Summary
Previous studies revealed that the promoters for driving both Cas9 and sgRNAs are quite important for efficient genome editing by CRISPR/Cas9 in plants. Here, we report our results of targeted genome editing using the maize dmc1 gene promoter combined with the U3 promoter for Cas9 and sgRNA, respectively. Three loci in the maize genome were selected for targeting. The T0 plants regenerated were highly efficiently edited at the target sites with homozygous or bi‐allelic mutants accounting for about 66%. The mutations in T0 plants could be stably transmitted to the T1 generation, and new mutations could be generated in gametes or zygotes. Whole‐genome resequencing indicated that no off‐target mutations could be detected in the predicted loci with sequence similarity to the targeted site. Our results show that the dmc1 promoter‐controlled (DPC) CRISPR/Cas9 system is highly efficient in maize and provide further evidence that the optimization of the promoters used for the CRISPR/Cas9 system is important for enhancing the efficiency of targeted genome editing in plants. The evolutionary conservation of the dmc1 gene suggests its potential for use in other plant species.
Keywords: maize, targeted genome editing, CRISPR/Cas9, dmc1 promoter
Introduction
The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR‐associated 9 (Cas9) system is derived from a bacterial immune system and has been widely used for targeted genome editing (Cong et al., 2013; Hsu et al., 2014; Mali et al., 2013). Unlike the two formerly developed sequence specific nucleases (SSNs), zinc finger nucleases (ZFNs) and transcription activator‐like effector nucleases (TALENs), the CRISPR/Cas9 is an RNA‐guided endonuclease to generate DNA double‐strand breaks (DSBs) at the target sites of the genome (Gaj et al., 2013). Usually two pathways, including nonhomologous end‐joining (NHEJ) or homology‐directed repair (HDR), are induced to repair the DSBs. The NHEJ pathway is error‐prone and frequently generates indels at the repair sites, while HDR could be adopted for precise sequence replacement or insertion when a template DNA is provided (Puchta and Fauser, 2014).
In 2013, targeted genome editing by CRISPR/Cas9 was first reported in mammalian cell lines (Cong et al., 2013; Mali et al., 2013). Later this system has been widely used in various species (DiCarlo et al., 2013; Friedland et al., 2013; Gratz et al., 2013; Hwang et al., 2013; Jiang et al., 2013; Li et al., 2013a,c; Yu et al., 2013). In plants, CRISPR/Cas9‐mediated genome editing was first simultaneously reported in three model species (Li et al., 2013b; Nekrasov et al., 2013; Shan et al., 2013). Since then, genome editing in plants has been reported by several other groups (Brooks et al., 2014; Feng et al., 2013; Liang et al., 2014; Miao et al., 2013; Xie and Yang, 2013; Xing et al., 2014). These reports provide evidence that the CRISPR/Cas9 could be used for targeted genome editing in plants; however, the editing efficiency is relatively low.
To enhance the efficiency of CRISPR/Cas9‐mediated genome editing in plants, improvements need to be made. In vertebrates, the Cas9 (mRNA or protein) and sgRNAs could be simultaneously injected into the zygotic cells. Bi‐allelic mutations for multiple sites in the genome took place before the first cell division, thus generating multigene bi‐allelic mutants (Friedland et al., 2013; Hwang et al., 2013; Li et al., 2013a,c). In this way, the dose of Cas9 and sgRNAs could be controlled to improve the targeting possibility. However, plant zygotic cells are difficult to be transformed directly due to technological limitations. At present, tissue culture is necessary for transformation of most plant species, which is preferentially mediated by Agrobacterium. Thus, for most plants, callus cells are exposed to gene editing reagents for initial targeting. The targeted editing efficiency in calli cells directly determines the proportion of targeted mutant output. Studies in rice and other species suggest the factors that could influence the targeting efficiency include the codons used for Cas9 translation, promoters used for both the sgRNAs and Cas9 gene expression and so on (Ma et al., 2015; Xie et al., 2015; Xu et al., 2014; Zhang et al., 2014). For example, the Ubiquitin gene promoter apparently works better than the CaMV 35S promoter in rice. In Arabidopsis, the CaMV 35S promoter used for Cas9 generated only chimeras in the T1 generation without homozygotes or bi‐allelic mutants (Feng et al., 2013, 2014; Xing et al., 2014). However, an egg cell‐specific promoter used for Cas9 could generate triple mutants in the T1 generation (Wang et al., 2015).
Maize is one of the most important crops in the world. Traditionally, genetic study of maize depends on chemical mutagenesis using ethyl methane sulphonate (EMS) or transposon tagging to generate mutants. Both are time consuming for mutant screening and the required generations of backcrossing (Nannas and Dawe, 2015). Efficient targeted genome editing in maize could simplify the mutant creation process. Targeted genome editing by CRISPR/Cas9 in maize protoplasts was first reported in 2014 (Liang et al., 2014). Thereafter, another group reported the generation of mutants of the ZmHKT gene in the maize B73 inbred line (Xing et al., 2014). The maize ubiquitin1 gene promoter and rice U3 or wheat U3 promoters were used for a maize codon‐optimized Cas9 (zCas9) and sgRNAs, respectively. The high efficiency of CRISPR/Cas9‐mediated targeted mutation in maize was also reported for three targeted alleles when using biolistic‐mediated transformation in one report (Svitashev et al., 2015). In another two reports, the combination of ubiquitin1 gene promoter and maize U6 promoter, or CaMV 35S promoter and maize U3 promoter for Cas9 and sgRNAs generated mutations at 13% or 2% in T0 generation seedlings when using Agrobacterium‐mediated Cas9/sgRNA delivery; most seedlings are chimeras (Feng et al., 2016; Zhu et al., 2016). More recently, when using the maize ubiquitin1 gene promoter and two rice U6 promoters for a rice codon‐optimized Cas9 and sgRNAs, mutations generated by a combination of two sgRNAs targeted at one gene were reported up to 70%, with bi‐allelic mutations at a frequency of 22%–58% for four genes (Char et al., 2016). Most recently, a high frequency of targeted mutation for one site in the LG1 gene in maize ZC01 inbred line was reported (Li et al., 2017). In most reports, Cas9 is driven by the maize ubiquitin1 promoter and indeed proved to be more efficient than CaMV 35S. However, with the use of different promoters for sgRNA expression, the targeting efficiency varied significantly from each other, although the same promoter was used for Cas9. These studies revealed that the mutation efficiency of CRISPR/Cas9 system largely depends on both the expressions of Cas9 and sgRNAs. Thus, screening and testing of new combinations of promoters for both Cas9 and sgRNA will improve targeted genome editing efficiency in plant species.
Here, we report our results of using a maize dmc1 promoter‐controlled (DPC) CRISPR/Cas9 system and generating highly efficient targeted genome editing in maize Hi‐II germplasm. Targeting a locus with one sgRNA, the T0 plants regenerated were high‐efficiently edited at the target sites with homozygous or bi‐allelic mutants accounting for about 66%, with the remainder heterozygous or mosaic mutants. Mutations in the T0 plants could be stably transmitted to the next generation, and at the same time, new mutations would be generated. Whole‐genome resequencing detected no mutations at the predicted off‐target sites.
Results
Dmc1 promoter‐controlled (DPC) CRISPR/Cas9‐mediated targeted editing of zb7 in maize T0 generation
In our previous work, the 35S promoter was used for driving the Cas9 (human codon‐optimized) expression in maize. A screen of over one hundred mutant lines only produced one plant with the mutant phenotype. Most of the regenerated plants were mosaic with a low mutation ratio (Feng et al., 2016). In this work, our purpose was to choose a meiosis‐specific promoter for increasing the mutation efficiency and at the same time avoiding the mosaicism. Thus, we chose the promoter of the maize dmc1 gene, which was reported to be expressed specifically in meiocytes (Klimyuk and Jones, 1997) and reasoned that gametes in T0 plants could be mutated and then recover homozygous or bi‐allelic mutants in the T1 generation at high frequency.
In our DPC CRISPR/Cas9 system, the expression of Cas9 gene is controlled by the maize dmc1 gene promoter and nos terminator, and the sgRNA is driven by the maize U3 promoter (Figure 1; Leader et al., 1994). We also checked the efficiency of targeted gene editing by U3 and U6 promoters in protoplasts (Zhu et al., 2016); the results revealed that the maize U3 promoter works better than U6 for the sites examined (Figure S1). To make the later analysis simpler, the maize zb7 gene was chosen as a target gene due to its albino phenotype when completely knocked out (Figure 2a; Lu et al., 2012). For targeting the zb7 gene, the DPC CRISPR/Cas9 constructs were transformed into maize Hi‐II immature embryos by Agrobacterium in two batches, and in total, 10 bialaphos‐resistant calli (transgene‐positive calli) were obtained. Although we supposed that the dmc1 promoter‐controlled Cas9 gene should be specifically expressed in reproductive tissue, we checked the expression of the Cas9 gene in the transgene‐positive calli. Total RNA was extracted from calli of the first batch (three of the four transgene‐positive calli) and was used for expression analysis. To our surprise, the Cas9 gene had a relatively high expression level in the calli (Figure S2a). To further test whether the dmc1 promoter is active in calli, we compared the expression level of dmc1 gene in different tissues. The results indicated that the dmc1 gene was highly expressed in callus and tassel, but weakly expressed in leaf and root (Figure S2b). These results prompted us to further identify whether the target locus was edited in calli. We then extracted genomic DNA from the four calli of the first batch and for each one we produced three independent sample duplicates. Because there was a restriction site coincident with the Cas9‐cutting site, by RFLP (restriction fragment length polymorphism) assay of the PCR amplicons involving the target site, we were again surprised to find that the zb7 gene was edited in all four calli, with homozygous or bi‐allelic mutations present in three of them (Figure 2b). In some calli, the mutation types of the three duplicate samples were different and indicated the editing occurred at different stages. In the second batch of transformations, six transgene‐positive calli were obtained, homozygous or bi‐allelic mutations could also be detected. To confirm the mutations in calli revealed by RFLP assays, the PCR products of the target site were directly subjected to Sanger sequencing. The sequencing results showed mono‐allelic, di‐allelic or multi‐allelic mutations in calli, and the mutation types were mostly small insertions and deletions (indels; Figure 2c). The pattern of mutation sequences in calli also indicated that mutations occurred at different stages during calli development.
Figure 1.

Schematic illustration of DPC CRISPR/Cas9 expression T‐DNA. Maize dmc1 gene and U3 promoters were used for Cas9 and sgRNA, respectively.
Figure 2.
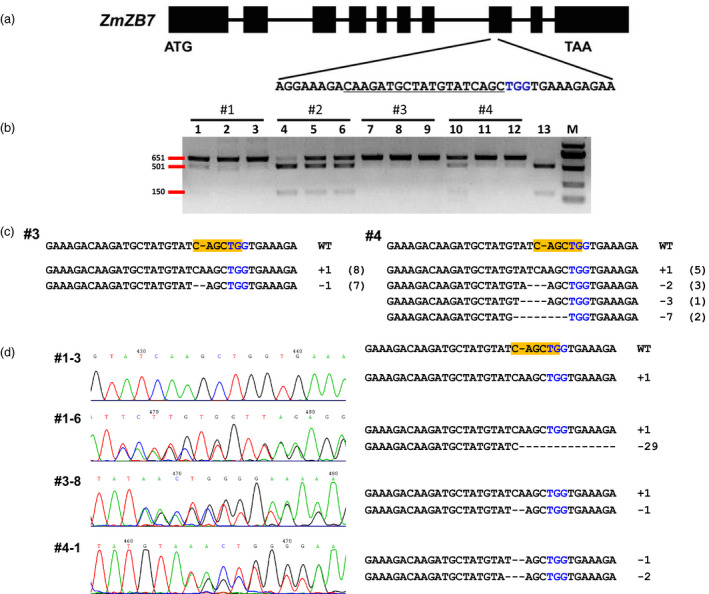
Identification of mutations in the zb7 gene by RFLP assay and Sanger sequencing. (a) Schematic illustration of the target site in the zb7 gene. Black box indicates exons, while the lines between them represent introns. Underlined sequence was selected for targeting; nucleotides marked in blue represent PAM (protospacer adjacent motif). (b) RFLP assay of the genomic DNA of four transgene‐positive calli (#1–#4) in the first batch. For each callus, three independent samples were collected and used. Lane 1–3, calli #1; lane 4–6, calli #2; lane 7–9, calli #3; lane 10–12, calli #4; lane 13, control (wild‐type DNA amplicons digested with Pvu II). M, DNA marker. Primer pair zb7‐F/zb7‐R was used for PCR amplification, Pvu II was used for digestion. (c) Mutation analysis of two transgene‐positive calli by cloning followed by Sanger sequencing. #3, #4 were the two calli sample with homozygous or bi‐allelic mutations by RFLP assay. (d) Sanger sequencing results of mutations in regenerated seedlings. #1–3, #1–6, #3–8, #4–1 were selected seedlings used for analysis. Left, sequencing chromatograph and right, the edited sequences at the target site. The yellow box indicates the Pvu II site.
The transgene‐positive calli were then cultured for regeneration. For the two batches, in total, 237 seedlings were regenerated from the ten calli. There were four kinds of phenotypes observed among all the seedlings, including albino, chlorosis, chimeric and wild type (Figure 3a–h), with the ratio for each one 51%, 15%, 19% and 15%, respectively (Table S1). The genotyping results indicated that the genotype corresponds to the phenotype. In albino seedlings, the mutations in both the two alleles were found to cause frameshift during translation, thus completely knocking out the gene. In chlorotic seedlings, bi‐allelic mutations were detected with a 3 bp deletion and a 11 bp deletion for the two alleles, respectively. Because the 3 bp deletion only led to a glutamine deletion at the protein level but not a frameshift, this kind of mutated allele may act as a weak allele and still retain partial function (Figure S3a). In fact, the chlorotic seedling could only grow for 3–4 weeks before it perished; the albino seedlings can just survive 2 weeks. In chimeric seedlings, a high ratio of mutant alleles could be detected, and in wild‐type seedlings, mono‐allelic or low‐ratio of mutant alleles was detected. The phenotypes of the seedlings were also consistent with the genotyping results of the calli. For example, in event (transgene‐positive calli) #4, seedlings regenerated include multiple phenotypes; in event #3, all seedlings regenerated were albino (Figure 3a,b and Table S1). In total, 66% of regenerated seedlings had a homozygous or bi‐allelic mutant phenotype (Table S1). The genotypes of the seedlings were also confirmed by RFLP assay of more than 10 seedlings selected from events #1–#4 (Figure S4). To further analyse the mutation types in the seedlings, the plants chosen for RFLP analysis were also selected for direct sequencing of the PCR products spanning the targeted site. The results revealed that all the albino seedlings were homozygous or bi‐allelic mutants, with most of the remainder being heterozygous or mosaic mutants (Table S2). Considering that the albino and chlorotic seedlings are homozygous or bi‐allelic mutants, the ratio of homozygous or bi‐allelic mutants in each event and total events was counted. For the target site in the zb7 gene, the T0 plants regenerated were edited high efficiently with homozygous or bi‐allelic mutants accounting for about 66% (Table 1).
Figure 3.

Phenotypes of selected T0 seedlings created by DPC CRISPR/Cas9 at the zb7 gene. (a, b) Seedling regenerated from two independent transgene‐positive calli. (c–h) Selected seedlings show different representative phenotypes. (c) and (d), albino. (e), chlorosis. (f) and (g), chimera. (h), control (wild‐type Hi‐II seedling). Bars = 1 cm.
Table 1.
Genotype statistics of transgenic T0 plants targeted by DPC CRISPR/Cas9 system at the zb7 gene
| Transgenic events | Homozygous & bi‐allelic mutants | Heterozygous & mosaic mutants | Total | Ratio of homozygous & bi‐allelic mutants |
|---|---|---|---|---|
| #1 | 29 | 10 | 39 | 74% |
| #2 | 1 | 19 | 20 | 5.0% |
| #3 | 41 | 0 | 41 | 100% |
| #4 | 18 | 3 | 21 | 86% |
| #5 | 9 | 0 | 9 | 100% |
| #6 | 16 | 2 | 18 | 89% |
| #7 | 19 | 0 | 19 | 100% |
| #8 | 1 | 13 | 14 | 7% |
| #9 | 16 | 12 | 28 | 57% |
| #10 | 6 | 22 | 28 | 21% |
| Total | 156 | 81 | 237 | 66% |
DPC CRISPR/Cas9‐mediated targeted editing of another two sites in the T0 generation
To test whether the DPC CRISPR/Cas9 system could generally mediate high‐efficiency genome editing in maize, we then chose another two target sites for targeting. One target site is located in the 3rd exon of the maize zyp1 gene, which is reported to encode the central element protein of the synaptonemal complex (SC) and be functional in meiosis (Figure 4a; Barakate et al., 2014). The DPC CRISPR/Cas9 construct for targeting this site was transformed into maize immature embryos by Agrobacterium. In total, 26 seedlings, which were regenerated from three independent calli lines, were subjected to analysis. The RFLP assay indicated that 12 seedlings showed homozygous or bi‐allelic mutations (Figure 4b). We then directly subjected the PCR products for Sanger sequencing. The results showed that 23 seedlings were homozygous or bi‐allelic mutants (Table S3). The reason that the RFLP results differ with the Sanger sequencing is due to the mutations not disrupting the restriction site (Figure 4c). Most mutations are small insertions and deletions; mutations altering protein sequence but not causing a frameshift were identified. For this target site, the regenerated T0 plants were edited efficiently with 23 of 26 seedlings homozygous or bi‐allelic mutants.
Figure 4.
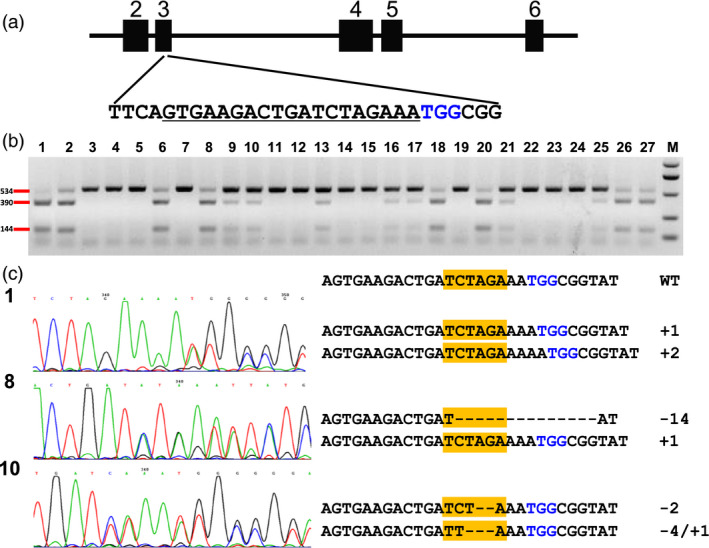
Identification of mutations in the zyp1 gene by RFLP assay and Sanger sequencing. (a) Schematic illustration of the target site in the zyp1 gene. Black box indicates exons, while the lines between them represent introns. Underlined sequence was selected for targeting; nucleotides marked in blue represent PAM. (b) RFLP assay of the genomic DNA of transgenic seedlings. Lane 1, control (wild‐type DNA amplicons digested with XbaI). Lane 2–27, 26 random selected transgenic seedlings. M, DNA marker. Primer pair zyp1‐F/zyp1‐R was used for PCR amplification, XbaI was used for digestion. (c) Sanger sequencing results of three seedlings. Left shows sequencing chromatograph, and right shows the edited sequences at the target site. Nucleotides marked in blue represent PAM. The yellow box indicates the XbaI site.
Another target site is located in the exonic region of the gene smc3, which has a putative role for chromosome structural maintenance and proved to be essential for embryo development (Figure 5a; Liu et al., 2002). The DPC CRISPR/Cas9 construct for targeting this gene was transformed into maize immature embryos by Agrobacterium. The transformation efficiency for the construct targeting this site is much lower (0.15%) compared with other constructs under the same conditions (1.5%). Only two transgene‐positive calli were obtained after a longer culture time (2 weeks) for selection due to the weak growth of the calli. The significantly lower transformation efficiency for this site suggested that knockout of this gene in maize calli may be lethal, implying that homozygous or bi‐allelic mutations occurred in most early transformed calli cells. To confirm that targeted mutation occurred in this site, we analysed the two transgene‐positive calli by genotyping. The results revealed that both the two calli are chimeras with multiple mutant alleles; the ratio of the mutant alleles in the two calli is 66.7% and 50%, respectively (Figure 5b,c).
Figure 5.
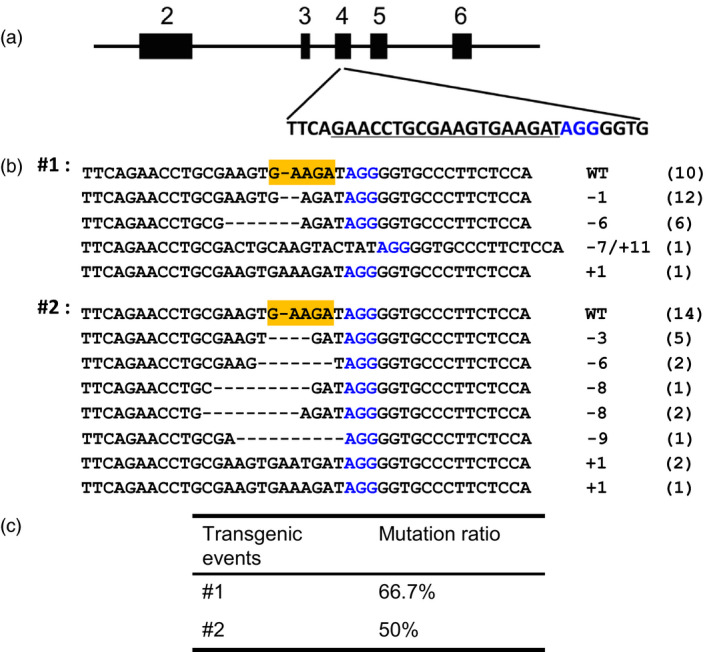
Mutation analysis of the smc3 site targeted by DPC CRISPR/Cas9 system in transgenic calli. (a) Schematic illustration of the target site in the smc3 gene. Black box indicates exons, while the line between them introns. Underlined sequence was selected for targeting; nucleotides marked in blue represent PAM. (b) Mutation analysis of the two transgene‐positive calli by cloning followed by Sanger sequencing. Sequence of the target site was amplified by PCR with primer pair smc3‐F/smc3‐R and cloned. Thirty clones were subjected to Sanger sequencing for each callus. The mutation types and number for different clones are shown on the right. (c) A table showing the ratio of mutant alleles in the two calli.
Because a pretest of the activity of sgRNAs is helpful, we also tested whether the DPC CRISPR/Cas9 system can generate genome editing in protoplasts. The results showed that all the three targeted loci chosen could be edited. By Sanger sequencing of the site‐specific PCR amplicons, indels at the target loci were detected (Figure S5a,b).
Transmission of the mutations to the next generation and generation of new mutations
As the homozygous or bi‐allelic mutants for the zb7 gene can only survive a few weeks, the mutant alleles could not be transmitted. The mosaic seedlings with high mutation ratio also grow weakly and do not survive. To check whether the mutations created by the DPC CRISPR/Cas9 system could be transmitted to the offspring and new mutations could be generated, we planted the heterozygous seedlings and mosaic seedlings with low mutation ratios and performed crosses among them (Table S4). In five crosses among heterozygous plants and mosaic plants with low mutation ratios (<10%), immature embryos of the hybrid ears were cultured, and the genotype of T1 seedlings was analysed by RFLP assay and Sanger sequencing (Table S4). In crosses 1, 2 and 3, the mutant alleles of the T0 generation were transmitted to the T1 generation (Table S4). The results indicated that the mutations in T0 plants generated by the DPC CRISPR/Cas9 system could be stably inherited. In all five crosses, new alleles could be detected in T1 seedlings. The new alleles were also detected in Cas9‐free T1 seedlings (crosses 3 and 5), suggesting that mutations occurred in gametes of the T0 plant, which is consistent with our assumption. However, new alleles could also be generated in zygotes or early‐stage embryos. Most homozygous or bi‐allelic mutants recovered in the T1 generation showed an albino phenotype (Figure 6a,b). Interestingly, one plant shows a zebra phenotype, which mimics the phenotype of a mutant reported previously (Lu et al., 2012; Figure 6c,d). From examination in detail, we found that this zb7 mutant protein has one tyrosine deletion combined with a substitution (glutamine to lysine; Figure S3b). In conclusion, the mutant alleles generated by the DPC CRISPR/Cas9 system could be stably transmitted to the next generation and furthermore, the gametes or early‐stage embryos in T0 plants could be edited.
Figure 6.
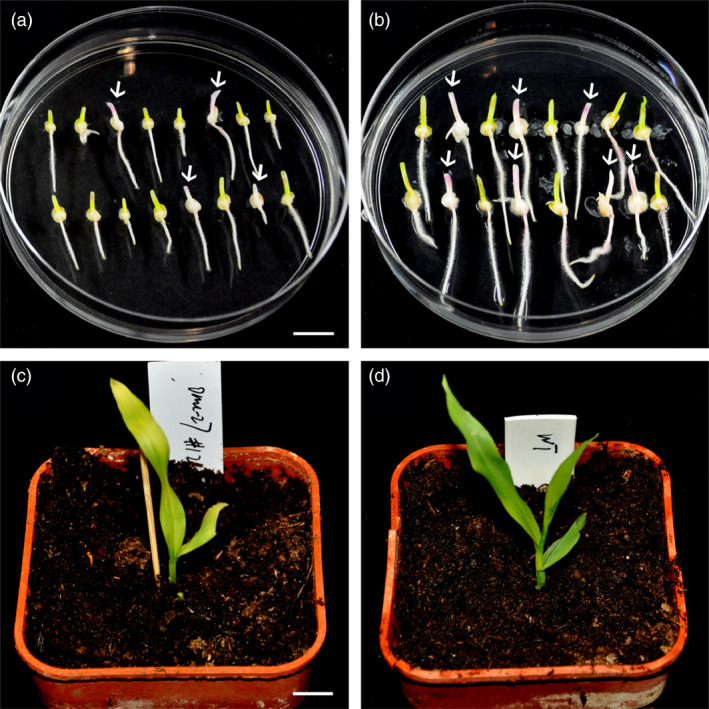
Phenotypes of the T1 seedlings by targeting the zb7 gene using DPC CRISPR/Cas9 system. (a, b) Culture of immature embryos from two sibling ears of T0 transgenic plants. Arrows indicate albino seedlings. (c) One seedling shows a zebra phenotype which mimics a previously reported zb7 mutant. (d) Control (regenerated wild‐type Hi‐II seedling). Bars = 1 cm.
Off‐target analysis
In addition to the editing efficiency, the specificity is also of concern when using CRISPR/Cas9 for targeted genome editing. In mammals, off‐target mutations were reported to take place in loci even with up to five nucleotide mismatch (Fu et al., 2013; Hsu et al., 2013; Pattanayak et al., 2013). To verify the specificity of using the DPC CRISPR/Cas9 system for targeted genome editing in maize, two Cas9‐positive T0 mutant plants with the zb7 gene edited and one control T0 plant without Cas9 were chosen for whole‐genome resequencing (Table S5). Approximately 11–14× average depth reads for each sample were sequenced with the Illumina Hiseq 2500 system (Table S5) and were examined for off‐target mutation. The resequencing data of Cas9‐positive and wild‐type plants were compared with the maize reference genome sequence. We first predicted the potential off‐target sites using online tools (Bae et al., 2014), over 1000 potential off‐target sites were identified with up to five mismatches. These potential off‐target sites were then checked with the resequencing data. The results show that no mutations were detected in these potential off‐target sites (Table S6). In contrast, the expected mutations in the target site of zb7 were easily identified for the two Cas9‐positive T0 plants (Figure S6). The number of indels and SNPs was revealed genome‐wide. The slight difference of the number of indels and SNPs between the Cas9‐positive mutant samples and control sample was likely due to sequencing depth. (Table S7). The predicted off‐target sites for another two sites were also analysed by T7EI assay (Figure 7 and Table S8). No mutations could be detected for the potential off‐target sites.
Figure 7.
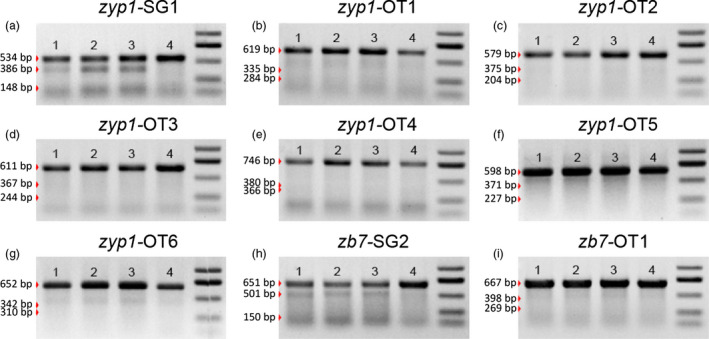
Off‐target analysis of the two loci by T7EI assay. (a) Target site in zyp1 gene (zyp1‐SG1 in Table S8). (b–g) Predicted off‐target sites (zyp1‐OT1 to zyp1‐OT6 in Table S8). (h) Target site in zb7 gene (zb7‐ SG2 in Table S8). (i) Predicted off‐target sites (zb7‐ OT1 in Table S8). For all sites, lanes 1–3 were three calli sample with homozygous or bi‐allelic mutation at the target sites and lane 4 is a control (wild‐type DNA sample used).
Discussion
To date, the CRISPR/Cas9 system has been widely adopted for genome editing in both monocots and dicots (Puchta, 2016). However, the efficiency of targeted genome editing is varied among species and different laboratories. Based on the reported results, the proper promoters used for both Cas9 and sgRNA are important for genome editing (Ma et al., 2016). We report here the DPC CRISPR/Cas9 system is highly efficient for targeted genome editing in maize. In this system, Cas9 and sgRNA are driven by the maize dmc1 promoter and U3 promoter, respectively. The mutation efficiency in transgene‐positive calli is 100%. Our results uncovered a new maize promoter that could be used for highly efficient genome editing in maize, thus providing further evidence that the efficiency of genome editing could be enhanced by screening and testing of promoters for Cas9 and sgRNA.
Previously the dmc1 gene was thought to be specifically expressed in reproductive tissue, and the dmc1 promoter would be meiosis specific. This gene is widely conserved in eukaryotes (Kagawa and Kurumizaka, 2010). By expression analysis, our results revealed that the dmc1 promoter could drive Cas9 to be highly expressed in callus, at least in maize. This was also confirmed by expression analysis of the endogenous dmc1 gene in different maize tissues including callus. The dmc1 gene has the highest expression level in tassel, but is also highly expressed in callus. These data could probably throw light on how the DPC CRISRP/Cas9 system can mediate targeted genome editing so efficiently.
It had been reported that the mutation efficiency of the vectors using ubiquitin promoter‐driven Cas9 combined with U6‐driven sgRNAs was more than 77% for three target sites when the constructs were delivered using biolistic‐mediated transformation (Svitashev et al., 2015). The biolistic transformation usually led to high copy number of transgenes integrated into the genomic region, not to mention the higher initial copy numbers of the plasmids delivered into the recipient cells. The high mutation efficiency may more or less benefit from the high expression level of the Cas9 gene caused by the high copy number of the transgene. The very similar constructs used by another group only generated a low mutation efficiency when using Agrobacterium for plasmid delivery (Zhu et al., 2016). Recently another group reported high‐efficiency targeted genome editing in maize Hi‐II germplasm using Agrobacterium‐mediated transformation (Char et al., 2016). The ubiquitin promoter is also used for driving Cas9 expression, and two sgRNAs driven by rice U6 promoters together were used for each gene to be targeted. The bi‐allelic mutant ratio is 21.3%–57.4% for the four genes targeted. Our results show that using the dmc1 promoter‐driven Cas9 combined with one sgRNA, the transgene‐positive calli were edited at the target loci at an efficiency of 100% with bi‐allelic mutations present in up to 66% of the regenerated seedlings. Thus, with a pretest of the activity of the sgRNAs in protoplasts, homozygous mutants for a specific gene could be obtained in the T0 generation at high efficiency using the DPC CRISPR/Cas9 system with one sgRNA. High‐efficiency multiplex genome editing in T0 generation is expected.
In most reports, sites in the gene coding region were chosen for applying the CRISPR/Cas9 to perform targeted genome editing, and usually the outcome is gene knockout. Recently, one group reported the results of editing the promoter region of a gene and generated quantitative trait variation with improved traits in tomato (Rodriguez‐Leal et al., 2017). By randomly selecting a target site in the zb7 gene, we obtained two kinds of mutant alleles with the coding sequence altered that show interesting phenotypes. Our results provide evidence that editing of the gene coding region without causing frameshift is probably also be useful for improving crop traits. It is also suggested that functional genomics could be explored deeper by altering the coding sequence of a gene rather than just making a knockout.
Except for the on‐target efficiency for CRISPR/Cas9‐mediated genome editing, another concern is the off‐target frequency. In mammals, the off‐target mutation frequency of the CRISPR/Cas9 system has been proven to be high when similar sites exist in the genome, especially sites with the same PAM‐proximal seed sequences. Off‐target mutation occurred in loci with even five mismatches. We conducted whole‐genome resequencing to detect off‐target mutation in the edited T0 plants. The results indicated no off‐target mutations occurred in the predicted sites with three or more mismatches.
In our experiments, using the dmc1 promoter‐controlled Cas9 system, high‐efficiency genome editing at the target site was identified in maize. The editing efficiency could be further improved by using more sgRNAs for one target site or by optimizing the criteria for sgRNA design (Dang et al., 2015; Doench et al., 2016; Wong et al., 2015). It is worth mentioning that previous studies have indicated that a maize codon‐optimized Cas9 could boost the mutation efficiency compared with a human codon‐optimized Cas9 (Xing et al., 2014). As the Cas9 used in our DPC CRISPR/Cas9 system is human codon‐optimized, we suggest that the targeted genome editing efficiency could be further increased using a plant or maize codon‐optimized Cas9. The higher genome editing efficiency could also largely benefit genome‐wide targeted mutagenesis in maize. Lastly, although at present we tested this system in maize, due to the evolutionary conservation of the dmc1 gene, we suggest that the dmc1 promoter‐controlled CRISPR/Cas9 system may be effective for other plant species.
Methods
Constructs and bacterial strains
The maize dmc1 promoter was amplified using primer pair dmcp‐F and dmcp‐R (Table S10) with B73 genomic DNA as template. The plasmid 35S‐Cas9‐SK (Feng et al., 2013) was digested with the restriction enzyme XhoI (NEB) to release the 35S promoter. Both the products were run in an agarose gel and then purified using a DNA purification Kit (TianGen). The purified linear fragments were assembled using the ‘Easygeno Assembly Cloning Kit’ (TianGen) to generate Dmc1‐Cas9‐SK. This plasmid was further digested by XmaI and EcoRI to release the Dmc1‐Cas9‐Nos cassette; the released cassette was then inserted into the binary vector pTF101.1 to make pDmc1‐Cas9 (Figure S7). The construction of the sgRNA cassette was described previously (Feng et al., 2016). The sgRNA cassette was amplified using primer pair sgRNA‐F and sgRNA‐R (Table S10), and then assembled into pDmc1‐Cas9 with the ‘Easygeno Assembly Cloning Kit’ (TianGen). The bacterial strains used in this study include DH5α (Escherichia coli) for conventional cloning and EHA105 (Agrobacterium) for maize transformation.
Agrobacterium‐mediated maize transformation
Maize Hi‐II germplasm was used as the transformation receptor. The seeds were planted in the experimental field in Beijing from May to September. The immature embryos were used for Agrobacterium‐mediated transformation following previous protocols (Frame et al., 2002). Tissue culture was performed in growth chambers in the dark. The regenerated plants were grown in chambers with a 16‐h light/8‐h dark condition at 25°C.
DNA extraction, PCR verification and genotyping
Genomic DNA of the maize calli, seedlings and protoplasts was extracted with Plant Genomic DNA Kit (TianGen Biotech., Beijing, China). The transgenic analysis was performed by PCR with specific primer pair Cas9‐F/Cas9‐R (Table S10). The PCR amplicons spanning the target sites were either subjected to direct Sanger sequencing, or cloned into the pMD19‐T vector (Takara Biomedical Technology, Beijing, China) and followed by Sanger sequencing. The chromatogram files were analysed by online tools (Liu et al., 2015).
RFLP assay and T7EI assay
Genome DNA that covered each target site was amplified from independent samples. Primers used were listed in Table S10. For RFLP assay, about 500 ng purified PCR products of each sample was digested with the corresponding enzyme (NEB, Ipswich, MA), the reaction products were analysed with 1.0% agarose gel electrophoresis. For the T7EI assay, PCR products obtained from the transgenic samples were mixed with the respective product derived from wild type, denatured (95°C for 5 min) and reannealed (ramp down from 95°C to 80°C at 5°C/min, from 80°C to 25°C at 5°C/2 min, hold on at 4°C). The reannealed products were then used to detect mutation with T7 endonuclease I (NEB, Ipswich, MA). The reaction products were analysed with 1.0% agarose gel electrophoresis.
Maize protoplast transformation
Maize B73 inbred seeds were germinated in the dark at 30°C for 3 days and then moved to a dark chamber at 25°C for a week. The fresh leaves of the seedlings were used to isolate mesophyll protoplasts. Maize protoplast transformation was carried out according to previously reported methods (Feng et al., 2016).
Whole‐genome resequencing and data analysis
Total DNA was isolated from the leaves of two Cas9‐positive transgenic plants and one wild‐type plant for genome resequencing. The genome sequence libraries were constructed according to the standard Illumina protocols and were applied to HiSeq 2500 system (Illumina, San Diego, CA). The whole‐genome resequencing data sets generated in this study are available from the NCBI BioProject (https://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA416839. The SNPs and indels were identified using the GATK Best Practices (Van der Auwera et al., 2013). The potential off‐target sites were identified with Cas‐OFFinder web tool with default parameters (Bae et al., 2014).
Conflict of interest
The authors declare competing financial interests: the authors have filed a patent application based on the results reported in this article.
Supporting information
Figure S1 Comparison of the mutation efficiency between the two maize polIII promoters.
Figure S2 Expression analysis of the Cas9 and dmc1 gene.
Figure S3 Detailed mutation analysis of the plants with chlorotic and zebra phenotypes.
Figure S4 Genotyping of the transgenic T0 plants by RFLP assay.
Figure S5 Mutation analysis of the three sites targeted by DPC CRISPR/Cas9 system in protoplasts.
Figure S6 Integrative Genomics Viewer (IGV) snapshots of the target site in the zb7 gene.
Figure S7 DNA sequence of the pDmc1‐Cas9 binary vector.
Table S1 Phenotype statistics of transgenic T0 plants targeted by DPC CRISPR/Cas9 system.
Table S2 Genotyping of the T0 plants by direct‐sequencing of the PCR products.
Table S3 Genotype statistics of transgenic T0 plants targeted by DPC CRISPR/Cas9 system at zyp1 gene.
Table S4 Genotyping of the T1 plants by RFLP assay and sanger sequencing at the zb7 gene.
Table S5 Summary of the re‐sequencing quality of different maize transgenic lines.
Table S6 Summary of the putative off‐target examination.
Table S7 Summary of the mutations of the whole genome re‐sequencing of maize transgenic lines and control.
Table S8 Predicted CRISPR/Cas9 off‐target sites of the two gene zyp1 and zb7.
Table S9 Maize genes targeted in this study.
Table S10 Primers used in this study.
Acknowledgements
We thank Professor Jiankang Zhu (Chinese Academy of Sciences) for kindly providing the 35S‐Cas9‐SK vector. We thank Prof. Kan Wang (Iowa State University) for sharing the pTF101.1 plasmid. This work is supported by the National Key Research and Development Program of China (2016YFD0102003), the State Key Program of National Natural Science Foundation of China (31630049) and the Ministry of Science and Technology (MOST) Project (2014ZX0801006B).
References
- Bae, S. , Park, J. and Kim, J.‐S. (2014) Cas‐OFFinder: a fast and versatile algorithm that searches for potential off‐target sites of Cas9 RNA‐guided endonucleases. Bioinformatics, 30, 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakate, A. , Higgins, J.D. , Vivera, S. , Stephens, J. , Perry, R.M. , Ramsay, L. , Colas, I. et al. (2014) The synaptonemal complex protein ZYP1 is required for imposition of meiotic crossovers in barley. Plant Cell, 26, 729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, C. , Nekrasov, V. , Lippman, Z.B. and Van Eck, J. (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR‐associated 9 system. Plant Physiol. 166, 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Char, S.N. , Neelakandan, A.K. , Nahampun, H. , Frame, B. , Main, M. , Spalding, M.H. , Becraft, P.W. et al. (2016) An Agrobacterium‐delivered CRISPR/Cas9 system for high‐frequency targeted mutagenesis in maize. Plant Biotechnol. J. 15, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, L. , Ran, F.A. , Cox, D. , Lin, S. , Barretto, R. , Habib, N. , Hsu, P.D. et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang, Y. , Jia, G. , Choi, J. , Ma, H. , Anaya, E. , Ye, C. , Shankar, P. et al. (2015) Optimizing sgRNA structure to improve CRISPR‐Cas9 knockout efficiency. Genome Biol. 16, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo, J.E. , Norville, J.E. , Mali, P. , Rios, X. , Aach, J. and Church, G.M. (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR‐Cas systems. Nucleic Acids Res. 41, 4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G. , Fusi, N. , Sullender, M. , Hegde, M. , Vaimberg, E.W. , Donovan, K.F. , Smith, I. et al. (2016) Optimized sgRNA design to maximize activity and minimize off‐target effects of CRISPR‐Cas9. Nat. Biotechnol. 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Z. , Zhang, B. , Ding, W. , Liu, X. , Yang, D.L. , Wei, P. , Cao, F. et al. (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23, 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Z. , Mao, Y. , Xu, N. , Zhang, B. , Wei, P. , Yang, D.L. , Wang, Z. et al. (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas‐induced gene modifications in Arabidopsis . Proc. Natl Acad. Sci. USA, 111, 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, C. , Yuan, J. , Wang, R. , Liu, Y. , Birchler, J.A. and Han, F. (2016) Efficient targeted genome modification in maize using CRISPR/Cas9 system. J. Genet. Genomics, 43, 37–43. [DOI] [PubMed] [Google Scholar]
- Frame, B.R. , Shou, H. , Chikwamba, R.K. , Zhang, Z. , Xiang, C. , Fonger, T.M. , Pegg, S.E.K. , et al. (2002) Agrobacterium tumefaciens‐mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 129, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland, A.E. , Tzur, Y.B. , Esvelt, K.M. , Colaiacovo, M.P. , Church, G.M. and Calarco, J.A. (2013) Heritable genome editing in C. elegans via a CRISPR‐Cas9 system. Nat. Methods, 10, 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. , Foden, J.A. , Khayter, C. , Maeder, M.L. , Reyon, D. , Joung, J.K. and Sander, J.D. (2013) High‐frequency off‐target mutagenesis induced by CRISPR‐Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj, T. , Gersbach, C.A. and Barbas, C.F. III . (2013) ZFN, TALEN, and CRISPR/Cas‐based methods for genome engineering. Trends Biotechnol. 31, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz, S.J. , Cummings, A.M. , Nguyen, J.N. , Hamm, D.C. , Donohue, L.K. , Harrison, M.M. , Wildonger, J. et al. (2013) Genome Engineering of Drosophila with the CRISPR RNA‐Guided Cas9 Nuclease. Genetics, 194, 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, P.D. , Scott, D.A. , Weinstein, J.A. , Ran, F.A. , Konermann, S. , Agarwala, V. , Li, Y. et al. (2013) DNA targeting specificity of RNA‐guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, P.D. , Lander, E.S. and Zhang, F. (2014) Development and applications of CRISPR‐Cas9 for genome engineering. Cell, 157, 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, W.Y. , Fu, Y.F. , Reyon, D. , Maeder, M.L. , Tsai, S.Q. , Sander, J.D. , Peterson, R.T. et al. (2013) Efficient genome editing in zebrafish using a CRISPR‐Cas system. Nat. Biotechnol. 31, 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W.Y. , Bikard, D. , Cox, D. , Zhang, F. and Marraffini, L.A. (2013) RNA‐guided editing of bacterial genomes using CRISPR‐Cas systems. Nat. Biotechnol. 31, 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa, W. and Kurumizaka, H. (2010) From meiosis to postmeiotic events: uncovering the molecular roles of the meiosis‐specific recombinase Dmc1. FEBS J. 277, 590–598. [DOI] [PubMed] [Google Scholar]
- Klimyuk, V.I. and Jones, J.D. (1997) AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: characterization, transposon‐induced allelic variation and meiosis‐associated expression. Plant J. 11, 1–14. [DOI] [PubMed] [Google Scholar]
- Leader, D.J. , Connelly, S. , Filipowicz, W. and Brown, J.W. (1994) Characterisation and expression of a maize U3 snRNA gene. Biochim. Biophys. Acta, 1219, 145–147. [DOI] [PubMed] [Google Scholar]
- Li, D.L. , Qiu, Z.W. , Shao, Y.J. , Chen, Y.T. , Guan, Y.T. , Liu, M. , Li, Y. et al. (2013a) Heritable gene targeting in the mouse and rat using a CRISPR‐Cas system. Nat. Biotechnol. 31, 681–683. [DOI] [PubMed] [Google Scholar]
- Li, J.F. , Norville, J.E. , Aach, J. , McCormack, M. , Zhang, D. , Bush, J. , Church, G.M. et al. (2013b) Multiplex and homologous recombination‐mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Teng, F. , Li, T.D. and Zhou, Q. (2013c) Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR‐Cas systems. Nat. Biotechnol. 31, 684–686. [DOI] [PubMed] [Google Scholar]
- Li, C. , Liu, C. , Qi, X. , Wu, Y. , Fei, X. , Mao, L. , Chen, B. et al. (2017) RNA‐guided Cas9 as an in vivo desired‐target mutator in maize. Plant Biotechnol. J. 15, 1566–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Z. , Zhang, K. , Chen, K. and Gao, C. (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics, 41, 63–68. [DOI] [PubMed] [Google Scholar]
- Liu, C.M. , McElver, J. , Tzafrir, I. , Joosen, R. , Wittich, P. , Patton, D. , Van Lammeren, A.A. et al. (2002) Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J. 29, 405–415. [DOI] [PubMed] [Google Scholar]
- Liu, W. , Xie, X. , Ma, X. , Li, J. , Chen, J. and Liu, Y.G. (2015) DSDecode: a web‐based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol. Plant, 8, 1431–1433. [DOI] [PubMed] [Google Scholar]
- Lu, X.M. , Hu, X.J. , Zhao, Y.Z. , Song, W.B. , Zhang, M. , Chen, Z.L. , Chen, W. et al. (2012) Map‐based cloning of zb7 encoding an IPP and DMAPP synthase in the MEP pathway of maize. Mol. Plant, 5, 1100–1112. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Zhang, Q. , Zhu, Q. , Liu, W. , Chen, Y. , Qiu, R. , Wang, B. et al. (2015) A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant, 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Zhu, Q. , Chen, Y. and Liu, Y.G. (2016) CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol. Plant, 9, 961–974. [DOI] [PubMed] [Google Scholar]
- Mali, P. , Yang, L. , Esvelt, K.M. , Aach, J. , Guell, M. , DiCarlo, J.E. , Norville, J.E. et al. (2013) RNA‐guided human genome engineering via Cas9. Science, 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, J. , Guo, D. , Zhang, J. , Huang, Q. , Qin, G. , Zhang, X. , Wan, J. et al. (2013) Targeted mutagenesis in rice using CRISPR‐Cas system. Cell Res. 23, 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannas, N.J. and Dawe, R.K. (2015) Genetic and genomic toolbox of Zea mays . Genetics, 199, 655–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov, V. , Staskawicz, B. , Weigel, D. , Jones, J.D. and Kamoun, S. (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA‐guided endonuclease. Nat. Biotechnol. 31, 691–693. [DOI] [PubMed] [Google Scholar]
- Pattanayak, V. , Lin, S. , Guilinger, J.P. , Ma, E. , Doudna, J.A. and Liu, D.R. (2013) High‐throughput profiling of off‐target DNA cleavage reveals RNA‐programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta, H. (2016) Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Curr. Opin. Plant Biol. 36, 1–8. [DOI] [PubMed] [Google Scholar]
- Puchta, H. and Fauser, F. (2014) Synthetic nucleases for genome engineering in plants: prospects for a bright future. Plant J. 78, 727–741. [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Leal, D. , Lemmon, Z.H. , Man, J. , Bartlett, M.E. and Lippman, Z.B. (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480. [DOI] [PubMed] [Google Scholar]
- Shan, Q.W. , Wang, Y.P. , Li, J. , Zhang, Y. , Chen, K.L. , Liang, Z. , Zhang, K. et al. (2013) Targeted genome modification of crop plants using a CRISPR‐Cas system. Nat. Biotechnol. 31, 686–688. [DOI] [PubMed] [Google Scholar]
- Svitashev, S. , Young, J.K. , Schwartz, C. , Gao, H. , Falco, S.C. and Cigan, A.M. (2015) Targeted mutagenesis, precise gene editing, and site‐specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169, 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera, G.A. , Carneiro, M.O. , Hartl, C. , Poplin, R. , Del Angel, G. , Levy‐Moonshine, A. , Jordan, T. et al. (2013) From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics, 43, 11.10.1‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.P. , Xing, H.L. , Dong, L. , Zhang, H.Y. , Han, C.Y. , Wang, X.C. and Chen, Q.J. (2015) Egg cell‐specific promoter‐controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, N. , Liu, W. and Wang, X. (2015) WU‐CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 16, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, K. and Yang, Y. (2013) RNA‐guided genome editing in plants using a CRISPR‐Cas system. Mol. Plant, 6, 1975–1983. [DOI] [PubMed] [Google Scholar]
- Xie, K. , Minkenberg, B. and Yang, Y. (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA‐processing system. Proc. Natl Acad. Sci. USA, 112, 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, H.L. , Dong, L. , Wang, Z.P. , Zhang, H.Y. , Han, C.Y. , Liu, B. , Wang, X.C. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, R. , Li, H. , Qin, R. , Wang, L. , Li, L. , Wei, P. and Yang, J. et al. (2014) Gene targeting using the Agrobacterium tumefaciens‐mediated CRISPR‐Cas system in rice. Rice, 7, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Z.S. , Ren, M.D. , Wang, Z.X. , Zhang, B. , Rong, Y.K.S. , Jiao, R. and Gao, G. (2013) Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila . Genetics, 195, 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zhang, J. , Wei, P. , Zhang, B. , Gou, F. , Feng, Z. , Mao, Y. et al. (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. [DOI] [PubMed] [Google Scholar]
- Zhu, J. , Song, N. , Sun, S. , Yang, W. , Zhao, H. , Song, W. and Lai, J. (2016) Efficiency and inheritance of targeted mutagenesis in maize using CRISPR‐Cas9. Plant Physiol. 43, 25–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Comparison of the mutation efficiency between the two maize polIII promoters.
Figure S2 Expression analysis of the Cas9 and dmc1 gene.
Figure S3 Detailed mutation analysis of the plants with chlorotic and zebra phenotypes.
Figure S4 Genotyping of the transgenic T0 plants by RFLP assay.
Figure S5 Mutation analysis of the three sites targeted by DPC CRISPR/Cas9 system in protoplasts.
Figure S6 Integrative Genomics Viewer (IGV) snapshots of the target site in the zb7 gene.
Figure S7 DNA sequence of the pDmc1‐Cas9 binary vector.
Table S1 Phenotype statistics of transgenic T0 plants targeted by DPC CRISPR/Cas9 system.
Table S2 Genotyping of the T0 plants by direct‐sequencing of the PCR products.
Table S3 Genotype statistics of transgenic T0 plants targeted by DPC CRISPR/Cas9 system at zyp1 gene.
Table S4 Genotyping of the T1 plants by RFLP assay and sanger sequencing at the zb7 gene.
Table S5 Summary of the re‐sequencing quality of different maize transgenic lines.
Table S6 Summary of the putative off‐target examination.
Table S7 Summary of the mutations of the whole genome re‐sequencing of maize transgenic lines and control.
Table S8 Predicted CRISPR/Cas9 off‐target sites of the two gene zyp1 and zb7.
Table S9 Maize genes targeted in this study.
Table S10 Primers used in this study.


