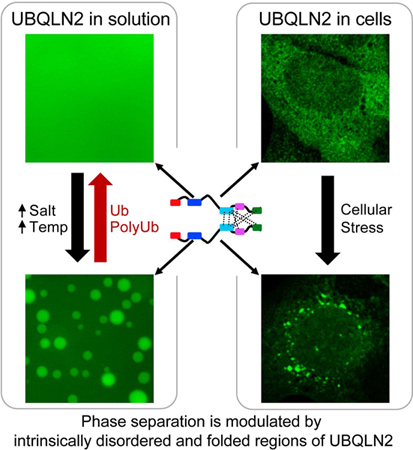
Summary
Under stress, certain eukaryotic proteins and RNA assemble to form membraneless organelles known as stress granules. The most well-studied stress granule components are RNA-binding proteins that undergo liquid-liquid phase separation (LLPS) into protein-rich droplets mediated by intrinsically-disordered low-complexity domains (LCDs). Here we show that stress granules include proteasomal shuttle factor UBQLN2, a LCD-containing protein structurally and functionally distinct from RNA-binding proteins. In vitro, UBQLN2 exhibits LLPS at physiological conditions. Deletion studies correlate oligomerization with UBQLN2’s ability to phase separate and form stress-induced cytoplasmic puncta in cells. Using NMR spectroscopy, we mapped weak, multivalent interactions that promote UBQLN2 oligomerization and LLPS. Ubiquitin or polyubiquitin binding, obligatory for UBQLN2’s biological functions, eliminates UBQLN2 LLPS, thus serving as a “switch” between droplet and disperse phases. We postulate that UBQLN2 LLPS enables its recruitment to stress granules where its interactions with ubiquitinated substrates reverse LLPS to enable shuttling of clients out of stress granules.
Keywords: Ubiquilin-2, stress granules, multivalent interactions, ubiquitin, protein quality control, liquid- liquid phase separation (LLPS), NMR spectroscopy, ligand-induced phase transition, amyotrophic lateral sclerosis (ALS)
eTOC Blurb
Ubiquilins (UBQLNs) are important shuttle proteins for cellular protein quality control machinery. In this issue of Molecular Cell, Dao et al. show that UBQLN2 colocalizes with stress granules in vivo and undergoes liquid-liquid phase separation at physiological conditions in vitro. Ubiquitin binding induces a transition that reverses UBQLN2 phase separation.
Graphical Abstract
Introduction
Liquid-liquid phase separation (LLPS) may promote the assembly of membraneless organelles and formation of cytoplasmic inclusions in neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) (Mitrea and Kriwacki, 2016; Protter and Parker, 2016; Taylor et al., 2016). LLPS is driven by multivalent, weak interactions involving globular, folded protein domains (Banani et al., 2016; Li et al., 2012), low-complexity, intrinsically-disordered regions (Uversky, 2016) and/or scaffolding DNA or RNA molecules (Aumiller et al., 2016). ALS-linked RNA-binding proteins (RBPs) with low-complexity, disordered regions (e.g., hnRNPA1, FUS, TDP-43) undergo LLPS into protein-containing droplets in solution (Conicella et al., 2016; Molliex et al., 2015; Patel et al., 2015). Inside cells, LLPS is linked to the formation of dynamic membraneless compartments, including stress granules (SGs), P bodies, and nucleoli (Brangwynne et al., 2009; Hyman et al., 2014; Protter and Parker, 2016). Many ALS-linked RBPs localize to SGs, which form when translation initiation is limited as part of a protective mechanism to sequester proteins and mRNA. Under normal conditions, SGs dissipate once the stress condition is removed. Importantly, disease mutations in RBPs disrupt SG assembly and disassembly and/or promote aggregates (Kim et al., 2013; Molliex et al., 2015).
SGs are heterogenous in composition and can contain not only RBPs but also protein quality control (PQC) components, including HSP70 chaperone proteins, valosin-containing protein (VCP), and ubiquitin (Ub) (Buchan et al., 2013; Jain et al., 2016; Kwon et al., 2007; Walters and Parker, 2015). SGs are dynamic signaling compartments (Mahboubi and Stochaj, 2017). Therefore, disruption of SG dynamics can lead to disease states. Defects in several PQC mechanisms involving VCP, or autophagy, impair proper SG assembly (Buchan et al., 2013; Ramaswami et al., 2013; Seguin et al., 2014).
Ubiquilins (UBQLNs) are adaptor proteins involved in PQC mechanisms including proteasomal degradation and autophagy, as well as stress response (Kleijnen et al., 2000; Mah et al., 2000; N’Diaye et al., 2009; Rothenberg et al., 2010; Yun Lee et al., 2013). Humans have four known paralogs of UBQLN (−1, −2, −3 and −4). Traditionally considered proteasomal shuttle proteins, UBQLNs contain an N-terminal ubiquitin-like domain (UBL) that interacts with proteasomal subunits, and a C-terminal ubiquitin-associating domain (UBA) that interacts with Ub and polyubiquitin chains. Both domains have been well characterized structurally and functionally (Walters et al., 2004; Zhang et al., 2008). In their central regions, UBQLNs contain multiple STI1-like domains that bind chaperone proteins (HSP70), autophagy components (LC3), and ubiquitinated substrates (Hjerpe et al., 2016; Kaye et al., 2000; Kurlawala et al., 2017; Yun Lee et al., 2013). The central region is also implicated in UBQLN dimerization, leading to formation of homodimers (e.g. UBQLN2-UBQLN2) or heterodimers (e.g. UBQLN1- UBQLN2) (Ford and Monteiro, 2006; Itakura et al., 2016). UBQLNs are implicated in many neurodegenerative disorders, and UBQLN-containing inclusions have been identified in ALS, Parkinson’s, Alzheimer’s, and Huntington’s diseases (Deng et al., 2011; Mori et al., 2012; Rutherford et al., 2013). UBQLN2 is found in cytoplasmic inclusions of ALS patients and mouse models post-mortem (Deng et al., 2011; Le et al., 2016). UBQLN2 also interacts with the ALS- linked SG components hnRNPA1 and TDP-43 and may regulate their expression levels (Deng et al., 2011; Gilpin et al., 2015).
UBQLN2 is expressed in various human tissues, with highest expression levels in the nervous system (Wu et al., 1999). Notably, UBQLN2 contains a unique proline-rich (Pxx) segment containing most familial ALS missense mutations, including residues P497, P506, P509, P525, among others (Deng et al., 2011). These mutations interfere with UBQLN2 trafficking of ubiquitinated substrates to the proteasome (Chang and Monteiro, 2015; Deng et al., 2011; Xia et al., 2014) and disrupt interactions with HSP70 (Hjerpe et al., 2016; Teyssou et al., 2017). However, no structural information exists on this central region (residues 100–570), and the disease-causing mechanisms of the mutations in the Pxx region remain unknown.
Here, we show that UBQLN2 colocalizes with SGs under different cellular stress conditions in vivo, and undergoes LLPS in vitro. Using biophysical techniques including NMR, we demonstrate that oligomerization mediated by the STI1-II, Pxx and UBA domains is a pre- requisite to promote UBQLN2 LLPS. Importantly, we show that UBQLN2 LLPS is eliminated entirely by non-covalent Ub or polyubiquitin (polyUb) binding. We propose a molecular model for how Ub disrupts multivalent interactions important for UBQLN2 LLPS. Identification of Ub as a modulator of LLPS behavior suggests that interaction between polyUb-tagged substrates and UBQLN2 may also disrupt LLPS, and enable UBQLN2-mediated trafficking of ubiquitinated substrates from SGs or other membraneless organelles to PQC systems, including the proteasome.
Results
UBQLN2 Is Recruited to Stress Granules
SGs form in response to proteotoxic stress and ubiquitinated species accrue here (Seguin et al., 2014). Moreover, UBQLNs, which have important roles in PQC, can also mediate cellular stress responses (Ko et al., 2002; Lim et al., 2009). To test if UBQLN2 is recruited to SGs, we investigated UBQLN2 distribution in response to stress. In unstressed U2OS cells, endogenous UBQLN2 showed diffuse staining in the cytoplasm. However, in response to four stressors (arsenite [oxidative stress], heat shock, puromycin [translation inhibition], and sorbitol [osmotic stress]), UBQLN2 accumulated in cytoplasmic puncta positive for SG marker eIF4G1 (Figure 1A). Quantitative assessment showed that nearly all SGs contained UBQLN2 (Figure 1B). These observations on endogenous UBQLN2 were recapitulated in HeLa cells (data not shown).
Figure 1. UBQLN2 is recruited to SGs and undergoes LLPS.
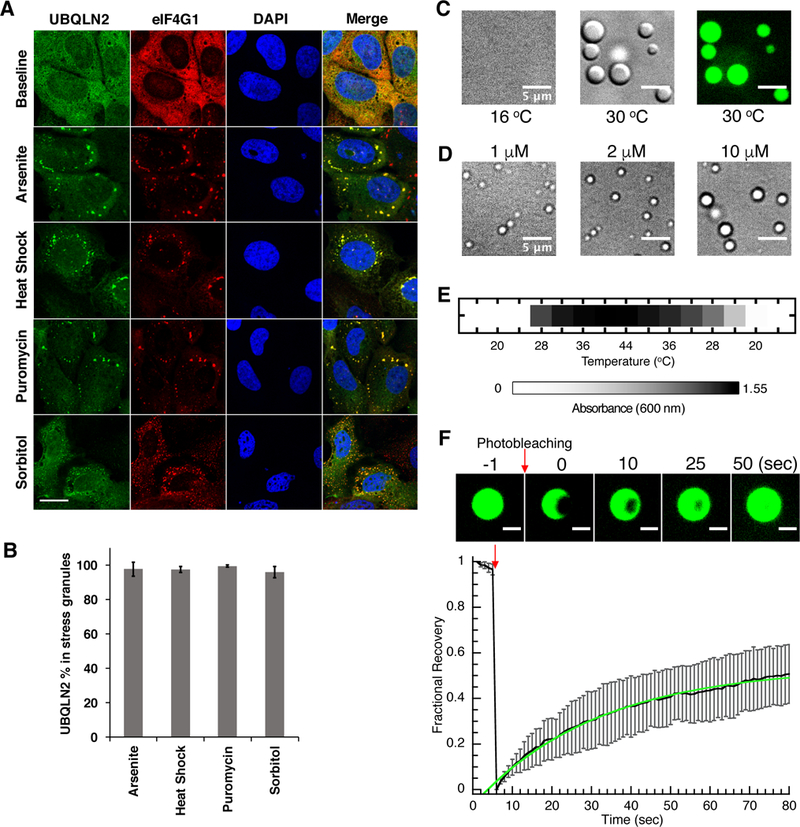
(A) Immunostaining for endogenous UBQLN2 in U2OS cells shows that UBQLN2 is diffuse in cytoplasm, but form puncta under four stress conditions tested. UBQLN2 colocalizes with eIF4G1, a SG marker. DAPI is used to stain nuclei. (B) Quantitation of UBQLN2 colocalization in SGs with error bars reflecting standard deviation from data in triplicate. (C) DIC and fluorescence microscopy shows 50 μM protein in 20 mM NaPhosphate, 200 mM NaCl, pH 6.8 phase separating into micron-sized droplets at 30oC but not at 16oC. (D) DIC microscopy shows UBQLN2 LLPS at physiological protein concentrations at 37oC in pH 7.4 buffer consisting of 150 mM KCl, 20 mM NaPhosphate, 1 mM DTT and 150 mg/mL Ficoll. (E) UBQLN2 LLPS is observed by measuring A600 as a function of temperature. (F) FRAP of UBQLN2 droplets. (Upper) Fluorescence images of partial droplet photobleaching experiments. (Lower) Black curve is an average of FRAP recovery curves from 6 separate droplets. Error bars represent the standard deviation. Green curve is a single exponential fit to the data. See also Figure S2.
UBQLN2 Undergoes LLPS at Physiological Conditions
Many SG proteins undergo LLPS in vitro, a process mediated by multivalent interactions involving low-complexity, intrinsically disordered regions and/or folded domains. Notably, UBQLN2 contains low-complexity segments predicted to be disordered (Figure S1). To test if UBQLN2 phase separates, we expressed and purified full-length (FL) UBQLN2. UBQLN2 solutions became turbid at physiological temperatures (37°C) and salt concentrations (200 mM NaCl, 20 mM sodium phosphate, pH 6.8), but clarified when cooled to <16°C. Using differential interference contrast (DIC) microscopy, we determined that turbidity results from the presence of micron-sized droplets rich in UBQLN2, as demonstrated by fluorescent imaging using a DyLight-488 fluorophore conjugated to UBQLN2 (Figure 1C). Imaging revealed that these droplets were dynamic: droplets fused with each other, creating larger droplets as they wetted the surface of the glass well over time (Movie S1). To mimic cellular environment, we monitored droplet formation at 37oC in pH 7.4 buffer consisting of 150 mM KCl, 20 mM NaPhosphate, 1 mM DTT and 150 mg/mL Ficoll (as molecular crowding agent). We observed droplets at 1 μM UBQLN2, within the estimated intracellular concentration of UBQLN2 of 1.25 μM in U2OS cells (Figure 1D and Methods), reinforcing the connection between our in vitro and in vivo observations of LLPS and SGs, respectively.
To quantify UBQLN2 LLPS, we used a spectrophotometric assay as a proxy for droplet formation (Figure 1E). We found that absorbance (A600) values increased with increasing temperatures. LLPS was generally reversible, as the A600 values for the same sample decreased when temperature was lowered. Droplets were observed at physiological temperature and ionic strength (200 mM NaCl). UBQLN2 also phase separated more readily with increasing protein and NaCl concentrations (Figure S2A).
To ascertain the mobility of UBQLN2 molecules inside these droplets, we monitored fluorescence recovery after photobleaching (FRAP) of droplets (Figure 1F). After photobleaching a small portion of a droplet, its fluorescence signal recovered relatively quickly, with a characteristic recovery time of 31.5 seconds, within the range observed for many RBPs (Lin et al., 2015). These data are consistent with the observation that UBQLN2 droplets are dynamic.
Mapping the Sequence Determinants of UBQLN2 LLPS
We next aimed to identify the domain(s) responsible for mediating UBQLN2 LLPS. We created UBQLN2 deletion constructs in which the STI1-II (residues 379–486), Pxx (residues 487–538), or UBA (residues 577–624) domains were removed (Figure 2B). Based on observations made for the deletion constructs as described below, we constructed C-terminal constructs 379–624 (which contained the entire C-terminal STI1-II region, Pxx and UBA domains) and 487–624 (which contained only the Pxx and UBA domains). We also created 450– 624 since residues 450–490 are predicted to be ordered (Figure S1).
Figure 2. UBLQN2 LLPS is modulated by its different domains, oligomerization states, temperature, protein and salt concentrations.
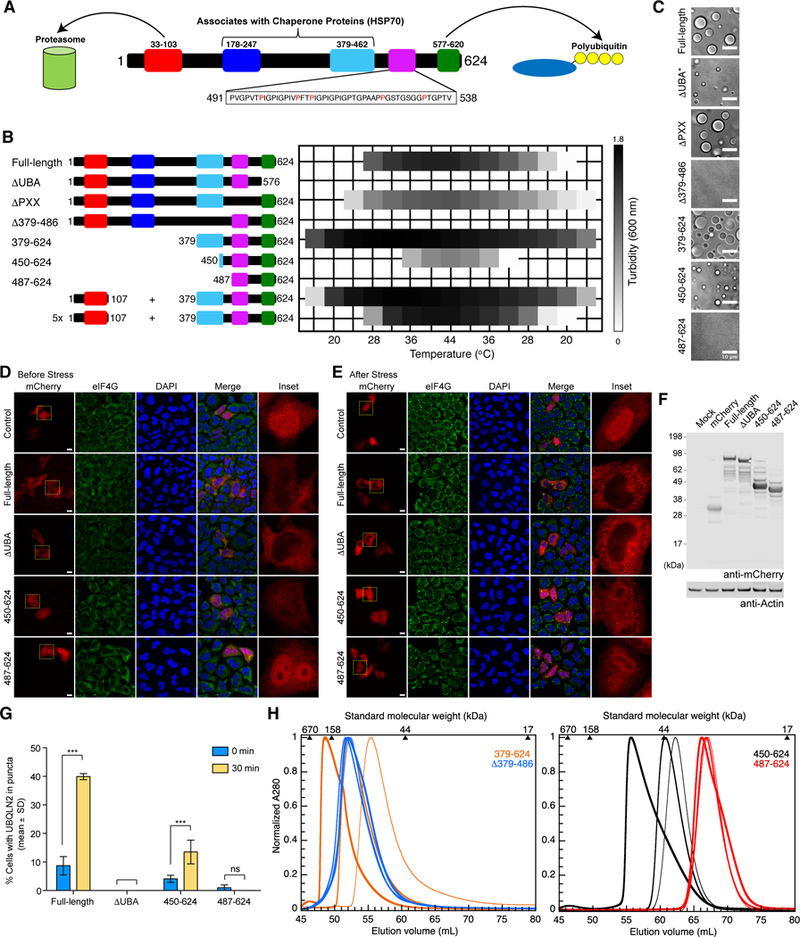
(A) Domain architecture of UBQLN2. The proline-rich repeat (Pxx) region of UBQLN2 harbors most familial ALS mutations (red). (B) Turbidity assays as a function of temperature comparing LLPS of different UBQLN2 constructs using 50 µM protein in 20 mM NaPhosphate, 200 mM NaCl, pH 6.8. The last two assays monitored the turbidity of solution consisting either 50 or 250 µM of the UBL domain (1–107) and 50 µM of UBQLN2 379–624. (C) DIC microscopy shows solutions of 50 μM constructs in 20 mM NaPhosphate, 200 mM NaCl, pH 6.8 after incubation at 37oC for 10 minutes. *ΔUBA microscopy was obtained at 500 mM NaCl since no droplets were observed at 200 mM NaCl. (D,E) HeLa cells were transfected with mCherry or mCherry-tagged UBQLN2 as indicated. Twenty-four hours post-transfection, cells were stimulated with 0.5 mM sodium arsenite for 30 min, and immunostained with anti-eIF4G and DAPI. Arrows indicate UBQLN2-positive puncta. Cells at pre- (D) and 30 min post-arsenite treatment (E) are shown. Scale bar: 10 µm. (F) Western blot analysis of mCherry-tagged UBQLN2 constructs shows comparable expression. Actin was blotted as a loading control. (G) Quantification of (D) and (E). The percentage of transfected cells with UBQLN2-positive puncta is plotted. ***p<0.001 two- way ANOVA, Sidak’s multiple comparisons test, n=3 biological repeats. Error bars reflect standard deviation. (H) SEC of UBQLN2 Δ379–486 (blue), 379–624 (orange), 450–624 (black) and 487–624 (red) at 10 µM (thinnest line), 100 µM (medium-thick), and 500 µM (thickest) protein concentrations. For UBQLN2 Δ379–486 construct, highest concentration used was 200µM. See also Figures S1, S2, S3 and S4.
We conducted spectrophotometric assays for all constructs under identical protein concentration and solution conditions (Figure 2B). To verify that any changes to A600 reading indeed result from LLPS and not other events such as protein aggregation or fibril formation, we also performed DIC microscopy (Figure 2C). UBQLN2 ∆UBA did not phase separate under these conditions, but did so at significantly higher salt and protein concentrations. More striking was the lack of LLPS for constructs ∆379–486 and 487–624 under all conditions tested, supporting a role for the STI1-II domain in mediating LLPS. UBQLN2 450–624, which contains a portion of the STI1-II domain, did undergo LLPS, albeit to a reduced degree, suggesting that this construct represents the minimum length required for LLPS. Supporting this idea, UBQLN2 450–624 did show enhanced LLPS with increasing protein and salt concentrations (Figure S2B). Furthermore, UBQLN2 450–624 and FL proteins colocalized in the same droplets (Figure S3A and Movie S2), suggesting similar or identical mechanism for LLPS of these constructs.
To test if our phase separation observations could be recapitulated in vivo, we transiently transfected four mCherry-labeled UBQLN2 constructs (FL UBQLN2, ∆UBA, 450–624, 487–624) into HeLa cells. Exogenously-expressed mCherry-UBQLN2 was diffuse in the cytoplasm (Figure 2D). When cells were exposed to arsensite stress, only FL UBQLN2 and UBQLN2 450–624 formed cytoplasmic puncta (Figure 2E). Moreover, even at slightly higher expression level than FL UBQLN2 (Figure 2F), UBQLN2 450–624 formed less puncta (Figures 2E, 2G), consistent with its reduced propensity to undergo LLPS in vitro. We noted that a portion of exogenously-introduced UBQLN2 colocalized with SG markers, whereas some exogenous UBQLN2 formed puncta that did not colocalize with SG marker. We surmise that UBQLN2 puncta formation and SG colocalization are highly sensitive to UBQLN2 expression levels, as others have reported UBQLN2 puncta when UBQLN2 was overexpressed, even in the absence of added cellular stress (Deng et al., 2011; Hjerpe et al., 2016).
Other deletion constructs showed modified LLPS behavior. In comparison to FL UBQLN2, removal of the Pxx region reduced turbidity, whereas LLPS of UBQLN2 379–624 occurred more readily. One major difference between FL and 379–624 constructs is the absence of the UBL domain in the latter. The UBL and UBA domains interact weakly (with a Kd of ~ 175 μM) (Nguyen et al., 2017). Removing the UBA-UBL interactions in UBQLN2 379–624 could enable the UBA domain to fully participate in promoting LLPS. Indeed, when we added the UBL domain in trans to UBQLN2 379–624, we observed reduction of LLPS (Figure 2B). In the presence of 5 times molar excess of UBL, UBQLN2 379–624 phase separated to a similar extent as FL UBQLN2. The UBL and UBA domains, as parts of the same molecule, can interact much more easily in FL UBQLN2 which exists as a dimer (Hjerpe et al., 2016).
Taken together, these data demonstrate that the STI1-II, Pxx, as well as the folded UBL and UBA domains all contribute to and modulate UBQLN2 LLPS. Specifically, we found that the STI1-II domain is necessary to drive LLPS, and that the UBA domain modulates STI1-II- mediated LLPS, as deletion of the UBA domain or binding of the UBL to the UBA domain substantially reduced the propensity of UBQLN2 to phase separate.
Oligomerization of UBQLN2 May Promote LLPS
Recent work showed that UBQLN1 dimerizes via the C-terminal portion of its STI1-II (Kurlawala et al., 2017). To examine the role of the STI1-II domain of UBQLN2 in oligomerization, we performed size-exclusion chromatography (SEC) of our deletion constructs under non-phase separating conditions (Figure 2H). UBQLN2 487–624 eluted at a volume consistent with a monomeric protein with very little concentration dependence between 10 µM and 500 µM, as confirmed by small angle neutron scattering (Figure S4, Table S1) and NMR (see below). In contrast, UBQLN2 450–624 eluted earlier with increasing protein concentration, consistent with concentration-dependent oligomerization. This effect was also evident for UBQLN2 379–624, which contains the full STI1-II region, but not for UBQLN2 ∆379–486. These results suggest that residues 379–486, which contain the STI1-II domain, mediate UBQLN2 oligomerization behavior. Furthermore, since the STI1-II domain is required for UBQLN2 LLPS, these findings suggest that oligomerization is a prerequisite for LLPS.
NMR Reveals That UBQLN2 C-Terminus is Mostly Disordered
We next sought to achieve an atomic-level understanding of UBQLN2 LLPS using NMR. For these experiments, we chose UBQLN2 450–624 (Figure 3) and UBQLN2 487–624 (Figure S5) since the former phase separates whereas the latter does not. Moreover, the molecular weights of these proteins are ideal for NMR (e.g. 450–624 is 18 kDa). The use of a larger C- terminal construct (e.g., UBQLN2 379–624) was not feasible given that only a portion of resonances in this protein were visible, likely due to its large size (Figure 2H, Figures S6B, and S6C). To justify the use of UBQLN2 450–624 for structural studies, we overlaid 1H-15N HSQC NMR spectra of FL and 450–624 constructs to show that many peaks superposed, indicating similar chemical and physical environments for many residues (Figure S3B).
Figure 3. NMR spectra for UBQLN2 450–624.
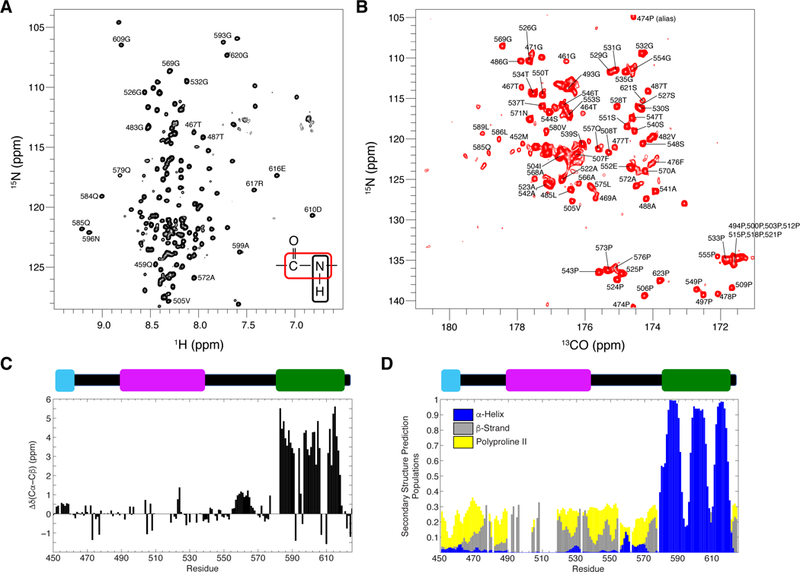
(A) 1H-15N TROSY-HSQC and (B) 15N-13CO (HACA)CON spectra of UBQLN2 450–624 at 10oC, pH 6.8, 20 mM NaPhosphate. (C) Residue-level secondary structure determination using Cα and Cβ secondary chemical shifts at 10oC. (D) Secondary structure prediction from δ2D calculations using all backbone chemical shift data. See also Figure S3 and S5.
1H-15N spectra of UBQLN2 450–624 revealed a concentration of amide resonances between 8.0 – 8.5 ppm in 1H dimension, consistent with a largely disordered construct (Figure 3A). The 15N-13C CON spectra reveal proline residues not visible in 1H-15N spectra as prolines do not contain amide 1H resonances (Figure 3B). After assigning the resonances (see Methods), we analyzed the secondary structure content of UBQLN2 450–624 on a residue-by- residue level using 13C chemical shifts for Cα and Cβ resonances (Figure 3C). The deviation of Cα and Cβ chemical shifts from random coil values provides a robust measurement of secondary structure propensity, with positive and negative values representing α-helical and β- strand population, respectively (Wishart et al., 1992). These data demonstrate that much of UBQLN2 450–624 is disordered. However, the UBA domain exhibits significant α-helical propensity, consistent with its known structure. Residues 450–460, located in the STI1-II domain, exhibit slight α-helical propensity and are predicted to be somewhat ordered according to PONDR-FIT calculations (Figure S1A). Noticeably, although predicted to be disordered, residues 550–570, consisting of hydrophobic and polar residues, exhibit some α-helical propensity, which is also observed in the shorter UBQLN2 487–624 construct (Figure S5C). Using a combination of backbone amide, Cα, Cβ, and CO chemical shifts, we predicted secondary structure population using the δ2D algorithm (Figure 3D) (Camilloni et al., 2012). These calculations confirmed the UBA domain to be α-helical. The remaining resonances are generally random-coiled, but have some β-strand or polyproline II propensity. The latter is consistent with the high proline content in this region of UBQLN2.
Multivalent Interaction Sites Exist Among STI1-II, Pxx, and UBA Domains
We next used NMR to characterize the interactions that drive UBQLN2 LLPS. Since UBQLN2 487–624 did not phase separate, whereas UBQLN2 450–624 did, we compared NMR spectra for both of these constructs under identical non-phase separating conditions (200 µM protein, pH 6.8, 25°C) (Figure 4A). Several well-de fined peaks in UBQLN2 487–624 were attenuated or broadened beyond detection in UBQLN2 450–624, specifically residues 505–508 of the Pxx region, 559–571 of the putative α-helical region, as well as 592–594 and 616–619 of the UBA domain. These observations indicated that residues 450–486 may interact with other sequence-distant parts of the protein, including the Pxx region and areas immediately upstream of, and including the UBA domain.
Figure 4. NMR identifies putative multivalent interactions in UBQLN2 450–624.
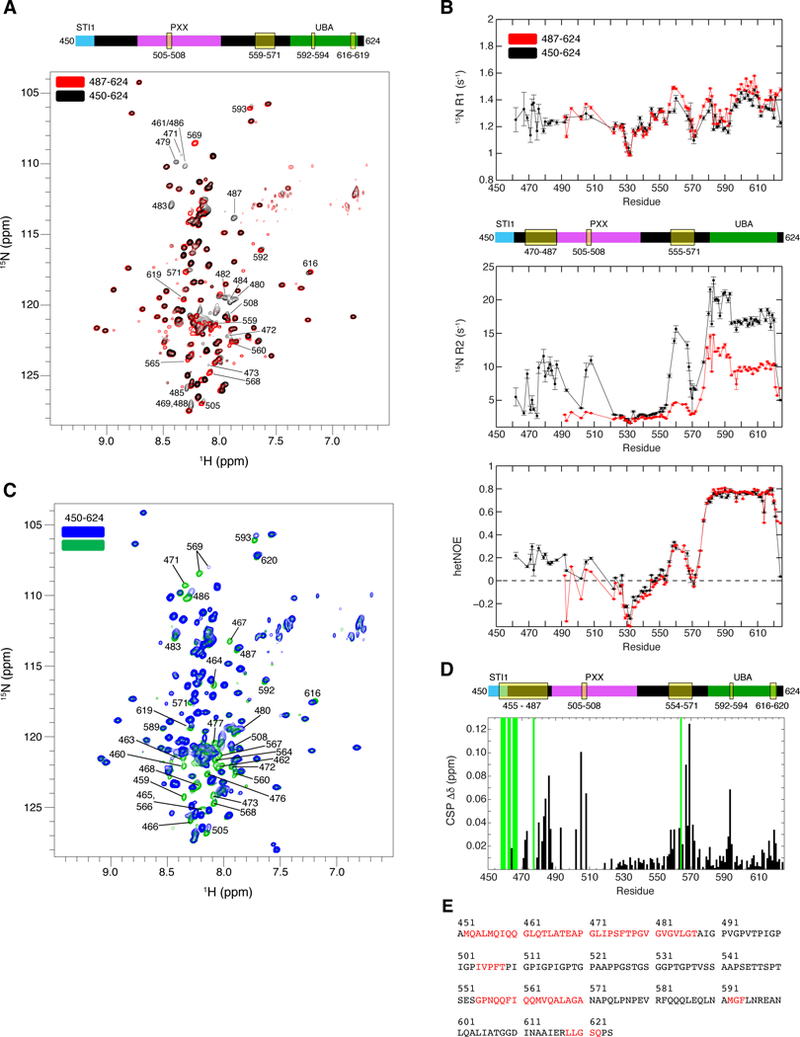
(A) Comparison of 1H-15N TROSY-HSQC spectra of 200 µM UBQLN2 450–624 and UBQLN2 487–624. Significant broadening was observed for residues identified in domain map above spectrum. (B) 15N R1 and R2 relaxation rates, and {1H}−15N hetNOE values for UBQLN2 450–624. Error bars represent standard errors of the experimental values. (C) Comparison of 1H-15N TROSY-HSQC spectra of UBQLN2 450–624 at 45 µM (green) and 600 µM (blue) protein concentrations. Contours are identical although 45 µM sample was collected with 4x scans as 600 µM sample. (D) CSPs represent residue-specific chemical shift differences between low (45µM) and high (600 µM) protein concentrations. Green bars mark resonances only visible at 45µM. Domain map marks residues that exhibit concentration-dependent peak broadening or significant CSPs. All spectra were collected at 25oC in pH 6.8 buffer under non-LLPS conditions. (E) UBQLN2 sequence whose amide resonances (red) exhibit elevated 15N R2 rates or concentration-dependent broadening or CSPs. See also Figure S6.
To quantitate 15N backbone dynamics of UBQLN2 on a residue-by-residue basis, we used standard R1, R2, and hetNOE experiments. R1 and R2 relaxation rates monitor backbone dynamics on ps-ns and µs-ms timescales, while hetNOE values reflect fast ps-ns dynamics. We performed these experiments on both UBQLN2 450–624 and 487–624 constructs (Figure 4B). HetNOE values for residues 450–580 are all below 0.3, indicative of significant ps-ns dynamics expected for intrinsically-disordered segments, particularly for residues 520–550 in both UBQLN2 450–624 and UBQLN2 487–624 constructs. Backbone R2 relaxation rates for UBQLN2 487–624 reinforced the notion that residues 487–580 were generally intrinsically disordered; R2 rates were 2–4 s-1, similar to the intrinsically disordered region in FUS in the non-phase separated state (Burke et al., 2015). In contrast, R1, R2 and hetNOE rates for UBA resonances in both UBQLN2 450–624 and UBQLN2 487–624 were consistent with well-ordered structure (Figure 4B, Table S2).
Peak broadening in NMR spectra of UBQLN2 450–624 indicated backbone dynamics occurring on an NMR millisecond timescale (Figure 4A). In UBQLN2 450–624, we noticed elevated R2 relaxation rates for residues 470–487, 505–508 in the Pxx region, and 555–570 in the putative α-helix upstream of the UBA domain (Figure 4B); these elevated R2 rates were not present in UBQLN2 487–624. Therefore, we hypothesized that these observations were related to the propensity for UBQLN2 450–624 to oligomerize at higher protein concentration (Figure 2H). To test for concentration-dependent effects, we collected NMR spectra of UBQLN2 450– 624 at concentrations between 45 µM and 600 µM (Figure 4C) under non-phase separating conditions. We observed that peaks corresponding to residues 450–470 were only visible at low concentration. A detailed residue-by-residue map between the chemical shifts at low and high protein concentration revealed that many peaks exhibited chemical shift perturbations (CSPs), specifically for residues 470–487, 505–508, 554–571, 592–594, and 616–620 (Figures 4D, 4E, Figure S6A). These highlighted regions correlate with the spectral observations made when comparing UBQLN2 450–624 and 487–624, and elevated R2 relaxation rates (Figure 4A, 4B). We also collected and analyzed NMR spectra of UBQLN2 379–624 and observed that resonances corresponding to residues 379–480, 505–508, and 550–570 were generally broadened beyond detection or exhibited significant CSPs (Figures S6B and S6C). Taken together, our data suggest that increases in protein concentration perturb multiple resonances across UBQLN2.
In summary, our NMR data, with corroboration by LLPS and SEC, strongly suggest that oligomerization mediated by residues in the STI1-II domain promotes UBQLN2 LLPS. Only the constructs that contain residues 379–486 (or some segment of this region) phase separate, and only these constructs exhibit concentration-dependent behavior via SEC. NMR measurements as a function of protein concentration and 15N relaxation data point towards residues 450–486 interacting with sequence-distant components of UBQLN2 in a weak, multivalent manner, including the Pxx region (residues 505–508) where most ALS-linked mutations of UBQLN2 reside, as well as residues in the region immediately upstream of the UBA domain (residues 550–570) and containing the UBA domain (residues 592–594, 616–620). We postulate that our NMR data identified the residues involved in interactions that drive UBQLN2 oligomerization and LLPS. Taken together, our data suggest that both folded domains (UBA) and intrinsically disordered regions of UBQLN2 are critical to mediating LLPS.
Ubiquitin Binding Disrupts UBQLN2 LLPS
UBQLN2 contains the domain architecture of archetypal proteasome shuttle proteins (Figure 2A). The UBA domain interacts directly with Ub molecules of ubiquitinated substrate proteins and can direct them to the proteasome by interaction of the UBL domain with proteasomal subunits (Walters et al., 2002). To see if Ub binding affects UBQLN2 LLPS, we added Ub to one region of a sample well containing UBQLN2 droplets and imaged the results over time. Remarkably, UBQLN2 droplets disassembled as Ub diffused through the well (Figure 5A, Movie S3), suggesting that Ub binding eliminated UBQLN2 LLPS.
Figure 5. Ubiquitin binding eliminates UBQLN2 LLPS.
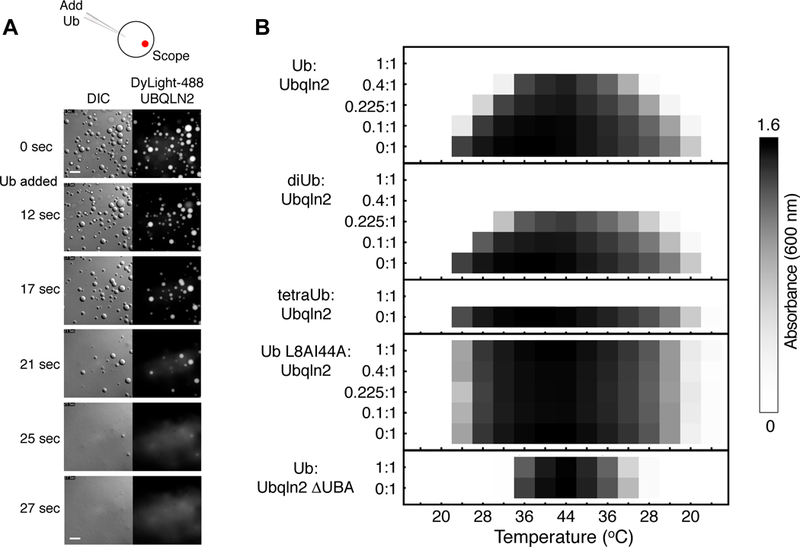
(A) DIC and fluorescence images showing disassembly of UBQLN2-protein droplets after Ub addition. (B) Turbidity assay as a function of temperature for mixtures containing both Ub and UBQLN2 in different molar ratios. For assays containing FL UBQLN2, solution consisted of 50 µM protein and 0–50 µM Ub (or diUb, tetraUb, L8AI44A Ub variant) in pH 6.8 buffer with 300 mM NaCl. For assay containing UBQLN2 ΔUBA, solution consisted of 75 µM protein and 0 or 75 µM Ub in a pH 6.8 buffer with 500 mM NaCl.
To quantify the effects of Ub binding on LLPS, we conducted spectrophotometric assays of Ub-UBQLN2 mixtures at different molar ratios (Figure 5B). LLPS decreased with increasing amounts of Ub, leading to complete elimination of LLPS when Ub:UBQLN2 ratio reached 1:1. We next monitored the effects of polyUb chains on UBQLN2 LLPS. As K48-linked chains are the most abundant type of polyUb chain in cells and its tagging of substrates signal for their proteasomal degradation, we enzymatically assembled K48-linked diUb and tetraUb. We observed that UBQLN2 LLPS was completely eliminated in the presence of either diUb or tetraUb (Figure 5B).
To test if specific binding between Ub and UBQLN2 eliminates LLPS, we repeated these assays using a Ub mutant (L8AI44A) that does not bind UBA domains (Castañeda et al., 2016). Addition of L8AI44A Ub had no effect on UBQLN2 LLPS, indicating that Ub binding to the UBA of UBQLN2 was required to disrupt LLPS. We next added WT Ub to UBQLN2 ∆UBA and found no effect on LLPS, indicating that specific binding of Ub to the UBA domain of UBQLN2 modulates LLPS and can indeed eliminate such behavior.
NMR titration of Ub into UBQLN2 450–624 indicated that Ub binds specifically to the UBA domain with a Kd of 3 ± 1 µM, as resonances for residues 450–580 were unaffected (Figure 6A). Relaxation data for the Ub-bound complex confirmed that Ub bound to the UBA domain only (Figure 6C). Binding affinity and CSPs were very similar to those reported for Ub binding to the isolated UBQLN1 UBA domain, which has 98% sequence identity to that of UBQLN2 (Zhang et al., 2008). In contrast, L8AI44A Ub bound very weakly, if at all, to UBQLN2 (Figure 6A).
Figure 6. Ub binds specifically to UBQLN2 UBA.
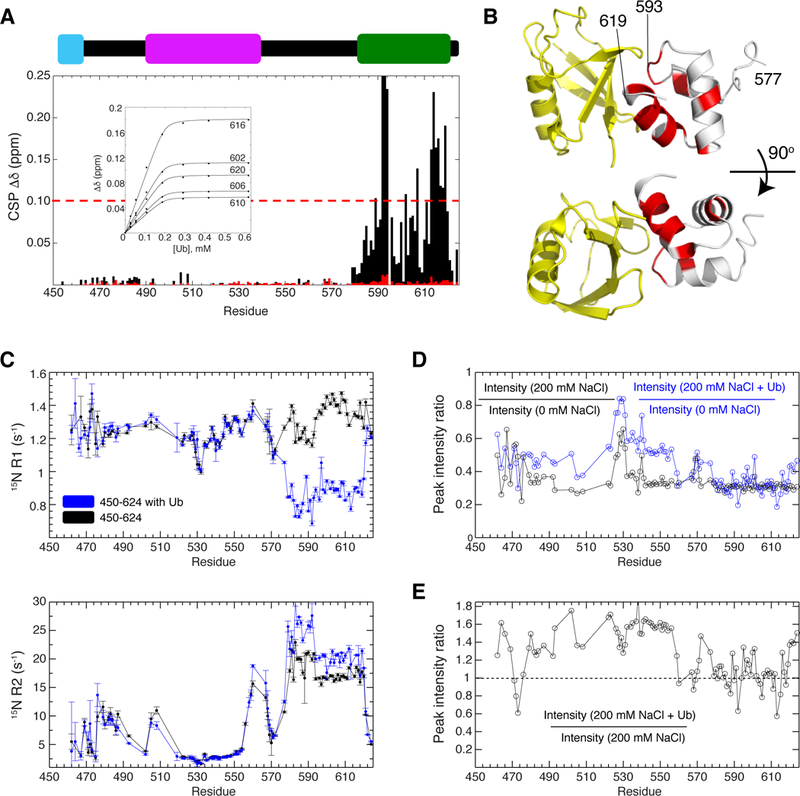
(A) CSPs for residues in UBQLN2 450–624 at the titration endpoint with Ub (black) or L8AI44A Ub (red). Inset contains sample titration curves; line represents fit to single-site binding model. (B) CSPs were mapped onto UBA (gray, residues with CSPs ≥ 0.1 ppm in red, and Ub (yellow) using PDB 2JY6. (C) 15N R1 and R2 relaxation rates for UBQLN2 450–624 in the absence (black) and presence of Ub (blue). (D) Peak intensities decreased after salt-induced LLPS (black). Peak intensities are partially recovered upon Ub-mediated elimination of UBQLN2 LLPS (blue). (E) Peak intensities increase across UBQLN2 after Ub was added except for the UBA, where Ub binds. See also Figure S7.
Mapping of the Ub binding site on UBQLN2 indicated that Ub binds to one face of the UBA domain (Figure 6B), with the largest CSPs observed for residues 592–594 and 610–620, the same residues exhibiting concentration-dependent CSPs in the absence of Ub (Figures 4D and4E). Interestingly, the UBL domain also binds the UBA domain at identical sites where Ub binds, albeit much more weakly (Nguyen et al., 2017). Therefore, we suspect that Ub binding to UBQLN2 eliminates at least two multivalent interaction sites, i.e., residues 592–594 and 610– 620, thus reducing the propensity for UBQLN2 to oligomerize and phase separate in the presence of Ub.
NMR analysis of phase separating RBPs FUS and TDP-43 revealed that the phase- separated state often contains broad resonances due to increased viscosity and restricted mobility of the protein in the liquid droplets (Burke et al., 2015; Conicella et al., 2016). We induced LLPS of 200 µM samples of UBQLN2 450–624 with 200 mM NaCl at 25°C (Figure S2B) and recorded NMR spectra immediately. We found that chemical shifts of residues in UBQLN2 450–624 were largely identical in the absence and presence of 200 mM NaCl (Figure S7A), consistent with prior observations in both FUS and TDP-43. However, generally across all residues, we observed a 70% reduction of peak intensity for the phase-separated 200 µM UBQLN2 450–624 sample (Figure 6D). Salt can cause loss of sensitivity of NMR signals. To approximate the peak intensity reduction as a result of salt-induced loss of sensitivity, we used a 200 µM sample of Ub under identical conditions, and observed only a 45–50% reduction of peak intensity upon addition of 200 mM NaCl (data not shown). Thus, 20–25% of the reduction of peak intensity is most likely due to LLPS. Indeed, upon the addition of Ub, which eliminates LLPS, peak intensities increased approximately 1.5-fold (150%) across many UBQLN2 residues, with the exception of the UBA domain, which remained unchanged (Figure 6E). The latter occurred due to Ub binding to the UBA domain. These observations are consistent with Ub binding specifically to the UBA domain and eliminating UBQLN2 LLPS.
These data suggest a model of how Ub binding modulates UBQLN2 LLPS. NMR data suggest that LLPS is driven by multivalent interactions across UBQLN2, involving residues 455– 487, 505–508, 554–571, 592–594, and 616–620 (Figure 4D). Interestingly, many of these residues are polar or hydrophobic (Figure 4E). Ub binds specifically to the UBA domain with the largest CSPs occurring at residues 592–594 and 615–619. Therefore, Ub binding disrupts only those UBQLN2 multivalent interactions involving the UBA domain, leading to elimination of LLPS (Figure 7).
Figure 7. Model for Ub-mediated elimination of UBQLN2 LLPS.
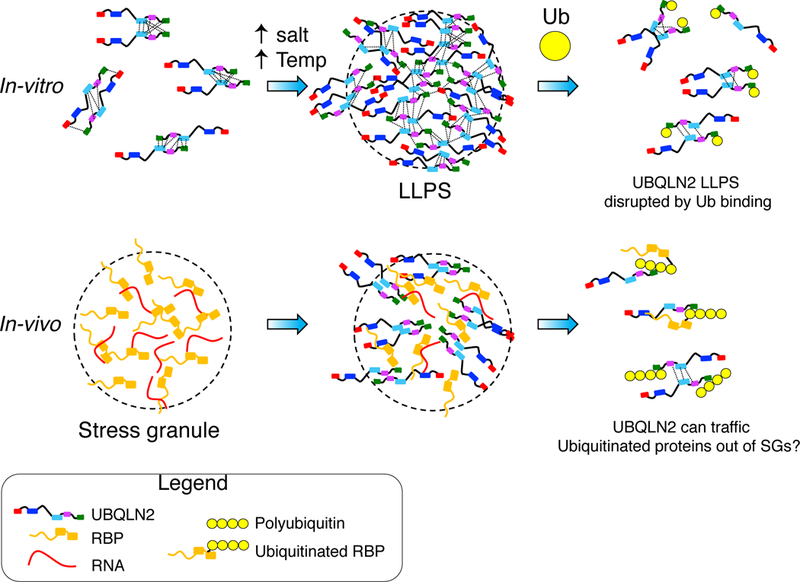
UBQLN2 undergoes LLPS under physiological conditions. Ub binding disrupts LLPS by interfering with multivalent interactions (dotted lines) involving the UBA domain. Inside cells, UBQLN2 LLPS promotes colocalization with SGs, whereby interaction with Ub or ubiquitinated substrates reverses LLPS and may shuttle clients out of stress granules.
Discussion
Recent studies have illuminated links between LLPS, SGs, and cytoplasmic inclusions characteristic of many neurodegenerative diseases (Kim et al., 2013; Molliex et al., 2015; Patel et al., 2015; Ramaswami et al., 2013). SG persistence or alterations in SG dynamics may be causative in ALS and other neurodegenerative disorders (Li et al., 2013; Monahan et al., 2016). Here we show that UBQLN2, a protein identified in inclusions of familial and sporadic ALS patients, colocalizes with SGs under different cellular stressors. Furthermore, we demonstrate that, at physiological protein concentration, temperature and ionic strength, UBQLN2 undergoes LLPS which correlates with UBQLN2’s ability to form stress-induced puncta inside cells. We show that specific non-covalent Ub binding can eliminate UBQLN2 LLPS entirely. Our observations further strengthen the established link between LLPS and SGs. Our biophysical studies using NMR identify the hydrophobic and polar multivalent interactions that promote UBQLN2 LLPS and provide a mechanism by which non-covalent interactions between Ub and UBQLN2 disrupt and eliminate LLPS.
A close link has been established between PQC components and SGs (Buchan et al., 2013; Jain et al., 2016; Walters et al., 2015). Chaperones such as heat shock proteins (HSPs) are integral components of SGs, and inhibition or reduced expression of HSP70 can lead to significant delays in SG disassembly and clearance (Cherkasov et al., 2013; Walters et al., 2015). Recently, UBQLN2 was shown to interact with HSP70, providing a molecular basis for how UBQLN2 can be recruited to SGs in vivo (Hjerpe et al., 2016). Since both UBQLN2 and HSPs are important for protein clearance, defects in either of these proteins or their interactions can result in disrupted SG dynamics and lead to disease states.
Ub is a common component of both SGs and pathological inclusions in eukaryotic cells (Deng et al., 2011; Kwon et al., 2007). Ubiquitination can occur in nuclear speckles, another type of membraneless organelle with liquid properties (Marzahn et al., 2016). Therefore, ubiquitination and other post-translational modifications (Jayabalan et al., 2016; Ohn et al., 2008) of SG components could regulate SG assembly, dynamics, and disassembly. For example, HDAC6, a deacetylase that contains a ZnF Ub-binding domain, is critical for SG formation, which is impaired in cells expressing a non-Ub-binding mutant of HDAC6 (Kwon et al., 2007). Alternatively, the membraneless organelle may provide a compartment for ubiquitination to occur by facilitating interactions between ubiquitinating enzymes and substrates.
Our in vitro observation that Ub and polyUb disrupt UBQLN2 LLPS suggests that UBQLN2 can be used to modulate the composition of SGs or other membraneless organelles through LLPS (Figure 7). Importantly, specific binding between Ub and UBQLN2 reverses UBQLN2 LLPS, an example of polyphasic linkage whereby ligand binding stabilizes or destabilizes specific phases (Posey et al., 2018; Wyman and Gill, 1980). In this case, Ub shifts the UBQLN2 LLPS transition such that the UBQLN2-dilute phase is stabilized (or that the UBQLN2-dense phase is destabilized). PolyUb binding may reduce propensity for UBQLN2 to oligomerize (Ford and Monteiro, 2006), akin to how UBQLN2 ΔUBA exhibits significantly reduced LLPS. Inside cells, we suggest that LLPS promotes UBQLN2 colocalization with SGs, as we observed with endogenous UBQLN2 in U2OS and HeLa cells. Moreover, given the observed elimination of UBQLN2 LLPS upon interaction with Ub or polyUb in vitro, we hypothesize that polyUb interactions with UBQLN2 result in reversal of LLPS, elimination of UBQLN2 from SGs with concomitant extraction of the polyubiquitinated substrate from SGs. This could provide a mechanism by which UBQLN2 can traffick polyUb-tagged substrates from SGs (or other membraneless organelles) to proteasomal or autophagic PQC systems (Figure 7).
Consistent with prior observations, we found that UBQLN2 oligomerizes in a concentration-dependent manner. Our SEC and NMR data show that oligomerization relies on at least a portion of the STI1-II domain, consistent with other reports suggesting that UBQLNs self-associate via their STI1 regions (Ford and Monteiro, 2006; Kurlawala et al., 2017). Recently, the latter half of STI1-II was found to be essential for mediating UBQLN1 dimerization (Kurlawala et al., 2017), consistent with our UBQLN2 findings. The STI1-II regions of both UBQLN1 and UBQLN2 are very similar, exhibiting 92% identity and similar order/disorder propensity (Figure S1A).
Importantly, UBQLN2’s concentration-dependent oligomerization correlated with the propensity for UBQLN2 to phase separate, since only those UBQLN2 constructs that contained complete or partial STI1-II domains exhibited LLPS. Dimerization or other higher-order oligomerization mediates LLPS for other proteins in membraneless organelle assembly, including homodimerization of G3BP1 in SG assembly (Tourrière et al., 2003) and SPOP oligomerization in localization to nuclear speckles (Marzahn et al., 2016). Therefore, we propose that UBQLN2 oligomerization promotes weak multivalent interactions that can drive UBQLN2 self-assembly into dynamic phase-separated liquid droplets, particularly as the local concentration of UBQLN2 may be increased in the cell in response to stress. We postulate that UBQLN2 LLPS could be further modulated through interactions with other binding partners via UBQLN2 STI1 regions (Kurlawala et al., 2017; Suzuki and Kawahara, 2016).
UBQLN2 phase separates readily at physiological temperature and ionic strength. However, unlike RNA-binding proteins FUS, TDP-43, and hnRNPA1, UBQLN2 does not have an overabundance of Asn/Gly/Gln/Tyr amino acids found in the LCDs of these proteins. Instead, LLPS appears driven by the C-terminal STI1-II region (residues 379–486), which is devoid of ionizable residues, but is high in hydrophobic amino acid content, particularly Ala, Leu, Met, Pro and Gln. Our NMR-identified multivalent interactions (Figure 4E) suggest that UBQLN2 LLPS is likely to be driven by multiple hydrophobic and polar interactions, consistent with the following observations. First, salt increases the propensity of UBQLN2 LLPS, as salt strengthens hydrophobic and polar interactions through increased electrostatic screening. Second, UBQLN2 LLPS is characteristic of LCST (lower critical solution temperature) polymers that have high hydrophobic amino acid content in the presence of prolines (Quiroz and Chilkoti, 2015). Third, we observed that UBQLN2 droplets dispersed with the addition of 1,6-hexanediol (data not shown), an aliphatic alcohol known to disrupt weak, hydrophobic interactions in phase- separated systems (Patel et al., 2007). Lastly, Ub binds to the UBA domain of UBQLN2 via at least two sets of hydrophobic interactions (residues 592–594 and 610–620). We hypothesize that Ub binding can outcompete UBQLN2 self-interactions and disrupt the contacts necessary to maintain UBQLN2 LLPS. Together, these metrics support the idea that UBQLN2 LLPS is driven by multivalent hydrophobic and polar interactions across multiple domains of UBQLN2.
UBQLN2 is the only ubiquilin paralog to contain the Pxx region, where most ALS-linked mutations reside. Although indirect evidence suggests that mutations in this region alter proteasomal degradation of proteins (Chang and Monteiro, 2015; Osaka et al., 2016), and impact the ability of UBQLN2 to interact with HSP70 chaperone proteins and associated cargo (Hjerpe et al., 2016; Teyssou et al., 2017), no known function has yet been assigned to this region. We originally reasoned that the Pxx region was important for mediating UBQLN2 LLPS given how its Pro and Gly sequence composition is similar to elastin-like peptides that phase separate (Muiznieks and Keeley, 2010). However, UBQLN2 phase separates without the Pxx region, albeit at reduced levels. Our NMR data show that the Pxx region dynamics (residues 505–508) is indeed perturbed with increasing protein concentration, suggesting that the Pxx region does contribute towards multivalent interactions. Therefore, we propose that while the STI1-II region drives LLPS through oligomerization of UBQLN2, the Pxx region tunes this behavior. This proposal is supported by recent studies showing that LLPS of designed elastin peptides is tuned by sequence modulation (Quiroz and Chilkoti, 2015).
Together, the multiple interactions among the STI, Pxx, and UBA domains and connecting regions as well as the interactions between the UBA and UBL domains offer several ways to modulate LLPS. We have demonstrated that Ub binding to the UBA is one way to eliminate LLPS. We propose that ALS mutations in the Pxx region may be another method by which UBQLN2 LLPS is altered, and future studies will address this hypothesis.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Carlos Castañeda (cacastan@syr.edu).
Experimental Model and Subject Details
Bacterial Culture
Unlabeled proteins were expressed in either NiCo21 (DE3) or Rosetta 2 (DE3) pLysS cells in Luria-Bertani (LB) broth. Uniformly 15N or 15N/13C labeled proteins were expressed in M9 minimal media with 15N ammonium chloride and 13C-glucose as the sole nitrogen and carbon sources, respectively. Cells were induced with IPTG and harvested after 12–16 hours at 37°C. Cell pellets were frozen, lysed and cleared by centrifugation at 22,000 × g for 20 min at 4°C.
Mammalian Cell Culture and Stable Cell Lines
HeLa (ATCC: CCL-2) and U2OS (ATCC: HTB-96) cells were maintained in GlutaMAX high- glucose DMEM (Gibco, 10566) with 10% fetal bovine serum and 1% penicillin/streptomycin. HeLa and U2OS cells were treated with the following stressors: 30 min of 0.5 mM sodium arsenite; 2 h of 43°C heat shock; 1 h of 2 mg/mL pu romycin, or 1 h 0.6 M sorbitol. HeLa and U2OS cells were authenticated by Short Tandem Repeat (STR) profiling. Both cell lines were derived from female.
Methods Details
Immunofluorescence of Endogenous UBQLN2
U2OS cells were seeded in 8-well glass slides (Millipore, PEZGS0816). After stress, cells were fixed using 4% formaldehyde at room temperature for 10 min and then washed 3X in phosphate-buffered saline (PBS). For immunostaining, fixed cells were blocked for 1 h in 5% normal goat serum and 0.25% Triton X-100 in PBS and were incubated in the same solution for 1 h at room temperature or overnight at 4°C with pr imary antibody (mouse anti-UBQLN2 [Ls- Bio NBP1–85639, 1:100] or rabbit anti-eIF4G1 [Santa Cruz, sc-11373, 1:200]), washed 3X in PBS, and incubated 1 h with secondary antibody (Alexa Fluor 488, 555 goat anti-mouse or anti- rabbit IgG [H+L], Invitrogen), washed 3X in PBS, and mounted on microscope slides using mounting media with DAPI to stain nuclei (ProLong Gold Antifade Reagent, Invitrogen P36834). Immunostaining was observed and imaged using a LSM510 (Zeiss) confocal microscope with a 63x objective. Representative images were compiled using Photoshop.
Estimation of UBQLN2 Concentration in Cells
We estimated UBQLN2 concentration using the PaxDb database (Wang et al., 2012). The underlying principle is that absolute protein abundance can be estimated by mass spectrometric analysis of whole cell/tissue lysate. In this database, human UBQLN2 has an abundance of ~250 ppm in U2OS cells according to published proteomics data (Geiger et al., 2012). As the sum of all protein molecules are estimated to be ~3 million per cubic micron in human cells (Milo, 2013), there are ~750 UBQLN2 molecules (1.25 × 10–21 moles) per cubic micron (1 × 10–15 liter). Thus, the concentration of UBQLN2 is ~1.25 µM in the cells.
Immunofluorescence of mCherry-tag UBQLN2
HeLa cells were seeded on 8-well glass slides (Millipore). Cells were transfected 24 hours after seeding using FuGene 6 (Promega) with mCherry-UBQLN2-Full-Length (wild-type), mCherry- UBQLN2-ΔUBA, mCherry-UBQLN2–450-624, or mCherry-UBQLN2–487-624 construct. 24 hr post transfection, cells were stressed with 500 µM sodium arsenite (Sigma-Aldrich) for 30 min. Cells were then fixed with 4% paraformaldehyde (Electron Microscopy Sciences), permeabilized with 0.5% Triton X-100, and blocked in 5% bovine serum albumin (BSA). Primary antibody used was against eIF4G (sc-11373; Santa Cruz). For visualization, the appropriate host-specific Alexa Fluor 488 (Molecular Probes) secondary antibody was used. Slides were mounted using Prolong Gold Antifade Reagent with DAPI (Life Technologies). Images were captured using a Leica TCS SP8 STED 3X confocal microscope (Leica Biosystems) with a 63x objective. Three independent experiments were performed.
Subcloning, Protein Expression, Purification
Ubiquitin (WT and L8AI44A) were expressed and purified as detailed elsewhere (Beal et al., 1996; Castañeda et al., 2016). The gene encoding mouse E1 was a kind gift from Jorge Eduardo Azevedo (Addgene plasmid 32534, (Carvalho et al., 2011)). E1 was expressed in Escherichia coli NiCo21 (DE3) cells in Luria-Bertani (LB) broth at 16oC overnight. GST-E2–25K in pGEX-4T2 was expressed in Escherichia coli Rosetta 2 (DE3) pLysS cells in Luria-Bertani (LB) broth at 16°C overnight. Bacteria were pellete d, frozen, then lysed via freeze/thaw method in 50 mM Tris, 1 mM EDTA (pH 8), 1 mM PMSF, 1 mM MgCl2, and 0.2 mg/mL DNase I. E1 was purified via Ni2+ chromatography. GST-E2–25K was purified via GST chromatography. Both E1 and GST-E2–25K were concentrated and buffer exchanged into 50 mM Tris and 1 mM EDTA (pH 8) and stored at −80oC for subsequent use in the production of K48-linked diubiquitin (Ub2) and tetra-ubiquitin (Ub4), as described in (Raasi and Pickart, 2005). Briefly, ubiquitin WT was reacted with ~1000 nM E1 and ~10 uM GST E2–25K in the presence of 10 mM ATP, 0.3 mM TCEP in Tris buffer at pH 8 for 3 hours at 37oC. This procedure generates polyubiquitin (polyUb) chains of different length. Ub2 and Ub4 were isolated from other polyUb chains using cation exchange chromatography at pH 4.5 with a HP SP column (GE Healthcare).
The gene encoding human UBQLN2 was a kind gift from the laboratory of Dr. Peter Howley (Addgene plasmid 8661). The gene was subcloned into pET24b (Novagen) using NEBuilder HiFi DNA assembly kit (NEB) with or without a C-terminal His tag. Deletion constructs were made using Phusion Site-Directed Mutagenesis Kit (Thermo Scientific). A tryptophan codon was added to the end of constructs 379–624, 450–624, and 487–624 to facilitate determination of protein concentration. The UBQLN2 UBL domain (construct 1–107) and 487–624W construct were expressed in Escherichia coli NiCo21 (DE3) cells in Luria-Bertani (LB) broth at room temperature overnight. The rest of the UBQLN2 constructs were expressed in Escherichia coli Rosetta 2 (DE3) pLysS cells in Luria-Bertani (LB) broth at 37°C overnight.
Bacteria were pelleted, frozen, then lysed in 50 mM Tris, 1 mM EDTA (pH 8), 1 mM PMSF, 1 mM MgCl2, and 0.2 mg/mL DNase I. Constructs that exhibited LLPS behaviors and therefore did not contain affinity tags for purification were purified via a “salting out” process. Briefly, NaCl was added to the cleared lysate to the final concentration of 0.5 M-1 M. UBQLN2 droplets were pelleted and then resuspended in 20 mM NaPhosphate, 0.5 mM EDTA (pH 6.8). The process was repeated to further remove protein and nucleic acid contaminants. Leftover NaCl was removed through HiTrap desalting column (GE Healthcare). Protein samples that needed further purification were subjected to size exclusion chromatography on a Superdex 75 16/600 column (GE Healthcare). Constructs that did not undergo LLPS (487–624, ∆379–486 and 1–107) were purified via Ni2+ chromatography, dialyzed into 20 mM NaPhosphate and 0.5 mM EDTA (pH 6.8), concentrated, and then subjected to size exclusion chromatography. Protein samples for NMR spectroscopy were produced in M9 minimum media supplemented with 15N ammonium chloride and 13C glucose as appropriate for the experiment. Purified proteins were frozen at −80°C.
Fluorescent Labeling
Full-length UBQLN2 and UBQLN2 450–624 constructs were fluorescently labelled with Dylight- 488 and DyLight-650 NHS Ester (Thermo Scientific), respectively, according to the manufacturer’s instructions. Mole dye per mole protein ratio for all samples was determined to be between 0.6 and 1.2.
Spectrophotometric Absorbance Measurements
Protein samples were prepared by mixing determined amounts of UBQLN2, NaCl, and ubiquitin (when appropriate) stocks and buffer to achieve desired concentrations of each component. Absorbance at 600 nm as a function of temperature was monitored on a Beckman DU-640 UV/Vis spectrophotometer and recorded after 2 min of reaching temperatures every 4°C. Absorbance values were reported after subtracting the optical density of buffer. Data were collected in duplicate (some with n=10), and representative traces are presented.
DIC/Fluorescence Imaging of Phase Separation
UBQLN2 full-length and 450–624 constructs were prepared to contain 50 µM protein (spiked with fluorophore-labeled UBQLN2 (Dylight-488 or Dylight-650), 1:100 molar ratio) in 20 mM NaPhosphate, 200 mM NaCl, and 0.5 mM EDTA (pH 6.8). Samples were added to MatTek glass bottom dishes that had been incubated with 3% BSA to reduce rapid coating of protein droplets onto the glass surface (Lin et al., 2015). Phase separation was imaged on a Leica DiM8 STP800 (Leica, Bannockburn, IL) equipped with a Lumencor SPECTRA X (Lumencor, Beaverton, Or), and Hamamatsu ORCAflash 4.0 V2 CMOS C11440–22CU camera using a 100×/1.4 N.A. HC Pl Apo objective to visualize the formation and fusion of UBQLN2 droplets over time. Images were taken every 4 seconds for up to 3 minutes to visualize these events. Exposures were 50 ms each on all channels (DIC, GFP, mCherry). To observe the effect of ubiquitin on UBQLN2 droplets, we began a time-lapse of a phase-separated UBQLN2 sample before gently adding a small volume of highly concentrated ubiquitin (to a final molar ratio of 1:1) to an area removed from where the camera was imaging and watched the droplets disappear as ubiquitin diffused into the sample.
Fluorescence Recovery After Photobleaching
FRAP experiments were performed on a PerkinElmer Ultraview VoX Spinning Disc Confocal system on a Nikon Eclipse Ti-E microscope using a Hammamatsu C9100–50 EMCCD camera and a 100×1.4 N.A. PlanApo objective. Images were acquired using the 488 nm laser at a 100 ms exposure. Images were taken every 1 s over 80 s. Images from each dataset were analyzed in ImageJ as 16-bit stacks after the bleach images were removed. The ImageJ FRAP Calculator Macro plug-in was used to generate FRAP curves for the images, measuring fluorescence intensity over time. The data points were then copied into Kaleidagraph where they were fitted to a single-exponential equation to determine recovery time.
Analytical Size Exclusion Chromatography
Purified UBQLN2 constructs at different concentrations (see above) were subjected to chromatography over a Superdex 75 HiLoad 16/600 column (GE Healthcare) to analyze concentration-dependent activity. Experiments were conducted at ambient temperatures at 1 mL/min in pH 6.8 buffer containing 20 mM NaPhosphate, 0.5 mM EDTA, with no added NaCl. Standard molecular weights were determined by subjecting a sample of Gel Filtration Standard (BioRad #1511901) over the column at the same conditions.
Small Angle Neutron Scattering
Samples of UBQLN2 constructs in D2O buffer (pD 6.8 20 mM NaPhosphate, 0.5 mM EDTA, with no added NaCl) were collected as previously described at 25°C (Castañeda et al., 2013). Data were reduced using the IGOR program. Sample-to-detector distances of 5 m and 1.5 m were used to cover the range 0.01 Å-1 ≤ q ≤ 0.4 Å-1, where q = 4π sin(θ)/λ, for scattering angle 2θ and neutron wavelength λ. Samples were prepared with approximately the same concentrations used for NMR measurements (Table S1). Expected I(0) value determined using small angle scattering calculator (Sarachan et al., 2013).
NMR Experiments
NMR experiments were performed at 10°C or 25°C on a Bruker Avance III 800 MHz spectrometer equipped with TCI cryoprobe. Proteins were prepared in 20 mM NaPhosphate buffer (pH 6.8), 0.5 mM EDTA, 0.02% NaN3, and 5% D2O. All NMR data were processed using NMRPipe (Delaglio et al., 1995) and analyzed using CCPNMR 2.4.2 (Vranken et al., 2005).
NH and CON NMR Spectra
1H-15N TROSY-HSQC experiments were acquired using spectral widths of 15 and 26 ppm in the direct 1H and indirect 15N dimensions, and corresponding acquisition times of 200 ms and 47 ms. Centers of frequency axes were 4.7 and 116 ppm for 1H and 15N dimensions, respectively. 1H-15N TROSY spectra were processed and apodized using a Lorentz-to-Gauss window function with 15 Hz line sharpening and 20 Hz line broadening in the 1H dimension, while 15N dimension was processed using a cosine squared bell function. Chemical shift perturbations (CSPs) were quantified as follows: ∆δ = [(∆δH)2 + (∆δN/5)2]1/2 where ∆δH and ∆δN are the differences in 1H and 15N chemical shifts, respectively. 15N-13CO spectra were collected using the (HACA)CON 13C-detect pulse program (Bastidas et al., 2015). Spectra were acquired with 16 or 32 transients using spectral widths of 40 and 36 ppm in the 13CO and 15N dimensions, respectively, with corresponding acquisition times of 64 ms and 47 ms. Centers of 15N and 13CO frequency axes were 122 ppm and 175 ppm, respectively.
NMR Chemical Shift Assignments
We determined resonance assignments using a combination of traditional 1H-detect and 13C- detect triple-resonance experiments (Bastidas et al., 2015) on 13C/15N samples containing 200µM UBQLN2 450–624 or 400 µM UBQLN2 487–624 proteins in pH 6.8 buffer (see above). Standard 1H-detect Bruker Topspin 3.2 pulse sequences for HNCO, HN(CA)CO, HNCACB and CBCA(CO)NH experiments using optimized parameter sets were used to assign non-proline resonances. 13C-detect experiments enabled the observation of the 25 proline residues that account for 14% of all amino acids in UBQLN2 450–624. To assign amide backbone proline and neighboring resonances through 15N-13CO correlations, we used the (HACA)N(CA)CON-IPAP and (HACA)N(CA)NCO-IPAP pulse programs (Bastidas et al., 2015). Additional 13C-detect experiments include standard Bruker Topspin 3.2 pulse sequences (H)CBCACON and (H)CBCANCO. Acquisition times for 1H-detect experiments were 13–18 ms, 21 ms, 5–9 ms, and 120 ms, in the indirect 15N dimensions, indirect 13CO, indirect 13Cα/Cβ dimensions, and direct 1H dimensions, respectively. Acquisition times for 13C-detect experiments were 15 ms, 10 ms, and 7 ms in the indirect 15N, indirect 13CO, and indirect 13Cα/Cβ dimensions, respectively. Spectral widths were generally 12 ppm in 13CO (for 1H-detect experiments), 40 ppm in 13CO (for 13C- detect experiments), 27 ppm in indirect 15N (for 1H-detect experiments), 36 ppm in indirect 15N (for 13C-detect experiments), and 65 ppm in indirect 13Cα/Cβ (for both 1H and 13C-detect experiments). Non-uniform sampling (NUS) was employed for all 1H-detect triple resonance experiments and (HACA)N(CA)NCO-IPAP. Experiments were acquired with 25%−30% sampling using the Poisson Gap sampling method (Hyberts et al., 2010). Spectra were processed using NMRPipe and employed standard apodization parameters and linear prediction in the indirect dimensions. Using these experiments, we successfully assigned backbone resonances (H, N, Ca, CO) for 90% of all residues. Nearly all backbone resonances were visible at 10°C in pH 6.8 buffer, and these were the initial conditions that we used for collecting NMR assignments of this protein. For Cα and Cβ secondary shift calculations, random coil chemical shifts for UBQLN2 constructs were determined at https://spin.niddk.nih.gov/bax/nmrserver/Poulsen_rc_CS/ using default parameters at sample temperature and pH 6.8 (Kjaergaard and Poulsen, 2011). Bioinformatic order/disorder prediction of UBQLN2 sequences were performed using PONDR- FIT (Xue et al., 2010).
15N Relaxation Experiments
Longitudinal (R1) and transverse (R2) 15N relaxation rates, and {1H}−15N steady-state heteronuclear Overhauser enhancement (hetNOE) were measured for UBQLN2 samples (200µM) using established interleaved relaxation experiments and protocols (Castañeda et al., 2016; Hall and Fushman, 2003). Relaxation inversion recovery periods for R1 experiments were 4 ms (x 2), 700 ms (x 2), and 1100 ms (x 2), using an interscan delay of 2.5 s. Total spin-echo durations for R2 experiments were 8 ms (x 2), 48 ms, 64 ms, 88 ms, 112 ms (x 2), and 200 ms (x 2) using an interscan delay of 2.5 s. Heteronuclear NOE experiments were acquired with an interscan delay of 4.5 s. All relaxation experiments were acquired using spectral widths of 12 and 24 ppm in the 1H and 15N dimensions, respectively, with corresponding acquisition times of 110 ms and 31 ms. Spectra were processed using squared cosine bell apodization in both 1H and 15N dimensions. Relaxation rates were derived by fitting peak heights to a mono- exponential decay using RELAXFIT (Fushman et al., 1997). The overall rotational diffusion tensors were determined for the UBA domain using the program ROTDIF (Walker et al., 2004) and only for residues in well-defined secondary structure elements. The ratio ρ of relaxation rates was determined for each residue as ρ = (2R2’/R1’ – 1)-1, where R1’ and R2’ are modified R1 and R2 values with the high-frequency contributions subtracted (Fushman et al., 2004).
NMR Titration Experiments
Unlabeled ligand (Ub or L8AI44A Ub) was titrated into 200 µM samples of 15N UBQLN2, and the binding was monitored by recording 1H-15N TROSY-HSQC spectra as a function of ligand concentration. At any titration point, the observed CSP can be represented as ∆δ = ∆δmax * [L]/([L] +Kd), where ∆δmax is the difference in the chemical shift between the free and fully bound states for a given amide resonance, Kd is the dissociation constant, and [L] is the molar concentration of the free ligand. Data fitting for each amide was performed using in-house Matlab program, assuming a single-site (1:1 stoichiometry) binding model. Only residues with CSP > 0.05 ppm at the titration endpoint were considered for Kd determination. Reported Kd values were averages of residue-specific Kd values with errors reflecting standard deviation of these values.
Quantification and Statistical Analysis
Analysis of UBQLN2 Colocalization in SGs
Colocalization analysis of UBQLN2 in stress granules was performed by measuring how many stress granules were also positive for UBQLN2 in three independent experiments in different stress conditions (see above). About 30 cells were measured per stress condition. Statistical analysis was performed using Prism 6. An ordinary two-way ANOVA followed by Sidak’s multiple comparisons test was chosed to compare the effect of two factors (time and construct). No method was used to determine strategies for randomization, inclusion and exclusion of any data, and whether the data met assumptions of the statistical approaches. No data point was excluded.
FRAP Experiments
Average FRAP curve was generated by averaging FRAP data from 6 separate droplets of similar size. FRAP curve error bars represent the standard deviation of the FRAP data. FRAP recovery times were derived by fitting a single exponential fit to the data using Kaleidagraph. No data point was excluded.
NMR Chemical Shift Perturbations
Where applicable, chemical shift perturbations (CSPs) were quantified as follows: ∆δ = [(∆δH)2 + (∆δN/5)2]1/2 where ∆δH and ∆δN are the differences in 1H and 15N chemical shifts, respectively, for the same residue in UBQLN2 at different protein concentrations, or in the absence and presence of ubiquitin. Only those resonances with well-defined peak positions were chosen for analysis.
NMR Relaxation
As described above, relaxation rates were derived by fitting peak heights to a mono-exponential decay using RELAXFIT (Fushman et al., 1997). Errors in 15N R1 and R2 relaxation rates were determined using 500 Monte Carlo trials in RELAXFIT. Errors in hetNOE measurements were determined using the standard error propagation formula. Errors in rotational diffusion tensor were determined using ROTDIF (Walker et al., 2004).
Data and Software Availability
NMR chemical shift assignments for UBQLN2 450–624 have been deposited in the Biological Magnetic Resonance Data Bank (BMRB, http://www.bmrb.wisc.edu/) with accession number 27339.
Supplementary Material
Highlights.
UBQLN2 forms stress-induced puncta and colocalizes with stress granules in cells.
UBQLN2 undergoes liquid-liquid phase separation under physiological conditions.
Phase separation is promoted by multivalent interactions across UBQLN2 domains.
Ubiquitin or polyubiquitin eliminates phase separation behavior of UBQLN2.
Acknowledgements
This work was supported by ALS Association grants 17-IIP-369 and 18-IIP-400 to C.A.C. and Syracuse University startup funds. H.H. was supported by NIH R00 GM107355. J.P.T was supported by NIH grant R35NS097974, The Packard Center for ALS Research at the Johns Hopkins University, the ALS Association, the Clinical Research in ALS and related disorders for Therapeutic Development (CReATe) Consortium, the American-Lebanese-Syrian Associated Charities, and the Howard Hughes Medical Institute. We acknowledge a Springboard Fellowship from Target ALS to R.M.K. Data collected on a Bruker 800 MHz NMR magnet was supported by NIH shared instrumentation grant 1S10OD012254. We thank Junmin Peng for estimating intracellular UBQLN2 concentrations. We thank Susan Krueger for assistance in collecting SANS data. Access to the NGB 30m SANS was provided by the Center for High Resolution Neutron Scattering, a partnership between the National Institute of Standards and Technology and the National Science Foundation under Agreement No. DMR-1508249. We thank SUNY- Upstate for access to the Perkin Elmer Spinning Disk confocal microscope. We thank Rohit Pappu, James Hougland, and Ananya Majumdar for critical discussions, and Natalia Nedelsky for editorial assistance.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aumiller WM, Pir Cakmak F, Davis BW, and Keating CD (2016). RNA-Based Coacervates as a Model for Membraneless Organelles: Formation, Properties, and Interfacial Liposome Assembly. Langmuir ACS J. Surf. Colloids 32, 10042–10053. [DOI] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, and Rosen MK (2016). Compositional Control of Phase-Separated Cellular Bodies. Cell 166, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas M, Gibbs EB, Sahu D, and Showalter SA (2015). A primer for carbon-detected NMR applications to intrinsically disordered proteins in solution. Concepts Magn. Reson Part A 44, 54–66. [Google Scholar]
- Beal R, Deveraux Q, Xia G, Rechsteiner M, and Pickart C (1996). Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc. Natl. Acad. Sci. U. S. A 93, 861–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, and Hyman AA (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis R-M, Taylor JP, and Parker R (2013). Eukaryotic Stress Granules Are Cleared by Autophagy and Cdc48/VCP Function. Cell 153, 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke KA, Janke AM, Rhine CL, and Fawzi NL (2015). Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol. Cell 60, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilloni C, De Simone A, Vranken WF, and Vendruscolo M (2012). Determination of Secondary Structure Populations in Disordered States of Proteins Using Nuclear Magnetic Resonance Chemical Shifts. Biochemistry 51, 2224–2231. [DOI] [PubMed] [Google Scholar]
- Carvalho AF, Pinto MP, Grou CP, Vitorino R, Domingues P, Yamao F, Sá-Miranda C, and Azevedo JE (2011). High-Yield Expression in Escherichia coli and Purification of Mouse Ubiquitin-Activating Enzyme E1. Mol. Biotechnol 51, 254–261. [DOI] [PubMed] [Google Scholar]
- Castañeda CA, Kashyap TR, Nakasone MA, Krueger S, and Fushman D (2013). Unique structural, dynamical, and functional properties of K11-linked polyubiquitin chains. Structure 21, 1168–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda CA, Dixon EK, Walker O, Chaturvedi A, Nakasone MA, Curtis JE, Reed MR, Krueger S, Cropp TA, and Fushman D (2016). Linkage via K27 Bestows Ubiquitin Chains with Unique Properties among Polyubiquitins. Structure 24, 423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, and Monteiro MJ (2015). Defective Proteasome Delivery of Polyubiquitinated Proteins by Ubiquilin-2 Proteins Containing ALS Mutations. PLoS ONE 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, and Bukau B (2013). Coordination of translational control and protein homeostasis during severe heat stress. Curr. Biol. CB 23, 2452–2462. [DOI] [PubMed] [Google Scholar]
- Conicella AE, Zerze GH, Mittal J, and Fawzi NL (2016). ALS Mutations Disrupt Phase Separation Mediated by α-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain. Structure 24, 1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995). NMRPIPE - A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6, 277– 293. [DOI] [PubMed] [Google Scholar]
- Deng H-X, Chen W, Hong S-T, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, et al. (2011). Mutations in UBQLN2 cause dominant X-linked juvenile and adult onset ALS and ALS/dementia. Nature 477, 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford DL, and Monteiro MJ (2006). Dimerization of ubiquilin is dependent upon the central region of the protein: evidence that the monomer, but not the dimer, is involved in binding presenilins. Biochem. J 399, 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushman D, Cahill S, and Cowburn D (1997). The main-chain dynamics of the dynamin pleckstrin homology (PH) domain in solution: analysis of 15N relaxation with monomer/dimer equilibration. J. Mol. Biol 266, 173–194. [DOI] [PubMed] [Google Scholar]
- Fushman D, Varadan R, Assfalg M, and Walker O (2004). Determining domain orientation in macromolecules by using spin-relaxation and residual dipolar coupling measurements. Prog Nucl Magn Reson Spectrosc 44, 189–214. [Google Scholar]
- Geiger T, Wehner A, Schaab C, Cox J, and Mann M (2012). Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics MCP 11, M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin KM, Chang L, and Monteiro MJ (2015). ALS-linked mutations in ubiquilin-2 or hnRNPA1 reduce interaction between ubiquilin-2 and hnRNPA1. Hum. Mol. Genet 24, 2565–2577. [DOI] [PubMed] [Google Scholar]
- Hall JB, and Fushman D (2003). Characterization of the overall and local dynamics of a protein with intermediate rotational anisotropy: Differentiating between conformational exchange and anisotropic diffusion in the B3 domain of protein G. J. Biomol. NMR 27, 261– 275. [DOI] [PubMed] [Google Scholar]
- Hjerpe R, Bett JS, Keuss MJ, Solovyova A, McWilliams TG, Johnson C, Sahu I, Varghese J, Wood N, Wightman M, et al. (2016). UBQLN2 Mediates Autophagy- Independent Protein Aggregate Clearance by the Proteasome. Cell 166, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyberts SG, Takeuchi K, and Wagner G (2010). Poisson-Gap Sampling and Forward Maximum Entropy Reconstruction for Enhancing the Resolution and Sensitivity of Protein NMR Data. J. Am. Chem. Soc 132, 2145–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, and Jülicher F (2014). Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol 30, 39–58. [DOI] [PubMed] [Google Scholar]
- Itakura E, Zavodszky E, Shao S, Wohlever ML, Keenan RJ, and Hegde RS (2016). Ubiquilins Chaperone and Triage Mitochondrial Membrane Proteins for Degradation. Mol. Cell 63, 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, and Parker R (2016). ATPase- Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 164, 487– 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayabalan AK, Sanchez A, Park RY, Yoon SP, Kang G-Y, Baek J-H, Anderson P, Kee Y, and Ohn T (2016). NEDDylation promotes stress granule assembly. Nat. Commun 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye FJ, Modi S, Ivanovska I, Koonin EV, Thress K, Kubo A, Kornbluth S, and Rose MD (2000). A family of ubiquitin-like proteins binds the ATPase domain of Hsp70-like Stch. FEBS Lett 467, 348–355. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang Y-D, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. (2013). Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaergaard M, and Poulsen FM (2011). Sequence correction of random coil chemical shifts: correlation between neighbor correction factors and changes in the Ramachandran distribution. J. Biomol. NMR 50, 157–165. [DOI] [PubMed] [Google Scholar]
- Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, and Howley PM (2000). The hPLIC Proteins May Provide a Link between the Ubiquitination Machinery and the Proteasome. Mol. Cell 6, 409–419. [DOI] [PubMed] [Google Scholar]
- Ko HS, Uehara T, and Nomura Y (2002). Role of Ubiquilin Associated with Protein- disulfide Isomerase in the Endoplasmic Reticulum in Stress-induced Apoptotic Cell Death. J. Biol. Chem 277, 35386–35392. [DOI] [PubMed] [Google Scholar]
- Kurlawala Z, Shah PP, Shah C, and Beverly LJ (2017). The STI and UBA Domains of UBQLN1 are Critical Determinants of Substrate Interaction and Proteostasis. J. Cell. Biochem 118, 2261–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S, Zhang Y, and Matthias P (2007). The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev 21, 3381–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le NTT, Chang L, Kovlyagina I, Georgiou P, Safren N, Braunstein KE, Kvarta MD, Dyke AMV, LeGates TA, Philips T, et al. (2016). Motor neuron disease, TDP-43 pathology, and memory deficits in mice expressing ALS–FTD-linked UBQLN2 mutations. Proc. Natl. Acad. Sci 201608432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, and Gitler AD (2013). Stress granules as crucibles of ALS pathogenesis. J Cell Biol 201, 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PJ, Danner R, Liang J, Doong H, Harman C, Srinivasan D, Rothenberg C, Wang H, Ye Y, Fang S, et al. (2009). Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J. Cell Biol 187, 201–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DSW, Rosen MK, and Parker R (2015). Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell 60, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah AL, Perry G, Smith MA, and Monteiro MJ (2000). Identification of Ubiquilin, a Novel Presenilin Interactor That Increases Presenilin Protein Accumulation. J. Cell Biol 151, 847–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahboubi H, and Stochaj U (2017). Cytoplasmic stress granules: Dynamic modulators of cell signaling and disease. Biochim. Biophys. Acta BBA - Mol. Basis Dis 1863, 884–895. [DOI] [PubMed] [Google Scholar]
- Marzahn MR, Marada S, Lee J, Nourse A, Kenrick S, Zhao H, Ben‐Nissan G, Kolaitis RM, Peters JL, Pounds S, et al. (2016). Higher‐order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J 35, 1254–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R (2013). What is the total number of protein molecules per cell volume? A call to rethink some published values. BioEssays News Rev. Mol. Cell. Dev. Biol 35, 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea DM, and Kriwacki RW (2016). Phase separation in biology; functional organization of a higher order. Cell Commun. Signal 14, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan Z, Shewmaker F, and Pandey UB (2016). Stress granules at the intersection of autophagy and ALS. Brain Res 1649, Part B, 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori F, Tanji K, Odagiri S, Toyoshima Y, Yoshida M, Ikeda T, Sasaki H, Kakita A, Takahashi H, and Wakabayashi K (2012). Ubiquilin immunoreactivity in cytoplasmic and nuclear inclusions in synucleinopathies, polyglutamine diseases and intranuclear inclusion body disease. Acta Neuropathol. (Berl.) 124, 149–151. [DOI] [PubMed] [Google Scholar]
- Muiznieks LD, and Keeley FW (2010). Proline Periodicity Modulates the Self-assembly Properties of Elastin-like Polypeptides. J. Biol. Chem 285, 39779–39789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N’Diaye E-N, Kajihara KK, Hsieh I, Morisaki H, Debnath J, and Brown EJ (2009). PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep 10, 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen K, Puthenveetil R, and Vinogradova O (2017). Investigation of the adaptor protein PLIC-2 in multiple pathways. Biochem. Biophys. Rep 9, 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, and Anderson P (2008). A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol 10, 1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka M, Ito D, and Suzuki N (2016). Disturbance of proteasomal and autophagic protein degradation pathways by amyotrophic lateral sclerosis-linked mutations in ubiquilin 2. Biochem. Biophys. Res. Commun 472, 324–331. [DOI] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. (2015). A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077. [DOI] [PubMed] [Google Scholar]
- Patel SS, Belmont BJ, Sante JM, and Rexach MF (2007). Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129, 83–96. [DOI] [PubMed] [Google Scholar]
- Posey AE, Ruff KM, Harmon TS, Crick SL, Li A, Diamond MI, and Pappu RV (2018). Profilin reduces aggregation and phase separation of huntingtin N-terminal fragments by preferentially binding to soluble monomers and oligomers. J. Biol. Chem jbc.RA117.000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, and Parker R (2016). Principles and Properties of Stress Granules. Trends Cell Biol 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz FG, and Chilkoti A (2015). Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mater 14, 1164–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasi S, and Pickart CM (2005). Ubiquitin Chain Synthesis. In Ubiquitin-Proteasome Protocols, pp. 47–55. [DOI] [PubMed] [Google Scholar]
- Ramaswami M, Taylor JP, and Parker R (2013). Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 154, 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg C, Srinivasan D, Mah L, Kaushik S, Peterhoff CM, Ugolino J, Fang S, Cuervo AM, Nixon RA, and Monteiro MJ (2010). Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum. Mol. Genet 19, 3219–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford NJ, Lewis J, Clippinger AK, Thomas MA, Adamson J, Cruz PE, Cannon A, Xu G, Golde TE, Shaw G, et al. (2013). Unbiased screen reveals ubiquilin-1 and −2 highly associated with huntingtin inclusions. Brain Res 1524, 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarachan KL, Curtis JE, and Krueger S (2013). Small-angle scattering contrast calculator for protein and nucleic acid complexes in solution. J. Appl. Crystallogr 46, 1889–1893. [Google Scholar]
- Seguin SJ, Morelli FF, Vinet J, Amore D, De Biasi S, Poletti A, Rubinsztein DC, and Carra S (2014). Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ 21, 1838–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, and Kawahara H (2016). UBQLN4 recognizes mislocalized transmembrane domain proteins and targets these to proteasomal degradation. EMBO Rep 17, 842–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Brown RH Jr, and Cleveland DW (2016). Decoding ALS: from genes to mechanism. Nature 539, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyssou E, Chartier L, Amador M-D-M, Lam R, Lautrette G, Nicol M, Machat S, Da Barroca S, Moigneu C, Mairey M, et al. (2017). Novel UBQLN2 mutations linked to Amyotrophic Lateral Sclerosis and atypical Hereditary Spastic Paraplegia phenotype through defective HSP70-mediated proteolysis. Neurobiol. Aging 58, 239.e11–239.e20. [DOI] [PubMed] [Google Scholar]
- Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, and Tazi J (2003). The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol 160, 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Uversky VN (2016). Intrinsically disordered proteins in overcrowded milieu: Membrane- less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol 44, 18–30. [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, and Laue ED (2005). The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins Struct. Funct. Bioinforma 59, 687–696. [DOI] [PubMed] [Google Scholar]
- Walker O, Varadan R, and Fushman D (2004). Efficient and accurate determination of the overall rotational diffusion tensor of a molecule from 15N relaxation data using computer program ROTDIF. J. Magn. Reson 168, 336–345. [DOI] [PubMed] [Google Scholar]
- Walters RW, and Parker R (2015). Coupling of Ribostasis and Proteostasis: Hsp70 Proteins in mRNA Metabolism. Trends Biochem. Sci 40, 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KJ, Kleijnen MF, Goh AM, Wagner G, and Howley PM (2002). Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 41, 1767–1777. [DOI] [PubMed] [Google Scholar]
- Walters KJ, Goh AM, Wang Q, Wagner G, and Howley PM (2004). Ubiquitin family proteins and their relationship to the proteasome: a structural perspective. Biochim. Biophys. Acta BBA - Mol. Cell Res 1695, 73–87. [DOI] [PubMed] [Google Scholar]
- Walters RW, Muhlrad D, Garcia J, and Parker R (2015). Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA 21, 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Weiss M, Simonovic M, Haertinger G, Schrimpf SP, Hengartner MO, and von Mering C (2012). PaxDb, a database of protein abundance averages across all three domains of life. Mol. Cell. Proteomics MCP 11, 492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD, and Richards FM (1992). The Chemical-Shift Index - A Fast And Simple Method For The Assignment Of Protein Secondary Structure Through Nmr- Spectroscopy. Biochemistry 31, 1647–1651. [DOI] [PubMed] [Google Scholar]
- Wu A-L, Wang J, Zheleznyak A, and Brown EJ (1999). Ubiquitin-Related Proteins Regulate Interaction of Vimentin Intermediate Filaments with the Plasma Membrane. Mol. Cell 4, 619–625. [DOI] [PubMed] [Google Scholar]
- Wyman J, and Gill SJ (1980). Ligand-Linked Phase Changes in a Biological System: Applications to Sickle Cell Hemoglobin. Proc. Natl. Acad. Sci. U. S. A 77, 5239–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Yan LH, Huang B, Liu M, Liu X, and Huang C (2014). Pathogenic mutation of UBQLN2 impairs its interaction with UBXD8 and disrupts endoplasmic reticulum- associated protein degradation. J. Neurochem 129, 99–106. [DOI] [PubMed] [Google Scholar]
- Xue B, Dunbrack RL, Williams RW, Dunker AK, and Uversky VN (2010). PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta BBA - Proteins Proteomics 1804, 996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun Lee D, Arnott D, and Brown EJ (2013). Ubiquilin4 is an adaptor protein that recruits Ubiquilin1 to the autophagy machinery. EMBO Rep 14, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Raasi S, and Fushman D (2008). Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J. Mol. Biol 377, 162–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


