Abstract
The microencapsulation process of bacteria has been used for many years, mainly in the food industry and, among the different matrixes used, sodium alginate stands out. This matrix forms a protective wall around the encapsulated bacterial culture, increasing its viability and protecting against environmental adversities, such as low pH, for example. The aim of the present study was to evaluate both in vitro and in vivo, the capacity of the encapsulation process to maintain viable lactic acid bacteria (LAB) strains for a longer period of time and to verify if they are able to reach further regions of mouse intestine. For this purpose, a recombinant strain of LAB (L. lactis ssp. cremoris MG1363) carrying the pExu vector encoding the fluorescence protein mCherry [L. lactis MG1363 (pExu:mCherry)] was constructed. The pExu was designed by our group and acts as a vector for DNA vaccines, enabling the host cell to produce the protein of interest. The functionality of the pExu:mCherry vector, was demonstrated in vitro by fluorescence microscopy and flow cytometry after transfection of eukaryotic cells. After this confirmation, the recombinant strain was submitted to encapsulation protocol with sodium alginate (1%). Non-encapsulated, as well as encapsulated strains were orally administered to C57BL/6 mice and the expression of mCherry protein was evaluated at different times (0–168 h) in different bowel portions. Confocal microscopy showed that the expression of mCherry was higher in animals who received the encapsulated strain in all portions of intestine analyzed. These results were confirmed by qRT-PCR assay. Therefore, this is the first study comparing encapsulated and non-encapsulated L. lactis bacteria for mucosal DNA delivery applications. Our results showed that the microencapsulation process is an effective method to improve DNA delivery, ensuring a greater number of viable bacteria are able to reach different sections of the bowel.
Keywords: recombinant Lactococcus lactis, sodium alginate encapsulation, mCherry reporter protein, pExu vector, DNA delivery
Introduction
The intestinal microbiota is composed of a huge diversity of ecological niches, encompassing more than 50 genera of bacteria (Holdeman et al., 1976) with different characteristics: harmful, beneficial, or both. Among the bacteria with beneficial activity, the LAB can be highlighted. These are Gram-positive, saccharolytic, rod shaped, non-sporulating bacteria with low-GC content genomes, that could reside in the large intestine (Salminen et al., 1998). The majority of LAB have a ‘Generally Recognized as Safe’ (GRAS) status according to the United Sates Food and Drug Administration (US FDA) and fulfill of the Qualified Presumption of Safety (QPS) notion criteria developed by the European Food Safety Authority (EFSA).
Some strains are classified as probiotic, promoting them to be beneficial for consumer health (Sanders et al., 2013). This intrinsic advantage of some LAB, as well as the easiness to design them in recombinant lactic acid bacteria (rLAB) makes them a good alternative to systemic delivery systems compared with other mucosal delivery vehicles such as liposomes and attenuated pathogens. For these reasons, over the past two decades, LAB has been intensively studied as potential carriers of compounds with therapeutic or prophylactic effects at mucosal surface (Wells and Mercenier, 2008; Bermúdez-Humarán, 2009; Zurita-Turk et al., 2014; Souza et al., 2016; Mancha-Agresti et al., 2017). In this context, plasmid-cured strains of L. lactis ssp. lactis IL1403 and L. lactis ssp. cremoris MG1363, whose genomes were sequenced in 1999/2001 and 2007, respectively, are the strains most commonly used (Bolotin et al., 2001; Wegmann et al., 2007; Linares et al., 2010) for production of recombinant molecules in mucosal vaccination (Wells and Mercenier, 2008). In addition, interest in genus Lactobacillus has also increased (Li et al., 2007; Hongying et al., 2014; Allain et al., 2016). In DNA delivery system the eukaryotic host cells express antigens encoded by the vaccine. Epitopes exposed by the recombinant protein (antigen) are very similar to those in their native form (Běláková et al., 2007), consequently they are presented to the immune system in an analogous form found in nature.
Oral administration of rLAB producing/coding therapeutics proteins have the benefit of direct proteins delivery in situ, avoiding degradation either by enzymatic action or adverse conditions encountered in the stomach such as severe acid challenges. Also, rLAB could be protect against stress elicited by the exposure to high bile and salt conditions of the intestine faced during transit through the gastrointestinal tract (Drouault et al., 1999). On the other hand, studies have shown that the percentage of viable bacteria that reaches the gut may be considerably lower compared to the number of administered bacteria (Charteris et al., 1998; Cook et al., 2012), due to above mentioned stress conditions.
To circumvent this drawback, bacterial microencapsulation technique using polymer matrix offers adequate conditions to reduce bacterial loss, in the gastrointestinal tract, as well as in food products (Fávaro-Trindade and Grosso, 2002). The most widely used matrix for microencapsulation is sodium alginate, a non-toxic and biocompatible polymer which is able to establish a versatile matrix which protects active components, as well as probiotic microorganisms sensible to pH, heat, oxygen, and other factors (Goh et al., 2012). Some studies report an increase of up to 80–95% survival of encapsulated probiotics with alginate beads (Sheu and Marshall, 1993; Jankowski et al., 1997; Krasaekoopt et al., 2003). Lower bacteria population reduction during exposure to simulated gastric environment and bile solution (Picot and Lacroix, 2004) are some features improved by encapsulation of probiotics in alginate. Encapsulation in alginate of Bifidobacterium longum as well as, Lactobacillus rhamnosus improved survival at acid pH (pH 2.0) up to 48 h and also, death rate decreased proportionately with increased alginate concentrations (2–4%) and bead size (Lee and Heo, 2000; Goderska et al., 2003).
In previous work, our group developed a new vector (pExu – Extra Chromosomal Unit) to be used as a DNA delivered by Lactococcus lactis and Lactobacillus strains. We showed eGFP expression (Enhanced Green Fluorescent Protein), by enterocytes in the duodenal region between 12 up to 72 h after oral administration of the recombinant strain L. lactis (pExu:egfp) (Mancha-Agresti et al., 2016). Nevertheless, tissue auto fluorescence is in the same GFP protein wavelength, hindering tissue analysis and eGFP expression detection in the lower bowel region. Nowadays, there are many fluorescent proteins available (Shaner et al., 2005; Ai et al., 2008; Day and Davidson, 2009). Among the best red members in the fruit series, the RFP (Red Fluorescent Protein) mCherry, can be highlighted. This protein is obtained from successive modifications of the Discosoma ssp. fluorescent protein DsRed, and it has longest excitation and emission wavelengths (587/610 nm, respectively) with shortest maturation time (Patterson et al., 2001; Shaner et al., 2004; Jach et al., 2006) in comparison with other RFPs. Proteins that emit fluorescence in red zone are more desirable for cellular studies than those which emit in green or blue zones because red light presents less dispersion allowing deep tissue penetration (Nienhaus and Wiedenmann, 2009), therefore increasing sensibility (Deliolanis et al., 2008).
The aim of this study was to encapsulate the L. lactis ssp. cremoris MG1363 strain carrying the pExu vector encoding RFP mCherry gene in alginate matrix and evaluate potential delivery across gut portions after oral administration by comparing non-encapsulated and encapsulated bacteria.
Materials and Methods
Bacterial Strains and Plasmids
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli Top10 strain was grown in Luria-Bertani (LB) medium (Acumedia, Lansing, MI, United States) at 37°C with vigorous shaking. L. lactis ssp. cremoris MG1363 was grown statically at 30°C in M17 medium (Sigma-Aldrich, St. Louis, MO, United States) supplemented with 0.5% glucose (w/v) (Labsynth, São Paulo, Brazil) (G-M17). The Erythromycin antibiotic (Sigma-Aldrich) was added at the indicated concentration as necessary; 500 μg/mL for E. coli and 125 μg/mL for L. lactis. Pure cultures of bacteria were kept as stock cultures in 40% glycerol (v/v) (Sigma-Aldrich) for E. coli and 25% glycerol (v/v) for L. lactis at −80°C.
Table 1.
Bacterial strains and plasmids used in this work.
| Bacterial strain | Characteristics | Source |
|---|---|---|
| Escherichia coli TOP10 | E. coli K-12-derived strain; F-mcrA 1 (mrr-hsdRMS-mcrBC) 8 80lacZ1M15 1lacX74 nupG recA1 araD139 1 (ara-leu)7697 galE15 galK16 rpsL (StrR) endA1 λ | Invitrogen |
| Escherichia coli TOP10 (pTP:mCherry) | E. coli TOP10 carrying the pTP:mCherry plasmid; Kmr | This work |
| Escherichia coli TOP10 (pExu:mCherry) | E. coli TOP 10 carrying the pExu:mCherry plasmid; Eryr | This work |
| Lactococcus lactis MG1363 | L. lactis ssp. cremoris | Gasson, 1983 |
| Lactococcus lactis MG1363 (pExu:mCherry) | L. lactis MG1363 carrying the pExu:mCherry plasmid; Eryr | This work |
| Plasmids | Characteristics | Source |
| Zero Blunt® TOPO® | Cloning vector (KmR, ccdB gene fused to the C-terminus of the LacZα fragment) | Invitrogen |
| pXJM19:mCherry | ori colE1, oricg, ptac, mCherry, Cmr | Ott et al., 2012 |
| pTP:mCherry | TOPO® vector with the mCherry ORF; Kmr | This work |
| pExu:mCherry | pCMV/Eryr/RepA/RepC/mCherry | This work |
Plac: promoter of operon lac; Kmr: gene that confers resistance to kanamycin; pUc ori: origin of replication; ccdB: lethal gene; ZeoR: gene resistance to zeomycin, ori ColE1: origin of replication for E. coli; Amp: gene that confers ampicillin resistance; pCMV: cytomegalovirus promoter; Ery: gene that confers erythromycin resistance; RepD and RepE: origins of prokaryotic replication for L. lactis and Lactobacillus; egfp: coding sequence of enhancer fluorescent green protein; Cm: gene that confers resistance to chloramphenicol; mCherry: coding sequence of red fluorescent monomeric protein.
DNA Manipulations
The protocols of DNA manipulation were performed following Sambrook protocols (Green and Sambrook, 2012) with some modifications. For plasmid DNA extraction from L. lactis, to prepare the protoplasts, TES buffer (25% sucrose, 1 mM EDTA, and 50 mM Tris-HCl pH 8) containing lysozyme (Sigma- Aldrich) (10 mg/mL) was added for 1 h at 37°C. Restriction enzymes were used as recommended by suppliers. Transformation of L. lactis was performed by electroporation (2,400 V, 25 μF capacitance and 200 Ω of resistance). L. lactis transformants were plated on G-M17 agar plates containing erythromycin and were counted after 48 h incubation at 30°C, whereas E. coli transformants were plated onto LB agar plates containing the required antibiotic for 24 h at 37°C.
pExu:mCherry and L. lactis ssp. cremoris MG1363 (pExu:mCherry)
The ORF of reporter gene mCherry was amplified by PCR technique using Hot Start® high-fidelity DNA Polymerase (Qiagen, Hilden, Germany). Oligonucleotides primers used for mCherry were: 5′ ggcGCGGCCGCAATGGTGAGCAAGGGCGAGG 3′ (forward) and 5′ ggcCTCGAGTTACTTGTACAGCTCGTCCATGC 3′ (reverse). Artificial restriction sites of NotI and XhoI enzyme and also, customized Kozak sequence were added in the forward oligonucleotides. Purified PCR product and pExu plasmid (Mancha-Agresti et al., 2016) were digested with the same restriction enzymes, as recommended by suppliers. The digested vector and insert were purified by IllustraTM GFXTM PCR DNA kit and then were ligated with T4 DNA ligase (Invitrogen, Carlsbad, CA, United States) and transformed into E. coli Top10, creating the E. coli Top10 (pExu:mCherry) strain. Sequence analysis was done to confirm the insert integrity using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, United States) and ABI3130 sequencing equipment. Lastly, pExu:mCherry was transformed into competent cells of L. lactis ssp. cremoris MG1363 strain generating L. lactis MG1363 (pExu:mCherry) strain, used in this study.
mCherry Production by Eukaryotic Cells in vitro
Chinese hamster ovarian cell line [Flp-InTM-CHO (Invitrogen)] (CRL 12023)-ATCC was used to evaluate the mCherry expression. For this intent, CHO cells were cultured in complete Nutrient Mixture F12 Ham media (Sigma-Aldrich), supplemented with 10% fetal bovine serum (Gibco-Thermo Scientific, Waltham, MA, United States), 1% L-glutamine (Sigma-Aldrich), and 1% HEPES (Sigma-Aldrich). Cells were seeded at 1 × 106 in a 6-well plate and at 90–95% of confluence, CHO cells were transfected with 4 μg of pExu:mCherry vector or no plasmid (negative control) using LipofectamineTM 2000 (Invitrogen), according to supplier’s recommendation. Eukaryotic cells protein expression was checked by epifluorescent microscope (Zeiss Axiovert 200, filter 585/42 nm, Oberkochen, Germany) and by flow cytometry (BD FACSCantoTM, BD Bioscience, Franklin Lakes, NJ, United States). Duplicate transfection assays were performed.
Bacterial Doses and Microencapsulation
To prepare the doses of L. lactis (pExu:mCherry), on the first day, the culture was grown at 30°C in M17 medium (Sigma-Aldrich) supplemented with 0.5% glucose (Labsynth) (GM17) and 125 μg/mL of erythromycin (Sigma-Aldrich). On the second day, a 1/20 dilution was performed until the culture reaches OD600 nm = 1.0. Then 2 mL, which corresponds to one dose, centrifuged for 10 min at 4°C at 4,000 rpm. Supernatant was discarded, and the pellet was washed twice with PBS (0.01 M) and then it was resuspended in 30 μL of sterile PBS, immediately frozen in ultra-freezer −80°C until use. At the moment of gavage, sterile PBS (0.01 M) was added (∼70 μL) to the prepared doses to reach a final volume of 100 μL. The final quantities of administrated bacteria were 1014 CFU/dose. This concentration was calculated after adequate dilution (10−14), where colonies were counted by pour plate method (G-M17 agar-medium with erythromycin).
For encapsulated doses, after washing the pellet with PBS (0.01 M), bacteria culture [1014 colony forming unit (CFU)] was mixed with 50 μL of sterile PBS (0.01 M)and 50 μL of 1% of Sodium Alginate Solution matrix as described by Krasaekoopt et al. (2003). For this procedure, alginate solution was prepared and sterilized by autoclaving (121°C for 15 min). The mixture was homogenized carefully and this homogeneous solution (Bacteria+PBS+1% of sodium alginate solution) was extruded through a 21-gauge nozzle into a sterile 3% CaCl2 solution magnetically stirring (a cross-linking solution), forming beads (encapsulated bacteria) by contact of both solutions. Afterward, beads were harvested by filtering using Cell Strainer 40 μm Nylon (Corning, NY, United States) and CaCl2 remnants were removed by pipetting. Then, they were transferred to a sterile micro tube and stored in ultra-freezer −80°C until use. Before gavage, ∼100 μL of sterile PBS was added to frozen encapsulated doses.
Encapsulation Efficiency and Viability After Freezing
To determine the encapsulation efficiency (EE), the beads were mixed with 100 μL of 2% (w/v) sterile sodium citrate buffer (pH = 8.36) during 10 min to dissociate the capsules. After this time, the mixture was vigorously homogenized and subsequently the entrapped viable bacteria, after adequate dilution (10−10), were counted by pour plate method (G-M17 agar-medium). Non-encapsulated bacteria were also submitted to the same treatment to guarantee the same experimental condition. This process was performed during 5 days after doses production to see viability of beads after freezing. Encapsulation efficiency after freezing was calculated by the following equation:
∗ CFU is a number of colony forming unit on the agar plate.
Efficiency was verified for freshly encapsulated and non-encapsulated bacteria (day 0) and for doses conditioned at −80°C (days 1 to 5). For the last condition, single doses were unfrozen and plated each day.
Viability of Encapsulated Bacteria to Artificial Juice
To verify the viability of entrapped bacteria, stomach environment was simulated. In a 20 mL solution containing sodium chloride 0.9% (w/v) and 0.3% of pepsin (w/v) (Sigma-Aldrich) (pH = 2), 1014 CFU (one dose) of encapsulated and non-encapsulated bacteria was added and incubated for 2 h at 37°C, with shaking at 50 rpm (Chávarri et al., 2010). Afterward, both suspensions were centrifuged and the supernatant was discarded (the encapsulated solution was filtered using Cell Strainer 40 μm Nylon-corning to avoid capsule loss during discard of the supernatant). Then, the bacterial content was washed twice with PBS (0.01 M) and centrifuged in order to remove the remaining acid solution. Finally, pellets were ressuspended with 100 μL of sodium citrate buffer (pH = 8.36) and after that, 100 μL of this mixture was assayed to pour plate method. This experiment was performed in duplicate. The survival (%) of free and encapsulated bacteria was calculated by spread plate count on G-M17 agar after incubation at 30°C for 48 h as shown in the following equation:
| (1) |
Mice
Conventional 4 to 6-week-old (25–30 g) male and female C57BL/6 mice, were obtained from Centro de Bioterismo (CEBIO) of Universidade Federal de Minas Gerais (UFMG, Brazil). Procedures and manipulation of animals followed the rules of Ethical Principles in Animal Experimentation, accepted by the Ethics Committee on Animal Experiments (Protocol # 114/2010, CETEA/UFMG/Brazil). All animals were maintained in collective cages in an environmentally controlled room with a 12-h light/dark cycle and given free access to water and food ad libitum.
Mice Handling: Gavage of Recombinant L. lactis MG1363 Strain (Encapsulated and Non-encapsulated) Into C57BL/6 Mice
Mice were split into three groups as follows: (i) PBS group, (ii) non-encapsulated L. lactis ssp. cremoris MG1363 (pExu:mCherry), and (iii) encapsulated L. lactis MG1363 (pExu:mCherry). Two independent experiments were performed with 24 mice in each group (72 mice/experiment). The animals were separated according to each evaluated time (three animals for each time). The recombinant strains (1014 CFU bacterial suspensions in a final volume of 100 μL of PBS) were orally administered by gavage. The administration was performed in one go (at zero time) and, at the following intervals: 12, 24, 48, 72, 96, 120, 144, and 168 h (post-gavage), animals were euthanized by cervical dislocation and intestinal sections of duodenum, jejunum, ileum and colon were analyzed by confocal microscopy and qRT-PCR. Images were captured using Zeiss LSM 540 META inverted confocal laser-scanning microscope and analyzed by Zeiss LSM Image Browser software.
mRNA Extraction and Real-Time PCR (qRT-PCR)
Samples of intestinal sections (duodenum, jejunum, ileum, and colon) at 24 h post-gavage, were collected to perform qRT-PCR assay to confirm mCherry expression in enterocytes after oral gavage. We even collected intestinal samples (duodenum, jejunum, ileum, and colon) at different times post-gavage (12, 48, 72, 96, 120, 144, and 168 h). Samples from each gut portion were collected after mice euthanasia and stored at −20°C in RNA later (Invitrogen) until RNA extraction. Total RNA was obtained using TRIzol reagent (Invitrogen), in compliance with manufacturer instructions. The quality and quantity of RNA samples were evaluated in agarose gel electrophoresis, and also through spectrophotometer analysis on NanoDrop® 2000 spectrophotometer (Thermo Scientific) taking into account absorbance ratios of 280/260 and 260/230 nm. Extracted RNA samples were treated with DNAse I (Invitrogen) which was subsequently deactivated. The complementary deoxyribonucleic acid (cDNA) synthesis was performed using 1 μg of RNA and ImProm-II reverse transcriptase (Promega, Madison, WI, United States) in compliance with its manual.
Quantitative reverse transcription PCR (qRT-PCR) was performed using iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, United States) and gene specific-primers for mCherry open reading frame (ORF). Housekeeping gene Actb was used as reference for normalization (Giulietti et al., 2001). Experimental approach was optimized by adjusting concentrations of primers for optimal specificity and efficiency (Table 2). Amplification reactions were performed in final volume of 10 μL, using 5 μL of SYBR green supermix and 10 ng of cDNA (5 ng/μL). The PCR cycle parameters were as follows: initial denaturation at 95°C for 30 s, annealing/extension at 60°C for 60 s, 40 cycles of 95°C for 15 s followed by a dissociation stage for recording the melting curve. Expression levels in control group were used as calibration data (i.e., animals which received PBS). Results were shown graphically as fold changes in gene expression, using the means and standard deviations of target gene expression amount in accordance with Hellemans et al. (2007). Data were analyzed according to the relative expression using 2−ΔΔCt method (Hellemans et al., 2007).
Table 2.
Targets and primers concentration of qRT-PCR performed in this study.
| RNA target | Primer sequence (5′–3′) |
|||
|---|---|---|---|---|
| Forward | Reverse | Amplicon size (pb) | Primer (μmol l−1) | |
| mCherry | CACTACGACGCTGAGGTCAA | GTGGGAGGTGATGTCCAACT | 97 | 0.25 |
| β-Actin | AGAGGGAAATCGTGCGTGAC | CAATAGTGATGACCTGGCCGT | 138 | 0.25 |
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 6. Statistical significance between the groups was calculated using a Student’s t-test. A 95% confidence limit was significant at a value of P < 0.05.
Results
Recombinant Strain of L. lactis – L. lactis MG1363 (pExu:mCherry)
The mCherry ORF (711bp) (GenBank No. AST15061.1) was successfully cloned into the pExu vector (Mancha-Agresti et al., 2016). This vector has an eukaryotic unit harboring the cytomegalovirus mammalian promoter (pCMV), the polyadenylation signal of the bovine growth hormone (BGH), and a prokaryotic region containing RepD/RepE replication origins for L. lactis and also OriColE1 replication origin for E. coli. The resistance marker Erythromycin (Ery), was used to select recombinant strain. pExu:mCherry vector construction was successfully confirmed by molecular techniques: PCR, enzymatic digestion (NotI and XhoI), and also DNA sequencing. pExu:mCherry was transformed into L. lactis ssp. cremoris MG1363 wild type strain generating L. lactis MG1363 (pExu:mCherry) recombinant strain.
Eukaryotic Cells Are Able to Express the mCherry Protein
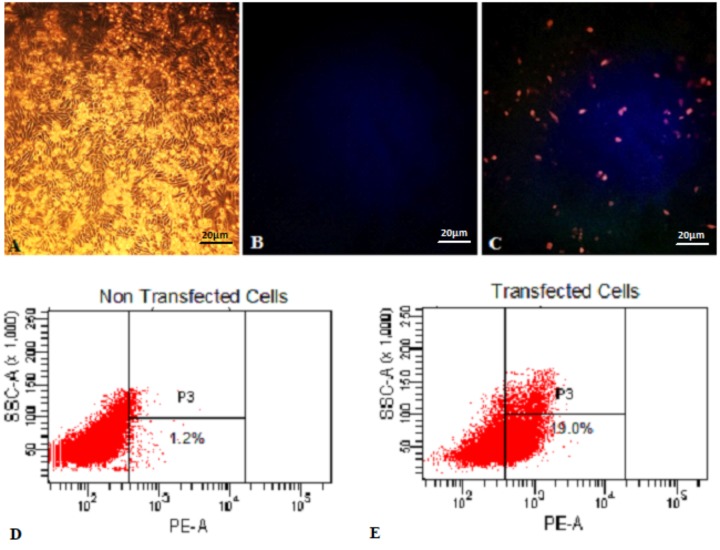
Functionality of pExu:mCherry was established after transfection of CHO cells in two independent assays. In the first one, fluorescence microscopy analysis showed that transfected eukaryotic cells were able to express the reporter protein. In contrast, non- transfected cells showed no fluorescence, as expected. In Figure 1, the pictures A, B, and C show the eukaryotic cell expressing or not the fluorescent protein. Images were captured by fluorescence microscopy (Figures 1A–C).
FIGURE 1.
Expression of mCherry protein by Eukaryotic cells transfected with pExu:mCherry plasmid evaluated by Fluorescence Microscopy and Flow Cytometry. (A) Light field capture of non-transfected CHO cells, (B) non-transfected CHO cells, (C) transfected CHO cells with pExu:mCherry (20X), (D) Dot plot of non-transfected cells, (E) Dot plot transfected cells with pExu:mCherry. The fluorescence images were obtained using the epifluorescent microscope (Zeiss Axiovert 200, filter 585/42 nm). The dot plot graph shows the cell count on the y axis and the PE-A filter on the x axis examined through BD FACSDivaTM Software-BD-biosciences.
In the second one, the percentage of expressing cells was evaluated by flow cytometry. After 48 h, 19% of transfected cells with pExu:mCherry showed ability to express the reporter protein. No expression was observed in the non-transfected cells, and also in cells transfected with pExu:empty (data not shown). In Figure 1 pictures D and E show the dot plot of non-transfected and transfected cells respectively. These results confirmed functionality of pExu:mCherry plasmid in eukaryotic cells.
Encapsulation Efficiency in 1% (w/v) Alginate and Loss of Viability After Encapsulation and Freezing of L. lactis MG1363 (pExu:mCherry)
At first, alginate concentration for encapsulation was standardized by testing three different concentrations (0.5, 1, and 2% w/v) with 3% CaCl2 (w/v). Results obtained at this stage showed that the uniformity and spherical beads at 0.5% (w/v) of alginate concentration were not satisfactory due to low viscosity and consequently, less number of binding sites for Ca2+ ions (cross-linkage) (Chandramouli et al., 2004; Lotfipour et al., 2012). With 2% (w/v) alginate concentration, beads were too viscous to be extruded from the 21G needle. Thus, the 1% (w/v) alginate concentration presented the best conditions for encapsulation.
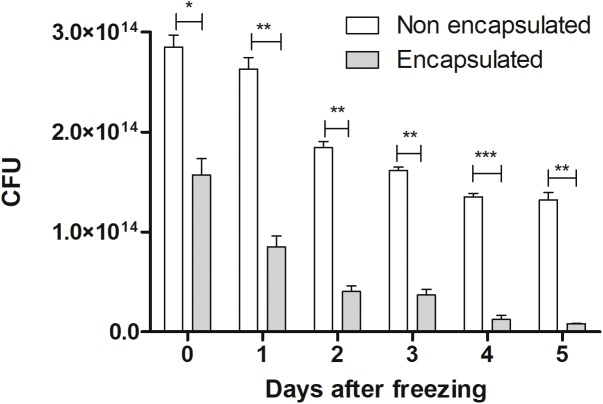
Regarding efficiency of encapsulation, our results showed that at the time of doses preparation it had 2.8 × 1014 CFU. After the encapsulation process (represented as 0 time), we observed a decrease in the number of CFU (1.6 × 1014), being about 60% efficiency, in comparison with non-encapsulated ones. It was possible to observe that both strains (encapsulated and non-encapsulated) lost viability during freezing days, in equivalent form, as demonstrated in Figure 2. Therefore, in this study, only recent confectioned doses were used in oral administration to mice.
FIGURE 2.
Cellular viability: viability of non-encapsulated and encapsulated cells of L. lactis MG1363 (pExu:mCherry) from 0 to 5 days after freezing (–80°C). The 0 represents the number of CFU by plate count immediately after the encapsulation process. ∗P < 0.05, ∗∗P < 0.001, and ∗∗∗P < 0.0001.
In artificial gastric juice, our results showed that the number of cultivable cells observed corresponds to 4.5 × 104 CFU and 4.7 × 104 CFU for non-encapsulated and encapsulated, respectively. Regarding initial doses in non-encapsulated bacteria (2.8 × 1014 CFU) and encapsulated (1.6 × 1014 CFU), we can observe that the percentage survival of non-encapsulated bacteria was 25.3%, while the survival of encapsulated strain was 32.9%. The bacterial survival showed around an 8% increase in encapsulated ones. These results lead to the conclusion that encapsulated bacteria has slightly more resistance to survive in artificial gastric juice, demonstrating that encapsulate process can be successfully used in oral administration strategies.
mRNA and Protein Expression in Mice Bowel
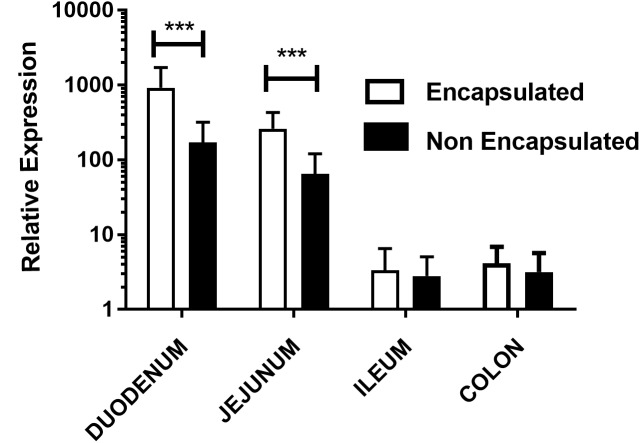
In order to investigate the expression of mCherry protein by eukaryotic cells in vivo, we evaluated mRNA expression of mCherry in different parts of the bowel, after gavage. Our results showed that animals which received encapsulated bacteria were able to express 4.34x fold increase in duodenum and 3.03x fold in jejunum portion when compared with non-encapsulated bacteria. However, no significant differences were seen in ileum and colon portions as shown in Figure 3. When the kinetics qRT-PCR experiments were performed, it was observed the same behavior as in confocal analysis. Animals which received the encapsulated doses have higher relative expression in duodenum and in jejunum sections while in ileum and colon the relative expression was lower. Nevertheless, in the colon no statistical differences were observed. The same behavior was observed in the relative expression of non-encapsulated bacteria, where the levels of expression in the duodenum and in the jejunum sections were higher compared to ileum and colon ones (Supplementary Figures 1, 2).
FIGURE 3.
Relative expression of mCherry protein in mouse intestinal cells: evaluation of mCherry protein expression by epithelial cells from different sections of the gut of mice treated orally with, non-encapsulated or encapsulated, L. lactis MG1363 (pExu:mCherry), by the qRT-PCR technique. Axis y shows the relative expression, and axis x shows the gut sections. ∗∗∗P < 0.0001.
Intestinal Cells of Mice Orally Administered With Encapsulated L. lactis MG1363 (pExu:mCherry) Showed mCherry Expression in Distant Regions of the Bowel
After oral administration of encapsulated and non-encapsulated L. lactis MG1363 (pExu:mCherry) to C57BL/6 mice at different times (12, 24, 48, 72, 96, 120, 144, and 168 h), the expression of mCherry protein by eukaryotic cells was investigated. The eukaryotic cells of the duodenum and jejunum were able to express mCherry protein until 3 days post-gavage with non-encapsulated bacteria as was shown in previous report with eGFP protein (Mancha-Agresti et al., 2016), while encapsulated bacteria was able to keep the protein expression for 7 and 4 days in duodenum and jejunum, respectively. When we investigated the ileum and colon sections, there was no expression found after gavage with non-encapsulated bacteria. Nevertheless, ileum of animals which received the encapsulated L. lactis expressed the recombinant protein between 2 and 4 days after gavage, as shown in Figure 4. In the colon section no expression was found (data not shown). The confocal microscopy showed two important aspects. First, there was more expression of mCherry protein by eukaryotic cells when encapsulated L. lactis was administered, as were shown in qRT-PCR results. The other aspect is the encapsulated strain is able to reach distant parts of the bowel, as ileum, for example (Figure 4).
FIGURE 4.
Evaluation of mCherry protein expression by mouse intestinal cells. Comparison between the expression obtained in the different intestinal sections (duodenum, jejunum, and ileum) from the delivery of the vector pExu:mCherry by non-encapsulated and encapsulated bacteria. The expression was evaluated between 12 and 168 h after mice gavage. The images were obtained using Zeiss LSM 510 META inverted confocal laser-scanning microscope and the images were analyzed by Zeiss LSM Image Browser software.
Discussion
Lactococcus lactis is commonly studied as a host for heterologous protein production and as a delivery platform for therapeutic molecules (Morello et al., 2008; Bahey-El-Din et al., 2010). Different strategies have been proposed in order to improve the delivery in the gut, including the membrane fragilization (Tao et al., 2011) or the expression of invasin proteins in the bacteria membrane (Innocentin et al., 2009). Even though these strategies have drawbacks, many reports showed delivery efficiency when using the DNA vaccine platform, by both oral (Zurita-Turk et al., 2014; Pereira et al., 2015; Souza et al., 2016) or nasal administration (Mancha-Agresti et al., 2017).
The eGFP expression by enterocytes, after mice oral administration of non- encapsulated L. lactis MG1363 (pExu:egfp), was successfully reported by our group (Mancha-Agresti et al., 2016). It was also reported that delivery by this recombinant strain only occurred in the duodenum section between 12 and 72 h after gavage. The eGFP protein is the marker of choice for many biological studies. However, there are limitations to its use, both in vivo and in vitro experiments. For example, GFP may lose its direct fluorescence during tissue fixation, as well as for subsequent processing which leads to commercial use of antibodies for immunostaining for GFP detection. Also, GFP expression shows substantial biological variability, even among similar cell types in a single animal (Anderson et al., 2005; Brazelton and Blau, 2005). In addition, GFP protein could have diminished brightness, variability, or loss of fluorescence due to auto-fluorescence of the cells in green wavelengths, and also a delay between protein synthesis and fluorescence development (Cubitt et al., 1995; Aspiras et al., 2000; Shaner et al., 2005). Even more, conventional fluorescent proteins, such as GFP, emit in visible spectrum, where light absorption by tissue is strong (Deliolanis et al., 2008), therefore, its sensibility is low.
In order to improve our previous results and circumvent the drawbacks of egpf or gfp reporter gene, we decided to evaluate another reporter, the mCherry, using the same system, the pExu vector delivered by L. lactis ssp. cremoris MG1363. The mCherry is a monomeric, non-toxic protein and doesn’t need any stress induction to host organisms expressed at high levels (Carroll et al., 2010). It is highly resistant to photobleaching, consequently losing less fluorescence overtime (Viegas et al., 2007). An important issue to highlight about mCherry in comparison with other red-shifted fluorescent proteins, is their fluorescence emission beyond the 600 nm spectra, where light absorption by tissues is substantially reduced, this showcases enhanced detection sensitivity in whole body imaging applications by at least two orders of magnitude over GFP (Deliolanis et al., 2008).
A previous study using mCherry reporter gene was conducted by van Zyl et al. (2015) to analyze, after mice gavage, the migration and colonization in the gastrointestinal tract (GIT) of two recombinant LAB: Enterococcus mundtii ST4SA and Lactobacillus plantarum 423. This study reports mCherry as a promising reporter system for living cell imaging studies in vitro, in vivo, and ex vivo. They showed that both bacteria predominantly colonized the cecum and colon, and also persisted in the gut for at least 24 h (van Zyl et al., 2015). Another study used mCherry ORF cloned in prCR12 vector (heterologous expression of mCherry protein), transfected in three strains of Lactobacillus (L. plantarum Lp90, L. plantarum B2, and L. fermentum Pbcc11.5) showed great strategy in estimating gut colonization in zebrafish model using probiotics. Another interesting point highlighted by these authors is to avoidance of animal sacrifice (Russo et al., 2015).
Acid gastric secretion, a major defense mechanism against the majority of ingested microorganisms, is the first stress confronted by orally administered bacteria (Marteau et al., 1997). Once L. lactis has overcome the gastric barrier, they need to envisage the enteric secretions encountered in the duodenum region such as mucus, lysozyme, and phospholipase A2 (Peeters and Vantrappen, 1975; Harwig et al., 1995), defensins secreted by Paneth cells (Mallow et al., 1996) and also bile salt and pancreatic juices. Due to these adversities, high cell mortality is detected in duodenum (Drouault et al., 1999). Divergence may occur once distinct bacterial species demonstrate different reductions in viability, as shown in a study comparing survival of Bifidobacterium bifidum, Lactobacillus acidophilus, Lactobacillus bulgaricus, and Streptococcus thermophilus in the gastric compartment (Marteau et al., 1997).
To prevent this situation, and consequently increase L. lactis viability in duodenum region and also reach further regions from the intestine, we investigated the encapsulation technique, that has been extensively studied and standardized in in vitro experiments (Sheu and Marshall, 1993; Rowley et al., 1999; Krasaekoopt et al., 2003; Desai and Jin Park, 2005; Urbanska et al., 2007; Nedovic et al., 2011).
Once the focus of our group is the use of LAB as DNA vaccine platform in the treatment of different gut disease, like inflammatory bowel disease (IBD), an important issue concern how the encapsulation doses could be affected by the freezing. Demonstrating that bacteria remains viable after freezing could help the development of new therapeutical strategies, for example, preparation of encapsulate doses in one moment and administrate them in another one. Even more, in developing countries this application could help to solve logistic issues, for example frozen doses could be transport further distances without big damage. In order to provide support for in vivo studies, we initially performed in vitro assays to verify the efficiency of encapsulation process with alginate matrix and the viability of bacteria doses for 5 days after freezing.
In contrast with Kailasapathy (2006) studies, in which EE of Lactobacillus acidophilus and Bifidobacterium lactis was tested after storage, we unexpectedly verified a decrease in the CFU of L. lactis ssp. cremoris MG1363 doses soon after the encapsulation process when compared with non-encapsulated. However, when these bacteria were submitted to low pH conditions, simulating gastric environment, the viability of encapsulated bacteria increased around 8% when compared with non-encapsulated. The important issue of this result is to highlight that encapsulation matrix was able to protect the bacteria against these astringent conditions.
Our findings are in accordance with Mortazavian et al. (2007) who tested three different pH (extreme, intermediate, and normal), in extreme and normal conditions they showed that encapsulated L. acidophilus, with 2% sodium alginate, had slightly higher percentages of survival than non-encapsulated, being 17.4 and 16%, respectively (Mortazavian et al., 2007). Also, an increase of 9.52 and 7.52% was described in the survival of encapsulated bacteria (L. acidophilus) with again 2% of sodium alginate at pH = 3.0 for 3 h in 2 and 1% of bile salt respectively (Chandorkar et al., 2018). These authors highlighted that microencapsulation is a competent technique able to protect probiotics against gastrointestinal environment. All this data supports the idea that encapsulated techniques improve the survival of probiotic bacteria against unfavorable conditions.
This finding was corroborated by our in vivo results where the expression of mCherry protein in the duodenum was higher for encapsulated bacteria and was also detected in ileum. The qRT-PCR provided quantitative results that were in line with those obtained in confocal microscopy, specially, in duodenum and jejunum portions, evidencing higher expression of mCherry protein in animals which received encapsulated doses. Although fluorescence had been observed by confocal microscopy at ileum portion, no significant differences were observed in qRT-PCR assay for both, ileum and colon.
Microencapsulation process shows to protect bacteria doses against environment through the GIT. Our hypothesis is that the alginate matrix protects the bacteria against the adversities (stomach and duodenum), allowing their movement across the bowel, being the bacteria able to deliver the plasmid in further sections, as jejunum, ileum, and also slightly in colon.
To our knowledge, this is the first study describing the comparison of encapsulated vs. non-encapsulated L. lactis using DNA delivery strategy, in vivo. Compared with our previous results (Mancha-Agresti et al., 2016) microencapsulation technique enables higher and longer recombinant protein expression in the gut. Furthermore, this process increases bacteria survival allowing further segments of the gut such as ileum to be reached. The methodology of encapsulation adopted in this work is able to enhance the potential for mucosal delivery by LAB. These findings are uplifting for mucosal therapy for different kinds of bowel diseases.
Author Contributions
NC-R, MD, CC, LJ, and PM-A conceived the project. NC-R, MD, PM-A, and SCS wrote the original draft of the manuscript. NC-R, MD, PM-A, SL, AN, and VA wrote, reviewed and edited the manuscript. VA obtained the funding and supervised the project.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported with grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02398/full#supplementary-material
References
- Ai H., Olenych S. G., Wong P., Davidson M. W., Campbell R. E. (2008). Hue-shifted monomeric variants of Clavularia cyan fluorescent protein: identification of the molecular determinants of color and applications in fluorescence imaging. BMC Biol. 6:13. 10.1186/1741-7007-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain T., Mansour N. M., Bahr M. M. A., Martin R., Florent I., Langella P., et al. (2016). A new lactobacilli in vivo expression system for the production and delivery of heterologous proteins at mucosal surfaces. FEMS Microbiol. Lett. 363:fnw117. 10.1093/femsle/fnw117 [DOI] [PubMed] [Google Scholar]
- Anderson D. A., Wu Y., Jiang S., Zhang X., Streeter P. R., Spangrude G. J., et al. (2005). Donor marker infidelity in transgenic hematopoietic stem cells. Stem Cells 23 638–643. 10.1634/stemcells.2004-0325 [DOI] [PubMed] [Google Scholar]
- Aspiras M. B., Kazmerzak K. M., Kolenbrander P. E., McNab R., Hardegen N., Jenkinson H. F. (2000). Expression of green fluorescent protein in Streptococcus gordonii DL1 and its use as a species-specific marker in coadhesion with Streptococcus oralis 34 in saliva-conditioned biofilms in vitro. Appl. Environ. Microbiol. 66 4074–4083. 10.1128/AEM.66.9.4074-4083.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahey-El-Din M., Gahan C. G. M., Griffin B. T. (2010). Lactococcus lactis as a cell factory for delivery of therapeutic proteins. Curr. Gene Ther. 10 34–45. 10.2174/156652310790945557 [DOI] [PubMed] [Google Scholar]
- Běláková J., Horynová M., Křupka M., Weigl E., Raška M. (2007). DNA vaccines: are they still just a powerful tool for the future? Arch. Immunol. Ther. Exp. 55 387–398. 10.1007/s00005-007-0044-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez-Humarán L. G. (2009). Lactococcus lactis as a live vector for mucosal delivery of therapeutic proteins. Hum. Vaccin. 5 264–267. 10.4161/hv.5.4.7553 [DOI] [PubMed] [Google Scholar]
- Bolotin A., Wincker P., Mauger S., Jaillon O., Malarme K., Weissenbach J., et al. (2001). The complete genome sequence of the lactic acid bacterium. Genome Res. 11 731–753. 10.1101/gr.169701.There [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazelton T. R., Blau H. M. (2005). Optimizing techniques for tracking transplanted stem cells in vivo. Stem Cells 23 1251–1265. 10.1634/stemcells.2005-0149 [DOI] [PubMed] [Google Scholar]
- Carroll P., Schreuder L. J., Muwanguzi-Karugaba J., Wiles S., Robertson B. D., Ripoll J., et al. (2010). Sensitive detection of gene expression in mycobacteria under replicating and non-replicating conditions using optimized far-red reporters. PLoS One 5:e9823. 10.1371/journal.pone.0009823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandorkar V. V., Shukla N. G., Gomashe A. V. (2018). Microencapsulation of Lactobacillus acidophilus for its survival under high acid and high bile salt concentrations. Int. J. Curr. Res. 10 63818–63820. [Google Scholar]
- Chandramouli V., Kailasapathy K., Peiris P., Jones M. (2004). An improved method of microencapsulation and its evaluation to protect Lactobacillus spp. in simulated gastric conditions. J. Microbiol. Methods 56 27–35. 10.1016/j.mimet.2003.09.002 [DOI] [PubMed] [Google Scholar]
- Charteris W. P., Kelly P. M., Morelli L., Collins J. K. (1998). Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J. Appl. Microbiol. 84 759–768. 10.1046/j.1365-2672.1998.00407.x [DOI] [PubMed] [Google Scholar]
- Chávarri M., Marañón I., Ares R., Ibáñez F. C., Marzo F., del Carmen Villarán M. (2010). Microencapsulation of a probiotic and prebiotic in alginate-chitosan capsules improves survival in simulated gastro-intestinal conditions. Int. J. Food Microbiol. 142 185–189. 10.1016/j.ijfoodmicro.2010.06.022 [DOI] [PubMed] [Google Scholar]
- Cook M. T., Tzortzis G., Charalampopoulos D., Khutoryanskiy V. V. (2012). Microencapsulation of probiotics for gastrointestinal delivery. J. Control. Release 162 56–67. 10.1016/j.jconrel.2012.06.003 [DOI] [PubMed] [Google Scholar]
- Cubitt A. B., Heim R., Adams S. R., Boyd A. E., Gross L. A., Tsien R. Y. (1995). Understanding, improving and using green fluorescent proteins. Trends Biochem. Sci. 20 448–455. 10.1016/S0968-0004(00)89099-4 [DOI] [PubMed] [Google Scholar]
- Day R. N., Davidson M. W. (2009). The fluorescent protein palette: tools for cellular imaging. Chem. Soc. Rev. 38 2887–2921. 10.1039/b901966a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deliolanis N. C., Kasmieh R., Wurdinger T., Tannous B. A., Shah K., Ntziachristos V. (2008). Performance of the red-shifted fluorescent proteins in deep-tissue molecular imaging applications. J. Biomed. Opt. 13:44008. 10.1117/1.2967184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai K. G. H., Jin Park H. (2005). Recent developments in microencapsulation of food ingredients. Dry. Technol. 23 1361–1394. 10.1081/DRT-200063478 [DOI] [Google Scholar]
- Drouault S., Corthier G., Ehrlich S. D., Corthier R. (1999). Survival, physiology, and lysis of Lactococcus lactis in the digestive tract. Appl. Environ. Microbiol. 65 4881–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fávaro-Trindade C. S., Grosso C. R. F. (2002). Microencapsulation of L. acidophilus (La-05) and B. lactis (Bb-12) and evaluation of their survival at the pH values of the stomach and in bile. J. Microencapsul. 19 485–494. 10.1080/02652040210140715 [DOI] [PubMed] [Google Scholar]
- Gasson M. J. (1983). Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulietti A., Overbergh L., Valckx D., Decallonne B., Bouillon R., Mathieu C. (2001). An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 25 386–401. 10.1006/meth.2001.1261 [DOI] [PubMed] [Google Scholar]
- Goderska K., Zybala M., Czarnecki Z. (2003). Characterisation of microemcapsulated Lactobacillus rhamnosus LR7 strain. Pol. J. Food Nutr. Sci. 12 21–24. [Google Scholar]
- Goh C. H., Heng P. W. S., Chan L. W. (2012). Alginates as a useful natural polymer for microencapsulation and therapeutic applications. Carbohydr. Polym. 88 1–12. 10.1016/j.carbpol.2011.11.012 [DOI] [Google Scholar]
- Green M. R., Sambrook J. (2012). Molecular Cloning: A Laboratory Manual, 4th Edn. New York, NY: Cold Spring Harbor. [Google Scholar]
- Harwig S. S., Tan L., Qu X. D., Cho Y., Eisenhauer P. B., Lehrer R. I. (1995). Bactericidal properties of murine intestinal phospholipase A2. J. Clin. Invest. 95 603–610. 10.1172/JCI117704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:R19. 10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdeman L. V., Good I. J., Moore W. E. (1976). Human fecal flora: variation in bacterial composition within individuals and a possible effect of emotional stress. Appl. Environ. Microbiol. 31 359–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongying F., Xianbo W., Fang Y., Yang B., Beiguo L. (2014). Oral immunization with recombinant Lactobacillus acidophilus expressing the adhesin Hp0410 of Helicobacter pylori induces mucosal and systemic immune responses. Clin. Vaccine Immunol. 21 126–132. 10.1128/CVI.00434-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocentin S., Guimaraes V., Miyoshi A., Azevedo V., Langella P., Chatel J.-M., et al. (2009). Lactococcus lactis expressing either Staphylococcus aureus fibronectin-binding protein A or Listeria monocytogenes internalin a can efficiently internalize and deliver DNA in human epithelial cells. Appl. Environ. Microbiol. 75 4870–4878. 10.1128/AEM.00825-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jach G., Pesch M., Richter K., Frings S., Uhrig J. F. (2006). An improved mRFP1 adds red to bimolecular fluorescence complementation. Nat. Methods 3 597–600. 10.1038/nmeth901 [DOI] [PubMed] [Google Scholar]
- Jankowski T., Zielinska M., Wysakowska A. (1997). Encapsulation of lactic acid bacteria with alginate/starch capsules. Biotechnol. Tech. 11 31–34. 10.1007/BF02764447 [DOI] [Google Scholar]
- Kailasapathy K. (2006). Survival of free and encapsulated probiotic bacteria and their effect on the sensory properties of yoghurt. LWT Food Sci. Technol. 39 1221–1227. 10.1016/j.lwt.2005.07.013 23021757 [DOI] [Google Scholar]
- Krasaekoopt W., Bhandari B., Deeth H. (2003). Evaluation of encapsulation techniques of probiotics for yoghurt. Int. Dairy J. 13 3–13. 10.1016/S0958-6946(02)00155-3 11139021 [DOI] [Google Scholar]
- Lee K. Y., Heo T. R. (2000). Survival of Bifidobacterium longum immobilized in calcium alginate beads in simulated gastric juices and bile salt solution. Appl. Environ. Microbiol. 66 869–873. 10.1128/AEM.66.2.869-873.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-G., Tian F.-L., Gao F.-S., Tang X.-S., Xia C. (2007). Immune responses generated by Lactobacillus as a carrier in DNA immunization against foot-and-mouth disease virus. Vaccine 25 902–911. 10.1016/j.vaccine.2006.09.034 [DOI] [PubMed] [Google Scholar]
- Linares D. M., Kok J., Poolman B. (2010). Genome sequences of Lactococcus lactis MG1363 (revised) and NZ9000 and comparative physiological studies. J. Bacteriol. 192 5806–5812. 10.1128/JB.00533-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfipour F., Mirzaeei S., Maghsoodi M. (2012). Preparation and characterization of alginate and psyllium beads containing Lactobacillus acidophilus. ScientificWorldJournal. 2012:680108. 10.1100/2012/680108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallow E. B., Harris A., Salzman N., Russell J. P., DeBerardinis R. J., Ruchelli E., et al. (1996). Human enteric defensins. Gene structure and developmental expression. J. Biol. Chem. 271 4038–4045. 10.1074/jbc.271.8.4038 [DOI] [PubMed] [Google Scholar]
- Mancha-Agresti P., de Castro C. P., dos Santos J. S. C., Araujo M. A., Pereira V. B., LeBlanc J. G., et al. (2017). Recombinant invasive Lactococcus lactis carrying a DNA vaccine coding the Ag85A antigen increases INF-γ, IL-6, and TNF-α cytokines after intranasal immunization. Front. Microbiol. 8:1263. 10.3389/fmicb.2017.01263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancha-Agresti P., Drumond M. M., Rosa do Carmo F. L., Santos M. M., Coelho dos Santos J. S., et al. (2016). A new broad range plasmid for DNA delivery in eukaryotic cells using lactic acid bacteria: in vitro and in vivo assays. Mol. Ther. Methods Clin. Dev. 4 83–91. 10.1016/j.omtm.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteau P., Minekus M., Havenaar R., Huis in’t Veld J. H. (1997). Survival of lactic acid bacteria in a dynamic model of the stomach and small intestine: validation and the effects of bile. J. Dairy Sci. 80 1031–1037. 10.3168/jds.S0022-0302(97)76027-2 [DOI] [PubMed] [Google Scholar]
- Morello E., Bermúdez-Humarán L. G., Llull D., Solé V., Miraglio N., Langella P., et al. (2008). Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J. Mol. Microbiol. Biotechnol. 14 48–58. 10.1159/000106082 [DOI] [PubMed] [Google Scholar]
- Mortazavian A., Hadi Razavi S., Reza Ehsani M., Sohrabvandi S. (2007). Principles and methods of microencapsulation of probiotic microorganisms. Iran. J. Biotechnol. 5 1–18. [Google Scholar]
- Nedovic V., Kalusevic A., Manojlovic V., Levic S., Bugarski B. (2011). An overview of encapsulation technologies for food applications. Procedia Food Sci. 1 1806–1815. 10.1016/j.profoo.2011.09.265 [DOI] [Google Scholar]
- Nienhaus G. U., Wiedenmann J. (2009). Structure, dynamics and optical properties of fluorescent proteins: perspectives for marker development. ChemPhysChem 10 1369–1379. 10.1002/cphc.200800839 [DOI] [PubMed] [Google Scholar]
- Ott L., McKenzie A., Baltazar M. T., Britting S., Bischof A., Burkovski A., et al. (2012). Evaluation of invertebrate infection models for pathogenic corynebacteria. FEMS Immunol. Med. Microbiol. 65 413–421. 10.1111/j.1574-695X.2012.00963.x [DOI] [PubMed] [Google Scholar]
- Patterson G., Day R. N., Piston D. (2001). Fluorescent protein spectra. J. Cell Sci. 114 837–838. [DOI] [PubMed] [Google Scholar]
- Peeters T., Vantrappen G. (1975). The Paneth cell: a source of intestinal lysozyme. Gut 16 553–558. 10.1136/gut.16.7.553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira V. B., Saraiva T. D. L., Souza B. M., Zurita-Turk M., Azevedo M. S. P., De Castro C. P., et al. (2015). Development of a new DNA vaccine based on mycobacterial ESAT-6 antigen delivered by recombinant invasive Lactococcus lactis FnBPA+. Appl. Microbiol. Biotechnol. 99 1817–1826. 10.1007/s00253-014-6285-3 [DOI] [PubMed] [Google Scholar]
- Picot A., Lacroix C. (2004). Encapsulation of bifidobacteria in whey protein-based microcapsules and survival in simulated gastrointestinal conditions and in yoghurt. Int. Dairy J. 14 505–515. 10.1016/j.idairyj.2003.10.008 [DOI] [Google Scholar]
- Rowley J. A., Madlambayan G., Mooney D. J. (1999). Alginate hydrogels as synthetic extracellular matrix materials. Biomaterials 20 45–53. 10.1016/S0142-9612(98)00107-0 [DOI] [PubMed] [Google Scholar]
- Russo P., Iturria I., Mohedano M. L., Caggianiello G., Rainieri S., Fiocco D., et al. (2015). Zebrafish gut colonization by mCherry-labelled lactic acid bacteria. Appl. Microbiol. Biotechnol. 99 3479–3490. 10.1007/s00253-014-6351-x [DOI] [PubMed] [Google Scholar]
- Salminen S., Bouley C., Boutron-Ruault M. C., Cummings J. H., Franck A., Gibson G. R., et al. (1998). Functional food science and gastrointestinal physiology and function. Br. J. Nutr. 80(Suppl. 1), S147–S171. 10.1079/BJN19980108 [DOI] [PubMed] [Google Scholar]
- Sanders M. E., Guarner F., Guerrant R., Holt P. R., Quigley E. M., Sartor R. B., et al. (2013). An update on the use and investigation of probiotics in health and disease. Gut 62 787–796. 10.1136/gutjnl-2012-302504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N. G., Palmer A. E., Tsien R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22 1567–1572. 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Steinbach P. A., Tsien R. Y. (2005). A guide to choosing fluorescent proteins. Nat. Methods 2 905–909. 10.1038/nmeth819 [DOI] [PubMed] [Google Scholar]
- Sheu T. Y., Marshall R. T. (1993). Microentrapment of lactobacilli in calcium alginate gels. J. Food Sci. 58 557–561. 10.1111/j.1365-2621.1993.tb04323.x [DOI] [Google Scholar]
- Souza B. M., Preisser T. M., Pereira V. B., Zurita-Turk M., de Castro C. P., da Cunha V. P., et al. (2016). Lactococcus lactis carrying the pValac eukaryotic expression vector coding for IL-4 reduces chemically-induced intestinal inflammation by increasing the levels of IL-10-producing regulatory cells. Microb. Cell Fact. 15:150. 10.1186/s12934-016-0548-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L., Pavlova S. I., Ji X., Jin L., Spear G. (2011). A novel plasmid for delivering genes into mammalian cells with noninvasive food and commensal lactic acid bacteria. Plasmid 65 8–14. 10.1016/j.plasmid.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanska A. M., Bhathena J., Prakash S. (2007). Live encapsulated Lactobacillus acidophilus cells in yogurt for therapeutic oral delivery: preparation and in vitro analysis of alginate–chitosan microcapsules. Can. J. Physiol. Pharmacol. 85 884–893. 10.1139/Y07-057 [DOI] [PubMed] [Google Scholar]
- van Zyl W. F., Deane S. M., Dicks L. M. T. (2015). Use of the mCherry fluorescent protein to study intestinal colonization by Enterococcus mundtii ST4SA and Lactobacillus plantarum 423 in mice. Appl. Environ. Microbiol. 81 5993–6002. 10.1128/AEM.01247-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas M. S., Martins T. C., Seco F., do Carmo A. (2007). An improved and cost-effective methodology for the reduction of autofluorescence in direct immunofluorescence studies on formalin-fixed paraffin-embedded tissues. Eur. J. Histochem. 51 59–66. [PubMed] [Google Scholar]
- Wegmann U., O’Connell-Motherway M., Zomer A., Buist G., Shearman C., Canchaya C., et al. (2007). Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189 3256–3270. 10.1128/JB.01768-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. M., Mercenier A. (2008). Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat. Rev. Microbiol. 6 349–362. 10.1038/nrmicro1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Turk M., Del Carmen S., Santos A. C. G., Pereira V. B., Cara D. C., Leclercq S. Y., et al. (2014). Lactococcus lactis carrying the pValac DNA expression vector coding for IL-10 reduces inflammation in a murine model of experimental colitis. BMC Biotechnol. 14:73. 10.1186/1472-6750-14-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.