Abstract
Recently lncRNAs have been implicated in the sub-compartmentalization of eukaryotic genome via genomic targeting of chromatin remodelers. To explore the function of lncRNAs in the maintenance of active chromatin, we characterized lncRNAs from the chromatin enriched with H3K4me2 and WDR5 using chromatin RNA immunoprecipitation (ChRIP). Significant portion of these enriched lncRNAs were arranged in antisense orientation with respect to their protein coding partners. Among these, 209 lncRNAs, commonly enriched in H3K4me2 and WDR5 chromatin fractions, were named as active chromatin associated lncRNAs (active lncCARs). Interestingly, 43% of these active lncCARs map to divergent transcription units. Divergent transcription (XH) units were overrepresented in the active lncCARs as compared to the inactive lncCARs. ChIP-seq analysis revealed that active XH transcription units are enriched with H3K4me2, H3K4me3 and WDR5. WDR5 depletion resulted in the loss of H3K4me3 but not H3K4me2 at the XH promoters. Active XH CARs interact with and recruit WDR5 to XH promoters, and their depletion leads to decrease in the expression of the corresponding protein coding genes and loss of H3K4me2, H3K4me3 and WDR5 at the active XH promoters. This study unravels a new facet of chromatin-based regulation at the divergent XH transcription units by this newly identified class of H3K4me2/WDR5 chromatin enriched lncRNAs.
INTRODUCTION
High throughput global transcriptome analyses have identified that majority of the human genome is dynamically transcribed to encode a complex range of lncRNAs (1). Pathway-based and phenotype-based screening approaches have been the prominent strategies for the identification of functional lncRNAs (2–4). Mechanisms by which these functional lncRNAs execute key regulatory roles in diverse biological processes have been found to be dynamic and context specific (5–7). They have been shown to function as the regulators of transcription and translation, as stabilizers of transcripts and proteins, as scaffolds facilitating the assembly of protein complexes and aid in the recruitment of the transcription factors/chromatin regulators to specific genomic regions (8). From the previous evidence, it is apparent that many examples ascribe a function of lncRNAs in guiding epigenetic effector complexes, in cis or trans, to catalyze epigenetic modifications in a sequence specific fashion across the genome (9,10). These functional precedents by several lncRNAs increase the potential of lncRNAs and chromatin modifying enzymes to collectively constitute a regulatory network with the requisite complexity to delineate dynamic epigenetic landscapes.
Chromatin, which fine-tunes gene expression in a spatio-temporal fashion is organized into active and inactive chromatin compartments with distinct epigenetics codes. Active chromatin is characterized by the presence of histone marks such as H3K4me1, H3K4me2, H3K4me3, H3K36me3 and H3K27ac (11–13). These modifications have been shown to be specifically enriched in the active chromatin regions such as promoters, enhancers and actively transcribed regions. Of these, H3K4me2 and H3K4me3 are promoter specific chromatin marks and the rest are used to define different types of enhancers (active, poised and inactive) (14,15). Inactive chromatin on the contrary is marked by histone modifications such as H3K27me3 and H3K9me3 (16). These active and inactive chromatin-specific histone codes are defined by chromatin remodelers in a tissue and developmental stage dependent manner (17). Although the functional role of various chromatin remodelers in defining different epigenetic states is well researched but how chromatin remodelers are targeted to specific genomic portions has not been investigated enough.
Recent evidences suggest that lncRNAs constitute an important feature in the targeting of chromatin modifiers across the genome (18,19). Thus, there is a growing interest to address the functional contribution of lncRNAs in defining the chromatin sub-compartmentalization of the eukaryotic genome. Consistent with this notion, recent investigations have characterized the lncRNAs enriched in interphase chromatin, repressive chromatin and mitotic chromosomes (20–22). Intergenic10 lncRNA, purified from interphase chromatin, shown to positively regulate the neighboring genes in cis while MEG3 lncRNA, purified from repressive chromatin, regulates its target genes in trans via triplex formation. Nevertheless, both these lncRNAs have been shown to regulate their target genes at transcriptional level through modulating chromatin structure (20,21).
In order to understand how lncRNAs maintain chromatin structures at the gene regulatory regions, one needs to understand the mode of action of lncRNAs both from repressive and active chromatin compartments. Active and inactive chromatin-specific chromatin modifiers WDR5 and EZH2, respectively, have been shown to collaborate with lncRNAs for their targeting to various regions across the genome (23,24). WDR5, which is required for the maintenance of H3K4me1, H3K4me2 and H3K4me3 chromatin modifications, has been shown to interact with lncRNA for its stabilization. The binding of lncRNAs to WDR5 confers its chromatin localization whereas mutant WDR5 lacking RNA binding fail to localize to chromatin and gets rapidly degraded (25). Interestingly, WDR5 has been shown to specifically interact with H3K4me2 and nucleate the assembly of MLL-SET1 complexes to catalyze the transition of H3K4me2 to H3K4me3 (26,27). However, no study has addressed the possible role of lncRNAs in the functional interactions between WDR5 and H3K4me2. Although both H3K4me2 and H3K4me3 have concordant enrichment patterns (28,29), H3K4me2 seem to be enriched at specific genomic regions. For example, H3K4me2 marks have been shown to define transcription factor binding regions (30) and also marks for tissue-specific gene regulation (31). Thus, exploring the possible role of lncRNA mediated interactions between WDR5 and H3K4me2 may reveal hitherto unknown role of H3K4me2/lncRNA/WDR5 nexus in the establishment of active chromatin structures.
To this end, we have used an optimized chromatin RNA immunoprecipitation method (ChRIP) as described previously (21,32) to characterize lncRNAs associated with different chromatin compartments. In this study, we have characterized lncRNAs from the chromatin enriched with both H3K4me2 and WDR5 to understand their potential role in the chromatin-dependent gene regulation.
MATERIALS AND METHODS
Breast cancer cell lines
BT-549 (CLS-300312; CLS cell line service, Germany), MDA-MB-231 (ACC-732; American Type Culture Collection, USA) and Hela (ATCC CCL-2) cell lines were cultured in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen) and 1% antibiotic cocktail (Pen-Strep, Invitrogen) at 37°C with 5% CO2 supply.
Chromatin RNA Immuno-Precipitation
Chromatin RNA immunoprecipitation (ChRIP) was performed as per the published protocol (32) using antibodies anti-H3K4me2 (Abcam: ab7766), anti-WDR5 (Abcam: ab56919) and anti-IgG (Millipore; 12-370). In brief, BT-549 cells were treated with 5 μg/μl of Actinomycin D (ActD) (Sigma) for 40 min to inhibit nascent transcription. ActD treated cells were first cross-linked (20 × 106 cells were fixed in 20 ml volume) with 1% formaldehyde (Sigma-) at room temperature (RT) for 10 min and the crosslinking reaction was quenched with Glycine at 125 μM of final concentration (Sigma-) at RT for 5 min. These cells were additionally cross-linked with UV light on ice. The later steps in the ChRIP protocol were strictly followed according to the published protocol. Ten percent input was used to calculate the percentage of transcript bound to chromatin in the H3K4me2 and WDR5 pull-down chromatin fractions as compared to the negative control IgG. Fold enrichment was normalized to GAPDH.
UV crosslinked RNA immunoprecipitation (UV-RIP)
BT-549 cells treated with Actinomycin-D for 1 h were cross linked using ultra violet wavelength of light, washed with 1× PBS (3×) and re-suspended in modified RIPA lysis buffer (150 mM NaCl, 50 mM Tris, 0.5% sodium deoxycholate, 0.2% SDS, 1% NP-40) supplemented with RNase inhibitors (Ambion, AM2694) and protease inhibitors. Cell suspension was sonicated using a Bioruptor Pico Ultra sonicator for 3 × 30 s cycles. 10 μl of DNase (Promega, M610A) was added to sonicated material, incubated at 37°C for 10 min, and spun down at max speed for 10 min at 4°C. Protein-A Dyna-beads were washed and pre-incubated with 4 μg of WDR5 antibody for minimum of 2 h at 4°C. Lysate and beads were incubated at 4°C for 2 h. Beads were washed 3× using wash buffer 1 (1 × PBS, 0.1% SDS, 0.5% NP-40) followed by 2× using a wash buffer 2 (5 × PBS, 0.1% SDS, 0.5% NP-40) and crosslinking was reversed with NaCl and proteins were digested with 5 μl proteinase-K (Invitrogen, PN100005393) at 55°C for 1 h. RNA was extracted using TRIzol Reagent (Ambion, AM9738).
Chromatin oligo affinity precipitation (ChOP)
The ChOP assay was performed as described before by Akhade et al. (33) with some modifications. BT-549 cells (20 × 106) were treated with 5μg/μl ActD for 1 h and the ActD treated cells were cross-linked using 1% formaldehyde for 10 min at room temperature followed by quenching (using 0.125 M glycine). The cross-linked cell pellet was obtained by centrifugation at 1000g at 4°C for 10 min. Cells were resuspended in 2 ml of buffer A (3 mM MgCl2, 10 mM Tris–HCl, pH 7.4, 10 mM NaCl, 0.5% v/v NP-40, 0.5 mM PMSF and 100 units/ml RNasin) and incubated on ice for 20 min. Nuclei were harvested by centrifugation and resuspended in 1.2 ml of buffer B (50 mM Tris–HCl, pH 7.4, 10 mM EDTA, 0.5% Triton X-100, 0.1% SDS, 0.5 mM PMSF and 100 units/ml RNasin) and incubated on ice for 40 min. An equal volume of buffer C (15 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5 mM PMSF and 100 units/ml RNasin) was then added and incubated on ice for 15 min. Samples were sonicated using a Bioruptor sonicator (Diagenode) for 45 cycles (30 s on, 30 s off at High Pulse). Nine different oligos complimentary to FOXD3-AS1 and 15 different oligos complimentary to both HOXC13-AS and GATA6-AS1 were pooled with a final concentration of 10 μM and then used for the RNA pull down. As a control, a pool of nine oligos in reverse orientation targeting to the FOXD3-AS1 and 15 oligos in reverse orientation to both the HOXC13-AS and GATA6-AS1 were used. The oligos were added to the chromatin solution along with yeast tRNA (100 μg/ml), salmon sperm DNA (100 μg/ml), BSA (400 μg/ml) and incubated overnight at 4°C. Samples were then incubated with streptavidin agarose beads for 3 h followed by two washes of low salt buffer (20 mM Tris–HCl, pH 7.9, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% TritonX-100, 0.5 mM PMSF and 50 units/ml RNasin), two washes of high salt buffer (20 mM Tris–HCl, pH 7.9, 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100, 0.5 mM PMSF and 50 units/ml RNasin) and one wash of 1× PBS. The beads were then incubated with elution buffer (1% SDS, 100 mM NaHCO3) for 45 min with intermittent mixing at 65°C followed by Proteinase K (80 μg/ml) digestion and DNA was isolated using phenol chloroform extraction. The fold enrichment over input was calculated considering the percentage of input chromatin used for ChOP and the Ct values obtained for the target from Input DNA and ChOP DNA as follows. Fold enrichment over input = % of input × 2[Ct(input) − Ct(ChOP)].
We checked the chromatin enrichment of the active XH lncCARs at the promoters of their partner protein coding genes with primers spanning –1 kb to +1.7 kb from the transcription start site (TSS) and also beyond transcription termination site (TTS) of the respective genes. The location of the primers with respect to TSS and TTS are mentioned in Dataset S9.
ChRIP library preparation and RNA sequencing
Since RNA yield from each ChRIP pull-down was not sufficient for library preparation to perform high-throughput RNA-sequencing using SOLiD (Applied Biosystem) sequencing platform, we had to pool the purified RNA for each antibody from three independent biological pull-down ChRIP experiments. The RNA sequencing library preparation for SOLiD high throughput RNA sequencing was done following standard library preparation protocol using SOLiD Total RNA-Seq Kit. The RNA sequencing was done at Uppsala Genome Center, Sweden.
RNA interference (RNAi)
We randomly chose ten XH loci with antisense lncRNAs and sense protein coding partner genes, encoding transcription factors. We custom designed five small interfering RNAs (siRNAs) for each lncRNA and transfected them along with control siRNA (Invitrogen) in both BT-549 and MDA-MB-231 cell lines. We could downregulate five out of ten lncRNAs by at least by one siRNA in one of the two cell lines. We downregulated mRNAs of protein coding partner genes such as HOXC13 and GATA6 using esiRNA pool (Sigma) while FOXD3 was depleted using two siRNAs (Sigma). We used esiRNA against EGFP as a negative control. We transfected 50 pmol of each siRNAs per 70 000–80 000 cells. For esiRNA, we had used 1.4 μg per 70 000–80 000 cells. We transfected siRNAs using RNAi Max (Invitrogen) or Lipofectamine 3000 (Invitrogen). We found RNAi Max to be more efficient in downregulating the active XH lncCARs and for protein coding mRNAs, both the transfection reagents were equally efficient. For WDR5 depletion we used three independent commercially available siRNAs from Invitrogen in the same way as described above for other genes. All siRNA sequences are provided in Dataset S9.
Subcellular fractionation
Subcellular fractionation was performed as prescribed earlier with following modifications(21). Briefly, approximately 1 × 106 BT-549 cells were washed with ice cold 1× PBS and resuspended in 175 μl of cold cytoplasmic lysis buffer (50 mM Tris–HCl pH 8.0; 140 mM NaCl; 1.5 mM MgCl2; 0.5% NP-40; supplied with RNase inhibitor) and incubated for 5 min on ice. Cytoplasmic fraction was separated as supernatant by centrifugation at 500g at 4°C for 4 min. Nuclear pellet was re-suspended in cold PBS and pelleted with high-speed centrifugation (5000 rpm) for 1 min. RNA was extracted from these two fractions using the TRIzol reagent (Life Technologies).
Chromatin immuno precipitation (ChIP)
ChIP was performed using kit for transcription factors (C01010055) and histone modifications (C01010073) as per manufacture's (Diagenode) recommendations using anti-H3K4me2 (Abcam: ab7766), anti-WDR5 (Abcam: ab56919), anti-H3K4me3 (Millipore: CS200554) and control IgG (Millipore; 12-370) antibodies. Briefly, BT-549 cells (2 × 106) were seeded in 10cm petri dish and transfected with 500 pmol of negative control or 500 pmol of siRNAs Cells treated with control and target-specific siRNA were harvested, and equal number of cells were cross-linked with 1% formaldehyde at room temperature for 5 min. Cross-linking was quenched using 125 μM of Glycine. Cells were lysed in lysis buffer using the manufacturer's recommendation and was sonicated for 30 cycles (30 s on, 30 s off in Diagenode Bioruptor Plus) to obtain fragment size of 100–300 bp. 3 μg of antibody was used per 7–10 μg of sonicated chromatin and binding reaction was carried out overnight. The steps following overnight incubation was performed as per the manufacturer's recommendations. ChIP DNA and Input was finally eluted in 250 μl volume. 10% of input from all the reactions was used for calculating the fold enrichment. 5 μl of purified DNA was used as template in qPCR reactions. Fold enrichment was normalized to the Input. A couple of primer pairs spanning the 1.5 kb protein coding gene promoter region was used to calculate H3K4me2, H3K4me3 and WDR5 enrichment. Primer pair spanning upstream non-promoter region was used as a negative control to determine the specificity of antibodies used (Refer primer sequence table from Dataset S9).
For ChIP sequencing using BT-549 cells with H3K4me2, H3K4me3 and WDR5 antibodies, we used the same ChIP kit that we used for ChIP-qPCR experiments with suggested scale up as per as manufacturer's recommendations.
LNA modified antisense GapmeRs treatment
We synthesized two antisense LNA GapmeRs (LNAs) each for GATA6-AS1, HOXC13-AS and FOXD3-AS1 along with negative control LNA (Refer oligo sequence table from Dataset S9). To downregulate GATA6-AS1, HOXC13-AS and FOXD3-AS1 lncRNAs, 20 pmol of LNA was transfected per 70 000–80 000 cells using RNAi Max (Life Technologies) for 48 h.
Quantitative RT-qRCR analysis
Total RNA was isolated from cells using Relia RNA isolation kit (Promega) following manufacturer's directions. Reverse transcription of RNA was carried out using InpromtII transcriptase (Promega). For detecting mature XH lncCARs and their protein coding gene pairs, exon-spanning primers were designed distal to the promoter. Gene expression was normalized to GAPDH. Error bars in RT-PCR analysis are standard deviations of mean expression relative to GAPDH from three independent experiments. Primer sequences used are given in Dataset S9. The level of nascent transcripts was measured using 5-ethynyl uridine (EtU) labeling and biotin tagging by Click-iT chemistry following protocol described in previous study (34).
Active XH lncCAR overexpression
Full length transcripts of HOXC13-AS and FOXD3-AS1 were amplified from total cDNA of BT-549 cell line using primers containing restriction enzyme recognition sequences for HindIII and NotI as described in Dataset S9. Both the amplified full-length transcripts were digested with HindIII and NotI and each transcript was cloned separately in HindIII/NotI linearized pcDNA3.1 (Invitrogen). HOXC13-AS and FOXD3-AS1 pcDNA3.1 constructs were transiently transfected into BT-549 cells using RNAi Max (Invitrogen) and cells were harvested for RNA isolation after 2 and 5 days post-transfection. RNA isolation and cDNA preparations were done as meontined above to check for the expression of ecopically expressed active XH CARs and also the expression of their respective protein coding genes.
Western Blotting
BT-549, MDA-MB231 and Hela cells from the outlined experiments were harvested, washed once with PBS and then lysed in lysis buffer containing 20 mM Tris, pH 8.0, 250 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.5% Triton X-100, 1 mM PMSF and 1× Protease inhibitor cocktail for 45 min at 4°C. The protein lysates were separated on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane. The membrane, after blocking in 5% skim milk (in PBS), was incubated with primary antibody overnight at 4°C. The membrane was then washed thrice in PBS–Tween (PBST) followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibody. After three washes in PBST, the blot was subjected to chemiluminescence using luminol as the substrate. The antibodies were used at the following dilutions: WDR5 (1:1000), α-Tubulin (1:5000), HOXC13 (Abcam: ab188043, 1:1000), FOXD3 (Cell Signaling #2019, 1:1000), FLAG (Sigma F7425, 1:1000), HRP-conjugated secondary antibody (1:5000).
Formaldehyde crosslinked RNA immunoprecipitation (f-RIP)
HeLa cells were transfected with 2 μg/ml of either FLAG-HA WDR5 WT or FLAG-HA WDR5 F266A plasmids. After 48 h of transfections cells were harvested. Approximately 1% of harvested cells were used for western analysis to detect induction of FLAG-HA WDR5 using anti-FLAG antibody (Sigma F7425). Roughly 12 × 106 cells were cross-linked with 1% formaldehyde at room temperature for 5 min. Cross-linking was quenched using 125 μM of Glycine, washed three times with 1× cold PBS and re-suspended in modified RIPA lysis buffer (150 mM NaCl, 50 mM Tris, 0.5% sodium deoxycholate, 0.2% SDS, 1% NP-40) supplemented with RNase inhibitors (Ambion, AM2694) and protease inhibitors. Cell suspension was sonicated using a Bioruptor Pico Ultra sonicator for 15 × 30 s cycles. 10 μl of DNaseI (Promega, M610A) was added to sonicated material, incubated at 37°C for 10 min, and spun down at max speed for 10 min at 4°C. Protein-A Dyna-beads were washed and pre-incubated with 4 μg of anti-FLAG antibody for 2 h at 4°C. Lysate and beads were incubated at 4°C for 2 h. Beads were washed three times with wash buffer 1 (1 × PBS, 0.1% SDS, 0.5% NP-40) and two times with wash buffer 2 (5 × PBS, 0.1% SDS, 0.5% NP-40) and cross-links were reversed and proteins were digested with 5 μl proteinase-K (Invitrogen, PN100005393) at 55°C for 1 h. RNA was extracted using TRIzol reagent (Ambion, AM9738).
Computational analysis for characterizing active chromatin associated RNAs (active CARs)
Sequencing, alignment and quantification of ChRIP-seq
The ChRIP samples for WDR5, H3K4me2 and nuclear RNA (input) were sequenced using Applied Biosystems SOLID platform by preparing strand specific library protocol according to the manufacturer's recommendations. Obtained sequencing reads (25–75 nucleotides long) were aligned to the reference hg19 genome using Lifescope 2.5 [http://www.lifetechnologies.com/lifescope], splice junction-aware aligner from AB SOLID with default parameters (number of mismatches allowed: 2). The alignment files were further processed for quality filtering (mapping quality, MAPQ ≥ 30; maximum mapping quality in SOLID, MAPQ = 50) using SAMtools (35) to avoid multiple mapped reads and obtained 6 259 178, 6 112 539 and 8 084 998 mapped reads in WDR5, H3K4me2 and nuclear RNA (Input) samples, respectively. This constitute 28%, 34.8% and 38% of total mapped reads from WDR5, H3K4me2 and nuclear input RNA. Later featureCounts (36) from Subread package was used to quantify expression (number of exonic reads mapped per transcript) of lncRNAs and protein coding genes (PCGs) using GENCODE (V19, equivalent to Ensembl GRCh37.74) annotation (37) in individual samples (-s 1 -t exon -Q 30). Obtained reads were normalized by Reads per Kilobase per Million (RPKM) normalization (https://github.com/santhilalsubhash/rpkm_rnaseq_count). Log-fold changes were calculated from RPKM values by considering Input sample as control (log2 [H3K4me2/Input] and log2 [WDR5/Input]). We used log-fold change >1 (fold-change > 2) to assign H3K4me2 and WDR5 associated lncRNAs and PCGs.
Neighboring gene analysis and classification
We looked for the genomic arrangement of H3K4me2 and WDR5 enriched transcripts in relation to the neighboring protein coding genes and lncRNAs based on first hit in a window of 50 kb regions using BEDtools (38) closest' (increasing >50 kb did not change the results significantly). We categorized the pairs into two major classes such as antisense (X) and sense (S) pairs based on the orientation towards their partners. There are few transcripts among the H3K4me2 and WDR5 transcripts having both antisense and sense partners (ambiguous paring) and to avoid any bias these transcripts were counted in both antisense and sense classification. Among antisense partners, there were H3K4me2 or WDR5 associated lncRNAs having protein coding genes as partners (pPCG), H3K4me2 or WDR5 associated lncRNAs having other lncRNAs as partners (pLncRNA), H3K4me2 or WDR5 associated PCGs having other lncRNAs as partners (pLncRNA) and H3K4me2 or WDR5 associated PCGs having other PCGs as partners (pPCG).
Within the antisense pattern, we found further four sub-patterns. For example, in H3K4me2 or WDR5 lncRNAs we found: XH, head-to-head divergent transcripts; XT, tail-to-tail; XI, lncRNA-Inside (lncRNA within a protein-coding gene); XO, lncRNA-Out (protein coding gene within a lncRNA). The other categories include XH-dist, if the promoters of head-to-head are separated by >2 kb and within 50 kb window. And the sense pattern was divided into two categories such as sense (within 2 kb distance) and sense-dist, if the genes are separated by >2 kb distance within 50 kb window. Similar classifications were also done for protein coding genes enriched in H3K4me2 and WDR5. In addition to the above analysis, we used non-chromatin enriched RNAs as control groups or background. Non-chromatin enriched RNAs were selected if they have at least one read (minimum expression) assigned in any of ChRIP samples used in this study (H3K4me2, WDR5 or nuclear input samples).
Functional enrichment analysis of neighboring protein coding genes
Gene enrichment analysis of neighboring protein coding genes to H3K4me2 and WDR5 lncRNAs was performed using GeneSCF v1.1 (39) and Gene Ontology processes were considered significant if there is at least 5 genes associated with the process and having P-value <0.05. We have performed enrichment analysis using different Gene Ontology annotations such as, Biological Process (BP) and Cellular Component (CC). There were 14 493 protein coding genes showing minimal expression in at least one sample (H3K4me2, WDR5 or nuclear input RNA ChRIP sample) were considered as background set.
For protein coding genes enriched in H3K4me2 (H3K4me2 PCGs) and WDR5 (WDR5 PCGs) ChRIP pull-downs, we performed gene enrichment analysis for H3K4me2 and WDR5 PCGs as well as for their partner protein coding genes.
ChIP-seq data processing and analysis
Initial quality checks for ChIP-seq was performed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to check for adapters. Obtained clean reads were aligned to hg19 genome with bowtie aligner (40). The duplicate sequences or reads from the samples were removed using SAMtools and picard (http://broadinstitute.github.io/picard/) utility. The blacklisted regions of hg19 genome (http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/) were removed using ‘bamutils’ from NGSUtils. To obtain log2-ratio of H3K4me2 and H3K4me3 sample over control/input sample, we used bamCompare from deepTools suite (41). For visualization of enrichments, deepTools utilities computeMatrix and plotProfile were used. In addition, we have analyzed ENCODE (42) ChIP-seq dataset H3K4me2 (ENCFF000BJF) and corresponding control sample (ENCFF000BHF) from human mammary epithelial cells (HMEC) and GEO (GSE55279) (43) dataset of WDR5 ChIP-seq from LnCaP cellline and corresponding control samples with the same approach used for BT-549 ChIP-seq analysis. All the ChIP-seq line plots were smoothened using ggplot2.
RESULTS
Characterization of H3K4me2 associated RNAs
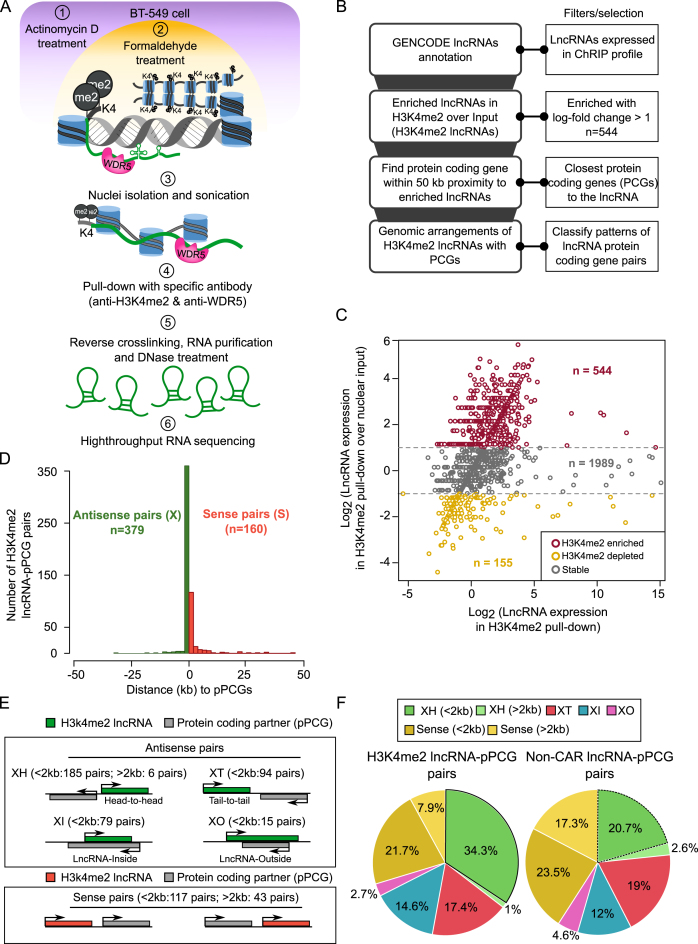
Previously, using ChRIP, we have characterized the chromatin association property of the human KCNQ1OT1 antisense lncRNA (44,45). Using a modified ChRIP protocol followed by high-throughput sequencing (ChRIP-seq), later we identified lncRNAs that are associated with repressive chromatin on a global scale (21). Here we used this modified ChRIP-seq protocol to characterize H3K4me2 enriched chromatin associated lncRNAs in breast cancer cell line BT-549 treated with Actinomycin D (ActD) to prevent any co-transcriptional crosslinking of lncRNAs to chromatin. The efficacy of the transcriptional arrest by ActD was tested by purifying nascent transcripts, prior and after ActD treatment, following 5-ethynyl uridine (EtU) labeling and biotin tagging by Click-iT chemistry (Supplementary Figure S1A) (34). We found that ActD treatment for 1 hour could significantly block de novo transcription of protein coding genes (GAPDH and HOXC13), ribosomal RNAs (45S, 28S and 18S) and lncRNAs (RP4-792G4.2 alias FOXD3-AS1 and HOXC-AS5 alias HOXC13-AS) (Supplementary Figure S1A). Following cross-linking with formaldehyde, the cells were sonicated and the chromatin was immunoprecipitated using antibodies against H3K4me2. From the H3K4me2 immunopurified chromatin, collected from three independent biological replicates, RNA was isolated, DNaseI-treated and then subjected to high-throughput sequencing along with nuclear input RNA (Figure 1A). We have considered annotation from GENCODE (V19) (37) and the transcripts which are expressed in either nuclear input or H3K4me2 ChRIP purified samples were used for further analysis (Figure 1B, see Dataset S1). Among these expressed transcripts, 544 lncRNAs (H3K4me2 lncRNAs) and 2293 protein coding genes (H3K4me2 PCGs) were enriched by two-fold in the H3K4me2 ChRIP compared to nuclear input (Figure 1C, see Dataset S2).
Figure 1.
Characterizing H3K4me2 enriched lncRNAs. (A) ChRIP experimental workflow to identify lncRNAs bound to chromatin enriched with H3K4me2 and WDR5 in BT-549 cell line. (B) Computational approach used for finding chromatin-associated RNAs and enriched patterns of genomic organization with respect to nearest protein coding genes. (C) Scatter plot showing the enrichment of H3K4me2 lncRNAs over input. X-axis denotes log transformed expression values of H3K4me2 lncRNAs and Y-axis denotes log-fold changes of lncRNAs in H3K4me2 ChRIP sample over nuclear input sample. (D) Histogram shows distribution of H3K4me2 lncRNAs with antisense (X) and sense (S) pattern with respect to their protein coding partner (pPCGs). ‘n’ denotes number of lncRNA-pPCG pairs in each pattern. (E) Genomic organization of H3K4me2 lncRNAs (green bars) with respect to nearest protein coding genes (grey bars). XH: where lncRNA shows Head-to-Head arrangement with nearby protein coding gene. XT: lncRNA in Tail-to-Tail arrangement with protein coding gene, XI: lncRNA located inside a protein coding gene, XO: lncRNA located outside and covers the entire protein coding gene. Sense pairs means lncRNAs in the same orientation as protein coding gene. The notion of greater or less than 2 kb means that the partner genes (pPCGs) are located within 2 kb (<2 kb) or away from 2 kb (> 2 kb) but within 50 kb window with respect to H3K4me2 lncRNAs. (F) Distribution of different patterns of genomic arrangements as described in (E) for H3K4me2 lncRNAs and non-CAR lncRNAs.
Next, we characterized these H3K4me2 enriched RNAs based on their genomic arrangements with respect to the nearest transcripts, which either partner with lncRNAs (pLncRNAs) or partner with protein coding genes (pPCGs). Based on this, H3K4me2 associated RNAs were categorized as: (i) H3K4me2 lncRNAs paired with protein coding genes (lncRNA-pPCG), (ii) H3K4me2 lncRNAs paired with other lncRNAs (lncRNA-pLncRNA), (iii) H3K4me2 PCGs paired with lncRNAs (PCG-pLncRNA), (iv) H3K4me2 PCGs paired with other protein coding genes (PCG-pPCG). Non-chromatin enriched RNAs (non-CARs) that are expressed in BT-549 were used as background (please see methods section). Non-CARs contain both lncRNAs (non-CAR lncRNAs) and protein coding genes (non-CAR PCGs). The genomic distribution of H3K4me2 enriched RNAs can be broadly classified into two types: transcripts that are transcribed either in antisense direction (designated ‘X’) or in sense direction (designated ‘S’) to their partners. Interestingly, ∼70% of the H3K4me2 lncRNAs and H3K4me2 PCGs were organized in an antisense orientation to their partner PCGs (pPCGs) and partner LncRNAs (pLncRNAs), respectively, suggesting that antisense organization is prominent among H3K4me2 enriched RNAs (Figure 1D and Supplementary Figure S1B).
H3K4me2 lncRNAs have non-random genomic distribution
The antisense organization of the H3K4me2 RNAs was further sub-classified into four categories based on a previous study (Figure 1E, see methods) (46). About 34.3% of the H3K4me2 lncRNAs have divergent organization (XH category: head-to-head) to their partner PCGs (H3K4me2 lncRNAs-pPCGs) as compared to 20.7% of non-CAR lncRNAs paired with coding genes (non-CAR lncRNAs-pPCGs) (hypergeometric P-value (XH) < 5.8E–112) (Figure 1F). This divergent organization was also not prominent in the case of other enriched pairs such as lncRNA–pLncRNA pairs (15.9%), PCG–pLncRNA pairs (21.4%) and PCG–pPCG pairs (15.6%) (Supplementary Figure S1C). These observations suggest that there is an overrepresentation of XH categories exclusively among the H3K4me2 lncRNAs (Figure 1F and Dataset S3). We name these H3K4me2 lncRNAs with XH transcription units as H3K4me2 XH lncRNAs. Interestingly, the other H3K4me2 lncRNAs from sense and other antisense (XT, XI and XO) categories located within a 2 kb window to the protein coding gene did not show any significant change in their distribution patterns compared to the non-CARs lncRNAs (Figure 1F), suggesting that enrichment of the H3K4me2 XH lncRNAs is not due to mere genomic proximity to protein coding genes.
H3K4me2 XH lncRNAs transcription units are associated with transcription factors
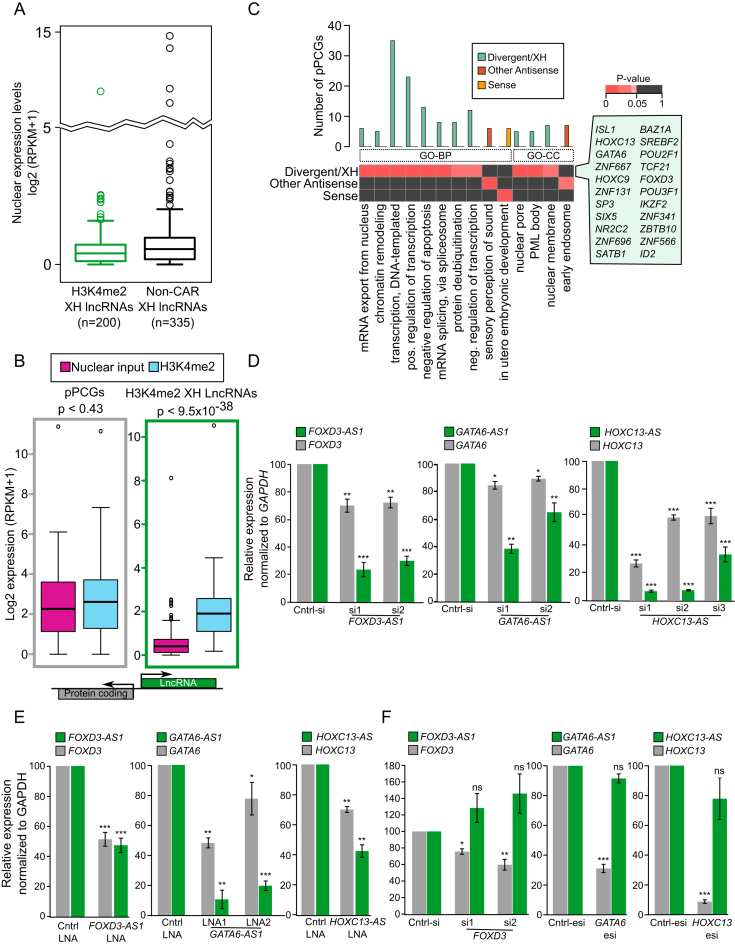
We next addressed whether the enrichment of H3K4me2 XH lncRNAs in our ChRIP pull-downs is not due to their higher expression in this cell line. For this we first compared the nuclear expression levels of H3K4me2 XH lncRNAs and non-CAR lncRNAs with the similar genomic organization (non-CAR XH lncRNAs) and we found both were expressed at similar levels in nuclear input (Figure 2A). Secondly, we compared the nuclear expression and chromatin enrichment of H3K4me2 XH lncRNAs and their protein coding partners (pPCGs). Although H3K4me2 XH lncRNAs have lower expression compared to pPCGs, the lncRNAs were found to be significantly enriched in our H3K4me2 ChRIP (P-value <3e–36) (Figure 2B). Taken together, these observations indicate that the chromatin enrichment of H3K4me2 XH lncRNAs is independent of their expression level. Functional enrichment analysis with the protein-coding gene partners of H3K4me2 XH lncRNAs revealed biological process (BP) terms related to transcription process (such as DNA-templated transcription, positive and negative transcriptional regulation) and apoptosis as the top enriched functions. However, these terms were not enriched with the coding partners (pPCGs) of other enriched antisense (XT + XI + XO) and sense categories of H3K4me2 lncRNAs (Figure 2C). Furthermore, these transcription factor related terms were also absent in H3K4me2 XH PCGs and non-CARs XH categories (Dataset S4). To verify whether the enrichment of the transcription related biological process terms is due to ActD-induced expression, we measured the steady state level of H3K4me2 lncRNAs and their coding partner (pPCGs) by qRT-PCR and they were found to be either unchanged or decreased. Moreover, we did not observe upregulation of the apoptosis related genes after ActD treatment (Supplementary Figure S2A). Also, cellular component (CC) terms for coding partner (pPCGs) of XH lncRNAs were enriched for distinct subcellular structures such as nuclear membrane, nuclear-pore and PML body (Figure 2C). Our results indicate that H3K4me2 XH lncRNAs may control important biological functions through regulating the expression of their coding partners (pPCGs) that encode important transcription factors (Figure 2C, Dataset S4). Though the other categories of H3K4me2 lncRNAs and H3K4me2 PCGs may have important regulatory functions, we have focused on understanding the function of overrepresented H3K4me2 XH lncRNAs category in the ChRIP pull-downs.
Figure 2.
H3K4me2 XH lncRNAs regulate the transcriptional activity of their protein coding partners. (A) Nuclear expression levels of H3K4me2 XH lncRNAs and non-CAR XH lncRNAs. (B) Expression of H3K4me2 XH lncRNA-protein coding pairs (pPCGs) in nuclear input and H3K4me2 ChRIP samples. The P-value denotes the significance of difference between nuclear input and H3K4me2. (C) Gene ontology based functional enrichment analysis of protein coding genes that are partners (pPCGs) to H3K4me2 lncRNAs: divergent (XH), other antisense (XT, XI, and XO) and sense lncRNAs. The bar graph shows number of genes in corresponding ontology term and the heatmap represents the significance of individual terms (P-values obtained using GeneSCF). The highlighted box, by the heatmap, is listed with different transcription factors from the enriched transcription related terms. (D) Gene expression analysis of three transcription factors (FOXD3, GATA6 and HOXC13) using RT–qPCR analysis following downregulation of their neighboring H3K4me2 XH lncRNAs (FOXD3-AS1, GATA6-AS1 and HOXC13-AS) with siRNA in BT-549 cells. LncRNA expression is in green bars while transcription factors expression is in grey. Data represent the mean ± SD. of two independent biological experiments. * P ≤ 0.05, ** P ≤ 0.01 and *** P ≤ 0.001. (E) RT–qPCR analysis of FOXD3, GATA6 and HOXC13 gene expression after strand-specific downregulation of FOXD3-AS1, GATA6-AS1 and HOXC13-AS respectively, using LNA. Data represent the mean ± SD. of two independent biological experiments. * P ≤ 0.05, ** P ≤ 0.01 and *** P ≤ 0.001. (F) Gene expression analysis of three H3K4me2 XH lncRNAs (FOXD3-AS1, GATA6-AS1 and HOXC13-AS) using RT–qPCR analysis following downregulation of their respective transcription factor partners with siRNA or esiRNA in BT-549 cells. LncRNAs expression is depicted as green bars while transcription factors expression is in grey bars. Data represent the mean ± SD. of two independent biological experiments. * P ≤ 0.05, ** P ≤ 0.01 and *** P ≤ 0.001. ns denotes non-significant.
H3K4me2 XH lncRNAs regulate nearby protein coding genes at the transcriptional level
Consistent with their chromatin enrichment, the H3K4me2 XH lncRNAs were found to be enriched in the nuclear fraction. Nuclear enrichment of the H3K4me2 lncRNAs from other categories (XT, XI and XO) were also very similar (Supplementary Figure S2B). Nuclear and chromatin enrichment of the H3K4me2 XH lncRNAs is suggestive of their transcriptional regulatory potential.
To address the functional role of some of these H3K4me2 XH lncRNAs in regulating their coding partners (pPCGs), we selected ten XH loci which encode transcription factors. We could downregulate five out of ten selected H3K4me2 XH lncRNAs, with at least by one siRNA, in BT-549 and MDA-MB-231 breast cancer cell lines. Downregulation of H3K4me2 XH lncRNAs FOXD3-AS1, GATA6-AS1, HOXC13-AS, POU2F1-AS (known as RP11-277B15.3) and HOXC-AS2 significantly affected the expression of their coding partner (pPCGs) FOXD3, GATA6, HOXC13, POU2F1 and HOXC9, respectively (Figure 2D and Supplementary Figure S3A). Unlike siRNAs, locked nucleic acid (LNA) modified antisense oligonucleotide gapmers degrade target RNA by recruiting RNaseH in a strand specific manner thereby providing an alternative method of strand specific gene silencing (47). Downregulation of FOXD3-AS1, GATA6-AS1 and HOXC13-AS using antisense LNA GapmeRs also resulted in the downregulation of their respective protein coding genes (Figure 2E). However, downregulation of all the five coding pairs (pPCGs) did not affect the expression of their respective XH lncRNAs (Figure 2F and Supplementary Figure S3B). Additionally, knockdown of the H3K4me2 XH PCGs did not show any consistent regulatory patterns in the transcriptional activity of their lncRNA partners (pLncRNAs). The pLncRNAs were showing increased, decreased or unaltered expression patterns following H3K4me2 XH PCGs knockdown (Supplementary Figure S3C). These observations suggest that H3K4me2 XH lncRNAs, investigated above, per se regulate their protein-coding gene partners (pPCGs) at the transcriptional level.
Characterization of WDR5-enriched chromatin interacting lncRNAs
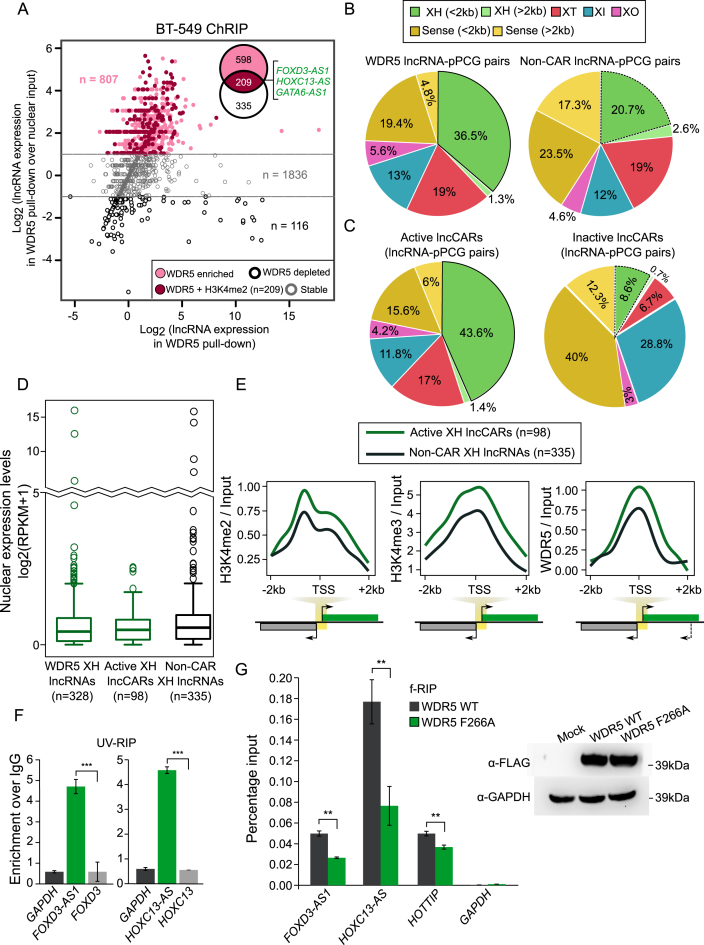
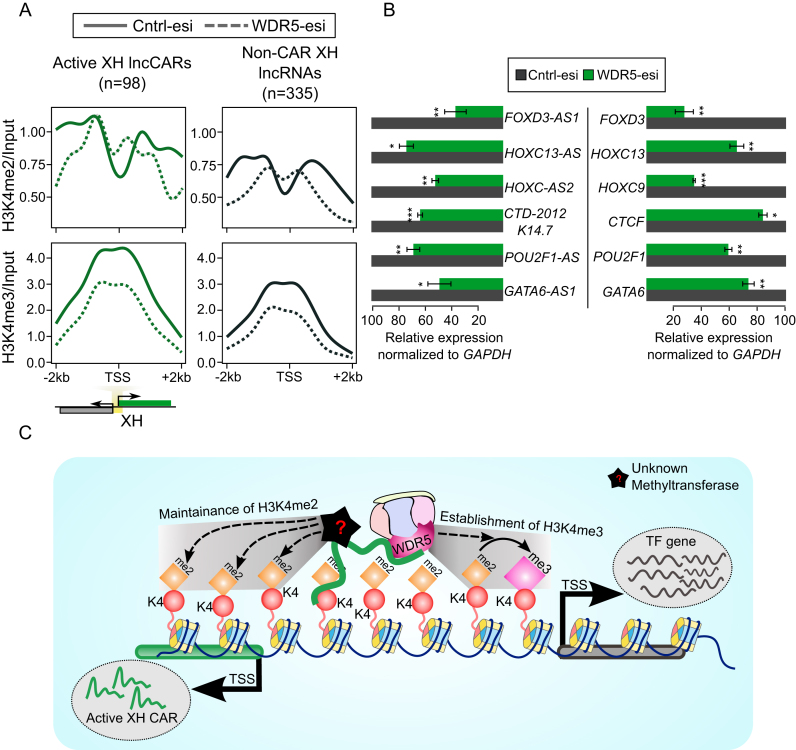
The role of some of the H3K4me2 XH lncRNAs in transcriptional regulation prompted us to look into the possible mechanism by which they carry out target gene regulation. Previously, it was proposed that WDR5, a critical component of active chromatin remodeling complexes such as MLL/SET1, binds directly to H3K4me2 and helps in the conversion of H3K4me2 to H3K4me3 (27). Hence using ChRIP we purified WDR5-enriched chromatin interacting RNAs in BT-549 cells (Figure 1A). Using the same approach as described earlier for characterizing the H3K4me2 RNAs, we identified 807 lncRNAs and 4381 protein coding genes to be significantly enriched in the WDR5 chromatin pull-down as compared to nuclear input (WDR5 lncRNAs and WDR5 PCGs) (Figure 3A and Dataset S5). Like in H3K4me2 enriched RNAs, >70% of the WDR5 enriched RNAs were also organized in an antisense orientation with their neighboring partners (Supplementary Figure S4A). 36.5% of the WDR5 lncRNAs have a XH organization as compared to the non-CAR lncRNAs (20.7%) and WDR5 PCGs paired with lncRNAs (20.6%) (Figure 3B, Supplementary Figure S4B, Dataset S6). Since both H3K4me2 and WDR5 lncRNAs exhibited similar genomic distribution i.e., enrichment of XH category, we analysed how many of the H3K4me2 XH lncRNAs were enriched with WDR5. We found that 209 of the H3K4me2 lncRNAs (around 50% of them, hypergeometric P-value < 2.08E–19) were also enriched in the WDR5 lncRNAs. We have termed these 209 lncRNAs as active chromatin associated lncRNAs (Active lncCARs) (Figure 3A). Active lncCARs were also organized in an antisense orientation with their neighboring partners (Supplementary Figure S4C). Interestingly, the distribution of XH lncRNAs in the active lncCARs (active XH lncCARs) is even more overrepresented (43.6%) compared to their distribution in the H3K4me2 lncRNAs (34.3%) and WDR5 lncRNAs (36.5%) (Figures 1F and 3B–C, Supplementary Figure S4D and Dataset S7).
Figure 3.
XH lncRNAs are overrepresented in active lncCARs. (A) Scatter plot showing enrichment of WDR5 lncRNAs over nuclear input. The X-axis denotes log transformed expression and Y-axis denotes log-fold change of WDR5 lncRNAs over nuclear input. Venn diagram, in the Scatter plot, shows 209 lncRNAs that are commonly enriched in both H3K4me2 and WDR5 ChRIP pulldowns. (B) Distribution of different patterns of genomic arrangements as described in Figure 1E for WDR5 lncRNAs and non-CAR lncRNAs. (C) Distribution of different patterns of genomic arrangements as described in Figure 1E for active lncRNA (ChRIP pulldown using H3K4me2 and WDR5 antibodies) and inactive lncRNA (ChRIP pulldown using EZH2 and H3K27me3 antibodies). (D) Nuclear expression levels of WDR5 XH lncRNAs, active XH lncCAR (WDR5 and H3K4me2) and non-CAR XH lncRNAs. (E) Enrichment of H3K4me2, H3K4me3, WDR5 ChIP-seq signals over active XH lncCAR (in green, n = 98) and non-CAR XH lncRNA (in black, n = 335) promoters (±2 kb) in BT-549 cells. The signals presented in the plots represent log2ratio between ChIP and input sample. TSS denotes the transcription start site of lncRNAs. (F) RT-qPCR analysis of WDR5 UV-RIP. The Y-axis shows fold enrichment relative to IgG. The significance of FOXD3-AS1 and HOXC13-AS enrichment in WDR5 RIP is calculated compared to the enrichment of FOXD3 and HOXC13 mRNAs, respectively. ***P ≤ 0.001. (G) Left panel: f-RIP of WDR5 WT and WDR5 F266A using FLAG antibody. The Y-axis shows percentage of Input. The significance of decrease in FOXD3-AS1, HOXC13-AS and positive control HOTTIP enrichment in WDR5 F266A RIP is calculated compared to their respective enrichments in WDR5-WT. GAPDH was used as negative control. Right panel: Expression of FLAG tagged WDR5 WT and WDR5 F266A mutant in HeLa cells by western blot. Data are shown as mean ± SD. **P ≤ 0.01.
Next, we investigated whether overrepresentation of XH lncRNAs is limited to active CARs or they are overrepresented among the other subtypes of chromatin interacting lncRNAs. Toward this, we have used our previously published repressive or inactive chromatin associated lncRNAs (inactive lncCARs) dataset, characterized using anti-H3K27me3 and anti-EZH2 ChRIP pull-downs in BT-549 cells (21). We found that the distribution of sense and antisense lncRNAs in the inactive lncCARs is in contrast to that seen in the active lncCARs. For example, XH transcription units were underrepresented among the inactive lncCARs 8.6%, which are far below in comparison to the XH transcription units among the active lncCARs 43.6%. This indicates that XH transcription units were more specifically enriched in the active lncCARs compared to the inactive lncCARs (Figure 3C). This comparison also revealed another interesting observation that sense category lncRNAs (within 2kb) showed overrepresentation (40%) in the inactive lncCARs compared to the sense active lncCARs (15.6%) (Figure 3C). This again suggest that specific enrichment of XH category in active lncCARs is independent of their proximity to nearby coding partners (pPCGs).
We then compared the nuclear expression levels of XH lncRNAs among WDR5 lncRNAs, active lncCARs and non-CAR lncRNAs. All the three categories of XH lncRNAs are expressed at similar levels (Figure 3D). Similar to H3K4me2 XH lncRNAs, active XH lncCARs, despite having similar nuclear expression, were significantly chromatin enriched compared to their pPCGs in both WDR5 and H3K4me2 pull-downs (Supplementary Figure S5A). This again suggests that chromatin enrichment of active XH lncCARs is independent of their expression level. Functional enrichment analysis with the protein coding partners (pPCGs) of the WDR5 XH lncRNAs and active XH lncCARs revealed similar biological terms related to transcription that were obtained with the H3K4me2 XH lncRNAs such as transcription DNA-templated, negative regulation of transcription. (Supplementary Figure S5B-C, Dataset S8).
Active XH transcription units have distinct H3K4 methylation patterns
Given the specific enrichment of divergent XH transcription units in our WDR5/H3K4me2 pull-downs, we wanted to check whether active XH transcription units (transcription units associated with active XH lncCARs) were enriched with differential H3K4 methylation patterns as compared to non-chromatin associated RNAs having divergent or XH patterns (non-CAR XH lncRNAs and non-CAR XH protein coding genes or PCGs). Toward this, we generated H3K4me2, H3K4me3 and WDR5 ChIP-seq data in BT-549 cell line. Interestingly, H3K4me2, H3K4me3 and WDR5 ChIP-seq signals were more enriched at the promoter regions (±2 kb from TSS) of active lncCARs having XH pattern (active XH lncCARs) compared to the non-CAR XH lncRNAs but this differential enrichment was absent between active pcCARs with XH pattern (active XH pcCARs) and non-CAR protein coding genes with XH pattern (non-CAR XH PCGs) (Figure 3E and Supplementary Figure S5D). These observations are consistent with enrichment of lncRNAs from divergent loci in our ChRIP pull-downs. Additionally, HMEC (epithelial derived breast tissue) H3K4me2 ChIP-seq data from ENCODE, and LnCaP WDR5 ChIP-seq data from Kim et al. (43) showed similar enrichment patterns for active XH lncCARs compared to active XH pcCARs and non-CAR XH lncRNAs (Supplementary Figure S5E).
Active XH lncCARs interact with WDR5
Out of five validated H3K4me XH lncRNAs (Figure 2D and Supplementary Figure S3A), four XH lncRNAs (FOXD3-AS1, HOXC13-AS, GATA6-AS1 and HOXC-AS2) were part of active XH lncCARs. To gain more insights into the mechanism of this positive regulation of gene expression by these active XH lncCARs, we performed UV cross-linked RNA immunoprecipitation (UV-RIP) with WDR5 antibody and looked for the enrichment of active XH lncCARs FOXD3-AS1, HOXC13-AS and GATA6-AS1 using ActD treated BT-549 cells. We found that these active XH lncCARs were significantly enriched in the WDR5 antibody pull-down, whereas their divergent coding partner mRNAs were not enriched (Figure 3F, and Supplementary Figure S6A). Additionally, we overexpressed FLAG WDR5 wild type (WT) and FLAG WDR5 F266A mutant (WDR5 mutant lacking RNA binding motifs (25) vectors in Hela cells. Comparable levels of FLAG WDR5 in WT and F266A was confirmed by Western (Figure 3G). We performed formaldehyde cross-linked RIP (fRIP) using antibodies against FLAG epitope and found that FOXD3-AS1 and HOXC13-AS active XH lncCARs interact with WT WDR5 but their interaction with F266A mutant WDR5 was compromised, suggesting that the interaction between the active XH lncCARs and WDR5 is mediated via RNA binding motif of WDR5 (Figure 3G).
Active XH lncCARs are targeted to their divergent transcription units
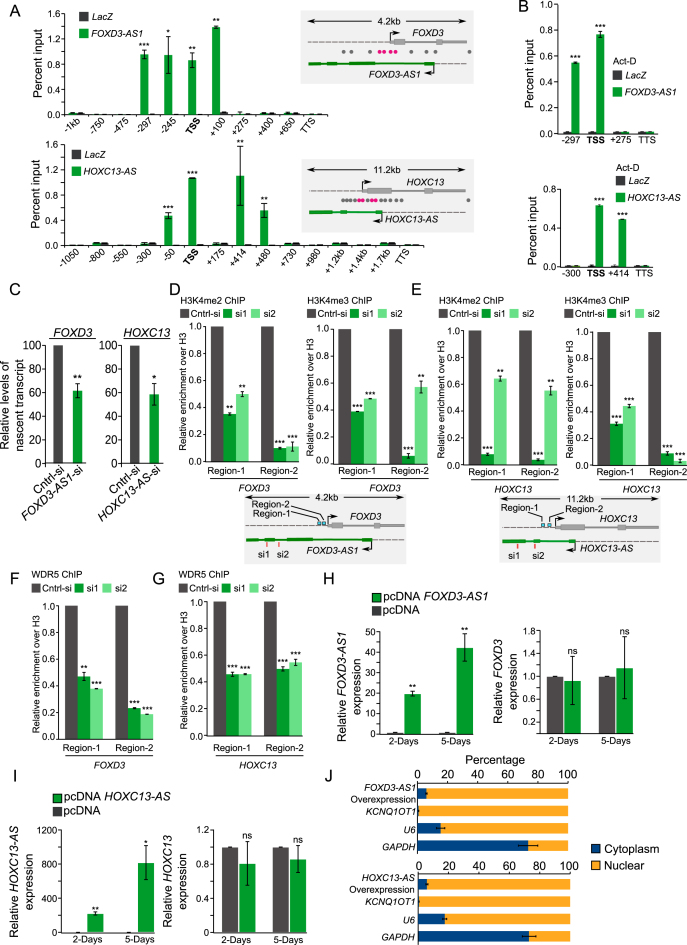
To further understand the mechanism(s) by which transcriptional competence is maintained at the active XH promoters, we performed chromatin oligo affinity precipitation (ChOP) assay to investigate if these active XH lncCARs are targeted to these divergent promoters. ChOP assay was carried out using biotinylated antisense oligos targeting FOXD3-AS1, HOXC13-AS and GATA6-AS1. We found that all the three active XH lncCARs were efficiently enriched in our ChOP pull-downs as determined by RT-qPCR (Data not shown). We then scanned for the enrichment of FOXD3-AS1, HOXC13-AS and GATA6-AS1 across the FOXD3, HOXC13 and GATA6 gene loci, respectively, with qPCR primers selectively amplifying regions spanning upstream and downstream of TSS. Enrichment of antisense lncRNAs, FOXD3-AS1, HOXC13-AS and GATA6-AS1 were found in the promoters of the corresponding coding partners. None of these enriched regions were overlapping with TSS of active XH lncCARs suggesting specific enrichment of these lncRNAs over the TSS of the coding partners (Figure 4A and Supplementary Figure S6B). The promoter targeting of all three active XH lncCARs were independent of the process of transcription, as ChOP carried after ActD treatment revealed that active XH lncCARs were still retained at their respective target region (Figure 4B and Supplementary Figure S6C). Downregulation FOXD3-AS1 and HOXC13-AS also resulted in decreased nascent transcription of partner protein coding genes (Figure 4C). Taken together, these results indicate that active XH lncCARs are targeted to their respective coding partner promoters to maintain their expression.
Figure 4.
Active XH lncCARs promote transcription of their protein coding partners. (A and B) Promoter targeting of active XH lncCARs detected by ChOP. A) qPCR analysis of ChOP pull-downs, performed using FOXD3-AS1 and HOXC13-AS antisense probes, with primers over TSS (transcription start sites), regions spanning upstream and downstream of TSS and over transcription termination sites (TTS) of FOXD3 and HOXC13 genes in BT-549 cells. The ChOP pull-down with LacZ antisense oligo, used as a negative control (grey bars). Specific enrichment pattern for each of the XH lncCARs is depicted in green bars. The schematic in the right of the bar graph represents the genomic locations of the respective primers (grey: not enriched and rosetta: showing enrichment). Data are shown as mean ± SD (n = 2 biological replicates). *P < 0.05, **P ≤ 0.01 and ***P ≤ 0.001. (B) Similar qPCR analysis of ChOP pull-downs using FOXD3-AS1 and HOXC13-AS antisense probes upon ActD treatment in BT-549 cells. Data are shown as mean ± SD (n = 2 biological replicates). *P < 0.05, **P ≤ 0.01 and ***P ≤ 0.001. (C) RT-qPCR analysis to detect relative levels of nascent FOXD3 and HOXC13 transcripts by Click-iT at 48 hours after the knockdown of FOXD3-AS1 and HOXC13-AS. Bar graph depicts the level of nascent protein coding transcripts upon removal of their respective active XH lncCARs. Data represent the mean ± SD of two independent biological experiments. * if P ≤ 0.05 and ** if P ≤ 0.01. (D and E) ChIP-qPCR analysis of the enrichment of H3K4me2 and H3K4me3 at the promoter region of FOXD3 (D) and HOXC13 (E) upon down regulation of active XH lncCAR FOXD3-AS1 and HOXC13-AS respectively. Fold enrichment of H3K4me2 and H3K4me3 was normalized to histone H3 and IgG. After normalization the ChIP data was represented as relative enrichment compare to control siRNA.The locations of ChIP primers used are depicted in schematic in the bottm of each bar graph. Data are shown as mean ± SD (n = 3 biological replicates). **P ≤ 0.01 and ***P ≤ 0.001. (F and G) ChIP-qPCR analysis of enrichment of WDR5 at the promoter region of FOXD3(F) and HOXC13 (G) upon downregulation of active XH lncCAR FOXD3-AS1 and HOXC13-AS respectively. Fold enrichment of WDR5 was normalized to histone H3 and IgG. After normalization the ChIP data was represented as relative enrichment compare to control siRNA. The ChIP primers used in the experiment are same as described in panel D–E. Data are shown as mean ± SD (n = 3 biological replicates). **P ≤ 0.01 and ***P ≤ 0.001. (H and I) FOXD3-AS1 and HOXC13-AS overexpression: RT–qPCR analysis of FOXD3 expression, upon ectopic overexpression of FOXD3-AS1 (H) and HOXC13-AS transcript (I) respectively, at Day 2 and Day 5 post transfection in BT-549 cell line. pcDNA empty vector transfection used as a control. Data represent the mean ± SD. of two independent biological experiments. P-values has been denoted as * if P ≤ 0.05, ** if P ≤ 0.01. ns denotes non-significant. (J) Cell fractionation in BT-549 cells show distribution of the overexpressed FOXD3-AS1 and HOXC13-AS transcripts. Data represent the mean ± SD of two independent biological experiments. GAPDH serves as positive control for cytoplasmic fraction, U6 and KCNQ1OT1 serves as positive control for nuclear fraction.
Active XH lncCARs are required for the maintenance of H3K4me2 and the establishment of H3K4me3 at their XH transcription units
To further dissect the mechanism of active lncCARs’ mediated regulation of transcriptional competence at divergent XH transcription units, we selected two of the active XH lncCAR loci HOXC13-AS and FOXD3-AS1. As mentioned earlier (Figure 2D and E), both siRNA and LNA mediated downregulation of active XH lncCARs FOXD3-AS1 and HOXC13-AS resulted in reduced expression of FOXD3 and HOXC13. We also observed decreased protein level of the FOXD3 and HOXC13 after downregulation of their respective XH lncRNA partners (Supplementary Figure S6D). Further we looked for the distribution of active chromatin marks i.e. H3K4me2 and H3K4me3 at the promoter regions of both HOXC13 and FOXD3 genes upon downregulation of HOXC13-AS and FOXD3-AS1 (Supplementary Figure S6E), respectively. Downregulation of both the active XH lncCARs resulted in decreased H3K4me2 and also H3K4me3 occupancy at the FOXD3 (Figure 4D) and HOXC13 (Figure 4E) promoters regions. We next investigated whether WDR5 occupancy at the FOXD3 and HOXC13 promoters was affected following FOXD3-AS1 and HOXC13-AS downregulation. Downregulation of the active XH lncCARs also resulted in significant decrease in the WDR5 occupancy at the HOXC13 and FOXD3 promoters regions (Figure 4F and G).
To understand if these XH lncCARs can act in trans to activate their divergent protein coding partner genes, we transiently overexpressed full-length cDNAs of FOXD3-AS1 and HOXC13-AS in BT-549 cell line for 2 days and 5 days. Transient overexpression of both FOXD3-AS1 and HOXC13-AS failed to upregulate the expression of their partner genes FOXD3 and HOXC13, respectively (Figure 4H and I), although the overexpressed transcripts were targeted to the nucleus (Figure 4J) indicating that they possibly need to be transcribed in cis as in the case of Xist and Kcnq1ot1 lncRNAs (5,45,48).
We then sought to know whether the maintenance of H3K4me2 and H3K4me3 at active XH transcription units is WDR5 dependent. To address this, we generated ChIP-seq profile for both H3K4me2 and H3K4me3 in control and in transiently depleted WDR5 cells. However, WDR5 depletion (Supplementary Figure S6F) resulted in loss of H3K4me3 but not H3K4me2 at the active XH transcription units. In contrast, both H3K4me2 and H3K4me3 loss was observed in non-CAR XH lncRNA promoters (Figure 5A). Transient depletion of WDR5 also led to downregulation of both active the XH lncCARs and their coding partners (Figure 5B). Downregulation of FOXD3-AS1 and HOXC13-AS results in the loss of both H3K4me2 and H3K4me3 over the promoters of the respective coding partners, whereas WDR5 loss affects only H3K3me3 over the active XH lncRNA loci, suggest that active XH transcription units can maintain H3K4me2 independent of WDR5 and that WDR5 is required for the establishment of H3K4me3 via the conversion of H3K4me2 into H3K4me3 (Figure 5C).
Figure 5.
H3K4me2 and H3K4me3 occupancy upon WDR5 depletion. (A) H3K4me2 and H3K4me3 ChIP-seq signals at the promoter regions (±2 kb) of active XH lncCARs (Green) and non-CAR XH lncRNA (Black) upon WDR5-esi depletion. The signals presented in the plots represent log2ratio between ChIP pull-down and input sample. TSS denotes the transcription start site of active XH lncCARs or non-CAR XH lncRNAs. The solid lines in the plot represents H3K4me2 and H3K4me3 signal in control esiRNA transfected cells and dotted lines represents WDR5 esiRNA transfection. (B) Normalized relative expression of active XH lncCARs (left panel) and their respective partner protein coding genes (right panel) upon removal of WDR5 by esiRNA as compared to that in control esiRNA transfection. Data represent the mean ± SD of two independent biological experiments. * P ≤ 0.05, ** P ≤ 0.01 and *** P ≤ 0.001. (C) Model depicting transcriptional regulation of divergent gene loci by active XH lncCARs. Active XH lncCARs transcribed from divergent transcription units are targeted to specific regions around TSS of partner protein coding gene that are enriched with H3K4me2, through their chromatin interaction property. Promoter targeting of these active XH lncCARs helps in maintaining H3K4me2 in WDR5 independent manner. XH lncCAR-dependent recruitment of WDR5 at the active XH transcription units helps in the conversion of H3K4me2 into H3K4me3, thereby establishing transcriptionally competent chromatin at the divergent transcription units. However, the establishment and maintenance of H3K4me2 levels at these transcription units are probably mediated by a different mechanism.
DISCUSSION
Accumulated evidence in the recent past suggested a functional nexus between lncRNA and DNA or chromatin modifying enzymes in the stringent regulation of both DNA and histone modifications (49–51). However, in most cases the mechanisms underlying their targeting to specific genomic regions is not clear. Several recent investigations tried to address this issue by identifying lncRNAs that interact with active and repressive (19,25) chromatin modifiers on a global scale. However, the functional significance of these lncRNAs in the chromatin-level gene regulation has however not been addressed in detail. In the current investigation we attempt to address this void by performing ChRIP assay with antibodies against active chromatin-specific histone mark H3K4me2 and its reader and H3K4-specifi methyltransferase WDR5 to identify active chromatin associated lncRNAs on a global scale in breast cancer cell line BT-549. ChRIP assay in combination with ActD treatment has identified robustly and functionally related chromatin bound lncRNAs. However, our assay might lead to loss of enrichment of short-lived lncRNAs or the RNAs which function by co-transcriptional association with chromatin.
The most prominent observation from this current investigation is that the purified lncRNAs from H3K4me2 and WDR5 defining active chromatin are enriched with antisense transcripts with overrepresented divergent transcription units (XH, head-to-head). This enrichment of XH transcription units appears to be unique to active lncCARs, as they were underrepresented in the inactive lncCARs purified using H3K27me3 and EZH2. Functional enrichment analysis revealed that the majority of the protein-coding partners of active XH lncCARs encode transcription factors. These observations indicate that active XH lncCARs may involve in specific transcriptional regulation of XH transcription units. Previous investigations have shown that lncRNA expression from the XH promoters correlates with active transcription in cis. The maintenance of transcriptionally competent chromatin at the divergent transcription units appeared to be dependent on both lncRNAs per se (46,52) as well as their act of transcription (53,54). Functional validation of the selected active XH lncCARs using siRNAs and LNAs, indicate that these transcripts per se positively regulate the transcription of protein-coding gene. In certain instances, however, siRNA or RNAi dependent silencing has been shown to involve transcriptional repression by modifying the chromatin structures (55,56). According to recent evidence, siRNA targeting the first exon only interfere with the transcriptional process, whereas the siRNAs that target downstream exons target transcript itself (57). The siRNAs used in the current study map to second or third exons of the analyzed active XH lncCAR, and hence they should primarily interfere with the transcript itself but not with the transcriptional process. LNA modified antisense GapmeRs preferentially degrade nuclear RNAs through RNaseH dependent mechanism (47), which further rule out the possibility that our active XH lncCAR knockdown approaches interfere with transcriptional process. Furthermore, enrichment of active XH lncCARs in the ActD treated ChOP assay is also in line our notion that active XH lncCARs act at the transcript level. In addition, the maintenance of active chromatin at these XH transcriptional units is specifically carried out by active XH lncCARs as the downregulation of protein-coding genes did not affect the transcription of the active XH lncCARs. This suggests that the lncRNA component of these XH transcription units harbors the transcriptional regulatory function but not their coding partners. Unlike transcription factors, which require a consensus motif for binding to target regions, lncRNAs have been shown to exploit multiple mechanisms in their targeting to genomic loci. For example, they interact with target regions via associating with chromatin regulatory proteins or through formation of RNA-DNA triplexes or RNA-DNA hybrids (10,21,58). It would be interesting to investigate the mechanism by which active XH lncCARs are targeted specifically to the promoters of their protein coding partners. In our ChRIP assay we have also identified a significant number of the protein-coding RNAs as chromatin bound (pcCARs). Unlike active XH lncRNAs, knockdown of protein-coding RNAs did not reveal any consistent regulatory pattern. Further systematic studies are required to explore the chromatin-based functions of protein-coding RNAs.
The possibility of trans regulatory role for these XH lncRNAs cannot be ruled out as XH lncRNAs have earlier been shown to act in trans (59,60). However, though transiently overexpressed of FOXD3-AS1 and HOXC13-AS show nuclear localization but has no seeming effects on expression levels of their protein-coding counter parts, indicating that active XH lncCARs need to be transcribed in cis in order to activate neighboring protein coding genes. This observation is consistent with Xist-dependent cis regulation of gene activity on the inactive X chromosome, where Xist per se is required in cis to regulate chromosome-wide transcriptional silencing on the inactive X-chromosome. Ectopic expression of Xist from the autosome had no effect on X chromosomes rather Xist transgene locally inactivated neighboring genes (48,61). Similar cis-acting RNA-dependent transcriptional silencing mechanisms have also been characterized for another lncRNA Kcnq1ot1 (5,45).
We show that active XH lncCARs interacts directly with WDR5 which might be critical for the establishment and/or maintenance of active chromatin at these active XH transcription units. WDR5 has been implicated in the establishment of global H3K4me1, -me2 and -me3 patterns, and our ChIP-seq data is also in concordance with the previous studies on the role of WDR5 in the establishment of global H3K4me1, -me2 and -me3 (27,62). According to ChIP-seq analysis, active XH lncCAR transcription units showed significant enrichment of H3K4me2, H3K4me3 and WDR5 compared to non-CAR XH or active XH pcCAR transcription units. Depletion of WDR5 results in the loss of transcription of both active XH lncCARs and their partner protein coding genes. However, we observed loss of only H3K4me3 but not H3K4me2 at the active XH transcription units upon WDR5 depletion. This observation particularly emphasises that WDR5 exploits different mechanisms in a promoter-specific manner in the establishment of H3K4me2 and H3K4me3. Whereas the depletion of active XH lncCARs results in loss of both H3K4me3 and H3K4me2 at active XH transcription units. Based on these observations we propose that the targeting of WDR5 to the active XH transcription units and the conversion of H3K4me2 to H3K4me3 requires XH lncCARs, whereas, the maintenance of H3K4me2 marks at active XH transcription units appears to be WDR5 independent (Figure 5C). This unravels a new facet of chromatin-based regulation at the active divergent XH transcription units by active XH lncCARs.
In sum, the current study together with earlier investigations on chromatin interacting lncRNAs, would form strong basis for more investigations to explore the functional role of lncRNAs in the maintenance of chromatin signatures at the various regulatory regions across the genome.
DATA AVAILABILITY
All sequencing data used in this study can be found at European Nucleotide Archive under projects ‘PRJEB20225’ (H3K4me2 and WDR5 ChRIP) and ‘PRJEB7307’ (nuclear/Input). The normalized read count data (RPKM) for ChRIP profiles are provided as supplement in ‘Dataset S1.zip’.
Supplementary Material
ACKNOWLEDGEMENTS
The computations for the dataset used in the current study were performed on resources provided by Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) high-performance computing (HPC) which is part of Swedish National Infrastructure for Computing (SNIC). FLAG-HA WDR5 WT and FLAG-HA WDR5 F266A plasmids were a kind gift from Dr. Howard Y. Chang.
Author Contributions: C.K., K.M., S.S. and T.M. conceptualized the work, analyzed the experimental data. T.M. and K.M. have generated HK4me2 and WDR5 ChRIP material. KM has generated both wild type and WDR5 depleted H3K4me2, H3K4me3 and WDR5 ChIP-seq data with suggestions from M.K. K.M. carried out all the wet-lab experiments except those performed by V.S.A. V.S.A. performed all the ChOP assays, western blots and transient overexpression experiments. S.S. has performed all the bioinformatics analysis and prepared the manuscript figures. K.M., C.K. and S.S. wrote the manuscript with vital inputs from T.M.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Knut and Alice Wallenberg Foundation [KAW 2014.0057]; Swedish Foundation for Strategic Research [RB13-0204]; Swedish Cancer Research foundation [Cancerfonden: Kontrakt no. 150796]; Swedish Research Council [2017-02834]; Barncancerfonden [PR2016-0057]; Ingabritt Och Arne Lundbergs forskningsstiftelse and LUA/ALF (to C.K.). Funding for open access charge: Knut and Alice Wallenberg Foundation (KAW) [KAW 2014.0057].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J. et al. . An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017; 543:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ali M.M., Akhade V.S., Kosalai S.T., Subhash S., Statello L., Meryet-Figuiere M., Abrahamsson J., Mondal T., Kanduri C.. PAN-cancer analysis of S-phase enriched lncRNAs identifies oncogenic drivers and biomarkers. Nat. Commun. 2018; 9:883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Loewer S., Cabili M.N., Guttman M., Loh Y.H., Thomas K., Park I.H., Garber M., Curran M., Onder T., Agarwal S. et al. . Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010; 42:1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lopez-Pajares V., Qu K., Zhang J., Webster D.E., Barajas B.C., Siprashvili Z., Zarnegar B.J., Boxer L.D., Rios E.J., Tao S. et al. . A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev. Cell. 2015; 32:693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mohammad F., Pandey G.K., Mondal T., Enroth S., Redrup L., Gyllensten U., Kanduri C.. Long noncoding RNA-mediated maintenance of DNA methylation and transcriptional gene silencing. Development. 2012; 139:2792–2803. [DOI] [PubMed] [Google Scholar]
- 6. Mondal T., Juvvuna P.K., Kirkeby A., Mitra S., Kosalai S.T., Traxler L., Hertwig F., Wernig-Zorc S., Miranda C., Deland L. et al. . Sense-Antisense lncRNA pair encoded by locus 6p22.3 determines neuroblastoma susceptibility via the USP36-CHD7-SOX9 regulatory axis. Cancer Cell. 2018; 33:417–434. [DOI] [PubMed] [Google Scholar]
- 7. Geisler S., Coller J.. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013; 14:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Akhade V.S., Pal D., Kanduri C.. Long noncoding RNA: genome organization and mechanism of action. Adv. Exp. Med. Biol. 2017; 1008:47–74. [DOI] [PubMed] [Google Scholar]
- 9. Kanduri C. Long noncoding RNAs: lessons from genomic imprinting. Biochim. Biophys. Acta. 2016; 1859:102–111. [DOI] [PubMed] [Google Scholar]
- 10. Rinn J.L., Chang H.Y.. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012; 81:145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fischle W., Wang Y., Allis C.D.. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 2003; 15:172–183. [DOI] [PubMed] [Google Scholar]
- 12. Nightingale K.P., Gendreizig S., White D.A., Bradbury C., Hollfelder F., Turner B.M.. Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J. Biol. Chem. 2007; 282:4408–4416. [DOI] [PubMed] [Google Scholar]
- 13. Margueron R., Trojer P., Reinberg D.. The key to development: interpreting the histone code. Curr. Opin. Genet. Dev. 2005; 15:163–176. [DOI] [PubMed] [Google Scholar]
- 14. Zhou V.W., Goren A., Bernstein B.E.. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011; 12:7–18. [DOI] [PubMed] [Google Scholar]
- 15. Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J.. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011; 470:279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gardner K.E., Allis C.D., Strahl B.D.. Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 2011; 409:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ho L., Crabtree G.R.. Chromatin remodelling during development. Nature. 2010; 463:474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gomez J.A., Wapinski Orly L., Yang Yul W., Bureau J.-F., Gopinath S., Monack Denise M., Chang Howard Y., Brahic M., Kirkegaard K.. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the Interferon-γ locus. Cell. 2013; 152:743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T.. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010; 40:939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mondal T., Rasmussen M., Pandey G.K., Isaksson A., Kanduri C.. Characterization of the RNA content of chromatin. Genome Res. 2010; 20:899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., Mitra S., Mohammed A., James A.R., Hoberg E. et al. . MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015; 6:7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meng Y., Yi X., Li X., Hu C., Wang J., Bai L., Czajkowsky D.M., Shao Z.. The non-coding RNA composition of the mitotic chromosome by 5′-tag sequencing. Nucleic Acids Res. 2016; 44:4934–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A. et al. . A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011; 472:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaneko S., Son J., Shen S.S., Reinberg D., Bonasio R.. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 2013; 20:1258–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Y.W., Flynn R.A., Chen Y., Qu K., Wan B., Wang K.C., Lei M., Chang H.Y.. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife. 2014; 3:e02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trievel R.C., Shilatifard A.. WDR5, a complexed protein. Nat. Struct. Mol. Biol. 2009; 16:678–680. [DOI] [PubMed] [Google Scholar]
- 27. Wysocka J., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D.. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005; 121:859–872. [DOI] [PubMed] [Google Scholar]
- 28. Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z.. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837. [DOI] [PubMed] [Google Scholar]
- 29. Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D.. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007; 39:311–318. [DOI] [PubMed] [Google Scholar]
- 30. Wang Y., Li X., Hu H.. H3K4me2 reliably defines transcription factor binding regions in different cells. Genomics. 2014; 103:222–228. [DOI] [PubMed] [Google Scholar]
- 31. Pekowska A., Benoukraf T., Ferrier P., Spicuglia S.. A unique H3K4me2 profile marks tissue-specific gene regulation. Genome Res. 2010; 20:1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mondal T., Subhash S., Kanduri C.. Chromatin RNA immunoprecipitation (ChRIP). Methods Mol. Biol. 2018; 1689:65–76. [DOI] [PubMed] [Google Scholar]
- 33. Akhade V.S., Arun G., Donakonda S., Rao M.R.. Genome wide chromatin occupancy of mrhl RNA and its role in gene regulation in mouse spermatogonial cells. RNA Biol. 2014; 11:1262–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meryet-Figuiere M., Alaei-Mahabadi B., Ali M.M., Mitra S., Subhash S., Pandey G.K., Larsson E., Kanduri C.. Temporal separation of replication and transcription during S-phase progression. Cell Cycle. 2014; 13:3241–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S . The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 37. Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D. et al. . GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006; 7(Suppl. 1):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Subhash S., Kanduri C.. GeneSCF: a real-time based functional enrichment tool with support for multiple organisms. BMC Bioinformatics. 2016; 17:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Consortium E.P. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011; 9:e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim J.-Y., Banerjee T., Vinckevicius A., Luo Q., Parker J.B., Baker M.R., Radhakrishnan I., Wei J.-J., Barish G.D., Chakravarti D.. A role for WDR5 in integrating threonine 11 phosphorylation to lysine 4 methylation on histone H3 during androgen signaling and in prostate cancer. Mol. Cell. 2014; 54:613–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chiesa N., De Crescenzo A., Mishra K., Perone L., Carella M., Palumbo O., Mussa A., Sparago A., Cerrato F., Russo S. et al. . The KCNQ1OT1 imprinting control region and non-coding RNA: new properties derived from the study of Beckwith-Wiedemann syndrome and Silver-Russell syndrome cases. Hum. Mol. Genet. 2012; 21:10–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pandey R.R., Mondal T., Mohammad F., Enroth S., Redrup L., Komorowski J., Nagano T., Mancini-Dinardo D., Kanduri C.. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008; 32:232–246. [DOI] [PubMed] [Google Scholar]
- 46. Luo S., Lu J.Y., Liu L., Yin Y., Chen C., Han X., Wu B., Xu R., Liu W., Yan P. et al. . Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell. 2016; 18:637–652. [DOI] [PubMed] [Google Scholar]
- 47. Kurreck J., Wyszko E., Gillen C., Erdmann V.A.. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002; 30:1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heard E., Mongelard F., Arnaud D., Chureau C., Vourc’h C., Avner P.. Human XIST yeast artificial chromosome transgenes show partial X inactivation center function in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:6841–6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Di Ruscio A., Ebralidze A.K., Benoukraf T., Amabile G., Goff L.A., Terragni J., Figueroa M.E., De Figueiredo Pontes L.L., Alberich-Jorda M., Zhang P. et al. . DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013; 503:371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mohammad F., Mondal T., Guseva N., Pandey G.K., Kanduri C.. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010; 137:2493–2499. [DOI] [PubMed] [Google Scholar]
- 51. Merry C.R., Forrest M.E., Sabers J.N., Beard L., Gao X.H., Hatzoglou M., Jackson M.W., Wang Z., Markowitz S.D., Khalil A.M.. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum. Mol. Genet. 2015; 24:6240–6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sigova A.A., Mullen A.C., Molinie B., Gupta S., Orlando D.A., Guenther M.G., Almada A.E., Lin C., Sharp P.A., Giallourakis C.C. et al. . Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Anderson K.M., Anderson D.M., McAnally J.R., Shelton J.M., Bassel-Duby R., Olson E.N.. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016; 539:433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Engreitz J.M., Haines J.E., Perez E.M., Munson G., Chen J., Kane M., McDonel P.E., Guttman M., Lander E.S.. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016; 539:452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weinberg M.S., Villeneuve L.M., Ehsani A., Amarzguioui M., Aagaard L., Chen Z.X., Riggs A.D., Rossi J.J., Morris K.V.. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006; 12:256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim D.H., Villeneuve L.M., Morris K.V., Rossi J.J.. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006; 13:793–797. [DOI] [PubMed] [Google Scholar]
- 57. Stojic L., Niemczyk M., Orjalo A., Ito Y., Ruijter A.E.M., Uribe-Lewis S., Joseph N., Weston S., Menon S., Odom D.T. et al. . Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions. Nat. Commun. 2016; 7:10406–10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Postepska-Igielska A., Giwojna A., Gasri-Plotnitsky L., Schmitt N., Dold A., Ginsberg D., Grummt I.. LncRNA Khps1 regulates expression of the Proto-oncogene SPHK1 via Triplex-Mediated changes in chromatin structure. Mol. Cell. 2015; 60:626–636. [DOI] [PubMed] [Google Scholar]
- 59. Musahl A.S., Huang X., Rusakiewicz S., Ntini E., Marsico A., Kroemer G., Kepp O., Orom U.A.. A long non-coding RNA links calreticulin-mediated immunogenic cell removal to RB1 transcription. Oncogene. 2015; 34:5046–5054. [DOI] [PubMed] [Google Scholar]
- 60. Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E. et al. . Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007; 129:1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee J.T., Strauss W.M., Dausman J.A., Jaenisch R.. A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell. 1996; 86:83–94. [DOI] [PubMed] [Google Scholar]
- 62. Sims R.J. 3rd, Reinberg D.. Histone H3 Lys 4 methylation: caught in a bind. Genes Dev. 2006; 20:2779–2786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data used in this study can be found at European Nucleotide Archive under projects ‘PRJEB20225’ (H3K4me2 and WDR5 ChRIP) and ‘PRJEB7307’ (nuclear/Input). The normalized read count data (RPKM) for ChRIP profiles are provided as supplement in ‘Dataset S1.zip’.