Abstract
Patients with active lupus have altered T cells characterized by low DNA methyltransferase levels. We hypothesized that low DNA methyltransferase levels synergize with low methionine levels to cause greater overexpression of genes normally suppressed by DNA methylation. CD4+ T cells from lupus patients and controls were stimulated with PHA then cultured in custom media with normal or low methionine levels. Oxidative stress was induced by treating the normal CD4+ T cells with peroxynitrite prior to culture. Methylation sensitive gene expression was measured by flow cytometry. Results showed low methionine levels caused greater overexpression of methylation sensitive genes in peroxynitrite treated T cells relative to untreated T cells, and in T cells from lupus patients relative to T cells from healthy controls. In conclusion, low dietary transmethylation micronutrient levels and low DNA methyltransferase levels caused either by oxidative stress or lupus, have additive effects on methylation sensitive T cell gene expression.
Keywords: lupus, diet, oxidative stress, T-lymphocytes
Introduction
Systemic lupus erythematosus (SLE) is an incompletely understood autoimmune disease that causes significant morbidity and mortality in those affected. While the etiology of lupus remains unknown, genetic and environmental factors play critical roles. Familial clustering as well as the association of numerous single nucleotide polymorphisms (SNPs) with SLE indicate a genetic requirement for the development of this disease [1]. However, an environmental component is also required, as evidenced by the incomplete concordance of lupus in monozygotic twins [2] and the chronic relapsing course of the disease. Exogenous agents associated with lupus onset and flares include certain drugs [3] as well as environmental agents that cause oxidative stress such as ultraviolet (UV) light [4], cigarette smoke [5], silica [6], and infections [7]. Lupus flares are characterized by continuing oxidative stress, caused by superoxide (O2-) combining with nitric oxide (NO) to form peroxynitrite (ONOO-) which nitrates tyrosines in serum proteins during the flares [8]. However, it is unclear how oxidizing agents alter the immune system to cause the flares in genetically predisposed individuals.
DNA methylation is an epigenetic mechanism that regulates gene expression. The drugs procainamide and hydralazine, as well as UV light, cause lupus-like autoimmunity in mice and likely humans at least in part by inhibiting T cell DNA methylation [3, 4]. Previous work demonstrated that treating CD4+ T cells with DNA methylation inhibitors such as 5-azacytidine causes hypomethylation and overexpression of immune genes including CD11a, CD70, KIR and perforin at the mRNA and protein levels [9], and that similar epigenetically altered CD4+ T cells are sufficient to cause lupus-like autoimmunity in animal models [3]. Further, the same genes are also hypomethylated and overexpressed by CD4+ T cells from patients with active lupus, again at the mRNA and protein levels [10-13].
The mechanisms causing T cell DNA demethylation in lupus patients are unclear. The replication of DNA methylation patterns during mitosis requires S-adenosylmethionine (SAM), the methyl donor, and DNA methyltransferase 1 (Dnmt1), the enzyme that transfers the methyl group from SAM to dC bases in DNA CpG pairs. Therefore, environmental factors such as diet, which provides the methyl donors, and agents that decrease Dnmt1 enzymatic activity may synergize to demethylate DNA in dividing T cells, causing overexpression of the methylation-sensitive immune genes that contribute to lupus flares [14]. T cell Dnmt1 levels are regulated in part by the extracellular receptor kinase (ERK) pathway [15], and we have reported that methylation sensitive T cell genes are overexpressed in transgenic C57BL/6 × SJL mice with an inducible T cell ERK pathway signaling defect. Inhibiting T cell ERK pathway signaling in these mice is sufficient to cause lupus-like autoimmunity. Further, the disease severity can be modulated in these mice by varying dietary transmethylation micronutrients in their chow when Dnmt1 levels are low, but not when the levels are normal [15].
CD4+ T cells from patients with active lupus also have impaired ERK pathway signaling. This causes a decrease in Dnmt1 levels during mitosis that contributes to T cell DNA hypomethylation in these patients [16]. This signaling defect is caused by nitration of protein kinase C delta (PKCδ) [17], an upstream regulator of ERK pathway signaling [18], and the nitration is caused by environmental agents that stimulate inflammation such as UV light, cigarette smoke, silica, and infections. The PKCδ inactivation prevents Dnmt1 upregulation to copy DNA methylation patterns when T cells divide [18]. Lupus patients with active disease also have other markers of ongoing oxidative stress including elevated levels of serum protein carbonyls [19] and urinary F2-isoprostane [20] as well as elevated 3-nitrotyrosine levels [21].
Since oxidative stress is increased [21] and T cell Dnmt1 levels are decreased in lupus patients [16], and low dietary transmethylation micronutrient levels also decrease lymphocyte DNA methylation [22], we hypothesized that the expression of methylation-dependent genes by T cells from patients with active lupus may be more sensitive to low micronutrient levels than normal T cells, causing demethylation and overexpression of the methylation-sensitive genes that contribute to disease flares. We therefore cultured CD4+ T cells from lupus patients and healthy controls in custom media containing a range of folate or methionine levels to test if CD4+ lupus T cells are more sensitive to low transmethylation micronutrient levels than CD4+ T cells from healthy controls. We also tested if gene expression by normal CD4+ T cells exposed to oxidative stress in vitro becomes more sensitive to low transmethylation micronutrient levels than untreated cells.
Materials and Methods
Subjects
Healthy subjects were recruited by advertising. Lupus patients (35 women, 2 men, average age 40, range 21-79 years) with active and inactive disease were recruited from the University of Michigan outpatient rheumatology clinic. Thirty of the patients were Caucasian, six African American and one was Hispanic. Lupus patients met the American College of Rheumatology's revised criteria for SLE [23]. Disease activity was assessed using the systemic lupus erythematosus disease activity index (SLEDAI) [24]. The mean activity was 3, ranging from 0-12. Patients receiving cyclophosphamide or methotrexate were excluded because of effects on transmethylation reactions. Healthy controls (n=6, average age 37 range 21-53 years) were recruited by advertising. Transmethylation micronutrient levels were measured in the University of Michigan MLabs and Department of Pathology Specimen Processing Laboratories, and Ralston Analytical Laboratories. The University of Michigan Institutional Review Board for Human Subject Research reviewed and approved this HUM00008181 study.
Chemicals and reagents
RPMI 1640, RPMI 1640 without folate and RPMI 1640 without methionine, as well as purified folate and methionine, were purchased from Invitrogen Life Technologies. Custom media, containing normal serum levels of the transmethylation micronutrients, was created using RPMI 1640 without vitamin B6, methionine, folate, vitamin B12, riboflavin, and choline, all purchased from Invitrogen Life Technologies. Zinc was also obtained from Invitrogen. Homocysteine was purchased from Sigma. Fetal bovine serum was dialyzed against Hank's Balanced Salt Solution (HBSS) using SpectraPor dialysis tubing with a molecular weight cutoff of 2,000 to remove micronutrients, then filter sterilized. Ficoll-Paque Plus was purchased from GE Healthcare; phytohemagglutinin was purchased from Remel; recombinant human IL-2 was purchased from Peprotech; and peroxynitrite was purchased from Calbiochem. Antibodies were obtained from BD Pharmingen, Beckman Coulter and R & D Systems. Cytofix/Cytoperm Kits were purchased from BD Biosciences. Standardization particles were purchased from Bangs Laboratories. Magnetic beads were from Miltenyi Biotech, the SV Total RNA Isolation System was from Promega Corporation, and the QuantiTect SYBR Green RT-PCR kit was purchased from Qiagen.
T cell isolation and culture
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by density gradient centrifugation on Ficoll-Paque, cultured in standard RPMI 1640/10% fetal bovine serum and stimulated with 1 μg/ml phytohemagglutinin (PHA) for 24 h. CD4+ T cells were then separated by positive selection using magnetic beads (Miltenyi Biotec), washed, and cultured for 72 h with 10 ng/ml IL-2 in either RPMI 1640/10% fetal bovine serum (FBS) or custom RPMI 1640 without vitamin B6, methionine (Met), folate, vitamin B12, riboflavin, and choline as the base. Vitamin B6, Met, folate, vitamin B12, riboflavin, and choline were added back at concentrations adjusted to physiologic levels as shown in Table 1. Homocysteine and zinc were also added at physiologic concentrations (Table 1). As described above, the fetal bovine serum for the custom media was dialyzed against Hank's Balanced Salt Solution (HBSS) using SpectraPor dialysis tubing with a molecular weight cutoff of 2,000 to remove additional transmethylation micronutrients, then filter sterilized. Where indicated the stimulated cells were treated with 20 or 25 μM ONOO- prior to culture.
Table 1. Media transmethylation micronutrient concentrations.
| Component | RPMI 1640 | Normal Serum Levels | Custom Media |
|---|---|---|---|
| Methionine (μM) | 101 | 10-42 | 33 |
| Choline (μM) | 21.4 | 7-20 | 10 |
| Folic acid (ng/ml) | 1000 | 3-17.5 | 7 |
| B6 (μg/L) | 1000 | 5-50 | 22 |
| B2 (μg/L) | 200 | 3-15 | 6 |
| B12 (pg/ml) | 5000 | 211-911 | 350 |
| Zn (μg/ml) | 0 | 0.55-1.5 | 0.5 |
| Homocysteine (μM) | 0 | 5-15 | 9 |
Flow cytometry
Purified CD4+ T cells were stained with fluorochrome-conjugated antibodies to human CD11a and CD70. KIR expression was measured using a cocktail of antibodies to KIR proteins including CD158b1/b2, j; CD158i; NKB1; CD158b; CD158a, h; and KIR2DL4/CD158d. Intracellular staining was performed using anti-human perforin antibodies and the Cytofix/Cytoperm Kit (BD Biosciences). The cells were then fixed and analyzed on a FACSCalibur flow cytometer (BD Biosciences) 24 hours later to allow for cell equilibration after contraction due to fixing. Isotype and single-color positive controls were included in each experiment. Standardization particles for fluorescein isothiocyanate (FITC), phytoerythrin (PE) and PE-Cy5 were run during each experiment for instrument calibration.
Statistical Analysis
To compare micronutrient levels in serum from lupus patients to normal population values, the means of the lupus patients' levels were compared to the midpoint of the normal range, using a one-sample t-test. For non-normally distributed variables, the median of the micronutrient value was compared to the midpoint of the normal range using a non-parametric Wilcoxon sign rank test. The percentage of patients who were out of the normal range (either above or below the normal range) was assessed using a one-sample binomial proportion, and a 95% confidence interval was computed.
Results
Initial studies measured transmethylation micronutrient levels in the serum of 28-35 patients with mildly active lupus (mean SLEDAI 3). Table 2 compares the mean levels of methionine, choline, folate, zinc, homocysteine and vitamins B6, B2, and B12 in serum from the lupus patients relative to population means. Overall, serum zinc, methionine and vitamin B6 levels were significantly (p<0.05) lower in the lupus patients relative to the population controls while homocysteine was increased as reported by others [25]. The number of SLE patients examined with highly active lupus was too small to draw any correlations with disease activity.
Table 2. Transmethylation micronutrient levels in lupus patients.
| Nutrient | Reference (Mean/Range) | Lupus (Mean/Range | p-value |
|---|---|---|---|
| Zinc (μg/ml) | 0.86 (0.55-1.50) | 0.71 (0.5-0.9 | 0.0001 |
| B6 (μg/L) | 22.5 (5-50) | 16.48 (2-57) | 0.021 |
| Homocysteine (μM) | 9.5 (5-15) | 11.37 (6-25) | 0.013 |
| Methionine (μM) | 26 (10-40) | 23.09 (13-47) | 0.012 |
| Folate (ng/ml) | 7.1 (1.5-22) | 18.27 (6.4->24) | N.S. |
| B12 (pg/ml) | 500 (211-911) | 569.8 (220-1770_ | N.S. |
| B2 (μg/L) | 6.0 (3-5) | 9.0 (1-78) | N.S. |
| Choline (ppm) | 225.4 (194-261) | 225 (201-260) | N.S. |
We then used custom media to test the effects of low transmethylation micronutrient concentrations on CD70, KIR and perforin expression by PHA stimulated CD4+ T cells from healthy subjects. Normal RPMI 1640 tissue culture media is enriched for transmethylation micronutrients. We therefore used custom media in which the transmethylation micronutrient concentrations were adjusted to normal human serum levels (Table 1). PBMCs from 9 healthy subjects were stimulated with PHA and cultured in the custom media described above, then CD70, KIR and perforin expression were measured by flow cytometry. Figure 1 shows that the custom media caused significant increases in KIR and perforin protein expression in CD4+ T cells, and significant increases in CD70, KIR and perforin protein in CD8+ T cells. The increases in CD70 and perforin expression were confirmed at the protein level in both CD4+ and CD8+ T cells.
Figure 1. Low transmethlyation micronutrient levels increase CD70, KIR and perforin expression by CD4+ and CD8+ T cells from healthy subjects.
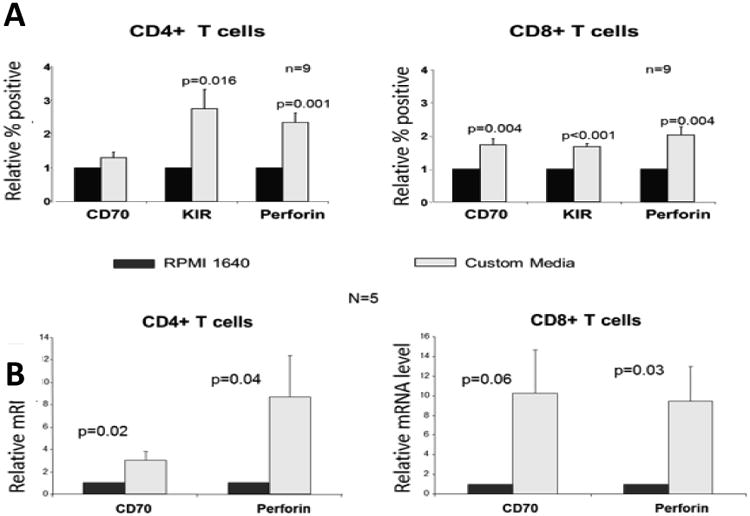
(A) PBMCs were isolated from 9 healthy donors, stimulated with PHA and cultured in either standard RPMI 1640 or custom media with low transmethylation micronutrient levels as described in “Materials and Methods”. The cells were then stained with fluorochrome conjugated antibodies to CD4 or CD8 and analyzed by flow cytometry. (B) PBMC from 5 healthy subjects were similarly stimulated with PHA, cultured in either complete RPMI 1640 or the custom media then separated into CD4+ and CD8+ subsets as in panel A. Total RNA was then extracted and analyzed by real-time RT-PCR. Results are shown relative to expression by T cells cultured in the complete RPMI 1640. Error bars represent SEM.
We next investigated whether gene expression by T cells from lupus patients was more sensitive to transmethylation micronutrient levels than T cells from healthy individuals. PBMCs from 9 female lupus patients (average age: 44.1 yrs; average SLEDAI: 6) and 9 age- and sex-matched controls (average age: 41.4 yrs.) were stimulated with PHA and cultured in standard RPMI 1640, which contains 101μM methionine or custom RPMI 1640 containing normal physiologic (30μM), or low (5μM) concentrations of methionine. The cells were then stained with antibodies to CD4 and CD70, KIR or perforin, and then analyzed by flow cytometry.
Decreasing media methionine levels did not significantly affect C70 expression by CD4+ T cells from the controls (Fig. 2A). However, decreasing media methionine levels caused a significant increase in CD70 expression by CD4+ T cells from the lupus patients (P=0.026). The overall increase in CD70 expression was also greater in T cells from the lupus patients relative to controls (P=0.002 by ANOVA). Figure 2B shows that low methionine levels also increased KIR expression on CD4+ T cells from both the lupus patients and controls, but KIR expression by CD4+ T cells from the lupus patients was significantly greater than by T cells from controls (p=0.032). Figure 2C similarly shows the effects of methionine restriction on perforin expression by PHA stimulated CD4+ T cells from healthy controls and lupus patients. Perforin was also overexpressed by significantly greater numbers of T cells from the lupus patients than the controls when methionine levels were low (p=0.001).
Figure 2. Low methionine levels cause overexpression of CD70, KIR and perforin by CD4+ T cells from lupus patients.
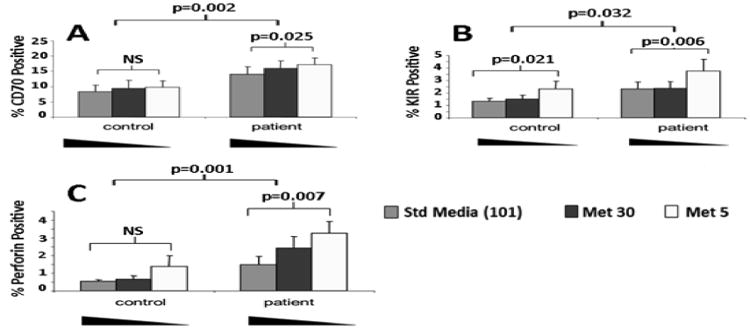
PBMCs from 9 female lupus patients (average age: 44.1 yrs.; average SLEDAI: 6) and 9 age- and sex-matched controls (average age: 41.4) were stimulated with PHA and cultured in RPMI 1640 with 101 μM, 30μM, or 5μM methionine. The cells were then stained with antibodies to CD4 and (A) CD70, (B) KIR or (C) perforin and analyzed by flow cytometry. The significance of the differences in gene expression both within and between subject groups was determined by ANOVA Error bars represent SEM.
The interactions between oxidative stress and transmethylation micronutrient deficiencies were then tested on methylation-sensitive T cell gene expression. As noted above, environmental agents that cause oxidative stress, such as UV light and infections, can trigger lupus flares, and treating dividing CD4+ T cells with oxidizing agents demethylates the DNA [26]. The reactive oxygen species inactivate PKCδ, preventing Dnmt1 upregulation during mitosis, inhibiting methylation of DNA in the daughter cells and making them autoreactive [26], and the oxidized, autoreactive cells are sufficient to cause lupus-like autoimmunity in mouse models [27]. This process suggests a mechanism by which oxidative stress could trigger flares in lupus patients. Since low Dnmt1 levels increase the sensitivity of T cell gene expression to low transmethylation micronutrient levels [28], we hypothesized that normal CD4+ T cells exposed to oxidizers such as ONOO- would overexpress methylation sensitive genes as reported [26], and that gene expression would further increase when the oxidized cells are cultured in media with low transmethylation micronutrient levels. To test this, PBMCs from healthy controls were stimulated with PHA, treated with 0, 20 or 25 μM ONOO- then similarly cultured in media containing 101 μM, 30 μM or 15 μM methionine (Met). Effects on CD70 and KIR expression were again measured by flow cytometry. Figures 3a and 3b show that low Met levels synergize with increased ONOO- concentrations to further increase CD70 and KIR expression. The effects of low folate levels on T cell gene expression were similarly tested by stimulating PBMC with PHA, treating with 0, 20 or 25 μM ONOO-, then culturing the cells in media containing 2270 nM, 40 nM or 10 nM folate. Figures 4a and 4b show that low folate levels also synergize with increased exposure to ONOO- mediated oxidative stress to increase CD70 and KIR expression.
Figure 3. Peroxynitrite and low methionine concentrations synergize to increase CD70 and KIR expression.
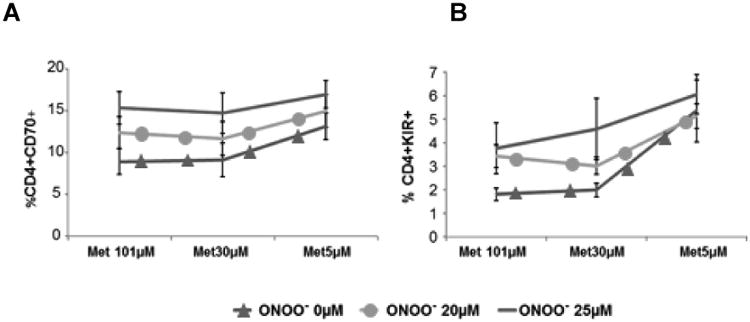
Peroxynitrite-treated CD4+ T cells were cultured in media containing low methionine concentrations then effects on CD70 and KIR expression were measured by flow cytometry. (A) CD70 (p<0.001 for overall Met effect; p<0.001 for overall peroxynitrite effect; n=6) and (B) KIR (p<0.001 for overall folate effect; p<0.01 for overall peroxynitrite effect; n=5). Repeated measures ANOVA was used to analyze relationship between methionine and peroxynitrite concentrations. Methionine and peroxynitrite concentration plus their interaction was included as the within-subjects effects. Post-hoc tests with Bonferroni correction for multiple comparisons were used to examine specific effects at each given concentration. Error bars represent SEM.
Figure 4. Peroxynitrite and low folate levels synergize to increase CD4+ T cell CD70 and KIR expression.
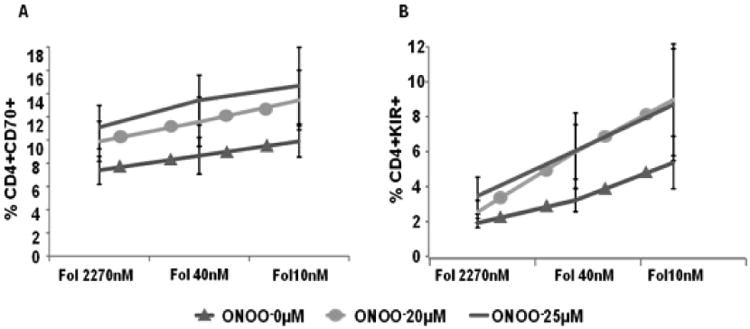
Peroxynitrite-treated CD4+ T cells were cultured in media containing low folate concentrations then expression of (A) CD70 (p<0.001 for overall folate effect; p<0.001 for overall peroxynitrite effect) or (B) KIR (p<0.001 for overall folate effect; p<0.05 for overall peroxynitrite effect) were measured by flow cytometry. n=6. Repeated measures ANOVA was used to analyze relationship between folate and peroxynitrite concentrations. Folate and peroxynitrite concentration plus their interaction was included as the within-subjects effects. Post-hoc tests with Bonferroni correction for multiple comparisons were used to examine specific effects at each given concentration. Error bars represent SEM.
Together these results indicate that oxidative stress and low transmethylation micronutrient levels can synergize to increase expression of methylation sensitive T cell genes in vitro when Dnmt1 levels are low, and likely in lupus patients when their T cell Dnmt1 levels are low.
Discussion
Environmental agents that cause inflammation such as sun exposure, infections, smoking, and silica are associated with lupus onset and flares, but how these agents trigger the flares is unclear. Lupus-inducing drugs such as hydralazine and procainamide, as well as reactive oxygen species such as O2- and ONOO-, generated by inflammation, inhibit CD4+ T cell DNA methylation by decreasing Dnmt1 enzymatic activity levels thereby increasing methylation sensitive gene expression [26], and T cells epigenetically altered with hydralazine, procainamide, H2O2 or ONOO- are sufficient to cause lupus-like autoimmunity in murine models [27]. Similar epigenetically altered T cells with low Dnmt1 levels and increased methylation sensitive gene expression are found in patients with active lupus, which is characterized by increased serum protein nitration [8], suggesting that the altered cells may contribute to the flares. However, the mechanism by which inflammation decreases Dnmt1 levels to inhibit DNA methylation in lupus T cells has been unclear.
Dnmt1 levels increase as T cells enter mitosis by signals transmitted through the ERK and JNK pathways [29]. Dnmt1 then binds the replication fork and “reads” CG pairs. Where the parent strand is methylated, Dnmt1 catalyzes transfer of the methyl group from S-adenosylmethionine (SAM) to the corresponding dC in the daughter strand, thereby replicating methylation patterns in the daughter cell. Previous studies have shown that impaired ERK pathway signaling decreases Dnmt1 levels in stimulated T cells from patients with active lupus, causing DNA hypomethylation in the daughter cells [30]. The signaling defect was traced to impaired PKCδ signaling function due to nitration, caused by ONOO- generated during inflammatory responses [17]. However, as noted above, the transmethylation reaction is also dependent on SAM levels, and SAM levels depend on methionine, an essential amino acid derived from the diet [31]. This interaction between dietary transmethylation micronutrient levels, Dnmt1 levels and autoimmunity was confirmed in vivo by restricting dietary methyl donors in lupus-prone transgenic mice with an inducible T cell ERK pathway signaling defect. The dietary restriction caused only a mild lupus-like disease when signaling was intact and Dnmt1 levels were normal, and more severe disease when ERK pathway signaling was disrupted, decreasing Dnmt1 levels [28].
The present study extends these reports using in vitro models to test if the expression of methylation sensitive genes by CD4+ T cells from lupus patients is more sensitive to dietary transmethylation micronutrient restriction than CD4+ T cells from healthy controls. Standard cell culture media often contains supraphysiologic levels of transmethylation micronutrients thereby masking epigenetic effects. Culturing PHA stimulated CD4+ T cells from healthy subjects and lupus patients in custom media in which methionine levels could be varied demonstrated that low methionine levels increased methylation sensitive gene expression in the lupus T cells relative to the T cells from healthy controls, consistent with the hypothesis. It should be noted that the KIR genes and perforin are normally silenced in CD4+ T cells by only DNA methylation. Inhibiting DNA methylation in CD4+ T cells activates expression of perforin as well as the entire the KIR gene family through demethylation of regulatory elements. Transfection of methylated and unmethylated KIR promoter constructs into normal T cells confirmed that promoter demethylation was necessary and sufficient to activate KIR gene expression [12]. Perforin expression is similarly suppressed primarily by DNA methylation in normal CD4+ T cells, and activated by DNA methylation inhibition [13].
Together, these studies indicate that low dietary transmethylation micronutrient levels can synergize with low T cell Dnmt1 levels, caused by oxidative stress, to increase methylation sensitive gene expression. These studies thus suggest that oxidative stress and a methyl-poor diet could combine to increase lupus flare severity in human lupus patients. The anti-oxidant N-acetylcysteine has already been shown to be beneficial in lupus [32], and dietary transmethylation micronutrient supplementation may also be beneficial.
Acknowledgments
The authors thank Ms. Stacy Fry for her expert administrative assistance.
This work was supported by a Merit grant from the Department of Veteran's Affairs and the Lupus Insight Prize from the Lupus Foundation of America, the Alliance for Lupus Research and the Lupus Research Institute. Donna Ray was supported by the National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases grant number T32AR007080.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ghodke-Puranik Y, Niewold TB. Immunogenetics of systemic lupus erythematosus: A comprehensive review. J Autoimmun. 2015;64:125–136. doi: 10.1016/j.jaut.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Block SR. A brief history of twins. Lupus. 2006;15:61–64. doi: 10.1191/0961203306lu2263ed. [DOI] [PubMed] [Google Scholar]
- 3.Yung R, Richardson B. Drug-Induced Lupus Mechanisms. In: Lahita R, Tsokos GC, Buyon JP, Takao K, editors. Systemic Lupus Erythematosus. Elsevier; 2011. pp. 385–404. [Google Scholar]
- 4.Lehmann P, Holzle E, Kind P, Goerz G, Plewig G. Experimental reproduction of skin lesions in lupus erythematosus by UVA and UVB radiation. J Am Acad Dermatol. 1990;22:181–187. doi: 10.1016/0190-9622(90)70020-i. [DOI] [PubMed] [Google Scholar]
- 5.Harel-Meir M, Sherer Y, Shoenfeld Y. Tobacco smoking and autoimmune rheumatic diseases. Nat Clin Pract Rheumatol. 2007;3:707–715. doi: 10.1038/ncprheum0655. [DOI] [PubMed] [Google Scholar]
- 6.Parks CG, Cooper GS, Nylander-French LA, Sanderson WT, Dement JM, Cohen PL, Dooley MA, Treadwell EL, St Clair EW, Gilkeson GS, Hoppin JA, Savitz DA. Occupational exposure to crystalline silica and risk of systemic lupus erythematosus: a population-based, case-control study in the southeastern United States. Arthritis Rheum. 2002;46:1840–1850. doi: 10.1002/art.10368. [DOI] [PubMed] [Google Scholar]
- 7.Cooper GS, Gilbert KM, Greidinger EL, James JA, Pfau JC, Reinlib L, Richardson BC, Rose NR. Recent advances and opportunities in research on lupus: environmental influences and mechanisms of disease. Environ Health Perspect. 2008;116:695–702. doi: 10.1289/ehp.11092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahsan H. 3-Nitrotyrosine: A biomarker of nitrogen free radical species modified proteins in systemic autoimmunogenic conditions. Hum Immunol. 2013;74:1392–1399. doi: 10.1016/j.humimm.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 9.Richardson B. Primer: epigenetics of autoimmunity. Nat Clin Pract Rheumatol. 2007;3:521–527. doi: 10.1038/ncprheum0573. [DOI] [PubMed] [Google Scholar]
- 10.Oelke K, Lu Q, Richardson D, Wu A, Deng C, Hanash S, Richardson B. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 2004;50:1850–1860. doi: 10.1002/art.20255. [DOI] [PubMed] [Google Scholar]
- 11.Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J Immunol. 2005;174:6212–6219. doi: 10.4049/jimmunol.174.10.6212. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Kuick R, Hanash S, Richardson B. DNA methylation inhibition increases T cell KIR expression through effects on both promoter methylation and transcription factors. Clin Immunol. 2009;130:213–224. doi: 10.1016/j.clim.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Q, Wu A, Ray D, Deng C, Attwood J, Hanash S, Pipkin M, Lichtenheld M, Richardson B. DNA methylation and chromatin structure regulate T cell perforin gene expression. J Immunol. 2003;170:5124–5132. doi: 10.4049/jimmunol.170.10.5124. [DOI] [PubMed] [Google Scholar]
- 14.Ray D, Richardson B. Toxicoepigenomics in lupus. In: Sahu S, editor. Toxicology and Epigenetics. John Wiley & Sons, Ltd; 2012. [Google Scholar]
- 15.Sawalha AH, Jeffries M, Webb R, Lu Q, Gorelik G, Ray D, Osban J, Knowlton N, Johnson K, Richardson B. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9:368–378. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng C, Kaplan MJ, Yang J, Ray D, Zhang Z, McCune WJ, Hanash SM, Richardson BC. Decreased Ras-mitogen-activated protein kinase signaling may cause DNA hypomethylation in T lymphocytes from lupus patients. Arthritis Rheum. 2001;44:397–407. doi: 10.1002/1529-0131(200102)44:2<397::AID-ANR59>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 17.Gorelik G, Fang JY, Wu A, Sawalha AH, Richardson B. Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J Immunol. 2007;179:5553–5563. doi: 10.4049/jimmunol.179.8.5553. [DOI] [PubMed] [Google Scholar]
- 18.Gorelik GJ, Yarlagadda S, Richardson BC. Protein kinase Cdelta oxidation contributes to ERK inactivation in lupus T cells. Arthritis and rheumatism. 2012;64:2964–2974. doi: 10.1002/art.34503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan PE, Sturgess AD, Davies MJ. Increased levels of serum protein oxidation and correlation with disease activity in systemic lupus erythematosus. Arthritis and rheumatism. 2005;52:2069–2079. doi: 10.1002/art.21130. [DOI] [PubMed] [Google Scholar]
- 20.Avalos I, Chung CP, Oeser A, Milne GL, Morrow JD, Gebretsadik T, Shintani A, Yu C, Stein CM. Oxidative stress in systemic lupus erythematosus: relationship to disease activity and symptoms. Lupus. 2007;16:195–200. doi: 10.1177/0961203306075802. [DOI] [PubMed] [Google Scholar]
- 21.Wang G, Pierangeli SS, Papalardo E, Ansari GA, Khan MF. Markers of oxidative and nitrosative stress in systemic lupus erythematosus: correlation with disease activity. Arthritis and rheumatism. 2010;62:2064–2072. doi: 10.1002/art.27442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacob RA, Gretz DM, Taylor PC, James SJ, Pogribny IP, Miller BJ, Henning SM, Swendseid ME. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr. 1998;128:1204–1212. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- 23.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 24.Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum. 1992;35:630–640. doi: 10.1002/art.1780350606. [DOI] [PubMed] [Google Scholar]
- 25.Haque S, Bruce IN. Therapy insight: systemic lupus erythematosus as a risk factor for cardiovascular disease. Nature clinical practice Cardiovascular medicine. 2005;2:423–430. doi: 10.1038/ncpcardio0270. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Gorelik G, Strickland FM, Richardson BC. Oxidative stress, T cell DNA methylation, and lupus. Arthritis & rheumatology. 2014;66:1574–1582. doi: 10.1002/art.38427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strickland FM, Li Y, Johnson K, Sun Z, Richardson BC. CD4(+) T cells epigenetically modified by oxidative stress cause lupus-like autoimmunity in mice. J Autoimmun. 2015;62:75–80. doi: 10.1016/j.jaut.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strickland FM, Hewagama A, Wu A, Sawalha AH, Delaney C, Hoeltzel MF, Yung R, Johnson K, Mickelson B, Richardson BC. Diet influences expression of autoimmune-associated genes and disease severity by epigenetic mechanisms in a transgenic mouse model of lupus. Arthritis Rheum. 2013;65:1872–1881. doi: 10.1002/art.37967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Gorelik GJ, Strickland FM, Richardson BC. Decreased ERK and JNK signaling contribute to gene overexpression in “senescent” CD4+CD28- T cells through epigenetic mechanisms. J Leukoc Biol. 2010;87:137–145. doi: 10.1189/jlb.0809562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng C, Lu Q, Zhang Z, Rao T, Attwood J, Yung R, Richardson B. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–756. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- 31.Ables GP, Hens JR, Nichenametla SN. Methionine restriction beyond life-span extension. Ann N Y Acad Sci. 2016;1363:68–79. doi: 10.1111/nyas.13014. [DOI] [PubMed] [Google Scholar]
- 32.Lai ZW, Hanczko R, Bonilla E, Caza TN, Clair B, Bartos A, Miklossy G, Jimah J, Doherty E, Tily H, Francis L, Garcia R, Dawood M, Yu J, Ramos I, Coman I, Faraone SV, Phillips PE, Perl A. N-acetylcysteine reduces disease activity by blocking mammalian target of rapamycin in T cells from systemic lupus erythematosus patients: a randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 2012;64:2937–2946. doi: 10.1002/art.34502. [DOI] [PMC free article] [PubMed] [Google Scholar]


