Abstract
Rationale:
The monitoring of TB treatments in clinical practice and clinical trials relies on traditional sputum-based culture status indicators at specific time points. Accurate, predictive, blood-based protein markers would provide a simpler and more informative view of patient health and response to treatment.
Objective:
We utilized sensitive, high throughput multiplexed ion mobility-mass spectrometry (IM-MS) to characterize the serum proteome of TB patients at the start of and at 8 weeks of rifamycin-based treatment. We sought to identify treatment specific signatures within patients as well as correlate the proteome signatures to various clinical markers of treatment efficacy.
Methods:
Serum samples were collected from 289 subjects enrolled in CDC TB Trials Consortium Study 29 at time of enrollment and at the end of the intensive phase (after 40 doses of TB treatment). Serum proteins were immunoaffinity-depleted of high abundant components, digested to peptides and analyzed for data acquisition utilizing a unique liquid chromatography IM-MS platform (LC-IM-MS). Linear mixed models were utilized to identify serum protein changes in the host response to antibiotic treatment as well as correlations with culture status end points.
Results:
A total of 10,137 peptides corresponding to 872 proteins were identified, quantified, and used for statistical analysis across the longitudinal patient cohort. In response to TB treatment, 244 proteins were significantly altered. Pathway/network comparisons helped visualize the interconnected proteins, identifying up regulated (lipid transport, coagulation cascade, endopeptidase activity) and down regulated (acute phase) processes and pathways in addition to other cross regulated networks (inflammation, cell adhesion, extracellular matrix). Detection of possible lung injury serum proteins such as HPSE, significantly downregulated upon treatment. Analyses of microbiologic data over time identified a core set of serum proteins (TTHY, AFAM, CRP, RET4, SAA1, PGRP2) which change in response to treatment and also strongly correlate with culture status. A similar set of proteins at baseline were found to be predictive of week 6 and 8 culture status.
Conclusion:
A comprehensive host serum protein dataset reflective of TB treatment effect is defined. A repeating set of serum proteins (TTHY, AFAM, CRP, RET4, SAA1, PGRP2, among others) were found to change significantly in response to treatment, to strongly correlate with culture status, and at baseline to be predictive of future culture conversion. If validated in cohorts with long term follow-up to capture failure and relapse of TB, these protein markers could be developed for monitoring of treatment in clinical trials and in patient care.
Keywords: Ion mobility spectrometry, Proteomics, Tuberculosis, Antibiotic treatment
1. Introduction
Tuberculosis (TB) is the leading cause of death from a single infectious agent [1]. According to the WHO, an estimated 10.4 million people fell ill with TB in 2016 with most of the incidents concentrated in developing countries [1]. Sputum-based culture remains the gold standard for monitoring response to treatment in clinical practice as well as in clinical trials [2–4]. Despite its long history in TB drug development, 8-week culture status and culture conversion as a measure of bacterial burden has limitations. First, the reliance on sputum is limiting given sputum production decreases rapidly on treatment. Additionally, analyses of performance of treatment culture conversion for predicting long term outcomes have shown variable results [5–7]. As a consequence, there has been a call for the development of new quantitative, non-sputum, nonculture-based TB biomarkers. If proven accurate and predictive, a blood-based biomarker signature would facilitate middle and phase 2 development for new TB drugs and regimens, as well as provide information on individual response to treatment.
Recent technologic advances in mass spectrometry (MS) based proteomics now provide multiple, robust tools that allow for high-accuracy, high-throughput analysis of clinical samples [8,9]. As a result, a better depth of coverage with accurate quantification can be obtained for a global proteome in complex clinical specimens. To date, discovery-based proteomics has primarily concentrated on investigating protein biomarkers of infection for early diagnosis of TB [10]. Early surface-enhanced laser desorption/ionization (SELDI) platforms compared serum samples from active TB patients with uninfected controls to identify possible discriminatory protein profiles, with recent studies involving a more in-depth separation [11,12]. Additionally, several liquid chromatography-mass spectrometry (LC-MS/MS) based platforms have been used as well to perform this comparison, providing a complimentary list of differentiated host proteins [13,14]. Achkar et al., presented a more comprehensive diagnostic study using LC-MS/MS and multiple-reaction monitoring (MRM) based assay to identify panels of host proteins that can distinguish active TB from latent infections as well as other respiratory disorders [15]. Longitudinal studies of TB treatment and response have also been conducted using aptamer array-based methods applied to small cohorts of patients [16–18].
In the present study we evaluate treatment response through the use of a sensitive MS platform, with the addition of ion mobility separations (IM) to an LC-MS platform, providing a more robust and higher throughput discovery analysis pipeline capable of larger scale clinical study investigations [19]. We assayed a large collection of serum samples collected at baseline and after 8 weeks of treatment from participants enrolled in a phase 2 clinical trial comparing rifapentine to rifampin, as part of combination therapy [20]. In this study, we sought to provide an unbiased characterization of the serum proteome in TB patients undergoing treatment, evaluating associations with clinical and microbiologic characteristics, comparing changes in protein biomarkers relative to culture status at time points 2, 4, 6 and 8 weeks during treatment, with the objective of identifying a serum protein signature indicative of treatment effect.
2. Materials and methods
2.1. Study population
CDC TB Trials Consortium (TBTC) Study 29 (ClinicalTrials.gov Identifier NCT00694629) is a prospective, multicenter, open-label Phase 2 clinical trial comparing the antimicrobial activity and safety of standard daily regimen containing rifampin, to that of the experimental regimen with daily rifapentine (10 mg/kg/dose), both given with isoniazid, pyrazinamide and ethambutol to adults with smear positive, culture-confirmed pulmonary TB. The primary efficacy endpoint of the trial was the proportion of patients, by regimen, having negative sputum cultures at completion of 8 weeks (40 doses) of treatment. All TB treatment was administered 5 days/week for 8 weeks and directly observed. All participants underwent HIV testing. Cultures were performed using both Lowenstein-Jensen (LJ) solid media (inoculum volume, 0.2 mL) and BACTEC mycobacterial growth indicator tube (MGIT, Becton Dickinson and Co, Franklin Lakes, New Jersey) liquid media (inoculum volume, 0.5 mL) with the MGIT 960 system, and assessed for presence of M. tuberculosis. Additional information regarding the design, conduct, and results of TBTC Study 29 has been published [20].
2.2. Selection of participants
Out of a total of 531 participants in the parent trial, we included 289 consecutively enrolled protocol-correct participants from CDC-TB Trials Consortium clinical trials sites in North America, South Africa, Uganda, and Spain, for this sub-study. The 289 participants included in the proteomic analyses had sputum smears positive for acid fast bacilli at baseline and were culture positive for drug-susceptible pulmonary TB. Detailed demographic, clinical, radiographic and microbiologic data were collected using CDC-TBTC developed case report forms as part of the parent clinical trial.
2.3. Sample processing
Serum was collected, processed and stored at baseline (time of enrollment), and after 8 weeks (40 doses) as previously described [20], in a standardized manner according to a manual of operating procedures. The investigators conducting LC-IM-MS assays were blinded to all patient characteristics until after assay results were submitted to the TBTC Data and Coordinating Center. Individual human serum samples were partitioned and depleted of 14 specific highly abundant proteins using a ProteomeLab™ 12.7 × 79.0-mm human IgY14 LC10 affinity LC column (Beckman Coulter, Fullerton, CA), which was performed using the manufacturer’s instructions to disrupt protein-protein binding and capture of additional proteins. Further processed of the flow-through portion of the depletion included automated protein isolation, denaturation, tryptic digestion, and peptide isolation [19].
2.4. LC-IM-MS analysis
Analysis of processed serum samples was performed on an in-house built instrument that couples a 1-m ion mobility separation with an Agilent 6224 TOF MS upgraded to a 1.5 m flight tube [19] coupled with LC and including a fully automated in-house built 4-column HPLC system equipped with in-house packed capillary columns [21]. A 60-min gradient with shorter columns (30 cm long with same dimensions and packing) was used with the IM-MS platform and data were collected from 100 to 3200 m/z. This unique platform has been previously described in detail [19,22], and has been optimized for higher throughput larger scale clinical sample analysis as it provides the benefit of the inclusion of an orthogonal ion mobility separations for depth of coverage coupled with a reduced LC separations component for faster overall analysis times (60 min per sample).
2.5. Data processing
Identification and quantification of the detected peptide peaks was performed utilizing the Accurate Mass and Time (AMT) tag approach [23]. Briefly, three pooled human serum samples were created (baseline, 8 weeks negative culture status, 8 weeks positive culture status) and divided into 24 fractions each using HPLC high pH fractionation. LC-MS/MS was performed on each fraction using the LTQ Orbitrap Velos MS and a reverse phase low pH LC separation [19]. A subsequent AMT tag database was created as previously described [24,25]. All MS/MS data can be obtained at ProteomeXchange under identifier PXD009029. Multiple in-house developed informatics tools (publicly available http://omics.pnl.gov/software) were used to process the LCMS data and correlate the resulting LC-MS features to an AMT tag database that contained accurate mass, LC separation elution time, and IMS drift time information for peptide tags generated from serum proteins. Among the tools used were algorithms for peak-picking and for determining isotopic distributions and charge states [26]. Further downstream data analysis incorporated all possible detected peptides into a visualization program, VIPER, to correlate LC-MS features to the peptide identifications in the AMT tag database [27].
2.6. Statistical analysis
Peptide values were log2 transformed and normalized for occurrence filtering and outlier removal as previously described [19]. Protein values were generated from the processed peptide level data utilizing the R roll-up approach as previously described [19]. All analyses were performed in the R environment, version 3.3.3. Linear mixed models were fit using the lme4 package in R. Contrasts were estimated using the package lsmeans in R (R Project for Statistical Computing, http://cran.r-project.org). Pathway enrichment statistics were calculated using a log-rank test on proteins that were ordered by significance level within each contrast. To determine which demographic, clinical, radiographic and microbiologic variables may drive protein expression during model building, univariate regressions or ANOVAs were assessed. Redundancy amongst variables was assessed using Spearman’s correlations of the variables themselves, as well as correlations calculated using protein abundance values, which shows the cross correlations of relevant clinical and demographic variables as well as bar plots, after only site/region adjustment, showing the number of significant proteins associated with each variable based upon the serum proteome results. Once the primary drivers of protein expression were determined, separate linear mixed models were fit to investigate the influence of culture conversion on protein expression in response to treatment (with adjustment for baseline disease severity). Baseline disease severity was determined by chest radiograph classification of cavitation status. All models adjusted for patient and study site as random effects, and the following fixed effects: geographic region, age, body mass index, sex, and HIV status.
Criteria to identify significantly different proteins included p-value < 0.05, minimum ± 0.1 log2 abundance change between comparisons and quantified in at least 50 patient pairs per protein. Samples without quantitative protein information were treated as missing data and not included for that particular protein analysis. Immunoglobulin constant region proteins and other immunoaffinity depleted proteins were excluded as described previously [19]. Pathway mapping was performed utilizing GO annotations extracted from DAVID version 6.8 (https://david.ncifcrf.gov/) using the ontology for biological process (GOTERM_BP_DIRECT). Bonferroni correction for only those GO term pathways that had a p value < 0.05 were selected. Cytoscape version 3.5.1 was used to create the interconnections of the pathway networks.
2.7. Ethics statement
Study 29 was approved by both CDC and local institutional review boards, and written informed consent was obtained from all study participants for collection of serum for TB-related research. The institutional review board at University of California, San Francisco approved this biomarker study (UCSF IRB Number: 12–10360).
3. Results
3.1. Study population and serum proteins identified
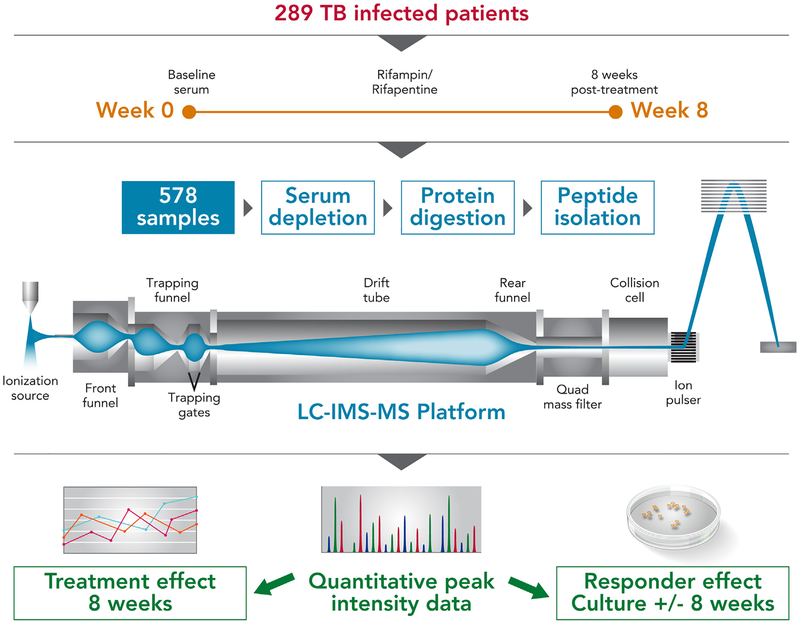
A schema of the proteome study is provided in Fig. 1. Table 1 shows the clinical metadata of the 289 study participants from whom serum was collected and LC-IM-MS high-throughput analysis applied. In total, 10,137 peptides corresponding to 872 proteins were identified, quantified, and used for statistical analysis across the longitudinal patient cohort (Supplemental Table 1).
Fig. 1.
Schematic workflow of the experimental design and analysis procedure. Utilization of the LC-IM-MS platform generated peptide level peak intensity data for each of the 578 samples (289 baseline samples and 289 8-week samples) from which protein level data was inferred. Protein level data was the basis to investigate the dynamic changes in the serum proteome after 8 weeks of treatment.
Table 1.
Demographic distribution of analyzed patient population.
| Category | No. of Patients (% of Total) | No. Sputum Converted (% of Category) | Median days to conversion | |||
|---|---|---|---|---|---|---|
| Liquid media at 8 wks | Solid media at 8 wks | Smear_WHO_8 wks | Solid media | Liquid media | ||
| All Subjects | 289 (100%) | |||||
| Gender | 190 (66%) | 227 (79%) | 206 (71%) | |||
| Female | 92 (32%) | 69 (75%) | 81 (88%) | 75 (81%) | 41.5 (13, 84) | 42 (13, 140) |
| Male | 197 (68%) | 121 (61%) | 146 (74%) | 131 (66%) | 42 (13, 168) | 56 (13, 168) |
| Age | 188 (65%) | 226 (78%) | 205 (71%) | |||
| 0 – 20 | 16 (5%) | 13 (81%) | 15 (94%) | 14 (87%) | 28 (14, 56) | 42 (14, 85) |
| 21 – 40 | 181 (63%) | 115 (64%) | 143 (79%) | 130 (72%) | 42 (14, 168) | 56 (13, 168) |
| 41 – 60 | 75 (26%) | 49 (65%) | 57 (76%) | 50 (67%) | 42 (13, 119) | 50 (13, 124) |
| 61 – | 17 (6%) | 11 (65%) | 11 (65%) | 11 (65%) | 29 (14, 84) | 29 (14, 112) |
| Body Mass Index | 168 (58%) | 227 (79%) | 178 (62%) | |||
| BMI < = 18.5 | 84 (29%) | 46 (55%) | 67 (80%) | 50 (60%) | 42 (13, 140) | 56 (13, 140) |
| BMI > 18.5 | 205 (71%) | 122 (60%) | 160 (78%) | 128 (62%) | 42 (13, 168) | 56 (13, 168) |
| Enrollment region | 190 (66%) | 227 (79%) | 206 (71%) | |||
| N. America (NA) | 105 (36%) | 84 (80%) | 83 (79%) | 76 (72%) | 35 (13, 119) | 42 (13, 124) |
| Spain (ESP) | 21 (7%) | 16 (76%) | 14 (67%) | 19 (90%) | 42 (14, 87) | 42 (14, 87) |
| S. Africa (AF) | 48 (17%) | 34 (71%) | 37 (77%) | 36 (75%) | 42 (13, 168) | 56 (13, 168) |
| Uganda (UGA) | 115 (40%) | 56 (49%) | 93 (81%) | 75 (65%) | 42 (14, 140) | 61 (28, 140) |
| HIV Status | 190 (66%) | 257 (89%) | 206 (71%) | |||
| Negative | 273 (94%) | 181 (66%) | 213 (78%) | 194 (71%) | 42 (13, 168) | 56 (13, 168) |
| Positive | 16 (6%) | 9 (56%) | 14 (87%) | 12 (75%) | 42 (14,82) | 56 (17, 112) |
| Baseline Chest X-Ray | 189 (66%) | 226 (78%) | 205 (71%) | |||
| No cavity | 101 (35%) | 70 (69%) | 81 (80%) | 80 (79%) | 42 (13, 141) | 49 (13, 140) |
| Cavity ≤ 4 cm | 78 (27%) | 53 (68%) | 60 (77%) | 57 (73%) | 42 (13, 168) | 56 (13, 168) |
| Cavity > 4 cm | 109 (38%) | 66 (60%) | 85 (78%) | 68 (62%) | 42 (13, 140) | 56 (13, 119) |
| Baseline MGIT960 | 149 (52%) | 215 (74%) | 161 (56%) | |||
| TTD < = 5 days | 77 (27%) | 25 (32%) | 42 (55%) | 35 (45%) | 42 (14, 168) | 59 (14, 168) |
| TTD > 5 days | 191 (66%) | 124 (65%) | 135 (70%) | 126 (66%) | 42 (13, 141) | 55 (13, 117) |
| Study Arm | 190 (66%) | 227 (79%) | 206 (71%) | |||
| Rifampin | 137 (47%) | 90 (66%) | 108 (79%) | 101 (74%) | 28 (13, 140) | 56 (13, 140) |
| Rifapentine | 152 (53%) | 100 (66%) | 119 (78%) | 105 (69%) | 42 (13, 168) | 56 (13, 124) |
BMI = Body Mass Index; TTD = Time to Detection
3.2. Treatment effect on serum protein abundances
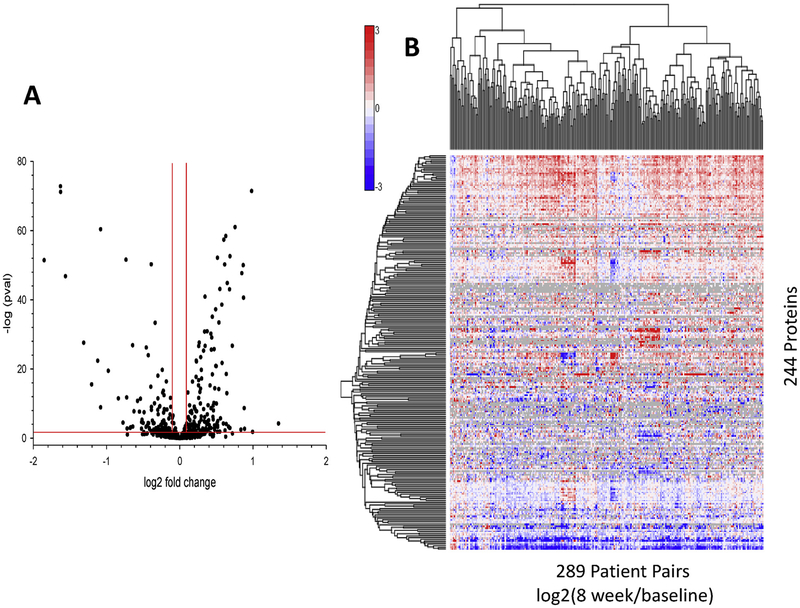
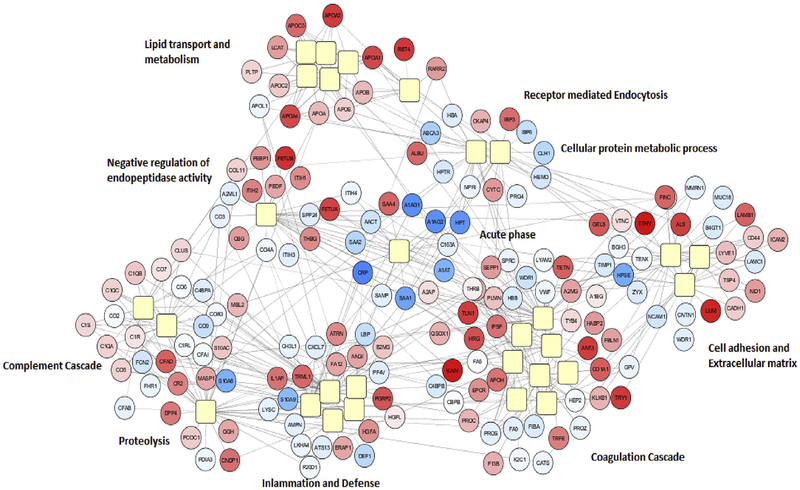
A linear mixed model, fit to investigate the outcome associated with treatment effect of each serum protein abundance over time, identified 244 serum proteins deemed significantly changing after 8 weeks of treatment (Fig. 2A and Supplemental Table 2). A heat map of the 244 serum proteins identified, where values represent the quantitative change in protein abundance between baseline and 8 weeks per protein, is shown in Fig. 2B. Observed are subsets of proteins which are quantitatively differentiated in the serum due to either up (red) or down (blue) regulation upon treatment. The majority of protein alterations are upregulated upon treatment (138 out of 244). Network and pathway interaction mapping of these treatment related proteins identified interconnections of multiple pathway networks and their response after 8 weeks of treatment (Fig. 3). In addition to acute phase proteins, pathway networks emerged for lipid transport and metabolism; complement cascade; proteolysis; coagulation cascade; inflammation and defense; and cell adhesion and extracellular matrix, among others.
Fig. 2.
Visual representation of the serum proteins differential abundant due to 8 weeks of antibiotic treatment. A) Volcano plot of all proteins analyzed via linear mixed model to determine differential abundance due to antibiotic treatment at 8 weeks. Red lines correspond to thresholds of acceptance (pval < 0.05 and > ±0.1 log2 fold change). B) Heat map of 244 proteins representing quantitative changes in protein abundances between baseline and 8 weeks from 289 patients. Red magnitude representing increase in abundance with treatment, blue magnitude representing reduced serum abundance upon treatment. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
Protein based pathway mapping of the patient response in the serum to antibiotic treatment. The GO annotations of 244 significantly altered proteins were extracted from DAVID and after corrected p-value filtering was visualized in Cytoscape to create the interconnected pathway network. Yellow squares: Pathways, spatial proximity indicates similarity in pathways; Red circles: proteins upregulated following 8 weeks of therapy; Blue circles: proteins downregulated following 8 weeks of therapy. Color intensity represents degree of fold change, darker signifying larger fold change. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Serum protein signatures and culture conversion
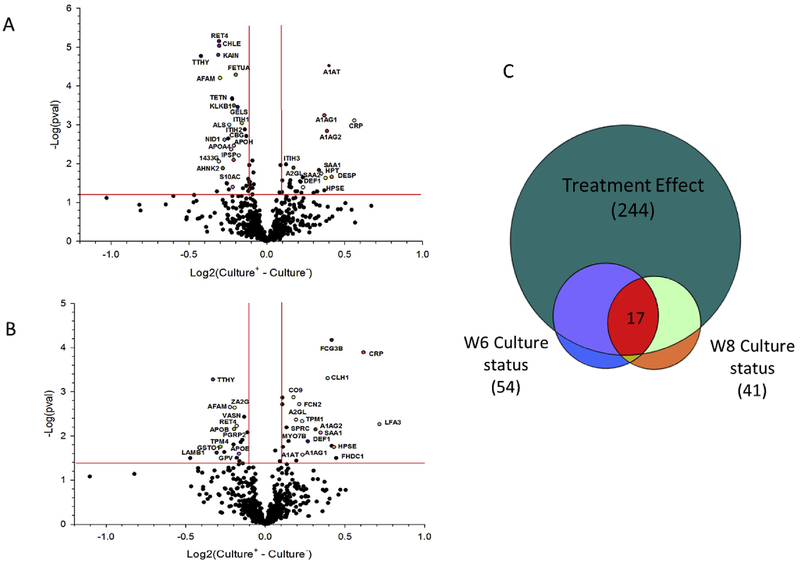
By utilizing the same comparison model described above, patient serum protein results were divided and analyzed based upon either positive or negative culture status for a particular time point to determine protein significance. Looking at week 2 culture status stratifications, CRP and many other highly downregulated proteins (SAA1, A1AG1) have reduced fold change differences for patients who have quickly converted at week 2 compared to week 8 stratifications where such proteins have similar or greater fold change differences between positive and negative stratification (Supplemental Table 3). Investigation of patients who remain culture positive after 8 weeks of treatment utilized the same model as previous but fit the outcome to culture status stratification at either 6 or 8 weeks of treatment. Fig. 4A and B shows the volcano plots of the week 8 and week 6 culture results respectively. Overall, we identified 41 and 54 proteins for week 8 and week 6 culture status respectively whose baseline to end of intensive phase biomarker changes were significantly differing between culture-negative and culture-positive patients. Fig. 4C shows the overlap between each comparison and the original 244 treatment effect serum proteins. Whereas these culture-status comparisons are statistically less significant, the 17 proteins which are consistently discriminatory for both week 6 and 8 culture status stratifications provide a promising list to explore further as predictive markers of culture conversion. (Fig. 4A and B and Supplemental Table 4).
Fig. 4.
Plots and diagram of proteins significantly altered based upon culture status at week 6 and week 8 treatment. Volcano plot showing log2 values calculated from the difference of fold change from baseline to week 8 for culture positive (culture+) patients compared to culture negative (culture-) patients at (a) Week 6 culture status and B) Week 8 culture status based upon a linear mixed model. Proteins labeled are those most significant and found in common across both week 6 and week 8 results. C) Venn diagram representing overlap between the 244 treatment serum proteins and discriminatory proteins for week 6 and week 8 culture status stratification.
3.4. Baseline serum proteins predictive of culture status
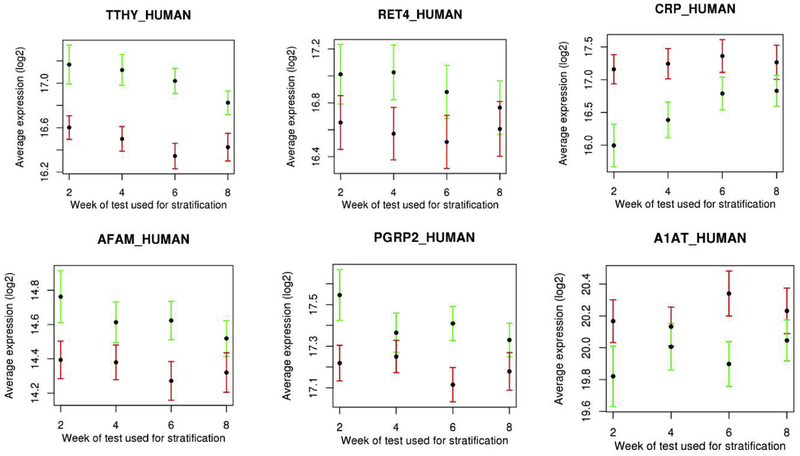
Seventeen proteins were identified with concentrations at baseline that were associated with culture conversion status with a p < 0.001 at any time point stratification (Supplemental Table 5). Across all culture time points, these seventeen proteins were most discriminatory for the week 6 culture status. The top six baseline proteins that discriminate culture status at subsequent time points are shown in Fig. 5, and all overlap with those identified as significant in fold change comparisons from baseline to week 8.
Fig. 5.
Plots of 6 serum proteins measured at baseline with highest significance in discriminating subsequent culture status across all culture time points. Black dots represent the average log2 protein abundance for that protein, red error bars (standard deviation) represent positive culture status patients, where green error bars represent patients with negative culture status. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
In this study, we identified a network of interconnected proteins and pathways representing the host response to TB treatment over 8 weeks that were differentially expressed in relation to culture conversion status. A total of 244 proteins were significantly altered in response to 8 weeks of intensive phase therapy. Pathway and network comparisons identify proteins that during treatment are up regulated (coagulation cascade, endopeptidase activity, lipid transport), with other proteins down regulated (acute phase), in addition to other cross regulated networks (inflammation, cell adhesion, extracellular matrix) (see Supplemental Tables 1 and 2). Across pathways and network comparisons, we discuss below selected proteins specifically associated with lung injury, bacterial load, and/or pulmonary bacterial infection in the published literature, in addition to proteins that are noted to be unique in terms of proteomic discovery outputs. The most significant changes between baseline and at 8-weeks of treatment occurred for proteins that were upregulated at the time of diagnosis and decreased significantly after treatment, predominated by acute phase reactants. Numerous proteins from coagulation and complement cascade pathways were also differentially expressed after treatment, including anti-coagulation proteins endothelial protein C receptor (EPCR) [28], plasma serine protease inhibitor (IPSP), alpha-2-macroglobulin (A2MG), coagulation factor XII (FA12) [29] and hyaluronan-binding protein 2 (HABP2), which aids in fibrinolysis [30] and prevention of fibrin clots formation by acting as an inhibitor of coagulation [29]. Additional coagulation-associated proteins included antithrombin III (ANT3) which participates in inhibition of several procoagulants [31], phosphatidylethanolamine-binding protein 1 (PEBP1) a serine protease inhibitor that inhibits thrombin, and Beta-2-glycoprotein 1 (APOH) and vitamin K-dependent protein C (PROC) that degrades factor V and VIII [32], further preventing formation of clots. Coagulation associated proteins that decreased with treatment were fibrinogen alpha chain (FIBA), factor IX (FA9), platelet factor 4 variant (PF4V) and vitamin K dependent protein S (PROS). Finally, we observed down-regulation of Ficolin-2 (FCN2), which acts as an opsonin that facilitates bacterial elimination via the complement cascade [33].
Several proteins involved in immune modulation and airway defense were also identified as differentially expressed, notably two antibacterial proteins that decreased with treatment: neutrophil defensin1 (DEF1) and platelet basic protein (CXCL7). Ashitani et al. reported an increased concentration of alpha-defensins in plasma of patients with active TB where the concentration of defensins reduced after 8 weeks of treatment [34]. Kalita et al. reported that human neutrophil peptides (HNP)s in conjugation with anti-TB drugs resulted in significant clearance of bacterial load [35] where the lower concentration post treatment is likely due to a decreased bacterial load. CXCL7, produced by human phagocytes, has been proposed to have a significant influence on the immune response against mycobacterial infection [34] and LYSC has been known to have bacteriolytic properties - both proteins decreased significantly during treatment. We also identified down-regulation of protein ATP-binding cassette sub-family A member 3 (ABCA3), which acts in the formation of pulmonary surfactant by transporting lipids [36]. Pulmonary surfactants provide alveolar stability by lowering surface tension at the air-liquid interface and also serve a direct anti-microbial role [37]. The presence of ABCA3 in the blood serum is interesting, and the down regulation may provide a blood marker of decreasing bacterial load. Two other proteins, cystatin C (CYTC), a potent inhibitor of the proteolytic activity of cysteine cathepsins that drive lung fibrosis [38–40], and selenoprotein P (SEPP1) reported to have antioxidant properties [15], were upregulated after 8 weeks of treatment.
Cavitation in pulmonary TB results in degradation of structural fibers and extracellular matrix (ECM) protein components [41], which can provide altered protein signatures in serum. We identified several likely ECM protein markers for further investigation. Fibrillar type I collagen in ECM is responsible for providing the tensile strength in lung tissues [41]. It has been reported that Mtb induces an increased expression as well as secretion of matrix metalloproteinase (MMP-1), which has been reported to increase following bacterial infection [42]. Metalloproteinase inhibitor-1 (TIMP-1) is an inhibitor of MMP-1 [42], and in our study, we found TIMP-1 decreased following treatment in conjunction with increasing MMP secretion. As cavitation resolves during treatment, upregulation of matrix and cell adhesion proteins that are involved in the repair process of the ECM of lung tissue is expected. Indeed, we found matrix proteins upregulated after treatment, including neural cell adhesion molecule L1-like protein (NCHL1) which facilitates cell-cell or cell-matrix interactions [43], lumican (LUM) known to regulate ECM’s function such as tissue hydration and collagen fibrillogenesis [44], fibronectin (FINC), which binds to integrins promoting cellular adhesion [45], laminin subunit beta-1 (LAMB1), an ECM protein, collagen alpha-1 chain (CO1A1), a major structural components of the lung [42], as well as gelsolin (GELS) and talin (TLN1) both involved in cell-matrix interactions [18,46].
Several proteases and peptidases were differentially regulated on treatment with a majority of peptidases and proteases up-regulated. Fetuin A (FETUA) enhances phagocytosis of apoptotic cells, and has been reported to decrease in whole blood samples of patients with active TB [47]. Olivier et al., found that both Fetuin A and Fetuin B (FETUB) are negative markers of acute phase reaction. We saw a similar trend, as both these peptidases were downregulated in patients at baseline and increased significantly post treatment [48]. Inter-alpha-trypsin inhibitor heavy chain (ITIH-H1 and H2) is known to contribute to extracellular matrix stability by covalently binding to hyaluronic acid [49]. This is in line with their upregulation post treatment. N-acetylmuramoyl-L-alanine amidase (PGRP2) was also up-regulated, likely due to its role in digesting biologically active bacterial peptidoglycan into inactive fragments [50]. In contrast, heparanase (HPSE) was down-regulated post-treatment. Heparanase is an endoglycosidase which cleaves heparan sulfate proteoglycans for extracellular matrix degradation and remodeling [51]. Among other functions, recent reports have it associated with acute lung injury [52], where heparanase was associated with loss of heparin sulfate in glycocalyx degradation, leading to neutrophil adherence and inflammation. Inhibition of heparanase prevented neutrophil adhesion and attenuated sepsis induced acute lung injury in mice [52]. In humans, heparanase was observed higher in abundance in lung biopsies with alveolar damage as well as higher plasma heparan sulfate degradation activity [53]. Identification and quantification of heparanase in the serum is significant as it groups with the acute phase markers of infection, and possibly provides a lung specific marker reflecting lung tissue damage and recovery.
Consistent with other reports [33], several lipid transport proteins were significantly upregulated after treatment. Hypocholesterolemia has been reported in TB patients [54], and it has been proposed that Mtb may derive carbon and energy from cholesterol [55]. Other lipoproteins with anti-inflammatory properties, namely platelet-activating factor acetyl hydrolase (PAFA) and phosphatidylcholine-sterol acyltransferase (LCAT) were also up-regulated with treatment. Binding proteins also provide indication of nutritional status and were down-regulated at the time of diagnosis. Examples include corticosteroid binding protein (CBG), transthyretin (TTHY), serotransferrin (TRFE), insulin-like growth factor-binding protein 3 (IGBP3) [56], albumin (ALBU) and tetranectin (TETN) and afamin (AFAM). Retinol binding protein (RET4) increased expression post-treatment might be due to retinoic acid’s ability to induce monocyte differentiation [42]. This helps in inhibition of bacterial proliferation in macrophages, thus further reducing the bacterial load [42]. TETN binds plasminogen, increasing its activity and resulting in more plasmin formation promoting fibrinolytic activity [57]. Afamin as a vitamin E binding protein is reduced at baseline likely due to a known decrease in concentration of antioxidants in TB infections [58]. Conversely, upregulation of Thyroxine binding globulin (THBG) upon treatment may be attributed to the hepatic effect of the use of rifamycins in the regimens studied in the parent trial [59]. Regarding drug effects, we found sex hormone-binding globulin (SHBG) to be significantly increased on treatment. Rifamycins have been shown to increase the expression of SHBG [60]. This is proposed to be due to the induction of cytochrome P450 mediated monoxygenase activity, which is believed to be a rate limiting step in the synthesis of testosterone and estrogen from cholesterol through pregnanolone [60]. As a result of the increase synthesis of these androgens, SHBG expression increases proportionally as well.
Utilizing culture status conversion data through 8 weeks, the change in protein serum concentrations over this timeframe provided mechanistic insights when correlating how quickly (or slowly) patients recover from TB disease. We observed a consistent set of proteins that represent a signature of a slower response to treatment (Fig. 4C). We focused on proteins which were consistently discriminatory for both 6 and 8 weeks of culture status stratification as these two time points represent the inflection point where the majority of patients convert from positive to negative. Interestingly, regardless of the time point stratification, the majority of these proteins overlap with those previously identified in the overall treatment effect changes, 17 proteins, implying they are associated with treatment or disease resolution. Each of these proteins has the same directionality of change regardless of time point stratification, see Supplemental Table 4. This suggests an overall delay in recovery response for those patients remaining culture positive at week 6 through week 8. Specifically, all acute phase proteins in this list, CRP, A1AT, A1AG1, A1AG2, SAA1, have lower fold changes for culture positive patients compared to culture negative patients at both week 6 and week 8, Supplemental Table 3. These proteins are the most reduced in abundance due to treatment and that reduction is attenuated for those still culture positive at these later time points. Similarly, proteins heparanase (HPSE) and DEF1 were also highly downregulated in treatment and showed diminished downregulation in culture positive individuals. The changes in heparanase abundance, which infers correlation with lung injury, has the potential to provide a unique specific serum marker of lung injury/and recovery in Mtb patients. Conversely, lipid transport proteins (APOA4, APOH), binding proteins (TTHY, AFAM, RET4), as well as protease PGRP2 were seen as upregulated overall due to treatment and disease resolution but this upregulation was diminished in patients still culture positive at 6 and 8 weeks. Though these proteins consistently correlated with disease severity and resolution, how predictive they are in regard to relapse cases will need further investigation.
An evaluation of baseline protein values in the serum identified a core set of proteins which correlated well with future culture conversion. Many of these proteins overlap with our treatment effect analyses, suggesting that for these particular proteins, the initial protein abundance at baseline appears to relate to their change in abundance in regard to patient culture status conversion, and hence can provide an important initial baseline indicator of treatment efficacy. Baseline values of proteins TTHY, AFAM, CRP, and PGRP2 showed the greatest differentiation, however we did see variations in predictive ability across the week stratifications. CRP differentiated best in early time points but gradually reduced out to week 8. Interestingly, TTHY, AFAM, and PGRP2 showed increased discrimination at week 6. It is somewhat unclear as to why, however it does appear to be driven by a drop in the remaining culture positive patients. Week 6 also had the most balanced stratification between culture positive/negative patients. The Study 29 serum sample set used in our study was also evaluated using directed immunoassays for a pre-selected panel of biomarkers, focusing primarily on biomarkers of inflammation and immune response [61]. We saw overlap in markers that were significantly modulated by treatment (CRP, SAA, MMP-1, TIMP-1) and in the biomarkers associated with early conversion (CRP). The non-directed MS approach used in the current study, however, was able to identify additional pathways than were found using the directed approach.
Our study has limitations. First, long-term outcomes of interest (relapse versus durable cure) were not a component of the parent phase2 trial, and our analyses were focused on biomarker associations with baseline severity and microbiologic indicators of treatment effect only. An evaluation of the most promising biomarkers we discovered should be pursued as part of clinical trials with long-term follow-up. Second, we were limited in our ability to explore the dynamics of biomarker change given two time points for serum collection. An assessment of the best performing markers in our study, across additional time points would improve our understanding of the predictive performance of these biomarkers. Third, the parent trial allowed for TB treatment prior to the randomization visit when serum was collected and study drugs initiated. This may have impacted baseline biomarker levels, however, in sensitivity analyses limited to participants with no pre-treatment, the biomarkers with the strongest associations with baseline severity and treatment effect remained the same.
In sum, we provide a comprehensive host serum protein dataset reflective of TB treatment effect. A core set of core serum proteins (TTHY, AFAM, CRP, RET4, SAA1, PGRP2, among others) were found to change significantly in response to treatment, to strongly correlate with culture status, and at baseline to be predictive of subsequent culture conversion. The protein signatures identified here have the potential to be informative of disease resolution throughout the treatment time course. If validated in cohorts of patients with longer treatment and follow-up, the proteins identified may serve as blood-based markers of treatment effect with applications in clinical care and clinical trials.
Supplementary Material
Acknowledgements
We thank Drs. Susan Dorman, Jason Stout, Neil Schluger as protocol co-Chairs and the TBTC Study 29 protocol team for their support of embedding biomarker studies into the clinical trial. We thank Drs. Karen Dobos and Nicole Kruh-Garcia for input on biomarker interpretations. We thank M. Engle for assistance in sample handling, testing and data processing. Funding for recruitment, enrollment, and clinical and laboratory follow-up of TBTC Study 29 participants was provided by the United States Centers for Disease Control and Prevention. The biomarker assays and PN were supported by the National Institutes of Health through National Institute of Allergy and Infectious Diseases funding (1R01AI104589: TB Surrogate Markers for Assessing Response to Treatment (TB SMART Study)), and by the Centers for Disease Control and Prevention, TB Trials Consortium Contract 200–200932597 (P.N.). Portions of this research were supported by NIH NIGMS GM103493. Work was performed in the Environmental Molecular Sciences Laboratory, a U. S. Department of Energy Office of Biological and Environmental Research national scientific user facility located at Pacific Northwest National Laboratory in Richland, Washington. Pacific Northwest National Laboratory is operated by Battelle for the U.S. Department of Energy under Contract No. DE-AC05–76RLO 1830.
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.tube.2018.07.005.
References
- [1].World Health Organization. Global tuberculosis report. Geneva, Geneva, Switzerland: World Health Organization; 2017. [Google Scholar]
- [2].Falzon D, Schunemann HJ, Harausz E, Gonzalez-Angulo L, Lienhardt C, Jaramillo E, et al. World Health Organization treatment guidelines for drug-resistant tuberculosis, 2016 update. Eur Respir J 2017;49(3). 10.1183/13993003.02308-2016. Epub 2017/03/24 PubMed PMID: ; PubMed Central PMCID: PMCPMC5399349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nahid P, Dorman SE, Alipanah N, Barry PM, Brozek JL, Cattamanchi A, et al. Official American thoracic society/centers for disease control and prevention/infectious diseases society of America clinical practice guidelines: treatment of drug-susceptible tuberculosis. Clin Infect Dis 2016;63(7):e147–95. 10.1093/cid/ciw376. Epub 2016/08/16 PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nahid P, Saukkonen J, Mac Kenzie WR, Johnson JL, Phillips PP, Andersen J, et al. CDC/NIH Workshop. Tuberculosis biomarker and surrogate endpoint research roadmap. Am J Respir Crit Care Med 2011;184(8):972–9. 10.1164/rccm.201105-0827WS. Epub 2011/07/09 PubMed PMID: ; PubMed Central PMCID: PMC3208659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Horne DJ, Royce SE, Gooze L, Narita M, Hopewell PC, Nahid P, et al. Sputum monitoring during tuberculosis treatment for predicting outcome: systematic review and meta-analysis. Lancet Infect Dis 2010;10(6):387–94. 10.1016/S1473-3099(10)70071-2. Epub 2010/06/01 PubMed PMID: ; PubMed Central PMCID: PMC3046810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wallis RS, Peppard T, Hermann D. Month 2 culture status and treatment duration as predictors of recurrence in pulmonary tuberculosis: model validation and update. PLoS One 2015;10(4):e0125403 10.1371/journal.pone.0125403. Epub 2015/04/30 PubMed PMID: ; PubMed Central PMCID: PMCPMC4414505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Phillips PPJ, Mendel CM, Nunn AJ, McHugh TD, Crook AM, Hunt R, et al. A comparison of liquid and solid culture for determining relapse and durable cure in phase III TB trials for new regimens. BMC Med 2017;15(1):207. 10.1186/s12916-017-0955-9. Epub 2017/11/25 PubMed PMID: ; PubMed Central PMCID: PMCPMC5701316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schubert OT, Rost HL, Collins BC, Rosenberger G, Aebersold R. Quantitative proteomics: challenges and opportunities in basic and applied research. Nat Protoc 2017;12(7):1289–94. 10.1038/nprot.2017.040. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [9].Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, et al. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell 2016;166(3):755–65. 10.1016/j.cell.2016.05.069. PubMed PMID: ; PubMed Central PMCID: PMCPMC4967013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Agranoff D, Fernandez-Reyes D, Papadopoulos MC, Rojas SA, Herbster M, Loosemore A, et al. Identification of diagnostic markers for tuberculosis by proteomic fingerprinting of serum. Lancet 2006;368(9540):1012–21. doi: S0140–6736(06)69342–2 [pii] 10.1016/S0140-6736(06)69342-2. Epub 2006/09/19 PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu J, Jiang T, Wei L, Yang X, Wang C, Zhang X, et al. The discovery and identification of a candidate proteomic biomarker of active tuberculosis. BMC Infect Dis 2013;13(1):1–11. 10.1186/1471-2334-13-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Song SH, Han M, Choi YS, Dan KS, Yang MG, Song J, et al. Proteomic profiling of serum from patients with tuberculosis. Annal Lab Med 2014;34(5):345–53. 10.3343/alm.2014.34.5.345. PubMed PMID: ; PubMed Central PMCID: PMC4151002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Xu D, Li Y, Li X, Wei LL, Pan Z, Jiang TT, et al. Serum protein S100A9, SOD3, and MMP9 as new diagnostic biomarkers for pulmonary tuberculosis by iTRAQ-coupled two-dimensional LC-MS/MS. Proteomics 2015;15(1):58–67. 10.1002/pmic.201400366. Epub 2014/10/22 PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [14].Xu DD, Deng DF, Li X, Wei LL, Li YY, Yang XY, et al. Discovery and identification of serum potential biomarkers for pulmonary tuberculosis using iTRAQ-coupled twodimensional LC-MS/MS. Proteomics 2014;14(2–3):322–31. 10.1002/pmic.201300383. Epub 2013/12/18 PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [15].Achkar JM, Cortes L, Croteau P, Yanofsky C, Mentinova M, Rajotte I, et al. Host protein biomarkers identify active tuberculosis in HIV uninfected and Co-infected individuals. EBioMedicine 2015;2(9):1160–8 10.1016/j.ebiom.2015.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nahid P, Bliven-Sizemore E, Jarlsberg LG, De Groote MA, Johnson JL, Muzanyi G, et al. Aptamer-based proteomic signature of intensive phase treatment response in pulmonary tuberculosis. Tuberculosis (Edinb). 2014;94(3):187–96. 10.1016/j.tube.2014.01.006. Epub 2014/03/19 PubMed PMID: ; PubMed Central PMCID: PMCPMC4028389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].De Groote MA, Nahid P, Jarlsberg L, Johnson JL, Weiner M, Muzanyi G, et al. Elucidating novel serum biomarkers associated with pulmonary tuberculosis treatment. PLoS One 2013;8(4):e61002 10.1371/journal.pone.0061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ochsner U, Green L, De Groote M, Sterling D, Ostroff R, Janjic N. Biomarkers of pulmonary tuberculosis identified in multiplexed proteomic assay (SOMAscan) of human serum (MPF2P.808). J Immunol 2014;192(1 Supplement). 67.7. [Google Scholar]
- [19].Baker ES, Burnum-Johnson KE, Jacobs JM, Diamond DL, Brown RN, Ibrahim YM, et al. Advancing the high throughput identification of liver fibrosis protein signatures using multiplexed ion mobility spectrometry. Mol Cell Proteomics 2014;13(4):1119–27. 10.1074/mcp.M113.034595. PubMed PMID: ; PubMed Central PMCID: PMC3977189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dorman SE, Goldberg S, Stout JE, Muzanyi G, Johnson JL, Weiner M, et al. Substitution of rifapentine for rifampin during intensive phase treatment of pulmonary tuberculosis: study 29 of the tuberculosis trials consortium. J Infect Dis 2012;206(7):1030–40. 10.1093/infdis/jis461. Epub 2012/08/02 PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [21].Livesay EA, Tang K, Taylor BK, Buschbach MA, Hopkins DF, LaMarche BL, et al. Fully automated, four-column capillary LC-MS system for maximizing throughput in proteomic analyses. Anal Chem 2008;80(1):294–302. 10.1021/ac701727r. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Baker ES, Livesay EA, Orton DJ, Moore RJ, Danielson WF 3rd, Prior DC, et al. An LC-IMS-MS platform providing increased dynamic range for high-throughput proteomic studies. J Proteome Res 2010;9(2):997–1006. 10.1021/pr900888b. Epub 2009/12/17 PubMed PMID: ; PubMed Central PMCID: PMC2819092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zimmer JS, Monroe ME, Qian WJ, Smith RD. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom Rev 2006;25(3):450–82. 10.1002/mas.20071. Epub 2006/01/24 PubMed PMID: ; PubMed Central PMCID: PMCPMC1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 1994;5(11):976–89 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- [25].Kim S, Gupta N, Pevzner PA. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J Proteome Res 2008;7(8):3354–63. 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA, Smith RD. Decon2LS: an open-source software package for automated processing and visualization ofhigh resolution mass spectrometry data. BMC Bioinf 2009;10:87. 10.1186/1471-2105-10-87. Epub 2009/03/19. PubMed PMID: ; PubMed Central PMCID: PMC2666663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Monroe ME, Tolic N, Jaitly N, Shaw JL, Adkins JN, Smith RD. VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics (Oxford, England) 2007;23(15):2021–3. 10.1093/bioinformatics/btm281. Epub 2007/06/05. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [28].Esmon CT, Xu J, Gu J-m, Qu D, Laszik Z, Ferrell G, et al. Endothelial protein C receptor. Thromb Haemostasis 1999;82(8):251–8. [PubMed] [Google Scholar]
- [29].Choi-Miura NH, Yoda M, Saito K, Takahashi K, Tomita M. Identification of the substrates for plasma hyaluronan binding protein. Biol Pharm Bull 2001;24(2):140–3. Epub 2001/02/24. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [30].Konings J, Hoving LR, Ariëns RS, Hethershaw EL, Ninivaggi M, Hardy LJ, et al. The role of activated coagulation factor XII in overall clot stability and fibrinolysis. Thromb Res 2015;136(2):474–80 10.1016/j.thromres.2015.06.028. [DOI] [PubMed] [Google Scholar]
- [31].Mintz PD, Blatt PM, Kuhns WJ, Roberts HR. Antithrombin III in fresh frozen plasma, cryoprecipitate, and cryoprecipitate-depleted plasma. Transfusion 1979;19(5):597–8. Epub 1979/09/01. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [32].Koedam JA, Meijers JC, Sixma JJ, Bouma BN. Inactivation of human factor VIII by activated protein C. Cofactor activity of protein S and protective effect of von Willebrand factor. J Clin Invest 1988;82(4):1236–43. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang C, Wei L-L, Shi L-Y, Pan Z-F, Yu X-M, Li T-Y, et al. Screening and identification of five serum proteins as novel potential biomarkers for cured pulmonary tuberculosis. Sci Rep 2015;5:15615. 10.1038/srep15615www.nature.com/articles/srep15615#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gonzalez-Cortes C, Diez-Tascon C, Guerra-Laso JM, Gonzalez-Cocano MC, Rivero-Lezcano OM. Non-chemotactic influence of CXCL7 on human phagocytes. Modulation of antimicrobial activity against L. pneumophila. Immunobiology 2012;217(4):394–401. 10.1016/j.imbio.2011.10.015. Epub 2011/11/22 PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [35].Kalita A, Verma I, Khuller GK. Role of human neutrophil peptide-1 as a possible adjunct to antituberculosis chemotherapy. J Infect Dis 2004;190(8):1476–80. 10.1086/424463. Epub 2004/09/21 PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [36].Ban N, Matsumura Y, Sakai H, Takanezawa Y, Sasaki M, Arai H, et al. ABCA3 as a lipid transporter in pulmonary surfactant biogenesis. J Biol Chem 2007;282(13):9628–34. 10.1074/jbc.M611767200. [DOI] [PubMed] [Google Scholar]
- [37].Han S, Mallampalli RK. The role of surfactant in lung disease and host defense against pulmonary infections. Annal Am Thorac Soc 2015;12(5):765–74. 10.1513/AnnalsATS.201411-507FR. Epub 2015/03/06 PubMed PMID: ; PubMed Central PMCID: PMCPMC4418337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kramer L, Turk D, Turk B. The future of cysteine cathepsins in disease management. Trends Pharmacol Sci 2017;38(10):873–98. 10.1016/j.tips.2017.06.003. Epub 2017/07/03 PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [39].Randers E, Kornerup K, Erlandsen EJ, Hasling C, Danielsen H. Cystatin C levels in sera of patients with acute infectious diseases with high C-reactive protein levels. Scand J Clin Lab Invest 2001;61(4):333–5. Epub 2001/07/24. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [40].Chapman HA Jr., Reilly JJ Jr., Yee R, Grubb A. Identification of cystatin C, a cysteine proteinase inhibitor, as a major secretory product of human alveolar macrophages in vitro. Am Rev Respir Dis 1990;141(3):698–705. 10.1164/ajrccm/141.3.698. Epub 1990/03/01 PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [41].Elkington PT, Emerson JE, Lopez-Pascua LD, O’Kane CM, Horncastle DE, Boyle JJ, et al. Mycobacterium tuberculosis up-regulates matrix metalloproteinase-1 secretion from human airway epithelial cells via a p38 MAPK switch. J Immunol (Baltimore Md 1950) 2005;175(8):5333–40. Epub 2005/10/08. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [42].Ong CW, Elkington PT, Friedland JS. Tuberculosis, pulmonary cavitation, and matrix metalloproteinases. Am J Respir Crit Care Med 2014;190(1):9–18. 10.1164/rccm.201311-2106PP. Epub 2014/04/10 PubMed PMID: ; PubMed Central PMCID: PMCPMC4226026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jacobs R, Malherbe S, Loxton AG, Stanley K, van der Spuy G, Walzl G, et al. Identification of novel host biomarkers in plasma as candidates for the immunodiagnosis of tuberculosis disease and monitoring of tuberculosis treatment response. Oncotarget 2016. 10.18632/oncotarget.11420. Epub 2016/08/25 PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Matsuda Y, Yamamoto T, Kudo M, Kawahara K, Kawamoto M, Nakajima Y, et al. Expression and roles of lumican in lung adenocarcinoma and squamous cell carcinoma. Int J Oncol 2008;33(6):1177–85. Epub 2008/11/21. PubMed PMID: . [PubMed] [Google Scholar]
- [45].Wang C, Li Y-Y, Li X, Wei L-L, Yang X-Y, Xu D-D, et al. Serum complement C4b, fibronectin, and prolidase are associated with the pathological changes of pulmonary tuberculosis. BMC Infect Dis 2014;14(1):52. 10.1186/1471-2334-14-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Calderwood DA, Ginsberg MH. Talin forges the links between integrins and actin. Nat Cell Biol 2003;5(8):694–7. [DOI] [PubMed] [Google Scholar]
- [47].Tanaka T, Sakurada S, Kano K, Takahashi E, Yasuda K, Hirano H, et al. Identification of tuberculosis-associated proteins in whole blood supernatant. BMC Infect Dis 2011;11(1):1–10. 10.1186/1471-2334-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Olivier E, Soury E, Ruminy P, Husson A, Parmentier F, Daveau M, et al. Fetuin-B, a second member of the fetuin family in mammals. Biochem J 2000;350 Pt 2:589–97. Epub 2000/08/19. PubMed PMID: ; PubMed Central PMCID: PMCPMC1221288. [PMC free article] [PubMed] [Google Scholar]
- [49].Hamm A, Veeck J, Bektas N, Wild PJ, Hartmann A, Heindrichs U, et al. Frequent expression loss of Inter-alpha-trypsin inhibitor heavy chain (ITIH) genes in multiple human solid tumors: a systematic expression analysis. BMC Canc 2008;8:25. 10.1186/1471-2407-8-25. Epub 2008/01/30 PubMed PMID: ; PubMed Central PMCID: PMCPMC2268946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang Z-M, Li X, Cocklin RR, Wang M, Wang M, Fukase K, et al. Human peptidoglycan recognition protein-L is an N-Acetylmuramoyl-L-alanine amidase. J Biol Chem 2003;278(49):49044–52. 10.1074/jbc.M307758200. [DOI] [PubMed] [Google Scholar]
- [51].Bame KJ. Heparanases: endoglycosidases that degrade heparan sulfate proteoglycans. Glycobiology 2001;11(6). 10.1093/glycob/11.6.91R.91R-8R. [DOI] [PubMed] [Google Scholar]
- [52].Morris A, Wang B, Waern I, Venkatasamy R, Page C, Schmidt EP, et al. The role of heparanase in pulmonary cell recruitment in response to an allergic but not non-allergic stimulus. PLoS One 2015;10(6):e0127032 10.1371/journal.pone.0127032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Schmidt EP, Yang Y, Janssen WJ, Gandjeva A, Perez MJ, Barthel L, et al. The pulmonary endothelial glycocalyx regulates neutrophil adhesion and lung injury during experimental sepsis. Nat Med 2012;18(8):1217–23 http://www.nature.com/nm/journal/v18/n8/abs/nm.2843.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Perez-Guzman C, Vargas MH. Hypocholesterolemia: a major risk factor for developing pulmonary tuberculosis? Med Hypotheses 2006;66(6):1227–30. 10.1016/j.mehy.2005.12.041. Epub 2006/02/28 PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [55].Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci USA 2008;105(11):4376–80. 10.1073/pnas.0711159105. Epub 2008/03/13 PubMed PMID: ; PubMed Central PMCID: PMCPMC2393810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Le Bouc Y, Bellocq A, Philippe C, Perin L, Garabedian M, Fouqueray B, et al. Insulinlike growth factors and their binding proteins in pleural fluid. Eur J Endocrinol 1997;137(5):467–73. Epub 1997/12/24. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [57].Rodríguez-Flores E, Campuzano J, Aguilar D, Hernández-Pando R, Espitia C. The response of the fibrinolytic system to mycobacteria infection. Tuberculosis 2012;92(6):497–504 10.1016/j.tube.2012.07.002. [DOI] [PubMed] [Google Scholar]
- [58].Seyedrezazadeh E, Ostadrahimi A, Mahboob S, Assadi Y, Ghaemmagami J, Pourmogaddam M. Effect of vitamin E and selenium supplementation on oxidative stress status in pulmonary tuberculosis patients. Respirology (Carlton, Vic) 2008;13(2):294–8. 10.1111/j.1440-1843.2007.01200.x. Epub 2008/03/15 PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- [59].Hill AR, Schmidt MFJ, Schussler GC. Rapid changes in thyroid function tests upon treatment of tuberculosis. Tuberculosis 1995;76(3):223–9. 10.1016/S0962-8479(05)80009-7. [DOI] [PubMed] [Google Scholar]
- [60].Brodie MJ, Boobis AR, Gill M, Mashiter K. Does rifampicin increase serum levels of testosterone and oestradiol by inducing sex hormone binding globulin capacity? Br J Clin Pharmacol 1981;12(3):431–3. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Sigal GB, Segal MR, Mathew A, Jarlsberg L, Wang M, Barbero S, et al. Biomarkers of tuberculosis severity and treatment effect: a directed screen of 70 host markers in a randomized clinical trial. EBioMedicine 2017;25:112–21. 10.1016/j.ebiom.2017.10.018. Epub 2017/11/05 PubMed PMID: ; PubMed Central PMCID: PMCPMC5704068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.