Abstract
Sensing of nucleic acids for molecular discrimination between self and non-self is a challenging task for the innate immune system. RNA acts as a potent stimulus for pattern recognition receptors including in particular human Toll-like receptor 7 (TLR7). Certain RNA modifications limit potentially harmful self-recognition of endogenous RNA. Previous studies had identified the 2′-O-methylation of guanosine 18 (Gm18) within tRNAs as an antagonist of TLR7 leading to an impaired immune response. However, human tRNALys3 was non-stimulatory despite lacking Gm18. To identify the underlying molecular principle, interferon responses of human peripheral blood mononuclear cells to differentially modified tRNALys3 were determined. The investigation of synthetic modivariants allowed attributing a significant part of the immunosilencing effect to the 2′-O-methylthymidine (m5Um) modification at position 54. The effect was contingent upon the synergistic presence of both methyl groups at positions C5 and 2’O, as shown by the fact that neither Um54 nor m5U54 produced any effect alone. Testing permutations of the nucleobase at ribose-methylated position 54 suggested that the extent of silencing and antagonism of the TLR7 response was governed by hydrogen patterns and lipophilic interactions of the nucleobase. The results identify a new immune-modulatory endogenous RNA modification that limits TLR7 activation by RNA.
INTRODUCTION
A limited set of germline-encoded pattern recognition receptors is capable of detecting the broad variety of invading pathogens due to the recognition of highly conserved pathogen-associated molecular patterns (PAMPs). Besides cell wall components of bacteria, nucleic acids serve as an important PAMP of viruses as well as of bacteria to activate the innate immune system within minutes. DNA and RNA can either be recognized by endosomal Toll-like receptors (TLR) or cytosolic receptors including retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5) and activation of the cGAS –STING axis. Endosomal TLRs like TLR3 and TLR7 are known to sense double-stranded RNA (dsRNA) and single-stranded RNA (ssRNA), respectively (1–4). Upon stimulation of TLR3 or TLR8 in myeloid dendritic (mDCs) cells and macrophages or TLR7 in plasmacytoid dendritic cells (pDCs) type-I interferons are secreted. Several groups consistently reported that, in isolated peripheral blood mononuclear cells (PBMCs), the release of interferon-α (IFN-α) upon stimulation with RNA is attributed to pDCs only (1,5–9).
Previous studies described activation of TLR7 by tRNAs although those RNAs are intrinsically folded and therefore show double-stranded structures. For example, the D-loop and T-loop of canonical tRNAs form very stable tertiary interactions which are reinforced by post-transcriptional modification of key nucleotides (10). Interestingly, one such modification was found to silence the TLR7-mediated response of pDCs toward tRNA of various origins. The highly conserved 2′-O-methylation of guanosine at position 18 is present in ‘self’ as well as ‘non-self’ tRNA isoacceptors (8,11). Given that the residues G18-G19 are buried deep inside the tRNA structure where they are structurally interacting with Ψ55 –C56 (10), TLR7 seems to be able to access residues that are not a priori single-stranded. Yet, it is still unclear if recognition of Gm18 by TLR7 is based on an intact tRNA which would imply tRNA unfolding and presumably base-flipping or Gm18 might exhibit its antagonism only after degradation.
Recent crystal structures of the ssRNA sensors TLR7 and TLR8 could only demonstrate the binding of a single nucleotide to a binding site 1 (first) and a tri- or dinucleotide at binding site 2 (second) (12,13). So far, no structure of full-length RNA bound to TLR7 is available, however, triggering TLR7 seems to require a minimum RNA length of ∼20 nt. This piece of biochemical evidence suggests that both binding sites might communicate via the continuous chain of a bound RNA (14–16) in turn might trigger TLR7 dimerization. However, because of the degradation of RNAs during the crystallization process, the role of chain connectivity between nucleotides addressing both sites remains unresolved. Of note, inhibition of TRL7 signaling could be achieved with a 2′-O-methylated oligoribonucleotide (17,18) at a minimal length of 9 nt. In addition, small activating molecules such as R848 demonstrate a preference for the first site, illustrating that TLR7 activation can proceed upon binding to site 1 alone (14), which involves an altered signaling cascade (19). The activity of R848 also underscores the importance of hydrophobic interactions within TLR7 (20) and highlights the limit of our understanding with respect to structure–function relationships for activation and inhibition of TLR7 by small molecules, but even more, for RNAs.
The identification of Gm18 as an immunosuppressive modification within tRNAs was initially stimulated by work from Kariko and Weissman (21), who showed that incorporation of certain RNA modifications in in vitro transcribed RNA reduced immune-stimulation. Yet, those approaches were done in artificial RNA and considered neither frequency nor position of endogenous, native RNA modifications (22,23). In previous publications, we investigated tRNA as activator of TLR7 for several reasons, namely (i) they comprise a substantial fraction (8–10%) of cellular total RNA, (ii) unlike ribosomal RNA, tRNAs are not tightly bound by proteins, and therefore could be easily accessible for TLR recognition and (iii) bacterial and mammalian tRNAs are extremely similar in both sequence and structure. Of note, tRNAs synthesized by in vitro transcription strongly activated TLR7 regardless of the sequence's evolutionary origin, be it prokaryotic or eukaryotic. While isolated eukaryotic tRNAs showed limited activation of TLR7, most isolated bacterial tRNA species were stimulatory (8). This demonstrated a post-transcriptional modification dependent ‘self’ and ‘non-self’ discrimination by TLR7. The non-stimulatory bacterial tRNATyr was found to contain Gm18 which was identified as an antagonist of TLR7 and therefore discussed as an immune evasion mechanism (8,17,18,24). Moreover, this work for the first time identified a native inhibitory modification within its natural context. This effect of Gm18 on TLR7 was simultaneously described by the Bauer group (11).
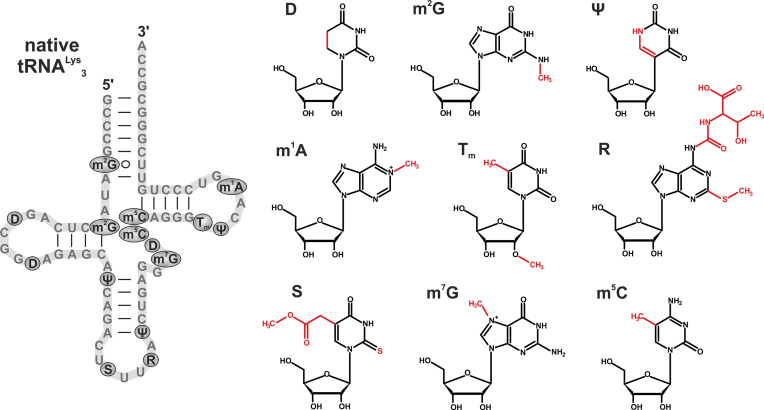
In the course of this project, we observed further non-stimulatory eukaryotic tRNAs, in particular, mammalian tRNALys3, which is depicted along with its post-transcriptional modifications in Figure 1. This tRNA, which holds a special interest in the HIV field (8,22,25–27), appeared to avoid immunostimulation by a novel modification-based principle, given that it does not contain Gm.
Figure 1.
RNA modifications in mammalian tRNALys3. Left part: native tRNALys3 and its corresponding modifications (illustrated as gray circles). Right part: all mentioned modifications and their structural formulas. D = Dihydrouridine, m2G = N2-methylguanosine, Ψ = Pseudouridine, m1A = 1-methyladenosine, Tm = m5Um = 2′-O-methylthymidine/5,2′-O-dimethyluridine, R = t6A = 2-methylthio-N6-threonylcarbamoyladenosine, S = mcm5s2U = 5-methoxycarbonylmethyl-2-thiouridine, m7G = 7-methylguanosine, m5C = 5-methylcytidine (according to modomics RNA modification database (40)).
To identify the immunosilencing modification pattern, we used a Colicin D-based molecular surgery approach. Colicin D is toxic to infected Escherichia coli cells and acts as a so-called tRNase which cleaves the four isoaccepting tRNAsArgin vivo, causing an arrest of protein synthesis (28). Here, we adapted its use in vitro and found that Colicin D also cleaved mammalian tRNALys3 at a precise position in the anticodon loop. This was exploited to synthesize tRNALys3 modivariants (23). Based on the latter, we could identify that 2′-O-methylthymidine (Tm) is, largely, responsible for the decline of immune response observed in tRNALys3-treated PBMCs.
MATERIALS AND METHODS
Cleavage of unmodified/native tRNALys3 by Colicin D
In a final volume of 20 μl, 200 pmoles of unmodified or native RNA were incubated in a solution of Buffer A (final: 5 mM Tris·HCl pH 7.8, 5 mM MgCl2, 25 mM KCl) or Buffer C (final: 5 mM HEPES·KOH pH 7.8, 0.5 mM DTT) and 0.8 μg Colicin D (prepared according to (28)) for 1 h at 37°C. The resulting RNA fragment mixture was used without further purification for the hybridization step.
Hybridization of DNA oligonucleotide to RNA
Hybridization buffer (final: 150 mM KCl, 75 mM HEPES pH 7.0) and an equimolar amount of complementary DNA oligonucleotide (listed below) were added to the fragment mixture. The solution was then incubated for 20 min at room temperature and after that, the DNA/RNA hybrid, as well as the remaining unhybridized RNA fragment were separated via 10% native polyacrylamide gel electrophoresis (PAGE). Bands containing hybrid and fragment were cut out of the gel and agitated in 0.3 M NaOAc overnight at room temperature. In the next step, the RNA was precipitated after addition of two volumes absolute EtOH and glycogen. Followed by resuspension in RNase-free water.
DNA oligonucleotide used for hybridization
| Name | Description | Sequence (5′-3′) | supplier |
|---|---|---|---|
| MH770 | DNA oligo for hybridization to 3′ fragment | TGGCGCCCGAACAGGGACTTGAACCCTGGACCCT | IBA |
DNase treatment and purification of RNA fragments
To obtain the purified, dephosphorylated native fragment, the DNA/RNA hybrid solution was incubated with 0.1 U/μl FastAP (Thermo Scientific) and 1.25 U/μl DNaseI (Thermo Scientific) for 1 h at 37°C. In the next step, the RNA was isolated via phenol-chloroform extraction and EtOH precipitation with 0.3 M NaOAc. The concentration of the received fragment in RNase-free water was then checked by UV absorbance.
Phosphorylation/dephosphorylation of RNA
To remove remaining 2′3′-cyclic phosphates and to get ligation-competent RNA strands (5′ phosphate necessary), 200 pmoles of the purified native fragments or unmodified commercial RNAs were treated with KL-Buffer (final: 10 mM Tris·HCl pH 7.4, 2 mM MgCl2), 5 mM DTT, 5 mM adenosine triphosphate (ATP) and 0.75 U/μl T4-Polynucleotide kinase (Thermo Scientific) for 1 h at 37°C. The reaction mixture was used directly for the subsequent splint ligation.
Splint ligations using tRNA fragments
Unmodified RNAs (see below) were ligated to the corresponding native fragment (Colicin D treatment) or the unmodified 5′ tRNA fragment (Tm modivariant synthesis) to receive a complete tRNALys3 modivariant. For this purpose, prephosphorylated RNA strands were mixed with a 2× molar excess (Colicin D treatment) or an equimolar amount (Tm modivariant synthesis) of the remaining tRNA parts and an equimolar amount of DNA splint in KL-Buffer. After that, the resulting solution was heated for 4 min to 75°C and then slowly cooled down to room temperature for 15 min. In the next step, 1.5 U/μl T4 DNA-Ligase (Thermo Scientific) and 70 ng/μl RNA Ligase 2 (e.g. Thermo Scientific) were added and the mixture was incubated for 24 h at room temperature. At last, the DNA splint was digested (2.5 U/μl DNaseI, 2.5 h at 37°C) and the ligation product was purified using 10% denaturing PAGE (10% acrylamide, 50% urea). The concentration was again checked by UV absorbance.
Nucleic acids used for splint ligation
| Name | Description | Sequence (5′-3′) | supplier |
|---|---|---|---|
| MH810 | tRNALys3 DNA splint (5′ biotin-, 3′ fluorescein-labeled) | AAATGGCGCCCGAACAGGGACTTGAACCCTGGACCCTCAGATTAAAAGTCTGATGCTCTACCGACTGAGCTATCCGGGC | IBA |
| MH823 | Unmodified 5′ fragment of tRNALys3 (RNA#2) | GCCCGGAUAGCUCAGUCGGUAGAGCAUCAGACUUUUAA | biomers |
| MH830 | Unmodified 3′ fragment of tRNALys3 | UCUGAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGCCA | biomers |
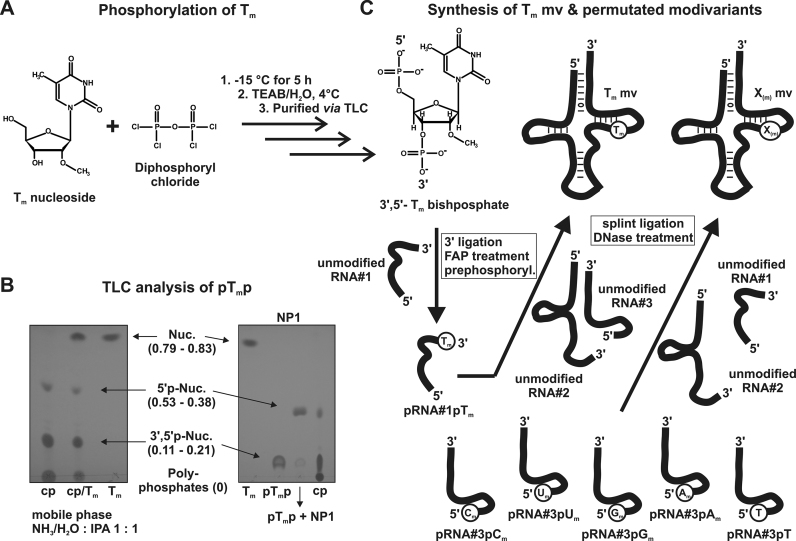
Synthesis of 2′-O-methylthymidine bisphosphate (pTmp)
The 5′-3′ bisphosphorylation of the Tm nucleoside was performed as previously described (29). In short, the nucleoside (2 mmoles) was added to a cooled solution of pyrophosphorylchloride (20 mmoles) under an argon atmosphere. Then, the solution was stirred for 5 h at −15°C and after hydrolysis, the evaporated crude product was purified via thin layer chromatography (mobile phase: IPA:conc. NH3·H2O 1:1). In the next step, the bisphosphate was eluted in 0.5 M NH4OAc and used as a minimal substrate (30) for mv synthesis. At last, the nucleotide-concentration was determined by UV absorbance. Assignment of product-spots was achieved according to literature (31).
To assure the production of the bisphosphate, an aliquot of pTmp (50 pmoles) was treated with 0.3 U Nuclease P1 (Sigma Aldrich) in 20 μl reaction solution (22.5 mM NH4OAc pH 5.0, 20 μM ZnCl2) for 30 min at 37°C. Conversion of the bisphosphate to the 5′ monophosphate indicated the presence of pTmp (verified by UV shadowing).
Ligation of pTmp to unmodified RNA and subsequent 3′ dephosphorylation
In a final volume of 30 μl, 1000 pmoles MH847 (unmodified RNA, sequence, see below) were incubated together with a 10× molar excess of pTmp and 1.6 U/μl RNA Ligase 1 (Thermo Scientific) in the corresponding buffer overnight at 37°C. After that, 1 U FastAP and FAP Buffer were added and the resulting solution (volume: 40 μl) was treated for 1 h at 37°C. At last, the resulting MH847pTm RNA strand was PAGE purified (see above).
Phosphorylation of MH847pTm and MH848 (tRNALys3 subfragments) and modivariant synthesis
Ligation-competent MH847pTm, as well as MH848, were obtained by 5′ phosphorylation using T4-Polynucleotide kinase (see phosphorylation/dephosphorylation of RNA). The received phosphorylated RNA strands were used without further purification for the Tm modivariant synthesis (see Splint ligations using tRNA subfragments).
Nucleic acids used for Tm mv synthesis
| Name | Description | Sequence (5′-3′) | supplier |
|---|---|---|---|
| MH847 | tRNALys3 subfragment (RNA#1) | UCUGAGGGUCCAGGG | biomers |
| MH848 | tRNALys3 subfragment (RNA#3) | UCAAGUCCCUGUUCGGGCGCCA | biomers |
Synthesis of permutated modivariants
Permutated modivariants were synthesized by using the RNA strands listed below. Here, RNA subfragments were 5′ phosphorylated and then subjected to splint ligations as previously described.
Nucleic acids used for synthesis of permutated modivariants
| Name | Description | Sequence (5′-3′) | supplier |
|---|---|---|---|
| MH862 | tRNALys3 subfragment (modified RNA#3) | Cm UCAAGUCCCUGUUCGGGCGCCA | biomers |
| MH863 | tRNALys3 subfragment (modified RNA#3) | Am UCAAGUCCCUGUUCGGGCGCCA | biomers |
| MH864 | tRNALys3 subfragment (modified RNA#3) | Um UCAAGUCCCUGUUCGGGCGCCA | biomers |
| MH865 | tRNALys3 subfragment (modified RNA#3) | Gm UCAAGUCCCUGUUCGGGCGCCA | biomers |
| MH866 | tRNALys3 subfragment (modified RNA#3) | rT UCAAGUCCCUGUUCGGGCGCCA | IBA |
Isolation and stimulation of human immune cells
Human PBMCs were isolated from heparinized blood of healthy donors upon informed consent and approval by the local ethics committee by standard Ficoll-Hypaque density gradient centrifugation (Ficoll 1.078 g/ml). PBMCs were resuspended in complete medium prepared of RPMI 1640 (Biochrom, Berlin, Germany) supplemented with 2% heat-inactivated human serum (1 h, 56°C). For stimulation experiments, RNA was encapsulated with DOTAP (N-[1-(2, 3-dioleoyloxy)propyl]-N, N, N-trimethylammonium-205 methylsulfate) (Carl Roth GmbH Karlsruhe, Germany) at a ratio of 3 μl DOTAP per 1 μg of RNA in Opti-MEM reduced serum medium (Life Technologies) and incubation for 10 min at room temperature. DOTAP encapsulation is necessary to deliver RNA into PBMCs. For transfection experiments, cells were stimulated with RNA at final concentrations of 500, 250 and 125 ng/ml. Where indicated, methylated or unmethylated oligoribonucleotides (ORNs) and native tRNALys3 were mixed with bacterial RNA prior encapsulation with DOTAP. As a positive control, PBMCs were stimulated with bacterial RNA (see below) and TLR7/8-agonist R848 (1 μg/ml) (Invivogen, San Diego, USA). All stimulations were performed in duplicate wells per individual donor at a density of 2 × 105 cells/well PBMCs in a 96-well flat bottom plate. Cells were incubated in a humidified 5% CO2 atmosphere at 37°C for 20 h. Cell-free supernatants were analyzed by sandwich ELISA for secretion of IFN-α (Affymetrix eBioscience, Frankfurt, Germany) according to the manufacturer’s protocol. Where indicated, values were normalized to cytokine production induced by stimulation with bacterial RNA to account for donor variation.
Nucleic acids used for stimulation of human immune cells
| Name | Description | Sequence (5′-3′) | supplier |
|---|---|---|---|
| Gm18 | 26mer GmG - RNA | GUGGGGUUCCCGAGC Gm GCCAAAGGGA | biomers |
| Unmod. | 26mer GG - RNA | GUGGGGUUCCCGAGCGGCCAAAGGGA | biomers |
Preparation of bacterial RNA from Staphylococcus aureus
Staphylococcus aureus ATCC 25923 was grown in Luria-Bertani (LB) Medium (Merck, Darmstadt, Deutschland) at 37°C until mid-log phase. Bacteria were harvested and treated with lysozyme (40 mg/ml, 20 min at 37°C) before RNA isolation using TRIzol reagent (Thermo Fisher, Waltham, USA) according to the manufacturer’s protocol. RNA underwent further purification using RNeasy mini kit (QIAGEN, Venlo, Netherlands) including an on-column DNase treatment. The purity and amount of bacterial RNA preparations were validated by NanoDrop measurement. Only RNA preparations with ratios 260/280 and 260/230 > 1.8 were used for stimulation.
Statistical analysis
Data were analyzed using GraphPad Prism 6.05 (GraphPad Software Inc.). Mean plus SEM were plotted and significant differences were assessed by two-way ANOVA including multiple comparison tests or linear regression model. P-values are indicated by ns (not significant, P > 0.05), * (P ≤ 0.05), ** (P ≤ 0.01), *** (P ≤ 0.001). The dose-response model was calculated by R software 3.4.1 with best fitting model for the corresponding RNA. Unmodified GG ORN: log-logistic function (three parameters), native Lys3: Weibull function (two to four parameters), modified GmG ORN: log-logistic function (two parameters).
RESULTS
Naturally occurring RNA modifications in tRNALys3 decrease, but do not inhibit immunestimulation in PBMCs
As an in-depth follow-up analysis of our initial work on bacterial tRNATyr (8) a side-by-side comparison of IFN-α secretion from PBMCs after transfection with native, fully modified tRNALys3 and synthesized unmodified tRNALys3 was performed. Bacterial RNA (bRNA) and the small molecule agonist R848 served as positive controls for TLR7 activation and non-treated PBMCs as negative control to exclude auto-activation of pDCs.
The IFN-α emission from PBMCs is a commonly used readout for TLR7 activation, which is validated by a chain of evidence, including the following elements. Depletion of pDCs from PBMCs results in complete loss of IFN-α secretion from the latter (8), and isolated pDCs show emission similar to PBMCs (not shown). Therefore the interferon response is clearly limited to pDCs. Ablation of the response upon inhibition of endosomal maturation by chloroquine treatment (8) argues for endosomal recognition and excludes cytosolic recognition. Among the TLR RNA receptors, pDCs express only TLR7 but not TLR3 or TLR8 (9) leading to IFN-α emission (1,7) as reviewed in (14,32). Of note, dendritic cells from TLR7 knockout mice did not show any response to RNA stimulation (33).
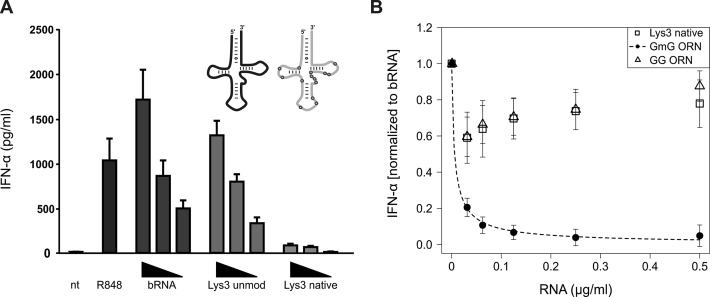
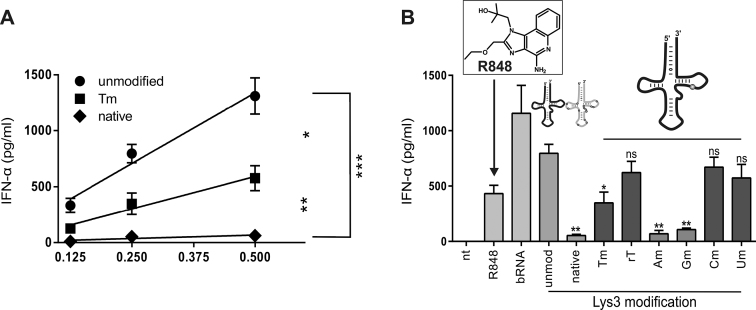
RNA preparations were titrated in three different concentrations on isolated PBMCs. To account for donor variation, a known problem in this type of assay, three independent experiments were performed in duplicate wells. Highest IFN-α levels were observed for bRNA from S. aureus. A synthetic unmodified tRNALys3 showed nearly similar efficiency, whereas the fully modified (native) tRNALys3 induced only minor IFN-α levels (Figure 2A and Supplementary Figure S1A).
Figure 2.
Native tRNALys3 and its immunostimulatory activity. (A) Human PBMCs were stimulated with DOTAP encapsulated bacterial RNA, an unmodified Lys3 tRNA and native tRNALys3 (see Supplementary Figure S1A) at three different concentrations (0.5, 0.25, 0.125 μg/ml). R848 (1 μg/ml) served as a positive control for TLR7 activation. IFN-α secretion was measured in cell-free supernatants after 20 h by ELISA. Shown bars represent the stimulation of 3–6 individual donors in duplicate wells +SEM. (B) To verify the antagonistic effect of native tRNALys3 human PBMCs of three individual donors were co-transfected in duplicates with a constant amount of bacterial RNA (0.5 μg/ml) and different concentrations of the indicated RNAs (0.5, 0.25, 0.125, 0.0625, 0.03125 μg/ml). Previously described GmG and GG ORNs served as positive and negative control for a trans-inhibitory modification, respectively. To account for donor variation data was normalized to bacterial RNA. Dose-response model was calculated by R software as described in statistical analysis.
At first glance, these results appeared similar to those obtained from the previous comparison of native and unmodified tRNATyr from E. coli (8). tRNATyr had been identified as an antagonist for TLR7, which even inhibited immunostimulation when co-delivered in combination with an otherwise immunostimulatory tRNA. To investigate the immunomodulating properties of tRNALys3 in more detail and in particular to study, whether tRNALys3 exerts inhibitory effects as observed for tRNATyr, we designed an assay to discriminate between immunosilencing and antagonistic abilities. In this assay, changes of the IFN-α secretion in response to constant amounts of immunostimulating bRNA combined with different concentrations of tRNALys3 were measured. As a positive control for TLR7 inhibition, the combination of bRNA with a Gm-containing 26mer (GmG ORN, mimicking the inhibitory activity of tRNATyr) was analyzed: whereas pure bRNA induced IFN-α secretion, the addition of the GmG ORN containing a single modified Gm led to complete suppression of TLR7 activation even at moderate concentrations (Figure 2B). As specificity control for inhibition of TLR7, we used an unmodified 26mer (GG ORN) which did not decrease bRNA induced IFN-α levels. Further details of the assay validation were previously published in Gehrig et al. (8) and Schmitt et al. (17).
Importantly, the addition of tRNALys3 did not decrease IFN-α levels induced by bRNA, indicating that the immune-modulatory effects of modifications within tRNALys3 were only sufficient to prevent TLR7 activation in cis (Figure 2A) but not acting inhibitory in trans to another RNA (Figure 2B). These findings demonstrate that tRNALys3 is non-stimulatory, though not acting as a TLR7 antagonist and thus differs from the previously characterized Gm18 modified tRNAs with respect to modulation of TLR7. Given that unmodified tRNALys3 is fully stimulatory, the modified nucleotides in tRNALys3 (Figure 1) are obvious mediators of the non-stimulatory (cis-silencing) effect.
Molecular surgery employing the tRNase Colicin D reveals reduced immunostimulation of the 3′ part of native tRNALys3
To examine the immunomodulatory contribution of post-transcriptional modifications in native tRNALys3, we synthesized tRNALys3 -modivariants using a molecular surgery procedure (8). Modivariants are RNA species that are identical in sequence but differ in post-transcriptional modifications. Their comparative testing allows to conclude on the influence on TLR7-mediated immunostimulation of a single or a subset of modifications. Modivariants can be synthesized by joining modified and unmodified fragments using biochemical methods that are frequently described as ‘cut-and-paste’ or ‘molecular surgery’ techniques (9,32,34). We have previously described the generation of RNA fragments from tRNAs that were well defined in terms of length and modification content by a cleavage reaction mediated by catalytic DNA, so-called DNAzymes (8,35). While the cleavage target site of DNAzymes can be programmed (23), their cleavage efficiency turned out to be impaired by the presence of multiple RNA modifications, which is a typical situation for eukaryotic tRNAs.
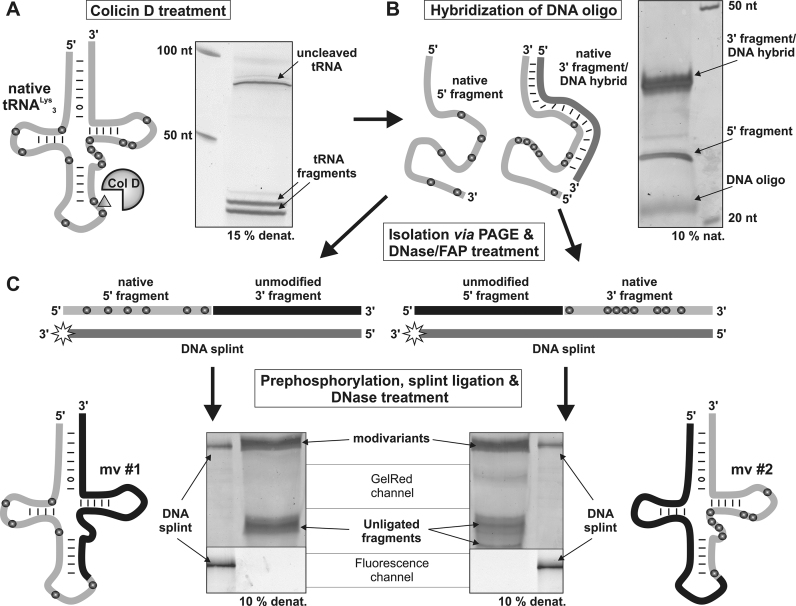
We therefore turned to an alternative approach, making use of the enzymatic activity of Colicin D (28). This tRNase was reported to specifically cleave tRNAArg isoacceptors between positions 38 and 39 in the anticodon-loop (28) in vivo. Interestingly, we observed that fully modified mammalian tRNALys3 was also a target of Colicin D cleavage in vitro, which resulted in two well-defined tRNA fragments (Figure 3A). Of note, the cleavage between position 38 and 39 resulted in two RNA fragments of similar size, leading to separation problems by denaturing PAGE. To address this issue, we hybridized a complementary DNA oligonucleotide to the cleaved 3′ fragment. On a native PAGE, the resulting DNA/RNA hybrid (Figure 3B) was considerably less mobile than the 5′-fragment. This procedure enabled simultaneous separation and isolation of both fragments, which were then submitted to treatment with DNase and alkaline phosphatase, followed by phenol–chloroform extraction. Subsequently, the corresponding RNAs were treated with T4-PNK and ATP to remove any cyclic 3′-phosphates and to obtain 5′-phosphorylated, ligation-competent strands (Figure 3C). Native tRNA fragments were then combined with equimolar amounts of a 5′-phosphorylated, synthetic, unmodified RNA, and hybridized to a full-length cDNA to yield a ligation-competent complex. These nicked DNA–RNA hybrids served as substrates for T4-DNA ligase, and after ligation, the DNA splint was removed enzymatically, with typical isolated yields between 40–60% (36). At last, both obtained modivariants were purified via denaturating PAGE and subjected to immunostimulation tests with PBMCs.
Figure 3.
Colicin D-based molecular surgery and preparation of Lys3-modivariants. (A) Treatment of native tRNALys3 (gray line, modifications indicated as gray dots) with Colicin D generated two fragments with 38 nt length (see PAGE analysis). Differences in migration distance were caused by 2′-3′-cyclic phosphates at the 3′ end of the 5′ fragment. (B) Due to similar fragment lengths, a DNA oligonucleotide (darker gray) was hybridized to the 3′ fragment. The corresponding band shift of the double-stranded RNA/DNA hybrid enabled separation and isolation of both tRNA fragments. RNAs were then treated with DNaseI and FastAP. (C) Prephosphorylated 5′ and 3′ fragments were subsequently annealed together with unmodified RNAs (black lines) on splint DNA (indicated by the darker gray line, fluorescence marker displayed as a star) which contains a 3′ fluorescein residue. Ligation and DNase treatment yielded mv#1 and mv#2 as shown in the respective PAGE analysis. Evaluation of the fluorescence channel revealed successful digestion of DNA splint in modivariant preparations.
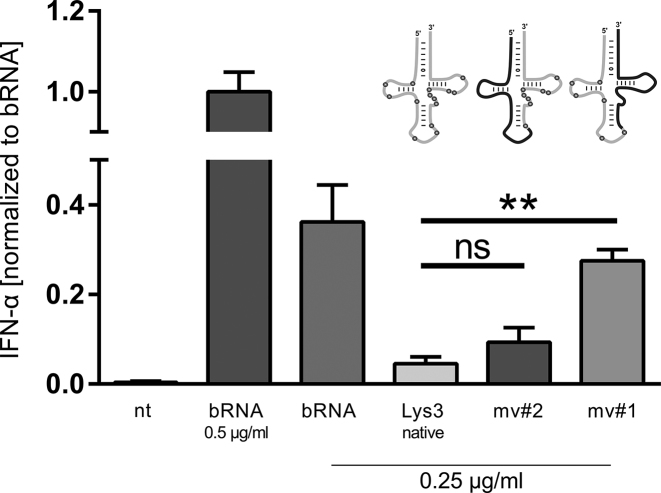
As depicted in Figure 4, bacterial RNA and modivariant 1 (mv#1, which included the native 5′ fragment and its modifications) induced comparable IFN-α secretion, indicating that none of the modifications in the 5′ fragment was sufficient to abolish TLR7 stimulation. In contrast, fully modified tRNALys3 and modivariant 2 (mv#2, which contains the native 3′ fragment) displayed clearly reduced immunostimulatory activity. Taken together, these results indicate that a single modified residue or the combination of multiple modifications in the 3′ part of native tRNALys3 caused decreased immunostimulation.
Figure 4.
Immunostimulatory activity tests of Lys3-modivariants. Human PBMCs were stimulated with DOTAP encapsulated RNA species at indicated concentrations. Splint-ligated mv#1 and mv#2 containing half part of native tRNALys3 and unmodified RNAs as well as native tRNALys3 were tested on three individual donors in duplicate wells. IFN-α levels were measured in cell-free supernatants 20 h post-transfection. To account for donor variation results were normalized to the amount of IFN-α induced by 0.5 μg/ml bRNA. Each bar illustrates mean +SEM.
Tm-modivariant displays decreased immunostimulatory activity
Based on this latter observation, we further inspected the underlying sequence context of the 3′ part of tRNALys3, which harbors the following RNA modifications (depicted in Figure 1): Ψ39, m7G46, D47, m5C48, m5C49, Tm54, Ψ55 and m1A58. So far, none of these modifications has been associated with reduced PBMC-stimulation when present in tRNAs. Yet, synthetic RNAs containing high amounts of Ψ and m5C exhibited low immunostimulation toward TLR7 and other RNA sensors (21,37,38). Bacterial tRNAs containing Ψ, m7G, D, t6A or T were stimulatory toward TLR7 (8), as was yeast tRNAAsp which contains m5C (not shown).
Among the modifications present in the mv#2, we focused on Tm which, in addition to the ubiquitous T (m5U) modification, also featured a 2′-O-methylated ribose moiety. The latter also occurs in Gm, which had been described as an inhibitory modification in recent studies (8,18). However, in various preceding permutation studies, inhibition had remained restricted to ribose- methylated purines, while the corresponding pyrimidines were innocuous.
Thus, we synthesized a tRNALys3-Tm modivariant containing Tm at position 54 in an otherwise unmodified tRNALys3 context. The first step of this synthesis included the bisphosphorylation of a commercially available Tm nucleoside with diphosphoryl chloride (Figure 5A). Successful preparation of the bisphosphorylated nucleoside (pTmp) was verified by thin-layer chromatography and nuclease P1 conversion of the bisphosphate to the 5′-monophosphate (Figure 5B). According to literature (30), nucleoside bisphosphates are minimal substrates for T4 RNA ligase 1, which selectively catalyzes the attachment of 5′,3′-bisphosphates to the 3′ end of single-stranded RNAs. Accordingly, pTmp was ligated to a synthetic unmodified RNA fragment (RNA#1) of tRNALys3 (Figure 5C) with about 25% isolated yield. After subsequent 3′-dephosphorylation and 5′-prephosphorylation by T4-PNK, pRNA#1pTm was used in a splint ligation together with the remaining RNA strands (#2 and #3) to yield the desired Lys3-Tm modivariant (Supplementary Figures S1B and C). Isolated yields were typically 15–25%, with ∼60% ligation efficiency per ligation site and some 30–50% loss during isolation of the final RNA constructs (36).
Figure 5.
Synthesis of Tm mv & permutated modivariants. (A) Reaction scheme describing the conversion of Tm to the corresponding bisphosphate by diphosphoryl chloride addition (B) Left part: TLC analysis of crude product (cp) showed the formation of 5′-monophosphate, 3′-5′-bisphosphate and polyphosphates (according to (31)). The Tm nucleoside (Tm) was not detectable which indicated complete conversion. Co-spotting (cp/Tm) of crude product and nucleoside is displayed in the middle lane. Right part: Isolated bisphosphate (pTmp) was treated with nuclease P1 (pTmp + NP1). The changed Rf value revealed the formation of the 5′-monophosphate out of the bisphosphate which proved pTmp-synthesis. (C) For the synthesis of the Tm modivariant, the respective bisphosphate was attached to an unmodified RNA#1 (part of tRNALys3) using RNA ligase 1. Further reaction steps then yielded the ligation-competent and Tm-modified RNA#1 which was subsequently ligated to RNAs #2 and #3 producing the desired modivariant. Xm modivariants were created the same way using commercially synthesized RNAs which were already attached to X(m) modifications.
The presence of Tm within the modivariant was verified via LC-MS/MS analysis (see supplementary information). As displayed in the corresponding diagram (Supplementary Figure S2), each modivariant molecule contained one Tm residue. The correct position of the Tm modification was furthermore confirmed by RiboMethSeq (39) (results are shown in Supplementary Figure S3).
The obtained modivariant was subsequently tested in comparison to the fully modified tRNALys3, which itself showed hardly any stimulation, and in comparison to a completely unmodified modivariant of tRNALys3. The introduction of the Tm modification efficiently reduced the immunostimulatory activity of the otherwise unmodified full-length tRNA by almost 50%. Of note, the native tRNALys3, was even less stimulatory than the Tm modivariant, indicating an immunosilencing effect of further modifications (Figure 6A).
Figure 6.
Immunostimulation of single modified tRNAs. (A) Isolated PBMCs were stimulated with indicated tRNAs encapsulated with DOTAP at a concentration of 0.5, 0.25 and 0.125 μg/ml. Cell-free supernatants were collected after 20 h and IFN-α release was detected by ELISA. Linear regression was calculated by GraphPad Prism. (B) Transfection of single modified tRNAs incorporating indicated modifications at position 54 of tRNA sequence at a final concentration of 0.25 μg/ml. R848 and bRNA were used as a positive control for TLR7 stimulation. IFN-α secretion was compared to unmodified tRNA control. All stimulations were performed on three individual donors in duplicate wells. Shown are mean + SEM.
Analysis of Xm-modivariants suggests a synergistic effect of nucleobase- and ribose-modifications on IFN-α secretion
To better understand details of Tm function in the context of tRNALys3, we permutated the nucleobase at position 54 as previously described (18). The starting material for these permutations were commercially available RNA oligonucleotides with 2′-O-methylcytidine (Cm), 2′-O-methyluridine (Um), 2′-O-methyladenosine (Am) or 2′-O-methylguanosine (Gm) at their 5′ ends. This allowed straightforward splint ligations utilizing the remaining unmodified RNAs #1 and #2 (see above and Supplementary Figures S1B and C).
Subsequent immunostimulation tests of these mutant/modivariants demonstrated significant differences between 2′-O-methylated purines and pyrimidines (Figure 6B). The modified pyrimidines Cm and Um induced IFN-α levels comparable to unmodified tRNAs. In contrast, the purine derivatives Am and Gm decreased the immunostimulatory activity of RNAs to levels which were comparable to those of native tRNALys3.
As a further observation of considerable interest, the presence of ribothymidine (T, rT) without ribose methylation did only slightly affect stimulation, similar to Um (Figure 6B). While single methylation neither of the ribose nor of the base of U54 was at all effective in decreasing the stimulation of TLR7, the combination of both showed a strong synergy, amounting to the observed 50% decrease in comparison to unmodified tRNALys3. The above observations may have implications for the recognition mode by TLR7, namely that Tm presumably binds to the same site and in a similar orientation as do Gm and Am.
As a final observation, this remarkable synergistic effect of Tm within tRNALys3 can only partially explain the observed low stimulatory potential on PBMC. Indeed, our results also imply a contribution of the remainder of the modifications shown in Figure 1, either single or as an ensemble.
DISCUSSION
A new molecular tool for biochemical manipulation of RNA in vitro
As indicated by our recent studies (8), ‘self’ tRNA is guarded against TLR7 activation by RNA modifications, such as Gm18, Gm34 and one or more hitherto unknown modifications in other tRNAs, including in particular tRNALys3, the object of the present work. Given that eukaryotic tRNAs may contain up to 25% modified nucleotides (40), the need to create modivariants containing selected defined modifications became even more imperative than in previous studies of bacterial tRNA, which are generally less modified (40). However, it turned out that the very same modifications impede our previously used DNAzyme approach for site-specific tRNA cleavage. The subsequent development of Colicin D, a ribotoxin with tRNase activity (28), as a new tool in the repertoire of methods often subsumed as ‘molecular surgery’ or ‘cut-and-paste’ of RNA biochemistry is an added bonus. Indeed, we anticipate its continued application in further studies of eukaryotic tRNAs, since preliminary data indicate, that the restriction of tRNase activity of Colicin D to tRNAArg and tRNALys can be overcome by increasing enzyme levels. So far, we succeded in conducting controlled cleavage of a yeast tRNAPhe transcript. Also, yeast total tRNA could be digested to >90%, leaving significant amounts of defined ∼30mer tRNA fragments (data not shown), suggesting that in vitro conditions for controlled cleavage of for any tRNA shaped molecule can be developed.
Distinction between minor and major determinants of immune suppression
Colicin D-based molecular surgery gave access to two modivariants whose testing identified a major effect caused by the modifications situated in the 3′-part. Among those modifications present, several occur in bacterial and other microbial tRNAs that had previously been found to be stimulatory. These modifications were considered as highly unlikely candidates for the observed effect.
The remainder consist of m1A58 and Tm54. Given the previous findings on ribose-methylated modifications, we turned our attention to the synthesis of a modivariant containing Tm54 as sole modified nucleotide. In this context, Tm decreased the interferon response by 50%. While this is a pronounced effect, it is considerably weaker than what was typically observed for Gm. Of note, since the fully modified tRNALys3 evokes almost no response, it is clear that the remainder of modifications, i.e. the ensemble except Tm, also has a contributing effect. Further studies may identify additional contributions of single modifications, but we would like to point out some of our results with synthetic modifications in this context. We recently reported a generic ‘shielding’ effect of a number of synthetic RNA modifications with no discernible common denominator in terms of chemical structure (41). The conclusion from those studies might also apply to heavily modified eukaryotic tRNA. An RNA carrying multiple and especially bulky modifications might escape detection by an altogether altered set of functional groups that mask the presence of typical RNA features. In the case at hand, the anticodon of tRNALys3 would be shielded by two bulky modifications (ms2t6A and mcm5s2U). Furthermore, modifications in the anticodon stem as well as in the D-loop are so numerous that they approach a density previously investigated in in vitro transcripts, which contained high levels of, e.g. Ψ or m5C (21,37,38). With regard to a fast immune response toward foreign RNA, the incorporation of bulky, and therefore likely unstimulatory, but not inhibitory modifications within human tRNAs may be crucial. Antagonistic effects of self-RNA would block the receptor and therefore impede an adequate immune response against invading pathogens.
Tm as a major determinant of immunosilencing
Our results indicate that Tm at position 54 is responsible for much of the interesting effect observed in tRNALys3. Contrary to Gm18, tRNALys3 reveals no dominant inhibitory effect when delivered in trans with a stimulatory RNA. In fact, our findings imply some kind of ‘RNA muting’ and the data indicate that some 50% of this ‘muting’ is due to the presence of Tm. As already discussed, this implies that other modifications within the RNA are also necessary for the full effect of tRNALys3. Of note, Tm therefore classifies differently from previously identified immune-modulatory Gm and Am modifications, the latter also suppressing in trans. In a previous study, we identified a minimal tri-nucleotide motif within RNA which is necessary and sufficient to antagonize TLR7 and TLR8 (17). A key finding of this study was the relevance of the nucleobase downstream the 2′-O-methylated nucleotide for immunosuppression. In case of the native tRNALys3, Tm is followed by pseudouridine whereas in the described modivariants Xm was followed by a uridine. The effect of Tm might potentially be modulated by synergistic effects with further modifications in the defined sequence context of the native tRNA.
TLR7 reportedly responds strongly to single-stranded RNA rich in guanosine and uridine residues (14). Supported by x-ray structures of TLR7 and TLR8 (12,13), recent reports postulated separate binding sites for both types of nucleosides in murine TLR7. Furthermore, RNA hydrolysis was discussed as an intrinsic part of TLR-signalling (12,42). In this backdrop, the current results raise multiple questions. Importantly, we are primarily assessing the evasion/inhibition of TLR7 signaling, rather than the signaling itself. It is therefore not at all clear if Tm and Gm mediate their TLR7-silencing effect through the same binding site that mediates TLR7-activation by U and G residues. It is furthermore open if immunesilencing by ribose-methylated nucleotides requires binding of RNA to one, two or even yet additional sites not yet identified by structural biology. Own data suggest that Gm modifications act as competitive antagonist inhibiting signaling by stimulatory RNA (24).
Progress toward a pharmacophore for TLR7 inhibition?
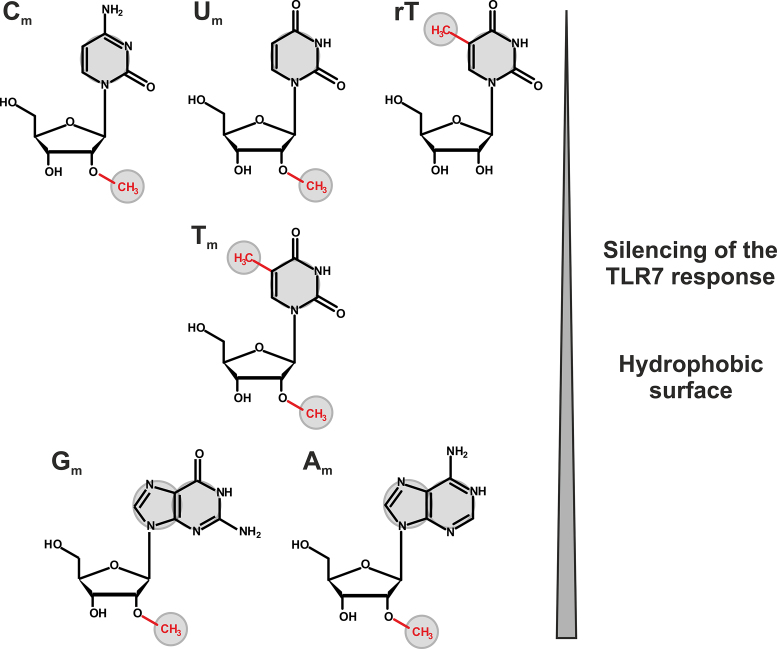
Pure biochemical data of the type presented here cannot unambiguously determine the number and structure of effective binding sites. However, we have added an interesting piece of data by comparing ribose-modified nucleosides Cm, Um, Tm, Am and Gm at the precisely same position of a tRNA scaffold (n.b.: the effects of Am and Gm do not differ significantly in Figure 6B, but previous studies showed Gm to be more potent in detailed dose-titration studies (17)). Furthermore, the sequence context in which the 2′-O-methylated nucleobase occurs is crucial for the antagonistic effect of an RNA. If Gm is followed by cytidine, no dominant inhibitory effect is detectable. Immunostimulation decreases from Cm to Gm, suggesting that the binding site is suited to optimally recognize features of a purine residue, and of course the 2′-O-methyl group. The synergistic effect of the two post-transcriptional methylations resulting in Tm thereby creates structure bearing similarities to Gm. These similarities concern the hydrogen pattern on the Watson–Crick face, but more importantly, the C5-methylation increases the hydrophobic surface of the base, bringing it closer to that of a purine ring (Figure 7). We therefore speculate that Tm mediates its effect by binding to the same site as Gm, using a hydrophobic interaction, potentially π-stacking, in addition to hydrogen bridges involving O6G/O4U as donor and NH1G/NH3U as acceptors. This view is supported by a recent study on the related TLR8, where stimulation was efficiently inhibited by a 7-deaza-8-azainosine structure, a purine whose hydrogen pattern on the Watson–crick face shows common features with the above (43). This study also indicates a direction for follow-up research on the present topic.
Figure 7.
Correlation between the hydrophobic surface of RNA modifications and their immunosilencing potential. Modifications in the upper part possess only small hydrophobic surfaces (indicated by gray circles) in comparison to purine compounds (Gm and Am). The latter also reveal the highest immunosilencing potential.
Biology of the Tm modification
Several lines of evidence disencourage the notion, that unmodifed tRNAs as used in this study, are recognized because of their failure to properly fold into the typical threedimensional L-shape of tRNAs. In particular, unmodified yeast tRNAPhe and mammalian tRNALys3 had previously been subject to structural comparison with their fully modified counterparts (44–47). Based on the minor differences reported, Tm might have subtle impact on local structure at best, making impact on TLR-recognition implausible. Also, in previous unpublished experiments, we found some TLR7 inhibtion by Gm in fragments of tRNATyr, and similarly, low TLR7 response for Tm containing fragments compared to unmodified fragments. These observations argue for the overall tRNA structure to be irrelevant in TLR/ recognition. Indeed, the concept that TLR7 inspects small unstructured stretches of RNA is supported by TLR7 inhibtion of Gm-containing RNA oligomers as short as 9 nt (17).
From the biological point of view, it might be possible that Tm serves as one of several modifications that differentiate between foreign- and self-RNAs. Whereas ample literature is available concerning the T54 modification (10,48), little is known concerning Tm beyond its identification (40,49). In the threedimensional structure of a canonical tRNA, T54/Tm54 are engaged in tertiary interactions that involve also G18/Gm18, and both residues are thus in close spatial proximity (10,48,49). So far, no tRNAs have been reported to simultaneously contain both Gm and Tm (40). Interestingly, the ensemble of modifications in tRNALys3 (Figure 1) is involved in HIV replication in multiple ways. For example, mcm5s2U34 is involved in the annealing process of the tRNA as primer for HIV reverse transcriptase (47,50), and m1A58 causes an arrest of reverse transcription that is mandatory for HIV replication (51). Given that tRNALys3 is selectively packaged into the HIV-virion (52,53), the presence of Tm in this tRNA might contribute to lowering an interferon response, although a tRNA containing Gm would arguably have a stronger effect. Of note, it was reported that pDCs of chronically HIV-infected patients displayed a reduced IFN production on a per-cell basis after treatment with TLR7 agonists (54). However, at this point, further interpretation is to be approached with great caution.
Part of our future research will be aimed at the contribution to immunoevasion of modifications other than Tm. We note that, while tRNA modifications in E. coli, yeast and mammalian mitochondria (40,55) have been mapped with high precision, knowledge on mammalian cytosolic tRNA modification patterns remains very sparse, and this constitutes a significant drawback in this extremely vibrant field, which currently develops intrinsic connections to medicinal biology.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Lilia AYADI (IMoPA, UMR7365 CNRS-UL) for excellent technical assistance for library preparation and sequencing and Dr Sébastien BOUTIN (Department of Infectious Diseases, University Hospital Heidelberg) for help with statistical analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
ANR-DFG Joint Grant [HTRNAMod to Y.M., M.H.; ANR-13-ISV8-0001/HE 3397/8-1]; DFG Grants [DA 592/5-1 to A.D., HE 3397/9-1, HE 3397/ 13-1 (SPP1784) to M.H.]. Funding for open access charge: Johannes Gutenberg-University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Diebold S.S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C.. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004; 303:1529–1531. [DOI] [PubMed] [Google Scholar]
- 2. Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M. et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006; 314:994–997. [DOI] [PubMed] [Google Scholar]
- 3. Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A.. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001; 413:732–738. [DOI] [PubMed] [Google Scholar]
- 4. Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J. et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006; 441:101–105. [DOI] [PubMed] [Google Scholar]
- 5. Siegal F.P., Kadowaki N., Shodell M., Fitzgerald-Bocarsly P.A., Shah K., Ho S., Antonenko S., Liu Y.J.. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999; 284:1835–1837. [DOI] [PubMed] [Google Scholar]
- 6. Yu H.R., Huang H.C., Kuo H.C., Sheen J.M., Ou C.Y., Hsu T.Y., Yang K.D.. IFN-alpha production by human mononuclear cells infected with varicella-zoster virus through TLR9-dependent and -independent pathways. Cell Mol. Immunol. 2011; 8:181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klonowska-Szymczyk A., Wolska A., Robak T., Cebula-Obrzut B., Smolewski P., Robak E.. Expression of toll-like receptors 3, 7, and 9 in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Mediat. Inflamm. 2014; 2014:381418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gehrig S., Eberle M.E., Botschen F., Rimbach K., Eberle F., Eigenbrod T., Kaiser S., Holmes W.M., Erdmann V.A., Sprinzl M. et al. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J. Exp. Med. 2012; 209:225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hornung V., Rothenfusser S., Britsch S., Krug A., Jahrsdorfer B., Giese T., Endres S., Hartmann G.. Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 2002; 168:4531–4537. [DOI] [PubMed] [Google Scholar]
- 10. Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006; 34:721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jockel S., Nees G., Sommer R., Zhao Y., Cherkasov D., Hori H., Ehm G., Schnare M., Nain M., Kaufmann A. et al. The 2′-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J. Exp. Med. 2012; 209:235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tanji H., Ohto U., Shibata T., Taoka M., Yamauchi Y., Isobe T., Miyake K., Shimizu T.. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat. Struct. Mol. Biol. 2015; 22:109–115. [DOI] [PubMed] [Google Scholar]
- 13. Zhang Z., Ohto U., Shibata T., Krayukhina E., Taoka M., Yamauchi Y., Tanji H., Isobe T., Uchiyama S., Miyake K. et al. Structural analysis reveals that Toll-like receptor 7 is a dual receptor for guanosine and Single-Stranded RNA. Immunity. 2016; 45:737–748. [DOI] [PubMed] [Google Scholar]
- 14. Dalpke A., Helm M.. RNA mediated Toll-like receptor stimulation in health and disease. RNA Biol. 2012; 9:828–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S., Noronha A., Manoharan M., Akira S., de Fougerolles A. et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005; 11:263–270. [DOI] [PubMed] [Google Scholar]
- 16. Vollmer J., Tluk S., Schmitz C., Hamm S., Jurk M., Forsbach A., Akira S., Kelly K.M., Reeves W.H., Bauer S. et al. Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. J. Exp. Med. 2005; 202:1575–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmitt F.C.F., Freund I., Weigand M.A., Helm M., Dalpke A.H., Eigenbrod T.. Identification of an optimized 2′-O-methylated trinucleotide RNA motif inhibiting Toll-like receptors 7 and 8. RNA. 2017; 23:1344–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaiser S., Rimbach K., Eigenbrod T., Dalpke A.H., Helm M.. A modified dinucleotide motif specifies tRNA recognition by TLR7. RNA. 2014; 20:1351–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Colak E., Leslie A., Zausmer K., Khatamzas E., Kubarenko A.V., Pichulik T., Klimosch S.N., Mayer A., Siggs O., Hector A. et al. RNA and imidazoquinolines are sensed by distinct TLR7/8 ectodomain sites resulting in functionally disparate signaling events. J. Immunol. 2014; 192:5963–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu H., Jin H., Sun L., Zhang L., Sun G., Wang Z., Yu Y.. Toll-like receptor 7 agonists: chemical feature based pharmacophore identification and molecular docking studies. PLoS One. 2013; 8:e56514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kariko K., Buckstein M., Ni H., Weissman D.. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005; 23:165–175. [DOI] [PubMed] [Google Scholar]
- 22. Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S., Lipford G., Wagner H., Bauer S.. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004; 303:1526–1529. [DOI] [PubMed] [Google Scholar]
- 23. Eigenbrod T., Keller P., Kaiser S., Rimbach K., Dalpke A.H., Helm M.. Recognition of specified RNA modifications by the innate immune system. Methods Enzymol. 2015; 560:73–89. [DOI] [PubMed] [Google Scholar]
- 24. Rimbach K., Kaiser S., Helm M., Dalpke A.H., Eigenbrod T.. 2′-O-Methylation within bacterial RNA acts as suppressor of TLR7/TLR8 activation in human innate immune cells. J. Innate Immun. 2015; 7:482–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kishimoto N., Onitsuka-Kishimoto A., Iga N., Takamune N., Shoji S., Misumi S.. The C-terminal domain of glyceraldehyde 3-phosphate dehydrogenase plays an important role in suppression of tRNA(Lys3) packaging into human immunodeficiency virus type-1 particles. Biochem. Biophys. Rep. 2016; 8:325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lepelley A., Louis S., Sourisseau M., Law H.K., Pothlichet J., Schilte C., Chaperot L., Plumas J., Randall R.E., Si-Tahar M. et al. Innate sensing of HIV-infected cells. PLoS Pathog. 2011; 7:e1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doyle T., Goujon C., Malim M.H.. HIV-1 and interferons: who's interfering with whom. Nat. Rev. Microbiol. 2015; 13:403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tomita K., Ogawa T., Uozumi T., Watanabe K., Masaki H.. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. PNAS. 2000; 97:8278–8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barrio J.R., Barrio M.C., Leonard N.J., England T.E., Uhlenbeck O.C.. Synthesis of modified nucleoside 3′,5′-bisphosphates and their incorporation into oligoribonucleotides with T4 RNA ligase. Biochemistry. 1978; 17:2077–2081. [DOI] [PubMed] [Google Scholar]
- 30. England T.E., Uhlenbeck O.C.. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry. 1978; 17:2069–2076. [DOI] [PubMed] [Google Scholar]
- 31. Murphy P.J. Organophosphorus Reagents: a Practical Approach in Chemistry. 2004; Oxford: Oxford University Press. [Google Scholar]
- 32. Eigenbrod T., Dalpke A.H.. Bacterial RNA: An underestimated stimulus for innate immune responses. J. Immunol. 2015; 195:411–418. [DOI] [PubMed] [Google Scholar]
- 33. Eberle F., Sirin M., Binder M., Dalpke A.H.. Bacterial RNA is recognized by different sets of immunoreceptors. Eur. J. Immunol. 2009; 39:2537–2547. [DOI] [PubMed] [Google Scholar]
- 34. Sakurai M., Ohtsuki T., Watanabe Y., Watanabe K.. Requirement of modified residue m1A9 for EF-Tu binding to nematode mitochondrial tRNA lacking the T arm. Nucleic Acids Res. Suppl. 2001; 1:237–238. [DOI] [PubMed] [Google Scholar]
- 35. Hengesbach M., Meusburger M., Lyko F., Helm M.. Use of DNAzymes for site-specific analysis of ribonucleotide modifications. RNA. 2008; 14:180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kurschat W.C., Muller J., Wombacher R., Helm M.. Optimizing splinted ligation of highly structured small RNAs. RNA. 2005; 11:1909–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kariko K., Weissman D.. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr. Opin. Drug Discov. Dev. 2007; 10:523–532. [PubMed] [Google Scholar]
- 38. Kariko K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D.. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008; 16:1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marchand V., Pichot F., Thuring K., Ayadi L., Freund I., Dalpke A., Helm M., Motorin Y.. Next-Generation Sequencing-Based RiboMethSeq protocol for analysis of tRNA 2′-O-Methylation. Biomolecules. 2017; 7:E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crecy-Lagard V., Ross R., Limbach P.A., Kotter A. et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018; 46:D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hellmuth I., Freund I., Schloder J., Seidu-Larry S., Thuring K., Slama K., Langhanki J., Kaloyanova S., Eigenbrod T., Krumb M. et al. Bioconjugation of small molecules to RNA impedes its recognition by Toll-Like receptor 7. Front. Immunol. 2017; 8:312–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Geyer M., Pelka K., Latz E.. Synergistic activation of Toll-like receptor 8 by two RNA degradation products. Nat. Struct. Mol. Biol. 2015; 22:99–101. [DOI] [PubMed] [Google Scholar]
- 43. Hu T., Suter S.R., Mumbleau M.M., Beal P.A.. TLR8 activation and inhibition by guanosine analogs in RNA: importance of functional groups and chain length. Bioorg. Med. Chem. 2018; 26:77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sampson J.R., Uhlenbeck O.C.. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. PNAS. 1988; 85:1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Isel C., Lanchy J.M., Le Grice S.F., Ehresmann C., Ehresmann B., Marquet R.. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J. 1996; 15:917–924. [PMC free article] [PubMed] [Google Scholar]
- 46. Gotte M., Marquet R., Isel C., Anderson V.E., Keith G., Gross H.J., Ehresmann C., Ehresmann B., Heumann H.. Probing the higher order structure of RNA with peroxonitrous acid. FEBS Lett. 1996; 390:226–228. [DOI] [PubMed] [Google Scholar]
- 47. Isel C., Marquet R., Keith G., Ehresmann C., Ehresmann B.. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J. Biol. Chem. 1993; 268:25269–25272. [PubMed] [Google Scholar]
- 48. Motorin Y., Helm M.. tRNA stabilization by modified nucleotides. Biochemistry. 2010; 49:4934–4944. [DOI] [PubMed] [Google Scholar]
- 49. Benas P., Bec G., Keith G., Marquet R., Ehresmann C., Ehresmann B., Dumas P.. The crystal structure of HIV reverse-transcription primer tRNA(Lys,3) shows a canonical anticodon loop. RNA. 2000; 6:1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Auxilien S., Keith G., Le Grice S.F., Darlix J.L.. Role of post-transcriptional modifications of primer tRNALys,3 in the fidelity and efficacy of plus strand DNA transfer during HIV-1 reverse transcription. J. Biol. Chem. 1999; 274:4412–4420. [DOI] [PubMed] [Google Scholar]
- 51. Renda M.J., Rosenblatt J.D., Klimatcheva E., Demeter L.M., Bambara R.A., Planelles V.. Mutation of the methylated tRNA(Lys)(3) residue A58 disrupts reverse transcription and inhibits replication of human immunodeficiency virus type 1. J. Virol. 2001; 75:9671–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kleiman L., Cen S.. The tRNALys packaging complex in HIV-1. Int. J. Biochem. Cell Biol. 2004; 36:1776–1786. [DOI] [PubMed] [Google Scholar]
- 53. Kleiman L., Jones C.P., Musier-Forsyth K.. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 2010; 584:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kaushik S., Teque F., Patel M., Fujimura S.H., Schmidt B., Levy J.A.. Plasmacytoid dendritic cell number and responses to Toll-like receptor 7 and 9 agonists vary in HIV Type 1-infected individuals in relation to clinical state. AIDS Res. Hum. Retroviruses. 2013; 29:501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suzuki T., Suzuki T.. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014; 42:7346–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.