Abstract
Controversy surrounds the molecular identity of mitochondrial K+ channels that are important for protection against cardiac ischemia–reperfusion injury. Although KNa1.2 (sodium-activated potassium channel encoded by Kcn2) is necessary for cardioprotection by volatile anesthetics, electrophysiological evidence for a channel of this type in mitochondria is lacking. The endogenous physiological role of a potential mito-KNa1.2 channel is also unclear. In this study, single channel patch clamp of 27 independent cardiac mitochondrial inner membrane (mitoplast) preparations from wild-type (WT) mice yielded 6 channels matching the known ion sensitivity, ion selectivity, pharmacology, and conductance properties of KNa1.2 (slope conductance, 138 ± 1 pS). However, similar experiments on 40 preparations from Kcnt2−/− mice yielded no such channels. The KNa opener bithionol uncoupled respiration in WT but not Kcnt2−/− cardiomyocytes. Furthermore, when oxidizing only fat as substrate, Kcnt2−/− cardiomyocytes and hearts were less responsive to increases in energetic demand. Kcnt2−/− mice also had elevated body fat, but no baseline differences in the cardiac metabolome. These data support the existence of a cardiac mitochondrial KNa1.2 channel, and a role for cardiac KNa1.2 in regulating metabolism under conditions of high energetic demand.—Smith, C. O., Wang, Y. T., Nadtochiy, S. M., Miller, J. H., Jonas, E. A., Dirksen, R. T., Nehrke, K., Brookes, P. S. Cardiac metabolic effects of KNa1.2 channel deletion and evidence for its mitochondrial localization.
Keywords: patch clamp, Slick, Slo2.1, bithionol
Numerous strategies for protection of the heart and other organs against ischemia–reperfusion (IR) injury are thought to require activation of K+ channels in the mitochondrial inner membrane [for a review, see Smith et al. (1)], including ischemic preconditioning, volatile anesthetic preconditioning (APC), and pharmacologic cardioprotection by K+ channel activators, such as NS-11021, bithionol (BT), and diazoxide (2–4). Concurrently, several K+ channels have been reported in mitochondria, including ATP-activated (KATP) (5), small-conductance Ca2+-activated (SK) (6, 7), and splice variants of large-conductance Ca2+ activated (BK) (8, 9). However, in only a small number of cases has the molecular (genetic) identity of specific mitochondrial channels involved in cardioprotection been proposed (3, 9–12). There are no K+ channels in the MitoCarta database of verified mitochondrial proteins (13), and no canonical mitochondrial target sequences have been identified in any K+ channel genes (1). As such, no clear mechanisms are known for the K+ channel targeting to mitochondria.
Mammalian sodium-activated potassium (KNa) channels are encoded by 2 genes: Kcnt1 (KNa1.1; Slack/SLO2.2) (14) and Kcnt2 (KNa1.2; Slick/SLO2.1) (15). Both KNa1.1 and KNa1.2 channels play important neurologic roles in the termination of seizure progression in epilepsy (16, 17). Although KNa1.2 expression has been reported in cardiac tissue (15, 18) and a generic KNa channel activity has been demonstrated in the cardiac cell membrane (19), notably the Kcnt2−/− mice have no cardiac phenotype (3, 18). Thus relatively little is known regarding the physiological role of KNa1.2 channels in the heart, including their subcellular location. In an earlier study, we showed that KNa1.2 is essential for cardiac APC, with hearts from Kcnt2−/− mice incapable of being protected against IR injury by isoflurane (3). In addition, we showed that the KNa opener BT is cardioprotective in wild-type (WT) mice but not in Kcnt2−/− mice (3, 20). Further, both BT and isoflurane activated a K+ flux in cardiac mitochondria isolated from WT mice but not in those from Kcnt2−/− mice (3). These observations led us to hypothesize that there are KNa1.2 channels in cardiac mitochondria.
There is considerable evidence that mitochondrial K+ channel activity can lessen the impact of IR injury (1). Less is known about the endogenous physiological roles of mitochondrial K+ channels (21)—in particular, mitochondrial KNa channels. Na+ enters mitochondria predominantly via an Na+/Ca2+ exchanger (NCLX) or an Na+/monocarboxylate transporter (MCT), with minor contribution from an Na+/H+ exchanger (22). Under conditions of elevated mitochondrial Na+ uptake (Ca2+ overload or cytosolic Na+ overload driving excessive NCLX activity or increased cytosolic Na+-lactate driving Na+ uptake via the Na+/monocarboxylate transporter), matrix Na+ may attain levels that can activate KNa channels (22–24). K+ influx via the KNa would dissipate mitochondrial membrane potential, thereby relieving a driving force for both Ca2+ uptake and NCLX activity. In effect, a mitochondrial KNa channel would be predicted to act as a safety valve to prevent ionic imbalance bought about by excessive Na+ uptake.
Using electrophysiological techniques (mitoplast patch clamp), we demonstrated that mitochondria contain a K+ channel that matches the known ion sensitivity, ion selectivity, pharmacology, and conductance properties of KNa1.2. Bioenergetic studies of WT and Kcnt2−/− hearts and cardiomyocytes also revealed a potential role for cardiac KNa1.2 in the metabolic response to a high energy demand.
MATERIALS AND METHODS
Animals
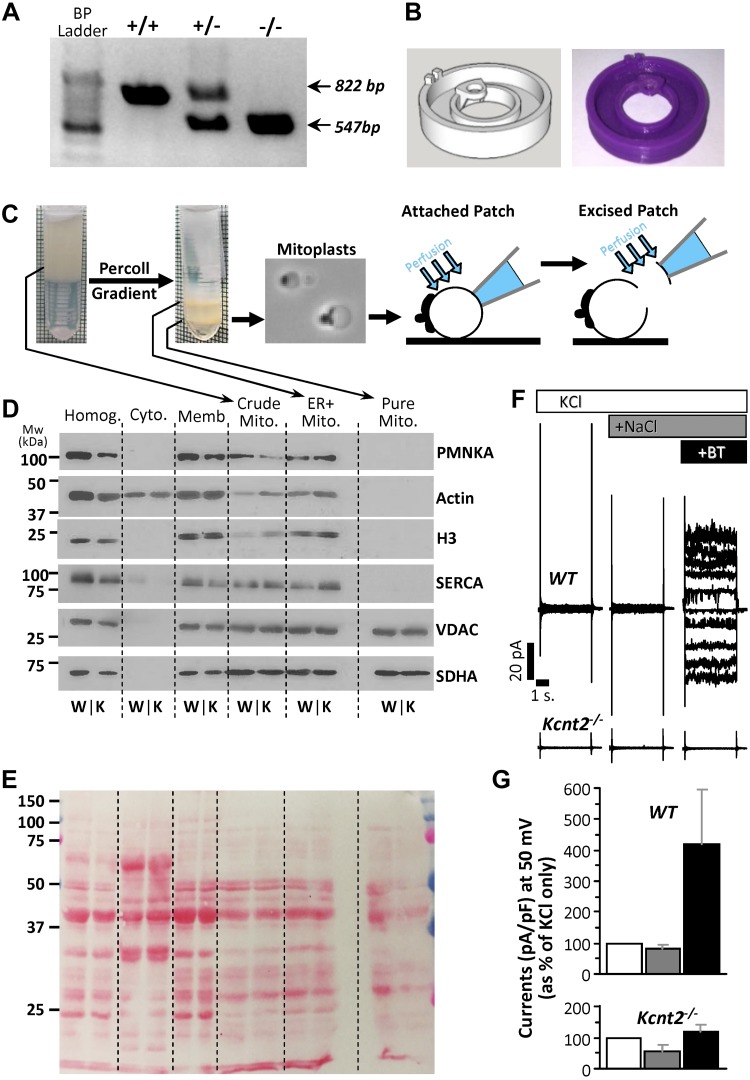
Male and female mice were housed in an Association for Assessment and Accreditation of Laboratory Animal Care–accredited pathogen-free facility with water and food available ad libitum. All procedures were locally approved and in accordance with the Guide for the Care and Use of Laboratory Animals [National Institutes of Health (NIH), Bethesda, MD, USA]. Mice were on a C57BL/6J background for >6 generations and periodically backcrossed to fresh stocks. Mice were bred from Kcnt2+/− parents, and male and females were separated, but otherwise littermate WT and Kcnt2−/− progeny were maintained in the same cages. Mice were genotyped by tail-clip PCR (Fig. 1A), with DNA extraction by a DNeasy Kit (Qiagen, Hilden, Germany) and genotyping by a KAPA2G Kit (Kapa Biosystems, Wilmington, MA, USA). Primers used were (5′→3′) forward-5′-AGGCAGCCATAGCTTTAGAGA-3′ and reverse 5′-CTCCTCATCGTGTGGTCCTA-3′, yielding amplicons at 822 and 547 bp for WT and Kcnt2−/− samples, respectively. Male and female mice were used for the experiments, and data are reported for the sexes separately or combined. Because the same personnel handled the mice and performed the experiments, the studies were not blinded to genotype.
Figure 1.
Kcnt2−/− mice, mitoplast purification and attached patch clamp. A) Example PCR analysis of tail clip genotyping of WT, heterozygous, and Kcnt2−/− mice. Amplicon expected for the WT (822 bp) and the knockout (547 bp) alleles. B) Custom 3-D–printed micro chamber for patch-clamp analysis of mitoplasts with computer model (left) and final product (right). C) Schematic depicting mitochondrial purification, mitoplast preparation, and attached patch and excised patch configuration. D) Western blot analysis of proteins from different cellular fractions during mitochondrial purification. Crude Mito, mitochondria-enriched fraction; Cyto, cytosol; ER+Mito, upper band following Percoll; [H3], Histone; Homog, homogenate; Memb, crude membrane; Pure Mito, lower band following Percoll. PMNKA, plasma membrane Na+/K+-ATPase; SERCA, sarcoendoplasmic reticulum Ca2+-ATPase; VDAC, voltage-dependent anion channel. E) Ponceau Red–stained membrane from blot in D for the loading control. (An empty lane was inserted between the ER+mit and Pure Mito samples.) F) Representative recordings from attached patch clamp of WT and Kcnt2−/− mitoplasts, with perfusion of 150 mM KCl (white), after addition of 40 mM NaCl (dark gray), and after addition of 10 µM BT (black). G) Quantitation of traces normalized to pA/pF and shown as percentage increase in current at a holding potential of 50 mV (same color scheme as F). Data are means ± sd (n = 4–10 independent mitoplast preparations).
Patch–clamp assessment of mitochondrial inner membranes
After anesthesia (tribromoethanol 200 mg/kg, i.p.), the heart from each 8–12-wk-old mouse was rapidly excised, washed, and chopped in ice-cold mitochondrial isolation medium [MIM, mM: 300 sucrose, 20 Tris, and 2 EGTA (pH 7.35)] at 4°C. All steps were performed on ice. Tissue was homogenized (Tissumizer; IKA, Inc., Wilmington, NC, USA) then centrifuged at 700 g, 5 min. Supernatants were saved and pellets rehomogenized and recentrifuged. Pooled supernatants were then centrifuged at 10,000 g for 10 min. The crude mitochondrial pellet was suspended in 0.2 ml MIM and layered over 1.75 ml of 30% osmotically balanced Percoll (MilliporeSigma, Burlington, MA, USA), in a round-bottomed microcentrifuge tube and centrifuged at 14,000 g for 1 h. Two mitochondrial layers were apparent (Fig. 1C), of which the lower (purified mitochondria) was washed twice by centrifugation. The mitochondrial pellet (∼25 µl) was suspended in 0.5 ml swelling buffer [mM: 30 KCl, 20 [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), and 1 EGTA, (pH 7.2)] for 15 min. Centrifugation (1000 g, 30 s) afforded a mitoplast pellet, resuspended in ∼20 µl MIM for immediate use in patch-clamp studies (n = 27 WT and 40 Kcnt2−/− mice).
For attached mitochondrial inner membrane (mitoplast) patch–clamp experiments, mitochondrial inner membranes (mitoplasts) were diluted 1:100 in a bath solution [mM: 150 KCl, 20 HEPES, 1 EGTA (pH 7.2)], and a 10-µl drop was placed on a glass coverslip attached to a custom 3-dimensional printed microchamber (Fig. 1B) (the stereolithography file is deposited at https://3dprint.nih.gov/discover/3dpx-008253). Electrodes (40–100 MΩ) (Sutter Instruments, Novato, CA, USA) were filled with a pipette solution [mM: 150 KCl, 0.025 NaCl, 20 HEPES, and 1 EGTA (pH 7.2)]. The bath was exchanged with buffer additionally containing 40 mM NaCl or 40mM NaCl with 10 µM BT (from stock in DMSO, final DMSO concentration, <0.01% v/v). For excised patch experiments, mitoplasts were diluted 1:100 in patch seal buffer [mM: 60 KCl, 80 K-gluconate, 40 LiCl, 0.025 NaCl, 0.1 CaCl2 [calculated free), 20 HEPES, and 1 EGTA (pH 7.2)]. Electrodes were filled with pipette solution (mM: 125 KCl, 15 K-gluconate, 15 LiCl 0.025 NaCl, 20 HEPES, and 1 EGTA). Mitoplasts were identified by their round shape and the presence of a cap structure (Fig. 1C). After formation of high-resistance seals, patches were excised and inside-out currents were recorded with an Axopatch 200B amplifier and Clampex10 software (Molecular Devices, Sunnyvale, CA, USA). All holding potentials reported are those applied to the patch pipette interior. The electrical connection was made using Ag/AgCl electrodes and an agar 2 M KCl salt bridge at the ground electrode. (Not all channels yielded currents at all potentials, and seal integrity was often compromised at the extremes of this range). Data were digitized and recorded at 10 kHz and filtered through an 8-pole low-pass 2 kHz filter. Patches were recorded under flow (0.1 ml/min) of: 1) Ca2+ free patch seal buffer with 0.076 mM sucrose for osmotic balance as above; 2) with LiCl replaced with 40 mM NaCl; and 3) further addition of 2.5 µM BT (from stock in DMSO, final DMSO concentration, <0.01% v/v). Although the study of K+ channels typically employs gluconate salts to exclude the possibility that measured conductances are related to Cl− channels, we used Cl− salts in this study because KNa1.2 activity is strongly enhanced in the presence of Cl− (15). All buffers were filtered (0.22 µm) immediately before use. Single channel analysis was performed with a Clampfit 10.0 single-channel search (Molecular Devices). Dwell times were calculated with the single-channel search with threshold crossing.
Cardiomyocyte isolation and respiration measurements
Primary adult ventricular cardiomyocytes from male and female mice were isolated by collagenase perfusion in the presence of 10 mM 2,3-butanedione monoxime (BDM), as we previously described (3). BDM was absent from all subsequent steps. Cells were step-wise rendered tolerant to 1.8 mM Ca2+, and the final pellet suspended in 1 ml MEM (supplemented with 1.8 mM, CaCl2 2.5% fetal bovine serum, and penicillin/streptomycin; 11095-080; Thermo Fisher Scientific, Waltham, MA, USA). Cell viability and yield were determined by Trypan blue and a hemocytometer. Only preparations with >85% viable rod-shaped cells were used for experiments. Cells were seeded at 2000/well on Seahorse XF96 V3-PS plates (Agilent Technologies, Santa Clara, CA, USA) and equilibrated for 1 h. Minimum essential medium was replaced with unbuffered DMEM (pH 7.4) containing various carbon sources (mM: 5 glucose, 0.1 palmitate, 4 glutamine, 5 galactose, 5 lactate, and 1 pyruvate) and either 10 mM 2-deoxyglucose or 20 µM etomoxir (MilliporeSigma), as detailed in the Results. All conditions with palmitate had 0.1 mM l-carnitine. Oxygen consumption rates (OCRs) were measured with an XF96 extracellular flux analyzer (n = 5 WT and 4 Kcnt2−/− mice).
Ex vivo heart perfusion
Hearts from male and female mice were perfused in constant flow (4 ml/min.) Langendorff mode as previously described (3). Krebs-Henseleit buffer (KH; in mM: 118 NaCl, 4.7 KCl, 25 NaHCO3, 1.2 MgSO4, 1.2 KH2PO4, and 2.5 CaCl2, gassed with 95%/5% O2/CO2, 37°C) was supplemented with 5 mM glucose or 0.1 mM bovine serum albumin–conjugated palmitate. Left ventricular pressure was measured via a water-filled, transducer-linked left ventricular balloon. Left ventricular and coronary root pressures were monitored and digitally recorded (Dataq, Akron, OH, USA). After equilibration hearts were treated with isoproterenol (100 nM final) for 5 min (n = 7 WT and Kcnt2−/− mice).
Electron microscopy
Hearts from male and female mice were fixed in 4% paraformaldehyde+2.5% glutaraldehyde in Millonig’s phosphate buffer [0.2 M NaH2PO4/Na2HPO4, and 0.5% NaCl (pH 7.4)]. One millimeter cubes were processed and digitally photographed on a 7650 electron microscope at ×12,000 (Hitachi, Chiyoda, Japan). Analysis of images was performed with ImageJ software (NIH). Mitochondrial areas and densities were placed into 8 or 5 bins, respectively, and the resulting histograms were fitted to a single gaussian. The form factor was calculated as 1/[(4π.area)/(perimeter2)], and the aspect ratio was calculated as major axis/minor axis (n = 5; WT and Kcnt2−/− mouse hearts, 5 fields of view/heart, and 50 mitochondria/field of view).
Metabolomics
Hearts from male mice were perfused as above in KH buffer supplemented with glucose plus palmitate for 20 min, then freeze clamped with Wollenberger tongs in liquid N2 and ground to powder. Samples representing ∼50% of each heart (50 mg) were shipped to Metabolon (Research Triangle Park, NC, USA) on dry ice, extracted by standard procedures, and analyzed by liquid chromatography–tandem mass spectrometry and gas chromatography MS/MS (Global Metabolomics solution; Metabolon, Inc.) to measure the relative steady-state abundance of metabolites (n = 7/WT and Kcnt2−/− group).
Data for each run were median normalized. Overall, 527 metabolites were identified, of which 26 (4.9%) were removed because of insufficient replicates, yielding 7014 theoretical individual data points (501 data points × 7 mice/group × 2 groups). A further 229 outlying data points (>1 sd) were removed, representing 3.3% of the data. Missing values were imputed as weighted medians (25). Metabolomic data were analyzed with Metaboanalyst software (26). In a separate series of experiments, WT and Kcnt2−/− hearts were perfused in KH buffer supplemented with fat as the only carbon source, adenine nucleotide levels (ATP, ADP, and AMP) were measured (27), and the energy charge was calculated as (ATP+1/2ADP)/(ATP+ADP+AMP).
Immunoblot analysis
The sample protein was determined by the Folin-Phenol (Lowry) assay. Nonmitochondrial samples were diluted 2 times in Laemmli sample loading buffer and incubated at 95°C for 1 min, whereas mitochondrial samples were diluted 2 times in sample loading buffer containing 5 times the standard concentration of SDS and incubated at 25°C for 30 min. Samples were separated by SDS-PAGE (10% gels) and transferred to nitrocellulose, followed by probing with antibodies as recommended by manufacturer’s protocols. Detection employed horseradish-linked secondary antibodies with enhanced chemiluminescence (GE Healthcare, Pittsburgh, PA, USA) (n = 4 mice/genotype).
Body composition, fasting glucose response, and electrocardiogram
Body weight from male and female mice was measured once a week, from weaning (3 wk) to 25 wk of age, by weighing nonanesthetized mice placed in a small plastic container on a digital scale. (For both sexes and for each genotype, n = 14). Eighty-four-day-old (12 wk) WT and littermate Kcnt2−/− male mice were anesthetized as described above, and body fat content was measured with dual-energy X-ray absorptiometry (DEXA) scanning (Lunar PIXImus densitometer; GE Healthcare) (n = 14 male WT and Kcnt2−/− mice; n = 8 female WT and Kcnt2−/− mice). Blood glucose was measured with a True2Go glucose meter with TrueTest glucose strips (Trividia Health, Fort Lauderdale, FL, USA). Mice were unfed overnight in cleaned cages with access to water and cotton bedding (for both sexes and for each genotype, N = 14). Alternatively, after anesthesia, electrocardiograms were recorded by 3 needle electrodes and an EKG amplifier (Harvard Apparatus, Cambridge, MA, USA). EKG measurements were averaged for each animal from 10 different segments of the trace, each containing R1-S1-T1-P2-Q2-R2 waves (n = 4 WT and 5 Kcnt2−/− mice).
Quantitative PCR analysis
mRNA was extracted from heart homogenates with acid phenol/Trizol, according to the Direct-zol RNA MiniPrep Kit R2050 (Zymo Research, Irvine, CA, USA), as described by Toledo-Arena et al. (28). cDNAs were prepared with an iScript Kit (170-8891; Bio-Rad, Hercules, CA, USA). qPCR analysis was performed with a PrimePCR regulation of lipid metabolism-PPAR M96 predesigned 96-well panel for use with Sybr Green (10031585; Bio-Rad) (n = 3 WT and 3 Kcnt2−/− mice).
Replicates and statistics
For samples comparing WT and Kcnt2−/−, n = 1 is 1 animal (i.e., biologic replicates). Significant differences between the WT and Kcnt2−/− groups were determined with 2-way ANOVA with a Bonferroni correction for multiple testing, followed by post hoc paired or unpaired Student’s t tests. A value of P < 0.05 indicated significance.
RESULTS
Kcnt2 knockout
Figure 1A shows an example of results of genotyping on WT and Kcnt2−/− mice, indicating the expected amplicons. The deletion targets exon 22 of the genomic open reading frame, and results in a complete loss of both mRNA and protein (18), as we have also confirmed in the heart (3).
Cardiac mitochondria contain a KNa1.2 channel
To investigate cardiac mitochondrial K+ channels, we performed electrophysiologic studies on Percoll-purified isolated mitochondrial inner membranes (mitoplasts) from hearts of WT or Kcnt2−/− mice (Fig. 1C). Mitochondrial enrichment was verified by Western blot analysis for mitochondrial proteins, and purity was confirmed by Western blot analysis for nonmitochondrial membrane proteins (Fig. 1D, E). Mitoplasts were readily identified under light microscopy (Fig. 1C) and high-resistance seals (2–10 GΩ) were formed with the inner membrane opposite the outer membrane cap. In an initial series of experiments using attached mitoplast patch configuration, channel activity in WT was not activated by NaCl alone, but manifested upon further addition of the KNa activator BT (3). No activation by NaCl or BT was seen in Kcnt2−/− mitoplasts (Fig. 1F, G). The absence of activation by NaCl alone in WT mitoplasts suggest that the channel’s ion-sensing site may be on the matrix side of the inner membrane and thus was inaccessible to Na+ from the bath perfusion on this time scale. To investigate such a channel, an excised mitoplast patch was used, affording access to the matrix side of the inner membrane (Fig. 1C).
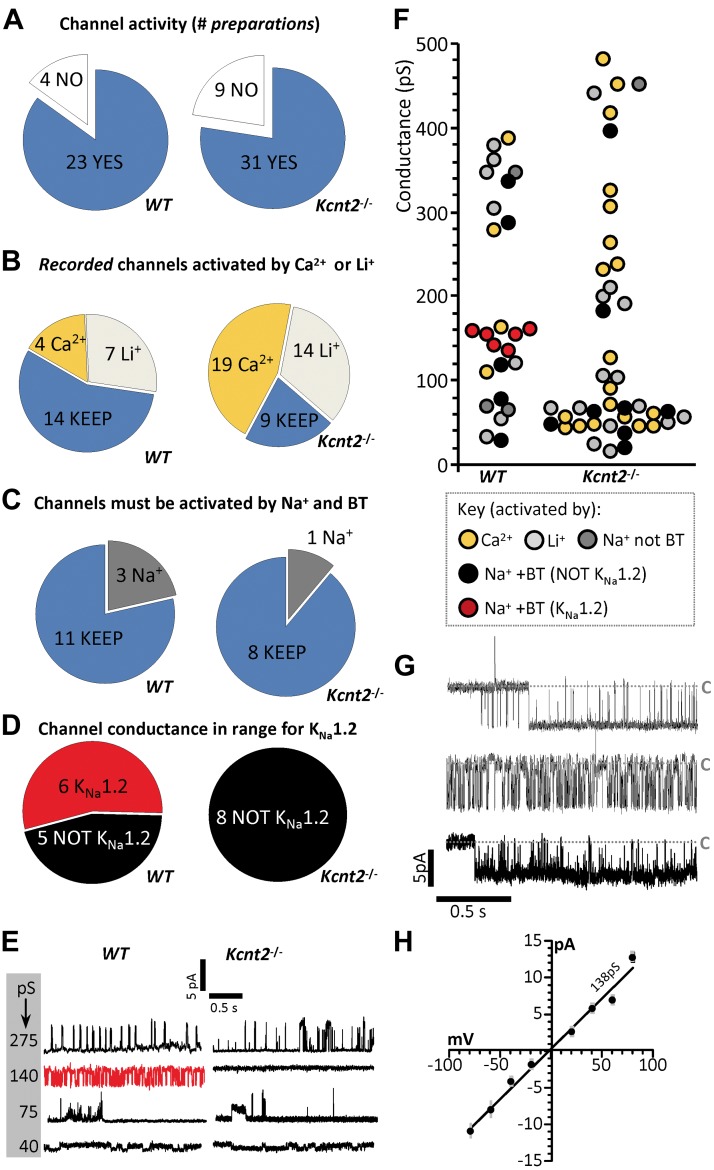
Channel activity was observed in a high proportion of independent preparations obtained from both WT (85%) and Kcnt2−/− (76%) hearts (Fig. 2A), consistent with previous reports that mitochondria contain numerous K+ channels (1, 29). In some cases, more than 1 channel was observed in a single preparation. yielding 25 recorded conductances in WT and 42 in Kcnt2−/−preparations. A multiple-level screen was performed to triage recordings containing channels other than KNa1.2 (Fig. 2B–D). The bath recording solution was sequentially switched from initial (containing 100 µM CaCl2), to 40 mM LiCl, then 40 mM NaCl, and finally 40 mM NaCl+2.5 µM BT. In each condition, channel activity was monitored at holding potentials from −100 to +100 mV.
Figure 2.
Mitochondria contain a KNa1.2 channel. A) Preparations from WT (left) and Kcnt2−/− (right) mitoplasts, sorted by channels observed (blue) or no channels observed (white). B) Exclusion of Ca2+-activated (gold) or Li+-activated (light gray) channels. In some preparations, more than 1 channel was observed. C) Selection of channels activated by Na+ and BT, with exclusion of channels activated by Na+ alone and subsequently blocked by BT (dark gray). B, C) Channels carried over to the subsequent screening step are shown in blue. D) Selection of channels with peak conductance matching that reported for KNa1.2 (red). E) Example traces of channels observed in WT and Kcnt2−/− preparations with a variety of peak conductances. Red trace indicates a channel assigned as KNa1.2. F) Peak conductance of channels observed from all traces. B–F) Color key shown at base of F. Channels with conductance >500 pS are omitted for clarity. G) Example of 2 s recordings from 3 KNa1.2 channels observed in WT mitoplasts (i.e., red points in Fig. 1F) at −40 mV holding potential. Gray dashed line labeled C: closed states. H) Channel current vs. voltage plot of peak unitary conductances of all KNa1.2 channels from WT mitoplast recordings. Average slope conductance was 138 ± 1 pS.
First, patches exhibiting channel activity in Ca2+ alone were discarded, as this is indicative of KCa channel activity (Fig. 2B, gold). Next, we discarded patches exhibiting activity after the switch to LiCl, as this is indicative of Na+-conductance (30) (Fig. 2B, light gray). Next, patches exhibiting channel activity in the presence of activating levels of NaCl (40 mM) and NaCl plus BT (2.5 µM) were considered potential KNa1.2 candidates (Fig. 2C, blue). Recordings exhibiting channel activity in the presence of Na+ but not after addition of BT, were also discarded (Fig. 2C, dark gray). Finally, single-channel unitary conductance was compared to the values expected for KNa1.2 (∼140 pS) (15, 30, 31), and those patches exhibiting appropriate conductance were considered to be KNa1.2 channels worthy of further analysis (Fig. 2D, red).
Figure 2E shows representative recordings observed in mitoplast patches from WT and Kcnt2−/− mice, exhibiting a variety of unitary conductances, with the knockouts having an absence of activity in the conductance range expected for KNa1.2. Quantitation of all conductances showed a cluster of 6 channels in WT mitoplasts that passed all screens (Fig. 2F red data points), with no channels of similar conductance observed in Kcnt2−/− mitoplasts. The Kcnt2−/− recordings contained proportionally more Na+- and BT-activated channels than WT in the small-conductance range (Fig. 2F, black symbols in 20–80 pS range, 6/42 Kcnt2−/− mice vs. 2/25 WT). A similar observation was made for the mitochondrial BK channel, wherein loss of channels with the expected conductance in a knockout was accompanied by an increase in small-conductance openings (12). Figure 2G shows representative examples of single-channel activity for 3 (of 6) Na+- and BT-activated conductances recorded at a holding potential of −40 mV in WT mitoplasts. As reported for KNa1.2 (15), these channels showed rapid flickering between open and closed states. The average unitary slope conductance was 138 ± 1 pS (Fig. 2H, peak conductance graph for all 6 channels). In our buffer conditions, reversal potential for channels conducting Na+ or Cl− was predicted to be −23 or −8 mV, respectively. K+ was the only other ion present and the channel current crossed 0 at −2 mV, and this finding indicates that K+ was the predominant conducting ion. The data in Fig. 2 demonstrate that WT cardiac mitochondria contained a K+ channel with the ion selectivity, ion sensitivity, pharmacology, and conductance properties of KNa1.2 that are absent in mitochondria from Kcnt2−/− mice.
Electrophysiological characterization of mitochondrial KNa1.2
Single-channel characteristics are useful for classifying channels. Of the 2 KNa channels; KNa1.1 (Kcnt1/Slack/SLO2.2) and KNa1.2 (Kcnt2/Slick/SLO2.1), KNa1.1 has been described in more detail at the single-channel level. Where possible, we compared the characteristics of the channels measured in this study to those published for KNa1.2 (15, 30, 31). If this was not possible, a comparison to KNa1.1 was included, given that both KNa channels share similar characteristics, and it is known that KNa1.1 is not present in the heart (3, 18).
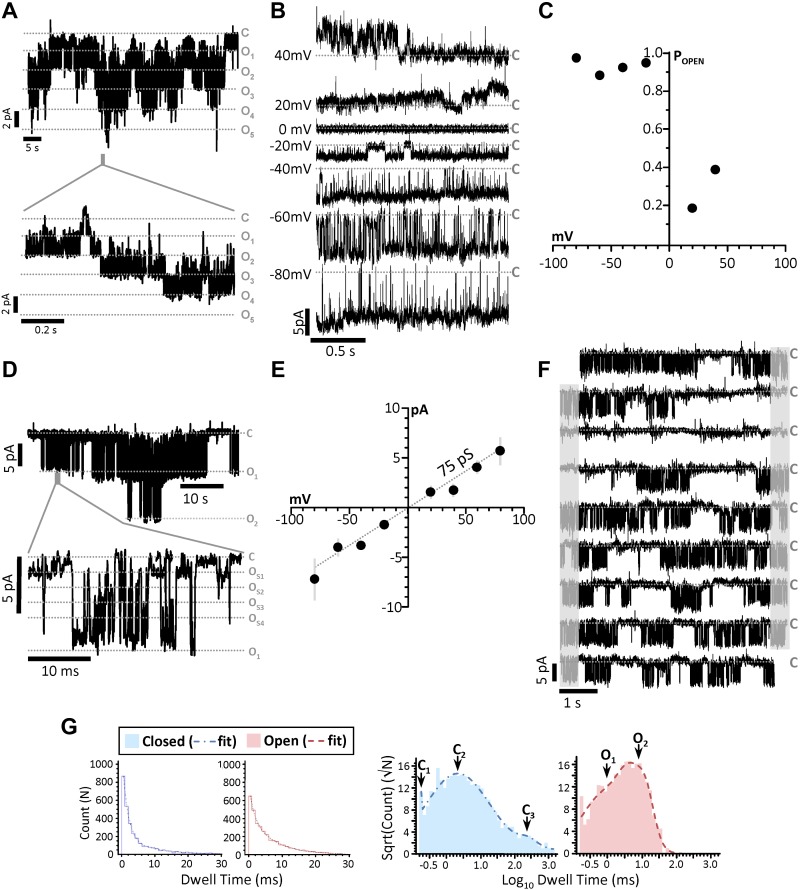
A detailed single-channel analyses of all 6 assigned KNa1.2 patches was not possible because of the presence of 2 or more identical channels within some patches (Fig. 3A), which is in agreement with previous reports that KNa1.2 channels cluster in neuronal plasma membranes (32). Examination of all suitable WT recordings (representative example, Fig. 3B) revealed a higher open probability at negative potentials (Fig. 3C). In addition, as previously reported for KNa1.2 (15, 33), numerous distinct subconductances were apparent between 35 and 140 pS (Fig. 3D). This behavior is a characteristic of KNa1.1 channels (34, 35), and although it has been observed in KNa1.2 mutants (33), this is the first observation of such in WT KNa1.2 channels in endogenous membranes. The average slope conductance when considering all subconductances was 75 pS (Fig. 3E), which agreed well with the average chord conductance of 74.8 ± 6.8 pS (assuming reversal potential of 0 mV). These data indicate that smaller subconductance states predominate in the active channel current.
Figure 3.
Single-channel characteristics of mitochondrial KNa1.2. A) Compressed traces from the recording of a patch containing 5 mitochondrial KNa1.2 channels (holding potential, −20 mV), and a time-expanded trace for the region highlighted by the gray bar in the trace above. Gray dashed lines: closed (C) and multiple open (O) states. B) Traces from a single KNa1.2 channel at holding potentials of 40 to −80 mV. C) Channel open probability plot from the channel shown in B. D) Representative trace of a recording with 2 channels on a compressed time scale (holding potential −40 mV) and a time-expanded trace for the region highlighted by the gray bar in the trace above, showing multiple subconductance states within the channel peak conductance (e.g., OS1). E) Current–voltage relationship of all 6 mito-KNa1.2 channels showing average current at each holding potential. The decreased slope conductance (compared to Fig. 2H showing peak unitary conductance) indicates that subconductances averaging 75 pS dominated the average current during the recordings. F) Continuous trace (45 s) of a single channel. Gray shading: portions of each trace (right) that are repeated (left) on the next line. Gray dashed lines labeled C: closed states. G) Log binned channel closed and open dwell-time peaks and frequency of closed dwell times plotted against their duration. Table inset shows calculated area and time constants (τ).
Open and closed channel dwell times were also calculated from continuous recordings at −40 mV (representative 45 s example, Fig. 3F). The frequency of channel open and closed dwell times was fitted to the sum of multiple simple exponentials, constituting 94.6% of the total area under the curve, and subsequent log-binned histograms revealed peaks representing 3 closed times and 2 open times (Fig. 3G). The longest closed time (τ3 = 262 ms) represents periods of channel closure between bursts of activity, which is characteristic of KNa1.2 (15).
KNa1.2 channel activation uncouples cardiomyocyte oxidative phosphorylation
We next sought to determine the impact of cardiac KNa1.2 channel activity on cardiomyocyte bioenergetics. Using Seahorse XF analysis, we measured OCRs of cardiomyocytes from WT and Kcnt2−/− mice. Cells from both genotypes had similar viability and rod-shaped morphology (Fig. 4A). The KNa opener BT (2.5 µM) significantly stimulated OCR in oligomycin-treated cardiomyocytes from WT mice, but not those from Kcnt2−/− mice (WT, 370 ± 48 maximum OCR; Kcnt2−/−, 154 ± 32 maximum OCR) (Fig. 4B). The marginal but persistent effect of BT in Kcnt2−/− cells is consistent with the observation of small conductances activated by Na+ and BT in Kcnt2−/− mitoplast patches (Fig. 2F).
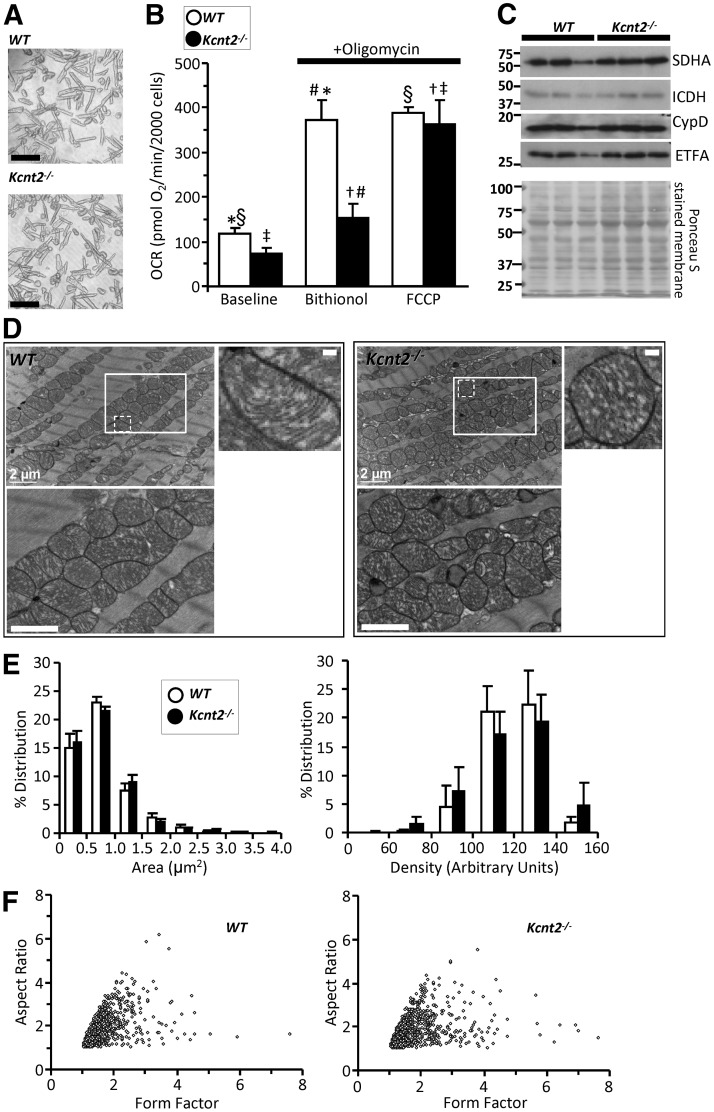
Figure 4.
Kcnt2−/− Cardiomyocyte bioenergetics and mitochondrial ultrastructure. A) Representative images of isolated cardiomyocytes from WT and Kcnt2−/− hearts. Scale bars, 100 µm. B) OCR of isolated cardiomyocytes measured with addition of oligomycin (1 µg/ml) and either 2.5 µM BT (KNa opener) or 500 nM FCCP (mitochondrial uncoupler). Statistics were measured using 2-way ANOVA with Bonferroni correction and post hoc t test. Bars with the same symbol are significantly different from each other (n = 4–5). Data are means ± sem. P < 0.05. C) Western blots from WT and Kcnt2−/− heart homogenates showing levels of mitochondrial proteins (SDHA, isocitrate dehydrogenase, cyclophilin D, and electron transfer flavoprotein subunit α) and Ponceau stain loading control. D) Representative transmission electron microscopic images of fixed heart slices. Bottom left panels: insets boxes at higher magnification. Scale bar, 1 µm. Top right panels: increased magnification of single mitochondria from WT and Kcnt2−/− mice with mitochondrial ultrastructure visible (i.e., cristae folds, outer and inner membrane contacts). Scale bar, 200 nm. E) Binned histogram of mitochondrial area or mitochondrial density, obtained from image analysis (n = 5). Data are means ± sd for each bin. F) Form-factor/aspect-ratio scatterplot. E, F) Data obtained from 1250 mitochondria, 25 fields of view, and 5 WT or Kcnt2−/− hearts.
In the mitochondrial K+ cycle (36), K+ entry into the organelle activates a mitochondrial K+/H+ exchanger, such that mitochondrial K+ channel activity can uncouple oxidative phosphorylation. The much larger BT-induced respiratory stimulation in WT vs. Kcnt2−/− cardiomyocytes is most likely related to mitochondrial uncoupling. As an additional control, the bona fide mitochondrial uncoupler carbonylcyanide-p-trifluoromethoxyphenylhydrazone (FCCP) elicited similar maximum respiration rates in cardiomyocytes from both genotypes (WT, 387 ± 15 maximum OCR; Kcnt2−/−, 364.2 ± 53 maximum OCR), rendering it unlikely that the differential effect of BT in WT vs. Kcnt2−/− cells is related to an underlying difference in overall bioenergetic capacity. Consistent with this result, Western blot analysis for several mitochondrial marker enzymes [succinate dehydrogenase complex, subunit A (SDHA), isocitrate dehydrogenase, cyclophilin-D, and electron transfer flavoprotein, subunit α] suggested no difference in mitochondrial mass or content between WT and Kcnt2−/− hearts (Fig. 4C).
Loss of KNa1.2 mildly impacts cardiac mitochondrial ultrastructure
An important function of the mitochondrial K+ cycle is the regulation of organelle volume (37, 38), and KNa1.2 activity is known to be sensitive to osmolarity (39, 40). Thus, we hypothesized that mitochondria from Kcnt2−/− may exhibit ultrastructural changes. Electron microscopic analysis of hearts from WT and Kcnt2−/− mice (Fig. 4D–F) revealed that mitochondria had similar areas (WT, 0.9 ± 0.30 μm2; Kcnt2−/−, 0.89 ± 0.43 μm2; mean ± sd; n = 5) and matrix density (WT, 134 ± 17; Kcnt2−/−, 135 ± 24; mean ± sd; n = 5). The distribution of these parameters (Fig. 4E) suggested a small shift toward increased area and density in Kcnt2−/− vs. WT, but it was not statistically significant. In addition, no difference in either form factor or aspect ratio was observed between the genotypes (Fig. 4F), indicating that KNa1.2 deficiency does not alter mitochondrial fission or fusion (41, 42). Together, the data in Fig. 4 suggest that, although activation of KNa1.2 can uncouple respiration, loss of the channel does not impact cardiac mitochondrial structure or content.
KNa1.2 is necessary for cardiac respiratory reserve capacity when oxidizing fat
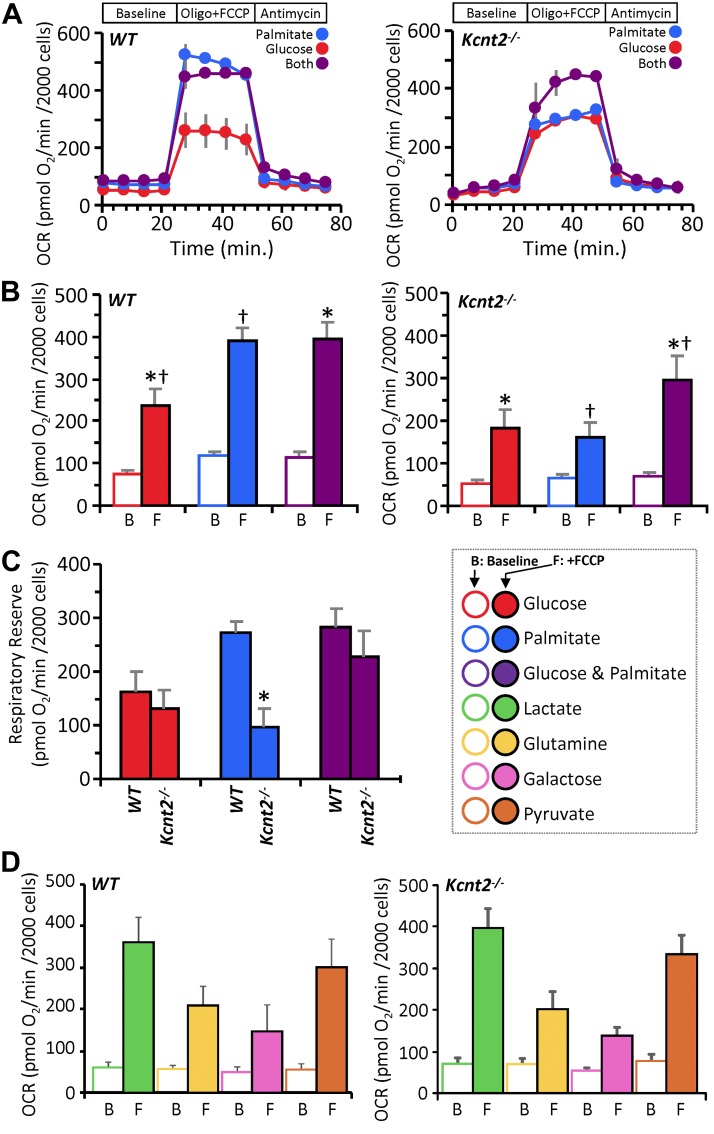
In an effort to further understand the bioenergetic effects of KNa1.2 deficiency, we compared metabolic substrate preferences in cardiomyocytes isolated from WT and Kcnt2−/− mice. Myocytes were incubated with 1) glucose alone (with etomoxir to inhibit fatty acid β-oxidation), 2) palmitate alone (with 2-deoxyglucose to inhibit glycolysis), or 3) glucose+palmitate. The response to uncoupling by FCCP (500 nM) was used to determine respiratory reserve (RR) capacity under each substrate condition (Fig. 5A–C).
Figure 5.
KNa1.2 loss impacts cardiac metabolic substrate choice under stress. A) Representative OCR traces of isolated WT or Kcnt2−/− cardiomyocytes, incubated in the presence of different metabolic substrates. Data from a single XF plate are shown (mean ± sd of 12 wells/substrate or genotype). Timeline above traces shows the OCR was measured at baseline, then with the addition of the ATP synthase inhibitor oligomycin (1 µg/ml) plus the mitochondrial uncoupler FCCP (500 nM), and finally with the mitochondrial complex III inhibitor antimycin A (5 µM). B) Group OCR averages of baseline (B) and FCCP-uncoupled (F) cardiomyocytes under substrate conditions as defined above. Statistics were measured using 2-way ANOVA with Bonferroni correction and post hoc Student’s t test. Data are mean ± sem, for n = 4 Kcnt2−/− or 5 WT, independent cardiomyocyte preparations. Bars with the same symbol are significantly different from one another. P < 0.05. C) RR capacity calculated from the data in B (i.e., uncoupled minus baseline OCR). Mean ± sem (n = 4–5). *P < 0.05 between genotypes. Color key for metabolic substrates used in all panels is shown to the right of C. D) OCR of WT and Kcnt2−/− cardiomyocytes metabolizing different substrates. Empty bars, baseline (B); filled bars, FCCP uncoupled (F). Data are means ± sem from 4 to 5 independent cardiomyocyte preparations.
Cardiomyocytes from WT and Kcnt2−/− cells exhibited a similar baseline OCR under all substrate conditions (Fig. 5B). In WT cells, a robust uncoupling response to FCCP was seen under all conditions, and notably, the uncoupling response with palmitate alone (3.3-fold) was equal to that seen when both substrates were present (3.3-fold). However, in Kcnt2−/− cells, the uncoupling response with palmitate alone (2.4-fold) was significantly blunted compared with that seen when both substrates were present (4.3-fold) (comparison between blue and purple bars in left and right panels of Fig. 5B). The additional OCR induced over baseline by FCCP was used to calculate RR capacity. Figure 5C shows that the RR of WT and Kcnt2−/− cells was similar in the glucose alone and the glucose+palmitate conditions. However, with palmitate alone, Kcnt2−/− cells exhibited a significant RR deficit relative to WT. No such RR deficit was observed in myocytes from Kcnt2−/− mice respiring on lactate, glutamine, galactose, or pyruvate (Fig. 5D), indicating that the Kcnt2−/− RR deficit is specific to fat oxidation.
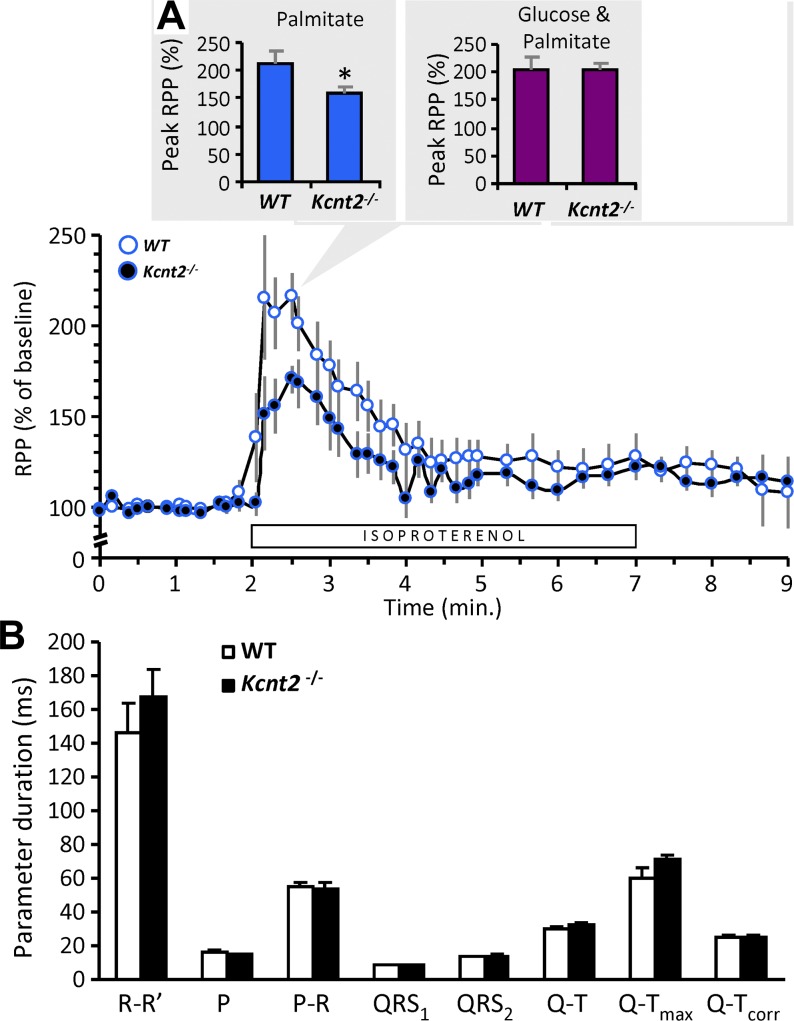
To test the physiological relevance of this RR deficit, the ability of perfused hearts to respond to increased metabolic demand was tested. Hearts from WT and Kcnt2−/− mice were perfused with palmitate as the sole carbon source, whereas stimulating workload by addition of the β-adrenergic agonist isoproterenol (100 nM). Hearts from Kcnt2−/− mice showed a significantly reduced functional response to isoproterenol, relative to WT (WT, 213 ± 20% increase in rate × pressure product (RPP), vs. Kcnt2−/− 159 ± 13%; mean ± sem, n = 7) (Fig. 6A). When RPP was broken into its component parameters, a greater difference in the effect of isoproterenol was observed on left-ventricular developed pressure than on heart rate (peak/baseline left-ventricular developed pressure: WT, 135 ± 9% vs. Kcnt2−/− 114 ± 4%; peak/baseline heart rate: WT, 162 ± 22% vs. Kcnt2−/− 140 ± 9%). However, consistent with the isolated cardiomyocyte OCR data (Fig. 5C), no difference in the isoproterenol-induced functional response was observed when the perfusion buffer was supplemented with glucose and palmitate (Fig. 6A, inset). Together, these data suggest that loss of KNa1.2 results in an impaired ability to respond to increased metabolic demand when oxidizing only fat. Consistent with previous reports (3, 18) no EKG differences were observed in Kcnt2−/−mice (Fig. 6B), suggesting that loss of KNa1.2 per se does not affect cardiac contractile function.
Figure 6.
Loss of KNa1.2 impacts whole heart substrate choice under stress. A) Cardiac function data (heart rate × pressure product, RPP) for isolated perfused hearts from WT or Kcnt2−/− mice, perfused with Krebs-Henseleit (KH) buffer containing palmitate as the sole metabolic substrate. Bar below the traces indicates duration of 100 nM isoproterenol infusion. Graph shows RPP as a percentage of the average baseline value for 1 min. before isoproterenol infusion. Inset: comparison of the peak response to isoproterenol under this substrate condition (palmitate only, blue). Adjacent inset (right): the peak response to isoproterenol from a separate series of perfusions in which the KH buffer contained both palmitate and glucose as substrates (purple). Data are means ± sem (n = 7). *P < 0.05 between genotypes. B) EKG parameters obtained in vivo from tribromoethanol-anesthetized WT and Kcnt2−/− mice. R–R′, distance between R waves of each beat (i.e., 1/HR); P, P-wave duration; P–R, interval between P and R waves; QRS1 & QRS2, diameter of QRS complex (different calculation algorithms); Q–T, interval between Q and peak of T wave; Q–Tmax, interval between Q and end of T wave; Q–Tcorr, QT interval corrected for heart rate. Data are mean ± sd (n = 4–5 animals).
Whole-animal metabolic differences in Kcnt2−/− mice
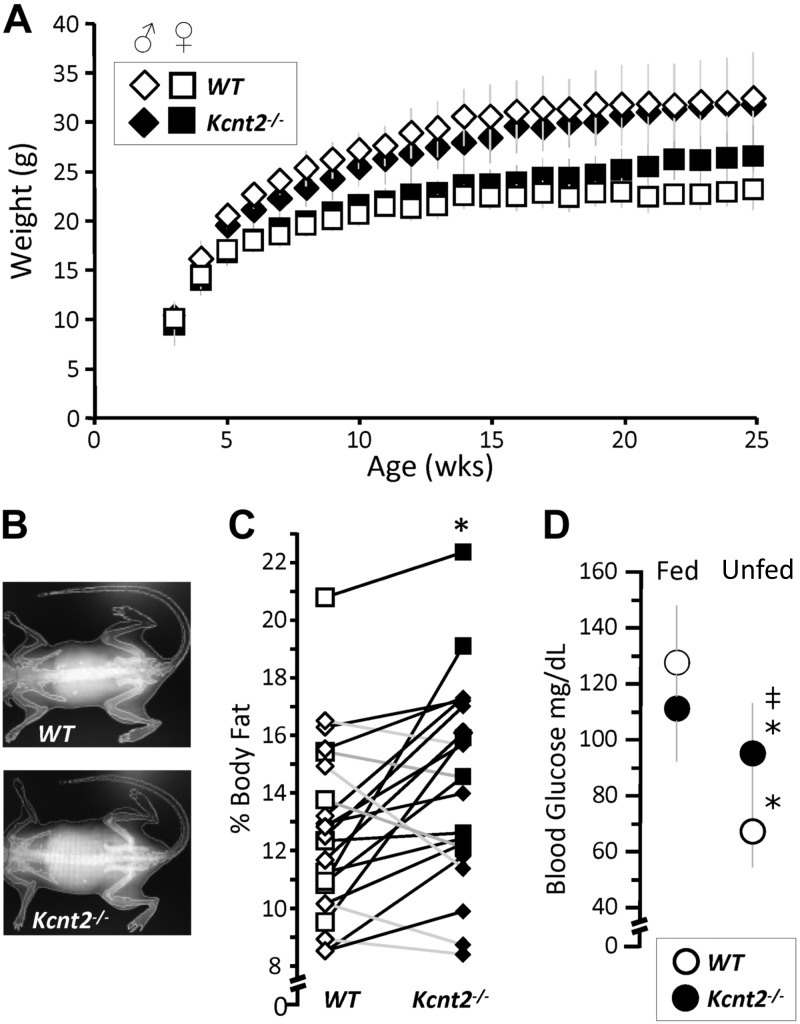
Because the heart is a major fat-burning organ, we hypothesized that the fat-specific RR capacity deficit in Kcnt2−/− would be accompanied by metabolic perturbations at the whole-animal level. No significant differences in weight gain were observed between WT and Kcnt2−/− mice over 25 wk (Fig. 7A). The percentage of body fat content determined by DEXA analysis (Fig. 7B) revealed a small difference in average fat content between genotypes (WT, 12.6 ± 3% vs. Kcnt2−/− 14.6 ± 4%), but that was statistically significant between paired littermates with (Fig. 7C). In addition, whereas WT mice showed an expected drop in blood glucose after food was withheld overnight (15 h), no such decrease was seen in Kcnt2−/− mice (Fig. 7D), suggesting elevated gluconeogenesis in response to unfed state in Kcnt2−/−, which is consistent with a potential increased reliance on glucose oxidation as a compensatory response to the fat-specific RR defect under stress conditions.
Figure 7.
Loss of KNa1.2 impacts whole body metabolic phenotype. A) Body weights of WT and Kcnt2−/− male (diamond) and female (square) mice from weaning (3 wk) to 25 wk of age. Data are mean ± sd (n = 12). B) Representative DEXA images from WT and Kcnt2−/− mice. C) Percentage of body fat measured by DEXA scan of WT (white symbols) and Kcnt2−/− (black symbols) littermates (pairs are indicated by connected data points) (n = 14). WT, 8 Kcnt−/−. *P < 0.05 between genotypes by paired Student’s t test. D) Left: blood glucose levels measured in WT and Kcnt2−/− mice at baseline (5 pm, fed) and following a 15-h withdrawal from food (8 am, unfed). Data are mean ± sd (n = 12). *P < 0.05 between fed and unfed states within a genotype, ‡P < 0.05 between genotypes.
Metabolomic and expression profiling of Kcnt2−/− hearts
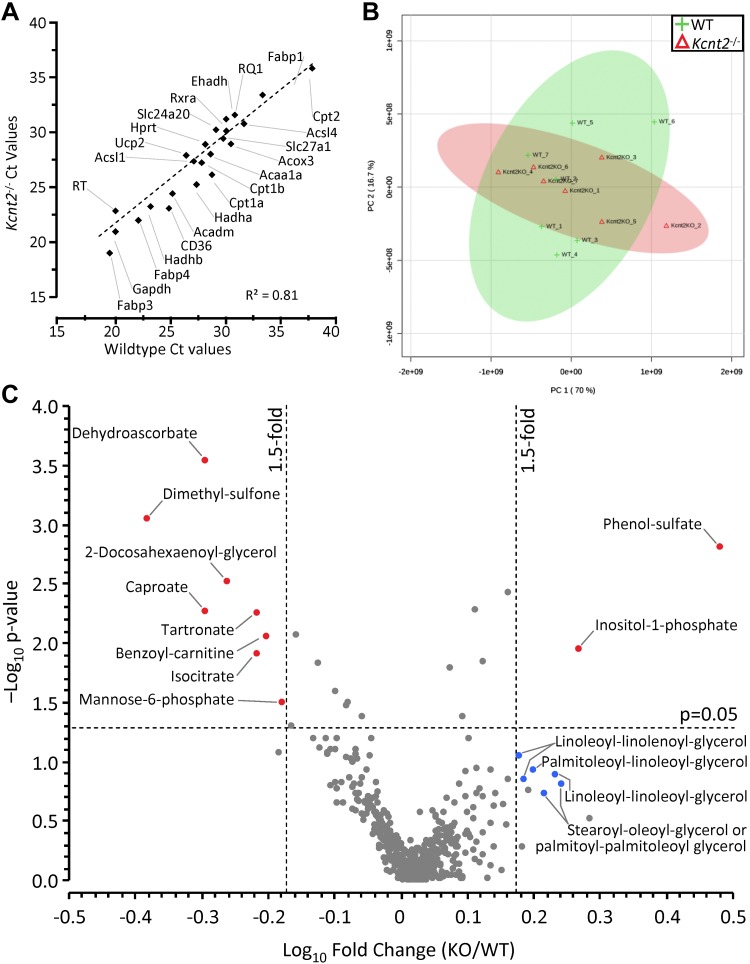
To investigate the molecular underpinnings of the fat-specific RR defect in Kcnt2−/− hearts, expression of metabolic regulatory genes was measured using a predesigned qPCR array (Fig. 8A and Table 1). No differences were observed between WT and Kcnt2−/− hearts, suggesting that the fat-specific RR defect is not related to a remodeling of metabolism at an expression level. Separately, a small but nonsignificant decrease in energy charge (ATP+1/2ADP)/(ATP+ADP+AMP) was observed in Kcnt2−/− mice (WT, 0.87 ± 0.05 vs. Kcnt2−/−, 0.77 ± 0.03; mean ± sem, n = 5–6), suggesting that energy-sensing metabolic regulators such as AMPK may be altered. However, Western blot analysis revealed no difference in AMPK phosphorylation between WT and Kcnt2−/− hearts (data not shown). Together with data in Figs. 4 and 5, these findings suggest that KNa1.2 deficiency does not induce large-scale remodeling of cardiac mitochondrial metabolism. Rather, KNa1.2 loss specifically affects cardiac fat oxidation only under conditions of high-energy demand such as uncoupling or β-adrenergic stimulation.
Figure 8.
Kcnt2−/− cardiac expression profiling and metabolomics. A) qPCR Ct values for 27 metabolically important genes (Table 1) in WT and Kcnt2−/− hearts (n = 3 independent RNA preparations/genotype). Data are mean, errors are omitted for clarity. B) WT and Kcnt2−/− hearts were perfused with KH buffer containing palmitate+glucose and freeze clamped for metabolomic analysis by liquid chromatography-MS/MS. Graph shows principle component analysis of 501 cardiac metabolites. The first and second principal components contributed 86.7% of the overall metabolic character. Shaded ovals overlaying the graph indicate 95% confidence intervals for WT (lime) and Kcnt2−/− (salmon pink) samples. C) Volcano plot of the metabolic profile of Kcnt2−/− vs. WT hearts. Dashed lines: P = 0.05 cut off (y axis) and 1.5-fold change cut off (x axis). Each point represents a single metabolite, and each point represents means (n = 7 mice). Errors are omitted for clarity. Red: metabolites meeting fold-change and P-value criteria; blue: additional metabolites discussed in the text. Diamonds: males; circles: data from both sexes combined.
TABLE 1.
Gene names and symbols for genes targeted by BioRad PrimePCR qPCR assay kit
| Gene name | Gene symbol |
|---|---|
| Acetyl-coenzyme A acyltransferase 1A | Acaa1a |
| Carnitine palmitoyltransferase 1a, liver | Cpt1a |
| Hydroxyacyl-coenzyme A dehydrogenase/3-ketoacyl-coenzyme A thiolase/enoyl-coenzyme A hydratase (trifunctional protein), α subunit | Hadha |
| Acyl-coenzyme A dehydrogenase, medium chain | Acadm |
| Carnitine palmitoyltransferase 1b, muscle | Cpt1b |
| Hydroxyacyl-coenzyme A dehydrogenase/3-ketoacyl-coenzyme A thiolase/enoyl-coenzyme A hydratase (trifunctional protein), β subunit | Hadhb |
| Glyceraldehyde-3-phosphate dehydrogenase | Gapdh |
| Acyl-Coenzyme A oxidase 3, pristanoyl | Acox3 |
| Carnitine palmitoyltransferase 2 | Cpt2 |
| Hypoxanthine guanine phosphoribosyl transferase | Hprt |
| Acyl-CoA synthetase long-chain family member 1 | Acsl1 |
| Enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | Ehhadh |
| Retinoid X receptor α | Rxra |
| PrimePCR DNA contamination control assay | gDNA |
| Fatty acid binding protein 1, liver | Fabp1 |
| Solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 | Slc25a20 |
| PrimePCR positive control assay | PCR |
| Acyl-CoA synthetase long-chain family member 4 | Acsl4 |
| Solute carrier family 27 (fatty acid transporter), member 1 | Slc27a1 |
| PrimePCR RNA quality assay | RQ1 |
| Fatty acid binding protein 3, muscle and heart | Fabp3 |
| CD36 antigen | Cd36 |
| Fatty acid binding protein 4, adipocyte | Fabp4 |
| Uncoupling protein 2 (mitochondrial, proton carrier) | Ucp2 |
| PrimePCR reverse transcription control assay | RT |
Genes are those shown in Fig. 8A.
Finally, to understand the effects of KNa1.2 deficiency on cardiac metabolism at a systems level, an unbiased metabolomics analysis was performed. Principal component analysis showed no significant difference in the fundamental character of metabolism between WT and Kcnt2−/−hearts at baseline (Fig. 8B). A volcano plot for all 501 metabolites measured revealed only 10 were significantly altered (>1.5-fold vs. WT; P < 0.05) (Fig. 8C). Of these metabolites, notable changes were an increase in phenol-sulfate and decrease in dimethyl-sulfone, potentially indicating perturbations in aryl-sulfotransferase activity and sulfur metabolism. Dehydroascorbate was significantly lower in Kcnt2−/− hearts, potentially indicating lower oxidative load. In addition, inositol-1-phosphate was significantly elevated, and a cluster of diacylglycerol metabolites was also elevated (although no individual diacylglycerol approached significance), suggesting enhanced PLC activity in Kcnt2−/−. Overall, the comparatively minor nature of metabolomic perturbations in Kcnt2−/− hearts at baseline is consistent with the notion that the impact of KNa1.2 loss is limited to fat oxidation under conditions of high energy demand.
DISCUSSION
A plethora of studies have identified mitochondrial K+ channels at the phenomenological level [reviewed in refs. (1, 43)], and several studies have linked these channels mechanistically to protection against IR injury (1, 44–46). However, surprisingly few examples exist of bona fide mitochondrial K+ channels that are: 1) identified at the molecular (genetic) level, 2) characterized with robust electrophysiology, and 3) linked to any specific mitochondrial channel function phenotype [examples include KCa1.1 (11, 12), KATP (10), and SK3 (47)]. Although the phenomenon of a cell surface KNa channel was first characterized in hearts 30 yr ago, the field of KNa research rapidly transitioned to study of these channels in brain (19). As such, very little is known about the role of KNa in cardiomyocytes, including potential subcellular localization. In patch–clamp studies of isolated cardiac mitochondrial inner membranes, we recorded a K+ channel matching the known characteristics of KNa1.2 (ion sensitivity, ion selectivity, pharmacology, and conductance) in mitoplasts from WT mice, that was absent in those from Kcnt2−/− mice (Figs. 1–3). This is the first report of a KNa channel in mitochondria.
Cardiac mitochondria are reported to contain several cation and anion channels (48, 49), and mitoplast recordings were subjected to a rigorous screening process to ensure that conductances assigned as KNa1.2 did not originate from other channels. Bath:pipette ratios of the major ion components (K+, Na+, and Cl−) enabled reversal potentials to be differentiated. The Na+ surrogate Li+ was used to screen out any Na+-activated currents that were in fact related to Na+ conductance. In addition, any conductances activated by Ca2+ were dismissed, to ensure patched membranes were free of KCa channels. Inner membrane anion channels and intracellular chloride channel variety exhibit conductances (∼100 pS: inner membrane anion channels, ∼8 pS: chloride channel-4) sufficiently distinct from KNa1.2 that, in combination with the observed reversal potential, we are confident do not contribute to assigned KNa conductances. Similarly, mitoKATP channels (∼10 pS) would not be mistaken for KNa1.2. Finally, KNa1.2 activation is significantly enhanced by Cl− in the presence of activating Na+, hence Cl− salts were used to maximize the probability of detecting these channels (15). Of 25 recordings in WT mitoplasts, this strategy yielded 6 channels, a 24% success rate. Application of this rate to the 42 recordings in Kcnt2−/− mitoplasts predicts 10 such channels, but we observed none.
Several known electrophysiological characteristics of KNa1.2 channels were also observed in our assigned KNa1.2 mitoplast recordings, including: 1) multiple subconductance states (Fig. 3D) (15, 35), 2) rapid flickering between open and closed states (Fig. 2G) (15), 3) burst operation with prolonged closed times between bursts (Fig. 3F, G) (15, 50), and 4) multiple channels within the same patch is consistent with the hypothesis that these channels form clusters in membranes, although could also arise from the random presence of 2 or more channels in a single patch (Fig. 3A) (35). In combination with the absence of such conductances in Kcnt2−/− mitoplasts, these characteristics strengthen the conclusion that the genetic origin of the channels herein designated mito-KNa1.2, is the Kcnt2 gene product. Whole-mitoplast attached patch experiments (Fig. 1E) suggest that the orientation of the channel in the mitochondrial inner membrane is the same as that reported for plasma membrane KNa1.2—namely, with the C-terminal Na+-sensing site on the inside of the membrane (15, 51).
Patches from WT and Kcnt2−/− recordings contained channels with a similar overall range of conductances. However, the distribution of these conductances between genotypes was shifted. In particular, channels with unitary conductances in the range of 20–80 pS were more frequently observed in Kcnt2−/− (Fig. 2F). A similar observation was recently made with a cardiac-specific knockout of the mito-BK channel (12), raising the intriguing possibility that loss of 1 mitochondrial K+ channel may lead to compensatory up-regulation of other channels, to maintain K+ homeostasis. Given the importance of the mitochondrial K+ cycle for the regulation of organelle volume (36), such compensatory K+ fluxes may account for the persistence of a small but nonsignificant respiratory uncoupling effect of BT in Kcnt2−/− cardiomyocytes (Fig. 4B), as well as the relatively minor effect of KNa1.2 loss on mitochondrial ultrastructure (Fig. 4D–F). An alternative to compensatory expression between different mitochondrial K+ channels, is the possibility they may form heteromers. Individual KNa1.1 (Kcnt1) subunits can combine with KCa1.1 [potassium calcium–activated channel subfamily M α1 gene (Kcnma1)] subunits to form functional heterotetramers of intermediate conductance and activation properties (14). Heterotetramers of KNa1.1 with KNa1.2 have also been demonstrated (35). However, the existence of KCa1.1/KNa1.2 heterotetramers has not been reported.
A key aspect of these studies was the discovery that loss of cardiac KNa1.2 results in a unique metabolic phenotype—namely, a specific defect in cardiac fat oxidation only under conditions of high energy demand. An important caveat is that we cannot definitively rule out the possibility that the metabolic phenotype of Kcnt2−/− is related to loss of the channel at the plasma membrane. However, this is true of all proteins that are expressed at both the plasma membrane and the mitochondria. Despite the development of anti-KNa antibodies, fluorescent tags, and KNa channel–knockout animals, the channel’s existence at the cardiac plasma membrane has not been confirmed using any of these methods during the intervening time period. In addition, the Kcnt2−/− mouse is a global knockout, and it is possible that KNa1.2 loss from other cell types contributes to the phenotypes observed in our study. KNa1.2 is broadly expressed in most regions of the brain and CNS, with similar expression levels in cardiac tissues and ovaries, although its relative expression is an order of magnitude lower in all other tissues measured (18). Currently, there are no reports exploring the function of KNa channels in cell types, other than neurons and cardiac myocytes. Because the metabolic phenotype we observed was acutely activated in isolated hearts and in cardiac myocytes, the most likely origin is cardiac KNa1.2. Coupled with definitive data herein demonstrating existence of KNa1.2 in cardiac mitochondria, and previous reports of a BT-activated mitochondrial K+ channel that is absent in isolated mitochondria from both C. elegans Slo2−/− (52) and mouse Kcnt2−/− (3), we assert that the most likely explanation of the mitochondrial/metabolic phenotype in Kcnt2−/− hearts is loss of the channel from cardiac mitochondria.
The metabolic phenotype of Kcnt2−/− (Figs. 5 and 6) was manifest under conditions corresponding to classic bioenergetic “state 3,” wherein the mitochondrial membrane potential is consumed and generates ATP. If the in situ reversal potential of mitochondrial KNa1.2 is above 0, then channel opening could readily occur under conditions of high energetic demand when potential is lowered. In addition, the mitochondrial membrane potential is known to flicker in vivo (53), such that transient depolarization events may activate mito-KNa1.2. These properties may render mitochondrial KNa1.2 function important under conditions of stress, such as tissue ischemia. Given the previously reported requirement of KNa1.2 for cardioprotection by APC (3), it is notable that the mitochondrial membrane potential depolarizes and that cytosolic Na+ levels also increase during ischemia (54, 55). Together, these observations suggest that mito-KNa1.2 channels may be activated by acute perturbations in mitochondrial energy demand or under stress conditions, such as ischemia. Such properties may explain why the metabolic phenotype of Kcnt2−/− is evident only under stress.
A potential confounding factor in these studies was the use of BDM to inhibit contraction during the first stage only of cardiomyocyte isolation. BDM has been shown to affect ion channels including the KV channels responsible for transient outward K+ current Ito, Ca2+/Na+ exchanger, l-type Ca2+ channel, and skeletal muscle ryanodine receptor (56–59). However BDM’s half-maximum inhibitory concentration for transient outward current and l-type Ca2+ channels are higher than the 10 mM used in our protocol, and the cardiac ryanodine receptor was found to be BDM insensitive. Although the half-maximum inhibitory concentration of Ca2+/Na+ exchanger for BDM was 2.4 mM, myocytes were successfully rendered Ca2+ tolerant and incubated in BDM-free medium after the first stage. As such, any effect of BDM on the channels being studied here is probably insignificant.
The data in Fig. 7 revealed additional chronic effects of KNa1.2 loss on bioenergetics that resulted in altered body fat content and fasting glucose metabolism. Because of the presence of a mitochondrial K+/H+ exchanger, activation of a mitochondrial K+ channel would be expected to decrease the mitochondrial ΔpH, thus uncoupling oxidative phosphorylation and stimulating OCR, as shown in Fig. 4B. Mild mitochondrial uncoupling by mito-KNa1.2 channel activators may represent a novel therapeutic avenue for obesity/diabetes/metabolic syndrome. It is therefore notable that the antihelminthic drug niclosamide, which activates KNa channels (60), has long been known to uncouple mitochondria (61) and was recently shown to confer benefits in a mouse high-fat diet model of diabetes (62). Furthermore, a recent case report highlighted a patient with a KNa1.2 mutation (Q270E) who had migrating focal seizures that were nonresponsive to the ketogenic diet typically used to treat such symptoms (63). Cardiac metabolomics also revealed a potential up-regulation of PLC signaling in the Kcnt2−/− heart (Fig. 8C). KNa1.2 channels are known to interact with the PLC substrate PIP2 (64), which raises the possibility that loss of KNa1.2 results in perturbation of PIP2/PLC signaling. An important PLC downstream target is PKCε, which is known to play a role in development of insulin resistance in response to a high-fat diet (65). In addition to mitochondrial uncoupling, mito-KNa1.2 channel activators may confer metabolic benefits via a PIP2/PLC/PKCε signaling axis. A deeper investigation of the relationship between mito-KNa1.2 activity and metabolic regulation is thus warranted.
ACKNOWLEDGMENTS
The authors thank Christopher Lingle (Washington University, St. Louis, MO, USA) for providing founders for the Kcnt2−/− mice; Kathleen Kinally (Emeritus, New York University, New York, NY, USA) for technical support in performing mitochondrial patch-clamp experiments; Dana Godfrey [University of Rochester Medical Center (URMC) Musculoskeletal Center] for support with the DEXA analyses; and Karen Bentley (URMC electron microscopy core) for support obtaining electron microscopic images. This work was funded by U.S. National Institutes of Health General Medical Research Grant R01-GM087483 (to P.S.B. and K.N.), National Heart, Lung and Blood Institute Grant R01-HL071158 (to P.S.B.), and National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant R01-AR-059646 (to R.T.D.). The authors declare no conflicts of interest.
Glossary
- APC
anesthetic preconditioning
- BDM
2,3-butanedione monoxime
- BT
bithionol
- DEXA
dual-energy X-ray absorptiometry
- FCCP
carbonylcyanide-p-trifluoromethoxyphenylhydrazone
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- IPC
ischemic preconditioning
- IR
ischemia–reperfusion
- KNa
sodium-activated potassium
- KH
Krebs-Henseleit
- MIM
mitochondrial isolation medium
- MS/MS
tandem mass spectrometry
- NCLX
Na+/Ca2+ exchanger
- OCR
oxygen consumption rate
- qPCR
quantitative PCR
- ROS
reactive oxygen species
- RR
respiratory reserve
- SDHA
succinate dehydrogenase complex, subunit A
- Slack
KNa1.1 (formerly Slo2.2; aka KCa4.1)
- Slick
KNa1.2 (formerly Slo2.1; aka KCa4.2)
- WT
wild type
- XF
extracellular flux
AUTHOR CONTRIBUTIONS
C. O. Smith, K. Nehrke, and P. S. Brookes designed the research; C. O. Smith, Y. T. Wang, S. M. Nadtochiy, and J. H. Miller performed the experiments; Y. T. Wang developed software for analysis of Langendorff traces; C. O. Smith analyzed the data; C. O. Smith and P. S. Brookes wrote the paper; and E. A. Jonas and R. T. Dirksen contributed new analytical tools, training, and critical resources.
REFERENCES
- 1.Smith C. O., Nehrke K., Brookes P. S. (2017) The Slo(w) path to identifying the mitochondrial channels responsible for ischemic protection. Biochem. J. 474, 2067–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentzen B. H., Nardi A., Calloe K., Madsen L. S., Olesen S. P., Grunnet M. (2007) The small molecule NS11021 is a potent and specific activator of Ca2+-activated big-conductance K+ channels. Mol. Pharmacol. 72, 1033–1044 [DOI] [PubMed] [Google Scholar]
- 3.Wojtovich A. P., Smith C. O., Urciuoli W. R., Wang Y. T., Xia X. M., Brookes P. S., Nehrke K. (2016) Cardiac Slo2.1 is required for volatile anesthetic stimulation of K+ transport and anesthetic preconditioning. Anesthesiology 124, 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garlid K. D., Paucek P., Yarov-Yarovoy V., Murray H. N., Darbenzio R. B., D’Alonzo A. J., Lodge N. J., Smith M. A., Grover G. J. (1997) Cardioprotective effect of diazoxide and its interaction with mitochondrial ATP-sensitive K+ channels. Possible mechanism of cardioprotection. Circ. Res. 81, 1072–1082 [DOI] [PubMed] [Google Scholar]
- 5.Inoue I., Nagase H., Kishi K., Higuti T. (1991) ATP-sensitive K+ channel in the mitochondrial inner membrane. Nature 352, 244–247 [DOI] [PubMed] [Google Scholar]
- 6.Dolga A. M., Netter M. F., Perocchi F., Doti N., Meissner L., Tobaben S., Grohm J., Zischka H., Plesnila N., Decher N., Culmsee C. (2013) Mitochondrial small conductance SK2 channels prevent glutamate-induced oxytosis and mitochondrial dysfunction. J. Biol. Chem. 288, 10792–10804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nabbi R., Gadicherla A. K., Kersten J. R., Stowe D. F., Lazar J., Riess M. L. (2014) Genetically determined mitochondrial preservation and cardioprotection against myocardial ischemia-reperfusion injury in a consomic rat model. Physiol. Genomics 46, 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siemen D., Loupatatzis C., Borecky J., Gulbins E., Lang F. (1999) Ca2+-activated K channel of the BK-type in the inner mitochondrial membrane of a human glioma cell line. Biochem. Biophys. Res. Commun. 257, 549–554 [DOI] [PubMed] [Google Scholar]
- 9.Singh H., Lu R., Bopassa J. C., Meredith A. L., Stefani E., Toro L. (2013) MitoBK(Ca) is encoded by the Kcnma1 gene, and a splicing sequence defines its mitochondrial location [published correction in Proc Natl Acad Sci (2013) 110, 18024]. Proc. Natl. Acad. Sci. USA 110, 10836–10841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster D. B., Ho A. S., Rucker J., Garlid A. O., Chen L., Sidor A., Garlid K. D., O’Rourke B. (2012) Mitochondrial ROMK channel is a molecular component of mitoK(ATP). Circ. Res. 111, 446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soltysinska E., Bentzen B. H., Barthmes M., Hattel H., Thrush A. B., Harper M. E., Qvortrup K., Larsen F. J., Schiffer T. A., Losa-Reyna J., Straubinger J., Kniess A., Thomsen M. B., Brüggemann A., Fenske S., Biel M., Ruth P., Wahl-Schott C., Boushel R. C., Olesen S. P., Lukowski R. (2014) KCNMA1 encoded cardiac BK channels afford protection against ischemia-reperfusion injury. PLoS One 9, e103402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frankenreiter S., Bednarczyk P., Kniess A., Bork N., Straubinger J., Koprowski P., Wrzosek A., Mohr E., Logan A., Murphy M. P., Gawaz M., Krieg T., Szewczyk A., Nikolaev V. O., Ruth P., Lukowski R. (2017) cGMP-elevating compounds and ischemic conditioning provide cardioprotection against ischemia and reperfusion injury via cardiomyocyte-specific BK channels. Circulation 136, 2337–2355 [DOI] [PubMed] [Google Scholar]
- 13.Calvo S. E., Clauser K. R., Mootha V. K. (2016) MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44(D1), D1251–D1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joiner W. J., Tang M. D., Wang L. Y., Dworetzky S. I., Boissard C. G., Gan L., Gribkoff V. K., Kaczmarek L. K. (1998) Formation of intermediate-conductance calcium-activated potassium channels by interaction of Slack and Slo subunits. Nat. Neurosci. 1, 462–469 [DOI] [PubMed] [Google Scholar]
- 15.Bhattacharjee A., Joiner W. J., Wu M., Yang Y., Sigworth F. J., Kaczmarek L. K. (2003) Slick (Slo2.1), a rapidly-gating sodium-activated potassium channel inhibited by ATP. J. Neurosci. 23, 11681–11691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawasaki Y., Kuki I., Ehara E., Murakami Y., Okazaki S., Kawawaki H., Hara M., Watanabe Y., Kishimoto S., Suda K., Saitsu H., Matsumoto N. (2017) Three cases of KCNT1 mutations: malignant migrating partial seizures in infancy with massive systemic to pulmonary collateral arteries. J. Pediatr. 191, 270–274 [DOI] [PubMed] [Google Scholar]
- 17.Gururaj S., Palmer E. E., Sheehan G. D., Kandula T., Macintosh R., Ying K., Morris P., Tao J., Dias K. R., Zhu Y., Dinger M. E., Cowley M. J., Kirk E. P., Roscioli T., Sachdev R., Duffey M. E., Bye A., Bhattacharjee A. (2017) A de novo mutation in the sodium-activated potassium channel KCNT2 alters ion selectivity and causes epileptic encephalopathy. Cell Reports 21, 926–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Espinosa P. L., Wu J., Yang C., Gonzalez-Perez V., Zhou H., Liang H., Xia X. M., Lingle C. J. (2015) Knockout of Slo2.2 enhances itch, abolishes KNa current, and increases action potential firing frequency in DRG neurons. Elife 4, e10014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kameyama M., Kakei M., Sato R., Shibasaki T., Matsuda H., Irisawa H. (1984) Intracellular Na+ activates a K+ channel in mammalian cardiac cells. Nature 309, 354–356 [DOI] [PubMed] [Google Scholar]
- 20.Yang B., Gribkoff V. K., Pan J., Damagnez V., Dworetzky S. I., Boissard C. G., Bhattacharjee A., Yan Y., Sigworth F. J., Kaczmarek L. K. (2006) Pharmacological activation and inhibition of Slack (Slo2.2) channels. Neuropharmacology 51, 896–906 [DOI] [PubMed] [Google Scholar]
- 21.Szabo I., Zoratti M. (2014) Mitochondrial channels: ion fluxes and more. Physiol. Rev. 94, 519–608 [DOI] [PubMed] [Google Scholar]
- 22.Murphy E., Eisner D. A. (2009) Regulation of intracellular and mitochondrial sodium in health and disease. Circ. Res. 104, 292–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nita I. I., Hershfinkel M., Lewis E. C., Sekler I. (2015) A crosstalk between Na+ channels, Na+/K+ pump and mitochondrial Na+ transporters controls glucose-dependent cytosolic and mitochondrial Na+ signals. Cell Calcium 57, 69–75 [DOI] [PubMed] [Google Scholar]
- 24.Bay J., Kohlhaas M., Maack C. (2013) Intracellular Na+ and cardiac metabolism. J. Mol. Cell. Cardiol. 61, 20–27 [DOI] [PubMed] [Google Scholar]
- 25.Aittokallio T. (2010) Dealing with missing values in large-scale studies: microarray data imputation and beyond. Brief. Bioinform. 11, 253–264 [DOI] [PubMed] [Google Scholar]
- 26.Xia J., Wishart D. S. (2016) Using metaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinformatics 55, 14.10.1–14.10.91 [DOI] [PubMed] [Google Scholar]
- 27.Nadtochiy S. M., Urciuoli W., Zhang J., Schafer X., Munger J., Brookes P. S. (2015) Metabolomic profiling of the heart during acute ischemic preconditioning reveals a role for SIRT1 in rapid cardioprotective metabolic adaptation. J. Mol. Cell. Cardiol. 88, 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toledo-Arana A., Dussurget O., Nikitas G., Sesto N., Guet-Revillet H., Balestrino D., Loh E., Gripenland J., Tiensuu T., Vaitkevicius K., Barthelemy M., Vergassola M., Nahori M. A., Soubigou G., Régnault B., Coppée J. Y., Lecuit M., Johansson J., Cossart P. (2009) The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956 [DOI] [PubMed] [Google Scholar]
- 29.Szewczyk A., Jarmuszkiewicz W., Kunz W. S. (2009) Mitochondrial potassium channels. IUBMB Life 61, 134–143 [DOI] [PubMed] [Google Scholar]
- 30.Dryer S. E., Fujii J. T., Martin A. R. (1989) A Na+-activated K+ current in cultured brain stem neurones from chicks. J. Physiol. 410, 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaczmarek L. K. (2013) Slack, slick and sodium-activated potassium channels. ISRN Neurosci. 2013, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim G. E., Kronengold J., Barcia G., Quraishi I. H., Martin H. C., Blair E., Taylor J. C., Dulac O., Colleaux L., Nabbout R., Kaczmarek L. K. (2014) Human slack potassium channel mutations increase positive cooperativity between individual channels. Cell Reports 9, 1661–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H., Kronengold J., Yan Y., Gazula V. R., Brown M. R., Ma L., Ferreira G., Yang Y., Bhattacharjee A., Sigworth F. J., Salkoff L., Kaczmarek L. K. (2009) The N-terminal domain of Slack determines the formation and trafficking of Slick/Slack heteromeric sodium-activated potassium channels. J. Neurosci. 29, 5654–5665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaczmarek L. K., Aldrich R. W., Chandy K. G., Grissmer S., Wei A. D., Wulff H. (2017) International union of basic and clinical pharmacology. C. Nomenclature and properties of calcium-activated and sodium-activated potassium channels. Pharmacol. Rev. 69, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim G. E., Kaczmarek L. K. (2014) Emerging role of the KCNT1 Slack channel in intellectual disability. Front. Cell. Neurosci. 8, 209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garlid K. D. (1996) Cation transport in mitochondria--the potassium cycle. Biochim. Biophys. Acta 1275, 123–126 [DOI] [PubMed] [Google Scholar]
- 37.Bednarczyk P., Wieckowski M. R., Broszkiewicz M., Skowronek K., Siemen D., Szewczyk A. (2013) Putative structural and functional coupling of the mitochondrial BKCa channel to the respiratory chain. PLoS One 8, e68125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Checchetto V., Teardo E., Carraretto L., Leanza L., Szabo I. (2016) Physiology of intracellular potassium channels: a unifying role as mediators of counterion fluxes? Biochim. Biophys. Acta. 1857, 1258–1266 [DOI] [PubMed] [Google Scholar]
- 39.Tejada M. A., Hashem N., Calloe K., Klaerke D. A. (2017) Heteromeric Slick/Slack K+ channels show graded sensitivity to cell volume changes. PLoS One 12, e0169914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tejada M. A., Stople K., Hammami Bomholtz S., Meinild A. K., Poulsen A. N., Klaerke D. A. (2014) Cell volume changes regulate slick (Slo2.1), but not slack (Slo2.2) K+ channels. PLoS One 9, e110833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Picard M., White K., Turnbull D. M. (2013) Mitochondrial morphology, topology, and membrane interactions in skeletal muscle: a quantitative three-dimensional electron microscopy study. J. Appl. Physiol. 114, 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bugger H., Chen D., Riehle C., Soto J., Theobald H. A., Hu X. X., Ganesan B., Weimer B. C., Abel E. D. (2009) Tissue-specific remodeling of the mitochondrial proteome in type 1 diabetic akita mice. Diabetes 58, 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laskowski M., Augustynek B., Kulawiak B., Koprowski P., Bednarczyk P., Jarmuszkiewicz W., Szewczyk A. (2016) What do we not know about mitochondrial potassium channels? Biochim. Biophys. Acta. 1857, 1247–1257 [DOI] [PubMed] [Google Scholar]
- 44.Testai L., Rapposelli S., Martelli A., Breschi M. C., Calderone V. (2015) Mitochondrial potassium channels as pharmacological target for cardioprotective drugs. Med. Res. Rev. 35, 520–553 [DOI] [PubMed] [Google Scholar]
- 45.Ertracht O., Malka A., Atar S., Binah O. (2014) The mitochondria as a target for cardioprotection in acute myocardial ischemia. Pharmacol. Ther. 142, 33–40 [DOI] [PubMed] [Google Scholar]
- 46.Tano J. Y., Gollasch M. (2014) Hypoxia and ischemia-reperfusion: a BiK contribution? Am. J. Physiol. Heart Circ. Physiol. 307, H811–H817 [DOI] [PubMed] [Google Scholar]
- 47.Stowe D. F., Gadicherla A. K., Zhou Y., Aldakkak M., Cheng Q., Kwok W. M., Jiang M. T., Heisner J. S., Yang M., Camara A. K. (2013) Protection against cardiac injury by small Ca(2+)-sensitive K(+) channels identified in guinea pig cardiac inner mitochondrial membrane. Biochim. Biophys. Acta 1828, 427–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ponnalagu D., Singh H. (2016) Anion channels of mitochondria. Handb. Exp. Pharmacol. 240, 71–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leanza L., Biasutto L., Managò A., Gulbins E., Zoratti M., Szabò I. (2013) Intracellular ion channels and cancer. Front. Physiol. 4, 227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang B., Desai R., Kaczmarek L. K. (2007) Slack and Slick K(Na) channels regulate the accuracy of timing of auditory neurons. J. Neurosci. 27, 2617–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomson S. J., Hansen A., Sanguinetti M. C. (2015) Identification of the intracellular Na+ sensor in Slo2.1 potassium channels. J. Biol. Chem. 290, 14528–14535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wojtovich A. P., Sherman T. A., Nadtochiy S. M., Urciuoli W. R., Brookes P. S., Nehrke K. (2011) SLO-2 is cytoprotective and contributes to mitochondrial potassium transport. PLoS One 6, e28287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Reilly C. M., Fogarty K. E., Drummond R. M., Tuft R. A., Walsh J. V., Jr. (2003) Quantitative analysis of spontaneous mitochondrial depolarizations. Biophys. J. 85, 3350–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green D. R., Kroemer G. (2004) The pathophysiology of mitochondrial cell death. Science 305, 626–629 [DOI] [PubMed] [Google Scholar]
- 55.Lesnefsky E. J., Moghaddas S., Tandler B., Kerner J., Hoppel C. L. (2001) Mitochondrial dysfunction in cardiac disease: ischemia–reperfusion, aging, and heart failure. J. Mol. Cell. Cardiol. 33, 1065–1089 [DOI] [PubMed] [Google Scholar]
- 56.Xiao Y. F., McArdle J. J. (1995) Activation of protein kinase A partially reverses the effects of 2,3-butanedione monoxime on the transient outward K+ current of rat ventricular myocytes. Life Sci. 57, 335–343 [DOI] [PubMed] [Google Scholar]
- 57.Watanabe Y., Iwamoto T., Matsuoka I., Ohkubo S., Ono T., Watano T., Shigekawa M., Kimura J. (2001) Inhibitory effect of 2,3-butanedione monoxime (BDM) on Na(+)/Ca(2+) exchange current in guinea-pig cardiac ventricular myocytes. Br. J. Pharmacol. 132, 1317–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eisfeld J., Mikala G., Varadi G., Schwartz A., Klöckner U. (1997) Inhibition of cloned human L-type cardiac calcium channels by 2,3-butanedione monoxime does not require PKA-dependent phosphorylation sites. Biochem. Biophys. Res. Commun. 230, 489–492 [DOI] [PubMed] [Google Scholar]
- 59.Tripathy A., Xu L., Pasek D. A., Meissner G. (1999) Effects of 2,3-butanedione 2-monoxime on Ca2+ release channels (ryanodine receptors) of cardiac and skeletal muscle. J. Membr. Biol. 169, 189–198 [DOI] [PubMed] [Google Scholar]
- 60.Biton B., Sethuramanujam S., Picchione K. E., Bhattacharjee A., Khessibi N., Chesney F., Lanneau C., Curet O., Avenet P. (2012) The antipsychotic drug loxapine is an opener of the sodium-activated potassium channel slack (Slo2.2). J. Pharmacol. Exp. Ther. 340, 706–715 [DOI] [PubMed] [Google Scholar]
- 61.Weinbach E. C., Garbus J. (1969) Mechanism of action of reagents that uncouple oxidative phosphorylation. Nature 221, 1016–1018 [DOI] [PubMed] [Google Scholar]
- 62.Tao H., Zhang Y., Zeng X., Shulman G. I., Jin S. (2014) Niclosamide ethanolamine-induced mild mitochondrial uncoupling improves diabetic symptoms in mice. Nat. Med. 20, 1263–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Madaan P., Jauhari P., Gupta A., Chakrabarty B., Gulati S. (2018) A quinidine non responsive novel KCNT1 mutation in an Indian infant with epilepsy of infancy with migrating focal seizures. Brain Dev. 40, 229–232 [DOI] [PubMed] [Google Scholar]
- 64.De los Angeles Tejada M., Jensen L. J., Klaerke D. A. (2012) PIP2 modulation of Slick and Slack K+ channels. Biochem. Biophys. Res. Commun. 424, 208–213 [DOI] [PubMed] [Google Scholar]
- 65.Samuel V. T., Liu Z. X., Wang A., Beddow S. A., Geisler J. G., Kahn M., Zhang X. M., Monia B. P., Bhanot S., Shulman G. I. (2007) Inhibition of protein kinase Cepsilon prevents hepatic insulin resistance in nonalcoholic fatty liver disease. J. Clin. Invest. 117, 739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]