Supplemental Digital Content is available in the text
Keywords: ANRIL, coronary artery disease, myocardial infarction, polymorphism, rs4977574
Abstract
Background:
Several studies have shown that ANRIL polymorphism may be associated with the risk of coronary artery disease (CAD). However, these studies do not provide a clear consensus in Asian population. Thus, this meta-analysis was aimed to evaluate the relationship between the common variant rs4977574 in ANRIL and CAD risk in Asian population.
Methods:
We conducted a systematic literature search of PubMed, Embase and the Cochrane Library and 2 Chinese databases. A total of 12,005 subjects from 6 independent studies were included. The pooled odds ratio (OR) and their corresponding 95% confidence intervals (CIs) were used to assess the association between rs4977574 and CAD using random effects model.
Results:
A significant association was observed between rs4977574 and CAD risk under the allelic (OR: 1.18, 95% CI: 1.04–1.34, P = .010), recessive (OR: 1.27, 95% CI: 1.01–1.60, P = .04), dominant (OR: 1.28, 95% CI: 1.13–1.44, P = .002), homozygous (OR: 1.46, 95% CI: 1.15–1.86, P = .002), and heterozygous model (OR: 1.17, 95% CI: 1.07–1.28, P = .0004), especially in the Chinese subgroup and the myocardial infarction (MI) subgroup (P < .05).
Conclusion:
The ANRIL polymorphism rs4977574 is associated with CAD risk in Asian population. The rs4977574 with G allele may confer to a higher risk of CAD, especially MI.
1. Introduction
Coronary artery disease (CAD) is a chronic multifactorial disease and a leading contributor to global morbidity and mortality.[1] Both genetic and environmental factors are involved in the pathogenesis of CAD; genetic factors could explain 30% to 60% of the variations in CAD risk.[2,3] To date, several common variants on chromosome 9p21.3 associated with CAD have been identified by genome-wide association studies (GWASs).[4–6]ANRIL, also known as CDKN2BAS and a potential candidate gene at chromosome 9p21.3 that encodes a large antisense non–coding RNA, is located adjacent to the CDKN2A-CDKN2B gene cluster.[4,7,8] The promoter region of ANRIL affects the transcription of CDKN2A and CDKN2B by binding to zinc-finger proteins.[9]ANRIL is expressed in cardiac tissues, which may affect development of CAD via regulating vascular cell proliferation with reduced expression of CDKN2A and CDKN2B.[10]ANRIL knockdown in vascular smooth muscle was shown to change the expression of genes involved in extracellular matrix remodeling, suggesting that ANRIL plays a role in vascular structure and function.[11]
Several GWASs have revealed that the single nucleotide polymorphism (SNP) rs4977574 of ANRIL was associated with increased CAD risk in different ethnicities.[12–19] A meta-analysis by Huang et al of 23 studies among 36,452 cases and 39,781 controls showed a strong association between rs4977574 and the risk of CAD (P < .0001, odds ratio [OR] = 1.27, 95% CI 1.22–1.31). A meta-analysis of 13 case-control studies involving 6796 cases and 9956 controls by Wang et al found that ANRIL rs2383207 polymorphism was associated with a significantly increased risk of CAD (OR = 1.47; 95% CI 1.33–1.62), including Caucasians (OR = 1.51; 95% CI 1.28–1.77) and Asians (OR = 1.42; 95% CI 1.26–1.61). These findings implicate ANRIL in CAD development.
However, the associations between rs4977574 and the risk of CAD are likely to be varied across different ethnicities. To date, most of the studies on ANRIL rs4977574 polymorphism focused on Caucasians,[17,20,21] and few studies are available on Asians. Huang et al carried out a case-control study of 590 Chinese CAD patients and 482 non-CAD patients subjects and found that ANRIL rs4977574 polymorphism was associated with a significantly increased risk of CAD in females (χ2 = 10.29, P = .003, OR = 2.14, 95% CI 1.31–2.77).[22] Additional studies on Chinese, Japanese and people from other Asian countries also suggested that ANRIL rs4977574 polymorphism was associated with an increased CAD risk,[16,17,21,23–24] but hitherto, no meta-analysis of ANRIL rs4977574 polymorphism and CAD risk is available. In the present study, we carried out a meta-analysis of the overall effect of ANRIL polymorphism rs4977574 (A >G) on CAD risk in Asian subjects.
2. Methods
2.1. Literature search
We conducted a systematically literature search of PubMed, Embase, and the Cochrane Library as well as 2 Chinese databasesWanfang and CNKI (China National Knowledge Infrastructure) for publications up to April 1, 2018 by using the following keywords: “rs4977574” AND (“polymorphism” OR “variant”) AND (“CAD” OR “CAD” OR “myocardial infarction [MI]” OR “coronary heart disease” OR “CAD” OR “MI”). The publications were limited to English and Chinese. All eligible studies and their references were evaluated for their eligibility for inclusion. Only original researches were included. Case reports, editorials and reviews were excluded.
2.2. Inclusion criteria
Publications were eligible if
-
(1)
the study examined the association ofrs4977574 with CAD including MI;
-
(2)
it was a case-control study;
-
(3)
the OR together with 95% confidence interval (CI) was obtained.
Major exclusions were
-
(1)
non-Asian population;
-
(2)
publications other than English or Chinese;
-
(3)
duplicated publications based on the same data.
2.3. Data extraction
We extracted the following characteristics: first author, publication year, region, genotyping, number of genotypes, and total number of cases and controls. All data were extracted from each literature by 2 authors (BX and ZF) independently. Disagreement was resolved by consensus of these 2 authors. If they could not reach a consensus, the result was reviewed by a third author (SH).
2.4. Quality assessment
To assess the quality of each study, the Newcastle–Ottawa scale (NOS) was used to evaluate all the observational studies. NOS score ranges from 4 to 9. If a study score was higher than 7, it was a high-quality study. Two investigators (BX and ZF) independently assessed the quality of these studies, and the third investigator (SH) resolved the disagreement between the first 2 investigators.
2.5. Ethical statement
All results and analyses were from previous published studies; thus, no ethical approval and patient consent are required.
2.6. Statistical analysis
We assessed the strength of the association between rs4977574 and CAD risk by ORs together with 95% CIs. Z test was used to determine the significance of the pooled ORs, and P values <.05 were considered significant. We evaluated the association between rs4977574 and CAD risk under the allelic (distribution of G allelic frequency of ANRIL gene polymorphism), recessive (GG versus AG+AA), dominant (GG+AG versus AA), homozygous (GG versus AA) and heterozygous (AG versus AA) genetic models. The heterogeneity of the ORs was checked by Q test or I2. A significant heterogeneity across studies was considered if P was less than .05 or I2 was greater than 50%. When a significant heterogeneity exists, we pooled ORs with the random-effects model. Otherwise, the fixed-effects model was selected.[25] Sensitivity analyses were performed to test the reliability of results. Hardy–Weinberg equilibrium (HWE) was assessed using Fisher exact test and significance was defined as P < .05. Potential publication bias was measured by the funnel plot and funnel plot asymmetry was assessed by Egger test with significance set at P < .05. All statistical tests were performed with STATA 11.0 (StataCorp, College Station, Texas) and Review Manager 5.0.
3. Results
3.1. Eligible studies
The study flowchart is shown in Figure 1. Thirteen relevant articles were identified from our initial search and 6 articles met the inclusion criteria. In total, data were extracted from 5683 CAD patients and 6322 controls. The study subjects came from China, Japan and Iran, and were categorized into Chinese or non-Chinese Asians (Table 1). Several studies carried out subgroup analysis of different genetic ethnicities. Seven studies were excluded; among them, 3 papers reported data of Caucasians and 4 papers did not report genotype data. By quality assessment with NOS, 3 studies scored higher than or equal to 7 and were considered high quality and 2 studies scored 6and were close to the high-quality standard (Table S1).
Figure 1.
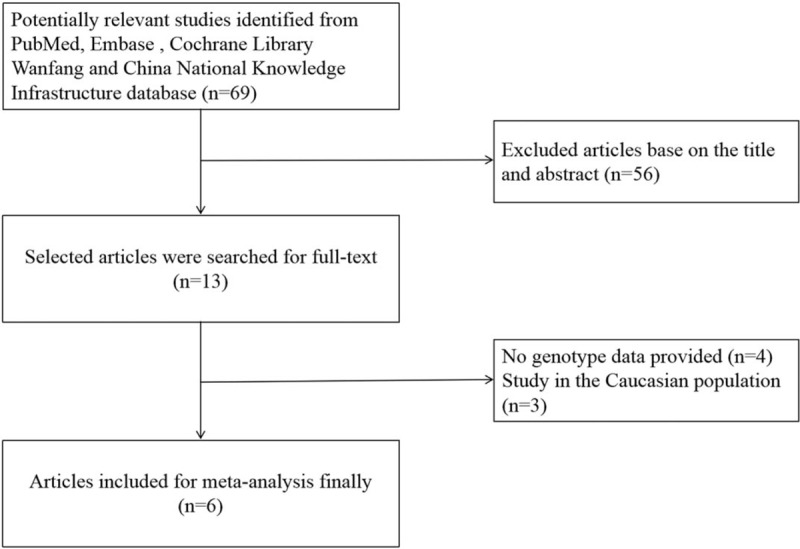
The study flowchart.
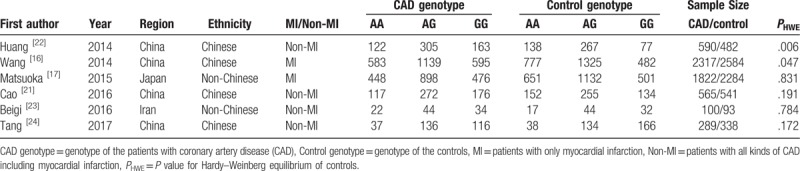
Table 1.
Characteristics of included studies on association between the ANRIL polymorphism rs4977574 and CAD.
3.2. Meta-analysis
Overall, significant associations were found between rs4977574 and CAD risk when all studies were pooled into the meta-analysis under the allelic model (OR = 1.18, 95% CI 1.04–1.34, P = .010), the dominant model (OR = 1.28, 95% CI 1.13–1.44, P < .0001), the recessive model (OR = 1.27, 95% CI 1.01–1.60, P = .04),the homozygous model (OR = 1.46, 95% CI 1.15–1.86, P = .002), and the heterozygous model (OR = 1.17, 95% CI 1.07–1.28, P = .0004) (Figs. 2 and 3). Under the dominant and heterozygous genetic model, no heterogeneity was detected in the whole population (Pheterogeneity = .18 and .67, respectively). In addition, the whole population displayed significant heterogeneity under another 3 genetic models (Pheterogeneity < .05). When Chinese subjects were analyzed as a subgroup, heterogeneity was still detected under the allelic, recessive and homogeneity models (Pheterogeneity < .05).In contrast, no heterogeneity was shown under all the genetic models when non-Chinese Asians were analyzed as a subgroup (Pheterogeneity < .05) (Figs. 2 and 3). For patients with MI as a subgroup, no heterogeneity was detected under all the genetic models (Pheterogeneity > .05). In contrast, no heterogeneity was demonstrated under only the dominant and heterozygous models when non-MI CAD patients were analyzed as a subgroup (Pheterogeneity > .05) (Supplementary Figure S1–5).
Figure 2.
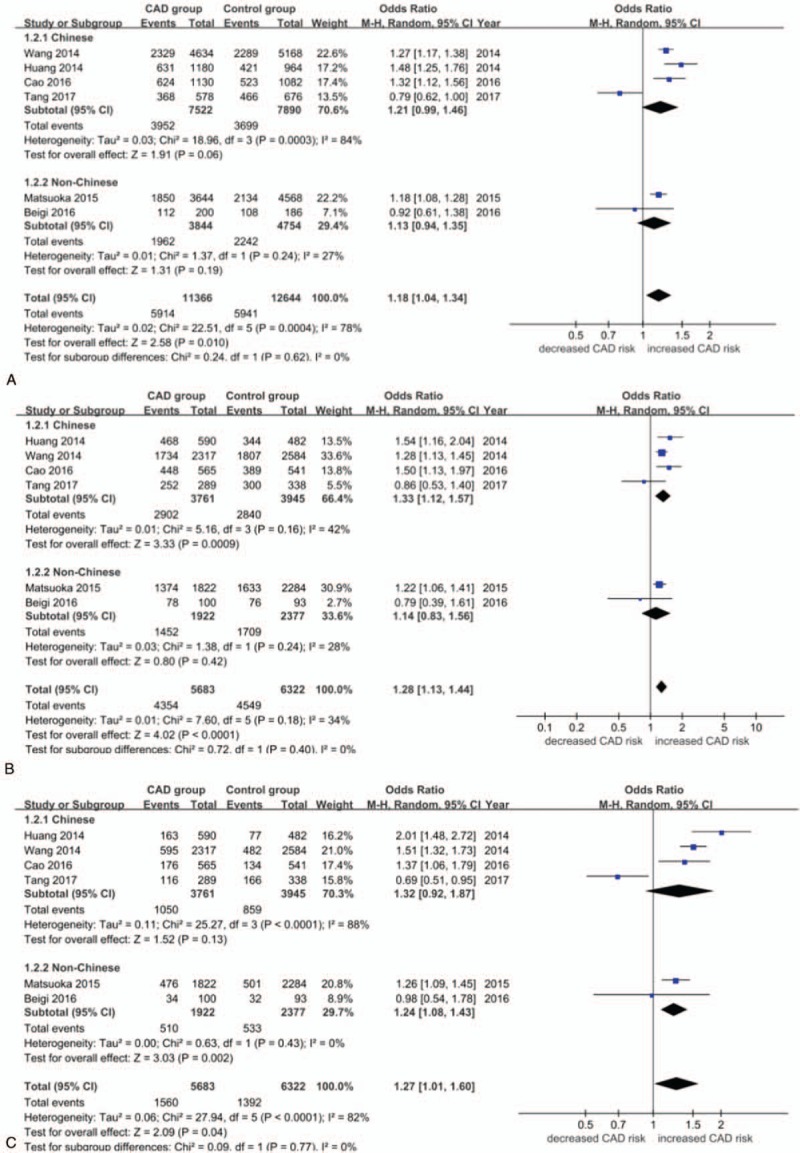
A. Forest plot (random effects model) for the association between ANRIL polymorphism rs4977574 and CAD stratified by ethnicity under the allelic genetic model (distribution of G allelic frequency of ANRIL gene polymorphism). G, guanine;CAD, coronary artery disease. B. Forest plot (random effects model) for the association between ANRIL polymorphism rs4977574 and CAD stratified by ethnicity under the dominant genetic model (GG+AG versus AA).A, adenine; G, guanine;CAD, coronary artery disease.C. Forest plot (random effects model) for the association between ANRIL polymorphism rs4977574 and CAD stratified by ethnicity under the recessive genetic model (GG versus AA+AG).A, adenine; G, guanine.CAD, coronary artery disease. CAD = coronary artery disease.
Figure 3.
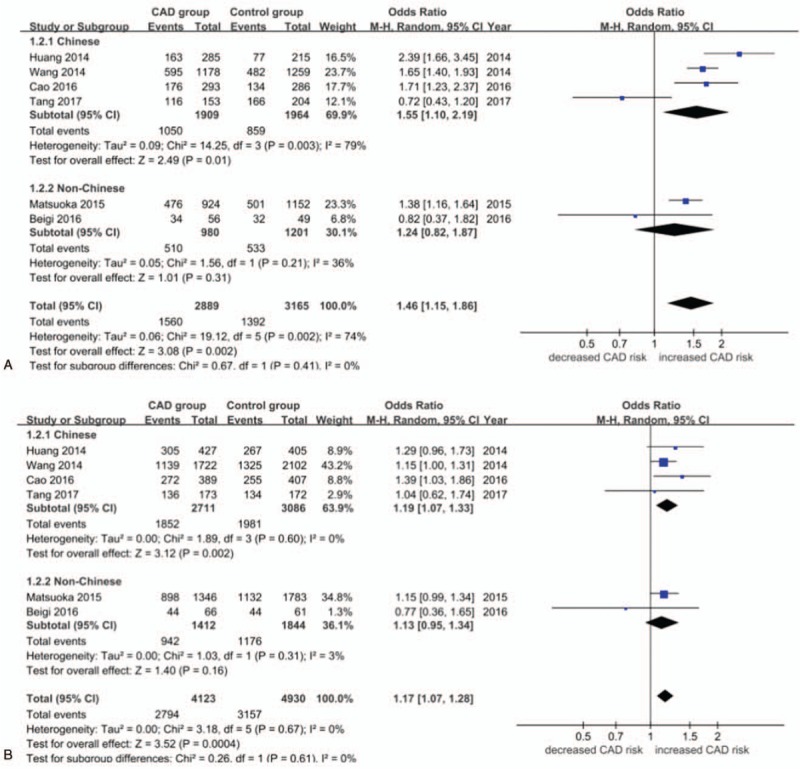
A. Forest plot (random effects model) for the association between ANRIL polymorphism rs4977574 and CAD stratified by ethnicity under the homozygous genetic model (GG versus AA).A, adenine; G, guanine; CAD, coronary artery disease. B. Forest plot (random effects model) for the association between ANRIL polymorphism rs4977574 and CAD stratified by ethnicity under the heterozygous genetic model (AG versus AA).A, adenine; G, guanine; CAD, coronary artery disease. CAD = coronary artery disease.
In the Chinese subgroup, a significant association between rs4977574 and CAD risk was found under the dominant model (OR = 1.33, 95% CI = 1.12–1.57, P = .0009), the homozygous model (OR = 1.55, 95% CI 1.10–2.19, P = .01), and the heterozygous model (OR = 1.19, 95% CI 1.07–1.33, P = .002). In the non-Chinese Asian subgroup, a significant association between rs4977574 and CAD risk was found under only the recessive model (OR = 1.24, 95% CI 1.08–1.43, P = .002) (Table S2).
In the MI subgroup, a significant association between rs4977574 and CAD risk was found under the allelic model (OR = 1.23, 95% CI 1.14–1.32, P < .00001), the recessive model (OR = 1.38, 95% CI 1.16–1.65, P = .0004), the dominant model (OR = 1.25, 95% CI 1.14–1.38, P < .00001), the homozygous model (OR = 1.51, 95% CI 1.27–1.80, P < .00001), and the heterozygous model (OR = 1.15, 95% CI 1.04–1.27, P = .006). In the non-MI CAD subgroup, a significant association between rs4977574 and CAD risk was found under only the heterozygous model (OR = 1.25, 95% CI 1.04–1.51, P = .02) (Table S3).
3.3. Sensitivity analysis and publication bias
Sensitivity analysis revealed that the significance of ORs was not materially changed by omitting any single study in the current meta-analysis. The shape of funnel plot and Egger test (P = .352) showed no evidence of obvious asymmetry under the allelic model (Fig. 4).
Figure 4.
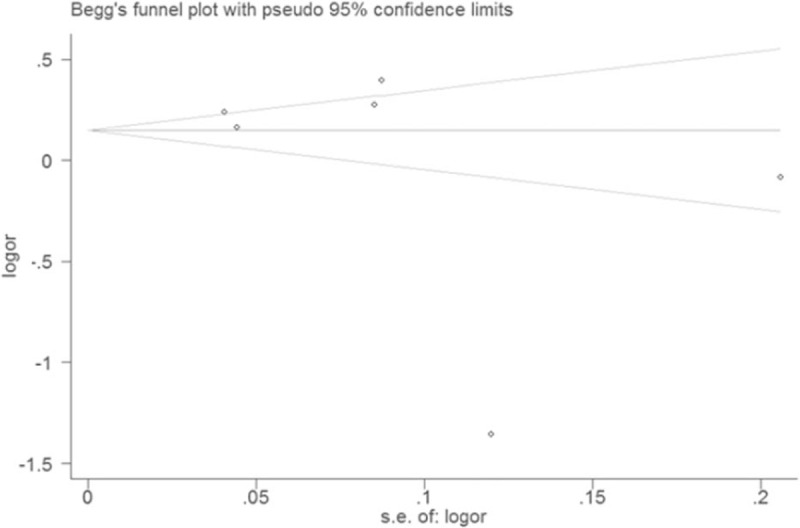
Funnel plot for eligible studies of association between ANRIL polymorphism rs4977574 and CAD under an allelic genetic model (distribution of G allelic frequency of ANRIL gene polymorphism). CAD = coronary artery disease, G = guanine, OR = odds ratio.
4. Discussion
Though several studies have demonstrated that rs4977574 of ANRIL is associated with increased CAD risk in different ethnicities, no meta-analysis of ANRIL rs4977574 polymorphism and CAD risk is available in Asian populations. Our current meta-analysis revealed significant correlation between rs4977574 and CAD risk under the allelic, recessive, dominant, homozygous, and heterozygous genetic models in the whole Asian population. When the Chinese subjects were analyzed as a subgroup, rs4977574 was significantly associated with CAD under the recessive, dominant, homozygous, and heterozygous genetic models. Moreover, the association strength grew higher with higher ORs under different models. When MI patients were analyzed as a subgroup, rs4977574 was also significantly associated with CAD under the allelic, recessive, dominant, homozygous, and heterozygous genetic models. In contrast, when non-MI CAD patients were analyzed as a subgroup, 4977574 was not significantly associated with CAD under the allelic, recessive, dominant, and homozygous genetic models, except for the heterozygous genetic model. The results suggested that ANRIL polymorphism rs4977574 with G-allele is associated with a higher CAD risk in Asians, especially in Asians of Chinese descent. Moreover, 4977574 with G-allele may be more evidently correlated with MI susceptibility when compared with other types of CAD.
Several common variants of ANRIL have been reported to associate with MI in European whites and Hispanic populations. The rs4977574 in ANRIL was first found to be associated with CAD in European and American-Caucasian population.[13,14]
Although Huang et al investigated the association between rs4977574 and CAD in a meta-analysis, the majority of the researches in their included studies were derived from subjects of European ancestry. Over the recent 3 years, an increasing number of GWASs were conducted in Asian countries, especially in China. So this meta-analysis was designed to summarize the latest findings from these studies. The results demonstrated a significant association between rs4977574 and CAD in Asians, which is consistent with previous studies in European countries. MI is the most severe entity of CAD with most severe atherosclerosis, which may lead to sudden death. Our meta-analysis proved that rs4977574 was associated with susceptibility of MI.
ANRIL, encoding a large antisense non–coding RNA, is adjacent to the CDKN2A-CDKN2B locus. CDKN2A-CDNK2B, coding for 2 cyclin-dependent kinase inhibitors, play an important role in regulation of the cell cycle, suggesting that these 2 genes may be involved in the pathogenesis of atherosclerosis. A functional analysis conducted by Jar Inova et al found risk allele within the 9p21.3 locus enhancer, which alters the transcriptional activity of ANRIL and then regulates cellular proliferation by interfering with adjacent protein-coding genes.[10] Congrains et al found abnormal expression of CDKN2A/B and reduced cell growth when ANRIL was knocked down in vascular smooth muscle cell.[26] So the genetic variants of ANRIL influence atherosclerosis process including thrombogenesis, vascular remodeling or repair, and plaque stability through altering ANRIL expression and modulating cell proliferation.[10,26]ANRIL changes the expression of these coding-related genes by multiple mechanisms including RNA interference, gene silencing, chromatin remodeling, or DNA methylation.[27] Besides, ANRIL is expressed in multiple cell types such as endothelial cells, smooth muscle cells, and inflammatory cells stimulated by atherosclerosis.[28]
The study also has several limitations. First, specific data such as genotype frequency were not obtained from some studies performed in Asian populations. In fact, the included studies in this meta-analysis only included 3 ethnicities, Chinese, Japanese and Iranians. They are not representative of all Asians. Chinese is also a broad category as it may include many minority groups apart from Han Chinese. The studies representative of minority groups is limited. Second, some included studies were not in accordance with HWE. Third, our search strategy limited publications to English and Chinese which may lead to some eligible studies being not taken into account.
5. Conclusion
We carried out this meta-analysis to identify the role of ANRIL polymorphism rs4977574 in the development of CAD in Asian population. More specifically, G allele may confer to a higher susceptibility of CAD, especially MI. It may be an effective prediction tool of CAD combined with environmental factors in the future. However, more functional studies are warranted for providing more evidence.
Author contributions
Conceptualization: Xiangjun Yang.
Data curation: Bing Xu, Zhen Fang, Shenghu He.
Formal analysis: Zhen Fang.
Funding acquisition: Xiangjun Yang.
Investigation: Bing Xu, Zhen Fang.
Methodology: Bing Xu.
Software: Shenghu He.
Supervision: Shenghu He, Xiangjun Yang.
Writing – original draft: Zhen Fang.
Writing – review & editing: Junhong Wang.
Supplementary Material
Footnotes
Abbreviations: CAD = coronary artery disease, CI = confidence interval, HWE = Hardy–Weinberg equilibrium, MI = myocardial infarction, OR = odds ratio.
BX and ZF contributed equally to this work.
This study was supported by grants from the National Natural Science Foundation of China (Grant no. 81770327), the Science and Technology Special Program for Clinical Medicine of Jiangsu Province (Grant no. BL2014050),
Jiangsu Province Commission of Health and Family Planning Research Project (Grant No.Z2017010), Subei People's Hospital Project (Grant No. yzucms201721).
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Lopez AD, Mathers CD, Ezzati M, et al. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet 2006;367:1747–57. [DOI] [PubMed] [Google Scholar]
- [2].Marenberg ME, Risch N, Berkman LF, et al. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med 1994;330:1041–6. [DOI] [PubMed] [Google Scholar]
- [3].Girelli D, Martinelli N, Peyvandi F, et al. Genetic architecture of coronary artery disease in the genome-wide era: implications for the emerging “golden dozen” loci. Semin Thromb Hemost 2009;35:671–82. [DOI] [PubMed] [Google Scholar]
- [4].McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science 2007;316:1488–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med 2007;357:443–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Willer CJ, Sanna S, Jackson AU, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 2008;40:161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Broadbent HM, Peden JF, Lorkowski S, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet 2008;17:806–14. [DOI] [PubMed] [Google Scholar]
- [8].Szpakowicz A, Pepinski W, Waszkiewicz E, et al. Polymorphism of 9p21.3 locus is associated with 5-year survival in high-risk patients with myocardial infarction. Plos One 2013;8:e72333. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [9].Rodriguez C, Borgel J, Court F, et al. CTCF is aDNA methylation-sensitive positive regulator of the INK/ARF locus. Biochem Biophys Res Commun 2010;392:129–34. [DOI] [PubMed] [Google Scholar]
- [10].Jarinova O, Stewart AF, Roberts R, et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol 2009;29:1671–7. [DOI] [PubMed] [Google Scholar]
- [11].Congrains A, Kamide K, Katsuya T, et al. CVD-associated non-coding RNA, ANRIL, modulates expression of atherogenic pathways in VSMC. Biochem Biophys Res Commun 2012;419:612–6. [DOI] [PubMed] [Google Scholar]
- [12].Schunkert S, König IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011;43:333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zheng Y, Li Y, Huang T, et al. Sugar-sweetened beverage intake, chromosome 9p21 variants, and risk of myocardial infarction in Hispanics. Am J Clin Nutr 2016;103:1179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Saade S, Cazier JB, Ghassibe-Sabbagh M, et al. Large scale association analysis identifies three susceptibility loci for coronary artery disease. PloS One 2011;6:e29427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].AbdulAzeez S, Al-Nafie AN, Al-Shehri A, et al. Intronic polymorphisms in the CDKN2B-AS1 gene are strongly associated with the risk of myocardial infarction and coronary artery disease in the saudi population. Int J Mol Sci 2016;17:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang Y, Wang L, Liu X, et al. Genetic variants associated with myocardial infarction and the risk factors in Chinese population. PloS One 2014;9:e86332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Matsuoka R, Abe S, Tokoro F, et al. Association of six genetic variants with myocardial infarction. Int J Mol Med 2015;35:1451–9. [DOI] [PubMed] [Google Scholar]
- [18].Hamrefors V, Hedblad B, Hindy G, et al. Smoking modifies the associated increased risk of future cardiovascular disease by genetic variation on chromosome 9p21. PloS one 2014;9:e85893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Qi L, Parast L, Cai T, et al. Genetic susceptibility to coronary heart disease in type 2 diabetes: 3 independent studies. J Am Coll Cardiol 2011;58:2675–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lee JY, Lee BS, Shin DJ, et al. A genome-wide association study of a coronary artery disease risk variant. J Hum Genet 2013;58:120–6. [DOI] [PubMed] [Google Scholar]
- [21].Cao XL, Yin RX, Huang F, et al. Chromosome 9p21 and ABCA1 genetic variants and their interactions on coronary heart disease and ischemic stroke in a Chinese Han population. Int J Mol Sci 2016;17:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huang Y, Ye H, Hong Q, et al. Association of CDKN2BAS polymorphism rs4977574 with coronary heart disease: a case-control study and a meta-analysis. Int J Mol Sci 2014;15:17478–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Beigi HSS, Ghaderian SMH, Doosti A. Investigation of the association between rs4977574 A >G polymorphismin ANRIL gene and coronaryartery disease in Iranian population. Cardiovascular 2015;9:139–44. [Google Scholar]
- [24].Tang O, Lv J, Cheng Y, et al. The correlation between 9p21 chromosome rs4977574 polymorphism genotypes and the development of coronaryartery heart disease. Cardiovasc Toxicol 2016;17:1–5. [DOI] [PubMed] [Google Scholar]
- [25].Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539–58. [DOI] [PubMed] [Google Scholar]
- [26].Congrains A, Kamide K, Oguro R, et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis 2012;220:449–55. [DOI] [PubMed] [Google Scholar]
- [27].Amaral PP, Dinger ME, Mercer TR, et al. The eukaryotic genome as an RNA machine. Science 2008;319:1787–9. [DOI] [PubMed] [Google Scholar]
- [28].Shanker J, Arvind P, Jambunathan S, et al. Genetic analysis of the 9p21.3 CAD risk locus in Asian Indians. Thromb Haemost 2014;111:960–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


