Abstract
The global temperatures are increasing. This increase is partly due to methane (CH4) production from ruminants, including dairy cattle. Recent studies on dairy cattle have revealed the existence of a heritable variation in CH4 production that enables mitigation strategies based on selective breeding. We have exploited the available heritable variation to study the genetic architecture of CH4 production and detected genomic regions affecting CH4 production. Although the detected regions explained only a small proportion of the heritable variance, we showed that potential QTL regions affecting CH4 production were located within QTLs related to feed efficiency, milk-related traits, body size and health status. Five candidate genes were found: CYP51A1 on BTA 4, PPP1R16B on BTA 13, and NTHL1, TSC2, and PKD1 on BTA 25. These candidate genes were involved in a number of metabolic processes that are possibly related to CH4 production. One of the most promising candidate genes (PKD1) was related to the development of the digestive tract. The results indicate that CH4 production is a highly polygenic trait.
Introduction
The increase in the global temperature has a serious impact on the environment and humans. Some of these consequences may exceed adaptive capacities of some species, lead to water supplies shortage, melt the glaciers and increase sea level as well as trigger extreme climatic events1. The estimated global temperature increase in 2010 due to greenhouse gas (GHG) and aerosol emissions was 0.81 °C in relation to the pre-industrial era. The 0.11 °C of this increase was contributed by methane (CH4) emissions from direct livestock emissions2. Most of livestock CH4 emissions are caused by ruminants3,4. The CH4 emissions from ruminants are mostly due to enteric fermentation.
In ruminants the enteric fermentation is a consequence of a normal digestive process. One of their stomachs, the rumen, is inhabited by rumen microorganisms, enabling digestion of feed that contains high amounts of fiber. One of the by-products of this digestive process converting the feed provided to the ruminants by the microorganisms is CH4.
The CH4 consists of carbon. When carbon is lost from the body it may no longer be used by the animal as a source of energy. Therefore, apart from its environmental impact, CH4 emission in ruminants has also a potential negative impact on the profitability of animal production5,6. Due to those potential consequences of CH4 emissions from ruminants, mitigation strategies are under investigations. Optional strategies range from adjusting the management to nutritional treatments7. These strategies may have a high impact on CH4 production. An additional strategy of mitigating CH4 production might involve selective breeding for lower emitters. Such a strategy could be possible in case of the existence of genetic variation in CH4 production and a favorable genetic association of CH4 and traits in the current breeding goals.
Recent genetic studies on dairy cattle revealed that while most of the variation in CH4 production is due to non-genetic factors (i.e. feed, management and other environmental factors), the genetic component (i.e. genetic variance) in CH4 production also exists8–13. However, information on the extent of genetic control over CH4 production and the genetic architecture of the trait is generally scant. For example, Manzanilla-Pech et al.14 performed GWAS on different methane phenotypes in beef cattle and validated the results on dairy cattle, whereas Van Engelen15 performed GWAS on Holstein cows using phenotypes predicted from milk and breath analyses.
Only lately the technology for measuring CH4 production both on a large scale and on individual animals has become available. Among others, high throughput measuring techniques are based on breath analyses, since approx. 90% of enteric CH4 is released during eructation events and by breathing5. Nonetheless, collection of such phenotypic records is still challenging and data sets are limited. This technique is promising as it is non-invasive, based on infra-red analyses of breath samples and measurements can easily be taken during milking or feeding13,16–19. The most common application of these techniques is in combination with automatic milking systems (AMS) enabling a relatively long measurement period (duration of the milking); additionally, several observations per cow per day may be collected from a large number of animals. This type of measurement set up enables collection of large volumes of data, which is a prerequisite for genetic analyses.
To our knowledge, to date no reports are available on genome-wide association analyses based on direct measurements of daily CH4 production in dairy cattle. Therefore, the objective of this study was to undertake a genome-wide association study using CH4 phenotypes measured by breath analyzers to unravel the genomic regions controlling CH4 production from dairy cattle.
Results
Detected SNPs
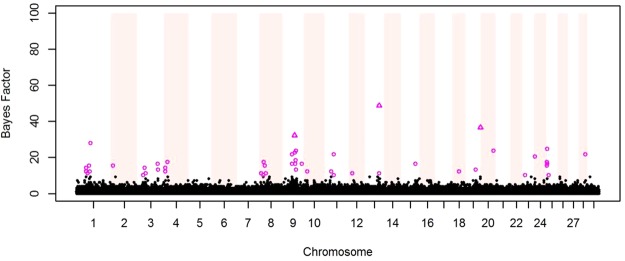
The genetic variance for daily CH4 production was estimated independently for each level of 2nd order Legendre polynomials. As the first parameter explains most of the variation, only SNP detected with it will be presented and discussed in this study. The GWAS performed on daily CH4 production indicated 50 SNPs with BF > 10 associated with CH4 production in dairy cattle (Fig. 1). Those SNPs were located on 18 different BTA (Tables 1 and 2). From detected SNPs, three had a BF above 30, which is defined as “very strong” association20. On BTA 1, 4, 9, 13 and 25 analysis in Haploview21 indicated six potential candidate QTL regions (Fig. 2). For those regions and two single SNP associations on BTA 9 and 20, a total of 130 candidate genes (protein-coding and non-coding RNA) were located with BIOMART22 (Table 1).
Figure 1.
Results of genome-wide association study for raw phenotypic methane production. Pink triangles indicate SNPs with Bayesian Factor (BF) >= 30, pink circles SNPs with 10 =< BF < 30 and black dots non-significant SNPs.
Table 1.
Candidate QTL regions and single SNPs detected for methane production with Bayesian Factor (BF) above 30, their position in base pairs, minor allele frequency (MAF), number of candidate genes and percentage of total genetic variance explained by them.
| BTA | SNP name | Position (bp) | MAF | BF | Candidate QTL | Allele subs. effecta | Number of candidate genes | Total genetic var. expl. (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | BTA89822nors | 46223040 | 0.491 | 14.33 | Yes | 0.113 | 12 | 0.006 |
| 1 | BTA89820nors | 46321775 | 0.488 | 12.26 | 0.193 | |||
| 4 | Hapmap39581BTA70101 | 9203380 | 0.497 | 12.26 | Yes | 0.226 | 14 | 0.003 |
| 4 | ARSBFGLNGS109843 | 9615916 | 0.430 | 14.33 | 0.251 | |||
| 9 | BTB00395654 | 60102040 | 0.353 | 32.29 | — | 0.132 | 1 | 0.003 |
| 9 | ARSBFGLNGS36482 | 64262480 | 0.365 | 16.41 | Yes | 0.201 | 9 | 0.005 |
| 9 | BTB01673493 | 64291804 | 0.365 | 16.41 | 0.172 | |||
| 9 | Hapmap42513BTA33276 | 66997852 | 0.215 | 23.76 | Yes | 0.183 | 7 | 0.005 |
| 9 | Hapmap27624BTA154889 | 67122449 | 0.318 | 13.29 | 0.158 | |||
| 13 | BPI1 | 67833218 | 0.402 | 11.22 | Yes | 0.144 | 21 | 0.003 |
| 13 | ARSBFGLNGS103635 | 67888763 | 0.467 | 48.68 | 0.152 | |||
| 20 | ARSBFGLNGS109784 | 909076 | 0.407 | 36.61 | — | 0.129 | 1 | 0.003 |
| 25 | ARSBFGLNGS61709 | 1086505 | 0.432 | 17.45 | Yes | 0.277 | 65 | 0.004 |
| 25 | ARSBFGLNGS103099 | 1127441 | 0.395 | 15.37 | 0.238 | |||
| 25 | ARSBFGLBAC43143 | 1184038 | 0.395 | 24.82 | 0.101 | |||
| 25 | Hapmap29768BTC016149 | 1205232 | 0.346 | 16.41 | 0.321 |
aAllele substitution effects were estimated as , where is the genetic variance explained by the SNP, and p and q are the frequencies of the two alleles76.
Table 2.
Suggestive SNPs detected for methane production with Bayesian Factor 10 < BF < 30, their position in base pairs, minor allele frequency (MAF) and percentage of total genetic variance explained by them.
| BTA | SNP name | Position (bp) | MAF | BF | Allele sub. effecta | Total genetic var. expl. (%) |
|---|---|---|---|---|---|---|
| 1 | ARSBFGLNGS94761 | 53656600 | 0.416 | 11.22 | 0.088 | 0.003 |
| 1 | ARSBFGLNGS3821 | 61286751 | 0.337 | 15.37 | 0.177 | |
| 1 | BTB01665387 | 63061634 | 0.437 | 12.26 | 0.231 | |
| 1 | ARSBFGLNGS4572 | 67212088 | 0.381 | 28.01 | 0.179 | |
| 2 | Hapmap44041BTA23382 | 10617894 | 0.266 | 15.37 | 0.200 | 0.001 |
| 3 | Hapmap33584BTA141202 | 30922247 | 0.128 | 10.19 | 0.235 | 0.075 |
| 3 | Hapmap44183BTA105889 | 37602383 | 0.428 | 14.33 | 0.172 | |
| 3 | ARSBFGLNGS38388 | 43476846 | 0.421 | 11.22 | 0.170 | |
| 3 | ARSBFGLNGS98870 | 98587436 | 0.428 | 16.41 | 0.114 | |
| 3 | Hapmap39765BTA62582 | 99317016 | 0.266 | 13.29 | 0.225 | |
| 4 | BTA72259nors | 20510260 | 0.360 | 17.45 | 0.172 | 0.001 |
| 8 | Hapmap26798BTA82382 | 11398105 | 0.191 | 11.22 | 0.207 | 0.012 |
| 8 | BTB00863195 | 23634451 | 0.449 | 10.19 | 0.088 | |
| 8 | ARSBFGLNGS39902 | 24288969 | 0.404 | 17.45 | 0.114 | |
| 8 | Hapmap52006BTA77999 | 29628947 | 0.449 | 15.37 | 0.133 | |
| 8 | BTB01356348 | 34847992 | 0.280 | 11.22 | 0.185 | |
| 9 | UAIFASA4057 | 50279445 | 0.245 | 16.41 | 0.197 | 0.005 |
| 9 | BTB00392496 | 50899854 | 0.322 | 21.65 | 0.155 | |
| 9 | BTB01520203 | 62539556 | 0.383 | 22.70 | 0.166 | |
| 9 | Hapmap58377rs29014990 | 66292441 | 0.353 | 18.50 | 0.179 | |
| 9 | Hapmap42705BTA85041 | 99135245 | 0.196 | 16.41 | 0.191 | |
| 10 | BTA59410nors | 17730891 | 0.215 | 12.26 | 0.194 | 0.002 |
| 11 | ARSBFGLNGS27959 | 22465305 | 0.128 | 12.26 | 0.161 | 0.004 |
| 11 | BTB01397452 | 33167082 | 0.428 | 21.65 | 0.156 | |
| 11 | BTB01641011 | 33771048 | 0.486 | 10.19 | 0.117 | |
| 12 | BTA31817nors | 22219373 | 0.241 | 11.22 | 0.289 | 0.001 |
| 15 | ARSBFGLNGS86665 | 67556240 | 0.227 | 16.41 | 0.196 | 0.003 |
| 18 | ARSBFGLNGS14182 | 33602408 | 0.323 | 12.26 | 0.144 | 0.001 |
| 19 | Hapmap48676BTA18047 | 47374363 | 0.490 | 13.29 | 0.094 | 0.003 |
| 20 | ARSBFGLBAC36856 | 63407185 | 0.356 | 23.76 | 0.129 | 0.001 |
| 23 | Hapmap61132rs29019650 | 11907305 | 0.402 | 10.19 | 0.158 | 0.001 |
| 24 | ARSBFGLBAC31288 | 4273189 | 0.178 | 20.60 | 0.242 | 0.002 |
| 25 | ARSBFGLNGS114786 | 7952738 | 0.400 | 10.19 | 0.141 | 0.002 |
| 28 | BTB00987935 | 35294673 | 0.400 | 21.65 | 0.169 | 0.005 |
aAllele substitution effects were estimated as , where is the genetic variance explained by the SNP, and p and q are the frequencies of the two alleles76.
Figure 2.
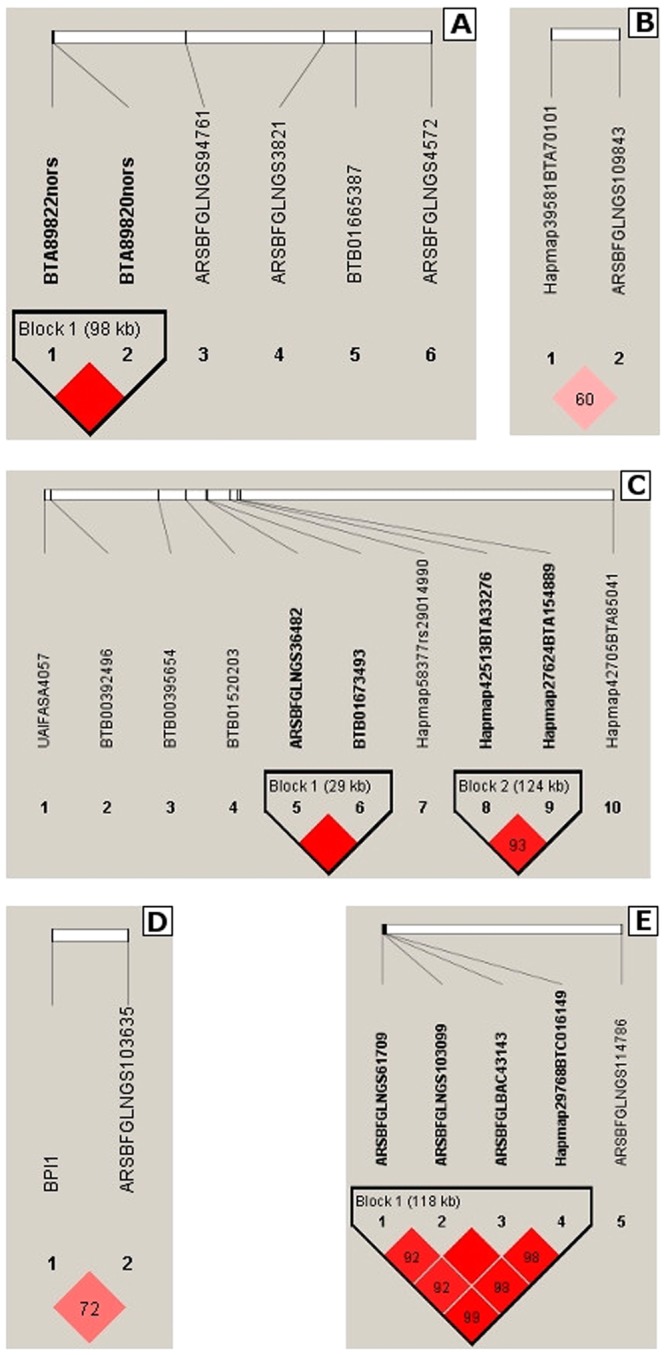
Results of the linkage disequilibrium (LD) analysis for significant SNPs detected on Bos Taurus autosomes (BTA) for raw phenotypic methane production. (A) BTA 1, (B) BTA 4, (C) BTA 9, (D) BTA 13, (E) BTA 25. Each square contains a value for r2 between neighboring SNP.
The three SNP detected for raw phenotypes with BF > 30 and six possible candidate QTL regions explained 0.032% of the total genetic variance (Table 1), whereas the remaining SNPs with 10 < BF < 30 explained 0.122% of this variance (Table 2). Overall this gives a very low result of 0.154% of the total genetic variance explained by detected SNPs.
Bioinformatics analysis of detected regions
Out of 130 candidate genes for CH4 production, 46 remained for a further GO Term analysis as known and non-ambiguous genes. For possible candidate genes, 428 different GO Terms were described: 82 cellular component terms, 251 biological process terms and 95 molecular function terms. Based on the GO Terms, five candidate genes were selected as the most promising: CYP51A1 on BTA 4, PPP1R16B on BTA 13, and NTHL1, TSC2, and PKD1 on BTA 25 (Table 3).
Table 3.
GO Terms for most promising candidate genes detected for methane production in dairy cattle.
| Gene | BTA | Position | Type of GO Term | GO Term name |
|---|---|---|---|---|
| CYP51A1 | 4 | 9306414–9323252 | Biological process | lipid metabolic process |
| steroid metabolic process | ||||
| PPP1R16B | 13 | 68258627–68366080 | Biological process | establishment of an endothelial barrier |
| positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | ||||
| NTHL1 | 25 | 1590252–1595934 | Biological process | metabolic process |
| TSC2 | 25 | 1596730–1626967 | Cellular component | Lysosome |
| PKD1 | 25 | 1627978–1666088 | Biological process | blood vessel development |
| nitrogen compound metabolic process | ||||
| digestive tract development |
Based on Cow QTLdb20, 52 QTLs involved in production and reproduction traits were selected as potentially playing a role in daily CH4 production in cows. Those QTLs were clustered into five groups: feed efficiency, milk related, body size and health status (see Table 4).
Table 4.
Previously detected QTLs within the identified genomic regions potentially related to methane production.
| Group of traits | Trait | BTA |
|---|---|---|
| Feed efficiency | Residual feed intake | 4 |
| Feed conversion ratio | 4 | |
| Average daily gain | 4 | |
| Body size | Height (mature) | 4 |
| Chest depth | 9 | |
| Body weight (mature) | 9 | |
| Milk | Milk fat yield | 9; 25 |
| Milk protein yield | 1; 13; 20 | |
| Milk yield | 13 | |
| Milk energy yield | 9 | |
| cis-Vaccenic acid content | 20 | |
| Docosatetraenoic acid content | 9 | |
| Eicosapentaenoic acid content | 9 | |
| Linoleic acid content | 1 | |
| Milk alpha-casein percentage | 1 | |
| Milk capric acid percentage | 13 | |
| Milk caproic acid percentage | 13 | |
| Milk caprylic acid percentage | 13 | |
| Milk myristoleic acid percentage | 13 | |
| Milk palmitoleic acid percentage | 1; 13 | |
| Oleic acid content | 1 | |
| Polyunsaturated fatty acid content | 1 | |
| Health status | Somatic cell score | 4; 9 |
| Clinical mastitis | 9 | |
| Immunoglobulin G level | 4; 20 | |
| Infectious bovine keratoconjunctivitis susceptibility | 1; 20 | |
| M. paratuberculosis susceptibility | 20 |
Discussion
To our knowledge this is the first GWAS on direct measurements of daily CH4 production performed in dairy cattle. So far one GWAS on direct measurements of CH4 production was performed in beef cattle with validation on dairy cattle14 and for CH4 intensity15. Another GWAS study on dairy cattle23 used predicted CH4 following the formula proposed by Dijkstra et al.24. Thus very little is still known on the actual genomic architecture of CH4 production in dairy cattle. Our results provide more insight into the genomic architecture of CH4 production thanks to the identification of genomic regions involved in the control of this trait and revealed genomic relationships between CH4 production and other traits.
Methane emission may be expressed in several ways depending on the aim of a given study25–28. First of all, when the total CH4 emitted by cows is of interest, the CH4 production phenotype expressed in g/d or l/d may be used16–18,28. When the goal is to minimize the amount of CH4 emitted from the supplied unit of feed (i.e. dry matter intake) in order to maximize feed conversion, the CH4 yield26 is the trait of interest (CH4 produced per kg of dry matter intake). Another way of expressing emission is CH4 intensity26, where produced CH4 is expressed per unit of product (milk or meat). Similarly to the residual feed intake, CH4 may be expressed as a difference between predicted and measured CH4 emission (i.e. residual CH4 emission)14,26,29. For our analyses we have decided to use the phenotype applied most widely in the literature and the least influenced by other traits not strictly related to CH4 emission itself (e.g. dry matter intake, milk production, live weight). Another reason is related with the fact that when calculating our CH4 production phenotype, we account for body weight, physiological status and milk production as described in Pszczola et al.13 following Madsen et al.28. Therefore, calculations of CH4 yield or CH4 intensity may have resulted in some potential overestimation of CH4 emissions due to double counting.
Selected candidate regions
Based on the bioinformatics analysis of detected regions for CH4 production in dairy cattle, five most promising candidate genes were selected based on GO Term analysis (Table 3). The first of them, CYP51A1 (BTA4: 9,306,414-9,323,252) located within the region of a candidate QTL on BTA 4, is a member of the cytochrome P450 family 51 subfamily A. Based on GO Terms this gene is involved in two biological processes that could potentially affect CH4 production in dairy cattle. Those GO Terms are the lipid metabolic process and the steroid metabolic process30,31, which are confirmed by CYP51A1 and its family members being involved in the synthesis of cholesterol, steroids and other lipids32. Lipids (i.e. fatty acids) were previously reported to be related to CH4 production, including several studies that used fatty acids present in milk to predict CH4 production24,33–39.
The second gene, namely PPP1R16B (BTA13: 68,258,627-68,366,080), a protein phosphatase 1 regulatory subunit 16B, is located within the candidate QTL region on BTA 13. For this gene two biological processes were found in GO Terms analysis that could link it to CH4 production. One of them, the establishment of the endothelial barrier, e.g. in the intestine, is defined as “… specific and selective control over the passage of water and solutes, thus allowing formation and maintenance of compartments that differ in fluid and solute composition”40. The other, the positive regulation of blood vessel endothelial cells30,40,41. The biological processes involving PPP1R16B suggest that this gene could affect the digestive process by controlling the passage of water within the intestine and providing blood vessels to the endothelial cells of the intestine. Being part of such processes, PPP1R16B could affect efficient use of feed and in this way control the amount of by-products (including CH4) produced during the process of digestion.
The three other genes were all located within the largest detected candidate QTL region on BTA 25, comprising of four SNPs. The first of the genes, NTHL1, nth like DNA glycosylase 1, is located at 1,590,252-1,595,934 bp. Its GO Term is the metabolic process, which includes protein synthesis and gradation31,40. The process involving this gene suggests that NTHL1 may affect digestive processes and consequently also a number of their by-products, e.g. CH4, being released post feeding.
The second of the above-mentioned genes, TSC2 (BTA25:1,596,730-1,626,967), tuberous sclerosis 2, is the only candidate gene with a GO Term related to a cellular component, in that case lysosome31,40. Moreover, TSC2 has been very well studied in humans, as its mutation causes tuberous sclerosis and its product is believed to be a tumor suppressor32. In the case of dairy cattle the location of the TSC2 gene in lysosome, which contains hydrolytic enzymes and takes part in energy metabolism, suggest that it could be involved in digestion processes and degradation of metabolites, this may affect CH4 produced by a cow.
The last of the candidate genes on BTA 25 is PKD1, encoding polycystein 1, a transient receptor potentially involved in channel interacting (1,627,978-1,666,088 bp). Three biological processes were assigned to it in GO Term analysis, i.e. blood vessel development40, nitrogen compound metabolic process31,40,42,43 and the digestive tract development40. All three GO Terms indicate that PKD1 is involved in digestion processes either directly by affecting the development of the digestive tract, or possibly also blood vessels around it as well as metabolic processes of a nitrogen compound. All those functions, in general, indicate that PKD1 might be involved in emissions of greenhouse gases, not only CH4 but also nitrogen related.
To confirm that the candidate genes detected in this study are the actual causative mutations affecting CH4 further functional studies such as gene expression or sequencing of the region of highest interest are required. However, this was outside the scope of this paper.
Potential Quantitative Trait Loci
Next to the search for candidate genes, we have also looked for previously detected QTLs for traits potentially related to CH4 production. Those QTLs were clustered in four groups of similar traits: feed efficiency, milk related, body size and health status (see Table 4). It has to be noted that in this study the estimation of CH4 production included an equation, in which fat-protein-corrected milk, live weight and pregnancy status are taken into account, and some of the found relationships may be present due to this fact. Alternatively, CH4 concentration (expressed in ppm) could be used for the association study. At this moment, however, CH4 production is the most widely reported trait in genetic studies regarding reduction of enteric CH4 emissions. For this reason we restricted our study to this trait.
Firstly, the comparison indicated an overlap between the genomic regions controlling the CH4 production and QTLs for feed efficiency traits (e.g. residual feed intake, feed conversion ratio, average daily gain; Table 4). The relationship between diet composition and CH4 production44 or the effect of additives reducing emission45–49 or dry matter intake50–52 is well known. It is anticipated that increased CH4 production leads to the loss of energy provided with feed5,6, and therefore more efficient cows should produce less CH4. Jentsch et al.53 showed that greater feed ingestion results in higher total CH4 production; however, CH4 production per kg dry matter intake decreases. Pickering et al.54 also reported the presence of a correlation between CH4 production and intake, while studies of55–57 showed that selection for cows with a low residual intake (efficient ones) results in lower CH4 production. Unfortunately, in this study no data was available on individual feed intake of cows and therefore we were not able to verify this statement empirically.
Secondly, regions controlling CH4 production were also overlapping with QTLs for traits describing various aspects of milk production (e.g. milk yield, milk protein and fat yield, milk composition; Table 4). The relationship between milk composition and CH4 production is particularly plausible because of common biochemical pathways between CH4, acetate and butyrate58. Furthermore, earlier studies showed that it is possible to use milk fatty acid composition to predict CH4 production24,33,35–39.
Thirdly, it was found that height, chest depth and body weight of the cow were genetically controlled by the same regions as potential QTLs for the CH4 production. Body characteristics such as body weight were earlier shown to be related to CH4 production52,59,60. Heavier cows are usually bigger and have a larger rumen capacity and a lower passage rate61, which leads to greater CH4 production52.
Finally, the QTLs detected previously for the health status of the cow (e.g. mastitis, somatic cell score, immunoglobulin G level) were also found in regions overlapping with SNPs detected in this study for CH4 production. Thus reports on the relationship between the health status of the animal and the direct CH4 production are limited. Zetouni et al. (2008) showed a negative genetic correlation on the health of the cows and methane production and a very low positive genetic correlation with udder health62. Elliott-Martin et al. (1997), based on breath analyses, indicated that CH4 could be used to diagnose ketosis. Moreover, the health status of the animal is known to affect other traits such as dry matter intake or production, and therefore is likely to affect CH4 production. It is likely that a sick animal produces less methane due to a lower milk production; however, methane intensity (i.e. the amount of methane produced per kg of milk) would increase. Next to QTLs related to traits indicating the health status of the cow also QTLs indicating susceptibility to illness were found in the regions important to CH4 production.
Based on the several traits mentioned above that share the genetic background with CH4 production, it may be suggested that some of the detected regions in this study have a pleiotropic effect. This knowledge is very beneficial especially in the case of production traits controlled by the same regions as CH4 production (i.e. assumed to be genetically correlated), which could serve as indicator traits for enteric CH4 production and eliminate difficult and time-consuming phenotyping. Our findings mostly match the study of Negussie et al.63, who reviewed literature on potential indirect traits for measuring CH4 production. Further evaluation of genetic relationships between CH4 and other traits is necessary to confirm relationships revealed by our study and before inclusion of CH4 to the breeding program can be made.
Power of the experimental design
The Bayesian method selected to perform GWAS for CH4 production allows for good distinctions between SNP with large and small effects on a trait, as in each iteration a different combination of SNPs is given a large effect. Thus detected SNPs give a valuable indication for the genomic regions potentially involved in CH4 production in dairy cattle. This was confirmed also by bioinformatics post-analysis of detected regions with the functions of selected candidate genes and QTLs for other traits detected within those regions. However, the total genetic variance explained by significant SNPs was very low. This could be due to several possible reasons, i.e. (1) a low number of animals used in the study, (2) the accuracy of the collected phenotypes, and (3) the polygenic nature of the studied trait.
Firstly, it should be noted that the analyzed dataset was relatively small, and therefore the power of the GWAS design was too low to detect a majority of SNPs associated with CH4 production. Taking into account the heritability of this trait at 0.2713, a higher number of genotyped animals would be needed to obtain a higher percentage of genetic variance explained by the detected SNP. Therefore, the analyses of a larger dataset (for both phenotypic and genomic data) may shed light on more specific SNPs with large effects However, generating a large data set by one project is difficult due to related costs (measuring and genotyping). Therefore, generating such a dataset by combining phenotypic observations and genotypes from various experiments could be a solution producing more reliable results in the future.
Secondly, to obtain reliable GWAS results reliable phenotypes are needed. In our study we used a technique that measures CH4 at the AMS during milking. To verify the accuracy of the sensor used in this study we validated the used sensor against sensors used in Respiration Chambers (the standard CH4 measuring technique). This comparison showed a high similarity between results generated by the two sensors when used in the AMS64. There are no studies comparing the performance of sensors used in the present study when installed in the Respiration Chamber. Several factors could lead to inaccuracies in the collected measurements such as occasional wind in the area of AMS or cows’ head movement. These factors were not controlled in this study. To account for these arguably random effects we measured CH4 for the individuals in the long period of time (i.e. resulting in multiple observations per animal). The average repeatability of the analyzed phenotype was 0.25 as reported in Pszczola et al.13.
Thirdly, the greater data set and increased accuracy of the measuring method could not have been enough to explain more genetic variation if the analyzed trait was highly polygenic. In previous studies using the same methodology, but larger data sets, only 0.83% of the genetic variance was explained by SNP in GWAS on litter size in pigs65 and 9.5% in GWAS on teat number in pigs66. Based on the presented results it seems that CH4 production is also a highly polygenic trait and many different regions are involved in its regulation. It might not be, therefore, possible to detect all of them using GWAS.
As CH4 production turned out to be a very polygenic trait in application to breeding practice, it may be more advisable to use the genomic prediction approach without specifying particular SNPs as being more important than others (e.g. genomic BLUP). In fact, de Haas et al.67, Lassen et al.68 and Wilson et al.69 performed genomic prediction type analyses while searching for correlated traits. The biggest challenge for the performance of genomic prediction with sufficient, reasonable or high accuracy of the estimated genotypic values is to create an adequately large reference population, which is likely to require cooperation between several countries.
Conclusions
This study aimed at detecting genomic regions affecting CH4 production in dairy cattle and showed that SNPs associated with the trait of interest may be detected. However, CH4 data collection poses a challenge, leading to a lower power of the experimental design and prevented detection of a high number of SNPs with a large effect on CH4 production. Consequently, only a small proportion of the genetic variance was explained by the SNPs. Nonetheless, the candidate QTL region on BTA 25, where three candidate genes were identified, may be considered as a genomic region regulating CH4 production in dairy cattle. Furthermore, the comparison of the QTL regions affecting CH4 production with previously reported QTLs indicated common genomic regions between CH4 production and traits related to feed efficiency, milk related, body size and health status. The found candidate genes were also involved in a number of metabolic processes possibly related to CH4 production. One of the most promising candidate genes (PKD1) was related to the development of the digestive tract being the environment inhabited by methanogens and the site for methane production. In general, all the evidence shows that CH4 production is a polygenic trait.
Methods
All research was approved by the Local Ethical Committee for Experiments on Animals in Poznan, Poland (Decision Number: 64/2012) and performed in accordance with the “Act on the protection of animals used for scientific purpose” of the Republic of Poland, which complies with the European Union Legislation for the protection of animals used for scientific purposes.
Phenotypes
The observations on CH4 production [g/d] used in this study were obtained from Pszczola et al.13, where all the detailed information on farms, measuring set-up and data processing can be found.
In short, animals available for this study were 287 Polish Holstein-Friesian cows kept on two commercial farms in Poland. This was a subset of 483 cows phenotyped for CH4 production and analyzed in Pszczola et al.13, of which 287 were genotyped. The CH4 production was measured repeatedly on Farm1 during two periods: from 2014/12/02 to 2016/02/03, and from 2016/06/01 to 2016/09/17, and on Farm2 from 2016/02/05 to 2016/03/14. Cows were milked repeatedly during the experiment, in total 25,872 CH4 production observations were collected for the genotyped animals.
The CH4 production was measured using a non-invasive Fourier Transform Infrared Spectroscopy breath analyzer (GASMET 4030; Gasmet Technologies Oy, Helsinki, Finland) during milking in AMS (Lely Astronaut A4). Concentrations of CH4 and CO2 measured during milking were converted to daily CH4 production in grams per day [g/d] following Madsen et al.28 and Pedersen et al.70. This calculation took into account the concentrations of CH4 and CO2, fat-protein corrected milk, live weight and duration of the pregnancy. Multiple daily outputs per cow were corrected for the diurnal variation in CH4 and averaged per cow per day.
Genotypes
Cows were genotyped with the Illumina BovineSNP50 v2.0 BeadChip (Illumina Inc., San Diego, CA) at the Cattle Genetics Laboratory of the Polish Federation of Cattle Breeders and Dairy Farmers. Ear tissue samples used to extract DNA were collected in the course of a routine procedure within the breeding program. The genotyped SNPs were processed with following quality control checks: (1) being in Hard-Weinberg equilibrium, (2) having the minor allele frequency above 0.05, (3) not being monomorphic, and (4) having a call rate of above 0.95. Six cows were removed as they had the call rate below 0.9. After quality control and removing SNPs located on sex chromosomes and chromosome 0 (unassigned), 39,680 SNPs remained for the genome-wide association analysis.
Genome-wide association
To identify regions of the genome affecting CH4 production, a multi-SNP genome-wide association analysis was performed with the application of the Bayesian Variable Selection method71. The method allows for a simultaneous estimation of the effects of all markers used in the analysis. The analysis was performed with the Bayz software72 on daily CH4 production using the model developed by Pszczola et al.13. The model was:
where CH4 stands for the daily CH4 production levels of a cow; µ is an n-vector equal to the mean; Xb is the design matrix of fixed effects of year-week of measurement and cow’s lactation number (levels 1 or 2+) fitted within the general lactation curve, which was modeled using 3rd order Legendre polynomials; and e is an n-vector of random residual effects assumed to be normally distributed . The Lk is a vector of individual random animal effect, which was modeled using 2nd order Legendre polynomials. The mapping of marker effects is constructed as a hierarchical model on random animal effects73. Firstly, the model accounts for genetic variance only. Secondly, at the next level the model allows disentangling permanent environmental (Note: this accounted for repeated observations of daily CH4 production per cow.) and genetic variances independently for each level of 2nd order Legendre polynomials. Here the Zu is a matrix with dimensions n by p, with n being the number of genotypes and p being the number of SNP coded as 0, 1, 2 copies of a specific allele vector; βijk is a p-vector with the random effects of markers; and εijk accounts for the permanent environmental effect assumed to be normally distributed .
For the marker effect the Bernoulli distribution was applied:
where for the first distribution it is assumed that the SNPs have a small effect (); whereas in the second distribution the SNPs are assumed to have a large effect, which explains a large part of variance () of analyzed traits. In this study, a prior of π1 = 0.001 was selected, thus on average only 1 in 1,000 SNPs was in the second distribution in each cycle. This resulted in only ~38 SNPs per cycle to have a large effect on the traits. The posterior means were calculated with 500k MCMC iterations with burn-in of 5k iterations to secure that all the SNPs were used65,66,74. Selecting a stringent prior provides a more precise distinction between SNPs with large and small effects on the trait66,75. If the SNP was not genotyped for a certain animal then Bayz assigned an average genotype to that position.
Identification of significant SNPs
The Bayes Factor (BF) was calculated for each SNP to determine the significant associations:
where π1 and π0 are the prior probabilities and is the posterior probability of the fraction of times the SNP was in the distribution with a large effect. Following the definitions of Kass and Raftery20, the SNPs with BF > 30 are described as a “very strong” association and with BF > 150 as “decisive”. The variance explained by significant SNPs was estimated as a fraction of the total genetic variance explained by all SNPs.
To confirm the potential QTL regions, also the linkage disequilibrium (LD) measured by r2 was estimated in Haploview21 between the SNPs detected on one BTA and not further from each other than 500 kbp. The candidate gene search was performed with the BIOMART software available in Ensembl Bos Taurus UMD 3.132 by entering the position of a possible QTL region or one of the most significant SNPs with ±500 kbp. To limit the number of QTLs to the most promising as candidate genes for daily CH4 production the BIOMART database was also used to study Gene Ontology Terms (GO Terms) of those QTLs. Furthermore, the Cow QTL database of the Animal Genome project20 was used to find previously detected QTLs within the most promising regions detected here for daily CH4 production. This was done analogically as for the candidate gene search, i.e. by entering the position of a possible QTL region or one of the most significant SNPs with ±500 kbp.
Acknowledgements
The authors would like to thank Luc Janss (Aarhus University) for valuable comments regarding the application of Bayz to dairy cattle data. This project was financed by the Polish National Center for Science (NCN OPUS grant no. 2013/09/B/NZ9/03179). MP and ESK acknowledge the financial support of the Polish Ministry of Science and Higher Education (grant no. 666/2014 and 1021/STYP/12/2017). Part of conducted research and publication of this manuscript was made possible by the statutory funding No. 508.534.01.6 of the Faculty of Veterinary Medicine and Animal Science Poznan University of Life Sciences, Poland; Department of Genetics and Animal Breeding. This study is partly based on the knowledge gained from the networks of COST Action FA1302 “Large-scale methane measurements on individual ruminants for genetic evaluations”.
Author Contributions
M.P. and E.S.K. designed the study and wrote the manuscript. M.P. was responsible for the collection of the phenotypic data, data editing, performing the statistical analysis of phenotypes and part of the post-genome-wide association study analysis. E.S.K. performed the genome-wide association and post analysis of those results. S.M. was responsible for the editing of the genotypes and was a discussion partner with respect to genome-wide association analysis. T.S. took part in data collection and was a discussion partner with respect to the statistical analysis of the phenotypic data and genome-wide association analysis. T.S. and M.P. initialized the project. All authors have read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
M. Pszczola and E. Sell-Kubiak contributed equally.
Contributor Information
M. Pszczola, Email: mbee@up.poznan.pl
T. Strabel, Email: strabel@up.poznan.pl
References
- 1.Stern, N. Stern Review Report on the Economics of Climate Change (2006).
- 2.Reisinger A, Clark H. How much do direct livestock emissions actually contribute to global warming? Global change biology. 2018;24:1749–1761. doi: 10.1111/gcb.13975. [DOI] [PubMed] [Google Scholar]
- 3.Opio, C. et al. Greenhouse gas emissions from ruminant supply chains – A global life cycle assessment. Food and Agriculture Organization of the United Nations (FAO), Rome, 1–214 (2013).
- 4.Smith, P. et al. Agriculture, forestry and other land use (AFOLU) (2014).
- 5.Murray RM, Bryant AM, Leng RA. Rates of production of methane in rumen and large-intestine of sheep. Br. J. Nutr. 1976;36:1–14. doi: 10.1079/bjn19760053. [DOI] [PubMed] [Google Scholar]
- 6.Johnson, D. E. et al. In Atmospheric methane: sources, sinks, and role in global change Vol. 13 (ed Khalil, M.A.K.) Ch. 11, 199–229 (Springer, 1993).
- 7.Knapp JR, Laur GL, Vadas PA, Weiss WP, Tricarico JM. Invited review: Enteric methane in dairy cattle production: Quantifying the opportunities and impact of reducing emissions. J. Dairy Sci. 2014;97:3231–3261. doi: 10.3168/jds.2013-7234. [DOI] [PubMed] [Google Scholar]
- 8.Lassen J, Lovendahl P. Heritability estimates for enteric methane emissions from Holstein cattle measured using noninvasive methods. J Dairy Sci. 2016;99:1959–1967. doi: 10.3168/jds.2015-10012. [DOI] [PubMed] [Google Scholar]
- 9.Lassen J, Lovendahl P. Heritability for enteric methane emission from Danish Holstein cows using a non-invasive FTIR method. Adv. Anim. Biosci. 2013;4:280. [Google Scholar]
- 10.de Haas Y, et al. Genetic parameters for predicted methane production and potential for reducing enteric emissions through genomic selection. J. Dairy Sci. 2011;94:6122–6134. doi: 10.3168/jds.2011-4439. [DOI] [PubMed] [Google Scholar]
- 11.Yin T, Pinent T, Brügemann K, Simianer H, König S. Simulation, prediction, and genetic analyses of daily methane emissions in dairy cattle. J. Dairy Sci. 2015;98:5748–5762. doi: 10.3168/jds.2014-8618. [DOI] [PubMed] [Google Scholar]
- 12.van Engelen S, Bovenhuis H, Dijkstra J, van Arendonk J, Visker M. Short communication: Genetic study of methane production predicted from milk fat composition in dairy cows. J. Dairy Sci. 2015;98:8223–8226. doi: 10.3168/jds.2014-8989. [DOI] [PubMed] [Google Scholar]
- 13.Pszczola M., Rzewuska K., Mucha S., Strabel T. Heritability of methane emissions from dairy cows over a lactation measured on commercial farms1. Journal of Animal Science. 2017;95(11):4813–4819. doi: 10.2527/jas2017.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manzanilla-Pech CIV, et al. Genomewide association study of methane emissions in Angus beef cattle with validation in dairy cattle. Journal of Animal Science. 2016;94:4151–4166. doi: 10.2527/jas.2016-0431. [DOI] [PubMed] [Google Scholar]
- 15.Engelen, S. v. The genetic background of methane emission by dairy cows, Wageningen University (2018).
- 16.Lassen J, Lovendahl P, Madsen J. Accuracy of noninvasive breath methane measurements using Fourier transform infrared methods on individual cows. J. Dairy Sci. 2012;95:890–898. doi: 10.3168/jds.2011-4544. [DOI] [PubMed] [Google Scholar]
- 17.Garnsworthy PC, Craigon J, Hernandez-Medrano JH, Saunders N. On-farm methane measurements during milking correlate with total methane production by individual dairy cows. J. Dairy Sci. 2012;95:3166–3180. doi: 10.3168/jds.2011-4605. [DOI] [PubMed] [Google Scholar]
- 18.Negussie E, et al. Non-invasive individual methane measurement in dairy cows. animal. 2016;11:890–899. doi: 10.1017/S1751731116002718. [DOI] [PubMed] [Google Scholar]
- 19.Chagunda MGG, Ross D, Roberts DJ. On the use of a laser methane detector in dairy cows. Computers and Electronics in Agriculture. 2009;68:157–160. doi: 10.1016/j.compag.2009.05.008. [DOI] [Google Scholar]
- 20.Kass RE, Raftery AE. Bayes Factors. Journal of the American Statistical Association. 1995;90:773–795. doi: 10.1080/01621459.1995.10476572. [DOI] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Cow QTL data base, https://www.animalgenome.org/cgi-bin/QTLdb/BT/index (2017).
- 23.Van Engelen, S., Bovenhuis, H., Dijkstra, J., Van Arendonk, J. A. M. & Visker, M. H. P. W. Genome wide association studies for milk fatty acids as a basis for methane prediction. Proceedings, 4–6 (2005).
- 24.Dijkstra J, et al. Relationships between methane production and milk fatty acid profiles in dairy cattle. Animal Feed Science and Technology. 2011;166-67:590–595. doi: 10.1016/j.anifeedsci.2011.04.042. [DOI] [Google Scholar]
- 25.Alcock DJ, Hegarty RS. Potential effects of animal management and genetic improvement on enteric methane emissions, emissions intensity and productivity of sheep enterprises at Cowra, Australia. Animal Feed Science and Technology. 2011;166-167:749–760. doi: 10.1016/j.anifeedsci.2011.04.053. [DOI] [Google Scholar]
- 26.Herd RM, et al. Measures of methane production and their phenotypic relationships with dry matter intake, growth, and body composition traits in beef cattle 1,2. Journal of Animal Science. 2014;92:5267–5274. doi: 10.2527/jas.2014-8273. [DOI] [PubMed] [Google Scholar]
- 27.de Haas Y., Pszczola M., Soyeurt H., Wall E., Lassen J. Invited review: Phenotypes to genetically reduce greenhouse gas emissions in dairying. Journal of Dairy Science. 2017;100(2):855–870. doi: 10.3168/jds.2016-11246. [DOI] [PubMed] [Google Scholar]
- 28.Madsen J, Bjerg BS, Hvelplund T, Weisbjerg MR, Lund P. Methane and carbon dioxide ratio in excreted air for quantification of the methane production from ruminants. Livestock Science. 2010;129:223–227. doi: 10.1016/j.livsci.2010.01.001. [DOI] [Google Scholar]
- 29.Donoghue, K. A., Herd, R. M., Bird, S. H., arthur, P. F. & Hegarty, R. S. In Proc. Assoc. Advmt. Anim. Breed. Genet. (AAABG) Vol. 20 290–293 (Napier, New Zealand, 2013).
- 30.Gene Ontology Consortium – Term Geniehttp://geneontology.org/page/termgenie (2017).
- 31.Smith, A. D., Datta, S. P. & Smith, G. H. Oxford Dictionary of Biochemistry and Molecular Biology (1997).
- 32.Ensembl Bos Taurus UMD 3.1http://www.ensembl.org (2017).
- 33.Dehareng F, et al. Potential use of milk mid-infrared spectra to predict individual methane emission of dairy cows. Animal. 2012;6:1694–1701. doi: 10.1017/s1751731112000456. [DOI] [PubMed] [Google Scholar]
- 34.Vanrobays ML, et al. Changes throughout lactation in phenotypic and genetic correlations between methane emissions and milk fatty acid contents predicted from milk mid-infrared spectra. J. Dairy Sci. 2016;99:7247–7260. doi: 10.3168/jds.2015-10646. [DOI] [PubMed] [Google Scholar]
- 35.Chilliard Y, Martin C, Rouel J, Doreau M. Milk fatty acids in dairy cows fed whole crude linseed, extruded linseed, or linseed oil, and their relationship with methane output. J. Dairy Sci. 2009;92:5199–5211. doi: 10.3168/jds.2009-2375. [DOI] [PubMed] [Google Scholar]
- 36.Mohammed R, McGinn SM, Beauchemin KA. Prediction of enteric methane output from milk fatty acid concentrations and rumen fermentation parameters in dairy cows fed sunflower, flax, or canola seeds. J. Dairy Sci. 2011;94:6057–6068. doi: 10.3168/jds.2011-4369. [DOI] [PubMed] [Google Scholar]
- 37.Rutten MJM, Bovenhuis H, Hettinga KA, van Valenberg HJF, van Arendonk JAM. Predicting bovine milk fat composition using infrared spectroscopy based on milk samples collected in winter and summer. Journal of Dairy Science. 2009;92:6202–6209. doi: 10.3168/jds.2009-2456. [DOI] [PubMed] [Google Scholar]
- 38.Soyeurt H, et al. Estimating fatty acid content in cow milk using mid-infrared spectrometry. J. Dairy Sci. 2006;89:3690–3695. doi: 10.3168/jds.S0022-0302(06)72409-2. [DOI] [PubMed] [Google Scholar]
- 39.Soyeurt H, et al. Mid-infrared prediction of bovine milk fatty acids across multiple breeds, production systems, and countries. J. Dairy Sci. 2011;94:1657–1667. doi: 10.3168/jds.2010-3408. [DOI] [PubMed] [Google Scholar]
- 40.Gene Ontology Consortium - Amigo (2017).
- 41.You C, et al. Loss of CCM3 impairs DLL4-Notch signalling: implication in endothelial angiogenesis and in inherited cerebral cavernous malformations. Journal of cellular and molecular medicine. 2013;17:407–418. doi: 10.1111/jcmm.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.CHEBI Chemical Entities of Biological Interest - https://www.ebi.ac.uk/chebi/.
- 43.Smith, A. D., Datta, S. P. & Smith, G. H. Oxford Dictionary of Biochemistry and Molecular Biology (2000).
- 44.Hegarty RS. Current and emerging technologies for decreasing enteric methane emission from individual ruminants. Recent advances in Animal Nutrition. 2009;17:81–88. [Google Scholar]
- 45.Martin C, Rouel J, Jouany JP, Doreau M, Chilliard Y. Methane output and diet digestibility in response to feeding dairy cows crude linseed, extruded linseed, or linseed oil. Journal of Animal Science. 2008;86:2642–2650. doi: 10.2527/jas.2007-0774. [DOI] [PubMed] [Google Scholar]
- 46.Hulshof RBA, et al. Dietary nitrate supplementation reduces methane emission in beef cattle fed sugarcane-based diets. Journal of Animal Science. 2012;90:2317–2323. doi: 10.2527/jas2011-4209. [DOI] [PubMed] [Google Scholar]
- 47.Cieslak A, et al. Tannins from sanguisorba officinalis affect in vitro rumen methane production and fermentation. JAPS: Journal of Animal & Plant Sciences. 2016;26:54–62. [Google Scholar]
- 48.Szczechowiak J, et al. Rumen fermentation, methane concentration and fatty acid proportion in the rumen and milk of dairy cows fed condensed tannin and/or fish-soybean oils blend. Animal Feed Science and Technology. 2016;216:93–107. doi: 10.1016/j.anifeedsci.2016.03.014. [DOI] [Google Scholar]
- 49.Cieslak A, Szumacher-Strabel M, Stochmal A, Oleszek W. Plant components with specific activities against rumen methanogens. animal. 2013;7:253–265. doi: 10.1017/S1751731113000852. [DOI] [PubMed] [Google Scholar]
- 50.Mills JAN, et al. Alternative approaches to predicting methane emissions from dairy cows 1. Journal of Animal Science. 2003;81:3141–3150. doi: 10.2527/2003.81123141x. [DOI] [PubMed] [Google Scholar]
- 51.Ellis JL, et al. Prediction of methane production from dairy and beef cattle. J. Dairy Sci. 2007;90:3456–3466. doi: 10.3168/jds.2006-675. [DOI] [PubMed] [Google Scholar]
- 52.Moraes LE, Strathe AB, Fadel JG, Casper DP, Kebreab E. Prediction of enteric methane emissions from cattle. Global change biology. 2014;20:2140–2148. doi: 10.1111/gcb.12471. [DOI] [PubMed] [Google Scholar]
- 53.Jentsch W, et al. Methane production in cattle calculated by the nutrient composition of the diet. Archives of Animal Nutrition. 2007;61:10–19. doi: 10.1080/17450390601106580. [DOI] [PubMed] [Google Scholar]
- 54.Pickering, N. K. et al. Consensus methods for breeding low emitting animals - a White Paper prepared by the Animal Selection Genetics Genomics Network of the Livestock Research Group of the Global Research Alliance on agricultural greenhouse gases. http://www.asggn.org/publications,listing,95,mpwg-white-paper.html (2013).
- 55.Nkrumah JD, et al. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle. Journal of Animal Science. 2006;84:145–153. doi: 10.2527/2006.841145x. [DOI] [PubMed] [Google Scholar]
- 56.Hegarty RS, Goopy JP, Herd RM, McCorkell B. Cattle selected for lower residual feed intake have reduced daily methane production12. Journal of Animal Science. 2007;85:1479–1486. doi: 10.2527/jas.2006-236. [DOI] [PubMed] [Google Scholar]
- 57.Fitzsimons C, Kenny DA, Deighton MH, Fahey AG, McGee M. Methane emissions, body composition, and rumen fermentation traits of beef heifers differing in residual feed intake1. Journal of Animal Science. 2013;91:5789–5800. doi: 10.2527/jas.2013-6956. [DOI] [PubMed] [Google Scholar]
- 58.Demeyer, D. I. & Van Nevel, C. J. In Digestion and metabolism in the ruminant (eds McDonald, I. W. & Warner, A. C. I.) 366–382 (The University of New England Publishing Unit, 1975).
- 59.Yan T, Porter MG, Mayne CS. Prediction of methane emission from beef cattle using data measured in indirect open-circuit respiration calorimeters. animal. 2009;3:1455–1462. doi: 10.1017/S175173110900473X. [DOI] [PubMed] [Google Scholar]
- 60.Holter JB, Young AJ. Methane Prediction in Dry and Lactating Holstein Cows. J. Dairy Sci. 1992;75:2165–2175. doi: 10.3168/jds.S0022-0302(92)77976-4. [DOI] [PubMed] [Google Scholar]
- 61.Demment MW, Soest PJV. A Nutritional Explanation for Body-Size Patterns of Ruminant and Nonruminant Herbivores. Am. Nat. 1985;125:641–672. doi: 10.1086/284369. [DOI] [Google Scholar]
- 62.Zetouni L, Kargo M, Norberg E, Lassen J. Genetic correlations between methane production and fertility, health, and body type traits in Danish Holstein cows. J. Dairy Sci. 2018;101:2273–2280. doi: 10.3168/jds.2017-13402. [DOI] [PubMed] [Google Scholar]
- 63.Negussie E, et al. Invited review: Large-scale indirect measurements for enteric methane emissions in dairy cattle: A review of proxies and their potential for use in management and breeding decisions. J. Dairy Sci. 2017;100:2433–2453. doi: 10.3168/jds.2016-12030. [DOI] [PubMed] [Google Scholar]
- 64.Sypniewski, M., Strabel, T., Cieslak, A., Szumacher-Strabel, M. & Pszczola, M. In 69th Annual Meeting of the European Federation of Animal Science (Dubrovnik, Corcotia, 2018).
- 65.Sell-Kubiak, E. et al. Genome-wide association study reveals novel loci for litter size and its variability in a Large White pig population. BMC Genomics16, 10.1186/s12864-015-2273-y (2015). [DOI] [PMC free article] [PubMed]
- 66.Duijvesteijn N, Veltmaat JM, Knol EF, Harlizius B. High-resolution association mapping of number of teats in pigs reveals regions controlling vertebral development. BMC Genomics. 2014;15:542. doi: 10.1186/1471-2164-15-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haas YD, et al. Breeding and genetics symposium: Resilience and lessons from studies in genetics of heat stress. Journal of Animal Science. 2017;95:1780–1787. doi: 10.2527/jas2016.0953. [DOI] [PubMed] [Google Scholar]
- 68.Lassen J, Poulsen NA, Larsen MK, Buitenhuis AJ. Genetic and genomic relationship between methane production measured in breath and fatty acid content in milk samples from Danish Holsteins. Animal Production Science. 2016;56:298–303. doi: 10.1071/AN15489. [DOI] [Google Scholar]
- 69.Wilson AM, et al. An international effort to improve feed efficiency and reduce methane emissions in dairy cows through genomics. Journal of Animal Science. 2016;94:152. doi: 10.2527/jam2016-0318. [DOI] [Google Scholar]
- 70.Pedersen, S. et al. Carbon Dioxide Production in Animal Houses: A literature review. Agricultural Engineering InternationalX (2008).
- 71.George EI, McCulloch RE. Variable Selection via Gibbs Sampling. Journal of the American Statistical Association. 1993;88:881–889. doi: 10.1080/01621459.1993.10476353. [DOI] [Google Scholar]
- 72.Heuven HCM, Janss LLG. Bayesian multi-QTL mapping for growth curve parameters. BMC Proceedings. 2010;4:S12. doi: 10.1186/1753-6561-4-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heuven, H., GJM., R & L. Janss. In 10th World Congress on Genetics Applied to Livestock Production (WCGALP) 2011–2013 (2014).
- 74.Lopes MS, et al. Using markers with large effect in genetic and genomic predictions. Journal of Animal Science. 2017;95:59–71. doi: 10.2527/jas.2016.0754. [DOI] [PubMed] [Google Scholar]
- 75.Van Den Berg I, Fritz S, Boichard D. QTL fine mapping with Bayes C (pi): a simulation study. Genet. Sel. Evol. 2013;45(10):1186. doi: 10.1186/1297-9686-45-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weller, J. I. Quantitative trait loci analysis in animals (2009).