Abstract
Cystic fibrosis (CF) is a common genetic disease caused by mutations in the gene coding for cystic fibrosis transmembrane conductance regulator (CFTR). Although CF affects multiple organ systems, chronic bacterial infections and inflammation in the lung are the leading causes of morbidity and mortality in people with CF. Complementation with a functional CFTR gene repairs this defect, regardless of the disease-causing mutation. In this study, we used a gene delivery system termed piggyBac/adenovirus (Ad), which combines the delivery efficiency of an adenoviral-based vector with the persistent expression of a DNA transposon-based vector. We aerosolized piggyBac/Ad to the airways of pigs and observed widespread pulmonary distribution of vector. We quantified the regional distribution in the airways and observed transduction of large and small airway epithelial cells of non-CF pigs, with ∼30–50% of surface epithelial cells positive for GFP. We transduced multiple cell types including ciliated, non-ciliated, basal, and submucosal gland cells. In addition, we phenotypically corrected CF pigs following delivery of piggyBac/Ad expressing CFTR as measured by anion channel activity, airway surface liquid pH, and bacterial killing ability. Combining an integrating DNA transposon with adenoviral vector delivery is an efficient method for achieving functional CFTR correction from a single vector administration.
INTRODUCTION
Cystic fibrosis (CF) is an autosomal recessive disease affecting >70 000 people worldwide, with ∼1000 new cases of CF each year (Cystic Fibrosis Foundation, www.cff.org). CF affects multiple organ systems; however, lung disease is the leading cause of morbidity and mortality in people with CF. CF is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene which results in dysregulated Cl− and HCO3− anion transport in the airways. Reduced levels of Cl− and HCO3− in CF airways leads to a relatively acidic airway surface liquid (ASL) pH, impaired mucociliary transport (MCT), decreased bacterial killing ability, and increased mucus viscosity (1,2). There are >2000 CFTR mutations (www.genet.sickkids.on.ca/cftr/app) potentially associated with CF. Small molecule potentiators and correctors are therapeutic for a small percentage of CF patients with specific mutations (3–5), however there is still a great need to develop a universal treatment for people with CF. Gene therapy for CF is a promising strategy regardless of the disease-causing mutation. Importantly, CFTR delivery prior to the onset of lung disease could prevent CF-related lung complications.
CF lung disease develops following bacterial colonization and onset of inflammation (as reviewed in (6)). Chronic inflammation causes permanent structural damage of the airways (7). Defective inflammatory responses and innate immunity dysregulation have been linked to CFTR deficiency (8). Evidence suggests that CF lung disease involves large and small airways (9,10), however thick large airway walls and narrow lumens develop within the first few months of life (11–13). Restoring CFTR function to the appropriate airway cells in both the large and small airways is critical to prevent the onset or progression of CF-related lung disease.
To achieve persistent expression in airway cells, we created a genomic integrating hybrid vector termed piggyBac/adenovirus (Ad). The DNA transposon piggyBac is comprised of a two-part system including a transposon (i.e., terminal repeats flanking a gene of interest) and a transposase that provides catalytic activity to mediate genomic integration. Using piggyBac/Ad, we reported transposase-dependent transgene expression for one year in immunodeficient mice (the duration of the experiment) (14). Yant and colleagues (15) reported using helper-dependent adenovirus (HDAd), an Ad vector devoid of all viral coding genes, to deliver a Sleeping beauty (SB) transposon. In those studies, hepatic delivery of SB/HDAd to mice led to persistent expression for at least 22 weeks when co-delivered with the transposase following a partial hepatectomy (15). SB/HDAd reproducibly results in persistent expression in vivo (16,17). We selected the piggyBac transposon because it has a large carrying capacity, does not require a Flp-recombinase step, and does not leave a footprint following transposition (18). In our previous studies, we mapped piggyBac transposase-dependent integrations in HeLa cells in vitro and in the mouse genome in vivo. Delivery of piggyBac/Ad encoding CFTR to primary human CF airway epithelia in vitro persistently corrected the anion channel defect for at least 17 weeks (14). Thus, this hybrid vector system was validated in immortalized epithelial cells, primary cultures of airway epithelial cells, and in the lungs of a small animal model (14).
Pigs are similar to humans in lung anatomy, size, and physiology (19). CF pigs develop lung disease similar to humans with CF (20,21). Previous studies indicate that by 6 weeks of age CF pigs have airway inflammation and remodeling. While the airways of the newborn CF pigs are free of infection and inflammation, they exhibit a reduced ASL pH and bacterial eradication defect (1,20). Investigation of these disease characteristics and their physiologic underpinnings allowed for novel assay development and validation (22,23) that influenced our study design and the selection of endpoints.
In this study, we delivered piggyBac/Ad encoding GFP and Gaussia luciferase to the airways of newborn non-CF pigs and examined expression 5 days later. We determined vector distribution throughout the airways by quantifying the number of cells transduced and identified cell types transduced. Using ‘gut-corrected’ CF pigs (24), we delivered piggyBac/Ad-CFTR to the airways and measured phenotypic correction by electrophysiology, ASL pH, and bacterial killing ability. Here, we report quantification of whole lung vector distribution of piggyBac/Ad vector and phenotypic correction in a large animal CF model.
MATERIALS AND METHODS
Constructs and vector production
The hybrid piggyBac/Ad vector was designed with the piggyBac transposon terminal repeats (TRs) flanking an expression cassette, such as CMV driving eGFP-T2A-Gaussia Luciferase (gLuc) or F5Tg83 promoter (25) driving a codon optimized pig CFTR cDNA (accession no. KT184306) (22). The piggyBac/Ad constructs were modified from the hybrid vector previously described (14). Briefly, the eGFP-T2A-gLuc or CFTR cassette was cloned into the piggyBac/Ad vector using HindIII and SpeI. Ad-transposase (iPB7) was previously described (14). PiggyBac/Ad, Ad-transposase, Ad-CMV-eGFP, and Ad-CMV-mCherry were produced, purified, and titered by the University of Iowa Viral Vector Core (medicine.uiowa.edu/vectorcore). Ad serotype 5 was used in all studies.
In vivo viral vector delivery
CF pigs were generated by homologous recombination as previously described (26) and the ‘gut-corrected pigs’ were generated by somatic cell nuclear transfer cloning (24). Wild-type pigs (hereafter referred to as non-CF pigs) and CF pigs were acquired from Exemplar Genetics (Coralville, IA). All pigs received vector within one week of birth and in the non-CF pig studies, both male and female pigs were used. Vector titers ranged from 2 × 1010 to 2 × 1011 pfu/ml. The vector was mixed with lysophosphatidylcholine (LPC) for a final concentration of 0.1% LPC (27) in a total volume of 2 ml (∼2 × 1010 pfu/kg). Pigs were anesthetized using inhaled 2% isoflurane and vectors were aerosolized into pig airways by intratracheal instillation using the PennCentury® Microsprayer (Wyndmoor, PA). All pigs were housed for 3 or 5 days at the University of Iowa during the duration of the studies. All animal procedures were reviewed and approved by the University of Iowa Institutional Animal Care and Use Committee (IACUC), in accordance with the United States Department of Agriculture and National Institutes of Health guidelines.
Ussing chamber studies
Freshly excised tracheal explant tissues were mounted in Ussing chambers to measure their bioelectric properties (24). The apical and basolateral chambers were bathed in symmetrical Ringer's solution (135 mM NaCl, 5 mM HEPES, 0.6 mM KH2PO4, 2.4 mM K2HPO4, 1.2 mM MgCl2, 1.2 mM CaCl2, 5 mM dextrose), apical solution was replaced with a low Cl− solution, and CFTR Cl− current was measured as previously described (28).
Bacterial killing assay
Pigs were sedated intramuscularly with ketamine (20 mg/kg) and xylazine (2 mg/kg) (both Akorn Animal Health, Lake Forest, IL, USA) and maintained with intravenous propofol (Fresenius Kabi USA, Lake Zurich, IL, USA; 1 mg/kg) for the pH and bacterial killing assays. During anesthesia, oxygen levels, temperature, pulse, and respiratory rate were monitored. Pigs were euthanized using Euthasol (Virbac, Forth Worth, TX, USA; 90 mg/kg). The bacterial killing assay was developed by Pezzulo et al. (1). Briefly, S. aureus (strain SA43) was immobilized on gold electron microscopy grids through a streptavidin and biotin interaction. Bacterial-coated grids were placed on the tracheal surface of a sedated pig for 1 min and queried for cell death by a propidium iodide stain (Live/Dead Bacterial Viability Assay, Invitrogen, Carlsbad, CA, USA). Grids were imaged by confocal microscopy and analyzed using ImageJ (ImageJ, Schneider, CA, USA).
Immunofluorescence
Lung regions were fixed in 4% paraformaldehyde, placed in sequential sucrose solutions, and embedded in OCT for freezing. Tissues were sectioned using a microtome. GFP positive images were quantified using ImageJ. The percentage of manually counted GFP positive cells was calculated as a ratio to the number of DAPI positive cells. Frozen sections were probed with anti-acetyl tubulin at 1:200 dilution (Cell Signaling Technology, D20G3, K40, Danvers, MA) and anti-human cytokeratin 5, 8 (CK5, 8) polyclonal antibody at 1:5 dilution (US Biological Life Sciences, 168994, Salem, MA). Basal cells were detected by anti-nerve growth factor receptor (NGFR) (1:100 dilution) (ThermoFisher Scientific, 14-9400-80, Waltham, MA, USA) and anti-cytokeratin 14 (CK14) (1:100) (Abcam, ab9220, Cambridge, UK). CFTR was detected by immunohistochemistry and immunofluorescence using mouse monoclonal CFTR 769 from the UNC/CFF consortium (29–31). Slides were counterstained and mounted with Vectashield containing DAPI (Vector Laboratories, Burlingame, CA, USA).
Quantitative Real-Time PCR
Genomic DNA was purified using the DNeasy Genomic Isolation Kit (Qiagen, Germantown, MD, USA). RNA was purified using the RNeasy Lipid Tissue Mini Kit (Qiagen, Germantown, MD) with RNase-Free DNase Set (Qiagen, Germantown, MD) and cDNA libraries were generated using Superscript IV Vilo Master Mix (ThermoFisher, Waltham, MA). GFP genomic DNA and mRNA abundance was quantified by qPCR using SYBR green (Applied Biosystems, Foster City, CA). Primer sequences for GFP were: Forward 5′-GGCGACGGCCCCGTGCTGCTGC-3′; Reverse 5′-CACGAACTCCAGCAGGACCATG-3′. For genomic DNA, Pig ribosomal protein L4 (RPL4) was used as a genomic standard (32) (Forward: 5′-AGCGCTGGTCATGTCTAAAG-3′; Reverse: 5′-TTCCAGGCCTTAAGCTTATTA-3′. β-actin was used as a standard for mRNA expression: Forward: 5′ - CTGCGGCATCCACGAAAC - 3′ Reverse: 5′-GTGATCTCCTCCTGCATCCTGTC-3′.
Statistics
All statistically significant differences were calculated using one-way ANOVA in Graphpad Prism (GraphPad Software, La Jolla, CA, USA). Unless otherwise noted, all data are presented as a mean ± SE. P < 0.05 was considered statistically significant.
RESULTS
Distribution of piggyBac/Ad in pig pulmonary airways
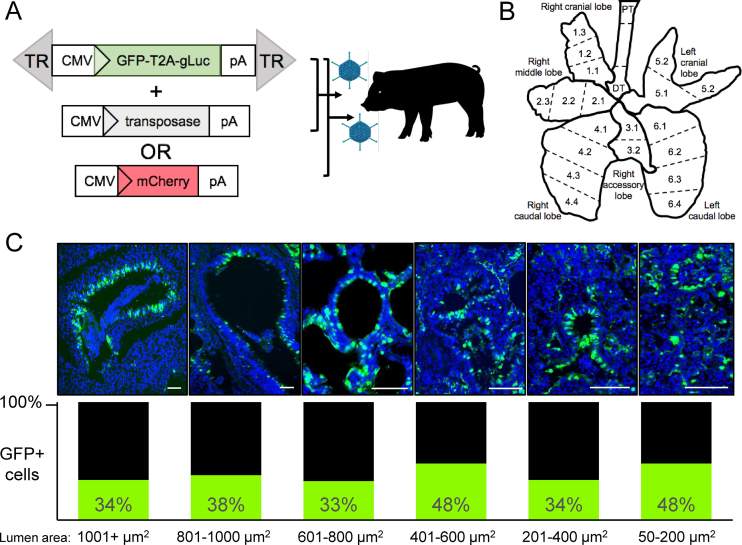
We performed a thorough analysis of the number and types of cells transduced in pig airways after transduction with the integrating hybrid piggyBac/Ad vector. This vector confers persistent gene expression in vitro and in vivo (14). The nonviral DNA transposon piggyBac is comprised of terminal repeats (TRs) that flank a transgene of interest. The GFP-T2A-Gaussia luciferase (gLuc) transposon cassette was cloned into the E1 region of Ad (Figure 1A). In the absence of the transposase, piggyBac/Ad is indistinguishable from first generation Ad5. In the presence of the transposase, the TRs and gene expression cassette can be integrated into the host genome. In this study, we aerosolized piggyBac/Ad-GFP-T2A-gLuc co-delivered with Ad-transposase or Ad-mCherry to the trachea of newborn non-CF pigs (Figure 1A). The Ad-mCherry served as a negative control for the transposase and as a means to measure how frequently two Ad vectors transduced the same cell. Five days post-transduction, serum, bronchoalveolar lavage (BAL), and lung tissues were collected. BAL was evaluated for neutrophil infiltration (Supplemental Figure S1A) and cytokine levels were measured in serum and BAL (Supplemental Figure S1B). Each of the six lung lobes were subdivided into two to four sections for whole-lung analysis. The trachea was divided into proximal and distal regions (Figure 1B). Tissue blocks were sectioned, slides were counterstained with DAPI, and imaged. We next quantified the number of GFP positive cells in large (>1001 μm2) and small (50 μm2) conducting airways using ImageJ. We consistently observed greater than 30% transduction in airways ranging from large to small (Figure 1C). GFP positive cells were identified in airways throughout all regions of all lobes. In addition, secreted gLuc expression was readily observed in both the BAL and serum (Supplemental Figure S2). We note that gLuc in the serum likely results from vector expression in alveolar cells and respiratory epithelium as previously observed (33–35).
Figure 1.
Transduced cells in conducting airways. (A) Ad5 carrying a piggyBac transposon with the terminal repeats (TRs) flanking a reporter gene cassette was co-delivered with Ad-transposase or Ad-mCherry and 0.1% LPC via intratracheal instillation to newborn non-CF pigs. 5 days post-delivery, lungs were collected for analysis. TR = piggyBac terminal repeats, pA = poly adenylation signal. (B) Schematic of pig lung indicates how tissues were portioned for analyses. PT = proximal trachea, DT = distal trachea. Each of the 6 lobes were divided into two to four regions, as indicated numerically. (C) Quantification of GFP positive cells in conducting airways of area sizes ranging from 50 to <1001 μm2 (n = 6 pigs). Airway sizes were calculated using ImageJ. Scale bar = 500 μm.
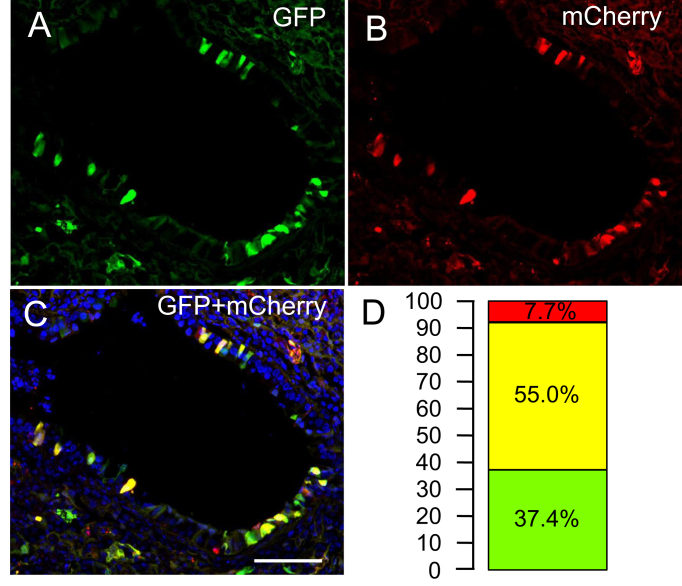
For transposition to occur, both components of the integrating piggyBac system must be in the same cell. As an indirect measure of how frequently the transposon and transposase are present in the same cell, we co-delivered piggyBac/Ad-GFP-T2A-gLuc with an Ad-mCherry vector. To measure the percentage of dual positive transduced cells in vivo, GFP (Figure 2A) and mCherry (Figure 2B) positive cells were quantified by counting. We observed that 55% of fluorescent cells were dual positive (Figure 2C, D). These data suggest that if an airway cell is permissive to Ad transduction, then at least two virions are likely to transduce the same cell.
Figure 2.
Dual positive GFP and mCherry cells in the airways. Individual channels of (A) GFP, (B) mCherry or (C) merged image of transduced airways. (D) Percentage of GFP, mCherry, and dual positive cells among transduced airways is indicated. Scale bar = 500 μm (n = 3).
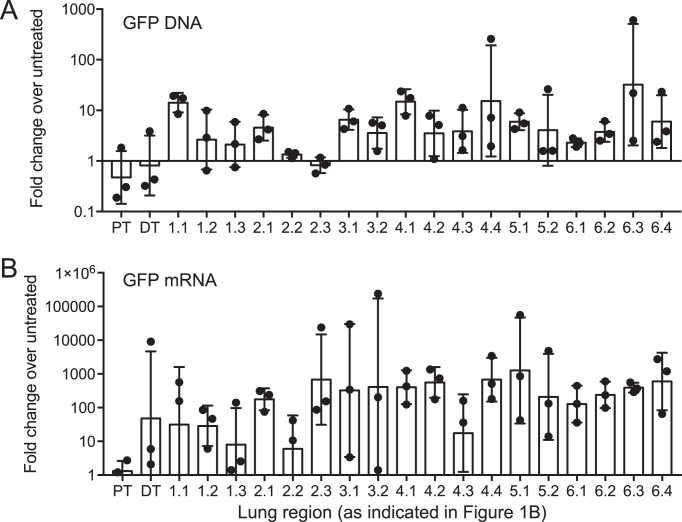
To determine the relative distribution of GFP throughout pig airways, we performed quantitative real-time PCR on genomic DNA or quantitative reverse-transcriptase PCR on mRNA isolated from 20 different regions of the lung from each pig (as indicated in Figure 1B). In general, we observed GFP genomic DNA and mRNA in all regions of each lung lobe (Figure 3A, B). These results, in combination with the fluorescence studies, suggest that aerosol delivery throughout the large and small airways of piggyBac/Ad to newborn pigs results in widespread airway distribution and expression.
Figure 3.
Distribution of GFP in pig lungs. (A) Quantitative real-time PCR of relative GFP levels in genomic DNA from each region of the pig lung 5 days post-delivery of piggyBac/Ad-GFP. GFP levels are normalized to the housekeeping gene RPL4 and fold changes are normalized relative to untreated animals (n = 3). (B) GFP mRNA expression levels from each region of the pig lung. Gene expression is normalized to the housekeeping gene pig β-actin, and fold changes are expressed relative to levels in untreated animals (n = 3). X-axis labels correspond to Figure 1B. Data are presented as the geometric mean ± SE.
Cell types transduced
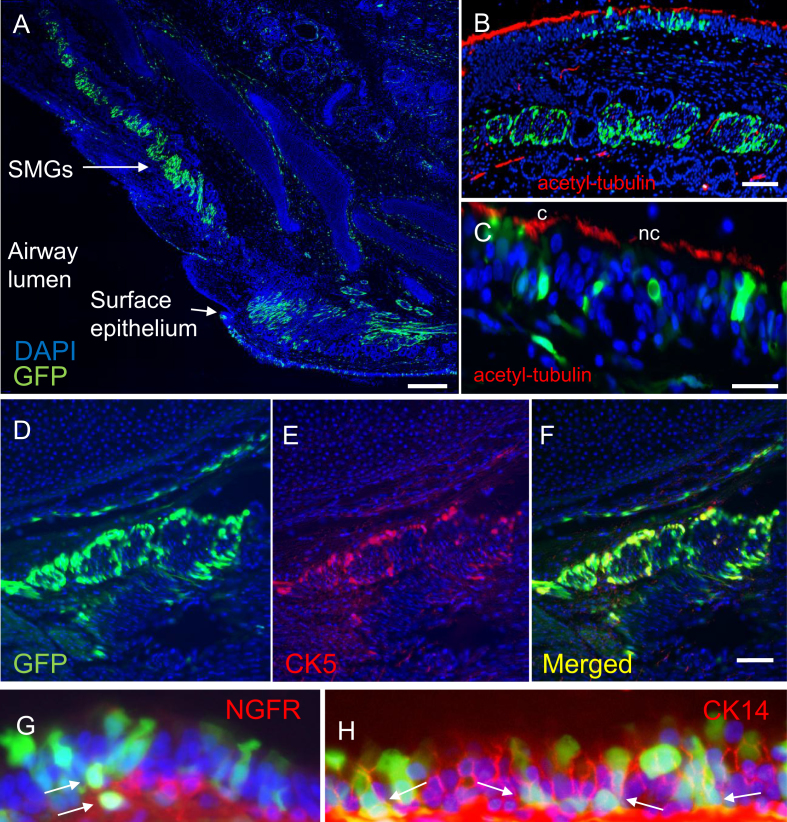
Previous reports show that Ad5 transduces all major cell types in the airways including ciliated cells, non-ciliated cells, goblet cells, basal cells, and submucosal glands (SMGs) (36). Here, we used immunofluorescence to visualize cell types transduced by piggyBac/Ad-GFP-T2A-gLuc. We observed GFP positive cells in the surface epithelium of the distal trachea as well as high levels of transduction in the SMGs along the trachea (Figure 4A). At a higher magnification, we found that both ciliated (c) (anti-acetyl α-tubulin) and nonciliated (nc) cells were positive for GFP (Figure 4B, C). Additionally, basal cells (anti-CK5) within the SMGs were GFP positive (Figure 4D, E, and F). GFP positive basal cells (anti-NGFR or anti-CK14) in the surface epithelium of the conductive airways were also observed (Figure 4G, H). As expected, piggyBac/Ad has wide cell tropism in the airways; however, the levels of SMG transduction were particularly remarkable.
Figure 4.
piggyBac/Ad-GFP transduces multiple cell types. Tissue from each region of the pig lung (as depicted in Figure 1B) was sectioned, counterstained with DAPI, and imaged. (A and B) piggyBac/Ad-GFP transduces submucosal glands (SMGs) throughout the trachea. (B and C) Tissues stained with acetyl-tubulin (red) show that piggyBac/Ad-GFP transduces ciliated (c) and non-ciliated (nc) cells at the airway surface. (D–F) piggyBac/Ad-GFP co-localizes with CK5 positive basal cells (red) within submucosal glands (n = 6). Scale bars for A = 250 μm; B–F = 500 μm. (G and H) piggyBac/Ad-GFP transduces basal cells at the airway surface. Arrows indicate GFP positive cells that co-localize with red (G) NGFR or (H) CK14 immunostaining. Images acquired at 63× magnification (n = 6).
Mapping integrations in vivo
Genomic integration may be a requirement for life-long expression of a therapeutic gene. Previously, we showed that piggyBac/Ad resulted in bona fide transposase-mediated integrations both in vitro and in mice in vivo (14). Here, we sequenced genomic DNA isolated from the distal trachea and mainstem bronchi of pigs following piggyBac/Ad delivery (37,38). As might be expected at 5 days post-delivery, the vast majority (>99.99%) of the piggyBac transposons mapped to the Ad genome. This indicates that at this early time point, episomal vector predominates and is a ready source of PCR amplification. However, our goal was to demonstrate that delivery of the piggyBac/Ad vector can support transposase-mediated integration in pig airways in vivo. As shown, we mapped 264 integrations that were widely distributed across all chromosomes (Supplemental Figure S3). Consistent with our previous observations (14,39), we observed a non-specific pattern of genomic integrations. There was no predilection of piggyBac to integrate near or away from DNase I hypersensitive sites, CpG islands, GC regions, or refSeq genes (Supplemental Figure S4).
Phenotypic correction of CF pigs
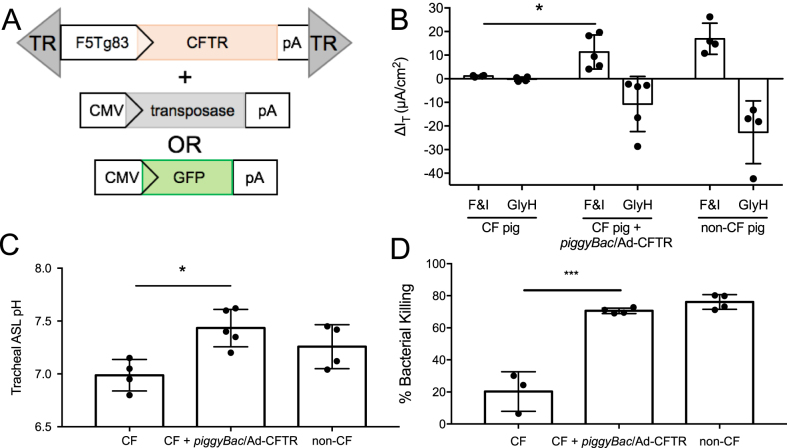
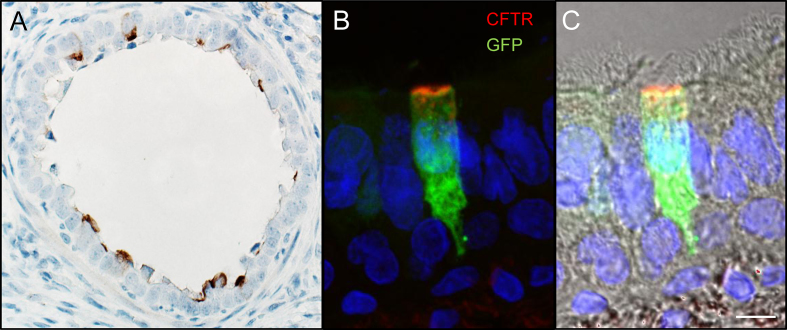
Functional correction in a large animal model is vital to validating piggyBac/Ad as a gene therapy vector. We next delivered piggyBac/Ad-CFTR with Ad-transposase or Ad-GFP (Figure 5A) by aerosol to five newborn CF pigs. Three days post-transduction, we measured phenotypic correction by several methods. In freshly excised tracheal tissues from pigs that received piggyBac/Ad-CFTR, we observed increased Cl− current in response to the cAMP agonists forskolin and IMBX (F&I) and decreased transepithelial Cl− current in response to the CFTR inhibitor GlyH-101 (GlyH). In contrast, these responses are nearly absent in tissues from untreated CF pigs (Figure 5B). In these same CFTR vector-treated animals, we also observed a restoration of ASL pH (Figure 5C) and bacterial killing ability (Figure 5D) to non-CF levels. Similar to reporter gene expression (Figure 1C), CFTR expression is readily detected in the conducting airways as determined by immunostaining (Figure 6A). In addition, CFTR was expressed at the apical membrane of conducting airway epithelia and co-localizes with GFP positive cells in vivo (Figure 6B, C). Based on these end-point metrics, delivery of CFTR by a piggyBac/Ad hybrid vector corrects several CFTR-dependent defects in vivo. In total, complementing CFTR using piggyBac/Ad in CF pigs rescued important phenotypic defects including Cl− current, ASL pH, and bacterial killing.
Figure 5.
piggyBac/Ad-CFTR corrects the CF pig phenotype in vivo. (A) piggyBac/Ad-CFTR was co-delivered with either Ad-CMV-transposase or Ad-CMV-GFP. (B) Anion channel current was measured in freshly excised tracheal tissues from naïve CF pigs, CF pigs that received piggyBac/Ad-CFTR (3 days post-delivery), or naïve non-CF pigs. Change in current was measured in response to F&I and GlyH. (C) Tracheal ASL pH was measured in vivo in a sedated pig. (D) Bacterial killing was quantified by immobilizing S. aureus on electron microscopy grids through streptavidin and biotin conjugation. Grids were placed on the tracheal surface of a sedated pig for 1 minute, stained with propidium iodide, and imaged by confocal microscopy. Live/dead bacteria were quantified using ImageJ (n = 4).
Figure 6.
CFTR delivered by piggyBac/Ad is expressed at the apical surface of airway cells. (A) Tissue sections were stained for CFTR using immunohistochemistry. Image taken at 40× objective. (B) piggyBac/Ad-CFTR was co-delivered with Ad-GFP to visualize co-transduction by two Ad vectors. Conducting airways show CFTR expression at the apical surface of airway epithelial cells. (C) CFTR and GFP immunofluorescence overlayed with transmitted light shows localization in ciliated cells at the airway surface. Scale bar = 10 μm (n = 3).
DISCUSSION
Here we present two methods for assessing gene transfer efficacy in the airways of a large animal model in vivo: quantifying the regional distribution of reporter gene expression and measuring phenotypic correction following therapeutic gene delivery. To investigate the deposition of piggyBac/Ad particles following aerosolization to non-CF pigs, we systematically divided pig lungs into twenty regions and assessed GFP expression in each sample. We found high levels of transduction of surface airway epithelia and submucosal glands by immunofluorescence and whole lung distribution at the DNA and mRNA levels. In CF pigs, we delivered piggyBac/Ad-CFTR and assayed for functional correction by quantifying the change in transepithelial current in response to cAMP agonists or CFTR inhibitor in freshly excised tracheal tissue. We also measured ASL pH and bacterial killing in the trachea. In total, we quantified vector distribution in pig airways and show phenotypic correction of a large animal CF model using a piggyBac/Ad vector carrying CFTR.
These studies are the first evidence to demonstrate vector distribution and transgene expression following vector delivery in a large animal model. We transduced cells at the airway surface and submucosal glands. CFTR localizes to the apical surface of airway epithelium to transport Cl− and HCO3− anions and is also highly expressed in the submucosal gland cells (40). CF lung disease involves the large and small airways, thus expressing CFTR in the appropriate cell types in both large and small airways will likely have functional consequences in delaying or preventing onset of lung disease. Here, we show widespread distribution throughout all lung lobes and in appropriate cell types in pig airways.
For this short-term (3–5 day) study, we delivered Ad vectors to newborn pigs. Although newborn CF pigs are free of inflammation and airway obstruction (20), the status of airway inflammation and responses to viral vectors may be quite different in older pigs. Future studies evaluating delivery and expression in older CF pigs will determine if delivering CFTR after onset of lung disease is as beneficial as delivering to newborn pigs. We note that in this study piggyBac transposon and transposase vectors were delivered separately. If they were delivered by a single vector, transposition efficacy would likely be increased and the vector load would be halved. Single vector delivery of piggyBac has been demonstrated by plasmid (41); however, generating a combined transposon/transposase viral vector is challenging because of unwanted transposition in the producer cells. Such transposition results in packaging of vector lacking the transposon. The use of inducible promoters to drive transposase expression in target cells is a potential strategy to overcome this challenge.
CF clinical trials using Ad vectors
Ad-based vectors were the first evaluated as a potential CF gene therapy vector in the lung. Since 1992, nine separate Phase 1 CF clinical trials using 1st or 2nd generation Ad-based vectors were approved (reviewed in (42)), with the last published in 2001 (43,44). In total, these clinical trials suggested that modest transduction efficiency, transient expression, and immune responses would preclude Ad from progressing as a candidate gene therapy vector. Since that time, much has been learned about Ad biology. For example, the receptor was assumed to be apically localized on airway cells simply because Ad is a respiratory virus. However, in 1997, the basolaterally localized Coxsackievirus and Adenovirus receptor (CAR) (45) was reported to be the principle cellular receptor (46). Considerable research has identified multiple strategies to facilitate receptor access for improved airway transduction efficiency (47–49). Vector co-administration with lysophosphatidylcholine (LPC), a natural airway surfactant, markedly increases viral transduction of Ad in the lung (27).
A major hurdle for Ad as a gene transfer vector for CF gene therapy is the innate and adaptive immune responses that result in transient transgene expression in the airways (50–54). Cytotoxic T cells target and eliminate transduced airway cells (55) and neutralizing antibodies prevent vector readministration (56,57). In this study at 5 days post-delivery, there was no difference in the percentage of polymorphonuclear leukocytes (PMNs) compared to naïve pigs (Supplemental Figure S1A). Adenoviral genes activate multiple signaling pathways that transiently induce expression of several proinflammatory cytokines, including TNF-α, IL-1β, IL-6, IFN-γ, (58) IL-2, IL-10, (59) and IL-8 (60). Transgene expression begins to decrease within 24 hours post-delivery(53) and cytokine levels are substantially decreased within 10 days of delivery (61). In our studies, we performed ELISAs at 5 days post-delivery on the BAL and serum (Supplemental Figure S1B). At 5 days post-Ad delivery, cytokine levels were no different than untreated controls; however, inflammation would likely have been observed at earlier timepoints (62).
Helper-dependent Ad
We previously validated piggyBac/Ad for expression and persistence in mice and showed that expression persisted in SCID mice for 1 year when co-delivered with the transposase. Moving forward, we will transition from a first generation Ad delivery vector to piggyBac/helper-dependent Ad (HDAd) vector carrying CFTR to validate persistence in a large animal model in vivo. HDAd retains a high transduction efficiency in the lung but lacks all viral coding genes, yielding a lower immunogenicity (63). Multiple studies highlight the efficacy of HDAd, particularly in a large animal pig model (64). Hu and colleagues evaluated the ability to readminister HDAd vectors in the lung without loss of gene expression. Persistent gene expression following readministration was attributed to blunted immune responses and reduced inflammation, as shown by decreased neutralizing antibody responses against Ad capsid proteins (64–67). HDAd delivery to the lungs also led to transgene expression in the submucosal glands (36). Transducing a progenitor cell population such as basal cells and submucosal glands is important to achieve persistent CFTR expression and repopulate the surface airway epithelium following cell turnover (68). In future studies using piggyBac/HDAd, we will monitor persistence using Gaussia luciferase (gLuc) as a sensitive reporter gene that can be measured in either BAL or serum (Supplemental Figure S2).
Current knowledge of vector integration patterns
Life-long gene expression from a single dose of a gene delivery vector likely requires genomic integration. Integrating vectors show promise for treating genetic diseases; however, there is inherent risk when introducing foreign DNA into a cell. Insertional mutagenesis may disrupt normal cell functions by inactivating an essential host gene or inappropriately causing expression of an undesirable gene. To date, malignant cell transformation after vector-mediated insertional mutagenesis has only been observed in three clinical entities including X-linked severe combined immunodeficiency, chronic granulomatous disease, and Wiskott-Aldrich syndrome, all of which occurred in conjunction with the use of first-generation murine leukemia virus (MLV)-based vectors harboring LTRs with strong enhancer/promoter sequences (69–73). Other reports suggest that, in T cells, piggyBac has an integration pattern similar to MLV. Specifically, biases towards DNase I hypersensitive sites, CpG islands, and transcription start sites were observed (74–76). However, the findings from this study and our previously published results (14,39) suggest that piggyBac integrates in a non-specific pattern. This may suggest that piggyBac integration patterns vary by cell type or delivery platforms.
Restoring CFTR in the appropriate airway cell types could slow or prevent the onset of lung disease in people with CF. Perhaps more importantly, transduction of progenitor cells with the capacity to repopulate the surface airway epithelium to continually express CFTR may prevent the need for repeat administration. In this study, we show transduction of multiple airway surface cell types known to express CFTR including NGFR and CK14 positive basal cells of surface airway epithelia and cytokeratin 5 positive cells in the submucosal glands that could have progenitor capacity. We show phenotypic correction in a CF pig model including anion channel activity, ASL pH, and bacterial killing ability. Additionally, we observed CFTR localized to the apical surface of the appropriate airway cells where anion transport is known to occur. Thus, we validated a hybrid piggyBac/Ad transposon system in a large animal model, including a relevant model that develops CF lung disease. Overall, these studies contribute an important advancement for developing an integrating vector for the preventative treatment of CF airway disease. Future studies will focus on long-term phenotypic correction.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Peter Taft, Nick Gansemer, Mallory Stroik, Brie Hilkin, Ni Li, Marisa Modjeski, Ana De La Torre, Emma Tighe, and Elizabeth Shirazi for their technical assistance in these studies. We thank our lab members who volunteer their time to feed and care for the animals during the duration of experiments. We thank the Iowa Office of Animal Resources and the animal caretakers. We thank Phil Karp and the In Vitro Models and Cell Culture Core for providing the primary cell cultures and the Viral Vector Core for vector production. We thank the Central Microscopy Research Facilities for providing secondary antibodies and the microtomes for sectioning. We thank Comparative Pathology Lab for immunostaining. Dr Fredric Bushman and his lab members Chris Nobles, Aoife Roche and John Everett at the University of Pennsylvania for their INSPIIRED sequencing and analysis. We thank Michael J. Welsh for his critical review of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [NIH P01 HL-51670, NIH P01 HL-091842, NIH R01 HL-133089, NIH R01 HL-105821]; Center for Gene Therapy of Cystic Fibrosis [NIH P30 DK-054759]; Cystic Fibrosis Foundation [SINN15XX0]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. P.B.M. is a founder of and holds equity in Talee Bio. The other authors have no conflicts of interests to declare.
REFERENCES
- 1. Pezzulo A.A., Tang X.X., Hoegger M.J., Abou Alaiwa M.H., Ramachandran S., Moninger T.O., Karp P.H., Wohlford-Lenane C.L., Haagsman H.P., van Eijk M. et al. Reduced airway surface pH impairs bacterial killing in the porcine cystic fibrosis lung. Nature. 2012; 487:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tang X.X., Ostedgaard L.S., Hoegger M.J., Moninger T.O., Karp P.H., McMenimen J.D., Choudhury B., Varki A., Stoltz D.A., Welsh M.J.. Acidic pH increases airway surface liquid viscosity in cystic fibrosis. J. Clin. Invest. 2016; 126:879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ramsey B.W., Davies J., McElvaney N.G., Tullis E., Bell S.C., Drevinek P., Griese M., McKone E.F., Wainwright C.E., Konstan M.W. et al. A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N. Engl. J. Med. 2011; 365:1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clancy J.P., Rowe S.M., Accurso F.J., Aitken M.L., Amin R.S., Ashlock M.A., Ballmann M., Boyle M.P., Bronsveld I., Campbell P.W. et al. Results of a phase IIa study of VX-809, an investigational CFTR corrector compound, in subjects with cystic fibrosis homozygous for the F508del-CFTR mutation. Thorax. 2012; 67:12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wainwright C.E., Elborn J.S., Ramsey B.W., Marigowda G., Huang X., Cipolli M., Colombo C., Davies J.C., De Boeck K., Flume P.A. et al. Lumacaftor-Ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N. Engl. J. Med. 2015; 373:220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stoltz D.A., Meyerholz D.K., Welsh M.J.. Origins of cystic fibrosis lung disease. N. Engl. J. Med. 2015; 372:1574–1575. [DOI] [PubMed] [Google Scholar]
- 7. Ranganathan S.C., Parsons F., Gangell C., Brennan S., Stick S.M., Sly P.D. Australian Respiratory Early Surveillance Team for Cystic, F. . Evolution of pulmonary inflammation and nutritional status in infants and young children with cystic fibrosis. Thorax. 2011; 66:408–413. [DOI] [PubMed] [Google Scholar]
- 8. Rieber N., Hector A., Carevic M., Hartl D.. Current concepts of immune dysregulation in cystic fibrosis. Int. J. Biochem. Cell Biol. 2014; 52:108–112. [DOI] [PubMed] [Google Scholar]
- 9. Tiddens H.A., Donaldson S.H., Rosenfeld M., Pare P.D.. Cystic fibrosis lung disease starts in the small airways: can we treat it more effectively. Pediatr. Pulmonol. 2010; 45:107–117. [DOI] [PubMed] [Google Scholar]
- 10. Armstrong D.S., Hook S.M., Jamsen K.M., Nixon G.M., Carzino R., Carlin J.B., Robertson C.F., Grimwood K.. Lower airway inflammation in infants with cystic fibrosis detected by newborn screening. Pediatr. Pulmonol. 2005; 40:500–510. [DOI] [PubMed] [Google Scholar]
- 11. Long F.R., Williams R.S., Castile R.G.. Structural airway abnormalities in infants and young children with cystic fibrosis. J. Pediatr. 2004; 144:154–161. [DOI] [PubMed] [Google Scholar]
- 12. Martinez T.M., Llapur C.J., Williams T.H., Coates C., Gunderman R., Cohen M.D., Howenstine M.S., Saba O., Coxson H.O., Tepper R.S.. High-resolution computed tomography imaging of airway disease in infants with cystic fibrosis. Am. J. Respir. Crit. Care Med. 2005; 172:1133–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stick S.M., Brennan S., Murray C., Douglas T., von Ungern-Sternberg B.S., Garratt L.W., Gangell C.L., De Klerk N., Linnane B., Ranganathan S. et al. Bronchiectasis in infants and preschool children diagnosed with cystic fibrosis after newborn screening. J. Pediatr. 2009; 155:623–628. [DOI] [PubMed] [Google Scholar]
- 14. Cooney A.L., Singh B.K., Sinn P.L.. Hybrid nonviral/viral vector systems for improved piggyBac DNA transposon in vivo delivery. Mol. Ther. 2015; 23:667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yant S.R., Ehrhardt A., Mikkelsen J.G., Meuse L., Pham T., Kay M.A.. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat. Biotechnol. 2002; 20:999–1005. [DOI] [PubMed] [Google Scholar]
- 16. Hausl M.A., Zhang W., Muther N., Rauschhuber C., Franck H.G., Merricks E.P., Nichols T.C., Kay M.A., Ehrhardt A.. Hyperactive sleeping beauty transposase enables persistent phenotypic correction in mice and a canine model for hemophilia B. Mol. Ther. 2010; 18:1896–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hausl M., Zhang W., Voigtlander R., Muther N., Rauschhuber C., Ehrhardt A.. Development of adenovirus hybrid vectors for Sleeping Beauty transposition in large mammals. Curr. Gene Ther. 2011; 11:363–374. [DOI] [PubMed] [Google Scholar]
- 18. Fraser M.J., Ciszczon T., Elick T., Bauser C.. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 1996; 5:141–151. [DOI] [PubMed] [Google Scholar]
- 19. Welsh M.J., Rogers C.S., Stoltz D.A., Meyerholz D.K., Prather R.S.. Development of a porcine model of cystic fibrosis. Trans. Am. Clin. Climatol Assoc. 2009; 120:149–162. [PMC free article] [PubMed] [Google Scholar]
- 20. Stoltz D.A., Meyerholz D.K., Pezzulo A.A., Ramachandran S., Rogan M.P., Davis G.J., Hanfland R.A., Wohlford-Lenane C., Dohrn C.L., Bartlett J.A. et al. Cystic fibrosis pigs develop lung disease and exhibit defective bacterial eradication at birth. Sci. Transl. Med. 2010; 2:29ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ostedgaard L.S., Meyerholz D.K., Chen J.H., Pezzulo A.A., Karp P.H., Rokhlina T., Ernst S.E., Hanfland R.A., Reznikov L.R., Ludwig P.S. et al. The {Delta}F508 mutation causes CFTR misprocessing and cystic fibrosis-like disease in pigs. Sci. Transl. Med. 2011; 3:74ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cooney A.L., Abou Alaiwa M.H., Shah V.S., Bouzek D.C., Stroik M.R., Powers L.S., Gansemer N.D., Meyerholz D.K., Welsh M.J., Stoltz D.A. et al. Lentiviral-mediated phenotypic correction of cystic fibrosis pigs. JCI Insight. 2016; 1:e88730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Steines B., Dickey D.D., Bergen J., Excoffon K.J., Weinstein J.R., Li X., Yan Z., Abou Alaiwa M.H., Shah V.S., Bouzek D.C. et al. CFTR gene transfer with AAV improves early cystic fibrosis pig phenotypes. JCI Insight. 2016; 1:e88728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stoltz D.A., Rokhlina T., Ernst S.E., Pezzulo A.A., Ostedgaard L.S., Karp P.H., Samuel M.S., Reznikov L.R., Rector M.V., Gansemer N.D. et al. Intestinal CFTR expression alleviates meconium ileus in cystic fibrosis pigs. J. Clin. Invest. 2013; 123:2685–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yan Z., Sun X., Feng Z., Li G., Fisher J.T., Stewart Z.A., Engelhardt J.F.. Optimization of recombinant Adeno-Associated Virus-Mediated expression for large transgenes, using a synthetic promoter and tandem array enhancers. Hum. Gene Ther. 2015; 26:334–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rogers C.S., Stoltz D.A., Meyerholz D.K., Ostedgaard L.S., Rokhlina T., Taft P.J., Rogan M.P., Pezzulo A.A., Karp P.H., Itani O.A. et al. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science. 2008; 321:1837–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parsons D.W., Grubb B.R., Johnson L.G., Boucher R.C.. Enhanced in vivo airway gene transfer via transient modification of host barrier properties with a surface-active agent. Hum. Gene Ther. 1998; 9:2661–2672. [DOI] [PubMed] [Google Scholar]
- 28. Chen J.H., Stoltz D.A., Karp P.H., Ernst S.E., Pezzulo A.A., Moninger T.O., Rector M.V., Reznikov L.R., Launspach J.L., Chaloner K. et al. Loss of anion transport without increased sodium absorption characterizes newborn porcine cystic fibrosis airway epithelia. Cell. 2010; 143:911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kreda S.M., Mall M., Mengos A., Rochelle L., Yankaskas J., Riordan J.R., Boucher R.C.. Characterization of wild-type and deltaF508 cystic fibrosis transmembrane regulator in human respiratory epithelia. Mol. Biol. Cell. 2005; 16:2154–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mall M., Kreda S.M., Mengos A., Jensen T.J., Hirtz S., Seydewitz H.H., Yankaskas J., Kunzelmann K., Riordan J.R., Boucher R.C.. The DeltaF508 mutation results in loss of CFTR function and mature protein in native human colon. Gastroenterology. 2004; 126:32–41. [DOI] [PubMed] [Google Scholar]
- 31. Meyerholz D.K., Lambertz A.M., Reznikov L.R., Ofori-Amanfo G.K., Karp P.H., McCray P.B. Jr, Welsh M.J., Stoltz D.A.. Immunohistochemical detection of markers for translational studies of lung disease in pigs and humans. Toxicol. Pathol. 2016; 44:434–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nygard A.B., Jorgensen C.B., Cirera S., Fredholm M.. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol. Biol. 2007; 8:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Auricchio A., O’Connor E., Weiner D., Gao G.P., Hildinger M., Wang L., Calcedo R., Wilson J.M.. Noninvasive gene transfer to the lung for systemic delivery of therapeutic proteins. J. Clin. Invest. 2002; 110:499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Siegfried W., Rosenfeld M., Stier L., Stratford-Perricaudet L., Perricaudet M., Pavirani A., Lecocq J.P., Crystal R.G.. Polarity of secretion of alpha 1-antitrypsin by human respiratory epithelial cells after adenoviral transfer of a human alpha 1-antitrypsin cDNA. Am. J. Respir. Cell Mol. Biol. 1995; 12:379–384. [DOI] [PubMed] [Google Scholar]
- 35. Engelhardt J.F. The lung as a metabolic factory for gene therapy. J. Clin. Invest. 2002; 110:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cao H., Machuca T.N., Yeung J.C., Wu J., Du K., Duan C., Hashimoto K., Linacre V., Coates A.L., Leung K. et al. Efficient gene delivery to pig airway epithelia and submucosal glands using helper-dependent adenoviral vectors. Mol. Ther. Nucleic Acids. 2013; 2:e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sherman E., Nobles C., Berry C.C., Six E., Wu Y., Dryga A., Malani N., Male F., Reddy S., Bailey A. et al. INSPIIRED: a pipeline for quantitative analysis of sites of new DNA integration in cellular genomes. Mol. Ther. Methods Clin. Dev. 2017; 4:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berry C.C., Nobles C., Six E., Wu Y., Malani N., Sherman E., Dryga A., Everett J.K., Male F., Bailey A. et al. INSPIIRED: quantification and visualization tools for analyzing integration site distributions. Mol. Ther. Methods Clin. Dev. 2017; 4:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li X., Burnight E.R., Cooney A.L., Malani N., Brady T., Sander J.D., Staber J., Wheelan S.J., Joung J.K., McCray P.B. Jr et al. piggyBac transposase tools for genome engineering. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E2279–E2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Engelhardt J.F., Yankaskas J.R., Ernst S.A., Yang Y., Marino C.R., Boucher R.C., Cohn J.A., Wilson J.M.. Submucosal glands are the predominant site of CFTR expression in the human bronchus. Nat. Genet. 1992; 2:240–248. [DOI] [PubMed] [Google Scholar]
- 41. Solodushko V., Bitko V., Fouty B.. Minimal piggyBac vectors for chromatin integration. Gene Ther. 2014; 21:1–9. [DOI] [PubMed] [Google Scholar]
- 42. Crystal R.G. Adenovirus: the first effective in vivo gene delivery vector. Hum. Gene Ther. 2014; 25:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Joseph P.M., O'Sullivan B.P., Lapey A., Dorkin H., Oren J., Balfour R., Perricone M.A., Rosenberg M., Wadsworth S.C., Smith A.E. et al. Aerosol and lobar administration of a recombinant adenovirus to individuals with cystic fibrosis. I. Methods, safety, and clinical implications. Hum. Gene Ther. 2001; 12:1369–1382. [DOI] [PubMed] [Google Scholar]
- 44. Perricone M.A., Morris J.E., Pavelka K., Plog M.S., O'Sullivan B.P., Joseph P.M., Dorkin H., Lapey A., Balfour R., Meeker D.P. et al. Aerosol and lobar administration of a recombinant adenovirus to individuals with cystic fibrosis. II. Transfection efficiency in airway epithelium. Hum. Gene Ther. 2001; 12:1383–1394. [DOI] [PubMed] [Google Scholar]
- 45. Bergelson J.M., Cunningham J.A., Droguett G., Kurt-Jones E.A., Krithivas A., Hong J.S., Horwitz M.S., Crowell R.L., Finberg R.W.. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997; 275:1320–1323. [DOI] [PubMed] [Google Scholar]
- 46. Walters R.W., Grunst T., Bergelson J.M., Finberg R.W., Welsh M.J., Zabner J.. Basolateral localization of fiber receptors limits adenovirus infection from the apical surface of airway epithelia. J. Biol. Chem. 1999; 274:10219–10226. [DOI] [PubMed] [Google Scholar]
- 47. Chu Q., St George J.A., Lukason M., Cheng S.H., Scheule R.K., Eastman S.J.. EGTA enhancement of adenovirus-mediated gene transfer to mouse tracheal epithelium in vivo. Hum. Gene Ther. 2001; 12:455–467. [DOI] [PubMed] [Google Scholar]
- 48. Wang G., Zabner J., Deering C., Launspach J., Shao J., Bodner M., Jolly D.J., Davidson B.L., McCray P.B. Jr. Increasing epithelial junction permeability enhances gene transfer to airway epithelia In vivo. Am. J. Respir. Cell Mol. Biol. 2000; 22:129–138. [DOI] [PubMed] [Google Scholar]
- 49. Kaplan J.M., Pennington S.E., St George J.A., Woodworth L.A., Fasbender A., Marshall J., Cheng S.H., Wadsworth S.C., Gregory R.J., Smith A.E.. Potentiation of gene transfer to the mouse lung by complexes of adenovirus vector and polycations improves therapeutic potential. Hum. Gene Ther. 1998; 9:1469–1479. [DOI] [PubMed] [Google Scholar]
- 50. Yang Y., Nunes F.A., Berencsi K., Furth E.E., Gonczol E., Wilson J.M.. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:4407–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Worgall S., Leopold P.L., Wolff G., Ferris B., Van Roijen N., Crystal R.G.. Role of alveolar macrophages in rapid elimination of adenovirus vectors administered to the epithelial surface of the respiratory tract. Hum. Gene Ther. 1997; 8:1675–1684. [DOI] [PubMed] [Google Scholar]
- 52. Hartman Z.C., Appledorn D.M., Amalfitano A.. Adenovirus vector induced innate immune responses: impact upon efficacy and toxicity in gene therapy and vaccine applications. Virus Res. 2008; 132:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nemerow G.R. A new link between virus cell entry and inflammation: adenovirus interaction with integrins induces specific proinflammatory responses. Mol. Ther. 2009; 17:1490–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zaiss A.K., Machado H.B., Herschman H.R.. The influence of innate and pre-existing immunity on adenovirus therapy. J. Cell. Biochem. 2009; 108:778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang Y., Su Q., Wilson J.M.. Role of viral antigens in destructive cellular immune responses to adenovirus vector-transduced cells in mouse lungs. J. Virol. 1996; 70:7209–7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang Y., Greenough K., Wilson J.M.. Transient immune blockade prevents formation of neutralizing antibody to recombinant adenovirus and allows repeated gene transfer to mouse liver. Gene Ther. 1996; 3:412–420. [PubMed] [Google Scholar]
- 57. Lei D., Lehmann M., Shellito J.E., Nelson S., Siegling A., Volk H.D., Kolls J.K.. Nondepleting anti-CD4 antibody treatment prolongs lung-directed E1-deleted adenovirus-mediated gene expression in rats. Hum. Gene Ther. 1996; 7:2273–2279. [DOI] [PubMed] [Google Scholar]
- 58. Otake K., Ennist D.L., Harrod K., Trapnell B.C.. Nonspecific inflammation inhibits adenovirus-mediated pulmonary gene transfer and expression independent of specific acquired immune responses. Hum. Gene Ther. 1998; 9:2207–2222. [DOI] [PubMed] [Google Scholar]
- 59. Zuckerman J.B., Robinson C.B., McCoy K.S., Shell R., Sferra T.J., Chirmule N., Magosin S.A., Propert K.J., Brown-Parr E.C., Hughes J.V. et al. A phase I study of adenovirus-mediated transfer of the human cystic fibrosis transmembrane conductance regulator gene to a lung segment of individuals with cystic fibrosis. Hum. Gene Ther. 1999; 10:2973–2985. [DOI] [PubMed] [Google Scholar]
- 60. Lyakh L.A., Koski G.K., Young H.A., Spence S.E., Cohen P.A., Rice N.R.. Adenovirus type 5 vectors induce dendritic cell differentiation in human CD14(+) monocytes cultured under serum-free conditions. Blood. 2002; 99:600–608. [DOI] [PubMed] [Google Scholar]
- 61. Teigler J.E., Iampietro M.J., Barouch D.H.. Vaccination with adenovirus serotypes 35, 26, and 48 elicits higher levels of innate cytokine responses than adenovirus serotype 5 in rhesus monkeys. J. Virol. 2012; 86:9590–9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Drazan K.E., Olthoff K.M., Wu L., Shen X.D., Gelman A., Shaked A.. Adenovirus-mediated gene transfer in the transplant setting: early events after orthotopic transplantation of liver allografts expressing TGF-beta1. Transplantation. 1996; 62:1080–1084. [DOI] [PubMed] [Google Scholar]
- 63. Muruve D.A., Cotter M.J., Zaiss A.K., White L.R., Liu Q., Chan T., Clark S.A., Ross P.J., Meulenbroek R.A., Maelandsmo G.M. et al. Helper-dependent adenovirus vectors elicit intact innate but attenuated adaptive host immune responses in vivo. J. Virol. 2004; 78:5966–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Croyle M.A., Chirmule N., Zhang Y., Wilson J.M.. “Stealth” adenoviruses blunt cell-mediated and humoral immune responses against the virus and allow for significant gene expression upon readministration in the lung. J. Virol. 2001; 75:4792–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Toietta G., Koehler D.R., Finegold M.J., Lee B., Hu J., Beaudet A.L.. Reduced inflammation and improved airway expression using helper-dependent adenoviral vectors with a K18 promoter. Mol. Ther. 2003; 7:649–658. [DOI] [PubMed] [Google Scholar]
- 66. Koehler D.R., Martin B., Corey M., Palmer D., Ng P., Tanswell A.K., Hu J.. Readministration of helper-dependent adenovirus to mouse lung. Gene Ther. 2006; 13:773–780. [DOI] [PubMed] [Google Scholar]
- 67. Cao H., Yang T., Li X.F., Wu J., Duan C., Coates A.L., Hu J.. Readministration of helper-dependent adenoviral vectors to mouse airway mediated via transient immunosuppression. Gene Ther. 2011; 18:173–181. [DOI] [PubMed] [Google Scholar]
- 68. Cao H., Ouyang H., Grasemann H., Bartlett C., Du K., Duan R., Shi F., Estrada M., Seigel K., Coates A. et al. Transducing airway basal cells with a helper-dependent adenoviral vector for lung gene therapy. Hum. Gene Ther. 2018; 29:643–652. [DOI] [PubMed] [Google Scholar]
- 69. Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E. et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003; 302:415–419. [DOI] [PubMed] [Google Scholar]
- 70. Howe S.J., Mansour M.R., Schwarzwaelder K., Bartholomae C., Hubank M., Kempski H., Brugman M.H., Pike-Overzet K., Chatters S.J., de Ridder D. et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008; 118:3143–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Boztug K., Schmidt M., Schwarzer A., Banerjee P.P., Diez I.A., Dewey R.A., Bohm M., Nowrouzi A., Ball C.R., Glimm H. et al. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N. Engl. J. Med. 2010; 363:1918–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stein S., Ott M.G., Schultze-Strasser S., Jauch A., Burwinkel B., Kinner A., Schmidt M., Kramer A., Schwable J., Glimm H. et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 2010; 16:198–204. [DOI] [PubMed] [Google Scholar]
- 73. Persons D.A., Baum C.. Solving the problem of gamma-retroviral vectors containing long terminal repeats. Mol. Ther. 2011; 19:229–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gogol-Doring A., Ammar I., Gupta S., Bunse M., Miskey C., Chen W., Uckert W., Schulz T.F., Izsvak Z., Ivics Z.. Genome-wide profiling reveals remarkable parallels between insertion site selection properties of the MLV retrovirus and the piggyBac transposon in primary human CD4(+) T cells. Mol. Ther. 2016; 24:592–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Galvan D.L., Nakazawa Y., Kaja A., Kettlun C., Cooper L.J., Rooney C.M., Wilson M.H.. Genome-wide mapping of PiggyBac transposon integrations in primary human T cells. J. Immunother. 2009; 32:837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Huang X., Guo H., Tammana S., Jung Y.C., Mellgren E., Bassi P., Cao Q., Tu Z.J., Kim Y.C., Ekker S.C. et al. Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and piggyBac transposons in human primary T cells. Mol. Ther. 2010; 18:1803–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.