Abstract
Maintenance of topological homeostasis is vital for gene expression and genome replication in all organisms. Similar to other circular genomes, also mitochondrial DNA (mtDNA) is known to exist in various different topological forms, although their functional significance remains unknown. We report here that both known type II topoisomerases Top2α and Top2β are present in mammalian mitochondria, with especially Top2β regulating the supercoiling state of mtDNA. Loss of Top2β or its inhibition by ciprofloxacin results in accumulation of positively supercoiled mtDNA, followed by cessation of mitochondrial transcription and replication initiation, causing depletion of mtDNA copy number. These mitochondrial effects block both cell proliferation and differentiation, possibly explaining some of the side effects associated with fluoroquinolone antibiotics. Our results show for the first time the importance of topology for maintenance of mtDNA homeostasis and provide novel insight into the mitochondrial effects of fluoroquinolones.
INTRODUCTION
Mitochondria are essential cell organelles, producing ATP and other key metabolites as well as regulating cell death and differentiation. These semi-autonomous organelles have their own genomes, which are in higher animals typically small double-stranded closed DNA circles. Similar to bacterial plasmids, mitochondrial DNA (mtDNA) can obtain various topological forms, including open circles, supercoils and catenanes consisting of interlocked circles of both forms (1–4). The topological conformation of DNA can influence protein interactions (5) and is known to regulate mtDNA transcription rate by influencing the promoter availability (6).
DNA topology is controlled by DNA topoisomerases, which are grouped in two classes with several subdivisions (7,8). Type I topoisomerases catalyze transient cleavage and ligation of a single DNA strand and relieve the topological stress experienced during transcription and replication (7). Both isoforms of Topoisomerase 1, Top1 and Top1mt, can relax both positive and negative supercoils, while Topoisomerase 3 with its isoforms Top3α and Top3β, catalyzes only the relaxation of negative supercoils. While type I topoisomerases create only single-strand nicks on DNA, type II topoisomerases are able to cleave and re-ligate both strands simultaneously in the presence of ATP and Mg2+. The only known eukaryotic type II topoisomerase is Topoisomerase 2, capable of relaxing positively and negatively supercoiled DNA as well as decatenating and catenating DNA (7). Most eukaryotes possess two isoforms of the enzyme: Top2α is primarily expressed in proliferating cells, whereas Top2β is present in all cell types (9,10). While Top2α is responsible for chromosome condensation and segregation in proliferating cells (11) and regulates the transcription especially of long genes (12), Top2β has a role in transcriptional activation and repression of specific nuclear gene loci (13,14). Eukaryotic Topoisomerase 2 can only relax supercoiled DNA, but the bacterial equivalent DNA gyrase is able to also introduce negative supercoils, which facilitates replication origin unwinding (8) and modulates promoter activity (15). Additionally, gyrases can relax negatively supercoiled DNA also in the absence of ATP.
So far, four topoisomerases have been identified to exist in mitochondria (16). The best characterized of these is Top1mt, which is also the only topoisomerase having a dedicated gene for the mitochondrial isoform. This enzyme has reduced DNA binding activity compared to the nuclear Top1 form (17) and appears to function in the topological regulation of mtDNA transcription (18,19). Also another type I topoisomerase, Top3α, has a mitochondrial version, which is expressed from the same gene as the nuclear protein, using an upstream alternative start codon creating a mitochondrial targeting sequence (20). As this targeting sequence is removed during the import of the protein, the resulting mitochondrial isoform is basically identical to the nuclear protein. The knockout of mitochondrial Top3α in flies leads to mtDNA instability, copy number depletion and premature ageing (21). In mammals, the enzyme has been recently suggested to be required for hemicatenane resolution and segregation of mtDNA (22).
In contrast to Top1mt and Top3α, mitochondrial type II topoisomerases have been more evasive. While both Top2α and Top2β (18) have been reported from the mitochondrial compartment, other studies have found no evidence of mitochondrial Top2 (22).
Considering the importance of topoisomerases in genome maintenance, the low number of studies dedicated to mitochondrial topoisomerases is surprising. Research is complicated by the fact that the gene products for both isoforms of Top2 as well as Top3α are shared between nucleus and mitochondria. Any manipulation of their coding sequence will simultaneously cause both nuclear and mitochondrial phenotypes, making the identification of the mitochondrial function difficult. This problem does not exist with Top1mt. Unlike its nuclear paralog (23) Top1mt is not essential for cell survival (24), but the loss of Top1mt results in accumulation of negatively supercoiled mtDNA species (18). This implies the protein to have a role in transcription and replication, as both processes result in the buildup of positive supercoils before the transcription/replication complex and negative behind it (8). As both negative and positive supercoiling need to be regulated as a part of the genome maintenance mechanisms, we hypothesized that additional topoisomerases should be involved in the process. We report here that the type IIA topoisomerase Top2β is required to relax positive mtDNA supercoiling in vivo and this relaxation is important for mtDNA replication initiation. The inhibition of mitochondrial Top2 by the fluoroquinolone antibiotic ciprofloxacin impairs mtDNA replication and transcription, thus having an impact also on cellular proliferation and differentiation. Besides providing a potential molecular mechanism for rare, but severe side effects of fluroquinolone antibiotics, our results help to understand the role of topological regulation in mtDNA maintenance and expression and provide novel evidence for mitochondrial retrograde signaling during cell differentiation.
MATERIALS AND METHODS
Cell culture
Cells were cultured in Dulbecco's Modified Eagle Medium supplemented with 10% FBS at 37°C in 8.5% CO2. C2C12 cell differentiation was induced upon confluency by changing the growth medium to DMEM containing 2% horse serum. Myotubes were harvested after 6 days in this differentiation medium unless mentioned otherwise.
Mitochondria extraction
Mitochondria were extracted from cultured cells and mouse tissues using differential centrifugation and purification by sucrose gradient as described in Goffart et al. (25). The purified mitochondrial layer was used for protein and nucleic acid extractions.
Protein extraction and Western blots
Proteins were extracted from cultured cells or purified mitochondria using TOTEX buffer, separated over 8% Tris/glycine or 4–12% Tris–Tricine SDS-PAGE, transferred to nitrocellulose membrane, incubated with antibodies and ECL-exposed as described in Herbers et al. (26).
| Antibody | Dilution | Source | Identifier |
|---|---|---|---|
| Rabbit-anti-Top2α | 1:1000 | Abgent | AP9248b |
| Rabbit-anti-Top2β | 1:1000 | Abcam | ab15565 |
| Rabbit-anti- Top2β | 1:1000 | Genetex | GTX102640 |
| Rabbit-anti-Top3α | 1:4000 | ProteinTech | 14525-1-AP |
| Rabbit-anti-Top1mt | 1:1000 | Bioss | bs-1211R |
| Rabbit-anti TFAM | 1:2000 | Abgent | AN1221 |
| Mouse-anti-HSP60 | 1:20 000 | Antibodies-online | ABIN361784 |
| Mouse-anti-vinculin | 1:10 000 | Sigma | V9264 |
| Rabbit-anti-Histone 2.1 | 1:2000 | Abcam | ab181973 |
| Rabbit anti-Tomm20 | 1:4000 | Sigma | HPA011562 |
| Goat-anti-rabbit IgG HRP | 1:10 000 | Antibodies-online | ABIN101744 |
| Goat-anti-mouse IgG HRP | 1:15 000 | Life Technologies | A16104 |
Floatation gradients
The colocalization of mitochondrial topoisomerases with mitochondrial nucleoids and known mtDNA interactors was analyzed by mitochondrial subfractionation and floatation gradient analysis as described (27). Briefly, mitochondria isolated form HEK293 cells were treated with digitonin to separate mitochondrial membrane-associated protein complexes in the pellet fraction from the supernatant fraction containing soluble mitochondrial proteins. Both fractions were separated over bottom-up flotation iodixanol gradients and their protein and mtDNA content analyzed by Western blot and Southern dot blot, using nts 14837–15367 as probe.
Transient transfection and immunocytochemistry
As the endogenous mitochondrial Top2β could not be localized by fluorescent microscopy, a mitochondrially targeted, myc-tagged version of human Top2β was overexpressed in HeLa cells. For this the human TOP2B-gene preceded by the mitochondrial targeting sequence of cytochrome C (kind gift of Dr Stefan Sobek) was equipped with a C-terminal myc-tag and cloned into pcDNA5 FRT/TO. The vector was introduced into Hela cells grown on coverslips using Turbofect according to the manufacturer's instructions. One day after transfection the cells were fixed with 3.3% PFA for 25 min at room temperature and permeabilized in PBS + 0.5% Triton-X, 10% FBS for 15 min. The slides were incubated with primary antibodies in PBS+ 0.1% Triton, 10% FBS for 1 h, washed in PBS, incubated with secondary antibodies for 1 h. After washing the cells were mounted and fluorescent staining analyzed with a Zeiss Axiovert/Axiocam fluorescent microscope.
siRNA knockdown
Cells were transfected in six-well plates with 25 pmol of siRNA or 12.5 + 12.5 pmol for combined transfections with two different siRNAs, using Lipofectamine RNAiMAX reagent according to the manufacturer's instructions. Protein and DNA were extracted 3 days after transfection. siRNAs used were Cat. No. AM4613 as negative siRNA control and Cat. No. 4390824 ID s14309 and ID s108 (ThermoFisher, Ambion) for knockdown of Top2α and Top2β, respectively.
DNA extraction
Total cell DNA was extracted as described in Goffart et al. (25), using proteinase K digest, phenol:chloroform extraction and ethanol precipitation. DNA samples were dissolved in TE buffer and digested with BglII (does not cleave human mtDNA) or KpnI (mouse) to facilitate solubilization.
RNA extraction and Northern blot analysis
RNA was extracted with TriReagent (Sigma) and analyzed by Northern blot as described in Herbers et al. (26). Mitochondrial RNA levels were quantified using ND2 and ND5 probes (nts 4470–5511 and 13 641–13 777 of human mtDNA, respectively) and normalized against 18S rRNA (nts 850–1347 bp of accession number NR_0032862) as a loading control.
Topology analysis
Topological forms of mtDNA were analyzed by agarose gel electrophoresis and Southern blotting as previously described (25). 1 μg of total DNA was separated over a 0.4% agarose gel in TBE, blotted and probed against nts 35–611 of human mtDNA or nts 14 783–15 333 of mouse mtDNA.
To investigate the direction of supercoiling in mtDNA 300 ng of total DNA or 100 ng of mtDNA isolated from HeLa cells was incubated for 10 min at RT with the indicated concentration of 0–100 μg/ml ethidium bromide in 20 mM HEPES pH 7.4. After addition of DNA loading dye the DNA samples were analyzed by gel electrophoresis and Southern blotting as above.
Neutral/Neutral two-dimensional agarose gel electrophoresis
For 2D agarose gels (2D-AGE), 5 μg of mtDNA or 10 μg of total DNA were digested with ClaI, HincII or MluI as indicated, separated by 2D agarose gel electrophoresis and blotted (25). The membranes were probed with 32P-labeled probes (nts 35–611 for human and 14 783–15 333 for mouse) and exposed to either an X-ray film or phosphor storage screen.
7S DNA quantification
Mitochondrial 7S DNA levels per mtDNA were quantified by Southern blotting (28) using 1 μg total DNA digested with HindIII and heated for 10 min at 65°C and probed against nts 16 177–40 of human mtDNA or nts 15 467–16 011 of mouse mtDNA. 7S DNA and mtDNA signal were quantified by phosphor storage screens using phosphorimager and the ratio of 7S per mtDNA signal calculated.
MtDNA copy number determination
Mitochondrial copy number per cell was determined from total DNA samples using quantitative PCR as described in Herbers et al. (26). For human mtDNA the primers HSmtDNA13456F (5′-ACC ATT GGC AGC CTA GCA TT-3′) and HSmtDNA13593R (5′-TGT CAG GGA GGT AGC GAT GA-3′) and the probe HSmtDNA 13546F (5′-FAM-ACA AAC GCC TGA GCC CTA- MGBNFQ-3′) were used using the identical PCR program.
Mitochondrial DNA damage measurement by long-range PCR
Integrity of mtDNA was determined by long-range Taqman from total DNA as described in Herbers et al. (26), but using a shorter amplicon in the long PCR. The primers used for the long mouse PCR were MMmtDNA3567F (5′-TTC GAG CAT CTT ATC CAC GC-3′) and MMmtDNA8496R (5′-ACC ATT TCT AGG ACA ATG GGC A-3′), the probe (16s-rRNA) 5′-FAM-TGA CCG TGC AAA GGT AGC AT-MGBNFQ-3′ and the elongation time 3 min.
For human mtDNA the primers used were HSmtDNA10131F (5′-ACC ACA ACT CAA CGG CTA CA -3′) and HSmtDNA14841R (5′-TTT CAT CAT GCG GAG ATG TTG GAT GG-3′) and the PCR program and evaluation identical.
In vitro topoisomerase assays
The in vitro activity of human Top2 on mtDNA were tested using purified Top2α (Topogen TG2000H) and Top2β (Inspiralis, Cat. No. HTB205) and isolated mtDNA of HEK293 cells. The proteins were diluted to the given unit in 20 μl of reaction buffer (50 mM Tris–HCl pH 7.5, 125 mM NaCl, 10 mM MgCl2, 5 mM DTT, 100 μg/ml albumin and 1 mM ATP), either without any inhibitor or in the presence of 50 ng/ml EtBr, 80 μg/ml ciprofloxacin or 3.4 μM doxorubicin. After 10 min incubation at room temperature 200 ng mtDNA in 10 μl reaction buffer was added and the samples were further incubated for 30 min at 37°C. The reaction was stopped by addition of 5 μl DNA loading dye (10 mM Tris-HCl pH 7.6, 0.03% bromophenol blue, 0.03% xylene cyanol FF, 60% glycerol, 60 mM EDTA), separated over a topology gel, blotted and probed as described above.
Catenated and supercoiled forms of mtDNA were identified by a treatment with 4 U Escherichia coli TopIV (Inspiralis, Cat. No. T4001) in 25 μl reaction buffer for 30 min at 37°C.
RESULTS
Top2α and Top2β are present in mitochondria and interacting with mtDNA
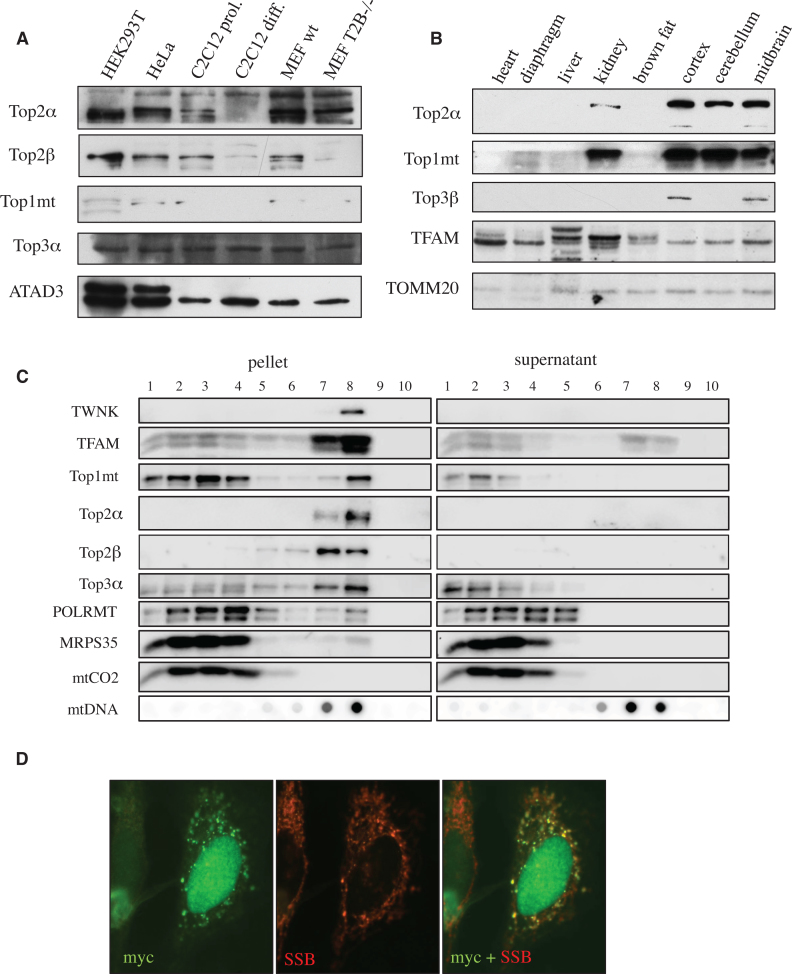
While the mitochondrial localization of Top1mt and Top3α is well established, the data for Top2α and Top2β are conflicting (18,22). We therefore analysed the levels of all four topoisomerases in purified mitochondria from various cell lines and mouse tissues using Western blot. Our results show that mitochondria of cultured human and mouse cells contain both Top2α and Top2β, with the size of the mitochondrial form being similar to the nuclear version (Figure 1A and Supplementary Figure S1A). The corresponding protein band was absent in Top2β-/- mouse embryonic fibroblasts demonstrating the specificity of the antibody. Top2α was not detectable in mitochondria of differentiated C2C12 myotubes or a range of post-mitotic mouse tissues, confirming that this isoform is expressed only in proliferating cells (Figure 1A and B). In contrast, Top2β was highly expressed in brain where also Top1mt and Top3α were strongly expressed (Figure 1B), and at much lower level in other tissues (Supplementary Figure S1B).
Figure 1.
Topoisomerase 2α and 2β co-localize with replicating mitochondrial DNA nucleoids. (A) Detection of topoisomerases in mitochondrial extracts of human and mouse cell lines by Western blot. Human cell lines: HEK293T and HeLa. Mouse cells: proliferating C2C12 myoblasts and differentiated C2C12 myotubes, mouse embryonic fibroblasts of wildtype and Top2β (–/–) knockout mice. ATAD3 is used as loading reference. Note that the used polyclonal antibodies might detect mouse and human proteins with different efficiencies. (B) Detection of topoisomerase Top2β, Top1mt and Top3α in mitochondrial extracts of various mouse tissues by Western blot. TFAM is used as indicator of mtDNA content, while TOMM20 serves as mitochondrial loading control. (C) Localization of mitochondrial topoisomerases in floatation gradients of mitochondrial extracts from HEK293T cells. The pellet fraction after digitonin treatment contains inner membrane-associated proteins and nucleoids, while the supernatant fraction contains soluble nucleoids and proteins without membrane-association. Top2α and Top2b co-localize well with the membrane-attached, replicating mtDNA nucleoids characterized by the presence of the mitochondrial helicase TWNK. Also the mitochondrial topoisomerases Top3a and Top1mt partially co-localize with this fraction, but a larger proportion is found in other fractions. (D) Mitochondrially targeted Top2β co-localizes with replicating nucleoids. HeLa cells transiently transfected with an expression vector for myc-tagged, mitochondrially targeted Top2β were immuno-stained with a monoclonal antibody against the myc-tag (green) and a polyclonal antibody against mtSSB (red) as a marker for mtDNA replication.
To analyse the mitochondrial role of the two Top2 isoforms, we investigated with which pools of mtDNA the enzymes are interacting. While the majority of mtDNA nucleoids is freely located in the mitochondrial matrix, replicating nucleoids are tightly associated with the inner membrane (27). These two mtDNA pools can be separated by gentle digitonin lysis followed by gradient centrifugation. In these gradients, both Top2α and Top2β specifically co-localized with the membrane-associated protein fractions containing the mitochondrial replisome, indicated by the replicative helicase TWNK and mitochondrial single-strand binding protein, mtSSB (Figure 1C). No association with non-replicating mtDNA or the mitochondrial RNA polymerase POLRMT was observed. Also Top3α was predominantly co-localizing with replicating mtDNA, but a considerable proportion of the protein was present in other fractions not containing mtDNA. While Top1mt was mainly membrane-associated, it showed only a partial co-localization with the membrane-attached mtDNA nucleoids. Instead, the majority of the protein was found together with POLRMT, in congruency with its role as a regulator of transcription (19).
We were unable to detect endogenous levels of mitochondrial Top2β in immunofluorescence staining of HeLa cells or to overexpress a tagged version of the wild-type Top2β protein. However, when a recombinant Top2β with a mitochondrial localization signal was expressed in the cells, the protein specifically co-localized with mtSSB, a marker of replicating mtDNA nucleoids (Figure 1D).
Top2 relaxes supercoiled mtDNA
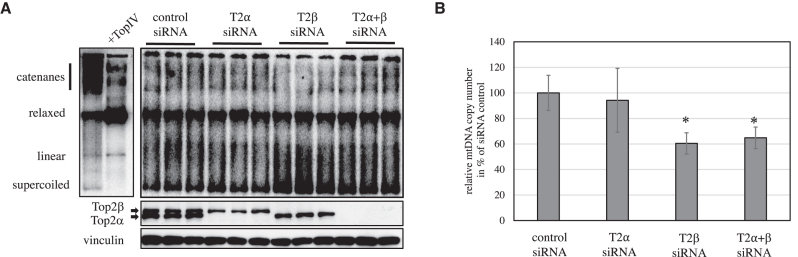
To further address the function of Top2 in mitochondria, we knocked down either Top2α, Top2β or both isoforms simultaneously in HeLa cells. siRNA treatment for 3 days was sufficient to reduce both proteins below detection level (Figure 2). The loss of Top2α did not have any effect on mtDNA topology or cellular mtDNA levels, while the knockdown of Top2β or both isoforms together resulted in noted accumulation of supercoiled mtDNA as well as reduction of cellular mtDNA levels (Figure 2B). Both isoforms of Top2 also relax supercoiled mtDNA in vitro and removed catenanes, while leaving dimeric mtDNA unaltered (Supplementary Figure S2B and C). These dimers represent either unicircular dimeric molecules or two monomeric molecules bound together by a four-way junction (2). An increase in linear mtDNA was observed at high enzyme concentrations.
Figure 2.
Knockdown of Top2β, but not Top2α affects mtDNA topology and replication. (A) mtDNA topology in HeLa cells after 3 days knockdown of Top2α, Top2β or Top2α and β simultaneously. The left panel shows the different topological forms of mtDNA. In vitro treatment with TopIV relaxes supercoiled mtDNA and removes catenanes, while it does not modify mtDNA multimers. The right panel shows the mtDNA topology after a 3 day siRNA knockdown of Top2a and b. Loss of Top2β, but not Top2α leads to accumulation of supercoiled mtDNA. Protein levels of Top2α and β after knockdown were determined by Western blot to confirm the reduction oin Top2 protein levels. (B) mtDNA copy number in HeLa cells after 3 days knockdown of Top2α, Top2β or Top2α and β simultaneously. Loss of Top2β alone or in combination with Top2α knockdown leads to a significant decrease of mtDNA (n = 3, ANOVA/Holm inference, * indicates P < 0.05, ns non-significant).
Ciprofloxacin and doxorubicin inhibit Top2 function in mitochondria
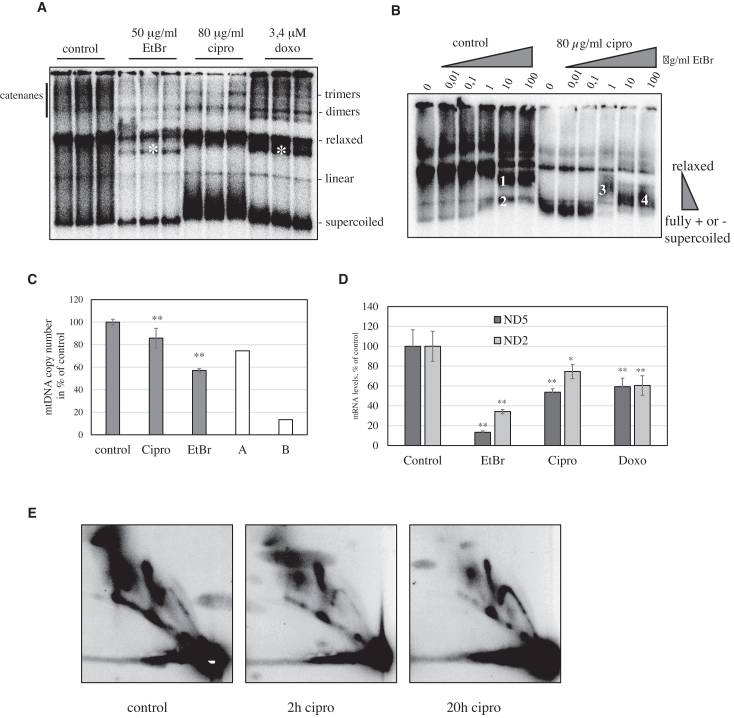
The relaxation activity of Top2 was completely inhibited in vitro by doxorubicin (Supplementary Figure S2B and C), but not etoposide or dexrazoxane. Also ciprofloxacin, an inhibitor of bacterial DNA gyrase and Top IV, blocked supercoil relaxation by Top2, confirming previous indications that the drug might target a broad spectrum of Type II topoisomerases (29). This inhibition was dose-dependent and less efficient than doxorubicin. In contrast to doxorubicin, a known DNA intercalator, ciprofloxacin did not alter the electrophoretic properties of mtDNA (Supplementary Figure S2A), indicating that the drug does not bind to DNA directly.
In agreement with the in vitro assay, also HeLa cells treated with ciprofloxacin or doxorubicin rapidly accumulated supercoiled mtDNA (Figure 3A). While ciprofloxacin also lead to the loss of mtDNA catenanes, doxorubicin-treated cells showed a small increase in multimeric forms without affecting the catenanes. As doxorubicin acts as a topoisomerase poison, stopping the catalytic reaction after strand cleavage (30), the resulting protein-DNA complexes lead to accumulation of mtDNA in the well of the separation gel. This effect was not observed with ciprofloxacin, indicating that the drug might inhibit mitochondrial Top2 in a different fashion or that the protein-DNA complexes are rapidly turned over.
Figure 3.
The topoisomerase inhibitors ciprofloxacin and doxorubicin inhibit Top2 relaxation of negative mtDNA supercoils, RITOLS mtDNA replication and transcription. (A) In vivo effects of ciprofloxacin, doxorubicin and ethidium bromide on mtDNA topology. HeLa cells were treated with 50 ng/ml ethidium bromide, 80 μg/ml ciprofloxacin or 3.4 μM doxorubicin for 24h and the topology of mtDNA compared to untreated control cells. Ciprofloxacin and doxorubicin both cause accumulation of supercoiled mtDNA. The intercalation of doxorubicin and ethidium bromide into intact relaxed circles leads to mild negative supercoiling and increased electrophoretic mobility (marked by *). The topology of nicked circles is not affected by intercalation. (B) Ciprofloxacin induces positive supercoiling in mtDNA. Ethidium bromide intercalation into DNA reduces positive supercoiling and increases negative supercoiling of non-nicked circular DNA. In mtDNA isolated from untreated HeLa cells in vitro incubation with rising amounts of ethidium bromide induces mild negative supercoiling in relaxed, non-nicked mtDNA molecules (1). At the same time fully supercoiled mtDNA migrates slightly slower during the electrophoresis, either due to reduced positive supercoiling or due to alteration in charge (2). In mtDNA isolated from ciprofloxacin-treated cells the abundant supercoiled molecules relax with low ethidium bromide intercalation (3), but supercoiling increases again with higher ethidium bromide concentrations (4), indicating the molecules to be initially positively supercoiled. (C) Effects of topoisomerase inhibitors on mtDNA copy number. HeLa cells were treated for 3 days with ethidium bromide, ciprofloxacin or doxorubicin and mtDNA copy number per nuclear DNA determined by quantitative PCR (n = 3, ANOVA/post-hoc Tukey HSD analysis, **P < 0,01). Doxorubicin cells died within 30 h of treatment and mtDNA levels could thus not be determined. The white bars represent the expected copy number decrease caused by a complete block of replication and dilution by cell division with the observed doubling time (A ciprofloxacin, doubling time 170 h; B ethidium bromide, doubling time 24.9 h). (D) Effects of topoisomerase inhibitors on mtDNA transcription. HeLa cells were treated with 50 ng/ml ethidium bromide, 80 μg/ml ciprofloxacin or 3.4 μM doxorubicin for 24 h and steady-state levels of mitochondrial transcripts were determined by Northern blot and normalized against 18S rRNA. Ciprofloxacin and doxorubicin both affect mitochondrial transcription, but less than ethidium bromide, that suppresses mitochondrial transcription completely at the applied concentration (n = 3, ANOVA/Holm post-hoc analysis, *P < 0.05, **P < 0.01 for treatments versus control). (E). Analysis of mtDNA replication processes. mtDNA of HEK293 cells treated with 80 μg/ml ciprofloxacin for 2 h and 20 h as well as untreated controls was digested with HincII, separated by two-dimensional agarose gel electrophoresis and probed for the 4 kb fragment containing the non-coding region. Already after 2h treatment the level of RITOLS replication intermediates decreased, and after 20 h the remaining replication intermediates represented nearly exclusively the COSCOFA-type replication mode. For detailed description of the various replication forms please see Supplementary Figure S6.
The rapid and dose-dependent accumulation of supercoiled mtDNA after ciprofloxacin treatment (Supplementary Figure S3) was more extreme than the changes caused by knockdown of Top2β. Ethidium bromide did not cause a comparable increase in supercoils, confirming that intercalation alone is not sufficient to alter mtDNA topology in the observed manner. To identify the precise topological conformation of ciprofloxacin treated and control mtDNA we used the intercalation of ethidium bromide, that introduces negative supercoiling on circular DNA (Figure 3B). Initially relaxed closed circles migrated faster due to a mild negative supercoiling caused by ethidium bromide. Supercoiled mtDNA in untreated cells showed a mild reduction in electrophoretic mobility caused by the reduced negative charge due to the intercalation of ethidium cation. In mtDNA from ciprofloxacin-treated cells, the separation of closed and nicked circles was observed, although the levels of both forms were drastically lower than in untreated mtDNA. The abundant supercoiled forms were instead converted to less supercoiled forms in the presence of 1 μg/ml ethidium bromide, indicating that they were originally positively supercoiled. Further incorporation of ethidium bromide at 10 and 100 μg/ml concentrations increased again the electrophoretic mobility due to negative supercoiling.
Inhibition of Top2β impairs mtDNA replication and transcription
The change in topology caused by the inhibition of mitochondrial Top2 was connected with an impairment of mtDNA replication. 7S DNA, the 650bp ssDNA strand incorporated at the D-loop region of mtDNA, was rapidly depleted upon ciprofloxacin, ethidium bromide and doxorubicin treatment (Supplementary Figure S5). While doxorubicin-induced cell death prevented the prolonged observation of mitochondrial DNA maintenance in proliferating cells, ciprofloxacin treatment reduced mtDNA copy number by 18% within 3 days (Figure 3C). As at the same time the growth rate of ciprofloxacin-treated cells was strongly reduced (doubling time 170.2 h versus 22.7 h in untreated controls (Supplementary Figure S4), the observed depletion reflects a nearly complete inhibition of mtDNA synthesis. The observed reduction in cell proliferation was connected to mitochondria as the growth rate of HeLa ρ0 cells lacking mtDNA was altered only after a longer incubation with ciprofloxacin (doubling time 19.9 h in ciprofloxacin-treated cells versus 22.9 h in controls during the first 24 h of treatment). As a comparison, ethidium bromide reduced mtDNA levels to ca. 50% after 3 days with only minor effects on cellular growth rate (24.9 h versus 21.8 h), indicating a milder impairment of mtDNA replication but a stronger dilution of mtDNA in cell divisions. Ciprofloxacin and doxorubicin reduced also mitochondrial transcription, although less efficient than ethidium bromide which reduced steady-state transcript levels to less than 40% within 24 h (Figure 3D).
To identify how ciprofloxacin affects mtDNA replication on molecular level, we analysed the mitochondrial replication intermediates of proliferating cells by 2D agarose gel electrophoresis (2DNAGE). The majority of the mtDNA replication intermediates from untreated HEK293 represents the standard strand-asynchronous replication mechanism (31), forming a typical pattern on 2D-AGE (for explanation see Supplementary Figure S6). Ciprofloxacin caused a strong reduction in these intermediates already after 2 h treatment (Figure 3E). After 20 h, this effect was clearly enhanced, with the strand-asynchronous intermediates being replaced by strand-coupled replication intermediates, a hallmark of mtDNA replication stalling (25,31–33).
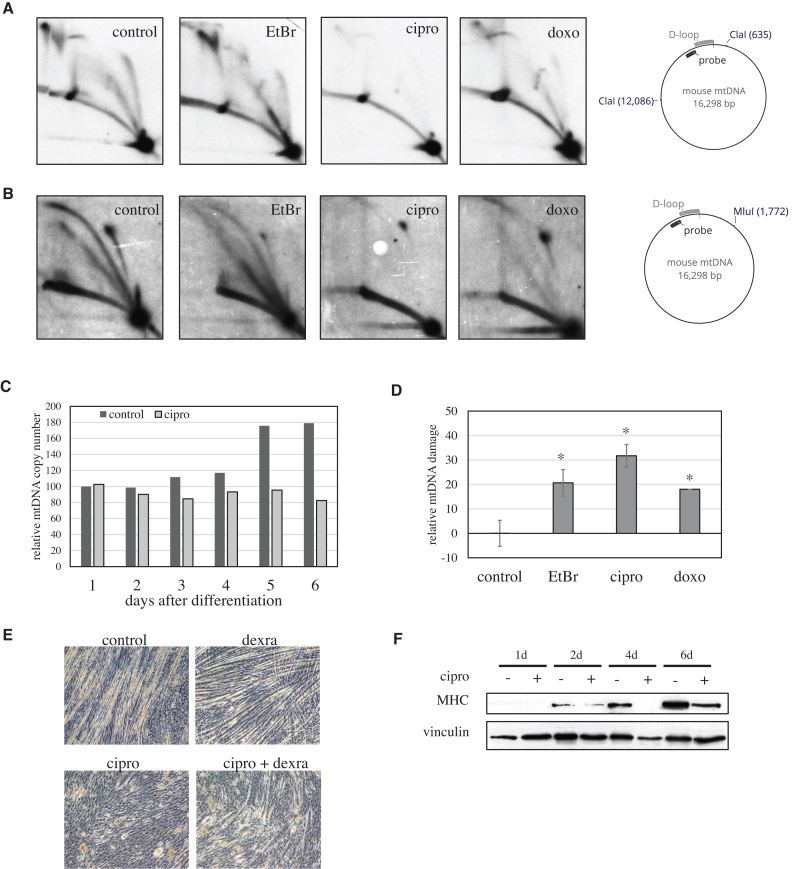
As ciprofloxacin has been described to have cytostatic effects on some cancer cell lines (34,35), we wanted to rule out the possibility that the observed inhibition of mtDNA maintenance is a secondary effect specific for proliferating cells. Thus, we investigated differentiated C2C12 myotubes that express Top2β as the only type II topoisomerase and do not depend on the function of nuclear Top2. Also in this cell type ciprofloxacin inhibited mtDNA replication. However, and in contrast to HEK293 cells, 2D-AGE showed a dramatic reduction of all non-linear mtDNA forms with ciprofloxacin (Figure 4A and B), while ethidium bromide did not have any obvious effects. Interference with progression of the mitochondrial DNA replication fork progression usually leads to replication stalling and accumulation of replication intermediates due to recurrent replication initiation (25,32,33). As ciprofloxacin reduced all replication intermediates, we hypothesized that this effect could be only caused by complete lack of replication initiation. This was confirmed by the strongest effect being observed on bubble-shaped replication intermediates arising during the early stages of replication close to the origin of heavy-strand replication (OH) (down to 18% of control), while the y-shaped intermediates arising from fork progression into the observed fragment were less reduced (down to 33% of control). Doxorubicin did not induce a rapid cell death in myotubes, enabling us to compare its effects with ciprofloxacin treatment more thoroughly. Interestingly, doxorubicin had a similar, but milder inhibitory effect on mtDNA replication than ciprofloxacin (Figure 4A and B).
Figure 4.
Ciprofloxacin disturbs mtDNA replication and mitochondrial physiology also in differentiated cells. (A) Replication patterns in the non-coding region of mtDNA in differentiated C2C12 myotubes after 24 h treatment with with 50 ng/ml ethidium bromide, 80 μg/ml ciprofloxacin or 3.4 μM doxorubicin. MtDNA digested with ClaI was analyzed by 2DNAGE using a probe detecting the non-coding region. While ethidium bromide and doxorubicin did not alter the pattern of replication intermediates, ciprofloxacin lead to the nearly complete loss of replicating molecules. (B) Overview over the replication processes of the whole mtDNA molecule using single-cut 2DNAGE and MluI restriction. Ethidium bromide reduces the abundance of mtDNA replication intermediates mildly, while doxorubicin has a stronger effect. Ciprofloxacin inhibits replication nearly completely. For detailed description of the various replication forms please see Supplementary Figure S5. (C) Ciprofloxacin abolishes the increase of mtDNA levels observed at later stages on C2C12 myotube maturation. (D) Treatment of already differentiated C2C12 cells with ethidium bromide, ciprofloxacin or doxorubicin for 3 days impaired the integrity of mtDNA damage as measured by long-range PCR (n = 3, Anova/Holm inference P < 0.05 for all treatments versus control). (E) In differentiating C2C12 myoblasts ciprofloxacin treatment impairs the fusion of myoblasts to multinuclear myotubes. Cirpofloxacin does also inhibit myotube formation in the presence of dexrazoxane, that itself does not inhibit differentiation. Light-field microscopy images with 100× magnification. (F) Ciprofloxacin also reduces the expression of myosin-heavy chain (MHC), a contractile protein expressed in differentiated muscle fibers, in differentiating C2C12 cells. Western blot of total cellular protein, Vinculin is used a s a loading control.
Ciprofloxacin impairs cellular physiology
The disturbance of mtDNA replication was also visible through the levels of mtDNA. Although post-mitotic C2C12 cells do not dilute mtDNA by cell division, the mtDNA levels rise during later stages of myotube maturation. This increase was completely abolished by ciprofloxacin (Figure 4C). While affecting mtDNA maintenance through different mechanisms, all three drugs had a negative effect on mtDNA integrity (Figure 4D). The impairment of mtDNA maintenance by ciprofloxacin not only disturbed cellular proliferation and the physiological increase of mtDNA copy number during muscle maturation, it also effectively impaired the fusion of confluent myoblasts to multinuclear myotubes (Figure 4E) and cell differentiation as indicated by the reduced expression of the heavy chain of Myosin II, a marker of differentiated skeletal muscle (Figure 4F). This effect is mediated by mitochondria, as simultaneous or separate inhibition of nuclear Top2 by dexrazoxane did not influence myotube formation (Figure 4E).
DISCUSSION
In the presented study, we confirmed previous findings reporting two isoforms of Topoisomerase 2, Top2α and β, in mitochondria (Figure 1A and B) (18). These isoforms are highly homologous and catalyse identical enzymatic reactions but have specialized roles in the maintenance of the nuclear genome. Top2α and β co-localized exclusively with the mitochondrial replication machinery in gradient fractionation of mitochondria (Figure 1C), suggesting a function in mtDNA maintenance in analogy to their nuclear role. Interestingly, none of the isoforms has an identified mitochondrial targeting signal and the bulk of both proteins is localized to the nucleus. This is typical for also other DNA maintenance factors present in both compartments. For example, the archaic primase–polymerase PrimPol does not have mitochondrial targeting signal and although it has a clear functional role in mtDNA maintenance (32), it is still unknown when and how the protein is targeted to mitochondria. Analysis of Top2β localization is complicated by the fact that the protein is prone to degradation in preparations and its high levels in the nucleus make the mitochondrially localized protein elusive in immunocytochemistry. However, a recombinant, mitochondrially targeted version specifically co-localizes only with replicating mitochondrial nucleoids (Figure 1D), confirming the results of the density gradient analysis.
While Top2α is essential for nuclear DNA maintenance, its knockdown did not have any effect on mtDNA topology or copy number, suggesting that it is not essential for mtDNA maintenance. Knockdown of Top2β in contrast had an immediate effect on mtDNA supercoiling (Figure 2), indicating that this isoform is more important for mitochondrial DNA maintenance. This also explains how mtDNA topology is regulated in post-mitotic cells not expressing Top2α. Curiously, Top2β is not essential for mtDNA maintenance under all circumstances, as several viable knockout cell lines and mouse models exist (14,36,37), suggesting that cells are able to adapt to the absence of Top2β, e.g. by recruitment of mitochondrial Top2α. Alternatively, Top1mt alone could control mtDNA supercoiling, although our data suggests that under normal conditions the two topoisomerases have a division of labour: Top1mt removes negative supercoils and Top2β positive supercoils (Figure 3B).
For successful mtDNA replication and segregation also the maintenance of catenane homeostasis is expected to be important. Catenane generation as well as resolution requires a double-strand cleavage reaction carried out by type II topoisomerases. Recently, a type I topoisomerase, Top3α was proposed to fulfill this function (22), which is possible if mtDNA forms exclusively hemicatenanes. This is unlikely as hemicatenane resolution can equally occur towards the inside of the DNA ring, generating a genuine catenane. In fact, a large fraction of high molecular weight mtDNA species are sensitive the type II decatenase TopIV (Figure 2A) (2). The same molecular forms were also modified or lost by Top2 knockdown and inhibition by ciprofloxacin (Figure 2A and Supplementary Figure S3A).
Ciprofloxacin caused a dramatic effect on mtDNA topology, blocking replication initiation, reducing copy number and inhibiting mitochondrial transcription (Figures 2B, 3A–E and 4A). Ciprofloxacin, the third most commonly used antibacterial antibiotic, stops the cleavage/re-ligation reaction of type II topoisomerases midway, generating double-strand breaks, persistent protein–DNA adducts and reduces also the overall enzyme activity (30). Its toxicity to mitochondria has been reported in various studies, suggesting a broad range of mechanisms including topoisomerase inhibition, oxidative stress, altered calcium handling and photosensitization (38–40). In our study, we observed ciprofloxacin to clearly reduce Top2 topoisomerase activity both in vitro and in vivo, but did not find any indication of increased mtDNA double-strand breaks (Figure 3A–C). However, ciprofloxacin did impair the overall mtDNA integrity in post-mitotic cells (Figure 4D). As our detection method (long-range PCR) does not distinguish between strand-breaks, abasic sites or base alterations inhibiting Taq polymerase, the observed effect might be caused by oxidative damage, which fluoroquinolones have been reported to induce in a variety of cell types (41,42).
Interestingly, while doxorubicin, another known type II topoisomerase inhibitor, was inhibiting Top2 function stronger in vitro (Supplementary Figure S2b-c), its effects on mtDNA replication in vivo were weaker (Figure 3A), suggesting that ciprofloxacin specifically accumulates in mitochondria or its activity might be enhanced by metabolic alterations in vivo. Doxorubicin induced mtDNA double-strand breaks in HEK293 (Supplementary Figure S3c), but not in HeLa cells (Figure 3C), demonstrating that the effects and efficiency of topoisomerase inhibition can also depend on the cell type. As ethidium bromide did not interfere with mtDNA topology at all in vivo (Figure 3A), it can be concluded that DNA intercalation alone is not sufficient to inhibit Top2. As of note, each drug, ethidium bromide, ciprofloxacin and doxorubicin improved the resolution of the different mtDNA forms on gel electrophoresis, probably due the inhibition of mitochondrial transcription, resulting in the clearance of heterogeneous RNA forms from the preparations.
The increase in supercoiled mtDNA due to the knockdown of Top2β or exposure to ciprofloxacin reveals interesting issues about their structure and potential function. These supercoils migrate slower and are less uniform in appearance than the supercoils present in untreated or ethidium bromide treated cells. Ethidium bromide is known to induce negative supercoiling of DNA due to its intercalation between the two strands of the double helix, causing twisting of the molecule (43). In bacteria supercoiling is an important genomic regulator, where negative supercoils are relaxed by the action of Topoisomerase I, while positive supercoils are relaxed by the type II topoisomerase DNA gyrase (44). Although eukaryotic Top1 and Top1mt as well as Top2 can relax both negative and positive supercoils, mitochondria appear to possess a similar functional specialization as bacteria, as the loss of Top1mt specifically results in accumulation of negatively supercoiled mtDNA (18), while we here show an accumulation of positively supercoiled mtDNA upon inhibition of Top2 (Figure 3B).
In bacterial systems negative supercoiling is required for replication origin recognition (45,46), with the binding of IHF or HU nucleoid proteins activating transcription and consequently replication origin activation. Also mitochondria have a functional equivalent to IHF and HU, the mitochondrial transcription factor A or TFAM, the main structural organizer of mtDNA nucleoids (47). Although not evolutionarily related, TFAM utilizes a similar mechanism of DNA bending and, assumedly, introduces negative supercoils to facilitate mtDNA transcription (48). In contrast to bacteria replication and transcription of mtDNA exclude each other (49), and this might be controlled by the topology of the respective mtDNA molecule and TFAM binding. TFAM is also essential for mtDNA copy number maintenance (50) and its overexpression results in accumulation of supercoiled mtDNA (3). If relaxation is required for mtDNA replication initiation, while negative supercoiling favours transcription (51), this could explain the loss of replication intermediates and 7S DNA in ciprofloxacin treated cells having positive supercoiled mtDNA (Figure 4 and Supplementary Figure S5). Both ciprofloxacin and doxorubicin also impaired mitochondrial transcription (Figure 3D), although less efficiently than ethidium bromide.
Ciprofloxacin did not only inhibit mtDNA synthesis, but also affected cellular growth and differentiation. The retardation of cell division was immediate and only present in cells with functional mitochondria, implying a retrograde signal from mitochondria to nucleus either caused by oxidative stress or impaired mtDNA replication. Similarly, the differentiation of myoblasts to multinucleated muscle fibres was heavily impaired upon inhibition of mitochondrial, but not nuclear Top2 (Figure 4E–F). This differentiation process does not require cellular proliferation, but is dependent on mitochondrial function (52), further supporting a signalling connection between mitochondria and nucleus. Interestingly, the knockout of Top2β has been reported to protect mice against the mitochondrial dysfunction and cardiomyopathy upon doxorubicin treatment, and a regulatory effect on mitochondrial biogenesis was proposed (53). Our data suggests that the negative effect of doxorubicin on cardiomyocytes is caused by mitochondrial Top2β poisoning and that in the absence of the enzyme the cells evade the deleterious consequences of this treatment.
The severe side effects of ciprofloxacin and other fluoroquinolones include tendinopathies such as tendon rupture, joint inflammation, muscle weakness, central and peripheral neuropathies, epilepsy and psychological symptoms such as depression. These symptoms have been proposed to be connected to enhanced oxidative stress (42,54,55), but the molecular mechanism remained unclear. The reduction of mtDNA copy number and mitochondrial transcription caused by the altered topology of mtDNA might result in severe dysregulation of the electron transport chain complexes, as known to occur under ciprofloxacin treatment (56), lead to respiratory chain dysfunction and cause the observed enhanced oxidative stress.
Ciprofloxacin has also been reported to interfere with physiologically significant cell differentiation processes, such as spermatogenesis (57), brain development (41), bone mineralization (58), as well as to induce renal toxicity and heart arrhythmia (59). While the molecular mechanisms of these adverse effects are yet unclear, mitochondria play a central role in all of these physiological processes, making mitochondrial impairment a likely culprit for the disturbed cellular physiology. Ciprofloxacin plasma concentrations in patients after a single oral administration reach normally around 2 μg/ml and after intravenous injection 6 μg/ml (60) and an up to 20-fold accumulation of ciprofloxacin in tissue has been reported (61). We found the inhibition of mtDNA maintenance starting from 40 μg/ml ciprofloxacin, a concentration that might be well exceeded in patients with altered pharmacokinetics due to obesity, age, renal impairment or simultaneous corticosteroid use, who experience the adverse side effects of fluoroquinolones most frequently (54,62). The fact that Top2β exists in brain mitochondria at far higher levels compared to other tissues, suggesting a specific demand for mtDNA topology regulation in neural cells, could indicate that the tissue has a higher sensitivity against inhibition that should be investigated.
As a conclusion, the maintenance of topological homeostasis in mitochondria is required for gene expression and replication of mtDNA as it is for other genomes. Similar to Top1mt, Top2 regulates mtDNA supercoiling, and their division of labor could be linked to differential association of these proteins between transcriptionally active versus replicating nucleoids. However, their functional redundancy might explain why neither Top1mt nor Top2β is essential for mtDNA maintenance. Although central in bacterial genome maintenance, the whole phenomena of DNA supercoiling and its functional implications are virtually unstudied in mitochondria and calls for future research. Our results identified mtDNA as a key target for ciprofloxacin-induced adverse effects through inhibition of Top2. As fluoroquinolone antibiotics are widely used and effective drugs against a number of important bacterial pathogens, their dosage, systemic enrichment and side-effects should be reviewed in the mitochondrial context, and their clinical use should be considered with great care.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Stefan Sobek and Prof. Fritz Boege for the donation of a mitochondrially targeted Top2β plasmid, Top2β –/– MEFs and helpful discussions as well as Kamila Puławska for her industrious measurement of growth curves.
Author Contributions: All experiments were conducted by A.H. and S.G. with the exception of mitochondrial topoisomerase expression analysis in mouse tissues (N.K.), mtDNA damage in proliferating cells (M.K.) and floatation gradient analysis (K.A. and J.G.). A.H., J.P. and S.G. planned the project and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academy of Finland [292822, 265314 to S.G.]; Finnish Cultural Foundation, North Karelia Regional fund [55172179 to A.H.]; Jane & Aatos Erkko Foundation [to J.P]; The Estonian Research Council [PUT610 to J.M.G]. Funding for open access charge: Jane ja Aaatos Erkon Säätiö.
Conflict of interest statement. None declared.
REFERENCES
- 1. Pohjoismaki J.L., Goffart S.. Of circles, forks and humanity: Topological organisation and replication of mammalian mitochondrial DNA. BioEssays. 2011; 33:290–299. [DOI] [PubMed] [Google Scholar]
- 2. Pohjoismaki J.L., Goffart S., Tyynismaa H., Willcox S., Ide T., Kang D., Suomalainen A., Karhunen P.J., Griffith J.D., Holt I.J. et al. . Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J. Biol. Chem. 2009; 284:21446–21457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pohjoismaki J.L., Wanrooij S., Hyvarinen A.K., Goffart S., Holt I.J., Spelbrink J.N., Jacobs H.T.. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 2006; 34:5815–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kolesar J.E., Wang C.Y., Taguchi Y.V., Chou S.H., Kaufman B.A.. Two-dimensional intact mitochondrial DNA agarose electrophoresis reveals the structural complexity of the mammalian mitochondrial genome. Nucleic Acids Res. 2013; 41:e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wei J., Czapla L., Grosner M.A., Swigon D., Olson W.K.. DNA topology confers sequence specificity to nonspecific architectural proteins. PNAS. 2014; 111:16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zollo O., Sondheimer N.. Topological requirements of the mitochondrial heavy-strand promoters. Transcription. 2017; 8:307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sobek S., Boege F.. DNA topoisomerases in mtDNA maintenance and ageing. Exp. Gerontol. 2014; 56:135–141. [DOI] [PubMed] [Google Scholar]
- 8. Bush N.G., Evans-Roberts K., Maxwell A.. DNA topoisomerases. EcoSal Plus. 2015; 6:doi:10.1128/ecosalplus.ESP-0010-2014. [DOI] [PubMed] [Google Scholar]
- 9. Turley H., Comley M., Houlbrook S., Nozaki N., Kikuchi A., Hickson I.D., Gatter K., Harris A.L.. The distribution and expression of the two isoforms of DNA topoisomerase II in normal and neoplastic human tissues. Br. J. Cancer. 1997; 75:1340–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gonzalez R.E., Lim C.U., Cole K., Bianchini C.H., Schools G.P., Davis B.E., Wada I., Roninson I.B., Broude E.V.. Effects of conditional depletion of topoisomerase II on cell cycle progression in mammalian cells. Cell Cycle. 2011; 10:3505–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Linka R.M., Porter A.C., Volkov A., Mielke C., Boege F., Christensen M.O.. C-terminal regions of topoisomerase IIalpha and IIbeta determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 2007; 35:3810–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thakurela S., Garding A., Jung J., Schubeler D., Burger L., Tiwari V.K.. Gene regulation and priming by topoisomerase IIalpha in embryonic stem cells. Nat. Commun. 2013; 4:2478. [DOI] [PubMed] [Google Scholar]
- 13. Haffner M.C., Aryee M.J., Toubaji A., Esopi D.M., Albadine R., Gurel B., Isaacs W.B., Bova G.S., Liu W., Xu J. et al. . Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat. Genet. 2010; 42:668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tiwari V.K., Burger L., Nikoletopoulou V., Deogracias R., Thakurela S., Wirbelauer C., Kaut J., Terranova R., Hoerner L., Mielke C. et al. . Target genes of Topoisomerase IIbeta regulate neuronal survival and are defined by their chromatin state. PNAS. 2012; 109:E934–E943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dorman C.J., Dorman M.J.. DNA supercoiling is a fundamental regulatory principle in the control of bacterial gene expression. Biophys. Rev. 2016; 8:209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pommier Y., Sun Y., Huang S.N., Nitiss J.L.. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016; 17:703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dalla Rosa I., Goffart S., Wurm M., Wiek C., Essmann F., Sobek S., Schroeder P., Zhang H., Krutmann J., Hanenberg H. et al. . Adaptation of topoisomerase I paralogs to nuclear and mitochondrial DNA. Nucleic Acids Res. 2009; 37:6414–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang H., Zhang Y.W., Yasukawa T., Dalla Rosa I., Khiati S., Pommier Y.. Increased negative supercoiling of mtDNA in TOP1mt knockout mice and presence of topoisomerases IIalpha and IIbeta in vertebrate mitochondria. Nucleic Acids Res. 2014; 42:7259–7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sobek S., Dalla Rosa I., Pommier Y., Bornholz B., Kalfalah F., Zhang H., Wiesner R.J., von Kleist-Retzow J.C., Hillebrand F., Schaal H. et al. . Negative regulation of mitochondrial transcription by mitochondrial topoisomerase I. Nucleic Acids Res. 2013; 41:9848–9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y., Lyu Y.L., Wang J.C.. Dual localization of human DNA topoisomerase IIIalpha to mitochondria and nucleus. PNAS. 2002; 99:12114–12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tsai H.Z., Lin R.K., Hsieh T.S.. Drosophila mitochondrial topoisomerase III alpha affects the aging process via maintenance of mitochondrial function and genome integrity. J. Biomed. Sci. 2016; 23:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nicholls T.J., Nadalutti C.A., Motori E., Sommerville E.W., Gorman G.S., Basu S., Hoberg E., Turnbull D.M., Chinnery P.F., Larsson N.G. et al. . Topoisomerase 3alpha is required for decatenation and segregation of human mtDNA. Mol. Cell. 2018; 69:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morham S.G., Kluckman K.D., Voulomanos N., Smithies O.. Targeted disruption of the mouse topoisomerase I gene by camptothecin selection. Mol. Cell. Biol. 1996; 16:6804–6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Douarre C., Sourbier C., Dalla Rosa I., Brata Das B., Redon C.E., Zhang H., Neckers L., Pommier Y.. Mitochondrial topoisomerase I is critical for mitochondrial integrity and cellular energy metabolism. PLoS One. 2012; 7:e41094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goffart S., Cooper H.M., Tyynismaa H., Wanrooij S., Suomalainen A., Spelbrink J.N.. Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum. Mol. Genet. 2009; 18:328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Herbers E., Kekalainen N.J., Hangas A., Pohjoismaki J.L., Goffart S.. Tissue specific differences in mitochondrial DNA maintenance and expression. Mitochondrion. 2018; doi:10.1016/j.mito.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 27. Rajala N., Gerhold J.M., Martinsson P., Klymov A., Spelbrink J.N.. Replication factors transiently associate with mtDNA at the mitochondrial inner membrane to facilitate replication. Nucleic Acids Res. 2014; 42:952–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hakonen A.H., Goffart S., Marjavaara S., Paetau A., Cooper H., Mattila K., Lampinen M., Sajantila A., Lonnqvist T., Spelbrink J.N. et al. . Infantile-onset spinocerebellar ataxia and mitochondrial recessive ataxia syndrome are associated with neuronal complex I defect and mtDNA depletion. Hum. Mol. Genet. 2008; 17:3822–3835. [DOI] [PubMed] [Google Scholar]
- 29. Jadhav A.K., Karuppayil S.M.. Molecular docking studies on thirteen fluoroquinolines with human topoisomerase II a and b. In Silico Pharmacol. 2016; 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anderson V.E., Gootz T.D., Osheroff N.. Topoisomerase IV catalysis and the mechanism of quinolone action. J. Biol. Chem. 1998; 273:17879–17885. [DOI] [PubMed] [Google Scholar]
- 31. Torregrosa-Munumer R., Goffart S., Haikonen J.A., Pohjoismaki J.L.. Low doses of ultraviolet radiation and oxidative damage induce dramatic accumulation of mitochondrial DNA replication intermediates, fork regression, and replication initiation shift. Mol. Biol. Cell. 2015; 26:4197–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Torregrosa-Munumer R., Forslund J.M.E., Goffart S., Pfeiffer A., Stojkovic G., Carvalho G., Al-Furoukh N., Blanco L., Wanrooij S., Pohjoismaki J.L.O.. PrimPol is required for replication reinitiation after mtDNA damage. PNAS. 2017; 114:11398–11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wanrooij S., Goffart S., Pohjoismaki J.L., Yasukawa T., Spelbrink J.N.. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007; 35:3238–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Herold C., Ocker M., Ganslmayer M., Gerauer H., Hahn E.G., Schuppan D.. Ciprofloxacin induces apoptosis and inhibits proliferation of human colorectal carcinoma cells. Br. J. Cancer. 2002; 86:443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kamat A.M., Lamm D.L.. Antitumor activity of common antibiotics against superficial bladder cancer. Urology. 2004; 63:457–460. [DOI] [PubMed] [Google Scholar]
- 36. Li Y., Hao H., Tzatzalos E., Lin R.K., Doh S., Liu L.F., Lyu Y.L., Cai L.. Topoisomerase IIbeta is required for proper retinal development and survival of postmitotic cells. Biol. Open. 2014; 3:172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maillet A., Tan K., Chai X., Sadananda S.N., Mehta A., Ooi J., Hayden M.R., Pouladi M.A., Ghosh S., Shim W. et al. . Modeling doxorubicin-induced cardiotoxicity in human pluripotent stem cell derived-cardiomyocytes. Sci. Rep. 2016; 6:25333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lowes D.A., Wallace C., Murphy M.P., Webster N.R., Galley H.F.. The mitochondria targeted antioxidant MitoQ protects against fluoroquinolone-induced oxidative stress and mitochondrial membrane damage in human Achilles tendon cells. Free Radic. Res. 2009; 43:323–328. [DOI] [PubMed] [Google Scholar]
- 39. Koziel R., Zablocki K., Duszynski J.. Calcium signals are affected by ciprofloxacin as a consequence of reduction of mitochondrial DNA content in Jurkat cells. Antimicrob. Agents Chemother. 2006; 50:1664–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ouedraogo G., Morliere P., Santus R., Miranda, Castell J.V.. Damage to mitochondria of cultured human skin fibroblasts photosensitized by fluoroquinolones. J. Photochem. Photobiol. B, Biol. 2000; 58:20–25. [DOI] [PubMed] [Google Scholar]
- 41. Dogan Z., Cetin A., Elibol E., Vardi N., Turkoz Y.. Effects of ciprofloxacin and quercetin on fetal brain development: a biochemical and histopathological study. J.Maternal-Fetal Neonatal Med. 2018; 10:1–9. [DOI] [PubMed] [Google Scholar]
- 42. Pouzaud F., Dutot M., Martin C., Debray M., Warnet J.M., Rat P.. Age-dependent effects on redox status, oxidative stress, mitochondrial activity and toxicity induced by fluoroquinolones on primary cultures of rabbit tendon cells. Comp Biochem. Phys. C. 2006; 143:232–241. [DOI] [PubMed] [Google Scholar]
- 43. Olavarrieta L., Martinez-Robles M.L., Sogo J.M., Stasiak A., Hernandez P., Krimer D.B., Schvartzman J.B.. Supercoiling, knotting and replication fork reversal in partially replicated plasmids. Nucleic Acids Res. 2002; 30:656–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu H.Y., Shyy S.H., Wang J.C., Liu L.F.. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988; 53:433–440. [DOI] [PubMed] [Google Scholar]
- 45. Lal A., Dhar A., Trostel A., Kouzine F., Seshasayee A.S., Adhya S.. Genome scale patterns of supercoiling in a bacterial chromosome. Nat. Commun. 2016; 7:11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Le Chatelier E., Janniere L., Ehrlich S.D., Canceill D.. The RepE initiator is a double-stranded and single-stranded DNA-binding protein that forms an atypical open complex at the onset of replication of plasmid pAMbeta 1 from Gram-positive bacteria. J. Biol. Chem. 2001; 276:10234–10246. [DOI] [PubMed] [Google Scholar]
- 47. Ngo H.B., Kaiser J.T., Chan D.C.. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat. Struct. Mol. Biol. 2011; 18:1290–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shi Y., Dierckx A., Wanrooij P.H., Wanrooij S., Larsson N.G., Wilhelmsson L.M., Falkenberg M., Gustafsson C.M.. Mammalian transcription factor A is a core component of the mitochondrial transcription machinery. PNAS. 2012; 109:16510–16515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Agaronyan K., Morozov Y.I., Anikin M., Temiakov D.. Mitochondrial biology. Replication-transcription switch in human mitochondria. Science. 2015; 347:548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Larsson N.G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G.S., Clayton D.A.. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998; 18:231–236. [DOI] [PubMed] [Google Scholar]
- 51. Fukuoh A., Ohgaki K., Hatae H., Kuraoka I., Aoki Y., Uchiumi T., Jacobs H.T., Kang D.. DNA conformation-dependent activities of human mitochondrial RNA polymerase. Genes Cells. 2009; 14:1029–1042. [DOI] [PubMed] [Google Scholar]
- 52. Wagatsuma A., Sakuma K.. Mitochondria as a potential regulator of myogenesis. Sci. World J. 2013; 2013:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang S., Liu X., Bawa-Khalfe T., Lu L.S., Lyu Y.L., Liu L.F., Yeh E.T.. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat. Med. 2012; 18:1639–1642. [DOI] [PubMed] [Google Scholar]
- 54. Stahlmann R., Lode H.. Safety Considerations of Fluoroquinolones in the Elderly An Update. Drug Aging. 2010; 27:193–209. [DOI] [PubMed] [Google Scholar]
- 55. Ilgin S., Can O.D., Atli O., Ucel U.I., Sener E., Guven I.. Ciprofloxacin-induced neurotoxicity: evaluation of possible underlying mechanisms. Toxicol. Mech. Methods. 2015; 25:374–381. [DOI] [PubMed] [Google Scholar]
- 56. Nadanaciva S., Dillman K., Gebhard D.F., Shrikhande A., Will Y.. High-content screening for compounds that affect mtDNA-encoded protein levels in eukaryotic cells. J. Biomol. Screen. 2010; 15:937–948. [DOI] [PubMed] [Google Scholar]
- 57. Khaki A., Heidari M., Novin M.G., Khaki A.A.. Adverse effects of ciprofloxacin on testis apoptosis and sperm parameters in rats. Iran. J. Reprod. Med. 2008; 6:71–76. [Google Scholar]
- 58. Holtom P.D., Pavkovic S.A., Bravos P.D., Patzakis M.J., Shepherd L.E., Frenkel B.. Inhibitory effects of the quinolone antibiotics trovafloxacin, ciprofloxacin, and levofloxacin on osteoblastic cells in vitro. J. Orthopaed. Res. 2000; 18:721–727. [DOI] [PubMed] [Google Scholar]
- 59. Liu X., Ma J.Y., Huang L., Zhu W.G., Yuan P., Wan R., Hong K.. Fluoroquinolones increase the risk of serious arrhythmias: A systematic review and meta-analysis. Medicine (Baltimore). 2017; 96:e8273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lipman J., Scribante J., Gous A.G., Hon H., Tshukutsoane S.. Pharmacokinetic profiles of high-dose intravenous ciprofloxacin in severe sepsis. The Baragwanath Ciprofloxacin Study Group. Antimicrob. Agents Chemother. 1998; 42:2235–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fabre D., Bressolle F., Gomeni R., Arich C., Lemesle F., Beziau H., Galtier M.. Steady-state pharmacokinetics of ciprofloxacin in plasma from patients with nosocomial pneumonia: penetration of the bronchial mucosa. Antimicrob. Agents Chemother. 1991; 35:2521–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hollenstein U.M., Brunner M., Schmid R., Muller M.. Soft tissue concentrations of ciprofloxacin in obese and lean subjects following weight-adjusted dosing. Int. J. Obesity. 2001; 25:354–358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.