Abstract
Fluorophore-linked, sequence-specific DNA binding reagents can visualize sequence information on a large DNA molecule. In this paper, we synthesized newly designed TAMRA-linked polypyrrole to visualize adenine and thymine base pairs. A fluorescent image of the stained DNA molecule generates an intensity profile based on A/T frequency, revealing a characteristic sequence composition pattern. Computer-aided comparison of this intensity pattern with the genome sequence allowed us to determine the DNA sequence on a visualized DNA molecule from possible intensity profile pattern candidates for a given genome. Moreover, TAMRA-polypyrrole offers robust advantages for single DNA molecule detection: no fluorophore-mediated photocleavage and no structural deformation, since it exhibits a sequence-specific pattern alone without the use of intercalating dyes such as YOYO-1. Accordingly, we were able to identify genomic DNA fragments from Escherichia coli cells by aligning them to the genomic A/T frequency map based on TAMRA-polypyrrole-generated intensity profiles. Furthermore, we showed band and interband patterns of polytene chromosomal DNA stained with TAMRA-polypyrrole because it prefers to bind AT base pairs.
INTRODUCTION
Direct visualization of individual DNA molecules is powerful because it allows us to understand biochemical events within the context of the DNA sequence (1). Although sequencing technology at the single nucleotide level has advanced dramatically and become less expensive, numerous unsolved biological problems remain that are limited by short read length and information loss within a large genome (2). The ultimate goal of DNA analysis would be to obtain sequence and epigenetic information directly from chromosomal DNA without fragmentation or amplification. Given these concerns, single large DNA molecules are a promising platform to overcome the limitations of current sequencing technology. Some key technological developments have focused on how to obtain genetic information by visualizing large DNA molecules. A pioneering approach, called Optical Mapping, digested a large, elongated DNA using sequence-specific restriction enzymes to make barcode-like patterns from a single DNA molecule (3,4); this platform has been a versatile tool for genome analysis, including as a guide for de novo sequence assembly (5) and genomic structural variation analysis in cancers (6).
Nanochannel-confined DNA analysis has spurred a variety of novel approaches because restriction enzyme digestion is inappropriate due to the random movements of cleaved DNA fragments in the nanochannel (7). As a first solution, Jo et al. developed an approach to label DNA backbones with red fluorescent dyes using a sequence-specific nicking and polymerase-filling reaction (8). Grunwald et al. developed another enzymatic biochemical approach to generate DNA physical map by using sequence-specific methyltransferase and fluorochrome labelling on the adenine of TCGA sequence (9). Alternatively, Reisner et al. reported a very different physical approach that generated a fluorescence intensity profile from large, partially melted DNA molecules, because elevating the temperature preferentially separates strands at AT-rich regions, causing YOYO-1 release from the DNA backbone and resulting in reduced fluorescence intensity (10). These approaches have the distinct advantage of providing more detailed sequence information because the signal comes directly from individual base pairs without any enzymatic biochemical reactions. Nyberg et al. reported another intensity profile approach using netropsin to block YOYO-1 intercalation by binding to AT-rich sequences (11). However, all of these methods have been based on YOYO-1, which often causes photo-induced DNA cleavage (12,13). DNA statically immobilized on the surface are relatively stable with YOYO-1 staining, but this issue is more problematic for dynamically moving DNA molecules because there are pulling forces to enhance the probability of DNA cleavage. For example, a tethered DNA molecule stretches by a microfluidic shear laminar flow that generates a pulling force to unravel coiled DNA (14–16). A nanofluidically confined DNA molecule should endure entropic pulling forces from thermally fluctuating local structures (17,18). These pulling forces would tear DNA double strands when both strands have photo-damaged nicks whose spacing is close enough for the pulling force to effect. Therefore, we speculate that it would be highly desirable to replace YOYO-1 with a novel DNA staining reagent that does not cause photo-cleavage. Furthermore, it would be more advantageous that sequence-specific binders like netropsin that could fluoresce alone rather than indirectly inhibiting fluorochrome binding.
To directly add fluorochromes to sequence-specific binders like netropsin, it is necessary to understand their chemical structure and DNA binding mechanism. Netropsin is a well-known DNA binding antibiotic polyamide (19). Its primary structure is a pyrrole dimer that forms hydrogen bonds in the minor-groove of the A/T base pair (20). Various polyamides containing pyrrole and imidazole have been widely investigated for biological and medical purposes because of their sequence-specific DNA binding characteristics (21,22). Moreover, fluorophore-linked pyrrole-imidazole polyamides have been utilized for a variety of biological applications, such as in vivo DNA targeting (23,24) and epigenetic analysis (25). In this context, we speculated that fluorophore-linked pyrrole-imidazole polyamides would be promising candidates for visualizing large elongated DNA molecules with sequence specificity, although they have never been applied for this purpose.
In this paper, we synthesized a newly designed fluorophore-linked polypyrrole, TAMRA-β2-Py4-β-Py4-Dp, to stain individual large DNA molecules with A/T-specificity. This reagent successfully allowed the visualization of specific fluorescence intensity patterns along DNA backbones that represented the A/T frequency. Importantly, this staining reagent overcame the undesirable features of YOYO-1, including DNA structural deformation and DNA cleavage. We demonstrated its use on tethered and surface-immobilized DNA molecules with novel, robust activity. Therefore, we applied it to large genomic DNA fragments from Escherichia coli cells. Using a Python program to compare intensity profiles based on the cross-correlation coefficient, we identified DNA fragments by aligning them with the genomic A/T frequency map. Finally, we used TAMRA-polypyrrole to stain large polytene chromosomal DNA to depict the band and interband patterns on a fluorescence microscope.
MATERIALS AND METHODS
Synthesis of TAMRA-pPy
Polyamides used in this study were synthesized as previously described elsewhere (23). Briefly, a computer-assisted Fmoc solid-phase synthesis of polyamides was performed from Fmoc-Py-oxime resin, and cleavage from the resin was performed with 1 ml of 3-(dimethylamino) propylamine (Dp) or 3,3′-diamino-N-methyldipropylamine treatment for 3 h at 55°C. More detailed procedure was included in SI ‘General Procedure of Solid-phase Synthesis of Polyamides.’ The crude products were purified by flash chromatography, and the collected fractions were lyophilized to collect the objective compounds. TAMRA-pPy was synthesized as follows: (i) 20% piperidine, DMF; (ii) Fmoc-Py-COOH, HCTU, DIEA, NMP; (iii) 20% piperidine, DMF; (iv) Fmoc-Py-COOH, HCTU, DIEA, NMP; (v) 20% piperidine, DMF; (vi) Fmoc-Py-COOH, HCTU, DIEA, NMP; (vii) 20% piperidine, DMF; (viii) Fmoc-β-Ala-COOH, HCTU, DIEA, NMP; (ix) 20% piperidine, DMF; (x) Fmoc-Py-COOH, HCTU, DIEA, NMP; (xi) 20% piperidine, DMF; (xii) Fmoc-Py-COOH, HCTU, DIEA, NMP; (xiii) 20% piperidine, DMF; (xiv) Fmoc-Py-COOH, HCTU, DIEA, NMP; (xv) 20% piperidine, DMF; (xvi) Fmoc-Py-COOH, HCTU, DIEA, NMP; (xvii) 20% piperidine, DMF; (xviii) Fmoc-β-Ala-COOH, HCTU, DIEA, NMP; (xix) 20% piperidine, DMF; (xx) Fmoc-β-Ala-COOH, HCTU, DIEA, NMP; (xxi) 20% piperidine, DMF; (xxii) 3-(dimethylamino) propylamine, 55°C, 3 h; (xxiii) 5-TAMRA NHS ester, DIEA, DMF (see Supplementary Figures S8–S16). TAMRA, carboxytetramethylrhodamine, has the absorption maximum at ∼546 nm and the emission at ∼580 nm: the quantum yield is 0.38, the extinction coefficient is 81 000 M−1 cm−1, and the fluorescence lifetime is 5.7 ns (26).
Microscopy
For single molecule observations, an inverted microscope (Olympus IX70, Tokyo, Japan) with 60× Olympus UPlanSApo oil immersion objectives illuminated with an LED light source (SOLA SM II light engine, Lumencor, Beaverton, OR, USA) was used. The light was passed through corresponding filter sets (Semrock, Rochester, NY, USA), to set the excitation and emission wavelengths. At the image plane, a maximum 140 mW/cm2 of light intensity was measured. Fluorescence images were taken with an electron-multiplying charge-coupled device digital camera (Evolve EMCCD, Roper Scientific, Tucson, AZ, USA) and stored in a 16-bit TIFF format using the software Micro-manager. For image processing and analysis, ImageJ with Java plug-ins and python programs developed in our lab were utilized.
Flow cell
A flow chamber was prepared by placing a custom-fabricated acrylic support on an acid-cleaned cover slip with a height of 100 μm formed by double-sided tape (16,17). The dimensions of the flow cell were approximately 5 × 10 × 0.1 mm (L × W × H). An NE-1000 syringe pump (New Era Pump Systems Inc., Wantagh, NY, USA) was used to control the buffer in the flow cell. After preparation, 40 μg/ml biotinylated bovine serum albumin were injected and incubated at room temperature for 10 min, followed by 20 μg/ml of Neutravidin in T50 solution (10 mM Tris, 50 mM NaCl, pH 8.0) with the same condition. Then, 1 μM of λ DNA overhang oligo (5′-p-GGGCGGCGACCT-Triethyleneglycol-biotin-3′) was loaded into the flow chamber and maintained at room temperature for 10 min. λ DNA, 200 U of T4 DNA ligase, and reaction buffer was added and incubated at room temperature for 30 min. After washing the residual enzyme mixture with 1 × TE (10 mM Tris, 1 mM EDTA, pH 8.0), the diluted TAMRA-polypyrrole solution flowed into the channels, resulting in visualization of the tethered DNA. Stained DNA molecules were visualized under a continuous flow of 1 × TE with the flow rate maintained at 100 μl/min.
Glass surface preparation
The brief experimental procedure used for glass surface preparation was as follows (15): The glass coverslips were soaked in piranha etching solution (30:70 v/v H2O2/H2SO4) for 3 h. After thorough rinsing with deionized water, the coverslips were sonicated for 30 min. To add positive charges to the surface, 400 μl N-trimethoxymethylsilylpropyl-N,N,N-trimethylammonium chloride in 50% methanol was added to 250 ml deionized water. The acid-cleaned coverslips were incubated in this solution overnight at 65°C, followed by ethanol rinsing. Positively charged glass surfaces were used within two weeks of preparation.
λ DNA preparation
λ DNA (NEB) was diluted to 5 ng/μl (0.16 nM, 7.76 μM as base pairs) with 1 × TE and mixed with 70 μM TAMRA-polypyrrole at a 1:1 volume ratio. The sample was incubated at room temperature for 15 min and diluted twenty times using 1 × TE with 4% β-mercaptoethanol (β-ME) as an anti-photobleaching reagent.
Yeast chromosome I DNA
Yeast S288c cells were grown for 1 day at 25°C, 220 rpm. The cells were harvested, and the OD600 was adjusted to 10.0. After mixing with 2% low gelling temperature agarose for a final concentration of 0.75%, the cells were lyticase-treated for 1 h at 37°C followed by washing the cell plugs in 1 × TE. Forty U of Proteinase K was added to a reaction tube and incubated at 42°C overnight. Then, gel plugs were washed several times in 1 × TE and subjected to pulse-field gel electrophoresis following the manufacturer's guide (Bio-Rad CHEF-DR® II): 1% low melting temperature agarose, 0.5 × TBE (Tris–borate–EDTA), 14°C, 60–120 s of switch time, with 24 h of running and a voltage gradient of 6 V/cm. After running, the gel was stained with SYBR Gold for 30 min, immediately extracted, and destained with 200 mM of NaCl and 300 mM of sodium acetate (pH 5.2) for 30 min. After repeating the destaining procedure and washing three times, the gel was used.
E. coli genomic DNA
Escherichia coli MG1655 cells were grown at 37°C, 220 rpm, until they reached an OD600 of 0.5 (27). After mixing with low melting temperature agarose for 0.75%, 16 U of proteinase K was added to a tube and incubated for 2 h at 42°C. The proteinase K reaction was repeated again with freshly prepared enzyme. Gel plugs were washed several times in 1 × TE, and then they were ready to use.
Genomic DNA preparation from gel piece
Gel pieces containing large DNA molecules were melted at 65°C for 15 min while the tube was carefully inverted. Then, the sample was digested with 2 U β-agarase at 42°C for 2 h. The melted DNA solution was mixed with 100 μM TAMRA-polypyrrole in a 1:1 volume ratio, diluted 20 times using 1 × TE with 4% β-ME solution, and observed. Each reagent was mixed with DNA and TAMRA-polypyrrole mixture in a 1:1 volume ratio, and entire solution was diluted to concentrations suitable for observation with a fluorescence microscope.
Polytene chromosome preparation
The Drosophila melanogaster polytene chromosomes were prepared by modifying the acid-fixation squash method (28). Briefly, dissected salivary glands from third larvae were fixed with a 1:2:3 ratio of propionic acid, deionized water, and acetic acid on a positively charged coverslip. Then, the cells were located between slide glass and the coverslip, followed by gentle spreading of the individual cells. After moving the coverslip back and forth on the slide glass, the spread cells were squashed. Fixation and cell spreading were done within 15 min. Then, we dipped the slide glass and coverslip into liquid nitrogen until the boiling stopped and immediately removed the coverslip from the slide glass. Finally, 2.5 mM TAMRA-polypyrrole with 4% β-ME was used to stain the polytene chromosomal DNA.
Python program
All five python codes were included in SI python program section, which also described how to use these programs. Five programs are ImageCompare.py that contains a library of functions, seq2map.py that converts a FASTA file into a A/T frequency in silico map, insilicoMapFolder.py that searches the best matching position for each DNA image in the folder by scanning through the entire genomic A/T frequency in silico map, sortView.py that compare images in the folder, and randomtiff.py that generate random intensity tiff images.
RESULTS AND DISCUSSION
Design and synthesis of TAMRA-β2-Py4-β-Py4-Dp for binding AT-rich sequences
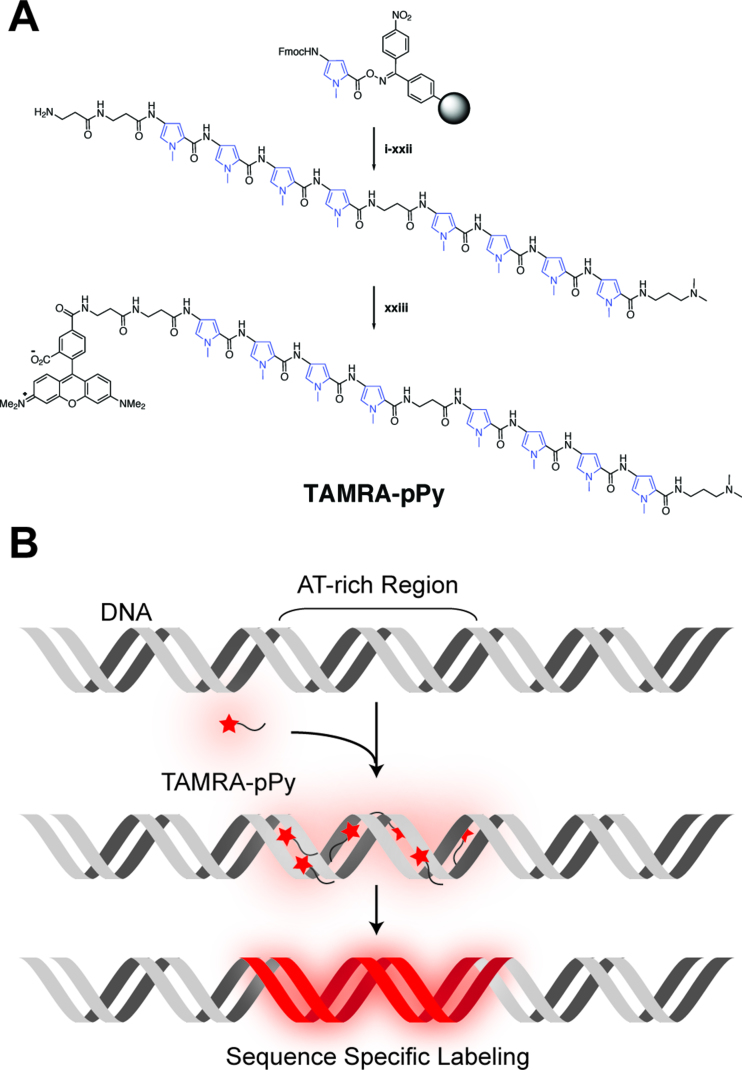
Figure 1 illustrates the molecular design and conceptual function of TAMRA-β2-Py4-β-Py4-Dp: 5-carboxytetramethylrhodamine-(β-alanine)2-(N-methylpyrrole)4-β-alanine-(N-methylpyrrole)4-(3-(dimethylamino) propylamine) (SI Scheme S1). The fluorophore TAMRA was at the N-terminus, and 3-(dimethylamino) propylamine (Dp) was at the C-terminus. Four consecutive pyrroles were repeated twice with a β-alanine residue to add structural flexibility. More detailed synthesis information is presented in the Materials and Methods and Supplementary Information, including NMR and mass spectra data to confirm the chemical structure. Before this design, we synthesized TAMRA-β2-Py4-Dp (SI Scheme S2), which contained four consecutive pyrroles deduced from the netropsin pyrrole–dimer structure (11). It was partially successful for visualizing AT-rich sequence clusters, although some problems remained unresolved (Supplementary Figure S1), including that staining did not detect many AT-rich regions, generating false negative data as well as unexpected false positive labels. In addition, many images were not bright enough to efficiently visualize the DNA backbone. Thus, we attempted to add another TAMRA to the C-terminus (TAMRA-β2-Py4-Dp-TAMRA: SI Scheme S3), but two TAMRA-linked polypyrroles were unable to bind double-stranded DNA at all, probably because two TAMRAs at both ends inhibited tetra-pyrrole binding to DNA due to steric hindrance. Finally, by adding one more tetra-pyrrole without a C-terminal TAMRA, we were able to successfully visualize λ DNA with sequence specificity as well as enhance the brightness, as shown in Figure 2.
Figure 1.
Design of a sequence-specific DNA staining reagent. (A) Synthetic scheme of TAMRA-β2-Py4-β-Py4-Dp (TAMRA-pPy) (see Materials and Methods, and SI for detailed chemical synthesis procedures). (B) Conceptual illustration of adenine (A) and thymine (T) base pair-rich regions specifically stained by TAMRA-polypyrrole.
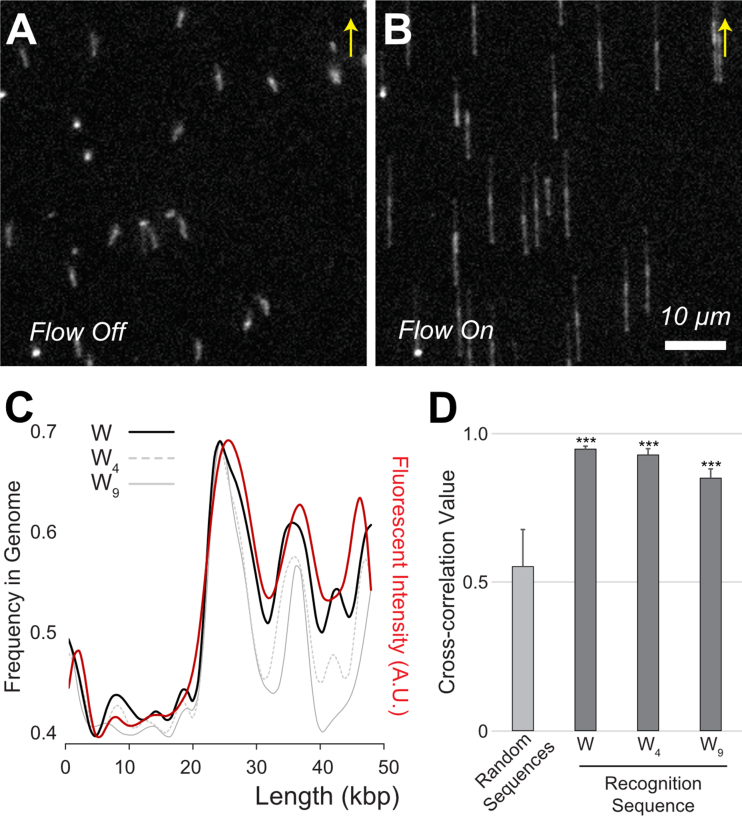
Figure 2.
TAMRA-pPy-stained λ DNA tethered on the surface of a flow cell. (A) Free-floating λ DNA (48.5 kb) molecules with a mushroom-like conformation in the flow off condition. (B) Fully elongated λ DNA molecules with a flow of 100 μl/min. Arrows indicate flow direction. Scale bar 10 μm. (C) Comparison of experimentally measured fluorescence intensity (red) with in silico sequence frequencies from the λ genome sequence. The black solid line represents A/T (W), the gray dotted line indicates four consecutive A/T (W4), and the grey solid line is W9. (D) Cross-correlation (cc) coefficient values calculated from the alignment of 20 molecular images with the genome using three kinds of binding sequences (W, W4 and W9). The control cc was obtained from 100 computer-generated random sequences (***P < 0.0001 for random-sequences paired t-test).
TAMRA-pPy stained tethered DNA molecules
Figure 2 shows TAMRA-polypyrrole-stained λ DNA (NCBI Reference Sequence: NC_001416.1) molecules tethered to the surface of a flow cell (16). Surface-tethered DNA molecules were free-floating with a mushroom-like conformation (Figure 2A) and were fully elongated at a flow of 100 μl/min (Figure 2B). In general, the surface-tethered DNA stretched uniformly, which is a distinct advantage over other DNA presentation methods, such as surface immobilization. In this experiment, the average DNA stretch was 14.03 ± 0.54 μm, corresponding to an 86% stretch compared with the full contour length of 16.3 μm (48 502 × 0.337 nm) (Supplementary Figure S2).
The measurement of the stretched DNA length has been a standard means to detect DNA’s structural deformation, particularly using optical/magnetic tweezers, atomic force microscope, and hydrodynamic stretching (29,30). Kundukad et al. reported a systematic study that they compared their result with previously reported eight papers. They concluded that the intercalation of YOYO-1 increases the contour length by the underwinding of DNA double helix (31). Therefore, the DNA stretching rates indicated the degree of dye-induced structural deformation such as double helix unwinding. In our previous study, we reported that YOYO-1-stained DNA exhibited severe structural deformations resulting in 21.8 μm λ DNA in a flow cell, which corresponds to 134% stretching compared with native λ DNA (21.8/16.3 μm), but our previous computer simulation using finite element analysis predicted that native DNA should stretch 84% in the same flow cell (32). This predicted value agrees relatively well with 14.03 μm (86%) observed for TAMRA-polypyrrole stained λ DNA in this report (Supplementary Figure S3). In addition, this stretching result is consistent with stretching values (13.4–14.6 μm) for various fluorescent DNA binding proteins to stain DNA molecules in the same flow cell that we reported earlier (32). Therefore, 86% DNA stretch suggests that TAMRA-polypyrrole does not cause considerable structural deformation (Supplementary Figure S3). This is likely because TAMRA-polypyrrole binds DNA via a hydrogen bonding interaction between a pyrrole and the minor groove, and TAMRA dyes remains far from the DNA backbone, unlike YOYO-1 intercalation (23). According to a previous X-ray crystallography study, pyrrole-imidazole polyamide bound dsDNA conserves native B-DNA structural features of 35° twist and 0.34 nm per residue (33).
Furthermore, the lack of backbone interaction is a significant advantage, because TAMRA-polypyrrole staining does not cleave DNA during repeated cycles of DNA elongation and relaxation in the flow cell (Figure 2). In contrast, YOYO-1 is well known to cause photo-induced DNA cleavage, which is a serious problem for tethered DNA (12,13). Supplementary Figure S4 illustrates the comparison of photo-induced effects between TAMRA-polypyrrole and YOYO-1 stained DNA molecules. After 30 min illumination, YOYO-1 stained DNA completely disappeared while TAMRA-polypyrrole stained DNA was not affected at all. We frequently observed YOYO-1-stained DNA cleavage and disappearance in the flow cell (15). For these reasons, TAMRA-polypyrrole has the important practical advantages of causing neither structural deformation nor DNA cleavage.
Elongated λ DNA backbones exhibit the same distinct florescent intensity patterns as shown in Figure 2B, because they are anchored at the 3′ end to the surface. The model of λ DNA is useful to depict specific intensity profile patterns, because the genome contains CG-rich (i.e. AT-poor) regions of up to 20 kb and AT-rich regions that are specifically located 20 kb from the end (48.5 kb) with three peaks. It is noteworthy that even CG-rich regions show dim but visible DNA backbones because some A/T bases are present in this region. Specifically, 14.03 μm of λ DNA corresponds to 52.6 pixels in the image. Thus, each pixel corresponds to 922 bp (48 502 bp/52.6 pixel). If we segmented the λ genome by 922 bp, the smallest AT content per pixel would be 39%. Therefore, even CG-rich regions should have at least 361 A/T base pairs out of 922 bp, which might be enough for DNA-bound TAMRA dyes to emit a dim signal for the DNA backbone. As a way to improve resolution, we also tested higher resolution camera, in which each pixel corresponded 231 bp. However, we only obtained lower cross-correlation values than the data of the low-resolution camera that we reported. Thus, simply increasing resolution may not be the solution to achieve high sensitivity. In addition, we tested a couple of extreme solutions such as pH 11 basic solution and salty buffer solution (1 × PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4), both of which detached fluorescent protein–DNA binding peptides from DNA in our previous report (34). However, we observed that TAMRA-polypyrrole DNA staining is successful in both the extreme solutions.
It is critical to further determine the actual recognition sequence of this reagent. A pyrrole residue is known to form hydrogen bond with each A/T base pair (W), and there are eight pyrroles in TAMRA-β2-Py4-β-Py4-Dp. If all of them linearly bound DNA, the recognition sequence would be W9, where the beta-alanine between the two Py4 units is known to recognize W (35). Another possibility is that tetra-pyrrole binds W4 and the other works as a linker. Alternatively, TAMRA-polypyrrole might randomly interact with any A/T (W) rather than the consecutive sequence. Accordingly, we identified three possible candidates as DNA binding sequences: W, W4 and W9. Figure 2C shows the comparison of the experimental fluorescence intensity profiles with the three candidates. According to this comparison, our experimental measurement agrees better with simple A/T frequency than W4 and W9. For a quantitative comparison, we calculated cross-correlation (cc) coefficients using a lab-made Python program, in which we utilized scipy.signal() functions to apply a low pass filter for reducing noises from the fluorescence intensity profile as well as the in silico A/T frequency map, and numpy.corrcoef() function to obtain their cc value (see SI ImageCompare.py). The calculation of cc coefficients for three cases show the highest cc value for simple A/T frequency (Figure 2D). Accordingly, in this report, we used only the A/T frequency from in silico genome sequences for the comparison of our experimental measurements.
TAMRA-pPy-stained DNA molecules immobilized on the surface
A number of approaches to visualize large DNA molecules have been developed, including surface immobilization (15,36,37), DNA tethering (16,38), microfluidic dynamic stretching (14,39) and nanochannel confinement (17,40). Surface immobilization is practically effective because the procedure is very simple (37). Moreover, images of immobilized DNA exhibit more intense fluorescence because stained fluorophores give off light for a longer period without movement. Nevertheless, surface immobilization generally has two shortcomings. First, the molecular orientations cannot be controlled due to random adsorption (41). Second, the DNA stretch is not uniform but is instead generally within a range, whereas tethered DNA stretches more uniformly (seeSupplementary Figure S2) (4). Neither of these shortcomings are issues for TAMRA-polypyrrole. As shown in Figure 3A, the sequence-specific patterns promptly revealed the orientations of randomly adsorbed DNA molecules, as indicated by the yellow arrows in the 5′→ 3′ direction. Moreover, sequence-specific patterns directly translate to the genomic map regardless of how long the DNA molecules stretch. The images of λ concatemers in Figure 3B show repeated λ monomer units with different stretching rates, but there is no ambiguity in interpreting these images.
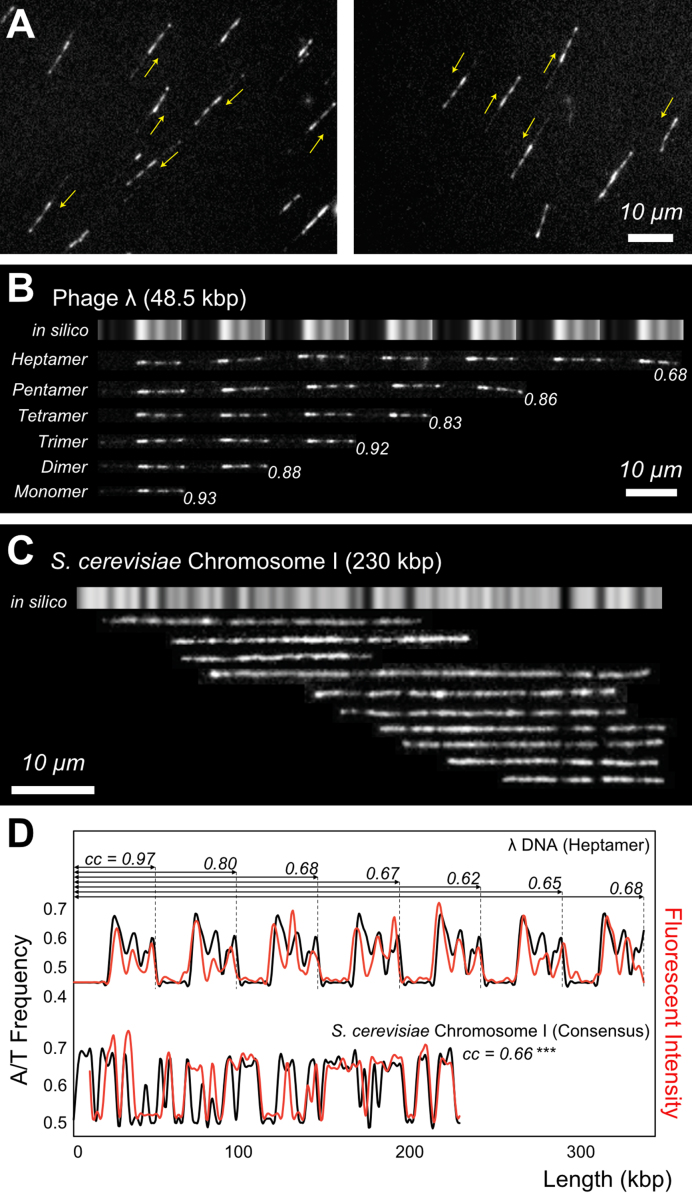
Figure 3.
TAMRA-pPy-stained λ DNA electrostatically immobilized on a surface (A) λ DNA on a positively charged surface; yellow arrows indicate the genome direction 5′→3′. (B) λ concatemers from monomer to heptamer. Gray bars represent the A/T frequency. (C) S. cerevisiae chromosome I DNA fragments aligned with the in silico A/T frequency map. Averaged cc value was 0.64 ± 0.14, whereas cc for computer-generated random images was 0.41 ± 0.06 (P < 0.003 for randomly-generated images paired t-test). (D) Cross-correlation analysis of λ DNA (heptamer in B) and S. cerevisiae chromosome I (consensus map of C): cc represents the cross-correlation coefficient for the range indicated by the arrows.
In addition to λ DNA and its concatemers, we applied TAMRA-polypyrrole to stain yeast (Saccharomyces cerevisiae S288c) chromosome I DNA (230 kb), which was prepared from a piece of dissected gel after pulsed-field gel electrophoresis. Because there was no possibility of confusion with other DNA molecules, these DNA fragments were aligned with an in silico A/T frequency genomic map. Figure 3C indicates that the fragment data agree relatively well with the genomic map. In particular, characteristic dark bands functioned as guidelines for assembling these DNA fragments.
Figure 3D illustrates quantitative analysis using intensity profiles and cc coefficients. From the comparison of the λ heptamer with its in silico map, it is noteworthy that the cc value generally decreases with increasing molecular length from 0.97 for the first monomer to 0.68 for the whole heptamer. As mentioned earlier, surface-immobilized DNA molecules stretch to different extents, which can reduce the cc for long DNA. The second profile graphs in Figure 3D represent the intensity profile of the consensus yeast chromosome DNA I map in Figure 3C compared with the in silico A/T frequency. The cc value is 0.66, which seems consistent with similarly sized λ concatemers. To evaluate the significance, we calculated the cc values for 100 computer-generated random sequences using the corresponding lengths as a control and obtained an average of 0.42 ± 0.08. This result demonstrates that our experimentally obtained cc is significantly higher than the random control.
DNA identification among the mixture of DNA fragments from E. coli genome
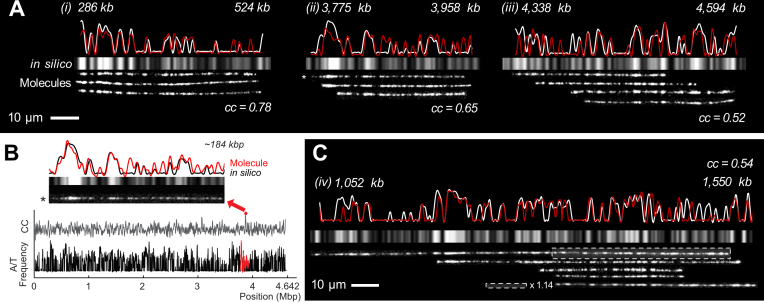
Figure 4 demonstrates examples of how to identify randomly generated DNA fragments obtained from the 4.64 Mb E. coli K-12 MG1655 genome (NCBI Reference Sequence: NC_000913.3). Currently, there is no approach to obtain the full length of 4.64 Mb genomic DNA. In general, a large genomic DNA is randomly broken into hundreds of kilo base pair (kb) fragments due to shear fluidic forces during sample preparation. We aligned the DNA fragments with the in silico bacterial genome map based on the cc values. Since genomic DNA molecules, hundreds kb long, stretched longer than λ DNA, we used 850 bp per pixel to generate the in silico map instead of 922 bp used for λ DNA in Figure 2. Then, we added other DNA fragments obtained from E. coli cells to support these alignments (Figure 4A). To find the primary alignment position for each fragment, we wrote a Python program to find the best matching position from the full genome as illustrated in Figure 4B (see SI insilicoMapFolder.py and Supplementary Figure S18). More specifically, the program read an in silico map and a DNA image as an intensity profile and smoothed the profiles with a low pass filter (42). Then, the programs scanned and compared two signal sets along the in silico map to find the best alignment position, returning a list of the highest cc values with the position and orientation of each aligned DNA molecule. As an intriguing example, Figure 4C exhibits 498 kb long DNA fragments. The first half of the image is well aligned to the reference map, but the second half does not exactly match the in silico map, which appears less stretched. This is an example of the different extents of DNA stretching on a surface, as mentioned above. Therefore, we extended the second half of the molecular image by a factor of 1.14, generating better alignment with the genomic map as well as the other DNA fragments. Quantitatively, this stretching enhanced the cc values from 0.48 to 0.54.
Figure 4.
Identification of bacterial genomic DNA fragments. (A) DNA fragments aligned with the bacterial genomic map. The white profiles represent the in silico map, and the red ones represent the consensus map of the experimental results. Grey bars represent the in silico A/T frequency. An asterisk indicates the image used in (B). (B) Example python program output to search alignment positions and draw graphs of mostly matched positions on the genome with its molecular image. (C) Alignment of 498 kb DNA fragment and stretch adjustment for the boxed region (×1.14) to properly align it with the in silico map and other DNA fragments. All of the cc values were from the consensus map (see Supplementary Figure S5 for statistical analysis).
Microscopic identification of DNA fragments from a given genome is a powerful tool for various biological applications. Recent advances in sequencing technology have provided a great deal of available sequence data. Therefore, it is rare to handle completely unknown DNA. Instead, most DNA analysis is resequencing rather than de novo sequencing. In this case, microscopic visualization of the A/T sequence frequency, as shown in Figure 4, can be an alternative tool to identify the sequence without performing sequencing. Moreover, this approach is particularly useful for analyzing chemically modified or damaged DNA backbones, because conventional sequencing methods can be ineffective on modified or damaged DNA (27,41,43). Therefore, microscopic DNA identification offers great promise for further genomic and biochemical studies on single DNA molecules.
Nonetheless, we also note the limitations of TAMRA-polypyrrole. Unfortunately, it only works for ∼40–60% AT content. λ DNA has an average 51% AT content that ranges from 39% to 67% with distinct fluctuations (Figures 2 and 3). Yeast chromosome I DNA has an average of 60% and a minimum of 40%, which produced characteristic dark bands (Figure 3). The E. coli genome has an average 49% AT content per pixel, which generates the well-matched patterns shown in Figure 4. However, DNA images from regions with higher or lower AT content are difficult to align to the A/T frequency map. For example, Figures 2 and 3 show very dim backbones of 39% AT content in λ DNA, which suggests the lower limit for detecting AT content. In another example, TAMRA-polypyrrole stained T4 DNA was stained too strongly to align the expected in silico pattern (Supplementary Figure S5). The T4 genome has an average AT content of 64%, ranging from 54% to 71%. This observation suggests the upper detection limit for this reagent.
TAMRA-pPy-stained polytene chromosomes
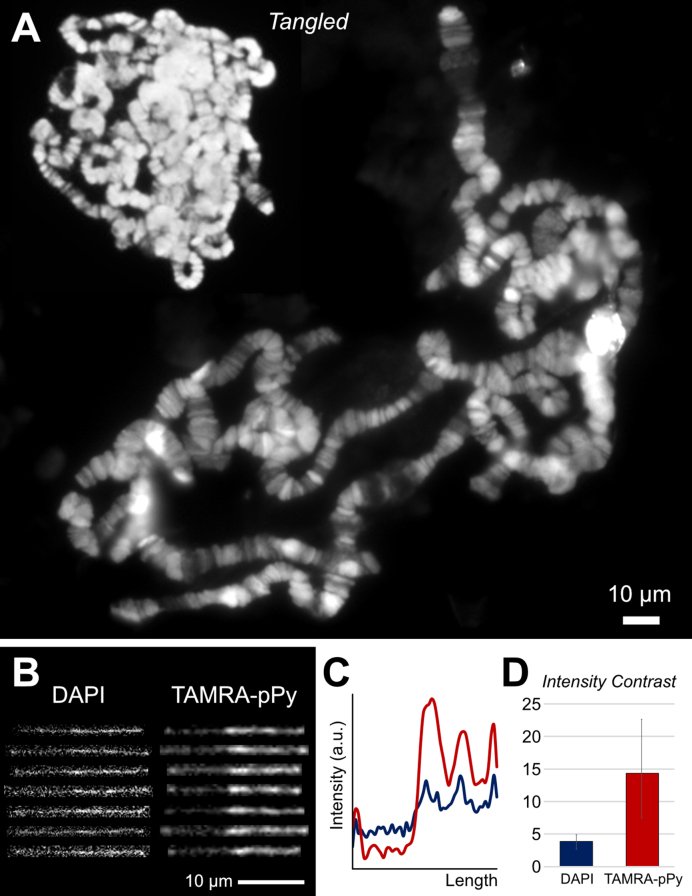
In addition to visualizing purified single DNA molecules, we sought to extend TAMRA-polypyrrole for visualizing chromosomal DNA. As an example, we stained the large polytene chromosomes from the D. melanogaster salivary gland. DAPI or Hoechst dyes are conventionally used to visualize polytene chromosomes on a fluorescence microscope (28). Interestingly, both dyes are known to bind the minor groove of double-stranded DNA with an A/T preference (44), which is similar to the characteristics of TAMRA-polypyrrole, which binds the minor groove of double-stranded DNA with A/T specificity. Figure 5A shows a typical TAMRA-polypyrrole image for tangled and spread polytene chromosomes, both of which clearly demonstrate band and interband patterns. As mentioned earlier, TAMRA-polypyrrole has the superb advantage of not damaging DNA. Furthermore, TAMRA is excited by yellowish-green wavelengths (550 nm), whereas DAPI and Hoechst require ultraviolet light sources or equivalent low wavelength light to excite the fluorophores, which can damage DNA structures. Importantly, TAMRA-polypyrrole-stained DNA exhibits a clear backbone compared with DAPI-stained DNA (Figure 5B) in terms of fluorescence intensity contrast (Figure 5C and D). Staining polytene chromosomal DNA with TAMRA-polypyrrole could demonstrate the band and interband patterns with a high-resolution, therefore, it is advantageous to study somatic genome instability (45), chromosomal organization of the genome, and protein immunolocalization (46). Accordingly, TAMRA-polypyrrole is a promising reagent for investigating large polytene structures as well as single large DNA molecules.
Figure 5.
(A) TAMRA-pPy-stained polytene chromosomes from D. melanogaster. Polytene chromosome samples were prepared via a conventional acid-wash protocol and exhibit bands and interbands in chromosomes on a fluorescence microscope. (B) Representative fluorescent λ DNA images stained with DAPI and TAMRA-polypyrrole. (C) Intensity profiles of DAPI and TAMRA-polypyrrole-stained λ DNA. Red line is TAMRA-polypyrrole, and blue line is DAPI staining. (D) Intensity contrast ratios for DAPI and TAMRA-polypyrrole. Bars represent the highest values over the lowest values for each DNA molecule. Error bars represent the minimum and maximum values for the images in (B).
CONCLUSION
In conclusion, our newly designed TAMRA-polypyrrole specifically stained AT-rich regions and exhibited distinct fluorescence intensity patterns on DNA backbones. TAMRA-polypyrrole staining generated specific patterns that allowed the interpretation of most DNA images to depict molecular direction or stretching rates. Moreover, a sequence-specific pattern provides an alternative method to determine the DNA sequence from a microscopic image of a DNA fragment if given the full sequence. Further, TAMRA-polypyrrole works better than DAPI and Hoechst dyes for staining large polytene chromosomal DNA to depict the band and interband patterns by fluorescence microscopy.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea (NRF) [2017R1A2B2012665, 2017R1E1A2A02080741, 2016M3A9B6947831, 2016R1A6A1A03012845, 2016K2A9A2A10005504 to K.J.]; Japan Society for the Promotion of Science (JSPS) KAKENHI [16H06356 to H.S. and 15J00928 to Y.K.]. Funding for open access charge: National Research Foundation of Korea.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lee J., Kim Y., Lee S., Jo K.. Visualization of large elongated DNA molecules. Electrophoresis. 2015; 36:2057–2071. [DOI] [PubMed] [Google Scholar]
- 2. Goodwin S., McPherson J.D., McCombie W.R.. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016; 17:333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schwartz D.C., Li X., Hernandez L.I., Ramnarain S.P., Huff E.J., Wang Y.K.. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science. 1993; 262:110–114. [DOI] [PubMed] [Google Scholar]
- 4. Dimalanta E.T., Lim A., Runnheim R., Lamers C., Churas C., Forrest D.K., de Pablo J.J., Graham M.D., Coppersmith S.N., Goldstein S.. A microfluidic system for large DNA molecule arrays. Anal. Chem. 2004; 76:5293–5301. [DOI] [PubMed] [Google Scholar]
- 5. Perna N.T., Plunkett G. 3rd, Burland V., Mau B., Glasner J.D., Rose D.J., Mayhew G.F., Evans P.S., Gregor J., Kirkpatrick H.A. et al. . Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001; 409:529–533. [DOI] [PubMed] [Google Scholar]
- 6. Gupta A., Place M., Goldstein S., Sarkar D., Zhou S.G., Potamousis K., Kim J., Flanagan C., Li Y., Newton M.A. et al. . Single-molecule analysis reveals widespread structural variation in multiple myeloma. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7689–7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Riehn R., Lu M.C., Wang Y.M., Lim S.F., Cox E.C., Austin R.H.. Restriction mapping in nanofluidic devices. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:10012–10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jo K., Dhingra D.M., Odijk T., de Pablo J.J., Graham M.D., Runnheim R., Forrest D., Schwartz D.C.. A single-molecule barcoding system using nanoslits for DNA analysis. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:2673–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grunwald A., Dahan M., Giesbertz A., Nilsson A., Nyberg L.K., Weinhold E., Ambjornsson T., Westerlund F., Ebenstein Y.. Bacteriophage strain typing by rapid single molecule analysis. Nucleic Acids Res. 2015; 43:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reisner W., Larsen N.B., Silahtaroglu A., Kristensen A., Tommerup N., Tegenfeldt J.O., Flyvbjerg H.. Single-molecule denaturation mapping of DNA in nanofluidic channels. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:13294–13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nyberg L.K., Persson F., Berg J., Bergstrom J., Fransson E., Olsson L., Persson M., Stalnacke A., Wigenius J., Tegenfeldt J.O. et al. . A single-step competitive binding assay for mapping of single DNA molecules. Biochem. Biophys. Res. Commun. 2012; 417:404–408. [DOI] [PubMed] [Google Scholar]
- 12. Tycon M.A., Dial C.F., Faison K., Melvin W., Fecko C.J.. Quantification of dye-mediated photodamage during single-molecule DNA imaging. Anal. Biochem. 2012; 426:13–21. [DOI] [PubMed] [Google Scholar]
- 13. Graneli A., Yeykal C.C., Prasad T.K., Greene E.C.. Organized arrays of individual DNA molecules tethered to supported lipid bilayers. Langmuir. 2006; 22:292–299. [DOI] [PubMed] [Google Scholar]
- 14. Jo K., Chen Y.L., de Pablo J.J., Schwartz D.C.. Elongation and migration of single DNA molecules in microchannels using oscillatory shear flows. Lab Chip. 2009; 9:2348–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim Y., Jo K.. Neutravidin coated surfaces for single DNA molecule analysis. Chem. Commun. 2011; 47:6248–6250. [DOI] [PubMed] [Google Scholar]
- 16. Lee S., Jo K.. Visualization of surface-tethered large DNA molecules with a fluorescent protein DNA binding peptide. J. Vis. Exp. 2016; 112:e54141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim Y., Kim K.S., Kounovsky K.L., Chang R., Jung G.Y., dePablo J.J., Jo K., Schwartz D.C.. Nanochannel confinement: DNA stretch approaching full contour length. Lab Chip. 2011; 11:1721–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kounovsky-Shafer K.L., Hernandez-Ortiz J.P., Potamousis K., Tsvid G., Place M., Ravindran P., Jo K., Zhou S.G., Odijk T., de Pablo J.J. et al. . Electrostatic confinement and manipulation of DNA molecules for genome analysis. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:13400–13405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Finlay A.C., Hochstein F.A., Sobin B.A., Murphy F.X.. Netropsin, a new antibiotic produced by a streptomyces. J. Am. Chem. Soc. 1951; 73:341–343. [Google Scholar]
- 20. Zimmer C., Wahnert U.. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog. Biophys. Mol. Biol. 1986; 47:31–112. [DOI] [PubMed] [Google Scholar]
- 21. Geierstanger B., Mrksich M., Dervan P., Wemmer D.. Design of a G.C-specific DNA minor groove-binding peptide. Science. 1994; 266:646–650. [DOI] [PubMed] [Google Scholar]
- 22. Wade W.S., Mrksich M., Dervan P.B.. Design of peptides that bind in the minor groove of DNA at 5′-(A,T)G(A,T)C(A,T)-3′ sequences by a dimeric side-by-side motif. J. Am. Chem. Soc. 1992; 114:8783–8794. [Google Scholar]
- 23. Kawamoto Y., Sasaki A., Chandran A., Hashiya K., Ide S., Bando T., Maeshima K., Sugiyama H.. Targeting 24 bp within telomere repeat sequences with tandem tetramer pyrrole–imidazole polyamide probes. J. Am. Chem. Soc. 2016; 138:14100–14107. [DOI] [PubMed] [Google Scholar]
- 24. Vaijayanthi T., Bando T., Pandian G.N., Sugiyama H.. Progress and prospects of pyrrole‐imidazole polyamide–fluorophore conjugates as sequence‐selective DNA probes. ChemBioChem. 2012; 13:2170–2185. [DOI] [PubMed] [Google Scholar]
- 25. Minoshima M., Bando T., Sasaki S., Fujimoto J., Sugiyama H.. Pyrrole-imidazole hairpin polyamides with high affinity at 5′-CGCG-3′ DNA sequence; influence of cytosine methylation on binding. Nucleic Acids Res. 2008; 36:2889–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Delgadillo R.F., Parkhurst L.J.. Spectroscopic properties of fluorescein and rhodamine dyes attached to DNA. Photochem. Photobiol. 2010; 86:261–272. [DOI] [PubMed] [Google Scholar]
- 27. Kang Y., Lee J., Kim J., Oh Y., Kim D., Lee J., Lim S., Jo K.. Analysis of alcohol-induced DNA damage in Escherichia coli by visualizing single genomic DNA molecules. Analyst. 2016; 141:4326–4331. [DOI] [PubMed] [Google Scholar]
- 28. Johansen K.M., Cai W., Deng H., Bao X., Zhang W., Girton J., Johansen J.. Polytene chromosome squash methods for studying transcription and epigenetic chromatin modification in Drosophila using antibodies. Methods. 2009; 48:387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Günther K., Mertig M., Seidel R.. Mechanical and structural properties of YOYO-1 complexed DNA. Nucleic Acids Res. 2010; 38:6526–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murade C.U., Subramaniam V., Otto C., Bennink M.L.. Force spectroscopy and fluorescence microscopy of dsDNA-YOYO-1 complexes: implications for the structure of dsDNA in the overstretching region. Nucleic Acids Res. 2010; 38:3423–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kundukad B., Yan J., Doyle P.S.. Effect of YOYO-1 on the mechanical properties of DNA. Soft Matter. 2014; 10:9721–9728. [DOI] [PubMed] [Google Scholar]
- 32. Lee S., Wang C., Song J., Kim D., Oh Y., Ko W., Lee J., Park J., Lee H.S., Jo K.. Investigation of various fluorescent protein-DNA binding peptides for effectively visualizing large DNA molecules. RSC Adv. 2016; 6:46291–46298. [Google Scholar]
- 33. Kielkopf C.L., White S., Szewczyk J.W., Turner J.M., Baird E.E., Dervan P.B., Rees D.C.. A structural basis for recognition of A center dot T and T center dot A base pairs in the minor groove of B-DNA. Science. 1998; 282:111–115. [DOI] [PubMed] [Google Scholar]
- 34. Lee S., Oh Y., Lee J., Choe S., Lim S., Lee H.S., Jo K., Schwartz D.C.. DNA binding fluorescent proteins for the direct visualization of large DNA molecules. Nucleic Acids Res. 2016; 44:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Urbach A.R., Dervan P.B.. Toward rules for 1: 1 polyamide: DNA recognition. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:4343–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matsumoto S., Morikawa K., Yanagida M.. Light microscopic structure of DNA in solution studied by the 4′,6-diamidino-2-phenylindole staining method. J. Mol. Biol. 1981; 152:501–516. [DOI] [PubMed] [Google Scholar]
- 37. Cai W., Aburatani H., Stanton V.P., Housman D.E., Wang Y.-K., Schwartz D.C.. Ordered restriction endonuclease maps of yeast artificial chromosomes created by optical mapping on surfaces. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:5164–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith S.B., Finzi L., Bustamante C.. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992; 258:1122–1126. [DOI] [PubMed] [Google Scholar]
- 39. Perkins T.T., Smith D.E., Chu S.. Single polymer dynamics in an elongational flow. Science. 1997; 276:2016–2021. [DOI] [PubMed] [Google Scholar]
- 40. Cao H., Yu Z., Wang J., Tegenfeldt J.O., Austin R.H., Chen E., Wu W., Chou S.Y.. Fabrication of 10 nm enclosed nanofluidic channels. Appl. Phys. Lett. 2002; 81:174–176. [Google Scholar]
- 41. Lee J., Park H.S., Lim S., Jo K.. Visualization of UV-induced damage on single DNA molecules. Chem. Commun. 2013; 49:4740–4742. [DOI] [PubMed] [Google Scholar]
- 42. Torche P.C., Muller V., Westerlund F., Ambjornsson T.. Noise reduction in single time frame optical DNA maps. PLoS One. 2017; 12:e0179041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee J., Kim Y., Lim S., Jo K.. Single-molecule visualization of ROS-induced DNA damage in large DNA molecules. Analyst. 2016; 141:847–852. [DOI] [PubMed] [Google Scholar]
- 44. Kapuscinski J. DAPI: a DNA-specific fluorescent probe. Biotech. Histochem. 1995; 70:220–233. [DOI] [PubMed] [Google Scholar]
- 45. Spradling A.C. Polytene chromosome structure and somatic genome instability. Cold Spring Harb. Symp. Quant. Biol. 2017; 82:033670. [DOI] [PubMed] [Google Scholar]
- 46. Zhimulev I.F., Belyaeva E.S., Vatolina T.Y., Demakov S.A.. Banding patterns in Drosophila melanogaster polytene chromosomes correlate with DNA-binding protein occupancy. Bioessays. 2012; 34:498–508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.