Abstract
Tunable induction of gene expression is an essential tool in biology and biotechnology. In spite of that, current induction systems often exhibit unpredictable behavior and performance shortcomings, including high sensitivity to transactivator dosage and plasmid take-up variation, and excessive consumption of cellular resources. To mitigate these limitations, we introduce here a novel family of gene expression control systems of varying complexity with significantly enhanced performance. These include: (i) an incoherent feedforward circuit that exhibits output tunability and robustness to plasmid take-up variation; (ii) a negative feedback circuit that reduces burden and provides robustness to transactivator dosage variability; and (iii) a new hybrid circuit integrating negative feedback and incoherent feedforward that combines the benefits of both. As with endogenous circuits, the complexity of our genetic controllers is not gratuitous, but is the necessary outcome of more stringent performance requirements. We demonstrate the benefits of these controllers in two applications. In a culture of CHO cells for protein manufacturing, the circuits result in up to a 2.6-fold yield improvement over a standard system. In human-induced pluripotent stem cells they enable precisely regulated expression of an otherwise poorly tolerated gene of interest, resulting in a significant increase in the viability of the transfected cells.
INTRODUCTION
Conditional systems for controlled modulation of gene expression are indispensable research tools in the life sciences (1,2). A typical configuration consists of a ligand-responsive transactivator or repressor that acts on a downstream gene of interest (3,4). Classically, the system’s performance is evaluated based on its sensitivity to the ligand concentration, its leakiness and its dynamic range (5). Other equally important performance considerations, such as the burden that the transactivator or repressor may impose on the host organism, unpredictable circuit response, and robustness features, have not been adequately addressed.
Previous work has highlighted how advanced functions and improved performance in endogenous circuits often emerge as properties of particular network topologies. For instance, robustness of biological function can often be attributed to the prevalence of feedback and feedforward loops (6–8). In a feedback loop, the gene product regulates the gene itself, either positively or negatively. Feedforward loops, on the other hand, are characterized by a master regulator that acts on a downstream gene both directly and indirectly through an auxiliary regulator (8). While some studies have investigated the realization of these regulatory strategies in naturally occurring motifs (9,10), others have focused on synthetically built regulatory modules, which exhibit features such as pulse generation (11), reduction of gene expression noise (12) and adaptation (13–16). Special attention has been devoted to RNA interference (RNAi) motifs (17), which have been involved in the regulation of differentiation, inflammation and many other processes (18,19). Such post-transcriptional repression can be selectively directed to a gene of interest by including suitable target sequences in its 3′ untranslated region (3′-UTR) (14,20,21).
Recently, cybergenetics has emerged as an active area of research where concepts from the field of control theory and cybernetics (22) are applied to the rational design of synthetic gene circuits that function to dynamically regulate cellular processes at the molecular scale (23–26). These cybergenetic circuits act as dynamic genetic control systems that typically employ feedback loops to achieve any one of several performance objectives such as robustness, disturbance rejection, tracking or adaptation. Recent examples of such genetic control systems in E. coli include (Gabriele, L., Aoki, S., Schweingruber, D. and Khammash, M. (2017) A synthetic integral feedback controller for robust tunable regulation in bacteria. bioRxiv, 170951; Kelly, L.C., Harris, A., Steel, H. et al. (2018) Synthetic negative feedback circuits using engineered small RNAs. bioRxiv, 184473; Huang, H.-H., Qian, Y. and Del Vecchio, D. (2018) A quasi-integral controller for adaptation of genetic modules to variable ribosome demand. bioRxiv, 336271). Here, we report on novel, rationally designed, cybergenetic gene expression controllers incorporating post-transcriptional feedback and feedforward regulation in mammalian cells. Guided by theoretical considerations, we reasoned that such regulation could improve the system's robustness to plasmid take-up and transactivator variability, and optimize the amount of cellular resources required to realize their function.
MATERIALS AND METHODS
We present here a brief summary of the computational and experimental methods. A more complete description is available in the Supplementary Materials and Methods.
Computational modeling
The genetic controllers were analyzed using dynamical models based on ordinary differential equations (ODE). The analytical steady-state solutions of the models were obtained using Mathematica version 8 (Wolfram Research). The numerical simulations were performed with MATLAB release 2014a (MathWorks). The models are discussed in detail in the Supplementary Text S1.
Plasmid construction
All recombinant DNA constructs were created using restriction-enzyme cloning, Gibson assembly and other standard molecular biology techniques. All plasmids were verified by sequencing. Cloning details are presented in the Supplementary Text S7.1.
Cell cultures and transient transfections
HEK293T, HeLa, CHO-K1 and human induced pluripotent stem cells (hIPSCs) were used in this study. Full details on specific culture procedures for each line are provided in the Supplementary Text S7.2. All cell lines were periodically screened for mycoplasma contamination using the Biotool Mycoplasma Detection Kit (cat. no. B39032) per manufacturer’s instructions. All DNA plasmids used in the transfections were purified with the ZymoPURE Plasmid Midiprep Kit (Zymo Research, cat. no. D4200), and further processed with the Norgen Biotek Endotoxin Removal Midi Kit (cat. no. 52200). The locked nucleic acid (LNA) inhibitor LNA-FF4 (miRCURY LNA Power Inhibitor) was purchased from Exiqon. Doxycycline (Dox) was purchased from Sigma-Aldrich (cat. no. D9891).
hIPSC viability assay
Calcein acetoxymethyl ester (calcein AM) was purchased from Sigma-Aldrich (cat. no. 56496-20X50UG). The supplied powder was resuspended in DMSO to obtain a 5 mM stock solution. This was then diluted in cell culture medium to 1 μM final concentration. Immediately before collection, the cells were incubated with the medium containing calcein AM for 15 min at 37°C, 100% humidity and 5% CO2. The medium was then removed and the cells detached with Accutase (Innovative Cell Technologies, Inc., cat. no. AT104). The resulting cell suspension was mixed with Spherotech AccuCount beads (cat. no. ACBP-100-10), which allowed for cell counting as described in the manufacturer’s protocol.
Flow cytometry
Approximately 48 h after transfection, the cells were collected and prepared as detailed in the Supplementary Text S7.2. The cell suspensions were analyzed on a LSRII Fortessa flow cytometer (BD Biosciences) equipped with the FACSDiva software program. mCitrine was measured with a 488 nm laser and 542/27 and 505 LP emission filters; DsRed with a 561 nm laser and 586/15 and 570 LP emission filters; Cerulean with a 445 nm laser and a 437/10 emission filter; iRPF670 with a 633 nm laser and a 670/14 emission filter. Data analysis procedures are detailed in the Supplementary Text S7.3.
RESULTS AND DISCUSSION
Study design
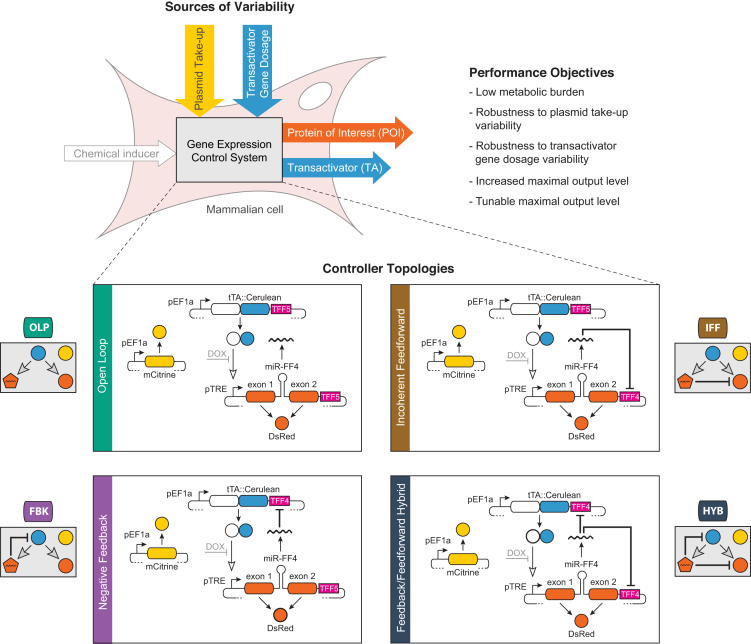
All of our controllers share the same genes, and they only differ in how the genes interact (Figure 1). A constitutively expressed mCitrine fluorescent protein serves as a reporter, generating a signal that is approximately proportional to the amount of plasmid taken up by the individual cells (13). The synthetic tetracycline-controller transactivator tTA-Advanced (referred to here as TA), fused to Cerulean, induces the expression of both a protein of interest (here the DsRed fluorescent protein, which we shall also refer to as POI) and an intronically encoded auxiliary repressor, consisting of a synthetic microRNA. The repressor can target the DsRed protein and the tTA::Cerulean simultaneously, separately, or neither, thus generating, respectively, the following regulation patterns (Figure 1): hybrid incoherent feedforward and negative feedback (HYB), incoherent feedforward only (IFF), negative feedback only (FBK), or no repression (open loop configuration, OLP). The latter recapitulates a standard inducible gene expression system. In all configurations, the POI level can be adjusted with Dox, which inhibits tTA::Cerulean activity.
Figure 1.
Study design. We introduce four cybergenetic circuits for controlling inducible gene expression in mammalian cells: an open loop circuit (OLP), a negative feedback controller circuit (FBK), an IFF controller circuit and a negative feedback/IFF hybrid controller circuit (HYB). We analyze their performance with respect to transactivator gene dosage and plasmid take-up variability (see Figure 2). Rounded rectangles denote coding sequences. Colored circles indicate fluorescent proteins of the corresponding color. The pentagon represents a post-transcriptional repressor. Solid pointed arrows denote gene expression, hollow pointed arrows show transcriptional activation, while blunt arrows denote post-transcriptional repression. Dashed lines represent variable circuit elements. Purple rectangles are micro-RNA targets.
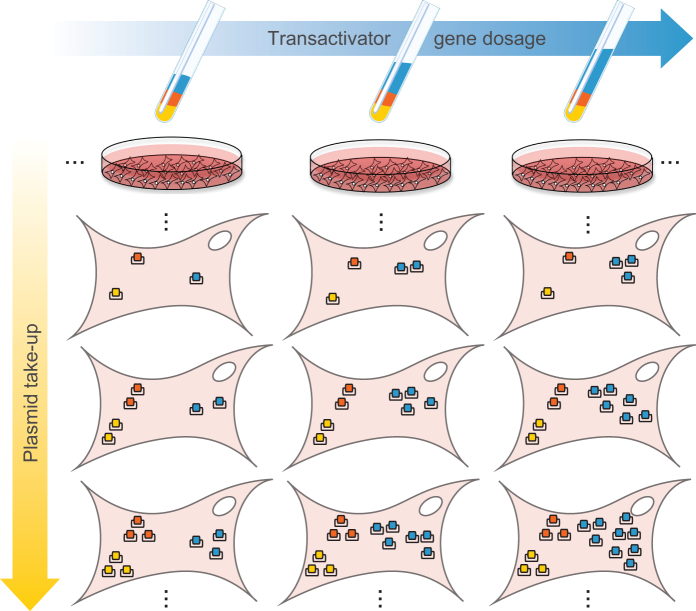
In inducible systems, the fully induced expression level of the regulated gene is of particular importance, since excess amounts of inducer are typically used in applications. We will refer to this quantity as maximal output level, or MOL, with the output in reference being the POI. Its major determinant is the amount of DNA delivered to the cell, usually in the form of plasmids or viral vectors. Other factors, such as the integration loci, the number of integrated copies and the cell’s metabolic state, also play a role. The large variability of MOL due to these factors leads to lengthy and labor-intensive screening of cell clones and embryos. In our controllers, the plasmid take-up and the other factors that generate cell-to-cell variation in the overall level of gene expression are captured by the constitutive mCitrine level. To study how differences in plasmid take-up affect MOL, we exploit transient transfection of DNA plasmids into mammalian cells, a process that typically results in large variability in the number of the plasmids internalized in the nuclei of individual cells (Figure 2). By observing the steady-state MOL of DsRed (which can be replaced by any gene of interest) as a function of mCitrine expression, the relationship between plasmid take-up and MOL can be captured. We refer to this curve as the plasmid-takeup/MOL function of a controller. Its significance lies in its ability to show how robust a circuit is. Robustness to plasmid take-up variation is indicated by the flatness of the curve, suggesting that MOL does not depend on plasmid take-up. Robustness to transactivator variability is reflected by the extent to which the curve is independendent of transactivator dosage. To assess this, we generate transactivator gene dosage variability by changing the relative amount of transactivator plasmid in the transfection samples (Figure 2). While we seek to implement controllers with such desirable input-output functions, we also aim to keep tTA levels as low as possible to minimize the consumption of cellular resources. As the tTA transactivator is a resource to be used sparingly, we consider the plasmid-takeup/TA relationship, which is characterized by the steady-state level of tTA::Cerulean as a function of mCitrine level. The slope of this curve indicates how much tTA::Cerulean is expressed per plasmid copy, and can be taken as a measure of resource utilization.
Figure 2.
Experimental setup. Mammalian transient transfection results in a large variability in plasmid take-up. Additionally, we generate transactivator gene dosage variability by changing the relative amount of transactivator plasmid in the transfection samples. Individual cells will on average take up plasmids in the same relative amount as those in the transfection samples.
Assessing the performance of a standard gene expression system
To quantitatively measure the performance of a standard gene expression system with the respect to the criteria outlined in Figure 1, we used a previously described toolbox of synthetic microRNAs (27) to construct three synthetic genes, two of which come in two different variants each. mCitrine driven by the constitutive human elongation factor 1 alpha promoter (PEF1α) serves as plasmid take-up reporter, while the tTA::Cerulean fusion (also driven by PEF1α) acts as transactivator. We further created a construct in which an intron containing the synthetic microRNA FF4 (miR-FF4) is embedded into the coding sequence of DsRed. We refer to this cassette as DsRed(FF4). Following splicing and RNA processing, the mature miR-FF4 becomes a repressor, and after translation, DsRed is the output gene of interest. DsRed(FF4) is driven by the synthetic promoter PTRE, consisting of seven tTA binding sites followed by a minimal cytomegalovirus promoter. Thus, tTA::Cerulean can simultaneously induce miR-FF4 and DsRed. Both the tTA::Cerulean and the DsRed(FF4) genes have been implemented with two different 3′-UTRs, containing three repeats of either the miR-FF4 target sequence or the miR-FF5 target sequence (also part of the toolbox mentioned above). The miR-FF5 targets are not recognized by miR-FF4, hence they provide a way of switching off the negative regulation. We refer to the targets as TFF4 and TFF5, respectively. We confirmed that miR-FF4 is functional in knocking down tTA::Cerulean as well as its own transcript (Supplementary Text S4 and Figures S3 and 4).
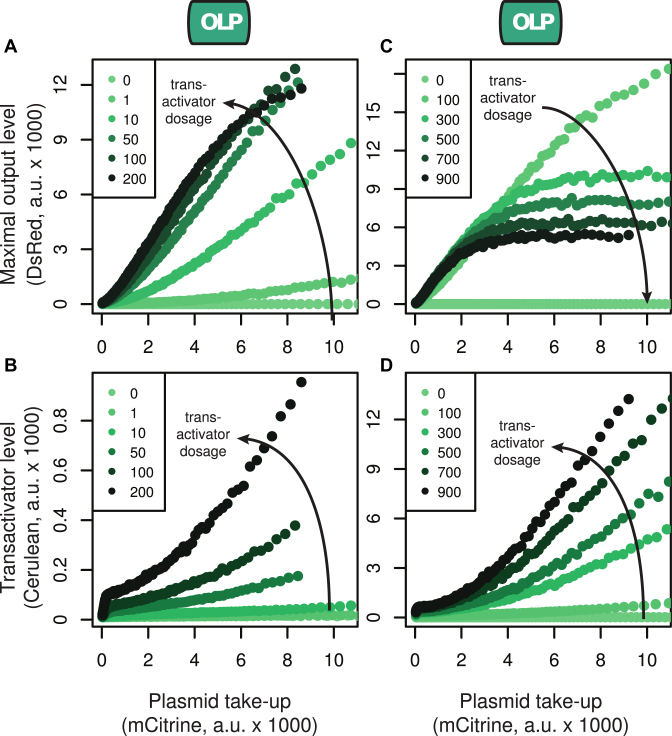
With the right fluorescent reporters in place, we could then move on to obtain the plasmid-takeup/MOL and plasmid-takeup/TA functions for the OLP controller. We co-transfected the reporter PEF1α-mCitrine with PEF1α-tTA::Cerulean-TFF5 and PTRE-DsRed(FF4)-TFF5. We performed a titration by keeping the amounts of mCitrine and DsRed plasmids fixed at 700 and 900 ng, respectively, while varying the amount of tTA::Cerulean plasmid. This procedure generates variability in both plasmid take-up and transactivator dosage (Figure 2). For dosages of PEF1α-tTA::Cerulean ranging between 0 and 200 ng, the levels of both Cerulean and DsRed increase with plasmid dosage for a given copy number (Figure 3A and B). Strikingly, however, if the tTA::Cerulean dosage is further increased, the DsRed level actually decreases as a function of tTA::Cerulean dosage for high copy numbers (Figure 3C and D). Our results suggest that in the OLP topology MOL is strongly affected by both plasmid take-up and transactivator dosage, and that tTA::Cerulean can reduce MOL when highly expressed. This effect is also displayed by the commercial tTA-Advanced (Supplementary Figure S5). We speculate that it is due to unnecessarily high levels of tTA expression causing burden on the host cells.
Figure 3.
Maximal output level (MOL) in the open loop controller as a function of plasmid take-up and transactivator dosage. The plasmids comprising the open loop controller were co-transfected using variable amounts of the tTA::Cerulean plasmid, thus varying the relative amount of tTA::Cerulean taken up by the individual cells. Plasmid-takup/MOL (top) and plasmid-takeup/TA (bottom) functions were obtained for low tTA::Cerulean dosages (A and B) and high tTA::Cerulean dosages (C and D). The numbers in the legends indicate the amounts (in nanograms) of tTA::Cerulean plasmid used in the corresponding transfection. All curves were obtained by pooling data from N = 3 replicates per condition, each of which had an average of ∼240 000 positively transfected cells (with above-background mCitrine expression).
Computational analysis of gene expression controllers
To gain a deeper understanding of how to build controllers with superior performance qualities, we created ODE models to calculate the plasmid-takeup/MOL and plasmid-takeup/TA functions (Supplementary Text S1). Our analysis shows that the plasmid-takeup/MOL function of the IFF and HYB controllers (Supplementary Equations S11 and 24) are of the following form:
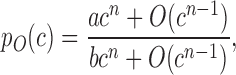 |
(1) |
where c is the plasmid copy number, pO is the steady-state MOL, a and b are constant parameters, n is in the range 1–3 and O(cn − 1) denotes terms containing powers of c of order n − 1 and lower. As the copy number increases, pO(c) approaches the constant value a/b. This indicates that our hybrid and IFF controllers are robust to plasmid take-up variation by adapting to the amount of their genetic template, similar to a previously reported IFF loop that used constitutively expressed output and negative regulator (13). In contrast, for the OLP and FBK systems (Supplementary Equations S4 and 17) we have:
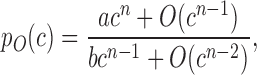 |
(2) |
indicating that MOL increases approximately linearly with the plasmid copy number. This finding also suggests that post-transcriptional feedback differs from transcriptional feedback, in which the output expression grows with the square root of the copy number (Supplementary Text S2). Analytical expressions of the plasmid-takeup/TA functions indicate that in the OLP and IFF circuits the transactivator level grows linearly with the copy number, while in the FBK and HYB controllers it remains asymptotically bounded (Supplementary Equations S6, 13, 19 and 26). This demonstrates that the FBK and HYB controllers are characterized by a lower resource utilization than the IFF and OLP systems. To better visualize the relevant curves, we solved our models numerically (Supplementary Text S3.1 and Table S1), and plotted MOL as a function of reporter protein (Supplementary Figure S1B). The simulations showed that MOL could be tuned by adjusting the binding affinity of the repressor for its targets. We also compared the plasmid-takeup/MOL (Supplementary Figure S1B) and plasmid-takeup/TA functions of the four circuits (Supplementary Figure S1C), and observed that the hybrid controller had the same plasmid-takeup/MOL function as the IFF topology and the same plasmid-takeup/TA function as the FBK topology. Hence, it can implement plasmid take-up adaptation while greatly reducing the transactivator level. Depending on the transactivator dose-response, this may also translate into an additional reduction in burden through a reduction in repressor levels (Supplementary Text S3.3 and Figure S2).
Implementation and characterization of synthetic gene expression controllers
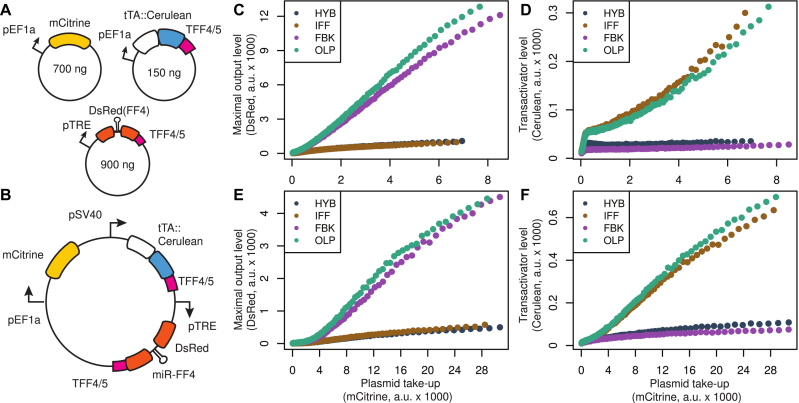
Next, we implemented the HYB, IFF and FBK genetic controllers, both as three separate plasmids (Figure 4A) and as single plasmids (Figure 4B), in order to experimentally characterize their plasmid-takeup/MOL and plasmid-takeup/TA functions. First, we co-transfected separate plasmids in the combinations required to generate the four controller topologies (Figure 4C and D). We also performed curve fitting and confirmed that the plasmid-takeup/MOL functions were consistent with the computational analysis (Supplementary Text S5 and Figure S6). The results indicate that: (i) the IFF motif displays genetic template adaptation, however its transactivator level grows quickly with plasmid copy number; (ii) the FBK motif can significantly reduce the transactivator level, but shows an approximately linear output expression without adaptation; (iii) the HYB motif combines the features of both mechanisms and shows plasmid copy number adaptation with greatly reduced transactivator level.
Figure 4.
Characterization of the cybergenetic circuits. (A) Implementation of the four controller topologies using separate plasmids. To avoid resource burden induced by overexpression of tTA::Cerulean, we used 150 ng of tTA::Cerulean plasmids in the transfection mixes, while the amounts of mCitrine and DsRed plasmids were 700 and 900 ng, respectively. (B) Implementation of the four controllers on a single plasmid each. (C–F) Experimental plasmid-takeup/MOL (left) and plasmid-takeup/TA (right) functions of the controllers on separate plasmids (top) or single plasmids (bottom). The colors indicate different topologies: hybrid feedback-feedforward (steel), feedback-only (lilac), feedforward-only (dark gold) and open loop (aqua). The curves were obtained by pooling data from N = 3 replicates per condition, each of which had an average of ∼460 000 positively transfected cells (with above-background mCitrine expression).
We then constructed single plasmids motivated by considerations of convenience and applicability. To achieve this, we needed to reduce the expression of tTA::Cerulean while keeping the dosage of the three parts identical. Hence, we replaced the promoter driving tTA::Cerulean with the Simian virus 40 promoter (PSV40), which was found to be ∼10-fold weaker than PEF1α in HEK cells (28). We assembled four plasmids, each containing one full synthetic controller. We transfected the single plasmids and repeated the characterization of the plasmid-takeup/MOL and plasmid-takeup/TA functions (Figure 4E and F). We found the same properties that were observed for the circuits on separate plasmids.
We verified the extent of gene expression variability induced by the transient transfection process by plotting the distributions of mCitrine expression by replicate for the four controller topologies (Supplementary Figure S9). These span 5 orders of magnitude. Given that mCitrine is a constitutively expressed reporter, this finding confirms that plasmid take-up variation is a major source of cell-to-cell variability in transient transfection experiments.
We further asked whether the four controllers differ in terms of cell-to-cell variability in the expression of the output and/or of the transactivator. To address this question, we calculated the Fano factor (or dispersion index) of the output and of the transactivator as a function of plasmid take-up (Supplementary Figure S10). We found that incoherent feedforward regulation results in a noticeable decrease in the Fano factor of the output, while negative feedback regulation significantly reduces the Fano factor of the transactivator. This is consistent with both intuition and previous reports that micro-RNA regulation can suppress gene expression noise (17).
Negative feedback regulation confers robustness to transactivator variability
Our FBK controller can achieve a similar MOL as the OLP system while using much less transactivator (Figure 4C–F). The computational analysis showed that this property requires a transactivator with a sharp dose-response curve, a frequently encountered feature (29) (Supplementary Text S3.4 and Figure S7). It also suggested that in the FBK controller the transactivator level remains bounded for large copy numbers. Therefore, we speculated that this topology could be used to stabilize MOL against fluctuations in the transactivator dosage. To verify that, we co-transfected the three plasmids comprising the FBK topology using variable amounts of the tTA::Cerulean plasmid. We found that MOL is essentially the same for all dosages, except for those with 0 and 100 ng tTA::Cerulean (Figure 5A and B). The first contained no transactivator and, as expected, expressed no Ds-Red output. The second expressed a lower level of Ds-Red output presumably because the dosage of tTA::Cerulean is such that PTRE is not operating at saturation.
Figure 5.
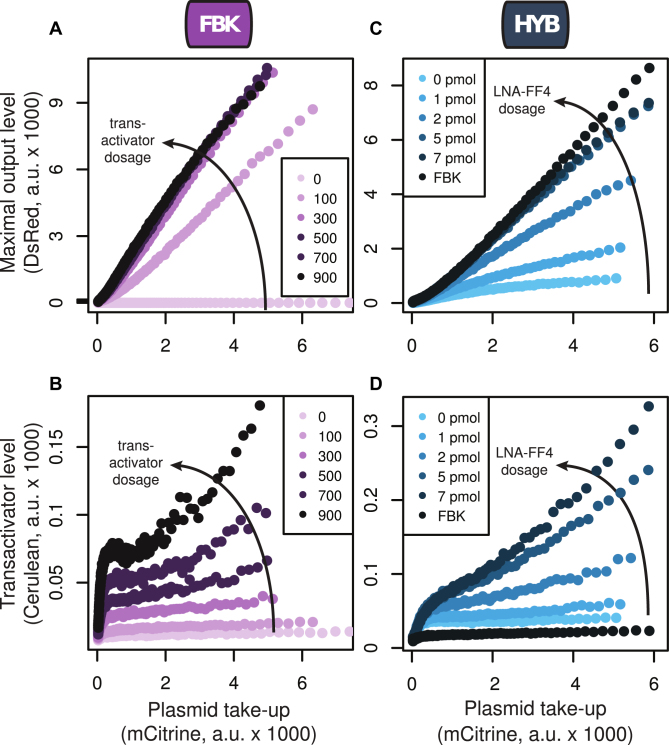
Robustness and tunability of MOL. (A and B) Plasmid-takeup/MOL (A) and plasmid-takeup/TA (B) functions for the transfections of the feedback controller with variable dosage of tTA::Cerulean. The numbers in the legends indicate the nanograms of tTA::Cerulean plasmid used in each transfection. (C and D) Plasmid-takeup/MOL (C) and plasmid-takeup/TA (D) functions for the transfections of the hybrid controller together with varying amounts of a miR-FF4 inhibitor. The numbers in the legend indicate the amount of inhibitor used in each transfection well. The black curve, denoted FBK, shows the results of a sample in which DsRed had FF5 targets instead of FF4 targets (thereby resulting in the feedback circuit). All curves were obtained by pooling data from N = 3 replicates per condition, each of which had an average of ∼600 000 positively transfected cells (with above-background mCitrine expression).
Output tuning
Our controllers contain tTA::Cerulean as a transactivator, therefore Dox can be used to adjust the Ds-Red output expression between 0 and the MOL of each topology. We performed a titration experiment and found that the FBK circuit displayed a 3.2-fold increase in Dox sensitivity, while in the IFF topology the sensitivity was decreased by 1.8-fold (Supplementary Text S6 and Figure S8). The response of the FBK circuit to Dox also showed a significantly higher Hill coefficient compared to the other topologies (Supplementary Table S2).
The computational analysis also uncovered a way to tune MOL by changing the miR-FF4 binding affinity for its targets (Supplementary Figure S1A). To test this, we co-transfected the three plasmids that comprise the hybrid topology together with varying amounts of an LNA antisense inhibitor against miR-FF4 (Figure 5C and D). We also included a control in which PTRE-DsRed(FF4)-TFF4 plasmid was replaced with PTRE-DsRed(FF4)-TFF5, thereby resulting in the FBK topology. We found that MOL increased with the dosage of the inhibitor, with 7 pmol resulting in essentially the same MOL as the FBK circuit (Figure 5C). The latter has a tTA::Cerulean level comparable to the 0 pmol dosage, which is expected since in both cases tTA::Cerulean has TFF4 targets.
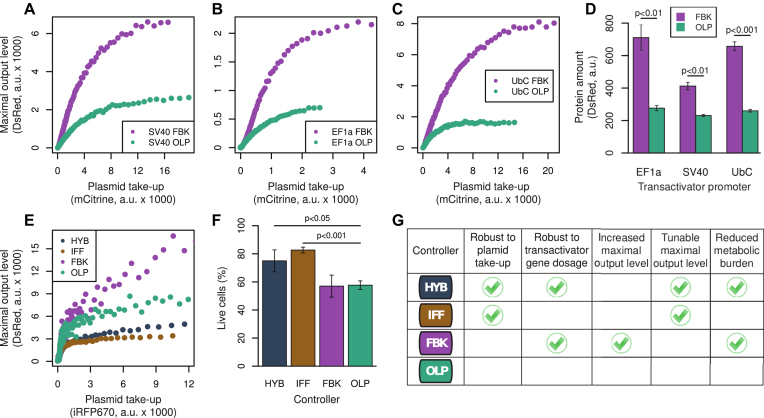
Negative feedback control improves output protein yield
An important consequence of the burden induced by tTA is that the FBK controller can achieve a higher MOL than the OLP system when the latter operates in a high tTA regime. To further investigate this aspect, we created additional versions of the FBK and OLP circuits, with different promoters driving tTA:Cerulean. In addition to PSV40, we used PEF1α and the human ubiquitin C promoter (PUbC). We then transfected these variants into suspension CHO-K1 cells, under conditions that are relevant for protein manufacturing. Specifically, we used polyethylenimine (PEI), an inexpensive reagent that can be also used for large-scale transfections. In all the tested cases the FBK controller resulted in a higher protein yield than the OLP system (Figure 6A–D). The effect was more pronounced in high-expressing cells, which could produce up to 4.6-fold more DsRed with the FBK circuit (Figure 6C). In the bulk populations of transfected cells, the DsRed amount obtained with the FBK topology was up to 2.6-fold that of the OLP topology (Figure 6D).
Figure 6.
The cybergenetic circuits as a platform for transgene expression. (A–C) Plasmid-takeup/MOL functions for the transfections in CHO-K1 cells with PEI. (D) Average DsRed levels in the populations of transfected CHO-K1. (E) Plasmid-takeup/MOL functions for the transfection in hIPSCs. (F) Percentage of transfected live hIPSCs as quantified by the calcein AM assay. In (A–C) and (E), the curves were obtained by pooling data from N = 3 replicates per condition, each of which had an average of ∼200 000 (A–C) or 28 000 (E) positively transfected cells (with above-background mCitrine or iRFP670 expression). In (D) and (F), the reported values are the means of N = 3 replicates, and the error bars denote standard deviation. The P-values were obtained using t-tests for the comparison of the means. (G) Summary of controller features.
In principle the same results could be obtained by carefully tuning tTA::Cerulean expression using genetic manipulation to find a suitable constitutive promoter. However, this approach would not only be much more complex and time-consuming, but also only valid for a specific cell line. Our data indicate that mammalian constitutive promoters do not have the same activity in all cell lines. In HEK293T cells, PSV40 ensures that the controllers operate in a low tTA regime, with the OLP and FBK circuits reaching similar MOL (Figure 4E). In contrast, in CHO-K1 cells PSV40 has a higher activity. As a result, the controllers operate in a high tTA regime, in which the burden effect becomes significant and the FBK controller achieves a higher MOL than the OLP system (Figure 6A).
Incoherent feedforward control enables defined transgene expression in sensitive cell lines
Our hybrid and IFF topologies display adaptation to gene dosage variability. Therefore, we reasoned that they could be useful in applications in which a protein of interest must be expressed at a well-defined level, above which toxicity effects to the host cells might become a concern. To investigate this, we tested our controllers in hIPSCs and measured the viability of the transfected cells with calcein acetoxymethyl ester (calcein AM), a green fluorescent dye that selectively stains live cells. To avoid spectral overlap with mCitrine, we used iRPF670 as plasmid take-up reporter. The circuits displayed the same behavior that was observed in other cell lines, including the burden effect of tTA (Figure 6E).
The results of calcein AM assay, however, suggest that hIPSCs do not tolerate high DsRed expression well (Figure 6F). This is consistent with previous studies such as (30), in which the authors reported lack of success in generating a stable ES cell line expressing DsRed. It has also been suggested that the toxicity is due to the poor solubility of DsRed, which is a tetramer in vivo (31). Our data show that controlling DsRed expression by means of incoherent feedforward regulation can help mitigate such toxic effects: cells transfected with the HYB or the IFF controllers were significantly more viable than those transfected with the FBK or the OLP controllers (Figure 6F).
CONCLUSION
Synthetic biology’s goal to program new functions into cells for useful applications is faced with numerous challenges, among which the often unpredictable behavior of biological parts and the erratic response of the cells when they are forced to produce exogenous material at high levels. By fluorescently tracking the individual parts of a standard inducible expression system in a novel way, we uncovered a number of previously unreported issues with this fundamental tool. We found that the standard expression system is not robust to transactivator variability and plasmid take-up. We also proved that it uses unnecessarily high transactivator levels, and that these can be detrimental if high expression of the gene of interest is desired. Additionally, our data suggest that particularly sensitive cell lines, such as hIPSCs, do not tolerate high-level transgene expression.
The negative effects of protein overexpression on cells are well-known and can result from a number of mechanisms including resource overload, stoichiometric imbalance, promiscuous interactions and pathway modulation (32). For the specific case of transcription factors, additional effects such as squelching can come into play as well (33). While our findings do not point at any of these phenomena specifically, they do highlight how crucial it is to keep the expression levels in transgenic constructs as finely regulated as those of the endogenous pathways. Our study represents a step in this direction, along with similar efforts in which advanced control architectures have been used to mitigate burden and improve robustness of gene expression (16,34).
We tackled sloppy component behavior by augmenting the standard induction system configuration with negative feedback and incoherent feedforward regulation, which significantly reduced transactivator levels, while conferring robustness to transactivator variability and plasmid take-up and allowing tunable programing of the fully induced expression of the gene of interest. We have thus arrived at a family of four cybergenetic gene expression circuits with different properties, which can be used in different situations based on the desired expression pattern of the output gene of interest (Figure 6G). We demonstrated their use in two proof-of-principle applications. In a transiently transfected culture of CHO-K1 cells, our feedback controller could eliminate the burden induced by tTA, resulting in up to a 2.6-fold increase in yield of the protein of interest. In hIPSCs that were transiently transfected with the controllers, the hybrid and IFF circuits could regulate the expression of the gene of interest to levels that were detectable, but low enough to mitigate the toxic effects caused by high-level expression. We anticipate that our controllers will find use in many similar situations that require reliable and flexible mammalian transgene expression.
Supplementary Material
ACKNOWLEDGEMENTS
We thank: Dr Laura Prochazka for the plasmid containing the insulator sequences; Dr Laura Prochazka and Mercedes Vazquez for assistance with the hIPSC cultures; Jiten Doshi for assistance with the CHO-K1 cell cultures; Timothy Frei for a plasmid expressing iRFP670; Dr Jörg Schreiber for a plasmid containing PUbC; Dr Benjamin Häfliger and Dr Nicolas Lapique for useful comments and discussion. We are especially grateful to Prof. Sai Reddy for the use of his laboratory’s cell culture facility, where all the transfection experiments reported in this manuscript were performed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme [743269 (CyberGenetics)]; European Union’s Horizon 2020 Research and Innovation Programme [766840 (COSY-BIO)]; Swiss National Science Foundation (31003A_149802). Funding for open access charge: ERC Horizon 2020 Research and Innovation Programme [743269].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hoppe P.S., Coutu D.L., Schroeder T.. Single-cell technologies sharpen up mammalian stem cell research. Nat. Cell Biol. 2014; 16:919–927. [DOI] [PubMed] [Google Scholar]
- 2. Ausländer S., Fussenegger M.. From gene switches to mammalian designer cells: present and future prospects. Trends Biotechnol. 2013; 31:155–168. [DOI] [PubMed] [Google Scholar]
- 3. Gossen M., Freundlieb S., Bender G., Muller G., Hillen W., Bujard H.. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995; 268:1766–1769. [DOI] [PubMed] [Google Scholar]
- 4. Cronin C.A., Gluba W., Scrable H.. The lac operator-repressor system is functional in the mouse. Genes Dev. 2001; 15:1506–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roney I.J., Rudner A.D., Couture J.-F., Kærn M.. Improvement of the reverse tetracycline transactivator by single amino acid substitutions that reduce leaky target gene expression to undetectable levels. Sci.Rep. 2016; 6:27697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005; 122:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Swiers G., Patient R., Loose M.. Genetic regulatory networks programming hematopoietic stem cells and erythroid lineage specification. Dev. Biol. 2006; 294:525–540. [DOI] [PubMed] [Google Scholar]
- 8. Alon U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 2007; 8:450–461. [DOI] [PubMed] [Google Scholar]
- 9. Mangan S., Itzkovitz S., Zaslaver A., Alon U.. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J. Mol. Biolo. 2006; 356:1073–1081. [DOI] [PubMed] [Google Scholar]
- 10. Kaplan S., Bren A., Dekel E., Alon U.. The incoherent feedforward loop can generate non-monotonic input functions for genes. Mol. Syst. Biol. 2008; 4:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Basu S., Mehreja R., Thiberge S., Chen M.-T., Weiss R.. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:6355–6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shimoga V., White J.T., Li Y., Sontag E., Bleris L.. Synthetic mammalian transgene negative autoregulation. Mol. Syst. Biol. 2013; 9:670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bleris L., Xie Z., Glass D., Adadey A., Sontag E., Benenson Y.. Synthetic incoherent feedforward circuits show adaptation to the amount of their genetic template. Mol. Syst. Biol. 2011; 7:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Strovas T.J., Rosenberg A.B., Kuypers B.E., Muscat R.A., Seelig G.. MicroRNA-based single-gene circuits buffer protein synthesis rates against perturbations. ACS Synth. Biol. 2014; 3:324–331. [DOI] [PubMed] [Google Scholar]
- 15. Bloom R.J., Winkler S.M., Smolke C.D.. Synthetic feedback control using an RNAi-based gene-regulatory device. J. Biol. Eng. 2015; 9:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Segall-Shapiro T.H., Sontag E.D., Voigt C.A.. Engineered promoters enable constant gene expression at any copy number in bacteria. Nat.Biotechnol. 2018; 36:352–358. [DOI] [PubMed] [Google Scholar]
- 17. Siciliano V., Garzilli I., Fracassi C., Criscuolo S., Ventre S., Di Bernardo D.. MiRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat. Commun. 2013; 4:2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aguda B.D., Kim Y., Piper-Hunter M.G., Friedman A., Marsh C.B.. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:19678–19683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marson A., Levine S.S., Cole M.F., Frampton G.M., Brambrink T., Johnstone S., Guenther M.G., Johnston W.K., Wernig M., Newman J. et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008; 134:521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xie Z., Wroblewska L., Prochazka L., Weiss R., Benenson Y.. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011; 333:1307–1311. [DOI] [PubMed] [Google Scholar]
- 21. Wroblewska L., Kitada T., Endo K., Siciliano V., Stillo B., Saito H., Weiss R.. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 2015; 33:839–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wiener N. Cybernetics or Control and Communication in the Animal and the Machine. 1961; Cambridge: MIT Press. [Google Scholar]
- 23. Briat C., Gupta A., Khammash M.. Antithetic integral feedback ensures robust perfect adaptation in noisy biomolecular networks. Cell Syst. 2016; 2:15–26. [DOI] [PubMed] [Google Scholar]
- 24. Lugagne J.-B., Carrillo S.S., Kirch M., Köhler A., Batt G., Hersen P.. Balancing a genetic toggle switch by real-time feedback control and periodic forcing. Nat. Commun. 2017; 8:1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Briat C., Zechner C., Khammash M.. Design of a synthetic integral feedback circuit: dynamic analysis and DNA implementation. ACS Synth. Biol. 2016; 5:1108–1116. [DOI] [PubMed] [Google Scholar]
- 26. Hsiao V., Swaminathan A., Murray R.M.. Control theory for synthetic Biology: Recent advances in system characterization, control design, and controller implementation for synthetic biology. IEEE Control Syst. 2018; 38:32–62. [Google Scholar]
- 27. Leisner M., Bleris L., Lohmueller J., Xie Z., Benenson Y.. Rationally designed logic integration of regulatory signals in mammalian cells. Nat. Nanotechnol. 2010; 5:666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lapique N., Benenson Y.. Digital switching in a biosensor circuit via programmable timing of gene availability. Nat. Chem. Biol. 2014; 10:1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prochazka L., Angelici B., Haefliger B., Benenson Y.. Highly modular bow-tie gene circuits with programmable dynamic behaviour. Nat. Commun. 2014; 5:4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hadjantonakis A.-K., Macmaster S., Nagy A.. Embryonic stem cells and mice expressing different GFP variants for multiple noninvasive reporter usage within a single animal. BMC Biotechnol. 2002; 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vintersten K., Monetti C., Gertsenstein M., Zhang P., Laszlo L., Biechele S., Nagy A.. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis. 2004; 40:241–246. [DOI] [PubMed] [Google Scholar]
- 32. Moriya H. Quantitative nature of overexpression experiments. Mol. Biol. Cell. 2015; 26:3932–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmidt S.F., Larsen B.D., Loft A., Mandrup S.. Cofactor squelching: artifact or fact. Bioessays. 2016; 38:618–626. [DOI] [PubMed] [Google Scholar]
- 34. Ceroni F., Boo A., Furini S., Gorochowski T.E., Borkowski O., Ladak Y.N., Awan A.R., Gilbert C., Stan G.-B., Ellis T.. Burden-driven feedback control of gene expression. Nat. Methods. 2018; 15:387–393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.