Abstract
Bladder cancer (BC) is one of the most common tumors in the urinary system. Noncoding RNAs are considered to take part in cellular phenotypes and are emerging as diagnostic and prognostic biomarkers of BC. The aim of this study is to investigate the clinical significance of neuroblastoma- associated transcript 1 (NBAT1) gene and its effects on malignant cellular phenotypes in BC. NBAT1 gene was low-expressed in BC tissues and cell lines and its low-expression was related with high pathological grade and metastasis of BC. Upregulation of NBAT1 gene depressed cell viability and invasiveness of KK47 and T24 cells and arrested KK47 and T24 cells at G1 stage. In addition, NBAT1 could target silence the expression of miR-21-5p in RNA-induced silencing complex-dependent manner. KK47 and T24 cells with miR-21-5p knockdown showed reduced cell viability, G1-stage arrest, and depressed invasiveness. MiR-21-5p mediates the regulatory effects of NBAT1 on malignant cellular phenotypes of BC cells. Moreover, SOCS6 gene was a target gene of miR-21-5p, and miR-21-5p modulated malignant cellular phenotypes of KK47 and T24 cells through targeted silencing of SOCS6. In conclusion, low-expression of NBAT1 is associated with the progress and metastasis of BC, and NBAT1 inhibits malignant cellular phenotypes through miR-21-5p/SOCS6 axis in BC. Our findings help to elucidate the tumorigenesis of BC, and future study will provide a novel therapeutic target for BC.
Introduction
Bladder cancer (BC) is a malignant tumor originating from bladder mucosa. In the urinary system, BC is the most common malignant tumor in China and the second most common tumor worldwide after prostatic cancer1,2. BC mainly included urothelial carcinoma, squamous cell carcinoma, and adenocarcinoma, as well as other rare types such as small cell carcinoma, carcinoid, malignant melanoma, and so on. Bladder urothelial carcinoma is the main type of BC, accounting for 95% of all BC.
BC can be divided into non-muscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer (MIBC) according to whether it invaded the muscular layer of the bladder wall. NMIBC, including Ta, T1, and Tis-stage BC, is also known as superficial BC. MIBC has intruded into the muscular layer of the bladder wall (T2–T4 stage) and is more likely to have lymphatic or distant metastasis. Despite the comprehensive treatment based on surgery, the recurrence rate of MIBC is high and the prognosis is poor3,4. Therefore, it is important and necessary to elucidate the underlying mechanism of BC growth and metastasis as well as find new therapeutic targets.
Noncoding RNAs (ncRNAs) consist of long noncoding RNAs (lncRNAs) and short noncoding RNA which include microRNAs, piwi-interacting RNAs, and short interfering RNAs. NcRNAs have become the focus of life science, especially oncology research in recent years. NcRNAs have been confirmed to participate in various complex diseases of human, especially malignant cancers5–7. It is well known that ncRNAs play important roles in tumorigenesis through modulating multiple important cellular biological phenotypes, such as cell proliferation, invasiveness, chemoresistance, and so on8–10. Recent studies found that ncRNAs were biomarkers for diagnosis and prognosis of some malignant cancers and might be new therapeutic targets in the future11,12.
Neuroblastoma-associated transcript 1 (NBAT1) gene is a newly identified functional lncRNA gene located at chromosome 6p22.3 and is identified and named by Pandey GK in the risk research of neuroblastoma in 201413. Heretofore, the research on NBAT1 and tumorigenesis is rare. Recent studies had found that NBAT1 gene downregulated and acted as a tumor suppressor gene in osteosarcoma and breast cancer14,15. However, the expression level and roles in BC remain unclear.
MiR-21-5p originates from 5′ end of pre-miR-21 which is mapped at chromosome 17q23.1. MiR-21-5p was confirmed to be highly expressed and plays its oncogene roles in a variety of tumors, including BC16–18. For example, miR-21-5p advanced migration and invasion of cervical carcinoma cells through targeting von Hippel-Lindau tumor suppressor (VHL) gene19. But the effects of miR-21-5p on malignant cellular phenotypes of BC are not very clear. Wu Y reported that formononetin could inhibit the invasiveness of BC cells and decrease the expression of miR-2120, but the correlation of miR-21-5p expression and the growth and metastasis of BC was not certain.
Suppressor of cytokine signaling 6 (SOCS6) gene is located at chromosome 18q22.2 and encodes a protein containing 535 amino acids. SOCS6 protein belongs to a suppressor of cytokine signaling family and is a cytokine-inducible negative regulator of cytokine signaling. SOCS6 gene has been proved to be a tumor suppressor gene in many malignant tumors, including prostate cancer, non-small-cell lung cancer, cervical cancer, and so on21–23. SOCS6 can control cell signal transduction by inducing ubiquitination degradation of signal protein24. There are no reports of SOCS6 and BC. Nevertheless, the function of SOCS6 in metastasis of BC is still unknown.
Together with the previous study that ncRNA NBAT1 could negatively modulate growth and metastasis of osteosarcoma cells through suppression of miR-2114, and SOCS6 is targeted by significantly downregulated miR-21-5p in secondary progressive multiple sclerosis25, this study will explore the clinical significance of NBAT1 expression, the role of NBAT1 in the regulation of cellular phenotypes, and its molecular mechanism in BC.
Methods
Clinical specimens
In total, 76 BC patients in this study were diagnosed and treated in the Department of Urology of Shengjing Hospital between July 2015 and May 2017. BC tissue and the corresponding normal bladder tissue (NBT) specimens were obtained through radical cystectomy and transurethral resection of bladder tumor. All the tissue specimens were confirmed by pathological diagnosis, and the clinical data were complete. All the patients were not treated with radiotherapy or chemotherapy before operation. This study got the approval of the Ethics Committee of Shengjing Hospital, and each participant provided an informed consent.
Cell lines and culture
Human normal bladder epithelial cell line (HCV29), BC cell line KK47 (low-grade NMIBC), and T24 (high-grade MIBC) were obtained from the Cell Resource Center of Chinese Academy of Medical Sciences (Beijing, China). Those cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (EXcell, Shanghai, China) in a 95% air/5% CO2 incubator at 37 °C.
Real-time quantitative PCR
Total RNA was extracted with TRNzol Universal reagent (Tiangen, Beijing, China) and reverse transcribed with lncRNA cDNA Synthesis Kit (Tiangen, Beijing, China). The expression of NBAT1 is examined with lncRNA qPCR Kit (Tiangen, Beijing, China) according to instructions, and primers of NBAT1 were 5′-ACTGAAACCCACAGAGATGAAG-3′ (sense) and 5′-CCCGTCATGTAGAGCAATATCC-3′ (antisense). The expression level of miR-21-5p was examined with Taqman Universal Master Mix II (Life Technologies, Carlsbad, CA, USA). The relative expression levels of NBAT1 and miR-21-5p were calculated using 2−ΔΔCT method after normalization with reference genes (β-actin and U6).
Cells transfection
The expression plasmid of NBAT1 (pUC-NBAT1) and its negative control (pUC-NC) were synthesized by Cyagen Biosciences Inc. (Santa Clara, USA). The plasmids were transfected into BC cells via HiPerFect reagent (QIAGEN, Hilden, Nordrhein-Westfalen, Germany) in a six-well culture plate according to instructions. The stable transfected cells were selected using GibcoTM Geneticin (Thermo Fisher Scientific, Waltham, MA, USA).
The agonist and antagonist of miR-21-5p (agomiR-21-5p and antagomiR-21-5p) and their negative control (agomiR-NC and antagomiR-NC) were purchased from GenePharma Co. (Shanghai, China). The expression plasmid and silence plasmid for SOCS6 (pEGFP-N1-SOCS6, pE-SOCS6; pSilencer3.1-SOCS6, pS-SOCS6) and their negative controls (pEGFP-N1, pE-NC; pSilencer3.1, pS-NC) were synthesized by Cyagen Co. (Santa Clara, CA, USA). HiPerFect reagent was applied to transfect transient microRNAs and plasmids were transiently transfected into cells.
Cell proliferation assay
The cell proliferation was examined using Enhanced Cell Counting Kit-8 (Beyotime, Beijing, China) according to instructions. Briefly, 2000 cells in 100 μl of medium were added into one pore of 96-well plates, addition of 10 μl enhanced CCK-8 solution to each pore, and incubated for 1 h. The absorbance value was detected with a Microplate Reader at 450 nm.
Flow cytometry assay
Cell Cycle Analysis Kit (Beyotime, Beijing, China) was used to examine the cell cycle (Beyotime, Beijing, China) in accordance with instructions. Cells were fixed for 2 h by 70% ethanol, centrifugated, and cleaned up. In total, 0.5 mL of propidium iodide (PI) staining solution was added and resuspended the cells, and then incubated for 30 min. The cell cycle was determined by the FACScan flow cytometry with Diva 8.0 software (Becton Dickinson, Franklin Lakes, NJ, USA).
Cell apoptosis was examined using Annexin V-FITC/PI apoptosis detection kit (Jiancheng, Nanjing, Jiangsu, China) according to instructions. In total, 2 × 105 cells were resuspended in 500 µL of binding buffer, added 5 µL of Annexin V-FITC, and 5 µl of PI in order, and incubated at 25 °C for 10 min. The apoptosis rate was detected and analyzed by FACScan flow cytometry with Diva 8.0 software. The apoptosis rate was the percent of cells with FITC-Annexin V positive/PI negative in the right lower quadrant.
Cell invasion assay
Cell invasiveness in vitro was assayed using Invasion Chamber with Matrigel Matrix (Corning, Corning, NY, USA). The lower chamber was added to 600 μL of DMEM medium containing 10% FB; the upper chamber was added to 100 μL of serum-free DMEM medium with 5 × 104 cells. Then the chamber was cultured at 37 °C for 48 h. Cells located at the upper side of the membrane were removed. The membrane was fixed with methanol and stained with Giemsa. Cells invaded to the lower side of the membrane were counted under a microscope in five randomly chosen fields and the average number was calculated.
Western blotting
Protein of cells was extracted using Protein Extraction Kit (Beyotime, Beijing, China), and quantified using Bradford Protein Assay Kit (Beyotime, Beijing, China). Totally, 30 μg of protein were processed, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene fluoride (PVDF) membrane, blocked with nonfat milk, hybridized with SOCS6 and cyclin D1 antibodies (ab197335 and ab134175, Abcam, Cambridge, MA, USA), and incubated with a secondary antibody. The PVDF membrane was treated with BeyoECL Plus reagent (Beyotime, China). Then, the bands were detected and analyzed by Image J software (NIH, Bethesda, MD, USA).
RNA pull-down assay
The biotinylated probes for NBAT1 and miR-21-5p (bio-NBAT1-W and bio-miR-21-5p-W, containing wild-type binding site) as well as their negative control (bio-NBAT1-M and bio-miR-21-5p-M, containing mutant binding site) were synthetized by GenePharma Co. (Shanghai, China). Probes were dissolved in the buffer and incubated with Dynabeads M-280 Streptavidin (Thermo Fisher Scientific, Waltham, MA, USA) for 10 min at 25 °C to form probe-coated beads. Those probe-coated beads were incubated with the lysates from KK47 and T24 cells, and eluted with the washing buffer. The pull- down RNAs were detected with qRT-PCR.
RNA immunoprecipitation (RIP) assay
RIP assay was operated using Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore Sigma, Burlington, MA, USA) according to previous literature26. Briefly, cells were lysed using RIP buffer and incubated with magnetic beads conjugated with human anti-Ago2 antibody or negative control normal IgG. Then the immunoprecipitated RNA was isolated and detected by qRT-PCR to demonstrate the presence of the binding targets.
Luciferase reporter assay
The luciferase reporter plasmids (pmiR-SOCS6-W, containing wild-type binding site; pmiR-SOCS6-M, containing mutant binding site) were synthesized by the Genescript Co. (Piscataway, NJ, USA). HEK-293T cells were co-transfected with the luciferase reporter plasmids and microRNAs, respectively. The Luciferase Reporter Kit (Beyotime, Beijing, China) was applied to detect the luciferase activity 48 h later in accordance with instructions.
Statistical analysis
The SPSS 22.0 (IBM, USA) was used for statistical analysis. All data were expressed as mean ± standard deviation and analyzed with one-way ANOVA and Student’s t test. If the P-value is less than 0.05, the difference was considered statistically significant.
Results
NBAT1 gene was low-expressed in BC and correlated with progress and metastasis of BC
The expression of NBAT1 gene in BC specimens was much higher than that in NBT specimens (Fig. 1a). Compared with HCV29 cells, NBAT1 gene was highly expressed in KK47 and T24 cells (Fig. 1b).
Fig. 1. NBAT1 was low-expressed in BC and its upregulation inhibited malignant cellular phenotypes of BC cells.
a The expression of NBAT1 gene in NBT and BC specimens. **P < 0.01 vs. NBT group. b The expression of NBAT1 gene in HCV29, KK47, and T24 cells. **P < 0.01 vs. HCV29 group, #P < 0.05 vs. KK47 group. c The expression of NBAT1 gene in KK47 and T24 cells. **P < 0.01 vs. pUC-NC group. d The cell viability of KK47 and T24 cells. **P < 0.01 vs. pUC-NC group. e The cell cycle of KK47 and T24 cells. *P < 0.05 vs. pUC-NC group. f The number of transmembrane KK47 and T24 cells. Scale bars represent 20 μm. **P < 0.01 vs. pUC-NC group. g The expression of cyclin D1 protein in KK47 and T24 cells. * P < 0.01 vs. pUC-NC group. h The cell apoptosis of T24 cells
Clinical parameters analysis showed that the decreased expression of NBAT1 was correlated with high pathological grade and smoking history, while it was independent of other parameters, including age and gender of BC patients (Table 1). Furthermore, the low-expression of NBAT1 was associated with the muscle invasion and lymph node metastasis of BC patients (Table 1), and the NBAT1 expression in T24 cells was much lower than that in KK47 cells (Fig.1b). These results revealed that NBAT1 gene was involved in the progress and metastasis of BC.
Table 1.
The correlation analysis of the expression of NBAT1 and miR-21-5p with the clinicopathological factors of 76 BC patients
| Pathological factors | Case | Relative NBAT1 expression | P-value | Relative miR-21-5p expression | P-value | |
|---|---|---|---|---|---|---|
| Age (years) | < 56 | 38 | 0.504 ± 0.168 | 0.647 | 3.689 ± 0.817 | 0.497 |
| ≥ 56 | 38 | 0.486 ± 0.172 | 3.811 ± 0.738 | |||
| Gender | Male | 50 | 0.484 ± 0.157 | 0.408 | 3.822 ± 1.032 | 0.387 |
| Female | 26 | 0.516 ± 0.163 | 3.613 ± 0.913 | |||
| Smoking history (more than 10 years) | No | 48 | 0.526 ± 0.174 | 0.043* | 3.613 ± 0.924 | 0.089 |
| Yes | 28 | 0.442 ± 0.166 | 3.986 ± 0.885 | |||
| Grade | Low grade | 36 | 0.548 ± 0.177 | 0.013 | 3.542 ± 0.735 | 0.033* |
| High grade | 40 | 0.447 ± 0.169 | 3.937 ± 0.842 | |||
| Muscle invasion | Negative | 41 | 0.552 ± 0.172 | 0.002** | 3.320 ± 0.823 | < 0.001** |
| Positive | 35 | 0.429 ± 0.165 | 4.257 ± 0.764 | |||
| Lymph node metastasis | Negative | 71 | 0.508 ± 0.186 | 0.027* | 3.670 ± 1.035 | 0.011* |
| Positive | 5 | 0.316 ± 0.132 | 4.890 ± 0.371 |
*P < 0.05, **P < 0.01
Upregulation of NBAT1 inhibited malignant cellular phenotypes of BC cells
Further, a series of gain-of-function assays was applied to examine the effect of NBAT1 on malignant cellular phenotypes of BC cells. First, KK47 and T24 cells were transfected with pUC-NBAT1 to upregulate the expression of NBAT1 (Fig.1c). And, the enhanced CCK8, flow cytometry, and cell invasion assays showed that upregulation of NBAT1 depressed cell viability of KK47 and T24 cells (Fig.1d), arrested KK47 and T24 cells at G1 stage (Fig.1e), and inhibited invasiveness of KK47 and T24 cells (Fig.1f). Meanwhile, western blotting assay showed that the expression of G1-stage checkpoint protein cyclin D1 was significantly increased when KK47 and T24 cells occurred in G1-stage block (Fig.1g). But, the cell apoptosis did not change significantly in the KK47 and T24 cells with NBAT1 enhancement (Fig.1h).
NBAT1 silenced specifically miR-21-5p expression of BC cells
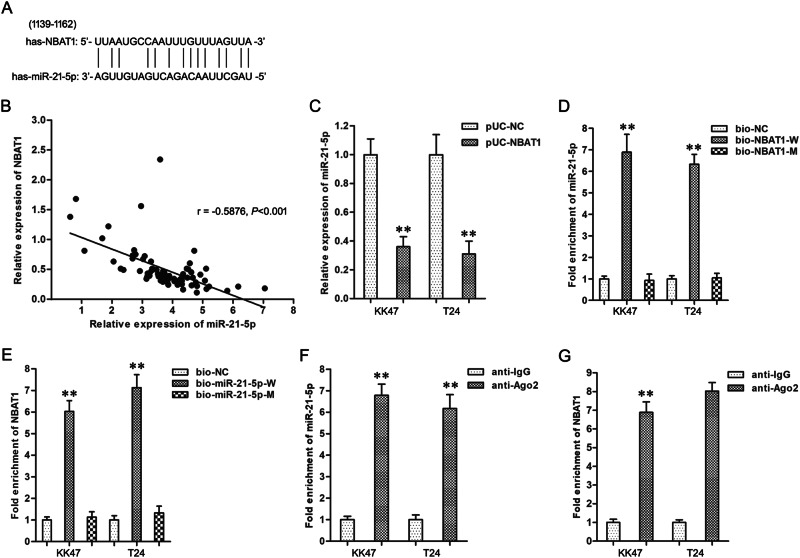
The bioinformatics analysis and previous literature14 forecasted the specific combination between NBAT1 and miR-21-5p (Fig.2a). First, the co-expression patterns analysis showed a negative correlation between NBAT1 and miR-21-5p in BC (Fig. 2b) (r = −0.5876, P < 0.001). Second, upregulation of NBAT1 significantly decreased the miR-21-5p expression in KK47 and T24 cells (Fig. 2c). Third, RNA pull-down assay verified that miR-21-5p was combined with bio-NBAT1-W probe but not bio-NBAT1-M probe (Fig. 2d); NBAT1 was specifically combined with bio-NBAT1-W probe correspondingly (Fig. 2e). Fourth, RIP experiment affirmed the enrichment of NBAT1 and miR-21-5p in the anti-Ago2 group (Fig. 2f, g), and both existed in a RNA-induced silencing complex (RISC). These findings elucidated NBAT1 that silenced specifically miR-21-5p expression of BC cells.
Fig. 2. NBAT1 silenced specifically miR-21-5p expression of in BC cells.
a The predicted miR-21-5p binding site in the NBAT1 sequence. Short vertical lines indicated complementary nucleotides. b The coexpression patterns analysis between NBAT1 and miR-21-5p in BC. c The expression of miR-21-5p gene in KK47 and T24 cells. **P < 0.01 vs. pUC-NC group. d Detection of miR-21-5p using qRT-PCR in the sample pulled down by biotinylated NBAT1 probe. **P < 0.01 vs. bio-NC group. e Detection of NBAT1 using qRT-PCR in the sample pulled down by biotinylated miR-21-5p probe. **P < 0.01 vs. bio-NC group. f Detection of miR-21-5p using qRT-PCR in RIP complex. **P < 0.01 vs. anti-IgG group. g Detection of NBAT1 using qRT-PCR in RIP complex. **P < 0.01 vs. anti-IgG group
miR-21-5p was high-expressed in BC and was necessary in NBT1-induced malignant cellular phenotypes of BC cells
The expression of miR-21-5p in BC was higher than that in NBT specimens (Fig. 3a). The expression of miR-21-5p in KK47 and T24 cells was highly expressed compared with HCV29 cells (Fig. 3b). Clinical parameters analysis displayed that the high-expression of miR-21-5p was correlated with high pathological grade, muscle invasion, and lymph node metastasis of BC patients (Table 1). In addition, the miR-21-5p expression in T24 cells was much higher than that in KK47 cells (Fig.1b). These results suggested that miR-21-5p also takes part in the progress and metastasis of BC.
Fig. 3. MiR-21-5p was high-expressed in BC and its knockdown inhibited malignant cellular phenotypes of BC cells.
a The expression of miR-21-5p gene in NBT and BC specimens. **P < 0.01 vs. NBT group. b The expression of miR-21-5p gene in HCV29, KK47, and T24 cells. **P < 0.01 vs. HCV29 group, #P < 0.05 vs. KK47 group. c The expression of miR-21-5p gene in KK47 and T24 cells. **P < 0.01 vs. pUC-NC group. d The cell viability of KK47 and T24 cells. **P < 0.01 vs. antagomiR-NC group. e The cell cycle of T24 cells. *P < 0.05 vs. antagomiR-NC group. f The number of transmembrane KK47 and T24 cells. Scale bars represent 20 μm. **P < 0.01 vs. antagomiR-NC group. g The expression of Cyclin D1 protein in KK47 and T24 cells. *P < 0.01 vs. antagomiR-NC group
AntagomiR-21-5p was transfected into T24 cells to depress the expression of miR-21-5p (Fig. 3c). And, the cellular phenotype detection confirmed that KK47 and T24 cells with miR-21-5p knockdown showed reduced cell viability (Fig. 3d), G1-stage arrest (Fig. 3e), and depressed invasiveness (Fig. 3f). Also, antagomiR-21-5p could decrease the expression level of G1-stage checkpoint protein cyclin D1 (Fig. 3g).
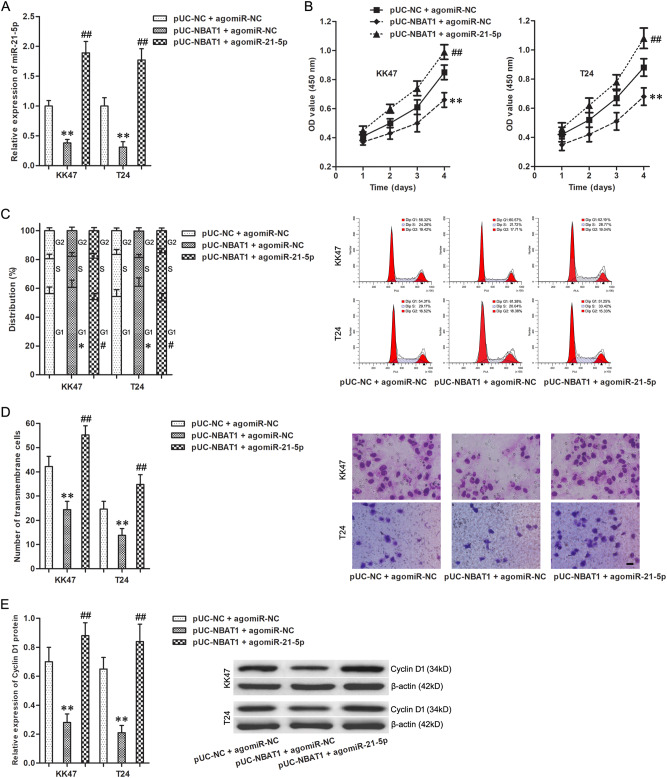
To clarify the effects of miR-21-5p in the regulation process of NBAT1 on malignant cellular phenotypes, agomiR-21-5p was transfected into T24 cells with stably high-expressed NBAT1 (Fig. 4a). Compared with the group of pUC-NBAT1 and agomiR-NC, pUC-NBAT1 and agomiR-21-5p group exhibited the stronger proliferation and invasiveness, as well as the weaker G1-stage arrest, and miR-21-5p enhancement rescued the inhibitory effect of NBAT1 upregulation on cellular phenotypes (Fig. 4b–e). In view of the above, miR-21-5p mediated the regulatory effects of NBAT1 on malignant cellular phenotypes in BC cells.
Fig. 4. MiR-21-5p mediated the regulatory effects of NBAT1 on malignant cellular phenotypes in BC cells.
a The expression of miR-21-5p gene in KK47 and T24 cells. **P < 0.01 vs. pUC-NC + agomiR-NC group; ##P < 0.01 vs. pUC-NBAT1 + agomiR-NC group. b The cell viability of KK47 and T24 cells. **P < 0.01 vs. pUC-NC + agomiR-NC group; ##P < 0.01 vs. pUC-NBAT1 + agomiR-NC group. c The cell cycle of KK47 and T24 cells. *P < 0.05 vs. pUC-NC + agomiR-NC group; #P < 0.05 vs. pUC-NBAT1 + agomiR-NC group. d The number of transmembrane KK47 and T24 cells. Scale bars represent 20 μm. **P < 0.01 vs. pUC-NC + agomiR-NC group; ##P < 0.01 vs. pUC-NBAT1 + agomiR-NC group. e The expression of Cyclin D1 protein in KK47 and T24 cells. **P < 0.01 vs. pUC-NC + agomiR-NC group; ##P < 0.01 vs. pUC-NBAT1 + agomiR-NC group
NBAT1 positively regulated SOCS6, which was a target gene of miR-21-5p in BC cells
SOCS6 gene might be a potential target for miR-21-5p predicted by TargetScan 7.1 (Fig. 5a). As shown in Fig. 5b, agomiR-21-5p significantly decreased the relative luciferase activity of HEK-293T cells co-transfected with pmiR-SOCS6-W, but agomiR-NC could not bring about this change; the relative luciferase activity did not change in the pmiR-SOCS6-M group wherever it was co-transfected with agomiR-21-5p or agomiR-NC. In addition, agomiR-21-5p depressed the expression of SOCS6 protein in KK47 and T24 cells, and antagomiR-21-5p showed the opposite result (Fig.5c). Accordingly, SOCS6 gene was a target gene of miR-21-5p in BC cells.
Fig. 5. SOCS6 was a target gene of miR-21-5p in BC cells.
a The predicted miR-21-5p binding site in the 3′UTR of SOCS6 mRNA. Short vertical lines indicated complementary nucleotides. b The relative luciferase reporter assay of HEK 293 T cells co-transfected with pmiR-SOCS6-W or pmiR-SOCS6-M and agomiR-21-5p or agomiR-NC. **P < 0.01 vs. pmiR-SOCS6-W + agomiR-NC group. c The expression of SOCS6 protein in KK47 and T24 cells. *P < 0.05 vs. agomiR-NC group, **P < 0.01 vs. agomiR-NC group; #P < 0.05 vs. antagomiR-NC group, ##P < 0.01 vs. antagomiR-NC group. d The expression of SOCS6 protein in KK47 and T24 cells. **P < 0.01 vs. pUC-NC + agomiR-NC group; ##P < 0.01 vs. pUC-NBAT1 + agomiR-NC group
Furthermore, western blotting assay showed that NBAT1 enhancement upregulated the expression of SOCS6 protein in KK47 and T24 cells, and agomiR-21-5p could rescue this upregulation (Fig. 5d), which proved that NBAT1 positively regulated the expression of SOCS6 through interacting with miR-21-5p.
Upregulation of SOCS6 inhibited malignant cellular phenotypes of BC cells
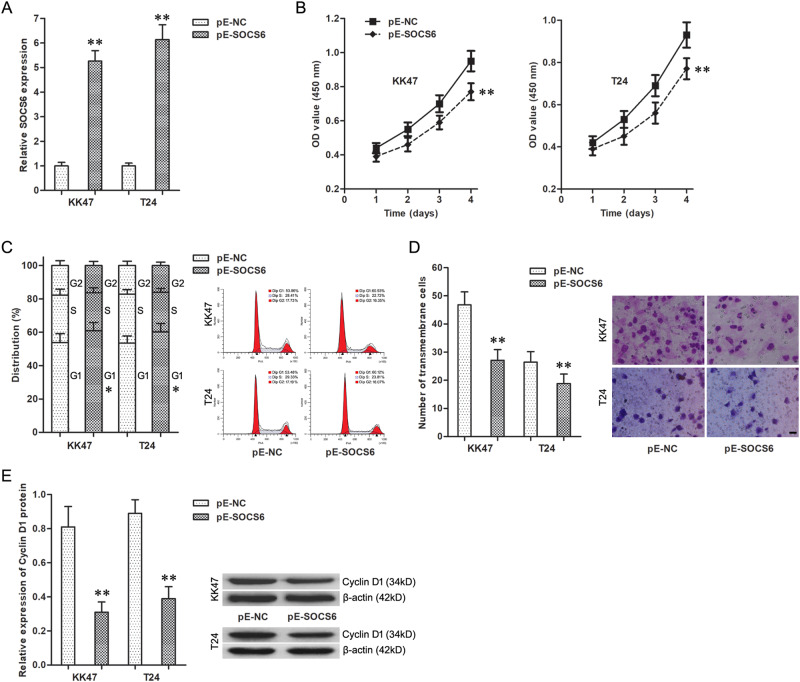
First, transfection with pE-SOCS6 significantly upregulated the expression of SOCS6 in KK47 and T24 cells (Fig. 6a). And, the enhanced CCK8, flow cytometry, and cell invasion assays showed that upregulation of SOCS6 depressed cell viability of KK47 and T24 cells (Fig. 6b), arrested KK47 and T24 cells at G1 stage (Fig. 6c), and inhibited invasiveness of KK47 and T24 cells (Fig. 6d). Meanwhile, western blotting assay showed that the G1-stage checkpoint protein cyclin D1 expression level was significantly increased when KK47 and T24 cells occurred in G1-stage block (Fig. 6e).
Fig. 6. Upregulation of SOCS6 inhibited malignant cellular phenotypes of BC cells.
a The expression of SOCS6 gene in KK47 and T24 cells. **P < 0.01 vs. pE-NC group. b The cell viability of KK47 and T24 cells. **P < 0.01 vs. pE-NC group. c The cell cycle of KK47 and T24 cells. *P < 0.05 vs. pE-NC group. d The number of transmembrane KK47 and T24 cells. Scale bars represent 20 μm. **P < 0.01 vs. pE-NC group. e The expression of Cyclin D1 protein in KK47 and T24 cells. *P < 0.01 vs. pE-NC group
MiR-21-5p modulated the malignant cellular phenotypes of BC cells by silencing SOCS6 expression
To determine whether the miR-21-5p-mediated malignant cellular phenotypes of BC cells were regulated by SOCS6, T24 cells were co-transfected with antagomiR-21-5p and pS-SOCS6 or pS-NC. Figure 7a demonstrated co-transfection with antagomiR-21-5p and pS-SOCS6 led to an obvious increase of SOCS6 expressions in T24 cells. The cellular phenotype detection found that, compared with antagomiR-21-5p and pS-NC group, the antagomiR-21-5p and pS-SOCS6 group displayed stronger proliferation and invasiveness as well as the weaker G1-stage arrest (Fig. 7b–e).
Fig. 7. MiR-21-5p modulated the malignant cellular phenotypes of BC cells by silencing SOCS6 expression.
a The expression of SOCS6 protein in KK47 and T24 cells. *P < 0.05 vs. antagomiR-NC + pS-NC group; ##P < 0.01 vs. agomiR-21-5p + pS-NC group. b The cell viability of KK47 and T24 cells. **P < 0.01 vs. antagomiR-NC + pS-NC group; ##P < 0.01 vs. agomiR-21-5p + pS-NC group. c The cell cycle of KK47 and T24 cells. *P < 0.05 vs. antagomiR-NC + pS-NC group; #P < 0.01 vs. agomiR-21-5p + pS-NC group. d The number of transmembrane KK47 and T24 cells. Scale bars represent 20 μm. **P < 0.01 vs. antagomiR-NC + pS-NC group; ##P < 0.01 vs. agomiR-21-5p + pS-NC group
These findings indicated that miR-21-5p modulated the malignant cellular phenotypes of BC cells by silencing SOCS6 expression.
Discussion
Accumulating research found that lncRNAs were involved in modulation of malignant cellular phenotypes in almost all malignant tumors and could be predictive biomarkers for metastasis and survival in various cancers. For instance, gastric cancer-associated transcript 3 (GACAT3) gene advanced cell proliferation and invasion of colorectal cancer cells;27 LINC00152 was a predictive biomarker of metastasis and survival in various cancers;28 our previous study found that growth arrest-specific 5 (GAS5) gene inhibited the malignant proliferation and doxorubicin resistance of BC, and was an independent prognostic biomarker for BC11.
Till now, there is no research on the expression level and functional role of NBAT1 in BC. In our study, NBAT1 gene was low-expressed in BC tissues and cell lines, and its low-expression was positively related with high pathological grade, lymphatic, and distant metastasis of BC, which suggested that NBAT1 gene was involved in the progress and metastasis of BC. Yang C reported that NBAT1 inhibited the growth and metastasis of osteosarcoma cells14. Hu P found that NBAT1 depressed the migration and invasion of breast cancer cells15. Nevertheless, it is still unknown whether NBAT1 takes part in the regulation of malignant cellular phenotypes in BC.
To verify the roles of NBAT1 on malignant cellular phenotypes, the expression of NBAT1 was upregulated in BC cells to carry out a series of gain-of-function assays. Upregulation of NBAT1 inhibited cell proliferation and invasiveness of BC cells and arrested them at G1 stage, which showed that NBAT1 enhancement restricted malignant cellular phenotypes of BC cells. Nevertheless, the underlying mechanism is unknown.
Little attention has been paid to the miR-21-5p-associated malignant cellular phenotypes of BC, and its roles of miR-21-5p are still unclear. Therefore, the correlation between the miR-21-5p expression and BC was examined, and the results found that its high-expression was related with progress and metastasis of BC patients. Furthermore, BC cells with miR-21-5p knockdown showed reduced cell viability and invasiveness as well as G1-stage arrest. Our previous research elucidated that miR-21-5p promoted invasion of renal cancer via TCF21–KISS1 pathway29.
Some lncRNAs could combine with microRNAs and form complex regulatory networks, and then modulate the expression and function of microRNAs30,31. In addition, noncoding RNAs ordinarily form ribonucleoprotein (RNP) complexes with their partner proteins to exert their functions and miRNAs assemble with argonaute (Ago) family proteins into the effector complex called RISC that mediates the target gene silencing32. For instance, X-inactive-specific transcript (XIST) could combine and silence miR-152 to exert its tumor-suppressive functions in glioblastoma33. Together with online bioinformatics databases and the published reference14, there may be the above lncRNA-microRNA regulatory model between NBAT1 and miR-21-5p.
Then, a series of experiments were carried out to test this hypothesis. First, there was a negative correlation between the expression of NBAT1 and miR-21-5p in BC tissues, and NBAT1 enhancement inhibited the expression of miR-21-5p in BC cells. Second, NBAT1 could specifically combine with miR-21-5p and downregulated the miR-21-5p expression. Third, miR-21-5p enhancement rescued the inhibitory effect of NBAT1 upregulation on cellular phenotypes. On these grounds, miR-21-5p mediated the regulatory effects of NBAT1 on malignant cellular phenotypes in BC cells.
It is well known that microRNAs exert their biological functions by targeted regulation of the expression of its target genes29,34; miR-21-5p might also regulate malignant cellular phenotypes through silencing its target gene. Subsequently, this conjecture was confirmed based on the following: (1) SOCS6 was confirmed to be a target gene of miR-21-5p; (2) upregulation of SOCS6 inhibited malignant cellular phenotypes of BC cells; (3) silencing of SOCS6 separately reversed the regulatory roles of miR-21-5p knockdown on malignant cellular phenotypes in BC cells. These findings verified that miR-21-5p modulated the malignant cellular phenotypes of BC cells by silencing SOCS6 expression. Similarly, Li ZB reported that miR-21 and miR-183 could simultaneously silence the expression of SOCS6, and then regulate cell viability and invasiveness of hepatocellular carcinoma cells35. Moreover, we found that NBAT1 positively regulated the expression of SOCS6 through interacting with miR-21-5p. Accordingly, NBAT1 inhibited malignant cellular phenotypes of BC through miR-21/SOCS6 axis.
Recent research found that miR-19 could activate JAK2/STAT3 signaling pathway via silencing SOCS6 and in osteosarcoma and promote osteosarcoma growth in vitro and in vivo, including the reduction of G1-S arrest and an increase of the S phase36. JAK2/STAT3 signaling pathway is one of the critical signaling pathways involved in the development and homeostasis in mammals and recent has been reported to involves in the pathogenesis of various cancers including BC37,38. Therefore, JAK2/STAT3 signaling pathway might also be involved in suppressing the modulation of NBAT1/miR-21/SOCS6 axis on malignant cellular phenotypes in BC.
In conclusion, low-expression of NBAT1 is associated with the progress and metastasis of BC, and NBAT1 inhibits malignant cellular phenotypes through miR-21-5p/SOCS6 axis in BC. Our findings help to elucidate the tumorigenesis of BC, and future study will provide a novel therapeutic target for BC.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (No. 81872067, No. 81172408, No. 30901480, and No. 81301834).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by E Candi
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41419-022-05485-2
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
12/7/2022
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41419-022-05485-2
References
- 1.Chen W, et al. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Martinez Rodriguez RH, Buisan Rueda O, Ibarz L. Med Clin (Barc) 2017. Bladder cancer: present and future; pp. 449–455. [DOI] [PubMed] [Google Scholar]
- 4.Sanli O, Lotan Y. Current approaches for identifying high-risk non-muscle invasive bladder cancer. Expert. Rev. AntiCancer Ther. 2018;18:223–235. doi: 10.1080/14737140.2018.1432358. [DOI] [PubMed] [Google Scholar]
- 5.Aryal, B. & Suárez, Y. Non-coding RNA regulation of endothelial and macrophage functions during atherosclerosis. Vascul. Pharmacol.10.1016/j.vph.2018.03.001 (2018). [DOI] [PMC free article] [PubMed]
- 6.Schmitt AM, Chang HY. Long noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao D, Zhang Y, Song L. MiR-16-1 targeted silences far upstream element binding protein 1 to advance the chemosensitivity to adriamycin in gastric cancer. Pathol. Oncol. Res7. 2018;24:483–488. doi: 10.1007/s12253-017-0263-x. [DOI] [PubMed] [Google Scholar]
- 8.Guo Y, et al. Long non-coding RNA CEBPA-AS1 correlates with poor prognosis and promotes tumorigenesis via CEBPA/Bcl2 in oral squamous cell carcinoma. Cancer Biol. Ther. 2018;19:205–213. doi: 10.1080/15384047.2017.1416276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang C, Tang W, Pan C, Hu X, Hong Y. Long non-coding RNA TUSC7 inhibits temozolomide resistance by targeting miR-10a in glioblastoma. Cancer Chemother. Pharmacol. 2018;81:671–678. doi: 10.1007/s00280-018-3522-y. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, et al. Silence of long noncoding RNA NEAT1 inhibits malignant biological behaviors and chemotherapy resistance in gastric cancer. Pathol. Oncol. Res. 2018;24:109–113. doi: 10.1007/s12253-017-0233-3. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, Guo Y, Song Y, Shang C. Long noncoding RNA GAS5 inhibits malignant proliferation and chemotherapy resistance to doxorubicin in bladder transitional cell carcinoma. Cancer Chemother. Pharmacol. 2017;79:49–55. doi: 10.1007/s00280-016-3194-4. [DOI] [PubMed] [Google Scholar]
- 12.Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol. Cancer. 2018;17:61. doi: 10.1186/s12943-018-0812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandey GK, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 14.Yang C, Wang G, Yang J, Wang L. Long noncoding RNA NBAT1 negatively modulates growth and metastasis of osteosarcoma cells through suppression of miR-21. Am. J. Cancer Res. 2017;7:2009–2019. [PMC free article] [PubMed] [Google Scholar]
- 15.Hu P, et al. NBAT1 suppresses breast cancer metastasis by regulating DKK1 via PRC2. Oncotarget. 2015;6:32410–32425. doi: 10.18632/oncotarget.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y, et al. MiR-21-5p, miR-34a, and human telomerase RNA component as surrogate markers for cervical cancer progression. Pathol. Res. Pract. 2018;214:374–379. doi: 10.1016/j.prp.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Shang C, Guo Y, Hong Y, Liu YH, Xue YX. MiR-21 up-regulation mediates glioblastoma cancer stem cells apoptosis and proliferation by targeting FASLG. Mol. Biol. Rep. 2015;42:721–727. doi: 10.1007/s11033-014-3820-3. [DOI] [PubMed] [Google Scholar]
- 18.Ohno R, et al. Both cancerous miR-21 and stromal miR-21 in urothelial carcinoma are related to tumour progression. Histopathology. 2016;69:993–999. doi: 10.1111/his.13032. [DOI] [PubMed] [Google Scholar]
- 19.Cai L, et al. MicroRNA-21-5p induces the metastatic phenotype of human cervical carcinoma cells in vitro by targeting the von Hippel-Lindau tumor suppressor. Oncol. Lett. 2018;15:5213–5219. doi: 10.3892/ol.2018.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, et al. Formononetin inhibits human bladder cancer cell proliferation and invasiveness via regulation of miR-21 and PTEN. Food Funct. 2017;8:1061–1066. doi: 10.1039/C6FO01535B. [DOI] [PubMed] [Google Scholar]
- 21.Yuan, D. et al. SOCS6 functions as a tumor suppressor by inducing apoptosis and inhibiting angiogenesis in human prostate cancer. Curr Cancer Drug Targets. 2018. 10.2174/1568009618666180102101442. [Epub ahead of print] [DOI] [PubMed]
- 22.Xue X, et al. MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget. 2016;7:84508–84519. doi: 10.18632/oncotarget.13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng L, Kong B, Zhao Y, Jiang J. miR-494 inhibits cervical cancer cell proliferation through upregulation of SOCS6 expression. Oncol. Lett. 2018;15:3075–3080. doi: 10.3892/ol.2017.7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabir NN, Sun J, Rönnstrand L, Kazi JU. SOCS6 is a selective suppressor of receptor tyrosine kinase signaling. Tumour Biol. 2014;35:10581–10589. doi: 10.1007/s13277-014-2542-4. [DOI] [PubMed] [Google Scholar]
- 25.Sanders KA, et al. Next-generation sequencing reveals broad down-regulation of microRNAs in secondary progressive multiple sclerosis CD4+ T cells. Clin. Epigenetics. 2016;8:87. doi: 10.1186/s13148-016-0253-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shang C, Guo Y, Hong Y, Xue YX. Long Non-coding RNA TUSC7, a Target of miR-23b, Plays Tumor-Suppressing Roles in Human Gliomas. Front. Cell. Neurosci. 2016;10:235. doi: 10.3389/fncel.2016.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou W, Wang L, Miao Y, Xing R. Novel long noncoding RNA GACAT3 promotes colorectal cancer cell proliferation, invasion, and migration through miR-149. Onco. Targets Ther. 2018;11:1543–1552. doi: 10.2147/OTT.S144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Jiaming, Yin Minuo, Huang Junming, Lv Zhengtao, Liang Shuang, Miao Xiaoao, Huang Fang, Zhao Yingchao. Long noncoding RNA LINC00152 as a novel predictor of lymph node metastasis and survival in human cancer: a systematic review and meta-analysis. Clinica Chimica Acta. 2018;483:25–32. doi: 10.1016/j.cca.2018.03.034. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Guo Y, Shang C, Song Y, Wu B. miR-21 downregulated TCF21 to inhibit KISS1 in renal cancer. Urology. 2012;80:1298–1302.e1. doi: 10.1016/j.urology.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Shuwen H, Qing Z, Yan Z, Xi Y. Competitive endogenous RNA in colorectal cancer: A systematic review. Gene. 2018;645:157–162. doi: 10.1016/j.gene.2017.12.036. [DOI] [PubMed] [Google Scholar]
- 31.Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell. Mol. Life Sci. 2018;75:67–484. doi: 10.1007/s00018-017-2626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi H, Tomari Y. RISC assembly: coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta. 2016;1859:71–81. doi: 10.1016/j.bbagrm.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Cheng Z, et al. Long non-coding RNA XIST promotes glioma tumorigenicity and angiogenesis by acting as a molecular sponge of miR-429. J. Cancer. 2017;8:4106–4116. doi: 10.7150/jca.21024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie D, Shang C, Zhang H, Guo Y, Tong X. Up-regulation of miR-9 target CBX7 to regulate invasion ability of bladder transitional cell carcinoma. Med. Sci. Monit. 2015;21:225–230. doi: 10.12659/MSM.893232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li ZB, Li ZZ, Li L, Chu HT, Jia M. MiR-21 and miR-183 can simultaneously target SOCS6 and modulate growth and invasion of hepatocellular carcinoma (HCC) cells. Eur. Rev. Med. Pharmacol. Sci. 2015;19:3208–3217. [PubMed] [Google Scholar]
- 36.Sun Z, Liu Q, Hong H, Zhang H, Zhang T. miR-19 promotes osteosarcoma progression by targeting SOCS6. Biochem. Biophys. Res. Commun. 2018;495:1363–1369. doi: 10.1016/j.bbrc.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Dodington DW, Desai HR, Woo M. JAK/STAT - emerging players in metabolism. Trends Endocrinol. Metab. 2018;29:55–65. doi: 10.1016/j.tem.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Wu Q, Zhao Y, Wang P. miR-204 inhibits angiogenesis and promotes sensitivity to cetuximab in head and neck squamous cell carcinoma cells by blocking JAK2-STAT3 signaling. Biomed. Pharmacother. 2018;99:278–285. doi: 10.1016/j.biopha.2018.01.055. [DOI] [PubMed] [Google Scholar]