Abstract
Synthetic RNA is widely used in basic science, nanotechnology and therapeutics research. The vast majority of this RNA is synthesized in vitro by T7 RNA polymerase or one of its close family members. However, the desired RNA is generally contaminated with products longer and shorter than the DNA-encoded product. To better understand these undesired byproducts and the processes that generate them, we analyze in vitro transcription reactions using RNA-Seq as a tool. The results unambiguously confirm that product RNA rebinds to the polymerase and self-primes (in cis) generation of a hairpin duplex, a process that favorably competes with promoter driven synthesis under high yield reaction conditions. While certain priming modes can be favored, the process is heterogeneous, both in initial priming and in the extent of priming, and already extended products can rebind for further extension, in a distributive process. Furthermore, addition of one or a few nucleotides, previously termed ‘nontemplated addition,’ also occurs via templated primer extension. At last, this work demonstrates the utility of RNA-Seq as a tool for in vitro mechanistic studies, providing information far beyond that provided by traditional gel electrophoresis.
INTRODUCTION
Transcription at its simplest is complex, but relatively straightforward: RNA polymerase binds its promoter sequence through interactions largely upstream of the start site of transcription, melts the DNA near the start site, initiates de novo (unprimed) synthesis of RNA utilizing the exposed template DNA, transitions through an unstable initial transcription (abortive) phase and then elongates the RNA faithfully until a termination event occurs or, in vitro, until the enzyme reaches the end of a linear template (1–6).
T7 RNA polymerase and other enzymes from related bacteriophages are highly processive DNA-dependent RNA polymerases, consisting of a single subunit (7,8). T7 RNA polymerase is highly specific for a relatively small consensus promoter sequence upstream of the start site (1,9) and transcribes the expected runoff RNA products with high incorporation fidelity (10). During initial transcription, prior to promoter release, RNA polymerase produces ‘contaminant’ abortive RNAs from 2 to 6 nts in length (11,12). Less widely addressed, but nevertheless well known, in vitro transcription reactions often produce RNAs shorter (n −i) or longer (n +i) than the template-encoded products (11,13).
It has also been reported that T7 RNA polymerase is able to synthesize RNA from single and double-stranded RNA, in the presence of the double-stranded DNA (dsDNA) promoter sequence (14). Additionally, T7 RNA polymerase can produce RNA in the absence of promoter DNA. Konarska and Sharp demonstrated that T7 RNA polymerase in some cases behaves like viral RNA-dependent RNA polymerases and self-replicates specific RNA sequences (RNA X) (15–17). Furthermore, it has been shown that synthesized runoff RNA can be extended to longer RNA products by RNA priming using RNA template-directed RNA synthesis (18). In principle, RNA extension can occur if the 3′ end of runoff RNA has sufficient complementarity to sit down in trans on a second RNA or fold back on itself in cis to form extendible inter- (19) or intramolecular (20) duplexes, respectively. The current work expands on this general result.
In this report, we develop an approach to augment classical gel electrophoresis with next generation RNA-Seq analysis of in vitro transcription reactions. This approach allows one to analyze RNA product lengths, as does gel electrophoresis. However, it also provides the sequence of each product, including the distribution of sequences within a given length of RNA. This is particularly important for n + i RNA products arising from events beyond the desired, template-encoded transcription.
In particular, these studies unambiguously confirm an earlier model in which RNA folds back on itself in cis to prime the self-templated extension (20). We also demonstrate that, as expected, production of such unintended products increases dramatically with overall RNA product yield, since the requisite rebinding of priming template RNA to T7 RNA polymerase increases as its concentration increases in the solution. We also demonstrate that this process is characterized by substantial heterogeneity, both in the priming structures and in the lengths of extensions. The data support a model in which the synthesis of longer extensions is distributive in nature, with extended products rebinding for further extension. At last, we provide evidence that even short extensions (n + 1, n + 2 etc.), previously assumed to be nontemplated (11,13,21), are largely templated through the same overall mechanism.
MATERIALS AND METHODS
In vitro transcription by T7 RNA polymerase
Template and nontemplate DNA oligonucleotides shown in Supplementary Table S1 were purchased from Integrated DNA Technologies (IDT). Most transcription reactions were carried out with constructs in which the nontemplate strand extends only to position +2, a common practice in the field (11,22). However, it is important to note that after the first round of transcription, the downstream template is unlikely to remain single stranded, as product RNA will bind and serve as the nontemplate strand in that region.
‘Low yield’ transcription reaction contained 2 μM each of template and nontemplate DNA, 0.5 μM T7 RNA polymerase, 0.6 mM guanosine triphosphate (GTP) and 0.4 mM each of cytidine triphosphate (CTP), adenosine triphosphate (ATP) and uridine triphosphate (UTP). Reactions were carried out in a buffer containing 15 mM magnesium acetate, 30 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 25 mM potassium glutamate, 0.25 mM ethylenediaminetetraacetic acid and 0.05% Tween-20 at 37°C for 5 min.
To be representative of common practices, ‘high yield’ transcription was performed using the HiScribe™ T7 High Yield RNA Synthesis Kit (New England BioLabs), in a transcription mixture containing 2 μM each of template and nontemplate DNA, 1.5 μl of T7 RNA Polymerase Mix™ for 20 μl reaction volume, 7.5 mM each of GTP, ATP, CTP and UTP, in the T7 Reaction Buffer (New England BioLabs). The reaction was incubated at 37°C for 4 h.
Primer extension on synthetic 24-mer RNA (purchased from IDT, sequence is shown in Supplementary Table S1) was performed under the above ‘low yield’ condition, with 25 μM RNA replacing template DNA. Reactions were carried out at 37°C for 5 min and for 4 h, as indicated.
RNase Inhibitor Murine (New England BioLabs) was added to each transcription mixture before and after the reaction, to inhibit any RNase activity. All reactions were stopped by heat inactivation at 70°C for 5 min.
Denaturing gel electrophoresis and radiochemical labeling of the synthetic RNA
Transcripts (expected size: 24 bases) were analyzed by 20% polyacrylamide, 7 M urea denaturing gel electrophoresis, and then visualized by SYBR™ Green II RNA Gel Stain (Invitrogen) or in some cases by radiochemical labeling of the 5′ end of the RNA with [γ-32P] ATP (PerkinElmer). For 5′ radiochemical labeling of the synthetic RNA, 50 μM of the synthetic RNA was incubated in the T4 polynucleotide kinase buffer, with [γ-32P] ATP (PerkinElmer) and 1 μl (10 Units) of T4 polynucleotide kinase (New England BioLabs) in a 10 μl reaction volume at 37°C for 30 min, and then heat inactivated at 65°C for 20 min. All gel electrophoresis experiments were repeated at least twice.
Library preparation for RNA-Seq
As outlined in Supplementary Figure S1, transcripts from all reactions were incubated at 37°C for 1 h with 5′ Pyrophosphohydrolase/RppH (New England BioLabs) to remove pyrophosphate from the 5′ end of tri-phosphorylated RNAs and to generate 5′ monophosphate RNAs. Transcripts were then incubated with Shrimp Alkaline Phosphatase/rSAP (New England BioLabs) at 37°C for 45 min to dephosphorylate the 5′ end of RNAs and to hydrolyze remaining NTPs from the reaction. Afterward, phosphatase activity was eliminated by heat inactivation at 65°C for 5 min. The 3′ ends of dephosphorylated RNAs were then ligated to 5′ pre-adenylated 3′ adapter with 3′ Biotin modification (the 3′ adapter sequence is shown in Supplementary Table S2), which was attached to Streptavidin Magnetic Beads (New England BioLabs) (23), by T4 RNA Ligase 2 truncated K227Q (New England BioLabs) (24). The 40 μl ligation reactions containing 20% (w/v) PEG 8000, 0.05 mg/ml bovine serum albumin, 50 mM NaCl and 2 μl of RNA Ligase 2 at 200 U μl−1 were incubated at 16°C overnight. After 3′ ligation, we performed three magnetic bead washing cycles to remove all unligated RNAs and NTPs. The 5′ ends of ligated product were then phosphorylated by T4 polynucleotide kinase (New England BioLabs) in a 40 μl reaction containing 2 μl of T4 polynucleotide kinase at 10 U μl−1 and 2 mM final concentration of ATP. The reactions were incubated at 37°C for 30 min, and then stopped by heat inactivation at 65°C for 20 min.
A 5′ adapter sequence, containing a barcode unique to each experiment (Supplementary Table S2), was then ligated to the 5′ mono-phosphorylated product by T4 RNA Ligase 1 (New England BioLabs) (24). The 40 μl reactions containing 10 μM 5′ adapter, 20% (w/v) PEG 8000, 1 mM ATP and 4 μl of enzyme at 10 U μl−1 were incubated at 16°C overnight. We then performed three bead washing cycles to remove all unligated 5′ adapter. At last, to remove ligated RNAs from the magnetic beads, we heated the reaction to 95°C for 5 min to denature the Biotin–Streptavidin interaction and isolated the supernatant from the beads.
Fully ligated RNAs were then reverse transcribed to complementary DNA (cDNA) using a primer that is specific to the ligated 3′ adapter (Supplementary Table S2). The reverse transcription reactions were performed in a 50 μl reaction volume containing 0.4 μM reverse transcriptase primer, 0.5 mM each deoxynucleoside triphosphate (dNTP), 10 mM dithiothreitol (DTT) and 2 μl ProtoScript® II Reverse Transcriptase (New England BioLabs) at 200 U μl−1 by incubation at 42°C for 1 h.
Amplification and quantification of the library
cDNA for each experiment was amplified by Phusion® High-Fidelity DNA Polymerase (New England BioLabs) using primers based on Illumina primers for TruSeq Small RNA Library, with a final concentration of 0.4 μM (primers were purchased from IDT and are shown in Supplementary Table S3). Note that different reverse primers with specific index reads were used for each experiment. Then we used ExoSAP-IT™ PCR Product Cleanup Reagent (Affymetrix) to remove excess primes and unincorporated nucleotides. We also performed G25 column clean up to remove salts. At last, the library was quantified by Qubit dsDNA HS Assay Kit (Invitrogen).
Next-generation sequencing and data analysis
The library was sequenced using an Illumina MiSeq sequencer with the MiSeq® Reagent Nano Kit v2 (Illumina), based on manufacturer’s instructions. Data analysis was carried out using code written in Python, including modules from Biopython (25), as shown in the flow chart of Supplementary Figure S1. Data from multiplexed sequencing were separated into individual experiments based on the barcode index reads. Only sequences with valid 5′ and 3′ adapters were analyzed. In most cases, sequences were further filtered for the expected GG sequence at positions +1 and +2. For 3′ end analyses, sequences were aligned to an internal expected sequence, although, mis-initiation was minor relative to the analyses. Counts at each step in processing are shown in Supplementary Table S4.
For RNA-Seq quantification of RNA products by length (Figures 1B, 3A and 5C), RNAs were simply binned by length (regardless of sequence). Note that this representation is the equivalent of a molar analysis and is not mass-weighted, as occurs in intercalator staining or [α-32P]NTP (nucleoside triphosphate) labeling.
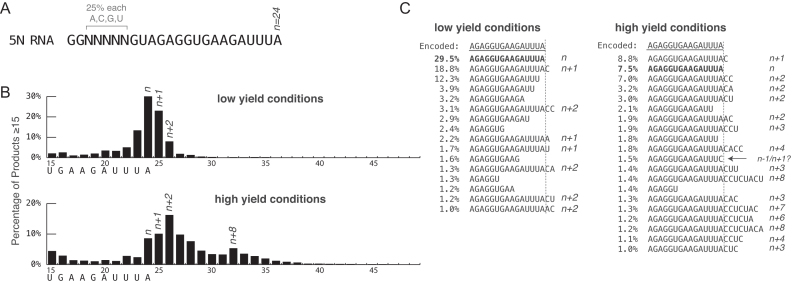
Figure 1.
Initial analysis of transcription reactions. (A) The RNA sequence encoded by the template DNA. Note that here, and forward, all sequences, including the output of RNA-Seq are shown as RNA. (B) RNA-Seq counting of RNAs ≥15 bases in length, synthesized under low and high yield conditions. Reported percentages are relative to that pool. (C) The most abundant RNA-Seq sequences from position +10 forward.
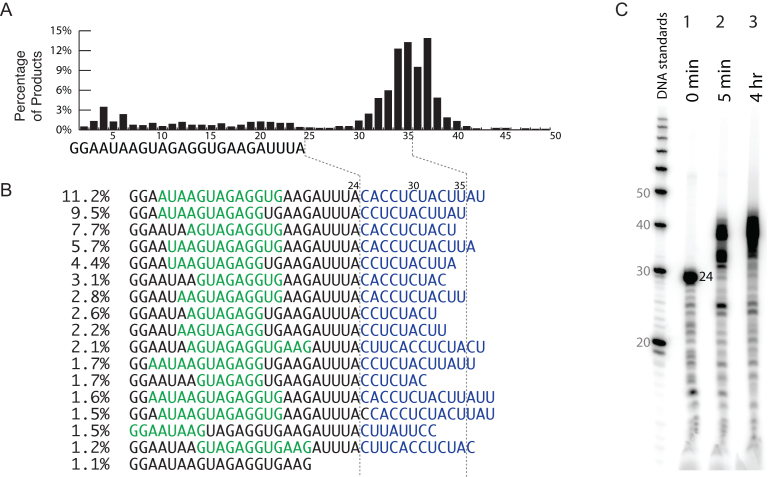
Figure 3.
Primer extension of synthetic RNA in the absence of promoter sequence. (A) RNA-Seq quantification of RNA products by length, for a 4 h incubation of synthetic RNA with T7 RNA polymerase, in the presence of 0.4 mM each NTP. (B) Most abundant RNA-Seq sequences. Extended RNA products show complementarity (blue: sequence beyond the synthetic RNA sequence, green: upstream sequence of the synthetic RNA that is complementary to the blue region). (C) Denaturing gel electrophoreses (20% denaturing urea) analysis of a parallel reaction using radiolabeled, synthetic RNA. Lane 1: Radiolabeled 24-base synthetic RNA. Lanes 2 and 3: Incubation of synthetic RNA with T7 RNA polymerase for 5 min and 4 h, respectively.
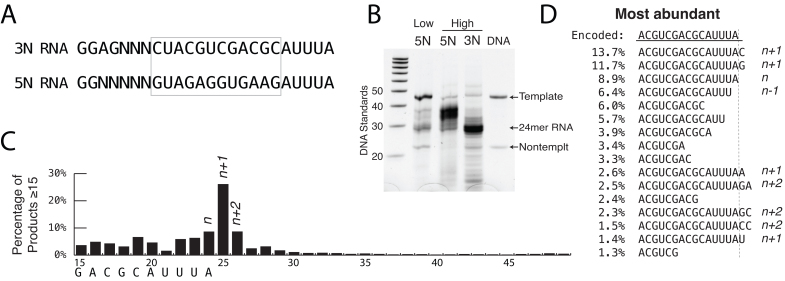
Figure 5.
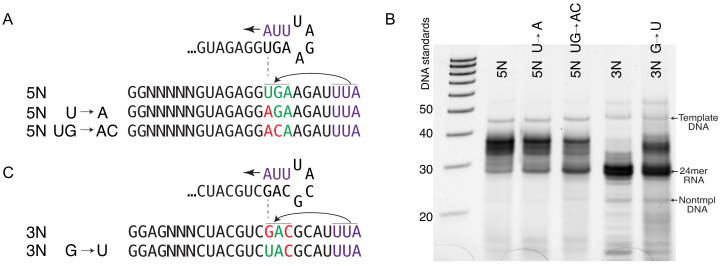
Cis primer extension depends on upstream sequence. (A) RNA sequences encoded by DNA templates. 3N RNA has same encoded 3′ end as the RNA from prior figures (5N RNA), but has a different upstream sequence. (B) Gel analysis of transcription profile for DNA templates encoding different internal sequences under the high yield conditions of Figure 1B. Twenty percent denaturing urea gel, stained with SYBR Green II RNA. Controls shown are DNA size standards and the promoter DNA strands used in transcription. (C) RNA-Seq counting of RNAs 15–49 bases in length, synthesized under high yield conditions for 3N RNA. (D) Most abundant RNA-Seq sequences from position +10 forward for 3N RNA.
The ‘high yield’ transcription reaction using DNA template 5N was replicated and comparisons (Supplementary Figures S2 and S3) with the initial experiment show excellent agreement.
RESULTS
In transcription from linear DNA, RNA polymerases are predicted to synthesize a defined full length (runoff) product (11). For decades, denaturing gel electrophoresis (11,18) has been the tool of choice in analyzing transcription products. Based on the known sequence of the DNA template and the lengths estimated from the gel, researchers have assigned bands in a gel to specific RNA products. However, it is not uncommon to observe products longer than the predicted runoff transcript (18–20). In this case, simple gel analyses provide no information on the nature of the products.
RNA-Seq analysis of in vitro transcription products
In order to determine the precise nature and distribution of in vitro synthesized transcription products, including sequences and relative abundances of individual sequences, we now analyze product RNAs by massively parallel next generation sequencing, RNA-Seq. Initial transcription reactions were carried out with a DNA template (5N template, Supplementary Table S1) encoding a 24-base runoff RNA, as shown in Figure 1A, using wild-type T7 RNA polymerase.
In order to analyze transcription profiles by RNA-Seq, the product RNA pool must be prepared for sequencing. After quenching each transcription reaction by heat inactivation at 70°C for 5 min, RNAs were processed without initial washing. As described in more detail in the ‘Materials and Methods’ section, to avoid self-ligation and formation of RNA dimer/multimers during the adapter ligation steps, the 5′ triphosphate was removed from RNAs using pyrophosphohydrolase and shrimp alkaline phosphatase, which also hydrolyzes substrate NTPs. RNAs were then ligated by their 3′ ends to a pre-adenylated 3′ adapter using an enzyme that is only able to use adapters pre-adenylated at their 5′ ends. Since this adapter is pre-bound to magnetic beads, after ligation the sample can be washed to new reaction conditions, removing all nucleic acids not ligated to the adapter. The previously dephosphorylated 5′ ends of ligated RNAs were then phosphorylated and subsequently ligated to a 5′ adapter carrying a barcode specific to each experiment. RNAs containing both adapters were then reverse transcribed and amplified by polymerase chain reaction, and the resulting cDNA was sequenced by next-generation sequencing (MiSeq-Illumina).
Low yield reaction conditions yield expected product profile
An initial transcription reaction was performed under conditions widely used in mechanistic studies of transcription (0.6 mM GTP, 0.4 mM each of ATP, CTP, and UTP, with 5 min incubation at 37°C). We will refer to this as ‘low yield’ conditions. The initial promoter-containing DNA template encodes a 24-base runoff transcript and includes a randomized (∼25% each of A, C, G and T in the DNA template) sequence from positions +3 to +7 (Figure 1A), the utility of which will become apparent later.
Analysis of the RNA-Seq length profiles for ‘low yield’ transcription, presented in Figure 1B, shows the expected runoff product, plus shorter and longer (n − 1, n, n + 1 etc.) products (see Supplementary Figure S2 for more detail). The data also reveal abortive RNAs (not shown) and products much longer than expected (for example, >30 bases, corresponding to n + 6 or longer). Although for this reaction, amounts of the latter are detectable, they are negligible in the bar graph. As expected, a 24-base runoff RNA is the most abundant product, however, substantial amounts of n − 1 (23 bases) and n + 1 (25 bases) products are also observed.
Counting specific sequences, rather than pooled RNA lengths, provides detail not available in gel electrophoresis. As shown in Figure 1C, low yield conditions, the n + 1 product containing an additional C is five times more abundant than those containing added A, G or U. Similarly, n + 2 products are observed, but the 16 possible n + 2 sequences are not observed uniformly. In comparing the data representations in Figure 1B and C, note that whereas there is one expected sequence for each major n −i product, the n + i products detailed in Figure 1C are spread across as many as 4i possible sequences.
High yield reaction conditions shift length distributions to longer RNA lengths
Preparative RNA reactions are often carried out for 4 h with NTP concentrations of 7.5 mM each. Under such ‘high yield’ conditions, using the same DNA template, the distribution of products skews significantly to longer RNA lengths. As shown in Figure 1B, high yield conditions, the expected length product (n = 24) is no longer the most abundant, but rather the most abundant RNA length is 26 bases (n + 2), with a wide range of longer RNA products. Analysis of the most abundant sequences, presented in Figure 1C shows that the n + 1 product containing an added C is slightly more abundant than the expected product, but the relative amounts of various n +i products increase substantially.
Casual analysis of the data reveals clearly that these longer RNA products are not at all random in their added sequences. For sequences ≥31 bases in length, in the ‘high yield’ reaction, 97% of the products show at least a 4-base window of reverse complementarity to upstream sequences (see Supplementary Figure S3). This is consistent with prior reports that RNA transcripts can be extended to longer RNA lengths as the result of primer extension using RNA (18–20), or the nontemplate strand (26,27), as the template.
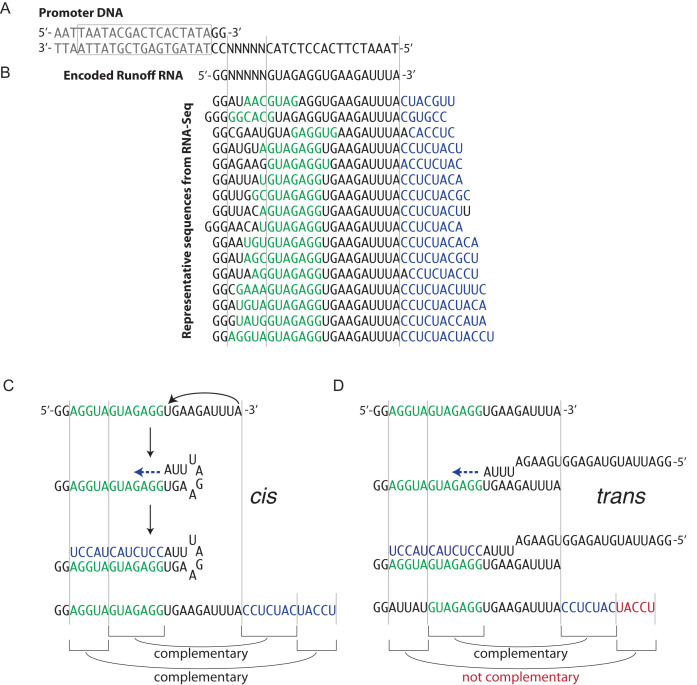
There have been conflicting reports in the literature regarding RNA primer extension with T7 RNA polymerase. The observed primer extension could in principle occur via a cis mechanism, with the 3′ end of the RNA looping back to sit down on its own upstream sequence (20), or via a trans mechanism, with the RNA binding to the equivalent region of a different RNA (18,19). The DNA template used here (Figure 2A), encodes a randomized sequence from position +3 through to position +7, allowing definitive resolution of the two models. The cis model predicts that each extended sequence should be the reverse complement of the corresponding sequence of the same RNA (self), while the trans model, in which templating is from a second RNA, predicts no such correlation. This is demonstrated by the collection of sequences shown in Figure 2B. In fact, as demonstrated in Supplementary Figure S3 for observed primer extension products that reach at least two bases into this randomized region, 75–80% have extensions that are the exact inverse complement of the randomized region of the same RNA (all other sequences, even those containing only one mismatch are scored as trans, suggesting that the actual percentage of cis originating RNAs is still higher). We conclude that primer extension occurs predominantly by RNA folding back on itself as the template in a cis (Figure 2C), rather than a trans (Figure 2D) mechanism.
Figure 2.
Representative depiction of RNA-Seq results from in vitro transcription reaction. (A) Transcription from partially single-stranded DNA template encoding a 24-base RNA transcript, which includes a randomized sequence from positions +3 to +7. The T7 promoter sequence is boxed and shown in gray. (B) Representative sequences of extended RNAs reveal complementarity (blue) to a sequence in the upstream region (green) within each RNA sequence. The cis (C) and trans (D) models for the formation of longer RNA products make distinct predictions for the expected downstream sequences corresponding to the upstream randomized regions. Longer RNA products shown in panel (B) are formed through cis primer extension.
Primer extension in cis could arise from nascent RNA rearranging within the active site to accommodate extension, or more simply, released RNA could rebind a vacant RNA polymerase. The latter model predicts that this type of primer extension will increase as runoff RNA accumulates in the reaction, as mass action will drive RNA rebinding and allow extension to compete more favorably with promoter-initiated transcription. The behavior observed in Figure 1 is fully consistent with this model.
Primer extension is promoter independent and competes with de novo initiation
To test whether primer extension occurs independent of promoter DNA, we (chemically) synthesized the expected 24-base RNA and incubated it with T7 RNA polymerase, in the absence of T7 promoter DNA. The synthetic RNA has same sequence as encoded by the DNA template above (with the only difference being that the sequence from position +3 to +7 is not randomized). The RNA-Seq results presented in Figure 3 clearly demonstrate extension of the synthetic RNA to longer RNA products, as illustrated both by RNA-Seq length counting (Figure 3A and B) and by gel analysis (Figure 3C). Indeed, in the absence of competing T7 promoter DNA, primer extension proceeds very efficiently and extends to greater lengths, as predicted by a distributive rebinding model in the absence of competition by promoter.
Analysis of the most abundant sequences presented in Figure 3B confirms that longer RNA products derive from primer extension using RNA as a template. Note also that this reaction was carried out under low yield transcription conditions, in particular, 0.4 mM each NTP and 15 mM magnesium acetate. This demonstrates that primer extension is not an artifact of high concentrations of these components in high yield transcription reactions, nor does it reflect differences in reaction conditions or enzyme preparation.
Although the RNA-Seq data reported are for the 4 h reaction, the gel data presented in Figure 3C reveal that at 5 min, essentially all of the RNA has been extended. Interestingly, at 5 min, the distribution of products skews shorter than at 4 h. Since the RNA concentration is 50-fold higher than the enzyme concentration, the intermediate length RNA products observed at 5 min that chase to longer products are not polymerase-bound intermediates. This argues that primer extension is distributive: polymerase extends one or a few bases, releases the RNA and then rebinds to continue extension.
Distribution of products reflects a range of mechanisms
The data in Figures 1 and 3 demonstrate significant heterogeneity in the primer extension products. The distribution of such products provides insights into the structural and energetic requirements of primer extension. To provide a more systematic analysis and noting that the DNA construct used (Figure 2A) contains a randomized initial transcription region, which is then copied in extension, we took the data from Figure 1 and ‘masked’ (N) both the bases from +3 to +7, and the corresponding, complementary downstream bases using the adjacent sequence (complementary to +8 to +13) for alignment. The analysis presented in Figure 4 compares the relative abundances of specific sequence types for both the transcription reaction in the high yield condition (Figure 1) and the primer extension reaction on synthetic RNA (Figure 3).
Figure 4.
Sequence distributions of primer extension products. Summaries of (A) 3342 sequences longer than 30 bases and containing at least six contiguous bases attributable to primer extension from the high yield transcription reaction of Figure 1 and (B) 1037 sequences, similarly filtered, from the synthetic RNA primer extension reaction of Figure 3. Sequences are grouped into profiles based on the initiation of primer extension. For the two largest profiles in each, individual sequences were counted and presented as a percentage of the total. For profiles representing <5% of the total, only summaries are provided (lower third of figure), and profiles representing <1% of each pool are not shown. In each, sequences in blue have reverse complementarity to sequences in green, indicating that the latter templates the former. In (iii), bases in red do not fit this pattern, but could be consistent with priming from n + 1 and n + 2 products. Conversely, the location marked by a delta symbol suggests priming from an n −1 product RNA. (C) Likely mechanisms for primer extension in profiles (i) and (ii).
The data in Figure 4A and B reveal that the same two sequence profiles represent more than half of the products in each of the two reactions. One of the profiles (i) represents 36 and 25% of products in promoter-driven transcription and in synthetic primer extension, respectively. The second profile (ii) represents 17 and 36% of products in promoter-driven transcription and in primer extension, respectively. Within each profile, there is heterogeneity in the lengths represented. Given the caveats of precisely quantifying RNA-Seq data, we prefer not to over-interpret these sub-data, but note simply the variations in both the locus of extension initiation and the range of the extension. The third most abundant profile, the top line in (iii), is the same in each, representing substantially fewer sequences in each (4.6 and 4.4%, respectively), and again, there is substantial variance in lengths (not shown). These three initiation profiles represent 58 and 65% of all products in promoter-driven transcription and in primer extension reaction, respectively. For each reaction, the remaining ≈40% of products show wide variation in initiation profiles and similar variations in lengths, as illustrated by those reported with higher than 1% frequencies of occurrence.
It is not possible from these data to know the precise structure of the RNA that first primes initiation, but in Figure 4C we propose likely hairpin structures. For the first sequence profile (i), a reasonable structure provides three good base pairs (remembering that GU is a stable pair in RNA), with a relatively small loop. However, for the second profile (ii), a similar construct, model (a) shows poor base pairing of the 3′ terminal base.
For the second sequence profile (ii), a similar mechanism (a) could be invoked, but would require extension from an AA mismatch (28). In the data presented in Figure 1C, there is substantial evidence of ‘nontemplated’ addition of 1 or 2 nts onto the expected runoff RNA, and in particular, addition of C is favored. A potential mechanism for adding C is shown in model (b) of profile (ii), where pairing further upstream generates a large and more stable duplex that allows for templated addition of C. If after addition, the complex dissociates (yielding an n + 1 product), the extended RNA could then rebind the enzyme with a smaller, differently positioned loop, yielding a good 3′ terminal base pair (29) and leading to the observed extension sequences. At last, note that in the mechanistic model for profile (i) and in the second step of model (b) in profile (ii) above, there are 8 bases between the bases forming the terminal base pair, suggesting a preferred size for efficiently extended priming structures.
Primer extension depends strongly on sequences upstream of the 3′ end
Although the above models are highly constrained by the sequences of the observed products, they make assumptions about pairing between downstream and upstream sequence elements. To examine this more directly, we compare transcription under high yield conditions from a DNA template (Supplementary Table S1, 3N template) encoding the same terminal sequence as above, but with a different upstream (internal) sequence (3N RNA in Figure 5A). Analysis both by gel electrophoresis and by RNA-Seq length counting, presented in Figure 5B and C, respectively (and in Supplementary Figure S4), shows a dramatic reduction in (long) primer extension products.
Interestingly, as before, significant amounts of n + 1 and n + 2 products are observed, as revealed in Figure 5C and D. This might suggest that short extensions are less sensitive to upstream sequence than are larger extensions. Note also that the 5N RNA is ‘C-less’ in the non-randomized region and n + 1 additions of G are barely detectable (Figure 1C). In contrast, the 3N RNA contains five internal C’s and addition of G is the second most abundant n + 1 product (Figure 5D). Together these results strongly suggest that n + 1 (and presumably n +i) additions are templated.
The primer initiating model presented in Figure 4C, profile (i) suggests a key role for pairing A at the 3′ end of RNA with the U, 9 bases upstream. The 3N RNA of Figure 5 has a G at that position and is observed to not support (long) primer extension. To confirm the importance of this particular pairing, we designed a new DNA template (Supplementary Table S1) identical to the template for 5N of Figure 1, but replacing (only) the U at position +15 of the encoded RNA with A, as shown in Figure 6A (5N U→A RNA). Gel electrophoretic analysis in Figure 6B (and Supplementary Figure S4) shows a significant reduction in the formation of longer RNA products for this RNA. A double mutant (5N UG→AC RNA), with a second modification expected to further disrupt the base pairing in profile (i) and also expected to disrupt base pairing in the second part of model (b) of profile (ii), dramatically reduces the amount of longer primer extension products (Figure 6B). Conversely, starting from the 3N template that shows low levels of primer extension, we designed a single base change variant that introduces pairing of the RNA 3′ end with the base 9 bases upstream (3N G→U RNA in Figure 6C). Consistent with the model, transcription from this template shows an increase in the formation of longer RNA products (Figure 6B).
Figure 6.
Transcription profiles (high yield conditions) from DNA templates containing internal base substitutions. (A) Encoded RNA sequences from templates, starting from the sequence in Figure 1 (5N RNA) with canonical pairing potential (in green and purple). Mutations were then targeted to disrupt pairing (in red). (B) Twenty percent denaturing gel analysis (as in Figure 5B) of RNA products from those templates. (C) Encoded RNA sequences as in (A), but starting from sequence 3N RNA (Figure 5) with poor pairing potential (in red). Mutations were then targeted to establish pairing (in green).
DISCUSSION
T7 RNA polymerase is widely used to produce RNA by in vitro transcription from linear DNA templates (11). It is known that such reactions often contain multiple byproducts (11,19,20), yet the sequence dependencies and the mechanistic origins of these byproducts have remained unclear. While for many decades, denaturing gel electrophoresis has been used to analyze transcription products, we now introduce a new approach to study transcription profiles of RNA polymerases generally: RNA-Seq of in vitro transcription products. In addition to the length profiles provided by electrophoresis, RNA-Seq provides the precise sequences and sequence distributions of each length.
Product runoff RNA loops back to prime from its own upstream sequence
In this study, we have focused on the formation of RNA products longer than the expected runoff. These products have been reported in the past and have been attributed to different mechanistic origins (19,20,27). The RNA-Seq results presented here confirm that longer RNA products arise from RNA primer extension (18). Specifically, the sequence beyond the expected runoff transcript is almost exclusively the reverse complement of a sequence in the upstream region of the encoded RNA. We further confirm that such products are formed predominantly by cis primer extension, in which the runoff RNA primes and transcribes from its own upstream sequence by looping back on itself.
It has been proposed previously that RNA polymerase, at the end of a runoff template, might switch to the nontemplate strand, giving the same reverse complement pattern to the appended sequence. Although transcription here was carried out with ‘partially single stranded templates’, in which the nontemplate strand is truncated at position +2, the first round of transcription could lay down RNA as a nontemplate strand for subsequent rounds. However, the strong correlation between RNA accumulation and primer extension argues convincingly that free RNA rebinds to the polymerase to initiate extension. The observation that synthetic RNA is very efficient in serving as a template and generates approximately the same extension profiles, which confirms the model.
The two most abundant initiation profiles from template 5N and from 24-mer RNA suggest that sequences with 8 or 10 intervening bases, which could form hairpins with a 2–3 base pair stem and a 4-base loop, could serve as initiator. The crystal structures of T7 RNA polymerase in its initiation (with and without promoter DNA) configuration confirm that the active site can accommodate ∼6–9 nts (30), suggesting such a hairpin structure could be formed within the initiation structure of the polymerase (see ‘initial transcription’ and ‘primer extension’ cartoons in Figure 7). Although the enzyme normally transitions from the initiation to the elongation configuration at a hybrid length of ∼8–9 bases (12), it is thought that complexes that do not transition correctly can extend RNAs only to hybrid lengths as long as 11–13 bases (31,32), consistent with the largest single addition lengths observed here.
Figure 7.
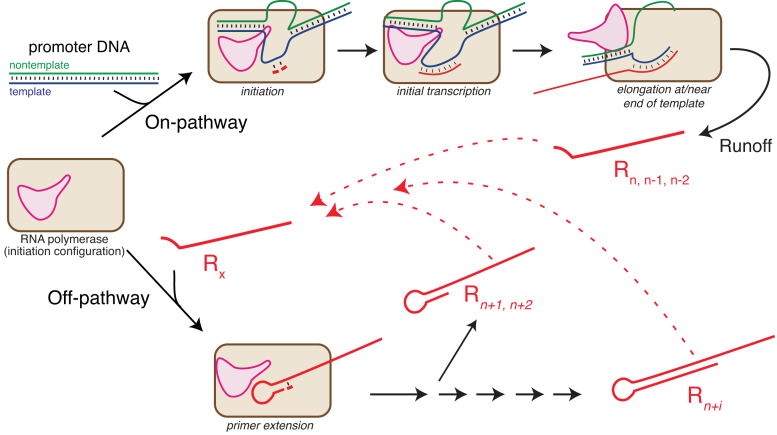
The ‘release and rebind’ model for the formation of longer RNA products by a cis primer extension mechanism. In the ‘On-pathway’ reaction, the promoter binding domain (pink) of T7 RNA polymerase binds promoter DNA and directs synthesis of the expected runoff RNA. In a competing ‘Off-pathway’ reaction, released RNA (Rx) rebinds to (a different) RNA polymerase and self-primes extension to longer RNA products in cis. RNAs shorter or longer than full length can function as Rx. Repeated rebinding/extension can (distributively) lead to still longer products.
Initiation of primer extension is heterogeneous and extension is distributive
These two profiles together account for slightly more than half of products ≥31 bases. The remaining products are spread across a variety of initiation possibilities. Many suggest initial priming at one position, followed by extension from another. Since n + 1 and n + 2 products begin to arise early in synthesis, it is not surprising that they can later rebind and prime further extension. Closer analysis of some of the minor sequences (see, for example, the third sequence in Figure 4A, profile (iii)) reveals apparent priming from n − 1 (and shorter) products as well.
The primer extension reaction of synthetic RNA shown in Figure 3C clearly demonstrates distributive synthesis. In other words, primer extended products are released and then rebind for further extension. Since the extension reaction does not compete with the promoter initiated transcription, distributive extension can grow each RNA to longer lengths. Note that in distributive synthesis, if rebinding is to the same site, the final sequence will be indistinguishable from that of a reaction that extends straight through (processive synthesis). Hence, this distributive behavior may be more prevalent than simple analysis of the transcription data suggests.
An important point arises from this distributive behavior. In traditional gel electrophoresis, the heterogeneity of the resulting products would yield a ‘smear’ at best, or each might drop below detection at worst. Thus, the nature of the 3′ end of the RNA could impact apparent yields of full length product.
Distributive primer extension may also have implications for early evolution, as it could provide a simple mechanism for rapidly increasing complexity in early RNAs. Conversely, later in evolution, the poly(U) tails that arise from hairpin-dependent termination and the poly(A) tails added during messenger RNA maturation could serve to limit formation of these now undesirable RNA species.
High yield transcription favors primer extension
The observations that primer extension is much more efficient in the absence of competing promoter DNA and that at low levels of RNA synthesis, primer extension is very inefficient relative to promoter-driven initiation indicate that (at least functional) rebinding of RNA is substantially weaker than (functional) promoter binding, as one would expect. Thus, primer extension increases dramatically as the runoff RNA accumulates under ‘high yield’ reaction conditions, as expected by a simple mass action model of binding. More specifically, efficient primer extension does not require the high concentrations of nucleoside triphosphates and/or magnesium typical of high yield reactions, as synthetic RNA in the absence of competing initiation is readily extended at much lower NTP and magnesium concentrations.
n + 1 and n + 2 ‘nontemplated’ products are actually templated
Under low yield conditions (template 5N), the expected length (n) product is the most abundant at 29%, but this drops to ∼7% under high yield conditions, confirming that longer products build off of shorter ones. RNAs of length n + 1 and n + 2 represent 23 and 7%, respectively, under low yield conditions, but represent 9 and 17%, respectively, under high yield conditions, suggesting that some n + 2 additions may be arising in a distributive manner, building on previously synthesized n + 1 RNAs.
Under both low and high yield conditions, the distribution of added nucleotides for these short lengths is not random, as 83% of n + 1 products contain added C. The predicted sequence of 5N RNA is ‘G-rich’ upstream of the terminal AUUUA-3′ sequence, which might explain the abundance of added C. While A and U are added at lower frequencies than C, we note that the predicted 5N RNA sequence is ‘C-less’ beyond the randomized region, and G is not observed in any n + 1 transcript more abundant than 1%. In contrast, the template for 3N RNA, which encodes the same 3′ terminal 5-base sequence, encodes five C’s upstream of that terminus, and added G represents 40% of the observed n + 1 products. Together, these data strongly argue that this well-known behavior previously termed ‘nontemplated addition’ in fact proceeds in a templated manner.
For decades, it has been assumed that ‘bad things’ happen when RNA polymerase sits around too long at the end of a linear DNA template. In retrospect, it should not be surprising that this is not the cause of n + i additions. The (hyper) forward translocation model for elongation complex dissociation predicts that RNA polymerases should become unstable as they approach the end of a template—the downstream barrier to forward translocation, melting of the bubble downstream of the active site, disappears as the polymerase nears the end of a template (33,34). Indeed, this instability is almost certainly the reason that RNA polymerases produce n −i products at the end of transcription. The notion that an RNA polymerase ‘waits around’ long enough to catalyze an unfavorable (nontemplated) reaction is unlikely, at best.
RNA-Seq: a new tool in mechanistic enzymology
The observed lack of G in the n + 1 products from the 5N RNA construct might have been ascribed to ligation bias (35). In other words, ligation of RNAs ending in …AUUUAG-3′ might be relatively inefficient, and so they would not appear or be under-represented in the RNA-Seq data set. The observation of an abundance of n + 1 products ending with G for the 3N RNA construct rules out this explanation, as both RNA sequences have the identical five encoded bases at their 3′ terminus. More broadly, however, one should always be aware of the potential for such artifacts in sequencing. In the current work, we have taken hypotheses generated by the RNA-Seq data and then tested them with both biochemical and follow-on RNA-Seq experiments.
Caution should be exercised in ‘horizontal’ quantitative comparisons, comparisons of the relative counts (intensities) of different RNA species within an experiment, as ligation bias against (or potentially for) structured RNAs could impact those comparisons (36). ‘Vertical’ quantitative comparisons of abundancies of the same RNA species across different conditions are much less likely to be influenced by such considerations and are particularly powerful.
While gel electrophoresis of RNA and DNA has been a staple of nucleic acid biochemistry for more than half a century, the complementary application of RNA-Seq approaches provides essential information not available from electrophoresis, or from even other sequencing approaches. Not only does RNA-Seq identify the most abundant sequence of any particular RNA length (and identify RNA lengths precisely), but also provides distributions of sequences, allowing powerful new insights into mechanism.
Summary
The model presented in Figure 7 summarizes our findings. The top ‘On-pathway’ process details expected runoff transcription, where the expected length (n) RNA is generated (this pathway also yields n − 1, n − 2 etc.). Rebinding of product RNA to polymerase initially competes poorly with ‘On-pathway’, promoter-initiated transcription. As product concentration grows, mass action drives ‘Off-pathway’ rebinding of product RNA to the polymerase, such that this process now competes favorably with ‘On-pathway’ initiation (the experiment with synthetic RNA only, in Figure 3, represents the extreme of ‘Off-pathway’ only).
Not only can the correct encoded product (n) rebind RNA polymerase, but also ‘Off-pathway’ n +i products can dissociate and then also rebind for further extension. Rebinding to the position from which each dissociated would be ‘silent’ in the sequencing results, but it is also possible, as evidenced here, that an n +i RNA product templated from one position could refold to prime at another position. More broadly, the current results explain why identical 3′ terminal sequences can yield very different patterns of ‘Off-pathway’ products.
DATA AVAILABILITY
Sequencing data have been deposited in the Small Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) with the BioProject accession code PRJNA486161
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [MCB-1516896]. Funding for open access charge: National Science Foundation [MCB-1516896].
Conflict of interest statement. None declared.
REFERENCES
- 1. Oakley J.L., Strothkamp R.E., Sarris A.H., Coleman J.E.. T7 RNA polymerase: promoter structure and polymerase binding. Biochemistry. 1979; 18:528–537. [DOI] [PubMed] [Google Scholar]
- 2. Martin C.T., Coleman J.E.. Kinetic analysis of T7 RNA Polymerase-Promoter interactions with small synthetic promoters. Biochemistry. 1987; 26:2690–2696. [DOI] [PubMed] [Google Scholar]
- 3. Cheetham G.M.T., Jeruzalmi D., Steltz T.A.. Structural basis for initiation of transcription from an RNA polymerase- promoter complex. Nature. 1999; 399:80–83. [DOI] [PubMed] [Google Scholar]
- 4. Yin Y.W., Steitz T.A.. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002; 298:1387–1395. [DOI] [PubMed] [Google Scholar]
- 5. Martin C.T., Mironova O. S.. Transcription: Initiation Wiley Encycl. Chem. Biol. 2008; 261:1076–1081. [Google Scholar]
- 6. Koh H.R., Roy R., Sorokina M., Tang G.Q., Nandakumar D., Patel S.S., Ha T.. Correlating transcription initiation and conformational changes by a Single-Subunit RNA polymerase with near Base-Pair resolution. Mol. Cell. 2018; 70:695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dunn J.J., Studier F.W., Gottesman M.. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 1983; 166:477–535. [DOI] [PubMed] [Google Scholar]
- 8. Chamberlin M., Ryan T.. 4 Bacteriophage DNA-dependent RNA polymerases. Enzymes. 1982; 15:87–108. [Google Scholar]
- 9. Sousa R., Patra D., Lafer E.M.. Model for the mechanism of bacteriophage T7 RNAP transcription initiation and termination. J. Mol. Biol. 1992; 224:319–334. [DOI] [PubMed] [Google Scholar]
- 10. Huang J., Brieba L.G., Sousa R.. Misincorporation by wild-type and mutant T7 RNA polymerases: Identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry. 2000; 39:11571–11580. [DOI] [PubMed] [Google Scholar]
- 11. Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C.. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987; 15:8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martin C.T., Muller D.K., Coleman J.E.. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry. 1988; 27:3966–3974. [DOI] [PubMed] [Google Scholar]
- 13. Krupp G. RNA synthesis: strategies for the use of bacteriophage RNA polymerases. Gene. 1988; 72:75–89. [DOI] [PubMed] [Google Scholar]
- 14. Arnaud-Barbe N., Cheynet-Sauvion V., Oriol G., Mandrand B., Mallet F.. Transcription of RNA templates by T7 RNA polymerase. Nucleic Acids Res. 1998; 26:3550–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Konarska M.M., Sharp P.A.. Replication of RNA by the DNA-dependent RNA polymerase of phage T7. Cell. 1989; 57:423–431. [DOI] [PubMed] [Google Scholar]
- 16. Konarska M.M., Sharp P.A.. Structure of RNAs replicated by the DNA-dependent T7 RNA polymerase. Cell. 1990; 63:609–618. [DOI] [PubMed] [Google Scholar]
- 17. Biebricher C.K., Luce R.. Template-free generation of RNA species that replicate with bacteriophage T7 RNA polymerase. EMBO J. 1996; 15:3458–3465. [PMC free article] [PubMed] [Google Scholar]
- 18. Cazenave C., Uhlenbeck O.C.. RNA template-directed RNA synthesis by T7 RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:6972–6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nacheva G.A., Berzal-Herranz A.. Preventing nondesired RNA-primed RNA extension catalyzed by T7 RNA polymerase. Eur. J. Biochem. 2003; 270:1458–1465. [DOI] [PubMed] [Google Scholar]
- 20. Triana-Alonso F.J., Dabrowski M., Wadzack J., Nierhaus K.H.. Self-coded 3′-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J. Biol. Chem. 1995; 270:6298–6307. [DOI] [PubMed] [Google Scholar]
- 21. Kao C., Zheng M., Rüdisser S.. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999; 5:1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maslak M., Martin C.T.. Kinetic analysis of T7 RNA polymerase transcription initiation from promoters containing single-stranded regions. Biochemistry. 1993; 32:4281–4285. [DOI] [PubMed] [Google Scholar]
- 23. Takita E., Kohda K., Tomatsu H., Hanano S., Moriya K., Hosouchi T., Sakurai N., Suzuki H., Shinmyo A., Shibata D.. Precise sequential DNA ligation on A solid substrate: solid-based rapid sequential ligation of multiple DNA molecules. DNA Res. 2013; 20:583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hafner M., Landgraf P., Ludwig J., Rice A., Ojo T., Lin C., Holoch D., Lim C., Tuschl T.. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008; 44:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B. et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009; 25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rong M., Durbin R.K., McAllister W.T.. Template strand switching by T7 RNA polymerase. J. Biol. Chem. 1998; 273:10253–10260. [DOI] [PubMed] [Google Scholar]
- 27. Mu X., Greenwald E., Ahmad S., Hur S.. An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 2018; 46:5239–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pomerantz R.T., Temiakov D., Anikin M., Vassylyev D.G., McAllister W.T.. A mechanism of nucleotide misincorporation during transcription due to template-strand misalignment. Mol. Cell. 2006; 24:245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zaher H.S., Unrau P.J.. T7 RNA polymerase mediates fast promoter-independent extension of unstable nucleic acid complexes. Biochemistry. 2004; 43:7873–7880. [DOI] [PubMed] [Google Scholar]
- 30. Cheetham G.M.T., Steitz T.A.. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999; 286:2305–2309. [DOI] [PubMed] [Google Scholar]
- 31. Ramírez-Tapia L.E., Martin C.T.. New insights into the mechanism of initial transcription: The T7 RNA polymerase mutant p266l transitions to elongation at longer RNA lengths than wild type. J. Biol. Chem. 2012; 287:37352–37361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Esposito E.A., Martin C.T.. Cross-linking of promoter DNA to T7 RNA polymerase does not prevent formation of a stable elongation complex. J. Biol. Chem. 2004; 279:44270–44276. [DOI] [PubMed] [Google Scholar]
- 33. Zhou Y., Navaroli D.M., Enuameh M.S., Martin C.T.. Dissociation of halted T7 RNA polymerase elongation complexes proceeds via a forward-translocation mechanism. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Santangelo T.J., Roberts J.W.. Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Mol. Cell. 2004; 14:117–126. [DOI] [PubMed] [Google Scholar]
- 35. Giraldez M.D., Spengler R.M., Etheridge A., Godoy P.M., Barczak A.J., Srinivasan S., De Hoff P.L., Tanriverdi K., Courtright A., Lu S. et al. Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat. Biotechnol. 2018; 36:746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fuchs R.T., Sun Z., Zhuang F., Robb G.B.. Bias in ligation-based small rna sequencing library construction is determined by adaptor and RNA structure. PLoS One. 2015; 10:e0126049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in the Small Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) with the BioProject accession code PRJNA486161