Abstract
Anabolism and catabolism are tightly regulated according to the cellular energy supply. Upon energy stress, ribosomal RNA (rRNA) biogenesis is inhibited, and autophagy is induced. However, the mechanism linking rRNA biogenesis and autophagy is unclear. Here, we demonstrate that the nucleolar protein NAT10 plays a role in the transition between rRNA biogenesis and autophagy. Under normal conditions, NAT10 is acetylated to activate rRNA biogenesis and inhibit autophagy induction. Mechanistic studies demonstrate that NAT10 binds to and acetylates the autophagy regulator Che-1 at K228 to suppress the Che-1-mediated transcriptional activation of downstream genes Redd1 and Deptor under adequate energy supply conditions. Upon energy stress, NAT10 is deacetylated by Sirt1, leading to suppression of NAT10-activated rRNA biogenesis. In addition, deacetylation of NAT10 abolishes the NAT10-mediated transcriptional repression of Che-1, leading to the release of autophagy inhibition. Collectively, we demonstrate that the acetylation status of NAT10 is important for the anabolism-catabolism transition in response to energy stress, providing a novel mechanism by which nucleolar proteins control rRNA synthesis and autophagy in response to the cellular energy supply.
INTRODUCTION
Under adequate nutrient and growth factor supply conditions, mammalian target of rapamycin (mTOR) is activated to promote cell growth by activating cell anabolism processes, which mainly include rRNA biogenesis and protein synthesis (1). When cells encounter energy stresses, such as deprivation of nutrients and/or growth factors, mTOR activity is inhibited, and autophagy is induced to maintain metabolic homeostasis and cell viability (2,3). Autophagy begins with the formation of autophagosomes, double-membrane structures that capture cargo from the cytoplasm and organelles. Autophagosomes then fuse with lysosomes to form autophagolysosomes, wherein cargo are degraded to generate the amino acids and nutrients necessary to provide cellular energy and support cell survival (4).
Redd1 and Deptor are inhibitors of mTOR signaling (5–7) and recent studies have shown that the transcription factor Che-1 (also known as AATF) induces autophagy by activating the transcription of Redd1 and Deptor upon energy stress (8). Che-1 plays a pivotal role in cell survival by promoting cell cycle progression, repressing apoptosis and activating autophagy (9), and the localization, stability and activity of Che-1 are tightly regulated by post-translational modifications under cellular stress (10–12). For instance, upon DNA damage, phosphorylation of Che-1 by ATM facilitates the binding of Che-1 to the promoters of genes involved in checkpoint activation, such as TP53 and p21, to promote their transcription, thus leading to cell cycle arrest and DNA repair (11,13). However, how Che-1 activity is regulated by post-translational modifications upon energy stress remains unclear.
Ribosome biogenesis, which consumes up to 80% of the cellular energy in proliferating cells, is a complex process that includes rRNA biogenesis and the assembly of ribosomal subunits (14,15). As an anabolic process, rRNA biogenesis is tightly controlled by the cellular energy status, which plays a critical role in ribosome biogenesis (16). Ribosomal RNA synthesis is activated to promote cell growth and proliferation under adequate nutrition conditions. Under energy stress, rRNA biogenesis is inhibited for the sake of saving energy (15), while autophagy is induced to produce energy and support cell survival upon energy deprivation. However, the crosstalk between rRNA synthesis and autophagy remains unclear.
The nucleolar protein N-acetyltransferase 10 (NAT10, also named hALP) was initially shown to activate telomerase by activating hTERT (human telomerase reverse transcriptase) (17). It has been found that NAT10 harbors RNA acetylase and lysine acetyltransferase activity. We previously identified NAT10 as a t-UTP that is required for rRNA biogenesis processes, including pre-rRNA transcription and 18S rRNA processing (18). NAT10 activates Pol I (RNA polymerase I) transcription by binding to UBF1 (upstream binding factor) and promoting UBF1 acetylation. Moreover, NAT10 is involved in rRNA processing by binding and acetylating rRNA (19–21). Recently, we found that the autoacetylation of NAT10 at K426 is required for its ability to activate rRNA biogenesis, and NAT10 is deacetylated in cells by Sirtuins (22). However, which member of the Sirtuin family regulates NAT10 acetylation and whether deacetylation of NAT10 regulates its biological function in rRNA biogenesis remain unknown.
NAT10 participates in multiple cellular biological processes via its lysine acetyltransferase activity, and the truncated recombinant NAT10 (amino acids 164–834) acetylates histones in vitro (17). Abolishment of NAT10-mediated tubulin acetylation at K40 by Remodelin ameliorates laminopathies (a group of rare genetic disorders caused by mutations in genes encoding proteins of the nuclear lamina) by correcting the nuclear architecture and reducing cellular senescence in cells and mice (23–25). Furthermore, we recently found that NAT10 acetylates p53 at K120 and contributes to p53 activation upon DNA damage (26,27). Thus, identification of NAT10 downstream substrates will provide evidence for exploring important biological functions of NAT10.
Sirt1, an important activator of autophagy, is activated by metabolic stresses, such as starvation, glucose withdrawal and energy deprivation (28–30). Overexpression of Sirt1 stimulates the formation of autophagosomes and elevates the basal levels of autophagy, while Sirt1 deficiency arrests autophagy in response to nutrient deprivation (31). Sirt1 regulates cell proliferation, DNA damage repair, cell survival and autophagy by deacetylating its downstream substrates, including histones, p53, FoxO1, β-catenin, Ku70, NF-κB, PTEN, ATG5, ATG7 and ATG8 (32–36). Thus, identification of Sirt1 downstream substrates will improve our understanding of the mechanisms underlying autophagy regulation.
In this study, we found that NAT10 is a downstream substrate of Sirt1 and that deacetylation of NAT10 controls the transition from rRNA biogenesis to the release of autophagy suppression in response to energy withdrawal. We have thereby elucidated the mechanism by which NAT10 regulates autophagy.
MATERIALS AND METHODS
Cell culture and transfection
The U2OS, HCT116 p53+/+, HCT116 p53−/−, HeLa, SW480 and HEK293T cell lines were maintained in DMEM supplemented with 10% fetal bovine serum. Cells were transfected with plasmid DNA or siRNA duplexes using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. In transient transfection experiments, plasmid DNA concentrations were maintained at a constant level with an empty vector. The HCT116 NAT10 Ctrl and HCT116 knockout (KO) cell lines were established by CRISPR–Cas9 genome editing technology in our laboratory (26) and maintained in DMEM.
Plasmids and antibodies
Flag- or GFP-tagged NAT10 and NAT10 mutants were cloned into the pCI-neo or pEGFP-C2 vector. Similarly, Flag-tagged Sirt1, Sirt6, Sirt7 and Che-1 were separately cloned into the pCI-neo vector. For in vitro GST pull-down assays, GST-NAT10, GST-Che-1 and GST-Sirt1 were separately cloned into the pGEX-4T1 vector. For protein purification, NAT10 was cloned into the pFast-Bac1 vector. All plasmids cloned with PCR inserts were confirmed by DNA sequencing. Mutant plasmids, including pCI-neo-Flag-NAT10 G641E, pCI-neo-Flag-NAT10 K461R, pCI-neo-Flag-Che-1 K138R, pCI-neo-Flag-Che-1 K145R, pCI-neo-Flag-Che-1 K228R, pCI-neo-Flag-Che-1 3KR, pGEX-4T1-Che-1 K138R, pGEX-4T1-Che-1 K145R, pGEX-4T1-Che-1 K228R, pGEX-4T1-Che-1 3KR and HA-Sirt1 H363Y, were obtained by mutagenesis using the QuickChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's protocol. The presence of mutations in the constructed plasmids was confirmed by DNA sequencing.
Anti-Sirt1 (B-10), anti-Che-1 (sc-81225), anti-p53 (DO-1), anti-actin (C-11) were purchased from Santa Cruz Biotechnology. Anti-LC3B (L7543), anti-Flag (F3165), anti-Flag (F1804) and anti-HA (H6908) were purchased from Sigma. Anti-acetylated lysine (clone 4G12) was purchased from Millipore. Anti-acetylated lysine (9441S) was purchased from Cell Signaling Technology. Anti-p62 (PM045) was purchased from Medical & Biological Laboratories International Corporation. Anti-Deptor (bs-8255R) was purchased from Beijing Biosynthesis Biotechnology. Anti-Redd1 (10638-1-AP) was purchased from Proteintech. Anti-NAT10 was a gift from Dr B. Zhang.
Coimmunoprecipitation assay
Cell lysates were prepared in Buffer A (25 mM Tris–Cl (pH 7.5), 150 mM KCl, 1 mM DTT, 2 mM EDTA, 0.5 mM PMSF and 0.2% Nonidet P-40) and used directly for immunoprecipitation. Antibodies were coupled with a 50% suspension of protein A-Sepharose beads (GE Healthcare) in IPP500 (500 mM NaCl, 10 mM Tris–Cl (pH 8.0), 0.2% Nonidet P-40), and the coupled beads were incubated with cell lysates for 2 h at 4°C. After washing, the precipitants were analyzed by Western blot using the indicated antibodies.
RT-qPCR analysis
Total RNA was extracted from cells using TRIzol reagent (Invitrogen), purified using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions, and electrophoresed on a denaturing agarose gel to examine the RNA integrity. Then, cDNA was synthesized from 2 μg of total RNA using the Superscript First-Strand Synthesis System (Invitrogen), and qPCR was performed as previously described (37) according to Minimal Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines. The sequences of the qPCR primers are shown in Supplementary Table S1.
In vitro acetylation and deacetylation assays
For in vitro acetylation, bead-bound His-NAT10 was incubated in buffer containing 50 mM Tris–Cl (pH 7.9), 10% glycerol, 0.1 mM EDTA, 1 mM PMSF, 10 mM sodium butyrate and 10 μM acetyl-CoA at 30°C for 1 h. After washing, bead-bound NAT10 was used as substrate for in vitro deacetylation assays or analyzed by immunoblot with an anti-acetyl-lysine antibody. For in vitro deacetylation, 0.25–1.0 μg of pre-acetylated His-NAT10 was incubated with 0.5 μg of Flag-tagged Sirt1 in the presence or absence of 1 mM NAD+ at 30°C for 1 h. Acetylation of NAT10 was monitored by immunoblot using an anti-acetyl-lysine antibody.
In-cell acetylation assay
The in-cell acetylation assay was performed as previously described (38,39). Briefly, cells were incubated with 1 μm trichostatin A (TSA) and 5 mM nicotinamide (NIA) for 6 h and then harvested. Cell extracts were prepared in whole cell lysis buffer (50 mM Tris–HCl (pH 7.8), 137 mM NaCl, 1 mM NaF, 1 mM NaVO3, 1% Triton X-100, 0.2% sarkosyl, 1 mM DTT and 10% glycerol) containing fresh protease inhibitors, 10 μm TSA and 5 mM NIA and then incubated with anti-acetyl lysine antibody-conjugated protein A-Sepharose beads at 4°C for 2 h. The beads were washed five times with BC100 buffer (50 mM Tris–HCl (pH 7.8), 100 mM NaCl, 0.2% Triton X-100 and 10% glycerol). The immunoprecipitants and total proteins were subjected to SDS-PAGE and analyzed by Western blot using the indicated antibodies.
Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde for 15 min and permeabilized using 0.2% Triton X-100 for 10 min at room temperature. After blocking with 10% goat serum, the cells were incubated with primary antibodies overnight at 4°C. After washing with PBS, a FITC-conjugated anti-rabbit antibody and a TRITC-conjugated anti-mouse antibody were added, and the samples were incubated for 1 h at room temperature. Finally, the cells were stained with DAPI to visualize the nuclei.
RESULTS
NAT10 is deacetylated by Sirt1 upon energy deprivation
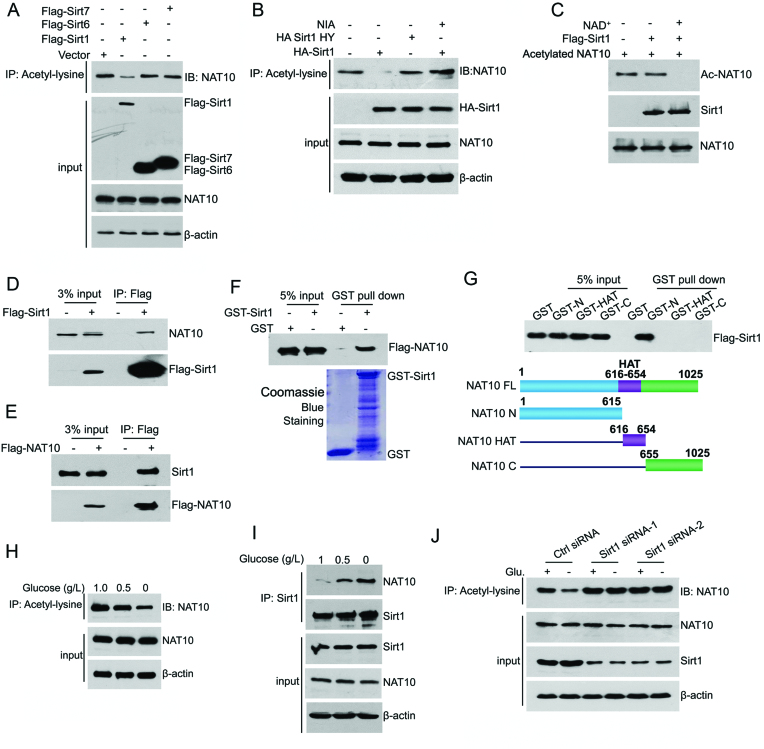
To determine which Sirtuin regulates NAT10 acetylation, the effects of three nuclear Sirtuins, Sirt1, Sirt6 and Sirt7 (40), on NAT10 acetylation were evaluated. As shown in Figure 1A, Sirt1 significantly decreased NAT10 acetylation levels, while neither Sirt6 nor Sirt7 affected NAT10 acetylation. Furthermore, the enzyme-dead Sirt1 mutant H363Y (HY) failed to deacetylate NAT10, indicating that NAT10 is deacetylated by Sirt1 (Figure 1B and Supplementary Figure S1A). In addition, in vitro deacetylation assays performed with purified Flag-Sirt1 and preacetylated His-NAT10 showed that purified Sirt1 prevented the acetylation of preacetylated His-NAT10 in the presence of NAD+ (Figure 1C). Together, these data demonstrated that NAT10 is a substrate of Sirt1.
Figure 1.
NAT10 is deacetylated by Sirt1 upon energy deprivation. (A) HCT116 cells were transfected with the indicated plasmids. Cell lysates were prepared and subjected to immunoprecipitation using an anti-acetyl-lysine antibody. The acetylation levels of NAT10 in the immunoprecipitants were evaluated by western blot using an anti-NAT10 antibody. (B) The indicated plasmids were coexpressed in HCT116 cells. Cell lysates were prepared and subjected to immunoprecipitation using an anti-acetyl-lysine antibody. The acetylation levels of NAT10 were evaluated by Western blot using an anti-NAT10 antibody. (C) Purified His-NAT10 was preacetylated and incubated with purified Flag-Sirt1 in the absence or presence of 1 mM NAD+. The acetylation levels of NAT10 were evaluated by Western blot with an anti-acetyl-lysine antibody. (D) Flag-Sirt1 was transfected into HCT116 cells. Immunoprecipitation was performed with an anti-Flag antibody, and NAT10 expression in the precipitants was determined by Western blot. (E) Flag-NAT10 was transfected into HCT116 cells. Immunoprecipitation was performed with an anti-Flag antibody, and Sirt1 expression in the precipitants was determined by Western blot. (F) A GST pull-down experiment was performed with GST-Sirt1 and purified Flag-NAT10. The binding of GST-Sirt1 to Flag-NAT10 was detected by immunoblotting with an anti-Flag antibody. (G) A GST pull-down experiment was performed using purified GST-NAT10 deletion mutants and purified Flag-Sirt1. The schematic diagram represents the GST-NAT10 deletion mutant constructs (lower panel). (H) HCT116 cells were cultured in medium containing 1.0 g/l, 0.5 g/l, or 0 g/l glucose for 18 h. The cells were harvested, and immunoprecipitation was performed using an anti-acetyl-lysine antibody. The acetylation levels of NAT10 were evaluated by Western blot using an anti-NAT10 antibody. (I) HCT116 cells were cultured in medium containing 1.0 g/l, 0.5 g/l, or 0 g/l glucose for 18 h. Immunoprecipitation was performed with an anti-Sirt1 antibody, and NAT10 expression in the precipitants was evaluated by immunoblotting. (J) HCT116 cells were transfected with the indicated siRNAs. Cells were cultured in glucose-rich or glucose-free medium for 18 h, harvested and then subjected to immunoprecipitation with an anti-acetyl-lysine antibody. The acetylation levels of NAT10 were evaluated by Western blot using an anti-NAT10 antibody on the immunoprecipitants. See also Supplementary Figure S1.
We next determined whether NAT10 interacts with Sirt1. Immunoprecipitation experiments showed that NAT10 interacted with Flag-Sirt1 in cells (Figure 1D and E). To determine whether NAT10 interacts directly with Sirt1, in vitro GST pull-down experiments were conducted. As shown in Figure 1F, purified Flag-NAT10 bound to GST-Sirt1 but not to GST alone, confirming that NAT10 directly interacts with Sirt1. Sirt1 specifically bound to the N-terminal domain of NAT10 (GST-N) instead of the HAT domain (GST-HAT) or the C-terminal domain (GST-C) (Figure 1G).
Under energy deficient conditions, Sirt1 facilitates cell survival by deacetylating its substrates in the nucleus (41,42). We thus explored whether Sirt1 deacetylates NAT10 upon glucose starvation. To this end, the acetylation levels of endogenous NAT10 were evaluated upon removal of glucose from the medium. As shown in Figure 1H, glucose deprivation reduced the acetylation levels of NAT10 without changing its total protein levels. To determine whether endogenous NAT10 binds Sirt1 in cells and how the NAT10-Sirt1 interaction is altered during energy stress, coimmunoprecipitation analysis was performed on the extracts of cells cultured in media containing different concentrations of glucose, revealing that endogenous NAT10 interacted with Sirt1 (Figure 1I). Notably, the interaction between endogenous Sirt1 and NAT10 was enhanced by glucose deprivation (Figure 1I and Supplementary Figure S1B). To determine whether the deacetylation of NAT10 during glucose deprivation was dependent on Sirt1, we silenced Sirt1 by siRNAs and evaluated NAT10 acetylation after removing glucose from the media. When Sirt1 was depleted, the NAT10 acetylation levels no longer changed under glucose deprivation (Figure 1J), indicating that NAT10 is deacetylated by Sirt1 upon glucose deprivation.
Deacetylation of NAT10 by Sirt1 leads to inhibition of NAT10-mediated rRNA biogenesis
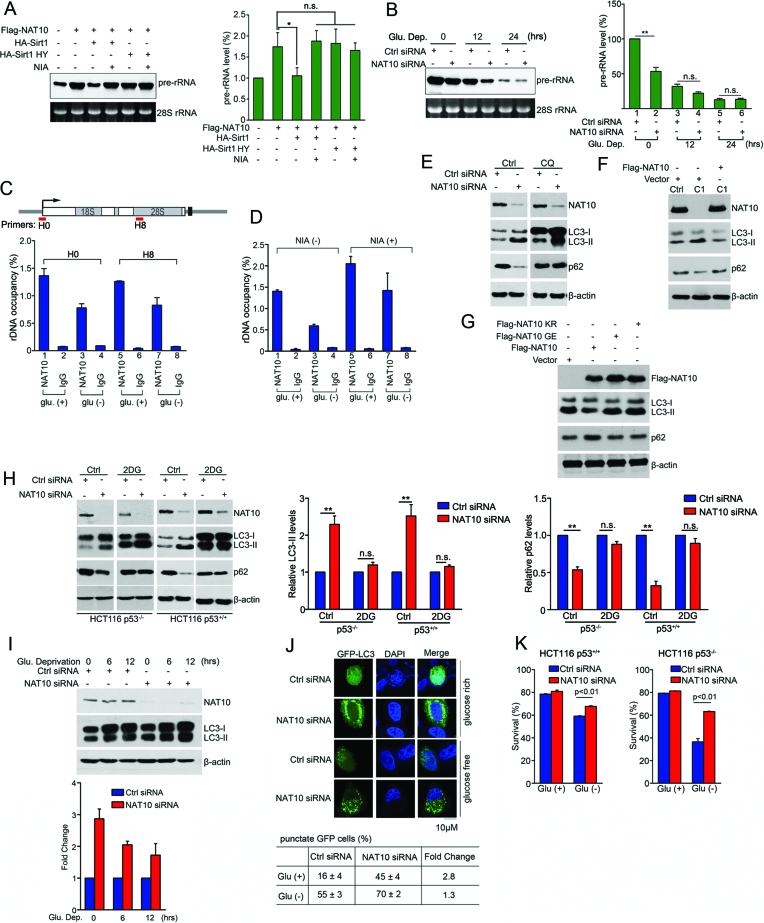
Since NAT10 has been shown to facilitate rRNA biogenesis (18–20), we further investigated whether deacetylation of NAT10 modulates its function in rRNA biogenesis in response to glucose starvation. First, we evaluated pre-rRNA levels by Northern blot and RT-qPCR as previously described (43) when Flag-NAT10 was ectopically expressed with or without ectopic HA-Sirt1. Flag-NAT10-activated pre-rRNA synthesis was inhibited by wild-type HA-Sirt1, and HA-Sirt1-mediated inhibition was blocked by NIA treatment (Figure 2A and Supplementary Figure S2A). However, HA-Sirt1 HY showed no effect on Flag-NAT10-activated pre-rRNA synthesis. These data demonstrated that Sirt1 inhibits NAT10-mediated rRNA biogenesis by targeting NAT10 for deacetylation. Then, we examined whether NAT10 regulates pre-rRNA levels upon the induction of energy stress. In glucose-rich cells, knockdown of NAT10 significantly decreased the pre-rRNA levels, and as expected, glucose deprivation gradually decreased the pre-rRNA levels over time (Figure 2B). However, depletion of NAT10 eventually had no effect on the pre-rRNA levels in HCT116 (Figure 2B and Supplementary Figure S2B), HeLa and U2OS cells (Supplementary Figure S2C and S2D) totally deprived of glucose. These data revealed that NAT10 fails to activate rRNA biogenesis under energy deprivation conditions. In addition, ChIP experiments showed that the interaction of NAT10 with rDNA was inhibited upon glucose deprivation (Figure 2C). Importantly, NIA treatment increased the NAT10 occupancy on rDNA in both glucose-rich and glucose-depleted cells (Figure 2D), indicating that deacetylation of NAT10 inhibits its function in rRNA biogenesis. These data suggested that Sirt1 deacetylates NAT10 to inhibit NAT10-mediated rRNA biogenesis, thus halting the energy-consuming ribosome biogenesis process to save cellular energy under energy stress.
Figure 2.
Deacetylation of NAT10 inhibits rRNA biogenesis and releases autophagy induction after glucose deprivation. (A) HCT116 cells were transfected with the indicated plasmids. After 24 h, pre-rRNA levels were determined by Northern blot (left) and RT-qPCR (right). Error bars indicate the SEM (n = 3). *P < 0.05. n.s., no significance. (one-way ANOVA). (B) HCT116 cells were transfected with the indicated siRNAs and cultured in glucose-free medium for the indicated amounts of time. The cells were harvested, and pre-rRNA levels were determined by Northern blot (left) and RT-qPCR (right). Error bars indicate the SEM (n = 3). *P < 0.05. n.s., no significance. (two-tailed t-test). (C) HEK293T cells were cultured in glucose-rich or glucose-free medium. rDNA occupancy by NAT10 was evaluated by ChIP experiment. Error bars indicate the SEM (n = 3). (D) HEK293T cells cultured in glucose-rich or glucose-free medium were treated with nicotinamide. rDNA occupancy by NAT10 was evaluated by ChIP experiment. Error bars indicate the SEM (n = 3). (E) HCT116 cells were transfected with the indicated siRNAs. After treatment with chloroquine (CQ, 100 μM), the cells were harvested, and Western blotting was performed to evaluate the indicated proteins. (F) HCT116 Ctrl or HCT116 NAT10 KO cells were transfected with the indicated plasmids. Cell lysates were subjected to immunoblot analysis using the indicated antibodies. (G) HCT116 cells were transfected with the indicated plasmids. Cell lysates were subjected to Western blot using the indicated antibodies. (H) HCT116 p53+/+ or HCT116 p53−/− cells were transfected with the indicated siRNAs. The cells were treated with 2-DG and harvested. Western blotting was performed to evaluate the indicated proteins. Densitometry scanning analyses of the LC3 and p62 bands standardized to β-actin are summarized from three independent experiments and shown on the right. Error bars indicate the SEM (n = 3). **P < 0.01. n.s., no significance. (two-tails t-test) (I) HCT116 cells were transfected with the indicated siRNAs and cultured in glucose-free medium for the indicated amounts of time. Western blotting was performed to evaluate the indicated proteins. Densitometry scanning analyses of LC3 bands standardized by β-actin are summarized from three independent experiments and shown in the lower panel. Error bars indicate the SEM (n = 3). (J) HCT116 cells transfected with the indicated siRNAs and GFP-LC3 were cultured in glucose-free medium for 18 h. GFP-LC3 distribution was assessed by fluorescence microscopy. The percentage of punctate GFP cells represents the number of cells with punctate GFP-LC3 among 100 GFP-LC3-expressing cells. Data represent the mean ± SD from three independent experiments performed in triplicate (lower table). Scale bars, 10 μM. (K) HCT116 cells were transfected with the indicated siRNAs and cultured in glucose-free medium for 18 h. The cell viability was measured by FACS analysis after Annexin V and propidium iodide (PI) double-staining. Error bars indicate the SEM (n = 3). P-values were calculated using the two-tailed t-test. See also Supplementary Figure S2.
Depletion of NAT10 facilitates autophagy induction and supports cell survival under energy stress
Given that Sirt1 deacetylates its substrates and activates autophagy under energy stress, we next asked whether NAT10 is involved in autophagy induction. Using accumulation of the autophagosomal marker LC3-II to reflect starvation-induced autophagic activity (44), we evaluated autophagy induction when NAT10 was knocked down, revealing that NAT10 depletion enhanced LC3-II accumulation (Figure 2E, left). In addition, this LC3-II accumulation was boosted by chloroquine (CQ), an inhibitor of lysosomal protein degradation, suggesting that NAT10 knockdown facilitated autophagosome formation (Figure 2E, right). Since the autophagy-specific substrate p62 is transported into autophagosomes and degraded in lysosomes (44), we further evaluated p62 levels. NAT10 depletion decreased the p62 level, and this effect was inhibited by CQ treatment, suggesting that NAT10 inhibits autophagy induction under normal conditions (Figure 2E). The NAT10 depletion-induced increase in LC3-II accumulation and decrease in p62 were also observed in p53-depleted cells (Supplementary Figure S2E). Together, these results suggest that NAT10 inhibits autophagy induction independent of p53. To rule out the off-targeting effect of NAT10 siRNAs, we analyzed the autophagy levels in Cas9-edited HCT116-NAT10 KO cells (22,26). LC3-II accumulation was increased in HCT116-NAT10 KO cells, while this effect was reversed by ectopic Flag-NAT10 expression, confirming that NAT10 inhibits autophagy under normal conditions (Figure 2F). Furthermore, ectopic Flag-NAT10 expression inhibited LC3-II accumulation and increased the p62 level, while the Flag-NAT10 KR or NAT10 GE mutants did not display these functions (Figure 2G), indicating that the autoacetylation and acetyltransferase activities of NAT10 are required for autophagy inhibition.
To further explore the role of NAT10 in glucose starvation-induced autophagy, we treated cells with 2-deoxy-glucose (2-DG) to block glycolysis. Interestingly, the NAT10 depletion-induced LC3-II accumulation and p62 decrease were significantly attenuated by 2-DG treatment in both p53-expressing and p53-null cells (Figure 2H). In addition, the NAT10 depletion-induced LC3-II accumulation was significantly reduced upon glucose withdrawal (Figure 2I), suggesting that inhibition of NAT10 facilitates autophagy induction in response to energy stress; similar results were observed in p53-null cells (Supplementary Figure S2F). Moreover, the observation of GFP-LC3 puncta showed that the NAT10 depletion-mediated autophagy induction decreased from 2.8-fold under normal conditions to 1.3-fold upon glucose deprivation (Figure 2J). These results indicated that NAT10 inhibits autophagy induction under energy-rich conditions, while NAT10-inhibited autophagy is released upon energy stress.
Since autophagy is induced to support cell survival in response to energy stress, we evaluated the effect of NAT10 depletion on cell survival upon glucose deprivation. NAT10 knockdown promoted the survival of cells grown in glucose-deprived conditions but had little effect on that of cells grown with an adequate glucose supply (Figure 2K). Taken together, these results demonstrated that NAT10 promotes rRNA biogenesis and inhibits autophagy under normal conditions, while deacetylation of NAT10 by Sirt1 halts rRNA biogenesis and facilitates autophagy induction under energy stress.
NAT10 interacts with Che-1
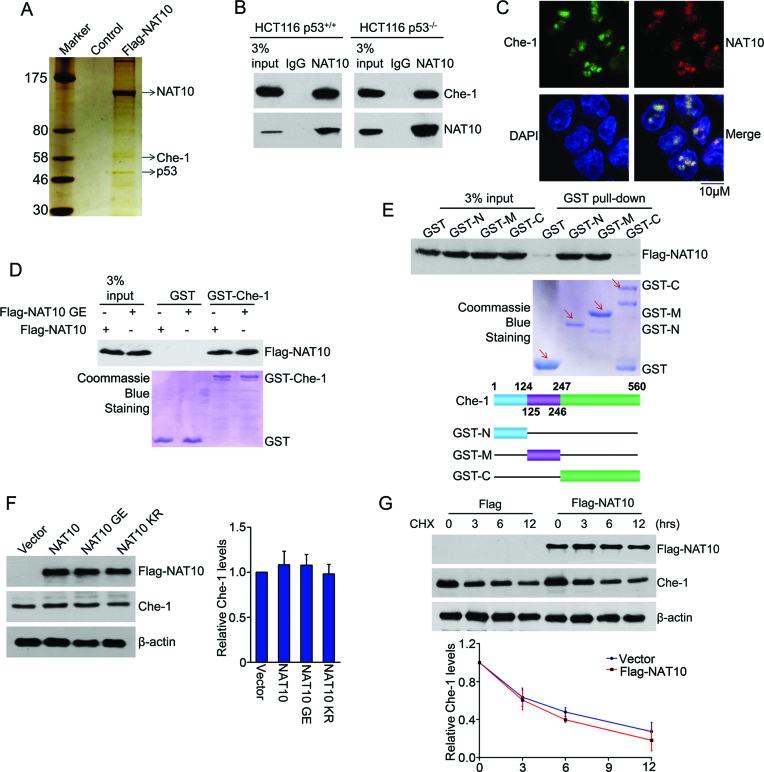
To uncover the mechanism by which NAT10 functions in autophagy induction, we purified a NAT10-specific immunocomplex and identified NAT10-interacting proteins (Figure 3A). Among various NAT10-interacting proteins identified by mass spectrometry, Che-1 is of particular interest due to its pivotal role in autophagy (45). Coimmunoprecipitation experiments confirmed the interaction between endogenous NAT10 and Che-1 in both HCT116 p53+/+ and HCT116 p53−/− cells (Figure 3B). Immunofluorescence staining showed that NAT10 colocalized with Che-1 in the nucleus (Figure 3C). To determine whether NAT10 directly interacts with Che-1, GST pull-down experiments were performed with purified Flag-NAT10 and GST-Che-1. Both Flag-NAT10 and Flag-NAT10 GE bound to GST-Che-1 in vitro (Figure 3D), indicating that the acetyltransferase activity of NAT10 is not required for its interaction with Che-1. Moreover, the NAT10-binding regions of Che-1 were narrowed down to the N-terminal domain (aa 1–124) and middle domain (aa 125–246) by GST pull-down experiments (Figure 3E). These results demonstrated that NAT10 interacts with Che-1 in cells and in vitro.
Figure 3.
NAT10 interacts with Che-1. (A) U2OS cells were transfected with Flag-NAT10. After 48 h, the cells were harvested, and whole cell extracts were immunoprecipitated with an anti-Flag antibody affinity resin. NAT10-binding proteins were resolved by SDS-PAGE, detected by silver staining and analyzed by mass spectrometry. (B) HCT116 cell lysates were immunoprecipitated with a control IgG or anti-NAT10 antibody. Immunoprecipitants were immunoblotted with the indicated antibodies. (C) HCT116 cells were fixed, and immunostaining was performed to determine the localization of Che-1 and NAT10. Immunofluorescence images were acquired with a confocal microscope. Scale bars, 10 μM. (D) GST pull-down assay was performed with purified Flag-NAT10 or Flag-NAT10 GE and GST-Che-1 proteins. Che-1-bound Flag-NAT10 was evaluated by Western blot using an anti-Flag antibody. The amounts of GST fusion proteins used in the GST pull-down assay are shown in the lower panel. (E) GST pull-down assay was performed with purified Flag-NAT10 and GST-Che-1 or its deletion mutants. Che-1-bound Flag-NAT10 was evaluated by Western blot using an anti-Flag antibody (upper). The amounts of GST fusion proteins used in the GST pull-down assays are shown (middle). The schematic diagram represents the GST-Che-1 deletion mutant constructs (lower). (F) HCT116 cells were transfected with the indicated plasmids. Twenty-four hours later, the cells were harvested, and proteins from the cell lysates were subjected to immunoblot analysis for the evaluation of Che-1 and NAT10. Beta-actin was evaluated as a loading control. Error bars indicate the SEM (n = 3) (right panel). (G) HCT116 cells were transfected with the Flag-NAT10 or Flag plasmids. Twenty-four hours later, the cells were treated with cycloheximide (CHX) and harvested at the indicated time points. Proteins from the cell lysates were subjected to immunoblot analysis for the evaluation of NAT10 and Che-1. Beta-actin was evaluated as a loading control (upper panels). Relative Che-1 levels standardized to β-actin at different time points are shown in the lower panel. Error bars indicate the SEM (n = 3).
We previously demonstrated that NAT10 promotes Mdm2 degradation via its atypical E3 ligase activity (26). Thus, we next wanted to know whether NAT10 regulates Che-1 protein stability. However, the Che-1 levels were not changed when Flag-NAT10, Flag-NAT10 GE or Flag-NAT10 KR was ectopically expressed (Figure 3F). In addition, the half-life of Che-1 was not altered by the expression of Flag-NAT10 (Figure 3G), confirming that Che-1 levels are not regulated by NAT10.
NAT10 inhibits the Che-1-dependent transcriptional activation of Deptor and Redd1 under energy-rich conditions
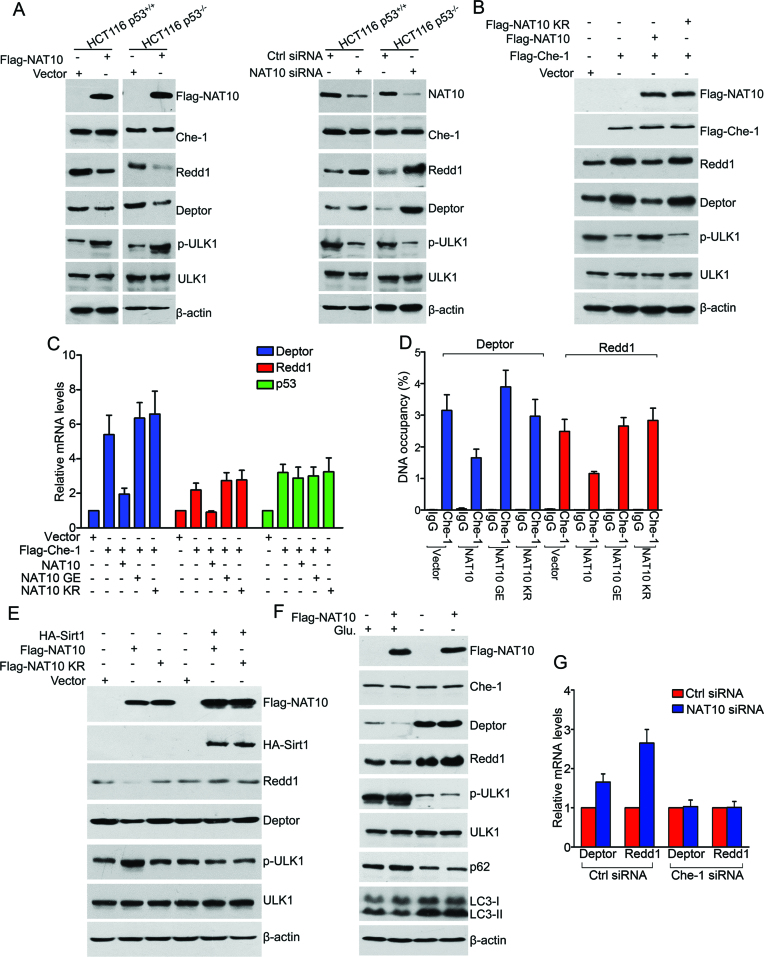
Che-1 enhances autophagy activation by transactivating the transcription of Deptor and Redd1, two important inhibitors of the mTOR pathway (45). We thus wanted to know whether NAT10 regulates autophagy induction by regulating the Che-1-mediated transcriptional activation of Deptor and Redd1. First, ectopically expressed Flag-NAT10 decreased Redd1 and Deptor levels in both HCT116 p53+/+ and HCT116 p53−/− cells without changing the Che-1 levels (Figure 4A, left and Supplementary Figure S3A), indicating that NAT10 inhibits the expression of Redd1 and Deptor. Consequently, phosphorylation of ULK1, a target of mTOR activation, was upregulated by ectopic Flag-NAT10 expression, indicating that NAT10 activates mTOR by inhibiting the expression of Redd1 and Deptor (Figure 4A, left). Accordingly, knockdown of NAT10 elevated Redd1 and Deptor levels and simultaneously inhibited the phosphorylation of ULK1 (Figure 4A, right); similar results were obtained in HCT116 NAT10 KO cells (Supplementary Figure S3B). To determine whether NAT10 inhibits the Che-1-dependent activation on Redd1 and Deptor, Flag-NAT10 was coexpressed with Flag-Che-1, and the expression levels of Redd1 and Deptor were evaluated. Flag-NAT10 suppressed the Che-1-activated expression of Redd1 and Deptor, while NAT10 KR failed to do so (Figure 4B), indicating that NAT10 suppresses the Che-1-mediated activation of Redd1 and Deptor, and autoacetylation of NAT10 is required for this function.
Figure 4.
NAT10 inhibits the Che-1-dependent transcriptional activation of Deptor and Redd1 under energy-rich conditions. (A) HCT116 cells were transfected with the indicated siRNAs or plasmids. Western blotting was performed to evaluate the indicated proteins. (B) HCT116 cells were transfected with the indicated plasmids. After 24 h, cell lysates were prepared and subjected to Western blot analysis for evaluation of the indicated proteins. (C) HCT116 cells were transfected with the indicated plasmids. After 24 h, total RNA was extracted and subjected to RT-qPCR for evaluation of the mRNA levels of the indicated genes. Error bars indicate the SEM (n = 3). (D) HCT116 cells were transfected with the indicated plasmids. The promoter occupancy of Deptor or Redd1 by Che-1 was analyzed by a ChIP assay. Error bars indicate the SEM (n = 3). (E) HCT116 cells were transfected with the indicated plasmids. After 24 h, the cells were harvested, and western blotting was performed to evaluate the indicated proteins. (F) U2OS cells were transfected with the indicated plasmids and cultured in medium with or without glucose for 36 h. The cells were harvested, and Western blotting was performed to evaluate the indicated proteins. (G) HCT116 cells were transfected with the indicated siRNAs. After 72 h, total RNA was extracted and subjected to RT-qPCR for evaluation of the Deptor and Redd1 mRNA levels. Error bars indicate the SEM (n = 3). See also Supplementary Figure S3.
To determine whether NAT10 regulates the Che-1-dependent transcriptional activation of Redd1 and Deptor, the mRNA levels of Redd1 and Deptor were evaluated by RT-qPCR when Flag-NAT10 was coexpressed with Flag-Che-1. The Flag-Che-1-induced elevation of Redd1 and Deptor mRNA levels was inhibited by the ectopic expression of Flag-NAT10 but not by Flag-NAT10 GE or Flag-NAT10 KR, indicating that NAT10 represses the transcriptional activation of Redd1 and Deptor by Che-1, and the acetyltransferase activity of NAT10 is required for this repression (Figure 4C). In addition, Flag-NAT10 failed to regulate the Che-1-mediated transcriptional activation of TP53, demonstrating that NAT10 specifically regulates the Che-1-mediated transcriptional activation of Redd1 and Deptor. We thereafter examined whether NAT10 affects the binding of Che-1 to Deptor and Redd1 promoters. ChIP assay results showed that Flag-NAT10 significantly decreased the binding of Che-1 to the Deptor and Redd1 promoters (Figure 4D), while neither NAT10 GE nor NAT10 KR lost this capability. Together, our data demonstrated that the acetyltransferase activity of NAT10 is required for its ability to inhibit the Che-1-dependent transcriptional activation of Redd1 and Deptor.
We further found that ectopically expressed HA-Sirt1 abolished the Flag-NAT10-induced inhibition of Deptor and Redd1 expression (Figure 4E). Collectively, we demonstrated that Sirt1 abrogates the NAT10-mediated inhibition of Che-1 transcriptional activity (the ability of Che-1 to transcriptionally activate Redd1 and Deptor). Significantly, Flag-NAT10 lost the ability to inhibit Deptor and Redd1 expression, activate ULK1 phosphorylation and suppress autophagy induction under glucose deprivation conditions (Figure 4F). Furthermore, when Che-1 was depleted by siRNA, knockdown of NAT10 failed to elevate the expression levels of Redd1 and Deptor (Figure 4G and Supplementary Figure S3C). Taken together, these data suggested that NAT10 inhibits Che-1 activity during autophagy induction under energy-rich conditions, while NAT10-mediated autophagy inhibition is suppressed by Sirt1 upon energy deprivation.
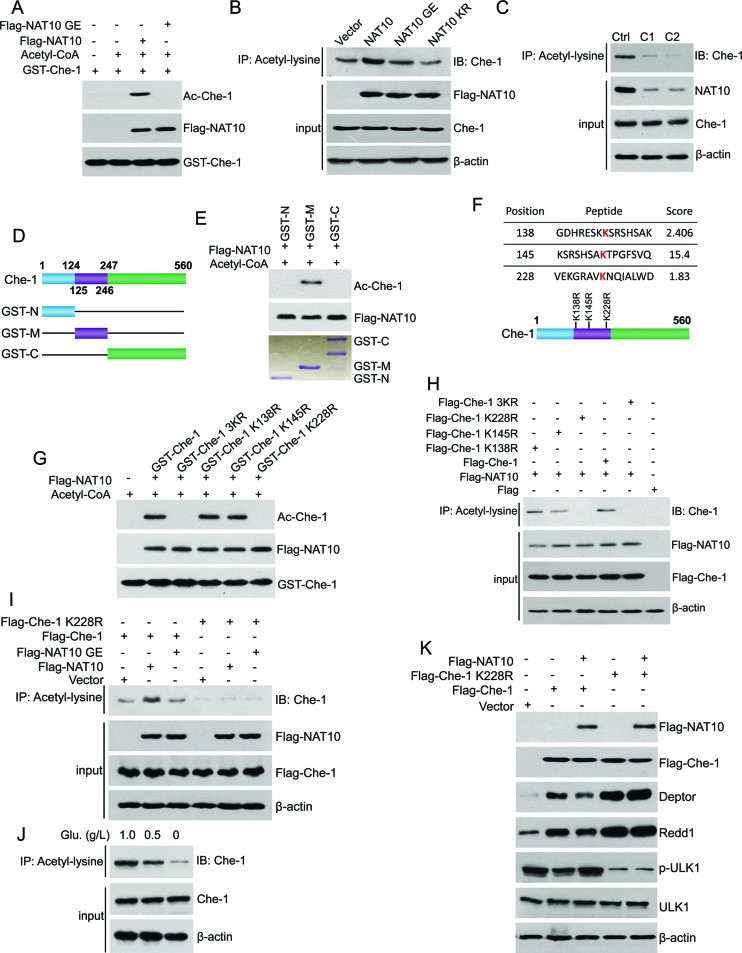
NAT10 acetylates Che-1 at K228
As acetyltransferase activity is required for the NAT10-mediated inhibition of Che-1, we hypothesized that NAT10 might acetylate Che-1. To this end, we performed in vitro acetylation assays using purified Flag-NAT10 from Sf9 insect cells and GST-Che-1 from E. coli bacterial cells. Che-1 was acetylated in the presence of both acetyl-CoA and Flag-NAT10 (Figure 5A). However, Flag-NAT10 GE lost the ability to acetylate Che-1. In addition, Flag-NAT10 increased the cellular acetylation levels of Che-1, while Flag-NAT10 GE and Flag-NAT10 KR failed to do so (Figure 5B). Moreover, NAT10 KR still bound to Che-1 as well as wild-type NAT10 (Supplementary Figure S4A and S4B), indicating that acetylation of NAT10 is essential for its ability to acetylate Che-1. To verify these results, we evaluated the acetylation levels of Che-1 in HCT116 NAT10 KO cells. The acetylation levels of Che-1 were dramatically decreased, while the Che-1 protein levels were not changed in HCT116 NAT10 KO cells (Figure 5C). Together, these data demonstrated that NAT10 functions as an acetyltransferase for Che-1, and acetylation of NAT10 is required for its ability to acetylate Che-1.
Figure 5.
NAT10 acetylates Che-1 at K228. (A) In vitro acetylation experiments were performed with GST-Che-1 and purified Flag-NAT10 or Flag-NAT10 GE as described in the Materials and Methods. Acetylation of Che-1 was evaluated by Western blot using an anti-acetyl-lysine antibody. (B) HCT116 cells were transfected with the indicated plasmids. Acetylation of Che-1 was evaluated by Western blot using an anti-Che-1 antibody on anti-acetyl-lysine antibody-specific immunoprecipitants. Inputs were evaluated by Western blot using the indicated antibodies. (C) Cell lysates were prepared from HCT116 Ctrl cells or HCT116 NAT10 KO cells (c1 or c2). Acetylation of Che-1 was evaluated by Western blot using an anti-Che-1 antibody on the anti-acetyl-lysine immunoprecipitants. The expression of NAT10 and Che-1 was evaluated by Western blot using the indicated antibodies. (D) The schematic diagram represents the Che-1 deletion mutant constructs. (E) In vitro acetylation experiments were performed with purified GST-Che-1-N, GST-Che-1-M or GST-Che-1-C fusion proteins and purified Flag-NAT10 as described in the Materials and Methods. Acetylation of Che-1 was evaluated by Western blot using an anti-acetyl-lysine antibody. (F) The potential acetylation sites on Che-1-M were predicted using the biocuckoo website. (G) In vitro acetylation experiments were performed with the purified GST-Che-1, GST-Che-1-3KR, GST-Che-1-K138R, GST-Che-1-K145R or GST-Che-1-K228R fusion protein and Flag-NAT10 as described in the Materials and Methods. Acetylation of Che-1 was evaluated by Western blot using an anti-acetyl-lysine antibody. (H) HCT116 cells were transfected with the indicated plasmids. Cells were harvested, and immunoprecipitation was performed to evaluate the level of Che-1 acetylation (upper). The expression levels of Flag-tagged proteins in the cell lysates were evaluated by Western blot using the indicated antibodies (lower). (I) U2OS cells were transfected with the indicated plasmids. Immunoprecipitation was performed using an anti-acetyl-lysine antibody to evaluate the level of Che-1 acetylation (upper). The expression levels of Flag-tagged proteins in the cell lysates were evaluated by Western blot using the indicated antibodies (lower). (J) HCT116 cells were cultured in medium containing different concentrations of glucose for 18 h. The acetylation levels of Che-1 in the anti-acetyl-lysine antibody-specific immunoprecipitants were evaluated using an anti-Che-1 antibody (upper). Total Che-1 levels were evaluated by western blot (lower). (K) HCT116 cells were transfected with the indicated plasmids. Cell lysates were prepared and subjected to Western blot for evaluation of the indicated proteins. See also Supplementary Figure S4.
To determine the sites of Che-1 acetylation catalyzed by NAT10, we performed in vitro acetylation experiments using purified GST-Che-1 deletion mutants that included the N-terminal fragment (aa 1–124), middle fragment (aa 125–246) and C-terminal fragment (aa 247–560) (Figure 5D). The middle fragment of Che-1 was acetylated by Flag-NAT10 (Figure 5E). Three potential acetylation sites in the middle portion of Che-1 were predicted at K128, K145 and K228 (Figure 5F). Therefore, in vitro acetylation experiments were performed with site-directed Che-1 mutants to determine the acetylation sites in Che-1. Substitution of lysine 228 with arginine in Che-1 (Che-1 K228R) and substitution of these three lysine residues with arginine (Che-1 3KR) totally abrogated the NAT10-mediated acetylation of Che-1 (Figure 5G), suggesting that NAT10 acetylates Che-1 at K228. Sequence alignment showed that K228 on Che-1 is highly conserved in various species (Supplementary Figure S4C). To confirm Che-1 acetylation at K228 by NAT10 in cells, Che-1 and its mutants were cotransfected with Flag-NAT10 into cells. Flag-Che-1, but not Flag-Che-1 K228R or Flag-Che-1 3KR, was acetylated in the presence of Flag-NAT10 (Figure 5H), further confirming that NAT10 acetylates Che-1 at K228. In addition, the Flag-NAT10 GE mutant failed to increase the Che-1 acetylation levels like Flag-NAT10 (Figure 5I). We thereafter evaluated the acetylation levels of Che-1 upon energy depletion. Significantly, acetylation of endogenous Che-1 gradually decreased upon energy deprivation in HCT116 cells (Figure 5J) and U2OS cells (Supplementary Figure S4D), while the total levels of Che-1 were not altered.
We further evaluated the role of NAT10-mediated Che-1 K228 acetylation in the Che-1-mediated activation of Deptor and Redd1. Flag-NAT10 inhibited the Che-1-mediated activation of Deptor and Redd1 but had no effect on Che-1 K228R-mediated transcription activation (Figure 5K). These results demonstrated that NAT10 inhibits Che-1 transcriptional activity by acetylating Che-1 at K228.
Taken together, our data demonstrated that NAT10 is autoacetylated and then acetylates Che-1 at K228 under energy-rich conditions. When cells sustain energy stress, NAT10 is deacetylated by Sirt1, and this deacetylation abolishes the ability of NAT10 to acetylate Che-1.
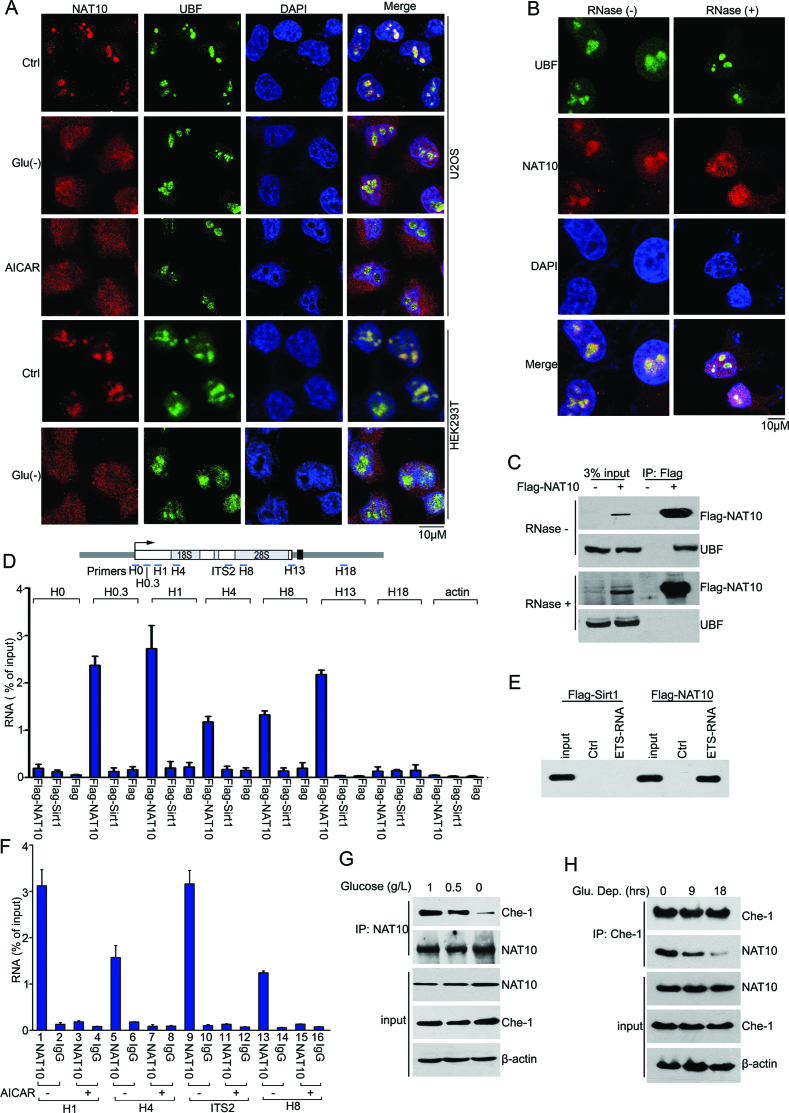
Glucose deprivation results in the release of NAT10 from nucleoli
We next investigated whether NAT10 remains in the nucleolus under glucose deprivation. Strikingly, NAT10 was excluded from the nucleoli upon glucose deprivation or AICAR (an energy depletion mimic) treatment, while NAT10 stayed predominantly in the nucleoli under normal conditions (Figure 6A). Interestingly, ectopic NAT10 KR remained in the nucleoli (Supplementary Figure S5A and S5B) under normal conditions and was excluded from the nucleoli under energy stress (Supplementary Figure S5C), indicating that translocation of NAT10 from the nucleoli to the nucleus was not due to its deacetylation. Because nucleolar proteins reside in the nucleolus by tethering to rRNA (46–48), we treated cells with RNase A and analyzed the localization of NAT10. RNase A treatment led to partial translocation of NAT10 from the nucleoli into the nucleoplasm, while UBF1 was retained in the nucleoli (Figure 6B), and immunoprecipitation analysis showed that NAT10 dissociated from UBF1 upon RNase A treatment (Figure 6C). These results suggested that RNA is required for the localization of NAT10 to the nucleoli.
Figure 6.
Nucleolar enrichment of NAT10 requires nascent pre-rRNA. (A) U2OS or HEK293T cells were cultured under normal conditions (ctrl), in glucose-deprived medium (Glu (-)) or in AICAR (2 mM)-containing medium for 36 h. Indirect immunofluorescence analysis was performed to determine the cellular localization of NAT10 (red) and UBF1 (green). Nuclei were stained with DAPI. Scale bar, 10 μM. (B) The cellular localization of NAT10 (red) and UBF1 (green) was visualized by indirect immunofluorescence after the cells were permeabilized and incubated with RNase (1 mg/ml, 10 min). Nuclei were stained with DAPI. Scale bar, 10 μM. (C) HEK293T cells transfected with Flag-NAT10 were cultured in the absence or presence of RNase (20 μg/ml). Immunoprecipitation was performed with an anti-Flag antibody. Flag-NAT10 and UBF1 in the immunoprecipitants were evaluated by immunoblot. (D) RNA immunoprecipitation (RIP) experiments were performed with cell lysates from HEK293T cells expressing Flag-NAT10 or Flag-Sirt1. Cell lysates were prepared after cells were exposed to UV light to crosslink the RNA with proteins and then incubated with anti-Flag beads. After elution with a Flag peptide and decrosslinking, coprecipitated RNA was purified and analyzed by RT-qPCR. The percentage of precipitated RNA relative to the total RNA is shown. Error bars indicate the SEM (n = 3). The positions of precipitated rRNA are shown in the upper panel. (E) Purified Flag-NAT10 or Flag-Sirt1 was incubated with streptavidin-coated Dynabeads (ctrl) or Dynabeads containing 5′ ETS RNA (+10/+389). Western blotting was performed to evaluate the RNA-bound proteins using an anti-Flag antibody. (F) HCT116 cells expressing Flag-NAT10 were treated with AICAR (2 mM) for 12 h. Flag-NAT10-RNA complexes were isolated and analyzed as described in D. Error bars indicate the SEM (n = 3). The positions of precipitated rRNA are shown in the upper panel of D. (G) HCT116 cells were cultured in medium containing 1.0 g/l, 0.5 g/l, or 0 g/l glucose for 18 h. Immunoprecipitation was performed with an anti-NAT10 antibody, and Che-1 expression in the precipitants was evaluated by immunoblot. (H) HCT116 cells were cultured in glucose-free medium for the indicated amounts of time. Immunoprecipitation was performed with an anti-Che-1 antibody, and NAT10 expression in the precipitants was evaluated by immunoblot. See also Supplementary Figure S5.
Since NAT10 participates in rRNA processing (18–20), we further analyzed the association of NAT10 with different regions of pre-rRNA in cells using RNA immunoprecipitation (RIP) assays. Pre-rRNA coprecipitated with NAT10 but was not associated with Sirt1 (Figure 6D), underscoring the specific binding of NAT10 to pre-rRNA. The association of NAT10 with rRNA was confirmed by an in vitro RNA pull-down assay performed using a biotin-labeled RNA fragment corresponding to the 5′ region (5′ ETS) of human pre-rRNA (from +10 to +389) (Figure 6E). Significantly, AICAR treatment diminished the binding of NAT10 to pre-rRNA (Figure 6F), suggesting that NAT10 departs from pre-rRNA and is excluded from nucleoli under energy stress.
We further performed immunoprecipitation experiments to evaluate whether the association of NAT10 with Che-1 is altered under energy stress conditions. The endogenous interaction between Che-1 and NAT10 was gradually decreased as the glucose concentration and duration of glucose deprivation in the medium decreased (Figure 6G, H and Supplementary Figure S5D). In addition, Che-1 was predominately retained in the nucleoli under glucose deprivation, while NAT10 was significantly translocated from the nucleoli to the nucleoplasm (Supplementary Figure S5E). Together, these results indicated that the Sirt1-dependent deacetylation of NAT10 prevents NAT10 from acetylating Che-1.
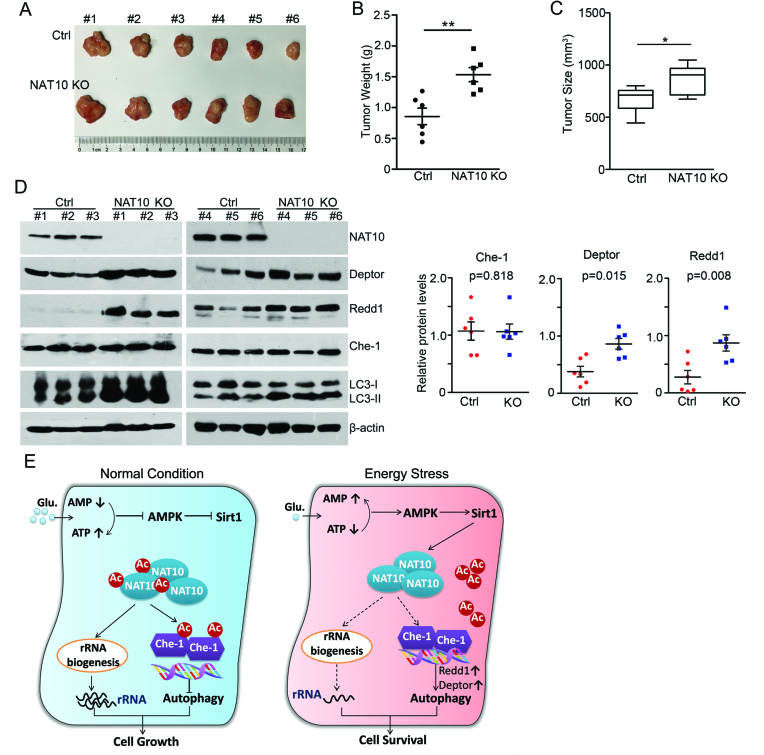
NAT10 regulates Che-1 activity in vivo
To further confirm the regulation of autophagy by NAT10 in vivo, we subcutaneously implanted HCT116 NAT10 KO cells into immunodeficient nude mice. The sizes and weights of tumor xenografts were then measured after the mice were sacrificed. The expression of Deptor and Redd1 in the tumor tissues was evaluated by Western blot. Ablation of NAT10 in HCT116 cells resulted in subtly increased tumor sizes and weights in the nude mice (Figure 7A–C). However, the expression levels of both Deptor and Redd1 were significantly increased in the tumor tissues generated from NAT10 KO cells, while Che-1 expression was not changed (Figure 7D). These data demonstrated that NAT10 inhibits the Che-1-mediated transcriptional activation of Deptor and Redd1 in vivo.
Figure 7.
NAT10 inhibits the expression of Redd1 and Deptor in mouse tumor xenografts. (A–C) NAT10 KO HCT116 cells were subcutaneously implanted into SCID mice. Tumors were dissected at the end of the experiment (A). The weights and sizes of the tumor xenografts are shown in B and C, respectively. Error bars indicate the SEM (n = 6). P-values were calculated using two-tailed unpaired Student's t-test. **P < 0.01, *P < 0.05. (D) Western blotting was performed on the tumor xenograft lysates to evaluate the levels of the indicated proteins (left). Densitometry scanning analyses of the Che-1, Deptor and Redd1 bands standardized to β-actin are summarized and shown (right). Error bars indicate the SEM (n = 6). (E) A working model explaining how NAT10 acetylation regulates rRNA biogenesis and autophagy upon glucose starvation.
DISCUSSION
Ribosome production is a major biosynthetic and energy-consuming process in cells, accounting for up to 80% of the energy consumption in proliferating cells (15). Under low energy levels, rRNA biogenesis is inhibited to save energy and maintain cell survival. Recently, substantial effort has been exerted to uncover the mechanism by which rRNA biogenesis is regulated upon energy stress. The protein complex eNoSC (energy-dependent nucleolar silencing complex) senses the cellular energy status and negatively controls rRNA transcription (43,46,49), and Sirt7 contributes to the repression of Pol I by deacetylating PAF53 upon stress (48). Moreover, SIRT7 is released from the nucleoli, resulting in hyperacetylation of U3-55k and attenuation of pre-rRNA processing (50). As a nucleolar acetyltransferase, NAT10 contributes to rRNA biogenesis by promoting UBF1 acetylation and facilitating 18S rRNA processing (18,19), and autoacetylation of NAT10 is required for NAT10’s ability to promote rRNA biogenesis (22). However, whether the acetylation of NAT10 is regulated during energy stress remains unknown.
Sirt1, a crucial target of AMPK (cellular energy sensor), is activated to maintain cell survival in response to metabolic stress (43,51–53) and regulates metabolic pathways by deacetylating its downstream substrates when cells encounter stress. For instance, Sirt1 targets FoxOs for deacetylation to promote cell survival upon energy stress and plays a protective role in ischemia by deacetylating PGC-1α (54,55). In the present study, we demonstrate that NAT10 interacts with Sirt1 in cells and in vitro, and importantly, the interaction of NAT10 with Sirt1 is enhanced by glucose starvation. NAT10 is deacetylated by Sirt1 in vitro and in cells under glucose deprivation. Deacetylated NAT10 fails to activate rRNA biogenesis and thus decreases energy consumption, which is beneficial for cell survival upon energy withdrawal. Therefore, deacetylation of NAT10 by Sirt1 contributes to halt rRNA biogenesis when cells undergo energy deprivation. We thus identify NAT10 as another important substrate of Sirt1 in energy stress response. Additionally, NAT10 is an ATP-dependent acetyltransferase responsible for 18S rRNA processing (19). Hence, NAT10-mediated rRNA processing should be inhibited by glucose starvation due to the absence of ATP.
Autophagy is inhibited under adequate energy supply conditions, and Che-1 is a key regulator of autophagy in that it activates the process by upregulating Deptor and Redd1 expression to inhibit mTOR activity (45). However, it is not fully understood how the function of Che-1 in autophagy induction is inhibited in energy-fed cells. We found that NAT10 interacts with Che-1 in cells and in vitro under adequate energy conditions. Interestingly, NAT10 inhibits the Che-1 transcriptional activation of Redd1 and Deptor but not that of TP53, suggesting the NAT10 specifically regulates autophagy-related downstream genes of Che-1. Consequently, NAT10 activates the mTOR pathway and represses autophagy induction. Interestingly, the enzyme-dead mutant Flag-NAT10 G641E and the autoacetylation-defective mutant Flag-NAT10 K462R lost the ability to inhibit Che-1 transcriptional activity, though they still bound to Che-1. These data indicate that both the acetyltransferase activity and acetylation of NAT10 at K462 are required for its ability to inhibit Che-1-mediated transcriptional activation in autophagy inhibition.
Given its pivotal role in cellular processes, Che-1 activity has been found to be controlled by post-translational modifications. Previous studies have demonstrated that Che-1 is phosphorylated and activated to protect cells from various cellular stresses (11,13). For instance, the MK2-mediated phosphorylation of Che-1 inhibits the transcription of p53-dependent proapoptotic genes, such as Puma, Bax and Bak, to protect cells from apoptosis (13). In addition, Che-1 transcriptional activity is activated by phosphorylation at residues S316, S320 and S321 during autophagy induction (56). However, how Che-1 activity is regulated by acetylation remains unknown. Here, we found that NAT10 acetylates Che-1 at K228 in energy-fed cells. Importantly, the Che-1 K228R mutant activated the expression of Deptor and Redd1 more efficiently than wild-type Che-1 (Figure 5K), indicating that acetylation of Che-1 at K228 attenuates its transcriptional activation of Redd1 and Deptor and consequently inhibits autophagy under adequate energy supply. Che-1 was previously shown to possess an inhibitory domain within residues 125–246 (57). Thus, acetylation at Che-1 K228 may be the critical post-translational modification within residues 125-246 for the negative regulation of Che-1. Flag-NAT10 inhibits the ability of wild-type Che-1 to transcriptionally activate its target genes but has no effect on Che-1 K228R-mediated transcriptional activation. Hence, the acetylation of Che-1 by NAT10 is critical for inhibiting Che-1 activity under adequate energy supply. We demonstrated that Che-1 transcriptional activity is suppressed by acetylation at K228. Whether Che-1 is acetylated by other acetytransferases, such as p300/CBP and TIP60, remains unknown. Further studies on Che-1 acetylation may provide more evidence for the functional regulation of Che-1 by post-translational modification.
Autophagy is induced to maintain metabolic homeostasis and cell viability upon sustained energy deprivation, and Che-1 acetylation should thus be inhibited under energy stress. In the present study, we demonstrated that the interaction between endogenous NAT10 and Che-1 is attenuated upon glucose deprivation. The dissociation of Che-1 from NAT10 prevents Che-1 from being acetylated by NAT10, and Che-1 is thus activated to induce autophagy. These observations demonstrated that the transcriptional activation of Redd1 and Deptor by Che-1 is regulated by NAT10 according to the cellular energy status. NAT10 is deacetylated by Sirt1 upon energy stress, but whether Sirt1 targets Che-1 for deacetylation when cells undergo energy stress remains unknown.
NAT10 activates rRNA biogenesis in nucleoli, and autoacetylation is required for NAT10-mediated rRNA synthesis (18–20,22). While it is possible that deficient acetylation could affect the nucleolar localization of NAT10, this is not the case since Flag-NAT10 KR and GFP-NAT10KR remain in the nucleolus. Interestingly, both endogenous NAT10 and ectopically expressed Flag-NAT10 KR translocated from the nucleolus to the nucleus when cellular RNA was digested. Thus, the nucleolar localization of NAT10 depends on the presence of RNA. This result is in accordance with previous studies demonstrating that some nucleolar proteins localize in the nucleolus by tethering to RNA (47,48). Since NAT10 is involved in rRNA biogenesis via its promotion of UBF1 acetylation and acetylation of rRNA in nucleoli (18–20), the exclusion of NAT10 from the nucleolus conversely abrogates the NAT10-elevated acetylation of UBF1 and NAT10-mediated rRNA acetylation, leading to attenuation of rRNA biogenesis. In summary, NAT10 maintains its acetylation level by autoacetylation, binding to UBF1 and enhancing rRNA biogenesis under adequate energy supply conditions. Under unfavorable growth conditions, NAT10 is deacetylated by Sirt1 and loses the ability to facilitate rRNA biogenesis, leading to decreased rRNA synthesis. The decreased rRNA level leads to the release of NAT10 from the nucleolus into the nucleoplasm.
In conclusion, our findings identified a critical role of NAT10 in the crosstalk between rRNA biogenesis and autophagy in response to energy stress. On the one hand, NAT10 is acetylated to activate rRNA biogenesis in proliferating cells. On the other hand, acetylated NAT10 binds to Che-1 and inhibits the Che-1-mediated transcriptional activation of Deptor and Redd1 to suppress autophagy induction. Under energy stress condition, NAT10-associated rRNA biogenesis is suppressed due to the deacetylation of NAT10 by Sirt1. Meanwhile, deacetylated NAT10 loses the ability to acetylate Che-1 and suppress autophagy, leading to autophagy induction and the facilitation of cell survival (Figure 7E).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr. Rui Yan (UC Berkeley) for editing the English in this manuscript. We thank Dr. Qihua He for assistance with confocal microscopy, Dr. Hounan Wu for flow cytometry assistance and Dr. Yan Gao for assisting in the mass spectrometric analysis. We thank Prof. Bo Zhang for providing us the anti-NAT10 antibody.
Author Contributions: X.L., X.D. and B.X. designed the project; X.L., S.C., C.Z. and Z.L. performed the experiments; X.L., B.X., J.L. and X.D. prepared the manuscript. All authors contributed and commented on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [81672735, 81621063, 81371868]; 973 Program [2013CB837201]; Innovation Team of Ministry of Education [IRT13001]. Funding for open access charge: National Natural Science Foundation of China [81672735, 81371868, 81621063].
Conflict of interest statement. None declared.
REFERENCES
- 1. Reiling J.H., Sabatini D.M.. Stress and mTORture signaling. Oncogene. 2006; 25:6373–6383. [DOI] [PubMed] [Google Scholar]
- 2. Zhou Y., Rucker E.B. 3rd, Zhou B.P.. Autophagy regulation in the development and treatment of breast cancer. Acta Biochim. Biophys. Sin.(Shanghai). 2016; 48:60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mizushima N., Klionsky D.J.. Protein turnover via autophagy: implications for metabolism. Annu. Rev. Nutr. 2007; 27:19–40. [DOI] [PubMed] [Google Scholar]
- 4. Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014; 24:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Neufeld T.P. TOR-dependent control of autophagy: biting the hand that feeds. Curr. Opin. Cell Biol. 2010; 22:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jung C.H., Ro S.H., Cao J., Otto N.M., Kim D.H.. mTOR regulation of autophagy. FEBS Lett. 2010; 584:1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao Y., Xiong X., Jia L., Sun Y.. Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis. 2012; 3:e386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Passananti C., Floridi A., Fanciulli M.. Che-1/AATF, a multivalent adaptor connecting transcriptional regulation, checkpoint control, and apoptosis. Biochem Cell Biol. 2007; 85:477–483. [DOI] [PubMed] [Google Scholar]
- 9. Iezzi S., Fanciulli M.. Discovering Che-1/AATF: a new attractive target for cancer therapy. Front. Genet. 2015; 6:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bruno T., Iezzi S., De Nicola F., Di Padova M., Desantis A., Scarsella M., Di Certo M.G., Leonetti C., Floridi A., Passananti C. et al. Che-1 activates XIAP expression in response to DNA damage. Cell Death Differ. 2008; 15:515–520. [DOI] [PubMed] [Google Scholar]
- 11. Bruno T., De Nicola F., Iezzi S., Lecis D., D’Angelo C., Di Padova M., Corbi N., Dimiziani L., Zannini L., Jekimovs C. et al. Che-1 phosphorylation by ATM/ATR and Chk2 kinases activates p53 transcription and the G2/M checkpoint. Cancer Cell. 2006; 10:473–486. [DOI] [PubMed] [Google Scholar]
- 12. Bacalini M.G., Di Lonardo D., Catizone A., Ciccarone F., Bruno T., Zampieri M., Guastafierro T., Calabrese R., Fanciulli M., Passananti C. et al. Poly(ADP-ribosyl)ation affects stabilization of Che-1 protein in response to DNA damage. DNA Repair (Amst.). 2011; 10:380–389. [DOI] [PubMed] [Google Scholar]
- 13. Hopker K., Hagmann H., Khurshid S., Chen S., Hasskamp P., Seeger-Nukpezah T., Schilberg K., Heukamp L., Lamkemeyer T., Sos M.L. et al. AATF/Che-1 acts as a phosphorylation-dependent molecular modulator to repress p53-driven apoptosis. EMBO J. 2012; 31:3961–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kusnadi E.P., Hannan K.M., Hicks R.J., Hannan R.D., Pearson R.B., Kang J.. Regulation of rDNA transcription in response to growth factors, nutrients and energy. Gene. 2015; 556:27–34. [DOI] [PubMed] [Google Scholar]
- 15. Schmidt E.V. The role of c-myc in cellular growth control. Oncogene. 1999; 18:2988–2996. [DOI] [PubMed] [Google Scholar]
- 16. Leary D.J., Huang S.. Regulation of ribosome biogenesis within the nucleolus. FEBS Lett. 2001; 509:145–150. [DOI] [PubMed] [Google Scholar]
- 17. Lv J., Liu H., Wang Q., Tang Z., Hou L., Zhang B.. Molecular cloning of a novel human gene encoding histone acetyltransferase-like protein involved in transcriptional activation of hTERT. Biochem. Biophys. Res. Commun. 2003; 311:506–513. [DOI] [PubMed] [Google Scholar]
- 18. Kong R., Zhang L., Hu L., Peng Q., Han W., Du X., Ke Y.. hALP, a novel transcriptional U three protein (t-UTP), activates RNA polymerase I transcription by binding and acetylating the upstream binding factor (UBF). J. Biol. Chem. 2011; 286:7139–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ito S., Horikawa S., Suzuki T., Kawauchi H., Tanaka Y.. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). J. Biol. Chem. 2014; 289:35724–35730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sharma S., Langhendries J.L., Watzinger P., Kotter P., Entian K.D., Lafontaine D.L.. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015; 43:2242–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kornprobst M., Turk M., Kellner N., Cheng J., Flemming D., Kos-Braun I., Kos M., Thoms M., Berninghausen O., Beckmann R. et al. Architecture of the 90S Pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell. 2016; 166:380–393. [DOI] [PubMed] [Google Scholar]
- 22. Cai S., Liu X., Zhang C., Xing B., Du X.. Autoacetylation of NAT10 is critical for its function in rRNA transcription activation. Biochem. Biophys. Res. Commun. 2017; 483:624–629. [DOI] [PubMed] [Google Scholar]
- 23. Larrieu D., Britton S., Demir M., Rodriguez R., Jackson S.P.. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science. 2014; 344:527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Balmus G., Larrieu D., Barros A.C., Collins C., Abrudan M., Demir M., Geisler N.J., Lelliott C.J., White J.K., Karp N.A. et al. Targeting of NAT10 enhances healthspan in a mouse model of human accelerated aging syndrome. Nat. Commun. 2018; 9:1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Larrieu D., Vire E., Robson S., Breusegem S.Y., Kouzarides T., Jackson S.P.. Inhibition of the acetyltransferase NAT10 normalizes progeric and aging cells by rebalancing the Transportin-1 nuclear import pathway. Sci. Signal. 2018; 11:eaar5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu X., Tan Y., Zhang C., Zhang Y., Zhang L., Ren P., Deng H., Luo J., Ke Y., Du X.. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep. 2016; 17:349–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Q., Liu X., Jin K., Lu M., Zhang C., Du X., Xing B.. NAT10 is upregulated in hepatocellular carcinoma and enhances mutant p53 activity. BMC Cancer. 2017; 17:605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bach M., Larance M., James D.E., Ramm G.. The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem. J. 2011; 440:283–291. [DOI] [PubMed] [Google Scholar]
- 29. Chang C., Su H., Zhang D., Wang Y., Shen Q., Liu B., Huang R., Zhou T., Peng C., Wong C.C. et al. AMPK-Dependent phosphorylation of GAPDH triggers Sirt1 activation and is necessary for autophagy upon glucose starvation. Mol. Cell. 2015; 60:930–940. [DOI] [PubMed] [Google Scholar]
- 30. Ha J., Guan K.L., Kim J.. AMPK and autophagy in glucose/glycogen metabolism. Mol. Aspects Med. 2015; 46:46–62. [DOI] [PubMed] [Google Scholar]
- 31. Lee I.H., Cao L., Mostoslavsky R., Lombard D.B., Liu J., Bruns N.E., Tsokos M., Alt F.W., Finkel T.. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:3374–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bosch-Presegue L., Vaquero A.. Sirtuin-dependent epigenetic regulation in the maintenance of genome integrity. FEBS J. 2015; 282:1745–1767. [DOI] [PubMed] [Google Scholar]
- 33. Feige J.N., Auwerx J.. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007; 17:292–301. [DOI] [PubMed] [Google Scholar]
- 34. Vinciguerra M., Fulco M., Ladurner A., Sartorelli V., Rosenthal N.. SirT1 in muscle physiology and disease: lessons from mouse models. Dis. Model Mech. 2010; 3:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meng X., Tan J., Li M., Song S., Miao Y., Zhang Q.. Sirt1: role under the condition of Ischemia/Hypoxia. Cell Mol. Neurobiol. 2017; 37:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang R., Xu Y., Wan W., Shou X., Qian J., You Z., Liu B., Chang C., Zhou T., Lippincott-Schwartz J. et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell. 2015; 57:456–466. [DOI] [PubMed] [Google Scholar]
- 37. Azkargorta M., Fullaondo A., Laresgoiti U., Aloria K., Infante A., Arizmendi J.M., Zubiaga A.M.. Differential proteomics analysis reveals a role for E2F2 in the regulation of the Ahr pathway in T lymphocytes. Mol. Cell. Proteomics. 2010; 9:2184–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yi J., Huang X., Yang Y., Zhu W.G., Gu W., Luo J.. Regulation of histone acetyltransferase TIP60 function by histone deacetylase 3. J. Biol. Chem. 2014; 289:33878–33886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fan W., Luo J.. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol. Cell. 2010; 39:247–258. [DOI] [PubMed] [Google Scholar]
- 40. Roth M., Chen W.Y.. Sorting out functions of sirtuins in cancer. Oncogene. 2014; 33:1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Feige J.N., Auwerx J.. Transcriptional targets of sirtuins in the coordination of mammalian physiology. Curr. Opin. Cell Biol. 2008; 20:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vaquero A., Reinberg D.. Calorie restriction and the exercise of chromatin. Genes Dev. 2009; 23:1849–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murayama A., Ohmori K., Fujimura A., Minami H., Yasuzawa-Tanaka K., Kuroda T., Oie S., Daitoku H., Okuwaki M., Nagata K. et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008; 133:627–639. [DOI] [PubMed] [Google Scholar]
- 44. Mizushima N., Yoshimori T.. How to interpret LC3 immunoblotting. Autophagy. 2007; 3:542–545. [DOI] [PubMed] [Google Scholar]
- 45. Desantis A., Bruno T., Catena V., De Nicola F., Goeman F., Iezzi S., Sorino C., Ponzoni M., Bossi G., Federico V. et al. Che-1-induced inhibition of mTOR pathway enables stress-induced autophagy. EMBO J. 2015; 34:1214–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mayer C., Schmitz K.M., Li J., Grummt I., Santoro R.. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 2006; 22:351–361. [DOI] [PubMed] [Google Scholar]
- 47. Todd M.A., Huh M.S., Picketts D.J.. The sub-nucleolar localization of PHF6 defines its role in rDNA transcription and early processing events. Eur. J. Hum. Genet. 2016; 24:1453–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen S., Seiler J., Santiago-Reichelt M., Felbel K., Grummt I., Voit R.. Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Mol. Cell. 2013; 52:303–313. [DOI] [PubMed] [Google Scholar]
- 49. Yuan X., Feng W., Imhof A., Grummt I., Zhou Y.. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol. Cell. 2007; 27:585–595. [DOI] [PubMed] [Google Scholar]
- 50. Chen S., Blank M.F., Iyer A., Huang B., Wang L., Grummt I., Voit R.. SIRT7-dependent deacetylation of the U3-55k protein controls pre-rRNA processing. Nat. Commun. 2016; 7:10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang L., Song T., Chen L., Kabra N., Zheng H., Koomen J., Seto E., Chen J.. Regulation of SirT1-nucleomethylin binding by rRNA coordinates ribosome biogenesis with nutrient availability. Mol. Cell Biol. 2013; 33:3835–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hardie D.G. The AMP-activated protein kinase pathway–new players upstream and downstream. J. Cell Sci. 2004; 117:5479–5487. [DOI] [PubMed] [Google Scholar]
- 53. Hardie D.G. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007; 8:774–785. [DOI] [PubMed] [Google Scholar]
- 54. Watroba M., Maslinska D., Maslinski S.. Current overview of functions of FoxO proteins, with special regards to cellular homeostasis, cell response to stress, as well as inflammation and aging. Adv. Med. Sci. 2012; 57:183–195. [DOI] [PubMed] [Google Scholar]
- 55. Fu B., Zhang J., Zhang X., Zhang C., Li Y., Zhang Y., He T., Li P., Zhu X., Zhao Y. et al. Alpha-lipoic acid upregulates SIRT1-dependent PGC-1alpha expression and protects mouse brain against focal ischemia. Neuroscience. 2014; 281:251–257. [DOI] [PubMed] [Google Scholar]
- 56. Alayev A., Doubleday P.F., Berger S.M., Ballif B.A., Holz M.K.. Phosphoproteomics reveals resveratrol-dependent inhibition of Akt/mTORC1/S6K1 signaling. J. Proteome Res. 2014; 13:5734–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ferraris S.E., Isoniemi K., Torvaldson E., Anckar J., Westermarck J., Eriksson J.E.. Nucleolar AATF regulates c-Jun-mediated apoptosis. Mol. Biol. Cell. 2012; 23:4323–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.