Abstract
Downstream stable mRNA secondary structures can stall elongating ribosomes by impeding the concerted movements of tRNAs and mRNA on the ribosome during translocation. The addition of a downstream mRNA structure, such as a stem-loop or a pseudoknot, is essential to induce -1 programmed ribosomal frameshifting (-1 PRF). Interestingly, previous studies revealed that -1 PRF efficiencies correlate with conformational plasticity of pseudoknots, defined as their propensity to form incompletely folded structures, rather than with the mechanical properties of pseudoknots. To elucidate the detailed molecular mechanisms of translocation and -1 PRF, we applied several smFRET assays to systematically examine how translocation rates and conformational dynamics of ribosomes were affected by different pseudoknots. Our results show that initial pseudoknot-unwinding significantly inhibits late-stage translocation and modulates conformational dynamics of ribosomal post-translocation complexes. The effects of pseudoknots on the structural dynamics of ribosomes strongly correlate with their abilities to induce -1 PRF. Our results lead us to propose a kinetic scheme for translocation which includes an initial power-stroke step and a following thermal-ratcheting step. This scheme provides mechanistic insights on how selective modulation of late-stage translocation by pseudoknots affects -1 PRF. Overall our findings advance current understanding of translocation and ribosome-induced mRNA structure unwinding.
INTRODUCTION
During elongation, the ribosome moves along the messenger RNA (mRNA) from 5′ to 3′ end and synthesizes proteins based on genetic information stored in the mRNA (1–4). Within each elongation cycle, the ribosome cycles between the post-translocation (POST) complex and the pre-translocation (PRE) complex and moves three nucleotide bases, a codon, at a time to maintain its reading frame. An elongation cycle starts with aminoacyl-tRNA (aa-tRNA) accommodation into the empty A-site of a POST complex and peptide transfer, resulting in formation of a PRE complex with a peptidyl-tRNA bound in the A-site and a deacylated tRNA bound in the P-site. Translocation, which is the movement of tRNA-mRNA complex on the ribosome catalyzed by elongation factor G (EF-G), leads to formation of a POST complex with a peptidyl-tRNA bound in the P-site and a deacylated tRNA bound in the E-site. Dissociation of the deacylated tRNA from the E-site completes an elongation cycle. Single-stranded mRNA is prone to forming secondary structures, including pseudoknots and stem-loops. Downstream mRNA structures have been shown to slow or even pause elongation, which can functionally couple with processes such as co-translational protein folding, protein modification and programmed ribosomal frameshifting (5–10).
Frameshifting occurs when the ribosome fails to maintain its reading frame, which usually leads to premature termination of elongation or production of a different polypeptide chain. Random frameshifting occurs at very low frequency (10−5) (11–13). In contrast, the efficiency of programmed ribosomal frameshifting (PRF) can be higher than 10−2 (14,15). –1 PRF, during which the reading frame shifts 1-nt backward, is a mechanism used by many viruses to synthesize two sets of proteins and to precisely regulate their relative production rates (16–18). Two essential elements are required to trigger –1 PRF. A hepta-nucleotide slippery sequence (X XXYYYZ, underlining denotes the 0 frame) and a downstream stimulatory secondary structure such as a pseudoknot or a stem-loop, usually located 5–8 nucleotides downstream of a slippery sequence (14,19). Previous studies have indicated that the –1 PRF-inducing secondary structure slows ribosomal elongation rate, and results in ribosome generation of a mechanical force which destabilizes codon:anticodon base pairs and induces frameshifting (6–8,20–22). Although pseudoknots with similar mechanical stabilities stimulate –1 PRF to quite different extents (2–28%) (23), -1 PRF efficiency does correlate with the conformational plasticity of pseudoknot (23,24). However, how pseudoknots with different conformational plasticities affect elongation rates and conformational dynamics of the ribosome has been unclear.
Here, we use single-molecule fluorescence resonance energy transfer (smFRET) assays (6,25) to determine the effects on elongation rates and conformational dynamics of a series of downstream mRNA pseudoknots on ribosomes carrying out two consecutive elongation cycles, which begin with ribosomes encountering and starting to unwind the pseudoknots. We find that –1 PRF efficiency strongly correlate with the rate of late-stage translocation in the first elongation cycle, and that pseudoknots modulate conformational dynamics of the POST complexes, whose transition rates also correlate with –1 PRF efficiencies. Together, our discoveries clarify how downstream pseudoknots with different conformational plasticity affect elongation, ribosome structural dynamics and –1 PRF.
MATERIALS AND METHODS
Reagents
Cyanine3 (NHS ester), Cyanine3 (maleimide), Cyanine5 (NHS ester) and Cyanine5 (maleimide) were purchased from Lumiprobe. Puromycin was purchased from Cayman Chemical. UltraPure™ 1M Tris–HCI pH 7.5 was obtained from Invitrogen. Other common materials and reagents were purchased from Sigma or Amresco.
Preparation of charged and labeled tRNA
Escherichia coli tRNAfMet was purchased from MP Biomedicals. Eschrichia coli tRNATyr (a mixture of isoacceptors) and tRNAArg (a mixture of isoacceptors) were purchased from Chemical Block (Moscow). Escherichia coli tRNAVal (a mixture of isoacceptors) was obtained from Sigma. tRNALys (anticodon: UUU, reading the AAA/AAG codon) and tRNAPhe (anticodon: GAA, reading the UUC/UUU codon) were separated from bulk E. coli tRNA according to previous studies (26,27).
Cy3/Cy5 labeled tRNALys and tRNAPhe were obtained through reaction of NHS ester with the primary aliphatic amino group of the 3-(3-amino-3-carboxypropyl)-uridine at position 47 (acp3U47) and purified with hydrophobic interaction chromatography on HPLC via a DeltaPak C4 Column (Waters, 300 Å, 15 μm, 3.9 mm × 300 mm) according to previous protocal (25,27). tRNAVal was labeled by Cy3-NHS and purified through the same procedure. Cy3-labeled tRNAArg was prepared using the reduction, charging, and labeling protocol as described (28). The charged tRNAArg was labeled through reaction of Cy3-hydrazide with the dihydrouridine residue in the D-loop region and purified by HPLC via a DeltaPak C4 Column (Waters, 300 Å, 15 μm, 3.9 mm × 300 mm).
Escherichia coli Phe, Lys, Tyr, Arg and Val tRNA synthetase, whose plasmids were constructed using vector pET-28a, were overexpressed in E. coli BL21 (DE3) cells and purified on a Ni-NTA column (Qiagen). Aminoacylation mixture containing 25 μM tRNA, 100 μM l-amino acid, 12.5 μM tRNA synthetase, 10 mM ATP, 3 mM dithiothreitol, 0.005 unit/μl Thermostable Inorganic Pyrophosphatase (NEB) in 100 mM Tris–HCl pH 7.8, 50 mM MgCl2, and 2.5 mM EDTA were kept at 37°C for 30 min. Aminoacyl tRNA was purified by phenol–chloroform extraction, Nap-5 column (GE) desalting, and ethanol precipitation as previous described (29,30). Partial separation of charged from uncharged tRNAs was achieved by reversed-phase HPLC on a DeltaPak C4 Column (Waters, 300Å, 15 μm, 3.9 mm × 300 mm). Aminoacyl tRNAs were precipitated with ethanol, dissolved in DEPC-treated water, and their concentrations were determined photometrically. Ternary complexes (aa-tRNA·EF-Tu·GTP, TCs) were formed by incubating 8 μM EF-Tu, 1 μM aminoacyl tRNA, 3 mM GTP, 1.3 mM phosphoenolpyruvate, and 5 μg/ml pyruvate kinase in TAM15 buffer (15 mM MgAc2, 50 mM Tris–HCl, pH 7.5, 30 mM NH4Cl, 70 mM KCl and 1 mM dithiothreitol) for 15 min at 37°C.
mRNA preparation
mRNAs for smFRET experiments were prepared via in vitro transcription and then annealed with a biotinylated DNA handle in order to achieve immobilization on slides. DNA fragments corresponding to mRNAs were synthesized into a pMV vector, which contains a T7 promoter. The DNA sequence in the coding region was confirmed by sequencing. The DNA construct was linearized by EcoR I and used as a template for transcription using the HiScribe™ T7 High Yield RNA Synthesis Kit (NEB). The transcripts were purified via phenol and chloroform extraction, followed by precipitation with 75% ethanol. The final RNA samples were dissolved in DEPC-treated water. The integrity and purity of the mRNAs were confirmed using agarose gel electrophoresis. All mRNA sequences were listed in Supplementary Table S1.
Initiation complex preparation
Ribosomes, initiation factors, elongation factors, and tRNAs were all from E. coli. Unlabeled 70S, 70S with Cy5-labeled L11 (70S-L11Cy5), 70S with Cy5-labeled L1 (70S-L1Cy5), initiation factors 1, 2 and 3, elongation factor G, and elongation factor Tu were prepared according to published procedures (29,31–33). Experiments were carried out in TAM15 buffer (15 mM MgAc2, 50 mM Tris–HCl, pH 7.5, 30 mM NH4Cl, 70 mM KCl and 1 mM dithiothreitol). To prepare initiation complexes, 70S, 70S-L11Cy5 or 70S-L1Cy5 ribosomes were incubated with mRNA, initiation factors, fMet-tRNAfMet and GTP in TAM15 buffer for 30 min at 37°C and purified by centrifugation through a 1.1 M sucrose cushion.
Preparation of PEG-passivated slides
PEG-passivated slides were prepared according to previous procedures with minor modifications (34). In brief, slides and coverslips were sonicated at 40°C in the order of acetone (10 min), 0.2 M KOH (20 min), and ethanol (10 min). Cleaned slides and coverslips were treated with amino-silane reagents (1 ml 3-aminopropyltriethoxysilane, 5 ml acetic acid and 94 ml methanol) at room temperature overnight and then incubated with polyethylene glycol (PEG, Laysan Bio, Inc., containing 20% (w/w) mPEG-Succinimidyl Valerate, MW 2000 and 1% Biotin-PEG-SC, MW 2000) in 0.1 M sodium bicarbonate (pH 8.3) for 3 h. Slides and coverslips were dried by clean N2, put in 50 ml falcon tubes, vacuum-sealed in food saver bags, and stored at –20°C.
smFRET Protocols
All smFRET studies were carried out at 25°C. All complex formations and single-molecule imaging were carried out in TAM15 buffer. An enzymatic oxygen scavenging system of 3 mg/ml glucose, 100 μg/ml glucose oxidase (Sigma-Aldrich), 40 μg/ml catalase (Roche), 1 mM cyclooctatetraene (COT, Sigma-Aldrich), 1 mM 4-nitrobenzylalcohol (NBA, Sigma-Aldrich), 1.5 mM 6-hydroxy-2,5,7,8-tetramethyl-chromane-2-carboxylic acid (Trolox, Sigma-Aldrich) was present in the final single-molecule imaging solutions to diminish fluorophore photobleaching and blinking.
Single-molecule fluorescence and FRET measurements were performed on a home-built objective-type TIRF microscope, based on a Nikon Eclipse Ti-E with an EMCCD camera (Andor iXon Ultra 897), and solid state 532 and 640 nm excitation lasers (Coherent Inc. OBIS Smart Lasers) which can be modulated using digital signals from the EMCCD camera. Fluorescence emission from the probes was collected by the microscope and spectrally separated by interference dichroic (T635lpxr, Chroma) and bandpass filters, ET585/65m (Chroma, Cy3) and ET700/75m (Chroma, Cy5), in a Dual-View spectral splitter (Photometrics, Inc., Tucson, AZ, USA). All smFRET movies were collected using Cell Vision software (Beijing Coolight Technology).
Collected movies were analyzed by a custom-made software program developed as an ImageJ plugin (http://rsb.info.nih.gov/ij). Fluorescence spots were fitted by a 2D Gaussian function within a 9-pixel by 9-pixel area, matching the donor and acceptor spots using a variant of the Hough transform (35). The background subtracted total volume of the 2D Gaussian peak was used as raw fluorescence intensity I. FRET efficiency is calculated as IA/ (IA+ID), where IA and ID are the Cy5 acceptor and Cy3 donor fluorescence intensity, respectively. IA and ID are subjected to background subtractions. Three or more replicates were performed for each experiments. Standard error of mean (SEM) was displayed as error bar and variation.
Immobilization of ribosomes
The sample flow chamber (∼7 μL) was formed between a PEG-coated slide and coverslip and held together by double-sided adhesive tape that served as spacers and borders of the flow chamber. Ribosomes were immobilized by hybridization of the 5′ of the mRNA with a 3′ biotinylated DNA handle (5′-CCCTGGTCCGGTGGTCCGCCTGCTGGTCCCTTTTTTTTTTTTTTTTTT-biotin-3′, underlined nucleotides base-paired with mRNA) that was bound via streptavidin to a PEG-coated chamber surface.
Dwell time experiments to examine real-time ongoing elongation
Single-molecule recording during ongoing elongation experiments began 10 s prior to injecting 10 nM labeled ternary complexes, 50 nM unlabeled ternary complexes, 4 μM EF-G, and 2 mM GTP into flow chambers containing immobilized initiation complexes, and was carried out without further washing for 10 min. Unless otherwise indicated, all dwell time experiments of ribosome translation on mRNAs with the non-slippery sequence were initiated from 70S initiation complexes. Dwell time experiments of ribosome on mRNAs with slippery sequence were initiated from pre-formed PRE-translocation complexes which contain Val and Lys codons in their P- and A-sites, respectively.
Fluctuation experiments to examine conformational dynamics of stalled ribosomes
Immobilized POST complexes were prepared by incubating immobilized initiation complexes with 50 nM unlabeled and labeled ternary complexes, 4 μM EF-G, and 2 mM GTP for 10 min to allow the ribosomes to translate to the designed POST complexes. Immobilized PRE complexes were formed by adding 50 nM cognate ternary complexes to immobilized POST complexes in the absence of EF-G and GTP. Unbound reagents were washed away before single-molecule recording.
Frameshifting efficiency measurement by single-molecule counting
Single-molecule counting was used to determinate the frameshifting efficiency induced by each secondary structure in the presence of slippery sequence A AAAAAG (0 frame: fMVKKF, –1 frame: fMVKKV). POSTV complex was first formed by mixing 1–5 nM biotinylated initiation complex, 200 nM Val ternary complex, 4 μM EF-G and 2 mM GTP for 10 min at 25°C and immobilized on the PEGylated surface. Mixture containing 50 nM Lys ternary complex, 50 nM Cy5 labeled Phe ternary complex, 50 nM Cy3 labeled Val ternary complex, 4 μM EF-G and 2 mM GTP was injected to flow chamber and incubated for 10 min to complete translocation and frameshifting. Numbers of Cy5 plots and Cy3 plots per field were recorded as X0 and X-1, respectively, by alternating excitation between 532 and 640 nm lasers. In the background experiments, immobilized POSTV complex was incubated with mixture containing no Lys ternary complex, 50 nM Cy5 labeled Phe ternary complex, 50 nM Cy3 labeled Val ternary complex, 4 μM EF-G and 2 mM GTP for 10 min. Then, numbers of Cy5 plots and Cy3 plots per field were recorded as B0 and B-1, respectively, which were at least 20 folds smaller than X0 and X-1. Frameshifting efficiency (E-1 PRF) was calculated via (X-1– B-1)/(X0 – B0 + X-1 – B-1).
Frameshifting efficiency measurement by an ensemble assay in solution
The efficiency of –1 PRF induced by each secondary structure in the presence of slippery sequence A AAA AAG was also determined by an ensemble assay in solution. POSTV complex was formed by mixing 100 nM initiation complex, 500 nM Val ternary complex, 4 μM EF-G and 2 mM GTP for 10 min at 25°C, and purified by centrifugation through a 1.1 M sucrose cushion. Purified POSTV complex was then incubated with 500 nM Lys ternary complex, 500 nM Cy5 labeled Phe ternary complex, 500 nM Cy3 labeled Val ternary complex, 4 μM EF-G and 2 mM GTP for 10 min at 25°C to complete translation and frameshifting. POSTV complex was incubated with the mixture without Lys ternary complex to serve as the background experiment. Ribosomes carrying Cy3 labeled fMVKKV-tRNAVal in the P-site (-1 frame) and ribosomes carrying Cy5 labeled fMVKKF-tRNAPhe in the P-site (0 frame) were purified through another 1.1 M sucrose cushion. Cy3 and Cy5 were excited by 532 and 640 nm, respectively, and their fluorescent signals were detected after passing LP550 and LP665 cut-off filters (SpectraMax), respectively. Signals captured from the background experiment were used as background signals, whose values were subtracted from Cy3 and Cy5 signals detected from frameshifting experiments. Concentrations of ribosomes carrying Cy3 labeled fMVKKV-tRNAVal (C-1) and ribosomes carrying Cy5 labeled fMVKKF-tRNAPhe (C0) were quantified by their fluorescence intensities through standard fluorescence curves of Cy3 labeled tRNAVal and Cy5 labeled tRNAPhe. Within the concentration range from 10 nM to 1 μM, fluorescence signals of Cy3 and Cy5 labeled tRNAs increased linearly with their concentrations. Ribosome samples were diluted so that their signals fell into the linear range of standard fluorescence curves. Frameshifting efficiency (E-1 PRF) was calculated via C-1/(C-1 + C0).
RESULTS
mRNA constructs and -1 PRF efficiency
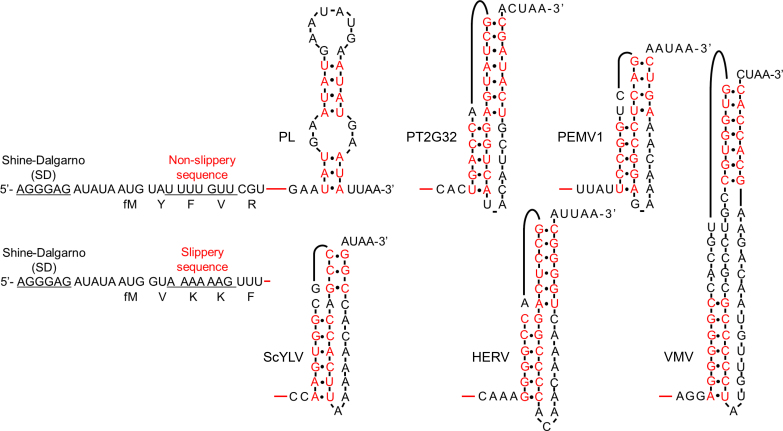
We used several RNA pseudoknots, denoted PT2G32, PEMV1, ScYLV, HERV and VMV (Figure 1), which were derived from the bacteriophage T2 gene 32 (36), pea enation mosaic virus-1 (37), sugarcane yellow leaf virus (38), human endogenous retrovirus-K10 (39), and Visna–Maedi retrovirus (40), respectively. Previous studies reported that these pseudoknots, which have similar unfolding energy barriers and unfolding forces, stimulate –1 PRF to different extents. To examine how they affect dynamics of elongation, which eventually leads to different frameshifting efficiencies, we placed them downstream from a common non-slippery sequence encoding fMYFVR or a common slippery sequence encoding fMVKKF (Figure 1 and Supplementary Table S1). An RNA sequence containing an unstable secondary structure (denoted PL) (6) was used as a control for comparison. The spacers between the common sequences and pseudoknots remained the same as the spacers between the slippery sequences and pseudoknots in their original viral sequences.
Figure 1.
mRNA constructs and their secondary structures. Two sets of mRNAs, one set contained a common non-slippery sequence (fMYFVR) and the other had a common slippery sequence (fMVKKF), were used in our experiments. fM, Y, F, V, R and K are abbreviations for initiator Methionine, Tyrosine, Phenylalanine, Valine, Arginine, and Lysine, respectively. The Shine-Dalgarno (SD) sequence, non-slippery sequence (U UUU GUU), and slippery sequence (A AAA AAG) were underlined. Pseudoknots PT2G32, PEMV1, ScYLV, HERV and VMV were placed downstream from the common mRNA sequences, whose connection sites were indicated by red lines. An unstable secondary structure PL was used as a control. Their full sequences were shown in Supplementary Table S1. Base pairs within secondary structures were indicated in red, whereas unpaired bases were indicated in black.
Although -1 PRF efficiencies have been reported for the pseudoknots we used, they were measured by different groups under different conditions (37–40). Here, we quantified the abilities of pseudoknots to induce –1 PRF of E. coli ribosomes under our experimental conditions. Translating our mRNAs with the common slippery sequence produces fMetValLysLysPhe (fMVKKF) in the 0 frame and fMetValLysLysVal (fMVKKV) if –1 PRF occurs. Using Cy5 labeled tRNAPhe and Cy3 labeled tRNAVal, the numbers of ribosomes translating in the 0 and -1 frames can be quantified by the numbers of Cy5 and Cy3 fluorescence spots, respectively, after elongating through the slippery sequence, permitting calculation of -1 frameshifting efficiency (Table 1). The mRNA containing the least stable structure (PL) caused the lowest frameshifting efficiency, 8 ± 1%. Frameshifting efficiencies induced by pseudoknots varied from 16 ± 2% to 61 ± 3%, presenting a similar trend as reported values (37–40). Because both UUU and UUC encode Phe. To examine the contribution of +1 frameshifting which might happen when the UUU codon is followed by a cytosine (C), we introduced single-base mutations in mRNAs containing ScYLV and HERV so that their codons after slippery sequences in the +1 frame codons did not encode Phe (Supplementary Table S2). Our results showed that, the -1 frameshifting efficiencies of these variants were the same as our original design, which indicated that the contribution of +1 frameshifting was neglectable (Supplementary Table S2). Furthermore, –1 frameshifting efficiencies obtained by single-molecule counting were validated by additional ensemble assay in solution. Similar frameshifting efficiencies were estimated by both single-molecule and ensemble assays (Table 1).
Table 1.
-1 PRF efficiency and dwell time of ribosomal complexes during ongoing elongation experiments
| Dwell time in the first cycle | Dwell time in the second cycle | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| –1 PRF efficiency (%)a | non-slippery sequence | slippery sequence | non-slippery sequence | ||||||
| mRNA structure | Single-molecule counting | Ensemble assay | PREFV (s) | POSTFV (s) | PREFV + POSTFV (s) | PREK1K2 (s) | POSTK1K2 (s) | PREVR (s) | POSTVR (s) |
| PL | 8 ± 1 | 10 ± 3 | 1.1 ± 0.1 | 2.9 ± 0.4 | 2.6 ± 0.4 | 1.6 ± 0.3 | 1.5 ± 0.3 | 1.3 ± 0.1 | 2.5 ± 0.3 |
| PT2G32 | 16 ± 2 | 19 ± 5 | 0.9 ± 0.2 | 4.6 ± 0.5 | 5.0 ± 0.5 | 2.3 ± 0.1 | 2.1 ± 0.2 | 1.4 ± 0.1 | 3.2 ± 0.4 |
| PEMV1 | 29 ± 3 | 23 ± 5 | 0.9 ± 0.1 | 5.7 ± 0.5 | 7.7 ± 0.6 | 2.3 ± 0.2 | 2.9 ± 0.2 | 1.5 ± 0.1 | 4.1 ± 0.4 |
| ScYLV | 61 ± 3 | 63 ± 7 | 0.7 ± 0.1 | 7.8 ± 0.3 | 9.6 ± 0.7 | 2.2 ± 0.3 | 4.0 ± 0.2 | 1.6 ± 0.1 | 3.8 ± 0.5 |
| HERV | 45 ± 2 | 48 ± 8 | 0.8 ± 0.1 | 8.6 ± 0.3 | 9.4 ± 0.8 | 2.0 ± 0.1 | 3.6 ± 0.1 | 1.5 ± 0.1 | 4.2 ± 0.6 |
| VMV | 44 ± 3 | 39 ± 3 | 0.9 ± 0.1 | 7.7 ± 0.8 | 9.1 ± 0.3 | 2.3 ± 0.3 | 3.2 ± 0.4 | 1.5 ± 0.1 | 3.3 ± 0.4 |
| Pearson correlation coefficientb | 0.73 (0.10) | 0.92* (0.01) | 0.93* (0.01) | 0.43 (0.39) | 0.97* (0.01) | 0.94* (0.01) | 0.66 (0.15) | ||
Mean ± SEM was calculated from three to four independent replicates.
a–1 PRF efficiency was measured in the presence of the slippery sequence (A AAA AAG).
bPearson correlation coefficient between dwell times of each ribosomal complex and -1 PRF efficiency measured by single-molecule counting. Corresponding P-value was listed within parentheses. * indicated significant correlation.
Because different experimental conditions were used, the numbers we obtained did not exactly match those obtained earlier using eukaryotic translation systems. For example, our measurements indicated that pseudoknot ScYLV displayed the highest efficiency to induce –1 PRF, whereas previous study reported that ScYLV caused moderate –1 PRF efficiency using rabbit reticulocyte lysate (Table 1 and Supplementary Table S1). The discrepancy might be caused by different buffer condition and difference between eukaryotic and prokaryotic ribosomes. Therefore, we used frameshifting efficiencies measured by us in the following correlation analysis to correlate with elongation rates captured under similar experimental conditions.
Design of the smFRET experiments using the non-slippery sequence
We applied several single-molecule FRET assays (6,25) to examine how downstream mRNA pseudoknots affect elongation rates and conformational dynamics of ribosomes in two consecutive elongation cycles with mRNAs containing the common non-slippery sequence encoding fMYFVR. Measurements started from the POSTF complex, in which intact pseudoknot structures engage the mRNA entrance located at the ribosomal 30S subunit. In the first elongation cycle, from POSTF to POSTV, ribosomes started to unwind pseudoknot structures. In the second cycle, from POSTV to POSTR, ribosomes unwound three more base pairs (Figure 2A).
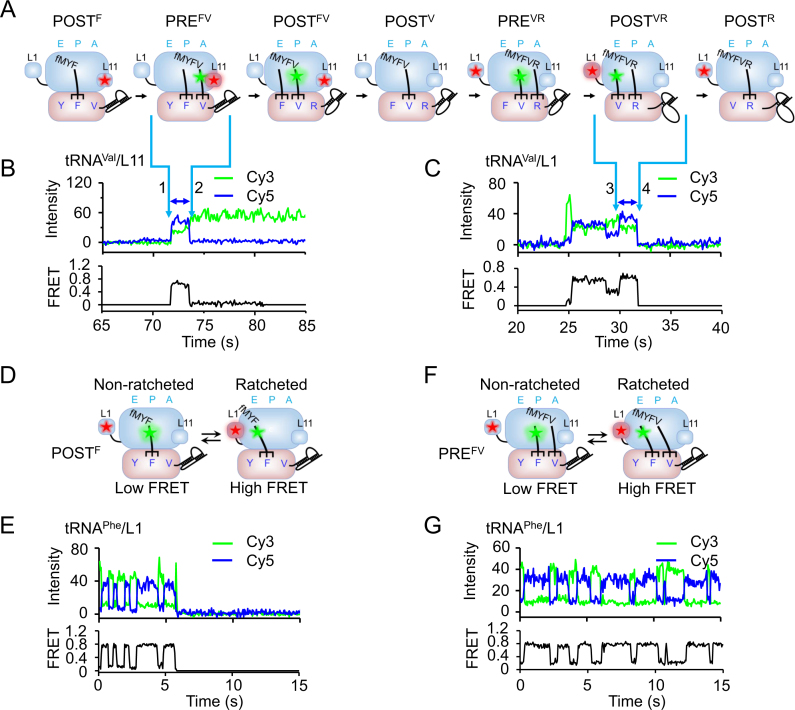
Figure 2.
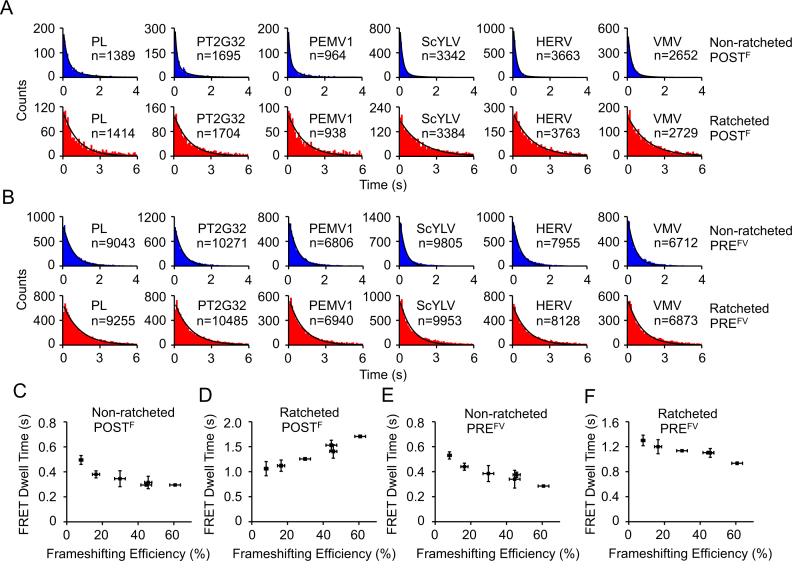
smFRET experimental design. (A) Schematic drawings of ribosomal complexes during two rounds of translational elongation cycles by adding Valine and Arginine to the growing peptide monitored by smFRET. Ribosomes engage downstream pseudoknots after formation of POSTF complexes and start to unwind pseudoknots in the following cycles. FRET pairs of Cy3-tRNAVal/Cy5-L11 and Cy3-tRNAVal/Cy5-L1 were drawn in the first and second cycles, respectively, to demonstrate our smFRET assays. (B) Typical real-time ribosome translation trace measured using Cy3-tRNAVal/Cy5-L11 FRET pair for ribosomes programmed with mRNAs containing the non-slippery sequence. Fluorescence of Cy3 (green) and Cy5 (FRET, blue) under 532 nm excitation were collected. Accommodation of Cy3-tRNAVal into the A-site led to spontaneous appearance of Cy3 and FRET signals (arrow 1). Translocation from PREFV to POSTFV caused decrease of FRET accompanied by increase of Cy3 signals (arrow 2). The high FRET state between arrows 1 and 2 corresponded to PREFV complex. (C) Typical real-time ribosome translation trace measured using Cy3-tRNAVal/Cy5-L1 FRET pair for ribosomes programmed with mRNAs containing the non-slippery sequence. According to our previous established assay (6), accommodation of Cy3-tRNAVal into the A-site led to appearance of Cy3 signals with a low FRET value. Movements of Cy3-tRNAVal to the P-site and arriving of the following Arg-tRNAArg into the A-site caused increase and decrease of FRET between Cy3-tRNAVal and Cy5-L1, respectively. Arriving of Cy3-tRNAVal into the E-site led to formation of the high FRET state (arrow 3). Dissociation of Cy3-tRNAVal from the E-site caused spontaneous disappearance of Cy3 and FRET signals (arrow 4). The last high FRET state between arrows 3 and 4 corresponded to POSTVR complex. (D–E) Schematic drawings (D and F) and example traces (E and G) using Cy3-tRNAPhe/Cy5-L1 FRET pair to capture conformational dynamics of POSTF (D and E) and PREFV (F and G) complexes. In both POSTF and PREFV complexes, ribosomes spontaneously transit between the non-ratcheted (low FRET) and ratcheted (high FRET) states, which was captured by smFRET as shown in E and G.
During undergoing active peptide synthesis, smFRET measurements of dwell time were carried out to measure the rates of specific elongation steps within each elongation cycle. These experiments measured smFRET between: (a) Cy3 labeled tRNA (Cy3-tRNA) and Cy5 labeled large-subunit protein L11 (Cy5-L11), near the A-site, to probe the movements of the A-site tRNA toward the P-site characterized by high FRET state between arrows 1 and 2 (an example of Cy3-tRNAVal/Cy5-L11 FRET pair was shown in Figure 2B) (29); (b) Cy3-tRNA and Cy5 labeled large-subunit protein L1 (Cy5-L1), to probe the dissociation of the E-site tRNA, which was characterized by the high FRET state between arrows 3 and 4 (an example of Cy3-tRNAVal/Cy5-L1 FRET pair was shown in Figure 2C) (6); (c) Cy5-tRNAPhe and Cy3-tRNAVal (Supplementary Figure S1) to determine dwell times of PREFV and POSTFV complexes.
In addition, when elongation was halted in the absence of elongation factors, smFRET between P-site Cy3-tRNA and Cy5-L1 was used to monitor spontaneous fluctuations between non-ratcheted (low FRET) and ratcheted (high FRET) states within both stalled PRE and POST complexes. Examples of Cy3-tRNAPhe/Cy5-L1 FRET pair were shown in Figure. 2D–G (41). Please notice that, for clarity, only three FRET pairs and four single-molecule trajectories were shown in Figure 2. Other FRET pairs were used and mentioned below. Unless otherwise indicated, measurements were performed with mRNAs containing the common non-slippery sequence (fMYFVR).
Elongation rates in the first pseudoknot-unwinding cycle
The first elongation cycle to unwind pseudoknots was from POSTF to POSTV. Cy3-tRNAVal/Cy5-L11 and Cy3-tRNAPhe/Cy5-L1 FRET pairs were used to capture dwell times of the PREFV and POSTFV, respectively, during ongoing elongation by subjecting immobilized initiation complexes containing either Cy5-L11 or Cy5-L1 labeled ribosomes, or unlabeled ribosomes to several rounds of elongation by injecting reaction mixtures containing Cy3-tRNAVal or Cy3-tRNAPhe ternary complexes, or Cy5-tRNAPhe and Cy3-tRNAVal ternary complexes, and other necessary components while recording single-molecule fluorescence signals.
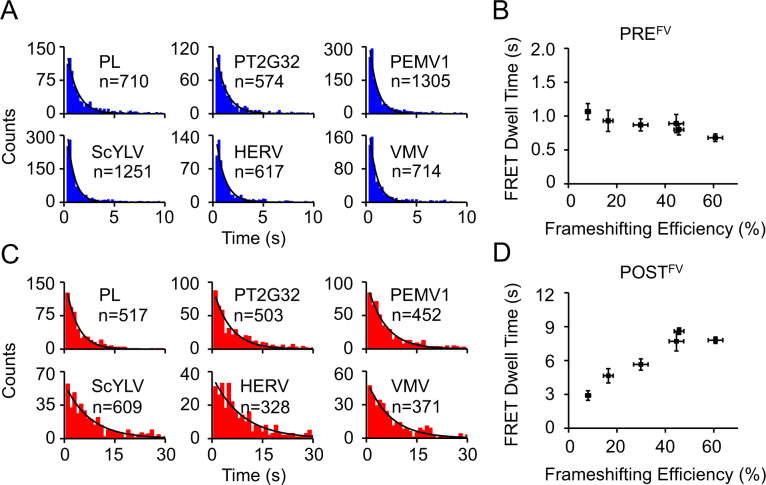
For the Cy3-tRNAVal/Cy5-L11 FRET pair, formation of PREFV caused by binding of Cy3-tRNAVal led to spontaneous appearance of Cy3 signal and FRET signal in Cy5 detection channel as indicated by arrow 1 in Figure 2b. Translocation of Cy3-tRNAVal from the A-site to the P-site led to decrease of FRET efficiency (E) from ∼0.6 (Supplementary Figure S2) to 0.1–0.2 as indicated by arrow 2 in Figure 2B (29). Average dwell times of PREFV were calculated from dwell time distributions of the high Cy3-tRNAVal/Cy5-L11 FRET state, whose values were in the range of 0.7–1.1 s and similar among different pseudoknots (Figure 3A, B and Table 1). These results are consistent with previous reports that most mRNA structures, except the ones with extreme high G–C base pairs, have minor effects on dwell times of PRE complexes (6).
Figure 3.
Elongation rates of ribosomes on non-slippery mRNAs with different pseudoknots during the first pseudoknot-unwinding cycle. (A) Dwell time distributions of PREFV complexes captured during ongoing elongation. (B) Plot of PREFV dwell time versus frameshifting efficiency. (C) Dwell time distributions of POSTFV complexes during ongoing elongation. (D) Plot of POSTFV dwell time versus frameshifting efficiency. n is the number of events. Dwell times were extracted by single exponential fitting of dwell time distributions. Error bars were standard errors.
For the Cy3-tRNAPhe/Cy5-L1 FRET pair, Cy3-tRNAPhe binding to, moving within, and dissociation from the ribosome led to four distinctive FRET states that occurred in the temporal sequence E = ∼0.1, ∼0.78, ∼0.2 and ∼0.78 (Supplementary Figure S3). Based on previous assignment (6), the last high FRET state, between arrows 1 and 2 in Supplementary Figure S3, was caused by FRET between Cy5-L1 and Cy3-tRNAPhe in the E-site, which were in close proximity to each other. Average dwell times of POSTFV quantified from dwell time distributions of the last high Cy3-tRNAPhe/Cy5-L1 FRET state were markedly increased by the presence of pseudoknots, which increased from 2.9 ± 0.4 s (PL) to 8.6 ± 0.3 s (HERV) (Figure 3C and Table 1). Interestingly and surprisingly, dwell times of POSTFV displayed a strong positive correlation with frameshifting efficiencies (Figure 3D and Table 1).
The Cy5-tRNAPhe/Cy3-tRNAVal FRET pair allowed measurement of dwell times of PREFV and POSTFV complexes (between arrows 1 and 2 in Supplementary Figure S1) which also displayed strong positive correlations with frameshifting efficiencies (Supplementary Figure S1D and Table 1). Based on results described above, such correlation should be mainly contributed by POSTFV complexes. In all, dwell times and correlation behaviors captured by different FRET pairs were consistent with each other.
A smaller set of data were obtained in the Philadelphia lab using a different non-slippery sequence encoding fMERFV, which gave tRNA/L11 dwell times for the PRE state encountering the unstable structure PL, and pseudoknots PT2G32 and VMV, independent of the frameshifting efficiency, and tRNA/L1 dwell times increasing with frameshifting efficiency from 2.5 ± 0.2 s to 5.9 ± 0.3 s (Supplementary Figure S4). Although a different non-slippery sequence was used, these results present the same behaviors in the first pseudoknot-unwinding cycle as the data measured in the Beijing lab.
Elongation rates in the first pseudoknot-unwinding cycle using mRNAs containing a slippery sequence
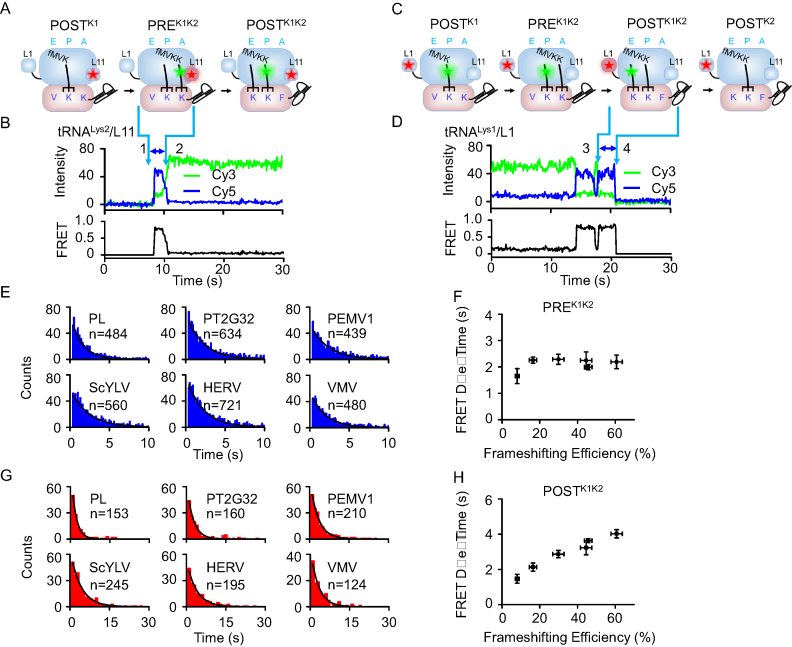
For measurements using mRNA encoding the common slippery sequence fMVKKF, K1 and K2 were used to represent the first and second lysines, respectively. Dwell times of PREK1K2 and POSTK1K2, which corresponded to PREFV and POSTFV in the non-slippery sequence, were measured by Cy3-tRNAK2/Cy5-L11 and Cy3-tRNAK1/Cy5-L1 FRET pairs, respectively (Figure. 4A–D). Consistent with results obtained with the non-slippery sequence, only dwell times of POSTK1K2 displayed a positive correlation with frameshifting efficiencies induced by pseudoknots, whereas PREK1K2 presented similar dwell times among different pseudoknots (Figure 4E–H and Table 1). Together, our results clearly indicated that, for both non-slippery and slippery sequences, the abilities of pseudoknots to induce –1 PRF strongly correlate with their abilities to stall elongating ribosomes in the POST complexes in the first pseudoknot-unwinding cycle.
Figure 4.
Elongation rates of ribosomes on slippery mRNAs with different pseudoknots sequence during the first pseudoknot-unwinding cycle. (A) Schematic drawings of ribosomal complexes from POSTK1 to POSTK1K2. (B) Typical real-time ribosome translation trace measured using Cy3-tRNALys2/Cy5-L11 FRET pair. Accommodation of Cy3-tRNALys2 into the A-site led to spontaneous appearance of Cy3 and FRET signals (arrow 1). Translocation from PREK1K2 to POSTK1K2 caused decrease of FRET accompanied by increase of Cy3 signals (arrow 2). The high FRET state between arrows 1 and 2 corresponded to PREK1K2 complex. (C) Schematic drawings of ribosomal complexes from POSTK1 to POSTK2. (D) Typical real-time ribosome translation trace measured using Cy3-tRNALys1/Cy5-L1 FRET pair. Arriving of Cy3-tRNALys1 into the E-site led to formation of the high FRET state (arrow 3). Dissociation of Cy3-tRNALys1 from the E-site caused spontaneous disappearance of Cy3 and FRET signals (arrow 4). The last high FRET state between arrows 3 and 4 corresponded to POSTK1K2 complex. (E) Dwell time distributions of PREK1K2 complexes during ongoing elongation. (F) Plot of PREK1K2 dwell time versus frameshifting efficiency. (G) Dwell time distributions of POSTK1K2 complexes during ongoing elongation. (H) Plot of POSTK1K2 dwell time versus frameshifting efficiency. n is the number of events. Dwell times were extracted by single exponential fitting of dwell time distributions. Error bars were standard errors.
Elongation rates in the first pseudoknot-unwinding cycle using mRNAs containing pseudoknots targeting prokaryotic ribosome
A pseudoknot, denoted copA, was identified as an efficient -1 PRF stimulator in E. coli copper transporter gene COPA (42). Using our single-molecule assays, we examined how copA and two of its variants (copA-S1 and copA-S2) induced –1 PRF and affected elongation rates in the first unwinding cycle (Supplementary Figure S5 and Supplementary Table S3). For both non-slippery and slippery sequences, the abilities of copA and its mutant variants to induce –1 PRF also correlate with their abilities to stall elongating ribosomes in POST complexes. Together, our results clearly demonstrated that copA, a pseudoknot targeting prokaryotic ribosome, exhibited the same behaviors as pseudoknots we tested above, most of which are from viral mRNAs and originally target eukaryotic ribosome.
Conformational dynamics of ribosomes in the first pseudoknot-unwinding cycle
Both ribosomal POST and PRE complexes have been shown to spontaneously fluctuate between two major conformational states (29,43–46). The Cy3-tRNAPhe/Cy5-L1 FRET pair was used to probe spontaneous transitions between the non-ratcheted (low FRET) and ratcheted (high FRET) states for both POSTF and PREFV (Figure 2D–G). Interestingly, we found that pseudoknots affected conformational dynamics of POSTF and PREFV to different extents (Figure 5 and Supplementary Table S4). For POSTF complex, pseudoknots decreased dwell times of the non-ratcheted state from 0.49 ± 0.04 s to 0.29 ± 0.01 s and increased dwell times of ratcheted state from 1.06 ± 0.14 s to 1.71 ± 0.01 s. Together, the chemical equilibrium was shifted towards the ratcheted POSTF state by ∼2.6 fold. On the other hand, for PREFV complex, pseudoknots decreased dwell times of non-ratcheted state from 0.53 ± 0.04 s to 0.28 ± 0.01 s and decreased dwell times of ratcheted state from 1.3 ± 0.1 s to 0.94 ± 0.02 s. Therefore, pseudoknots accelerated spontaneous transitions between the non-ratcheted and ratcheted PREFV states and moderately affected their equilibrium by ∼30%. Our observations suggest that there are relative movements between mRNA and 30S subunit during transitions between the non-ratcheted and ratcheted states, which are modulated by pseudoknots unwinding. It is plausible that the relative movements between mRNA and 30S subunit are caused by transitions of tRNAs between their canonical states and non-canonical chimeric states, which have been identified in several ratcheted ribosomal structures (47–49) and probed by single-molecule FRET assays (50–52). Furthermore, the abilities of pseudoknots to induce –1 PRF correlated with their abilities to affect conformational dynamics of POSTF and PREFV, two major ribosomal complexes in the first pseudoknot-unwinding cycle.
Figure 5.
Conformational dynamics of ribosomal complexes on non-slippery mRNAs with different pseudoknots in the first pseudoknot-unwinding cycle. (A) Dwell time distributions of the non-ratcheted and ratcheted POSTF states. (B) Dwell time distributions of the non-ratcheted and ratcheted PREFV states. (C–F) Plots of dwell times of non-ratcheted POSTF (C), ratcheted POSTF (D), non-ratcheted PREFV (E) and ratcheted PREFV (F) captured by smFRET under equilibrium conditions versus frameshifting efficiency. n is the number of events. Dwell times were extracted by single exponential fitting of dwell time distributions except the non-ratcheted POSTF, whose dwell times were extracted using double exponential decay. Error bars were standard errors.
We did not observe transitions between the non-ratcheted and ratcheted states during real-time ongoing elongation experiments, although dwell times of the non-ratcheted and ratcheted states of stalled POST and PRE complexes were shorter than dwell times of POST and PRE complexes during ongoing elongation. Our observations are consistent with previous reports that structural fluctuations observed on stalled ribosomes are suppressed during ongoing protein synthesis (52).
Elongation rates and conformational dynamics of ribosomes in the second pseudoknot-unwinding cycle
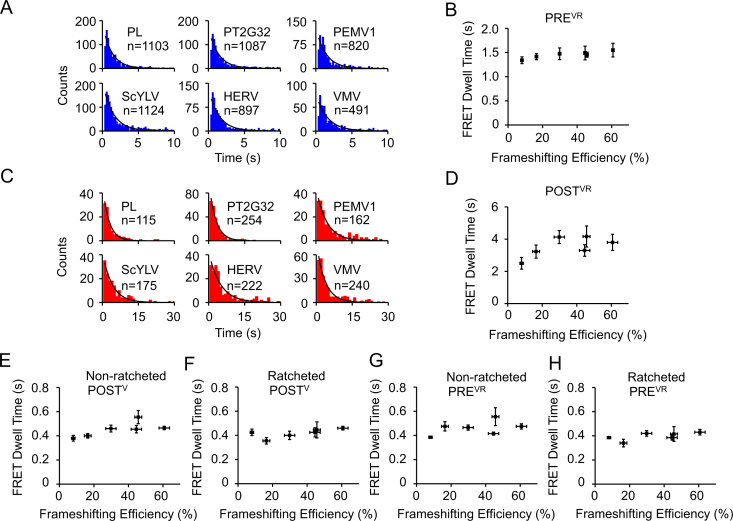
The second pseudoknot unwinding elongation cycle using mRNAs containing the common non-slippery sequence encoding fMYFVR results in conversion of POSTV to POSTR. We applied the same assays as mentioned above to extract elongation rates and conformational dynamics of ribosomes in this cycle. During ongoing elongation assays, dwell times of PREVR and POSTVR complexes, which were used to characterize elongation rates, were captured using Cy3-tRNAArg/Cy5-L11 (Supplementary Figure S6) and Cy3-tRNAVal/Cy5-L1 (Figure 2C) FRET pairs, respectively. Both the dwell times of PREVR and POSTVR complexes displayed no significant change among different pseudoknots (Figure 6A–D and Table 1), which was different from our findings in the early cycle. In addition, the Cy3-tRNAVal/Cy5-L1 FRET pair was used to capture spontaneous fluctuations between the non-ratcheted and ratcheted states for both stalled POSTV and PREVR complexes (Supplementary Figure S7 and Supplementary Table S4). Here dwell times displayed no significant change among different pseudoknots (Figure 6E–H); indeed, influence of pseudoknot on elongation rates and conformational dynamics of ribosomes in the second pseudoknot-unwinding cycle were mostly abolished.
Figure 6.
Conformational dynamics of ribosomes on non-slippery mRNAs with different pseudoknots in the second pseudoknot-unwinding cycle. (A) Dwell time distributions of PREVR complexes captured during ongoing elongation. (B) Plot of PREVR dwell time versus frameshifting efficiency. (C) Dwell time distributions of POSTVR complexes during ongoing elongation. (D) Plot of POSTVR dwell time versus frameshifting efficiency. (E–H) Plots of dwell times of non-ratcheted POSTV (E), ratcheted POSTV (F), non-ratcheted PREVR (G) and ratcheted PREVR (H) captured by smFRET versus frameshifting efficiency. n is the number of events. Dwell times were extracted by single exponential fitting of dwell time distributions. Error bars were standard errors.
DISCUSSION
Translocation impeded by pseudoknot
Translocation, defined as the concerted movements of tRNAs and mRNA on the ribosome, is a complex multi-step process (46,50,53–56). In previous work we used tRNA/L11 and tRNA/L1 FRET pairs to examine translocation in the presence of a downstream mRNA structure stem-loop or pseudoknot (6). Our results suggested that the dwell time of PRE complex captured by tRNA/L11 FRET mainly corresponds to tRNA movements in the large 50S ribosomal subunit, whereas the dwell time of POST complex captured by tRNA/L1 FRET should correspond to the movements of tRNAs and mRNA in the small 30S ribosomal subunit and subsequent dissociation of the E-site tRNA. Previous study also indicated that pseudoknot unwinding reduced rates of both early- and late-stage translocation. Based on previous established assays and discoveries, here we demonstrate that the extent to which pseudoknots impede translocation strongly correlates with their abilities to induce –1 PRF and their conformational plasticity. Our findings shed light on molecular mechanisms of translocation, mRNA structure unwinding, and –1 PRF, as discussed below.
Woodside and coworkers (23), using optical tweezers to examine the mechanical properties of various pseudoknots (including all pseudoknots listed in Table 1 and Figure 1), found that neither the unfolding energy barriers, nor the unfolding forces, nor the unfolding rates of pseudoknots could be correlated to –1 PRF efficiencies. Rather, –1 PRF efficiencies strongly correlated with pseudoknot conformational plasticity, i.e. with the propensity for pseudoknots to form alternative, incompletely folded structures. Here, using the same pseudoknots in combination with either a non-slippery sequence or a slippery sequence, we demonstrate that pseudoknot conformational plasticity also correlates with the ability to impede late-stage translocation in the first pseudoknot-unwinding cycle (up to three fold, Figures 3D and 4H). Our present results are consistent with previous proposals that motions between tRNAs and mRNA leading to frameshifting occur at the late stage of impeded translocation in the first structure-unwinding cycle (7,20,21,57). This empirically-driven conclusion raises two questions. Why is late-stage but not early-stage translocation sensitive to pseudoknot unwinding? Why is conformational plasticity a significant factor in pseudoknot impeded translocation?
Recent work by ourselves and others support a translocation model, in which movements of tRNAs and mRNA are initiated by a force-generating power-stroke step linked to EF-G dependent GTP hydrolysis followed by one or more thermal-ratcheting steps (55,58,59). There is also evidence that the ribosome unwinds mRNA structures using two distinct mechanisms, one converting free energy released from GTP hydrolysis (∼ –10 kcal/mol (60)) and/or peptide bond formation (∼ –8 kcal/mol (60)) to mechanically unwind mRNA structure, the other requiring no extra energy source and involving thermal ratcheting of the mRNA structure, which exists in rapid equilibrium between its closed and open forms (9). The free energy potentially available for mechanical unwinding during the early translocation stage force-generating power stroke greatly exceeds that needed to unwind three base pairs (2–6 kcal/mol (61)). In addition, depending on the length of the spacer between slippery sequence and pseudoknot, the ribosome may engage and start to unwind mRNA pseudoknot in the late translocation stage. Together, it is reasonable to expect that the rate of early-stage mechanical-unwinding step would be less sensitive to downstream structures than the rate of late-stage thermal-ratcheting step.
Translocation and -1 PRF modulated by conformational plasticity of pseudoknot
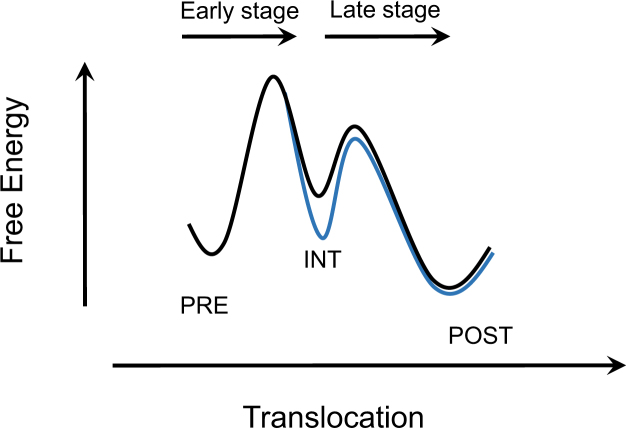
How does conformational plasticity of pseudoknots relate to their abilities to affect translocation? We proposed the following hypothesis to rationalize our findings. Optical tweezers measurements showed that, during unfolding of pseudoknots, the distance from the folded state to the transition state (the unfolding energy barrier) is only 1.6–2 nm, which corresponds to unwinding 1–2 base pairs (23). Therefore, in the first pseudoknot-unwinding cycle, the initial power-stroke step during translocation might lead the elongating ribosome to approach or to even pass the transition state of pseudoknot unfolding energy landscape (Figure 7, black curve). Transforming a partially unwound pseudoknot to an alternative more stable structure would reshape the unfolding energy landscape by stabling the intermediate (INT) state and introducing a higher unfolding energy barrier in the late stage (Figure 7, blue curve). Therefore, the late translocation stage after the initial power stroke could be further impeded by pseudoknots more prone to form stable alternative structures. In the presence of a slippery sequence, –1 PRF might provide an alternative and faster reaction pathway for the ribosome to overcome the energy barrier in the late translocation stage and to continue translation in the new -1 frame (20).
Figure 7.
Proposed energy landscape modulated by alternative folding of pseudoknot. The initial power-stroke step leads the elongating ribosome to approach or even pass the transition state of pseudoknot unfolding energy landscape (black curve). Rapidly transforming a partially unwound pseudoknot to an alternative more stable structure could stabilize the intermediate translocation state and introduce a higher energy barrier in the late stage (blue curve). INT denotes an intermediate state.
In summary, using several smFRET assays, we reveal that pseudoknots with different conformational plasticity inhibit the late-stage translocation in the first pseudoknot-unwinding cycle to different extents, which strongly correlate with their abilities to induce –1 PRF. We proposed a hypothesis to provide mechanistic insights on how pseudoknots with high conformational plasticity could reshape the energy landscape of translocation and selectively modulate the late-stage translocation by rapidly forming alternative incompletely folded structures during translocation. Furthermore, our findings also advanced current understanding of molecular mechanisms of translocation, –1 PRF and ribosome-induced mRNA structure unwinding.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31570754]; Tsinghua-Peking Joint Center for Life Sciences; Beijing Advanced Innovation Center for Structural Biology (to C.C.). National Institutes of Health [R35GM118139] (to Y.E.G.) and [GM080376] (to B.S.C.). Funding for open access charge: National Natural Science Foundation of China [31570754].
Conflict of interest statement. None declared.
REFERENCES
- 1. Schmeing T.M., Ramakrishnan V.. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009; 461:1234–1242. [DOI] [PubMed] [Google Scholar]
- 2. Agirrezabala X., Frank J.. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q. Rev. Biophys. 2009; 42:159–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moore P.B. How should we think about the ribosome. Annu. Rev. Biophys. 2012; 41:1–19. [DOI] [PubMed] [Google Scholar]
- 4. Voorhees R.M., Ramakrishnan V.. Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 2013; 82:203–236. [DOI] [PubMed] [Google Scholar]
- 5. Rodnina M.V. The ribosome in action: tuning of translational efficiency and protein folding. Protein Sci. 2016; 25:1390–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen C., Zhang H., Broitman S.L., Reiche M., Farrell I., Cooperman B.S., Goldman Y.E.. Dynamics of translation by single ribosomes through mRNA secondary structures. Nat. Struct. Mol. Biol. 2013; 20:582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim H.K., Liu F., Fei J., Bustamante C., Gonzalez R.L. Jr, Tinoco I. Jr. A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. PNAS. 2014; 111:5538–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Namy O., Moran S.J., Stuart D.I., Gilbert R.J., Brierley I.. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006; 441:244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Qu X., Wen J.D., Lancaster L., Noller H.F., Bustamante C., Tinoco I. Jr. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011; 475:118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Atkins J.F., Loughran G., Bhatt P.R., Firth A.E., Baranov P.V.. Ribosomal frameshifting and transcriptional slippage: from genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016; 44:7007–7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kurland C.G. Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 1992; 26:29–50. [DOI] [PubMed] [Google Scholar]
- 12. Parker J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989; 53:273–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Manickam N., Nag N., Abbasi A., Patel K., Farabaugh P.J.. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA. 2014; 20:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tinoco I. Jr, Kim H.K., Yan S.. Frameshifting dynamics. Biopolymers. 2013; 99:1147–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Advani V.M., Dinman J.D.. Reprogramming the genetic code: The emerging role of ribosomal frameshifting in regulating cellular gene expression. BioEssays. 2016; 38:21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farabaugh P.J. Programmed translational frameshifting. Microbiol. Rev. 1996; 60:103–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giedroc D.P., Cornish P.V.. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009; 139:193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brierley I., Gilbert R.J., Pennell S.. RNA pseudoknots and the regulation of protein synthesis. Biochem. Soc. Trans. 2008; 36:684–689. [DOI] [PubMed] [Google Scholar]
- 19. Caliskan N., Peske F., Rodnina M.V.. Changed in translation: mRNA recoding by -1 programmed ribosomal frameshifting. Trends Biochem. Sci. 2015; 40:265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Caliskan N., Katunin V.I., Belardinelli R., Peske F., Rodnina M.V.. Programmed -1 frameshifting by kinetic partitioning during impeded translocation. Cell. 2014; 157:1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caliskan N., Wohlgemuth I., Korniy N., Pearson M., Peske F., Rodnina M.V.. Conditional switch between frameshifting regimes upon translation of dnaX mRNA. Mol. Cell. 2017; 66:558–567. [DOI] [PubMed] [Google Scholar]
- 22. Chen J., Petrov A., Johansson M., Tsai A., O’Leary S.E., Puglisi J.D.. Dynamic pathways of -1 translational frameshifting. Nature. 2014; 512:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ritchie D.B., Foster D.A., Woodside M.T.. Programmed -1 frameshifting efficiency correlates with RNA pseudoknot conformational plasticity, not resistance to mechanical unfolding. PNAS. 2012; 109:16167–16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ritchie D.B., Soong J., Sikkema W.K., Woodside M.T.. Anti-frameshifting ligand reduces the conformational plasticity of the SARS virus pseudoknot. J. Am. Chem. Soc. 2014; 136:2196–2199. [DOI] [PubMed] [Google Scholar]
- 25. Fei J., Wang J., Sternberg S.H., MacDougall D.D., Elvekrog M.M., Pulukkunat D.K., Englander M.T., Gonzalez R.L. Jr. A highly purified, fluorescently labeled in vitro translation system for single-molecule studies of protein synthesis. Methods Enzymol. 2010; 472:221–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yokogawa T., Kitamura Y., Nakamura D., Ohno S., Nishikawa K.. Optimization of the hybridization-based method for purification of thermostable tRNAs in the presence of tetraalkylammonium salts. Nucleic Acids Res. 2010; 38:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peng S., Sun R., Wang W., Chen C.. Single-Molecule photoactivation FRET: A General and Easy-To-Implement approach to break the concentration barrier. Angew. Chem. 2017; 56:6882–6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pan D., Qin H., Cooperman B.S.. Synthesis and functional activity of tRNAs labeled with fluorescent hydrazides in the D-loop. RNA. 2009; 15:346–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen C.L., Stevens B., Kaur J., Cabral D., Liu H.Q., Wang Y.H., Zhang H.B., Rosenblum G., Smilansky Z., Goldman Y.E. et al. . Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol. Cell. 2011; 42:367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walker S.E., Fredrick K.. Preparation and evaluation of acylated tRNAs. Methods. 2008; 44:81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodnina M.V., Wintermeyer W.. GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. PNAS. 1995; 92:1945–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stevens B., Chen C., Farrell I., Zhang H., Kaur J., Broitman S.L., Smilansky Z., Cooperman B.S., Goldman Y.E.. FRET-based identification of mRNAs undergoing translation. PLoS One. 2012; 7:e38344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Subramanian A.R., Dabbs E.R.. Functional studies on ribosomes lacking protein L1 from mutant Escherichia coli. Eur. J. Biochem. 1980; 112:425–430. [DOI] [PubMed] [Google Scholar]
- 34. Roy R., Hohng S., Ha T.. A practical guide to single-molecule FRET. Nat. Methods. 2008; 5:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Illingworth J., Kittler J.. A survey of the hough transform. Comput. Vis. Graph. Image Process. 1988; 44:87–116. [Google Scholar]
- 36. Holland J.A., Hansen M.R., Du Z., Hoffman D.W.. An examination of coaxial stacking of helical stems in a pseudoknot motif: the gene 32 messenger RNA pseudoknot of bacteriophage T2. RNA. 1999; 5:257–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nixon P.L., Rangan A., Kim Y.G., Rich A., Hoffman D.W., Hennig M., Giedroc D.P.. Solution structure of a luteoviral P1-P2 frameshifting mRNA pseudoknot. J. Mol. Biol. 2002; 322:621–633. [DOI] [PubMed] [Google Scholar]
- 38. Cornish P.V., Hennig M., Giedroc D.P.. A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated -1 ribosomal frameshifting. PNAS. 2005; 102:12694–12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y., Wills N.M., Du Z., Rangan A., Atkins J.F., Gesteland R.F., Hoffman D.W.. Comparative studies of frameshifting and nonframeshifting RNA pseudoknots: a mutational and NMR investigation of pseudoknots derived from the bacteriophage T2 gene 32 mRNA and the retroviral gag-pro frameshift site. RNA. 2002; 8:981–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pennell S., Manktelow E., Flatt A., Kelly G., Smerdon S.J., Brierley I.. The stimulatory RNA of the Visna-Maedi retrovirus ribosomal frameshifting signal is an unusual pseudoknot with an interstem element. RNA. 2008; 14:1366–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fei J., Kosuri P., MacDougall D.D., Gonzalez R.L. Jr. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell. 2008; 30:348–359. [DOI] [PubMed] [Google Scholar]
- 42. Meydan S., Klepacki D., Karthikeyan S., Margus T., Thomas P., Jones J.E., Khan Y., Briggs J., Dinman J.D., Vazquez-Laslop N. et al. . Programmed ribosomal frameshifting generates a copper transporter and a copper chaperone from the same gene. Mol. Cell. 2017; 65:207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Korostelev A., Ermolenko D.N., Noller H.F.. Structural dynamics of the ribosome. Curr. Opin. Chem. Biol. 2008; 12:674–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Munro J.B., Sanbonmatsu K.Y., Spahn C.M.T., Blanchard S.C.. Navigating the ribosome's metastable energy landscape. Trends Biochem. Sci. 2009; 34:390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Frank J., Gonzalez R.L.. Structure and dynamics of a processive brownian motor: The translating ribosome. Annu. Rev. Biochem. 2010; 79:381–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fischer N., Konevega A.L., Wintermeyer W., Rodnina M.V., Stark H.. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010; 466:329–333. [DOI] [PubMed] [Google Scholar]
- 47. Zhou J., Lancaster L., Donohue J.P., Noller H.F.. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science. 2014; 345:1188–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramrath D.J., Lancaster L., Sprink T., Mielke T., Loerke J., Noller H.F., Spahn C.M.. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:20964–20969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brilot A.F., Korostelev A.A., Ermolenko D.N., Grigorieff N.. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:20994–20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wasserman M.R., Alejo J.L., Altman R.B., Blanchard S.C.. Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat. Struct. Mol. Biol. 2016; 23:333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Adio S., Senyushkina T., Peske F., Fischer N., Wintermeyer W., Rodnina M.V.. Fluctuations between multiple EF-G-induced chimeric tRNA states during translocation on the ribosome. Nat. Commun. 2015; 6:7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jamiolkowski R.M., Chen C., Cooperman B.S., Goldman Y.E.. tRNA fluctuations observed on stalled ribosomes are suppressed during ongoing protein synthesis. Biophys. J. 2017; 113:2326–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ermolenko D.N., Noller H.F.. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat. Struct. Mol. Biol. 2011; 18:457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pan D.L., Kirillov S.V., Cooperman B.S.. Kinetically competent intermediates in the translocation step of protein synthesis. Mol. Cell. 2007; 25:519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Holtkamp W., Cunha C.E., Peske F., Konevega A.L., Wintermeyer W., Rodnina M.V.. GTP hydrolysis by EF-G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J. 2014; 33:1073–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Belardinelli R., Sharma H., Peske F., Wintermeyer W., Rodnina M.V.. Translocation as continuous movement through the ribosome. RNA Biol. 2016; 13:1197–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yan S., Wen J.D., Bustamante C., Tinoco I. Jr. Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell. 2015; 160:870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu T., Kaplan A., Alexander L., Yan S., Wen J.D., Lancaster L., Wickersham C.E., Fredrick K., Noller H., Tinoco I. et al. . Direct measurement of the mechanical work during translocation by the ribosome. eLife. 2014; 3:e03406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen C., Cui X., Beausang J.F., Zhang H., Farrell I., Cooperman B.S., Goldman Y.E.. Elongation factor G initiates translocation through a power stroke. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:7515–7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Petrov A., Chen J., O’Leary S., Tsai A., Puglisi J.D.. Single-Molecule analysis of translational dynamics. Csh Perspect. Biol. 2012; 4:a011551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Freier S.M., Kierzek R., Jaeger J.A., Sugimoto N., Caruthers M.H., Neilson T., Turner D.H.. Improved free-energy parameters for predictions of rna duplex stability. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.