Abstract
Groups of proximate continental islands may conceal more tangled phylogeographic patterns than oceanic archipelagos as a consequence of repeated sea level changes, which allow populations to experience gene flow during periods of low sea level stands and isolation by vicariant mechanisms during periods of high sea level stands. Here, we describe for the first time an ancient and diverging lineage of the Italian wall lizard Podarcis siculus from the western Pontine Islands. We used nuclear and mitochondrial DNA sequences of 156 individuals with the aim of unraveling their phylogenetic position, while microsatellite loci were used to test several a priori insular biogeographic models of migration with empirical data. Our results suggest that the western Pontine populations colonized the islands early during their Pliocene volcanic formation, while populations from the eastern Pontine Islands seem to have been introduced recently. The inter-island genetic makeup indicates an important role of historical migration, probably due to glacial land bridges connecting islands followed by a recent vicariant mechanism of isolation. Moreover, the most supported migration model predicted higher gene flow among islands which are geographically arranged in parallel. Considering the threatened status of small insular endemic populations, we suggest this new evolutionarily independent unit be given priority in conservation efforts.
Introduction
Islands epitomize a suite of simplified conditions in spatially bounded areas, which make harbored populations an ideal model to investigate several evolutionary topics1–3. For example, oceanic islands contributed a lot to our empirical understanding of a variety of microevolutionary processes that drive diversification and eventually speciation4–7. In such a context, island size, physiography, proximity among islands and dispersal capabilities are expected to shape the genetic structure of a pioneer species that colonizes an oceanic archipelago through oversea dispersal. Conversely, islands situated on continental shelves have often been reconnected with the mainland after sea level drops during glacial phases8. This scenario creates allopatric conditions that change through time, enabling both vicariant and dispersal mechanisms to act on genetic diversity and divergence9,10. In addition, groups of small geographically proximate islands may comprise an even more complex phylogeographic setting compared to single continental islands because nearby islands may experience repeated cycles of isolation (during sea level transgression) and connection (during periods of low sea level stands). Indeed, a complex geological setting dominated by considerable changes of islands size might lay the foundation for a variety of different genetic outcomes as a result of changing demographic processes and recurrent periods of fragmentation and gene flow11–13.
With almost 5000 islands, the Mediterranean Basin provides an attractive geological history which helps to explain the insular biodiversity observed today14. During the Pleistocene, the basin was characterized by cyclic sea level drops, which caused the formation of transient land bridges connecting nearby islands to each other, as well as islands and the mainland. The Mediterranean insular biota contains numerous taxa formed in response to such eustatic fluctuations15–17.
Almost half of the 23 recognized species of Podarcis lizards are endemic to Mediterranean islands and they represent an ideal model to explore the evolution of genetic diversity on islands. Here, we focus on the Italian wall lizard Podarcis siculus which, aside from inhabiting the entire Italian Peninsula, is also widespread across the greater islands of Sicily, Corsica, and Sardinia and across all the main Italian archipelagos and smaller islands (Fig. 1a). The phylogeography of this species across the Italian Peninsula is very complex18 and a recent study19 revealed that long term processes, giving rise to a mosaic of allopatric lineages, combined with more recent demographic dynamics, have driven the evolutionary history of this species. Here, we examine insular genetic variation of P. siculus from the Pontine Islands using mitochondrial sequences (mtDNA), nuclear sequences (nuDNA), and microsatellite loci. The Pontine Archipelago is formed by two groups of islands. The western group, which lies 32 km from the Tyrrhenian Coast, includes the oldest island of Ponza and the nearby Gavi Islet, Zannone Island, and Palmarola Island. These islands are separated from the eastern Pontine Islands by 40 km of open sea; this eastern group of islands includes Ventotene and the Santo Stefano islands, located 50 km away from the mainland (Fig. 1b). On the basis of morphological traits, different subspecies have been described within this archipelago: Podarcis s. latastei (Bedriaga, 1879) on Ponza Island, Podarcis s. patrizii (Lanza, 1952) on Zannone Island, Podarcis s. lanzai (Mertens, 1952) on Gavi Islet, and Podarcis s. palmarolae (Mertens, 1967) on Palmarola Island.
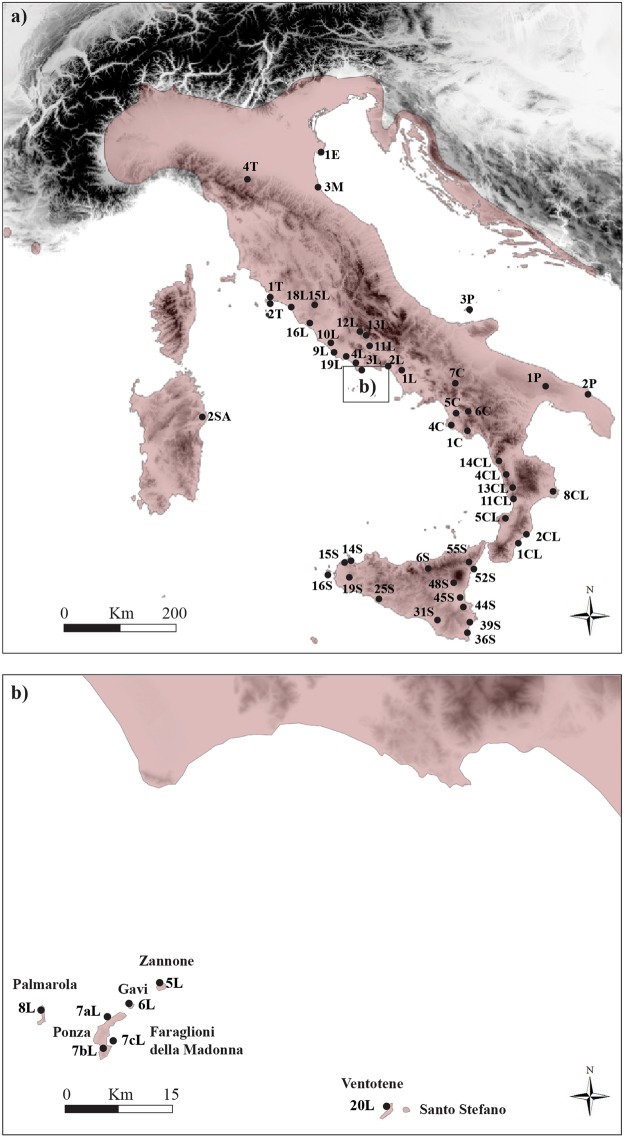
Figure 1.
(a) Map of the study area showing all sampling sites on the Italian Peninsula with the geographic distribution range of Podarcis siculus highlighted in red. (b) Map of the Pontine Archipelago with relative sampling locations.
This particular insular context provides ample opportunity to investigate several factors regarding the origin and evolution of intraspecific genetic diversity within a Mediterranean archipelago. First, the volcanic origin of the islands has been thoroughly investigated and relatively precise ages for their emergence have been reported20,21. Presumably, they never experienced any stable contact with the mainland. Second, the surrounding shallow waters indicate that the islands have been subjected to repeated episodes of fragmentation during high sea level stands as well as reunification when sea levels were lowest, during glacial phases. Finally, the western Pontine Islands have a spatial arrangement that allows testing different hypotheses of island biogeography, specifically regarding their compatibility with the observed patterns of genetic divergence and differentiation in this Mediterranean archipelago.
In this study we aim to (i) unveil the evolutionary history of Podarcis siculus from the Pontine Islands, and (ii) assess the influence of different scenarios of inter-island genetic diversification and gene flow by comparing models of island biogeography with genetic empirical data.
Results
Phylogenetic reconstruction and time of divergence
In the concatenated mtDNA alignment of 1701 bp (nd4 = 766 bp and cytb = 935 bp;156 sequences; accession numbers are reported in Supplementary Table S4), we found 100 haplotypes with 368 polymorphic sites and an overall mean distance of 0.062 among the main clades. Uncorrected pairwise p-distances are reported in Supplementary Table S5.
The phylogenetic inference returned the same topology under the two Bayesian approaches applied, revealing the presence of eight clades (Fig. 2a and Supplementary Fig. S1), seven of which had already been described in previous works18,19. However, a new diverging lineage (clade P in Fig. 2a,b) was discovered to which all individuals from the western Pontine Islands belong. The phylogenetic reconstruction indicated an early split separating the S, C1, and C2 clades from all the other clades (A1, A2, T1, T2, and P). Subsequently, a second cladogenetic event involved the separation of the Pontine clade P.
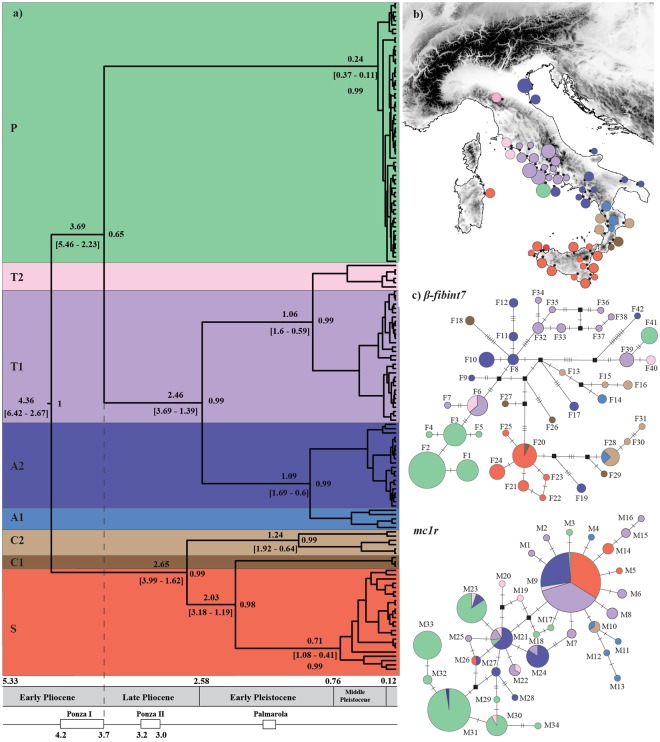
Figure 2.
(a) Chronogram based on mtDNA performed with beast. At each node, the posterior probabilities and the coalescence time with their relative 95% high posterior densities (HPD) are shown. Chronology of the geological epochs with the two principal volcanic episodes that formed the western Pontine Islands shown below. (b) Geographic distribution of the mtDNA clades at each sampled locality. (c) Statistical parsimony networks of the two nuclear gene fragments; circle sizes are proportional to allele frequencies. Alleles are colored based on the mtDNA clade assignment of the respective specimen.
The divergence time analysis carried out using beast showed an effective sample size over 200 for all parameters, indicating good chain mixing and adequate posterior estimates. The coefficient of variation in the average mutation rate estimates suggests a substantial heterogeneity among clades, supporting a non-clock-like model for the phylogeny (ucld.stdev parameter was 0.33 for cytb and 0.47 for nd4). According to our divergence time estimate, the Pontine clade (P) separated about 3.7 Mya (95% HPD: 5.5–2.2 Mya, Fig. 2a).
The final nuDNA alignment consisted of 252 phased sequences (600 bp) for mc1r and 164 sequences (728 bp) for β-fibint7 (accession numbers are reported in Supplementary Table S4). A test for recombination using the Pairwise Homoplasy Index (phi) did not find significant evidence for recombination in either of the two genes (p-value = 0.772 for mc1r; p-value = 0.559 for β-fibint7). The nuDNA did not show a phylogenetic pattern fully congruent with the mtDNA, but a certain level of accordance can be detected for the β-fibint7 fragments (Fig. 2c). In those specimens assigned to the new mtDNA clade P, all but one (F41) β-fibint7 allele form a monophyletic group. Moreover, all but three β-fibint7 alleles (F6, F20 and F28) were found only in specimens belonging to the same mtDNA clade. The mc1r alleles were typically shared among specimens of different mtDNA lineages. However, most alleles found in individuals of the western Pontine Islands are closely related at this locus and are mostly restricted to these islands, while they are very rare or absent elsewhere.
Among island structure and gene flow
The Podarcis siculus populations of the western Pontine Islands were characterized by 31 mtDNA haplotypes with 44 polymorphic sites. The number of haplotypes (H) and nucleotide (π) and haplotype (h) diversity are reported for each dataset and marker in Table 1.
Table 1.
Genetic diversity estimates for each Podarcis siculus insular population (PO: Ponza, GA: Gavi, PA: Palmarola, ZA: Zannone, FM: Faraglioni della Madonna).
| island | mtDNA | mc1r | β-fibint7 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | p.s. | H | h + sd | π + sd | n | p.s. | H | h + sd | π + sd | n | p.s. | H | h + sd | π + sd | |
| PO | 18 | 11 | 10 | 0.86 ± 0.06 | 0.001 ± 0.0002 | 36 | 10 | 6 | 0.69 ± 0.04 | 0.004 ± 0.0007 | 22 | 14 | 4 | 0.72 ± 0.06 | 0.006 ± 0.001 |
| FM | 5 | 6 | 4 | 0.90 ± 0.10 | 0.001 ± 0.0005 | 8 | 7 | 4 | 0.78 ± 0.11 | 0.004 ± 0.0012 | 6 | 12 | 3 | 0.73 ± 0.15 | 0.009 ± 0.002 |
| GA | 20 | 4 | 5 | 0.44 ± 0.13 | 0.0003 ± 0.0001 | 40 | 9 | 6 | 0.69 ± 0.04 | 0.004 ± 0.0003 | 14 | 1 | 2 | 0.52 ± 0.06 | 0.0007 ± 0.0001 |
| PA | 10 | 11 | 8 | 0.95 ± 0.05 | 0.001 ± 0.0002 | 24 | 9 | 7 | 0.79 ± 0.04 | 0.005 ± 0.0004 | 14 | 3 | 4 | 0.62 ± 0.11 | 0.001 ± 0.0008 |
| ZA | 8 | 9 | 7 | 0.96 ± 0.07 | 0.001 ± 0.0003 | 14 | 8 | 5 | 0.72 ± 0.08 | 0.005 ± 0.0006 | 12 | 2 | 3 | 0.54 ± 0.14 | 0.0008 ± 0.0002 |
| Tot | 61 | 43 | 31 | 0.87 ± 0.03 | 0.002 ± 0.0002 | 122 | 11 | 12 | 0.79 ± 0.02 | 0.005 ± 0.0002 | 68 | 15 | 6 | 0.68 ± 0.04 | 0.0040 ± 0.0009 |
n; number of individuals/alleles, p.s.; number of polymorphic sites, H; number of haplotypes, h; haplotype diversity, π; nucleotide diversity, sd; standard deviation.
The mtDNA parsimony network showed the presence of two main genetic assemblages (Fig. 3a), one of which is restricted to the Zannone Island, where all specimens carried haplotypes of that assemblage. This result was also confirmed by the DAPC analysis, which suggested K = 2 as the most reliable number of genetic clusters (Supplementary Fig. S2).
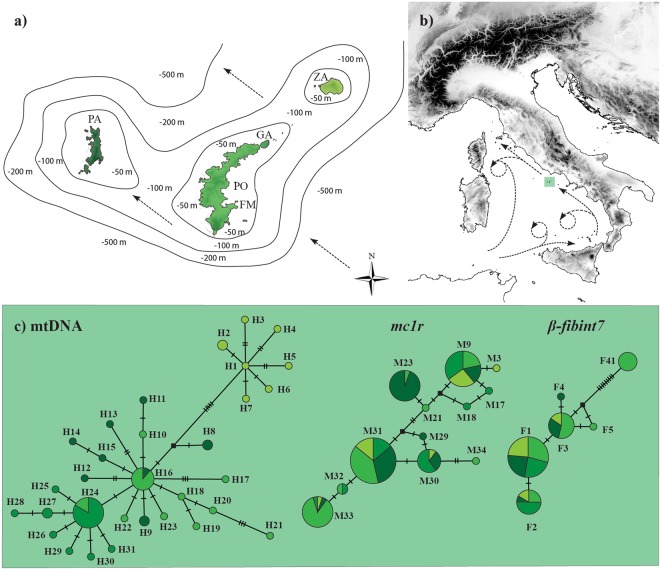
Figure 3.
(a) Map and bathymetry of the western Pontine Islands (PO: Ponza, GA: Gavi, PA: Palmarola, ZA: Zannone, FM: Faraglioni della Madonna). (b) Geographic location of the western Pontine Islands with the direction of the main marine currents in the Tyrrhenian Sea according to El-Geziry and Bryden78. (c) Statistical parsimony networks of the mtDNA (cytb + nd4) and nuDNA (mc1r and β-fibint7) fragments. Circles sizes are proportional to the haplotype frequencies and filled rectangles representing one mutation step. Haplotypes are colored by islands with different shades of green (for color code, see (c).
The nuDNA parsimony networks did not identify a concordant pattern for either mitochondrial or island repartition (Fig. 3a). The mc1r fragment showed the presence of 12 alleles and most of them were found in individuals across all island populations. Notably, of the 6 β-fibint7 alleles found, one allele (F4) was separated by 11 substitutions from the other 5 alleles. The DAPC carried out on nuclear sequences did not allow the identification of an optimal K value, returning a flat BIC curve (Supplementary Fig. S2).
All the 61 P. siculus specimens were genotyped at 11 polymorphic microsatellite loci, but the loci Pli 21 and Lv4a were excluded from further analyses because the former was monomorphic, while the latter might be influenced by positive selection. Genetic characteristics of the microsatellite loci are reported in Supplementary Table S6. Significant departures from the Hardy-Weinberg equilibrium (HWE) were observed in 2 out of 45 locus/population exact tests (Bonferroni correction applied: p < 0.001, see Supplementary Table S7). No linkage disequilibrium was detected between any pair of loci. Evidence of null alleles was found for several loci, but none was consistent across all populations, therefore we did not exclude them from further analysis. An indication of recent reduction in population size (bottlenecks) was obtained only for the Faraglioni della Madonna population by both the Wilcoxon test (P = 0.00684) and the mode-shift indicator, which revealed a distortion in the typical L-shape distribution of allele frequencies.
Both clustering analyses conducted on microsatellite data (DAPC and structure approaches) assigned K = 2 as the most likely number of genetic clusters among islands (Supplementary Figs S2 and S3). The first genetic cluster includes only the population from Gavi, while the second cluster includes all the remaining populations. Despite a further subdivision scheme, as suggested by the deltaK when K = 4, the individual assignment at the fourth cluster did not correspond to any specific island, suggesting a reliable genetic partition not higher than two (Supplementary Fig. S3). The presence of further differentiation among all P. siculus populations is suggested by FST analyses, where all comparisons yielded significant values (P < 0.005) (Bonferroni correction applied; Table 2).
Table 2.
FST values (below) and respective p-values (above) for each pair of populations (PO: Ponza, GA: Gavi, PA: Palmarola, ZA: Zannone, FM: Faraglioni della Madonna).
| ZA | GA | PO | FM | PA | |
|---|---|---|---|---|---|
| ZA | — | <0.00001 | 0.0048 | 0.0020 | 0.0010 |
| GA | 0.1144 | — | <0.00001 | <0.00001 | <0.00001 |
| PO | 0.0466 | 0.0655 | — | <0.00001 | 0.0010 |
| FM | 0.1859 | 0.1595 | 0.1081 | — | 0.0010 |
| PA | 0.0710 | 0.0807 | 0.0394 | 0.1347 | — |
Significant differences after Bonferroni correction (p < 0.005) are emboldened.
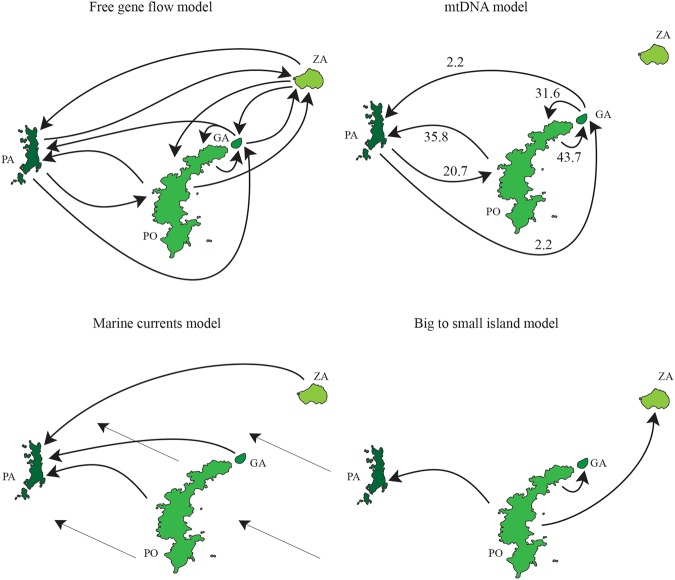
The analysis of current gene flow performed with BayesAss indicated no evidence of recent gene flow in any pairwise comparisons. The migration rate estimates were all below 0.07 with the exception of the migration estimate between the Zannone and Palmarola populations. This was detected above the background noise (M = 0.16). The Migrate-n analysis carried out on the microsatellite dataset under a full migration matrix model indicated the presence of asymmetric gene flow between island pairs, with higher migration rates detected between the island of Ponza and all the others (Fig. 4). The Bayes factor calculating the model probability using the log marginal likelihood of each generated migration scenario, indicated that the mtDNA migration model was the most reliable scenario for gene exchange.
Figure 4.
Models of migration among western Pontine Islands tested through Bayes factor analysis (PO: Ponza, GA: Gavi, PA: Palmarola, ZA: Zannone). Inferred migration rates are only shown for the most supported model.
Discussion
Dispersal plays a key role for the biodiversity of oceanic islands5,6. Conversely, studies on continental islands provided more complex evolutionary histories dominated by a variety of microevolutionary processes, such as vicariance, dispersal, and gene flow upon secondary contact11,15,22–24. Although the Pontine Archipelago could be referred as a particular case of oceanic islands system in the biogeographic sense of the term, it is located on continental shelf where changes in island size induced by sea level fluctuations may have been drastic. Such a special context provides a compelling setting to investigate the birth and the formation of intraspecific genetic diversity within archipelagos. Our results outline a tangled evolutionary history triggered by an ancient dispersal event of P. siculus from the mainland to islands, followed by local extinctions and intra-islands vicariant processes with gene flow.
On the origin of island genetic divergence
Phylogenetic reconstruction using two mitochondrial markers confirmed the complex phylogeographic pattern across the Italian Peninsula, with seven very divergent clades whose evolutionary histories have been described in detail in previous works18,19. In this study, we detect the presence of a new divergent 8th P. siculus clade corresponding to populations inhabiting the western Pontine Islands of Ponza, Gavi, Palmarola, and Zannone. On the contrary, the island of Ventotene of the eastern Pontine Island group showed haplotypes belonging to the A2 clade, indicating a recent introduction of P. siculus to this island, as already reported25.
According to our estimate of divergence time, the ancestry of the Pontine clade dates back to the early/late Pliocene transition (3.7 Mya; 95% HPD: 5.5–2.2 Mya). Although we used the same methodology, our estimates were slightly different to those obtained in a previous work19, but comparable when the HPD for each node is taken into account. Such a discrepancy could be due to the incorporation of an additional marker (nd4) highlighting how this analysis can be influenced by the reliability of the mutation rates. However, the present reconstruction is in line with the geological evolution of the western Pontine Islands (Fig. 2a). Indeed, on the basis of potassium-argon dating, the first volcanic event that accompanied the formation of the Ponza Island occurred between 4.2 and 3.7 Mya, followed by a quiescent period up to 3.2 Mya21. Although the estimated dates are associated with large uncertainty values of the coalescent process, our molecular clock suggests that the western Pontine populations colonized the archipelago early after their Pliocene volcanic formation. Furthermore, global changes in ice volume and records of terrestrial and marine sedimentary sequences suggest four glaciation events during the Pliocene, with one occurring about 3.6 Mya during the early/late Pliocene transition26. A consequence of global cooling is the sea-level retreat, which likely relaxed the separation of Ponza Island from the mainland facilitating a possible dispersal event. A similar explanation has also proposed to explain other ancient colonization events in the Mediterranean Basin15,27. It must be emphasized though, that the origin of these islands only puts an upper limit to the time since colonization and the age of the respective evolutionary lineages, as there is evidence that other vertebrate species colonized these islands much later28.
The genetic diversity of P. siculus from the Pontine Islands has been previously investigated using allozymes29. These authors were unable to detect any genetic differentiation of the western Pontine Island populations, neither with respect to the eastern Pontine Islands nor in comparison with the mainland populations. They subsequently suggested a possible human-mediated introduction in historical times. Our results from the nuDNA dataset clearly reject such a hypothesis, as allele frequencies of western Pontine Islands were significantly different from the other populations and several alleles of both mc1r and β-fibint7 fragments were private either for the western Pontine Islands or even for single islands. The population structure defined by β-fibint7 was more similar to the mtDNA, as all island alleles except one (F41) appear to form a monophyletic assemblage and no allele is shared with the mainland. On the contrary, most alleles of the mc1r gene occurred in specimens belonging to several mitochondrial clades. Such discrepancy is not surprising if we bear in mind that coding genes, such as mc1r, have lower mutation rates and are thus more prone to exhibit incomplete lineage sorting30,31. In addition, a recent study which reconstructed the phylogenetic relationship of 934 sequences of mc1r in nine Podarcis species revealed only slight differentiation, with some alleles shared even among different species32, corroborating our interpretation of incomplete lineage sorting.
Inter-island genetic diversification
On the basis of historical sea level changes from the Early Pleistocene, the Pontine Islands have probably been subjected to repeated cycles of connection (into a single paleo-island setting) and isolation33. Our inter-island diversification reconstruction is in line with such a scenario only if we consider a Middle Pleistocene divergence followed by different extent of genetic exchange during subsequent periods. The absence of an earlier divergence episode is interesting considering the ancestry of these populations and the fragmentation of the islands assumed for the Middle Pleistocene periods due to sea level elevation. At the same time, it is also difficult to imagine that migration processes have been maintained for such an extended period (from 3.7 to 0.2 Mya) to prevent further divergence. A more plausible explanation is that populations diverged in allopatry, i.e., when islands were disconnected. However, some of these ancestral island populations may have become extinct during strong marine transgressions, such that their present populations originate from a more recent episode of divergence. Such an interpretation is also supported by stratigraphic and paleontological evidence, which suggest that at least two principal episodes of strong marine transgression on the Tyrrhenian Coast occurred some 0.54 and 0.35 Mya ago, which prompted a sea level rise from 55 up to 120 m a.s.l.34. In addition, beach deposits from the Early Pleistocene are present at an elevation of 200–250 m a.s.l. on Monte Guardia (Ponza Island), while erosional surfaces from the Middle Pleistocene are widespread at 100–120 m a.s.l35. Therefore, considering the low altitude of some Pontine Islands (maximum altitude: Gavi = 60 m; Zannone = 170 m; Palmarola = 180 m; Ponza = 250 m), some of them might have been totally or partially submerged. Subsequent re-colonization from nearby islands during marine regressions may then have homogenized the genetic make-up among island populations.
According to our chronogram, the first episode of divergence within the Pontine Islands involved the separation of the Zannone Island population during the Mindel–Riss interglacial period (MIS 11, Fig. 2a). This period has been described as one of the longest warm periods of the Quaternary, inducing marked footprints of global sea level rise36. Under such vicariant mechanisms of divergence, taking into account similar distances and bathymetric depths among islands, we should expect also a similar extent of among islands genetic diversification followed by genetic admixture as a consequence of glacial inter-island reunification. Indeed, sea depth among islands never exceeds 100 m (Fig. 3b), which is almost the same magnitude of sea level drop during the last glacial maximum37,38.
Our multilocus dataset revealed a pattern of mito-nuclear discordance within the western Pontine Islands. The mtDNA showed the presence of two principal assemblages: one composed of individuals from Zannone Island and the other including individuals from Ponza, Gavi, and Palmarola islands. However, the nuDNA did not support strong population structure, neither when relating it to geography (different islands) nor to the mtDNA groups. Finally, the clustering analysis carried out with both DAPC and structure on the microsatellite dataset identified a K = 2 subdivision, revealing the Gavi Islet as a separate cluster. The lack of accordance between genes and genealogies represents a common issue, but can provide relevant insights into microevolutionary processes underlying the origin of small island system biodiversity39. In such an insular context, the causes of mito-nuclear discordance should be primarily a result of the different evolutionary trajectories of distinct markers. Indeed, the mtDNA has an inherently higher mutation rate and, due to its uniparental inheritance, is expected to reach fixation fourfold faster than the nuDNA genome because of its smaller effective population size and the associated higher susceptibility to demographic changes and genetic drift. On the contrary, the lack of congruence of the nuDNA dataset to both geographic settings and mtDNA differentiation is probably because genetic differentiation on these islands is rather recent, such that the sorting of the nuclear genealogies is not complete. Such a scenario has been repeatedly reported in many vertebrate species, including other Podarcis lizards30,31.
Although a principal trend of mito-nuclear discordance emerged when considering the mere genealogies and the current structure of the markers, our coalescent-based analysis of gene flow allowed us to partially explain this complexity. Indeed, the most supported migration scenario using microsatellite data, was the mtDNA migration model. This model suggests reduced gene flow between the Ponza and Zannone islands, and higher gene flow between the Ponza and Palmarola islands (Fig. 3b). Such a prediction is particularly plausible because these latter two islands have a parallel geographic arrangement. Therefore, the genetic exchange could have been even more pervasive during a low sea level stand, when islands shared a larger contact area. However, we cannot rule out that the reduced gene flow between the Ponza and Zannone populations is the outcome of demographic dynamics like “density blocking”40,41, or alternatively, is due to morphological or behavioral changes which occurred during the allopatric divergence.
Taxonomic and conservation implications
The genetic differentiation we found in the populations from the western Pontine Islands with respect to the mainland is comparable with those observed for many other Podarcis lizards species and over twice as high as between the insular endemic P. raffoneae and its sister taxon P. waglerianus24,42,43. On the other hand, populations from the eastern Pontine Islands seem to have colonized the Ventotene Island in recent times25. Considering the greater distance from the mainland and the deeper water in which the eastern Pontine Islands are located, the absence of a comparable ancient lineage is fairly surprising. Interestingly, in 1926 the herpetologist Robert Mertens described a particular phenotype endemic to Santo Stefano Island (P. siculus sanctistephani), which then became extinct and was replaced by the ordinary mainland phenotype during the first decades of the last century44,45. Such observations deserve special attention and may have several implications. First, it could be argued that an ancient lineage was also historically present on the eastern Pontine Islands, including Ventotene. Secondly, and perhaps more fascinating, it would be interesting to understand why the P. siculus sanctistephani became extinct. Currently, there is no trace of introgression in the current allochthonous population from the Santo Stefano Island25,29. In addition, a recent work focused on morphological variation of 374 individuals from the Pontine Islands and mainland, including the extinct population of Santo Stefano island, did not find intermediate phenotypes as could be expected during a complete admixture process46. If the local endemic P. siculus sanctistephani became extinct after the new colonizers and was indeed not hybridizing with later arriving conspecifics of the mainland type, there might have been a reproductive isolation mechanism, e.g. a behavioral or morphological change present in these island populations.
All these considerations point to a priority of the conservation of all the western Pontine Island populations, which should be referred to a different taxon. Indeed, the aforementioned example of possible population replacement underscores the vulnerability of endemic insular populations, suggesting that further introductions from the mainland could be deleterious and should be prevented in order to maintain the genetic integrity on this archipelago. In support of our conclusions, additional morphological, behavioral, and physiological studies would be needed to unravel whether the found genetic divergence is associated with ecological and/or phenotypic diversification on these islands.
Materials and Methods
Laboratory techniques and molecular data
A total of 156 specimens of P. siculus have been analyzed in this study: 63 new tail tissues were collected from six islands (Ponza, Faraglioni della Madonna, Gavi, Zannone, Palmarola, and Ventotene) while 93 individuals from 46 localities of the Italian Peninsula have been selected from a previous study19 (see Supplementary Table S1). The tail tissues were collected in accordance with relevant guidelines in full compliance with specific permits released by the Italian Environment Ministry (Prot. 00017879/PNM-09/09/2012) and no lizards were killed.
Genomic DNA was extracted according to the universal protocol of Salah47. We amplified one portion of the NADH dehydrogenase subunit 4 (nd4) with flanking regions (tRNASer, tRNAHis and tRNALeu) in all 156 collected samples. Additionally, three fragments including the cytochrome b (cytb), the coding melanocortin receptor 1 (mc1r), and the β-fibrinogen intron 7 (β-fibint7) were amplified only in the 61 specimens of the Pontine Islands, as the samples from the Italian Peninsula had been previously analyzed at these loci by Senczuk et al.19. Details on reaction buffer, primers, and cycling conditions are reported in Supplementary Table S2. The obtained amplified products were purified with Sure Clean (Bioline) and sequenced with an automated DNA sequencer by Macrogen (www.macrogen.com).
BioEdit 7.248 was used to evaluate the electropherograms, encode heterozygote positions using IUPAC ambiguity codes, and compute consensus alignments. The two mitochondrial fragments (mtDNA) were concatenated. For each nuclear gene (nuDNAs), we inferred the gametic phase using the coalescent-based Bayesian algorithm in phase 2.149,50. The β-fibint7 showed insertion/deletion (INDEL) polymorphisms in a region of poly-T and poly-G starting at 157 bp. Following the method described by Flot51, this region was used to determine the phase for sequences that were heterozygous for INDELs. The known phases were then implemented in the coalescent-based Bayesian reconstruction. Three independent runs were conducted with a burn-in at 1 × 103, 1 × 104 iterations, and thinning at each 100 steps. Finally, the occurrence of recombination events was assessed for each nuclear gene through the Pairwise Homoplasy Index (phi) test implemented in splitstree v. 452.
The 61 samples from the western Pontine Islands were also genotyped at 11 microsatellite loci. Details on primer and PCR conditions are reported in Supplementary Table S3. Microsatellite multiplex amplifications were performed and then compared with singleplex PCR to confirm that there were no apparent allele dropouts or allele size differences. Amplification products were run on an ABI 3130xl Genetic Analyzer and allele size was scored using peak scanner 1.0 (Applied Biosystems). Amplification and genotyping were repeated twice for some 15% of the total samples (10 lizards) to verify reproducibility in microsatellite scoring. Genepop 3.453 was used to assess deviations from Hardy-Weinberg equilibrium and linkage disequilibrium. The statistical significance levels were corrected for multiple comparisons using Bonferroni’s method (α = 0.05). lositan54 was used to identify candidate loci under selection. Before subsequent analyses, the presence of null alleles, large allele dropouts, and scoring errors was checked using micro-checker55.
Phylogenetic reconstruction and time of divergence
To infer phylogenetic relationships, the mtDNA dataset was analyzed using MrBayes v. 3.1.256. The best substitution model for each partition (i.e., HKY + I + G for nd4 and GTR + I + G for cytb) was selected using JModeltest57 under the Akaike Information Criterion (AIC). The analysis was run for 3 × 106 generations using four annealing chains, sampling every 1 × 104 and discarding the first 10% of computed trees as burn-in. Sequences of Podarics muralis retrieved from GenBank (accession numbers: HQ65293 for cytb, KF372413 for nd4) were used as an outgroup.
The software beast 1.8.158 was used to date the most recent common ancestors (TMRCA). In this analysis the selection of proper priors is a crucial component although it may be challenging. Because of the absence of fossil records in Podarcis lizards to calibrate the phylogeny, we used substitution rates previously estimated for both gene fragments12,43. We incorporated a lognormal prior distribution to the mean rate in order to constrain the 95% of the prior distribution between the upper and lower values estimated for Podarcis (μ = 0.0175, SD = 0.2 for cytb, and μ = 0.0115, SD = 0.5 for nd4). Another relevant issue is the consideration of how substitution rates can vary across branches assuming a clock-like or alternatively a relaxed-clock model. To resolve this issue, a preliminary run was performed using a lognormal relaxed-clock in order to account for branch heterogeneity by screening the ucld.stdev parameter, which provides information about whether evolutionary rates inferred from the data are clock-like. Finally, the choice of an outgroup can be difficult when the phylogeny is not fully resolved32. However, the use of a relaxed-clock allows estimates of the tree root even in the absence of a known outgroup59. beast was run for 1 × 108 generations sampling every 1 × 104 steps and the software tracer 1.660 was used to check parameter convergence. The final consensus tree was summarized from the stationary distribution (burn-in = 25%) in TreeAnnotator 1.8.158.
Furthermore, to better understand the relationship between mtDNA and nuDNA and the variation of the latter throughout the whole species distribution area, two nuDNA parsimony networks were built using the software popart61. For this analysis we removed 19 bp from the original β-fibint7 dataset (157–175), due to the presence of many INDELs polymorphisms in a region of poly-T and poly-G. Finally, uncorrected pairwise p-distances were calculated for each mtDNA clade obtained by the phylogenetic reconstruction using mega 7.062.
Among island structure and gene flow
To infer the genetic relationship between mitochondrial and nuclear haplotypes among island populations, we used the mtDNA and nuDNA dataset to construct statistical parsimony networks under the 95% probability connection limit using the software popart.
For the mtDNA and nuDNA datasets, the number of haplotypes (H) as well as nucleotide (π) and haplotype (h) diversity were estimated using DnaSP 5.163. The microsatellite genetic variability was estimated by calculating allele frequencies, expected (HE) and observed (HO) heterozygosities, number of alleles, and number of private alleles using Genetix 4.0564,65 and Fstat 2.9.3.266. All calculations were performed for the whole dataset and for each island. The software Bottleneck 1.2.0267 was used to assess the occurrence of recent bottlenecks; the tests were performed assuming the two-phase model (TPM).
To explore the genetic structure among island populations, we performed a discriminant analysis of principal component (DAPC) implemented in the R-package ‘Adegenet’68 using the mtDNA, nuDNA, and microsatellite datasets. For the microsatellite data we also performed a Bayesian clustering analysis implemented in structure 2.3.169 to infer the number of different genetic clusters (K), assuming the admixture model. Ten simulations of 1 × 105 iterations for each K (from K = 1 to K = 7) were run with a burn-in of 1 × 106. The best K value, based on the modal value ΔK70, was detected using the online software Structure Harvester 0.6.9371. In addition, Arlequin 3.572 was used to investigate the interpopulation level of genetic differentiation by calculating FST, according to Weir and Cockerham73.
In order to assess the presence of gene flow and whether such transfer was recent or historical we employed two different approaches. The software BayesAss 3.074 was used to estimate the presence of gene flow within the past few generations. Several preliminary runs were performed to adjust the acceptance rate of the mixing parameters (migration rates, allele frequencies, and inbreeding coefficients). The final analysis was carried out with 5 independent replicates running for 1 × 107 iterations, sampling every 100 steps and discarding the first 1 million generations as burn-in. Convergence was assessed by comparing the accordance of the posterior mean parameter estimates across independent runs and by checking the stationary of the trace file using the software tracer. To obtain an estimate of a more ancient signal of gene flow, we used the software migrate-n75,76. The program implements a coalescent-based approach to estimate the mutation scaled effective population size (θ = 4Neu) and the mutation scaled long-term migration rate (M = m/u, where m is the migration rate, and u is the mutation rate). We generated a set of plausible a priori biogeographic migration models, to test microsatellite empirical data: (i) the “free gene flow model”, which implies gene exchange among all islands, (ii) gene flow among all islands except for Zannone, as indicated by our mtDNA results (“mtDNA model”), (iii) gene flow in a south-east/north-west direction as could be supposed when considering the marine currents, favoring overseas dispersal (“marine currents model”), and (iv) gene flow from the bigger island to the smaller (“big to small island model”). Bayes factors were calculated for each model and used to test the more reliable scenario following the guideline of Kass and Raftery77. The Faraglioni della Madonna population was excluded from this analysis because of the small sample size (n < 5). After a preliminary tuning run to estimate θ and m parameters through FST to use as starting values, we performed four independent runs for each generated model using four parallel chains with static heating (temperatures 1.0, 1.5, 3.0 100,000.0) and Brownian motion microsatellite model.
Electronic supplementary material
Acknowledgements
The authors are very grateful to Paolo Colangelo, Flavia Annesi, Ignazio Avella, and Laura Gramolini for their advice and laboratory support. Thanks are also extended to all the collectors who have contributed to improving our sampling. We wish to give a special thanks to Dario D’ Eustacchio, who tragically passed away, for his precious suggestions and for his valuable help in the field. We thank Rebecca Nagel for her “native-speaker’s” corrections of our manuscript. We are grateful to the Italian Ministry for the Environment, Land and Sea for research permission (Prot. 00017879/PNM-09/09/2012). We also wish to thank the Editor and two anonymous reviewers for their useful suggestions. Funding was provided by the Italian Ministry of Education, University and Research (PRIN 2012 project) and by “Progetti di Ricerca di Università” to RC, German Academic Exchange Service (DAAD) fellowship to GS, and by the Institute of Biochemistry and Biology, University of Potsdam to RT.
Author Contributions
G.S. and R.C. designed the study. R.C. and R.T. provided funding. G.S., R.C., and E.D. performed fieldwork and sample collection. G.S., K.H., C.R., and E.D. obtained the molecular data. G.S., V.M., and C.R. analyzed the data. R.T. guided and contributed to discussions on the phylogeographical and microsatellite analyses. G.S. wrote the manuscript. All authors reviewed and approved the manuscript.
Data Availability
All DNA sequences used in this study will be submitted to GenBank upon acceptance.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33326-w.
References
- 1.MacArthur, R.H. and Wilson, E.O. The Theory of Island Biogeography, Princeton University Press (1967).
- 2.Carlquist, S. Island biology. Columbia University Press: New York & London. 660pp, 581, 5279 (1974).
- 3.Losos JB, Ricklefs RE. Adaptation and diversification on islands. Nature. 2009;457(7231):830–836. doi: 10.1038/nature07893. [DOI] [PubMed] [Google Scholar]
- 4.Wallace, A. R. Island Life: Or, The Phenomena and Causes of Insular Faunas and Floras, Including a Revision and Attempted Solution of the Problem of Geological Climates (Harper, 1881).
- 5.Cowie RH, Holland BS. Dispersal is fundamental to biogeography and the evolution of biodiversity on oceanic islands. Journal of Biogeography. 2006;33(2):193–198. doi: 10.1111/j.1365-2699.2005.01383.x. [DOI] [Google Scholar]
- 6.Heaney LR. Is a new paradigm emerging for oceanic island biogeography? Journal of Biogeography. 2007;34(5):753–757. doi: 10.1111/j.1365-2699.2007.01692.x. [DOI] [Google Scholar]
- 7.Whittaker RJ, Triantis KA, Ladle RJ. A general dynamic theory of oceanic island biogeography. Journal of Biogeography. 2008;35(6):977–994. doi: 10.1111/j.1365-2699.2008.01892.x. [DOI] [Google Scholar]
- 8.Rohling EJ, et al. Magnitudes of sea-level lowstands of the past 500,000 years. Nature. 1998;394(6689):162–165. doi: 10.1038/28134. [DOI] [Google Scholar]
- 9.Paulo OS, et al. The role of vicariance vs. dispersal in shaping genetic patterns in ocellated lizards in the western Mediterranean. Molecular Ecology. 2008;17:1535–1551. doi: 10.1111/j.1365-294X.2008.03706.x. [DOI] [PubMed] [Google Scholar]
- 10.Li JW, et al. Rejecting strictly allopatric speciation on a continental island: prolonged postdivergence gene flow between Taiwan (Leucodioptron taewanus, Passeriformes Timaliidae) and Chinese (L. canorum canorum) hwameis. Molecular Ecology. 2010;19(3):494–507. doi: 10.1111/j.1365-294X.2009.04494.x. [DOI] [PubMed] [Google Scholar]
- 11.Runemark A, Hey J, Hansson B, Svensson EI. Vicariance divergence and gene flow among islet populations of an endemic lizard. Molecular Ecology. 2012;21(1):117–129. doi: 10.1111/j.1365-294X.2011.05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salvi D, Schembri PJ, Sciberras A, Harris DJ. Evolutionary history of the Maltese wall lizard Podarcis filfolensis: insights on the ‘Expansion–Contraction’ model of Pleistocene biogeography. Molecular Ecology. 2014;23(5):1167–1187. doi: 10.1111/mec.12668. [DOI] [PubMed] [Google Scholar]
- 13.Reynolds RG, et al. Archipelagic genetics in a widespread Caribbean anole. Journal of Biogeography. 2017;44(11):2631–2647. doi: 10.1111/jbi.13072. [DOI] [Google Scholar]
- 14.Medail Frederic, Quezel Pierre. Hot-Spots Analysis for Conservation of Plant Biodiversity in the Mediterranean Basin. Annals of the Missouri Botanical Garden. 1997;84(1):112. doi: 10.2307/2399957. [DOI] [Google Scholar]
- 15.Brown RP, et al. Bayesian estimation of post-Messinian divergence times in Balearic Island lizards. Molecular Phylogenetics and Evolution. 2008;48(1):350–358. doi: 10.1016/j.ympev.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 16.Delicado D, Machordom A, Ramos MA. Living on the mountains: patterns and causes of diversification in the springsnail subgenus Pseudamnicola (Corrosella) (Mollusca: Caenogastropoda: Hydrobiidae) Molecular phylogenetics and evolution. 2013;68(3):387–397. doi: 10.1016/j.ympev.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 17.Rodríguez V, et al. Multilocus genetic diversity and historical biogeography of the endemic wall lizard from Ibiza and Formentera, Podarcis pityusensis (Squamata: Lacertidae) Molecular Ecology. 2013;22(19):4829–4841. doi: 10.1111/mec.12443. [DOI] [PubMed] [Google Scholar]
- 18.Podnar M, Mayer W, Tvrtković N. Phylogeography of the Italian wall lizard, Podarcis sicula, as revealed by mitochondrial DNA sequences. Molecular Ecology. 2005;14(2):575–588. doi: 10.1111/j.1365-294X.2005.02427.x. [DOI] [PubMed] [Google Scholar]
- 19.Senczuk G, Colangelo P, De Simone E, Aloise G, Castiglia R. A combination of long term fragmentation and glacial persistence drove the evolutionary history of the Italian wall lizard Podarcis siculus. BMC evolutionary biology. 2017;17(1):6. doi: 10.1186/s12862-016-0847-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bellucci F, Grimaldi M, Lirer L, Rapolla A. Structure and geological evolution of the island of Ponza, Italy: inferences from geological and gravimetric data. Journal of volcanology and geothermal research. 1997;79(1):87–96. doi: 10.1016/S0377-0273(97)00025-5. [DOI] [Google Scholar]
- 21.Cadoux A, Pinti DL, Aznar C, Chiesa S, Gillot PY. New chronological and geochemical constraints on the genesis and geological evolution of Ponza and Palmarola volcanic islands (Tyrrhenian Sea, Italy) Lithos. 2005;81(1):121–151. doi: 10.1016/j.lithos.2004.09.020. [DOI] [Google Scholar]
- 22.Meiri S. Oceanic island biogeography: Nomothetic science of the necdotal. Frontiers of Biogeography. 2017;9:1. doi: 10.21425/F59132081. [DOI] [Google Scholar]
- 23.Veith M, Kosuch J, Vences M. Climatic oscillations triggered post-Messinian speciation of Western Palearctic brown frogs (Amphibia, Ranidae) Molecular phylogenetics and evolution. 2003;26(2):310–327. doi: 10.1016/S1055-7903(02)00324-X. [DOI] [PubMed] [Google Scholar]
- 24.Lymberakis P, Poulakakis N. Three continents claiming an archipelago: the evolution of Aegean’s herpetofaunal diversity. Diversity. 2010;2(2):233–255. doi: 10.3390/d2020233. [DOI] [Google Scholar]
- 25.Biaggini M, et al. Low genetic differentiation between populations of Podarcis sicula (Reptilia, Lacertidae) from the Italian islands off the coast of Campania and the mainland. Belg. J. Zool. 2009;139(2):169–172. [Google Scholar]
- 26.De Schepper S, Gibbard PL, Salzmann U, Ehlers J. A global synthesis of the marine and terrestrial evidence for glaciation during the Pliocene Epoch. Earth-Science Reviews. 2014;135:83–102. doi: 10.1016/j.earscirev.2014.04.003. [DOI] [Google Scholar]
- 27.Carranza S, Harris DJ, Arnold EN, Batista V, Gonzalez De La Vega JP. Phylogeography of the lacertid lizard, Psammodromus algirus, in Iberia and across the Strait of Gibraltar. Journal of biogeography. 2006;33(7):1279–1288. doi: 10.1111/j.1365-2699.2006.01491.x. [DOI] [Google Scholar]
- 28.Avella I, Castiglia R, Senczuk G. Who are you? The genetic identity of some insular populations of Hierophis viridiflavus sl from the Tyrrhenian Sea. Acta. Herpetologica. 2017;12(2):209–214. [Google Scholar]
- 29.Capula M, Ceccarelli A. Distribution of genetic variation and taxonomy of insular and mainland populations of the Italian wall lizard. Podarcis sicula. Amphibia-Reptilia. 2003;24(4):483–495. doi: 10.1163/156853803322763945. [DOI] [Google Scholar]
- 30.Pinho C, Harris DJ, Ferrand N. Non-equilibrium estimates of gene flow inferred from nuclear genealogies suggest that Iberian and North African wall lizards (Podarcis spp.) are an assemblage of incipient species. BMC Evolutionary Biology. 2008;8(1):63. doi: 10.1186/1471-2148-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rato C, Carranza S, Perera A, Carretero MA, Harris DJ. Conflicting patterns of nucleotide diversity between mtDNA and nDNA in the Moorish gecko. Tarentola mauritanica. Molecular Phylogenetics and Evolution. 2010;56(3):962–971. doi: 10.1016/j.ympev.2010.04.033. [DOI] [PubMed] [Google Scholar]
- 32.Salvi D, Pinho C, Harris DJ. Digging up the roots of an insular hotspot of genetic diversity: decoupled mito-nuclear histories in the evolution of the Corsican-Sardinian endemic lizard Podarcis tiliguerta. BMC evolutionary biology. 2017;17(1):63. doi: 10.1186/s12862-017-0899-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emig, C. & Geistdoerfer, P. The Mediterranean deep-sea fauna: historical evolution, bathymetric variations and geographical changes. arXiv preprint q-bio/0507003 (2005).
- 34.Malatesta A, Zarlenga F. Evidence of Middle Pleistocene marine transgressions along the Mediterranean coast. Palaeogeography, Palaeoclimatology, Palaeoecology. 1988;68(2–4):311–315. doi: 10.1016/0031-0182(88)90048-X. [DOI] [Google Scholar]
- 35.Carrara C, Cremaschi M, Dai Pra G. Oscillazioni del livello marino, depositi e morfogenesi continentali nelle isole pontine (Italia Centrale) Geogr. Fis. Din. Quat. 1994;17:139–153. [Google Scholar]
- 36.Kukla G. Saalian supercycle, Mindel/Riss interglacial and Milankovitch’s dating. Quaternary Science Reviews. 2005;24(14):1573–1583. doi: 10.1016/j.quascirev.2004.08.023. [DOI] [Google Scholar]
- 37.Siddall M, et al. Sea-level fluctuations during the last glacial cycle. Nature. 2003;423(6942):853. doi: 10.1038/nature01690. [DOI] [PubMed] [Google Scholar]
- 38.Lambeck K, et al. Sea level change along the Italian coast during the Holocene and projections for the future. Quaternary International. 2011;232(1):250–257. doi: 10.1016/j.quaint.2010.04.026. [DOI] [Google Scholar]
- 39.Toews DP, Brelsford A. The biogeography of mitochondrial and nuclear discordance in animals. Molecular Ecology. 2012;21(16):3907–3930. doi: 10.1111/j.1365-294X.2012.05664.x. [DOI] [PubMed] [Google Scholar]
- 40.Waters JM, Fraser CI, Hewitt GM. Founder takes all: density-dependent processes structure biodiversity. Trends in Ecology & Evolution. 2013;28(2):78–85. doi: 10.1016/j.tree.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 41.Moritz C, et al. Multilocus phylogeography reveals nested endemism in a gecko across the monsoonal tropics of Australia. Molecular ecology. 2016;25(6):1354–1366. doi: 10.1111/mec.13511. [DOI] [PubMed] [Google Scholar]
- 42.Harris DJ, Pinho C, Carretero MA, Corti C, Böhme W. Determination of genetic diversity within the insular lizard Podarcis tiliguerta using mtDNA sequence data, with a reassessment of the phylogeny of Podarcis. Amphibia-Reptilia. 2005;26(3):401–407. doi: 10.1163/156853805774408676. [DOI] [Google Scholar]
- 43.Poulakakis N, Lymberakis P, Valakos E, Zouros E, Mylonas M. Phylogenetic relationships and biogeography of Podarcis species from the Balkan Peninsula, by Bayesian and maximum likelihood analyses of mitochondrial DNA sequences. Molecular Phylogenetics and Evolution. 2005;37(3):845–857. doi: 10.1016/j.ympev.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Bruno S. Catalogo sistematico, zoogeografico e geonemico dei Lacertidae di Corsica, Italia e Isole Maltesi. Natura Bresciana Ann. Mus. civ. Stor. nat., Brescia. 1992;19:39–95. [Google Scholar]
- 45.Lanza B, Corti C. Evolution of the knowledge on the Italian herpetofauna during the 20th century. Bollettino del museo civico di storia naturale di Verona. 1996;20(1993):373–436. [Google Scholar]
- 46.Senczuk Gabriele, Colangelo Paolo, Avramo Vasco, Castiglia Riccardo, Böhme Wolfgang, Corti Claudia. A study in scarlet: incipient speciation, phenotypic differentiation and conservation implications of the Podarcis lizards of the western Pontine Islands, Italy. Biological Journal of the Linnean Society. 2018;125(1):50–60. doi: 10.1093/biolinnean/bly091. [DOI] [Google Scholar]
- 47.Salah MA, Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997;25:4692–4693. doi: 10.1093/nar/25.22.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hall, T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. InNucleic acids symposium series (Vol. 41, No. 41, pp. 95–98). [London]: Information Retrieval Ltd., c1979-c2000 (1999).
- 49.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. The American Journal of Human Genetics. 2001;68(4):978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. The American Journal of Human Genetics. 2003;73(5):1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flot JF. SeqPHASE: a web tool for interconverting PHASE input/output files and FASTA sequence alignments. Molecular Ecology Resources. 2010;10:162–166. doi: 10.1111/j.1755-0998.2009.02732.x. [DOI] [PubMed] [Google Scholar]
- 52.Huson, D. H., & Bryant, D. Estimating phylogenetic trees and networks using SplitsTree 4. Manuscript in preparation, software available from www.splitstree.org (2005).
- 53.Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49(6):1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- 54.Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. Lositan: a workbench to detect molecular adaptation based on a F ST-outlier method. BMC bioinformatics. 2008;9(1):323. doi: 10.1186/1471-2105-9-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P. Micro‐checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Resources. 2004;4(3):535–538. [Google Scholar]
- 56.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic biology. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature methods. 2012;9(8):772–772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular biology and evolution. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS biology. 2006;4(5):e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rambaut, A., Suchard, M., Xie, D., & Drummond, A. (2014). Tracer v1. 6 http://beast. bio. ed. ac. uk. Tracer>(Online 2015, May 29).
- 61.Leigh JW, Bryant D. popart: full‐feature software for haplotype network construction. Methods in Ecology and Evolution. 2015;6(9):1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- 62.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular biology and evolution. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Librado P, Rozas J. DnaSPv5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 64.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire génome, populations, interactions, CNRS UMR. 1996;5000:1996–2004. [Google Scholar]
- 65.Belkhir, K., Borsa, P., Chikhi, L., Raufaste, N., & Bonhomme, F. Genetix 4.05, Population genetics software for Windows TM. Université de Montpellier II. Montpellier (2004).
- 66.Goudet, J. Fstat; a program to estimate and test gene diversities and fixation indices version 2.9. 3. http://www.unil.ch/izea/softwares/fstat.html (2001).
- 67.Piry S, Luikart G, Cornuet JM. Computer note. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. Journal of heredity. 1999;90(4):502–503. doi: 10.1093/jhered/90.4.502. [DOI] [Google Scholar]
- 68.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24(11):1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 69.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular ecology. 2005;14(8):2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 71.Earl DA, VonHoldt BM. Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conservation genetics resources. 2012;4(2):359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 72.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular ecology resources. 2010;10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 73.Weir BS, Cockerham CC. Estimating F‐statistics for the analysis of population structure. evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 74.Wilson GA, Rannala B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics. 2003;163(3):1177–1191. doi: 10.1093/genetics/163.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proceedings of the National Academy of Sciences. 2001;98(8):4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beerli P. How to use migrate or why are Markov chain Monte Carlo programs difficult to use. Population genetics for animal conservation. 2009;17:42–79. doi: 10.1017/CBO9780511626920.004. [DOI] [Google Scholar]
- 77.Kass RE, Raftery AE. Bayes factors. Journal of the american statistical association. 1995;90(430):773–795. doi: 10.1080/01621459.1995.10476572. [DOI] [Google Scholar]
- 78.El-Geziry TM, Bryden IG. The circulation pattern in the Mediterranean Sea: issues for modeller consideration. Journal of Operational Oceanography. 2010;3(2):39–46. doi: 10.1080/1755876X.2010.11020116. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All DNA sequences used in this study will be submitted to GenBank upon acceptance.