Summary
Starch is the main form of energy storage in higher plants. Although several enzymes and regulators of starch biosynthesis have been defined, the complete molecular machinery remains largely unknown. Screening for irregularities in endosperm formation in rice represents valuable prospect for studying starch synthesis pathway. Here, we identified a novel rice white‐core endosperm and defective grain filling mutant, ospk2, which displays significantly lower grain weight, decreased starch content and alteration of starch physicochemical properties when compared to wild‐type grains. The normal starch compound granules were drastically reduced and more single granules filled the endosperm cells of ospk2. Meanwhile, the germination rate of ospk2 seeds after 1‐year storage was observably reduced compared with wild‐type. Map‐based cloning of OsPK2 indicated that it encodes a pyruvate kinase (PK, ATP: pyruvate 2‐O‐phosphotransferase, EC 2.7.1.40), which catalyses an irreversible step of glycolysis. OsPK2 has a constitutive expression in rice and its protein localizes in chloroplasts. Enzyme assay showed that the protein product from expressed OsPK2 and the crude protein extracted from tissues of wild‐type exhibits strong PK activity; however, the mutant presented reduced protein activity. OsPK2 (PKpα1) and three other putative rice plastidic isozymes, PKpα2, PKpβ1 and PKpβ2, can interact to form heteromer. Moreover, the mutation leads to multiple metabolic disorders. Altogether, these results denote new insights into the role of OsPK2 in plant seed development, especially in starch synthesis, compound granules formation and grain filling, which would be useful for genetic improvement of high yield and rice grain quality.
Keywords: grain filling, amyloplast development, pyruvate kinase, starch biosynthesis, rice
Introduction
Starch is the essential carbohydrate storage material in plants. Because of its relevance as staple crop, rice has been used to study starch formation in seeds. Starch synthesis is a complex process involving a series of biosynthetic enzymes such as ADP‐Glc pyrophosphorylase (AGPase), starch synthase (SS), starch branching enzyme (SBE) and starch debranching enzyme (DBE) (Toyosawa et al., 2016). Mutation of those genes changes the appearance of the endosperm and alters the characteristics of storage starch. For instance, mutations in AGPase subunits create a wrinkled endosperm and significantly reduce the starch content (Akihiro et al., 2005; Lee et al., 2007; Tuncel et al., 2014; Wei et al., 2017). Loss‐of‐function mutations of the GBSSI gene produce a complete lack of endosperm amylose (Sato et al., 2002; Tian et al., 2009). Knockout of OsSSIIIa causes a white‐core floury endosperm and changes starch granules morphology, starch structure and gelatinization temperature (Ryoo et al., 2007). Mutation of BEIIb causes a chalkiness endosperm and has great influence on the properties of starch (Tanaka et al., 2004). Loss‐of‐function mutations of ISA1, encoding a starch DBE, cause a serious defect in amylopectin structure and a sugary endosperm (Kawagoe et al., 2005; Kubo et al., 1999). Besides starch biosynthesis enzymes, other factors associated to starch formation can also affect endosperm development in rice. For example, FLOURY ENDOSPERM4 (FLO4), encoding a pyruvate orthophosphate dikinase (PPDK; EC 2.7.9.1), was shown to regulate carbon flow from starch and lipid biosynthesis during seeding stage (Kang et al., 2005). Some other factors such as flo2, flo6, flo7, OsbZIP58, OsBT1 (ADP‐glucose transporter), RPBF (rice P‐box binding factor), OspPGM (plastidic phosphor‐glucomutase) and RSR1 (rice starch regulator1) have also been shown to influence starch synthesis and endosperm development in rice (Fu and Xue, 2010; Lee et al., 2016; Li et al., 2017; Peng et al., 2014; She et al., 2010; Wang et al., 2013; Yamamoto et al., 2006; Zhang et al., 2016). Mutants of SUBSTANDARD STARCH GRAIN4 (SSG4) and SSG6 show enlarged starch grains (SGs) in the rice endosperm (Matsushima et al., 2014, 2016). All these data evidence that the genetic basis of starch synthesis and grain development implies a complex regulatory network. Therefore, identification and characterization of different rice endosperm defective mutants are necessary for further understanding this process in rice.
Pyruvate kinase (PK, ATP: pyruvate 2‐O‐phosphotransferase, EC 2.7.1.40) is a key enzyme that regulates and adjusts the final step of the glycolysis pathway. It catalyses the irreversible transfer of the high‐energy phosphate group from phosphoenolpyruvate (PEP) to ADP to synthesize pyruvate and ATP (Ambasht and Kayastha, 2002; Valentini et al., 2000). The glycolytic pathway provides intermediates for other crucial metabolic reactions. The product pyruvate can be decarboxylated by pyruvate dehydrogenase complex (PDC) to generate acetyl‐CoA, which can be used as the initial substrate to participate in the tricarboxylic acid (TCA) cycle to generate ATP under aerobic condition or be converted into lactic acid or ethanol under hypoxia. Besides, acetyl‐CoA can be involved in fatty acid (FA) biosynthesis and the mevalonate pathway (Grodzinski et al., 1999). PEP, as the intermediate product in glycolysis pathway, can also be catalysed by phosphoenolpyruvate carboxylase (PEPC) to form oxaloacetic acid (OAA) which is required for amino acid synthesis (Schwender et al., 2004). Therefore, PK plays an important role in the cellular metabolic flux (Zhang et al., 2012). PKs in plant exist as cytosolic (PKc) and plastidic (PKp) isozymes (Plaxton, 1996). In most organisms, PKs are usually present as a homotetramer (Schramm et al., 2000), but can also be found in a monomer (Knowles et al., 1989) or in complexes as homodimer (Plaxton et al., 2002) or in different heteromeric forms (αnβn) (Andre et al., 2007; Plaxton, 1989). For example, PKc in castor oil seed germinating endosperm is existed as 240 kDa heterotetramers comprised by 56 and 57 kDa subunits (Hu and Plaxton, 1996). PKp purified from castor bean had been reported to exist as a 334 kDa α3β3 heterohexameric (63.5 and 54 kDa polypeptides considered as α‐subunit and β‐subunit, respectively) (Plaxton, 1991). There are 14 genes encoding putative PK isoforms in Arabidopsis (Initiative, 2000). At least six PKc potato genes were identified by Southern blot analyses (Oliver et al., 2008). Cotton has at least 19 PKc and 14 PKp genes (Zhang and Liu, 2016). In rice, at least 11 genes have been predicted to encode putative PK isozymes (Zhang et al., 2012). Thus, the component forms of PK in higher plant display abundant diversity.
In plants, a ‘bottom‐up’ allosteric regulatory mechanism regulates the glycolysis pathway where PEP is considered as a potent inhibitor for ATP‐ and PPi‐dependent phosphofructokinase (Plaxton and Podestá, 2006). Meanwhile, PK as an allosteric enzyme can be inhibited by glutamate and activated by aspartate (Hu and Plaxton, 1996; Smith et al., 2000). Allosteric activation by 6‐p‐gluconate (G6P) in Brassica napus PKp has been described (Andre et al., 2007). The major role of PKc is to produce a precursor and ATP for various synthetic metabolic pathways (Smith et al., 2000). Transgenic tobacco plants lacking the leaf PKc display decreased root elongation and delayed bud differentiation and flowering (Knowles et al., 1998). The cotton PKc gene GhPK6 is primarily expressed in cotton fibres, and its expression negatively correlates with the rate of fibre cell elongation (Zhang and Liu, 2016). Identification of the rice OsPK1 gene suggested that PKc might affect plant morphological development, causing dwarfism and panicle enclosure in rice (Zhang et al., 2012). On the other hand, PKp is thought to provide pyruvate and ATP to support biosynthetic pathways in plastids (Andre and Benning, 2007). Besides the defects in oil accumulation and delayed germination found in Arabidopsis pkp1 mutants (Andre et al., 2007; Baud et al., 2007), research on PKp is limited and defined roles in growth and development are unknown.
In this study, we identified a white‐core endosperm and defective grain filling mutant, ospk2. Map‐based cloning showed that OsPK2 encodes a plastidic PK enzyme that can interact with three other putative PKp subunits to form a polymer which affects starch synthesis, compound granule formation and grain weight during rice seed development. This deep dissection of OsPK2 will enhance our understanding of the genetic basis of endosperm starch synthesis and grain filling in rice and provides useful information for genetic improvement of high yield and grain quality.
Results
The rice ospk2 mutant displays defects in endosperm appearance and grain filling
A chalky endosperm and defective grain filling mutant, named ospk2, were identified from an ethyl methane sulphonate (EMS)‐mutagenized rice population. Compared to the wild‐type, the mature grains of ospk2 displayed an opaque endosperm and presented a white‐core in the interior whereas the peripheral endosperm was transparent (Figure 1a–c). Scanning electron microscopy (SEM) analysis of the endosperm transverse sections showed that wild‐type endosperm cells were filled with densely packed and irregularly polyhedral SGs (Figure 1d,e), whereas the ospk2 endosperm cells presented spherical and loosely packed SGs (Figure 1f,g). The grain filling rate in the ospk2 mutant was significantly slower than the wild‐type from 10 days after fertilization (DAF), resulting in a significantly reduced grain weight and grain yield per plant in ospk2 (Figures 1h,i and S1a). Quantification of seed size showed no significant difference in grain length but the grain width and the grain thickness of ospk2 were reduced compared with the wild‐type (Figure 1j–l). Furthermore, germination rates of fresh harvested seeds were not significantly different between wild‐type and ospk2, but the buds of ospk2 were weaker. However, the seed germination rates after 1 year of storage were strongly reduced in ospk2 compared with wild‐type (Figure S2). Besides these grain endosperm defects, no other obvious differences in plant architecture between wild‐type and the ospk2 mutant were observed (Figure S1b–g).
Figure 1.
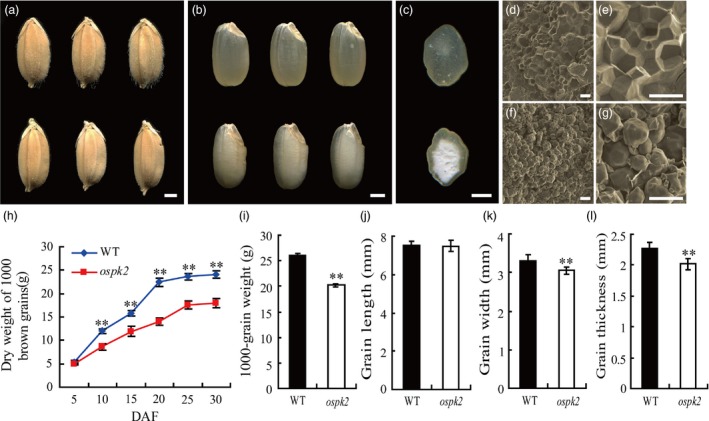
Phenotype of the ospk2 mutant. (a–b) Appearance comparison of seeds (a) and brown rice (b) of wild‐type (WT) (above) and ospk2 (below). (c) Transverse sections of WT (above) and ospk2 (below) brown rice. (d–g) Scanning electron microscopy images of transverse sections of the WT (d, e) and ospk2 mutant (f, g) grains. Scale bars, 1 mm in (a–c); 20 μm in (d, f) and 50 μm in (e, g). (h) Weight of dry grains of WT and ospk2 at various stages of grain filling. DAF, days after fertilization. (i–l) Weight of 1000‐grains (i), grain length (j), grain width (k) and grain thickness (l) of WT and ospk2. Data in (h–l) are means ± SD from three biological replicates, and each replication in (i) and (h, j–l) not less than 200 and 50 seeds, respectively. Asterisks in (i–l) indicate statistical significance between the WT and the mutant, as determined by a Student's t‐test (**P < 0.01).
The structure of starch compound granules is abnormal in ospk2 endosperm cells
Semithin sections of developing endosperms at 10 DAF were prepared to observe the structure of starch compound granules in wild‐type and ospk2. Fewer well‐developed compound granules and some immature starch granules were observed in the peripheral endosperm cells of both wild‐type and ospk2 (Figure 2a,e). In central endosperm cells of wild‐type seeds, several polyhedral and sharp‐edged starch granules congregate to form well‐developed amyloplasts (Figure 2a,c). In central endosperm cells of ospk2, however, abundant single, smaller, scattered and weakly stained starch granules were also observed (Figure 2f,g). The morphology of individual starch granules was also analysed using SEM. The starch granules in wild‐type endosperm appear polygonal with sharp edges and similar size, whereas in the mutant, they were smaller and with irregular shape (Figure 2d,h). Amyloplasts at earlier developmental stages (3, 6 and 9 DAF) were observed by transmission electron microscopy. Wild‐type amyloplasts were filled with polyhedral starch granules and the interior space was quickly occupied during grain development, eventually forming a typical complex structure (Figure 3a–c). Similar to wild‐type, there were few well‐developed amyloplasts in the endosperm cells of the ospk2 mutant (Figure 3d–f), however, the majority of the starch granules in ospk2 endosperm cells existed as small, single and disperse granules rather than aggregated (Figure 3g–i). This was consistent with our observations of the semithin sections (Figure 2f,g). These results suggest that OsPK2 affects the formation of starch compound granules in endosperm cells during grain development.
Figure 2.
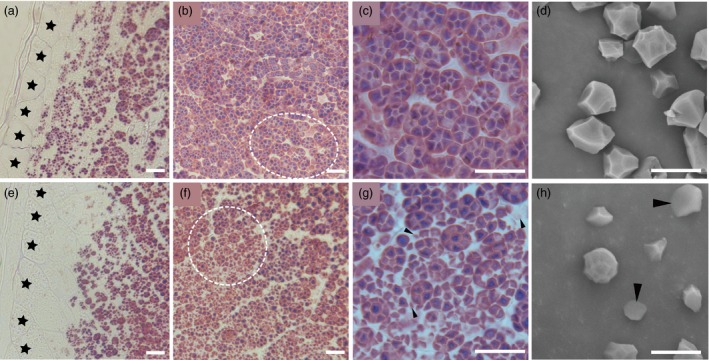
Starch granules formation in endosperm cells. (a–c) Semithin sections of wild‐type endosperm at 10 days after fertilization (DAF). (d) SEM analysis of starch or insoluble glucan granules purified from mature kernels of the wild‐type. (e–g) Semithin sections of ospk2 mutant endosperm at 10 DAF. (h) SEM analysis of starch or insoluble glucan granules purified from mature kernels of ospk2. (a, e) represent the peripheral region of endosperm of wild‐type and ospk2 mutant, respectively; (b, c) represent the central region of endosperm of wild‐type; (f, g) represent the central region of endosperm ospk2 mutant; (c, g) are enlarged sections of the white circle region from (b) and (f), respectively. Stars in (a, e) indicate the aleurone layer cells. Arrowheads in (g) indicate smaller, weak staining or abnormal starch granules in cytosol of ospk2. Arrowheads in (h) represent smaller or irregular shapes starch granules in ospk2. Scale bars: 10 μm in (a–h).
Figure 3.
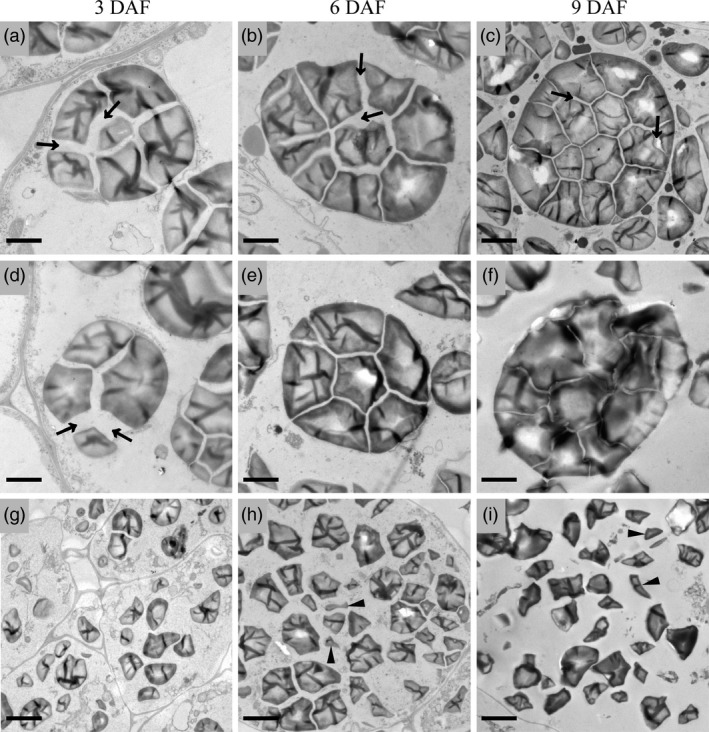
Electron micrographs depicting amyloplast development in endosperm cells of wild‐type (a–c) and ospk2 (d–i). (a, d, g) display an amyloplast in endosperm cells of wild‐type and ospk2 at 3 days after fertilization (DAF). (b, e, h) Amyloplast in endosperm cells of wild‐type and ospk2 at 6 DAF. (c, f, i) Amyloplast in endosperm cells of wild‐type and ospk2 at 9 DAF. (d–f) Represent few well‐developed amyloplasts in the endosperm cells of ospk2 mutant. (g–i) Represent the majority phenomenon of starch granules in ospk2 endosperm cells. Arrowheads in (a–d) show the stroma inside the amyloplast. Arrowheads in (h, i) indicate smaller, irregular shape, abnormal granules in ospk2. Bars: 2 μm (a–f), 5 μm (g–i).
Starch physicochemical properties in ospk2
Because the starch granules were abnormal in both mature and developing endosperm in the ospk2 mutant, the starch physicochemical properties were also examined. The total starch and amylose contents significantly decreased in the endosperm of ospk2 compared with wild‐type (Figure 4a,b). Nonetheless, the contents of protein and lipid in ospk2 were higher than these in wild‐type (Figure 4c,d). In the case of lipids, the contents of the C16:0 (palmitic acid), C18:0 (stearic acid), C18:1 (oleic acid), C18:2 (linoleic acid) and C22:0 (behenic acid) all increased (Figure S3). To further analyse the fine structure of amylopectin, its chain length distribution was determined. Compared to wild‐type, the proportion of short chains with degree of polymerization (DP) values between 6 and 12 decreased, whereas the proportion of intermediate chains with DP values between 13 and 25 was elevated in the ospk2 mutant (Figure 4e).
Figure 4.
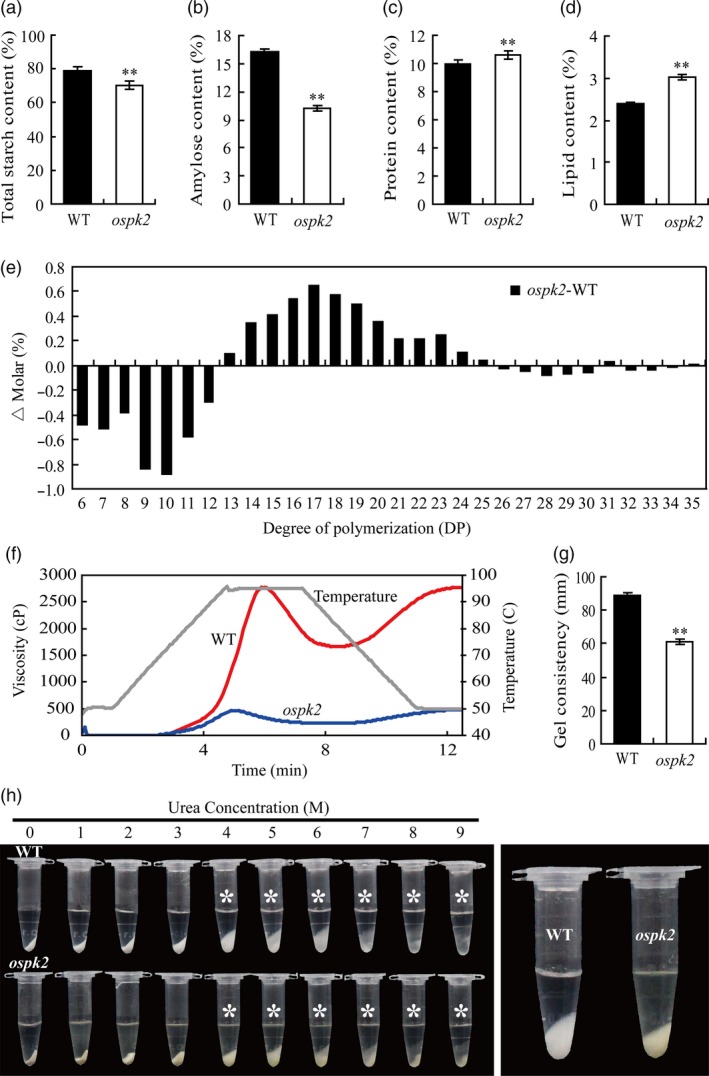
Grain characteristics and starch physicochemical characteristics in the ospk2 mutant. (a–d) The per cent contents of total starch, amylose, protein and lipid in endosperm of wild‐type (WT) and ospk2. (e) Differences in the amylopectin chain length distributions between the WT and ospk2. (f) Pasting properties of endosperm starch of WT (red line) and ospk2 mutant (blue line). The viscosity value at each temperature is the average of three replicates. The grey line indicates the temperature changes during the measurements. (g) The gel consistency of endosperm starch of WT and ospk2 mutant. (h) Gelatinization characteristics of starch in urea solutions. Starch powder of WT and ospk2 was mixed with varying concentrations (1–9 m) of urea solution. Asterisks indicate the starch of ospk2 endosperm is more difficult to gelatinize in 4–9 m urea solution than that of WT. The most significant difference was observed for 4 m urea (right panel). Values in (a–d, g) are means ± SD from three biological replicates. Asterisks indicate statistical significance between WT and mutant, as determined by a Student's t‐test (*P < 0.05; **P < 0.01).
The pasting properties of starch were analysed with a Rapid Visco Analyzer (RVA; Figure 4f). The RVA profile showed significant differences in pasting properties of the starch derived from wild‐type and ospk2. The viscosity of pasting starch in ospk2 was maintained at a very low level and did not show the distinct characteristics of the peak viscosity and breakdown when the temperature rose. When the temperature was lowered, the viscosity of pasting starch in wild‐type augmented quickly and showed a high final value, whereas that of ospk2 almost did not increase and the final viscosity was only 16.68% of that of wild‐type. The gelatinization properties of the SGs also were examined, and the result showed that SGs in ospk2 had a significantly shorter gel consistency than wild‐type (Figure 4g). Starch solubility in urea solutions was measured. Mixing powdered starch with varying concentrations (0–9 m) of urea solutions showed that the ospk2 starch was more difficult to gelatinize and exhibited significant different gelatinization characteristics in 4–9 m urea compared to that of wild‐type (Figure 4h). In brief, the physicochemical properties of the starch in ospk2 endosperm were significantly different from those in wild‐type.
Map‐based cloning and complementation of the ospk2 mutation
To identify the gene controlling the ospk2 phenotypes, map‐based cloning was conducted using an F2 mapping population that was generated by crossing the ospk2 mutant (japonica) with Nanjing11 (NJ‐11, indica). The mutation locus was first mapped on the short arm of chromosome 7 between the simple sequence repeat (SSR) markers RM21078 and RM3484. Over 978 individuals showing the ospk2 phenotypes were chosen from this F2 population. The ospk2 locus was further narrowed down to a 90.8‐kb region between InDel markers In18 and In20 located on the BAC clone OSJ1119_A04, which included nine putative open reading frames (ORFs, Figure 5a; Table S1). Sequence analysis of the ORFs revealed a single nucleotide substitution from a cytosine (C) to thymine (T) on the 4th exon in ORF4 (Os07g0181000) in the ospk2 mutant, which led a Serine296 replaced by a Leucine296 (Figure 5b). Os07g0181000 was predicted to encode a putative pyruvate kinase (PK) of about 63.58 kDa with 578 amino acids (http://smart.embl-heidelberg.de). Among a total of eleven Oryza sativa PK isozymes, OsPK2 (Os07g0181000, also named PKpα1) belongs to the α‐subunit type within the plastidic clade (Figure S4). Protein sequence alignment showed that the mutation site lies in a highly conserved Serine (S) residue on the PK domain among the OsPK2 homologues and that OsPK2 shares 74.33% identity with Arabidopsis AtPKp1 (Figure S5) which was reported as a plastidic PK involved in embryo development and control of carbon flux in maturing Arabidopsis seeds (Andre et al., 2007).
Figure 5.
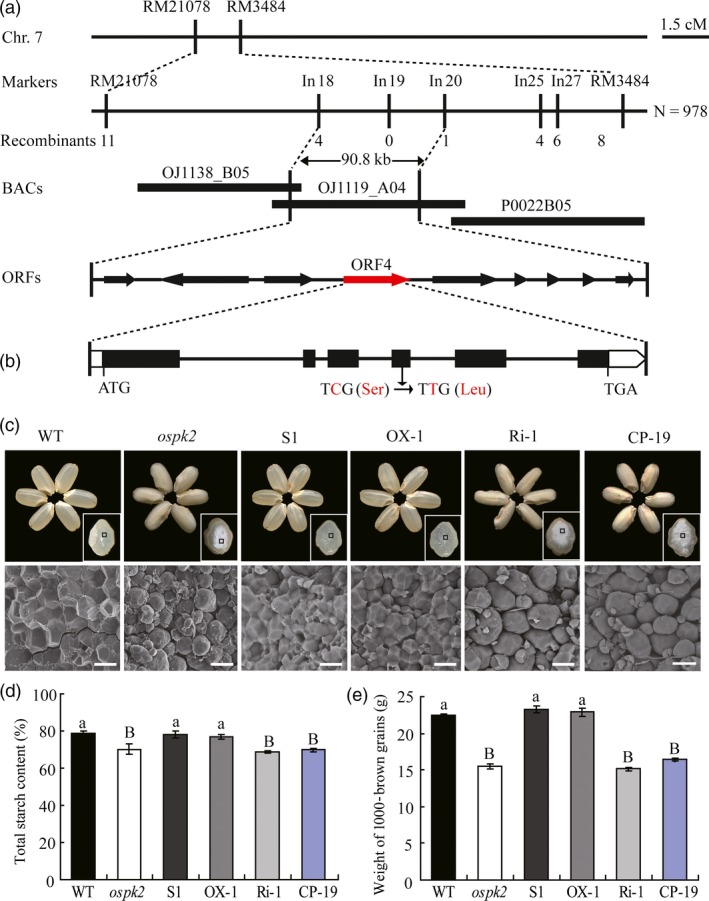
Map‐based cloning and complementation of the ospk2 mutant. (a) Fine mapping of the OsPK2 locus. The OsPK2 locus was mapped to a 90.8 kb region by markers In18 and In20 on chromosome 7 (Chr.7) which contained nine predicted genes. The molecular markers and the number of recombinants are shown. cM, centimorgan; ORF, open reading frame. (b) Gene structure and mutation site in OsPK2. Base pair change (C to T) detected in ospk2 in the 4th exon of Os07g0181000 causing a Ser‐296 to Leu‐296 change. ATG and TGA represent the start and stop codons, respectively. (c) Complementation of the ospk2 mutation in transgenic lines (S1) and overexpression of OsPK2 in ospk2 (OX‐1) showing the restored wild‐type seed appearance and normal starch granules in endosperm, whereas RNAi (Ri‐1) and CRISPR/Cas9 mediated editing (CP‐19) of OsPK2 in ZH11 background produce abnormal seed appearance and seeds became chalky. Bars: 20 μm. (d, e) Total starch content and 1000‐brown grains weight of grains in complementation (S1), overexpression (OX‐1), RNAi (Ri‐1) and CRISPR/Cas9 mediated editing lines (CP‐19). Data are shown as means ± SD from three biological replicates, each replication in (e) is not less than 200 seeds. Significant difference analysed by multiple‐comparison. Different letters indicate statistically significant difference (small letter, P < 0.05 and capital letter, P < 0.01).
To verify whether OsPK2 (Os07g0181000) is the gene responsible for the mutant phenotypes, a vector carrying the OsPK2 genomic sequence including its native promoter and another vector with the OsPK2 coding region driven by the UBIQUTIN1 promoter were constructed and introduced into the ospk2 mutant. The expression of OsPK2 in complementation and overexpression transgenic lines was significantly higher than that in ospk2 and wild‐type plants (Figure S6). Seeds harvested from independent complementation and overexpression positive transgenic T1 lines showed similar phenotypes to wild‐type (Figure 5c). The total starch content, 1000‐brown grain weight, grain length, width and thickness of brown rice of the complementation and overexpression lines achieved the same levels of those of wild‐type (Figures 5d,e and S7a–d). SGs in endosperm cells of these positive transgenic lines also presented the polyhedral and densely packed granule morphology, similar to those of wild‐type (Figure 5c). RNAi knock‐down plants (Ri) of OsPK2 (Os07g0181000) were generated in a wild‐type genetic background resulting in lines with significantly lower expression levels of OsPK2 than in wild‐type (Figure S6). The grains of those Ri lines displayed opaque endosperm (Figure 5c). The total starch content and grain weight, grain length, width and thickness of brown rice of Ri were all markedly decreased compared with wild‐type (Figures 5d,e and S7e–h). CRISPR/Cas9 technology was also applied to confirm that the phenotypic was due to the knockout of the OsPK2 gene (Figures 5c–e and S8). Therefore, it was deduced that the abnormal grain phenotypes and starch physicochemical properties observed in the mutant were due to the mutation detected in the OsPK2 gene.
Expression pattern, subcellular localization and enzyme assay of OsPK2
Temporal and spatial expression analysis showed that OsPK2 is constitutively expressed in all tested organs. Transcript levels of OsPK2 were higher in stems, panicles and leaves but less abundant in roots and developing grains. Compared to the wild‐type, OsPK2 transcription level in the mutant was much lower in most organs (Figure 6a). To confirm these results, a vector with the GUS reporter gene driven by the OsPK2 promoter was transformed into rice. Histochemical analysis of GUS activity in transgenic plants corroborated that OsPK2 presents a constitutive expression pattern (Figure 6b).
Figure 6.
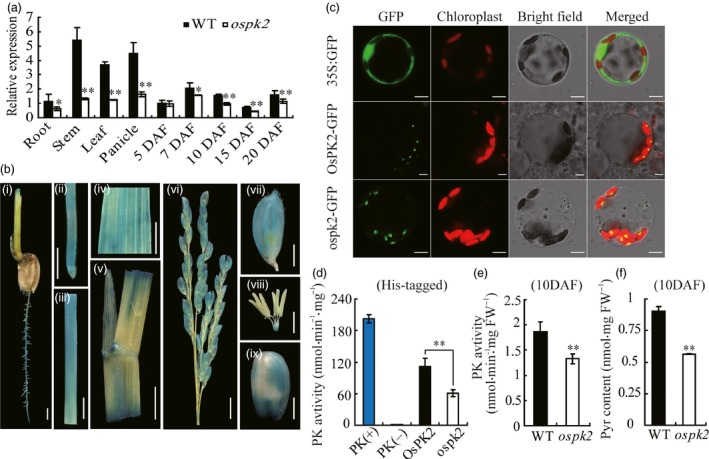
Expression pattern, subcellular localization and enzyme assay of OsPK2. (a) OsPK2 expression level in various tissues and in developing endosperms of the WT and ospk2. RNA was isolated from root, stem, leaf and panicle at the heading stage and developing endosperms at 5, 7, 10, 15 and 20 days after fertilization (DAF). Values are means ± SD from three biological replicates. Asterisks indicate statistical significance between wild‐type and mutant, as determined by a Student's t‐test (*P < 0.05, **P < 0.01). (b) GUS staining in root (i, ii), stem (iii), leaf (iv), leaf sheath (v), panicle (vi), glume (vii), spikelet (viii) and brown rice (ix) driven by the OsPK2 promoter. Bar: 2 mm (i, vii–ix); 1 cm (ii–v,); 5 mm (vi). (c) Subcellular localization of OsPK2 in rice protoplast cells. From the top panel to the bottom panel: free GFP used as control; OsPK2 full‐length coding region and GFP fusion protein (OsPK2‐GFP) and ospk2 full‐length coding region and GFP fusion protein (ospk2‐GFP). Forty‐eight hours after transformation, protoplast cells were observed using a confocal laser scanning microscope. GFP signals, chlorophyll autofluorescence, bright‐field images and the merged images of GFP and chlorophyll signals are shown in each panel. Bars: 5 μm. (d) Enzyme activity assay of OsPK2 and ospk2 protein expressed in baculovirus system and purified by His‐tag. PK activity was determined by the decreasing of NADH (measured the absorbance value at 340 nm for 2 min). (e) PK activity of fresh seeds (10 DAF) from wild‐type and ospk2. (f) Pyruvate content of seeds (10 DAF) from wild‐type and ospk2. Data in (d–f) are shown as mean ± SD from three biological replicates. Asterisks indicate statistical significance as determined by a Student's t‐test (*P < 0.05, **P < 0.01).
OsPK2 was predicted as a chloroplast‐targeted protein by the online tools Plant‐PLoc (http://www.csbio.sjtu.edu.cn/cgi-bin/PlantPLoc.cgi) and WoLF PSORT (http://wolfpsort.org). To verify the predicted subcellular localization and the length and position of the chloroplast‐targeting signal in OsPK2, GFP fusions driven by the CaMV 35S promoter, OsPK2‐GFP, ospk2‐GFP, OsPK21–451aa‐GFP, OsPK21–97aa‐GFP and OsPK298–578aa‐GFP were constructed and transiently expressed in rice protoplasts and tobacco leaves. The GFP control protein was widely present in the cytoplasm and nuclei; however, both OsPK2‐GFP and ospk2‐GFP fusion proteins were localized in chloroplasts, evidenced by the exclusive colocalization with the chlorophyll autofluorescence signal (Figures 6c and S9a,b). GFP signals of OsPK21–451aa‐GFP and OsPK21–97aa‐GFP fusion proteins were also targeted to the chloroplasts, while the signal of OsPK298–578aa‐GFP was not (Figure S9c–e). These results suggest that OsPK2 is a chloroplast‐located protein and that the 97‐amino acid N‐terminal sequence is essential for targeting OsPK2 to chloroplasts.
The proteins encoded by OsPK2 and ospk2 were expressed in baculovirus system and purified by His‐tag (Figure S10) and used to examine PK activity. The result showed that both OsPK2 and ospk2 present PK activity. However, the activity of ospk2 (~60.98 nmol/min/mg) was significantly lower than that of wild‐type (~112.09 nmol/min/mg) (Figure 6d). The PK activity was also detected in seeds (10 DAF) and leaves of wild‐type and ospk2, and the results indicated that the PK activity was also significantly decreased in the mutant (Figures 6e and S11a). Additionally, the content of pyruvate (product of PK) in seeds (10 DAF) and leaves was also significantly reduced in ospk2 compared with wild‐type (Figures 6f and S11b). All these results demonstrate that the protein encoded by OsPK2 has PK activity and a single amino acid substitution (Ser296 to Leu296) in the PK domain compromised its enzymatic activity.
OsPK2 can interact with other plastidic pyruvate kinases to form heteropolymers
There are at least eleven genes predicted to encode putative PK isoforms in rice (Zhang et al., 2012), among them, OsPK2 (PKpα1), Os03g0672300 (PKpα2, 53.48% amino acid sequence identity with OsPK2), Os01g0660300 (PKpβ1, 39.47% identity with OsPK2) and Os10g0571200 (PKpβ2, 39.43% identity with OsPK2) exist as plastidic PK isozymes (Baud et al., 2007). Transcription levels of the PK family were analysed in wild‐type and ospk2 mutant. The results showed that the expression of OsPK2 and PKpβ2 was significantly decreased, and that of PKpα2, PKpβ1 and two plant cytoplasmic PK isozyme genes, Os01g0276700 and Os11g0216000, were significantly increased in the ospk2 mutant (Figure S12). Usually, PKs exist as homotetramers, but are also present as monomers, homodimers, heterodimers, heterotetramers or heteropolymers (Andre et al., 2007; Knowles et al., 1989; Plaxton, 1989; Plaxton et al., 1990, 2002). Approaches to test recombinant subunits of rice PKps were applied in our study. The possible interactions of PKp subunits with OsPK2 were examined by yeast two‐hybrid analysis (Y2H) and bimolecular fluorescence complementation (BiFC). Results showed that three putative PKp subunits (PKpα2, PKpβ1 and PKpβ2) can interact with OsPK2 in yeast cells (Figure 7a). The BiFC assay in tobacco epidermal cells also confirmed that OsPK2 (PKpα1) can physically interact with PKpα2, PKpβ1 and PKpβ2 (Figure 7b). In addition, Y2H and BiFC revealed that PKpβ1 and PKpβ2 can interact with each other to form a heteromer (Figure 7a,b). Moreover, we found that OsPK2 (PKpα1) and PKpα2 can interact with themselves. However, pKpβ1 and PKpβ2 cannot interact with themselves and no interaction was detected between PKpα2 and pKpβ1 or PKpβ2 (Figures 7a,b and S13). These results suggest that OsPK2 can form polymeric protein complexes in vivo by interacting with other PKp.
Figure 7.
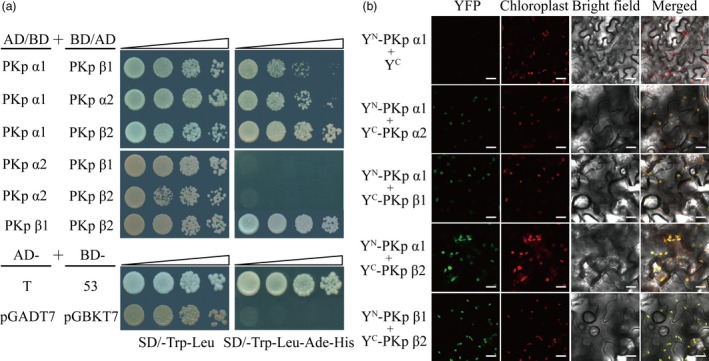
The interactions between OsPK2 and other PKps (a) yeast two‐hybrid assays showed interactions between OsPK2 (PKpα1, α1) and three putative pyruvate kinase plastidic isozymes, PKpα2 (α2), PKpβ1 (β1) and PKpβ2 (β2). Serial dilutions (10‐fold) of yeast cells expressing the indicated proteins were plated onto nonselective medium (SD/‐Leu/‐Trp) (left) or selective medium (SD/‐Leu/‐Trp/‐Ade/‐His) (right). The interactions between pGADT7‐T (T) and pGBKT7‐53 (53), pGADT7 (AD) and pGBKT7 (BD) were used as the positive and negative controls, respectively. (b) BiFC assay showed that OsPK2 (PKpα1, α1) can interact with PKpα2 (α2), PKpβ1 (β1) and PKpβ2 (β2), and PKpβ1 (β1) also can interact with PKpβ2 (β2) in the chloroplast of tobacco cells. Bars = 30 μm.
Expression of glycolysis‐, pyruvate metabolism‐, FA metabolism‐ and starch synthesis‐related genes was affected in the ospk2 mutant
Pyruvate kinase is a key enzyme involved in the glycolytic pathway, thus, the expression of ten putative enzyme genes related to glycolysis, pyruvate metabolism and FA metabolism was analysed in developing grains of wild‐type and ospk2. Compared to wild‐type, expression levels of OsFBP1 encoding fructose‐1,6‐bisphosphatase (FBPase, EC: 3.1.3.11), a key enzyme in gluconeogenesis or Calvin cycle converting fructose‐1,6‐bisphosphate to fructose‐6‐phosphate (F6P), and triose phosphate isomerase (TIM, EC: 5.3.1.1), which reversibly interconvert dihydroxyacetone phosphate (DHAP) and 3‐phosphoglyceraldehyde (GA3P), were significantly increased in the ospk2 mutant. The expression level of a gene encoding the putative plastidic pyruvate dehydrogenase (PDH, EC: 4.1.1.1, the dehydrogenase E1 component of PDC) which converts pyruvate to acetyl‐CoA was also significantly increased. Whereas no differences were observed for the expression of the gene encoding glucose‐6‐phosphate isomerase (GPI, EC: 5.3.1.9) that interconvert G6P and F6P (Figure 8a). Otherwise, expression levels of pyruvate metabolism‐related genes PEPC‐2 encoding PEP carboxylase, PEPCK for PEP carboxykinase and ATL‐2 for alanine transaminase were all significantly increased in ospk2. Meanwhile, expression of the genes that encode a chloroplastic NADP‐dependent malic enzyme (ME6, Os01g0188400) and OsAlaATL1 (Os10g0390500) was lower in the ospk2 mutant (Figure 8b). The expression of OsKASI, encoding a β‐ketoacyl‐[acyl carrier protein] synthase I which participates in FA synthesis, was significantly higher in the mutant. Genes for lipolytic enzymes such as THIS1 and Lipase‐II displayed lower expression in ospk2 (Figure 8c). The expression of starch synthesis‐related genes was also evaluated in 12 DAF grains. Results showed that transcript levels of three AGP genes (OsAGPL2, OsAGPS1 and OsAGPS2b), six amylopectin synthesis‐related genes (OsSSI, OsSSIVb, OsGBSSI, OsISA1, OsISA3 and OsPUL), Susy3 and pyruvate phosphate dikinase gene PPDKB were all significantly lower in ospk2 than in the wild‐type. Conversely, the transcript levels of OsBEIIb, OsSSIIIa, OsPHOL, OsISA2, Susy1 and Susy2 were markedly higher in ospk2 mutant (Figure 8d).
Figure 8.
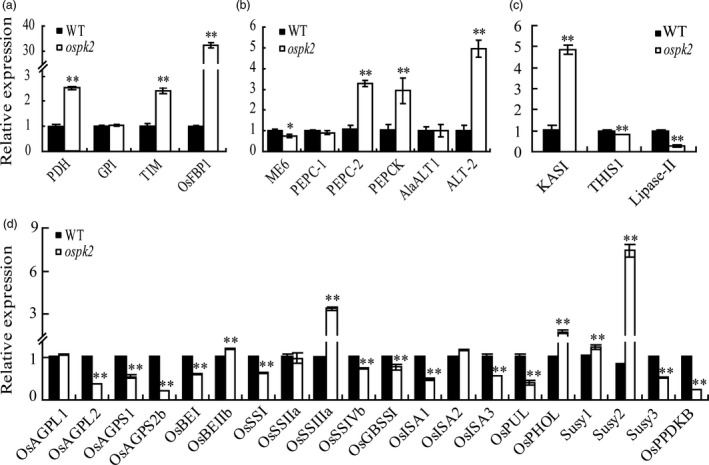
Expression analyses of genes related to glycolysis/gluconeogenesis, pyruvate/phosphoenolpyruvate metabolism, fatty acid metabolism and starch synthesis. (a) Enzyme genes of glycolysis/gluconeogenesis metabolism. PDH (Os04g0119400), a putative pyruvate dehydrogenase; GPI (Os06g0256500), glucose‐6‐phosphate isomerase; TIM (Os09g0535000), similar to the chloroplast precursor of triosephosphate isomerase and OsFBP1 (Os01g0866400), a putative fructose‐1, 6‐bisphosphatase. (b) Genes related to pyruvate/PEP metabolism. ME6 (Os01g0188400) encodes a putative chloroplastic NADP‐dependent malic enzyme; PEPC‐1 (Os02g0244700) and PEPC‐2 (Os01g0208700), putative phosphoenolpyruvate (PEP) carboxylases; PEPCK (Os10g0204400), a putative PEP carboxykinase; OsAlaAT1 (Os10g0390500) and ALT‐2 (Os07g0108300) encode alanine transaminases. (c) Genes involved in fatty acid synthesis and degradation. OsKASI (Os06g0196600) encodes a β‐ketoacyl‐[acyl carrier protein] synthase;. THIS1 (Os01g0751600) and Lipase‐II (Os07g0668700) as lipolytic enzymes. (d) Genes associated with starch metabolism, including AGP genes (OsAGPL1, OsAGPL2, OsAGPS1 and OsAGPS2b), starch branching enzyme genes (OsBEI, OsBEIIb), amylopectin synthesis genes (OsSSI, OsSSIIa, OsSSIIIa and OsSSIVb), amylose starch synthase gene (OsGBSSI), starch debranching enzyme genes (ISA1,ISA2,ISA3,PUL), sucrose synthase genes (Susy1, Susy2 and Susy3), OsPHOL and OsPPDKB. In (a–d), RNA was isolated from wild‐type (WT) and ospk2 grains of 12 days after fertilization. Expression levels are represented as relative to the corresponding genes in WT (set as reference value of 1) and data are shown as means ± SD from three biological replicates. Asterisks indicate statistical significance between the WT and the mutant, as determined by a Student's t‐test (*P < 0.05; **P < 0.01).
Discussion
OsPK2 can interact with other plastidic PKs to form heteropolymers
Pyruvate kinase isoforms vary in different tissues and subcellular components (Andre et al., 2007). In cytosol, PKc exists as homotetramer in both germinating cotyledons and developing endosperm of castor oil seeds (Podestá and Plaxton, 1994; Turner et al., 2005). In plastids, native enzyme PKp in developing endosperm of castor oil seeds was present as a heterohexamer (α2β2; Plaxton et al., 1990). PKp in Arabidopsis was as appeared to be a 460‐kDa heterooctamer (α4β4) containing four α subunits (59.6 kDa) and four β subunits (56.8 kDa; Andre et al., 2007). In this work, we identified and characterized OsPK2 encoding a PKp isozyme in rice. Subcellular localization analysis confirmed that the green fluorescent signals of OsPK2‐GFP were small dot‐like structures and only colocalized with the autofluorescent signals of chlorophyll (Figures 6c and S9), implying that OsPK2 localizes in the chloroplast nucleoids, similar to what was observed for AtPKp1 and AtPKp2 in Arabidopsis (Baud et al., 2007). In developing castor oil seeds, the N‐terminal 44 and 60 amino acids of PKp α‐ and β‐subunit contain their respective transit peptides which were responsible for their import to the leucoplast (Negm et al., 1995). Here, we found that the first 97 N‐terminal amino acids were necessary for the chloroplast location of OsPK2 (Figure S9). Y2H and BiFC assays indicated that OsPK2 (PKpα1) can interact with three other putative PKp subunits (PKpα2, PKpβ1 and PKpβ2) and that OsPK2 (PKpα1) and PKpα2 can also interact with themselves (Figures 7a,b and S13). These results suggest that the PKp in rice can exist as heteromeric complex. Meanwhile, BiFC assays also showed the green fluorescent signals of PKpα1&PKpα2, PKpα1&PKpβ1, PKpα1&PKpβ2 and PKpβ1&PKpβ2 overlapping with the chlorophyll autofluorescence signal in tobacco cells (Figure 7b), supporting the idea that PKpα2, PKpβ1 and PKpβ2 with OsPK2 (PKpα1) are all plastidic isozymes, consistent with the phylogenetic analysis of OsPK2 homologous proteins (Figure S4). Therefore, the enzyme complexes formed by OsPK2 subunit coordinated with three other subunits would fulfil PKp function in rice. Together, these results suggest that the PKp complex in rice might be more multifaceted and diverse than in other plants due to the multiple interactions between different subunits.
Mutation in OsPK2 leads to multiple disorders in metabolic processes
Pyruvate kinase catalyses the final step in glycolysis which produces ATP and NADH by breakdown glucose into pyruvate. Pyruvate can then be converted to acetyl‐CoA and NADH by the PDC which contains two distinct forms, mitochondrial (mtPDC) and plastidial (plPDC). In the cytosol, pyruvate produced by PKc is transported into mitochondria to undergo oxidative decarboxylation by mtPDC and offer carbon (acetyl‐CoA) to the TCA cycle (Smith et al., 2000). In plastids, the main role of PKp is to generate ATP and substrate (pyruvate) which can be catalysed by plPDC to synthesize acetyl‐CoA for FA biosynthesis (Andre et al., 2007). Pyruvate and acetyl‐CoA are also important products in the conversion of lipids to sugars via gluconeogenesis and the glyoxylate cycle. In addition, the intermediate PEP can be catalysed by PEPC to produce OAA which can enter the mitochondria to fuel the TCA pathway or amino acids synthesis. Malate produced by OAA in the cytosol can also enter the mitochondria (Plaxton and Leary, 2012; Sweetlove et al., 2010). Therefore, pyruvate is an important intermediate for sucrose/starch, lipid and amino acid metabolism. In our study, mutation in OsPK2 significantly reduced the PK activity in ospk2 (Figures 6e and S10b), which led to alteration of the expression of genes involved in multiple metabolic processes. For instance, the expression of TIM, OsFBP1 and PDH was significantly higher in ospk2 (Figure 8a). The transcription level of OsKASI encoding a FA synthesis enzyme was significantly higher in the mutant than in the wild‐type, while that of THIS1 and Lipase‐II gene encoding the lipolytic enzyme was lower in ospk2 (Figure 8c). In addition, expression of Susy1 and Susy2 was significantly higher in ospk2 (Figure 8d). These data imply that OsPK2 can directly or indirectly affect the TCA cycle, FA and sugar metabolism. In addition, the expression of starch synthesis‐related genes was also changed in ospk2 (Figure 8d). Therefore, mutation in OsPK2 leads to a variety of metabolic disorders in the ospk2 mutant.
OsPK2 affects normal starch/FA biosynthesis and seed germination rate in rice
The de novo synthesis of FA in plants occurs predominantly in the plastid, which requires acetyl‐CoA as substrate (Harwood, 1988). Import of metabolites from the cytosol to synthesize acetyl‐CoA is needed as acetyl‐CoA is not directly imported into plastids (Roughan et al., 1979; Weaire and Kekwick, 1975). In plastids, pyruvate can be converted to acetyl‐CoA and NADH by the plPDC (Baud et al., 2007; Harwood, 1988). In our study, expression of a PDH gene encoding the dehydrogenase E1 component of PDC was higher in ospk2 than in wild‐type (Figure 8a), which may lead to increased content of acetyl‐CoA and eventually increase FA content in the mutant (Figure S3). Synthesis of FA and starch in plastids requires abundant ATP. In fact, increased FA content will consume more ATP and the reduced PK activity in ospk2 would produce less ATP, resulting in less available ATP for starch synthesis. Meanwhile, expression levels of several genes involved in starch synthesis (such as AGPL2, AGPS1, AGPS2b and GBSSI) also were reduced in the mutant (Figure 8d). All these alterations may cause the lower starch content observed in ospk2 (Figure 4a,b). In addition, we also found that the germination rate of ospk2 seeds after 1‐year storage declined largely (Figure S2). This might be caused by the increased amount of FA in ospk2 (Figure S3). The lower germination rate was consistent with the results described for the mutation of AtPKp1 in Arabidopsis (Baud et al., 2007). Together, these results denote new insights into the role of OsPK2 in plant seed development, especially in starch/FA biosynthesis, compound granules formation and grain filling, and provide useful grounds for potential genetic improvement of high yield and grain quality in rice.
Materials and methods
Plant materials and growth conditions
The ospk2 mutant was created from O. sativa subsp. japonica cv. Zhonghua 11 (ZH11) by EMS mutagenesis. The F2 population for gene mapping was generated by crossing the ospk2 mutant with the indica rice cv. Nanjing11 (NJ11). Plants were grown under natural conditions at China National Rice Research Institute experimental fields in Fuyang, Hangzhou, China.
Map‐based cloning of the OsPK2 gene
To map the OsPK2 locus, white‐core endosperm seeds were selected from the F2 population produced from the cross between the ospk2 and NJ‐11. More than 170 polymorphic SSR markers evenly distributed over the whole genome were selected to identify markers linked to OsPK2. Further molecular markers were developed based on nucleotide polymorphisms between wild‐type and NJ‐11 in the corresponding regions (Table S2). To fine map the OsPK2 locus, 978 recessive individuals with the mutant phenotype were selected from the F2 population.
Microscopy analysis
The brown rice of wild‐type and the ospk2 mutant was cut transversely, and the ruptured transverse surface was coated with gold to prepare samples. Images were obtained with a HITACHI S‐3400N scanning electron microscope (Hitachi, Tokyo, Japan; http://www.hitachi‐ hitec.com). To examine the development of compound granules, transverse sections (approximately 1 mm in thickness) of wild‐type and ospk2 endosperms at 10 DAF were used to prepare semithin sections (800 nm). Samples were stained with I2‐KI for 5 s and subsequently examined under a light microscope (Nikon Eclipse 80i, Japan; http://www.nikon.com). All treatments of samples were followed as described previously (Peng et al., 2014). To observe the ultrastructure of amyloplasts, developing seeds (3–12 DAF) from wild‐type and ospk2 were analysed by transmission electron microscope (H‐7650; Hitachi, http://www.hitachi.com). Samples were treated as described by Takemoto et al. (2002).
Physicochemical properties of the starch in rice grains
The total starch content of the rice powder was measured using a starch assay kit (Megazyme, Wicklow, Ireland; http://www.megazyme.com). Amylose content was determined following the method depicted by Liu et al. (2009). Lipid and protein contents in the grains were measured according to the method described by Kang et al. (2005). Content of FA was determined by gas chromatography‐mass spectrometer (GC‐MS, Li et al., 2006). To determine the starch pasting properties, 3 g of milled rice power (0.5 mm or less, 14% moisture basis) was transferred into a container with 25 mL of distilled water. The sample was mixed and measured with a Rapid Visco Analyzer (RVA Techmaster; Newport Scientific, Narrabeen, Australia), applying the protocol described by the manufacturer. To determine the chain length distribution of amylopectin, 5 mg of rice flour was digested with Pseudomonas amyloderamosa isoamylase (Megazyme), then analysed using the capillary electrophoresis (PA800 plus pharmaceutical analysis system; Beckman Coulter, http://www.beckmancoulter.com). The swelling and gelatinization properties of endosperm starch in urea solution were measured according to the method described by Nishi et al. (2001). All parameters related to physicochemical properties included three biological replications.
RNA extraction and real‐time PCR analysis
Total RNA was extracted from different plant tissues (root, stem, leaf, panicle and seeds at 5, 7, 10, 15, 20 DAF) using the Trizol reagent (Life Technologies). A 2‐μg portion of total RNA was reverse‐transcribed into cDNA according to the manufacturer's instructions (ReverTra Ace qPCR RT Kit; Toyobo). qRT–PCR was performed using the SYBR Green Real‐time PCR Master Mix (Toyobo). PCR programme was as follows: 95 °C for 30 s, 40 cycles of 95 °C for 5 s, 60 °C for 35 s and 95 °C for 15 s. Gene expression was calculated by the 2−ΔΔCT method using the Ubiquitin gene (Os03g0234200) as internal control. The primer sequences used in this analysis are listed in Table S2.
Plasmid construction and rice transformation
For complementation of the ospk2 mutant, the wild‐type OsPK2 genomic fragment from ZH11, including its native promoter, was amplified by PCR (all primers used for plasmid construction are listed in Table S2) and then cloned into the binary vector pCAMBIA1300 to generate the complementation vector. A vector bearing the OsPK2 cDNA sequence driven by UBIQUTIN1 promoter was cloned into the binary vector pCAMBIA1390 to generate the overexpression vector. To obtain the RNAi vector, a 481 bp fragment from the OsPK2 cDNA from ZH11 was cloned into the binary vector LH‐FAD2‐1390RNAi under the control of the maize UBIQUTIN1 promoter. The CRISPR target of OsPK2 (GGCCGACCTGAGGGAGAAC) was cloned into a guide sgRNA (U3 gRNA) by restriction site (BsaI), and gateway recombination was used to incorporate gRNA into the pYLCRISPR/Cas9‐MH/B vector. These plasmids were introduced into Agrobacterium tumiefaciens strain EHA105. Complementation and overexpression vectors were transformed into ospk2, and the RNAi and the CRISPR/Cas9 vectors were transformed into wild‐type plants. The genotypes of the transgenic plants were determined by PCR amplification of the specific transgenic fragment.
Subcellular localization of OsPK2 protein
To verify the subcellular localization of OsPK2, the coding regions without a termination codon from wild‐type and ospk2 were amplified by PCR and cloned into the pAN580 and pCAMBIA1305‐GFP vectors to generate the OsPK2‐GFP, and ospk2‐GFP constructs. Other four vectors bearing the fragment of OsPK21–97aa, OsPK298−578aa and OsPK21–451aa were also constructed into pCAMBIA1305‐GFP. Stem and leaf sheath tissues from 12‐day‐old green rice seedlings were sliced into 0.5 mm long segments to prepare protoplasts. The OsPK2‐GFP, ospk2‐GFP and pAN580 vectors were transformed into rice protoplasts and incubated in dark at 28 °C for 2 days following previous description (Chen et al., 2006). The other constructs were introduced into Agrobacterium strain GV3101 which was then infiltrated into 3‐week‐old leaf epidermal cells of Nicotiana benthamiana (tobacco). After infiltration for 48 h, GFP fluorescence in tobacco leaves was observed with a LSM710 confocal laser scanning microscope (Carl Zeiss AG, Oberkochen, Germany).
GUS staining
The putative promoter region of OsPK2 (~2‐kb upstream of ATG) was amplified by PCR and cloned into the EcoRI/NcoI sites of pCAMBIA1305. The resultant construct was transformed into ZH11 calli, and independent lines of positive T1 transgenic progeny were used to detect GUS activity. Tissues were submerged in GUS staining solution (10 mm EDTA, 0.1% Triton X‐100, 1 mm 5‐bromo‐4‐chloro‐3‐indoyl‐ b‐D‐glucuronide, 100 mm sodium phosphate (pH 7.0), 2.5 mm K4Fe(CN)6 and 2.5 mm K3Fe(CN)6 at 37 °C for 12–15 h). After incubation, tissues were discoloured several times in pure ethyl alcohol.
Protein expression, enzyme assay and pyruvate content detection
To validate the PK activity of OsPK2, the full‐length CDS of OsPK2 and ospk2 were cloned into the Bac‐to‐Bac baculovirus expression vector PFAST‐BAC I (Thermo Fisher Scientific). Protein expression and purification by His‐tag using the baculovirus expression system followed the method depicted by King and Possee (1992). On the other hand, a PK crude enzyme solution was also extracted from fresh leaves and 10 DAF seeds of wild‐type and ospk2 according to a previous report (Baud et al., 2007). First, 0.1 g of plant material was ground in liquid nitrogen and homogenized with 1 mL extraction buffer consisting of 50 mm HEPES‐KOH buffer (pH 8.0), 100 mm KCL, 5 mm MgCl2, 20 mm NaF, 1 mm EDTA, 0.1% (v/v) Triton X‐100, 20% (v/v) glycerol, 5% (w/v) polyethylene glycol 8000, 1 mm 2,2′‐dithiodipyridine and 1% (w/v) insoluble poly(vinylpolypyrrolidone). The homogenate was centrifuged at 14 000 g for 10 min at 4 °C, and the supernatant was used for enzyme activity. Both expressed protein and crude enzyme solution extracted from fresh tissues were used to measure PK activity by the method of coupling reaction of pyruvate and the conversion of NADH to NAD+. PKp reaction solution contained 100 mm Hepes‐KOH (pH 8.25), 50 mm KCl, 10 mm MgCl2, 5% (w/v) polyethylene glycol 8000, 1 mm dithiothreitol, 2 mm PEP, 0.30 mm NADH, 2.5 mm ADP and 2 U/mL desalted rabbit muscle lactate dehydrogenase (Sigma, L1254; Sigma‐Aldrich Co., St. Louis, MO). About 0.5 U/mL PK from rabbit muscle (Sigma, P9136; Sigma‐Aldrich Co.) was used as the positive control, and the inactivated His‐OsPK2 by heat treated at 95 °C for 20 min served as negative control. The declined absorbance values at 340 nm of the reaction solution from 20 s to 2 min 20 s were analysed with a microplate spectrophotometer (Infinite 200 PRO; TECAN, Mannedorf, Switzerland) to calculate PK activity. Simultaneously, pyruvate content in leaf and 10 DAF seeds of wild‐type and ospk2 was determined by the 2,4‐dinitrobenzene hydrazine colorimetric method using the pyruvate assay kit (TC0750‐100T; Beijing Leagene Biotechnology Co., Ltd., http://www.leagene.com). First, 0.5 g of fresh leaves and 10 DAF seeds were ground in liquid nitrogen and homogenized with 9 mL of 8% trichloroacetic acid in ice for 30 min, then centrifuged at 4000 g for 10 min at 4 °C. About 1.5 mL of supernatant was mix well with 0.5 mL 0.1% 2,4‐dinitrobenzene hydrazine, then 2.5 mL of 2 m NaOH was added with adequate mixing. After 5 min, the absorbance value at 520 nm was recorded using a microplate spectrophotometer (Infinite200 PRO; TECAN).
Yeast two‐hybrid assay and BiFC assay
The coding region of OsPK2 without a termination codon was amplified by PCR and cloned into pGBKT7 and pGADT7 (Clontech, http://www.clontech.com). Coding regions of several rice PK genes were also cloned into the same vectors (Table S2). Yeast transformation and selecting procedures were accomplished according to the manufacturer's guidebook (Clontech). For BiFC assays, the full‐length cDNAs without a termination codon of Os03g0672300 (named PKpα2), Os01g0660300 (PKpβ1), Os10g0571200 (PKpβ2) and OsPK2 (Os07g0181000, also named PKpα1) were cloned into the binary vectors pSPYCE or pSPYNE to create the constructors pPKα2‐YFPC, pPKβ1‐YFPC, pPKβ2‐YFPC and pOsPK2–YFPN, respectively. The plasmids were transiently expressed in tobacco leaves following the method described by Waadt and Kudla (2008). A confocal laser scanning microscope (Zeiss LSM710) was used to detect YFP fluorescent signals after 48 h post‐transfection. Primers used in this assay are listed in Table S2.
Author contributions
All authors contributed to the design and implementation of the experimental strategy and participated in the data collection. P. H. and X. W. designed the experiments. Y. C., S. L., X. W., G. J., Z. S., Y. W., G. S., L. X., C. P. and S. T. carried out the experimental work. Y. C., S. L. and X. W. wrote the first draft of the manuscript which was critically revised and implemented by P. H., S. T. and J. X. All authors discussed the results and commented on the final version of the manuscript.
Conflict of interest
The authors declare that they have no competing financial interest.
Supporting information
Figure S1 Characteristics of adult wild‐type and ospk2 plants.
Figure S2 The germinability of fresh harvested and 1 year storage seeds of wild‐type and ospk2.
Figure S3 Fatty acid composition in mature seeds.
Figure S4 Phylogenetic analysis of OsPK2.
Figure S5 Amino acid sequence alignment between OsPK2 and other homologous proteins.
Figure S6 qRT‐PCR analysis of OsPK2 in transgenic plants.
Figure S7 Analysis of OsPK2 transgenic plants.
Figure S8 Analysis of OsPK2 CRISPR‐CAS9‐mediated editing.
Figure S9 Subcellular localization of OsPK2 and truncated OsPK2 in tobacco cells.
Figure S10 SDS‐PAGE and western‐blot analysis of his‐OsPK2 and his‐ospk2 purified from baculovirus expression system under native condition.
Figure S11 PK activity assay in fresh leaves.
Figure S12 qRT‐PCR analysis of genes encoding putative PK in rice.
Figure S13 Yeast two‐hybrid assays showed that both OsPK2 (PKpα1) and PKpβ1 can interact with itself.
Table S1 Gene products of the nine predicted ORFs in the fine mapping region.
Table S2 Primers used in this study.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2016YFD0101801), the National Natural Science Foundation of China (grants No. 31471472, 31501280) and by the National S&T Major Project of China (2016ZX08001006).
Contributor Information
Xiangjin Wei, Email: weixiangjin@caas.cn.
Peisong Hu, Email: peisonghu@126.com, Email: hupeisong@caas.cn.
References
- Akihiro, T. , Mizuno, K. and Fujimura, T. (2005) Gene expression of ADP‐glucose pyrophosphorylase and starch contents in rice cultured cells are cooperatively regulated by sucrose and ABA. Plant Cell Physiol. 46, 937–946. [DOI] [PubMed] [Google Scholar]
- Ambasht, P.K. and Kayastha, A.M. (2002) Plant pyruvate kinase. Biol. Plantarum, 45, 1–10. [Google Scholar]
- Andre, C. and Benning, C. (2007) Arabidopsis seedlings deficient in a plastidic pyruvate kinase are unable to utilize seed storage compounds for germination and establishment. Plant Physiol. 145, 1670–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre, C. , Froehlich, J.E. , Moll, M.R. and Benning, C. (2007) A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis . Plant Cell, 19, 2006–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud, S. , Wuillème, S. , Dubreucq, B. , de Almeida, A. , Vuagnat, C. , Lepiniec, L. , Miquel, M. et al (2007) Function of plastidial pyruvate kinases in seeds of Arabidopsis Thaliana . Plant J. 52, 405–419. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Tao, L. , Zeng, L. , Vega‐ Sanchez, M.E. , Umemura, K. and Wang, G.L. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein–protein interactions in rice. Mol. Plant Pathol. 7, 417–427. [DOI] [PubMed] [Google Scholar]
- Fu, F.F. and Xue, H.W. (2010) Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 154, 927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodzinski, B. , Jiao, J. , Knowles, V.L. and Plaxton, W.C. (1999) Photosynthesis and carbon partitioning in transgenic tobacco plants deficient in leaf cytosolic pyruvate kinase. Plant Physicol. 120, 887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood, J.L. (1988) Fatty acid metabolism. Annu. Rev. Plant Physiol. Mol. Biol. 39, 101–138. [Google Scholar]
- Hu, Z. and Plaxton, W.C. (1996) Purification and characterization of cytosolic pyruvate kinase from leaves of the castor oil plant. Arch. Biochem. Biophys. 333, 298–307. [DOI] [PubMed] [Google Scholar]
- Initiative, A.G. (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Kang, H.G. , Park, S. , Matsuoka, M. and An, G. (2005) White‐core endosperm floury endosperm‐4 in rice is generated by knockout mutations in the C‐type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J. 42, 901–911. [DOI] [PubMed] [Google Scholar]
- Kawagoe, Y. , Kubo, A. , Satoh, H. , Takaiwa, F. and Nakamura, Y. (2005) Roles of isoamylase and ADP‐glucose pyrophosphorylase in starch granule synthesis in rice endosperm. Plant J. 42, 164–174. [DOI] [PubMed] [Google Scholar]
- King, L.A. and Possee, R.D. (1992) The Baculovirus Expression System: A Laboratory Guide, 1st ed. Edinburgh: Chapman & Hall; 10.1007/978-94-011-2374-7. [DOI] [Google Scholar]
- Knowles, V.L. , Dennis, D.T. and Plaxton, W.C. (1989) Purification of a novel pyruvate kinase from a green alga. FEBS Lett. 259, 130–132. [Google Scholar]
- Knowles, V.L. , McHugh, S.G. , Hu, Z. , Dennis, D.T. , Miki, B.L. and Plaxton, W.C. (1998) Altered growth of transgenic tobacco lacking leaf cytosolic pyruvate kinase. Plant Physiol. Biochem. 116, 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, A. , Fujita, N. , Harada, K. , Matsuda, T. , Satoh, H. and Nakamura, Y. (1999) The Starch‐debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol. 121, 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.K. , Hwang, S.K. , Han, M. , Eom, J.S. , Kang, H.G. , Han, Y. Choi, S.B. et al (2007) Identification of the ADP‐glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.). Plant Mol. Biol. 65, 531–546. [DOI] [PubMed] [Google Scholar]
- Lee, S.K. , Eom, J.S. , Hwang, S.K. , Shin, D. , An, G. , Okita, T.W. and Jeon, J.S. (2016) Plastidic phosphoglucomutase and ADP‐glucose pyrophosphorylase mutants impair starch synthesis in rice pollen grains and cause male sterility. J. Exp. Bot. 67, 5557–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Beisson, F. , Pollard, M. and Ohlrogge, J. (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant‐to‐plant variation. Phytochemistry, 67, 904–915. [DOI] [PubMed] [Google Scholar]
- Li, S. , Wei, X. , Ren, Y. , Qiu, J. , Jiao, G. , Guo, X. , Tang, S. et al (2017) OsBT1 encodes an ADP‐glucose transporter involved in starch synthesis and compound granule formation in rice endosperm. Sci. Rep. 7, 40124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. , Ma, X. , Liu, S. , Zhu, C. , Jiang, L. , Wang, Y. , Shen, Y. et al (2009) Identification and characterization of a novel Waxy allele from a Yunnan rice landrace. Plant Mol. Biol. 71, 609–626. [DOI] [PubMed] [Google Scholar]
- Matsushima, R. , Maekawa, M. , Kusano, M. , Kondo, H. , Fujita, N. , Kawagoe, Y. and Sakamoto, W. (2014) Amyloplast‐localized SUBSTANDARD STARCH GRAIN 4 protein influences the size of starch grains in rice endosperm. Plant Physiol. 164, 623–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima, R. , Maekawa, M. , Kusano, M. , Tomita, K. , Kondo, H. , Nishimura, H. , Crofts, N. et al (2016) Amyloplast membrane protein SUBSTANDARD STARCH GRAIN6 controls starch grain size in rice endosperm. Plant Physiol. 170, 1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negm, F.B. , Cornel, F.A. and Plaxton, W.C. (1995) Suborganellar localization and molecular characterization of nonproteolytic degraded leukoplast pyruvate kinase from developing castor oil seeds. Plant Physiol. 109, 1461–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi, A. , Nakamura, Y. , Tanaka, N. and Satoh, H. (2001) Biochemical and genetic analysis of the effects of amylose‐extender mutation in rice endosperm. Plant Physiol. 127, 459–472. [PMC free article] [PubMed] [Google Scholar]
- Oliver, S.N. , Lunn, J.E. , Urbanczyk‐Wochniak, E. , Lytovchenko, A. , van Dongen, J.T. , Faix, B. , Schmalzlin, E. et al (2008) Decreased expression of cytosolic pyruvate kinase in potato tubers leads to a decline in pyruvate resulting in an in vivo repression of the alternative oxidase. Plant Physiol. 148, 1640–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, C. , Wang, Y. , Liu, F. , Ren, Y. , Zhou, K. , Lv, J. , Zheng, M. et al (2014) FLOURY ENDOSPERM 6 encodes a CBM48 domain‐containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant J. 77, 917–930. [DOI] [PubMed] [Google Scholar]
- Plaxton, W.C. (1989) Molecular and immunological characterization of plastid and cytosolic pyruvate kinase isozymes from castor‐oil‐plant endosperm and leaf. Eur. J. Biochem. 181, 443–451. [DOI] [PubMed] [Google Scholar]
- Plaxton, W.C. (1991) Leucoplast pyruvate kinase from developing castor oil seeds. Characterisation of the enzyme's degradation by a cysteine endopeptidase. Plant Physiol. 97, 1334–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton, W.C. (1996) The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 185–214. [DOI] [PubMed] [Google Scholar]
- Plaxton, W.C. and Leary, B.O. (2012) The central role of phosphoenolpyruvate metabolism in developing seeds In Seed development: OMICS technologies toward improvement of seed quality and crop yield (Agrawal G.K. and Rakwal R., eds), pp. 279–301. Dordrecht: Springer; 10.1007/978-94-007-4749-4 [DOI] [Google Scholar]
- Plaxton, W.C. and Podestá, F.E. (2006) The functional organization and control of plant respiration. Crit. Rev. Plant Sci. 25, 159–198. [Google Scholar]
- Plaxton, W.C. , Dennis, T.D. and Knowles, V.L. (1990) Purification of leucoplast pyruvate kinase from developing castor bean endosperm. Plant Physiol. 94, 1528–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton, W.C. , Smith, C.R. and Knowles, V.L. (2002) Molecular and regulatory properties of leucoplast pyruvate kinase from Brassica napus (rapeseed) suspension cells. Arch. Biochem. Biophys. 400, 54–62. [DOI] [PubMed] [Google Scholar]
- Podestá, F.E. and Plaxton, W.C. (1994) Regulation of cytosolic carbon metabolism in germinating Ricinus communis cotyledons. Planta, 194, 381–387. [Google Scholar]
- Roughan, P.G. , Holland, R. , Slack, C.R. and Mudd, J.B. (1979) Acetate is the preferred substrate for long‐chain fatty acid synthesis in isolated spinach chloroplasts. Biochem. J. 184, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo, N. , Yu, C. , Park, C.S. , Baik, M.Y. , Park, I.M. , Cho, M.H. , Bhoo, S.H. et al (2007) Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white‐core floury endosperm in rice (Oryza sativa L.). Plant Cell Rep. 26, 1083–1095. [DOI] [PubMed] [Google Scholar]
- Sato, H. , Suzuki, Y. , Sakai, M. and Imbe, T. (2002) Molecular characterization of Wx‐mq, a novel mutant gene for low‐amylose content in endosperm of rice (Oryza sativa L.). Breed. Sci. 52, 131–135. [Google Scholar]
- Schramm, A. , Siebers, B. , Tjaden, B. , Brinkmann, H. and Hensel, R. (2000) Pyruvate kinase of the hyperthermophilic crenarchaeote thermoproteus tenax: physiological role and phylogenetic aspects. J. Bacteriol. 182, 2001–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender, J. , Ohlrogge, J. and Shachar‐Hill, Y. (2004) Understanding flux in plant metabolic networks. Curr. Opin. Plant Biol. 7, 309–317. [DOI] [PubMed] [Google Scholar]
- She, K.C. , Kusano, H. , Koizumi, K. , Yamakawa, H. , Hakata, M. , Imamura, T. , Fukudai, M. et al (2010) A novel factor FLOURY ENDOSPERM 2 is involved in regulation of rice grain size and starch quality. Plant Cell, 22, 3280–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.R. , Knowles, V.L. and Plaxton, W.C. (2000) Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell cultures. Eur. J. Biochem. 267, 4477–4485. [DOI] [PubMed] [Google Scholar]
- Sweetlove, L.J. , Beard, K.F.M. , Nunes‐Nesi, A. , Fernie, A.R. and Ratcliffe, R.G. (2010) Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci. 15, 462–470. [DOI] [PubMed] [Google Scholar]
- Takemoto, Y. , Coughlan, S.J. , Okita, T.W. , Satoh, H. , Oqawa, M. and Kumamaru, T. (2002) The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol. 128, 1212–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, N. , Fujita, N. , Nishi, A. , Satoh, H. , Hosakam, Y. , Ugaki, M. , Kawasaki, S. et al (2004) The structure of starch can be manipulated by changing the expression levels of starch branching enzyme IIb in rice endosperm. Plant Biotechnol. J. 2, 507–516. [DOI] [PubMed] [Google Scholar]
- Tian, Z. , Qian, Q. , Liu, Q. , Yan, M. , Liu, X. , Yan, C. , Liu, G. et al (2009) Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl Acad. Sci. USA, 106, 21760–21765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyosawa, Y. , Kawagoe, Y. , Matsushima, R. , Crofts, N. , Ogawa, M. , Fukuda, M. , Kumamaru, T. et al (2016) Deficiency of starch synthase IIIa and IVb alters starch granule morphology from polyhedral to spherical in rice endosperm. Plant Physiol. 170, 1255–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncel, A. , Kawaguchi, J. , Ihara, Y. , Matsusaka, H. , Nishi, A. , Nakamura, T. , Kuhara, S. et al (2014) The rice endosperm ADP‐glucose pyrophosphorylase large subunit is essential for optimal catalysis and allosteric regulation of the heterotetrameric enzyme. Plant Cell Physiol. 55, 1169–1183. [DOI] [PubMed] [Google Scholar]
- Turner, W.L. , Knowles, V.L. and Plaxton, W.C. (2005) Cytosolic pyruvate kinase: subunit composition, activity, and amount in developing castor and soybean seeds, and biochemical characterization of the purified castor seed enzyme. Planta, 222, 1051–1062. [DOI] [PubMed] [Google Scholar]
- Valentini, G. , Chiarelli, L. , Fortin, R. , Speranza, M.L. , Galizzi, A. and Mattevi, A. (2000) The allosteric regulation of pyruvate kinase. J. Biol. Chem. 275, 18145–18152. [DOI] [PubMed] [Google Scholar]
- Waadt, R. and Kudla, J. (2008) In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc. pdb.prot4995. 10.1101/pdb.prot4995. [DOI] [PubMed] [Google Scholar]
- Wang, J.C. , Xu, H. , Zhu, Y. , Liu, Q.Q. and Cai, X.L. (2013) OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 64, 3453–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaire, P.J. and Kekwick, R.G. (1975) The synthesis of fatty acids in avocado mesocarp and cauliflower bud tissue. Biochem. J. 146, 425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, X. , Jiao, G. , Lin, H. , Sheng, Z. , Shao, G. , Xie, L. , Tang, S. et al (2017) GRAIN INCOMPLETE FILLING 2 regulates grain filling and starch synthesis during rice caryopsis development. J. Integr. Plant Biol. 59, 134–153. [DOI] [PubMed] [Google Scholar]
- Yamamoto, M.P. , Onodera, Y. , Touno, S.M. and Takaiwa, F. (2006) Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol. 141, 1694–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. and Liu, J.Y. (2016) Cotton cytosolic pyruvate kinase GhPK6 participates in fast fiber elongation regulation in a ROS‐mediated manner. Planta, 244, 915–926. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Xiao, W. , Luo, L. , Pang, J. , Rong, W. and He, C. (2012) Downregulation of OsPK1, a cytosolic pyruvate kinase, by T‐DNA insertion causes dwarfism and panicle enclosure in rice. Planta, 235, 25–38. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Ren, Y. , Lu, B. , Yang, C. , Feng, Z. , Liu, Z. , Chen, J. et al (2016) FLOURY ENDOSPERM 7 encodes a regulator of starch synthesis and amyloplast development essential for peripheral endosperm development in rice. J. Exp. Bot. 67, 633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Characteristics of adult wild‐type and ospk2 plants.
Figure S2 The germinability of fresh harvested and 1 year storage seeds of wild‐type and ospk2.
Figure S3 Fatty acid composition in mature seeds.
Figure S4 Phylogenetic analysis of OsPK2.
Figure S5 Amino acid sequence alignment between OsPK2 and other homologous proteins.
Figure S6 qRT‐PCR analysis of OsPK2 in transgenic plants.
Figure S7 Analysis of OsPK2 transgenic plants.
Figure S8 Analysis of OsPK2 CRISPR‐CAS9‐mediated editing.
Figure S9 Subcellular localization of OsPK2 and truncated OsPK2 in tobacco cells.
Figure S10 SDS‐PAGE and western‐blot analysis of his‐OsPK2 and his‐ospk2 purified from baculovirus expression system under native condition.
Figure S11 PK activity assay in fresh leaves.
Figure S12 qRT‐PCR analysis of genes encoding putative PK in rice.
Figure S13 Yeast two‐hybrid assays showed that both OsPK2 (PKpα1) and PKpβ1 can interact with itself.
Table S1 Gene products of the nine predicted ORFs in the fine mapping region.
Table S2 Primers used in this study.


