Abstract
For the opportunistic pathogen Acinetobacter baumannii, desiccation tolerance is thought to contribute significantly to the persistence of these bacteria in the healthcare environment. We investigated the ability of A. baumannii to survive rapid drying, and found that some strains exhibited a profoundly desiccation-resistant phenotype, characterized by the ability of a large proportion of cells to survive on a dry surface for an extended period of time. However, this phenotype was only displayed during the stationary phase of growth. Most interestingly, we found that drying resistance could be lost after extended cultivation in liquid medium. Genome sequencing of isolates that became drying-sensitive identified mutations in bfmR, which encodes a two-component response regulator that is important for A. baumannii virulence. Additionally, BfmR was necessary for the expression of stress-related proteins during stationary phase, and one of these, KatE, was important for long-term drying survival. These results suggested that BfmR may control stress responses, and we demonstrated that the ΔbfmR mutant was more sensitive to hydrogen peroxide, nutrient starvation, and increased osmolarity. We also found that cross-protection against drying could be stimulated by either starvation, which required BfmR, or increased osmolarity. These results imply that BfmR plays a role in controlling stress responses in A. baumannii which help protect cells during desiccation, and they provide a regulatory link between this organism’s ability to persist in the environment and pathogenicity.
Introduction
The gram-negative bacterium Acinetobacter baumannii is an opportunistic pathogen that primarily infects severely ill or wounded individuals, but it rarely causes disease outside of the healthcare setting [1]. These infections have become a growing concern in recent years due to the ability of this organism to develop multi-drug resistance [2]. In contrast to many other Acinetobacter species, which are mostly non-pathogenic and can be isolated from a wide variety of environmental sources, A. baumannii is most frequently found in association with hospitalized patients or in the nosocomial environment, where it can become a persistent contaminant [3, 4]. Sustained colonization of the hospital environment by A. baumannii can lead to outbreaks of infection, particularly in intensive care units and trauma centers [5–8]. This was clearly illustrated during U.S. military operations in Iraq and Afghanistan during 2001–2007, where more than 700 wounded soldiers were either infected or colonized with A. baumannii during passage through the military evacuation chain [9, 10]. One trait that is thought to aid in the survival of A. baumannii in the clinical environment is its ability to tolerate desiccation.
Desiccation is a common environmental stress that poses many challenges to bacterial cells. Water loss leads to decreased turgor pressure and biochemical changes that can damage cell membranes, and drying can also cause denaturation of intracellular proteins and conformational changes to DNA [11, 12]. Additionally, reactive oxygen species generated during desiccation can damage both proteins and DNA [11, 13]. In response, bacteria have developed a variety of mechanisms to mitigate the damage from drying. Some species can differentiate into dormant forms such as endospores or cysts, which are extremely desiccation-resistant, but this strategy requires a significant expenditure of time and resources [14, 15]. Other species have developed mechanisms that either help protect susceptible cellular components from damage, or that sequester water in an attempt to avoid dehydration. These mechanisms include the alteration of membrane composition or LPS modification to help stabilize membranes during drying, and the accumulation of compatible solutes such as trehalose, which can protect cytoplasmic and membrane constituents [11, 14]. Alternatively, exopolysaccharide production and biofilm formation can be a way to retain water in the local microenvironment and protect bacterial cells during dry conditions [16]. Yet while many bacterial species possess these capabilities, their ability to tolerate desiccation can vary widely. Drying survival times can range from minutes to years, and there can even be considerable variation between different strains of the same species [11]. Therefore, the molecular, biophysical, and regulatory processes that determine desiccation tolerance in non-spore forming bacteria remains a poorly understood aspect of bacterial physiology.
Compared to Escherichia coli and Pseudomonas aeruginosa, two other Gram-negative pathogens commonly found in the nosocomial environment, A. baumannii typically displays a greater ability to survive drying in laboratory tests, but the survival of different A. baumannii strains when dried can be variable, ranging from a few days to months [17–19]. Overall, laboratory studies have shown that many clinical A. baumannii strains can survive at least 20 days on dry surfaces, and some can survive 100 days or more [20–22]. A. baumannii has also been isolated from many different sources in the hospital environment, such as bed rails, pillows, mattresses, tables, and medical equipment [8]. These findings indicate that transfer of this pathogen via fomites is a major issue in clinical environments, and is most likely sustained by this organism’s ability to withstand desiccation and prolonged dry conditions. While this points to the importance of desiccation tolerance for this opportunistic pathogen, only a few specific factors that contribute to this property have been identified. A recent study showed that hepta-acylated lipid A, a component of the A. baumannii outer membrane, is important for drying survival [23]. Another study found that RecA, an important cellular factor for DNA repair, was required for cells to withstand desiccation [24]. Additional investigations have also suggested that biofilm formation or transition to a dormant state could be important for desiccation survival [25, 26]. While these studies have provided valuable insights, they do not provide a comprehensive understanding of A. baumannii desiccation tolerance.
We decided to further investigate this property of A. baumannii since it likely helps this opportunistic pathogen to come in contact with hospitalized individuals that are most susceptible to infection. We assessed the drying survival of a small group of A. baumannii strains, and confirmed that different strains appear to have distinct desiccation tolerance phenotypes. We also discovered that drying-tolerant strains can convert to a drying-sensitive phenotype during laboratory cultivation, and analysis of these strains led us to establish a role for the regulator BfmR in desiccation tolerance and other stress-related phenotypes.
Results
Some A. baumannii strains display an extremely desiccation-tolerant phenotype
Previous studies found that A. baumannii strains can vary widely in their ability to survive desiccation, with some only surviving a few days, and others being able to survive for weeks or months in a dried state [17]. To identify suitable strains for studying desiccation tolerance we tested the ability of a small group of A. baumannii strains to withstand drying. For these experiments we harvested cells from stationary phase broth cultures, washed the cells with water, and allowed small samples of washed cells to dry on a plastic surface (see Materials and methods). Bacterial samples were visibly dry in 30 min to 1 h under the conditions tested. This procedure allowed us to focus on the intrinsic properties of cells that promote desiccation tolerance, since media components can affect bacterial drying survival [17, 18]. Additionally, we believed that this method would reasonably simulate a bacterial contamination event that could occur in the nosocomial environment. We began by examining the maximum survival time of dried A. baumannii cells after incubation at ambient temperature and humidity (19–23°C, 25–61% RH). The results from these experiments showed that this group of A. baumannii strains displayed three different desiccation-tolerance phenotypes (Table 1). One widely studied strain, ATCC 19606, had low desiccation tolerance under these conditions, losing viability after a few days of drying. A second group of strains, including another commonly studied strain, ATCC 17978, had moderate resistance to drying, surviving for an average of 20–43 days. The maximum survival times for this group of strains were quite variable, and were likely influenced by fluctuations in relative humidity [17], which ranged from 25–61% based on regular monitoring of the ambient conditions. However, another set of strains consistently survived desiccation for 90 days or longer. We defined these strains as having a profoundly desiccation-tolerant phenotype.
Table 1. Maximum survival time of dried A. baumannii cells.
| Strain | Average survival time in days (mean ± SD)1 |
Average percent survival after 90 days |
|---|---|---|
| ATCC 19606 | 3 ± 1 | 0 |
| A118 | 20 ± 7 | 0 |
| ATCC 17978 | 34 ± 22 | 0 |
| ATCC 17904 | 22 ± 12 | 0 |
| AB09-002 | 43 ± 26 | 0 |
| AB09-003 | > 90 | 5% |
| ATCC 17961 | > 90 | 6% |
| AB5075 | ~ 90† | < 0.01% |
1 Washed A. baumannii cells were spotted on a smooth polystyrene surface and allowed to air dry. Dried cells were incubated at 19–23°C (mean 22°C), with a relative humidity of 25–61% (mean 46%). Results are from at least three separate experiments.
† Average survival was approximately 90 days, based on the following results from four independent experiments: > 90 days, > 90 days, 44 days, 90 days.
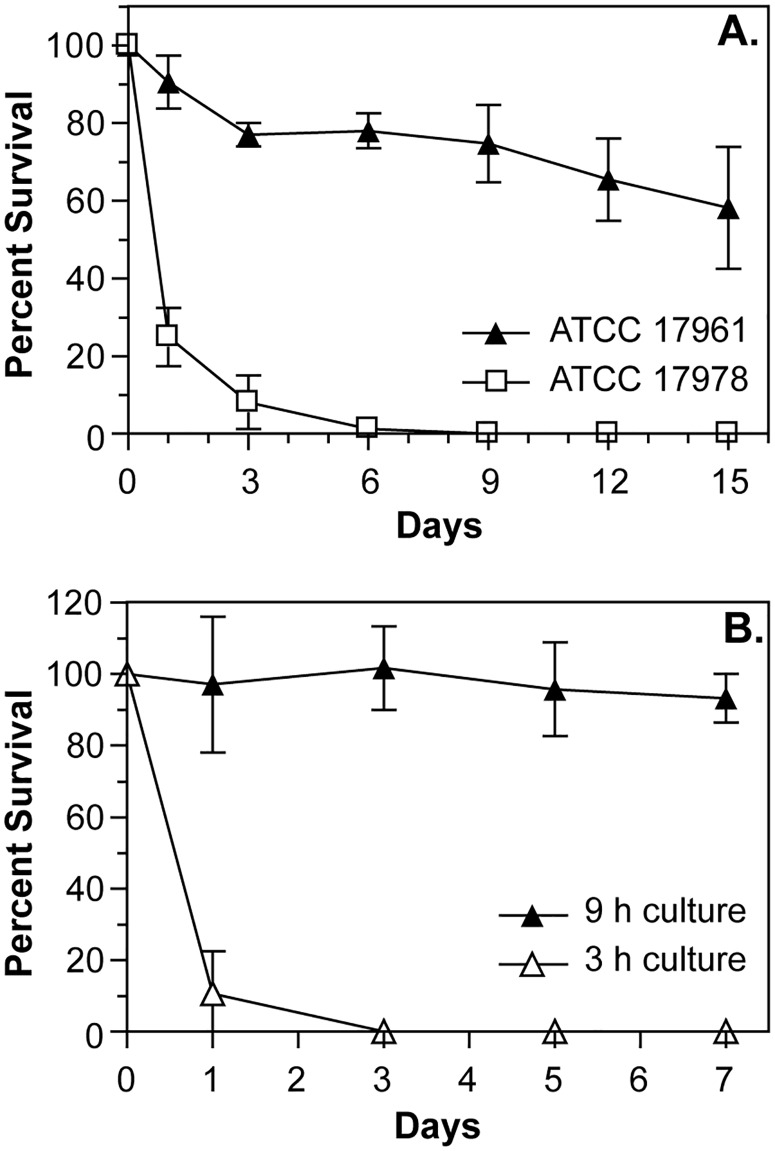
The difference between moderately and profoundly desiccation-tolerant strains was even more apparent when we examined the survival curves of A. baumannii cells during the first two weeks of drying. We performed these and all following experiments under conditions of controlled temperature and humidity (24–26°C and 79–81% RH) to minimize variations in survival caused by these factors. Moderately drying-resistant strains, represented by strain ATCC 17978, had a large decrease in the number of viable cells during the first week of desiccation, with only a small proportion (less than 1%) of the population being able to survive being dried for an extended period of time (Fig 1A). We observed similar patterns of survival for other moderately desiccation-tolerant strains A118, ATCC 17904, and AB09-002 (S1 Fig). In contrast, for profoundly desiccation-tolerant strains, represented by strain ATCC 17961, we saw only a slight decrease in the number of viable cells after the first few days of drying, followed by a slow decline in viability (Fig 1A). This survival pattern was also displayed by strains AB09-003 and AB5075 (S1 Fig). In addition, we observed similar trends in drying survival for strains ATCC 17978 and AB09-003 under conditions of lower relative humidity (25% RH, 24–26°C). These results showed that profound desiccation tolerance is a distinct phenotype of certain A. baumannii strains, characterized by the survival of a large proportion of the cells when dried for an extended period of time.
Fig 1. Desiccation survival phenotypes of A. baumannii cells.
(A) A. baumannii strains were grown overnight in LB medium, and then cells were washed with water and samples were dried on polystyrene. Dried cells were incubated at 25°C and 80% RH, and at the indicated times dried samples were suspended, and survival was assessed by CFU counts. (B) Cells from cultures of A. baumannii strain ATCC 17961 were harvested after either 3 h (logarithmic phase) or 9 h (stationary phase) of growth in LB medium, washed with water, dried, and then survival was assessed at the indicated times. The data presented are the mean ± SD from at least three independent experiments.
We also made another interesting observation about the desiccation-tolerant phenotype. In our initial experiments we tested the survival of cells harvested from the stationary phase of growth, but we saw large differences in drying survival when we tested cells taken from the logarithmic growth phase. Compared with cells that were grown in lysogeny broth (LB) for 9 h, cells from strain ATCC 17961 that grew for only 3 h had a greatly reduced ability to survive desiccation, and instead generated a survival curve that was more similar to strains with moderate to low desiccation tolerance (Fig 1B). Cells taken from logarithmic phase cultures of strains AB09-003 and AB5075 also had a greatly decreased ability to survive drying. These results implied that factors which are important for desiccation tolerance in A. baumannii are regulated in a growth phase-dependent manner.
Laboratory adaptation reveals a regulator that is required for desiccation tolerance
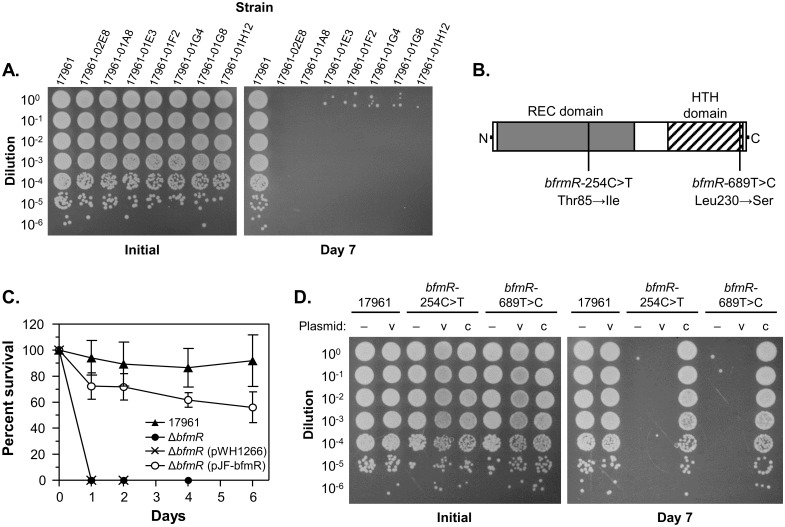
A previous study that examined the drying survival of a larger group of A. baumannii strains noted that more recent isolates tended to be more desiccation tolerant than laboratory strains [17]. Since bacteria are rarely, if ever, subjected to dry conditions in the laboratory, we hypothesized that desiccation resistance could be lost during laboratory cultivation, and that this could be a route to understanding the molecular mechanisms that allow A. baumannii to survive drying. To test this, we cultured the highly desiccation-resistant strain ATCC 17961 for five to seven passes in liquid medium, then isolated individual clones and screened them for drying survival. We then performed a more controlled test for drying survival on a subset of clones that appeared to have reduced desiccation tolerance, and were able to readily identify drying-sensitive isolates derived from each strain (Fig 2A). Unfortunately, our primary screening procedure produced a large number of false-positive results, so we were unable to accurately determine the frequency at which drying-sensitive clones arose in the population. However, we were able to isolate sensitive clones in experiments where only approximately 200 clones were screened, suggesting that desiccation resistance can be lost at a fairly high rate.
Fig 2. The two-component response regulator BfmR is required for desiccation tolerance.
A. baumannii strains were grown overnight in LB medium, and then cells were washed with water, dried, and assessed for desiccation tolerance at 25°C and 80% RH. (A) The survival of dried cells from the wild-type strain ATCC 17961 and derivatives isolated after extended culture in LB medium were tested. Samples from the initial inoculum and after 7 days of drying were diluted and spotted onto the surface of an LB plate. These data are representative of at least two independent experiments. (B) A representation of the BfmR protein showing the relative positions of the receiver domain (REC domain, gray box), the DNA-binding helix-turn-helix domain (HTH domain, hashed box), and the nucleotide and corresponding amino acid substitutions identified in the strains tested in panel (A). (C) A. baumannii strains ATCC 17961 (solid triangle), 17961-ΔbfmR (solid circle), 17961-ΔbfmR carrying vector plasmid pWH1266 (X), and 17961-ΔbfmR carrying an intact copy of bfmR on plasmid pJF-bfmR (open circle) were assessed for drying survival on the indicated days. The data presented are the mean ± SD from at least four independent experiments. (D) Cells from strains ATCC 17961, 17961-bfmR-254C>T, and 17961-bfmR-689T>C carrying either no plasmid (–), vector plasmid pWH1266 (v), or plasmid pJF-bfmR for complementation (c) were tested for survival after drying. Samples from the initial inoculum and after 7 days of drying were diluted and spotted onto the surface of an LB plate. These data are representative of at least two independent experiments.
Recently, two different mechanisms have been described to account for frequent phenotypic variations in A. baumannii. The first involves the loss of a large, self-transmissible plasmid during laboratory cultivation [27]. Strain 17961 did not appear to carry a large pAB3/pAB04-1-type plasmid as assessed by PCR screening, but it does harbor at least two other plasmids. Monitoring of these plasmids did not detect any difference in the number or size of plasmids present in the drying-sensitive derivatives of 17961, which indicates that the observed changes in desiccation tolerance were not due to plasmid loss. The second mechanism involves a cell density-dependent phase change that was first observed in strain AB5075 [28]. We viewed colonies of ATCC 17961 and the drying-sensitive derivatives under the appropriate conditions to observe phase change (growth on 0.5X LB, 0.8% agar), and we did not observe any changes in colony opacity, although we were able to see both opaque and translucent colony variants for our control strain AB5075. Additionally, we took two of our drying-sensitive isolates, cultured them repeatedly in LB medium, and repeated our screening procedure to see if we could detect any reversion to a drying-resistant phenotype. We did not isolate any desiccation-tolerant clones from these experiments. Together, these results strongly suggested that the variation in drying-resistance phenotypes that we observed was not the result of a phase change mechanism.
To identify any genetic changes that could be the cause of the change in desiccation tolerance, we analyzed the genome of strain ATCC 17961 and seven drying-sensitive derivatives of this strain by high-throughput DNA sequencing. First, we generated a draft genome sequence for strain 17961, which we have made available on the PATRIC website (http://www.patricbrc.org). The total length of the generated contigs was 4,057,452 bp, with a 39.2% GC content, which is similar to genomes of other A. baumannii strains. Next, we aligned sequencing reads from each of the drying-sensitive isolates to the parent genome and identified variations. For six of the isolates (17961-02E8, 17961-01E3, 17961-01F2, 17961-01G4, 17961-01G8, and 17961-01H12) we identified the same two single-nucleotide variations: insertion of a “T” residue in an intergenic region upstream from a gene encoding a predicted fimbrial protein precursor (17961 genome feature 470.2202.peg.2098), and substitution of a “C” residue for a “T” at bp 689 of the bfmR gene, leading to the conversion of BfmR Leu230 to Ser. These six isolates were collected during the same experiment, and likely represent the expansion of a single clone. In the seventh isolate (17961-01A8) we identified a single nucleotide substitution at bp 254 of bfmR (C to T), converting BfmR Thr85 to Ile. Since these analyses identified two independent, nonsynonymous mutations in bfmR, we decided to further investigate the role of bfmR in desiccation tolerance.
The bfmR gene encodes an OmpR/PhoB-family bacterial two-component response regulator. Like other members of this family, BfmR contains an N-terminal receiver domain, which can be phosphorylated at a conserved aspartate residue to control protein function, and a C-terminal helix-turn-helix DNA binding domain (Fig 2B). Phosphorylation of BfmR is likely controlled by the sensor histidine kinase BfmS, which is co-transcribed with bfmR [29], and which controls the expression of many of the same genes as BfmR [30]. Since it was unclear how each independent point mutation that we identified would affect the function of BfmR, we constructed an isogenic, in-frame bfmR deletion mutant in strain 17961. We also re-introduced the bfmR-254C>T and bfmR-689T>C point mutations onto the strain 17961 chromosome by allelic exchange. We then tested each of these newly constructed strains for the ability to survive desiccation. Compared to the wild-type strain, the bfmR mutant had a greatly decreased ability to survive drying, and we were unable to recover any viable cells from the ΔbfmR strain after more than four days of drying (Fig 2C). Desiccation tolerance could be restored to the bfmR deletion mutant by supplying an intact copy of bfmR on a multi-copy plasmid, but was not restored by the vector plasmid alone (Fig 2C). Similarly, each of the strains with point mutations in bfmR had a large decrease in survival after drying, and survival was restored by supplying a wild-type copy of bfmR to these strains (Fig 2D). These results implied that the bfmR-254C>T and bfmR-689T>C mutations inactivated BfmR, and indicated that BfmR is required for desiccation tolerance in strain 17961.
BfmR controls the expression of stress-related proteins during the stationary phase of growth
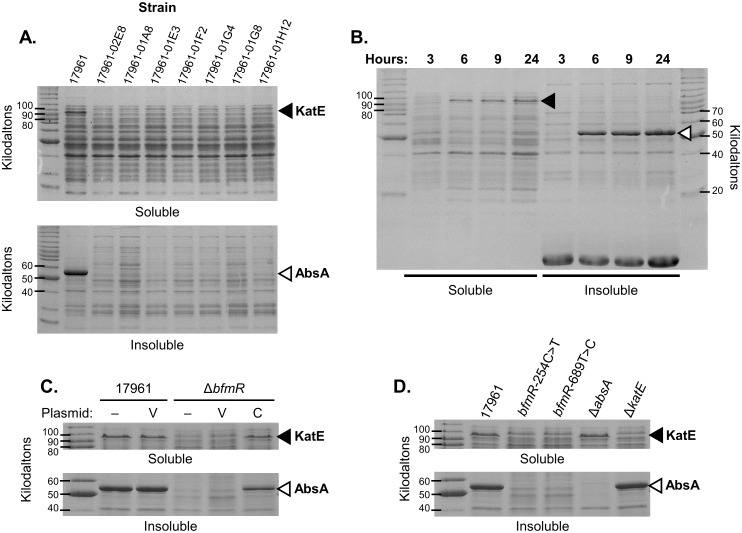
While analyzing the genome sequences of strain 17961 and drying-sensitive derivatives of this strain, we decided to also examine protein expression profiles to try to identify differences that could be important for desiccation tolerance. Cells for each strain were disrupted using a commercial lysis reagent (BugBuster protein extraction reagent, EMD, Millipore), and then we analyzed the soluble and insoluble fractions of each lysate by SDS-PAGE. Interestingly, we saw that the production of two different proteins appeared to be greatly reduced in the drying-sensitive isolates (Fig 3A). We additionally observed that the expression of these proteins was increased during the stationary phase of growth in strains 17961 (Fig 3B) and AB09-003 (data not shown). We excised the bands corresponding to each of these proteins from polyacrylamide gels and identified them by mass spectrometry. The protein present in the soluble fraction of lysates was identified as the KatE catalase, which aids in the detoxification of hydrogen peroxide. Consistent with our observations, another study showed that katE expression was greatly increased during the stationary phase of growth in A. baumannii [31]. The protein present in the insoluble fraction of lysates is an acidophilic repeat motif-containing protein that appears to be encoded in the genome of many A. baumannii strains, as assessed by BLAST (http://www.ncbi.nlm.nih.gov/blast). This protein is annotated as a stress-induced protein in some A. baumannii genomes due to the presence of a conserved N-terminal domain that is present in a variety of stress-related proteins in other bacterial species. Therefore we designated this protein as A. baumannii stress-related protein A (AbsA).
Fig 3. Identification of proteins that are differentially expressed in desiccation tolerant versus sensitive A. baumannii cells.
Lysates from A. baumannii cells were separated into soluble and insoluble fractions. Samples from each strain containing approximately equivalent amounts of total protein were analyzed by SDS-PAGE, and gels were stained with Coomasie-R250. The positions of KatE and AbsA are indicated by closed and open arrowheads, respectively. These data are representative of at least two independent experiments. (A) Samples from the wild-type strain ATCC 17961, and derivatives of strain 17961 that were isolated after extended culture in LB medium, were analyzed for protein expression. (B) Cells from strain ATCC 17961 were harvested after 3, 6, 9, and 24 h of growth in LB medium and analyzed. (C) Protein expression was analyzed in cells from strains ATCC 17961 and 17961-ΔbfmR carrying either no plasmid (–), vector plasmid pWH1266 (v), or plasmid pJF-bfmR for complementation (c). (D) Protein expression was examined in strains ATCC 17961, 17961-bfmR-254C>T, 17961-bfmR-689T>C, 17961-ΔabsA, and 17961-ΔkatE.
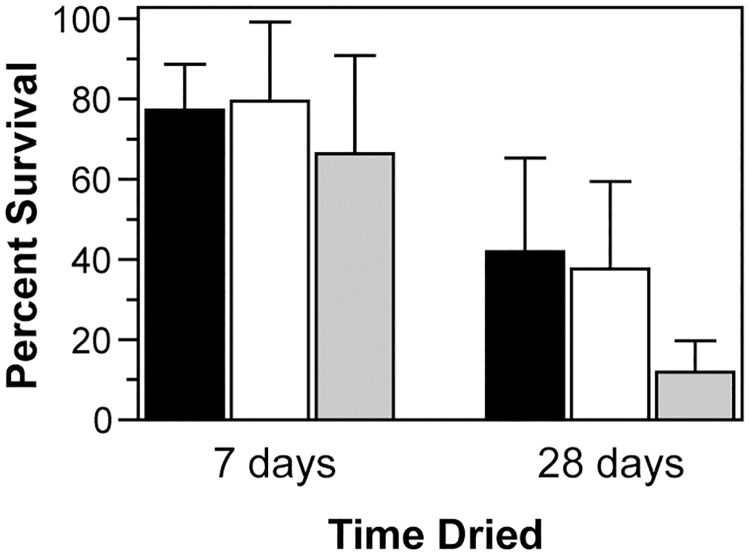
Since BfmR is expected to be a regulator of gene expression, we wondered if the decreased levels of KatE and AbsA observed in the drying-sensitive isolates from strain 17961 was a consequence of bfmR inactivation. To examine this possibility we analyzed cell lysates from the bfmR deletion mutant and the bfmR-254C>T and bfmR-689T>C mutant strains. We saw that KatE levels appeared to be lower in these strains, and AbsA levels appeared to be greatly reduced (Fig 3C and 3D). KatE and AbsA expression could be restored to the ΔbfmR strain by supplying an intact copy of bfmR on a plasmid, but not by the vector plasmid alone (Fig 3C). These results indicated that BfmR positively regulates the production of both KatE and AbsA. To test if the loss of either KatE or AbsA could explain the large decrease in desiccation tolerance displayed by the bfmR mutant, we constructed strains with in-frame deletions in either katE or absA. We observed that KatE appeared to be expressed in the ΔabsA mutant at a level similar to the wild-type, and that AbsA was produced at a wild-type level in the ΔkatE mutant (Fig 3D), implying that bfmR was not inactivated in these strains during laboratory manipulations, and that deletion of either of these genes did not affect expression of the other. Next, we tested the ability of these strains to survive desiccation. We saw that the ΔkatE and ΔabsA mutant strains survived just as well as the wild-type strain after seven days of drying (Fig 4). After 28 days of drying the ΔabsA mutant still survived as well as the wild-type, but there was a significant decrease in the viability of the ΔkatE mutant strain compared to strain 17961 (Fig 4, ΔkatE vs. 17961 at 28 days, P = 0.02 as determined by t-test). However, these results were not sufficient to explain the large decrease (>99%) in viability seen when ΔbfmR mutant cells were dried for only a few days (Fig 2C), showing that additional factors that contribute to desiccation tolerance must be absent in the ΔbfmR mutant.
Fig 4. Effects of KatE and AbsA on desiccation tolerance.
Dried cells from the wild-type strain ATCC 17961 (black bars), the absA deletion mutant 17961-ΔabsA (white bars), or the katE deletion mutant 17961-ΔkatE (gray bars) were assessed for survival after 7 and 28 days of desiccation at 25°C and 80% RH. [Note: For comparison, the bfmR deletion mutant had <1% survival after one day of drying (Fig 2C)]. Data represent the mean ± SD from at least five independent experiments.
BfmR is important for phenotypes associated with the general stress response
Although we did not identify a specific BfmR-controlled factor that was fully responsible for drying survival, the fact that we found BfmR controls two potentially stress-related proteins that were also upregulated during the stationary phase of growth caused us to consider a role for BfmR as a stress response regulator. In particular, we noted that katE expression in E. coli, despite its role in protecting cells from oxidative stress, is not stimulated by oxidative stress [32]. Instead, katE is upregulated during the stationary phase of growth as part of the general stress response [33]. In Gram-negative bacteria the general stress response involves the induction of a large number of genes that provide cross-protection against a variety of environmental stresses [34]. Considering this, we hypothesized that BfmR may coordinate multiple stress responses, allowing A. baumannii cells to combat the various stresses encountered during desiccation. To examine this, we tested the susceptibility of the wild-type and bfmR mutant strains to a variety of stressors.
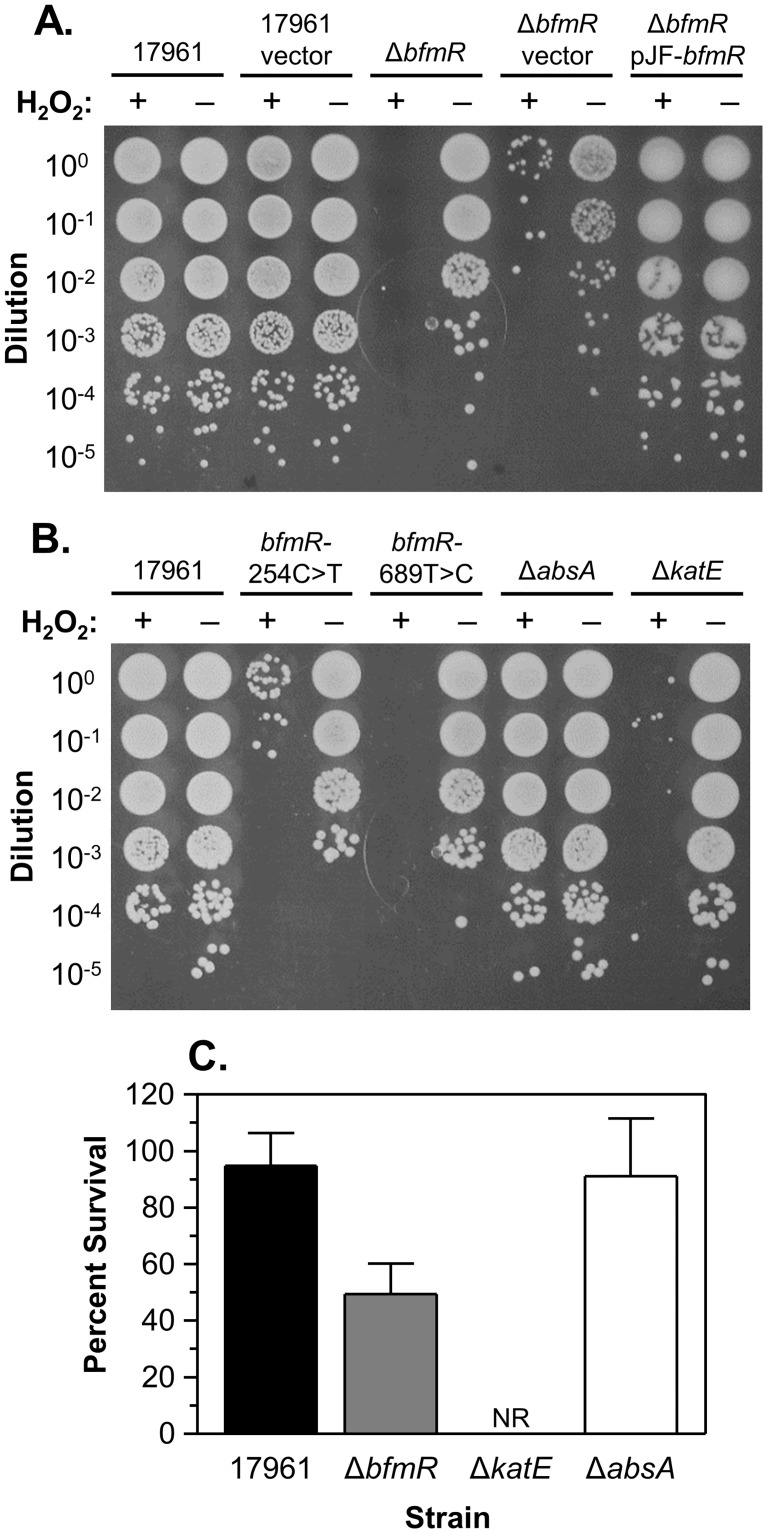
First, we analyzed the resistance of both dried and broth-cultured cells to hydrogen peroxide treatment. Dried cells of strain 17961 were resistant to H2O2 treatment, with samples showing only a slight decrease in viability after challenge with 1.5% H2O2 (0.49 M) for 45 sec (Fig 5A). In contrast, samples of the ΔbfmR, bfmR-254C>T, and bfmR-689T>C mutants had large decreases in survival after H2O2 treatment (Fig 5A and 5B). Resistance to hydrogen peroxide was restored in the ΔbfmR mutant carrying an intact copy of bfmR on a plasmid, but not with the vector plasmid alone (Fig 5A). Similarly, cells from the ΔbfmR mutant had a reduced ability to survive H2O2 treatment in broth culture as well. After 30 min in the presence of 10 mM H2O2, the ΔbfmR mutant had approximately a 50% decrease in survival compared with only a 5% decrease in the survival of the wild-type strain (Fig 5C). These results showed that BfmR is important for hydrogen peroxide resistance in A. baumannii strain 17961. Additionally, we tested the susceptibility of the ΔabsA and ΔkatE mutant strains to H2O2. The ΔabsA mutant strain had a similar level of resistance to H2O2 treatment as the wild-type strain, and as expected the ΔkatE mutant was highly sensitive to H2O2 both when dried and in liquid culture (Fig 5B and 5C, respectively). This demonstrated that KatE is critical for the hydrogen peroxide resistance of stationary-phase cells in A. baumannii strain 17961, and also suggests that the increased susceptibility of bfmR mutants to H2O2 treatment is due to decreased levels of KatE in these strains.
Fig 5. Sensitivity of A. baumannii cells to hydrogen peroxide.
(A) and (B) Dried bacterial samples were challenged with H2O2. The indicated A. baumannii strains were grown overnight in LB medium. Samples of washed cells were dried, and then treated with either water or 1.5% H2O2, as indicated, for 45 s. Each sample was then diluted and spotted onto the surface of an LB plate. These data are representative of at least two independent experiments. (C) Cells in liquid culture were challenged with H2O2. A. baumannii strains were grown overnight, and then diluted into fresh LB medium and challenged with 10 mM H2O2 for 30 min at 37°C. The designation “NR” indicates that no viable cells were recovered after the H2O2 challenge. The data presented are the mean ± SD from at least four independent experiments.
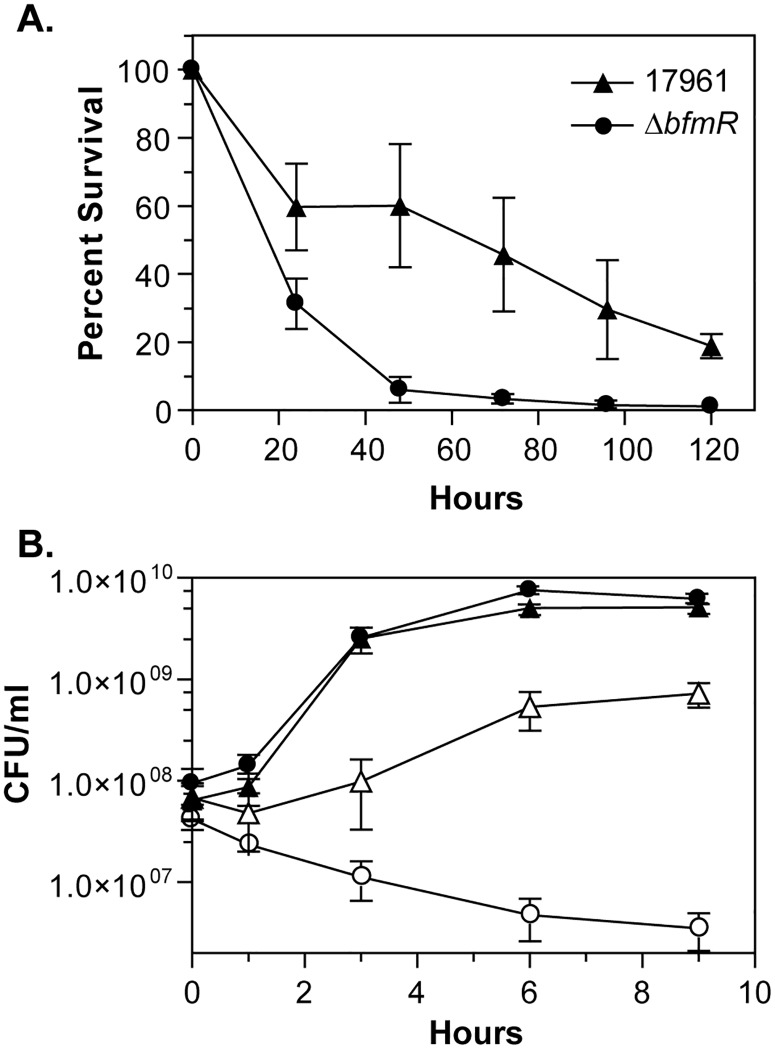
Next, we tested the ability of the wild-type and ΔbfmR mutant strains to survive during an extended period of nutrient deprivation. For each strain, cells from overnight cultures grown in LB medium were washed and incubated in phosphate-buffered saline for five days. During the first day, cultures of strain 17961 had an approximately 40% decrease in the number of viable cells, followed by a continued slow decline in viability over the next four days (Fig 6A). However, the ΔbfmR mutant strain had an approximately 94% decrease in survival during the first 48 hours of nutrient starvation (Fig 6A). These data showed that BfmR is necessary for optimal survival in the absence of nutrients. We also analyzed the growth of the ΔbfmR mutant strain under conditions of increased osmolarity. The wild-type and ΔbfmR mutant strains both grew equally well in LB medium without sodium chloride (Fig 6B). When strain 17961 was shifted from medium without NaCl to medium containing 0.6 M NaCl there was initially a small decrease in viability, but the cells were able to adapt to the high salt conditions and resume growth, although they did not grow to as high of a density as in media without NaCl (Fig 6B). Unlike the wild-type strain, the ΔbfmR mutant was not able to resume growth after being placed in media containing 0.6 M NaCl, and instead these cultures had a slow decrease in cell viability throughout the course of the experiment (Fig 6B), indicating that BfmR is required for cells to adapt to high salt conditions. Together, these results showed that BfmR is important for A. baumannii to survive under multiple stressful conditions.
Fig 6. BfmR is important for survival during nutrient limitation and conditions of high osmolarity.
(A) The survival of the wild-type strain ATCC 17961 (triangles) and the bfmR mutant 17961-ΔbfmR (circles) incubated at 37°C in PBS was assessed by counting the number of CFU recovered at each time point. The data presented represent the mean ± SD from three separate experiments. (B) The growth of the wild-type (triangles) and the ΔbfmR mutant strains (circles) were monitored at 37°C in the absence (closed symbols) or presence (open symbols) of 0.6 M NaCl. The data presented are the mean ± SD from at least four independent experiments.
Finally, we tested whether responses to other stresses can promote cross-protection against desiccation in A. baumannii. To do this, we challenged cells during the logarithmic phase of growth, when they are more sensitive to desiccation (Fig 1B), with either nutrient starvation or increased osmolarity, and then compared the ability of stressed and unstressed cells to survive drying. For starvation experiments, A. baumannii strains were grown in phosphate-buffered LB for 2.5 hours, and then samples of cultures were diluted 1 to 5 either into fresh medium or phosphate-buffered saline, which was at the same temperature, pH, and NaCl concentration as the original growth medium. One hour after this transfer, cells from each condition were tested for desiccation tolerance. Cells of strain 17961 that were sub-cultured in LB were very sensitive to drying, with few to no viable cells recovered after being dried for one day, but cells that were starved by sub-culture in buffer were much more resilient to desiccation (Fig 7A). We did not observe this effect in the ΔbfmR mutant strain, which was still highly sensitive to desiccation after starvation (Fig 7A). As expected, the ΔbfmR mutant carrying an intact copy of bfmR on a plasmid was protected from drying after starvation (Fig 7A). To test the effects of osmolarity, strain 17961 was initially grown in LB without NaCl for 2.5 hours, and then cells were diluted into pre-warmed LB medium containing varying amounts of NaCl. After one hour of incubation, cells from cultures containing 0.4–0.5 M NaCl had an increased ability to survive drying compared with cultures containing lower amounts of NaCl (Fig 5B). We were unable to clearly assess whether BfmR was necessary for this effect, since the viability of the ΔbfmR mutant greatly decreased after one hour of incubation with 0.5M NaCl. Nevertheless, these experiments indicated that responses to both starvation and increased osmolarity can provide cross-protection against drying in A. baumannii, and that BfmR is required for this effect in response to general nutrient starvation.
Fig 7. Nutrient starvation and increased osmolarity can stimulate cross-protection against desiccation.
(A) A. baumannii cells in the exponential phase of growth were diluted 1:5 either into fresh growth medium (LB) or phosphate-buffered saline (s). After one hour of incubation at 37°C, samples of cells were taken from each culture, washed with water, and dried for one day. “Vector” refers to the cloning vector pWH1266, and pJF-bfmR carries an intact copy of bfmR. (B) A. baumannii strain ATCC 17961 was grown in LB without NaCl, and then diluted 1:5 into LB supplemented with the indicated concentrations of NaCl. After one hour of incubation at 37°C, samples of cells were taken from each culture, washed with water, and dried. For both experiments, samples from the initial inoculum and after one day of drying were diluted and spotted onto the surface of an LB plate. The data presented are representative of at least two independent experiments.
Discussion
Desiccation tolerance is considered to be one of the abilities that has allowed A. baumannii to become a successful opportunistic pathogen in the nosocomial environment, but the molecular mechanisms that contribute to this trait are poorly understood. While many A. baumannii strains have been reported to survive drying for 20 days or longer [20], clinical isolates of A. baumannii can vary in their ability to survive desiccation similar to the group of strains analyzed in this study [21, 35]. Several mechanisms have been described to account for phenotypic variations in A. baumannii strains that occur during both culture in the laboratory and during infection, including plasmid loss, mobilization of genetic elements, and phase change, which helps to partly explain this organism’s naturally occurring variability [27, 28, 36]. For profoundly drying-resistant strains we found that a much larger proportion of the bacterial population survived drying for an extended period of time, suggesting that strains which exhibit this phenotype could have more opportunities for survival and spread in the hospital setting. However, we also observed that this phenotype could be lost after growth in liquid medium. A previous study noted that A. baumannii strains isolated from dry sources tended to survive desiccation better than strains isolated from wet sources [35], and together these findings suggest that desiccation can be an important selective pressure in the environment.
Through an analysis of strains that lost the ability to survive drying during laboratory culture we were able to identify BfmR as a regulator of the desiccation resistance phenotype. In A. baumannii, BfmR is part of a two-component regulatory system with the sensor histidine kinase BfmS, which appears to inactivate BfmR through phosphorylation [30]. Originally identified as having a role in controlling biofilm formation, bfmR and bfmS have subsequently been linked to several phenotypes, including formation of pili, motility, complement resistance, antibiotic susceptibility, and virulence [29, 30, 37–41]. BfmR appears to be similar to other response regulators in the OmpR/PhoB family, and based on structural data [39, 42] one of the naturally-occurring mutations that we found (in the “switch” threonine, Thr85) likely affected BfmR function by disrupting conformational changes in the protein that occur in response to phosphorylation. The effects of the other mutation we observed, in Leu230, are unclear, but both mutations appear to have altered BfmR’s regulatory activity, leading to increased sensitivity to desiccation (Fig 2). There is currently interest in developing compounds to target BfmR in order to treat A. baumannii infections [39, 42], and our findings provide evidence that this could be a potential strategy to control A. baumannii in the healthcare environment as well.
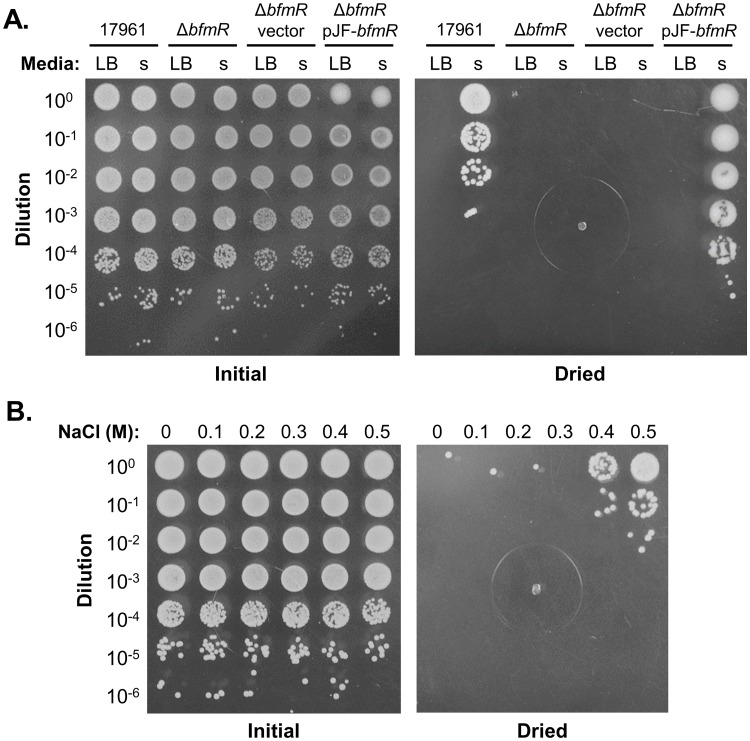
BfmR and BfmS were previously shown to influence phenotypes which could potentially affect drying survival. BfmR was originally identified as a regulator of biofilm formation [29], and another study found that A. baumannii strains that formed robust biofilms were more desiccation tolerant than strains that could not form biofilms [25]. However, in our studies broth-grown cells were washed thoroughly and then samples were dried rapidly, typically in approximately 45 minutes. This procedure provided little time for the cells to respond to the change in water availability and form a complex biofilm matrix. Therefore we believe that a difference in biofilm formation is an unlikely explanation for the significant drying survival defect of the bfmR mutant strains in our experiments (Fig 2), although biofilm formation likely plays a role in the persistence of A. baumannii in the nosocomial environment. Capsule production is another phenotype influenced by BfmRS that could contribute to desiccation tolerance, since exopolysaccharides are known to aid a variety of bacterial species in drying survival [11]. Strains with mutations that either delete or inactivate bfmS have increased capsular polysaccharide production, but this does not occur in bfmRS mutant strains, implying a role for BfmR in regulating capsule synthesis [30, 43]. However, another study found that inactivation of bfmR alone in two different A. baumannii strains did not have any effect on capsular polysaccharide production [39]. From these findings it appears that BfmR negatively regulates capsule production in response to phosphorylation by BfmS, but the exact role of BfmR in controlling capsule expression in wild-type strains is still somewhat unclear. While extracellular polysaccharides could certainly help protect A. baumannii cells during desiccation, the capsule layer surrounding laboratory-grown A. baumannii cells appears to be relatively thin, and a capsule is produced by both the extremely desiccation-tolerant strain ATCC 17961, and the more desiccation-sensitive strains ATCC 17978 and ATCC 19606 [43]. Based on this, we believe that differences in capsule production are not likely to be the only cause of the large differences in drying survival that we observed. Instead, we hypothesize that these differences are more likely due to the fact that BfmR appears to coordinate multiple stress responses.
Our decision to investigate the role of BfmR in additional stress responses was in response to our discovery that BfmR controlled the expression of two potential stress-related proteins, KatE and AbsA. Both of these proteins were maximally produced during the stationary phase of growth (Fig 3B), which coincided with the time of greatest desiccation tolerance (Fig 1B). Additionally, a recently reported transcriptome analyses of bfmR and bfmS mutant strains found that the BfmRS system controls a post-exponential growth phase-like pattern of gene expression in the moderately desiccation-resistant strain ATCC 17978 [30]. Several other bacterial species exhibit enhanced desiccation tolerance during stationary phase, and in both Escherichia coli and Pseudomonas fluorescens the general stress response regulator RpoS is necessary for maximal survival when dried [19, 44–46]. A role for general stress responses in desiccation tolerance makes sense given that drying can adversely affect many different cellular components [14]. General stress responses can provide protection against oxidative stress, starvation, and increased osmolarity [34], and we found that BfmR was important for defense against these stresses as well (Figs 5 and 6). Desiccation itself can also stimulate responses that help protect bacterial cells from both drying and other stresses [47], and A. baumannii was found to produce higher levels of molecular chaperones and proteins involved in protection from oxidative stress and antibiotic resistance after exposure to dry conditions [26], indicating that a broad range of responses is likely important for A. baumannii to survive and recover from drying. We additionally found that BfmR was required for starvation-induced cross-protection against drying, and that increased osmolarity can also trigger cross-protection against drying (Fig 7), supporting the notion that decreased desiccation tolerance in the bfmR mutant strains was due to a defect in the ability to mount an effective stress response. These results are intriguing since A. baumannii does not appear to encode an RpoS homolog, and together with the transcriptome data of Geisinger et al. [30], our findings imply that general stress responses in A. baumannii are coordinated by an alternative mechanism that includes the BfmRS two-component signaling system.
While we did not identify all of the BfmR-controlled factors that contribute to desiccation tolerance, we did find that the KatE catalase supports the long-term survival of A. baumannii cells when dried (Fig 4), which is a novel function for this protein. KatE was previously shown to be important for H2O2 degradation in A. baumannii and A. nosocomialis [31], and we found that KatE was necessary to protect dried cells from H2O2 treatment (Fig 5). KatE is also required for maximum long-term survival of E. coli cells in broth culture [33], and a proteomics analysis of A. baumannii cells under dry conditions found increased levels of KatE during desiccation [26], providing further support for KatE’s role in long-term drying survival. Other genes that are important for defense against oxidative stress, including sodC, ahp, and acnA, were found to be controlled by BfmRS according to transcriptome analyses [30], which also identified some genes that help to explain the susceptibility of the ΔbfmR mutant to other stresses as well. In particular, genes involved in the production of compatible solutes, including genes for the synthesis of trehalose and genes for the uptake and accumulation of glycine betaine, were up-regulated by BfmR [30]. Synthesis of compatible solutes is a strategy used by many organisms to protect against both osmotic stress and desiccation, and A. baumannii produces trehalose, mannitol, and glutamate in response to increased osmolarity caused by challenge with NaCl [48]. Decreased production of compatible solutes could be a potential reason for the inability of the ΔbfmR mutant to adapt to high salt condtions (Fig 6B). However, the specific contribution of these compounds to A. baumannii desiccation tolerance have not yet been defined.
Our current findings are especially interesting based on the potential link between environmental persistence and virulence in A. baumannii. Stress-related genes, including genes involved in DNA repair, osmotolerance, and nutrient acquisition, were identified in a screen for survival in Galleria mellonella larvae, an insect model of infection [41]. The bfmS gene was also identified in this screen [41], and both bfmR and bfmS have been shown to be important for virulence in mammalian infection models [40, 49]. Based on these studies and our current results, we expect that the BfmRS system directs stress responses that are essential for survival in the clinical environment and during the infectious process. However, other regulatory systems likely play a role in these responses as well. The regulators GigA and GigB were also identified as essential for growth in Galleria, and are important for resistance to heat and antibiotic-induced stress [50]. Therefore the coordination of A. baumannii stress responses may involve cross-talk between multiple regulatory systems. Overall, our results show that in addition to its established roles in virulence and biofilm formation, BfmR regulates processes that are important for surviving desiccation, which is an ability that allows A. baumannii to survive on surfaces in the hospital environment where it can come in contact with the individuals who are most susceptible to infection.
Materials and methods
Bacterial strains and growth conditions
The A. baumannii strains used in this study are listed in Table 2. Stocks of each strain containing 15% glycerol (v/v) were stored at -80°C, and bacteria were freshly plated to begin each experiment. For all experiments bacteria were cultured in lysogeny broth (LB; Lennox formulation), and cultures were incubated at 37°C with shaking at 260–280 rpm. When necessary to maintain plasmids cultures were supplemented with 5 μg/ml tetracycline.
Table 2. A. baumannii strains and plasmids used in this study.
| Strain or Plasmid | Description | Reference or source |
|---|---|---|
| ATCC 19606 | Clinical isolate from urine | ATCC |
| ATCC 17978 | Clinical isolate from spinal meningitis | ATCC |
| ATCC 17904 | Clinical isolate from urine | ATCC |
| ATCC 17961 | Clinical isolate from blood | ATCC |
| 17961-02E8 | Laboratory-adapted isolate derived from ATCC 17961 | This study |
| 17961-01A8 | Laboratory-adapted isolate derived from ATCC 17961 | This study |
| 17961-01E3 | Laboratory-adapted isolate derived from ATCC 17961 | This study |
| 17961-01F2 | Laboratory-adapted isolate derived from ATCC 17961 | This study |
| 17961-01G4 | Laboratory-adapted isolate derived from ATCC 17961 | This study |
| 17961-01G8 | Laboratory-adapted isolate derived from ATCC 17961 | This study |
| 17961-01H12 | Laboratory-adapted isolate derived from ATCC 17961 | This study |
| 17961-ΔbfmR | bfmR deletion mutant derived from strain ATCC 17961 | This study |
| 17961-bfmR-254C>T | ATCC 17961 derivative with a C-to-T substitution in bfmR at bp 254 | This study |
| 17961-bfmR-689T>C | ATCC 17961 derivative with a T-to-C substitution in bfmR at bp 689 | This study |
| 17961-ΔabsA | absA deletion mutant derived from strain ATCC 17961 | This study |
| 17961-ΔkatE | katE deletion mutant derived from strain ATCC 17961 | This study |
| A118 | Clinical isolate from blood | [51] |
| AB09-002 | Military isolate from wound culture | G. Plano, Univ. of Miami |
| AB09-003 | Military isolate from cerebrospinal fluid | G. Plano, Univ. of Miami |
| AB5075 | Military isolate from osteomyelitis | [52] |
| Plasmids | ||
| pEX18Ap | Suicide vector | [53] |
| pBfmR-suc | bfmR region on pEX18Ap | This study |
| pΔbfmR-suc | Suicide plasmid carrying bfmR deletion | This study |
| pBfmR-254C>T-suc | Suicide plasmid carrying bfmR with point mutation | This study |
| pBfmR-689T>C-suc | Suicide plasmid carrying bfmR with point mutation | This study |
| pΔkatE-suc | Suicide plasmid carrying katE deletion | This study |
| pΔabsA-suc | Suicide plasmid carrying absA deletion | This study |
| pWH1266 | Escherichia coli-A. baumannii shuttle plasmid | [54] |
| pJF-bfmR | bfmR cloned into pWH1266 | This study |
Desiccation tolerance assays
Cells were harvested from 1 ml samples of cultures by centrifugation, and then were washed twice with 1 ml of sterile deionized water (dH2O). Water was used to avoid additional osmotic stress that could occur from the concentration of salts or buffers during evaporation of the suspending solution. Cell suspensions in dH2O were adjusted to an optical density at 600 nm (OD600) of 1.0, and then 5 μl droplets of each adjusted suspension were deposited onto a plastic (polystyrene) surface with a micropipette. These samples were left uncovered in a biosafety cabinet at room temperature (21–25°C) until visibly dry, approximately 30 min to 1 h. For experiments to determine the maximum survival time of dried A. baumannii cells, dried samples were incubated in a dark drawer at ambient temperature and humidity (19–23°C, 25–61% RH), and conditions of temperature and humidity were monitored every other day with a Fisherbrand Traceable relative humidity/temperature meter (Thermo Fisher Scientific). For all other experiments, dried samples were incubated in the dark in a sealed chamber at 24–26°C and at an equilibrium relative humidity 79–81%, which was achieved by storing samples above a saturated solution of sodium chloride within the chamber. Conditions of temperature and humidity within the chamber were monitored with a Fisherbrand Traceable relative humidity/temperature meter (Thermo Fisher Scientific) several times a week. To determine the initial number of cells present, 5 μl samples of each washed and OD600-adjusted bacterial suspension were diluted to a volume of 20 μl with saline solution (0.9% sodium chloride), and were then diluted serially and plated onto LB agar. To determine viability after drying, 20 μl of saline was placed onto each dried sample, and the samples were incubated at room temperature for 5 min to rehydrate. The bacteria were suspended by pipetting and gentle scraping to remove the cells from the plastic surface, and the suspended cells were diluted serially in saline and plated onto LB agar. For qualitative assessments, 10 μl of each dilution was spotted onto the surface of the agar, and the plate was incubated at 37°C overnight. For quantitative assessments, dilutions were spread onto agar and the number of viable cells was determined by CFU counts. Percent survival was calculated by comparing the number of CFU recovered from each dried sample to the initial number of CFU present in 5 μl of washed and OD600-adjusted cells suspensions prior to drying. At least two dried samples per time-point were suspended and plated for quantitative assays. To determine the maximum survival time of desiccated samples, one dried sample was suspended every other day, and the entire volume was plated onto LB agar. Strains were considered no longer viable if no bacteria were recovered from two consecutive platings.
Screen for spontaneous mutants and genomic analysis
To isolate spontaneous desiccation-sensitive mutants, freshly plated bacteria were used to inoculate 10 ml of LB medium, and cultures were incubated at 37°C. Approximately every 24 h, 0.2 ml of each culture was used to inoculate 10 ml of LB, and this was continued for a total of five to seven passes in liquid medium. Samples from these cultures were diluted and plated to obtain isolated colonies, and then bacteria from single colonies was used to inoculate 150 μl of LB in 96-well plates. Cultures in 96-well plates were incubated at 37°C with shaking for 6 h. Next, 20 μl from each well was diluted into 100 μl dH2O, and 5 μl of these diluted samples was dried in the wells of 96-well round-bottom plates. The remainder of each 96-well plate culture was supplemented with glycerol to achieve a final concentration of 12%, and stored at -80°C. Dried samples were stored in the dark at ambient temperature and humidity for 20 days. After this, 120 μl of LB medium was placed onto each dried sample, and they were incubated at 37°C overnight. Wells were scored visually for growth to identify clones that had lost the ability to survive desiccation. Potentially drying-sensitive clones were then screened using our standard desiccation tolerance assay described above.
To determine the mutations present in desiccation-sensitive isolates that were identified through screening, chromosomal DNA from the parent strain ATCC 17961 and its derivatives was analyzed by next-generation DNA sequencing. Libraries for sequencing were prepared with a TruSeq DNA library prep kit (Illumina), and samples were sequenced using the Illumina MiSeq system by the Genomic Sciences Laboratory at North Carolina State University, Raleigh, NC. Sequencing reads were filtered and trimmed for quality, and adapter sequences were removed using Trimmomatic v0.32 [55]. DNA sequences were analyzed using resources available on the PATRIC website [56]. A draft genome assembly for strain ATCC 17961 was generated using the PATRIC genome assembly service with the “miseq” assembly strategy. The assembled contigs were reordered using the Mauve aligner [57], with the AB5075 genome sequence as a reference. The reordered genome sequence was then annotated with the PATRIC genome annotation service to complete the draft genome. Genetic variations in the drying-sensitive derivatives of strain 17961 were identified through the PATRIC variation analysis service by aligning Illumina sequencing reads to the 17961 genome using BWA-MEM [58], followed by variant calling using either FreeBayes [59] or SAMtools [60]. Variation analyses were performed using the default settings on the PATRIC website. Genetic variations that were identified in either repetitive or duplicated regions of the genome were ignored. Other potential variations were confirmed by PCR amplification and sequencing of the genomic region of interest.
Construction of plasmids and mutant strains
To construct plasmids for generating strains with mutations in bfmR, an approximately 2.5 kb DNA fragment containing the bfmR gene was amplified by PCR using chromosomal DNA from strain ATCC 17961 as a template. The oligonucleotide primers for this reaction were designed to add a PstI site to each end of the fragment (S1 Table). Both the DNA fragment and the vector plasmid pEX18Ap were digested with PstI, and the two were ligated together to produce pBfmR-suc. Next, plasmid pBfmR-suc acted as a template for PCR reactions using oligonucleotide primers with 5’-phosphate groups (S1 Table). The DNA fragments generated from these reactions were circularized by ligation to produce plasmids pΔbfmR-suc, pBfmR-254C>T-suc, and pBfmR-689T>C-suc. Plasmid pΔbfmR-suc carries a mutated allele of bfmR with an in-frame deletion that removes the DNA sequence coding for amino acids 12 to 237 of BfmR (95% of the protein sequence). Plasmids pBfmR-254C>T-suc and pBfmR-689T>C-suc carry mutant bfmR alleles with single nucleotide substitutions at either bp 254 (C to T) or bp 689 (T to C), respectively, of the bfmR coding sequence. DNA fragments carrying in-frame deletions in either absA or katE were generated by PCR as described previously [61]. These fragments were designed to remove the DNA sequence coding for amino acids 26 to 388 of AbsA (88% of the protein sequence) or 41 to 680 of KatE (90% of the protein sequence). The fragments were digested with the appropriate restriction enzyme (PstI for ΔabsA or EcoRI for ΔkatE), and then were ligated with pEX18Ap, which had been digested with the same enzyme, to produce plasmids pΔabsA-suc and pΔkatE-suc.
To construct a plasmid carrying the wild-type bfmR gene for genetic complementation, a DNA fragment that began 470 bp upstream from the bfmR translational start site and ended 17 bp downstream from the bfmR stop codon was amplified by PCR. The oligonucleotide primers for this amplification were designed to contain an EcoRI site at the upstream end of the fragment, and a PstI site at the downstream end (S1 Table). The DNA fragment was digested with these enzymes, and then ligated with pWH1266, which had been digested with the same enzymes, to produce plasmid pJF-bfmR.
Mutant strains of A. baumannii were generated using the procedures described by Hoang et al. [53], with some modifications. Plasmids carrying mutant alleles were transferred into A. baumannii via conjugation with Escherichia coli strain SM10. A. baumannii cells that integrated the plasmid sequence onto the chromosome were selected by plating onto LB supplemented with 150 μg/ml carbenicillin and 5 μg/ml chloramphenicol. Next, mutants were selected by plating carbenicillin-resistant isolates onto LB supplemented with 10% sucrose to resolve plasmid sequences from the chromosome. Potential mutant strains were confirmed by PCR amplification and DNA sequencing of the chromosomal region of interest. For complementation, replicative plasmids were transferred into A. baumannii strains by electroporation.
Protein analysis and identification
Cells were harvested from cultures by centrifugation and stored at -80°C until analysis. To extract soluble proteins, each cell pellet was suspended in an appropriate volume (10 μl per mg wet pellet weight) of BugBuster protein extraction reagent (EMD Millipore) supplemented with 2 mg/ml lysozyme and Benzonase nuclease (EMD Millipore), as directed by the manufacturer. Samples were incubated with gentle mixing for 20 min at room temperature, and then were centrifuged at 20,000 × g for 20 min, 4°C. The supernatant (soluble fraction) was collected, and solubilization buffer (50 mM Tris pH 6.8, 1 mM EDTA, 100 mM NaCl, 0.5% SDS) was added to each pellet at a volume equal to the amount of BugBuster reagent used for cell lysis. Pellets were incubated at 55°C for 15 min, and then homogenized to produce the insoluble fraction for analysis. Soluble and insoluble fractions were analyzed by SDS-PAGE on a 12% polyacrylamide gel, and gels were visualized by staining with Coomassie-R250. To identify proteins, individual bands were excised from the gels, and proteins within these gel slices were identified by mass spectrometry (performed by the Proteomics Core Facility at the University of California, Davis).
Hydrogen peroxide sensitivity assays
To assess the sensitivity of dried A. baumannii cells to hydrogen peroxide, cells were harvested from overnight cultures by centrifugation, and then were washed twice with sterile dH2O. Cell suspensions in dH2O were adjusted to an OD600 of 1.5, and then 5 μl samples were spotted onto a plastic surface. These samples were incubated uncovered in a biosafety cabinet until visibly dry, 1–2 h. Dried samples were treated with 10 μl of either dH2O or 1.5% H2O2 for 45 sec. Next, 10 μl of 1.5% sodium thiosulfate pentahydrate (STS) was added to each sample to neutralize H2O2, and bacterial cells were gently suspended by pipetting. Suspended samples were mixed with 180 μl of 1.5% STS, and then suspensions were diluted serially in 1.5% STS. Controls were also performed to ensure that STS did not affect bacterial viability. To assess survival, 10 μl of each dilution was spotted onto the surface of a LB agar plate, and the plate was incubated at 37°C overnight.
To measure the survival of A. baumannii cells after hydrogen peroxide challenge in broth culture, cells from overnight cultures were used to inoculate fresh LB medium, which was warmed to 37°C, to a density of approximately 1 × 108 CFU/ml. Samples of inoculated medium were supplemented with 10 mM H2O2, and were incubated at 37°C for 30 min. Samples taken both prior to and after H2O2 treatment were diluted serially in 1% STS and plated onto LB medium. The number of viable cells was determined by CFU counts, and percent survival was calculated by comparing the number of CFU recovered after treatment with H2O2 to the number present prior to treatment.
Nutrient deprivation and osmotolerance assays
For experiments to measure the survival of A. baumannii strains after extended nutrient starvation, cells from overnight cultures were used to inoculate 10 ml of LB to a density of approximately 6 × 106 CFU/ml. These cultures were incubated at 37°C for 6 h. Then 2 ml samples of each culture were washed twice with 2 ml of sterile phosphate-buffered saline (PBS). Washed cells were suspended in 25 ml PBS at a density of approximately 2.5–3 × 108 CFU/ml, and were incubated with shaking at 37°C. At various time-points samples were taken from the suspensions, diluted serially in PBS, and plated onto LB medium to determine the number of viable cells. Percent survival was calculated by comparing the number of CFU recovered at each time-point to the number of CFU present at T0.
To test for tolerance of changes in osmolarity, A. baumannii cells were grown overnight at 37°C in LB medium without sodium chloride. Cells from overnight cultures were subcultured in 37°C LB medium either without or with 0.6M NaCl at a density of 0.5–1 × 108 CFU/ml. Cultures were incubated at 37°C. At various time-points samples were taken from cultures, diluted serially in 0.5X PBS, and plated onto LB medium to determine CFU/ml.
Cross-protection assays
Cells from overnight cultures were used to inoculate fresh growth medium to an OD600 of 0.05, and cultures were incubated at 37°C for 2.5 h. Then 2.5 ml of this culture was diluted into 10 ml of either fresh growth medium or medium to provide a stress, which had been prewarmed to 37°C. For nutrient starvation, cells were cultured in LB buffered with 10 mM potassium phosphate, pH 7.0, and then diluted into either the same medium or into a buffer containing 10 mM potassium phosphate, pH 7.0, 5 g/l NaCl. To test the effects of osmolarity, cells were grown in LB without NaCl, and then diluted into LB supplemented with NaCl as indicated in the text. After transfer, cultures were incubated at 37°C for 1 h, and then cells were harvested from approximately 1.7 ml of each culture, washed with 1 ml dH2O, and suspended in 0.25–0.5 ml dH2O. Cell suspensions were adjusted to an OD600 of 1.0 with dH2O, and were assayed for desiccation tolerance as described above.
Statistical analysis
Statistical significance was determined using Student’s t-test where appropriate, with P < 0.05 considered significant. Analyses were performed using Microsoft Excel.
Accession numbers
Illumina sequencing reads from A. baumannii strain ATCC 17961, and the drying-sensitive isolates derived from strain ATCC 17961, were deposited in the NCBI Sequence Read Archive under BioProject PRJNA437968, accession numbers SRX3783010, SRX3783011, SRX3783012, SRX3783013, SRX3783014, SRX3783015, SRX3783016, and SRX3783017. The draft genome sequence for strain ATCC 17961 is available on the PATRIC website (http://www.patricbrc.org), Genome ID 470.2202.
Supporting information
(PDF)
A. baumannii strains were grown overnight in LB medium, and then cells were washed with water and samples were dried on polystyrene. Dried cells were incubated at 25°C and 80% RH and, at the indicated times, dried samples were suspended and survival was assessed by CFU counts. For data with error bars, the data represent the mean ± SD from at least three independent experiments. For data without error bars, the data are representative of at least two independent experiments.
(PDF)
Acknowledgments
We would like to thank G. Plano, Univ. of Miami, and P. Rather, Emory University, for providing A. baumannii strains and plasmids. We would also like to thank K. Tipton for helpful insights into genetic manipulation of A. baumannii, V. Vasquez-Rios for assistance with assay development, and S. Palethorpe, S. Lotlikar, and J. Coleman for critical reading of the manuscript.
Data Availability
Illumina sequencing reads from A. baumannii strain ATCC 17961, and the drying-sensitive isolates derived from strain ATCC 17961, were deposited in the NCBI Sequence Read Archive under BioProject PRJNA437968, accession numbers SRX3783010, SRX3783011, SRX3783012, SRX3783013, SRX3783014, SRX3783015, SRX3783016, and SRX3783017. The draft genome sequence for strain ATCC 17961 is available on the PATRIC website (http://www.patricbrc.org), Genome ID 470.2202.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. Clinical and Pathophysiological Overview of Acinetobacter Infections: a Century of Challenges. Clin Microbiol Rev. 2017;30(1):409–47. 10.1128/CMR.00058-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rice LB. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J Infect Dis. 2008;197(8):1079–81. 10.1086/533452 [DOI] [PubMed] [Google Scholar]
- 3.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538–82. 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2017;16:91 10.1038/nrmicro.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang SH, Sheng WH, Chang YY, Wang LH, Lin HC, Chen ML, et al. Healthcare-associated outbreak due to pan-drug resistant Acinetobacter baumannii in a surgical intensive care unit. J Hosp Infect. 2003;53(2):97–102. 10.1053/jhin.2002.1348 [DOI] [PubMed] [Google Scholar]
- 6.Wilks M, Wilson A, Warwick S, Price E, Kennedy D, Ely A, et al. Control of an Outbreak of Multidrug-Resistant Acinetobacter baumannii-calcoaceticus Colonization and Infection in an Intensive Care Unit (ICU) Without Closing the ICU or Placing Patients in Isolation. Infect Control Hosp Epidemiol. 2006;27(7):654–8. 10.1086/507011 [DOI] [PubMed] [Google Scholar]
- 7.Öncül O, Keskin Ö, Acar HV, Küçükardalı Y, Evrenkaya R, Atasoyu EM, et al. Hospital-acquired infections following the 1999 Marmara earthquake. J Hosp Infect. 2002;51(1):47–51. 10.1053/jhin.2002.1205 [DOI] [PubMed] [Google Scholar]
- 8.Fournier PE, Richet H, Weinstein RA. The Epidemiology and Control of Acinetobacter baumannii in Health Care Facilities. Clin Infect Dis. 2006;42(5):692–9. 10.1086/500202 [DOI] [PubMed] [Google Scholar]
- 9.Scott P, Deye G, Srinivasan A, Murray C, Moran K, Hulten E, et al. An outbreak of multidrug-resistant Acinetobacter baumannii-calcoaceticus complex infection in the US military health care system associated with military operations in Iraq. Clin Infect Dis. 2007;44(12):1577–84. 10.1086/518170 [DOI] [PubMed] [Google Scholar]
- 10.Moultrie D, Hawker J, Cole S. Factors Associated with Multidrug-Resistant Acinetobacter Transmission: An Integrative Review of the Literature. AORN J. 2011;94(1):27–36. 10.1016/j.aorn.2010.12.026 [DOI] [PubMed] [Google Scholar]
- 11.Potts M. Desiccation tolerance of prokaryotes. Microbiol Rev. 1994;58(4):755–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolfe J, Bryant G. Freezing, drying, and/or vitrification of membrane- solute-water systems. Cryobiology. 1999;39(2):103–29. 10.1006/cryo.1999.2195 [DOI] [PubMed] [Google Scholar]
- 13.França MB, Panek AD, Eleutherio EC. Oxidative stress and its effects during dehydration. Comp Biochem Physiol A Mol Integr Physiol. 2007;146(4):621–31. 10.1016/j.cbpa.2006.02.030 [DOI] [PubMed] [Google Scholar]
- 14.Lebre PH, De Maayer P, Cowan DA. Xerotolerant bacteria: surviving through a dry spell. Nat Rev Microbiol. 2017;15(5):285–96. 10.1038/nrmicro.2017.16 [DOI] [PubMed] [Google Scholar]
- 15.Berleman JE, Bauer CE. Characterization of cyst cell formation in the purple photosynthetic bacterium Rhodospirillum centenum. Microbiology. 2004;150(2):383–90. 10.1099/mic.0.26846-0 [DOI] [PubMed] [Google Scholar]
- 16.Chang WS, van de Mortel M, Nielsen L, Nino de Guzman G, Li X, Halverson LJ. Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J Bacteriol. 2007;189(22):8290–9. 10.1128/JB.00727-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jawad A, Heritage J, Snelling AM, Gascoyne-Binzi DM, Hawkey PM. Influence of relative humidity and suspending menstrua on survival of Acinetobacter spp. on dry surfaces. J Clin Microbiol. 1996;34(12):2881–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nocker A, Fernandez PS, Montijn R, Schuren F. Effect of air drying on bacterial viability: A multiparameter viability assessment. J Microbiol Methods. 2012;90(2):86–95. 10.1016/j.mimet.2012.04.015 [DOI] [PubMed] [Google Scholar]
- 19.Chen AI, Goulian M. A network of regulators promotes dehydration tolerance in Escherichia coli. Environ Microbiol. 2018;20(3):1283–95. 10.1111/1462-2920.14074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J Clin Microbiol. 1998;36(7):1938–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giannouli M, Antunes LC, Marchetti V, Triassi M, Visca P, Zarrilli R. Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect Dis. 2013;13:282 10.1186/1471-2334-13-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapartegui-González I, Lázaro-Díez M, Bravo Z, Navas J, Icardo JM, Ramos-Vivas J. Acinetobacter baumannii maintains its virulence after long-time starvation. PLoS One. 2018;13(8):e0201961 10.1371/journal.pone.0201961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boll JM, Tucker AT, Klein DR, Beltran AM, Brodbelt JS, Davies BW, et al. Reinforcing Lipid A Acylation on the Cell Surface of Acinetobacter baumannii Promotes Cationic Antimicrobial Peptide Resistance and Desiccation Survival. MBio. 2015;6(3):e00478–15. 10.1128/mBio.00478-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aranda J, Bardina C, Beceiro A, Rumbo S, Cabral MP, Barbé J, et al. Acinetobacter baumannii RecA protein in repair of DNA damage, antimicrobial resistance, general stress response, and virulence. J Bacteriol. 2011;193(15):3740–7. 10.1128/JB.00389-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Espinal P, Martí S, Vila J. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J Hosp Infect. 2012;80(1):56–60. 10.1016/j.jhin.2011.08.013 [DOI] [PubMed] [Google Scholar]
- 26.Gayoso CM, Mateos J, Méndez JA, Fernández-Puente P, Rumbo C, Tomás M, et al. Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacter baumannii. J Proteome Res. 2014;13(2):460–76. 10.1021/pr400603f [DOI] [PubMed] [Google Scholar]
- 27.Weber BS, Ly PM, Irwin JN, Pukatzki S, Feldman MF. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci USA. 2015;112(30):9442–7. 10.1073/pnas.1502966112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tipton KA, Dimitrova D, Rather PN. Phase-Variable Control of Multiple Phenotypes in Acinetobacter baumannii Strain AB5075. J Bacteriol. 2015;197(15):2593–9. 10.1128/JB.00188-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology. 2008;154:3398–409. 10.1099/mic.0.2008/019471-0 [DOI] [PubMed] [Google Scholar]
- 30.Geisinger E, Mortman NJ, Vargas-Cuebas G, Tai AK, Isberg RR. A global regulatory system links virulence and antibiotic resistance to envelope homeostasis in Acinetobacter baumannii. PLoS Pathog. 2018;14(5):e1007030 10.1371/journal.ppat.1007030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun D, Crowell SA, Harding CM, De Silva PM, Harrison A, Fernando DM, et al. KatG and KatE confer Acinetobacter resistance to hydrogen peroxide but sensitize bacteria to killing by phagocytic respiratory burst. Life Sci. 2016;148:31–40. 10.1016/j.lfs.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loewen PC, Switala J, Triggs-Raine BL. Catalases HPI and HPII in Escherichia coli are induced independently. Arch Biochem Biophys. 1985;243(1):144–9. [DOI] [PubMed] [Google Scholar]
- 33.Mulvey MR, Switala J, Borys A, Loewen PC. Regulation of transcription of katE and katF in Escherichia coli. J Bacteriol. 1990;172(12):6713–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hengge R. The General Stress Response in Gram-Negative Bacteria In: Storz G, Hengge R, editors. Bacterial Stress Responses, Second Edition Washington, DC: ASM Press; 2011. pp. 251–290. [Google Scholar]
- 35.Wendt C, Dietze B, Dietz E, Ruden H. Survival of Acinetobacter baumannii on dry surfaces. J Clin Microbiol. 1997;35(6):1394–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright MS, Jacobs MR, Bonomo RA, Adams MD. Transcriptome Remodeling of Acinetobacter baumannii during Infection and Treatment. MBio. 2017;8(2). 10.1128/mBio.02193-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clemmer KM, Bonomo RA, Rather PN. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology. 2011;157:2534–44. 10.1099/mic.0.049791-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liou ML, Soo PC, Ling SR, Kuo HY, Tang CY, Chang KC. The sensor kinase BfmS mediates virulence in Acinetobacter baumannii. J Microbiol Immunol Infect. 2014;47(4):275–81. 10.1016/j.jmii.2012.12.004 [DOI] [PubMed] [Google Scholar]
- 39.Russo TA, Manohar A, Beanan JM, Olson R, MacDonald U, Graham J, et al. The Response Regulator BfmR Is a Potential Drug Target for Acinetobacter baumannii. mSphere. 2016;1(3). 10.1128/mSphere.00082-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Umland TC, Schultz LW, MacDonald U, Beanan JM, Olson R, Russo TA. In vivo-validated essential genes identified in Acinetobacter baumannii by using human ascites overlap poorly with essential genes detected on laboratory media. MBio. 2012;3(4). 10.1128/mBio.00113-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gebhardt MJ, Gallagher LA, Jacobson RK, Usacheva EA, Peterson LR, Zurawski DV, et al. Joint Transcriptional Control of Virulence and Resistance to Antibiotic and Environmental Stress in Acinetobacter baumannii. MBio. 2015;6(6):e01660–15. 10.1128/mBio.01660-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Draughn GL, Milton ME, Feldmann EA, Bobay BG, Roth BM, Olson AL, et al. The Structure of the Biofilm-controlling Response Regulator BfmR from Acinetobacter baumannii Reveals Details of Its DNA-binding Mechanism. J Mol Biol. 2018;430(6):806–21. 10.1016/j.jmb.2018.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geisinger E, Isberg RR. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog. 2015;11(2):e1004691 10.1371/journal.ppat.1004691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemcke RM. THE CHANGES WITH AGE IN THE RESISTANCE OF ESCHERICHIA COLI TO DRYING UNDER ATMOSPHERIC CONDITIONS. J Appl Bacteriol. 1959;22(2):253–7. 10.1111/j.1365-2672.1959.tb00158.x [DOI] [Google Scholar]
- 45.Vriezen JAC, deBruijn FJ, Nüsslein K. Desiccation responses and survival of Sinorhizobium meliloti USDA 1021 in relation to growth phase, temperature, chloride and sulfate availability. Lett Appl Microbiol. 2006;42(2):172–8. 10.1111/j.1472-765X.2005.01808.x [DOI] [PubMed] [Google Scholar]
- 46.Stockwell VO, Loper JE. The sigma factor RpoS is required for stress tolerance and environmental fitness of Pseudomonas fluorescens Pf-5. Microbiology. 2005;151(9):3001–9. 10.1099/mic.0.28077-0 [DOI] [PubMed] [Google Scholar]
- 47.Gruzdev N, Pinto R, Sela S. Effect of desiccation on tolerance of Salmonella enterica to multiple stresses. Appl Environ Microbiol. 2011;77(5):1667–73. 10.1128/AEM.02156-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeidler S, Hubloher J, Schabacker K, Lamosa P, Santos H, Müller V. Trehalose, a temperature- and salt-induced solute with implications in pathobiology of Acinetobacter baumannii. Environ Microbiol. 2017;19(12):5088–99. 10.1111/1462-2920.13987 [DOI] [PubMed] [Google Scholar]
- 49.Wang N, Ozer EA, Mandel MJ, Hauser AR. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. MBio. 2014;5(3):e01163–14. 10.1128/mBio.01163-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gebhardt MJ, Shuman HA. GigA and GigB are Master Regulators of Antibiotic Resistance, Stress Responses, and Virulence in Acinetobacter baumannii. J Bacteriol. 2017;199(10). 10.1128/JB.00066-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramirez MS, Don M, Merkier AK, Bistue AJ, Zorreguieta A, Centron D, et al. Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J Clin Microbiol. 2010;48(4):1488–90. 10.1128/JCM.01264-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, et al. AB5075, a Highly Virulent Isolate of Acinetobacter baumannii, as a Model Strain for the Evaluation of Pathogenesis and Antimicrobial Treatments. MBio. 2014;5(3):e01076–14. 10.1128/mBio.01076-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212(1):77–86. [DOI] [PubMed] [Google Scholar]
- 54.Hunger M, Schmucker R, Kishan V, Hillen W. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene. 1990;87(1):45–51. [DOI] [PubMed] [Google Scholar]
- 55.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017;45:D535–D42. 10.1093/nar/gkw1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30(20):2843–51. 10.1093/bioinformatics/btu356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 2012:arXiv:1207.3907.
- 60.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farrow JM, Pesci EC. Two distinct pathways supply anthranilate as a precursor of the Pseudomonas quinolone signal. J Bacteriol. 2007;189(9):3425–33. 10.1128/JB.00209-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
A. baumannii strains were grown overnight in LB medium, and then cells were washed with water and samples were dried on polystyrene. Dried cells were incubated at 25°C and 80% RH and, at the indicated times, dried samples were suspended and survival was assessed by CFU counts. For data with error bars, the data represent the mean ± SD from at least three independent experiments. For data without error bars, the data are representative of at least two independent experiments.
(PDF)
Data Availability Statement
Illumina sequencing reads from A. baumannii strain ATCC 17961, and the drying-sensitive isolates derived from strain ATCC 17961, were deposited in the NCBI Sequence Read Archive under BioProject PRJNA437968, accession numbers SRX3783010, SRX3783011, SRX3783012, SRX3783013, SRX3783014, SRX3783015, SRX3783016, and SRX3783017. The draft genome sequence for strain ATCC 17961 is available on the PATRIC website (http://www.patricbrc.org), Genome ID 470.2202.