Abstract
Diversity-generating retroelements (DGRs) create unparalleled levels of protein sequence variation through mutagenic retrohoming. Sequence information is transferred from an invariant template region (TR), through an RNA intermediate, to a protein-coding variable region. Selective infidelity at adenines during transfer is a hallmark of DGRs from disparate bacteria, archaea, and microbial viruses. We recapitulated selective infidelity in vitro for the prototypical Bordetella bacteriophage DGR. A complex of the DGR reverse transcriptase bRT and pentameric accessory variability determinant (Avd) protein along with DGR RNA were necessary and sufficient for synthesis of template-primed, covalently linked RNA–cDNA molecules, as observed in vivo. We identified RNA–cDNA molecules to be branched and most plausibly linked through 2′-5′ phosphodiester bonds. Adenine-mutagenesis was intrinsic to the bRT-Avd complex, which displayed unprecedented promiscuity while reverse transcribing adenines of either DGR or non-DGR RNA templates. In contrast, bRT-Avd processivity was strictly dependent on the template, occurring only for the DGR RNA. This restriction was mainly due to a noncoding segment downstream of TR, which specifically bound Avd and created a privileged site for processive polymerization. Restriction to DGR RNA may protect the host genome from damage. These results define the early steps in a novel pathway for massive sequence diversification.
INTRODUCTION
Diversity-generating retroelements (DGRs) create massive protein sequence variation in ecologically diverse bacteria, archaea, and microbial viruses, including constituents of the human microbiome (1–7). An extraordinary prevalence of DGRs was recently identified in the microbial ‘dark matter’ (8). Variation by DGRs extends to at least 1020 possible sequences (9). The only parallel to this scale of variation occurs in vertebrate adaptive immune systems, in which, for example, 1014–16 sequences are possible in antibodies and T cell receptors (10,11). Strikingly, DGRs dedicate 103-fold less DNA but can achieve 104–6-fold greater variation than these adaptive immune systems. Massive protein sequence variation enables binding of novel targets, and is recognized to enable the immune system to adapt to dynamic environments.
A similar fitness benefit appears to be provided by the prototypical DGR of Bordetella bacteriophage (1). This DGR encodes the phage's receptor-binding protein Mtd (12,13), which is localized to the tip of phage tail fibers (Supplementary Figure S1A). Massive sequence variation in Mtd (∼1013) enables the phage to adapt to the loss of Bordetella surface receptors due to environmentally-responsive shifts in gene expression or immune-driven selection (4,14). For example, the Bordetella adhesin pertactin, which is expressed by the in vivo pathogenic or Bvg+ phase of Bordetella, can serve as a surface receptor for the phage (1,15). However, when Bordetella transitions to the ex vivo or Bvg– phase, expression of pertactin is lost. The loss of this phage receptor is counterbalanced by the DGR through the generation of Mtd variants, some of which have the ability to maintain phage infectivity by binding surface receptors expressed by Bvg–Bordetella (1,15).
DGRs diversify proteins through mutagenic retrohoming, which requires three fundamental components: a specialized reverse transcriptase and two nearly identical repeat regions, a variable region (VR) and a template region (TR) (Figure 1A). VR is located within the coding sequence of a diversified protein, such as mtd, and TR is proximally located in a noncoding region. Sequence information is transferred unidirectionally from TR to VR in a selectively error-prone manner focused on adenines in TR. Adenine-specific mutagenesis is a unique hallmark of DGRs. The process involves transcription of TR to produce TR-RNA, which is then reverse transcribed by the DGR reverse transcriptase to produce TR-cDNA (1,2,5,16–19) (Figure 1A). The TR-cDNA, which has specific errors due to the unfaithful copying of adenines, homes to and replaces VR through a mechanism that is as yet undefined. As a result of the adenine-specificity of mutagenesis, only amino acids that are encoded with adenines in TR are subject to diversification. The most frequently observed adenine-containing codon in TR is AAY, which encodes Asn. Adenine-mutagenesis of AAY can result in the encoding of 14 other amino acids, which cover the gamut of amino acid chemical character, but cannot result in a stop codon (12). Notably, adenine-encoded variable amino acids have been seen to form the solvent-exposed, ligand-binding sites of diversified proteins and to participate in ligand binding (9,12,15,20).
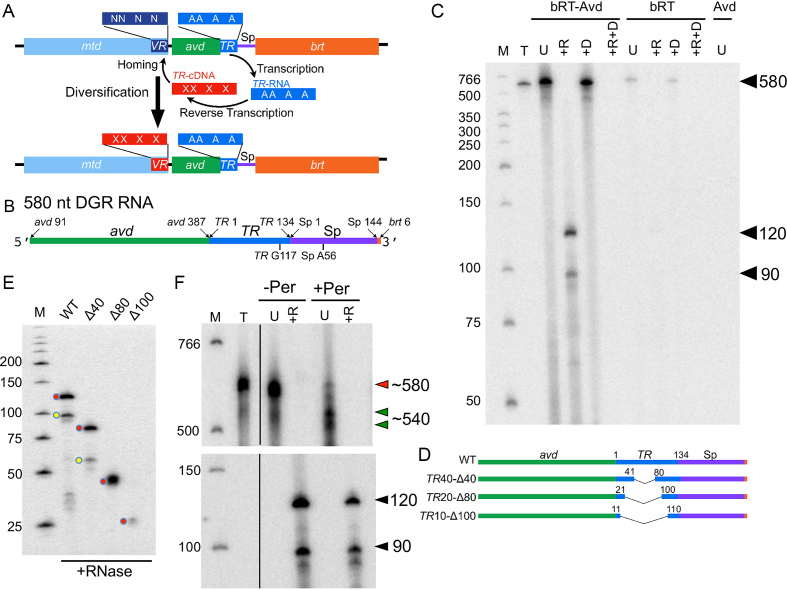
Figure 1.
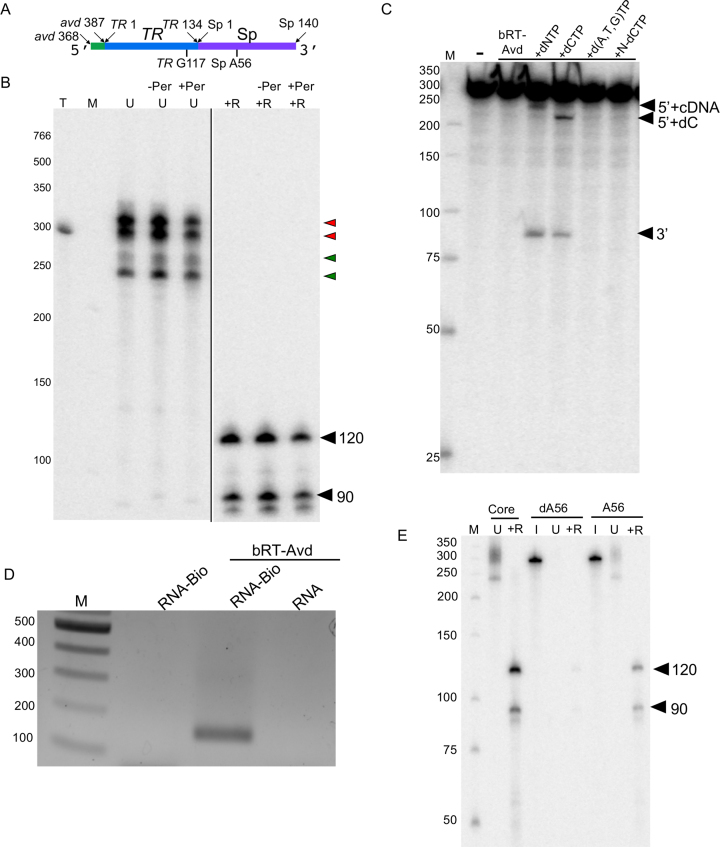
In vitro template-primed cDNA synthesis. (A) Bordetella bacteriophage DGR diversification of Mtd. mtd contains a variable region (VR), which encodes the receptor-binding site of the Mtd protein. Downstream of VR is the template region (TR). Adenines in TR (‘A’) are frequently replaced by another base in VR (‘N’). TR is transcribed to produce TR-RNA, which is then reverse transcribed to TR-cDNA. During this process, adenines in TR are mutagenized, as depicted by ‘X’ in TR-cDNA. Adenine-mutagenized TR-cDNA homes to and replaces VR, resulting in diversification of Mtd. bRT is the DGR reverse transcriptase, and avd the DGR accessory variability determinant. (B) Sequence elements of the 580 nt DGR RNA template used for reverse transcription reactions. (C) bRT-Avd, bRT, or Avd was incubated with the 580 nt DGR RNA and dNTPs, including [α-32P]dCTP, for 2h. Products resulting from the incubation were untreated (U), or treated with RNase (+R), DNase (+D), or both RNase and DNase (+R+D), and resolved by 8% denaturing polyacrylamide gel electrophoresis (PAGE). Lane T corresponds to internally-labeled 580 nt DGR RNA as a marker for the size of the template. The positions of the 580 nt band, and 120 and 90 nt cDNA bands are indicated. Nuclease-treated samples were loaded at twice the amount as untreated samples, here and throughout unless otherwise indicated. Lane M here and throughout corresponds to radiolabeled, single-stranded DNA molecular mass markers (nt units). (D) DGR RNA templates containing internal truncations in TR. (E) Radiolabeled cDNA products resulting from bRT-Avd activity for 2 h with intact (WT) or internally truncated 580 nt DGR RNA as template. Samples were treated with RNase and resolved by denaturing PAGE. The positions of the 120 and 90 nt cDNAs produced from intact template are indicated by red and yellow circles, respectively, as are positions of the correspondingly shorter cDNAs produced from truncated RNA templates. (F) Radiolabeled products resulting from bRT-Avd activity for 2 h with the 580 nt DGR RNA as template. Prior to reverse transcription, the RNA template was mock-treated (–Per) or treated with periodate (+Per). Products of the reaction were untreated (U) or treated with RNase (+R), and resolved by 4% (top) or 8% (bottom) denaturing PAGE. In the top gel, the red arrowhead indicates the ∼580 nt species, and the green arrowheads the several ∼540 nt species. In the bottom gel, the black arrowheads indicate the 120 and 90 nt cDNA products. The black vertical line within the gel indicates irrelevant lanes that were removed for display purposes. A 2-fold higher quantity was loaded for +Per samples than –Per samples.
In a number of retrotransposons, reverse transcription is primed by the DNA target of integration, thereby coupling cDNA synthesis to homing (21,22). However, recent in vivo studies show that cDNA synthesis by the Bordetella bacteriophage DGR does not require the target VR, and is instead primed by the DGR RNA template itself (19). Reverse transcription to produce TR-cDNA requires the Bordetella bacteriophage DGR reverse transcriptase bRT and the DGR accessory variability determinant (Avd) protein (1,19,23). The structure of bRT is not known, but it is predicted to resemble the reverse transcriptase domain of group II intron maturases (24–27). bRT appears to contain no additional functional domains. The structure of Avd is known, and it forms a pentameric and highly positively charged barrel (23). Avd interacts functionally with bRT (23).
To gain mechanistic insight into cDNA synthesis and adenine-mutagenesis by DGRs, we reconstituted reverse transcription by the Bordetella bacteriophage DGR in vitro. We found that a purified complex of bRT and Avd along with the DGR RNA template were necessary and sufficient for the synthesis of template-primed and covalently linked RNA–cDNA molecules, as observed in vivo (19). We showed that these RNA–cDNA molecules were branched due to a priming site located internally in the RNA, and linked most plausibly through 2′-5′ RNA–cDNA phosphodiester bonds. bRT alone had no reverse transcriptase activity, and required association with Avd for this function. Most significantly, we found that adenine-mutagenesis was intrinsic to the bRT-Avd complex, which displayed an unprecedented promiscuity in reverse transcribing adenines of either DGR or non-DGR RNA templates. In contrast, the processivity of bRT-Avd was strictly dependent on the RNA template, occurring only for the DGR RNA. This restriction was mainly due to a noncoding region downstream of TR, which specifically bound Avd and created a privileged site for processive polymerization by bRT-Avd. These results provide fundamental advances in understanding the mechanism of mutagenic retrohoming.
MATERIALS AND METHODS
Cloning, expression and purification of proteins
Bordetella bacteriophage bRT, with an N-terminal His6-tag followed by a PreScission protease cleavage site, and Avd were co-expressed using pET-28b (Novagen) and pACYCDuet-1 vectors (Novagen), respectively, in Escherichia coli BL21-Gold (DE3). Bacteria were grown with shaking at 37°C to OD600 0.6–0.8, and induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and then grown further overnight with shaking at 20°C. Bacteria were centrifuged and frozen at −80°C, and then thawed and re-suspended in 500 mM (NH4)2SO4, 50 mM HEPES, pH 8.0, 0.1% β-mercaptoethanol, 20% glycerol, 2 mM PMSF, protease inhibitor tablet(s) (Roche), DNase I (Ambion, 0.25 units/ml) and RNase A/T1 mix (Ambion, 0.2 μl/mL). Bacteria were lysed using an Emulsiflex C-5, and the lysate was clarified by centrifugation. His6-bRT/Avd from the supernatant was purified by Ni2+-NTA column chromatography. The His6-tag was cleaved with PreScission protease, and the bRT-Avd complex was further purified by gel filtration chromatography (Superdex 200). Two peaks were evident, with the first peak corresponding to the bRT-Avd complex and the second to bRT alone. Avd was expressed and purified as described previously (19).
RNA synthesis and purification
RNA was synthesized in vitro through runoff transcription reactions performed overnight at 37°C using 40 ng/μl DNA template, 4 mM NTPs (Promega), 0.5 units/μl RNase inhibitor (NEB), and 0.6 mg/mL T7 RNA polymerase in T7 buffer (3 mM MgCl2, 5 mM DTT, 2 mM spermidine, 40 mM Tris, pH 7.5). Products were extracted with phenol:chloroform:isoamyl alcohol (25:24:1 v/v, P:C:I) and ammonium acetate/ethanol (AA/E) precipitated. The pellet was resuspended in water and digested with 0.5 units/μl DNase I (Ambion) for 1 h at 37°C, and then P:C:I extracted and AA/E precipitated as above. RNA was further purified by denaturing polyacrylamide gel electrophoresis. RNA was recovered from the gel by diffusion overnight in 300 mM NaCl, 0.1% SDS, 1 mM EDTA, and extracted with P:C:I and chloroform:isoamyl alcohol (24:1 v/v, C:I), and AA/E precipitated. The pellet was washed with cold 80% ethanol, air dried for 10 min at RT, and resuspended in water. Internally-labeled RNA was produced similarly, except the NTP was 0.5 mM and 0.2 μCi/μl of [α-32P]UTP was included. Mutations were introduced into DNA templates by the QuikChange method (Agilent). Scrambled sequences in the Sp region are indicated in Supplementary Table S1. For the non-DGR RNA template, pET28a was digested with HindIII and used for in vitro runoff transcription from its T7 promoter.
Reverse transcription
Reactions were carried out in 20 μl containing 1.8 μM bRT-Avd, bRT or Avd, 100 ng/μl RNA template, 100 μM dNTPs (Promega) (or varying concentrations of certain dNTPs), 0.5 μCi/μl [α-32P]dCTP, 20 units RNase inhibitor (NEB) in 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 50 mM HEPES, pH 7.5, 10% glycerol for 2 h at 37°C. Oligodeoxynucleotide-primed reactions included 0.25–1 μM primer (Supplementary Table S2). Samples were digested with proteinase K (Ambion, 0.4 mg/ml) at 50°C for 30 min, and quenched with 2 mM EDTA. Samples were then desalted using a G-25 spin column, ammonium acetate/ethanol precipitated with 10 μg glycogen included with the ethanol (AA/E-glycogen). The pellet was resuspended in 10 μl water. Nuclease treatments were carried out on reverse transcription products (2 μl for RNase digestion and 1 μl for DNase digestion) in 10 μl containing 0.5 μl RNase A/T1 mix or 0.5 μl DNase I in RNase or DNase buffer. For digestion with UDG and RNase, 10 μl containing 2 μl of reverse transcription products, 5 units UDG (NEB) and 2 μl RNase A/T1 in UDG reaction buffer was incubated at 37°C for 20 min, and quenched by the addition of 10 μl denaturing loading dye.
Samples were heated for 5 min at 95°C in denaturing loading dye, resolved on denaturing polyacrylamide gels, and visualized by autoradiography using a Typhoon Trio (GE Healthcare Life Sciences). Quantification of band intensities was carried out using ImageQuant TL 8.1 (GE Healthcare Life Sciences). Background, as measured from a lane containing no sample in the gel being quantified, was subtracted from band intensities.
Reverse transcription reactions with HIV-1 RT were carried out as above, except in 10 μl and containing 10 units RNase inhibitor (NEB), 0.1 μCi/μl [α-32P]dCTP, 30 ng/μl RNA template, 1 μM PG117 primer (Supplementary Table S2) and 2 units of HIV-1 RT (Worthington Biochemical), and the reaction was carried out for 30 min.
Periodate treatment of RNA
RNA (200 μg) was incubated in 400 μl borax buffer, pH 9.5 (50 mM boric acid and 5 mM borax) and 50 mM sodium periodate at 30°C for 1 h in the dark, followed by addition of 40 μl glycerol, and quenching of the reaction at RT for 10 min. The sample was concentrated using a Speedvac to 50 μl, then diluted to 400 μl in borax buffer, pH 9.5, and incubated at 45°C for 90 min. The sample was finally purified through a desalting column (G-25) and a denaturing polyacrylamide gel, as above.
RT-PCR of RNA–cDNA molecules
RNA–cDNA molecules (200 ng) resulting from bRT-Avd activity on DGR RNA was annealed with in 13 μl with primer 1 (Supplementary Table S2) at 65°C for 5 min in the presence of 0.5 mM dNTPs and 5% DMSO. The sample was quenched on ice for 5 min and 200 units Super RT (BioBharati Life Sciences Pvt. Ltd), 20 units RNase inhibitor, and 5 mM DTT in RT buffer (75 mM KCl, 3 mM MgCl2, 50 mM Tris, pH 8.4) were added, yielding a 20 μl reaction. The reaction was incubated at 50°C for 90 min, and 70°C for 15 min. PCR amplification was then carried out with primers 1 and 2 (Supplementary Table S2), and PCR products were gel-purified and restriction digested, and cloned and sequenced.
Sequencing of 3′ ends of cDNAs
The cDNA portions of RNA–cDNA molecules (800 ng) resulting from bRT-Avd activity on DGR RNA were poly-dA tailed by incubation in 200 μl with 40 units of terminal deoxynucleotidyl transferase (TdT, NEB), 0.25 mM CoCl2, and 0.2 mM dATP in TdT buffer at 37°C for 45 min, and 70°C for 10 min. The sample was P:C:I and C:I extracted, and AA/E-glycogen precipitated. The pellet was re-suspended in water and used for PCR amplification using primers 3 and 4 (Supplementary Table S2). PCR products were gel-purified, restriction digested, and cloned and sequenced.
Complementary DNAs produced from non-DGR templates were poly-dA tailed, as above, except the reaction volume was 50 μl and 10 units of TdT were used, and the reaction was carried out at 37°C for 30 min, and 75°C for 20 min. The sample was treated as above, except that primers 5′-pET28b and 3′-pET28b were used for PCR amplification (Supplementary Table S2).
RNA cleavage by bRT-Avd
A 20 μl reaction consisting of 1 μg of [α-32P]UTP internally-labeled DGR RNA, 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 50 mM HEPES, pH 7.5, 20 units RNase inhibitor (NEB), 10% glycerol was incubated with 1.8 μM of bRT-Avd for 2 h at 37°C, in the presence or absence of 100 μM dNTPs. In the case of nonhydrolyzeable dCTP, 100 μM 2′-deoxycytidine-5′-[(α,β)-methyleno]triphosphate (Jena Bioscience) was used.
3′-Biotinylated DGR RNA
RNA was biotinylated using the Pierce™ RNA 3′ end biotinylation kit, according to manufacturer's instructions. Reverse transcription with bRT-Avd and 3′-biotinylated RNA was carried out as described above. After proteinase K treatment, 1 μl of the reaction was incubated with 5 μl Dynabeads M-280 (Invitrogen), which had been equilibrated in binding buffer (1 M NaCl, 50 mM Tris, pH 8, 0.1% Triton X-100 and 5% nuclease free-BSA). The sample was then incubated at RT for 20 min with constant shaking, and separated from solution magnetically. Beads were washed 10x with 1 mL binding buffer, and resuspended in 100 μl of 150 mM NaCl, 50 mM Tris, pH 8. PCR was carried out with 5 μl of the resuspended beads using primers 1 and 5 (Supplementary Table S2).
Hybrid molecules
RNA spanning Sp 56–140 (5′ phosphorylated and containing a 3′ dideoxy) with either adenosine or deoxyadenosine at Sp 56 was chemically synthesized (IDT) and gel-purified. Chemically synthesized molecules (12 μM) were incubated with in vitro transcribed and gel-purified RNA spanning avd 91-Sp 55 or avd 368-Sp 55 (3.4 μM) in the presence of 8 μM splint-1 DNA oligonucleotide (Supplementary Table S2), 8% DMSO, and 0.2× T4 RNA Ligase 1 (T4Rnl1) buffer in 250 μl. The splint was annealed to the RNA molecules by heating at 95°C for 3 min and cooling at 0.2°C/min to 20°C. A 250 μl solution consisting of 0.8× T4Rnl1 buffer, 2 mM ATP, 4% DMSO and 540 units T4Rnl1 was mixed with the annealed reaction. The reaction was incubated for 5 h at 37°C. The sample was then extracted with phenol:chloroform (P:C) and AA/E-glycogen precipitated. The pellet was resuspended in water and gel-purified. Hybrid molecules for cleavage experiments were prepared similarly, except the in vitro transcribed RNA was internally-labeled with [α-32P]UTP.
For detection, hybrid molecules (2 pmol) were treated with Shrimp Alkaline Phosphatase (2 units) in 20 μl cutsmart buffer for 30 min at 37°C, and 10 min at 65°C. The sample was purified using G-25 desalting column, and the RNA was 5′ 32P-labeled in a 40 μl reaction containing 4 μCi [γ-32P] ATP and 20 units of T4 polynucleotide kinase (NEB, PNK) in PNK buffer. The reaction was carried out at 37°C for 45 min, and 65°C for 20 min. The sample was purified using a G-25 desalting column.
Sequencing of RNA cleavage product
Gel-purified cleaved RNA (400 ng), 4 μM adapter DNA, and 5.2 μM splint-2 DNA (Supplementary Table S2) were annealed in 6% DMSO and 0.2× T4Rnl1 buffer in 25 μl by heating at 95°C for 3 min and cooling at 0.2°C/min to 20°C (Supplementary Table S2). A 25 μl solution consisting of 0.8x T4Rnl1 buffer, 2 mM ATP, 4% DMSO, and T4Rnl1 (60 units) was mixed with the annealed reaction, and the mixture was incubated for 5 h at 37°C. The sample was then P:C extracted and AA/E-glycogen precipitated. The pellet was resuspended in water and amplified by RT-PCR using primers 6 and 7 (Supplementary Table S2). RT-PCR products were gel-purified and restriction digested, and cloned and sequenced.
RNase protection assay and small RNA sequencing
A 50 μl reaction consisting of 100 ng of [α-32P]UTP internally-labeled RNA, 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 50 mM HEPES, pH 7.5, 10% glycerol was incubated with 1.8 μM of bRT, Avd, or bRT-Avd for 20 min at 37°C. RNase A/T1 mix (2 μl) was added and the incubation was carried out a further 30 min at 37°C. The reaction was stopped through P:C:I extraction and AA/E-glycogen precipitation.
For sequencing the protected RNA, the reaction was scaled up 5-fold with unlabeled RNA. Following purification, the protected RNA was dephosphorylated using alkaline phosphatase (NEB), and 5′-phosphorylated using T4 polynucleotide kinase (NEB), according to manufacturer's directions. This 5′-phosphorylated RNA sample was used for cDNA library preparation using the Illumina TruSeq small RNA library preparation kit according to manufacturer's directions, and sequencing was carried out at the Institute of Genomic Medicine, University of California, San Diego. Illumina small RNA sequencing output reads were trimmed with cutadapt 1.9.0 (28) using the adapter sequence TGGAATTCTCGGGTGCCAAG. Trimmed reads were aligned to a DGR reference sequence (corresponding to the DGR RNA) using bowtie2-align-s version 2.2.6 with default parameters, and unmapped reads were filtered using samtools (29,30). Reads that successfully mapped to the DGR were analyzed with a Python script to determine regions of the DGR sequence that were statistically enriched with mapped reads. Briefly, read start position counts were binned in ∼10 bp regions and tested for binomial enrichment within each bin with Bonferroni correction for the number of bins tested. Reads within each significantly enriched region were assembled through overlap consensus (31).
Quantification of dNTPs in Bordetella
Bordetella bronchiseptica ML6401 was grown overnight with shaking at 37°C in LB supplemented with streptomycin (20 μg/ml) and nicotinic acid (10 mM) to OD600 ∼0.4. One milliliter of bacterial culture was centrifuged, and the bacterial pellet was washed three times with 1 ml of ice cold PBS, centrifuged again and resuspended in 70% CH3OH. Internal standards (50 pmol 13C15N-dATP, 100 pmol 13C15N-dCTP, 50 pmol 13C15N-dGTP, 50 pmol 13C15N-TTP) were added to the samples, and the samples were stored overnight at −80°C. Samples were then thawed and centrifuged, and the supernatant was air-dried and stored at −20°C. Samples were reconstituted in 200 μl 2 mM NH3H2PO4 with 3 mM hexylamine, and 20 μl were injected for dNTP analysis by liquid chromatography–tandem mass spectrometry (LC–MS/MS). Intracellular dATP, dCTP, dGTP, TTP and dUTP were separated by Ultimate 3000 LC system (Thermo Scientific) using a Kinetex EVO C18 column (Phenomenex). An API5000 triple quadrupole mass spectrometer (ABsciex, MA, USA) was used for detection by multiple reaction monitoring. For estimation of nucleotide concentrations, a single cell volume of 4.5 fl determined for E. coli was used (32), based on the similarity between B. bronchiseptica and E. coli, both rod-shaped bacteria.
RESULTS
Reconstitution of cDNA synthesis
The bRT-Avd complex was co-expressed and co-purified, in apparent 1:5 stoichiometry, and incubated with in vitro transcribed and gel-purified DGR RNA (Figure 1B and Supplementary Figure S1B, C). This ∼580 nucleotide (nt) RNA contained TR (134 nt), and extensive upstream (297 nt from avd) and downstream sequences (144 nt of spacer between TR and brt, and 6 nt of brt). The extensive avd sequence was based on prior results showing that 300 nt upstream of TR are required for near wild-type retrohoming activity (23). Shorter lengths of avd, especially below 115 nt, lead to ∼500–1400-fold lower retrohoming activity. The downstream sequence was based on studies described below showing that nearly the entire length of the spacer region was required for cDNA synthesis. In reverse transcription reactions carried out in the absence of an exogenous primer and monitored through the incorporation of [α-32P]dCTP, we observed production of a major radiolabeled band with an apparent size similar to the template RNA, ∼580 nt (Figure 1C and Supplementary Figure S1D, E). This 580 nt band, however, was not entirely composed of cDNA, as it was digested by RNase into smaller bands, primarily ∼120 and ∼90 nt in length (both slightly smaller than the 134-nt TR), along with a number of minor bands (Figure 1C and Supplementary Figure S1F). These 120- and 90-nt bands corresponded to cDNAs as indicated by their sensitivity to DNase. All bands were sensitive to combined RNase and DNase digestion.
Evidence that these cDNAs corresponded to TR-cDNA came from templates deleted internally within TR (Figure 1D). A template containing an internal 40 nt deletion in TR resulted in cDNA products that were correspondingly 40 nt shorter than the 120 nt and 90 nt cDNAs, that is, ∼80 and ∼50 nt (Figure 1E). A template containing an internal 80 nt deletion in TR resulted in a single cDNA product which was correspondingly 80 nt shorter than the 120 nt cDNA (Figure 1E). A second shorter cDNA was not observed, suggesting that the reverse transcription termination site that led to the 90 nt cDNA was missing in the TR20-Δ80 construct. This supposition was confirmed by sequencing of cDNAs, as detailed below. A template containing an internal 100 nt deletion in TR resulted in a single weak cDNA product which was correspondingly 100 nt shorter than the 120 nt cDNA (Figure 1E). These results provided evidence for template-primed reverse transcription, which resulted in TR-cDNA molecules covalently linked to the template RNA, as observed in vivo (19).
Curiously, the 580 nt radiolabeled band resulting from reverse transcription was sensitive to RNase but did not appreciably change size upon DNase digestion (Figure C). To address this, we analyzed the 580 nt band on a low percentage gel. Several distinct species were resolved—a high-intensity ∼580 nt species (Figure 1F, red arrowhead) and several lower-intensity and smaller (∼540 nt) species (Figure 1F, green arrowheads). These smaller species were also visible upon overloading (Supplementary Figure S1D). The nature of these species was clarified by blocking the 3′-OH of the RNA through periodate treatment prior to reverse transcription (33,34). Most significantly, the amount of the 580 nt species was substantially diminished upon periodate-treatment, but the yields of the 90 and 120 nt TR-cDNAs were hardly changed (Figure 1F). These data indicate that the 580 nt species lacked TR-cDNAs, and instead consisted of the RNA template with one or just a few dNTPs added to its 3′ end in nontemplated fashion (3′ addition activity). DNase does not degrade further than tetranucleotides on average (35), explaining why the radiolabel on the 580 nt species survives DNase treatment. These data further suggested that covalently linked RNA–cDNA molecules were found in the several 540 nt species. The nature of these RNA–cDNA molecules, which migrate faster than the input 580 nt RNA template, is investigated below. bRT alone produced the 580 nt band but no cDNA (Figure 1C and Supplementary Figure S1G), indicating that it had the (presumably artifactual) 3′ addition activity but no reverse transcription activity. In support of this conclusion, when bRT alone was reacted with periodate-treated RNA, no radiolabeled products were formed (Supplementary Figure S1H). No radiolabeled products resulted from Avd alone, demonstrating that Avd had neither cDNA synthesis nor 3′ addition activity (Figure 1C and Supplementary Figure S1G).
Adenine-mutagenized TR-cDNA
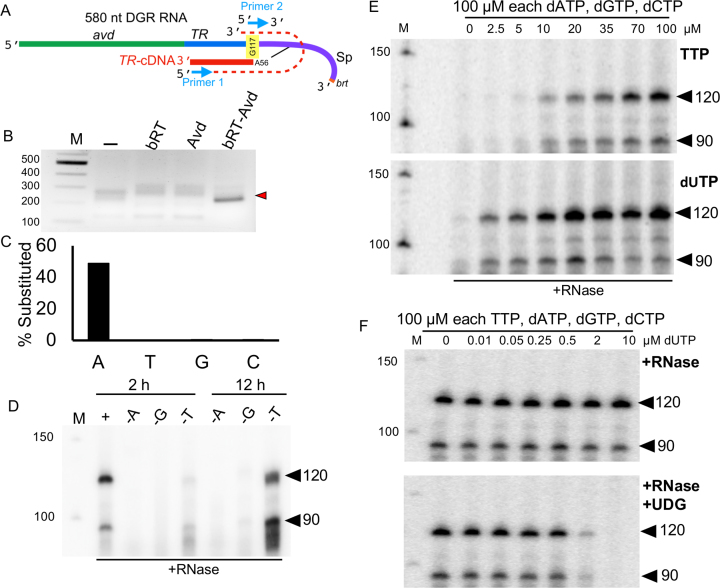
The sequence of cDNAs produced by bRT-Avd were identified from RNA–cDNA molecules that had been amplified by RT-PCR (Figure 2A, Supplementary Table S2). Amplicons were evident specifically for templates reacted with bRT-Avd, but not bRT or Avd alone, or no protein (Figure 2B). Sequencing of these amplicons showed that they contained TR, and remarkably ∼50% of TR adenines were substituted (Figure 2C and Supplementary Figure S2), identical to the ∼50% observed in vivo (1,4). Substitutions at other bases were detected, but at extremely low frequency (0–0.5%), as also reported in vivo (1), and rare single basepair deletions and insertions could be detected as well (0.1%). Adenine-specific misincorporation by bRT-Avd was confirmed by its ability to synthesize TR-cDNA in the absence of TTP, but not dATP or dGTP (Figure 2D and Supplementary Figure S3); dCTP was not omitted, as [α-32P]dCTP was used for radiolabeling. Synthesis excluding TTP resulted in greater production of the 90 nt product as compared to the 120 nt product and exhibited slower kinetics, most likely because misincorporation was required at every TR adenine.
Figure 2.
Adenine mutagenesis and template-priming. (A) Covalently-linked RNA–cDNA molecule. The linkage is to Sp A56 of the RNA, and the first nucleotide reverse transcribed is TR G117. The RT-PCR product resulting from primers 1 and 2 (blue arrows) is indicated by the dashed red line. (B) RT-PCR amplicons from 580 nt DGR RNA reacted with no protein (–), bRT, Avd, or bRT-Avd, separated on a 2% agarose gel and ethidium bromide-stained. The specific amplicon produced from reaction with bRT-Avd shown by the red arrowhead. (C) Percentage of substitutions in TR-cDNA determined by sequencing. (D) Radiolabeled 120 and 90 nt cDNA products, indicated by arrowheads, resulting from bRT-Avd activity with the 580 nt DGR RNA as template for 2 h (left) or 12 h (right). Either standard dNTPs (dATP, dGTP, dCTP, TTP), as indicated by ‘+’,were present in the reaction, or standard dNTPs excluding dATP (-A), dGTP (–G), or TTP (-T) were present. Products were treated with RNase, and resolved by denaturing PAGE. (E) Radiolabeled 120 and 90 nt cDNA products, indicated by arrowheads, resulting from bRT-Avd activity for 2 h with the 580 nt DGR RNA as template with varying TTP (top) or dUTP (bottom) concentrations. Products were treated with RNase, and resolved by denaturing PAGE. (F) Radiolabeled 120 and 90 nt cDNA products, indicated by arrowheads, resulting from bRT-Avd activity for 2 h with the 580 nt DGR RNA as template with varying dUTP concentrations. Products were either RNase-treated (top), or both RNase- and UDG-treated (bottom), and resolved by denaturing PAGE.
We also found that bRT-Avd was able to incorporate dUTP, but not UTP or dUMP, into TR-cDNA (Supplementary Figure S4A, B). Incorporation was assessed by scission of uracylated DNA upon digestion with uracil DNA glycosylase (UDG) and heating (36). bRT-Avd had an apparent ∼10-fold preference for dUTP over TTP (Figure 2E and Supplementary Figure S4C), and incorporated dUTP even when the dUTP concentration was 50-fold lower than that of competing dNTPs (Figure 2F and Supplementary Figure S4D). As validation of these assays, HIV-1 RT was shown to have a marginal preference for TTP over dUTP, as previously reported (37) (Supplementary Figure S4E–G). In Bordetella, however, dUTP was measured to be >200-fold lower in concentration than TTP (Supplementary Table S3), indicating that mutagenesis in vivo almost certainly proceeds through misincorporation of dATP, dGTP, or dCTP at template adenines rather than incorporation of dUTP followed by de-uracylation and repair (1,4).
Together, these results establish that bRT-Avd is sufficient for template-primed TR-cDNA synthesis from DGR RNA, and that bRT-Avd displays a vigorous promiscuity at template adenines but fidelity at other bases.
Priming from Sp 56
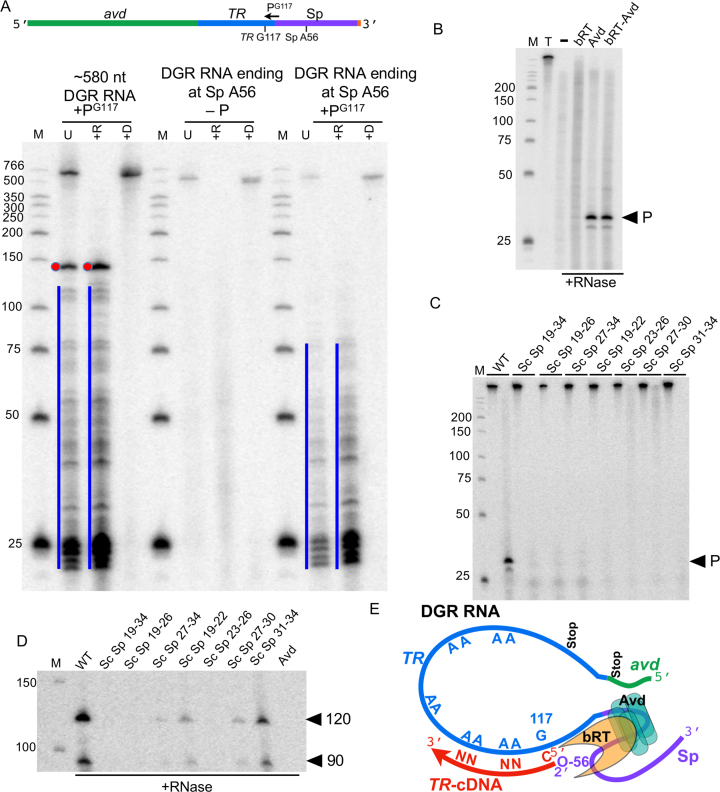
Sequencing of RT-PCR amplicons generated from RNA–cDNA molecules showed that immediately adjacent to TR was sequence from the noncoding spacer (Sp) region (Figure A and Supplementary Figure S2). TR and Sp sequences originated from different strands (i.e. sense versus antisense), signifying that Sp had folded back onto TR during priming (Figure 3A). The precise site of priming was ambiguous due to sequence degeneracy, but was clarified by the pattern of RNA substitutions in TR or Sp transferred to RNA–cDNA molecules (Supplementary Figure S5A and S5B). In short, we found that substitutions at Sp 55 and 56, but not TR 118 and 119, were transferred (Supplementary Figure S5C). These data showed that synthesis was primed from Sp A56, and that TR G117 was the first nucleotide reverse transcribed (Supplementary Figure S5D). TR G117 is located in a GC-rich region of the TR that is therefore not prone to adenine-mutagenesis. These assignments are identical to those derived from analysis of RNA–cDNA molecules produced in vivo (19).
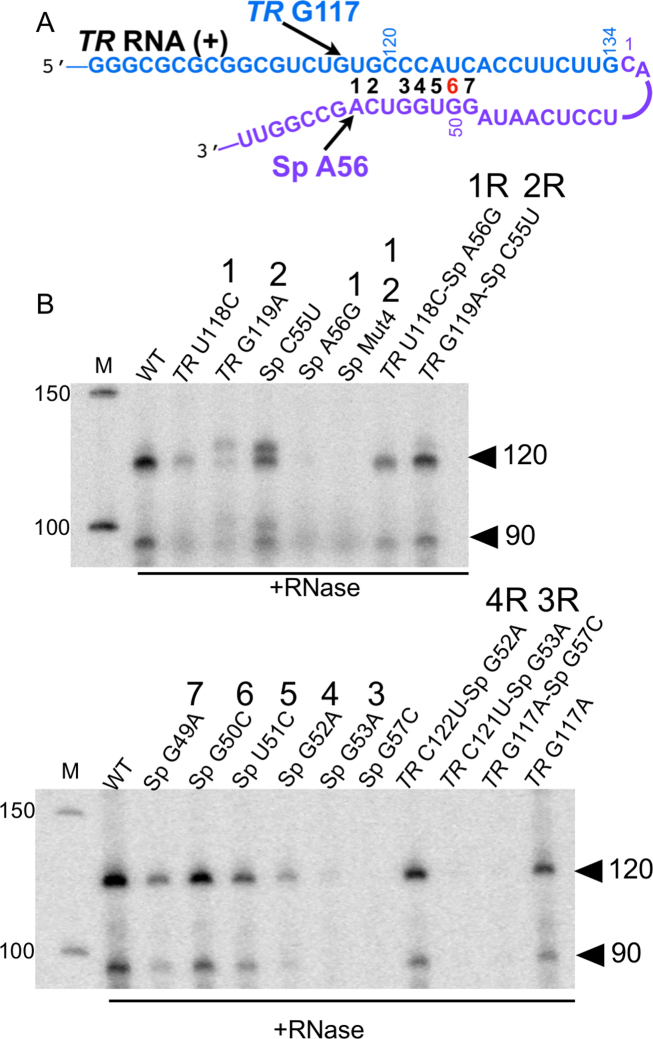
Figure 3.
TR-Sp interactions. (A) Complementarity between TR (blue) and Sp (purple) segments. Potential basepairs are numbered (wobble in red). (B) Radiolabeled 120 and 90 nt cDNA products, indicated by arrowheads, resulting from bRT-Avd activity for 2h with the WT or mutated 580 nt DGR RNA as template. Products were treated with RNase, and resolved by denaturing PAGE. Numbers over lane labels correspond to the basepair tested by the mutation, with ‘R’ referring to restoration of the basepair. Sp Mut4 corresponds to Sp 55-CAGC substituted with 55-GUCG.
The 3′ ends of cDNAs were identified by poly-dA tailing and PCR amplification (Supplementary Figure S6A). Amplicons were once again evident specifically from templates reacted with bRT-Avd, but not with bRT or Avd alone, or no protein (Supplementary Figure S6B). Three length classes of cDNAs, all adenine-mutagenized, were identified (Supplementary Figure S6C). The longest cDNAs were 118–122 nt, having terminated within the six 3′ bases of avd; these corresponded to the most abundant radiolabeled species, the 120 nt band (Figure 1C, Supplementary Figure S1F). The second longest were almost all 94–96 nt, having terminated between TR 22–24; these corresponded to the second most abundant radiolabeled species, the 90 nt band. Notably, codons permissive to adenine mutagenesis occur exclusively between TR 23–98, and thus both the 90 and 120 nt cDNAs carry the full potential of adenine-mutagenesis. Short cDNAs of 40–59 nt, for which synthesis had terminated between TR 59–78, were also found, and these corresponded to a minor population of radiolabeled cDNAs that became evident only after prolonged reaction times (Supplementary Figure S3). The relative proportion of these cDNAs did not change over time, indicating independent termination events rather than precursor-product relationships. The internally deleted TR20-Δ80 and TR10-Δ100 DGR RNAs (Figure 1D) lack the TR 22–24 termination site, explaining why they produced one and not two cDNA products.
TR-Sp interactions
Seven basepairs (bp), including a wobble, can be formed between sequences proximal to the initiation site in TR and the priming site in Sp (Figure 3A). Basepairing appeared to be important at three of these seven (bp 1, 2 and 4), as disruption led to greatly attenuated cDNA synthesis, and complementary substitutions rescued cDNA synthesis (Figure 3B and Supplementary Figure S7). For the complementary substitution at bp 1 (TR U118C-Sp A56G), Sp 56G was the priming nucleotide (Supplementary Figure S5C), indicating that the identity of the base at Sp 56 was not strictly essential to priming. For bp 2, which is adjacent to the priming site, disruption resulted in additional cDNA bands, which were due to non-templated insertion of four to seven nucleotides (Supplementary Figure S5C). Disruption of bp 3 caused a drastic reduction in cDNA synthesis, but a complementary substitution was ineffective at restoring synthesis, suggesting that guanine is required at Sp 53. Basepairs 5, 6 and 7 appeared not to be crucial, as disruptions affected cDNA synthesis only mildly. The in vitro results for bp 1 and 2 are similar to those found in vivo (19), including the addition of non-templated nucleotides. The other potential basepairs were not examined in vivo.
We asked whether the priming site could be shifted by introducing an additional basepair downstream of Sp 56, at Sp 57 (Sp G57C-TR G117). However, this nearly abrogated cDNA synthesis (Figure 3B and Supplementary Figure S7). Disruption of this artificial basepair (Sp G57C-TR G117A) did not rescue cDNA synthesis (TR G117A by itself was competent for cDNA synthesis), indicating a specific role for guanine at Sp 57.
In sum, these results provide evidence for limited basepairing between TR and Sp, and also suggest higher-order organization of the template, with specific bases required at certain positions.
RNA–cDNA linkage
The apparent ∼540 nt size of RNA-cDNA molecules produced by bRT-Avd was puzzling, given a 580 nt RNA template and 90 or 120 nt cDNAs. Two possibilities were examined. The first was that the RNA had been cleaved at Sp 56, thereby freeing its 3′-OH for priming. We asked whether a template truncated at Sp 56 was competent for cDNA synthesis, and found that it was not (Supplementary Figure S8). This result indicated that regions downstream of Sp 56 were essential for cDNA synthesis, but did not rule out cleavage at Sp 56. Cleavage was directly examined with an internally-labeled 580 nt DGR RNA incubated with bRT-Avd in the presence or absence of dNTPs. No cleavage products were detected, with as much as 10-fold overloading of the RNA (Supplementary Figure S9).
The second possibility examined was internal priming. To test this hypothesis, we biotinylated the 3′ end of the 580 nt DGR RNA, and used that as a template for reverse transcription by bRT-Avd (Figure 4A). Following reverse transcription, 3′-biotinylated RNA was isolated using streptavidin beads, and the presence of TR-cDNA accompanying the intact RNA was probed by PCR. A specific PCR amplicon of the correct size was evident for 3′-biotinylated RNA reacted with bRT-Avd, but not for 3′-biotinylated RNA reacted with no protein or unbiotinylated RNA reacted with bRT-Avd (Figure 4B). The PCR amplicon was confirmed to correspond to adenine-mutagenized TR-cDNA by sequencing (Supplementary Figure S10A), and input samples were confirmed to all contain RNA by RT-PCR (Supplementary Figure S10B); the two input samples reacted with bRT-Avd to contain TR-cDNA by PCR (Supplementary Figure S10C); and the two purified biotinylated samples to contain RNA by RT-PCR (Supplementary Figure S10B). We also verified that the RNA–cDNA molecules examined were in cis (part of the same molecule) and not in trans (two or more RNA molecules associated through a common TR-cDNA or other means). This was done by carrying out the bRT-Avd reverse transcription reaction with unbiotinylated RNA, inactivating bRT-Avd with proteinase K as in all experiments, and then adding an equal quantity of unreacted biotinylated RNA prior to isolation with streptavidin beads. We found no recruitment of unbiotinylated RNA–cDNA molecules by biotinylated RNA (Supplementary Figure S10D), demonstrating that the molecules examined were indeed in cis. These results indicated that the streptavidin beads had effectively and specifically captured the biotin at the 3′ end of the RNA, and that the captured and intact RNA contained TR-cDNA. This result also showed that internally located Sp 56 can serve as a priming site, with its 2′-OH being the most chemically plausible site for priming. RNA–cDNA linkage through a 2′-OH has been observed previously in retrons (38–41), in which a specific guanosine internally located in an RNA template primes cDNA synthesis by the retron reverse transcriptase, resulting in a branched RNA–cDNA molecule linked by a 2′-5′ phosphodiester bond.
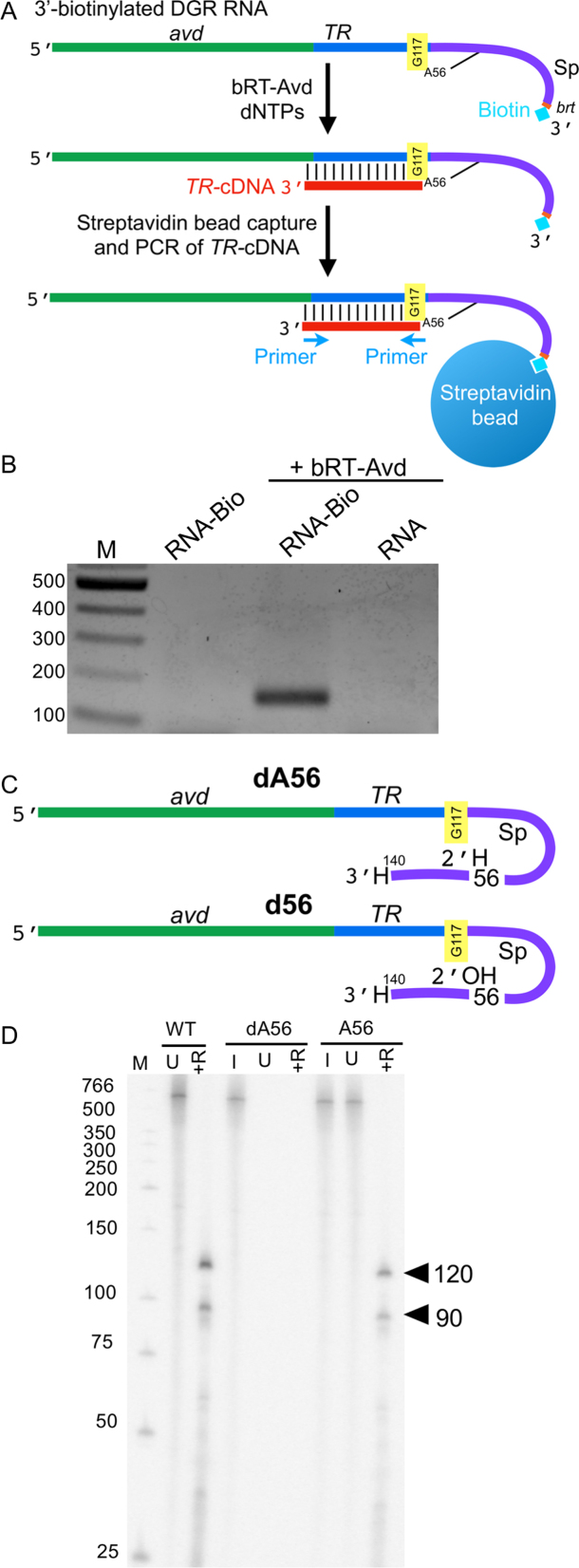
Figure 4.
Branched RNA–cDNA. (A) The 580 nt DGR RNA was biotinylated at its 3′ end and used as a template for reverse-transcription by bRT-Avd, after which biotinylated RNA was captured with streptavidin beads, and the presence of TR-cDNA was detected by PCR using the indicated primers. (B) The 580 nt DGR RNA was biotinylated at its 3′ end (RNA-Bio), and either reacted with no protein or used as a template for reverse transcription with bRT-Avd. The 580 nt DGR RNA in its unbiotinylated form (RNA) was also used as a template for reverse transcription with bRT-Avd. Samples were then purified using streptavidin beads, and the presence of TR-cDNA in the purified samples was assessed by PCR. Products from the PCR reaction were resolved on an agarose gel. (C) Hybrid dA56 580 nt DGR RNA containing deoxyadenosine at Sp 56 (indicated with H at 2′ position) and hybrid d56 580 nt DGR RNA containing adenosine at Sp 56 (indicated with OH at 2′). Both molecules terminate at Sp 140 and have a dideoxynucleotide at the 3′ end (indicated with H at 3′). (D) Radiolabeled products resulting from bRT-Avd activity for 12 h with 580 nt DGR RNA, hybrid 580 nt dA56, or hybrid 580 nt A56 DGR RNA as template. Products were untreated (U) or RNase-treated (+R), and resolved by denaturing PAGE. Separate samples of dA56 and A56 were 5′ 32P-labeled for visualization of input templates (I). The positions of the 120 and 90 nt cDNAs are indicated.
To test the importance of the 2′-OH of Sp 56 in priming, we used chemical synthesis and ligation to create a hybrid 580 nt DGR RNA with a deoxyadenosine at Sp 56 (dA56), and as a control, an equivalent molecule with ribonucleotides throughout (A56) (Figure 4C). For the efficiency of ligation, it was necessary to use a template that ended at Sp 140 and had a 3′ dideoxynucleotide. The latter precluded the 3′ addition activity of bRT. The A56 molecule supported cDNA synthesis, albeit less efficiently than fully in vitro transcribed DGR RNA (48%), which was likewise truncated at Sp 140 (Figure 4D). In contrast, an equivalent quantity of the dA56 hybrid molecule was incompetent for cDNA synthesis, demonstrating that the 2′-OH of Sp 56 was essential for priming. Taken together, these results support the conclusion that priming from the 580 nt DGR RNA occurs from Sp 56 without RNA cleavage, resulting in branched molecules linked by 2′-5′ RNA–cDNA phosphodiester bonds. Such RNA–cDNA branched molecules apparently septate through gels faster than the unreacted RNA template, as shown by the ∼540 nt species corresponding to RNA–cDNA molecules (Figure 1F).
While no RNA cleavage was evident for the 580 nt DGR RNA template, we did detect cleavage of a shorter template, called the ‘core DGR RNA’. This 294-nt template had just 20 nt of avd at its 5′ end and extended to Sp 140 at its 3′ end (Figure 5A). The core DGR RNA supported cDNA synthesis (Figure 5B), whereas a template lacking the entire avd sequence at the 5′ end or the entire Sp sequence at the 3′ end did not (Supplementary Figure S11A and B). Similarly, templates that contained only 10 nt of avd at the 5′ end or were deleted of 20 nt of Sp at the 3′ end did not support cDNA synthesis (Supplementary Figure S11C). These experiments showed that the extensive 300 nt sequence of avd upstream of TR required for near wild-type retrohoming activity (23) was almost entirely dispensable for cDNA synthesis. Notably, reverse transcription from the core DGR RNA resulted in ∼7-fold greater cDNA production (i.e., the 90 and 120 nt cDNAs) as compared to the 580 nt DGR RNA (Supplementary Figures S11C and S12A). We verified that cDNAs produced from the core DGR RNA were still primed from Sp 56 with TR G117 the first nucleotide reverse transcribed through sequencing of amplified RNA–cDNA molecules (Supplementary Figure S12B and C).
Figure 5.
Core DGR RNA. (A) Schematic of core DGR RNA. (B) Radiolabeled products resulting from bRT-Avd activity for 2 h with the core DGR RNA as template. Prior to the reverse transcription reaction, the RNA template was untreated (-Per) or treated with periodate (+Per). Products from the reaction were untreated (U) or treated with RNase (+R), and resolved by 6% denaturing PAGE. Lane T corresponds to internally-labeled core DGR RNA as a marker for the size of the template. Red arrowheads indicate radiolabeled product bands that migrate at the same position or slower than the core DGR RNA, and green arrowheads ones that migrate faster. The positions of the 120 and 90 nt cDNA bands are indicated. The two panels are from the same gel, with the black line indicating that intermediate lanes were removed. (C) Internally-labeled core DGR RNA was not incubated (–), or incubated with bRT-Avd alone or bRT-Avd with 100 μM standard dNTPs (+dNTP), 100 μM dCTP (+CTP), 100 μM dNTPs excluding dCTP (+d(A,T,G)TP), or 100 μM nonhydrolyzeable analog of dCTP (+N-dCTP) for 2 h. Incubation products were resolved by denaturing PAGE. The band corresponding to the 5′ fragment of the cleaved core RNA containing either a deoxycytidine alone (5′+dC) or cDNA (5′+cDNA), and the band corresponding to the 3′ fragment of the RNA are indicated. (D) The core DGR RNA was biotinylated at its 3′ end (RNA-Bio), and either reacted with no protein or used as a template for reverse transcription with bRT-Avd. The core DGR RNA in its unbiotinylated form (RNA) was also used as a template for reverse transcription with bRT-Avd. Samples were then purified using streptavidin beads, and the presence of TR-cDNA in the purified samples was assessed by PCR. Products from the PCR reaction were resolved on an agarose gel. (E) Radiolabeled products resulting from bRT-Avd activity for 12 h with core, hybrid core dA56, or hybrid core A56 DGR RNA as template. Products were untreated (U) or treated with RNase (+R), and resolved by denaturing PAGE. Separate samples of core dA56 and A56 were 5′ 32P-labeled for visualization of inputs (I). The positions of the 120 and 90 nt cDNAs are indicated.
Most interestingly, several RNA–cDNA bands were produced from the core DGR RNA, with some of these having a mobility greater than the unreacted template (Figure 5B). To delve into these RNA–cDNA molecules further, we asked whether the core DGR RNA had been cleaved. Upon incubation with bRT-Avd, cleavage of ∼10% of internally-labeled core DGR RNA was evident, but only in the presence of dNTPs (Figure 5C). Indeed, the initiating dNTP (i.e. dCTP, which basepairs to TR G117) was necessary and sufficient for cleavage, and hydrolysis of dCTP was required, as cleavage did not occur with a nonhydrolyzeable dCTP analog. Since the core and 580 nt DGR RNAs differed at both 5′ and 3′ ends, we examined cleavage of a 580 nt DGR RNA lacking just the 3′ end (3′Δ10). Cleavage was not evident (Supplementary Figure S12D), indicating that the length of the avd sequence at the 5′ end modulated cleavage.
The sizes of the cleavage products were consistent with bond breakage circa Sp 56. The larger cleavage product was ∼210 nt in length when dCTP was added and >210 nt when dNTPs were added, indicating that it included cDNA linked to Sp 56 and corresponded to the 5′ end of the core DGR RNA. The smaller cleavage product had a size of ∼80 nt regardless of whether dNTPs or dCTP alone were added, indicating that it did not include cDNA or the Sp 56 priming site, and corresponded to the 3′ end of the core DGR RNA. The 3′ cleavage product was purified, amplified by RT-PCR following ligation of an adaptor to its 5′ end, and cloned and sequenced. This showed that, surprisingly, Sp 58 and not Sp 57 was at its 5′ end, indicating that cleavage had occurred between Sp 57–58 (Supplementary Figure S12E and F). This 3′ cleavage product was also phosphorylated at its 5′ end prior to adaptor ligation to account for the possibility that some cleaved molecules may have lacked a 5′ phosphate. The sequencing results from this experiment were identical, showing that Sp 58 was at the 5′ end. We attempted a parallel method for sequencing the 3′ end of the larger cleavage product, but were unsuccessful, perhaps due to deoxycytidine at Sp 56 blocking adapter ligation. We conclude from these data that synthesis-dependent cleavage occurs between Sp 57–58, but cannot rule out the possibility that a second cleavage event occurs between Sp 56–57.
To determine whether cleavage was necessary for cDNA synthesis from the core DGR RNA, we used 3′-biotinylated core DGR RNA as a template for reverse transcription, as described above (Figure 4A). We once again captured the 3′-biotinylated RNA following reverse transcription with streptavidin beads, and asked whether TR-cDNA accompanying the intact RNA was present by PCR. We observed a specific amplicon of the correct size for 3′-biotinylated core DGR RNA reacted with bRT-Avd, but not for 3′-biotinylated RNA reacted with no protein or unbiotinylated RNA reacted with bRT-Avd (Figure 5D). The PCR amplicon was verified to be adenine-mutagenized TR-cDNA by sequencing (Supplementary Figure S13A), and the integrity of input and purified samples was verified, as described above for the 580 nt DGR RNA (Supplementary Figure S13B and C). We also confirmed that RNA–cDNA molecules were in cis (Supplementary Figure S13D). This result indicated that cleavage was not required for priming, as internally located Sp 56 could serve as the priming site. To verify the importance of the 2′-OH of Sp A56 for priming in the core DGR RNA, we created a hybrid molecule with dA56 and a control molecule with A56, as described above (Figure 4C). The A56 form of the core DGR RNA supported cDNA synthesis (50% of that from fully in vitro transcribed RNA). However, cDNA synthesis was severely attenuated from an equivalent quantity of the core dA56 hybrid molecule (Figure 5E), showing that the 2′-OH of Sp A56 was crucial for cDNA synthesis from the core DGR RNA. The 2′-OH of Sp A56 was also crucial for cleavage of the core DGR RNA, as the core A56 molecule was cleaved in the presence of bRT-Avd and dCTP, whereas dA56 was not (Supplementary Figure S12G). In this experiment, only the in vitro transcribed portion of the hybrid molecule was internally-labeled, explaining why only the larger of the two cleavage bands was visible for A56.
With these findings in hand, we sought to assign the RNA–cDNA bands from the core DGR RNA. To this end, the core DGR RNA was treated with periodate prior to reverse transcription to minimize the 3′ addition activity of bRT. Four RNA–cDNA bands were observed (Figure 5B). The very uppermost band (Figure 5B, top red arrowhead), at an apparent size of ∼330 nt, likely contained the 120 nt cDNA covalently linked to the ∼210 nt RNA that had been cleaved at Sp 57, and the next uppermost band, at an apparent size of ∼300 nt, likely contained the 90 nt cDNA linked to the cleaved RNA. The two lower bands (Figure 5B, green arrowheads), with greater mobility than the unreacted core DGR RNA, likely corresponded to branched RNA–cDNA molecules containing intact RNA, as seen with the 580 nt DGR RNA.
These results indicate that the core DGR RNA can be cleaved by bRT-Avd and that cleavage is coupled to cDNA synthesis. However, cleavage was not required for cDNA synthesis. The 2′-OH of internally located Sp 56 could serve as the priming site, leading to branched RNA–cDNA molecules.
Specificity to DGR RNA
We asked whether template-primed cDNA synthesis could also occur on a heterologous template. Using a ∼200 nt non-DGR template and no primer, we observed no cDNA synthesis and only 3′ addition activity (Supplementary Figure S14A). However, with the addition of an oligodeoxynucleotide (ODN) primer, bRT-Avd synthesized cDNAs, which were as expected not covalently attached to the RNA, but were quite short (∼5–35 nt).
We next asked whether template-priming was required for cDNA synthesis from the DGR RNA. When synthesis was primed with an ODN such that TR G117 was the first nucleotide reverse transcribed (Supplementary Figure S14B and C, PG117 primer), a 120 nt TR-cDNA and other extended TR-cDNAs were produced, along with some shorter ones. This ODN primer inhibited template-primed cDNA synthesis. In contrast, when synthesis was primed by ODNs that annealed to other locations on the DGR RNA, exclusively short ODN-primed cDNAs were produced, just as with the non-DGR template. Important insight into the organization of the DGR RNA template came from the observation that truncation of the DGR RNA at Sp 56 eliminated the synthesis of extended cDNAs primed by PG117. Only short cDNAs were produced from the truncated template (Figure 6A, PG117 primer). Deletion of the avd sequence at the 5′ end of the 580 nt DGR RNA greatly reduced but did not eliminate synthesis of extended cDNAs from primer PG117 (Supplementary Figure S14D), indicating that the avd sequence has a role in processive polymerization but a less major one than the Sp region. These results suggest that avd and Sp are organized such that TR G117 exists at a functionally privileged location that promotes the processivity of bRT-Avd and enables synthesis of extended cDNAs. In addition, these results showed that processivity from TR 117 was independent of the mechanism of priming, occurring when synthesis was primed either by the RNA template or an exogenous ODN.
Figure 6.
Specificity to DGR RNA. (A) Top, schematic of DGR RNA template and primer PG117. Bottom, radiolabeled products resulting from bRT-Avd activity for 2 h with intact 580 nt DGR RNA or DGR RNA truncated at Sp A56 as template. Reverse transcription reactions were carried out in the absence (-P) or presence of primer PG117. Reaction products were untreated (U), treated with RNase (+R), or treated with DNase (+D), and resolved by denaturing PAGE. The blue line indicates ODN-primed cDNA products. The red dot indicates ODN-primed 120 nt cDNA (cDNA + 20 nt primer for a 140 nt band). (B) Protection of internally-labeled 580 nt DGR RNA from RNase by bRT, Avd, or bRT-Avd, with products resolved by 15% denaturing PAGE. The protected band (P) is indicated. (C) RNase protection by Avd, as in panel B, carried out on internally-labeled wild-type 580 nt DGR RNA or 580 nt DGR RNA with scrambled (Sc) Sp sequences, with the first lane in each pair untreated and the second RNase-treated. Products were resolved by denaturing PAGE. The protected band (P) is indicated. (D) Radiolabeled products resulting from bRT-Avd activity for 2 h with the WT 580 nt DGR RNA or DGR RNA containing scrambled (Sc) Sp sequences as template. The last lane shows the activity of Avd alone for 2 h with the WT 580 nt DGR RNA as template. Products were treated with RNase, and resolved by denaturing PAGE. The positions of the 120 and 90 nt cDNAs indicated. (E) Model of processive polymerization of adenine-mutagenized cDNA by bRT-Avd/RNA ribonucleoprotein particle. The 2′-OH of Sp 56 serves as the priming site and forms a 2′-5′ phosphodiester bond with the cDNA. The first nucleotide reverse transcribed is TR 117. Adenines in TR are unfaithfully reverse transcribed by bRT-Avd (represented by ‘N’). The RNP promotes processive polymerization, which terminates at one of two stops in the DGR RNA.
Both bRT and Avd were required for ODN-primed cDNA synthesis from DGR and non-DGR templates, suggesting a role for Avd in catalysis. Sequencing of short ODN-primed cDNAs synthesized from the non-DGR RNA template revealed adenine-mutagenesis (Supplementary Figure S14E). This promiscuity extended to the ability of bRT-Avd to incorporate dUTP into cDNAs synthesized from a non-DGR template (Supplementary Figure S14F). These results indicate that adenine-mutagenesis, unlike synthesis of extended cDNAs, is not restricted by the RNA template and is instead intrinsic to bRT-Avd.
To determine the basis for this restriction to the DGR RNA, an RNase protection assay was carried out with internally-labeled 580 nt DGR RNA. A ∼30 nt band was protected by Avd and bRT-Avd, but not bRT alone (Figure 6B). Deep sequencing revealed that this band contained Sp 19–34 (Supplementary Figure S15A). The region downstream of Sp 56 was not required for protection by Avd (Supplementary Figure S15B). Scrambling the sequence of Sp 19–34, either in its entirety or as 4- or 8-nt segments, resulted in loss of RNase protection by Avd and bRT-Avd (Figure 6C and Supplementary Figure S15C, Supplementary Table S1). Synthesis of cDNA from these templates was also severely attenuated (Figure 6D and Supplementary Figure S15C). The exception was scrambled Sp 31–34, for which RNase protection was lost but cDNA synthesis maintained at one-third of wild-type. Apparently, Avd binding to this template was sufficiently weakened to permit access to RNase but not enough to drastically reduce cDNA synthesis, consistent with multiple Avd contact points to the RNA. The boundaries of the Avd binding site were probed by scrambling Sp 4–18 and Sp 35–44. The former eliminated RNase protection and severely attenuated cDNA synthesis, while the latter had little effect on either (Supplementary Figure S15E–G). Interestingly, with Sp 45–49 scrambled, the RNA was not protected by Avd but was protected by bRT-Avd, and cDNA synthesis was reduced to one-fifth of wild-type (Supplementary Figure S15G). This suggests that Avd forms weak contacts to this priming-proximal region, and these contacts are reinforced by bRT. Taken together, these results are consistent with Sp 4–30 forming the Avd binding site, and indicate that Avd, in addition to its role in catalysis, is required for targeting bRT-Avd to the Sp region of DGR RNA.
DISCUSSION
Adenine-mutagenesis is a hallmark feature of DGRs. We sought to identify the events involved in cDNA synthesis and adenine-mutagenesis through in vitro reconstitution of components of the Bordetella bacteriophage DGR. An essential step in accomplishing this was isolation of biochemically well-behaved bRT, which required its co-expression with Avd. A second essential step was the purification to homogeneity of in vitro transcribed RNA, without which no cDNA synthesis was evident. Once purified, bRT-Avd and DGR RNA recapitulated synthesis of template-primed RNA–cDNA molecules in vitro, as observed in vivo (19). The correspondence to in vivo observations included identical sites of priming (Sp 56) and initiation (TR 117) (19). This correspondence further included the level of cDNA synthesis from mutated DGR RNA templates (19). Specifically, substitutions that abrogate or decrease cDNA synthesis in vivo did the same in vitro (i.e., Sp A56G and Mut4 for the former, and TR U118C and G119A for the latter), and a substitution that has a mild effect in vivo also had a mild effect in vitro (Sp C55U). Most significantly, these trends in cDNA synthesis closely parallel retrohoming activity (19), providing evidence that the RNA–cDNA molecules observed in vitro and in vivo are on the path to sequence diversification.
Using the in vitro system, we discovered that the bRT-Avd complex was necessary and sufficient for adenine-mutagenesis. bRT by itself had no reverse transcription activity, and required the participation of Avd. Synthesis of adenine-mutagenized cDNA was intrinsic to the bRT-Avd complex, occurring for either DGR or non-DGR RNA templates, and occurring either when primed by the template or an exogenous ODN. Remarkably, misincorporation by bRT-Avd at template adenines occurred with near equal efficiency as proper incorporation. This promiscuity extended to dUTP incorporation, with bRT-Avd displaying an unprecedentedly strong preference for dUTP over TTP (37). In Bordetella, the dUTP concentration was measured to be sufficiently low that misincorporation rather than uridine incorporation was the likely pathway for adenine-mutagenesis. The proclivity to incorporate uridines likely reflects the extreme infidelity of bRT-Avd at template adenines, but may also have a role in diversification in organisms or conditions in which the dUTP concentration is substantially higher.
While the RNA template had no apparent role in adenine-mutagenesis, the processivity of bRT-Avd was strictly dependent on the RNA template. This was evident in experiments using exogenous primers. A primer that initiated synthesis from TR 117 (PG117) resulted in extended cDNA synthesis, but only when the RNA template included intact Sp. When Sp was truncated at Sp 56, only short cDNAs could be primed from TR 117 by PG117. This was remarkable as PG117 annealed to TR, quite distant from Sp 56. Likewise, only short cDNAs were synthesized when primed by an ODN from other locations of the DGR RNA or from non-DGR RNA templates. Loss of the avd sequence upstream of TR had a less dramatic effect than loss of Sp on synthesis primed by PG117, but still resulted in a substantial decrease in the synthesis of extended cDNA. These upstream and downstream sequences surrounding TR were essential for template-primed synthesis. Almost the entire Sp region was required for template-primed synthesis, as were 20 nt of avd. Taken together, these results suggest that the DGR RNA and bRT-Avd combine to form a ribonucleoprotein (RNP) particle that creates a privileged site for cDNA synthesis from TR 117 (Figure 6E). Recruitment of bRT-Avd by Sp 4–30 along with higher-order structure in Sp, and perhaps avd, is likely to position the 2′-OH of Sp 56 as the priming group and TR 117 as the templating base at the catalytic site of bRT. Without avd and Sp sequences surrounding TR, it is likely that no RNP forms or is incorrectly structured such that Sp 56 is unavailable for priming. This would explain why these surrounding sequences are absolutely required for template-primed synthesis of TR-cDNA. We further envision that the RNP stabilizes contacts between bRT-Avd and the RNA template, thereby supporting processive polymerization. While an exogenous primer that initiates from TR 117 can circumvent the need for template-priming from Sp 56, this would also explain why processive polymerization from such a primer is reliant on avd and Sp sequences surrounding TR.
Although processive cDNA synthesis can occur from this privileged site either through template- or ODN-primed mechanisms, template-priming provides a clear advantage in obviating the need for additional factors encoded by the DGR (e.g., primase) or the host (e.g., primer). The former may be important for the genomic economy of DGRs, and the latter for the horizontal spread of DGRs to disparate hosts. The existence of a privileged site in the DGR RNA for reverse transcription, beyond providing efficiency for the synthesis of TR-cDNA, is also likely to protect the host genome from damage. Genome damage due to retrotransposons, such as LINE and Ty elements, has been documented (42,43). Aberrant reverse transcription of cellular mRNAs by retrotransposon reverse transcriptases results in cDNAs that become incorporated into the genome as pseudogenes via DNA repair and recombination pathways (44). Thus, the functional necessity for the avd and Sp regions provides a mechanism for restricting bRT-Avd action to DGR RNA and excluding host RNAs, thereby curtailing the possibility of deleterious pseudogenes arising in host genomes from DGR activity.
Lastly, we found that bRT-Avd is able to carry out RNA cleavage in a manner that is coupled to reverse transcription. However, the functional importance of RNA cleavage remains obscure. We observed cleavage of the core DGR RNA between Sp 57–58, but not the 580 nt DGR RNA, which differs from the core DGR RNA primarily in the sequence length of avd upstream of TR. RNA cleavage occurs in group II introns as well, and is catalysed in large part by the RNA itself (45). The group II intron maturase protein has a minor role in RNA cleavage, but its reverse transcriptase active site is not required (45), suggesting different mechanisms for RNA cleavage in group II introns and DGRs. Cleavage has also been observed for DGR RNA isolated from Bordetella, and here too, cleavage required a catalytically active bRT (19). The implication from this study was that RNA cleavage frees the 3′-OH of Sp 56 to act as the priming site. However, no evidence in this study indicates that RNA cleavage is required for reverse transcription. Our results, on the other hand, provide evidence that RNA cleavage is not required for reverse transcription. By isolating RNA–cDNA molecules and probing for the presence of intact Sp, we found that the priming site at Sp 56 can be internally located within the RNA. We next showed that the 2′-OH of Sp 56, the most chemically plausible site for priming, was required for reverse transcription from both the core and 580 nt DGR RNA templates. This was done by constructing hybrid RNA molecules that had a deoxynucleotide specifically at Sp 56. In the case of the core DGR RNA, we also found that RNA cleavage was dependent on the 2′-OH of Sp 56. Taken together, these observations suggest that reverse transcription is primed by the 2′-OH of Sp 56, and that RNA cleavage between Sp 57–58 follows priming, explaining the dependency of cleavage on the 2′-OH of Sp 56. If RNA cleavage is not required for cDNA synthesis, what is its function? This remains an open question, but our evidence suggests that if RNA cleavage has a functional role, it is likely to be at a step following cDNA synthesis.
In summary, our studies set the stage for addressing the next steps in mutagenic retrohoming as well as elucidating the structural basis for the uniquely selective promiscuity of bRT-Avd.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Naorem and H. Guo for sharing results prior to publication. We also thank A. Robart, N. Toor, S. Joseph and G. Ghosh for discussion and helpful suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01 AI096838 to J.F.M. and P.G.]. Funding for open access charge: NIH [R01 AI096838].
Conflict of interest statement. J.F.M. is a cofounder, equity holder and chair of the scientific advisory board of AvidBiotics Inc., a biotherapeutics company in San Francisco. The remaining authors declare that they have no competing interests.
REFERENCES
- 1. Liu M., Deora R., Doulatov S.R., Gingery M., Eiserling F.A., Preston A., Maskell D.J., Simons R.W., Cotter P.A., Parkhill J. et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science. 2002; 295:2091–2094. [DOI] [PubMed] [Google Scholar]
- 2. Minot S., Grunberg S., Wu G.D., Lewis J.D., Bushman F.D.. Hypervariable loci in the human gut virome. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:3962–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ye Y. Identification of diversity-generating retroelements in human microbiomes. Int. J. Mol. Sci. 2014; 15:14234–14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guo H., Arambula D., Ghosh P., Miller J.F.. Diversity-generating retroelements in phage and bacterial genomes. Microb. Spectr. 2014; 2:doi:10.1128/microbiolspec.MDNA3-0029-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paul B.G., Bagby S.C., Czornyj E., Arambula D., Handa S., Sczyrba A., Ghosh P., Miller J.F., Valentine D.L.. Targeted diversity generation by intraterrestrial archaea and archaeal viruses. Nat. Commun. 2015; 6:6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nimkulrat S., Lee H., Doak T.G., Ye Y.. Genomic and metagenomic analysis of Diversity-Generating retroelements associated with treponema denticola. Front. Microbiol. 2016; 7:852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu L., Gingery M., Abebe M., Arambula D., Czornyj E., Handa S., Khan H., Liu M., Pohlschroder M., Shaw K.L. et al. Diversity-generating retroelements: natural variation, classification and evolution inferred from a large-scale genomic survey. Nucleic Acids Res. 2018; 46:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paul B.G., Burstein D., Castelle C.J., Handa S., Arambula D., Czornyj E., Thomas B.C., Ghosh P., Miller J.F., Banfield J.F. et al. Retroelement-guided protein diversification abounds in vast lineages of Bacteria and Archaea. Nat. Microbiol. 2017; 2:17045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Le Coq J., Ghosh P.. Conservation of the C-type lectin fold for massive sequence variation in a Treponema diversity-generating retroelement. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:14649–14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boehm T., McCurley N., Sutoh Y., Schorpp M., Kasahara M., Cooper M.D.. VLR-based adaptive immunity. Annu. Rev. Immunol. 2012; 30:203–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Litman G.W., Rast J.P., Fugmann S.D.. The origins of vertebrate adaptive immunity. Nat. Rev. Immunol. 2010; 10:543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McMahon S.A., Miller J.L., Lawton J.A., Kerkow D.E., Hodes A., Marti-Renom M.A., Doulatov S., Narayanan E., Sali A., Miller J.F. et al. The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat. Struct. Mol. Biol. 2005; 12:886–892. [DOI] [PubMed] [Google Scholar]
- 13. Dai W., Hodes A., Hui W.H., Gingery M., Miller J.F., Zhou Z.H.. Three-dimensional structure of tropism-switching Bordetella bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4347–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Melvin J.A., Scheller E.V., Miller J.F., Cotter P.A.. Bordetella pertussis pathogenesis: current and future challenges. Nat. Rev. Microbiol. 2014; 12:274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miller J.L., Le Coq J., Hodes A., Barbalat R., Miller J.F., Ghosh P.. Selective ligand recognition by a diversity-generating retroelement variable protein. PLoS Biol. 2008; 6:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doulatov S., Hodes A., Dai L., Mandhana N., Liu M., Deora R., Simons R.W., Zimmerly S., Miller J.F.. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004; 431:476–481. [DOI] [PubMed] [Google Scholar]
- 17. Simon D.M., Zimmerly S.. A diversity of uncharacterized reverse transcriptases in bacteria. Nucleic Acids Res. 2008; 36:7219–7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo H., Tse L.V., Barbalat R., Sivaamnuaiphorn S., Xu M., Doulatov S., Miller J.F.. Diversity-generating retroelement homing regenerates target sequences for repeated rounds of codon rewriting and protein diversification. Mol. Cell. 2008; 31:813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Naorem S.S., Han J., Wang S., Lee W.R., Heng X., Miller J.F., Guo H.. DGR mutagenic transposition occurs via hypermutagenic reverse transcription primed by nicked template RNA. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E10187–E10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Handa S., Paul B.G., Miller J.F., Valentine D.L., Ghosh P.. Conservation of the C-type lectin fold for accommodating massive sequence variation in archaeal diversity-generating retroelements. BMC Struct. Biol. 2016; 16:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cost G.J., Feng Q., Jacquier A., Boeke J.D.. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002; 21:5899–5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luan D.D., Korman M.H., Jakubczak J.L., Eickbush T.H.. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993; 72:595–605. [DOI] [PubMed] [Google Scholar]
- 23. Alayyoubi M., Guo H., Dey S., Golnazarian T., Brooks G.A., Rong A., Miller J.F., Ghosh P.. Structure of the essential diversity-generating retroelement protein bAvd and its functionally important interaction with reverse transcriptase. Structure. 2013; 21:266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qu G., Kaushal P.S., Wang J., Shigematsu H., Piazza C.L., Agrawal R.K., Belfort M., Wang H.W.. Structure of a group II intron in complex with its reverse transcriptase. Nat. Struct. Mol. Biol. 2016; 23:549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao C., Pyle A.M.. Crystal structures of a group II intron maturase reveal a missing link in spliceosome evolution. Nat. Struct. Mol. Biol. 2016; 23:558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stamos J.L., Lentzsch A.M., Lambowitz A.M.. Structure of a thermostable group II intron reverse transcriptase with Template-Primer and its functional and evolutionary implications. Mol. Cell. 2017; 68:926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011; 17:10–12. [Google Scholar]
- 29. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S. . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miller J.R., Koren S., Sutton G.. Assembly algorithms for next-generation sequencing data. Genomics. 2010; 95:315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Volkmer B., Heinemann M.. Condition-dependent cell volume and concentration of Escherichia coli to facilitate data conversion for systems biology modeling. PLoS One. 2011; 6:e23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neu H.C., Heppel L.A.. Nucleotide sequence analysis of polyribonucleotides by means of periodate oxidation followed by Cleavage with an amine. J. Biol. Chem. 1964; 239:2927–2934. [PubMed] [Google Scholar]
- 34. Tanner N.K., Cech T.R.. Guanosine binding required for cyclization of the self-splicing intervening sequence ribonucleic acid from Tetrahymena thermophila. Biochemistry. 1987; 26:3330–3340. [DOI] [PubMed] [Google Scholar]
- 35. Matsuda M., Ogoshi H.. Specificity of DNase I:Estimation of nucleosides present at the 5′-Phosphate terminus of a limit digest of DNA by DNase I. J. Biochem. 1966; 59:230–235. [PubMed] [Google Scholar]
- 36. Hayakawa H., Kumura K., Sekiguchi M.. Role of uracil-DNA glycosylase in the repair of deaminated cytosine residues of DNA in Escherichia coli. J. Biochem. 1978; 84:1155–1164. [DOI] [PubMed] [Google Scholar]
- 37. Kennedy E.M., Daddacha W., Slater R., Gavegnano C., Fromentin E., Schinazi R.F., Kim B.. Abundant non-canonical dUTP found in primary human macrophages drives its frequent incorporation by HIV-1 reverse transcriptase. J. Biol. Chem. 2011; 286:25047–25055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Furuichi T., Dhundale A., Inouye M., Inouye S.. Branched RNA covalently linked to the 5′ end of a single-stranded DNA in Stigmatella aurantiaca: structure of msDNA. Cell. 1987; 48:47–53. [DOI] [PubMed] [Google Scholar]
- 39. Furuichi T., Inouye S., Inouye M.. Biosynthesis and structure of stable branched RNA covalently linked to the 5′ end of multicopy single-stranded DNA of Stigmatella aurantiaca. Cell. 1987; 48:55–62. [DOI] [PubMed] [Google Scholar]
- 40. Elfenbein J.R., Knodler L.A., Nakayasu E.S., Ansong C., Brewer H.M., Bogomolnaya L., Adams L.G., McClelland M., Adkins J.N., Andrews-Polymenis H.L.. Multicopy Single-Stranded DNA directs intestinal colonization of enteric pathogens. PLos Genet. 2015; 11:e1005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shimamoto T., Inouye M., Inouye S.. The formation of the 2′,5′-phosphodiester linkage in the cDNA priming reaction by bacterial reverse transcriptase in a cell-free system. J. Biol. Chem. 1995; 270:581–588. [DOI] [PubMed] [Google Scholar]
- 42. Derr L.K., Strathern J.N.. A role for reverse transcripts in gene conversion. Nature. 1993; 361:170–173. [DOI] [PubMed] [Google Scholar]
- 43. Esnault C., Maestre J., Heidmann T.. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000; 24:363–367. [DOI] [PubMed] [Google Scholar]
- 44. Maxwell P.H., Curcio M.J.. Retrosequence formation restructures the yeast genome. Genes Dev. 2007; 21:3308–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cui X., Matsuura M., Wang Q., Ma H., Lambowitz A.M.. A group II intron-encoded maturase functions preferentially in cis and requires both the reverse transcriptase and X domains to promote RNA splicing. J. Mol. Biol. 2004; 340:211–231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.