Abstract
Clustered regularly interspaced short palindromic repeats (CRISPRs) and CRISPR-associated (Cas) proteins provide microbial adaptive immunity against invading foreign nucleic acids. In type II-A CRISPR–Cas systems, the Cas1–Cas2 integrase complex and the subtype-specific Csn2 comprise the CRISPR adaptation module, which cooperates with the Cas9 nuclease effector for spacer selection. Here, we report the molecular organization of the Streptococcus pyogenes type II-A CRISPR adaptation module and its interaction with Cas9 via Csn2. We determined the crystal structure of S. pyogenes type II-A Cas2. Chromatographic and calorimetric analyses revealed the stoichiometry and topology of the type II-A adaptation module composed of Cas1, Cas2 and Csn2. We also demonstrated that Cas9 interacts with Csn2 in a direct and stoichiometric manner. Our results reveal a network of molecular interactions among type II-A Cas proteins and highlight the role of Csn2 in coordinating Cas components involved in the adaptation and interference stages of CRISPR-mediated immunity.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPRs) and CRISPR-associated (Cas) proteins constitute a microbial adaptive immune system against phages and other invading nucleic acids (1–5). CRISPRs are a class of repetitive genetic elements found in microbial genomes and consist of invariable ‘repeat’ sequences interspaced with variable ‘spacer’ sequences (6–8). Adjacent to the CRISPR loci lie genes encoding the Cas proteins that function in all three stages of CRISPR-mediated immunity: adaptation, expression and interference (9–11). During adaptation, short DNA fragments derived from the invading nucleic acids, called prespacers, are integrated between two repeat sequences in the CRISPR array as new spacers (12–14). In the expression stage, the CRISPR array is transcribed as a long precursor CRISPR RNA (crRNA), which is processed within the repeat sequences to produce mature CRISPR RNAs (9,15).During the interference stage, the crRNAs assemble with single or multiple Cas proteins to form the crRNA–effector complex that identifies and degrades the complementary sequences in re-invading foreign nucleic acids (15,16).
CRISPR–Cas systems can be classified into two major classes and further divided into six types or into more than 30 subtypes (17,18). The adaptation mechanism is fairly conserved among the different CRISPR–Cas systems. The universal Cas1 and Cas2 proteins comprise an integrase complex responsible for space acquisition (12–14). The interference mechanism varies significantly among CRISPR–Cas systems. The interference machinery of class 1 systems (types I, III and IV) is composed of multiple Cas proteins, whereas class 2 systems (types II, V and VI) rely on a single effector Cas protein for interference (15,16,19). In type II CRISPR–Cas systems, the multi-domain Cas9 functions as a sole effector protein that binds to the crRNA and cleaves target DNAs (16,20,21). Cas9 also requires a trans-activating crRNA (22,23), which base pairs with the repeat region of the crRNA to form a mature dual RNA hybrid for Cas9 guidance (24,25).
Among the three subtypes of type II CRISPR–Cas systems (20,21), the molecular mechanism of type II-A systems has been studied the most. As in type I systems, two Cas1 dimers and a single Cas2 dimer form a heterohexameric integrase complex in type II-A CRISPR–Cas systems (26). The type II-A Cas1–Cas2 complex is sufficient for in vitro spacer integration without the need for any additional factors (26,27). Type II-A Cas9 has attracted significant attention since Streptococcus pyogenes Cas9 with the engineered single-guide RNA (sgRNA) has been developed as the revolutionary genome editing tool (24,28–30). The structure and function of type II-A Cas9 have been characterized thoroughly (24,31–35). It has also been reported, in the type II-A systems of Streptococcus species, that Cas9 contributes not only to the interference stage, but also to the adaptation stage of CRISPR-mediated immunity (36–38). In addition to Cas1, Cas2 and Cas9, type II-A CRISPR–Cas systems also include Csn2, which is considered a subtype-specific signature Cas protein (17,18,20). Csn2 has been implicated in the adaptation stage of CRISPR-mediated immunity since it was shown to be required for the acquisition of new spacers during in vivo adaptation experiments (1,36,37). When the S. pyogenes type II-A Cas operon was expressed in Escherichia coli, the four Cas proteins co-purified (36), suggesting the formation of a multi-protein Cas complex in the subtype II-A CRISPR–Cas systems.
We previously reported the crystal structures of S. pyogenes Cas1 and Csn2 (39,40) and demonstrated that Cas1 interacts directly with Csn2 (41). In the present study, we further examined the structure and interaction of the four Cas proteins in type II-A CRISPR–Cas systems. The crystal structure of S. pyogenes type II-A Cas2 was solved. The stoichiometry and topology of a larger Cas1–Cas2–Csn2 complex (hereafter referred to as the ‘type II-A CRISPR adaptation module’) were determined based on size-exclusion chromatography (SEC) and isothermal titration calorimetry (ITC) analyses. We also assessed interactions between Cas9 and the Cas components of the adaptation module and found that Cas9 interacts with Csn2. The binding stoichiometry and affinity between Cas9 and Csn2 were characterized using SEC and surface plasmon resonance (SPR). These results more precisely establish an interaction network between the four type II-A Cas proteins and provide experimental evidence of how the Cas9 nuclease effector and Cas1–Cas2 integrase are connected in type II-A CRISPR–Cas systems.
MATERIALS AND METHODS
Cloning, expression and purification
The S. pyogenes cas1 and cas9 genes were cloned into pET28a vectors with a C-terminal (His)6 tag (Supplementary Table S1). The S. pyogenes cas2 and csn2 genes were cloned into pET28a and pHMGWA vectors, respectively, each of which contain an N-terminal (His)6-maltose binding protein (MBP) tag and a tobacco etch virus (TEV) protease cleavage site (Supplementary Table S1). E. coli BL21 (DE3) cells transformed with each individual construct were cultured in lysogeny broth (LB) medium at 37°C until the optical density at 600 nm reached 0.7. Protein expression was induced by the addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside, followed by incubation at 17°C for 16 h. Cells were harvested by centrifugation and resuspended in lysis buffer (300 mM NaCl, 10% (w/v) glycerol, 5 mM β-mercaptoethanol (BME) and 20 mM Tris–HCl (pH 8.0)).
After sonication and centrifugation, the supernatant was loaded onto a 5 ml HisTrap HP column (GE Healthcare, USA) that was pre-equilibrated with elution buffer (300 mM NaCl, 10% (w/v) glycerol, 5 mM BME, 30 mM imidazole, 20 mM Tris–HCl (pH 8.0)). After washing the column with elution buffer, the bound protein was eluted by applying a linear gradient of imidazole (up to 450 mM). The (His)6-MBP tag of Csn2 was cleaved with TEV protease and separated by using a 5 ml HisTrap HP column (GE Healthcare), whereas that of Cas2 was maintained due to the solubility issue. Cas1 was further purified using a 5 ml HiTrap Heparin HP column (GE Healthcare). All proteins were finally purified using a HiLoad 16/60 Superdex200 column (GE Healthcare) equilibrated with SEC buffer (200 mM NaCl, 10% (w/v) glycerol, 2 mM dithiothreitol (DTT), 20 mM Tris–HCl (pH 8.0)).
The Cas1–(His)6-MBP-Cas2 complex was generated by co-expression. The genes of Cas1 and Cas2 were cloned into pET21a, which does not contain a tag, and pET28a, which includes an N-terminal (His)6-MBP tag, respectively. Cas1 and (His)6-MBP-Cas2 were co-expressed in E. coli BL21 (DE3) cells containing both constructs. The expression was induced as described above for the individual Cas proteins. The Cas1–(His)6-MBP-Cas2 complex was purified in a three-step procedure using a HisTrap HP column, a HiTrap Heparin HP column and a HiLoad 16/60 Superdex200 column (GE Healthcare).
Crystallization and structure determination of Cas2
To determine the crystal structure of S. pyogenes Cas2, we generated a truncated cas2 construct, in which the first N-terminal five residues were replaced with a Ser-Gly-Ser-Gly-Ser segment, and the C-terminal tail (residues 92–113) was removed. This construct was cloned into a pET28a vector containing an N-terminal (His)6-MBP tag and a TEV protease cleavage site (Supplementary Table S1). The protein was expressed and purified as described above for Csn2 without the (His)6-MBP tag. Crystals were grown at 20°C by the sitting-drop vapor diffusion method from 14 mg/ml protein solution in buffer (300 mM NaCl, 5% (w/v) glycerol, 5 mM BME, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.0)) mixed with an equal amount of reservoir solution (22.0% (w/v) polyethylene glycol (PEG) 4000, 325 mM ammonium sulfate, 0.1 M sodium acetate (pH 4.6)). To determine the crystal structure using single-wavelength anomalous diffraction, the selenomethionyl protein was expressed in E. coli BL21 (DE3) cells grown in M9 medium supplemented with SeMet, as described previously (42). The selenomethionyl protein was purified as described above for the native protein, and its crystals were grown under similar conditions (26.5% (w/v) PEG 4000, 225 mM ammonium sulfate, 0.1 M sodium acetate (pH 3.6)). The native and selenomethionyl crystals were flash-frozen in liquid nitrogen without additional cryo-protective reagents.
Diffraction data were collected at the beamline 7A of the Pohang Accelerator Laboratory at 100 K. Diffraction images were processed with HKL2000 (43). Determinations of selenium positions, density modification and initial modeling of the selenomethionyl protein were done using PHENIX (44). The initial model of the selenomethionyl structure was used for phasing of the native structure in PHASER (45). The structure of the native protein was completed using alternate cycles of manual fitting in COOT (46) and refinement in PHENIX (44). The stereochemical quality of the final model was assessed using MolProbity (47).
Analytical SEC
Analytical SEC was performed on Superdex 200 10/300 GL columns (GE Healthcare). The columns were equilibrated with buffer (150 mM NaCl, 2 mM DTT, 20 mM Tris–HCl (pH 8.0)). To test complex formation, protein samples were incubated together in buffer (200 mM NaCl, 2 mM DTT, 20 mM Tris–HCl (pH 8.0)) at 4°C for 1 h and loaded onto one of the columns at a flow rate of 0.5 ml/min. Control experiments for single protein components were performed as references in the same column as their mixtures. A standard curve for the SEC column is provided in Supplementary Figure S1. Experiments with varying salt concentrations were performed by applying different amounts of NaCl to the column. Elution fractions were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by Coomassie staining.
ITC
The ITC experiments were performed at 25°C using a MicroCal iTC200 system (GE Healthcare). Proteins in a 200 μl sample cell were titrated with 20 consecutive 2 μl injections in buffer (150 mM NaCl, 10% (w/v) glycerol, 20 mM Tris–HCl (pH 8.0)). Origin software (OriginLab) was used for processing and analysis of the ITC titration data with a fitting model assuming multiple identical independent binding sites.
SPR
The SPR binding assays were performed using a CM5 chip on a Biacore T200 (GE Healthcare). The amine coupling ligand immobilization procedure was performed at a flow rate of 5 μl/min. The CM5 chip was activated with a mixture of 0.1 M N-hydroxysuccinimide and 0.4 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride at a ratio of 1:1 for 400 s. Subsequently, 60 μg/ml of Cas9 dissolved in 10 mM sodium acetate (pH 5.0) was injected for 30 s. The remaining activated carboxyl groups on the sensor chip surface were deactivated with 1 M ethanolamine (pH 8.5) for 400 s. The multi-cycle analysis was performed at a flow rate of 30 μl/min. Each concentration of Csn2 in running buffer (150 mM NaCl, 10 mM Tris–HCl (pH 8.0)) was injected for 600 s, followed by dissociation for 1200 s in a separate analysis cycle. The sensor chip surface was regenerated with 10 mM NaOH between cycles. Data were fit with the bivalent analyte model. The equilibrium dissociation constant (Kd) was determined based on kinetic rate constants calculated using Biacore T200 evaluation software (GE Healthcare).
RESULTS
The crystal structure of S. pyogenes Cas2 reveals conformational variability in type II-A Cas2 homologues
Among the four S. pyogenes type II-A Cas proteins, the crystal structures of Cas1, Csn2 and Cas9 have previously been reported (31–34,39,40). Thus, we aimed to determine the crystal structure of S. pyogenes type II-A Cas2, to fill the knowledge gap on the structure of individual S. pyogenes type II-A Cas proteins. Under our experimental conditions, the full-length Cas2 precipitated, and an N-terminal (His)6-MBP tag was required for solubilization. While we were not able to crystallize the (His)6-MBP-tagged full-length Cas2, the crystal structure was solved for its untagged, truncated form. In this construct, the first five N-terminal residues (Met-Ser-Tyr-Arg-Tyr) were replaced with a more flexible segment (Ser-Gly-Ser-Gly-Ser), and the C-terminal tail (residues 92–113) was eliminated (Supplementary Figure S2) since it was often unresolved in the crystal structures of other Cas2 homologues due to insufficient electron density (41,48,49). Data collection, phasing and refinement statistics are summarized in Table 1.
Table 1.
Data collection, phasing and refinement statisticsa
| Native | SeMet | |
|---|---|---|
| Space group | P212121 | P212121 |
| Unit cell parameters (Å) | a = 51.1, b = 75.6, c = 146.4 | a = 51.1, b = 76.4, c = 145.2 |
| Wavelength (Å) | 0.9793 | 0.9793 |
| Data collection statistics | ||
| Resolution range (Å) | 50.00–1.76 (1.82–1.76) | 50.00–2.00 (2.07–2.00) |
| Number of reflections | 56417 (5499) | 38716 (3813) |
| Completeness (%) | 99.7 (98.6) | 99.9 (100.0) |
| R merge b | 0.071 (0.452) | 0.099 (0.644) |
| Redundancy | 7.2 (6.5) | 7.2 (7.3) |
| Mean I/σ | 22.5 (3.5) | 12.0 (3.1) |
| Phasing statistics | ||
| f', f'' used in phasing | -8.0, 4.3 | |
| Figure of merit | 0.414 | |
| Refinement statistics | ||
| Resolution range (Å) | 47.96–1.76 | |
| R cryst c/Rfreed (%) | 19.7/23.3 | |
| RMSD bonds (Å) | 0.007 | |
| RMSD angles (deg) | 1.00 | |
| Average B-factor (Å2) | 42.0 | |
| Number of water molecules | 293 | |
| Ramachandran favored (%) | 99.2 | |
| Ramachandran allowed (%) | 0.6 | |
aValues in parentheses are for the highest resolution shell.
b R merge = ΣhΣ|Ii(h) − <I(h)> |/ ΣhΣiIi(h), where Ii(h) is the intensity of an individual measurement of the reflection and <I(h)> is the mean intensity of the reflection.
c R cryst = Σh||Fobs| − |Fcalc||/Σh|Fobs|, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively.
d R free was calculated as Rcryst using ∼5% of the randomly selected unique reflections that were omitted from structure refinement.
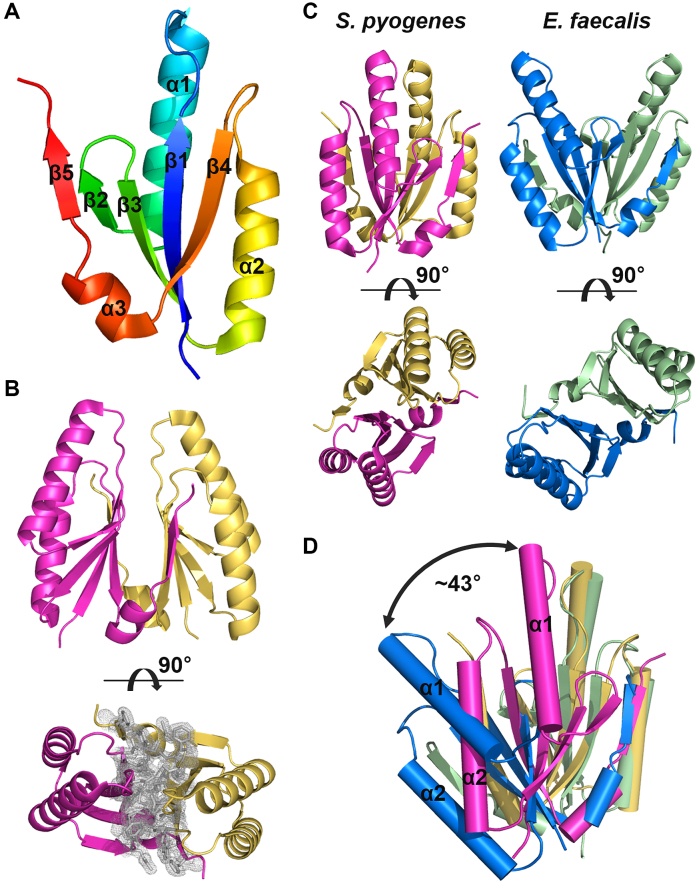
The asymmetric unit contains six Cas2 molecules forming three dimers with non-crystallographic pseudo-2-fold symmetry. The structures of the three Cas2 dimers are very similar, where the root mean square deviation (RMSD) values of the Cα atomic positions among them range from 0.7 to 1.3 Å. Therefore, we hereafter describe the single Cas2 dimer composed of chains A and B, which includes the most amino acid residues (residues 6–91) in the final model. The crystal structure of the S. pyogenes type II-A Cas2 revealed a topology and protomer fold that are essentially identical to those of other homologous structures, including Cas2 from the Enterococcus faecalis type II-A system (26,41,48,49). The S. pyogenes Cas2 protomer contains an N-terminal ferredoxin fold composed of a four-stranded antiparallel β-sheet (β1–4) and two α-helices (α1, α2) and a C-terminal segment that includes a short α-helix (α3) and a β-strand (β5) (Figure 1A). The C-terminal β5 strand in one protomer aligns with the β4 strand of the other protomer, extending the antiparallel β-sheet in the N-terminal ferredoxin fold (Figure 1B). Such β5 swapping creates a dimer interface that exists mostly between the two five-stranded antiparallel β-sheets (Figure 1B). The dimerization of S. pyogenes Cas2 buries 1355 Å2 of the total surface area and forms 22 hydrogen bonds.
Figure 1.
Crystal structure of Streptococcus pyogenes type II-A Cas2. (A) Protomer structure of S. pyogenes type II-A Cas2. Secondary structure elements are also indicated. (B) Dimeric structure of S. pyogenes type II-A Cas2. The 2mFobs − DFcalc map is contoured at 1.0 σ for residues at the interface. (C) Side-by side comparison of S. pyogenes and Enterococcus faecalis type II-A Cas2 structures. The C-terminal tails (residues 92–109) of E. faecalis Cas2 dimer are removed for clarity. (D) Structural alignment of S. pyogenes and E. faecalis type II-A Cas2 dimers based on only one of the two protomers. Note that the ∼43° rigid-body rotation between the two unaligned protomers.
Despite the conserved protomer fold and identical CRISPR subtype (Supplementary Figure S3), the S. pyogenes Cas2 displays a notable conformational difference from the E. faecalis Cas2 in terms of the relative protomer orientation at the dimer interface. In the S. pyogenes Cas2, the two protomers orient approximately parallel to each other, whereas the E. faecalis Cas2 dimerizes at a relatively tilted angle (Figure 1C) (26). When these two type II-A Cas2 structures are aligned based on only one of their two protomers, a significant structural deviation is noted in the positioning of the other protomer (Figure 1D). Between the two unaligned protomers, an ∼43° rigid-body rotation can be detected, and the RMSD value of the equivalent Cα atomic positions is 8.3 Å. The two conserved Asp13 residues at the tip of the β1 strands are further apart in the E. faecalis Cas2 dimer (9.6 Å) than in the S. pyogenes Cas2 structure (3.2 Å). Such structural variability was observed in type I-C Cas2 proteins, which also involve β5 swapping for dimerization, and hinge-bending conformational switching for catalytic activation was suggested (41,48,49). However, it is not clear whether the conformational heterogeneity found in the type II-A Cas2 structures is biologically relevant or simply an experimental artifact (see ‘Discussion’ section).
S. pyogenes Cas2 interacts with Cas1 through its C-terminal tail to form a heterohexameric complex
In E. coli type I-E and E. faecalis type II-A systems, Cas1 and Cas2 form an integrase complex, in which a single Cas2 dimer serves as a bridge between two distal Cas1 dimers (26,50). Perturbation of the Cas1–Cas2 interaction disrupts spacer acquisition in vivo (50), indicating that the complex formation is crucial for CRISPR adaptation. In the present study, we performed biochemical analyses to characterize the interaction between Cas1 and Cas2 in the S. pyogenes type II-A CRISPR–Cas system. We first examined complex formation with individually purified S. pyogenes type II-A Cas1 and Cas2. Since the untagged full-length Cas2 was soluble only at high salt concentrations above 500 mM, and was not stable under our experimental conditions requiring lower salt concentrations for binding assays, we hereafter describe the results obtained with the N-terminal (His)6-MBP-tagged Cas2 (Supplementary Figure S2), unless otherwise stated. In the analytical SEC, the pre-incubated S. pyogenes Cas1 and (His)6-MBP-Cas2 migrated together and had a shorter retention time than those of the individual proteins, implying formation of the S. pyogenes type II-A Cas1–(His)6-MBP-Cas2 complex (Figure 2A).
Figure 2.
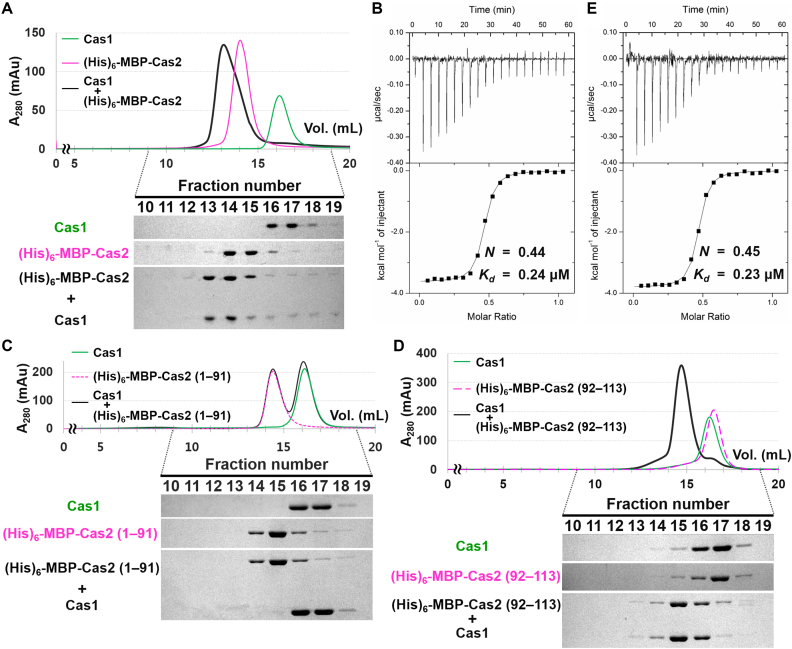
Cas2 interacts with Cas1 through its C-terminal tail. (A) Interaction between Cas1 and (His)6-MBP-Cas2 determined by analytical SEC. Individually purified Cas1 (20 μM) and (His)6-MBP-Cas2 (20 μM) samples were used. Elution fractions were analyzed by SDS-PAGE. Uncropped gel images are shown in Supplementary Figure S18. (B) ITC analysis showing the binding of (His)6-MBP-Cas2 to Cas1. (His)6-MBP-Cas2 (400 μM) was added consecutively to the chamber containing Cas1 (80 μM). The experimentally determined N and Kd values are also indicated. (C and D) The interactions between Cas1 and truncated Cas2 variants were tested in analytical SEC. Cas1 (40 μM) does not interact with the C-terminally truncated form (residues 1–91) of Cas2 (C), but forms a stable complex with the C-terminal tail (residues 92–113) of Cas2 (D). These two Cas2 variants (20 μM) were purified with an N-terminal (His)6-MBP tag. Elution fractions were analyzed by SDS-PAGE. Uncropped gel images are shown in Supplementary Figure S18. (E) ITC trace for the binding of the C-terminal tail of Cas2 to Cas1. The peptide corresponding to the C-terminal tail of Cas2 (residues 92–113) was commercially synthesized (BIONICS, Korea). The C-terminal tail of Cas2 (400 μM) was injected into Cas1 (80 μM). The experimentally determined N and Kd values are also indicated.
We then used ITC to quantitatively analyze the Cas1–Cas2 interaction (Figure 2B). (His)6-MBP-Cas2 bound to Cas1 with sub-micromolar affinity (Kd = 0.24 μM). The binding stoichiometry (N) of (His)6-MBP-Cas2 to Cas1 was calculated to be 0.44, indicating 2:1 binding between Cas1 and Cas2. This is consistent with the formation of a heterohexameric complex composed of one Cas2 dimer and two Cas1 dimers. When the (His)6-MBP-tagged Cas2 was expressed together with untagged Cas1 in E. coli cells, co-purification of the two proteins was observed (Supplementary Figure S4). They co-migrated as one peak in SEC, and the elution volume was identical to that of the Cas1–(His)6-MBP-Cas2 complex reconstituted by mixing the individually purified Cas1 and (His)6-MBP-Cas2 at a 2:1 ratio (Supplementary Figure S5). This confirms the formation of a stable and stoichiometric complex by S. pyogenes type II-A Cas1 and Cas2. We tried to remove the (His)6-MBP tag from the complex, but the untagged Cas2 precipitated even with the bound Cas1 at low salt concentrations below 500 mM.
As described above, we deleted the C-terminal tail (residues 92–113) of S. pyogenes type II Cas2 for its crystallization, since the equivalent region was disordered in several other homologous structures (41,48,49). However, in the crystal structures of the E. faecalis and E. coli Cas1–Cas2 complexes, the C-terminal tail of Cas2 is visible and mediates the Cas1–Cas2 interaction (26,50). To determine whether, and to what extent, the C-terminal tail contributes to the interaction with Cas1, we analyzed the binding of truncated S. pyogenes Cas2 variants (Supplementary Figure S2) to Cas1. In the analytical SEC, the C-terminally truncated variant (residues 1–91) of (His)6-MBP-Cas2 did not interact with Cas1 (Figure 2C), whereas the (His)6-MBP-tagged C-terminal tail of Cas2 co-eluted with Cas1 (Figure 2D). In the ITC experiment using a peptide corresponding to the C-terminal tail of Cas2, the stoichiometry (N) and dissociation constant (Kd) were determined to be almost identical to those obtained with the full-length Cas2 (Figure 2B and E). These results suggest that the C-terminal tail region of S. pyogenes type II-A Cas2 is primarily responsible for the interaction with Cas1 in the formation of the Cas1–Cas2 complex.
The Cas1–Cas2 complex interacts with Csn2 to form the type II-A CRISPR adaptation module
In type II-A CRISPR–Cas systems, Cas1, Cas2 and Csn2 are not necessary for the interference process but are required for spacer integration (1,36,37). We previously showed that, in the S. pyogenes type II-A system, Cas1 interacts directly with Csn2, and identified the binding interface and key residues for the complex formation (39). Mutations in these residues severely disturbed the Cas1–Csn2 interaction (39). Streptococcus pyogenes Csn2 did not interact with Xanthomonas campestris type I-F Cas1 (Supplementary Figure S6), suggesting that S. pyogenes Csn2 interacts specifically with S. pyogenes Cas1. In the present study, we also demonstrated complex formation between S. pyogenes type II-A Cas1 and Cas2. These observations raised a question of whether Cas1, Cas2 and Csn2 form a three-component Cas complex in type II-A CRISPR–Cas systems.
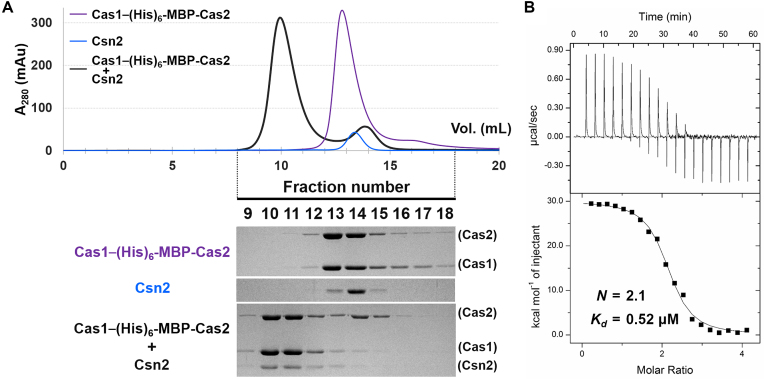
To verify the formation of the type II-A CRISPR adaptation module composed of Cas1, Cas2 and Csn2, we tested the interaction between the Cas1–(His)6-MBP-Cas2 complex and Csn2 in the S. pyogenes type II-A CRISPR–Cas system. In the analytical SEC, they co-migrated with a smaller elution volume than those of either the Cas1–(His)6-MBP-Cas2 complex or Csn2 tetramer alone (Figure 3A), indicating that Cas1, Cas2 and Csn2 form a larger three-component protein complex. We also performed the ITC experiment for quantitative analysis (Figure 3B). The binding stoichiometry (N) of the Cas1–(His)6-MBP-Cas2 complex per the Csn2 tetramer was 2.1, suggesting that two hexameric Cas1–Cas2 complexes interact with a single Csn2 tetramer, consistent with our previous observation that the Csn2 tetramer contains two symmetric binding sites for Cas1 (39).
Figure 3.
The Cas1–Cas2 complex and Csn2 comprise the type II-A CRISPR adaptation module. (A) The interaction between the Cas1–(His)6-MBP-Cas2 complex (10 μM) and Csn2 (40 μM) was assessed by analytical SEC. Elution fractions were analyzed by SDS-PAGE. Uncropped gel images are shown in Supplementary Figure S18. (B) ITC trace for the binding of the Cas1–(His)6-MBP-Cas2 complex to Csn2. Csn2 (50 μM) was titrated with the Cas1–(His)6-MBP-Cas2 complex (250 μM). The experimentally determined N and Kd values are indicated.
The dissociation constant (Kd) between the Cas1–(His)6-MBP-Cas2 complex and Csn2 was measured as 0.52 μM, whereas that between Cas1 and Csn2 was previously determined to be ∼5.0 μM (39). This indicates that Csn2 binds to the Cas1–Cas2 complex more strongly than to Cas1. We initially suspected that tighter Csn2 binding by the Cas1–(His)6-MBP-Cas2 complex could result from a direct interaction between (His)6-MBP-Cas2 and Csn2. However, they did not interact with each other in SEC (Supplementary Figure S7). Thus, Cas2 does not appear to possess intrinsic binding affinity to Csn2 and likely enhances the Cas1–Csn2 interaction indirectly. It is possible that Cas2 binding to Cas1 may aid in the formation and/or stabilization of the binding interface(s) for Csn2 in the Cas1 structure. It is important to note that both Cas2 and Csn2 interact with the N-terminal domain of Cas1 (26,39).
The Cas9–Csn2 interaction manifests a connection between the CRISPR adaptation and interference
In type II-A CRISPR–Cas systems, Cas9 was initially implicated in crRNA biogenesis and interference (1,22,24). It was later found that Cas9 also contributes to the adaptation of CRISPR immunity (36,37). Cas9 selects new spacer sequences during spacer acquisition and associates with other Cas proteins in the acquisition machinery when they are co-expressed in E. coli (36). A mutation in Cas9 increased the rate of spacer acquisition significantly (38). To better understand how Cas9 is connected with the type II-A CRISPR adaptation module at the molecular level, we investigated in vitro interactions between Cas9 and other Cas components of the S. pyogenes type II-A system, Cas1, Cas2 and Csn2.
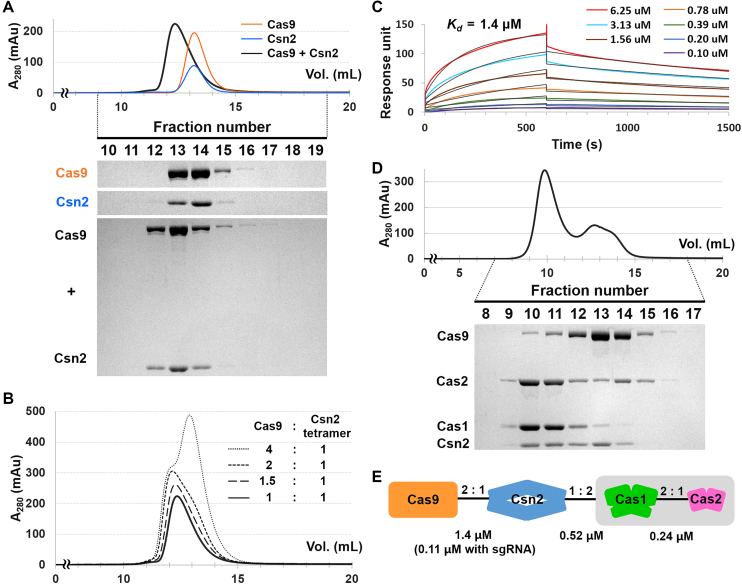
With individually purified proteins, we performed SEC to test the complex formation between Cas9 and each of the Cas components comprising the type II-A CRISPR adaptation module. In the SEC experiment, Cas9 did not interact with Cas1, (His)6-MBP-Cas2 or the Cas1–(His)6-MBP-Cas2 complex (Supplementary Figures S8–S10). In contrast, Cas9 interacted directly with Csn2 to form a stable complex (Figure 4A). The formation of the Cas9–Csn2 complex was perturbed by increasing the NaCl concentrations of the SEC buffer (Supplementary Figure S11), suggesting that electrostatic interactions play an important role in complex formation. To determine the binding stoichiometry of Cas9 to Csn2, we repeated the SEC analyses with increasing amounts of Cas9. Until the molar ratio between Cas9 and Csn2 reached ∼1:2, the elution volume of the Cas9–Csn2 complex decreased and its peak height increased (Figures 4B and Supplementary S12). This result indicates that a maximum of two monomeric Cas9 proteins bind to a single Csn2 tetramer. For the quantitative analysis of the binding affinity between Cas9 and Csn2, the SPR binding assays were performed since we were unable to obtain ITC data due to the precipitation of the proteins during the titration. In the SPR analysis, the dissociation constant (Kd) between Cas9 and Csn2 was determined to be 1.4 μM (Figure 4C). The sgRNA-loaded Cas9 exhibited higher binding affinity (Kd = 0.11 μM) to Csn2 than the apo-Cas9 (Supplementary Figure S13).
Figure 4.
Cas9 interacts with the type II-A CRISPR adaptation module via Csn2. (A) Interaction between Cas9 (10 μM) and Csn2 (40 μM) in analytical SEC. Elution fractions were analyzed by SDS-PAGE. Uncropped gel images are shown in Supplementary Figure S18. (B) Determination of the binding stoichiometry between Cas9 and Csn2 by analytical SEC. Csn2 (40 μM) was pre-incubated with increasing amounts of Cas9 (10, 15, 20 and 40 μM), and the interactions were tested by analytical SEC. The chromatogram for the interaction between Cas9 (10 μM) and Csn2 (40 μM) was reproduced from Figure 4A for comparison. SDS-PAGE analyses for the elution fractions are shown in Supplementary Figure S12. (C) SPR analysis of the Cas9–Csn2 interaction. Sensorgrams show the binding of increasing concentrations (0.10, 0.20, 0.39, 0.78, 1.56, 3.13 and 6.25 μM) of free Csn2 to immobilized Cas9. The calculated Kd value is also shown. (D) Test of the Csn2–Cas9–Cas1–Cas2 complex formation in analytical SEC. Separately purified Csn2 (40 μM), Cas9 (10 μM) and Cas1–(His)6-MBP-Cas2 complex (10 μM) were used. Elution fractions were analyzed by SDS-PAGE. Uncropped gel images are shown in Supplementary Figure S18. (E) Schematic representation of the type II-A Cas complex illustrating direct interactions between Cas9, Csn2, Cas1 and Cas2. The binding stoichiometry and Kd values are also shown.
Finally, we examined whether Cas9 and the Cas1–Cas2 complex bind to Csn2 simultaneously to form a larger, four-component Cas complex. When the separately purified Cas9, Csn2 and Cas1–(His)6-MBP-Cas2 complex were incubated together, they co-eluted in the analytical SEC with a shorter retention time than those for each individual component (Figure 4D, fractions 10 and 11). This result is consistent with the previous observation that S. pyogenes Cas9 associates with Cas1, Cas2 and Csn2 when expressed together in E. coli (36), suggesting the formation of the Cas9–Csn2–Cas1–Cas2 complex (Figure 4E). However, we also observed an unbound form of Cas9 (Figure 4D, fractions 13 and 14), and the elution volume of the four-component Cas complex is similar to that of the three-component complex composed of Cas1, (His)6-MBP-Cas2 and Csn2 (Figures 3A and 4D). Thus, it is still unclear to determine whether the four Cas components can from a stable complex based on our experimental data. Binding of the Cas1–(His)6-MBP-Cas2 complex to Csn2 may hinder the interaction between Csn2 and Cas9 if the binding interfaces for the Cas1–(His)6-MBP-Cas2 complex and Cas9 in Csn2 partially overlap. The (His)6-MBP tag, which was required for solubilizing Cas2 under our experimental conditions, could also interfere with the Cas9–Csn2 interaction. The dissociation constant (Kd) between Csn2 and the Cas1–(His)6-MBP-Cas2 complex is smaller than that between Csn2 and Cas9 (Figures 3B and 4C), implying that the Cas1–Cas2 complex binds more tightly to Csn2 than Cas9.
DISCUSSION
The type II-A CRISPR–Cas system from S. pyogenes has attracted significant attention, since its Cas9 protein is widely used as a core component of the CRISPR–Cas genome editing tool (24,28–30). Together with previous studies, our work expands on structural knowledge of S. pyogenes type II-A Cas proteins. We determined the crystal structure of S. pyogenes type II-A Cas2, which makes all of the individual Cas structures available in the S. pyogenes type II-A CRISPR–Cas system. We also revealed the network of molecular interactions among the type II-A Cas components in more detail. In particular, Cas9 interacts specifically with Csn2, implying that Csn2 is a natural binding partner of Cas9. The tetrameric Csn2 also binds to the Cas1–(His)6-MBP-Cas2 complex. These findings suggest the potential of Csn2 in the Cas9-based genome editing technology as a scaffold connecting Cas9 and other functional effectors. Notably, Csn2 exhibited calcium-dependent conformational changes in its tertiary and quaternary structures (40,51), based on which a regulatory mechanism could be developed.
Compared to the E. faecalis type II-A homologue, S. pyogenes type II-A Cas2 revealed a more lateral protomer arrangement at the dimer interface. Such conformational variability has also been recognized in type I-C Cas2 structures (41,48,49). Based on the structural analyses and other experimental data, hinge-bending conformational switching for nuclease activity was suggested for the type I-C Cas2 homologues (41,48,49). When the S. pyogenes Cas2 protomers were arranged as those in the E. faecalis dimer, severe steric clashes were not observed at the dimer interface (Supplementary Figure S14). However, it is unclear whether the conformational heterogeneity is relevant to the function of type II-A Cas2 in CRISPR-mediated immunity. Although some Cas2 proteins of various subtypes have been characterized as nucleases (41,48,49,52), other homologues, including S. pyogenes type II-A Cas2, did not exhibit nuclease activity (39,53). Moreover, active site mutants of Cas2 still support spacer acquisition in E. coli (50), indicating that the catalytic activity of Cas2 is dispensable during the adaptation stage. Given that a conformational change of Cas2 can induce significant structural rearrangement of the Cas1–Cas2 complex, we also explored whether different patterns of Cas2 dimerization were related to the preferred sizes of DNA substrates for Cas1–Cas2 integrase. However, the lengths of both repeat and spacer sequences are identical in the S. pyogenes and E. faecalis type II-A CRISPR arrays (Supplementary Figure S15), suggesting no obvious correlation between Cas2 conformation and the length of the integration substrate. Alternatively, the distinct conformational states of the two type II-A Cas2 structures may result from differences in experimental settings. We used a truncated form of S. pyogenes Cas2 for the structural study, whereas E. faecalis Cas2 was crystallized as a component of the DNA-bound Cas1–Cas2 complex (26). Note that E. coli Cas1 and Cas2 proteins undergo conformational changes upon complex formation and DNA binding (50,54).
In the SEC and ITC experiments (Figure 2), we demonstrated that the C-terminal tail of Cas2 is critical for its interaction with Cas1, consistent with the crystal structure of the E. faecalis type II-A Cas1–Cas2 complex (26), in which the C-terminal tail of Cas2 makes extensive contacts with Cas1. In the crystal structure, only one E. faecalis Cas1 protomer in a single dimer is involved in the interaction with Cas2 (26), suggesting that two Cas2 dimers simultaneously bind to one Cas1 dimer by interacting with each Cas1 protomer. However, in our ITC analysis, in which the short peptide corresponding to the C-terminal tail was used as a titrant (Figure 2E), the binding stoichiometry (N) of the Cas2 tail to Cas1 was determined to be ∼0.5, implying that only one C-terminal tail of Cas2 binds to a single Cas1 dimer. Considering the relatively small size of the peptide used in the ITC, it is unlikely that the second binding would be prevented by steric hindrance caused by the bound Cas2 tail at the first binding site. Rather, the Cas2 binding to one Cas1 protomer may induce a structural change in the other unbound Cas1 protomer and decrease the affinity of the second binding site for Cas2, similar to allosteric proteins displaying negative cooperativity. Although the crystal structure of the S. pyogenes Cas1 dimer alone exhibited a symmetric arrangement of its two protomers (39), a significant conformational difference was observed between the Cas2-bound and unbound Cas1 protomers in the E. faecalis structure (26). The superposition of the two E. faecalis Cas1 protomers revealed ∼33° hinge motion between the N- and C-terminal domains (26). Indeed, the C-terminal tail of the E. faecalis Cas2 interacts with residues from both domains of Cas1 (26), and the S. pyogenes Cas1 protomer exhibits greater conformational similarity to the Cas2-bound E. faecalis Cas1 protomer than the unbound one. These observations suggest that the conformational change may deform the potential Cas2 binding interface in the unbound Cas1 protomer.
The involvement of Csn2 in the adaptation process has been demonstrated, but its precise role therein remains unclear. In vivo deletion of csn2 genes inhibits new spacer insertion into CRISPR arrays upon bacteriophage challenge (36,37). However, in vitro, the type II-A Cas1–Cas2 complex alone catalyzes full-site spacer integration when provided with proper substrates, indicating the dispensability of Csn2 in spacer integration in vitro (26,27). Based on these observations, it was previously suggested that Csn2 may be involved in different step(s) of the adaptation process, such as the generation of prespacers (27). The connection between Csn2 and the prespacer substrate is also supported by the molecular organization of the S. pyogenes type II-A adaptation module, which we described herein. Considering the E. faecalis Cas1–Cas2 structure (26), and our previous finding that Csn2 interacts with the N-terminal domains of the Cas1 dimer (39), Csn2 is presumably in close proximity to the prespacer binding surface of the Cas1–Cas2 complex, but distant from the target DNA-binding site (Supplementary Figure S16). This suggests that Csn2 functions in molecular event(s) related to the prespacer substrate rather than the DNA target, such as the production and/or recruitment of prespacers. Csn2 exhibited the binding affinity for a prespacer in an electrophoretic mobility shift assay (Supplementary Figure S17). The double-stranded DNA end-binding and sliding properties of Csn2 (55) fit well with its proposed involvement with prespacers, which are essentially linear duplex DNAs.
Several other CRISPR–Cas systems contain conserved Cas components that participate in the CRISPR adaptation in addition to Cas1 and Cas2. The Sashital group recently reported that Bacillus halodurans type I-C Cas4 interacts tightly with Cas1 and processes prespacers (56). The integral roles of Cas4 in spacer acquisition have also been identified in the CRISPR–Cas systems of Synechocystis sp. 6803 (57) and Pyrococcus furiosus (58). In type I-F CRISPR–Cas systems, Cas3, the effector nuclease for target degradation is fused with Cas2 into a single polypeptide. This fusion protein forms a stable complex with Cas1, supporting the possibility that the type I-F Cas3 is also involved in the spacer acquisition process (59,60).
Our work also highlights the role of Csn2 in coordinating the functions of Cas9 and the Cas1–Cas2 complex within the type II-A CRISPR–Cas system. Although co-purification of the four S. pyogenes type II-A Cas proteins was reported (36), how the nuclease effector and the integrase complex are connected at the molecular level has not yet been determined. In the present study, we applied multiple experimental techniques to show that Cas9 interacts directly with Csn2, which forms a stable, stoichiometric adaptation module with the Cas1–Cas2 integrase complex. These results suggest that Csn2 mediates Cas9 involvement in the type II-A adaptation process. The overall architecture of the four-component type II-A Cas complex, including the binding interface between Cas9 and Csn2, and details of the prespacer generation mechanism remain to be determined.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data Bank under accession number 5ZYF.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff of the beamline 7A of the Pohang Accelerator Laboratory for their support with data collection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cooperative Research Program for Agricultural Science & Technology Development funded by Rural Development Administration [PJ013181]; Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology [NRF-2016R1D1A1A09916821]. Funding for open access charge: Seoul National University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P.. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007; 315:1709–1712. [DOI] [PubMed] [Google Scholar]
- 2. Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J.. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008; 321:960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wiedenheft B., Sternberg S.H., Doudna J.A.. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012; 482:331–338. [DOI] [PubMed] [Google Scholar]
- 4. Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015; 526:55–61. [DOI] [PubMed] [Google Scholar]
- 5. Hille F., Richter H., Wong S.P., Bratovic M., Ressel S., Charpentier E.. The biology of CRISPR-Cas: backward and forward. Cell. 2018; 172:1239–1259. [DOI] [PubMed] [Google Scholar]
- 6. Bolotin A., Quinquis B., Sorokin A., Ehrlich S.D.. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005; 151:2551–2561. [DOI] [PubMed] [Google Scholar]
- 7. Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Soria E.. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005; 60:174–182. [DOI] [PubMed] [Google Scholar]
- 8. Pourcel C., Salvignol G., Vergnaud G.. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005; 151:653–663. [DOI] [PubMed] [Google Scholar]
- 9. van der Oost J., Westra E.R., Jackson R.N., Wiedenheft B.. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 2014; 12:479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reeks J., Naismith J.H., White M.F.. CRISPR interference: a structural perspective. Biochem J. 2013; 453:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiang F., Doudna J.A.. The structural biology of CRISPR-Cas systems. Curr. Opin. Struct. Biol. 2015; 30:100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J.. CRISPR-Cas: adapting to change. Science. 2017; 356:eaal5056. [DOI] [PubMed] [Google Scholar]
- 13. Amitai G., Sorek R.. CRISPR-Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 2016; 14:67–76. [DOI] [PubMed] [Google Scholar]
- 14. Sternberg S.H., Richter H., Charpentier E., Qimron U.. Adaptation in CRISPR-Cas systems. Mol. Cell. 2016; 61:797–808. [DOI] [PubMed] [Google Scholar]
- 15. Mohanraju P., Makarova K.S., Zetsche B., Zhang F., Koonin E.V., van der Oost J.. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016; 353:aad5147. [DOI] [PubMed] [Google Scholar]
- 16. Nishimasu H., Nureki O.. Structures and mechanisms of CRISPR RNA-guided effector nucleases. Curr. Opin. Struct. Biol. 2017; 43:68–78. [DOI] [PubMed] [Google Scholar]
- 17. Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H. et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015; 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koonin E.V., Makarova K.S., Zhang F.. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017; 37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Plagens A., Richter H., Charpentier E., Randau L.. DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes. FEMS Microbiol. Rev. 2015; 39:442–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N., Yan W., Abudayyeh O.O., Gootenberg J.S., Makarova K.S., Wolf Y.I. et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017; 15:169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chylinski K., Makarova K.S., Charpentier E., Koonin E.V.. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014; 42:6091–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E.. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011; 471:602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chylinski K., Le Rhun A., Charpentier E.. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013; 10:726–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fonfara I., Le Rhun A., Chylinski K., Makarova K.S., Lecrivain A.L., Bzdrenga J., Koonin E.V., Charpentier E.. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014; 42:2577–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xiao Y., Ng S., Nam K.H., Ke A.. How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration. Nature. 2017; 550:137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wright A.V., Doudna J.A.. Protecting genome integrity during CRISPR immune adaptation. Nat. Struct. Mol. Biol. 2016; 23:876–883. [DOI] [PubMed] [Google Scholar]
- 28. Wright A.V., Nunez J.K., Doudna J.A.. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell. 2016; 164:29–44. [DOI] [PubMed] [Google Scholar]
- 29. Mali P., Esvelt K.M., Church G.M.. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013; 10:957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsu P.D., Lander E.S., Zhang F.. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014; 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jinek M., Jiang F., Taylor D.W., Sternberg S.H., Kaya E., Ma E., Anders C., Hauer M., Zhou K., Lin S. et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014; 343:1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O.. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014; 156:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Anders C., Niewoehner O., Duerst A., Jinek M.. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014; 513:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang F., Zhou K., Ma L., Gressel S., Doudna J.A.. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015; 348:1477–1481. [DOI] [PubMed] [Google Scholar]
- 35. Anders C., Jinek M.. In vitro enzymology of Cas9. Methods Enzymol. 2014; 546:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heler R., Samai P., Modell J.W., Weiner C., Goldberg G.W., Bikard D., Marraffini L.A.. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015; 519:199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wei Y., Terns R.M., Terns M.P.. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 2015; 29:356–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heler R., Wright A.V., Vucelja M., Bikard D., Doudna J.A., Marraffini L.A.. Mutations in Cas9 enhance the rate of acquisition of viral spacer sequences during the CRISPR-Cas immune response. Mol. Cell. 2017; 65:168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ka D., Lee H., Jung Y.D., Kim K., Seok C., Suh N., Bae E.. Crystal structure of streptococcus pyogenes Cas1 and its interaction with Csn2 in the type II CRISPR-Cas system. Structure. 2016; 24:70–79. [DOI] [PubMed] [Google Scholar]
- 40. Koo Y., Jung D.K., Bae E.. Crystal structure of Streptococcus pyogenes Csn2 reveals calcium-dependent conformational changes in its tertiary and quaternary structure. PLoS One. 2012; 7:e33401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ka D., Kim D., Baek G., Bae E.. Structural and functional characterization of Streptococcus pyogenes Cas2 protein under different pH conditions. Biochem. Biophys. Res. Commun. 2014; 451:152–157. [DOI] [PubMed] [Google Scholar]
- 42. Mark B.L., Vocadlo D.J., Knapp S., Triggs-Raine B.L., Withers S.G., James M.N.. Crystallographic evidence for substrate-assisted catalysis in a bacterial beta-hexosaminidase. J. Biol. Chem. 2001; 276:10330–10337. [DOI] [PubMed] [Google Scholar]
- 43. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 44. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 47. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nam K.H., Ding F., Haitjema C., Huang Q., DeLisa M.P., Ke A.. Double-stranded endonuclease activity in Bacillus halodurans clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas2 protein. J. Biol. Chem. 2012; 287:35943–35952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ka D., Hong S., Jeong U., Jeong M., Suh N., Suh J.Y., Bae E.. Structural and dynamic insights into the role of conformational switching in the nuclease activity of the Xanthomonas albilineans Cas2 in CRISPR-mediated adaptive immunity. Struct. Dyn. 2017; 4:054701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nunez J.K., Kranzusch P.J., Noeske J., Wright A.V., Davies C.W., Doudna J.A.. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat. Struct. Mol. Biol. 2014; 21:528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nam K.H., Kurinov I., Ke A.. Crystal structure of clustered regularly interspaced short palindromic repeats (CRISPR)-associated Csn2 protein revealed Ca2+-dependent double-stranded DNA binding activity. J. Biol. Chem. 2011; 286:30759–30768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Beloglazova N., Brown G., Zimmerman M.D., Proudfoot M., Makarova K.S., Kudritska M., Kochinyan S., Wang S., Chruszcz M., Minor W. et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J. Biol. Chem. 2008; 283:20361–20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Samai P., Smith P., Shuman S.. Structure of a CRISPR-associated protein Cas2 from Desulfovibrio vulgaris. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010; 66:1552–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang J., Li J., Zhao H., Sheng G., Wang M., Yin M., Wang Y.. Structural and mechanistic basis of PAM-Dependent spacer acquisition in CRISPR-Cas systems. Cell. 2015; 163:840–853. [DOI] [PubMed] [Google Scholar]
- 55. Arslan Z., Wurm R., Brener O., Ellinger P., Nagel-Steger L., Oesterhelt F., Schmitt L., Willbold D., Wagner R., Gohlke H. et al. Double-strand DNA end-binding and sliding of the toroidal CRISPR-associated protein Csn2. Nucleic Acids Res. 2013; 41:6347–6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee H., Zhou Y., Taylor D.W., Sashital D.G.. Cas4-Dependent prespacer processing ensures High-Fidelity programming of CRISPR arrays. Mol. Cell. 2018; 70:48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kieper S.N., Almendros C., Behler J., McKenzie R.E., Nobrega F.L., Haagsma A.C., Vink J.N.A., Hess W.R., Brouns S.J.J.. Cas4 facilitates PAM-Compatible spacer selection during CRISPR adaptation. Cell Rep. 2018; 22:3377–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shiimori M., Garrett S.C., Graveley B.R., Terns M.P.. Cas4 nucleases define the PAM, length, and orientation of DNA fragments integrated at CRISPR loci. Mol. Cell. 2018; 70:814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rollins M.F., Chowdhury S., Carter J., Golden S.M., Wilkinson R.A., Bondy-Denomy J., Lander G.C., Wiedenheft B.. Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E5113–E5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fagerlund R.D., Wilkinson M.E., Klykov O., Barendregt A., Pearce F.G., Kieper S.N., Maxwell H.W.R., Capolupo A., Heck A.J.R., Krause K.L. et al. Spacer capture and integration by a type I-F Cas1-Cas2-3 CRISPR adaptation complex. PProc. Natl. Acad. Sci. U.S.A. 2017; 114:E5122–E5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data Bank under accession number 5ZYF.