Abstract
Limbal epithelial stem cells (LESC) maintenance requires communication between stem cells and neighboring stromal keratocytes. Extracellular vesicles (EVs) are important for intercellular communication in various stem cell niches. We explored the regulatory roles of limbal stromal cell (LSC)-derived exosomes (Exos), an EV sub-population, in limbal epithelial cells (LEC) in normal and diabetic limbal niche and determined differences in Exo cargos from normal and diabetic LSC. Wound healing and proliferation rates in primary normal LEC were significantly enhanced upon treatment by normal Exos (N-Exos), but not by diabetic Exos (DM-Exos). Western analysis showed increased Akt phosphorylation in wounded LECs and organ-cultured corneas treated with N-Exos, compared to untreated wounded cells and DM-Exos treated fellow corneas, respectively. N-Exos treated organ-cultured corneas showed upregulation of putative LESC markers, keratin 15 (K15) and Frizzled-7, compared to the DM-Exos treated fellow corneas. By next generation sequencing, we identified differentially expressed small RNAs including microRNAs in DM-Exos vs. N-Exos. Overall, N-Exos have greater effect on LEC proliferation and wound healing than DM-Exos, likely by activating Akt signaling. The small RNA differences in Exos from diabetic vs. normal LSC could contribute to the disease state. Our study suggests that exosomes may serve as novel therapeutic tools for diabetic cornea.
Introduction
The homeostasis of corneal epithelium is a dynamic and complex process that plays a key role in the corneal transparency and visual function. Renewal of terminally differentiated central corneal epithelium, essential of transparency, is orchestrated through the differentiation and centripetal migration by the limbal epithelial stem cells (LESC). Thus, maintenance of LESC in their niche environment is critical for proper functioning of corneal epithelium. LESC are quiescent cells located at the corneal periphery in the corneoscleral limbus inside specific structures called palisades of Vogt1, and/or in the deeper limbal epithelial crypts and focal stromal projections2–4. Unlike central epithelial cells, LESC are in close contact with the cells in the underlying limbal stroma and the vasculature that surrounds the limbal crypts5.
It is well established that limbal stromal cells (LSC), or keratocytes, support limbal epithelial cell (LEC) homeostasis through their secreted soluble factors6–9. Central corneal stroma promotes cell proliferation and differentiation, whereas limbal stroma helps maintaining cell stemness10. Therefore, the stem cell maintenance and function in normal and diseased states of the cornea involve complex interactions of various intrinsic and extrinsic factors between all the resident cell types as well as infiltrating cells from the circulation in the limbal stromal microenvironment, or niche. Any damage to LESC or limbal stromal niche due to the external insults or diseases such as diabetes may lead to pathological state of altered vision, and in severe cases of LESC loss may lead to limbal stem cell deficiency (LSCD) and blindness.
Recent studies have shown the important roles of extracellular vesicles (EVs), in addition to direct cell-cell contact or secreted molecules, in communication between the surrounding cells and ECM in stem cell maintenance and activation11. There are different types of EVs including exosomes, microvesicles, ectosomes or shedding vesicles, which differ by their subcellular origin, type of release, and size, and are secreted by most cells and contain mRNA, microRNA, DNA and protein cargo mediating physiological intercellular crosstalk12. An EV sub-population, exosomes (Exos), are small endosomal membrane-bound vesicles about 50–200 nm, with a range of nucleic acids and proteins contents, which differ with the cell and tissue of origin12. They exert their effects by fusion to the target cells and transferring their cargo, which may include bioactive molecules such as proteins, lipids, mRNAs and miRNAs12. The important roles of exosomes have been shown in pathological conditions, such as cancer13, inflammation14, cardiovascular diseases15,16, diabetes17,18, as well as in wound healing19,20. Exosome-like vesicles were described between central corneal epithelial cells and the stroma during wound healing after epithelial debridement of mouse cornea21.
In this study, we characterized both healthy or normal (N) and diabetic (DM) human limbal LSC-derived Exos and examined their roles in survival, migration and proliferation of LEC in normal and diabetic corneas. Furthermore, next generation sequencing (NGS) was performed to identify distinct miRNA players and investigate the effect of diabetes on LSC-derived Exo population. Our study indicates that normal LSC-derived Exos (N-Exos) have a greater potential in cell proliferation and wound healing than diabetic LSC-derived Exos (DM-Exos). Using NGS analysis, we have also documented differences in Exo cargos derived from normal and diabetic limbal keratocytes.
Results
Primary limbal stromal cells (keratocytes) characterization
Corneal stromal cells isolated from the limbal region were characterized based on their morphology and specific marker expression. The morphology was typical of LSC22,23, that is, elongated, spindle-shaped with long spreading cellular processes, which was maintained at confluence and at later passages (Supplementary Fig. S1a). Immunocytochemistry showed positive staining for lumican, keratocan and aldehyde dehydrogenase 3 (ALDH3), which are specific keratocyte markers (Supplementary Fig. S1b). The staining for myofibroblast marker α-SMA, and corneal epithelial marker keratin 12 (K12) was negative (Supplementary Fig. S1b). Flow cytometry further confirmed the expression of lumican and ALDH3 (Supplementary Fig. S1c). Western blot analysis also showed lumican expression in both normal and diabetic keratocytes (Supplementary Fig. S1d).
Characterization of normal and diabetic human limbal LSC-derived Exos
Exos derived from conditioned media of at least three biological replicates of each normal and diabetic keratocytes were characterized by several analytic approaches. The typical cup shape EV morphology was detected by transmission electron microscopy (TEM, Fig. 1a). EV size was measured using TEM and NanoSight technology (Fig. 1a,b) with the size ranging between 50 and 200 nm (mean size 159.3 nm for normal EVs and 145.3 nm for diabetic EVs). We used common exosome markers to characterize normal and diabetic human LSC-derived EVs by flow cytometry and western blotting (Fig. 1c,d). Flow cytometry using BD LSR II instrument (BD Biosciences, San Jose, CA), showed that both normal and diabetic EVs were positive for both CD63 and CD81, with no significant difference (using Student’s t-Test, n = 3, p > 0.05) between two groups (Fig. 1c). Western analysis of EVs showed positive bands for CD63 and heat-shock protein (HSP)70 (Fig. 1d). The data suggested that the majority of our isolated EVs were exosomes.
Figure 1.
Characterization of normal and DM human LSC-derived Exos. (a) Representative TEM images showing a range of exosomal size from 50–200 nm and typical doughnut shape morphology in both N (n = 3) and DM (n = 3) human LSC-derived Exos. (b) size distribution of LSC-derived EVs was determined by NanoSight LM10. Histogram shows particle size distribution typical of exosomes. (c) Flow cytometry was performed on normal (healthy)- and DM-EVs using anti-CD63-coated beads. Vesicles were immunostained against CD63 (red) and CD81 (red) and compared with appropriate isotype control (blue), n = 3. (d) Western blot showed expression of typical exosomal markers HSP70 and CD63 in both N (n = 3) and DM (n = 3) vesicles.
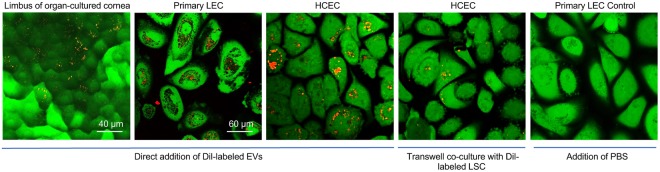
Internalization of Exos by cultured cells and organ-cultured corneas
To determine whether human limbal epithelial cells are the targets for LSC-derived Exos, Dil red fluorescent dye was used to label Exos. Primary LEC were treated with 10 and 25 μg/ml Dil-labeled LSC-derived Exos for 24 h to optimize the internalization of Exos by cultured cells (Supplementary Fig. S2). After incubating the labeled Exos (25 μg/ml or 3 × 108/ml; this dose was chosen for all other experiments) with organ-cultured corneas, primary LEC and telomerase immortalized human corneal epithelial cells (HCEC) for 24 h, the uptaken Exos were observed in limbal region of organ-cultured corneas (Fig. 2) and in perinuclear region of both cell types, LEC and HCEC, by confocal microscopy (Fig. 2), demonstrating the internalization of labeled Exos by live cells. Similar results were observed in co-culture system where Dil-labeled human LSC were seeded onto the 0.4 µm inserts in transwell system with HCEC as a recipient (Fig. 2).
Figure 2.
Dil-labeled normal human LSC-derived EVs can be internalized by human primary LEC, HCEC and organ-cultured corneas. Dil-labeled LSC-derived Exos (25 μg/ml or 3 × 108/ml) were incubated (direct addition of Dil-labeled Exos) with organ-cultured corneas, primary LEC and HCEC for 24 h, no Exo was added to the control. Dil-labeled normal keratocytes were co-cultured up to 48 h on trans-wells. The cells were stained with calcein-AM, which exhibits green fluorescence and demonstrates live cells and their uptake of EVs.
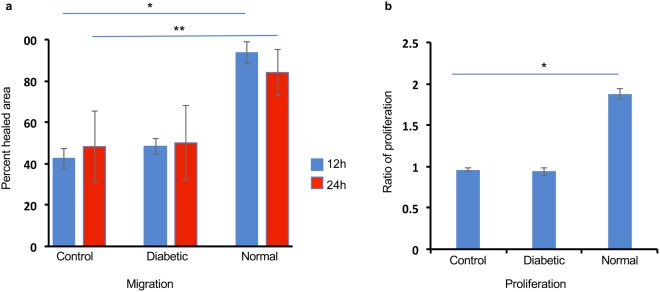
Exos derived from normal but not DM LSC enhance cell proliferation and wound healing rate in normal LEC in vitro
Scratch-wounded cultured LEC treated with N-Exos increased wound healing rate compared to untreated control cells (Fig. 3a). In addition, LEC treatment with N-Exos for 24 h increased cell proliferation rate compared to untreated control cells (Fig. 3b). Interestingly, DM-Exos didn’t exert significant effects on proliferation or migration rates in treated LEC compared to control cells (Fig. 3a,b). Additionally, immunostaining of normal primary LECs treated with N-Exos showed a similar increase in Ki-67 compared to those treated with DM-Exos or untreated control cells (Supplementary Fig. S3). DM-Exo treatments decreased the expression of Ki-67 compared to untreated control cells (Supplementary Fig. S3).
Figure 3.
Normal EVs increase migration and proliferation of normal primary LEC. (a) LEC were scratch-wounded and incubated with 25 μg/ml N/DM LSC-derived EV, and wound closure was quantified using ImageJ software at 12 and 24 hr after wounding. Cell migration and wound closure were significantly enhanced in normal primary LEC treated with N-Exos compared to the cells treated with DM-Exos or PBS/untreated control cells. DM-Exos treatments did not change the wound healing rate compared to control. (b) MTS proliferation assay showed increase in proliferation rate in LEC treated with N-Exos vs. those treated with DM-Exos or untreated control cells. DM-Exo treatments did not change the proliferation rate compared to control. The bar graph represents average ± SEM of pooled values of three independent triplicate assays and compared to untreated control cells (negative control). **p < 0.01, *p < 0.05 by paired two-tailed t test.
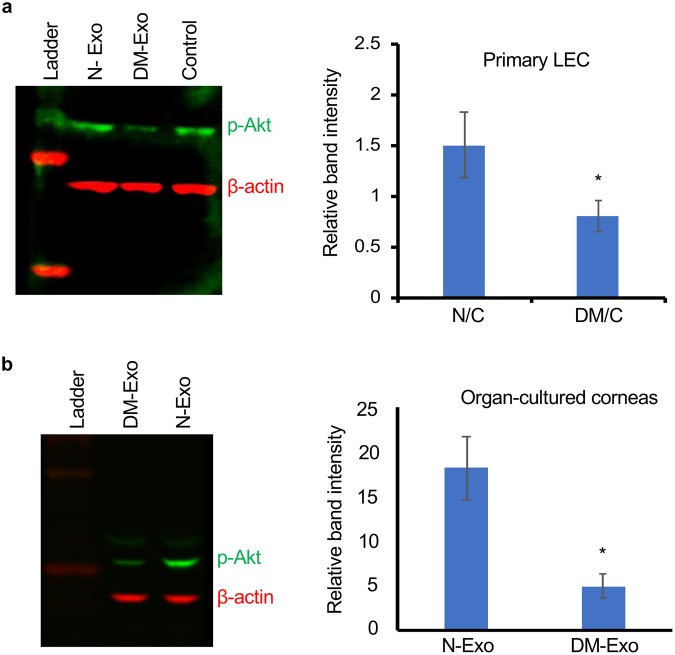
Effects of Exos on activation of wound healing-related signaling molecules in primary LEC and organ-cultured corneas
To investigate the effect of normal and diabetic Exos on signaling pathways during wound healing, LEC cultures and organ-cultured corneas were wounded and treated with either N-Exos or DM-Exos. Western analysis of wound healing related signaling molecules showed significantly increased expression of p-Akt in wounded LEC incubated with N-Exos compared to control untreated cells, whereas DM-Exos treated wounded cells showed significantly decreased expression of p-Akt compared to control untreated cells (Fig. 4a). Similarly, wounded organ-cultured corneas incubated with N-Exos showed increased level of p-Akt protein levels in comparison with the fellow corneas incubated with DM-Exos (Fig. 4b). However, there were no significant differences in levels of p-p38 among any treatments both in LEC cultures and organ-cultured corneas (Supplementary Fig. S4).
Figure 4.
Western blot analysis of p-Akt expression in wounded LEC and organ-cultured corneas treated with normal or DM LSC-derived Exos. (a) Total extracted protein from wounded primary LEC treated with N/DM Exos and untreated cells (control) was separated on gradient SDS-PAGE gels, transferred to nitrocellulose and probed with antibodies to p-Akt. Normal-Exo treatment increases protein levels of p-Akt vs. control (PBS/untreated) and DM-Exo treated cells. (b) Western analysis showed increases p-Akt expression in wounded organ-cultured corneas treated with N-Exos compared to the fellow corneas treated with DM-Exos. Antibody to β-actin was used as equal loading control and for semi-quantitation. All experiments were performed in triplicate. *p < 0.05 by paired two-tailed t test.
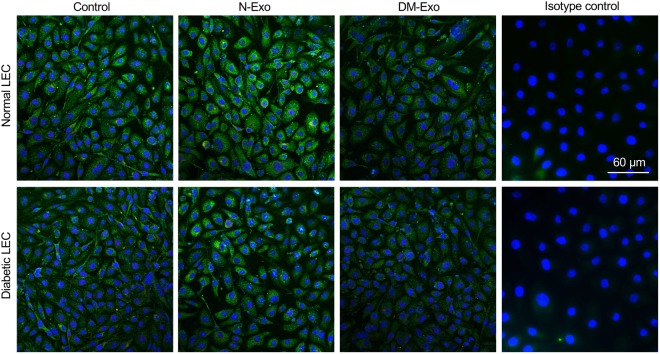
Effects of Exos on LESC markers
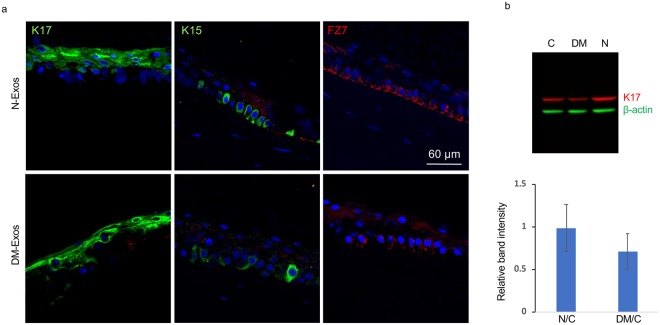
The effects of N-Exos and DM-Exos on the expression of putative LESC markers were examined in primary normal LEC cultures and in organ-cultured corneas by immunostaining and western blot. Our data showed increased staining of putative stem cell marker, K15, in LEC treated with N-Exos compared to DM-Exo treated cells or untreated control LEC (Fig. 5). In addition, normal organ-cultured corneas treated with N-Exos showed upregulation, whereas corneas treated with DM-Exos showed downregulation of putative LESC markers K15, frizzled-7 (FZ7), and to some degree K17 (Fig. 6a). Western blot analysis showed N-Exo treatment increased, while the DM-Exo treatment decreased K17 protein expression level in primary LEC compared to control, which did not reach significance (Fig. 6b).
Figure 5.
Effect of N and DM Exos on the expression of putative LESC marker, K15, in normal and DM primary LEC. Both normal and DM LEC treated with N-Exos increased K15 expression in comparison with untreated control or DM-Exo treated cells. DM-Exo treated cells showed no change in K15 expression level in comparison to control cells. IgG2aκ isotype control antibody (Thermo Fisher Scientific) was used as negative control. The experiments were performed at least in triplicate and same exposure time was used for each set of compared stained sections, and the assessment was done by more than one observer.
Figure 6.
Effect of N- or DM-Exos on LESC marker expression in normal organ-cultured corneas and primary LEC. (a) Normal Exo treatment in normal organ-cultured corneas led to increased expression of putative LESC markers, K15 and FZ7, and no significant change in K17 protein level compared to fellow corneas treated with DM-Exos (immunofluorescent staining of limbal corneal sections). The same exposure time was used for each set of compared stained sections, and the assessment was done by more than one observer. (b) Western analysis shows that N-Exos treatment increased, whereas DM-Exo treatment decreased K17 protein expression level in primary LEC compared to control treated cells, which did not reach significance. Antibody to β-actin was used as equal loading control and for semi-quantitation.
Distinct small RNA profile of normal and diabetic LSC-derived Exos
We conducted a comprehensive analysis of small RNAs including microRNA expression in normal and diabetic LSC-derived Exos by NGS. On average, 24.4 million reads were obtained per sample and the average reads for the vast majority of the data had Q score greater than Q30. Mapping of the sequencing reads aligned to the abundant sequences such as poly A, ribosomal RNA and the mitochondrial chromosomes, as well as to the reference genome including fragments of mRNA and lncRNA transcripts, to small RNA database, and to miRBase. A total of 772 and 337 small RNAs including piRNA, snoRNA and Y_RNA were identified in all samples with the average threshold of more than one Taq Per Million (TPM) > 1 and TPM > 10, respectively (Supplementary Dataset S1).
The number of known miRNAs was calculated after mapping the data and counting relevant entries in miRBase 20. A total of 297 and 150 known miRNAs were identified in all samples with the average threshold of more than one Taq Per Million (TPM) > 1 and TPM > 10 respectively (Supplementary Dataset S2).
The IsomiR analysis was performed for each sample based on the occurrence of count variants for each detected miRNA. These variants were identified by changes in start or stop position, or occurrence of mutations within the read. IsomiRs were not included in differential expression analysis.
A set of 312 small RNAs including miRNAs was identified as differentially expressed in DM-Exos vs. N-Exos with the raw p < 0.05 and fold change of greater than 2 (Supplementary Dataset S3). Of these 312 small RNAs and miRNAs 219 showed a false discovery rate (FDR)-adjusted p < 0.05 (Supplementary Dataset S3). miRNA analysis revealed 66 (54 upregulated and 12 downregulated) and 10 (5 upregulated and 5 downregulated) differentially expressed miRNA in DM-Exos vs. N-Exos with the raw p < 0.05 and FDR < 0.05 respectively. Table 1 shows the 20 most differentially expressed miRNAs, and a full list of differentially expressed miRNAs is given in the Supplementary Dataset S4. The Volcano plot shows a quick visual identification of miRNAs with large-magnitude changes, which are also statistically significant (Fig. 7). The plot is constructed by plotting the p-value (−log10) on the y-axis, and the expression fold change between the two experimental groups (N-Exos vs. DM-Exos) on the x-axis. The top of the plot (high statistical significance) and the extreme left or right (strongly down- and up-regulated respectively) are the two regions of interest (Fig. 7).
Table 1.
List of the 20 most significantly differentially expressed microRNAs and annotation.
| Names | Log FC | Log CPM | P Value | FDR | Diabetic | Normal |
|---|---|---|---|---|---|---|
| has-miR-4516 | −8.716 | 6.458 | 1.81E-09 | 6.02E-07 | 0.92 | 216.8 |
| has-miR-184 | 6.534 | 15.435 | 5.08E-08 | 8.47E-06 | 76887.86 | 827.6 |
| has-miR-4449 | −7.263 | 4.216 | 9.35E-06 | 0.001037 | 0.23 | 52.23 |
| has-miR-4461 | −5.725 | 3.523 | 6.95E-05 | 0.005783 | 0.5 | 32.87 |
| has-miR-7704 | −3.610 | 5.939 | 0.000195 | 0.011698 | 11.4 | 135.75 |
| has-miR-3195 | −4.076 | 4.882 | 0.000211 | 0.0698 | 4.15 | 71.59 |
| has-miR-708-5p | 9.756 | 5.546 | 0.00028 | 0.013308 | 70.76 | 0 |
| has-miR-200b-3p | 7.604 | 8.513 | 0.00075 | 0.031223 | 622.97 | 2.25 |
| has-miR-200c-3p | 12.523 | 8.152 | 0.001051 | 0.038903 | 483.32 | 0 |
| has-let-7c-5p | 2.676 | 12.326 | 0.001335 | 0.044449 | 8032.43 | 1257.32 |
| has-miR-23b-3p | 4.107 | 8.846 | 0.002746 | 0.076447 | 757.39 | 46.62 |
| has-miR-146a-5p | −2.747 | 14.633 | 0.002755 | 0.076447 | 7372.85 | 49485.89 |
| has-miR-103a-3p | 3.359 | 9.607 | 0.004341 | 0.107733 | 1258.86 | 125.21 |
| has-miR-1304-3p | 8.507 | 4.447 | 0.004885 | 0.107733 | 30.04 | 0 |
| has-miR-99a-5p | 2.882 | 14.769 | 0.005247 | 0.107733 | 44332.22 | 6011.66 |
| has-miR-142-5p | 8.507 | 4.803 | 0.005273 | 0.107733 | 39.82 | 0 |
| has-miR-500a-3p | 3.641 | 4.945 | 0.0055 | 0.107733 | 42.81 | 2.25 |
| has-miR-27b-5p | 3.154 | 5.813 | 0.006391 | 0.113713 | 82.84 | 6.75 |
| has-miR-107 | 4.005 | 5.472 | 0.006785 | 0.113713 | 64.51 | 5.18 |
| has-miR-655-3p | −3.512 | 2.974 | 0.006996 | 0.113713 | 1.84 | 25.22 |
| has-miR-125b-3p | 2.948 | 7.837 | 0.007332 | 0.113713 | 352.81 | 48.87 |
Log fold change (log FC) between diabetic and normal Exos, raw p-values, Benjamini-Hochberg FDR corrected p-values as well as the average TMM values per group.
Figure 7.

Volcano plot displaying differentially expressed miRNAs between normal and diabetic exosomes. The Y-axis corresponds to the mean expression value of log 10 (p-value), and the X-axis displays the log2 fold change, N-Exos vs. DM-Exos.
Gene ontology enrichment and pathway analyses
Gene ontology (GO) enrichment analysis was performed to identify GO terms that are significantly associated with differentially expressed miRNAs in DM-Exos vs. N-Exos identified to their target genes. Using miRSearch, we mapped the differentially expressed miRNAs (Supplementary Dataset S4) to their target genes and investigated whether specific GO terms were associated with these miRNAs. First, a standard Fisher’s test was used to investigate enrichment of terms between the two test groups. Second, the Elim method was used for more conservative approach by incorporating the topology of the GO network to compensate for local dependencies between GO, which can mask significant GO terms. Comparisons of the predictions from these two methods can highlight truly relevant GO terms. The top 20 most significant GO terms for the target genes found to be differentially expressed in DM- vs. N-Exos and their corresponding annotation for Biological process (BP) are shown in Table 2. Complete GO enrichment analysis of the comparisons of related terms, Molecular functions (MF) and Cellular components (CC) analyses in addition for BP is presented in Supplementary Dataset S5.
Table 2.
List of the top 20 most significant GO terms for the target genes found to be differentially expressed in DM-Exos vs. N-Exos and their corresponding annotation for Biological process (BP).
| GO ID | Term | P value |
|---|---|---|
| GO: 0045471 | Response to ethanol | 5.30-E-05 |
| GO: 0042593 | Glucose homeostasis | 0.00016 |
| GO: 0045616 | Regulation of keratinocyte differentiation | 0.00018 |
| GO: 0051150 | Regulation of smooth muscle cell differentiation | 0.00025 |
| GO: 0060017 | Parathyroid gland development | 0.00048 |
| GO: 0007219 | Notch signaling pathway | 0.00054 |
| GO: 0060560 | Developmental growth involved in morphogenesis | 0.00056 |
| GO: 0043406 | Positive regulation of MAP kinase activity | 0.00073 |
| GO: 0007157 | Heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | 0.00075 |
| GO: 0000083 | Regulation of transcription involved in G1/S transition of mitotic cell cycle | 0.00096 |
| GO: 0051897 | Positive regulation of protein kinase B signaling | 0.00098 |
| GO: 0030261 | Chromosome condensation | 0.00121 |
| GO: 0001843 | Neural tube closure | 0.00124 |
| GO: 0021846 | Cell proliferation in forebrain | 0.00138 |
| GO: 0050680 | Negative regulation of epithelial cell proliferation | 0.00149 |
| GO: 0005979 | Regulation of glycogen biosynthetic process | 0.00151 |
| GO: 0001755 | Neural crest cell migration | 0.00154 |
| GO: 0045598 | Regulation of fat cell differentiation | 0.0017 |
| GO: 0000077 | DNA damage checkpoint | 0.00176 |
| GO: 0050714 | Positive regulation of protein secretion | 0.00177 |
Discussion
Recent studies have shown that, beside cell-cell contact and soluble factors, EVs constitute an important mechanism for cell-cell communication both in physiological and pathological conditions11,24. Further, EVs play an important role in different stem cell niches such as the mesenchymal25, cancer26, cardiac27, neurogenic28,29 and bone marrow niches30. However, their roles in adult limbal niche remain to be elucidated.
The close spatial arrangement and known communication between putative LESC niche structures and stromal keratocytes31 may suggest that EVs may have a role in LEC-LSC crosstalk. There are few studies on corneal EVs/Exos21,32 and none in stem cell-enriched limbal area and in disease state such as corneal diabetes. In the present study, we isolated and characterized EV sub-population, exosomes, derived from both normal and diabetic human limbal stromal keratocytes. As confirmed by several analytic approaches, they were in the 50–200 nm size range and positive for CD63, CD81 and HSP70 (exosome markers) suggesting that the majority of our isolated EVs were exosomes. To examine the possible role of Exos in LSC-LEC communications, we showed that exogenous Dil-labeled Exos were uptaken by HCEC and primary LECs in vitro and limbal region of ex vivo organ-cultured corneas (Fig. 2). Additionally, endogenous Exos released from Dil-labeled LSC were uptaken by primary LECs in co-culture system, suggesting that Exos are involved in paracrine activity of LSC and LEC in limbal niche.
Recent studies have revealed that Exos can affect many biological processes such as cell proliferation, differentiation, angiogenesis, cell migration and wound healing, through their cargo transfer from the originating cells to the recipient cells33–35. In order to confirm the role of Exos in LEC-LSC crosstalk in limbal niche, we performed functional analysis of Exos derived from LSC on their recipient cells, LEC. Our study demonstrated that epithelial healing was significantly promoted in wounded primary LEC when incubated with N-Exos compared to untreated wounded cells. However, LEC treated with DM-Exos didn’t show any significant changes in wound healing rate compared to control, untreated wounded cells. Similar studies have previously explored the role of Exos in cell repair and wound healing in other cell types such as in skin20,33,36,37, skeletal38 and cardiac39,40 muscle. A very recent study has shown that Exos derived from human amniotic epithelial cells promoted wound healing and inhibited scar formation in skin36. Similarly, a study by Zieske’s group has documented the communication between epithelial cells and keratocytes as well as endothelial cells by Exos secreted by mouse corneal epithelial cells in vitro that may suggest their involvement in corneal wound healing21.
Our study has also shown that proliferation rate is significantly enhanced in primary LEC when incubated with N-Exos compared to untreated cells. Interestingly, LEC treated with DM-Exos showed less or not any significant changes in proliferation compared to control cells. These data show greater potential of normal Exos in stimulating cell proliferation and wound healing than diabetic Exos. This may suggest that there is a difference in exosomes’ cargos derived from normal and diabetic LSC, which might contribute to the disease state. Furthermore, we observed upregulation of wound healing-related signaling molecule, p-Akt, in wounded LEC and organ-cultured corneas treated with N-Exos compared to untreated wounded cells and DM-Exo treated fellow corneas, respectively. It may be suggested that N-Exos cargos may contain signaling molecules such as p-Akt or its upstream signaling molecules and/or specific miRNAs35,41,42 that regulate signaling pathways in their wounded target cells and could actively regulate migration and proliferation in recipient LEC. Additionally, the HSP70 expression in exosomes may promote cell motility beside its other roles such as assisting in proper folding and preventing the aggregation of proteins43.
In our study, we assessed for the first time the role of Exos in LESC survival and maintenance. Immunostaining of LEC treated with N-Exos showed upregulation of putative LESC marker, K15, in comparison to untreated cultured cells or DM-Exo treated cells. In addition, organ-cultured corneas treated with N-Exos showed upregulation of putative LESC markers, K15 and FZ7, compared to the organ-cultured fellow corneas treated with DM-Exos. These data further suggest that LSC-derived Exos may contribute to LSC-LEC crosstalk and maintenance of LESC. Downregulation of both K15 and FZ7 by DM-Exos suggest an important difference in exosome cargos derived from normal and diabetic LSC contributing to the disease state. These data are in line with our previous results on downregulation of a number of putative LESC markers in human diabetic corneas44. In fact, the mechanism of this effect may be related to exosomes secreted by diabetic LSC that may not support normal LESC maintenance.
In all types of EVs including exosomes, miRNAs have been found in large amounts, which may exert various effects in recipient cells due to their key regulatory roles in gene expression45. Thus, we performed comparative exosomal small RNA profiling using NGS analysis (Exiqon) for both normal and DM LSC-derived Exos, which could help us reveal the mechanism of exosomal function in normal and diabetic limbal niche. We quantitatively identified the spectrum of small RNAs (including miRNAs) profiles of N and DM LSC-derived Exos and those miRNAs that abnormally expressed in DM-Exos of diabetic corneas (Supplementary Dataset S3). The top differentially expressed miRNA, miR-4516, has been shown to inhibit skin keratinocyte migration by targeting fibronectin/integrin α9 signaling46. Paradoxically, it was significantly downregulated in DM-Exos although diabetes induces corneal wound healing impairment. As fibronectin is not significantly changed in diabetic corneas, miR-4516 in the cornea may work on other targets, e.g., on MMP-2 that is elevated in the diabetic corneas, presumably in the corneal stroma47. Alternatively, downregulation of miR-4516, may present an attempt of the diabetic stromal cells to preserve normal wound healing, which may be eventually counteracted by other miRs that become elevated in diabetes, for instance, miR-146a that inhibits wound healing-stimulating EGFR48. Interestingly, miR-184, the most abundantly expressed miRNA in central corneal epithelium49,50 is among the top differentially expressed miRNAs and is significantly upregulated in DM-Exos. Its key role in corneal epithelial homeostasis has been shown in several studies50–52. It has been suggested to function in corneal angiogenesis by targeting VEGF and Akt signaling50, regulate the transition from proliferation to early differentiation52, while its abnormal expression or mutation resulted in impaired homeostasis leading to corneal diseases such as familial severe keratoconus51. Therefore, its upregulation in DM-Exos may lead to the alteration of limbal niche homeostasis resulting in diabetic disease state such as altered proliferation and differentiation53,54. Both miR-200b-3p and miR-200c-3p, which are significantly upregulated in DM-Exos, have been shown to inhibit growth and motility55,56, which may contribute to slow wound healing and migration of the diabetic epithelium. Interestingly, miR-146a is another differentially expressed miRNA in DM vs. normal LSC-Exos with the potential of targeting and downregulating signaling molecules and putative stem cell markers, K15 and FZ7, as we have shown previously57.
We also identified the spectrum of small RNAs including, piRNA, tRNA, snRNA and Y_RNAs, in N and DM LSC-derived Exos, and the abnormally expressed small RNAs in DM-Exos. These categories of small RNAs are involved with both epigenetic and post-transcriptional gene silencing, splicing and DNA replication. Further studies are required to determine the functions of the differentially expressed small RNAs including miRNAs in limbal niche.
To identify differentially expressed miRNA target genes that might be responsible for the diabetic corneal abnormalities, gene ontology (GO) and pathway analysis were performed to determine the underlying mechanisms. Biological function, cellular component, and molecular function were the most related terms in the GO analysis. Interestingly, the most significant GO terms include insulin receptor signaling, cell cycle regulators, signaling molecules, and negative regulation of epithelial cell proliferation, which have been shown to be dysregulated in diabetes54. In addition, significant GO terms such as Notch signaling pathway and regulation of keratinocyte differentiation of differentially expressed miRNA target genes suggest the alteration of LESC function observed in diabetic cornea54. These data suggest a disturbed LSC-LEC communication in the diabetic cornea, which may lead to disease alterations.
In conclusion, this is the first study showing the Exos’ role in limbal niche in LSC-LEC communications in healthy and diabetic corneas. We have documented Exos’ influence on a key signaling molecule involved in migration and proliferation and putative LESC marker in in vitro and ex-vivo organ-cultured corneas. Further, we identified both normal and DM LSC-derived Exos’ cargos and differentially expressed small RNAs including miRNAs in DM-Exos, which may have roles in disease state. Further studies are required to determine their functions in normal and diabetic cornea. The presented data may contribute to better understanding of the complexity of the limbal niche in physiological and pathological conditions. This would help us develop more effective therapeutic approaches by targeting the niche and its cellular components for the treatment of corneal diseases such as diabetic keratopathy.
Materials and Methods
Human specimens
Age-matched human autopsy healthy and diabetic corneas (Supplementary Table S1) were received from National Disease Research Interchange (NDRI, Philadelphia, PA) in Optisol storage medium; donor identity was withheld by the supplier. NDRI has a human tissue collection protocol approved by a managerial committee and subject to National Institutes of Health oversight. In all cases the required informed consent from donors next of kin specifying the use of postmortem tissue for research was obtained by NDRI-affiliated eye banks. The work reported here was covered by approved Cedars-Sinai Medical Center IRB protocols EX-1055 and Pro00019393. Corneas were harvested within 5 hours of donor death and reached our laboratory within 24 h of death, and studies were conducted in accordance with approved guidelines.
Isolation and maintenance of primary limbal epithelial and stromal cells
Primary limbal epithelial and stromal cells were dissociated from the age-matched autopsy normal and diabetic limbal rims. LEC containing LESC were isolated from corneoscleral rims by Dispase/Trypsin digestion to dissociate LEC from stroma, and were maintained and characterized as previously described49,57. After removal of epithelial cells by enzymatic digestion, corneal stroma was chopped and kept in 1 mg/ml collagenase type IV solution at 37 °C overnight9. LSC were filtered through a 70 μm mesh, washed and re-suspended in complete culture medium (CCM) [DMEM/F12 supplemented with B27, N2, 1% antibiotic/antimycotic and 5 ng/ml basic fibroblast growth factor (FGF2), PeproTech Inc, Rocky Hills, NJ], plated at 8 × 103 cells/cm2 and kept in the incubator at 37 °C and 5% CO2. Cells were passaged after 70–80% confluence using TrypLE express (Thermo Fisher Scientific, Waltham, MA).
Maintenance of human organ-cultured cornea
Corneal organ cultures were established as described in details58 and were maintained in Dulbecco’s Modified Eagle’s Medium (Thermo Fisher Scientific) with 1X insulin-transferrin-selenite (Sigma-Aldrich), 1X non-essential amino acids, and 1X antibiotic/antimycotic mix (Thermo Fisher Scientific).
Isolation of EVs/Exos from primary LSC culture supernatants
Normal and diabetic LSC were cultured in serum free CCM. EVs were prepared from conditioned media of normal and diabetic LSC using ultracentrifugation and ExoQuick-TC (SBI, Palo Alto, CA) precipitation as described previously59. In brief, an initial spin was performed at 10,000 × g at room temperature for 10 min for each sample to remove cells and debris, followed by filtration through 0.2 µm filter to remove cell debris and particles larger than 200 nm. The resulting cell free medium was concentrated by ultrafiltration using Amicon® Ultra-15 membrane with molecular weight cutoff of 3,000 Daltons (Millipore, St. Louis, MO). EVs were precipitated in Exo-Quick-TC (SBI), exosome isolation reagent, following the manufacturer’s protocol. Briefly, concentrated cell culture supernatant and Exo-Quick-TC reagent were added proportional to the starting sample volume, mixed and incubated at 4 °C for up to an hour and then centrifuged at 10,000 × g for 30 minutes at room temperature to precipitate Exo pellets. The Exo pellet was re-suspended in appropriate buffer and stored at −80 °C immediately after isolation until further analysis.
Transmission electron microscopy (TEM)
Isolated Exos (1 µg) were resuspended in phosphate-buffered saline (PBS) and the suspension was adsorbed on carbon-formvar 300 mesh grids for 30 min, fixed with 2% glutaraldehyde, washed and stained with 2% uranyl acetate (UA). The grids were dragged on a piece of filter paper to remove the excess of UA, allowed to dry and examined on a JEOL 100CX electron microscope at 60 kV. Images were collected on type 4489 EM film, and the negatives scanned to create digital files.
NanoSight particle size analysis by dynamic light scattering
Exos were suspended in PBS and analyzed in real time using dynamic light scattering measurements with NanoSight LM10-HS instrument equipped with a laser (638 nm) and Nanoparticle Tracking Analysis software version 2.3, Build 0033 (NanoSight, Westborough, MA). Post-acquisition settings were based on the manufacturer’s recommendations and kept constant between the samples. Each video was analyzed to obtain particle size distribution profiles and concentration measurements.
Flow cytometry
Flow cytometry was performed as per manufacturer’s instructions. Briefly, the harvested and fixed keratocytes from passage 2–3 (1.0 × 106 cells/ml) were incubated with unconjugated primary antibodies (Supplementary Table S2), rabbit anti-lumican (aa64–91) and mouse anti-ALDH3 (1B6) followed by incubating with secondary antibodies (Li-Cor Biosciences, Lincoln, NE) and measured with a flow cytometer BD LSR II instrument (BD Biosciences, San Jose, CA). Pooled Exos from keratocytes (1 × 108 per 10 µl of Dynabeads®) were incubated overnight at 4 °C with Dynabeads® magnetic beads (Invitrogen, Carlsbad, CA) coated with primary monoclonal antibody specific for the CD63 (Supplementary Table S2) membrane antigen. To detect the presence of specific antigens like CD81 or CD63, the exosome-coated beads were incubated with PE-CD63 (H5C6) and APC-CD81(5A6) antibodies (Supplementary Table S2) for 45 min at RT on a sample shaker (1000 rpm) followed by isolation buffer (PBS with 0.1% BSA, filtered through 0.2 µm filter) washes. Complexes were resuspended in isolation buffer and subjected to flow cytometry using BD LSR II instrument (BD Biosciences), where at least 50,000 events were collected, and results were analyzed by Flowjo software.
Western blot analysis and immunostaining
Western blot was performed as described previously48. Briefly, treated cells or Exos were lysed and suspended in Tris-glycine sample buffer with proteinase inhibitor EDTA-free cocktail. Equal amounts of lysates were subjected to SDS-PAGE and transferred onto nitrocellulose membranes. Blots were blocked followed by incubation with primary antibodies (Supplementary Table S2) overnight at 4 °C. IRDye LiCor secondary antibodies (Li-Cor Biosciences) were used for protein detection with LiCor Odyssey CLX imaging system (Li-Cor Biosciences). Quantification of protein bands was done with Image Studio. Immunostaining was performed as described previously57. Isotype control (IgG2a kappa) was used as a negative control for K15 antibody.
Exos cellular uptake
Cellular uptake of Exos was followed using confocal microcopy. LSC- derived Exos were labeled with Dil fluorescent dye (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate; Thermo Fisher Scientific) that labels the plasma membrane, according to the manufacturer’s instructions. Briefly, Exos were incubated with Dil dye for 1 h at RT in the dark followed by two washes in PBS. Next, 10 μg/ml and/or 25 μg/ml Dil-labeled Exos in a total volume of 100 μl were diluted in respective medium as described previously36 and were added to N or DM (N/DM) LEC cultures or organ-cultured corneas for 24–48 h and washed prior to staining with 10 μM calcein-AM (Thermo Fisher Scientific) at 37 °C for 30 min in the dark. Cells or organ-cultured corneas were briefly washed and examined under a Zeiss LSM-780 confocal microscope (BioSciences, Jena, Germany). Control group was cultured in respective medium with added 100 μl of PBS.
Co-culture assay
Dil fluorescent dye was added to limbal keratocytes and allowed to incubate at 37 °C for 30 min. Cells were washed and transferred for co-culturing with LEC in 24-well plate with cell culture porous membrane inserts. Dil-labeled keratocytes were seeded onto the 0.4 µm inserts which allow transport of EVs but no cells. After 48 hours the inserts were removed and LECs were labeled with calcein-AM for 30 min at 37 °C and imaged with confocal microscopy.
In-vitro MTS proliferation and wound healing assays
Proliferation assay was performed as described previously49. Briefly, HCEC were seeded on 96-well plates at 5,000/well and 25 μg N/DM LSC-derived Exos were added to the basal medium without growth factors. After 24 h, proliferation was measured using MTS assay (CellTiter 96 Aqueous One Solution Cell Proliferation Assay, Promega, Madison, WI). Scratch wound assay was performed either on treated HCEC or LEC with 25 μg N/DM-Exos as described48. Briefly, treated LEC or HCEC at confluence were scratch wounded using the pipette tip and photographed at time 0. The wounds were allowed to heal and photographed every 6 h. All images were then analyzed using ImageJ software. The percent area healed was calculated with reference to time 0. Wound healing in organ-cultured corneas was performed as published57. A 5 mm wound in central cornea was created using a disk soaked in n-heptanol and incubated with Exos (3.5 × 108) and wound closure was monitored over time.
Small RNA Next Generation Sequencing (NGS)
Exo RNA isolation
Total RNA within Exos was isolated by miRCURY RNA Isolation Kit–Cell & Plant (Exiqon, Woburn, MA) according to the manufacturer’s instructions. The concentration of total RNA was determined by NanoDrop spectrophotometer (Thermo Fisher Scientific) and their quality was analyzed using 2100 Bioanalyzer (Agilent, Santa Clara, CA). NGS was performed by Exiqon (Denmark).
Small RNA library preparation
The library preparation was done using the NEBNext® Small RNA Library preparation kit (New England Biolabs). Sequencing libraries were generated by ligation of adapters to the small RNAs extracted from 100 ng of total RNA for each sample followed by reverse transcription and PCR amplification (15 cycles) and purification of small RNA libraries. QC for the generated libraries was performed using either Bioanalyzer 2100 (Agilent, Carpinteria, CA) or TapeStation 4200 (Agilent) and they were size sorted using the LabChip XT (Perkin Elmer, Inc, San Jose, CA) aiming to select the fraction with the size corresponding to microRNA libraries (~145 nt). The library pools were quantified using the qPCR KAPA Library Quantification Kit (KAPA Biosystems, Wilmington, MA).
Small RNA and miRNA sequencing
The library pool was sequenced on a NextSeq 500 sequencing instrument according to the manufacturer’s instructions. Raw data were de-multiplexed and FASTQ files for each sample were generated using the bcl2fastq software (Illumina Inc., San Diego, CA). FASTQ data were checked using the FastQC tool60. RNA adapters were trimmed off and the resulting reads were mapped to miRBase and small RNA database.
Data analysis workflow
Adapters were removed from the sequences using cutadapt61. Bowtie2 version 2.2.262 was used to map against the human reference genome (GRCh37). No mismatches were allowed for mapping against miRbase 20, one mismatch in the first 32 bases of the read and no indels were allowed for mapping to the genome.
NGS statistical analysis
For the statistical analyses presented in this study, the trimmed mean of M-values normalization method (TMM normalization) was used63, in addition to TPM normalization, which compensate for sample specific effects caused by the variation in library size/sequencing depth between samples. The differential expression analysis was done using TMM in the EdgeR statistical software package64. For gene ontology enrichment analysis, the TopGO R package65, two different statistical tests were used and compared. First, a standard Fisher’s test was used to investigate enrichment of terms between the two test groups. Second, the ‘Elim’ method takes a more conservative approach by incorporating the topology of the GO network to compensate for local dependences between GO, which can mask significant GO terms. Comparisons of the predictions from these two methods can highlight truly relevant GO terms.
Statistical analysis
Experiments were analyzed by Student’s t-test for two groups, or ANOVA for three or more groups with p < 0.05 considered significant, using Prism6 (GraphPad Software, San Diego, CA).
Electronic supplementary material
Acknowledgements
We would like to thank Kavita Patel, Adam Poe and Ruchi Shah for their assistance with immunostaining. Presented in part at the Association for Research in Vision and Ophthalmology (ARVO) annual meeting, May 2018, Honolulu, Hawaii. Funding sources: NIH R01 EY025377, EY013431, Board of Governors Regenerative Medicine Institute & Biomedical Sciences, Cedars-Sinai Medical Center.
Author Contributions
M.S., A.L., designed the experiments. M.S., A.L., M.K., performed the experiments and analyzed the data. M.S., A.L., A.V.L., M.K., wrote the manuscript. All authors contributed to the editing the manuscript.
Data Availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33169-5.
References
- 1.Davanger M, Evensen A. Role of the pericorneal papillary structure in renewal of corneal epithelium. Nature. 1971;229:560–561. doi: 10.1038/229560a0. [DOI] [PubMed] [Google Scholar]
- 2.Dua HS, Shanmuganathan VA, Powell-Richards AO, Tighe PJ, Joseph A. Limbal epithelial crypts: a novel anatomical structure and a putative limbal stem cell niche. Br. J. Ophthalmol. 2005;89:529–532. doi: 10.1136/bjo.2004.049742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shortt AJ, et al. Characterization of the limbal epithelial stem cell niche: novel imaging techniques permit in vivo observation and targeted biopsy of limbal epithelial stem cells. Stem Cells. 2007;25:1402–1409. doi: 10.1634/stemcells.2006-0580. [DOI] [PubMed] [Google Scholar]
- 4.Molvaer RK, et al. Interactive 3D computer model of the human corneolimbal region: crypts, projections and stem cells. Acta Ophthalmol. 2013;91:457–462. doi: 10.1111/j.1755-3768.2012.02446.x. [DOI] [PubMed] [Google Scholar]
- 5.Li W, Hayashida Y, Chen YT, Tseng SC. Niche regulation of corneal epithelial stem cells at the limbus. Cell Res. 2007;17:26–36. doi: 10.1038/sj.cr.7310137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li DQ, Tseng SC. Three patterns of cytokine expression potentially involved in epithelial fibroblast interactions of human ocular surface. J Cell Physiol. 1995;163:61–79. doi: 10.1002/jcp.1041630108. [DOI] [PubMed] [Google Scholar]
- 7.Higa K, et al. Aquaporin 1-positive stromal niche-like cells directly interact with N-cadherin-positive clusters in the basal limbal epithelium. Stem Cell Res. 2013;10:147–155. doi: 10.1016/j.scr.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Dziasko MA, et al. Localisation of epithelial cells capable of holoclone formation in vitro and direct interaction with stromal cells in the native human limbal crypt. PLoS One. 2014;9:e94283. doi: 10.1371/journal.pone.0094283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basu S, et al. Human limbal biopsy-derived stromal stem cells prevent corneal scarring. Sci Transl Med. 2014;6:266ra172. doi: 10.1126/scitranslmed.3009644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espana EM, et al. Stromal niche controls the plasticity of limbal and corneal epithelial differentiation in a rabbit model of recombined tissue. Invest. Ophthalmol. Vis. Sci. 2003;44:5130–5135. doi: 10.1167/iovs.03-0584. [DOI] [PubMed] [Google Scholar]
- 11.Théry C. Exosomes: secreted vesicles and intercellular communications. F1000 Biology Reports. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 13.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013;32:623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carney EF. Chronic kidney disease: Key role of exosomes in albumin-induced inflammation. Nat. Rev. Nephrol. 2018;14:142. doi: 10.1038/nrneph.2018.6. [DOI] [PubMed] [Google Scholar]
- 15.Aliotta JM, et al. Exosomes induce and reverse monocrotaline-induced pulmonary hypertension in mice. Cardiovasc Res. 2016;110:319–330. doi: 10.1093/cvr/cvw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai RC, Chen TS, Lim SK. Mesenchymal stem cell exosome: a novel stem cell-based therapy for cardiovascular disease. Regen. Med. 2011;6:481–92. doi: 10.2217/rme.11.35. [DOI] [PubMed] [Google Scholar]
- 17.Ying W, et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171:372–384. doi: 10.1016/j.cell.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 18.Pardo F, Villalobos-Labra R, Sobrevia B, Toledo F, Sobrevia L. Extracellular vesicles in obesity and diabetes mellitus. Mol. Aspects Med. 2018;60:81–91. doi: 10.1016/j.mam.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Zhang B, et al. HucMSC-exosome mediated-Wnt4 signaling is required for cutaneous wound healing. Stem Cells. 2015;33:2158–2168. doi: 10.1002/stem.1771. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, et al. Exosomes secreted by human adipose mesenchymal stem cells promote scarless cutaneous repair by regulating extracellular matrix remodeling. Sci. Rep. 2017;7:13321. doi: 10.1038/s41598-017-12919-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han KY, Tran JA, Chang JH, Azar DT, Zieske JD. Potential role of corneal epithelial cell-derived exosomes in corneal wound healing and neovascularization. Sci. Rep. 2017;7:40548. doi: 10.1038/srep40548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abd Ghafar N, et al. Phenotypic characterization of culture expanded rabbit limbal corneal keratocytes. Cell Tissue Bank. 2014;15:25–34. doi: 10.1007/s10561-012-9360-y. [DOI] [PubMed] [Google Scholar]
- 23.Ljubimov AV, Saghizadeh M. Progress in corneal wound healing. Prog. Retin. Eye Res. 2015;4:17–45. doi: 10.1016/j.preteyeres.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 25.Baglio SR, et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 2015;6:127. doi: 10.1186/s13287-015-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu S, Cao H, Shen B, Feng J. Tumor-derived exosomes in cancer progression and treatment failure. Oncotarget. 2015;6:37151–37168. doi: 10.18632/oncotarget.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellison-Hughes GM, Madeddu P. Exploring pericyte and cardiac stem cell secretome unveils new tactics for drug discovery. Pharmacol Ther. 2017;171:1–12. doi: 10.1016/j.pharmthera.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bátiz LF, et al. Exosomes as Novel Regulators of Adult Neurogenic Niches. Front Cell Neurosci. 2015;9:501. doi: 10.3389/fncel.2015.00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luarte A, Bátiz LF, Wyneken U, Lafourcade C. Potential Therapies by Stem Cell-Derived Exosomes in CNS Diseases: Focusing on the neurogenic niche. Stem Cells Int. 2016;2016:5736059. doi: 10.1155/2016/5736059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reagan MR, Rosen CJ. Navigating the bone marrow niche: translational insights and cancer-driven dysfunction. Nat. Rev. Rheumatol. 2016;12:154–168. doi: 10.1038/nrrheum.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Notara M, Shortt AJ, Galatowicz G, Calder V, Daniels JT. IL6 and the human limbal stem cell niche: a mediator of epithelial-stromal interaction. Stem Cell Res. 2010;5:188–200. doi: 10.1016/j.scr.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Han KY, Dugas-Ford J, Seiki M, Chang JH, Azar DT. Evidence for the Involvement of MMP14 in MMP2 Processing and Recruitment in Exosomes of Corneal Fibroblasts. Invest. Ophthalmol. Vis. Sci. 2015;56:5323–5329. doi: 10.1167/iovs.14-14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B, et al. Human umbilical cord mesenchymal stem cell exosomes enhance angiogenesis through the Wnt4/ β-catenin pathway. Stem Cells Transl. Med. 2015;4:513–522. doi: 10.5966/sctm.2014-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang S, et al. Umbilical cord-derived mesenchymal stem cell-Derived exosomal microRNAs suppress myofibroblast differentiation by inhibiting the transforming growth factor-β/SMAD2 pathway during wound healing. Stem Cells Transl. Med. 2016;5:1425–1439. doi: 10.5966/sctm.2015-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun, Y. et al. Platelet-derived exosomes affect the proliferation and migration of human umbilical vein endothelial cells via miR-126. Curr. Vasc. Pharmacol. [Epub ahead of print] (2018). [DOI] [PubMed]
- 36.Zhao B, et al. Exosomes derived from human amniotic epithelial cells accelerate wound healing and inhibit scar formation. J Mol Histol. 2017;48:121–132. doi: 10.1007/s10735-017-9711-x. [DOI] [PubMed] [Google Scholar]
- 37.Than UTT, Guanzon D, Leavesley D, Parker T. Association of Extracellular Membrane Vesicles with Cutaneous Wound Healing. Int. J. Mol. Sci. 2017;18:E956. doi: 10.3390/ijms18050956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jalabert A, et al. Exosome-like vesicles released from lipid-induced insulin-resistant muscles modulate gene expression and proliferation of beta recipient cells in mice. Diabetologia. 2016;59:1049–1058. doi: 10.1007/s00125-016-3882-y. [DOI] [PubMed] [Google Scholar]
- 39.Barani B, Rajasingh S, Rajasingh J. Exosomes: Outlook for future cell-free cardiovascular disease therapy. Adv. Exp. Med. Biol. 2017;998:285–307. doi: 10.1007/978-981-10-4397-0_19. [DOI] [PubMed] [Google Scholar]
- 40.Xu JY, Chen GH, Yang YJ. Exosomes: A rising star in falling hearts. Front Physiol. 2017;8:494. doi: 10.3389/fphys.2017.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clark DJ, Fondrie WE, Yang A, Mao L. Triple SILAC quantitative proteomic analysis reveals differential abundance of cell signaling proteins between normal and lung cancer-derived exosomes. J. Proteomics. 2016;133:161–169. doi: 10.1016/j.jprot.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 42.Dai G, et al. Colorectal cancer cell-derived exosomes containing miR-10b regulate fibroblast cells via the PI3K/Akt pathway. Bull Cancer. 2018;105:336–349. doi: 10.1016/j.bulcan.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 43.Gastpar R, et al. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res. 2005;65:5238–5247. doi: 10.1158/0008-5472.CAN-04-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saghizadeh M, et al. Alterations of epithelial stem cell marker patterns in human diabetic corneas and effects of c-met gene therapy. Mol. Vis. 2011;17:2177–2190. [PMC free article] [PubMed] [Google Scholar]
- 45.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/S0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 46.Chowdhari S, Sardana K, Saini N. miR-4516, a microRNA downregulated in psoriasis inhibits keratinocyte motility by targeting fibronectin/integrin α9 signaling. Biochim Biophys Acta. 2017;1863:3142–3152. doi: 10.1016/j.bbadis.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 47.Saghizadeh M, et al. Overexpression of matrix metalloproteinase-10 and matrix metalloproteinase-3 in human diabetic corneas: a possible mechanism of basement membrane and integrin alterations. Am. J. Pathol. 2001;158:723–34. doi: 10.1016/S0002-9440(10)64015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Funari VA, et al. Differentially expressed wound healing-related microRNAs in the human diabetic cornea. PLoS One. 2013;8:e84425. doi: 10.1371/journal.pone.0084425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kulkarni M, et al. Genome-wide analysis suggests a differential microRNA signature associated with normal and diabetic human corneal limbus. Sci. Rep. 2017;7:3448. doi: 10.1038/s41598-017-03449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park JK, et al. miR-184 exhibits angiostatic properties via regulation of Akt and VEGF signaling pathways. FASEB J. 2017;31:256–265. doi: 10.1096/fj.201600746R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hughes AE. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am. J. Hum. Genet. 2011;89:628–33. doi: 10.1016/j.ajhg.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagosa S. microRNA-184 induces a Commitment Switch to Epidermal Differentiation. Stem Cell Reports. 2017;9:1991–2004. doi: 10.1016/j.stemcr.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saghizadeh M, Dib CM, Brunken WJ, Ljubimov AV. Normalization of wound healing and stem cell marker patterns in organ-cultured human diabetic corneas by gene therapy of limbal cells. Exp. Eye Res. 2014;129:66–73. doi: 10.1016/j.exer.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ljubimov AV. Diabetic complications in the cornea. Vision Res. 2017;139:138–152. doi: 10.1016/j.visres.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li D, et al. The microRNAs miR-200b-3p and miR-429-5p target the LIMK1/CFL1 pathway to inhibit growth and motility of breast cancer cells. Oncotarget. 2017;8:85276–85289. doi: 10.18632/oncotarget.19205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maolakuerban N, et al. MiR-200c-3p inhibits cell migration and invasion of clear cell renal cell carcinoma via regulating SLC6A1. Cancer Biol. Ther. 2018;19:282–291. doi: 10.1080/15384047.2017.1394551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winkler MA, Dib C, Ljubimov AV, Saghizadeh M. Targeting miR-146a to treat delayed wound healing in human diabetic organ-cultured corneas. PLoS One. 2014;9:e114692. doi: 10.1371/journal.pone.0114692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kramerov AA, Saghizadeh M, Ljubimov AV. Adenoviral gene therapy for diabetic keratopathy: Effects on wound healing and stem cell marker expression in human organ-cultured corneas and limbal epithelial cells. J Vis Exp. 2016;110:e54058. doi: 10.3791/54058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ibrahim AG, Cheng K, Marbán E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Reports. 2014;2:606–619. doi: 10.1016/j.stemcr.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 61.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 62.Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bioconductor, http://www.bioconductor.org.
- 65.Alexa, A. & Rahnenfuhrer, J. TopGO: Enrichment Analysis for Gene Ontology. R package version 2.32.0 (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).