Abstract
Objective
ABCC8 encodes sulfonylurea-receptor-1, a key regulatory protein of cerebral edema in many neurological disorders including traumatic brain injury (TBI). Sulfonylurea-receptor-1 inhibition has been promising in ameliorating cerebral edema in clinical trials. We evaluated whether ABCC8 tag-single-nucleotide-polymorphisms predicted edema and outcome in TBI.
Methods
DNA was extracted from 485 prospectively enrolled severe TBI patients. 410 were analyzed after quality control. ABCC8 tag-single-nucleotide-polymorphisms were identified (Hapmap, r2 ≥0.8, minor-allele frequency>0.20) and sequenced (iPlex-Gold, MassArray). Outcomes included radiographic edema, intracranial-pressure (ICP), and 3-month Glasgow Outcome Scale (GOS) score. Proxy-single-nucleotide-polymorphisms, spatial-modeling, amino-acid topology, and functional predictions were determined using established software-programs.
Results
Wild-type rs7105832 and rs2237982 alleles and genotypes were associated with lower average ICP (β=−2.91 p=0.001; β=−2.28 p=0.003), and decreased radiographic edema (OR=0.42 p=0.012; OR=0.52 p=0.017). Wild-type rs2237982 also increased favorable 3-month GOS (OR=2.45 p=0.006); this was partially mediated by edema (p=0.03). Different polymorphisms predicted 3-month outcome: variant-rs11024286 increased (OR=1.84, p=0.006) and wild-type rs4148622 decreased (OR=0.40 p=0.01) odds of favorable outcome. Significant tag and concordant proxy SNPs regionally span introns/exons 2–15 of the 39-exon gene.
Conclusions
This study identifies four ABCC8 tag-SNPs associated with cerebral edema and/or outcome in TBI, tagging a region including 33 polymorphisms. In polymorphisms predictive of edema, variant alleles/genotypes confer increased risk. Different variant-polymorphisms were associated with favorable outcome potentially suggesting distinct mechanisms. Significant polymorphisms spatially clustered, flanking exons encoding the sulfonylurea-receptor-site and transmembrane-domain-0/loop-0 (juxtaposing the channel-pore/binding-site). This builds a foundation for developing future strategies that may guide individualized care, treatment response, prognosis, and patient selection for clinical trials.
Keywords: ABCC8, sulfonylurea receptor 1 / Sur-1, traumatic brain injury / TBI, cerebral edema, polymorphisms
INTRODUCTION
In the developing era of precision medicine, genetic factors are increasingly recognized as important contributors to traumatic brain injury (TBI) variability and outcome[1,2]. Other than a few outcome-association studies of aquaporin-4 and interleukins, there is limited genetic evaluation of pathways affecting cerebral edema (CE) after severe TBI (sTBI)[2,3]. However, clinical evaluation and management of sTBI patients largely focuses on CE- a key prognosticator of unfavorable outcomes and frequent cause of herniation, morbidity and mortality[4]. The lack of preventive or targeted treatments against CE likely reflects challenges associated with disease heterogeneity and complexity of mechanisms underlying CE formation, which may have contributory genetic influences.
Several processes are implicated in CE including blood-brain-barrier disruption, cell volume regulation, oncotic gradients, and inflammation[5]. Genetic variations in such targets may influence transcriptional regulation, protein expression, structure or function. This could direct individual differences in timing and degree of CE, dominance of specific pathways, therapeutic response likelihood, and outcome. A priori identification of specific polymorphisms could provide valuable prognostic and predictive enrichment strategies to directing patient-care and clinical trials[6,7]. One such pathway involves sulfonylurea-receptor-1 (Sur1)- transient-receptor potential cation-channel member-4 (Trpm4). Polymorphisms in ABCC8 (encoding Sur1) especially warrant exploration given this pathway’s unique attributes and implications for targeted therapies.
Sur1, a transmembrane protein initially known for regulating a potassium-channel modulating insulin secretion (Kir6.2), is now also thought to play an important role in CE regulation[8]. In central nervous system (CNS) injury, Sur1 is upregulated and associates with Trpm4 (a non-selective cation channel) causing depolarization, water influx, oncotic edema, and cell death[8]. Unlike other known mechanisms underlying CE, CNS Sur1-Trpm4 is not normally expressed but is upregulated after injury[8,9]. This pathway harbors an exciting potential therapeutic target to prevent/decrease CE: an existing anti-diabetes medication (glyburide) inhibits channel opening with promising results in clinical trials of stroke[10,11]. Sur1 is expressed in human contusions[12] and cerebrospinal-fluid (CSF) Sur1 levels are a potential biomarker of CE in TBI[13]. ABCC8 single-nucleotide-polymorphisms (SNPs) in whole-exome analysis have been associated with CE in TBI[3]; however intrinsic to whole-genome exomic studies, SNP identification was limited to existing partial gene coverage, likely resulting in missed relevant SNPs (particularly intronic).
We examine effects of coding and non-coding sequence variability on CE and sTBI outcome across the entire ABCC8 gene using a targeted, candidate-gene approach to genotype tag-SNPs and capture a substantial proportion of common sequence variations[14]. We hypothesized that ABCC8 tag-SNPs are associated with CE and sTBI outcome. We also explored whether CE mediated SNP associations with outcome.
METHODS
Study Design
Subjects were prospectively enrolled through the Brain-Trauma Research Center protocol after informed consent from surrogate decision-makers. Inclusion criteria were age 16–80 years, sTBI with a Glasgow Coma Scale (GCS) score ≤ 8 on presentation and an external ventricular drain (EVD) per standard care (at our institution all sTBI patients have dual monitors placed including intraparenchymal multimodal monitors in addition to EVDs). Brain death on presentation and pregnancy were excluded. 485 subjects were consecutively enrolled between 2000–2014. The University of Pittsburgh Institutional Review Board approved the study.
ABCC8 Tag and proxy SNP identification
23 ABCC8 tag-SNPs were identified using HapMap with the tagger-algorithm pairwise approach, r2 ≥0.8 and minor allele frequency >0.20. After eliminating tag-SNPs that failed quality control, 15 were included in the final analysis (Supplementary Methods-A). Proxy-SNPs associated with tag-SNPs based on LD (r2 ≥0.8) were identified using SNP annotation and proxy search (SNAP, Broad Institute, Version 2.2).
DNA collection and Genotyping
DNA was obtained and extracted from white blood cells by the simple-salting out method as per our previously published report[3]. If blood was unavailable, since SNPs are constant per individual (regardless of time/source) DNA was extracted from excess CSF containing white blood cells in the ventriculostomy bag[3]. Genotyping using iPlex-Gold was performed with an Agena Compact MassArray with Nanodispenser by the University of Pittsburgh Genomics Research Core. Genotype analysis was performed using Typer-4.0. Assay. After quality control (Supplementary Methods-B), 75/485 samples were excluded, yielding 410 samples for final analysis.
Clinical Data Collection
Three previously established outcome measures were investigated[3,13].
Edema on initial head CT (binary)[15]: Consistent with NINDS common data element variables this includes size of ventricles, basilar cisterns, transfalcine herniation, and loss of gray/white matter differentiation – all of which are associated with raised ICP after TBI[15,16]. Determinations were per official radiologist reports.
Hourly ICP (continuous) – subcategorized into average and peak ICP (typically 5 days of neuromonitoring at our institution).
3-month Glasgow Outcome Scale (GOS) score (binary): favorable (GOS≥4) versus unfavorable (GOS<4).
Spatial relationship modeling between ABCC8 regions and Sur1 structure
Chromosomal locations were determined using the University of California, Santa Cruz genome browser, human genome assembly-38. Putative and canonical topology for human Sur1 amino-acid sequence was obtained from PROTTER version-1.0[17]. Peptide sequences encoded by specific exons and residue overlap splice sites were identified by Ensembl (release-87)[18]. ProtAnnot identified Sur1 protein annotations in the context of ABCC8 genomic sequence[19]. The established 3-dimensional electron microscopy structure of Sur1-Kir6.2 was obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank[20]. University of California, San Francisco Chimera software was used to generate the 3-dimensional structure of Sur1 without the associated channel Kir6.2[21].
Functional category of ABCC8 tag/proxy SNPs
Predicted SNP functional effects were investigated using F-SNP to explore potentially clinically significant implications of the genetic variation[22]. SNPs were evaluated for established clinical significance via PubMed, Embase, and ClinVar searches (Supplementary Methods-C).
Statistical Analysis
Analysis of variance (ANOVA, F-test) and Fisher’s exact test compared outcome variables between genotypes of each tag-SNP. Between-group comparisons were adjusted using Bonferroni’s method. The exact test assessed Hardy-Weinberg Equilibrium (HWE). Univariate linear and logistic regression models evaluated independent relationships between ABCC8 tag-SNPs and continuous or categorical outcomes. Multivariate regression models were developed with clinically relevant variables (age, gender, initial GCS-score) to control for confounders. Odds ratios, β-coefficients, and 95% confidence intervals (CI) were based on modes of inheritance: homozygous-wild-type (AA), heterozygous (Ab), homozygous-variant (bb), as well as by an allele difference model (A vs b). Multiple comparisons for the regression models were adjusted using the established Benjamin-Yekutieli (B-Y) method[3,14,23] yielding a significance threshold of p=0.01064. The Sobel-Goodman test determined mediation effects[24]. Analyses were performed using Stata 14.0(StataCorp, TX). Pairwise LD between tag-SNPs was determined using a java-based linkage disequilibrium plotter (JLIN) and LDlink for proxy-SNPs[2,3],[25]. R-package Haplo.Stats version-1.7.7 was used for haplotype/risk-allele construction (Supplementary Table-1, Supplementary Results).
RESULTS
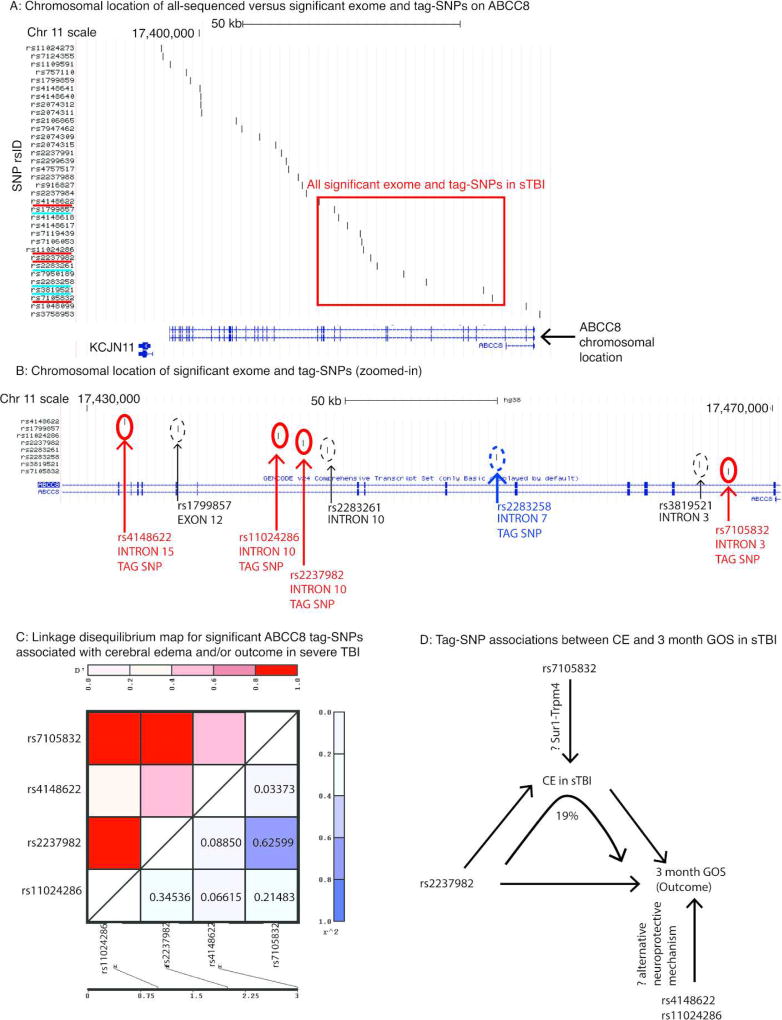
Table 1 summarizes demographic and clinical characteristics. Four tag-SNPs (rs7105832, rs2237982, rs11024286 and rs4148622) had significant associations with CE and/or outcome in univariate and multivariate single-locus analyses. Table 2 shows relative genotype frequencies and descriptive statistics of variation in CE and 3-month GOS between genotypes. Although the 15 analyzed tag-SNPs were distributed along the length of ABCC8, rs7105832, rs2237982, rs11024286 and rs4148622 were clustered between introns 3–15 (Figure 1A–B, Figure 2). Tag-SNPs were in HWE and no significant LD was noted (r2 <0.8, Figure 1C). This regional clustering is consistent with previously identified ABCC8 SNPs associated with CE in sTBI in whole-exome analysis (rs1799857, rs2283261, rs2283258, rs3819521)[3].
TABLE 1.
PATIENT CHARACTERISTICS
| Variable (n=410) | Mean (SD, Range) |
|---|---|
| Age | 38.3 (16.7, 16–78) |
| Initial GCS | 5.83 (1.48, 3–9) |
| Average ICP | 10.6 (4.8, 4–45) |
| Peak ICP | 27.6 (15.2, 9–105) |
| Frequency n (%) | |
| Gender (M) | 315 (77.8%) |
| Race | |
| • White | 371 (90.5%) |
| • African American | 29 (7.1%) |
| • Asian | 2 (0.5%) |
| • Native American | 2 (0.5%) |
| • Unknown | 6 (1.4%) |
| Primary Injury | |
| • Intraparenchymal hemorrhage +/− intraventricular hemorrhage | 135 (33.3%) |
| • Epidural | 29 (7.1%) |
| • Subdural | 133 (32.4%) |
| • Diffuse Axonal | 30 (7.3%) |
| • Subarachnoid | 72 (17.6%) |
| • None | 6 (1.5%) |
| • Unknown | 5 (1.2%) |
| Edema noted on acute CT (Y) | 163 /369 (44.2%) |
| Decompressive Craniotomy (Y) | 131 /405 (32.4%) |
| • Day 0 | 86 /131 (65.6%) |
| • Day 1 | 18 /131 (13.7%) |
| • Days 2–5 | 16 /131 (12.2) |
| • > 5 Days | 11 /131 (8.4%) |
| Favorable Outcome GOS ≥ 4 (Y) | 126/ 410 (30.7%) |
TABLE 2.
TAG-SNP GENOTYPE FREQUENCIES IN THE SEVERE TBI POPULATION
| SNP Details |
Variable Measured | Homozygous Wild-Type |
Heterozygous | Homozygous Variant | p-value (ANOVA /Fisher) |
|---|---|---|---|---|---|
|
| |||||
| rs7105832 (Intron 3) Wild-type A Variant C | SNP Frequency in sTBI population (%) | 160 (42.9%) | 171 (45.8%) | 42 (11.3%) | |
| • Average ICP, Mean (SD) | 10 ± 3 mmHg | 10 ± 4 mmHg | 13 ± 7.5 mmHg | 0.003* | |
| • Peak ICP, Mean (SD) | 28.8 ± 17 mmHg | 24.2 ± 10.3 mmHg | 31.9 ± 19 mmHg | 0.020* | |
| • Acute CT Edema, Y frequency (%) | 68/150 (45.3%) | 57/155 (36.8%) | 25/40 (62.5%) | 0.012* | |
| • Favorable GOS ≥ 4, Y frequency (%) | 53/160 (33.1%) | 53/171 (31%) | 9/42 (21.4%) | 0.358 | |
|
| |||||
| rs2237982 (Intron 10) Wild-type C Variant T | SNP Frequency in sTBI population (%) | 133 (33.17%) | 192 (47.88%) | 76 (18.95%) | |
| • Average ICP, Mean (SD) | 9.9 ± 2.9 mmHg | 10.3 ± 3.8 mmHg | 12.4 ± 7.8 mmHg | 0.0107* | |
| • Peak ICP, Mean (SD) | 29.2 ± 17.5 mmHg | 24.8 ± 11 mmHg | 31.6 ± 18.3 mmHg | 0.0190* | |
| • Acute CT Edema, Y frequency (%) | 54/120 (45%) | 66/176 (37.5%) | 38/67 (56.7%) | 0.0204* | |
| • Favorable GOS ≥ 4, Y frequency (%) | 43/133 (32.3%) | 66/192 (34.4%) | 13/76 (17.1%) | 0.015* | |
|
| |||||
| rs11024286 (Intron 10) Wild-type G Variant A | SNP Frequency in sTBI population (%) | 187 (47.10%) | 160 (40.3%) | 50 (12.59%) | |
| • Average ICP, Mean (SD) | 11.2 ± 5.9 mmHg | 10.2 ± 3.7 mmHg | 9.8 ± 2.9 mmHg | 0.2207 | |
| • Peak ICP, Mean (SD) | 28.6 ± 15.2 mmHg | 26.9 ± 14.7 mmHg | 26 ± 17.5 mmHg | 0.6108 | |
| • Acute CT Edema, Y frequency (%) | 74/169 (43.8%) | 64/147 (43.6%) | 19/44 (43.2%) | 1.0 | |
| • Favorable GOS ≥ 4, Y frequency (%) | 44/187 (23.5%) | 60/160 (37.5%) | 16/50 (32%) | 0.017* | |
|
| |||||
| rs4148622 (Intron 15) Wild-type G Variant A | SNP Frequency in sTBI population (%) | 224 (55.86%) | 141 (35.16%) | 36 (8.98%) | |
| • Average ICP, Mean (SD) | 10.6 ± 4.6 mmHg | 10.8 ± 5.5 mmHg | 9.9 ± 3.5 mmHg | 0.7402 | |
| • Peak ICP, Mean (SD) | 26.7 ± 13.1 mmHg | 29.4 ± 19.1 mmHg | 24.7 ± 9.7 mmHg | 0.3162 | |
| • Acute CT Edema, Y frequency (%) | 90/206 (43.7%) | 59/129 (45.8%) | 10/28 (35.7%) | 0.636 | |
| • Favorable GOS ≥ 4, Y frequency (%) | 70/224 (31.3%) | 35/141 (24.82%) | 18/36 (50%) | 0.016* | |
Figure 1. ABCC8 tag-SNPs associated with cerebral edema (CE) and/or outcome in severe TBI (sTBI).
A: Graphical display of the chromosomal location of all sequenced versus significant ABCC8 polymorphisms plotted on the University of California Santa Crux (UCSC) genome browser (Assembly version GRCh38/hg38, scale-bar for 50 kilo-basepairs shown). A schematic of the 39-exon ABCC8 gene is on the X-axis. Exons (separated by introns) are marked with vertical lines/blocks. Exon #1 is on the right extreme and exon #39 is on the left extreme of the gene schematic. The vertical axis lists all of the single nucleotide polymorphisms (SNPs) by rsID sequenced in this study as well as a prior exome analysis. Significant tag-SNPs identified in this study (rsID solid red underline) were rs7105832, rs2237982, rs11024286, and rs4148622. SNPs identified in the whole exome analysis as being associated with edema are also shown for comparison (rsID solid blue underline). The chromosomal location of all significant exome and tag-SNPs in sTBI is highlighted (box).
B: Magnified display of the chromosomal location for significant tag-SNPs in sTBI (scale-bar for 10 kilobasepairs shown). Tag-SNP location marked with red circles, labeled with the appropriate rsID). Direct comparison with significant SNPs identified from a previously published whole-exome analysis (dashedcircles) demonstrates consistent geographical clustering between introns 3–15.
C: Linkage disequilibrium plot including r2 values between the four significant tag-SNPs were generated using the Java LINkage (JLIN) disequilibrium plotter program. The results show that none of the r2 values between any of the four tag-SNPs are ≥0.8.
D: Summary schematic of tag-SNPs associated with CE (rs7105832), 3-month GOS (rs4148622, rs11024286), or both (rs2237982). The diagram illustrates that 19% of the effect of rs2237982 on 3-month GOS is mediated via CE, however 81% is not. Moreover, both rs4148622 and rs11024286 are associated with 3-month GOS but not CE. This gives rise to the hypothesis that the association with CE presumably mediated via the Sur1-Trpm4 pathway, may be mechanistically distinct from Sur1 mediated neuroprotection/effect on outcome.
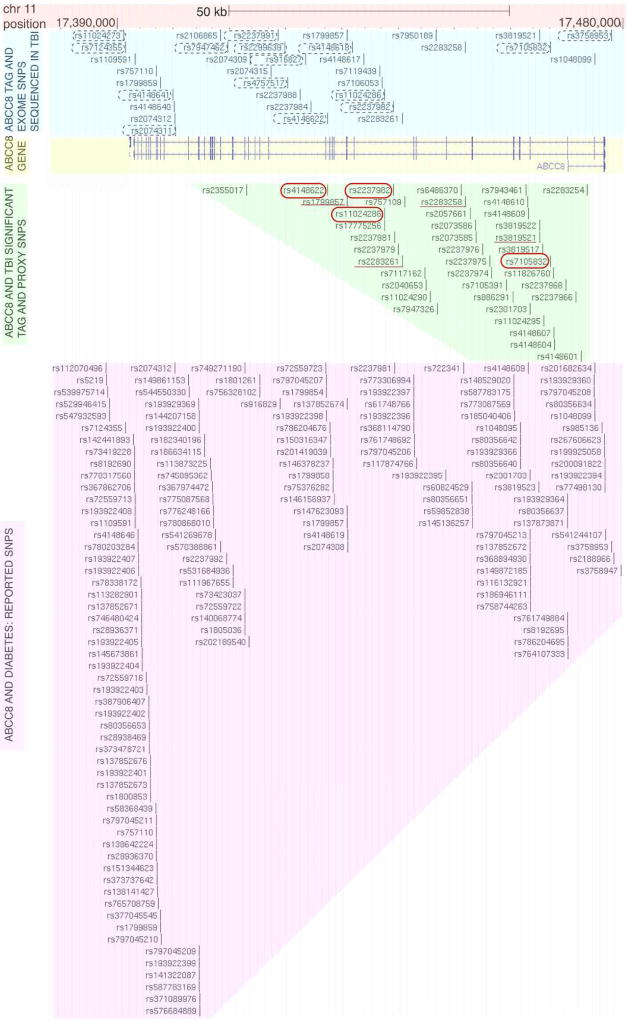
Figure 2. Spatial comparison of ABCC8 tag and proxy SNPs associated with cerebral edema (CE) and/or outcome in severe TBI versus previously reported SNPs implicated in glucose metabolism disorders.
This is a graphical display of the chromosomal location of ABCC8 polymorphisms plotted on the UCSC genome browser (Assembly version GRCh38/hg). Base-pair position on chromosome 11 is numbered and a scale-bar for 50 kilo-basepairs is shown (shaded orange). A schematic of the 39-exon ABCC8 gene is shaded in yellow where exons (separated by introns) are marked with vertical blue lines/blocks. Exon #1 is on the right extreme, and exon #39 is on the left extreme of the illustrated gene. The position and identification of all sequenced SNPs in sTBI is marked (shaded blue), with the 15 tag-SNPs specifically sequenced in this study circled (black dashed-outline) showing coverage across the entire gene. The position and rsIDs of all significant SNPs associated with CE and/or outcome in sTBI is depicted in the shaded-green region- they cluster primarily between introns 2–15 with a single exception (a proxy-SNP rs2355017 at intron 20 tagged by rs4148622 with incomplete LD r2 =0.83). Significant tag-SNPs from this study are circled (solid-red outline), and significant SNPs from the previously published whole-exome study are underlined (solid red). The remaining SNPs are proxy-SNPs associated with the significant tag-SNPs with r2 ≥0.8. The position and rsIDs of reported clinically significant SNPs associated with disorders of glucose metabolism are depicted in the shaded-pink region: more than 66% cluster downstream of intron 15 in contrast with the polymorphisms significant in sTBI.
rs7105832 is associated with CE measures
CE measures were different between rs7105832 genotypes (Table 1, average ICP p=0.003, peak ICP p=0.02, acute CT edema p=0.012). A higher proportion of homozygous-variant patients had acute CT edema (CC=62.5%) versus heterozygotes (CA=36.8%) and homozygous-wild-type (AA=45.3%). After Bonferroni’s correction, homozygous-variant had higher average (13.0±7.5 mmHg) and peak (31.9±19.0 mmHg) ICP versus heterozygotes (average ICP 10.0±4, p=0.004; peak ICP 24.2±10.3, p=0.045) and homozygous-wild-type (average ICP 10.0±3, p=0.004; peak ICP 28.8±17 p=0.099). Univariate and multivariate regression analyses (Table 3) demonstrated that homozygous-variant rs7105832 was an independent predictor of increased average ICP (β=2.88 univariate, β=2.73 multivariate), surviving the B-Y correction (p=0.001). The allele-difference model was consistent: wild-type allele was protective for lower average ICP (β=-2.9, p=0.001).
TABLE 3.
ABCC8 TAG-SNP ASSOCIATION WITH CEREBRAL EDEMA IN SEVERE TBI
| SNP | Model | Average ICP | Peak ICP | Acute CT Edema | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | Univariate | Multivariate | ||||||||
| β coeffici ent (95% CI) |
p | β coeffic ient (95% CI) |
p | β coefficie nt (95% CI) |
p | β coefficie nt (95% CI) |
p | Odds Ratio (95% CI) |
p | Odds Ratio (95% CI) |
p | ||
| rs2237982# | Genotype (reference CC) | ||||||||||||
| • CT | 0.34 (−1.07–1.75) | 0.63 | 0.53 (−0.83–1.89) | 0.44 | −4.4 (−8.87–0.07) | 0.05 | −3.72 (−8.03–0.60) | 0.09 | 1.47 (0.81–2.70) | 0.20 | 0.73 (0.45–1.17) | 0.19 | |
| • TT | 2.48 (0.77–4.19) | 0.005* | 2.16 (0.49–3.83) | 0.012 | 2.37 (−3.05–7.80) | 0.39 | 1.89 (−3.42–7.20) | 0.48 | 2.64 (1.03–6.74) | 0.042 | 1.56 (0.85–2.86) | 0.16 | |
|
| |||||||||||||
| Allele Difference | |||||||||||||
| • C | −2.28 (−3.77–−0.79) | 0.003* | −1.84 (−3.31–−0.38) | 0.014 | −4.98 (−9.74– −0.22) | 0.04 | −4.09 (−8.76–0.58) | 0.086 | 0.52 (acute) (0.30–0.89) | 0.017 | 0.53 (0.31–0.92) | 0.023 | |
| • T | 1.01 (−0.32–2.35) | 0.13 | 1.03 (−0.25–2.31) | 0.11 | −2.26 (−6.49–1.97) | 0.29 | −2.0 (−6.07–2.08) | 0.34 | 1.67 (acute) (0.94–2.97) | 0.083 | 0.90 (0.58–1.41) | 0.65 | |
|
| |||||||||||||
| rs7105832 | Genotype (reference AA) | ||||||||||||
| • AC | −0.06 (−1.25–1.14) | 0.92 | 0.23 (−0.90–1.26) | 0.69 | −4.59 (−8.8—0.37) | 0.033 | −3.94 (−7.98–0.10) | 0.056 | 0.70 (acute) (0.44–1.11) | 0.13 | 0.68 (0.43–1.09) | 0.108 | |
| • CC | 2.88 (1.12–4.64) | 0.001* | 2.73 (1.07–4.40) | 0.001* | 3.14 (−3.04–9.32) | 0.32 | 2.72 (−3.21–8.65) | 0.37 | 2.01 (acute) (0.98–4.11) | 0.056 | 1.95 (0.95–4.01) | 0.068 | |
|
| |||||||||||||
| Allele Difference | |||||||||||||
| • A | −2.91 (−4.57–−1.26) | 0.001* | −2.62 (−4.19–1.05) | 0.001* | −5.37 (−11.2–0.5) | 0.07 | −4.6 (−10.2–1.03) | 0.11 | 0.42 (acute) (0.21–0.82) | 0.012 | 0.42 (0.21–0.84) | 0.014 | |
| • C | 0.63 (−0.51–1.78) | 0.28 | 0.83 (−0.25–1.90) | 0.13 | −2.77 (−6.76–1.23) | 0.17 | −2.36 (−6.17–1.45) | 0.22 | 0.88 (0.57–1.34) | 0.54 | 0.86 (0.56–1.32) | 0.48 | |
rs2237982 is also associated with outcome in severe TBI.
rs2237982 is associated with CE measures and 3-month outcome
CE measures and 3-month GOS were different between rs2237982 genotypes (average ICP p=0.0107, peak ICP p=0.0190, acute CT edema p=0.0204, 3-month GOS p=0.015; Table 2). Homozygous-variants (TT) had higher average ICP in univariate analysis (β=2.48, p=0.005) and presence of wild-type allele (C) was protective (β=-2.28, p=0.003, Table 3). Homozygous-variant genotype decreased odds of favorable outcome (OR 0.43, p=0.010) and concordantly presence of a wild-type allele increased odds of favorable outcome by 2.45 (p=0.006), withstanding the B-Y correction in the multivariate model (OR=2.62, p=0.005, Table 4). The protective effect of rs2237982 on 3-month GOS was partially mediated through CE (Sobel-Goodman proportion of total effect mediated=0.198, p=0.03, Figure 1D).
TABLE 4.
ABCC8 TAG-SNP ASSOCIATION WITH OUTCOME IN SEVERE TBI
| SNP | Model | 3 month GOS | |||
|---|---|---|---|---|---|
| Univariate Odds ratio (95% CI) |
p | Multivariate Odds ratio (95% CI) |
p | ||
| rs11024286 | Genotype (reference GG) | ||||
| • GA | 1.95 (1.22–3.10) | 0.005* | 2.57 (1.53–4.31) | 0.000* | |
| • AA | 1.53 (0.77–3.02) | 0.22 | 1.94 (0.93–4.04) | 0.077 | |
|
| |||||
| Allele Difference | |||||
| • G | 0.91 (0.48–1.72) | 0.77 | 0.81 (0.41–1.60) | 0.55 | |
| • A | 1.84 (1.19–2.86) | 0.006* | 2.40 (1.47–3.89) | 0.000* | |
|
| |||||
| rs2237982 | Genotype (reference CC) | ||||
| • CT | 1.10 (0.69–1.75) | 0.70 | 1.08 (0.65–1.79) | 0.76 | |
| • TT | 0.43 (0.21–0.87) | 0.01* | 0.40 (0.19–0.83) | 0.014* | |
|
| |||||
| Allele Difference | |||||
| • C | 2.45 (1.29–4.63) | 0.006* | 2.62 (1.34–5.15) | 0.005* | |
| • T | 0.87 (0.56–1.37) | 0.56 | 0.84 (0.52–1.36) | 0.48 | |
|
| |||||
| rs4148622 | Genotype (reference GG) | ||||
| • GA | 0.73 (0.45–1.17) | 0.19 | 0.66 (0.40–1.11) | 0.12 | |
| • AA | 2.2 (1.08–4.48) | 0.03 | 2.40 (1.01–4.77) | 0.047 | |
|
| |||||
| Allele Difference | |||||
| • G | 0.40 (0.20–0.81) | 0.01* | 0.39 (0.18–0.83) | 0.015 | |
| • A | 0.94 (0.61–1.44) | 0.78 | 0.87 (0.55–1.39) | 0.57 | |
Multivariate model controls for age, sex and initial GCS
rs11024286 and rs4148622 are associated with 3-month outcome
Genotype distributions (Table 2) and descriptive statistics for both rs11024286 and rs4148622 showed that favorable 3-month GOS was different between genotypes (p=0.017 and p=0.016 respectively). Table 4 presents univariate and multivariate results. Unlike tag-SNPs associated with CE where variant-alleles conferred increased risk, for both tag-SNPs associated with 3-month GOS, variant-alleles conferred protection against unfavorable outcome, suggesting a distinct neuroprotective pathway. Rs11024286 heterozygotes had increased odds of favorable outcome (multivariate OR 2.57, p=0.000) versus homozygous-wild-type(GG). Variant-allele (A) was an independent predictor of favorable outcome (multivariate OR 2.40, p=0.000). For rs4148622, wild-type-allele (G) demonstrated decreased odds of favorable outcome (OR 0.010) however this did not retain multivariate significance (p=0.015).
Predicted functional implications of ABCC8 tag/proxy-SNPs
Thirty-three proxy-SNPs were in LD (r2 ≥0.8) with the significant tag-SNPs (rs7105832, rs2237982, rs11024286 and rs4148622). Supplementary Table 2 summarizes association strengths/LD, chromosomal locations, predicted functional category, functional significance (FS) score[22], and known clinical implications[3,26–28]. Consistent with known SNP distributions in the human genome, significant SNPs were in non-coding portions of ABCC8 with predicted effects on transcriptional regulation or splice site alterations. The significant proxy-SNPs, while intronic, have LD extending across coding regions/exons (Supplementary Figure 1, Supplementary Table 2). One of the significant proxy-SNPs rs2283261 (r2 =1.0 with rs2237982) had been previously evaluated in a smaller untargeted analysis where it was independently identified as a predictor of CE [3]. In type-2 diabetes, rs2237981 (a proxy-SNP identified in this study r2 =1.0 with rs2237982, intron-10), had a trend towards predicting gliclazide responsiveness (p=0.045) without altering the amino-acid sequence of Sur1[27] and rs4148609 (r2 =0.97 with rs7105832 intron-4) had a modest interaction with metformin treatment[28].
Gene-protein spatial model of significant ABCC8 tag/proxy-SNPs
ABCC8 has 39 exons. All significant tag-SNPs and majority (32/33) proxy-SNPs were upstream of intron 15 (Supplementary Table 2, Figure 1A–B, 2). This geography is consistent with clustering of significant SNPs from a prior whole-exome analysis in sTBI (Figure 1A–B)[3] but in contrast with SNPs reported in glucose-metabolism disorders- most of which (64.1%) regionally cluster towards introns/exons 16–39 (Figure 2).
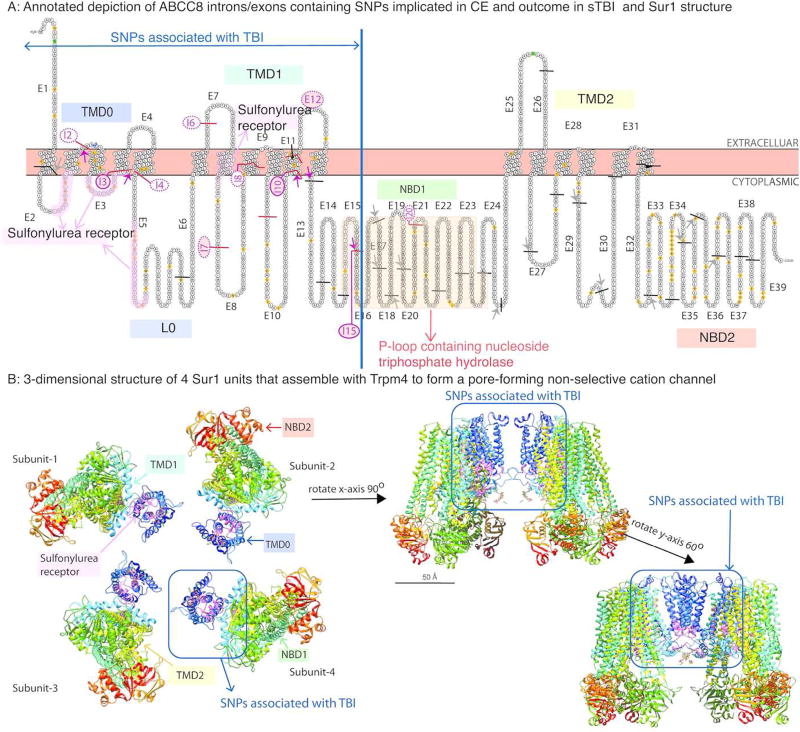
Combining human Sur1 putative and canonical amino-acid topology with ABCC8 exons, we determined that exons between introns 1–15 encode transmembrane-domain-0 (TMD0, exons 1–4), loop-0 (L0, exons 5–6), transmembrane-domain-1 (TMD1, exons 6–12), part of nucleotide-binding-domain-1 (NBD1, exons 13–15), and the sulfonylurea receptor motif of the protein (exons 2,3, and 7; Figure 3). A 3-dimensional model demonstrates that these domains predominantly contribute to the Trpm4 binding site and channel-pore (Figure 3B). Conversely, exons contained between introns 16–39 encode majority of the P-loop containing nucleoside triphosphate hydrolase, tranemembrane-domain-2 (TMD2), and nucleotide-binding-domain-2(NBD2).
Figure 3. Spatial relationships between ABCC8 gene regions and Sur1 protein structure.
A: This is an annotated depiction of ABCC8 introns/exons containing SNPs implicated in CE and outcome in sTBI as they relate to specific locations in the Sur1 protein. The diagrammatic representation of Sur1 was generated using PROTTER software (UniProt ID Q09428). Exons (E) are numbered and separated by solid lines (black line for exons separated by introns that do not contain significant tag/proxy SNPs, magenta line for exons separated by introns that harbor significant tag/proxy SNPs associated with CE or outcome in sTBI). These interspersed introns (I) that contain significant tag/proxy SNPs are numbered and categorized into those harboring significant tag/exomic-SNPs (shaded magenta circle) versus those containing significant proxy-SNPs (unshaded magenta circle). Sulfonylurea-receptor-1 contains 3 transmembrane domains and 2 nucleotide binding domains. Different domains of the protein are labeled: TMD0=transmembrane-domain 0, L0=Loop 0, TMD1=transmembrane-domain 1, NBD1=nucleotide binding domain 1, TMD2=transmembrane-domain 2, NBD2=nucleotide binding domain 2. The sulfonylurea receptor binding site is shaded in light pink and labeled. The p-loop containing nucleoside triphosphate hydrolase is shaded in yellow and labeled. Residue overlap splice sites separated by introns containing a significant tag/proxy SNP are marked (short magenta-arrows). Non-significant residue overlap splice sites in other regions of the protein are also shown (short gray-arrows). All significant tag, and 32/33 significant proxy-SNPs associated with CE+/− outcome in sTBI are located in the gene region that contains exons encoding TMD0-L0, the sulfonylurea receptor binding site, TMD1, and a small portion of NBD1.
B: The 3-dimensional structure of Sur1 subunits that assemble with Trpm4 to form a pore-forming non-selective cation channel was obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB-PDB) based on work from Li et al (Cell, 2017) using cryoelectron-microscopy to elucidate the structure of the pancreatic Kir6.2-Sur1 complex. University of California, San Francisco (UCSF) Chimera software was used to generate the 3-dimension structure of Sur1 in this figure without the associated Kir6.2 channel. The panel on the left illustrates 4 Sur1 subunits. TMD0, TMD1, TMD2, NBD1, NBD2 and the sulfonylurea subunit are labeled. 1 Sur1 subunit is then rotated 90° on the X axis and 60° on the Y axis to better illustrate the orientation of TMD0/TMD1 and the sulfonylurea receptor binding site since these domains are encoded by sequences in ABCC8 regions that are in spatial proximity to significant tag/proxy SNPs associated with CE+/− outcome in sTBI.
Introns-2, 3, 4,10 and 15 all contain significant tag and associated proxy-SNPs (Supplementary Table 2), and separate exons with residue overlap splice sites where 1–2 nucleotides are encoded in the exon preceding the intron, and the remaining nucleotide(s) are in the following exon (Figure 3A). Thus, an intermediary intronic SNP may be of particular functional or regulatory significance to the encoded protein.
DISCUSSION
In sTBI, CE is a predictor of unfavorable outcome and can account for up to 50% of mortality[4]. CE also represents a frequent, often life-threatening concern in other neurological disorders including stroke, intracranial-hemorrhage, cardiac-arrest, and subarachnoid-hemorrhage. Sur1 plays a key regulatory role in all of these disorders[3,8,10–12]. Sur1-Trpm4 inhibition is also independently linked with reduced tissue loss, white matter preservation, and improved cognition- independent of effects on CE[8,29]. Genetic variation may influence the degree to which Sur1-mediated CE or neurodegeneration predominates over other pathways, and whether (or how) individual patients respond to targeted inhibition. This has potential implications for prognostic and predictive enrichment strategies for sTBI and beyond: ABCC8 polymorphisms could guide risk-stratification/prognostication and may become valuable patient-selection tools for clinical trials evaluating efficacy of targeted inhibition with treatments like glyburide. This is an important consideration in the context of many failed TBI trials attributed to disease heterogeneity and dearth of informative biomarkers[13,30]. It is also immediately relevant given Sur1 expression in human contusions, its potential as a biomarker of CE in human sTBI, and the promising results of CE reduction in recent clinical trials assessing glyburide/Sur1-inhibition in stroke[10–13].
This study lays some groundwork for developing the predictive and prognostic utility of ABCC8 genetic variability in sTBI with three central findings:
Four ABCC8 tag-SNPs were independent predictors of CE (rs7105832), outcome (rs11024286, rs4148622), or both (rs2237982). These were markers of 33 proxy-SNPs.
The effect of rs2237982 on outcome was partially mediated through CE. Eight proxy-SNPs were in complete LD with rs2237982 (r2 =1.0) suggesting ~24.3% (9/37) of the identified SNPs would also be associated with both CE and outcome.
Significant tag/associated-proxy SNPs clustered upstream of intron 15, with predicted functional consequences related to splicing or transcriptional regulation. The interspersed exons (in regions of LD with tag and proxy SNPs) encode specific Sur1 regions (TMD0-L0, TMD1 and part of NBD1) that contain the sulfonylurea-receptor site and the Trpm4 binding-site/channel-pore.
ABCC8 polymorphisms, CE, and sTBI outcome
Sur1 is a key regulator of CE[8]. An exomic study in sTBI identified four ABCC8 SNPs (rs3819521, rs2283258, rs2283261, rs1799857) where variant-alleles increased risk of CE[3]. These led to the hypothesis that a targeted analysis of ABCC8 tag-SNPs would detect variants associated with both CE and outcome, and that CE would mediate this relationship. However, only variant-rs2237982 was associated with both increased measures of CE and unfavorable 3-month GOS. Rs2237982 was in complete LD (r2 =1) with eight proxy-SNPs and partial LD (r2 =0.97) with two proxy-SNPs (Supplementary Table 2), suggesting that these may similarly be associated with both CE and outcome. Statistically, 19.8% of rs2237982’s effect of on outcome was mediated by CE. Variant-rs7105832 was solely associated with increased CE measures, and variant-rs11024286 and rs4148622 were associated with improved neurological outcome but not CE. This suggests potential alternate neuroprotective mechanisms (Figure 1D) that have been posited by some preclinical studies[8,29].
Spatial location and potential significance of ABCC8 polymorphisms
Unlike glucose-metabolism disorders, where ~2/3rd of reported clinically significant SNPs are between introns 16–39 (adjacent to KCJN11 encoding Kir6.2), 4/4 (100%) of the tag-SNPs and 32/33 (97%) proxy-SNPs associated with CE and/or sTBI outcome occur between introns 2–15 (Figure 2). This is also true for the four previously identified SNPs associated with CE (Figure 1B)[3]. This portion of DNA is interspersed between sequences encoding critical regions of Sur1 encompassed by LD (Supplementary Figure 1, Figure 3): a) the sulfonylurea-receptor site which binds sulfonylureas/medications like glyburide and regulates channel opening and b) TMD0-L0 domain which juxtaposes the channel pore and thought to be the Sur1-Trpm4 binding site[8,20]. One of the proxy-SNPs identified in this study (rs2237981, r2 =1.0 with tag-SNP rs2237982) is implicated in predicting sulfonylurea responsiveness[27]. None of the significant tag-SNPs/correlated-proxy-SNPs in this study occurred in the ABCC8 region containing exons 25–39, ultimately destined for translation into TMD2/NBD2.
Given the different spatial proclivities of polymorphisms in glucose-metabolism disorders vs. sTBI, it is not surprising that only 5 of the 37 tag/proxy SNPs (Supplementary Table 2) have been previously reported. Few SNPs in similar locations (introns 3,4, 7,10,15) have been associated with aberrant splicing in hyperinsulinism[31,32]. The four significant tag-SNPs (rs7105832, rs2237982, rs11024286, rs4148622) are harbored in introns-3, 10 and 15. These introns are flanked by exons with residue overlap splice sites i.e. 1–2 nucleotides are encoded in the preceding exon and the remaining nucleotide(s) are in the following exon. A SNP in the connecting intron may therefore have important splicing, functional or regulatory consequences. Using F-SNP software and FS-score, the functional significance for all tag and 29/33 proxy-SNPs was predicted as ‘changed’, particularly with respect to transcriptional regulation[22]. FS-scores were between 0–0.208; most intronic SNPs (even those known to be disease causing) have an FS-score <0.5 with a median of 0.176 in neutral SNPs[22].
Although the predicted functional implications of the tag/proxy-SNPs predominantly relate to transcriptional function, the spatial locations described above also suggest a possible role involving the Sur1-Trpm4 interaction. These may not be mutually exclusive. Moreover, ABCC8 has >20 known transcripts (isoforms)[33]- thus, in addition to potential effects on activation/enhancement/repression, these polymorphisms may regulate the type of splice variant/transcript produced which, in turn, may affect the Sur1-Trpm4 interaction. The true functional implications of the tag/proxy-SNPs currently remain unestablished and need further exploration in biological models (in vitro and/or in vivo).
Limitations
The functional significance of the identified tag/proxy-SNPs is unknown, therefore potential biologic causality between ABCC8 genetic variation and CE/outcome in sTBI remains to be elucidated which was beyond the scope of this study. This was a single-center population prone to selection bias. Regional demographics resulted in a 90% Caucasian population. Due to quality control, we excluded eight tag-SNPs from the analysis, potentially underestimating the number of significant polymorphisms. A validated measure like ‘Therapeutic Intensity Level’ accounting for escalating treatments against CE that could influence ICP or CT-findings would have been useful, however was unavailable. While an important limitation, these maneuvers would reduce detected measures of CE and decrease the likelihood of false-positive results. We did not evaluate effects of TRPM4 polymorphisms on CE/outcome- our focus was on ABCC8 since Sur1 is the key regulatory protein of the channel contributing to CE. Exploring distinct neuroprotective pathways regulated by ABCC8/Sur1 also merits investigation of potentially relevant genes including TRPM4, KCJN11 (encoding Kir6.2), MMP-9, TNFA, and CASP-3.
Our report represents one of the largest polymorphisms studies in sTBI[2,3,34,35]. Nevertheless, our sample size is small for genetic studies. However, it is a candidate-gene targeted (not genome-wide) approach with lower sample-size requirements and reduced likelihood of false positive relationships after correcting for multiple comparisons and retaining statistical significance[14].
Only recently have exciting multi-center initiatives such as Transforming Research and Clinical Knowledge in TBI (TRACK-TBI) begun developing the required biomarker and tissue repositories necessary for future precision medicine and genetic analyses[36]. After 4 years with 11 enrolling sites in TRACK-TBI, currently there are approximately 294 sTBI patients enrolled in this multi-center consortium (with a significant overlap with this study’s cohort). To place our cohort in context, recent publications from TRACK TBI reporting important genetic contributions to TBI outcome have had between 93–220 subjects[34,37–39]. A European multi-center initiative (Genetic Associations in Neurotrauma) was unfortunately not funded but is crucial to endeavors researching the role of genetics in TBI pathophysiology and outcome. Pilot data such as ours may be important to developing and funding essential multi-center resources. Our results need validation/replication in such multi-center cohorts. Nonetheless, effect-sizes were large and robust to adjusting for multiple comparisons and confounding variables in multivariate-regression models. Moreover, there is a strong pathophysiologic basis for ABCC8 variability to influence Sur1 expression/regulation, the presence and degree of CE, response to targeted therapy, and outcome in sTBI.
CONCLUSIONS
Identification of polymorphisms in various genes characterizes treatment-responders in many fields of medicine, guiding precision medicine, and changing clinical practice[6,7]. Our results give credence to the hypothesis that ABCC8 polymorphisms (rs7105832, rs2237982, rs4148622, rs11024286 and proxy-SNPs) are important in the equation between genetic variability, CE and sTBI outcome. Spatial clustering of significant ABCC8 polymorphisms occurs around critical regions of DNA interspersed between sequences encoding the sulfonylurea receptor and a transmembrane-domain close to the channel-pore that binds Trpm4. Consequently, it is possible that these may differentially impact sulfonylurea (e.g. glyburide) sensitivity in individual patients. Although consistent with known pathophysiology, our findings need to be validated. To our knowledge, this is the first report exploring the association between tag-SNPs across the entire ABCC8 gene as they correlate with both CE and outcome. Our findings are relevant in the context of reports demonstrating Sur1-overexpression in human contusions, and recent clinical-trials suggesting that Sur1-inhibition may reduce CE in susceptible patients[10–12]. Understanding the impact of ABCC8 variability on Sur1 mediation of CE and outcome in sTBI warrants further investigation given its potential to guide patient risk-stratification, characterization of treatment responders, development of novel gene-based/targeted therapies, and patient-selection for future precision-medicine based clinical trials.
Supplementary Material
Acknowledgments
Dr. Ruchira M. Jha, MD MS discloses funding by the following NIH grants: T-32HL007820, NCATS KL2-TR000146, NCATS KL2-TR001856 and NINDS K23NS101036.
Dr. Theresa A. Koleck, PhD discloses NIH funding NINR F31NR014590 and T32NR007969.
Dr. Ava M. Puccio, PhD discloses funding by the following NIH grant: NINR R00 NR013176.
Dr. Jessica S. Wallisch, MD discloses funding by the following NIH grant NICHD T32-HD040686
Dr. Patrick M. Kochanek, MD discloses funding by the following NIH grant: 1R01NS087978-01 titled 2',3'-cAMP in Traumatic Brain Injury.
Dr. Yvette P. Conley, PhD discloses funding by the following NIH grant: NINR R01NR013342.
Footnotes
Statistical work contributors:
Ruchira M. Jha, MD MS, University of Pittsburgh
Theresa A. Koleck, PhD, University of Pittsburgh
Seo-Young Park, PhD, University of Pittsburgh
Author contributions:
Ruchira M. Jha was involved in study concept, design, data analysis and interpretation and manuscript generation. Theresa A. Koleck was involved with statistical analysis and haplotype generation and analysis. Ava M. Puccio, David O. Okonkwo, and Benjamin E. Zusman were involved in acquisition of data and patient samples. Seo-Young Park was involved in statistical analysis and review. Jessica Wallisch, Philip E. Empey, Lori A. Shutter and Robert S.B. Clark were involved in content expertise and critical revision of the manuscript. Patrick M. Kochanek was involved in study concept, content expertise, supervision and critical review of the manuscript. Yvette P. Conley was involved in study concept, design, data acquisition and interpretation, supervision and critical revision of the manuscript.
Author Financial Disclosures:
Dr. David O. Okonkwo, MD PhD reports no disclosures.
Dr. Seo-Young Park, PhD reports no disclosures.
Mr. Benjamin E. Zusman, BS reports no disclosures.
Dr. Robert S.B. Clark, MD reports no disclosures.
Dr. Lori A. Shutter, MD reports no disclosures.
Dr. Philip E. Empey, PharmD PhD reports no disclosures.
Bibliography
- 1.Weaver SM, Portelli JN, Chau A, et al. Genetic polymorphisms and traumatic brain injury: the contribution of individual differences to recovery. Brain Imaging Behav. 2014;8:420–34. doi: 10.1007/s11682-012-9197-9. [DOI] [PubMed] [Google Scholar]
- 2.Dardiotis E, Paterakis K, Tsivgoulis G, et al. AQP4 tag single nucleotide polymorphisms in patients with traumatic brain injury. J Neurotrauma. 2014;31:1920–6. doi: 10.1089/neu.2014.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jha RM, Puccio AM, Okonkwo DO, et al. ABCC8 Single Nucleotide Polymorphisms are Associated with Cerebral Edema in Severe TBI. Neurocrit Care. 2017;26:213–24. doi: 10.1007/s12028-016-0309-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah S, Kimberly WT. Today’s approach to treating brain swelling in the neuro intensive care unit. Semin Neurol. 2016;36:502–7. doi: 10.1055/s-0036-1592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marmarou A. A review of progress in understanding the pathophysiology and treatment of brain edema. Neurosurg Focus. 2007;22:E1. doi: 10.3171/foc.2007.22.5.2. [DOI] [PubMed] [Google Scholar]
- 6.SEARCH Collaborative Group. Link E, Parish S, et al. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;359:789–99. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 7.Shuldiner AR, O’Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–57. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simard JM, Woo SK, Schwartzbauer GT, et al. Sulfonylurea receptor 1 in central nervous system injury: a focused review. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2012;32:1699–1717. doi: 10.1038/jcbfm.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woo SK, Kwon MS, Ivanov A, et al. The sulfonylurea receptor 1 (Sur1)-transient receptor potential melastatin 4 (Trpm4) channel. J Biol Chem. 2013;288:3655–67. doi: 10.1074/jbc.M112.428219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheth KN, Elm JJ, Molyneaux BJ, et al. Safety and efficacy of intravenous glyburide on brain swelling after large hemispheric infarction (GAMES-RP): a randomised, double-blind, placebo-controlled phase 2 trial. Lancet Neurol. 2016;15:1160–9. doi: 10.1016/S1474-4422(16)30196-X. [DOI] [PubMed] [Google Scholar]
- 11.Kimberly WT, Battey TWK, Pham L, et al. Glyburide is associated with attenuated vasogenic edema in stroke patients. Neurocrit Care. 2014;20:193–201. doi: 10.1007/s12028-013-9917-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez-Valverde T, Vidal-Jorge M, Martínez-Saez E, et al. Sulfonylurea Receptor 1 in Humans with Post-Traumatic Brain Contusions. J Neurotrauma. 2015;32:1478–87. doi: 10.1089/neu.2014.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jha RM, Puccio AM, Chou SH-Y, et al. Sulfonylurea Receptor-1: A Novel Biomarker for Cerebral Edema in Severe Traumatic Brain Injury. Crit Care Med. 2017;45:e255–64. doi: 10.1097/CCM.0000000000002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–91. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 15.Miller MT, Pasquale M, Kurek S, et al. Initial Head Computed Tomographic Scan Characteristics Have a Linear Relationship with Initial Intracranial Pressure after Trauma. The Journal of Trauma: Injury, Infection, and Critical Care. 2004;56:967–73. doi: 10.1097/01.TA.0000123699.16465.8B. [DOI] [PubMed] [Google Scholar]
- 16.Eisenberg HM, Gary HE, Aldrich EF, et al. Initial CT findings in 753 patients with severe head injury. A report from the NIH Traumatic Coma Data Bank. J Neurosurg. 1990;73:688–98. doi: 10.3171/jns.1990.73.5.0688. [DOI] [PubMed] [Google Scholar]
- 17.Omasits U, Ahrens CH, Müller S, et al. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30:884–6. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- 18.Yates A, Akanni W, Amode MR, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–6. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mall T, Eckstein J, Norris D, et al. ProtAnnot: an App for Integrated Genome Browser to display how alternative splicing and transcription affect proteins. Bioinformatics. 2016;32:2499–501. doi: 10.1093/bioinformatics/btw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li N, Wu J-X, Ding D, et al. Structure of a Pancreatic ATP-Sensitive Potassium Channel. Cell. 2017;168:101–110. e10. doi: 10.1016/j.cell.2016.12.028. [DOI] [PubMed] [Google Scholar]
- 21.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 22.Lee PH, Shatkay H. An integrative scoring system for ranking SNPs by their potential deleterious effects. Bioinformatics. 2009;25:1048–55. doi: 10.1093/bioinformatics/btp103. [DOI] [PubMed] [Google Scholar]
- 23.Narum SR. Beyond Bonferroni: Less conservative analyses for conservation genetics. Conserv Genet. 2006;7:783–7. doi: 10.1007/s10592-005-9056-y. [DOI] [Google Scholar]
- 24.MacKinnon DP, Lockwood CM, Hoffman JM, et al. A comparison of methods to test mediation and other intervening variable effects. Psychol Methods. 2002;7:83–104. doi: 10.1037/1082-989X.7.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–7. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knüppel S, Rohde K, Meidtner K, et al. Evaluation of 41 candidate gene variants for obesity in the EPIC-Potsdam cohort by multi-locus stepwise regression. PLoS One. 2013;8:e68941. doi: 10.1371/journal.pone.0068941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Y, Mao G, Ren X, et al. Ser1369Ala variant in sulfonylurea receptor gene ABCC8 is associated with antidiabetic efficacy of gliclazide in Chinese type 2 diabetic patients. Diabetes Care. 2008;31:1939–44. doi: 10.2337/dc07-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jablonski KA, McAteer JB, de Bakker PIW, et al. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes. 2010;59:2672–81. doi: 10.2337/db10-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel AD, Gerzanich V, Geng Z, et al. Glibenclamide reduces hippocampal injury and preserves rapid spatial learning in a model of traumatic brain injury. J Neuropathol Exp Neurol. 2010;69:1177–90. doi: 10.1097/NEN.0b013e3181fbf6d6. [DOI] [PubMed] [Google Scholar]
- 30.Schwamm LH. Progesterone for traumatic brain injury–resisting the sirens’ song. N Engl J Med. 2014;371:2522–2523. doi: 10.1056/NEJMe1412951. [DOI] [PubMed] [Google Scholar]
- 31.Nestorowicz A, Glaser B, Wilson BA, et al. Genetic heterogeneity in familial hyperinsulinism. Hum Mol Genet. 1998;7:1119–28. doi: 10.1093/hmg/7.7.1119. [DOI] [PubMed] [Google Scholar]
- 32.Flanagan SE, Clauin S, Bellanné-Chantelot C, et al. Update of mutations in the genes encoding the pancreatic beta-cell K(ATP) channel subunits Kir6.2 (KCNJ11) and sulfonylurea receptor 1 (ABCC8) in diabetes mellitus and hyperinsulinism. Hum Mutat. 2009;30:170–80. doi: 10.1002/humu.20838. [DOI] [PubMed] [Google Scholar]
- 33.Zerbino DR, Achuthan P, Akanni W, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–61. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winkler EA, Yue JK, McAllister TW, et al. COMT Val 158 Met polymorphism is associated with nonverbal cognition following mild traumatic brain injury. Neurogenetics. 2016;17:31–41. doi: 10.1007/s10048-015-0467-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osier ND, Bales JW, Pugh B, et al. Variation in PPP3CC Genotype Is Associated with Long-Term Recovery after Severe Brain Injury. J Neurotrauma. 2017;34:86–96. doi: 10.1089/neu.2015.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yue JK, Vassar MJ, Lingsma HF, et al. Transforming research and clinical knowledge in traumatic brain injury pilot: multicenter implementation of the common data elements for traumatic brain injury. J Neurotrauma. 2013;30:1831–44. doi: 10.1089/neu.2013.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yue JK, Pronger AM, Ferguson AR, et al. Association of a common genetic variant within ANKK1 with six-month cognitive performance after traumatic brain injury. Neurogenetics. 2015;16:169–80. doi: 10.1007/s10048-015-0437-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winkler EA, Yue JK, Ferguson AR, et al. COMT Val158Met polymorphism is associated with posttraumatic stress disorder and functional outcome following mild traumatic brain injury. J Clin Neurosci. 2017;35:109–16. doi: 10.1016/j.jocn.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yue JK, Winkler EA, Rick JW, et al. DRD2 C957T polymorphism is associated with improved 6-month verbal learning following traumatic brain injury. Neurogenetics. 2017;18:29–38. doi: 10.1007/s10048-016-0500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.