Abstract
Mutations in SPOP, the gene most frequently point-mutated in primary prostate cancer, are associated with a high degree of genomic instability and deficiency in homologous recombination repair of DNA but the underlying mechanisms behind this defect are currently unknown. Here we demonstrate that SPOP knockdown leads to spontaneous replication stress and impaired recovery from replication fork stalling. We show that this is associated with reduced expression of several key DNA repair and replication factors including BRCA2, ATR, CHK1 and RAD51. Consequently, SPOP knockdown impairs RAD51 foci formation and activation of CHK1 in response to replication stress and compromises recovery from replication fork stalling. An SPOP interactome analysis shows that wild type (WT) SPOP but not mutant SPOP associates with multiple proteins involved in transcription, mRNA splicing and export. Consistent with the association of SPOP with transcription, splicing and RNA export complexes, the decreased expression of BRCA2, ATR, CHK1 and RAD51 occurs at the level of transcription.
INTRODUCTION
The recent advances in whole-genome and exome sequencing of tumors have provided new insights into the genomic alterations underlying prostate cancer (PCa). Interestingly, the gene encoding Speckle Type POZ protein (SPOP) is the most frequently point-mutated in localized prostate tumors. Heterozygous missense mutations in SPOP were found in 8–15% of localized prostate tumors (1–3). SPOP is the substrate binding subunit of a Cullin3 E3 ubiquitin ligase complex and nearly all cancer-associated mutations reside in proximity of the substrate binding cleft of its MATH domain, suggesting a loss of function phenotype. It is thought that SPOP binding to its targets promotes their polyubiquitylation and proteasomal degradation. Therefore, cancer-associated SPOP mutations are thought to have a dominant negative effect that leads to increased stabilization of target proteins (4). Since the discovery of SPOP mutations in PCa, a diverse array of SPOP targets for ubiquitylation have been found. Examples include DEK, TRIM24, ERG, androgen receptor (AR) and its co-activator SRC-3 (4–9). Several of these factors are closely linked to PCa development, supporting a role for SPOP as a tumor suppressor. Interestingly, a recent study revealed that localized prostate tumors with SPOP mutations harbor an exceptionally high amount of genomic rearrangements when compared with other tumor subtypes, indicating that SPOP mutant tumors have a high degree of genomic instability (10). However, the mechanisms through which SPOP promotes chromosome stability remain poorly understood.
Chromosome instability is a hallmark of cancer and is associated with both poor prognosis and drug resistance and two of the major sources of genome instability are DNA double-strand breaks (DSBs) and replication stress. A general consensus view of DSB repair is that it involves a choice between two major pathways; non-homologous end-joining (NHEJ), which is considered error-prone, and homologous recombination (HR), which allows for potential error-free repair (11). A key step in HR is loading of RAD51 onto single-stranded DNA (ssDNA) filaments previously generated by 5′ to 3′ DNA end resection and coated by replication protein A (RPA) (12). Many factors are involved in RAD51 loading, but particularly crucial roles are played by BRCA1 and BRCA2 (13).
DNA replication stress, which is characterized by DNA synthesis slow down and/or replication fork stalling is another significant challenge to genome stability and has a central role in the generation of structural and numerical chromosome alterations (14). A number of mechanisms are in place to prevent replication stress and the critical involvement of HR in promoting replication fork progression has recently become an object of more intense research. Indeed components of the core HR machinery such as RAD51, BRCA1 and BRCA2 all play important roles in preventing replication stress by protecting stalled forks from breakage as well as facilitating restart of broken replication forks (15–17). Another critical pathway for preventing and reacting to replication stress is the ATR-CHK1 kinase cascade. Uncoupling of the MCM replicative helicase from DNA polymerase as occurs upon replication fork stalling leads to recruitment and activation of ATR by the exposed, RPA-coated ssDNA at the stalled fork (18). ATR activation not only promotes stability locally at the stalled fork, but phosphorylation of its key target CHK1 leads to diffusion of CHK1 into the nucleoplasm where it inhibits new replication origin firing on a global level. The concerted actions of ATR and CHK1 serve to coordinate fork progression and origin firing at the local and global level respectively, balancing the demand for limiting replication proteins such as RPA (19–21).
Here, we investigate whether SPOP regulates the response to replication stress. We show that SPOP knockdown impairs RAD51 foci formation and leads to spontaneous replication stress and aberrant cell cycle progression. Importantly, these phenotypes are associated with reduced transcriptional expression of several key DNA repair and replication factors including BRCA2, RAD51, CHK1 and ATR. Analysis of the SPOP interactome reveals its association with multiple proteins involved in transcription, mRNA splicing and export, suggesting a role of SPOP in promoting the transcription of these repair factors.
MATERIALS AND METHODS
Cell culture
U2OS cells were grown in DMEM (high glucose and Glutamax) supplemented with 10% fetal bovine serum and antibiotics (Penicillin, streptomycin), all from GIBCO (31966047, 10500064, 15070063). Prostate cancer cell lines LnCaP, C4-2b, PC3 and 22rv1 were cultured in RPMI (61870010) supplemented with 10% fetal bovine serum and antibiotics (Penicillin, streptomycin), all from GIBCO. For laser-induced DNA damage, the cells were maintained in CO2 independent media (GIBCO) for the duration of the damage-induction and recovery. For transfection of siRNA (20 nM concentration), all cells were reverse-transfected using Optimem and Lipofectamine RNAimax (Invitrogen, 13778150) according to the manufacturer's instructions.
Immunofluorescence
Cells were seeded on 13 mm glass coverslips (VWR, 631-0148) while reverse transfected when applicable. The samples were washed once in ice-cold PBS and fixed with 4% formaldehyde in PBS for 15 min at RT. When pre-extraction was performed in order to remove the non-chromatin-bound fraction, this was carried out prior to fixation using 0.2% Triton in PBS for 90 sec. The samples were permeabilized in 0.5% Triton in PBS for 15 min prior to incubation with primary antibodies for 1–2 h depending on the antibody. The samples were subsequently incubated with secondary fluorescence-coupled antibodies (Alexa Fluor) for 1 h and finally stained with DAPI at 1 μg/ml for 5 min before being mounted on glass slides with Prolong Diamond mounting medium (Invitrogen, P36961). The coverslips were washed at least 3 times after each antibody incubation as well as after DAPI staining. Incubations with antibodies were done in 3% BSA in PBS-T (0.01% Tween-20 in PBS) and all washes were performed with PBS-T. When Click-it reactions were performed as for EdU incorporation, this was done prior to the primary antibody incubation according to the manufacturer's instructions (Invitrogen, C10638).
Colony assays
Cells were transfected and 24 h post transfection trypsinized, counted and reseeded at 300 cells/well in 6-well format. At 48 h post transfection, the cells were treated with replication stress-inducing agents. After treatment, the cells were washed three times and allowed to recover for 12 days. The cells were then washed once in PBS, left to dry and stained with cell staining solution (0.5% w/v Crystal Violet, 25% v/v methanol). The plates were washed three times in deionized water and colonies were counted manually and normalized to the untreated sample of each siRNA, respectively.
Laser microirradiation
Laser microirradiation was performed essentially as described previously (22). Briefly, cells were plated on glass coverslips, sensitized to laser damage by 24 h incubation with 10 μM BrdU and then transferred to CO2 independent media for equilibration and damage induction with a custom-designed PALM MicroBeam equipped with a 355 nm UV-A pulsed laser. The cells were left to recover for the indicated time and then fixed and processed for immunofluorescence staining.
QIBC
Quantitative image-based cytometry (QIBC) was performed as previously described (19). In brief, images used for QIBC were obtained using a motorized Olympus IX-81 wide-field microscope equipped with filters for DAPI, FITC, Cy3, and Cy5 fluorescent dyes, an MT20 Illumination system, and a digital monochrome Hamamatsu C9100 charge coupled device (CCD) camera. When foci analysis was performed, 40×/0.9 NA or 20×/0.75 NA objectives were used, whereas a 10×/0.4 NA objective was used when whole-nuclei mean intensities were the end point. Image acquisition was carried out using the ScanR acquisition software. Depending on magnification and cell density, 100–200 images were acquired per sample, allowing for the detection of at least 1000 cells per sample. The acquisition times were adjusted for nonsaturated conditions in 12-bit dynamic range. The ScanR analysis software was then utilized performing a dynamic background correction to all images and the DAPI signal was used to identify individual nuclei by an intensity-threshold-based mask. Within this mask, pixel intensities were analyzed for each channel to yield mean and total intensities for each nucleus. For foci analysis, the spot detection module of ScanR analysis was applied to detect individual foci. All data was exported into TIBCO Spotfire software, which was used to generate all scatterplots for figures.
Antibodies
53BP1 (Santacruz, SC-22760, WB, IF 1:500), Actin (Sigma, WB 1:5000), ATR (Cell Signaling, #2790, WB 1:500), BRCA1 (Santacruz SC-646 IF 1:200), BRCA2 (Abcam ab123491 WB 1:500), CHK1 (Cell Signaling #2360, WB 1:500), CHK1-pS345 (Cell signaling, #2348, WB 1:500), FLAG (Sigma, F1804, IF 1:400, WB 1:1000), H2AX-pS139 (Biolegend, 613401, IF 1:1000), H2AX-pS139 (Cell signaling #9718 IF 1:500), KU70 (Abcam Ab2172, WB 1:400), PRPF8 (Santacruz, SC30207, WB 1:400), RAD51 (Abcam ab213, IF 1:100, WB 1:500), RAP80 (Bethyl A300–763-A1 IF 1:200), RNA Polymerase II (Santacruz SC-899, WB 1:500), RPA70 (Abcam, ab79398, IF 1:500), SPOP (Proteintech 16750-1-AP, WB 1:1000), SSRP1 (Biolegend 10D1 WB 1:2000), THOC2 (Abcam ab129485).
DNA fiber labeling
Cells were pulse-labelled with 25 μM of CldU (Sigma-Aldrich C6891) for 20 min at 37°C, washed and incubated in fresh medium containing 250 μM of IdU (Sigma-Aldrich I7125) for 20 min at 37°C. Cells were harvested and DNA fibers prepared as described previously (23). CldU was detected with rat anti-BrdU (OBT0030, Serotec) and the AlexaFluor-568 anti-rat antibodies and IdU was detected with a mouse anti-BrdU (347580, Becton Dickinson) and the AlexaFluor-488 anti-mouse antibodies. Images of well spread DNA fibers were acquired using the LSM700 confocal microscope (Carl Zeiss), the 63×/1.4 oil immersion objective (Carl Zeiss) and LSM ZEN software. Analysis of double-labeled replication forks was performed manually using LSM ZEN software.
siRNA oligos
All siRNAs were from Ambion: Ctrl si (4390843), SPOP si#1 (s15955), SPOP si#2 (13369), SPOP si#3 (13447).
Cloning of SPOP and SPOP-F133V
The full length SPOP gene was cloned into pCPR0085, a modified version of pNIC-CH (GenBank EF199843) containing an C-terminal Tobacco Etch Virus (TEV) protease recognition site followed by a TwinsStrep-His6 tag using ligation-independent cloning. The SPOP-F133V mutant was created by QuickChange site-directed mutagenesis (Agilent 200523).
See Supplementary material for additional material and methods.
RESULTS
SPOP is required for proper response to replication fork stalling
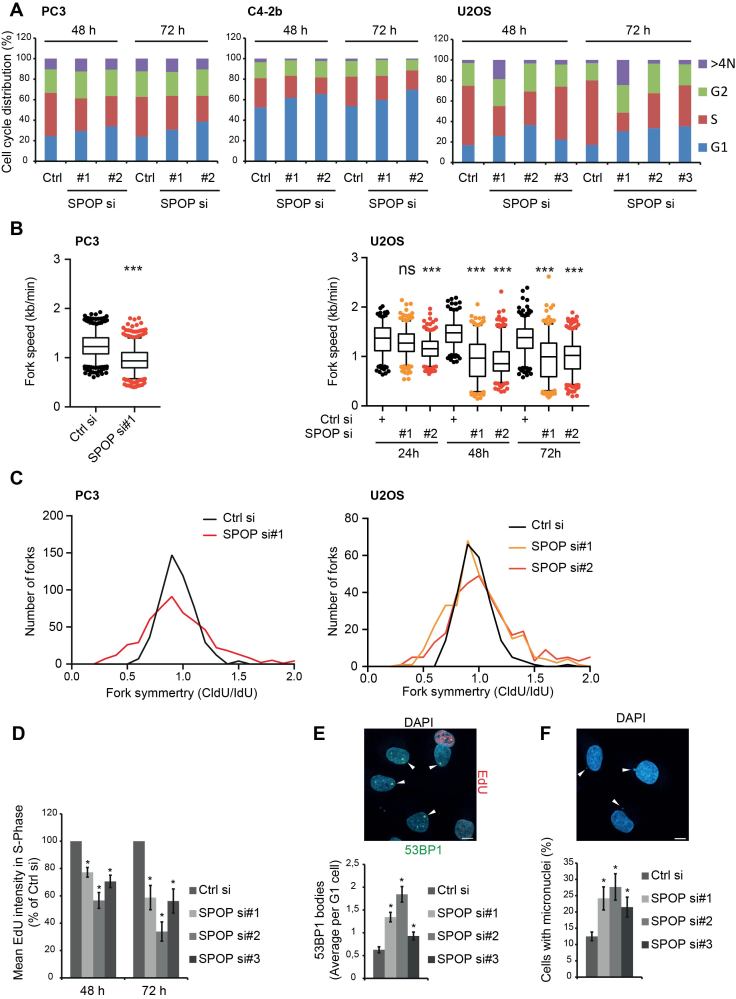
Since loss of SPOP function impairs HR-mediated DSB repair (10), we set out to investigate a role for SPOP in countering replication stress. To assess replication stress-induced RAD51 foci formation in S-phase we took advantage of Quantitative Image-based cytometry (QIBC), which allows for automated high-throughput detection and quantitation of cellular features such as DNA damage foci in individual cells of large populations of thousands of cells as a function of cell cycle status (19). We established efficient siRNA-mediated knockdowns of SPOP in prostate cancer cell lines C4-2b and PC3 as well as the non-prostate cell line U2OS (Figure 1A). We found that RAD51 foci formation in response to replication fork stalling induced by treatment with hydroxyurea (HU) or low dose camptothecin (CPT) was significantly reduced by SPOP knockdown (Figure 1B). We proceeded to investigate how the impaired response to replication stress would impact the ability of cells to recover from prolonged fork stalling. To specifically monitor the recovery of cells affected by replication stress, we labelled cells in S-Phase by a short EdU pulse and then stalled replication forks by addition of HU for 24 h. The cells were then washed and allowed to recover. The cells exposed to replication stress could then be followed over time by QIBC-derived gating of the EdU+ cells (Figure 1C–E). Replication fork stalling leads to accumulation of protective RPA on the exposed ssDNA, which counters fork breakage. Prolonged stalling does, however, lead to fork collapse characterized by hyper-phosphorylated γH2AX (19). As expected, we found that HU-induced fork stalling for 24 h lead to increased RPA accumulation on chromatin (Figure 1E). Interestingly, the proportion of EdU+ cells with hyper-phosphorylated γH2AX (γH2AX+), was increased in SPOP knockdown cells after 24 h of HU treatment, suggesting that replication forks in these cells are more prone to collapse (Figure 1C and E). Also, while ctrl siRNA transfected EdU+ cells recovered partially from the prolonged fork stalling, SPOP knockdown EdU+ cells instead displayed persistent γH2AX phosphorylation (Figure 1C and E). Colony assays further demonstrated that knockdown of SPOP significantly impaired relative clonal outgrowth after both short (2 h) and long (24 h) HU-induced replication fork stalling as well as persistent replication stress by low dose CPT for 24 h (Figure 1F). Taken together, these data demonstrate that depletion of SPOP impairs the formation of RAD51 foci in response to replication stress as well as the ability of cells to recover from replication fork stalling.
Figure 1.
SPOP knockdown impairs RAD51 foci formation in response to replication stress and impairs recovery from prolonged replication fork stalling. (A) C4-2b, PC3 and U2OS cells were transfected with siRNA and lysed 48 h post transfection. Whole cell extracts were analyzed by western blotting with the indicated antibodies. (B) C4-2b, PC3 and U2OS cells were transfected with ctrl siRNA or SPOP si#2 and treated with 2 mM HU or low dose CPT (50 nM) and pre-extracted and fixed after the indicated times. Samples were stained by immunofluorescence for RAD51, RPA70 and DAPI. QIBC analysis was performed to quantify RAD51 foci in S-Phase cells (RPA+) based on DAPI and chromatin-bound RPA70 staining. (C) U2OS cells were transfected with siRNA and pulsed for 30 min with EdU to label replicating cells. Replication fork progression was then blocked by treatment with HU for 24 h. HU was washed out and the cells were left to recover and resume replication for the indicated times. The samples were then fixed and analyzed by immunofluorescence for RPA70, EdU, γH2AX and DAPI. QIBC was performed in order to generate scatterplots for DAPI/EdU with γH2AX+ cells shown in red. An outline of the experiment is shown in (D). (E) Additional scatterplots were generated to show chromatin-bound RPA and γH2AX inside the EdU+ population (replicating cells at the time of HU-treatment). (F) U2OS cells were transfected with siRNA and subjected to colony assay following replication stress-inducing treatment with HU (2 mM for 2 or 24 h) or low dose CPT (10 nM for 24 h).The colony count was normalized to the respective untreated sample. Error bars represent SD, n = 3. Significance was determined by a two-tailed t test: *P < 0.05.
Loss of SPOP leads to spontaneous replication stress and aberrant cell cycle progression
Given that SPOP mutations are very early events in PCa progression (10), we speculated whether the lack of SPOP would be sufficient to elicit any spontaneous effects in unchallenged cells. Indeed, when the QIBC-derived cell cycle profiles of C4-2b, PC3 and U2OS cells were analyzed at 48 and 72 h after transfection with siRNA targeting SPOP, we observed a clear reduction in the amount of cells in S-Phase, while the G1 population increased (Figure 2A). We then performed DNA fiber labeling to analyze replication fork progression at the level of individual DNA molecules and found that SPOP knockdown significantly reduced replication fork progression in both PC3 and U2OS cells (Figure 2B) while also leading to increased fork arrest and collapse indicated by the analysis of fork symmetry (Figure 2C). In addition, quantification of replication stress associated phenotypes revealed that SPOP knockdown lead to a significant decrease in the mean EdU incorporation of S-phase cells as quantified by QIBC (Figure 2D). Furthermore, SPOP knockdown lead to a significant increase in G1 cells with 53BP1 bodies as well as increased micronuclei (Figure 2E and F), both of which are strongly associated with replication stress (24). These data clearly indicate that loss of SPOP function promotes spontaneous replication stress and genome instability.
Figure 2.
SPOP knockdown leads to replication stress and aberrant cell cycle progression. (A) C4-2b, PC3 and U2OS cells were transfected with siRNA. At the indicated time points, the cells were pulsed for 30 min with EdU and fixed for immunofluorescence analysis for DAPI and EdU. QIBC was performed and cell cycle profiles were derived from this. PC3 and U2OS cells were transfected with siRNA for the indicated times, incubated for 20 min with CldU and IdU, followed by DNA fiber preparation, staining, and quantification. Total fork speed was determined (B) and fork symmetry is shown for the 48 h time point (C). Significance was determined by a two-tailed t test: ***P < 10−14. U2OS cells were transfected with siRNA. At the indicated time points, the cells were pulsed for 30 min with EdU and fixed for immunofluorescence analysis for DAPI, EdU and 53BP1. QIBC was performed and cell cycle profiles were derived from this. The mean EdU intensities of cells in S-Phase were quantified by QIBC (D). QIBC analysis of the 72 h time points was used to quantify 53BP1 bodies in G1 cells (E) and micronuclei were scored manually (200 cells analyzed per sample) (F). Example pictures are shown above. Scale bars, 10 μm. Error bars represent SD, n = 3. Significance was determined by a two-tailed t test: *P < 0.05.
SPOP knockdown impairs RAD51 foci formation in response to DNA DSBs
To gain further insight into the mechanisms underlying the observed HR defects of SPOP deficient cells, we investigated the response of SPOP knockdown cells to DNA DSBs induced by ionizing radiation. Consistent with previous findings (10,25), SPOP knockdown had no impact on the formation of γH2AX foci, indicating that recognition of DSBs and the immediate activation of the DNA damage response are unaffected by SPOP knockdown (Supplementary Figure S1A and B). However, HR-associated RAD51 foci formation was significantly decreased by SPOP knockdown while 53BP1 foci persisted (Supplementary Figure S1C–F) indicating that repair of DNA DSBs in SPOP depleted cells was channeled toward NHEJ instead of HR as it was previously suggested (10).
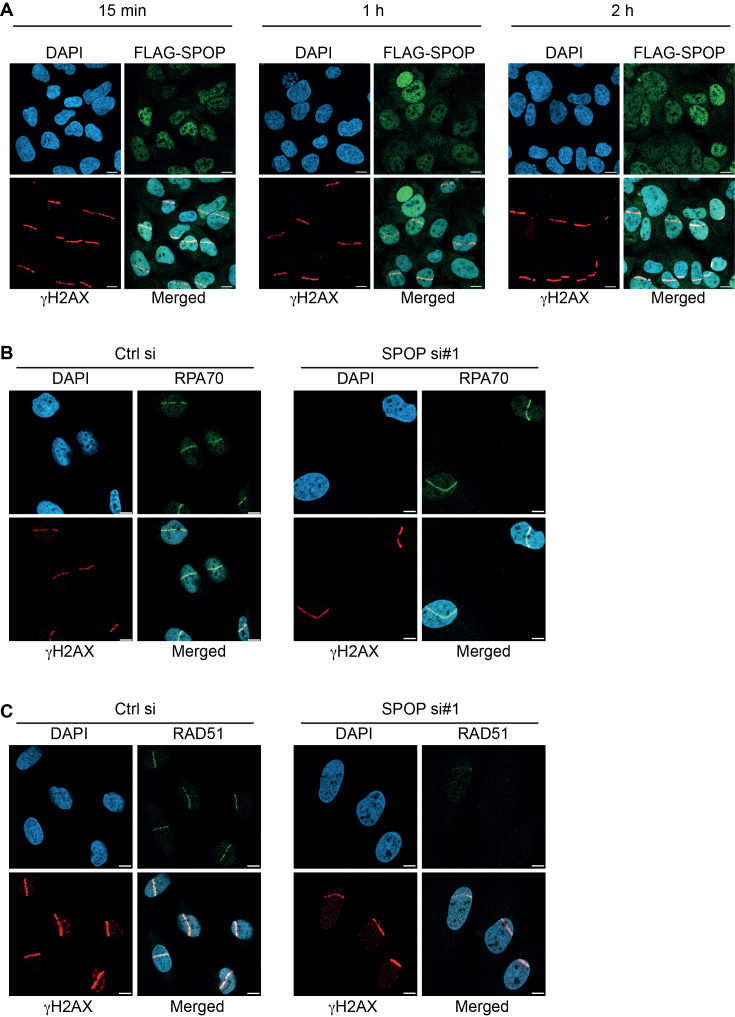
In order to further dissect the role of SPOP in the DNA damage response, we took advantage of laser-induced micro-irradiation to monitor recruitment of repair factors to DSBs (22). Using a FLAG-tagged, doxycycline-inducible SPOP, we found SPOP to be localized in a speckled, predominantly nuclear pattern as reported earlier (26). However, we did not observe any recruitment of SPOP to the site of damage at any of the time points (Figure 3A). No changes in the recruitment of components of the resection-promoting complex, BRCA1 and RAP80, to the γH2AX-marked sites of damage was observed upon SPOP knockdown (Supplementary Figure S2A and B). Recruitment of RPA, which coats ssDNA exposed by resection, was also observed to similar extents (Figure 3B), indicating that resection appears to proceed as normal in the absence of SPOP. However, as would be expected in light of results shown above, RAD51 recruitment to the DSBs was clearly reduced (Figure 3C). These results show that SPOP is not recruited actively to the sites of DSB-induction suggesting that the role of SPOP in promoting HR is likely indirect. Furthermore, the main defect of SPOP knockdown cells in recruiting RAD51 appears to reside downstream of DNA end resection at the level of RAD51 loading.
Figure 3.
Recruitment dynamics of SPOP and key effectors in DNA repair in response to laser-induced double-strand breaks. (A) U2OS cells expressing doxycycline-inducible FLAG-SPOP were microlaser irradiated, fixed at the indicated time points and stained by immunofluorescence for γH2AX and FLAG. U2OS cells were transfected with siRNA and microlaser irradiated, fixed after 1 h and stained by immunofluorescence for γH2AX, RPA70 (B) and RAD51 (C). Scale bars, 10 μm.
SPOP interactome reveals association with core transcription, splicing and mRNA export machinery
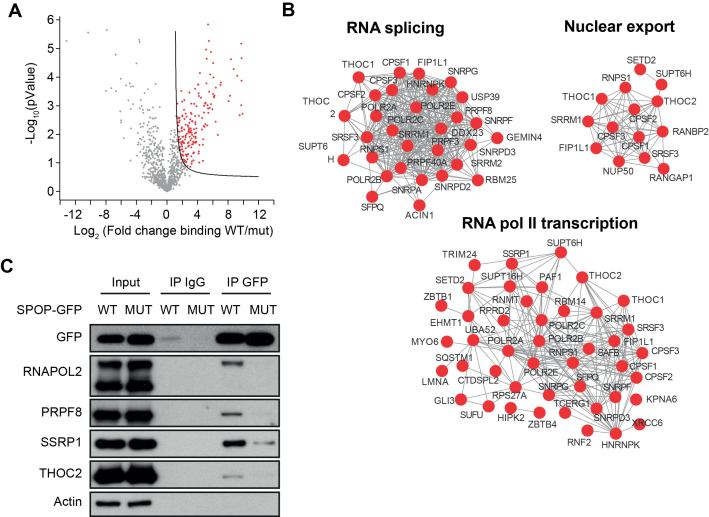
To find new clues as to how SPOP may impact on genome stability, we performed an extensive, mass spectrometry-driven analysis of the SPOP interactome. Recombinant, STREP-tagged WT SPOP was used in pull-downs assays on lysates from prostate cancer LnCaP cells. Since mutations in the SPOP MATH domain abrogates the binding of SPOP to target proteins, we used recombinant, STREP-tagged SPOP containing the MATH domain mutation F133V, as a negative control in the pull-downs. We identified 209 proteins showing significantly (FDR<0,05) increased association to WT than to F133V SPOP (Figure 4A). Among the proteins that preferentially bind to WT SPOP there were several known SPOP targets such as DAXX, SETD2, TRIM24, GLI3 and INF2 (4,27–30), validating our approach (Supplementary Table S1). Interestingly, gene ontology analysis performed on the WT-specific interactors revealed that these proteins were highly enriched (FDR<10-5) for factors involved in RNA polymerase II transcription, mRNA splicing, and nuclear export (Figure 4B). This included several subunits of RNA polymerase II as well as SSRP1 and SUPT16h, the two subunits of the FACT complex, which promotes transcriptional elongation. Also, components of the spliceosome were bound to SPOP including PRPF3, PRPF40A, and the essential core component PRPF8. In addition, THOC1 and THOC2, which are important parts of the TREX complex were associated with WT SPOP. To verify these interactions, we used U2OS expressing doxycycline-inducible GFP-tagged WT and F133V SPOP to co-immunoprecipitate selected proteins. Indeed, we verified the interactions of WT but not F133V SPOP with RNA polymerase 2, SSRP1, PRPF8, and THOC2 (Figure 4C). Interestingly, we noted that WT SPOP appeared to interact primarily with the slow-migrating, hyper-phosphorylated form of RNA polymerase II, which is commonly assumed to be actively elongating (Figure 4C). In order to investigate whether any of the identified proteins interacting with WT SPOP could be targeted for degradation for polyubiquitylation, we analysed the overall protein levels these proteins upon overexpression of both WT and F133V SPOP. However, none of the identified proteins were decreased in abundance upon overexpression of WT SPOP (Supplementary Figure S3) suggesting that they are not regulated on overall protein level by SPOP or may be indirect interactors as part of a bigger complex regulating transcription or mRNA processing.
Figure 4.
SPOP is in complex with the core transcription and RNA processing machinery. Lysates from LnCaP cells were subjected to pull-down assays using recombinant WT- or F133V mutant STREP-tagged SPOP. The pull-downs were performed in triplicate and analyzed by mass spectrometry. The WT-specific interactors are highlighted in a volcano plot (red color) (A). Gene ontology analysis was performed and a functional network of the most highly enriched protein within functional groups is shown (B). U2OS cells expressing doxycycline-inducible GFP-tagged WT- or F133V mutant SPOP were lyzed and subjected to IP using anti-GFP antibodies or unspecific IgG. The IPs were analyzed by western blotting using the indicated antibodies (C).
SPOP promotes transcriptional expression of DNA repair and replication factors and is required for proper activation of CHK1 in response to replication stress
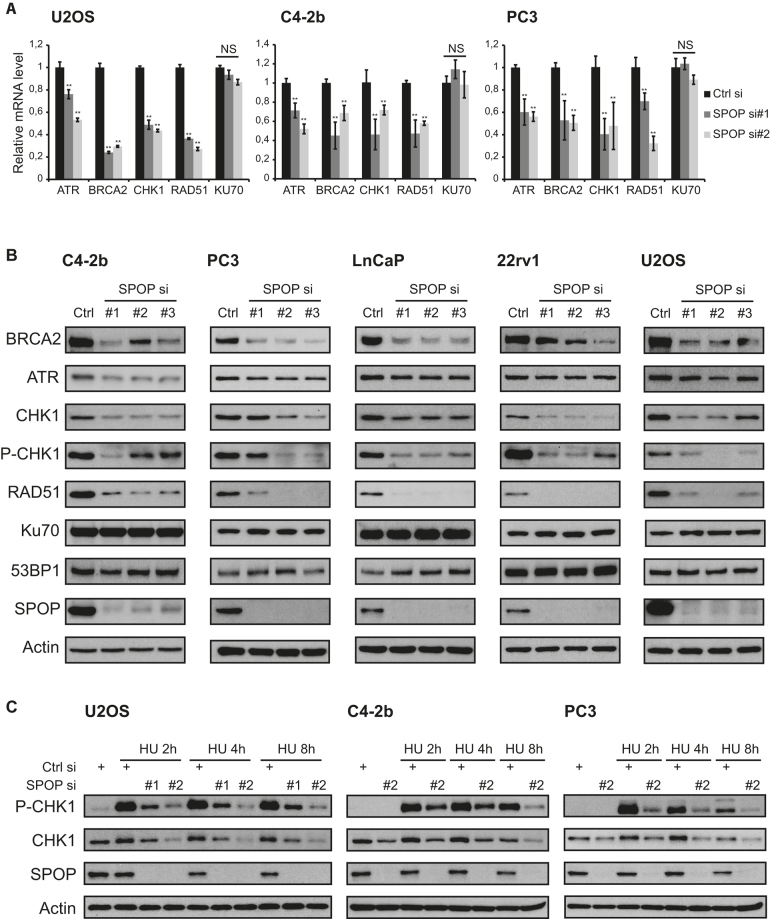
The association of SPOP with the transcription machinery led us to investigate whether the spontaneous replication stress and HR defects of SPOP depleted cells was associated with altered mRNA expression of a panel of key factors involved in DNA repair and the replication stress response. Interestingly, analysis of mRNA levels by qPCR showed a significant downregulation of RAD51, BRCA2, CHK1 and ATR in SPOP siRNA transfected U2OS, C4-2b and PC3 cells, whereas the NHEJ factor KU70 was unaffected (Figure 5A). Expanding on these results, we analysed additional DNA repair factors ATM, BRCA1, CHK2 and 53BP1, but these genes were either not affected by SPOP knockdown or inconsistently affected across the cell lines tested (Supplementary Figure S4). The transcriptional downregulation of RAD51, BRCA2, CHK1 and ATR translated into similar results when protein levels were examined by western blotting with RAD51, BRCA2, CHK1, and, to a lesser extent, ATR all showing decreased levels after knockdown of SPOP in prostate cancer cell lines C4-2b, PC3, LnCaP, and 22rv1 as well as U2OS (Figure 5B). The protein levels of KU70 and 53BP1 were, however, not affected by SPOP knockdown (Figure 5B). In addition, activated CHK1 as measured by phosphorylation of the ATR-dependent CHK1-S345 residue was suppressed by SPOP knockdown (Figure 5B). Activation of CHK1 in response to replication stress was also impaired as knockdown of SPOP lead to reduced levels of activated, phosphorylated CHK1-S345 in C4-2b and PC3 cells in response to HU treatment (Figure 5C). Similarly, replication stress-induced levels of CHK1-S345 in SPOP depleted U2OS cells were also clearly suppressed (Figure 5C).
Figure 5.
SPOP promotes transcriptional expression of DNA repair and replication factors and is required for proper activation of CHK1 in response to replication stress. (A) U2OS, C4-2b and PC3 cells were transfected with siRNA. At 48 h after transfection, mRNA was isolated, reverse-transcribed and analyzed by qPCR with the indicated primers. (B) C4-2b, PC3, LnCaP, 22rv1 and U2OS cells were transfected with siRNA and lysed 48 h post transfection. Whole cell extracts were analyzed by western blotting with the indicated antibodies (Note: P-CHK1 blots were exposed for a prolonged time to analyse P-CHK1 in unchallenged cells). (C) U2OS, C4-2b and PC3 cells were transfected with siRNA and treated with HU for the indicated times. Whole cell extracts were analyzed by western blotting with the indicated antibodies. Error bars represent SD, n = 3. Significance was determined by a two-tailed t test: *P < 0.05; **P< 0.01.
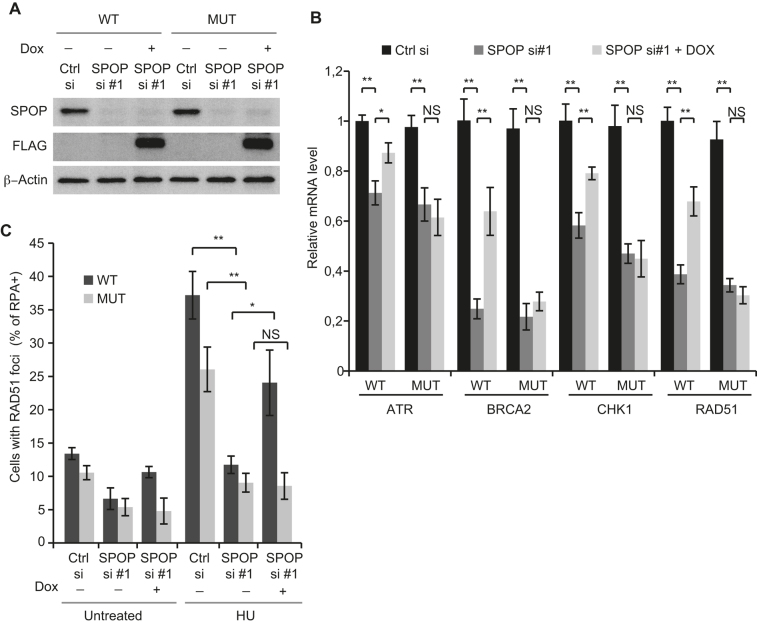
WT but not F133V mutant SPOP rescues transcriptional expression of DNA repair genes and replication stress induced RAD51 foci formation
In order to further investigate the role of the prostate cancer associated mutation in the context of replication stress and transcriptional expression of DNA repair factors, we established U2OS cells expressing inducible siRNA resistant FLAG-tagged WT and F133V mutated SPOP (Figure 6A). As expected, expression of WT FLAG-SPOP partially restored the mRNA levels of ATR, BRCA2, CHK1 and RAD51. However, interestingly, expression of F133V-FLAG-SPOP did not rescue the transcriptional expression of these genes (Figure 6B). To validate these results in the context of replication stress, we assessed RAD51 foci formation in response to replication fork stalling by HU. Confirming our previous results, we found that expression of WT-FLAG-SPOP significantly increased RAD51 foci formation in response to HU above the levels imposed by SPOP knockdown whereas expression of F133V-FLAG-SPOP did not rescue foci formation, indicating that the F133V mutation confers deficiency in promoting proper mRNA expression of DNA repair and replication factors to suppress replication stress (Figure 6C).
Figure 6.
SPOP knockdown-mediated repression of trancriptional expression of DNA repair genes and replication stress induced RAD51 foci formation are rescued by WT but not mutant SPOP. U2OS cells expressing siRNA resistant doxycycline-inducible FLAG-tagged WT- or F133V mutant SPOP were grown in the presence or absence of doxycycline and transfected with siRNA and lysed 48 h post transfection. Whole cell extracts were analysed by western blotting (A) with the indicated antibodies and mRNA was isolated, reverse-transcribed and analyzed by qPCR with the indicated primers (B). U2OS cells expressing siRNA resistant doxycycline-inducible FLAG-tagged WT- or F133V mutant SPOP were grown in the presence or absence of doxycycline and transfected with siRNA. At 48 h post transfection, the cells were treated with 2 mM HU for 24 h and then pre-extracted and fixed. Samples were stained by immunofluorescence for RAD51, RPA70 and DAPI. QIBC analysis was performed to quantify RAD51 foci in S-Phase cells (RPA+) based on DAPI and chromatin-bound RPA70 staining (C). Error bars represent SD, n = 3. Significance was determined by a two-tailed t test: *P < 0.05; **P< 0,01.
Taken together these results demonstrate that SPOP is required for sufficient expression of the key DNA replication and repair factors RAD51, BRCA2, CHK1 and ATR and that inability of SPOP knockdown cells to promote sufficient expression of these factors is likely due to defects at the level of transcription or mRNA processing. These findings describe a novel function of SPOP in regulating mRNA transcription, which seems to be specific for a subset of genes involved in DNA replication and repair.
DISCUSSION
Previous work by ourselves and others have demonstrated the importance of the DNA damage checkpoint barrier in preventing tumorigenesis (31) and recently we have characterized the progressive activation of these responses in PCa development (32). Although SPOP is broadly recognized as a tumor suppressor in prostate cancer and SPOP mutations are associated with an unusually high frequency of genomic rearrangements, the mechanisms by which SPOP promotes genome stability have so far evaded discovery. In the present study, we now demonstrate that the role of SPOP in promoting genomic stability is likely conferred by promoting expression of key factors involved in resistance to replication stress and HR-mediated DNA repair. Moreover, we show that these changes in protein levels are correlated with significant reductions in mRNA levels indicating that the underlying defects reside at the level of transcription. Our MS-driven profiling of SPOP WT-specific binding partners represents to our knowledge the first extensive interactome performed for SPOP. Interestingly, the protein networks associated with WT SPOP are highly enriched for components of the general mRNA transcription-, splicing- and processing machinery. This appears consistent with the well-established localization of SPOP to nuclear speckles (26), which are enriched in pre-mRNA splicing- and export factors (33). Indeed, a recent study establishing SETD2 as a target for polyubiquitylation by SPOP reports changes in pre-mRNA splicing upon SPOP knockdown (30).
The role of the general transcription and pre-mRNA processing machinery in promoting genome stability has been emphasized by recent studies detailing how interference with core processes such as transcriptional elongation (34), splicing (35,36), or mRNA export (37) appears to selectively affect transcripts encoding replication and HR factors but not NHEJ factors, yielding phenotypes reminiscent of SPOP knockdown. Since we find SPOP in complex with core transcription, spliceosome and mRNA export modules, it is tempting to speculate that the observed attenuation of RAD51, BRCA2, ATR, and CHK1 transcripts upon SPOP knockdown may be due to misregulation of these processes. We show that SPOP does not clearly localize to sites of induced DNA DSBs. This is consistent with an indirect role for SPOP in promoting genome stability by facilitating normal expression of repair factors although it appears to somewhat contradict earlier reports of partial colocalization with γH2AX foci (10,25). Our finding that cells depleted for SPOP display a concomitant decrease in levels of both RAD51 and BRCA2, which plays a key role in loading RAD51 onto ssDNA, provides novel mechanistic insight regarding the HR deficiencies and genomic instability of SPOP deficient tumors. Moreover, these findings are consistent with our data indicating that recognition and resection of DSBs appear to proceed as normal in SPOP knockdown cells, with the main HR defects residing at the level of RAD51 loading.
Our work also provides first evidence that spontaneous replication stress as result of SPOP knockdown is a general phenomenon as observed across different cell lines. In addition to spontaneous replication stress, the impact of SPOP knockdown on replication dynamics was also evident by the impaired ability to recover from prolonged replication fork stalling. Considering the well-established roles of BRCA2, RAD51, CHK1 and ATR in promoting fork progression and resistance to replication stress, it is not surprising that the observed suppression in the expression of all four genes is associated with reduced fork speed as well as an increase in 53BP1 bodies and micronuclei. BRCA2 and RAD51 both play important roles at the replication fork, where they promote fork progression and protect stalled forks from extensive resection (16,38,39). The ATR-CHK1 axis is also pivotal in the control of replication, acting on a global level to control origin firing, but also promoting the progression of individual forks (40–42). The inability of SPOP knockdown cells to recover from prolonged fork stalling is likely multi-facetted. Suppression of the ATR-CHK1 axis has been demonstrated to promote replication fork collapse in response to fork stalling (19) while decreased levels of RAD51 impairs the restart of collapsed forks (15). The contribution of modulation of AR dependent transcription by SPOP deficiency towards the replication stress phenotypes observed in this study could also be important to consider. On one hand, androgen signaling has been shown to promote expression of DNA repair genes (43) but on the other hand, AR is also known to promote expression of replication licensing factors such as CDC6 (44), the aberrant overexpression of which can lead to replication stress (45–47). However, since two of the cell lines employed here, U2OS and PC3, are androgen independent it is unlikely that aberrant androgen signaling underlies the replication stress observed upon SPOP deficiency.
Our finding that SPOP knockdown induces spontaneous replication stress thus expands the current understanding of the mechanisms underlying the high degree of genomic instability associated with SPOP mutation and it further strengthens the rationale for specialized treatment of cancers with SPOP loss of function with therapeutics aimed at DNA repair and replication defects. Recent data from cell culture experiments indicate that cells expressing mutant SPOP or transfected with SPOP siRNA are more sensitive to treatment with PARP inhibitors (10). Also, results from a recent clinical study show that patients with castration-resistant metastatic PCa harboring defects in DNA repair genes BRCA1, BRCA2 or ATM had a high response rate to PARP inhibitor treatment (48). The study does, however, not report on response rates for patients with SPOP mutation, but considering the intrinsic replication stress and HR defects described in this study, SPOP mutation could prove a useful biomarker for treatment stratification.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (49) partner repository with the dataset identifier PXD010527.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Protein Production Facility at the Novo Nordisk Foundation Center for Protein Research for production of SPOP and SPOP mutant and we thank MyungHee Lee for technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Danish Research Council [DFF 4004-00450, Flores-Morales]; Danish Cancer Society [R90-A6060-14-S2, Flores-Morales]; The Novo Nordisk Foundation Center for Protein Research is financially supported by the Novo Nordisk Foundation [NNF14CC0001]. Jiri Bartek and Apolinar Maya-Mendoza are funded by the Danish Cancer Society (R204/A12) and the Swedish Research Council. Funding for open access charge: Novo Nordisk Foundation Center for Protein Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Baca S.C., Prandi D., Lawrence M.S., Mosquera J.M., Romanel A., Drier Y., Park K., Kitabayashi N., MacDonald T.Y., Ghandi M. et al. . Punctuated evolution of prostate cancer genomes. Cell. 2013; 153:666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barbieri C.E., Baca S.C., Lawrence M.S., Demichelis F., Blattner M., Theurillat J.P., White T.A., Stojanov P., Van Allen E., Stransky N. et al. . Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012; 44:685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Le Gallo M., O’Hara A.J., Rudd M.L., Urick M.E., Hansen N.F., O’Neil N.J., Price J.C., Zhang S., England B.M., Godwin A.K. et al. . Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat. Genet. 2012; 44:1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Theurillat J.P., Udeshi N.D., Errington W.J., Svinkina T., Baca S.C., Pop M., Wild P.J., Blattner M., Groner A.C., Rubin M.A. et al. . Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science. 2014; 346:85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. An J., Ren S., Murphy S.J., Dalangood S., Chang C., Pang X., Cui Y., Wang L., Pan Y., Zhang X. et al. . Truncated ERG oncoproteins from TMPRSS2-ERG fusions are resistant to SPOP-mediated proteasome degradation. Mol. Cell. 2015; 59:904–916. [DOI] [PubMed] [Google Scholar]
- 6. An J., Wang C., Deng Y., Yu L., Huang H.. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014; 6:657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gan W., Dai X., Lunardi A., Li Z., Inuzuka H., Liu P., Varmeh S., Zhang J., Cheng L., Sun Y. et al. . SPOP promotes ubiquitination and degradation of the ERG oncoprotein to suppress prostate cancer progression. Mol. Cell. 2015; 59:917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Geng C., He B., Xu L., Barbieri C.E., Eedunuri V.K., Chew S.A., Zimmermann M., Bond R., Shou J., Li C. et al. . Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:6997–7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Groner A.C., Cato L., de Tribolet-Hardy J., Bernasocchi T., Janouskova H., Melchers D., Houtman R., Cato A.C., Tschopp P., Gu L. et al. . TRIM24 is an oncogenic transcriptional activator in prostate cancer. Cancer Cell. 2016; 29:846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boysen G., Barbieri C.E., Prandi D., Blattner M., Chae S.S., Dahija A., Nataraj S., Huang D., Marotz C., Xu L. et al. . SPOP mutation leads to genomic instability in prostate cancer. Elife. 2015; 4:e09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jeggo P.A., Pearl L.H., Carr A.M.. DNA repair, genome stability and cancer: a historical perspective. Nat. Rev. Cancer. 2016; 16:35–42. [DOI] [PubMed] [Google Scholar]
- 12. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Prakash R., Zhang Y., Feng W., Jasin M.. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 2015; 7:a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burrell R.A., McClelland S.E., Endesfelder D., Groth P., Weller M.C., Shaikh N., Domingo E., Kanu N., Dewhurst S.M., Gronroos E. et al. . Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013; 494:492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T.. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010; 37:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M.. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011; 145:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schlacher K., Wu H., Jasin M.. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012; 22:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saldivar J.C., Cortez D., Cimprich K.A.. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017; 18:622–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Toledo L.I., Altmeyer M., Rask M.B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J.. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013; 155:1088–1103. [DOI] [PubMed] [Google Scholar]
- 20. Petermann E., Woodcock M., Helleday T.. Chk1 promotes replication fork progression by controlling replication initiation. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:16090–16095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Syljuasen R.G., Sorensen C.S., Hansen L.T., Fugger K., Lundin C., Johansson F., Helleday T., Sehested M., Lukas J., Bartek J.. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 2005; 25:3553–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M.B., Bartek J., Lukas J.. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006; 173:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maya-Mendoza A., Olivares-Chauvet P., Kohlmeier F., Jackson D.A.. Visualising chromosomal replication sites and replicons in mammalian cells. Methods. 2012; 57:140–148. [DOI] [PubMed] [Google Scholar]
- 24. Lukas C., Savic V., Bekker-Jensen S., Doil C., Neumann B., Pedersen R.S., Grofte M., Chan K.L., Hickson I.D., Bartek J. et al. . 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 2011; 13:243–253. [DOI] [PubMed] [Google Scholar]
- 25. Zhang D., Wang H., Sun M., Yang J., Zhang W., Han S., Xu B.. Speckle-type POZ protein, SPOP, is involved in the DNA damage response. Carcinogenesis. 2014; 35:1691–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marzahn M.R., Marada S., Lee J., Nourse A., Kenrick S., Zhao H., Ben-Nissan G., Kolaitis R.M., Peters J.L., Pounds S. et al. . Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 2016; 35:1254–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kwon J.E., La M., Oh K.H., Oh Y.M., Kim G.R., Seol J.H., Baek S.H., Chiba T., Tanaka K., Bang O.S. et al. . BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J. Biol. Chem. 2006; 281:12664–12672. [DOI] [PubMed] [Google Scholar]
- 28. Chen M.H., Wilson C.W., Li Y.J., Law K.K., Lu C.S., Gacayan R., Zhang X., Hui C.C., Chuang P.T.. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009; 23:1910–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jin X., Wang J., Gao K., Zhang P., Yao L., Tang Y., Tang L., Ma J., Xiao J., Zhang E. et al. . Dysregulation of INF2-mediated mitochondrial fission in SPOP-mutated prostate cancer. PLos Genet. 2017; 13:e1006748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu K., Lei P.J., Ju L.G., Wang X., Huang K., Yang B., Shao C., Zhu Y., Wei G., Fu X.D. et al. . SPOP-containing complex regulates SETD2 stability and H3K36me3-coupled alternative splicing. Nucleic Acids Res. 2017; 45:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bartkova J., Horejsi Z., Koed K., Kramer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C. et al. . DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005; 434:864–870. [DOI] [PubMed] [Google Scholar]
- 32. Kurfurstova D., Bartkova J., Vrtel R., Mickova A., Burdova A., Majera D., Mistrik M., Kral M., Santer F.R., Bouchal J. et al. . DNA damage signalling barrier, oxidative stress and treatment-relevant DNA repair factor alterations during progression of human prostate cancer. Mol. Oncol. 2016; 10:879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Galganski L., Urbanek M.O., Krzyzosiak W.J.. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res. 2017; 45:10350–10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blazek D., Kohoutek J., Bartholomeeusen K., Johansen E., Hulinkova P., Luo Z., Cimermancic P., Ule J., Peterlin B.M.. The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011; 25:2158–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adamson B., Smogorzewska A., Sigoillot F.D., King R.W., Elledge S.J.. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012; 14:318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tanikawa M., Sanjiv K., Helleday T., Herr P., Mortusewicz O.. The spliceosome U2 snRNP factors promote genome stability through distinct mechanisms; transcription of repair factors and R-loop processing. Oncogenesis. 2016; 5:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wickramasinghe V.O., Savill J.M., Chavali S., Jonsdottir A.B., Rajendra E., Gruner T., Laskey R.A., Babu M.M., Venkitaraman A.R.. Human inositol polyphosphate multikinase regulates transcript-selective nuclear mRNA export to preserve genome integrity. Mol. Cell. 2013; 51:737–750. [DOI] [PubMed] [Google Scholar]
- 38. Daboussi F., Courbet S., Benhamou S., Kannouche P., Zdzienicka M.Z., Debatisse M., Lopez B.S.. A homologous recombination defect affects replication-fork progression in mammalian cells. J. Cell Sci. 2008; 121:162–166. [DOI] [PubMed] [Google Scholar]
- 39. Michl J., Zimmer J., Buffa F.M., McDermott U., Tarsounas M.. FANCD2 limits replication stress and genome instability in cells lacking BRCA2. Nat. Struct. Mol. Biol. 2016; 23:755–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maya-Mendoza A., Petermann E., Gillespie D.A., Caldecott K.W., Jackson D.A.. Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO J. 2007; 26:2719–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Petermann E., Maya-Mendoza A., Zachos G., Gillespie D.A., Jackson D.A., Caldecott K.W.. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol. Cell. Biol. 2006; 26:3319–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wilsker D., Petermann E., Helleday T., Bunz F.. Essential function of Chk1 can be uncoupled from DNA damage checkpoint and replication control. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20752–20757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goodwin J.F., Schiewer M.J., Dean J.L., Schrecengost R.S., de Leeuw R., Han S., Ma T., Den R.B., Dicker A.P., Feng F.Y. et al. . A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov. 2013; 3:1254–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jin F., Fondell J.D.. A novel androgen receptor-binding element modulates Cdc6 transcription in prostate cancer cells during cell-cycle progression. Nucleic Acids Res. 2009; 37:4826–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liontos M., Koutsami M., Sideridou M., Evangelou K., Kletsas D., Levy B., Kotsinas A., Nahum O., Zoumpourlis V., Kouloukoussa M. et al. . Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 2007; 67:10899–10909. [DOI] [PubMed] [Google Scholar]
- 46. Sideridou M., Zakopoulou R., Evangelou K., Liontos M., Kotsinas A., Rampakakis E., Gagos S., Kahata K., Grabusic K., Gkouskou K. et al. . Cdc6 expression represses E-cadherin transcription and activates adjacent replication origins. J. Cell Biol. 2011; 195:1123–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Galanos P., Vougas K., Walter D., Polyzos A., Maya-Mendoza A., Haagensen E.J., Kokkalis A., Roumelioti F.M., Gagos S., Tzetis M. et al. . Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat. Cell Biol. 2016; 18:777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mateo J., Carreira S., Sandhu S., Miranda S., Mossop H., Perez-Lopez R., Nava Rodrigues D., Robinson D., Omlin A., Tunariu N. et al. . DNA-Repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 2015; 373:1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al. . 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016; 44:D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (49) partner repository with the dataset identifier PXD010527.