Abstract
Transfer of genetic material from parents to progeny via fusion of gametes is a way to ensure flow of information from one generation to the next. Apart from the genetic material, gametes provide a rich source of other factors such as RNA and proteins which can control traits of the embryo. Non-coding RNAs are not only carriers of regulatory information but can also encode memory of events of parental life. Here, we explore the possibility of parental inheritance of non-coding RNAs, especially long non-coding RNAs. Meta-analysis of RNA-seq data revealed several non-coding RNAs present in zebrafish oocyte, sperm and 2cell-stage. The embryo is transcriptionally silent at this stage, we rationalize that all the RNAs detectable at 2cell-stage are deposited either by sperm or oocyte or both and thus inherited. In the inherited pool, we noticed a conserved lncRNA, Cyrano previously known for zebrafish brain development. Knockdown of inherited Cyrano by miR-7 without changing zygotic Cyrano altered brain morphology at 24 hpf and 48 hpf. This defect could be partially rescued by injecting full length Cyrano lncRNA or a mutant resilient to knock-down by miR-7. In future, there is ample scope to check the possibility of inherited lncRNAs as carriers of memory of parental life events and building blocks that set up an initial platform for development.
INTRODUCTION
An interesting facet of biology is that the entire body of a sexually reproducing diploid organism is derived from a single celled embryo formed by the union of gametes. This union brings numerous factors apart from genomic DNA such as mitochondria, proteins and mRNAs into the zygote (1). In fact, the zygote is transcriptionally incompetent during early cell divisions. Thus, early stages of embryonic development, such as dorso-ventral patterning, cell fate determination and cell division are largely under the control of these maternal products and to a smaller extent influenced by paternal products (2,3). For example, loss of maternal Runx2b, that specifies ventral fate in zebrafish embryos, results in dorsalization (4). Maternal factors not only control all steps until the embryo begins synthesis of its own gene products, but they also impact development well beyond zygotic genome activation. For instance, we have shown previously that miR-34 is maternally inherited in Drosophila and zebrafish and knocking down the maternal miR-34 while retaining zygotic miR-34 led to abnormal neural development in zebrafish (5). In another study, maternally supplied transcription factor REST has been shown to regulate multiple genes during embryonic development and impact behaviour. Loss of maternal REST makes larvae hyperactive with abnormal primary motor neuron architecture (6). Such important and long lasting effects are produced by these inherited factors that dynamically regulate target expression during early development. Regulation of gene expression is achieved in two layers. Firstly, at the transcriptional level, chromatin accessibility is controlled by chemically modifying DNA or histones to facilitate the binding of transcription factors. Secondly, at the post-transcriptional level, various events such as splicing, RNA editing, nuclear export of RNA, association of RNA to ribosomes and binding of RNA stabilizing and destabilizing factors are modulated. Long non-coding RNAs (lncRNAs) are versatile molecules which have emerged as novel players in all of the above steps of gene regulation (7–11). Thousands of lncRNAs have been identified in diverse species marking different developmental stages, only a fraction of them have been functionally studied. For example, 550 distinct long intergenic non-coding RNAs (lincRNAs), a class of lncRNAs, were detected in zebrafish embryos at 24 and 48 hpf (12). Further, systematic investigation of RNA-seq data generated from eight stages of early zebrafish development identified 1133 lncRNAs (13), speculating contribution of parentally provided, functionally unknown transcripts. Single-cell RNA sequencing analysis of cells from human pre-implantation embryos identified 8701 maternal lncRNAs (14) highlighting the need to explore their role in maternal cytoplasm.
Apart from maternally inherited factors, paternally inherited ones have also gained attention recently. Thousands of such factors including small non-coding RNAs have been detected in sperm owing to advancement in next generation sequencing technology. Small RNA deep sequencing of mouse mature sperm identified a novel class of abundantly expressed, tRNA derived small RNAs (tsRNAs) that are 29–34 nucleotides in length (15). Such sperm-borne tsRNAs are active players in intergenerational inheritance and have been implicated in paternal diet induced metabolic disorders in offspring (16,17). However, there is scope to systematically monitor parental inheritance of lncRNAs.
Here, we have explored the possibility of maternal and paternal inheritance of non-coding RNAs by meta-analysis of RNA-seq data of zebrafish sperm, oocyte and 2cell-stage. The zebrafish embryos are transcriptionally inactive at the 2cell-stage. Since many transcripts present in the gametes may not be retained in the fertilized embryo, we used expression in the gamete and at 2cell-stage as a criterion for identifying inherited lncRNAs. The transcripts detected only in sperm and 2cell-stage are paternally inherited, while those common to oocyte and 2cell-stage exclusively are maternally inherited RNA. Interestingly, the highly conserved lncRNA Cyrano (12) was expressed in a testis, oocyte and 2cell-stage; thus, it could be maternally and paternally inherited in zebrafish. Expression analysis of Cyrano from publicly available single cell RNA seq data (14) of human oocyte, zygote, 2cell-stage and 4cell-stage embryos showed that human Cyrano is a maternally inherited lncRNA as well. Cyrano has a conserved miR-7 binding site through which both interact and mutually repress each other in embryonic stem cells (18). Ectopic over-expression using in vitro synthesized mature miR-7 duplex RNA successfully degraded inherited Cyrano. This resulted in severe deformities in the opening of midbrain and hindbrain ventricles at 24 hpf embryos as well as altered brain architecture at 48 hpf. These defects could be partially rescued by co-injecting Cyrano lncRNA or a mutant of Cyrano wherein the miR-7 binding sites were deleted.
This is the first report that systematically identifies maternal as well as paternal non-coding RNAs in zebrafish. The impact of parental inheritance of lncRNAs is illustrated by prototypical Cyrano, loss of which leads to defects in brain development.
MATERIALS AND METHODS
Animal husbandry
Zebrafish (Danio rerio) stocks were maintained at the zebrafish facility of CSIR-Institute of Genomics and Integrative Biology (IGIB), New Delhi, India, according to standard procedures (19) with 13/11 h light/dark cycle. Zebrafish experiments were performed as per recommendations and guidelines from Institutional Animal Ethics Committee (IAEC) of the CSIR-IGIB, New Delhi, India. Embryos from wild type and [Tg(HuC: Kaede × her4.1: mCherry)] strains were used for experiments. A night before the experiment, male and female zebrafish were kept in mating cages. Embryos were collected at desired stages of development and unfertilized oocytes were obtained by squeezing the abdomen of gravid zebrafish females.
RNA-seq datasets and data analysis
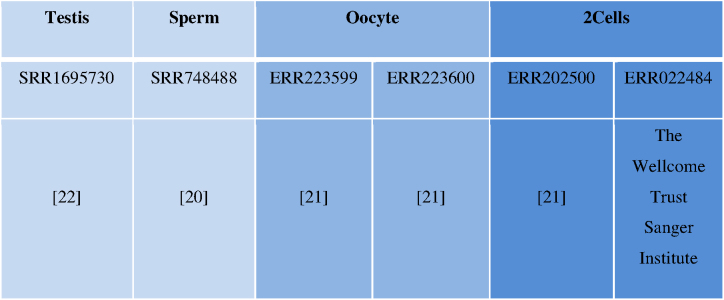
To check inheritance of non-coding RNAs, publically available transcriptome datasets of zebrafish testis, sperm, oocyte, 2cell-stage (20–22) were reanalysed (Details are in Table 1). To predict non-coding RNAs with higher confidence and overcoming inter-lab variation, two RNA-seq datasets were used as replicates for each stage.
Table 1.
RNA-seq datasets used in the study
|
Quality of raw reads from high throughput RNA sequencing of above samples was checked by FastQC. Trimmomatic was used to improve per base sequence quality and to remove over-represented sequences if any (23). TopHat v2.0.9 was used to align RNA-Seq reads to a zebrafish genome (Zv10) (24). Cuffquant was used further to quantify the expression. Transcripts were normalized by Cuffnorm. Then, from total expressed transcripts of each sample, Ensembl protein coding transcripts were discarded to get putative non-coding set of transcripts. As lncRNAs have arbitrary cut-off of 200 nucleotides (nt) for its length, all the transcripts with less than 200nt were discarded. Next, open reading frame (ORF) of remaining transcripts was checked by getORF (http://emboss.toulouse.inra.fr/cgi-bin/emboss) and transcripts having >100 amino acids ORF were discarded. In the end 1908 non-coding RNAs were retained after removing transcript with zero FPKM in all stages.
In vitro Transcription (IVT) for synthesis of microRNA (miRNA) mimics
DNA templates for IVT of miRNAs were synthesised by gap filing of overlapping oligonucleotides (Table 2). Two partially overlapping oligonucleotides (forward and reverse) were hybridized and extended by PCR. From this template sense and antisense control mimic/miRNAs were in vitro transcribed from T7 promoter (MEGAscript transcription kit, Thermo Fisher Scientific). The separately transcribed sense and antisense control mimic RNAs/miRNAs were further hybridized by boiling and then cooling at room temperature to make duplex control mimic/miRNA mimic.
Table 2.
Primers used in the study
| Primer name | Sequence | Experiment | Figure No. |
|---|---|---|---|
| ENSDART00000154527_F | GAAGCTCGCTTATCCAAATCCG | RT-PCR | 2 |
| ENSDART00000154527_R | GCCTACATCCCTCTCCCAAATC | RT-PCR | 2 |
| ENSDART00000168346_F | GCATCTTGCGGCAGAACACAT | RT-PCR | 2 |
| ENSDART00000168346_R | TGACCATACTCTGTCCAGCT | RT-PCR | 2 |
| ENSDART00000148229_F | CTTCAGGGCCATCAGAACATG | RT-PCR | 2 |
| ENSDART00000148229_R | CAGCACAATCATCAGCGGTTA | RT-PCR | 2 |
| Zf_ISH_cyrano_T7F | CGCTAATACGACTCACTATAGGGTAATCACTATTAGTTGATG | In situ hybridization | 3 |
| Zf_ISH_cyrano_SP6R | GCGATTTAGGTGACACTATAGAGTCACAACACTGGTCCACTC | In situ hybridization | 3 |
| ZF_cyrano_F | ACAAACCAAGACAGGCAGTGGCA | RT-PCR, qPCR | 3 |
| ZF_cyrano_R | TGCAACTCAATAGCACCCCGCT | RT-PCR, qPCR | 3 |
| ZF_cyrano_Exon1_F | TCACTCCAGTCCACATGTTTCC | RT-PCR, qPCR | 3 |
| ZF_cyrano_Intron1_R | GTACATCTCTACTCCCGCACAATG | RT-PCR, qPCR | 3 |
| ZF_sox11_F | AGAGCACGAAAGTTTCAAAGACTG | qPCR | 4 |
| ZF_sox11_R | TATCGGATCCTCTTCCTCCTC | qPCR | 4 |
| ZF_wnt8a_F | AATAGCGGACACGTTCAGCG | qPCR | 4 |
| ZF_wnt8a_R | TGTAGACATTCCCTGCCTTC | qPCR | 4 |
| ZF_gsc_F | ACGTCGGCACAAGAGAACAA | qPCR | 4 |
| ZF_gsc_R | TCCTCTGACGACGACCTTTTC | qPCR | 4 |
| ZF_miR-7_T7_Sense_F | GCTAATACGACTCACTATAGTGGAAGACTAGTGATTTTGTTGTAT | Anneal for IVT template | 4,5 |
| ZFmiR-7_Sense_R | ATACAACAAAATCACTAGTCTTC | Anneal for IVT template | 4,5 |
| ZFmiR-7_T7_Antisense _R | GCTAATACGACTCACTATAGACAACAAAATCACTAGTCTTCCA | Anneal for IVT template | 4,5 |
| ZF_miR-7_Antisense_F | TGGAAGACTAGTGATTTTGTTGT | Anneal for IVT template | 4,5 |
| Control _Sense_F | GCTAATACGACTCACTATAGCTCTAGGTTAAACTCCTGGTT | Anneal for IVT template | 4,5 |
| Control _Sense_R | AACCAGGAGTTTAACCTAG | Anneal for IVT template | 4,5 |
| Control _Antisense _F | GCTAATACGACTCACTATAGCCAGGAGTTTAACCTAGAG | Anneal for IVT template | 4,5 |
| Control _Antisense _R | CTCTAGGTTAAACTCCTGG | Anneal for IVT template | 4,5 |
| ZF_Cyrano_T7_F | |||
| TATAATACGACTCACTATAGATATCAGTAGCAGCACACGGGACCGAAAT | Full length Cyrano amplification and IVT | 5 | |
| ZF_Cyrano_full_R | GATCTAGACGCTTCTGTAGTGCATAGATCAGACTAAGCA | Full length Cyrano amplification | 5 |
| ZF_miR-7 resistantF_Cyrano | CAAACAAGTGACAAGCTGCTTGCTTTAGGG | miR-7 mutated Cyrano amplification | 5 |
| ZF_ miR-7 resistantR _Cyrano | CCCTAAAGCAAGCAGCTTGTCACTTGTTTG | miR-7 mutated Cyrano amplification | 5 |
miRNA mimic over-expression by microinjections
miRNA mimic was over-expressed at one-cell stage of zebrafish embryos by injecting 300 pg of in vitro synthesized RNA respectively. The embryos were monitored constantly and about 20 embryos per stage per sample were fixed for RNA isolation.
Zebrafish Cyrano amplification, IVT and rescue experiment
DNA template for IVT of full length ∼4.6 kb Cyrano was amplified by primers (Table 2). Flanking regions of miR-7 binding sites in Cyrano were amplified using PCR and then these two fragments were annealed to form miR-7 resistant Cyrano. Using these templates, wild type Cyrano and miR-7 resistant Cyrano was in vitro transcribed from T7 promoter (MEGAscript transcription kit, Thermo Fisher Scientific). At one cell stage of embryo, 300 pg of miR-7 mimic was injected with 6pg of either wild type Cyrano, or miR-7 resistant Cyrano transcript. The embryos were monitored for defects and death.
Quantitative real time PCR
Total RNA was isolated from embryos using TRizol® Reagent (Invitrogen). cDNA was synthesized by using gene specific reverse primer or random hexamer primers and M-MuLV reverse transcriptase at 42°C for 1 h. For quantitation of miRNA expression, poly(A) tail was attached to RNA by poly(A) polymerase and cDNA was synthesized by using universal adapter, M-MuLV reverse transcriptase at 42°C for 1 hour. Expression of mature miRNAs was measured by qPCR with miRNA specific primer using SYBR-green PCR master mix. The Real Time data analysis was done by using 2−ΔΔCTmethod.
RNA detection by in situ hybridization
For detection of RNA, probe sequence was amplified with specific primers (Table 2) and cloned in Topo2.1 vector (Invitrogen). T7 and SP6 promoter was used to synthesize digoxigenin labeled antisense RNA probes. Embryos were fixed at desired stages in 4% PFA then transferred to 100% methanol to dehydrate. Embryos were incubated in probe overnight, thereafter given stringent washes to remove non-specific binding. Embryos were further incubated in anti DIG antibody and was developed using the chromogenic agent NBT/BCIP.
Image acquisition and analysis
Images of in situ hybridized embryos were captured using Nikon microscope. Fluorescence images of zebrafish embryos were captured using Leica confocal microscope at 40x magnification from the randomly selected pool of embryos. Distance between two points was measured using ImageJ tool in the images of control or miR-7 mimic over-expressed embryos.
RESULTS
In sexually reproducing diploid organisms, each gamete contributes one copy of the genetic material in the form of DNA which remains inaccessible or silent for a few early cell divisions preceding embryonic development. During these early stages, the extrachromosomal factors set up a basic fate map of central nervous system, although the actual process of neurogenesis starts after two weeks of gestation in human. Next generation sequencing technology helped in identifying a hidden layer of non-coding RNAs in the transcriptome. However, their presence and relevance in the maternal factor pool is not much explored. We hypothesized that lncRNAs are parentally inherited during fertilization, and decided to explore their functions in zebrafish development.
Meta-analysis of RNA-seq data reveals parental inheritance of non-coding RNAs in zebrafish
For prediction of parentally inherited non-coding RNAs, RNA-seq data of sperm, oocyte and 2cell-stage were reanalyzed (details are in Table 1). Zebrafish embryos at 2cell-stage are transcriptionally inactive but rich in parentally inherited factors. This implies that, all the RNAs that are detected at 2cell-stage, were deposited by sperm,oocyte or both. RNA detected in only sperm and 2cell-stage were classified as paternally inherited RNA. Similarly, RNA found in oocyte and 2cell-stage only were thought to be maternally inherited.
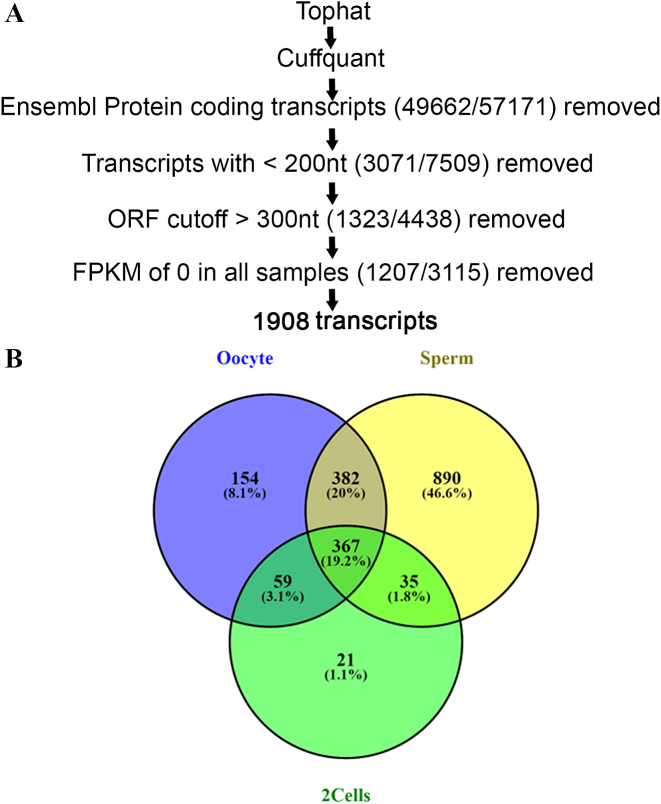
We checked the quality of RNA-seq raw reads of sperm, oocyte and 2cell-stage by FastQC and over-represented sequences were removed by trimmomatic. Remaining reads were aligned to zebrafish genome version 10 (Zv10) by Tophat. Then, Cuffquant was used to compute the transcript expression profile of 57171 transcripts obtained from the datasets mentioned above. We filtered out 49662 Ensembl protein coding transcripts to retain 7509 non-coding RNAs. As lncRNAs are, by convention longer than 200nt, all the transcripts shorter than 200nt were discarded. Next we checked the open reading frame (ORF) of remaining transcripts by getORF (http://emboss.toulouse.inra.fr/cgi-bin/emboss) and transcripts having >100 amino acids ORF were discarded. Further, removal of transcripts having zero FPKM in all samples retained 1908 transcripts (Figure 1A). Out of 1908 transcripts, 59 were exclusively maternally inherited and 35 were paternally inherited (Figure 1B).
Figure 1.
Overview of lncRNAs filtering pipeline and their distribution in various stages. (A) Systematic workflow for identification of lncRNAs. (B) Number of filtered non-coding RNAs in sperm, oocyte, 2cell-stage and common between them.
Validation of maternally inherited lncRNAs
To validate expression of systematically filtered lncRNAs, total RNA was isolated from zebrafish testis, oocyte and 2cell-stage and cDNA was synthesized. We used testis instead of sperm since sperm could not be extracted in amounts sufficient for RNA isolation. Further, expression of selected lncRNAs was checked by RT-PCR using these cDNAs. Interestingly, filtered lncRNAs ENSDART00000154527, ENSDART00000168346, ENSDART00000148229 which are antisense to Cacng2A, CDC6 and Kcnq5b, respectively, were found to be expressed only in oocyte and 2cell-stage (Figure 2A–C). They are maternally inherited non-coding RNAs; thus, validating the bio-informatics prediction.
Figure 2.
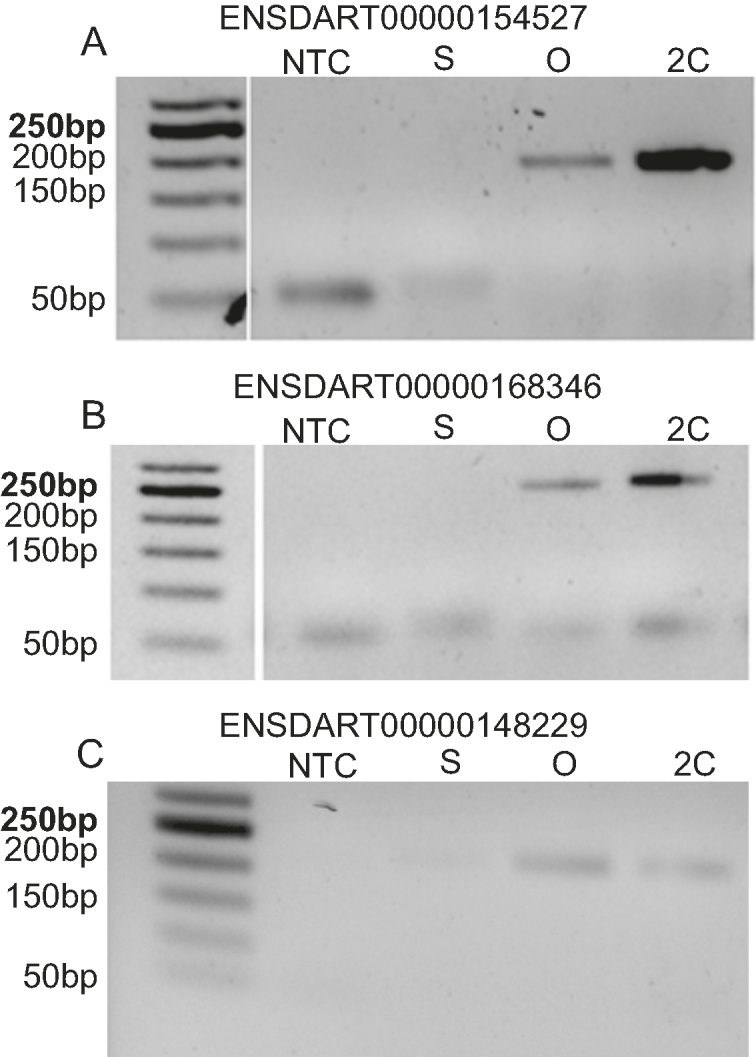
Validation of maternally inherited lncRNAs by RT-PCR. (A) ENSDART00000154527, (B) ENSDART00000168346, (C) ENSDART00000148229 which are antisense to Cacng2A, CDC6, Kcnq5b respectively. NTC-Non Template Control, S – testis, O – oocyte, 2C – 2cell-stage.
Conserved lincRNA Cyrano transcript gets inherited in zebrafish
Evolutionary conservation is frequently used as a criterion to narrow down important RNAs. The RNA can be conserved at its sequence, structure or synteny. Here, we checked sequence conservation of 1908 non-coding RNAs by BLAST and found that 70 zebrafish non-coding RNAs were conserved with human non-coding RNAs from Non-code database (Supplementary Figure S1A). These 70 transcripts include various classes of non-coding RNAs such as lincRNAs, antisense lncRNAs, processed transcripts etc (Supplementary Figure S1B) expressed across sperm, oocyte and/or 2cell-stages (Supplementary Figure S1C).
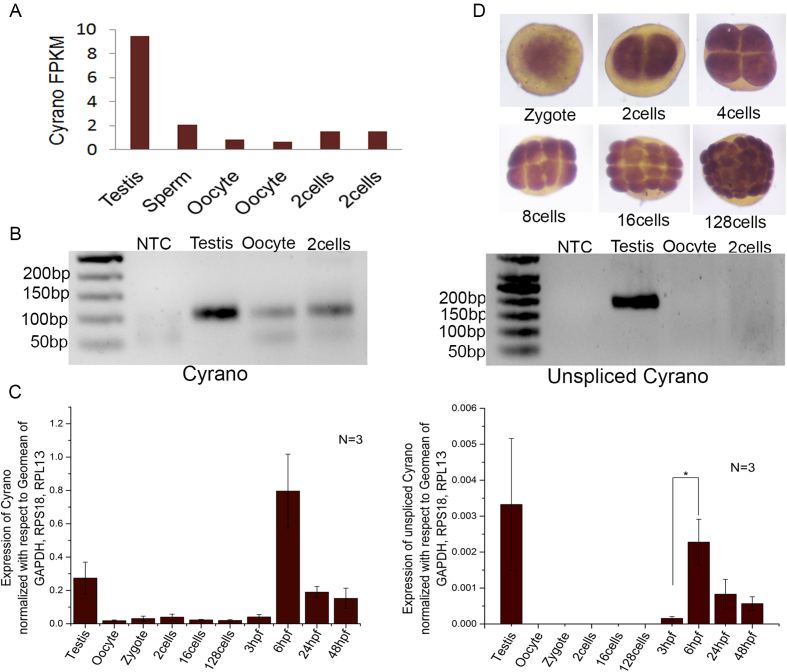
Interestingly, highly conserved lincRNA Cyrano (12) was found to be expressed in testis, oocyte and 2cell-stage thus it could be maternally and paternally inherited in zebrafish (Figure 3A). Cyrano expression from RNA-seq data corroborates with its expression obtained by RT-PCR (Figure 3B). Further, in order to unambiguously differentiate inherited Cyrano from zygotically expressed Cyrano, mRNA splicing is used as a handle. The maternally inherited proteome of zebrafish is rich in pre-mRNA splicing factors and ∼97.5% introns gets removed from maternally inherited RNAs (25). As expected, unspliced Cyrano transcripts were not detected in oocyte and 2cell-stage ruling out zygotic contribution to the inherited pool at 2cell-stage (Figure 3B). Expression of Cyrano and unspliced Cyrano was quantitated by qRT-PCR, which also detected the mature transcript but not unspliced Cyrano in testis, oocyte and 2cell-stage (Figure 3C). Zygotic Cyrano was first detected at 1000 cells stage (3 hpf) and later during gastrulation (6 hpf), both Cyrano and unspliced Cyrano were highly detected (Figure 3C). In situ hybridization also confirmed inherited Cyrano expression in the zygote (Figure 3D). Expression analysis of Cyrano from publicly available single cell RNA seq data (14) of human oocyte, zygote, 2cell-stage and 4cell-stage embryos found that Cyrano is maternally inherited (Supplementary Figure S2). Thus, sequence conservation as well as maternal inheritance in zebrafish and human made Cyrano as an interesting candidate to study further.
Figure 3.
Inheritance of conserved lncRNA Cyrano in zebrafish. (A) FPKM values of Cyrano expression in RNA-seq datasets of zebrafish testis, sperm, oocyte, 2cell-stage shows that Cyrano is present as early as 2cell-stage in zebrafish embryo. (B) RT-PCR validation of Cyrano and unspliced Cyrano expression in zebrafish testis, oocyte and 2cell-stage. (C) qPCR validation of Cyrano and unspliced Cyrano expression in zebrafish developmental stages. (D) Expression of Cyrano in zebrafish developmental stages checked by In situ hybridization. Error bar represents standard error of mean from N = number of independent biological replicates. *P value ≤0.05.
Loss of inherited Cyrano affects brain development
To investigate function of inherited Cyrano, it is important to selectively knock it down without affecting expression of zygotic Cyrano. In an earlier attempt to knockdown zebrafish Cyrano, Ulitsky et al. focused on total Cyrano not differentiating between inherited and zygotic Cyrano as inheritance of Cyrano was not known then (12).
Ulitsky et al. used two different types of morpholino antisense oligos (MOs) (12). One type of MO targets pre-RNA Cyrano splice site that disrupts RNA splicing. The other type of MO is complementary to the highly conserved site of Cyrano. After injection of splice-site MO at one cell stage zebrafish embryos, they observed reduced Cyrano transcript accumulation, whereas injection of conserved site MO did not affect transcript levels (12). As inherited Cyrano is already spliced, splice-site MO would be ineffective against inherited Cyrano. We rationalised that conserved site MO might remain stable several hours after injection in embryos and hence disrupt zygotic Cyrano functioning. Therefore, It appears that effects shown by Ulitsky et. al. were specific to zygotic Cyrano knockdown. So the function of inherited Cyrano remains elusive.
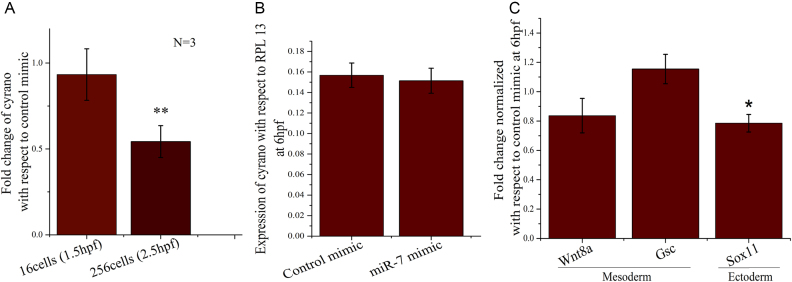
Cyrano has a conserved miR-7 binding site. Cyrano and miR-7 interact and mutually represses each other in embryonic stem cells (18). The ectopic over-expression of miR-7 may potentially degrade inherited Cyrano. Thus, we injected in vitro synthesized mature miR-7 duplex RNA in one cell stage of zebrafish. The concentration of miR-7 was optimised to degrade only inherited Cyrano but not zygotic Cyrano (Supplementary Figure S3). Over-expression of miR-7 (Supplementary Figure S3) and knockdown of inherited Cyrano were confirmed by qRT-PCR (Figure 4A). Inherited Cyrano successfully degraded at 256 cell-stage but zygotic expression at 6hpf was not changed (Figure 4A, B). Since brain development starts with ectoderm, we compared ectodermal markers to mesodermal markers after knockdown of inherited Cyrano. There was a small but significant change in the ectodermal marker sox11 but mesodermal markers wnt8a and goosecoid (gsc) remained unchanged upon knockdown of inherited Cyrano (Figure 4C).
Figure 4.
Knockdown of inherited Cyrano by ectopic over-expression of miR-7 mimic and its effect on germ layers. (A) 300 pg of In vitro synthesized miR-7 or control mimic injected at one cell stage of zebrafish embryos and inherited Cyrano level was checked by qPCR at 16cell-stage, 256cell-stage and (B) 6 hpf, (C) fold change of mesodermal (Wnt8a, Gsc) and ectodermal markers (Sox11) checked at 6hpf by qPCR in miR-7 mimic over-expressed conditions (normalized to control mimic; Fold change = 1). N = number of biological replicates. **P value ≤ 0.01, *P value ≤ 0.05.
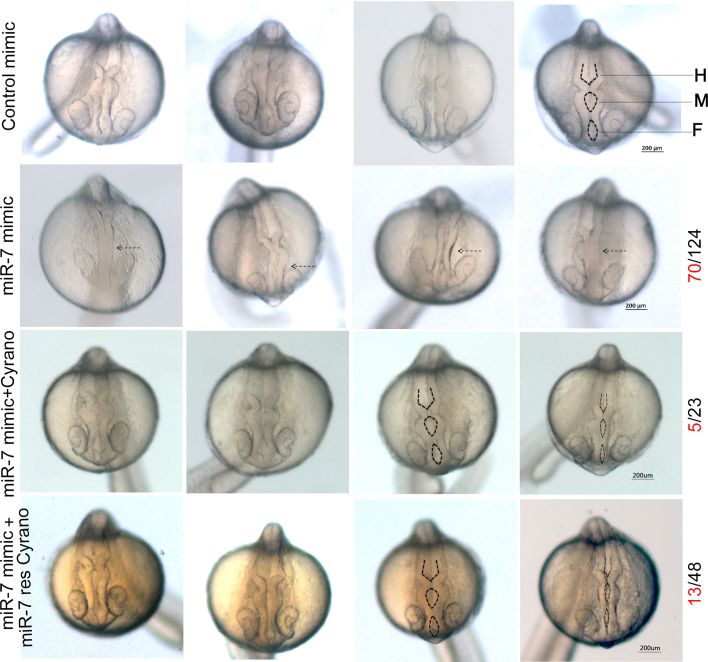
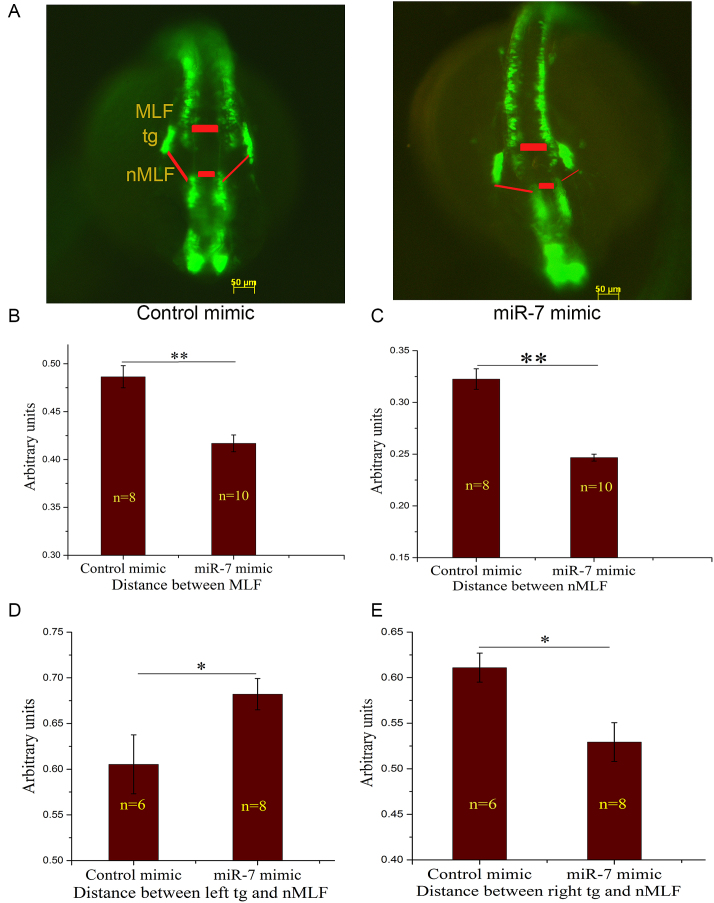
At 24hpf, the zebrafish brain is marked by completely open forebrain, midbrain and hindbrain ventricles. After knockdown of inherited Cyrano by miR-7 mimic, we identified severe deformities in the opening of midbrain and hindbrain ventricles (Figure 5) in 56% of the injected embryos. Since there was a possibility of disruption of other known or unknown targets of miR-7 that might produce the observed phenotypes, rescue of the defects by introduction of Cyrano was essential. Cyrano transcript partially rescued the miR-7 mediated defects, leaving a residual defect in 21% of the injected embryos. To rule out the possibility that the ventricle deformities were an indirect effect caused by ectopically injected Cyrano transcript sequestering miR-7 and allowing other miR-7 targets to escape, we designed a mutant Cyrano transcript. We made Cyrano that is impervious to miR-7 binding by deleting a 60 nucleotide region around the binding site of miR-7. The miR-7 resistant Cyrano, could also rescue the ventricle defects (73% of injected embryos) seen in the embryo injected with miR-7 alone. The residual 27% of defective embryos are perhaps due to some yet unknown miR-7 target. Cyrano at 6pg concentration was sufficient to rescue miR-7 mediated knockdown of inherited Cyrano at 24hpf (Figure 5). Thus, it was confirmed that knockdown of inherited Cyrano is the main cause of the phenotypes we observed at 24hpf. Further, to study the effect of knockdown of inherited Cyrano on brain development we used transgenic zebrafish HuC:kaede which labels mature neurons with GFP variant kaede. At 24hpf HuC:kaede zebrafish shows GFP labelled medial longitudinal fasciculus (MLF), nucleus of medial longitudinal fasciculus (nMLF) and trigeminal ganglion (tg). These brain structures make a network which takes signals from the eye, pass it on to midbrain where it gets processed and send back to the eye for eye movement. We found that the MLF-tg-nMLF architecture changed significantly after knockdown of inherited Cyrano (Figure 6A). Distance between left and right MLF was reduced in the inherited Cyrano knockdown embryos (Figure 6B). Similarly distance between left and right nMLF was reduced in the inherited Cyrano knockdown embryos (Figure 6C). This suggests an incomplete opening of midbrain and forebrain ventricles. Further, distance between left tg and nMLF increased significantly while right tg was located closer to the nMLF (Figure 6D, E). This suggests that, loss of inherited Cyrano disrupts the symmetric development of neuronal structures.
Figure 5.
Ectopic over-expression of miR-7 leads to defective ventricle opening and it is rescued by Cyrano. First panel shows control mimic injected zebrafish embryos, second panel with 300 pg of miR-7 mimic injected embryos, third panel has embryos injected with 6 pg of Cyrano transcript and 300 pg of miR-7 mimic and fourth panel has embryos injected with 6 pg of miR-7 resistant Cyrano transcript (miR-7 res Cyrano) along with 300 pg of miR-7 mimic imaged at 24 hpf. Defects were observed (shown by dotted arrow) in 70 out of 124 animals with miR-7 mimic injection and these defects were restricted to 5 out of 23 and 13 out of 48 animals after rescue with wild type Cyrano or miR-7 res Cyrano, respectively. Forebrain (F), Midbrain (M), Hindbrain (H) ventricles shown by dotted lines.
Figure 6.
Loss of inherited Cyrano by ectopic over-expression of miR-7 leads to defective brain development in 24 hpf embryos. (A) HuC: kaede zebrafish injected with control or miR-7 mimic imaged at 24 hpf to capture GFP labelled medial longitudinal fasciculus (MLF), nucleus of medial longitudinal fasciculus (nMLF) and trigeminal ganglion (tg). (B) Distance between MLF and (C) nMLF in control and miR-7 mimic over-expressed embryos measured, both the regions shown by red boxes in A. (D) Distance between left tg to nMLF and (E) right tg to nMLF in control and miR-7 mimic over-expressed embryos measured, as shown by red lines in A. Error bar represents standard error of mean, n = number of animals, **P value ≤ 0.01, *P value ≤ 0.05.
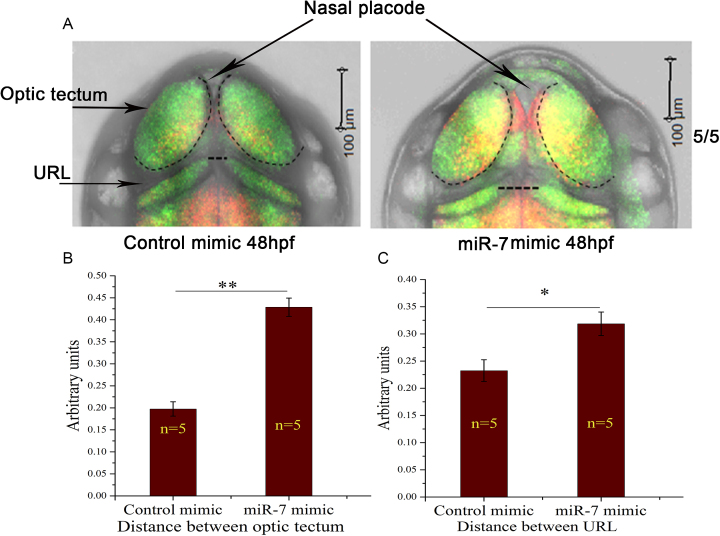
To study brain development after 48 hpf, we used double transgenic zebrafish embryos HuC:kaede X her4.1:mCherry which labels neural progenitors with mCherry in various niches and mature neurons with GFP. Inherited Cyrano knockdown showed enlargement of nasal placode that can be visualized using GFP labelled neurons at 48 hpf (Figure 7A). These embryos also showed increased distance between left and right optic tectum as well as distance between left and right upper rhombic lip (Figure 7B, C).
Figure 7.
Effect of loss of inherited Cyrano by ectopic over-expression of miR-7 in 48 hpf embryos. (A) Tg[HuC:kaede X her4.1:mCherry] zebrafish embryos injected with control or miR-7 mimic imaged at 48 hpf to capture optic tectum and Upper Rhombic Lip (URL). (B) Distance between left and right optic tectum and (C) URL in control and miR-7 mimic over-expressed embryos measured, both the regions marked by black dotted lines in A. Error bar represents standard error of mean, n = number of animals, **P value ≤ 0.01, *P value ≤ 0.05.
DISCUSSION
Neurodevelopment depends heavily on the genetic and epigenetic blueprint presented within gametes by parents. Non-coding RNAs are not only part of this blueprint in the gametes but they can also be important to decipher, implement and fine-tune it after the birth of the embryo. In this study, we systematically analysed RNA-seq data of zebrafish gametes and early developmental stages in order to find non-coding RNAs which are part of oocyte, sperm or both and sustain into the transcriptionally silent zygote. This reveals the previously underexplored inheritance of non-coding RNAs from both the gametes to the embryo. In future, an allele specific analysis may reveal the relative contribution of paternal and maternal non-coding transcripts. Genomes of vertebrate organisms encode thousands of lncRNAs of which only a fraction of it gets expressed in gametes and further deposited in the zygote. This suggests that inheritance of lncRNA is a highly selective process. Here, we report that one such conserved zebrafish lncRNA cyrano in part of the inherited pool, previously known for zebrafish brain development. We asked what can be the function of brain specific lncRNA in the gametes. To our surprise, we found that inheritance of Cyrano is also important for zebrafish brain development as knockdown of inherited Cyrano without changing zygotic Cyrano, altered brain morphology. It is well known that Cyrano interacts with several RNA binding proteins (RBPs), for example Cyrano sequesters HuR protein thus destabilizing cell cycle regulating cyclins A and D1 mRNAs (26). Cyrano is also known to interact with several mRNAs. Interaction of Cyrano with GAK mRNA in the cytoplasm reduces GAK mRNA half life and Cyrano knockdown elevates GAK and promotes aberrant mitoses (27). Further, Cyrano has miRNA binding sites and mutual repression of miR-7 and Cyrano is also known to be important for maintenance of self-renewing state of pluripotent cells (18). Considering the complexity of factors in the inherited pool it is highly likely that some of these Cyrano interacting factors are deposited in the zygote and required for proper function of Cyrano. For example, HuR is a well known protein in the deposited factor pool (28), that may interact with inherited Cyrano before the start of zygotic transcription. Overall, these studies, with our present observations point towards the possibility that inherited transcripts of Cyrano, with the help of other deposited factors might be regulating cell divisions during initial embryonic development.
The previous attempt to explore the single cell transcriptome of human pre-implantation embryo revealed >8000 maternally expressed lncRNAs (14), paving the way to explore their functions and increasing scope to study paternally inherited factors. Contribution of inherited factors from sperm was previously underestimated but surprisingly we found that sperm is also enriched with non-coding RNAs. Presence of these pre-made non-coding RNAs in gametes may serve as a link between parental experiences and postnatal life of the organism. Effects of maternal experiences during pregnancy such as stress, nutrition or infection were widely studied and shown to affect fetal growth, disease risk and resilience (29). However, maternally inherited transcriptome is shaped by maternal experiences just before or during ovulation and implantation. Thus reaching further, our study opens up new avenues to explore factors that modulate maternally deposited transcriptome formed during the critical period of ovulation and implantation. In future, it will be interesting to explore how stress, dietary factors or infection during ovulation and implantation changes the non-coding transcriptome of oocyte and thus subsequently inheritance of non-coding RNAs to zygote. In parallel, recent advancement in next generation sequencing technology has revealed the complex transcriptome and methylome of sperm (20), providing opportunity to explore the impact of paternal experiences in shaping up sperm transcriptome. Here, we project inherited non-coding RNAs as a signature of perturbations undergone by parents that warrants further detailed investigation in future.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the core imaging facility of CSIR-IGIB, New Delhi, India funded by CSIR Task Force project BSC0403 and the zebrafish facility. We thank Ashwani Kumar Choudhary, who created the tg(her4.1:mCherry) line which was used to derive the double transgenic line used here. We thank Hitoshi Okamoto and Tomomi Sato, RIKEN Brain Science Institute, Japan for providing tg(HuC:Kaede) line. MAS acknowledges to University Grant Commission (UGC), New Delhi, for fellowship.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Science and Technology (DST-SERB) [GAP0103]. Funding for open access charge: Department of Science and Technology (DST-SERB) [GAP0103].
Conflict of interest statement. None declared.
REFERENCES
- 1. Abrams E.W., Mullins M.C.. Early zebrafish development: It's in the maternal genes. Curr. Opin. Genet. Dev. 2009; 19:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pelegri F. Maternal factors in zebrafish development. Dev. Dyn. 2003; 228:535–554. [DOI] [PubMed] [Google Scholar]
- 3. Langdon Y.G., Mullins M.C.. Maternal and Zygotic Control of Zebrafish Dorsoventral Axial Patterning. Annu. Rev. Genet. 2011; 45:357–377. [DOI] [PubMed] [Google Scholar]
- 4. Flores M.V.C., Lam E.Y.N., Crosier K.E., Crosier P.S.. Osteogenic transcription factor Runx2 is a maternal determinant of dorsoventral patterning in zebrafish. Nat. Cell Biol. 2008; 10:346–352. [DOI] [PubMed] [Google Scholar]
- 5. Soni K., Choudhary A., Patowary A., Singh A.R., Bhatia S., Sivasubbu S., Chandrasekaran S., Pillai B.. miR-34 is maternally inherited in Drosophila melanogaster and Danio rerio. Nucleic Acids Res. 2013; 41:4470–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moravec C.E., Samuel J., Weng W., Wood I.C., Sirotkin H.I.. Maternal Rest/Nrsf regulates zebrafish behavior through snap25. J. Neurosci. 2016; 36:9407–9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilusz J.E., Sunwoo H., Spector D.L.. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009; 23:1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mercer T.R., Mattick J.S.. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013; 20:300–307. [DOI] [PubMed] [Google Scholar]
- 9. Kornienko A.E., Guenzl P.M., Barlow D.P., Pauler F.M.. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013; 11:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ng S.-Y., Johnson R., Stanton L.W.. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012; 31:522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iyengar B.R., Choudhary A., Sarangdhar M.A., Venkatesh K.V., Gadgil C.J., Pillai B.. Non-coding RNA interact to regulate neuronal development and function. Front. Cell. Neurosci. 2014; 8:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P.. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011; 147:1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pauli A., Valen E., Lin M.F., Garber M., Vastenhouw N.L., Levin J.Z., Fan L., Sandelin A., Rinn J.L., Regev A. et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012; 22:577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J. et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013; 20:1131–1139. [DOI] [PubMed] [Google Scholar]
- 15. Peng H., Shi J., Zhang Y., Zhang H., Liao S., Li W., Lei L., Han C., Ning L., Cao Y. et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012; 22:1609–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sharma U., Conine C.C., Shea J.M., Boskovic A., Derr A.G., Bing X.Y., Belleannee C., Kucukural A., Serra R.W., Sun F. et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016; 351:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J., Feng G.H., Peng H., Zhang X., Zhang Y. et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016; 351:397–400. [DOI] [PubMed] [Google Scholar]
- 18. Smith K.N., Starmer J., Miller S.C., Sethupathy P., Magnuson T.. Long Noncoding RNA Moderates MicroRNA Activity to Maintain Self-Renewal in Embryonic Stem Cells. Stem Cell Rep. 2017; 9:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. M. Westerfield The Zebrafish Book A Guide for the Laboratory Use of Zebrafish (Danio rerio). 2007; 5th ednEugene: University of Oregon Press. [Google Scholar]
- 20. Jiang L., Zhang J., Wang J.J., Wang L., Zhang L., Li G., Yang X., Ma X., Sun X., Cai J. et al. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 2015; 153:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harvey S.A., Sealy I., Kettleborough R., Fenyes F., White R., Stemple D., Smith J.C.. Identification of the zebrafish maternal and paternal transcriptomes. Development. 2013; 140:2703–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang L., Zhang Z., He S.. Both male-biased and female-biased genes evolve faster in fish genomes. Genome Biol. Evol. 2016; 8:3433–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. DespicV. D.M., Gu M., Krishnan J., Zhang J., Herzel L., Straube K., Gerstein M.B., Butter F., Neugebauer K.M. et al. Dynamic RNA-protein interactions underlie the zebrafish maternal-to-zygotic transition. Genome Res. 2017; 27:1184–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim J., Abdelmohsen K., Yang X., De S., Grammatikakis I., Noh J.H., Gorospe M.. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res. 2016; 44:2378–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim J., Noh J.H., Lee S.K., Munk R., Sharov A., Lehrmann E., Wang W., Abdelmohsen K., Gorospe M.. LncRNA OIP5-AS1/cyrano suppresses GAK expression to control mitosis. Oncotarget. 2017; 8:49409–49420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li X., Lu Y.C., Dai K., Torregroza I., Hla T., Evans T.. Elavl1a regulates zebrafish erythropoiesis via posttranscriptional control of gata1. Blood. 2014; 123:1384–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bale T.L. Epigenetic and transgenerational reprogramming of brain development. Nat. Rev. Neurosci. 2015; 16:332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.