Abstract
During HIV-1 assembly and budding, Gag protein, in particular the C-terminal domain containing the nucleocapsid domain (NCd), p1 and p6, is the site of numerous interactions with viral and cellular factors. Most in vitro studies of Gag have used constructs lacking p1 and p6. Here, using NMR spectroscopy, we show that the p1–p6 region of Gag (NCp15) is largely disordered, but interacts transiently with the NCd. These interactions modify the dynamic properties of the NCd. Indeed, using isothermal titration calorimetry (ITC), we have measured a higher entropic penalty to RNA-binding for the NCd precursor, NCp15, than for the mature form, NCp7, which lacks p1 and p6. We propose that during assembly and budding of virions, concomitant with Gag oligomerization, transient interactions between NCd and p1–p6 become salient and responsible for (i) a higher level of structuration of p6, which favours recruitment of budding partners; and (ii) a higher entropic penalty to RNA-binding at specific sites that favours non-specific binding of NCd at multiple sites on the genomic RNA (gRNA). The contributions of p6 and p1 are sequentially removed via proteolysis during Gag maturation such that the RNA-binding specificity of the mature protein is governed by the properties of NCd.
INTRODUCTION
In retroviruses, the Pr55Gag polyprotein precursor (Gag) is the only protein required for particle formation. It is composed of the structural protein domains (Figure 1A): the matrix (MA), the capsid (CA), a spacer peptide p2, the nucleocapsid domain (NCd), a hydrophobic peptide p1, and an acidic p6 domain. As part of Gag, the MA domain targets Gag to the plasma membrane; CA drives Gag multimerization during assembly; NCd recruits the viral genomic RNA (gRNA) into virions and facilitates the assembly process; and p6 recruits the endosomal sorting complex (ESCRT), which catalyses the membrane fission step to complete the budding process (1).
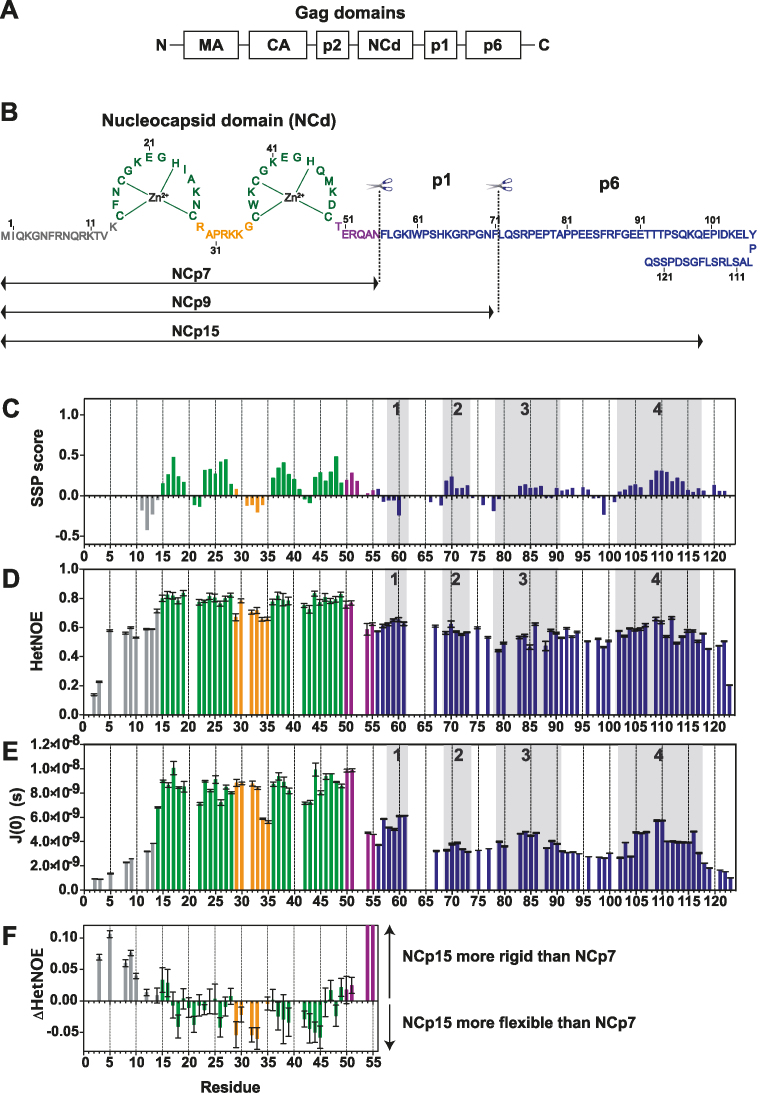
Figure 1.
Structural and dynamic characterization of NCp15 from NMR chemical shifts and 15N relaxation data. (A) Schematic of full-length Gag, (B) Sequence of the HIV-1 C-terminal domain of Gag (NCp15). The first cleavage by the HIV-1 protease first liberates the NCp15 protein, then NCp9 and finally the mature form of NCd called NCp7. The dashed line represents the two sites of protease cleavage present in NCp15. Residues are coloured in grey for residues in the N-terminal part of NCd, green for those in the zinc knuckles and orange for residues in the linker between the two zinc knuckles of NCd and purple for the C-terminal domain of NCd. Due to a limitation in space, the sequence of p6 is not drawn linearly, but this does not represent a fold back of p6 on itself, (C) Secondary structural propensities calculated by the SSP program (58). Cα, C β, CO and Hα were used as input data for the calculation of the SSP score. Positive values indicate the amount of α-helical conformation present along the sequence whereas negative values indicate extended or β-strand conformations. (D) 15N–{1H} NOE (HetNOE) values are indicative of the magnitude of local subnanosecond motions (high values: restricted motions; low values: high-amplitude motions). (E) Spectral densities J(0) extracted by spectral density mapping from 15N relaxation data (T1, T2, HetNOE) of NCp15. J(0) values are indicative of slow overall and segmental tumbling motions present in NCp15. The boxes in grey indicated the four regions of p1–p6 domains showing significant secondary structure propensities. (F) Differences of HetNOE within the NCd between NCp15 and NCp7 to probe the change in restrictions of motions of the backbone of the precursor and mature forms of the NCd.
Gag is present as monomers or dimers in the cytoplasm where Gag and gRNA first interact (2–6). Once targeted to the plasma membrane, Gag oligomerisation and the gRNA nucleate particle assembly (1). Both MA and NCd within Gag are able to bind to RNA in the cytoplasm (7–9). However, the NCd of Gag is the primary viral determinant that drives gRNA packaging by binding to a packaging Ψ signal, whereas MA selects a subset of tRNAs in the cytosol which regulates MA and Gag-membrane binding (8,10–12). Global changes in the RNA binding specificity of Gag regulate virion synthesis (12). Prior to, and after virion assembly and maturation, the NCd preferentially binds to the Ψ region at the level of SL3, one of the major determinants of gRNA encapsidation (6,13,14), and to Rev Response elements (RRE) of the gRNA (12). The RNA-binding properties of NCd in its mature forms (also called NCp7, Figure 1B) hence resemble that of unassembled Gag in the cytosol. During assembly and budding, this specificity transiently changes in a manner that facilitates genome packaging, i.e. the NCd within Gag non-specifically binds to many sites on the gRNA (12).
After assembly, the release step is mediated by the cellular ESCRT machinery, which is hijacked by Gag. p6 harbors motifs known as late (L) domains. Its primary L domain motif, P78TAP81, binds the ESCRT-I component TSG101 (15,16), which in turn recruits the ESCRT-III machinery enabling final pinching off of particles and subsequent recycling of the ESCRT components (reviewed in (17,18)). p6 also harbors an auxiliary L domain (L106YPLASL112) that engages the ESCRT pathway component ALIX, in addition to its primary PTAP-type L domain (19,20). For both TSG101 and ALIX, the L domains in p6 are the strongest interaction sites with Gag but, the NCd cooperates with p6 for their binding and to promote virus budding (21–27). p6 is also engaged in the binding of the retroviral protein Vpr via its conserved L112RSLFG117 motif (28–30). The coordination between protease activation, virion assembly and budding is critical and ensures the infectiousness of progeny virions (21,31).
During or shortly after budding of the particle from the cell surface, the viral protease cleaves the Gag polyprotein precursor to trigger HIV-1 maturation (31). Three different forms of NCd appear subsequently (Figure 1B). The first cleavage by the HIV-1 protease occurs between MA-CA-p2 and NCd-p1–p6, thereby liberating NCp15. The second cleavage frees NCp9 (NCd-p1) whereas the last one releases the NCd itself, which constitutes the final maturation form of NCd (NCp7). NCp7 exhibits the optimal RNA chaperone activity, essential notably during reverse transcription (32–39). NCp15 and NCp9 intermediates appear transiently but their lifetime is specifically programmed (31,40). The correct processing of the cleavage site between p1 and p6 is essential and NCp15-containing virions are non-infectious (41–43).
Although the structures of isolated domains of the C-terminal part of Gag, NCd or p6, have been extensively studied independently (30,44,45) or in complex with their partners (29,37,46–53), no structural data are available about the conformation of these domains when they are present together. Yet, p6 is an essential domain for assembly and budding. There is therefore a clear need for in-depth studies of the C-terminal part of Gag containing p1 and p6. The low solubility of full-length Gag and its extreme sensitivity to proteolysis (54) precludes this investigation. Only a Gag construct lacking p1 and p6 was amenable to an NMR dynamic study which showed that the structured domains within Gag (MA, CA and NCd) retain the same fold as their isolated counterparts. They reorient semi-independently from each other through unfolded and dynamic linkers that connect the structural domains (3). We can thus anticipate that the dynamic properties of a construct composed of NCd-p1–p6 (NCp15) are conserved in the context of Gag.
In the present study, we have explored the conformational landscape of NCd by a comprehensive NMR dynamic study using 15N relaxation measurements of NCd in its different stages of maturation by the HIV-1 protease (NCp7, NCp9 and NCp15). We also analyzed their interaction with the RNA stem-loop SL3 from the encapsidation signal. For the first time, we show that p1–p6 within NCp15 is disordered in solution and that short-lived α-helices are formed in p1–p6 upon long-range and transient contacts with the NCd, linking p6 to NCd and likely promoting the binding of p6 to its partners. Using NMR and microcalorimetry, we also demonstrate that this dynamical behavior changes the thermodynamics of the NCd interaction with SL3. Indeed, we uncover an entropic penalty to RNA-binding higher for the NCd precursors than for its mature form. We propose that during assembly and budding of virions, concomitant with Gag oligomerization, the transient interactions between NCd and p1–p6 become salient and responsible for (i) a higher level of p6 structuration favoring recruitment of its budding partners and (ii) a higher entropic penalty to NCd RNA-binding at specific sites leading to non-specific RNA-binding of NCd at a multitude of sites on gRNA necessary during assembly. The maturation by the protease cleaving off p6 and p1 from NCd eventually reverts the RNA-binding specificity of the mature NCd, ie the NCd no longer binds to multiple sites, but binds to discrete specific sites on gRNA containing unpaired guanine.
MATERIALS AND METHODS
Expression and purification of recombinant HIV-1 NCp7, NCp9 and NCp15
NCp7, NCp9 and NCp15 (HIV-1 strain NL4-3) were expressed without any tag either at their N- or C-termini in Escherichia coli from plasmids built into a pET-3a vector (Novagen). Mutants of NCp9 were generated by PCR amplifications using specific primers (55). NCp7 and NCp9 were overexpressed in E. coli BL21(DE3)pLysE strain while NCp15 was overexpressed in E. coli BL21(DE3)star. NCp7 was overexpressed isotopically 15N-labeled and 13C–15N-labeled as previously described (47,56). The same protocols were used for NCp9. For NCp15, the protocols of purification were modified since NCp15 presented a lower expression level and was more sensitive to proteolysis. Briefly, the cells were resuspended in a lysis buffer containing 50 mM Tris–Cl (pH 8.5), 10% glycerol, 100 mM NaCl, 10 mM EDTA, 10 mM β-mercaptoethanol, 1 mM PMSF, 5 μg/ml pepstatin A and one protease inhibitor tablet at 4°C. The cells were sonicated and then centrifuged at 45 000 g for 30 min at 4°C. The supernatant was collected and the nucleic acids were precipitated by adding 5% polyethyleneimine dropwise to a final concentration of 1%, and the mixtures were stirred for 45 min before centrifugation at 45 000 g for 30 min at 4°C. The supernatant was collected and loaded onto a 53 ml Q-Sepharose (HiLoad 26/40, HP, GE Healthcare) equilibrated in buffer A (50 mM Tris–Cl pH 8.5, 10% glycerol, 100 mM NaCl, 10 mM EDTA, 10 mM β-mercaptoethanol, 1 mM PMSF). The fractions containing NCp15 were pooled and the sample was supplemented with ammonium sulfate to a final concentration of 1 M before loading it onto a 53 ml phenyl-Sepharose (HiLoad 26/10, HP, GE Healthcare). The protein was eluted with a 500 ml gradient of 30–70% buffer B (50 mM Tris–Cl pH 8.5, 10% glycerol, 1 M ammonium sulfate, 5 mM EDTA, 10 mM β-mercaptoethanol). The fractions containing NCp15 were pooled and loaded onto a 320 ml Superdex75 (HiLoad 26/60, Prep Grade, GE Healthcare) equilibrated in buffer C (50 mM Tris–Cl pH 8.5, 10% glycerol, 100 mM NaCl, 10 mM β-mercaptoethanol, 0.1 mM ZnCl2). The eluted protein was pooled and loaded onto a column of 5 ml Heparin Sepharose (HiTrap Heparin, HP, GE Healthcare) equilibrated with buffer D (50 mM Tris–Cl pH 8.5, 100 mM NaCl, 10 mM β-mercaptoethanol, 0.1 mM ZnCl2). The protein was eluted with a 200 ml gradient of 15% to 30% buffer E (50 mM Tris–Cl pH 8.5, 1 M NaCl, 10 mM β-mercaptoethanol, 0.1 mM ZnCl2). The fractions containing NCp15 were pooled, concentrated and dialyzed against the NMR buffer (25 mM deutered sodium acetate pH 6.0, 25 mM NaCl, 0.1 mM ZnCl2 and 0.1 mM β-mercaptoethanol) using an amicon unit of 3 kDa (Millipore).
RNA sample preparation
The SL3 RNA stem–loop, in its extended version, 5′-GGACUAGCGGAGGCUAGUCC-3′ was purchased, deprotected and desalted, from Dharmacon Research®. The RNA sample was heated at 90°C for 5 min and cool on ice quickly for 10 min. It was then lyophilized and dissolved in the NMR buffer.
NMR experiments
NMR spectra of NCp7, NCp9 and NCp15, unless otherwise stated, were recorded at 10°C on Bruker 950 MHz spectrometer equipped with a cryogenic probe. NMR data on complexes with SL3 were recorded at 25°C and 950 MHz. Some data were also recorded on Bruker 600 or 700 MHz spectrometers, only the 600 MHz spectrometer being equipped with a cryogenic probe.
Analysis of NMR chemical shifts
Backbone chemical shift assignments of NCp7, NCp9 and NCp15 were performed using standard 3D NMR experiments (57). The backbone NMR chemical shifts of NCp15 were deposited to BMRB (entry code 26843). Combined NMR chemical shift perturbations of amide groups Δδ (in ppm) were derived from 1H and 15N chemical shift differences as Δδ (H,N) = √ [(Δ15N * WN)2 + (Δ1H * WH)2], where Δ = δ protein 1 - δ protein 2 (difference of chemical shifts between protein 1 and protein 2) and WH = 1 and WN = 1/6. A statistical analysis of these chemical shift perturbations is presented in Supplementary Figure S8BC.
Secondary structure propensity (SSP) scores were calculated using Cα, C β, CO and Hα chemical shifts, five-residue weighted averaging, and all other default parameters (58).
15N NMR relaxation experiments
The 15N backbone relaxation experiments were performed on 0.9–1 mM samples of 15N-uniformly labeled NCp15, NCp9 and NCp7. The T1, T2 and hetNOE 15N experiments have been performed using the standard Bruker pulse sequence libraries. All the experiments were recorded with a relaxation delay of 4s between two successive scans. The T1 data were collected using 20, 50 (repeat), 80, 100, 150, 200, 250, 300, 400 (repeat), 500, 700, 800, and 3000 ms for the values (13) of recovery delay. The T2-CPMG experiments were collected with 10 values of 16, 32, 64, 80, 128 (repeat), 160, 208 (repeat), 256, 320 and 400 ms. CPMG pulse trains were used with a 0.9 ms delay between successive 15N 180° pulses. The 15N-{1H} NOE (HetNOE) values were taken as the ratio between the intensities recorded with and without saturation of the amide protons. In all experiments, the points corresponding to the different relaxation delays were acquired in an interleaved manner.
The experiments were processed using NMRPipe (59) and SPARKY (60) was used to measure the intensities values of cross-peaks at the different relaxation delays. CurveFit (AG. Palmer Lab) and PYTHON (www.python.org), R (www.r-project.org) scripts were used to determine the R1 and R2 relaxation rates as well as the associated uncertainties from the single-exponential decay. Additionally, reduced power spectral density mapping for the determination of J(0) were used using the strategy designed by Farrow et al. (61) and PYTHON scripts to achieve the procedure. The uncertainties in the calculated spectral density values were obtained from Monte Carlo simulations using the errors deduced from the preceding fits of R1 and R2 values and the spectral noise errors for HetNOE experiments. Using these errors, 500 synthetic files were generated for each observable (T1, T2, HetNOE) and after extraction of the 500 J(0) values, these later were examined and the means and standard deviations extracted. The whole calculations were achieved using PYTHON and R scripts. A statistical analysis of the variations of HetNOE within the NCd between the three forms of the NC protein is presented in Supplementary Figure S8A.
Isothermal titration calorimetry (ITC)
Dissociation constants for NCp7, NCp9 and NCp15 binding to SL3 were recorded on a MicroCal® ITC200 system. SL3 RNA and the protein samples were extensively dialyzed against the ITC buffer (25 mM Na-acetate pH 6.0, 25 mM NaCl, 0.1 mM TCEP, 0.1% Tween, 0.1 mM ZnCl2) prior to titration. Heats of reaction were measured at 30°C for 15 injections of 2.54 μl of one NC protein (in the syringe at a concentration between 60 and 70 μM) into 280 μl of SL3 RNA (in the cell at a concentration of 5 μM). The heats of dilution were obtained by titrating the identical protein sample into a cell containing the ITC buffer and was subtracted from the raw data. The ITC data were analyzed with the software ORIGIN® using a single set of sites model. The standard errors were estimated from the data spread and from the uncertainty of the titrant concentration determination as previously described (62). A statistical analysis of the variations of the thermodynamic functions measured for the three complexes with SL3 is presented in Supplementary Figure S8D.
RESULTS
p1–p6 within NCp15 is largely disordered, but presents regions that transiently adopt α-helical structures
We explored the conformational landscape of NCd by a comprehensive NMR dynamic study of NCd in its different stages of maturation. We thus expressed without any tags, purified and assigned the NMR backbone resonances of NCp7, NCp9 and NCp15 proteins that correspond to the three states of maturation of NCd by the HIV-1 protease (BMRB entry number 26843, (63)).
The limited frequency dispersion of the resonances from p1–p6 (Supplementary Figure S1) within NCp15, is characteristic of an intrinsically disordered domain (IDD). The secondary structure propensity (SSP) score (58) calculated from NMR chemical shifts of NCp15 (Figure 1C) and the 15N-{1H} NOE (HetNOE) (Figure 1D and Supplementary Figure S2DE) support this trend, but highlight several regions in p1–p6 with a significant propensity to form secondary structures. Indeed, helical propensities of 17%, 14% and 11%, were estimated for regions 4, 2 and 3 respectively. The conversion of the measured 15N T1, T2 and HetNOE data into spectral densities at zero-frequency, J(0), gives information on the motional restriction at single residue resolution. Basically, higher J(0) values are a hallmark of regions displaying well-folded or transient secondary structures. These latter regions are characterized by the occurrence of J(0) maxima in their center (64,65). In p1–p6, J(0) maxima were observed for regions 1, 2, 3 and 4 (Figure 1E). p1–p6 within NCp15 is thus largely disordered, but presents regions that transiently adopt secondary structures, notably in regions known to interact with protein partners (TSG101 with region 3 and ALIX and Vpr with region 4 of p6).
The NMR data of the NCd within NCp15 (Figure 1 and Supplementary Figure S2) showed that the two zinc knuckles (ZKs) define two well-structured regions (high HetNOE and J(0) values) separated by a more flexible linker. Similar data were obtained for NCp7 (51) and NCp9 (Supplementary Figure S3), showing that the global folding and dynamics of the NCd is conserved in the three forms. However, the precursor NCp15 is more rigid in the N-terminal part and globally more flexible for residues of the ZKs and of the linker (Figure 1F). In addition, the J(0) values for residues of the NCd increased by 2.5 fold when comparing NCp15 to NCp7, related to the presence of the disordered p1–p6 tail (Supplementary Figure S3). The motions of the NCd and p1–p6 are thus coupled, suggesting that transient contacts between NCd and p1–p6 could occur.
Long-range transient interactions occur between p1 or p1–p6 and the NCd
As suggested by the dynamics of NCp15, we next investigated whether the NMR data could provide evidence of transient interactions between p1–p6 and NCd as sometimes observed between a structured domain and an unfolded tail of a protein (66).
Since p1–p6 is disordered within NCp15, when one compares the NMR chemical shifts of the NCd residues within NCp7 and NCp15, the chemical shift changes observed for NCd residues that are not sequentially close to the last C-terminal residue of NCp7, namely residue 55, are indicative of transient long-range interactions with the part of the protein that is absent in NCp7, therefore p1–p6. Globally, the presence of p1 in NCp9 leads to large chemical shift variations for K3, G4, F16, N17, C18, A25, K33 and K38 in NCd, whereas the presence of p6 in NCp15 causes additional chemical shift perturbations mainly for A25 and K38 (Figure 2AB and Supplementary Figure S4C). Such perturbations are also observed for residues L57, I60 and W61 of p1, which exhibited chemical shift variations in NCp15 compared to their chemical shifts in NCp9 (Figure 2A and Supplementary Figure S4A).
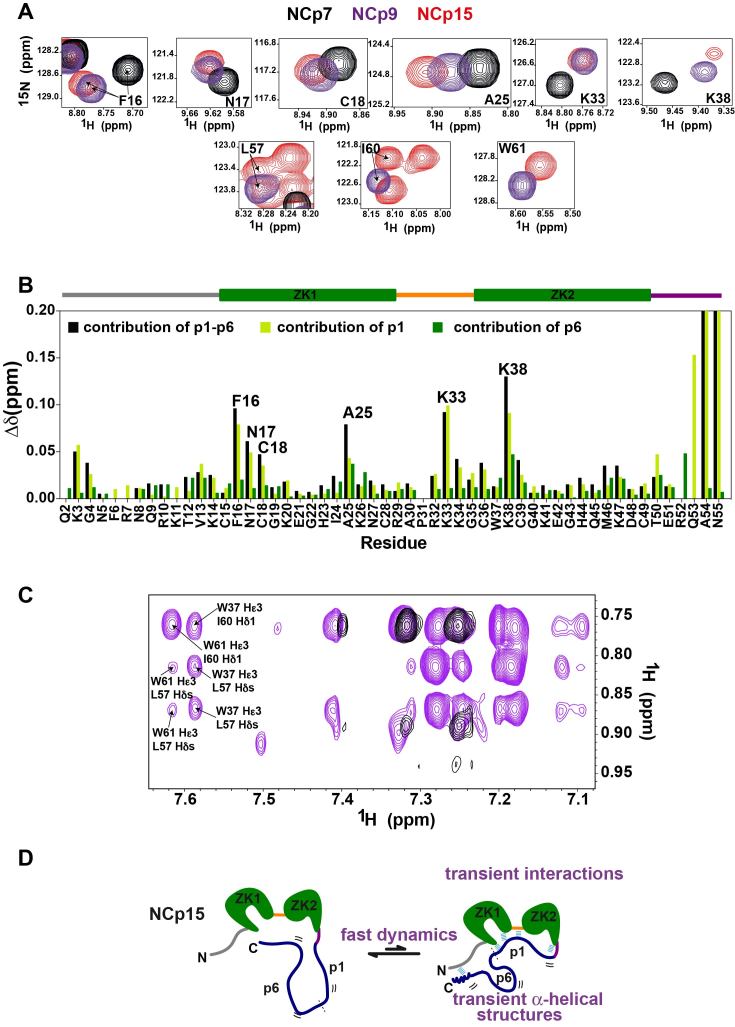
Figure 2.
Transient interactions between p1–p6 and NCd domains highlighted by NMR data measured on the precursor and mature forms of the NCd. (A) Peaks of F16, N17, C18, A25, K33 amide groups in NCp7 (black), in NCp9 (purple) and in NCp15 (red); and peaks of L57, I60 and W61 amide groups in NCp9 (purple) and in NCp15 (red), all extracted from 1H–15N BEST-TROSY. (B) Contributions of p1–p6 (black bars), p1(light green bars) and p6 (dark green bars) to NCd chemical shift perturbations measured for the amide groups. The p1–p6 (respectively p1) contributions were calculated as the difference in chemical shifts of NCd amide groups (combined 1H/15N shifts) between NCp15 (respectively NCp9) and NCp7, the p6 contributions were calculated as the difference in chemical shifts of NCd amide groups (combined 1H/15N shifts) between NCp15 and NCp9. The delimitation of the different regions in NCd is indicated as a drawing placed above the graphs using the color code of Figure 1A. (C) 2D NOESY (150 ms) measured in D2O on NCp9 (in purple) showing NOEs between proton Hϵ3 of W37 and protons of methyl groups of L57 and I60, the same region of a 2D NOESY recorded in the same conditions on NCp7 is displayed in black. (D) Schematic summarizing the interactions between NCd, p1 and p6 highlighted by the NMR chemical shifts and the dynamics analysis of the NCd in NCp7, NCp9 and NCp15.
To determine whether any persistent inter-residue contact was detectable between p1 or p1–p6 and NCd, we measured 3D 15N- and 13C-edited NOESY-HSQCs. Only intra-residue and nearest-neighbour inter-residue 1H–1H NOEs were observed in the p1 and p1–p6 domains, and no long-range NOEs were detected between p1 and NCd or p1–p6 and NCd. These data confirm that there is no evidence of any persistent structural element within p1 or p1–p6, and show that there is no long-lived interaction between p1–p6 and NCd. Weak 1H–1H NOEs could be detected and assigned in 2D (1H,1H)-NOESY spectra of NCp9 recorded in D2O with mixing times of 150 or 300 ms in order to obtain enhanced sensitivity for the weak NOEs (Figure 2C). Indeed, the Hϵ3 proton of W37 exhibits cross-peaks with both methyl groups of L57 and with the δ1 methyl group of I60, demonstrating proximity between W37 in NCd and L57 and I60 in p1. The introduction of the L57A, I60A and W61A point mutations in NCp9 confirmed the assignment of these NOEs (Supplementary Figure S5). These NOEs are also observable in 2D (1H,1H)-NOESY spectra of NCp15 measured in D2O (Supplementary Figure S5A). Overall, these data show that p1 transiently interacts with a region of the NCd defined by F16, N17, A25, C18, A25, K33 and K38, both in NCp9 and NCp15 and that p6 reinforces these interactions.
Interactions between p6 and the NCd were not clearly revealed by chemical shift variations which implies that p6 interacts at multiple sites on the NCd like commonly observed for fuzzy complexes (67). Indeed, p1–p6 rapidly interconverts between an ensemble of conformations that are partially restrained by transient interactions with the NCd. This ensemble may involve binding of the flexible p1–p6 at one predominant site or at multiple sites on the NCd. In NCp15, the N-terminal residues are globally less flexible than in NCp7 (Figure 1F) and NCp9. This restriction in mobility is likely due to transient interactions between acidic residues of p6 and the N-terminal basic residues of NCd within NCp15. The NMR data collected for the three proteins are thus in agreement with p1 transiently interacting with the NCd at one site described by the NMR chemical shift perturbations, whereas the acidic p6 domain has multiple sites of interaction in the N-terminal tail which is unfolded and positively charged (Figure 2D).
Changes in fast dynamics observed for the different states of maturation of the nucleocapsid domain correlate with changes in entropy penalty for SL3 binding
We next investigated whether these transient interactions could impact the interaction of NCd with SL3, the major component of the gRNA encapsidation signal, by NMR and ITC. The NMR chemical shift perturbations measured on NCd amide groups upon binding to SL3 were very similar regardless of the maturation state of this domain (Figure 3A and Supplementary Figure S6). Therefore, SL3 is very likely positioned into the hydrophobic plateau of the NCd using the same specific interactions, independently of its maturation state. Moreover, p1–p6 did not show large chemical shift perturbations upon SL3 binding (Figure 3B), showing that they do not participate directly in SL3 binding. Overall, this shows that the NCd is involved in SL3 binding in both the precursor or mature forms of this domain and that the SL3-binding interface is virtually identical in the three complexes.
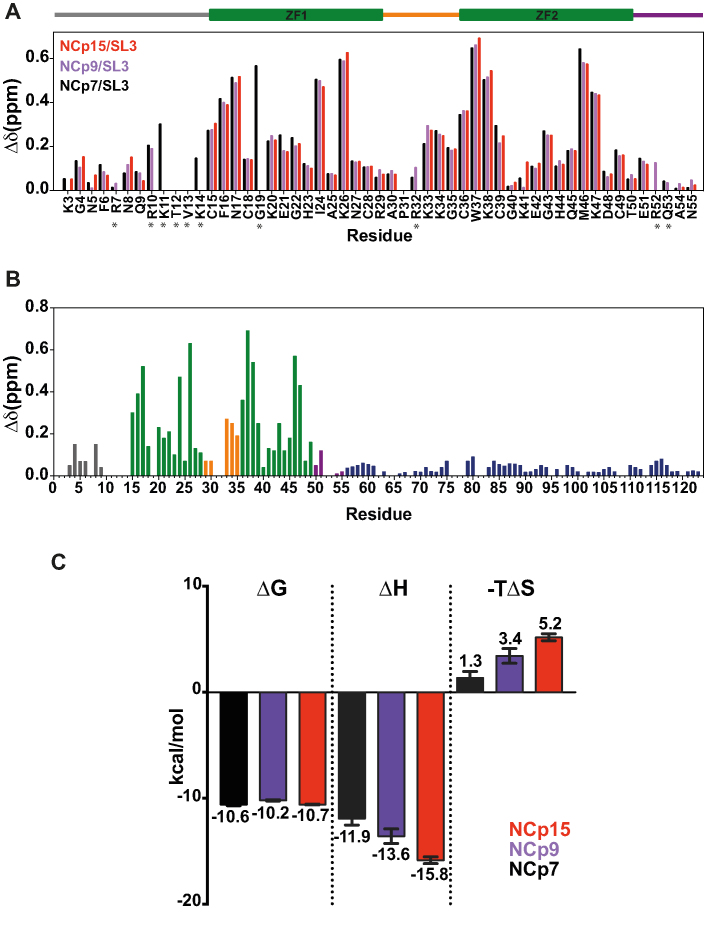
Figure 3.
NMR and ITC analysis of NCd/SL3 complexes. (A) Chemical shift perturbations upon SL3 binding for NCp7 (black bars), NCp9 (purple bars) and NCp15 (red bars) observed at the level of the NC domain. The delimitation of the different regions in NCd is indicated as a drawing placed above the graphs using the color code of Figure 1A. Stars indicate that we do not have the data for the three complexes due to peak superposition. (B) NMR chemical shift perturbations upon SL3 binding measured for NCp15 as a function of the sequence of NCp15. The color code for residues is that of Figure 1. (C) Thermodynamic components (ΔG: free energy, ΔH: enthalpy and –TΔS: entropy) displayed as bars extracted from the calorimetric titrations of SL3 RNA with NCp7 (in black), NCp9 (in purple) and NCp15 (in red). Values are reported as means ± standard error. The uncertainties on the fitted parameters were estimated from the data spread and from the uncertainty of the protein concentration determination (5%) (62).
We used ITC to gain insight into the thermodynamics of the SL3/NCd interaction (Figure 3C and Supplementary Figure S7). In agreement with the NMR structure of NCp7/SL3 complex (47), the stoichiometry of binding is 1 to 1 (Supplementary Figure S7B). The free energies of binding (ΔG) of NCp7, NCp9 and NCp15 to SL3 complexes were similar (≈-10 kcal/mol). However, the enthalpy and entropy contributions to the free energy of binding differed remarkably (Figure 3C). To be precise, in the constructs containing p1 or p1–p6, a higher -TΔS penalty was measured for SL3 binding (2 kcal/mol for NCp9 and 4 kcal/mol for NCp15 compared to NCp7) which was compensated by a higher enthalpy of interaction, showing that both p1 and p6 contribute to changes in the entropic penalty (Figure 3C). An entropy change could arise from a number of factors, the major ones being differences in solvent entropy or conformational entropy of binding that reflects the changes in dynamics between the free and bound states of interacting species (68). Although we cannot definitively rule out that differences in solvation upon SL3 binding contribute to this entropic penalty, it is reasonable to assume that the changes in solvent entropy upon ligand binding are similar in the three complexes, given that both the ligand and the ligand-binding interface are identical. Moreover, it was previously shown that the binding of NCd to nucleic acids (NA) notably results in a loss of motion of the NCd ZKs and linker (3). Therefore, the differences between the three complexes in the entropic contribution to RNA binding most probably arise from differences in the conformational entropy as previously observed in other complexes (68,69). Indeed, it was recently shown that changes in conformational entropy originate from enhanced pico to nanosecond internal motions and that fast timescale dynamics tune the entropic contribution of ligand affinity (68–71). In NCp15, transient interactions are responsible for a higher flexibility of the ZKs and the linker (Figure 1E). Changes in fast dynamics observed for the different states of maturation of the nucleocapsid domain correlate with changes in entropic penalty for SL3 binding. Therefore, the resulting increase in global flexibility of the ZKs and of the linker that occur in the fast timescale (ns–ps) do not change the affinity of NCd to SL3 but result in a conformational entropic penalty to SL3 binding by the precursors.
DISCUSSION
The C-terminal domain of Gag is a platform for molecular interactions involved in the binding of gRNA, ALIX, TSG101 and Vpr. In this work, we have studied the C-terminal part of Gag encompassed in NCp15 and we report the first comprehensive study that analyses the conformational landscape of each step of maturation of the NCd (NCp15, NCp9 and NCp7) together with their interactions with SL3. It was previously shown using NMR on a large HIV-1 Gag fragment containing MA-CA-p2-NCd, but lacking p1 and p6, that each folded domain (MA, CA and NCd) exhibit the same fold as their isolated counterparts and that they reorient semi-independently from each other through unfolded spacers that linked them together (3). Moreover, in presence of NA mimicking the gRNA, it was shown that a 1:1 Gag/NA complex is formed involving NCd and that a secondary weak interaction site involving MA can be observed at high concentration of NA (3) in agreement with the property of MA to bind NA. In cells, it was demonstrated that MA binds tRNAs (12). Globally, these previous studies demonstrate that the dynamic and binding properties of NCp15 that we uncover in this work are conserved in the context of Gag.
Our work thus provides a new picture of the structural and dynamic properties of the NCd during the assembly, budding and maturation of the virions (Figure 4). Indeed, we have demonstrated that p1–p6 within NCp15 is an IDD that experiences transient folding and long-range transient interactions with the NCd. p6 interacts at multiple sites within the positively-charged unstructured N-terminal part of NCp15 (Figure 4A). Interestingly, previous NMR studies on p6 or parts of p6 in various media (29,72) identified helices in the regions 3 and 4 where we uncovered transient helical propensities (Figure 1), but no significant amount of secondary structures was found for p6 in aqueous buffer. This strongly suggests that the transient folding of p6 occurs concomitantly to its short-lived contacts with the NCd. This behavior resembles that of multifunctional, intrinsically disordered proteins (IDPs) of HCV virus such as NS5A in which a network of electrostatic long-range interactions channels the transient folding of charged elements (73). The influence of the distribution of residues with opposite-charged residues on the conformations of IDPs appears to be a general feature of this class of proteins (74). Note also that similarly to the NS5A protein, p6 contains a large number of phosphorylation sites that could bring further negative charges (75–77). It is also interesting to underline the hydrophobic character of most of the residues of p1 involved in the transient contacts wih the NCd (L57, W61), in agreement with the fact that p1 is rather poor in charged residues. The synergy between hydrophobic and charged residues to mediate the transient contacts between NCd and p1–p6 is reminiscent to that observed within the intrinsically disordered DNA-binding inhibitory elements of the Ets-1 transcription factor (67).
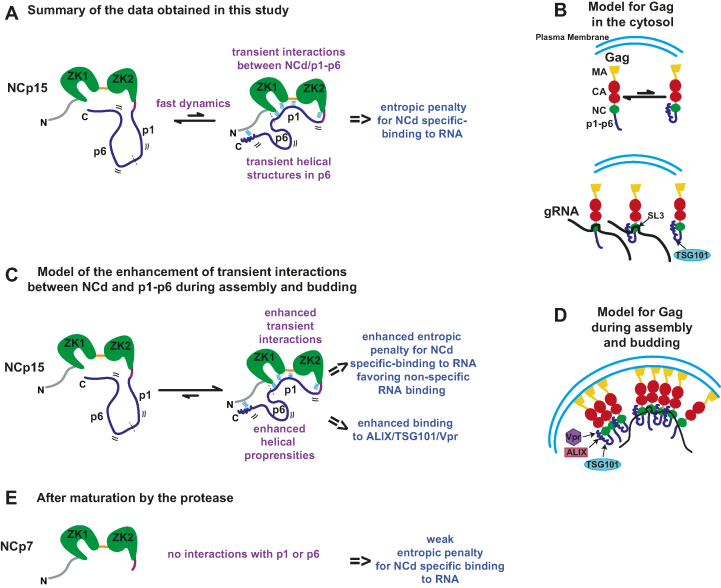
Figure 4.
Summary of the dynamic behaviour and RNA-binding properties observed for the C-terminal part of Gag (NCp15) and implications for its binding properties during virion genesis. (A) The fast dynamics motions observed in NCp15 are characterized by transient interactions between NCd and p1–p6 and transient helical folding of p6. These dynamical processes result in an entropic penalty for NCd specific-binding to RNA. (B) In the context of Gag in the cytosol, the C-terminal part of Gag is in equilibrium between the two forms, that can bind equally SL3 and the presence of helical folding can also promote binding to TSG101. (C) Transient interactions could be amplified during assembly and budding of virions due to the packing and oligomerization of Gag to the plasma membrane. The enhancement of the transient interactions between NCd and p1–p6 and the enhancement of helical propensities of p6 could have two non-related consequences: 1) an enhanced entropic penalty for NCd specific-binding to RNA that could prevent the binding of NCd to its preferred sites and favor its binding to non-specific sequences explaining previous results obtained in cell and 2) enhanced binding to partners of p6. We propose that this enhanced entropic penalty explains the specificity change observed for Gag/RNA interaction occuring in immature virion. (D) Model for Gag during assembly and budding taking into account hypothesis of (C). (E) After maturation by the protease, NCp7 is freed from p1 and p6 and from Gag oligomerization and can again binds to its specific RNA sequences.
In nucleocapsid proteins of retroviridae, interactions between ZK2 and residues just after this ZK were previously shown for MMTV (78) and MPMV (79). In both cases, the C-terminal extension that is organized, either as a reverse-turn like structure or as a β-hairpin, forms a compact structure with ZK2. In the case of HIV, the interactions are transient, but one could imagine that these interactions are a general feature shared by nucleocapsid proteins in retroviridae.
The long-range transient interactions of p1–p6 with the NCd lead to significant changes in the NCd dynamics (Figure 1E). Indeed, in the NCp15 precursor form, the N-terminal part of NCd is significantly less flexible than the mature NCp7 form whereas the two ZKs and the linker exhibit a higher flexibility. The resulting increase in global flexibility of the ZKs and of the linker within NCp15 do not change the affinity of NCd to SL3 but result in an entropic penalty to SL3 binding.
These results can have several implications for the understanding of the function of Gag and NCd throughout the viral replication cycle (Figure 4). Prior to virion assembly, Pr55Gag exists as a diffuse pool of monomers or low-order multimers in the cytoplasm of infected cells with its NCd bound to discrete sequences on the gRNA (i.e. Ψ and RRE). These binding sites strikingly also coincide with the major sites of NCd binding in mature virons; NCd being in the NCp7 form at this step (12). This result is in agreement with our study showing that the precursor NCd form (NCp15) is able to bind SL3 with the same affinity as the mature NCd form (NCp7). During assembly, as Gag molecules become more tightly packed, coincident with higher-order multimerization at the plasma membrane, Gag binds to multiple sites on the gRNA, independently of Ψ, and this loss of specificity facilitates genome packaging (12). To constrain thousands of Gag molecules into a pseudo-curved array induces a dramatic increase of local concentration of the C-terminal part of Gag consisting of NCd-p1–p6 at this step of the viral cycle. We speculate that this will enhance the transient interactions between p1–p6 and NCd leading to two non-related consequences (Figure 4C): (i) a higher level of p1–p6 folding and (ii) a higher entropic penalty for NCd binding to RNA. Regarding the first point, the regions of p6 exhibiting larger propensities to form transient helical conformations are located in the binding sites of TSG101, ALIX and Vpr. Therefore, the enhancement of these transient foldings during assembly and budding could help the recruitment of p6 partners (Figure 4D) that are essential during these steps of the viral cycle. This can explain how the NCd cooperate with p6 to recruit ESCRT proteins.
For point (ii), the entropic penalty could become so high that the enthalpy might no longer compensate for this increase, leading to a loss of affinity of the NCd in the precursors for its specific RNA binding sites and thus a redirection of NCd-binding to non-specific binding sites (Figure 4C, D). This could explain how the necessary non-specific binding to RNA genome is achieved at this step of the virion formation. This model is in agreement with a recent study that shows that oligomeric-capable Gag displayed ∼3-times stronger binding affinity for non-specific RNA motifs over the cytosolic specific target of NCd. This was not observed for oligomerization-impaired forms of Gag, and for Gag lacking p6 capable of high-order Gag oligomerization, showing that p6 has a role in facilitating the binding of non-specific RNA to oligomeric forms of Gag (80). The processing by the protease will cleave off p1 and p6 from the NCd allowing it to bind again to its specific targets (Figure 4E).
We have thus shed light on the fine regulation of the NCd dynamics and of its RNA-binding properties by p1–p6 and by the maturation process orchestrated by the protease. The concept that IDD could mediate and/or regulate protein-protein or protein-NA interactions through transient contacts with others structured regions of a protein is now supported by a number of reports (81–85). The use of a highly disordered domain in the fine regulation of the RNA recognition for the various steps of maturation of the NCd adds to the known repertoire of the unstructured protein's roles.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Ewen Lescop for helpful advice and technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
TGIR-RMN-THC FR3050 CNRS; SIDACTION, ANRS; National Cancer Institute; NIH [HHSN261200800001E to R.G.] (in part); A.M. is recipient of a SIDACTION fellowship to finance her last year of PhD; EQUIPEX CACSICE; LABEX Dynamo; Feder and Sesame Ile-de-France. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1. Freed E.O. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015; 13:484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kutluay S.B., Bieniasz P.D.. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010; 6:e1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Deshmukh L., Ghirlando R., Clore G.M.. Conformation and dynamics of the Gag polyprotein of the human immunodeficiency virus 1 studied by NMR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:3374–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mailler E., Bernacchi S., Marquet R., Paillart J.C., Vivet-Boudou V., Smyth R.P.. The Life-Cycle of the HIV-1 Gag-RNA complex. Viruses. 2016; 8:E248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hellmund C., Lever A.M.. Coordination of genomic RNA packaging with viral assembly in HIV-1. Viruses. 2016; 8:E192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bernacchi S., Abd El-Wahab E.W., Dubois N., Hijnen M., Smyth R.P., Mak J., Marquet R., Paillart J.C.. HIV-1 Pr55(Gag) binds genomic and spliced RNAs with different affinity and stoichiometry. RNA Biol. 2017; 14:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Datta S.A., Heinrich F., Raghunandan S., Krueger S., Curtis J.E., Rein A., Nanda H.. HIV-1 Gag extension: conformational changes require simultaneous interaction with membrane and nucleic acid. J. Mol. Biol. 2011; 406:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alfadhli A., Still A., Barklis E.. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J. Virol. 2009; 83:12196–12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rein A., Datta S.A., Jones C.P., Musier-Forsyth K.. Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem. Sci. 2011; 36:373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chukkapalli V., Oh S.J., Ono A.. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:1600–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shkriabai N., Datta S.A., Zhao Z., Hess S., Rein A., Kvaratskhelia M.. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry. 2006; 45:4077–4083. [DOI] [PubMed] [Google Scholar]
- 12. Kutluay S.B., Zang T., Blanco-Melo D., Powell C., Jannain D., Errando M., Bieniasz P.D.. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell. 2014; 159:1096–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paillart J.C., Shehu-Xhilaga M., Marquet R., Mak J.. Dimerization of retroviral RNA genomes: an inseparable pair. Nat. Rev. Microbiol. 2004; 2:461–472. [DOI] [PubMed] [Google Scholar]
- 14. Lu K., Heng X., Summers M.F.. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 2011; 410:609–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. VerPlank L., Bouamr F., LaGrassa T.J., Agresta B., Kikonyogo A., Leis J., Carter C.A.. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. U.S.A. 2001; 98:7724–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garrus J.E., von Schwedler U.K., Pornillos O.W., Morham S.G., Zavitz K.H., Wang H.E., Wettstein D.A., Stray K.M., Cote M., Rich R.L. et al. . Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001; 107:55–65. [DOI] [PubMed] [Google Scholar]
- 17. Votteler J., Sundquist W.I.. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013; 14:232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Engelenburg S.B., Shtengel G., Sengupta P., Waki K., Jarnik M., Ablan S.D., Freed E.O., Hess H.F., Lippincott-Schwartz J.. Distribution of ESCRT machinery at HIV assembly sites reveals virus scaffolding of ESCRT subunits. Science. 2014; 343:653–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Strack B., Calistri A., Craig S., Popova E., Gottlinger H.G.. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003; 114:689–699. [DOI] [PubMed] [Google Scholar]
- 20. Martin-Serrano J., Bieniasz P.D.. A bipartite late-budding domain in human immunodeficiency virus type 1. J. Virol. 2003; 77:12373–12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chamontin C., Rassam P., Ferrer M., Racine P.J., Neyret A., Laine S., Milhiet P.E., Mougel M.. HIV-1 nucleocapsid and ESCRT-component Tsg101 interplay prevents HIV from turning into a DNA-containing virus. Nucleic Acids Res. 2015; 43:336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weiss E.R., Gottlinger H.. The role of cellular factors in promoting HIV budding. J. Mol. Biol. 2011; 410:525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dussupt V., Javid M.P., Abou-Jaoude G., Jadwin J.A., de La Cruz J., Nagashima K., Bouamr F.. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 2009; 5:e1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dussupt V., Sette P., Bello N.F., Javid M.P., Nagashima K., Bouamr F.. Basic residues in the nucleocapsid domain of Gag are critical for late events of HIV-1 budding. J. Virol. 2011; 85:2304–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sette P., Dussupt V., Bouamr F.. Identification of the HIV-1 NC binding interface in Alix Bro1 reveals a role for RNA. J. Virol. 2012; 86:11608–11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sette P., O’Connor S.K., Yerramilli V.S., Dussupt V., Nagashima K., Chutiraka K., Lingappa J., Scarlata S., Bouamr F.. HIV-1 Nucleocapsid mimics the membrane adaptor syntenin PDZ to gain access to ESCRTs and promote virus budding. Cell Host Microbe. 2016; 19:336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. El Meshri S.E., Boutant E., Mouhand A., Thomas A., Larue V., Richert L., Vivet-Boudou V., Mely Y., Tisne C., Muriaux D. et al. . The NC domain of HIV-1 Gag contributes to the interaction of Gag with TSG101. Biochim. Biophys. Acta. 2018; 1862:1421–1431. [DOI] [PubMed] [Google Scholar]
- 28. Bachand F., Yao X.J., Hrimech M., Rougeau N., Cohen E.A.. Incorporation of Vpr into human immunodeficiency virus type 1 requires a direct interaction with the p6 domain of the p55 gag precursor. J. Biol. Chem. 1999; 274:9083–9091. [DOI] [PubMed] [Google Scholar]
- 29. Salgado G.F., Marquant R., Vogel A., Alves I.D., Feller S.E., Morellet N., Bouaziz S.. Structural studies of HIV-1 Gag p6ct and its interaction with Vpr determined by solution nuclear magnetic resonance. Biochemistry. 2009; 48:2355–2367. [DOI] [PubMed] [Google Scholar]
- 30. Salgado G.F., Vogel A., Marquant R., Feller S.E., Bouaziz S., Alves I.D.. The role of membranes in the organization of HIV-1 Gag p6 and Vpr: p6 shows high affinity for membrane bilayers which substantially increases the interaction between p6 and Vpr. J. Med. Chem. 2009; 52:7157–7162. [DOI] [PubMed] [Google Scholar]
- 31. Bendjennat M., Saffarian S.. The race against protease activation defines the role of ESCRTs in HIV budding. PLoS Pathog. 2016; 12:e1005657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tisne C. Structural bases of the annealing of primer tRNA(3Lys) to the HIV-1 viral RNA. Curr. HIV Res. 2005; 3:147–156. [DOI] [PubMed] [Google Scholar]
- 33. Tisne C., Roques B.P., Dardel F.. Heteronuclear NMR studies of the interaction of tRNA(Lys)3 with HIV-1 nucleocapsid protein. J. Mol. Biol. 2001; 306:443–454. [DOI] [PubMed] [Google Scholar]
- 34. Tisne C., Roques B.P., Dardel F.. The annealing mechanism of HIV-1 reverse transcription primer onto the viral genome. J. Biol. Chem. 2004; 279:3588–3595. [DOI] [PubMed] [Google Scholar]
- 35. Barraud P., Gaudin C., Dardel F., Tisne C.. New insights into the formation of HIV-1 reverse transcription initiation complex. Biochimie. 2007; 89:1204–1210. [DOI] [PubMed] [Google Scholar]
- 36. Barraud P., Paillart J.C., Marquet R., Tisne C.. Advances in the structural understanding of Vif proteins. Curr. HIV Res. 2008; 6:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sleiman D., Goldschmidt V., Barraud P., Marquet R., Paillart J.C., Tisne C.. Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions. Virus Res. 2012; 169:324–339. [DOI] [PubMed] [Google Scholar]
- 38. Sleiman D., Barraud P., Brachet F., Tisne C.. The Interaction between tRNA(Lys) 3 and the primer activation signal deciphered by NMR spectroscopy. PLoS One. 2013; 8:e64700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu T., Gorelick R.J., Levin J.G.. Selection of fully processed HIV-1 nucleocapsid protein is required for optimal nucleic acid chaperone activity in reverse transcription. Virus Res. 2014; 193:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mirambeau G., Lyonnais S., Gorelick R.J.. Features, processing states, and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biol. 2010; 7:724–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Coren L.V., Thomas J.A., Chertova E., Sowder R.C. 2nd, Gagliardi T.D., Gorelick R.J., Ott D.E.. Mutational analysis of the C-terminal gag cleavage sites in human immunodeficiency virus type 1. J. Virol. 2007; 81:10047–10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kafaie J., Dolatshahi M., Ajamian L., Song R., Mouland A.J., Rouiller I., Laughrea M.. Role of capsid sequence and immature nucleocapsid proteins p9 and p15 in Human Immunodeficiency Virus type 1 genomic RNA dimerization. Virology. 2009; 385:233–244. [DOI] [PubMed] [Google Scholar]
- 43. Lee S.K., Harris J., Swanstrom R.. A strongly transdominant mutation in the human immunodeficiency virus type 1 gag gene defines an Achilles heel in the virus life cycle. J. Virol. 2009; 83:8536–8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morellet N., Jullian N., De Rocquigny H., Maigret B., Darlix J.L., Roques B.P.. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992; 11:3059–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Summers M.F., Henderson L.E., Chance M.R., Bess J.W. Jr, South T.L., Blake P.R., Sagi I., Perez-Alvarado G., Sowder R.C. 3rd, Hare D.R. et al. . Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992; 1:563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Amarasinghe G.K., De Guzman R.N., Turner R.B., Chancellor K.J., Wu Z.R., Summers M.F.. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000; 301:491–511. [DOI] [PubMed] [Google Scholar]
- 47. De Guzman R.N., Wu Z.R., Stalling C.C., Pappalardo L., Borer P.N., Summers M.F.. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998; 279:384–388. [DOI] [PubMed] [Google Scholar]
- 48. Bourbigot S., Ramalanjaona N., Boudier C., Salgado G.F., Roques B.P., Mely Y., Bouaziz S., Morellet N.. How the HIV-1 nucleocapsid protein binds and destabilises the (-)primer binding site during reverse transcription. J. Mol. Biol. 2008; 383:1112–1128. [DOI] [PubMed] [Google Scholar]
- 49. Bazzi A., Zargarian L., Chaminade F., Boudier C., De Rocquigny H., Rene B., Mely Y., Fosse P., Mauffret O.. Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC. Nucleic Acids Res. 2011; 39:3903–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Morellet N., Demene H., Teilleux V., Huynh-Dinh T., de Rocquigny H., Fournie-Zaluski M.C., Roques B.P.. Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC). J. Mol. Biol. 1998; 283:419–434. [DOI] [PubMed] [Google Scholar]
- 51. Zargarian L., Tisné C., Barraud P., Xu X., Morellet N., René B., Mély Y., Fossé P., Mauffret O.. Dynamics of linker residues modulate the nucleic acid binding properties of the HIV-1 nucleocapsid protein fingers. PLoS One. 2014; 9:e102150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee S., Joshi A., Nagashima K., Freed E.O., Hurley J.H.. Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 2007; 14:194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Im Y.J., Kuo L., Ren X., Burgos P.V., Zhao X.Z., Liu F., Burke T.R. Jr, Bonifacino J.S., Freed E.O., Hurley J.H.. Crystallographic and functional analysis of the ESCRT-I /HIV-1 Gag PTAP interaction. Structure. 2010; 18:1536–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Campbell S., Rein A.. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 1999; 73:2270–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu H., Naismith J.H.. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008; 8:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee B.M., De Guzman R.N., Turner B.G., Tjandra N., Summers M.F.. Dynamical behavior of the HIV-1 nucleocapsid protein. J. Mol. Biol. 1998; 279:633–649. [DOI] [PubMed] [Google Scholar]
- 57. Salzmann M., Pervushin K., Wider G., Senn H., Wuthrich K.. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:13585–13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Marsh J.A., Singh V.K., Jia Z., Forman-Kay J.D.. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 2006; 15:2795–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 60. Goddard T.D., Kneller D.G.. SPARKY 3. San Francisco: University of Califormia. [Google Scholar]
- 61. Farrow N.A., Zhang O., Szabo A., Torchia D.A., Kay L.E.. Spectral density function mapping using 15N relaxation data exclusively. J. Biomol. NMR. 1995; 6:153–162. [DOI] [PubMed] [Google Scholar]
- 62. Tellinghuisen J., Chodera J.D.. Systematic errors in isothermal titration calorimetry: concentrations and baselines. Anal. Biochem. 2011; 414:297–299. [DOI] [PubMed] [Google Scholar]
- 63. Larue V., Catala M., Belfetmi A., Zargarian L., Mauffret O., Tisne C.. (1)H, (13)C and (15)N backbone and partial side-chain resonance assignments of the C-terminal domain of HIV-1 Pr55(Gag) encompassed in NCp15. Biomol. NMR Assign. 2018; 12:139–143. [DOI] [PubMed] [Google Scholar]
- 64. Brutscher B., Bruschweiler R., Ernst R.R.. Backbone dynamics and structural characterization of the partially folded A state of ubiquitin by 1H, 13C, and 15N nuclear magnetic resonance spectroscopy. Biochemistry. 1997; 36:13043–13053. [DOI] [PubMed] [Google Scholar]
- 65. Feuerstein S., Solyom Z., Aladag A., Favier A., Schwarten M., Hoffmann S., Willbold D., Brutscher B.. Transient structure and SH3 interaction sites in an intrinsically disordered fragment of the hepatitis C virus protein NS5A. J. Mol. Biol. 2012; 420:310–323. [DOI] [PubMed] [Google Scholar]
- 66. Lee G.M., Pufall M.A., Meeker C.A., Kang H.S., Graves B.J., McIntosh L.P.. The affinity of Ets-1 for DNA is modulated by phosphorylation through transient interactions of an unstructured region. J. Mol. Biol. 2008; 382:1014–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Desjardins G., Meeker C.A., Bhachech N., Currie S.L., Okon M., Graves B.J., McIntosh L.P.. Synergy of aromatic residues and phosphoserines within the intrinsically disordered DNA-binding inhibitory elements of the Ets-1 transcription factor. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:11019–11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Frederick K.K., Marlow M.S., Valentine K.G., Wand A.J.. Conformational entropy in molecular recognition by proteins. Nature. 2007; 448:325–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tzeng S.R., Kalodimos C.G.. Protein activity regulation by conformational entropy. Nature. 2012; 488:236–240. [DOI] [PubMed] [Google Scholar]
- 70. Liu X., Speckhard D.C., Shepherd T.R., Sun Y.J., Hengel S.R., Yu L., Fowler C.A., Gakhar L., Fuentes E.J.. Distinct Roles for Conformational Dynamics in Protein-Ligand Interactions. Structure. 2016; 24:2053–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chakrabarti K.S., Agafonov R.V., Pontiggia F., Otten R., Higgins M.K., Schertler G.F., Oprian D.D., Kern D.. Conformational selection in a Protein-Protein interaction revealed by dynamic pathway analysis. Cell Rep. 2016; 14:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fossen T., Wray V., Bruns K., Rachmat J., Henklein P., Tessmer U., Maczurek A., Klinger P., Schubert U.. Solution structure of the human immunodeficiency virus type 1 p6 protein. J. Biol. Chem. 2005; 280:42515–42527. [DOI] [PubMed] [Google Scholar]
- 73. Solyom Z., Ma P., Schwarten M., Bosco M., Polidori A., Durand G., Willbold D., Brutscher B.. The disordered region of the HCV protein NS5A: Conformational dynamics, SH3 binding, and phosphorylation. Biophys. J. 2015; 109:1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Das R.K., Pappu R.V.. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:13392–13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Muller B., Patschinsky T., Krausslich H.G.. The late-domain-containing protein p6 is the predominant phosphoprotein of human immunodeficiency virus type 1 particles. J. Virol. 2002; 76:1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Radestock B., Morales I., Rahman S.A., Radau S., Glass B., Zahedi R.P., Muller B., Krausslich H.G.. Comprehensive mutational analysis reveals p6Gag phosphorylation to be dispensable for HIV-1 morphogenesis and replication. J. Virol. 2013; 87:724–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Radestock B., Burk R., Muller B., Krausslich H.G.. Re-visiting the functional relevance of the highly conserved Serine 40 residue within HIV-1 p6(Gag). Retrovirology. 2014; 11:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Klein D.J., Johnson P.E., Zollars E.S., De Guzman R.N., Summers M.F.. The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle. Biochemistry. 2000; 39:1604–1612. [DOI] [PubMed] [Google Scholar]
- 79. Gao Y., Kaluarachchi K., Giedroc D.P.. Solution structure and backbone dynamics of Mason-Pfizer monkey virus (MPMV) nucleocapsid protein. Protein Sci. 1998; 7:2265–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tanwar H.S., Khoo K.K., Garvey M., Waddington L., Leis A., Hijnen M., Velkov T., Dumsday G.J., McKinstry W.J., Mak J.. The thermodynamics of Pr55Gag-RNA interaction regulate the assembly of HIV. PLoS Pathog. 2017; 13:e1006221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fuxreiter M., Tompa P.. Fuzzy complexes: a more stochastic view of protein function. Adv. Exp. Med. Biol. 2012; 725:1–14. [DOI] [PubMed] [Google Scholar]
- 82. Fuxreiter M. Fuzziness in protein interactions - a historical perspective. J. Mol. Biol. 2018; 430:2278–2287. [DOI] [PubMed] [Google Scholar]
- 83. Pufall M.A., Lee G.M., Nelson M.L., Kang H.S., Velyvis A., Kay L.E., McIntosh L.P., Graves B.J.. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005; 309:142–145. [DOI] [PubMed] [Google Scholar]
- 84. Milles S., Mercadante D., Aramburu I.V., Jensen M.R., Banterle N., Koehler C., Tyagi S., Clarke J., Shammas S.L., Blackledge M. et al. . Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell. 2015; 163:734–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Currie S.L., Lau D.K.W., Doane J.J., Whitby F.G., Okon M., McIntosh L.P., Graves B.J.. Structured and disordered regions cooperatively mediate DNA-binding autoinhibition of ETS factors ETV1, ETV4 and ETV5. Nucleic Acids Res. 2017; 45:2223–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.