Abstract
Exostosin-1 (EXT1) has been demonstrated to participate in the progression of many cancers. However, it has not been previously described in patients with hepatocellular carcinoma (HCC) without vascular invasion. In this study, we got the accurate data of EXT1 mRNA Z-score from the CBio data portal of The Cancer Genome Atlas (TCGA), which was used to express the level of EXT1 gene expression. We analyzed the EXT1 gene expression between HCC and normal liver tissue and compared the clinical significance of tumor tissue's EXT1 gene expression of HCC patients without vascular invasion based on data from TCGA database. The association between EXT1 gene expression and disease-free survival (DFS) was further analyzed. EXT1 gene copy number was also analyzed in this study. Univariate and multivariate analyses showed that high EXT1 gene expression group was significantly poorer than that of the low EXT1 gene expression group (P = .004). In addition, EXT1 gene expression was positively associated with α-fetoprotein (AFP), which is a well-known marker for HCC. There was a significant positive correlation between EXT1 copy number and upregulated EXT1 gene (P < .0001). In conclusion, upregulation of EXT1 could be an important indicator to the short DFS of HCC patients without vascular invasion. EXT1 gene copy number amplification is one of the mechanisms underlying the upregulation of EXT1.
Keywords: copy number, disease-free survival, EXT1, hepatocellular carcinoma, prognosis
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most common and malignant tumors in the world.[1] With the surgical technology being gradually improved and preoperation period of the treatment becoming better, safety of liver resection and survival rate have been raised, but the recurrence rate is still very high.[2,3] Although surgical resection offers the best chance of cure of HCC, the prognosis after surgery differs considerably among patients which have hampered both treatment and prognostic prediction. The molecules responsible for early stage HCC,[4] especially in patients with HCC without vascular invasion have not been fully elucidated.[5]
EXT1 is a protein that in humans is encoded by the EXT1 gene. This gene encodes an endoplasmic reticulum-resident type II transmembrane glycosyltransferase involved in the chain elongation step of heparan sulfate biosynthesis.[6] Mutations in this gene cause the type I form of multiple exostoses.[7] Furthermore, aberrant EXT1 expression is significantly associated with prognosis in breast cancer,[8] bile duct cancer,[9] and acute lymphoblastic leukemia cancer (ALL).[10] However, the prognostic value of EXT in patients with HCC without vascular invasion has not been clearly defined.
In the current study, we have taken advantage of Oncomine datasets[11] and The Cancer Genome Atlas (TCGA) database by the CBio data porta.[12] Then, we performed gene expression of EXT1 in HCC. We also investigated the clinical significance of EXT1 expression in HCC cases and assessed whether EXT1 expression was associated with the prognosis in patients with HCC without vascular invasion. We analyzed genomic copy number variation of EXT1 gene, which maybe one of the mechanisms underlying the upregulation of EXT1 which could be related to the prognosis.
2. Materials and methods
2.1. Ethical approval
This study was approved by the ethics committee of Wuxi No.9 People's Hospital affiliated to Soochow University, P.R. China. All methods were performed in accordance with the relevant guidelines and regulations.
2.2. Public dataset
Bioinformatic analysis was based on data from Oncomine datasets and TCGA database. Oncomine datasets (https://www.oncomine.org) is a public cancer microarray platform incorporating 392 independent microarray datasets, totaling >28,880 microarray experiments and spanning 41 cancer types. It unifies a large compendium of other published cancer microarray data, including Gene Expression Omnibus (GEO)[13] and Stanford Microarray Database (SMD),[14] and uniquely provides differential expression analyses comparing most major types of cancer with their respective normal tissues.
The Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov) database stores genomic and clinical data of a series of common cancers including HCC[15]; all data have been made public for analysis. Tissue and tumor specimens were obtained from patients with appropriate consent from institutional review boards under the guidelines of The Cancer Genome Atlas Research Network. Informed consent was obtained from all patients or their guardians, including the use of tissue samples for research purposes. The official TCGA report on HCC was released and the database was analyzed on the CBio data portal (http://www.cbioportal.org/). Copy number variance was determined by profiling HCC on Affymetrix SNP 6.0 arrays[16] and analysis by GISTIC 2.0.[17] Values: −2 = deep deletion; −1 = shallow deletion; 0 = neutral/no change (diploid); 1 = gain; 2 = amplification. Surgical resection of biopsy bio specimens was collected from patients diagnosed with HCC, and had not received prior treatment for their disease (ablation, chemotherapy, or radiotherapy). Cases were staged according to the American Joint Committee on Cancer (AJCC).[18]
2.3. Patients and analysis of clinical data
From Jan 1st, 1995 to Feb 16th, 2016, 363 TCGA-HCC cases by whole-exome sequencing and DNA copy number analyses were analyzed, there were 203 patients without vascular invasion (including macroscopic and microscopic vascular invasion) among these patients. The patients without vascular invasion included 134 men cases and 69 women cases with an average age of 60.1 years (from 16–85 years of age). A descriptive table of clinical features, histological features, and molecular features for the 203 cases cohort as well as a patient level summary are shown in tables below. We got the accurate data of EXT1 mRNA Z-score from the CBio data portal of TCGA-HCC, which was used to express the level of EXT1 gene expression. We analyzed the clinical significance of tumor tissue's EXT1 gene expression of HCC patients without vascular invasion. The association with EXT1 expression and disease-free survival (DFS) was also analyzed. We investigated EXT1 gene copy number of this study in comparison with its mRNA levels. The heat map data of EXT1 gene copy number and mRNA levels were collected using the TCGA database by the CBio data portal. t-test was used to evaluate the effect of EXT1 gene copy number on its mRNA levels.
2.4. Statistical analysis
The Kaplan–Meier survival analysis (log-rank test) was used to analyze HCC patients’ DFS time. The factors with P < .05 from the Kaplan–Meier analysis were enrolled in the Cox regression hazard model. Log-rank test was applied to compare the survival distribution of 2 groups. Categorical variables were compared using the chi-squared test or Fisher exact test. P values were 2 tailed. Statistical significance was accepted for P values <.05. The data were statistically analyzed with the software SPSS Statistics, version 18.0 (SPSS Inc., Chicago, IL). The tumor tissue's EXT1 mRNA expression and its copy number variation status in different HCC patients were examined by the GraphPad Prism7 software (GraphPad Software, Inc., La Jolla, CA). Data were presented with mean ± SD.
3. Results
3.1. EXT1 is upregulated in HCC
To explore the expression and significance of EXT1 in HCC, a study was performed to compare the gene expression between HCC and normal liver tissue using the Oncomine database. In this study, we chose the EXT1 expression data in Roessler Liver[19] (Hepatocellular Carcinoma vs Normal), including 21 normal liver tissues and 22 HCC tissues. Gene expression profiling was carried out on Affymetrix Gene Chip HG-U133A 2.0 (Affymetrix, Santa Clara, CA) arrays according to the manufacturer's protocol. Then, the gene expression (mRNA) data were plotted. We found that EXT1 was upregulated in HCC tissues compared with normal liver tissue (P = .002) (Fig. 1).
Figure 1.
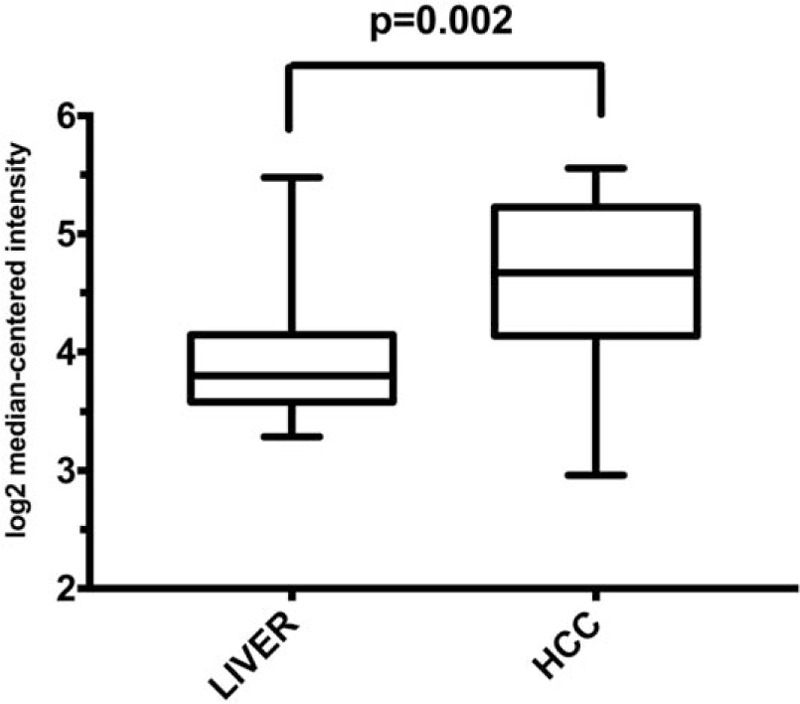
Expression of EXT1 mRNA levels in 21 normal liver tissues and 22 HCC tissues. The data were displayed as log 2 median-centered intensity, P = .002; t-test = 3.118; fold change = 1.548. EXT1 = (Exostosin-1), HCC = hepatocellular carcinoma.
3.2. Independent factors influencing prognosis of patients after HCC resection
To estimate the clinical significance of EXT1 expression in HCC patients without vascular invasion, EXT1 mRNA expression of the patients who underwent resection of HCC was analyzed. The mRNA Z-score value was used to express the level of EXT1 gene expression. According to the Z-score value, cases were divided into 2 groups, the high EXT1 expression group were defined as the Z-score value >2 while the low EXT1 expression group were defined as the Z-score value <2. The data of 203 HCC patients without vascular invasion was analyzed by Kaplan–Meier analysis. The DFS rates of 1, 2, and 5 years were 74.4%, 58.7%, and 38.0%, respectively; the median DFS time was 36.0 months. The overall survival rates of 1, 2, and 5 years were 88.4%, 77.0%, and 56.0%, respectively; the median overall survival time was 80.7 months. The results also showed that white people, men, no HBV infection, HCV infection, R1 hepatectomy, TNM stage (T2–T4), high EXT1 expression were important factors that had influenced the DFS time (P < .05). The results of Cox regression hazard model analysis showed that HCV infection, TNM stage (T2–T4), and high EXT1 expression were independent risk factors influencing DFS (P < .05) (Table 1).
Table 1.
The influencing factors of the DFS in HCC patients without vascular invasion.
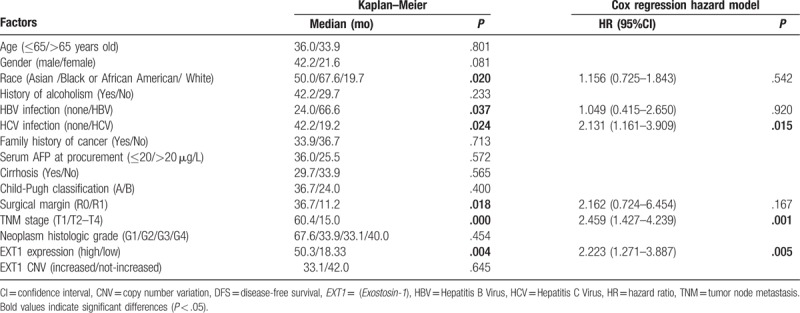
The results indicate that high EXT1 expression is an important and independent factor influencing the DFS. Therefore, we focused on this factor and performed further analyses.
3.3. The clinical significance of EXT1 expression in HCC patients without vascular invasion
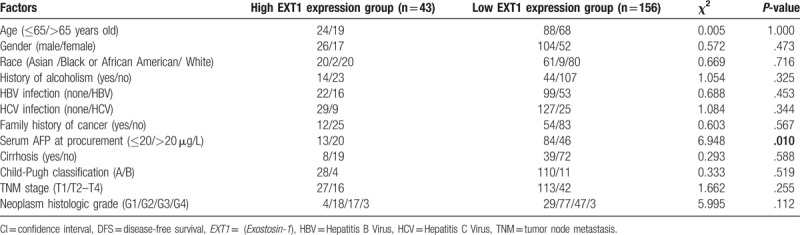
Then the correlations between EXT1 expression and clinicopathological characteristics of HCC were analyzed. As shown in Table 2, we found that the expression of EXT1 of HCC patients was positively associated with the corresponding α-fetoprotein (AFP) level (χ2 = 6.948, P = .010). As AFP is frequently used for the prognosis of HCC, the correlation indicated that EXT1 might be a potential biomarker of HCC.
Table 2.
Clinical pathological data in the high EXT1 expression group and the low EXT1 expression group.
The DFS of the patients in high and low EXT1 expression group. The results of Kaplan–Meier analysis showed that in the high EXT1 expression group, the median DFS was 18.3 months. The 1-, 2-, and 5-year DFS rates were 61.0%, 38.6%, and 0.0%, respectively. In the low EXT1 expression group, the median DFS was 50.3 months. The 1-, 2-, and 5-year DFS rates were 78.0%, 63.5%, and 44.1%, respectively. The analysis showed that there was statistically significant difference in the DFS between the 2 groups (P = .004). The DFS of the high EXT1 expression group was significantly shorter (Fig. 2). The result suggests that EXT1 gene expression is important to DFS of the HCC patients.
Figure 2.
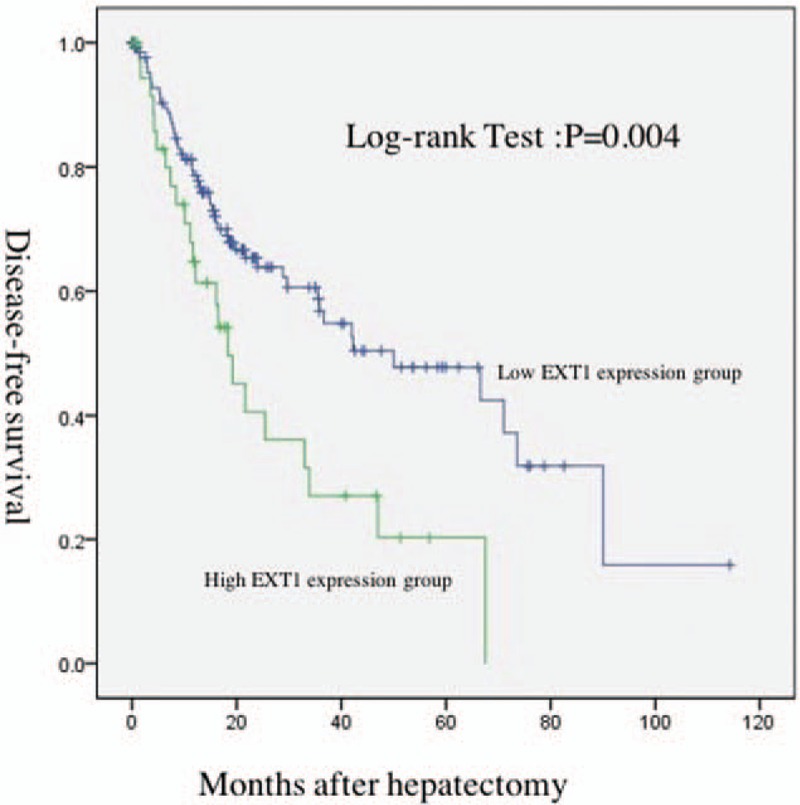
Comparison of DFS between high EXT1 expression group (green line) and low EXT1 expression group (blue line). Some patients’ EXT1 gene expression data or survival were missing, P = .004. DFS = disease-free survival, EXT1 = (Exostosin-1).
3.4. The expression of EXT1 correlates with the gene copy number in HCC patients without vascular invasion
To verify EXT1 gene copy number variation and their impact on expression of EXT1 gene in HCC, we used the CBio data portal to quarry the TCGA dataset. Overall, the average EXT1 mRNA Z-score level was 0.8341 (range, −1.4292–8.6263). Patients were divided into 2 groups: increased EXT1 gene copy number group (gene amplification), not increased EXT1 gene copy number group (including gene gain, deep deletion, shallow deletion, diploid). Increased EXT1 gene copy number group was found in 19.4% patients, EXT1 mRNA Z-score level was 2.300 ± 0.3765, n = 38. While in the group of not increased EXT1 gene copy number, EXT1 Z-score level was 0.4833 ± 0.1196, n = 158. As shown in Figs. 3 and 4, a positive correlation was found between EXT1 gene copy number and EXT1 mRNA (P < .0001).
Figure 3.
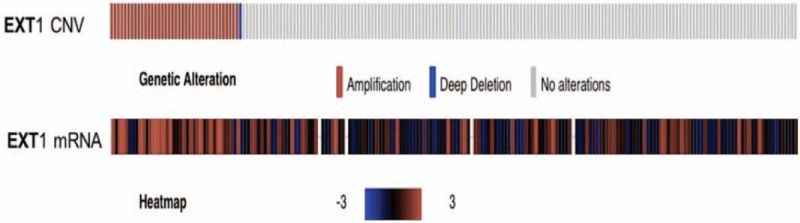
An expression heat map of frequent copy number amplification of EXT1 gene and mRNA in HCC patients. Some patients’ EXT1 gene expression or copy number data were missing. EXT1 = (Exostosin-1), HCC = hepatocellular carcinoma.
Figure 4.
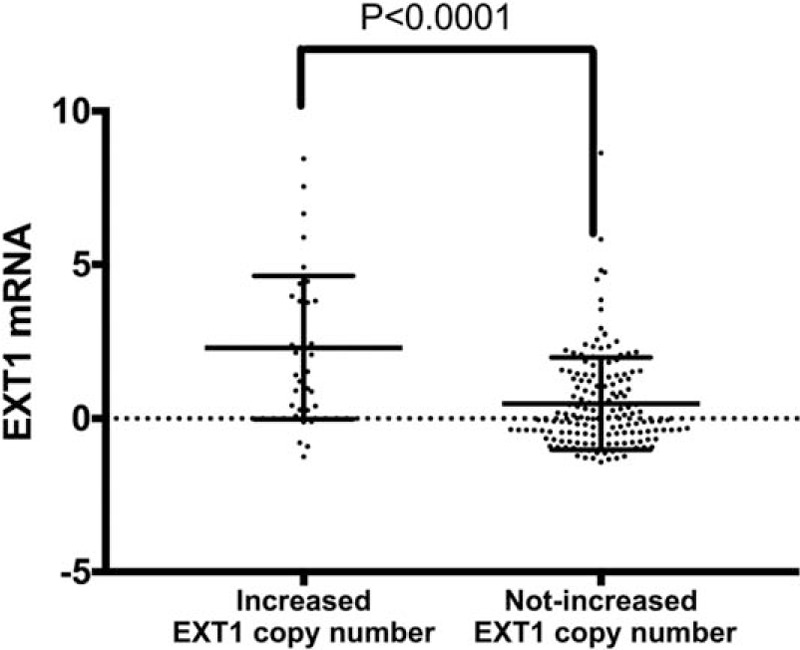
Increased EXT1 gene copy number correlates with increased mRNA levels in HCC patients without vascular invasion, P < .0001. EXT1 = (Exostosin-1), HCC = hepatocellular carcinoma.
4. Discussion
Years ago, techniques such as ultrasonography, computed tomography, and magnetic resonance imaging were not available. HCC was almost always diagnosed when the cancer produced symptoms. Years later, it became feasible to diagnose HCC at earlier stages, before symptoms developed.[20] Very early and early-stage HCC patients without both macrovascular and microvascular invasion can benefit from potentially curative treatments, such as surgical resection.[21] But long-term survival of early-stage HCC patients is still very low,[22] studies now aim to identify molecular markers for risk and determination of prognosis and treatment[23].
Now EXT1 gene has been used as an approach to identification of molecular biomarkers for cancer occurrence, classification, and prognosis. But until now little has been known about the role and molecular mechanism of EXT1 in HCC. In this study, we found that EXT1 gene expression was upregulated in some cases. These data suggested that EXT1 gene might participate in the tumorigenesis and metastasis of HCC. Moreover, EXT1 gene mRNA expression was significantly correlated with serum AFP. AFP has been a useful biomarker for HCC,[1] the significant positive correlation between AFP and EXT1 gene strongly suggested that EXT1 might be another potential biomarker of HCC.
We used the Oncomine data (Roessler Liver) to explore the possibility, we found that EXT1 was upregulated in HCC tissues compared with normal liver tissue. In fact, EXT1 was localized in the endoplasmic reticulum of fibroblast cell lines that exhibits glycosyltransferases activities involved in the chain elongation step of heparin sulfate (HS) biosynthesis. Expression of EXT1 may promote carcinogenesis via its glycosyltransferases activities to synthesize HS.[24] The increase in HS has been reported in many cancer types such as in HCC,[25] pancreatic cancer,[26] breast cancer.[27] Its role in tumor progression towards an aggressive phenotype, particularly invasiveness and preparation of metastatic niche, has been proposed. Consistent with these results, we believed that the upregulation of EXT1 is a consequence of the development of HCC.
Furthermore, we also showed the evidence for the correlation between EXT1 gene copy number and its expression level. EXT1 was always associated with diseases including hereditary multiple exostoses, an autosomal dominant bone disorder of its mutation[28,29] or, more recently, its expression levels.[30] The copy number of EXT1 gene or the correlation between EXT1 gene copy number and its expression level has not been previously examined. In this study, we found a positive correlation between EXT1 gene copy number and EXT1 mRNA (P < .0001). One possible mechanism is gene amplification.[31] Gene amplification can be defined as any duplication of a region of DNA that contains a gene. Gene amplification include retro transposition event, ectopic recombination, polyploidy, aneuploidy, and replication slippage. From the perspective of molecular genetics, gene copy amplification is one of many ways in which a gene can be overexpressed. If gene copy amplification occurred in the same reading frame in the coding portion of a gene, it may lead to a long chain of the amino acid, possibly creating increase of protein expression and if this occurred in the non-coding part of the gene, it may affect gene expression and regulation too.[32] For example, gene copy amplification and overexpression of c-myc, could be frequently observed in many cancers.[33]
But there were still some cases of upregulated EXT1 without increased gene copy number. There are some other mechanisms may contribute to EXT1 over-expression. Variants located in one or both EXT1 promoters might influence EXT1 expression. We searched the relevant documents, and found the EXT1 promoter was identified within a region of 123 bp and the promoter single nucleotide polymorphisms (SNP) rs3401643 which was lost with the presence of the C-allele resulting in a 56% increase in EXT1 promoter activity, that could explain part of the EXT1 over-expression.[34] Certain microRNA may also modulate EXT1 expression. A recent report described there is a negative correlation between miR-1303 and EXT1 expression in pediatric astrocytomas, which strongly suggested the regulation of EXT1 by this miRNA. EXT1 is also regulated by let-7g-3p. This miRNA had a similar expression pattern to that of miR-1303, which suggested a synergic action of these miRNAs over the regulation of EXT1 expression.[35] It is certain that some other mechanisms can affect the EXT1 mRNA as well as its gene copy number. These mechanisms can explain for some outlier value (cases). We recognize it is a limitation of not having a larger series of patients to explore whether EXT1 gene copy number as well as upregulated mRNA associated with HCC patients’ survival. In vitro or in vivo assays will also be done in our further study and will further illuminate factors affecting the expression of EXT1 and the prognosis of HCC. Animal models such as nude mice would also be selected to further demonstrate this discovery in this study.
Taken together, this study has identified upregulation of EXT1 could be an important indicator to the short DFS of HCC patients without vascular invasion. Our data also implied that EXT1 could be a useful target in patients with HCC without vascular invasion. EXT1 gene copy number amplification may be one of the mechanisms underlying the upregulation of EXT1.
Author contributions
Conceptualization: Sheng Dong, Yifeng Wu, Shigang Yu.
Data curation: Sheng Dong.
Investigation: Yinxi Yang.
Methodology: Shigang Yu, Yinxi Yang.
Project administration: Yinxi Yang.
Resources: Yinxi Yang.
Software: Yifeng Wu.
Supervision: Shurong Fan.
Validation: Sheng Dong, Lijun Lu, Shurong Fan.
Visualization: Sheng Dong, Lijun Lu, Shurong Fan.
Writing – original draft: Sheng Dong, Lijun Lu, Shurong Fan.
Writing – review & editing: Sheng Dong, Shurong Fan.
Footnotes
Abbreviations: AFP = alpha fetoprotein, AJCC = American Joint Committee on Cancer, ALL = acute lymphoblastic leukemia cancer, ALT = alanine aminotransferase, CI = confidence interval, DFS = disease-free survival, EXT1 =(Exostosin-1), GEO = Gene Expression Omnibus, HBV = Hepatitis B Virus, HCC = hepatocellular carcinoma, HCV = Hepatitis C Virus, HS = heparin sulfate, SMD = Stanford Microarray Database, TCGA = The Cancer Genome Atlas, TNM = tumor node metastasis.
The authors have no conflicts of interest to disclose.
References
- [1].Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69. [DOI] [PubMed] [Google Scholar]
- [2].Wang CH, Wey KC, Mo LR, et al. Current trends and recent advances in diagnosis, therapy, and prevention of hepatocellular carcinoma. Asian Pac J Cancer Prev 2015;16:3595–604. [DOI] [PubMed] [Google Scholar]
- [3].Sun CY, Sarna L. Symptom management in hepatocellular carcinoma. Clin J Oncol Nurs 2008;12:759–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Colombo M, Sangiovanni A. Treatment of hepatocellular carcinoma: beyond international guidelines. Liver Int 2015;35suppl:129–38. [DOI] [PubMed] [Google Scholar]
- [5].Hsieh CH, Wei CK, Yin WY, et al. Vascular invasion affects survival in early hepatocellular carcinoma. Mol Clin Oncol 2015;3:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Busse M, Feta A, Presto J, et al. Contribution of EXT1, EXT2, and EXTL3 to heparan sulfate chain elongation. J Biol Chem 2007;282:32802–10. [DOI] [PubMed] [Google Scholar]
- [7].Wuyts W, Van HW, De BK, et al. Mutations in the EXT1 and EXT2 genes in hereditary multiple exostoses. Am J Hum Genet 1998;62:346–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Manandhar S, Kim C, Su YO, et al. Abstract B21: Exostosin 1 regulates cancer cell stemness in breast cancer cells. Cancer Res 2016;76(15 suppl):B30. [Google Scholar]
- [9].Khoontawad J, Hongsrichan N, Chamgramol Y, et al. Increase of exostosin 1 in plasma as a potential biomarker for opisthorchiasis-associated cholangiocarcinoma. Tumour Biol 2014;35:1029–39. [DOI] [PubMed] [Google Scholar]
- [10].Daakour S, Hajingabo LJ, Kerselidou D, et al. Systematic interactome mapping of acute lymphoblastic leukemia cancer gene products reveal EXT-1 tumor suppressor as a Notch1 and FBWX7 common interactor. BMC Cancer 2016;16:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Edgar R, Lash A. The Gene Expression Omnibus (GEO): A gene expression and hybridization repository. National Center for Biotechnology Information. Nucleic Acids Res 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gollub J, Ball CA, Sherlock G. The Stanford Microarray Database. Clifton, New Jersey: Humana Press; 2006. [DOI] [PubMed] [Google Scholar]
- [15].Ally A, Balasundaram M, Carlsen R, et al. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017;169:1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mccarroll SA, Kuruvilla FG, Korn JM, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet 2008;40:1166–74. [DOI] [PubMed] [Google Scholar]
- [17].Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual, and the future of TNM. Ann Surg Oncol 2010;17:1471–4. [DOI] [PubMed] [Google Scholar]
- [19].Roessler S, Jia HL, Budhu A, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res 2010;70:10202–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dumitrescu CI, Gheonea IA, Săndulescu L, et al. Contrast enhanced ultrasound and magnetic resonance imaging in hepatocellular carcinoma diagnosis. Med Ultrason 2013;15:261–7. [DOI] [PubMed] [Google Scholar]
- [21].Thein HH, Isaranuwatchai W, Qiao Y, et al. Cost-effectiveness analysis of potentially curative and combination treatments for hepatocellular carcinoma with person-level data in a Canadian setting. Cancer Med 2017;6:2017–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chang HC, Lin YM, Yen AM, et al. Predictors of long-term survival in hepatocellular carcinomas: a longitudinal follow-up of 108 patients with small tumors. Anticancer Res 2013;33:5171–8. [PubMed] [Google Scholar]
- [23].Yim SH, Chung YJ. An overview of biomarkers and molecular signatures in HCC. Cancers (Basel) 2010;2:809–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mccormick C, Leduc Y, Martindale D, et al. The putative tumour suppressor EXT1 alters the expression of cell-surface heparan sulfate. Nat Genet 1998;19:158–61. [DOI] [PubMed] [Google Scholar]
- [25].Tátrai P, Egedi K, Somorácz A, et al. Quantitative and qualitative alterations of heparan sulfate in fibrogenic liver diseases and hepatocellular cancer. J Histochem Cytochem 2010;58:429–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Korc M. Pancreatic cancer associated stroma production. Am J Surg 2007;194(4 suppl):S84–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Koo CY, Sen YP, Bay BH, et al. Targeting heparan sulfate proteoglycans in breast cancer treatment. Recent Pat Anticancer Drug Discov 2008;3:151–8. [DOI] [PubMed] [Google Scholar]
- [28].Hong G, Guo X, Yan W, et al. Identification of a novel mutation in the EXT1 gene from a patient with multiple osteochondromas by exome sequencing. Mol Med Rep 2017;15:657–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu H, Song WU, Duan LI, et al. Identification of a novel EXT1 mutation in patients with hereditary multiple exostosis by exome sequencing. Oncol Rep 2015;33:547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shieh YE, Wells DE, Sater AK. Zygotic expression of Exostosin1 (EXT1) is required for BMP signaling and establishment of dorsal-ventral pattern in Xenopus. Int J Dev Biol 2014;58:27–34. [DOI] [PubMed] [Google Scholar]
- [31].Zhang J, Zhang J. Evolution by gene duplication. Trends Ecol Evol 2003;18:292–8. [Google Scholar]
- [32].Myers RH. Huntington's disease genetics. NeuroRx 2004;1:255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lewis LM, Edwards MC, Meyers ZR, et al. Replication Study: transcriptional amplification in tumor cells with elevated c-Myc. elife 2018;7:e30274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jennes I, Zuntini M, Mees K, et al. Identification and functional characterization of the human EXT1 promoter region. Gene 2012;492:148–59. [DOI] [PubMed] [Google Scholar]
- [35].Lópezaguilar JE, Velázquezflores MA, Simónmartínez LA, et al. Circulating microRNAs as biomarkers for pediatric astrocytomas. Arch Med Res 2017;48:323–32. [DOI] [PubMed] [Google Scholar]


