TATA box is underlined) (B); Sequence of sgRNA is underlined with terminator (C).
Figure S4 Relative expression of sgRNA under control of barley and rice U3 promoter (pHvU3 and pOsU3) in leaves of hvmorc1‐KO T2 homozygous mutants measured by RT‐PCR and normalized against Hygromycin gene.
Figure S5 SpCas9‐induced frame‐shift mutations in HvMORC1 leads to premature STOP codons Predicted HvMORC1 open reading frames (in red) with premature stop codons after Cas9 induced mutation (b‐d) compared to wt (A) using online tool (http://web.expasy.org/translate/).
Table S1 Oligonucleotide primers used in this study (restriction sites are underlined).
Appendix S1 Cloning manual for pMGE genome editing vectors.
Appendix S2 Annotated sequence files (Genbank) for pMGE vectors.
Supplementary File


