Abstract
Despite recent progress on synthetic transcription factor generation in eukaryotes, there remains a need for high-activity bacterial versions of these systems. In synthetic biology applications, it is useful for transcription factors to have two key features: they should be orthogonal (influencing only their own targets, with minimal off-target effects), and programmable (able to be directed to a wide range of user-specified transcriptional start sites). The RNA polymerase of the bacteriophage T7 has a number of appealing properties for synthetic biological designs: it can produce high transcription rates; it is a compact, single-subunit polymerase that has been functionally expressed in a variety of organisms; and its viral origin reduces the connection between its activity and that of its host's transcriptional machinery. We have created a system where a T7 RNA polymerase is recruited to transcriptional start sites by DNA binding proteins, either directly or bridged through protein–protein interactions, yielding a modular and programmable system for strong transcriptional activation of multiple orthogonal synthetic transcription factor variants in Escherichia coli. To our knowledge this is the first exogenous, programmable activator system in bacteria.
INTRODUCTION
Synthetic biology represents on ongoing effort to treat biology as an engineering science. First the individual biological parts such as cells, proteins, and metabolites must be understood by thorough characterization and standardization. Then these parts can be interfaced with other parts to reconstitute natural biological networks and their behaviors, or to generate novel ones. Experience shows, however, that native biological parts often prove too complex for an engineer to characterize and standardize. Additionally, even if a part is well characterized in one system it is currently not possible to predict that it will operate identically in another context. Thus scientists and engineers often devise simpler parts and networks that capture the desired natural behaviors.
When creating a regulatory circuit for use in a synthetic biological design, it is helpful if the components (such as the transcription factors we will discuss here) have two properties. They should be orthogonal: able to act on their own targets without exerting off-target effects on the targets of other synthetic components. And they should be programmable: capable of being directed to a wide range of user-specified targets rather than restricted to a limited set of naturally-occurring ones. An orthogonal, programmable set of regulatory components (such as transcription factors) offers designers in synthetic biology the capacity to create complex circuits where multiple inputs and outputs are processed in the same cell without interfering with one another.
In the initial phase of synthetic biology's development, there was a significant shortage of useful biological components; this became known the ‘parts/components problem’ (1,2). At the level of the regulation of transcriptional initiation, only a handful of well-characterized promoter/transcription factor combinations were practical in bacteria and eukaryotes, and the same few components played central roles in many synthetic circuit designs. In 2012, the development of synthetic transcription factors (sTFs) in yeast (3) presented a path towards resolving the issue: the sTFs could be programmed to initiate transcription from any location definable by the DNA binding sequences of zinc finger arrays (ZFAs). This work was soon expanded with other programmable DNA binding proteins (DBPs), including transcription activator–like effectors (TALEs) and dCas9 (4–10). There is now such a wide variety of parts based on sTFs that it is reasonable to conclude that eukaryotes no longer have a significant transcriptional parts problem.
A similarly wide range of synthetic transcriptional control options have not been developed for bacteria. This is at least partly due to the predominantly ‘on by default’ nature of transcription in bacteria: unlike in eukaryotes where transcription is generally off by default in the absence of RNAP recruitment by transcription factors, the bacterial RNAP holoenzyme recognizes its promoter without auxiliary transcription factors (11). This results in a situation where promoters are most effectively controlled by blocking or otherwise inhibiting the polymerase using repressor proteins, but limited ability exists to activate transcription directly with transcription factors. The inducible repressors AraC, LacI and TetR (controlled with arabinose, isopropyl β-d-1-thiogalactopyranoside (IPTG) and anhydrotetracycline (ATc), respectively) are still the repressible promoter systems seen most frequently in the literature, and inducible transcriptional activation in bacteria is most commonly achieved by de-repression (12).
Considerable effort has been directed to expanding the pool of orthogonal bacterial repressors through mutagenesis, bioinformatics-driven parts mining and directed evolution (13). This can provide repressors that bind new inducers (14–17) as well as bind new operator sites (18,19). This approach has been refined to the point that the design and construction of arbitrary logical operations in DNA can be automated (20).
However, as successful as repressor-based transcriptional regulation has been, direct transcriptional activation offers an important alternative mode of regulation in synthetic biology, with the potential to simplify the design and implementation of synthetic circuits. The active recruitment of transcription factors could also allow synthetic systems in bacteria to explore the more complex and modular modes of activation characteristic of eukaryotic systems. Two main approaches exist for synthetic transcriptional activation in bacteria: bacterial one/two-hybrid type systems and phage-based systems.
Bacterial hybrid systems use minimal bacterial promoters and replace the UP element (which recruits the C-terminal domain of the RNA polymerase alpha or omega subunits) with the DNA binding motif of the desired DNA-binding domain. The C-terminal domain is then replaced with a DNA-binding domain matching the DNA binding motif. A two hybrid version associates the DNA-binding domain with the RNAP alpha subunit through protein–protein interactions. Thus, the bacterial RNAP is recruited to the minimal promoter with the help of a DNA-binding domain. This approach has been successfully used with zinc finger arrays, TALEs, dCas9 and others for transcriptional activation (21–27). These systems also double as repressors when targeted to sterically block the RNAP binding at the promoter. Other successful non-hybrid approaches to activation use activating RNAs that de-repress RNAP elongation (28) and sigma factor-anti-sigma pairs (29,30). However, what limits the hybrid systems, with few exceptions, is their modest activity. Additionally, the use of the host RNAP in these systems makes it more challenging to produce large sets of orthogonal activators, and introduces potential interference between the host's native RNAP and the engineered hybrid system.
The second approach to bacterial activation consists of phage-based systems, employing proteins from bacteriophage viruses (especially RNA polymerases, but phage-derived transcription factors like the lambda repressor can also be used as activators) (11). RNA polymerases from bacteriophages transcribe so strongly that they form the basis of the majority of protein over-expressing bacterial cell lines. The most common phage RNAP system is the T7 RNAP, a single subunit RNAP that recognizes a single DNA motif between 17 and 22 bp long. The T7 RNA polymerase has appealing properties for synthetic biological applications: it can produce very high transcription rates (31,32) useful for example in amplification steps or high-gain feedback circuits (33); its compact single-unit nature makes it relatively easy to express; and it has been functionally expressed in a variety of contexts including both prokaryotes and eukaryotes (34–37).
Numerous successful efforts have been made to engineer the T7 RNAP to recognize different promoter sequences (38–42) as well as to generally enhance its activity (31,32). Further, T7 RNAP has been split to generate inducible (43,44) and logic gated operations (45–47). Combined, these efforts yield an impressive library of T7 RNAPs that can target orthogonal promoters with very high transcriptional activation. Furthermore, T7 RNAP can be made functional in eukaryotes, allowing it to act as a widely-applicable tool (34,35). However, the convenient single-subunit nature of the T7 RNAP also results in coupled binding and transcription initiation activities, preventing the relatively straightforward process of directing the polymerase to a specific start site by modifying the DNA binding behavior of a DNA-binding domain in isolation, as is possible in bacterial hybrid and eukaryotic systems. Whereas the same core polymerase can be fused or post-translationally dimerized to a DBP to define its activity in other systems, phage RNAPs must be re-engineered specifically for each new DNA binding motif, currently a time-consuming process.
Our goal in this work was to make the T7 RNAP programmable (like recruitment-based bacterial hybrid and eukaryotic systems), while retaining its strong transcriptional activity. Achieving this aim requires fusing a customizable DNA-binding domain to the T7 RNAP, either directly or through a protein–protein bridge. For programmability, this DNA-binding protein must be modifiable such that designers can select a wide range of DNA sequences as the binding target. To achieve the desired property of orthogonality, we require two system features: the activity of the T7 RNAP must depend strongly on its recruitment by the DNA-binding domain (preventing significant activity in the absence of the target DNA-binding sequence); and multiple DNA-binding domains must be sufficiently specific that they do not induce significant T7 RNAP recruitment on DNA sequences other than their own targets.
We have developed a solution to this design challenge, which we call the ‘iiT7 system’, where ‘ii’ stands for ‘initiation impaired’. The system, to be described below, consists of a set of DNA-binding proteins (zinc finger arrays), fused or bridged to either wild-type or mutated T7 RNA polymerases, along with a truncated T7 promoter that displays recruitment-dependent activity when placed adjacent to the DNA-binding targets of the zinc finger arrays. The iiT7 system represents a programmable transcriptional activator in Escherichia coli, the first such system of which we are aware.
MATERIALS AND METHODS
Cell culture
Escherichia coli strain DH5αZ1 (Expressys, Germany) was used in prototyping measurements and strain MG1655Z1 (a gift from Jeff Hasty, UCSD) was used for final measurements using leucine zippers. Cells were grown in EZ Rich Defined Media (Teknova, USA) with antibiotics where appropriate; 50 ug/ml carbenicillin, 50 ug/ml kanamycin, 25 ug/ml Zeocin.
DNA parts and manipulation
The reporter plasmid pOEGFP (this lab) was modified to contain the ZFA binding sites upstream of the T7 promoter using restriction digest of annealed oligonucleotides (Eurofins Operon, USA) between the NheI and AvaI sites. ‘Spacer’ distances are from the end of the NheI scar to the beginning of the ZFA binding site. The plasmid pSB3K3 (BioBricks, iGEM.org) was used to express T7 (BBa_I202079), ZFA-T7 and Leucine Zipper (LZ)-T7 constructs from a Lac inducible promoter (BBa_J04500) cloned in through the BioBrick standard assembly. Any further modifications to any plasmids were done with site directed mutagenesis (Q5 Site-Directed Mutagenesis Kit, NEB) or Gibson Assembly (Gibson Assembly® Cloning Kit and NEBuilder® HiFi DNA Assembly Master Mix, NEB). The plasmid pADCR4 (a gift from Jeff Hasty (48), UCSD) was used to express ZFA-LZ. ZFAs were provided by Keith Joung (Harvard) and Marcus Noyes (NYU), and variants were ordered as gBlocks (IDT, USA). Leucine zippers were ordered as gBlocks (IDT, USA) and derived from An3.5, Bn3 and Bn3.5 (49) due to their well characterized binding affinities and tunability. All inserts into plasmids were confirmed by DNA sequencing (TCAG, Toronto and Eurofins Operon, USA) prior to measurement. Sequences and descriptions can be found in the Supplementary Information.
Cell transformation and measurement
Escherichia coli cells were made chemically competent with standard procedures. Cells were transformed with a bulk heat shock method in a 96 well round bottomed plate (29,50). Briefly, 50 μl of chemically competent E. coli were incubated with 1 μl of 5 ng/μl per plasmid on ice for 30 min. Cells were then heat shocked at 44°C for 45 s and returned to ice for 2 min. 100μl of room temperature SOC was added per well, covered with a breathable membrane (VWR 60941-086) and the plate was incubated at 37°C for 2 h at 900 revolutions per minute (rpm). A 2 ml deep 96 well plate (VWR 89237-526) with a 6 mm borosilicate bead per well was prepared with 400 μl of EZ Rich Defined Media (Teknova M2105) per well as described and 30 μl of culture was added to each respective well to incubate overnight at 37°C at 600 rpm with appropriate antibiotics. Overnight cultures were diluted 1/50 into a new 2 ml deep 96 well plate containing 600 μl EZ Rich Defined Media per well and incubated for up to 7 hrs with appropriate antibiotics. Starting at 2 h, 100 μl aliquots were measured hourly in a black 96 well plate (Thermo 152036) in a Tecan M1000 Pro plate reader. Settings were as follows: 4 s of horizontal shaking; absorbance 600 nm, 4 s horizontal shaking, fluorescence (excitation = 485 nm, emission = 525 nm).
Data analysis
Data analysis was performed in Microsoft Excel. Fluorescence was normalized with the following formula ((Fsample – Fmedia)/(ODsample – ODmedia)) – ((Fcontrol – Fmedia)/(ODcontrol – ODmedia)). Errors bars are one standard deviation of three independently cultured samples. P-values for statistical significance were not calculated as only large effects were of interest in this investigation. Figures were generated from time points in which cell were in exponential growth (usually 3 or 4 h). Fluorescence Relative to WT T7 was calculated by dividing each well by the well containing T7 on its own pt7 promoter (this promoter is often denoted PT7 or pT7, but here we will use the convention that all DNA sequences, including promoters, are entirely in lower case, to distinguish them from the all-upper-case protein constructs). ‘Fold activity’ was calculated by dividing each T7 or ZFA variant's fluorescence output by the fluorescence output of wild-type T7 RNA polymerase (WT T7 RNAP) on the promoter corresponding to the same T7 or ZFA variant. This normalizes each T7 or ZFA variant to WT T7 RNAP on the modified promoter, providing the fold activity that the variant offers over WT T7 RNAP alone. This is a measure of the performance of the recruitment-based iiT7 system versus WT T7 RNAP, and uses the WT T7 RNAP as a proxy for off-target T7 constructs that would be present in a system containing multiple promoters.
RESULTS AND DISCUSSION
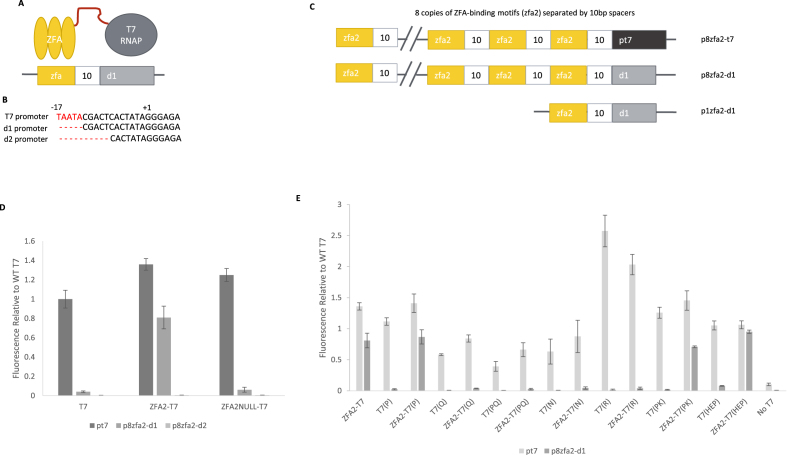
The initial version of the iiT7 system is shown schematically in Figure 1A: a zinc finger array (ZFA) is fused by a linker to the T7 RNA polymerase. Transcription is initiated by recruitment of the ZFA-T7 RNAP complex to the vicinity of a truncated T7 promoter, d1 (Figure 1B). The top DNA sequence represents the WT T7 promoter whereas the bottom d2 promoter represents additional deletion of the ‘specificity loop’ binding region. Later, we replace the direct linker with a protein–protein interaction ‘bridge’ (Figure 3A). Selection and testing of each element of the system will be discussed below. Table 1 offers a list of the labels we have used for the key protein constructs and promoters that appear below.
Figure 1.
The direct-fusion version of the iiT7 system. (A) A schematic of the direct-fusion iiT7 system. A zinc finger array (ZFA) is attached by a peptide linker to the T7 RNA polymerase. The DNA sequence binding target (zfa) of the ZFA serves to recruit the protein fusion to the vicinity of a truncated T7 promoter sequence, d1. (B) The promoter sequences of the wild-type T7 promoter (pt7), and two truncated versions of the promoter (d1 and d2). (C) Schematics of several promoters used for testing: p8zfa2-t7 incorporates 8 copies of the zfa2 binding sequence, each spaced 10 bp apart, followed by the wild-type pt7 promoter sequence; p8zfa2-d1 replaces the pt7 sequence with the truncated d1 sequence; and p1zfa2-d1 uses only a single copy of the zfa2 binding sequence before d1. (D) Expression from promoters pt7, p8zfa2-d1, and p8zfa2-d2, in the presence of the wild-type T7 RNAP (T7), the fusion ZFA2-T7 (WT T7 RNAP fused to ZFA2), and the fusion ZFA2NULL-T7 (WT T7 RNAP fused to a non-DNA-binding mutant of ZFA2). Values are normalized to WT T7 activity on its native pt7 promoter. [Error bars represent one standard deviation for an n of 3]. (E) Expression of a set of T7 RNAP mutants (labels described in the text) from promoters pt7 and p8zfa2-d1, normalized to WT T7 on pt7. Each mutant's normalized activity is shown on both promoters, when expressed on its own (e.g. T7(PQ)) and when fused to a zinc finger array (e.g. ZFA2-T7(PQ)). The T7-HEP mutant shows nearly wild-type activity on the p8zfa2-d1 promoter when recruited by in the ZFA2-T7(HEP) construct. [Error bars represent one standard deviation for an n of 3].
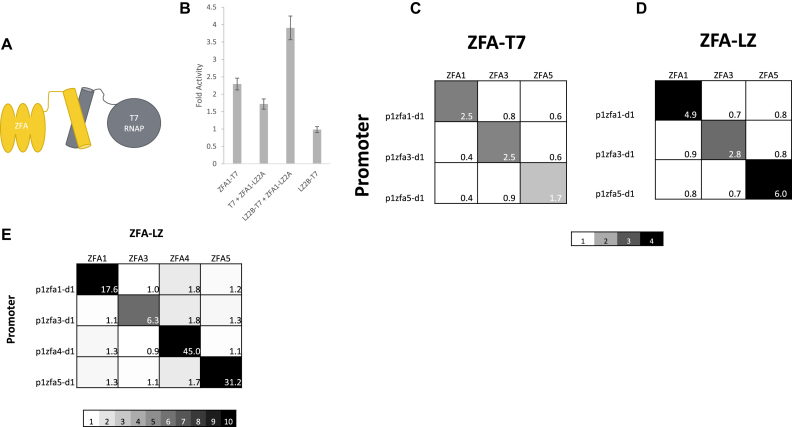
Figure 3.
The leucine zipper bridged version of the iiT7 system. (A) A schematic of the bridged iiT7 system. A zinc finger array (ZFA) is attached by a peptide linker to a leucine zipper (LZ), and a complementary LZ is attached to the wild-type T7 RNAP (WT T7). (B) Activities of various construct combinations on promoter p1zfa2-d1, normalized to WT T7 RNAP on the same promoter (fold activity): the direct fusion ZFA1-T7; the bridged pair ZFA1-LZ2A + LZ2B-T7; the unbridged pair ZFA1-LZ2A + WT T7; and the unrecruited LZ2B-T7. [Error bars represent one standard deviation for an n of 3]. (C) An orthogonality plot showing fold activities on p1zfa{1,3,5}-d1 for the fused iiT7 system (constructs ZFA{1,3,5}-T7), provided for direct comparison with the bridged system. (D) An orthogonality plot showing fold activities on promoters p1zfa{1,3,5}-d1 for the bridged iiT7 system (constructs ZFA{1,3,5}-LZ2A + LZ2B-T7). Data in panels (C) and (D) are with the target promoters on plasmid backbone pADCR4. (E) Improved orthogonality and fold activity in the bridged system when placing the target promoters on plasmid backbone pADCR5, which lowers the basal expression level. The 4 × 4 grid shows fold activities on promoters p1zfa{1,3,4,5}-d1, in the presence of the bridged pairs of constructs ZFA{1,3,4,5}-LZ2A + LZ2B-T7. Scale color range is used to emphasize orthogonality.
Table 1.
A list of labels used for the protein and DNA constructs examined here. Proteins and protein fusions are given labels entirely in upper-case letters (ZFA1, ZFA2-LZ2A, etc.) DNA sequences, including promoters, are labeled entirely in lower-case letters (d1, zfa2, etc.) All promoter sequences begin with the letter ‘p’ (p8zfa2-t7, etc.)
| Label | Type | Description |
|---|---|---|
| ZFA{1-5}; ZFA{1,2}NULL | Proteins | Zinc finger arrays. ZFA1: sTF 43-8 (3). ZFA2: sTF 43.8 (3) with a Zif268 (53) backbone. ZFA3: sTF 54.8 (3). ZFA4: Zif268 (53). ZFA5: 46698 (61). ZFA1NULL, ZFA2NULL: ZFA1/ZFA2 with mutations to abolish DNA-binding activity |
| WT T7 RNAP | Protein | The wild-type T7 RNA polymerase |
| T7(HEP) | Protein | WT T7 RNAP with mutations H772R, E775R, and P266L |
| ZFA{1-5}-T7; ZFA{1-5}-T7(HEP) | Protein fusions | ZFA{1-5} fused with linker GS(GGGS)2 to WT T7 RNAP; or fused with the same linker to T7(HEP) |
| Linker {1-3} | Peptide linkers | Linker 1: GS(GGGS). Linker 2: GS(GGGS)2. Linker 3: GS(GGGS)3 |
| zfa1, zfa2, zfa3, zfa4, zfa5 | DNA sequences | The DNA-binding target sequences of ZFA1 through ZFA5 |
| pt7 | Promoter | Wild-type T7 promoter sequence |
| d1 | DNA sequence | pt7 with bases -13 to -17 deleted |
| d2 | DNA sequence | pt7 with bases -8 to -17 deleted |
| p8zfa2-t7 | Promoter | 8 copies of zfa2 (each separated by 10-bp spacers), followed by pt7 |
| p{1,2,8}zfa2-d1; p{1,2,8}zfa2-d2 | Promoters | 1, 2, or 8 copies of zfa2 (each separated by 10-bp spacers), followed by d1; or followed by d2 |
| LZ1A/LZ1B, LZ2A/LZ2B, LZ3A/LZ3B | Proteins | Leucine zipper pairs. LZ1A/LZ1B: AN3/BN3. LZ2A/LZ2B: AN3.5/ BN3.5. LZ3A/LZ3B: AN4/BN4 (46) |
| ZFA{1-5}-LZ2A, LZ2B-T7 | Protein fusions | ZFAs 1 through 5 fused with linker GS(GGGS)2 to LZ2A; and LZ2B fused with the same linker to WT T7 RNAP. (And similarly for other LZ pairs.) |
Selection of DNA binding domains
To achieve recruitment at user-specified DNA sequences, our system requires a set of DNA binding domains that allow targeting to arbitrary DNA sequences. These domains must be able to fuse to the T7 RNAP while maintaining their own activity and not disrupting the T7 RNAP’s activity. Zinc fingers (ZFs) are a family of DNA binding proteins that are promising candidates: they are relatively small, with a simple structure that reduces the chances of misfolding or aggregation; they have been successfully employed in the design of eukaryotic synthetic transcription factors (3) and in bacterial hybrid screens in E. coli; and they can be designed to bind almost any DNA sequence. TALEs and CRISPR-Cas9 were also considered, but both are significantly larger and more complex, so at this stage we have focused on zinc fingers.
The best studied ZFs are the members of the C2H2 fold group which bind to both strands of DNA through their alpha helical region and can be fused into zinc finger arrays (ZFAs) to bind longer and more specific target sequences. Large libraries of arrays have been developed using methods such as modular assembly (51), OPEN (52) and CoDA (53), and can be searched through ZiFit (54,55). The ZFA sTF 43-8, a three fingered ZFA (henceforth denoted ZFA1; all our protein and protein fusion names will use only upper-case letters) was selected based on its high specificity as described in Khalil et al. (3). Additionally, a hybrid ZFA (denoted ZFA2) was constructed using the DNA binding helix from 43-8 and the backbone of the canonical ZFA Zif268 (56). While beta-hairpin backbones do differ between zinc fingers, the placement of the critical C2H2 residues are highly conserved. DNA-binding alpha helices can be grafted on to different ZF beta-hairpin backbones and still retain function (57,58).
Both ZFA1 and ZFA2 repressed expression approximately 4- to 5-fold in a repression assay where a tetracycline inducible promoter was modified by replacing the TetO elements with ZFA DNA binding motifs (Supplementary Figure S1). ‘Null’ variants of the zinc finger arrays (ZFA1NULL and ZFA2NULL) were produced by changing the DNA-binding amino acids in each finger of the ZFAs to alanine, abolishing the ZFAs DNA-binding activity as shown by the same repression assay (Supplementary Figure S1).
We next fused each of ZFA1 and ZFA2 to the T7 RNAP, resulting in two protein constructs: ZFA1-T7 and ZFA2-T7. The result is shown schematically in Figure 1A: a DNA-binding domain (ZFA, in this case ZFA1 or ZFA2) is fused by a peptide linker (here GS(GGGS)2, or GSGGGSGGGS) to the T7 RNA polymerase. Repeating the repression assay while expressing ZFA1-T7 and ZFA2-T7 showed that the fusion to the T7 RNAP did not affect the ability of the ZFAs to bind (Supplementary Figure S1). The activity of the ZFA1-T7 and ZFA2-T7 constructs on the wild-type pt7 promoter was comparable to the activity of the wild-type T7 RNAP on the same pt7 promoter (Supplementary Figure S2). In some cases the fusions showed slightly higher activity (20–40% higher) than the wild-type T7 RNAP on its own, indicating some unknown interaction between the ZFAs and the T7 RNAP.
Construction of the iiT7 prototype
To recruit the ZFA-T7 constructs to the vicinity of a transcriptional start site, we began by augmenting the wild-type pt7 promoter with upstream sequences matching the shared DNA-binding target sequence of ZFA1 and ZFA2 (which have common DNA-binding helices and differ only in their backbones). We placed eight copies of their DNA-binding target sequence (denoted zfa2; all DNA sequences, including promoters, will have names that are entirely in lower case) upstream of the pt7 promoter, with each zfa2 sequence separated by a 10 base-pair spacer. The spacer sequence was randomly generated, but checked for interference with other sequences in the other promoters or genes present in the system. This promoter is denoted p8zfa2-t7: eight copies of the DNA binding domain target site (zfa2), followed by the full pt7 promoter sequence, as shown in Figure 1C. Measuring GFP fluorescence from this promoter failed to show any enhanced activity by the ZFA{1,2}-T7 constructs from the p8zfa2-t7 promoter: instead, activity was similar to that obtained from the wild-type pt7 promoter (Supplementary Figure S2). We speculated that this was a saturation effect, wherein the expression from just the pt7 promoter was already so strong that it was not possible to increase it further through recruitment by the fused ZFAs.
We then explored making use of truncated versions of the pt7 promoter sequence, seeking variants that would continue to support transcriptional initiation by the T7 RNAP, while being sufficiently weakened to allow for increased activity in the presence of recruitment by the ZFAs. The fact that DNA binding is mechanistically distinct from promoter melting (59) (though WT T7 RNAP interactions require the free energy of binding for promoter melting (60)) suggests that it should be possible to alter the promoter region to reduce the T7 RNAP’s binding affinity while still retaining its ability to initiate transcription. Ideally, the ZFA should replace the binding function of the T7 RNAP’s AT loop, while being structurally dissociated enough through the linker that it does not hinder promoter clearance. Thus, in the truncated promoter sequence we denote ‘d1’, bases –17 to –13 were deleted from the wild-type pt7 sequence, removing the DNA motif that binds the T7 RNAP AT loop as well as the 5′ A specificity base (Figure 1B), which is expected to dramatically reduce WT T7 RNAP activity (61). A second, more severe truncation we denote ‘d2’ further removed bases –13 to –8, containing the entire specificity loop (Figure 1B). These truncated T7 promoter sequences were again combined with eight repeats of the zfa2 binding target sequences to create two new promoters, p8zfa2-d1 and p8zfa2-d2 (eight copies of the DNA-binding domain target sequence, followed by the d1 or d2 truncations of the pt7 promoter sequence); see Figure 1C.
Our initial testing of the d1 and d2 truncated promoter variants used a two plasmid system, with one plasmid expressing the ZFA2-T7 construct from the lactose inducible promoter BBa_J04500 while the reporter plasmid incorporated the p8zfa2-d1 or p8zfa2-d2 promoter expressing EGFP. The output of this system was normalized to the level of fluorescence (as a proxy for transcriptional activity) obtained by wild-type T7 RNAP acting on its own promoter in a second two-plasmid system expressing WT T7 RNAP from the same promoter and same plasmid backbone as the ZFA2-T7 construct, with a reporter plasmid expressing EGFP from the wild-type pt7 promoter. This ‘T7-normalized activity’ allowed us to measure how much transcriptional activity was being achieved through recruitment, compared to what the natural T7 system produced.
The complete iiT7 system consists of two key elements: the promoter (incorporating the ZFA target binding sequence as well as a truncated portion of the wild-type pt7 promoter); and a protein construct (consisting of a zinc finger array fused to the T7 RNAP). Figure 1D shows the result of measuring T7-normalized activity of the expression from three promoters (pt7, p8zfa2-d1, and p8zfa2-d2), in the presence of three protein constructs (wild-type T7 RNAP (T7), ZFA2-T7, and ZFA2NULL-T7, in which the DNA-binding ability of ZFA2 has been disrupted as described above). The wild-type T7 RNAP’s output from its wild-type promoter pt7 has, by definition, a T7-normalized activity of 1. WT T7 RNAP displays barely measurable activity on the p8zfa2-d1 promoter, and no activity above background on the p8zfa2-d2 promoter.
The ZFA2-T7 construct showed a T7-normalized activity of 1.4 on the wild-type pt7 promoter (Figure 1D), indicating a slight enhancement effect for the fusion construct when compared to the WT T7 RNAP, in the absence of any recruitment elements in the promoter. (We have not investigated the nature of this enhancement, though the T7-normalized activity of 1.2 shown by ZFA2NULL-T7 on the pt7 promoter suggests that the effect is not specifically related to the DNA-binding function of the ZFA.) Successful recruited activity is illustrated by the 0.8 T7-normalized activity of ZFA2-T7 on p8zfa2-d1 whereas the WT T7 RNAP shows near-zero activity from this same promoter, as does the nonbinding ZFA2NULL-T7 construct.
The p8zfa2-d2 promoter variant showed no transcriptional activity (Figure 1D) in the presence of any of the protein constructs, and no subsequent efforts were able to induce measurable expression from this more severe truncation of the pt7 promoter sequence. We therefore discontinued investigation of the d2 truncation variant, and all subsequent testing and optimization of the iiT7 system concentrated entirely on variants using the d1 truncation. The failure of this d2 truncation means that we were not able to achieve a version of the iiT7 system in which transcriptional initiation would be triggered entirely through recruitment to the DNA-binding target sites, without even a truncated pt7 promoter sequence; such a version may yet be possible, but would likely require extensive modification of the T7 RNAP itself. Our screens of T7 RNAP mutants (described in the next section) did not uncover any ability to initiate transcription from promoters based on the d2 truncation.
Optimizing the iiT7 system's activity
In an effort to increase the level of recruited activity seen with ZFA2-T7 acting on p8zfa2-d1, we screened a number of T7 RNA polymerase mutants that affected DNA binding in the specificity loop (N748, R756, and Q758) (39,40,62), two involved in the progression to elongation (P266L, K172L) (63) and two that were common variants in a screen for alternate promoter specificity (H772R, E775V) (41). Since each of the mutations is of a different amino acid, we will refer to T7 RNAP mutants using one or more parenthetical letters to indicate which of the above-listed mutations has been applied: T7(P) refers to WT T7 RNAP with the P266L mutation; T7(HEP) refers to WT T7 RNAP with the H772R, E775V, and P266L mutations; and so on. Figure 1E shows the result of testing a range of ZFA-mutant T7 RNAP protein constructs for activity on both the pt7 promoter and the p8zfa2-d1 promoter. Each T7 RNAP mutant was tested on its own as well as fused (with linker GS(GGGS)2) to the ZFA2 DNA-binding domain (yielding constructs like ZFA2-T7(P), and so on). A wide range of T7-normalized activities was obtained from the mutants and their fusion constructs, on both promoters, but of particular note is the behaviour of the T7(HEP) mutant (right-hand set of bars in Figure 1E). Without a fused ZFA domain, T7(HEP) shows wild-type activity on pt7, and near zero activity on p8zfa2-d1. The ZFA2-T7(HEP) construct continues to show wild-type activity on pt7, but now also shows close to wild-type activity (T7-normalized activity of 0.95) from p8zfa2-d1, demonstrating that a recruited mutant T7 RNAP can initiate transcription from the truncated d1 sequence nearly as strongly as WT T7 RNAP does from the wild-type, non-truncated pt7 promoter sequence.
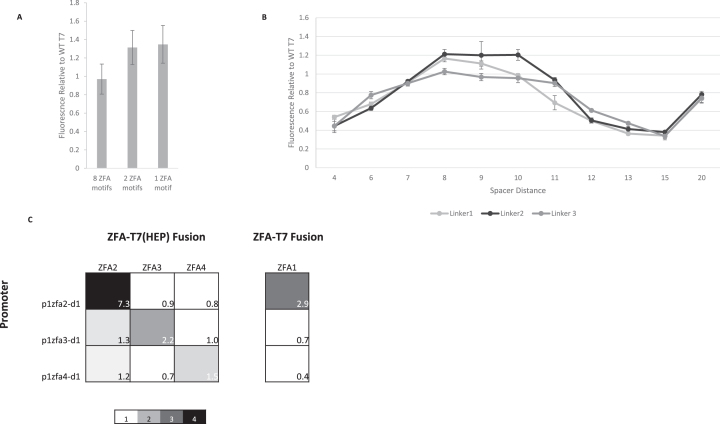
In an effort to simplify the design, we investigated the effect of varying the number of zfa2 binding sites upstream of the d1 sequence: we created promoters with one and two copies of the zfa2 sequence, called p1zfa2-d1 and p2zfa2-d1, respectively. Surprisingly, having fewer ZFA binding sequences does not reduce the recruited activity levels (Figure 2A) shown by the fusion ZFA2-T7(HEP). This simplification came at the cost of somewhat increased background expression (from p1zfa2-d1 in the absence of any T7 RNAP or fusion construct; see Supplementary Figure S3), but the greatly reduced complexity of the design was a compelling reason to select the single-zfa2 site version of the promoter (p1zfa2-d1) as the default for further development of the iiT7 system.
Figure 2.
Tuning and orthogonality in the direct-fusion iiT7 system. (A) Activities of the ZFA2-T7(HEP) fusion on three promoter variants incorporating 8, 2, or 1 copy of the zfa2 DNA-binding target sequences before the truncated d1 promoter sequence (p8zfa2-d1, p2zfa2-d1, and p1zfa2-d1, resp.) Values are normalized to WT T7 RNAP on its own pt7 promoter. Reducing the number of zfa2 sites does not reduce the activity of the promoters. [Error bars represent one standard deviation for an n of 3]. (B) Activities of the ZFA2-T7(HEP) fusion on versions of the p1zfa2-d1 promoter in which the length of the spacer sequence has been varied from 4 to 20 bp. The length of the linker fusing ZFA2 to T7(HEP) is also varied, creating ZFA2-T7(HEP) variants using Linker 1 (GS(GGGS)), Linker 2 (GS(GGGS)2), and Linker 3 (GS(GGGS)3). Activities are normalized relative to the fluorescence output of WT T7 RNAP on its native pt7 promoter. Spacer length has a strong effect on activity, while linker length has a much less substantial influence. [Error bars represent one standard deviation for an n of 3]. (C) An orthogonality plot, showing on-target and off-target activity levels for fusions ZFA{2,3,4}-T7(HEP), acting on promoters p1zfa{2,3,4}-d1. Values are given as ‘fold activities’: the ratio of observed fluorescence to the fluorescence measured with WT T7 RNAP acting on the same promoter. The fourth, separated column shows the reduced off-target activity obtained by the fusion ZFA1-T7 (using the wild-type, unmutated T7 RNAP). Scale color range is used to emphasize orthogonality.
We investigated the effect of varying both the number of bases in the spacer sequence and the length of the linker used to fuse ZFA2 to the wild-type T7 RNAP. (Complete spacer/linker tests with the ZFA2-T7(HEP) fusions were not conducted, but partial tests with this mutant showed very a very similar pattern to that seen in Figure 2B; see Supplementary Figure S12.) Figure 2B shows the result of varying the spacer length from 4 to 20 bases (as before, the spacer sequences are randomly chosen but subject to disqualification based on interference with other sequences in the system), measuring output from varying-spacer-length versions of the promoter p8zfa2-d1. Three different protein fusions were created, with linkers GS(GGGS) (Linker 1), GS(GGGS)2 (Linker 2), and GS(GGGS)3 (Linker 3) connecting the ZFA2 domain to WT T7 RNAP. The results suggest that spacer length has a strong influence on T7-normalized transcriptional activity, with the strongest outputs observed for spacers in the 8–10 bp range. All three linkers showed the same pattern as a function of spacer length, with relatively small differences between the linkers. On the basis of these results, we decided to fix a spacer length of 10 bases in our promoters and to employ Linker 2 (GS(GGGS)2) in our fusions, as the basis for further development of the design.
Orthogonality testing with additional ZFAs
The desired property of orthogonality means that we must be able to direct the iiT7 system to multiple user-defined targets, with different variants having minimal off-target effects. We tested the orthogonality of the iiT7 system by creating a set of new zinc finger arrays as the DNA-binding domains: 54.8 (3) (ZFA3), Zif268 (56) (ZFA4), and 46698 (64) (ZFA5). Applying the notation established above, each of these has its own target DNA binding sequence (zfa3, zfa4, and zfa5), and its own corresponding promoter (p1zfa3-d1, p1zfa4-d1 and p1zfa5-d1). For orthogonality, we would ideally have the fusions ZFA2-T7(HEP) through ZFA5-T7(HEP) showing high activity from their corresponding promoters, while showing little activity on off-target promoters (those not incorporating matching DNA binding sites). In a synthetic regulatory circuit we would have multiple protein constructs and multiple promoters present simultaneously, so it is important to measure the level of transcriptional activity induced by an unrecruited T7 RNA polymerase (e.g. in a system that included the p1zfa2-d1 promoter, the ZFA3-T7(HEP) construct could still induce some level of nonspecific activity from the zfa2-based promoter.) We will use the term ‘fold activity’ to denote ratio of a candidate ZFA-T7 construct's observed fluorescence (with baselines removed as described in the Methods section) to the fluorescence output from that same promoter in the presence of the wild-type T7 RNAP. Fold activity provides a ratio of recruited to unrecruited activity, in both on-target and off-target cases, and uses the WT T7 RNAP as a proxy for the effect of a generic off-target construct on a given promoter.
Figure 2C shows the results (expressed as fold activities) of testing each of the protein fusions ZFA2-T7(HEP), ZFA3-T7(HEP) and ZFA4-T7(HEP) against their own cognate promoters (p1zfa2-d1, p1zfa3-d1, and p1zfa4-d1) and against the promoters matching the other constructs. (Note that ZFA1 and ZFA2 have the same DNA binding target sequence, but the ZFA1-T7(HEP) construct failed to show good on-target activity on p1zfa2-d1 and we thus excluded it from the initial set of ZFAs; see Supplementary Figure S4). The 3 × 3 array in Figure 2C shows moderate orthogonality, with on-target fold activities ranging from 1.5 to 7.3, but with off-target activations of 1.2 and 1.3 too similar to the lowest on-target activation of 1.5. While the ZFA1-T7(HEP) construct did not activate successfully, a test of ZFA1-T7 (a fusion to the wild-type T7 RNAP), shown as a separate column in Figure 2C, produced improved results: an on-target fold activity of 2.9, with significantly lower off-target activations than those produced by ZFA2-T7(HEP). These initial results suggested that the system had the potential to generate orthogonal sets of transcription factors, but that it would require further development to improve both on-target and off-target performance.
Improved activity with leucine zippers
Searching for additional zinc finger arrays revealed that at least one of the candidate ZFAs (ZFA5) showed very strong binding to its target sites (through a repression assay, see Supplementary Figure S6) but failed to show significant recruited activation. We speculated that this was a result of ‘tethering,’ whereby the fused ZFA-T7 construct failed to escape the promoter after transcriptional initiation, because it was being physically constrained by the strong binding of the ZFA’s DNA binding domain. This hypothesis gained support when we produced a new (and improved) iiT7 system variant in which the bridge between the ZFAs and the T7 RNAP consisted of pairs of leucine zipper (LZ) domains, with protein–protein interactions that are intended to be less strong than the ZFA-DNA binding interactions (see Figure 3A). Leucine zippers (LZs) are small, coiled coil domains that in their simplest form are a pair of alpha helices. They associate naturally through hydrophobic core interactions that form primarily homodimers (or at least do not preferentially heterodimerize). However, recent efforts have generated a number of heterodimerizing LZs by mutating key hydrophobic residues into complementary charged residues between LZ pairs (49,65,66). Electrostatics between two charged LZs generate a strong attraction for the complementary charged partner, resulting in a strong preference for heterodimers (67). These LZs exist as small libraries with characterized binding constants spanning two orders of magnitude, providing the necessary tuning information, and have been used to assemble protein sensors and actuators (68,69). We selected the LZ pairs AN3/BN3 (we will label them LZ1A/LZ1B), AN3.5 and BN3.5 (LZ2A/LZ2B), and AN4/BN4 (LZ3A/LZ3B) (the superscripts refer to lengths of the domains, expressed as numbers of heptad repeats) (49). We will show results from all three of these pairs, but the intermediate-length leucine zipper pair LZ2A and LZ2B was selected for initial testing of the use of protein–protein interaction domains in the iiT7 system (this also represents the first instance of their in vivo use as far as we are aware). The LZ2A domain was fused to the C terminus of ZFA1, producing the construct ZFA1-LZ2A. A complementary fusion construct, LZ2B-T7(HEP), was generated by fusing the LZ2B domain to the N terminus of the T7(HEP) mutant RNA polymerase. Tests of the LZ2B-T7(HEP) construct had high on-target levels of activation, but the off-target activation was also found to be unacceptably high (see Supplementary Figure S9). Replacing the mutated T7(HEP) RNAP with the wild-type T7 RNAP, yielding the construct LZ2B-T7, reduced the off-target activity to reasonable levels (to be discussed below). The cause of this context-specific change in the behaviour of the T7(HEP) mutant compared to WT T7 is not currently understood. We chose to adopt the best-performing available version of the polymerase in this revised context, which turned out to be the wild-type T7 RNAP, and used that for further development of the iiT7 system. The new construct ZFA1-LZ2A was expressed from the plasmid pADCR4 under the control of the ptet promoter (usually rendered as Ptet or Ptet, but exclusively lower-case will continue to be used for DNA sequences).
Figure 3B shows tests and controls of the new system, with fusion and LZ-bridged constructs expressed in the presence of the promoter p1zfa1-d1. The direct fusion construct ZFA1-T7 shows recruited activation, as before, but the bridged combination of constructs ZFA1-LZ2A + LZ2B-T7 shows significantly higher fold activity than the original fused version. The construct LZ2B-T7 on its own (without the matching ZFA1-L2A construct) has a fold activity of 1, indicating wild-type levels of activity in the absence of the recruiting element. A spontaneous mutation in LZ2B-T7 disrupted LZ2A-LZ2B binding, and confirmed that this binding was required for specific recruited activity (Supplementary Figure S7). For reasons not currently understood, expressing WT T7 RNAP in the presence of the ZFA1-LZ2A construct (T7 + ZFA1-LZ2A) shows a fold activity well above 1.
Figure 3C and D provide a side-by-side comparison between the direct-fusion (Figure 3C) and leucine zipper bridged (Figure 3D) orthogonality plots for a set of three ZFAs and their corresponding promoters. Figure 3C shows the results of testing constructs ZFA1-T7, ZFA3-T7 and ZFA5-T7 against the set of promoters p1zfa1-d1, p1zfa3-d1 and p1zfa5-d1, yielding an orthogonality plot with moderate on-target fold activities (from 1.7 to 2.5), and an average off-target fold activity of 0.6 (ranging from 0.4 to 0.9). The leucine-zipper equivalents of these direct-fusion constructs are the following pairs of constructs: ZFA1-LZ2A + LZ2B-T7; ZFA3-LZ2A + LZ2B-T7 and ZFA5-LZ2A + LZ2B-T7 (note that only the ZFA constructs vary, the LZ2B-T7 construct is common to all pairs). Each of these pairs of constructs were co-expressed in the presence of same set of promoters (p1zfa1-d1, p1zfa3-d1, and p1zfa5-d1), leading to the orthogonality plot shown in Figure 3D. The on-target fold activities are all higher in the LZ-bridged version, with improvements ranging from slight to substantial (2.5 to 4.9; 2.5 to 2.8 and 1.7 to 6.0). The off-target activity also increased somewhat, with an average off-target fold activity of 0.8 (ranging from 0.7 to 0.9). It is interesting to note that what had been the poorest-performing activator (ZFA5-T7) in the direct-fusion version is now the best-performing activator (ZFA5-LZ2A + LZ2B-T7) in the LZ-bridged version. This result lends indirect support to the tethering hypothesis: ZFA5 showed strong binding to its target in repression assays, so its relatively poor activation upon recruitment through a direct fusion could be explained by a failure to allow promoter escape; this could be relieved by the LZ-bridged system, in which the strongly-bound ZFA5-LZ2A element can remain behind as the LZ2B-T7 element breaks the LZ2A-LZ2B association and proceeds to escape the promoter.
The performance of the system was further improved by placing all of the reporter promoters into a different plasmid backbone (pADCR5, see the spreadsheet provided in the Supplementary Information for full information); this new plasmid has the same antibiotic resistance but a reduced copy number, resulting in lower basal expression from the promoters. Figure 3E provides the final version of the iiT7 system's orthogonality results, showing fold activity values for constructs ZFA{1,3,4,5}-LZ2A, in each case co-expressed with LZ2B-T7 and tested against the full set of cognate promoters (p1zfa{1,3,4,5}-d1). On-target activations range from 6.3 to 45, with an average off-target fold activity of 1.3 (ranging from 0.9 to 1.8). This set of constructs represents a set of highly orthogonal transcriptional activators for use in genetic circuit designs in E. coli.
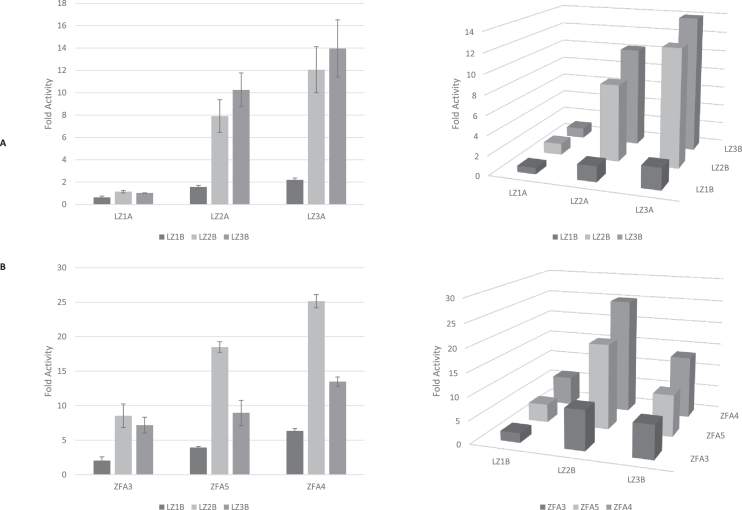
To explore how the choice of leucine zipper affected recruited activation levels, we created the remaining two additional leucine zipper bridged pairs of constructs, each fused to ZFA1: ZFA1-LZ1A + LZ1B-T7, and ZFA1-LZ3A + LZ3B-T7. The strength of the in vitro binding affinities of the LZ pairs increases with length (49), with the LZ1A-LZ1B pair having the weakest binding and LZ3A-LZ3B having the strongest. We examined the effect of varying the members of the LZ pairs, including ‘mismatched’ pairings, by testing co-expressed constructs representing the nine possible permutations of LZ pairs: ZFA1-LZ1A + LZ1B-T7; ZFA1-LZ1A + LZ2B-T7; ZFA1-LZ1A + LZ3B-T7; ZFA1-LZ2A + LZ1B-T7; ZFA1-LZ2A + LZ2B-T7; ZFA1-LZ2A + LZ3B-T7; ZFA1-LZ3A + LZ1B-T7; ZFA1-LZ3A + LZ2B-T7 and ZFA1-LZ3A + LZ3B-T7. The results are shown in Figure 4A (left) as bar graphs to allow display of the error bars, and the same data is also displayed (right) on a 3 × 3 grid to show the two-dimensional pattern. Within each choice of ‘A-side’ LZ domain (LZ1A, LZ2A or LZ3A), there is a trend: fold activity generally increases with the length of the paired ‘B-side’ LZ domain (LZ1B, LZ2B, or LZ3B). The same trend applies in the inverse: within each choice of B-side LZ domain, fold activity increases with the length of the paired A-side LZ domain. Varying the choice of LZ domain pairs offers designers of synthetic circuits a method of tuning the level of transcriptional activation in their implementation of the iiT7 system.
Figure 4.
Effects of varying leucine zipper pairs in the bridged iiT7 system. (A) With the ZFA element fixed at ZFA1, we show expression from promoter p1zfa1-d1 as fold activity (normalized to WT T7 RNAP on the same promoter), for the 9 permutations of constructs ZFA1-LZ{1,2,3} + LZ{1,2,3}B-T7. (left) The data presented as bar plots. [Error bars represent one standard deviation for an n of 3]. (right) The same data presented as a 2D plot. (B) With one LZ element fixed at LZ2A, fold activities from the nine permutations of promoters p1zfa{3,4,5}-d1 in the presence of constructs ZFA{3,4,5}-LZ2A + LZ{1,2,3}B-T7. (left) The data presented as bar plots. [Error bars represent one standard deviation for an n of 3]. (right) The same data presented as a 2D plot.
The above results kept the DNA-binding domain ZFA1 fixed and varied the LZ domains. We also explored the interactions between the choices of the ZFA and the LZ domains, by keeping one side of the LZ pairs fixed at LZ2A, while varying the ZFA (among ZFA3, ZFA4, and ZFA5) as well as varying the other side of the LZ pair (among LZ1B, LZ2B, and LZ3B). This led to the following nine permutations of pairs of constructs: ZFA3-LZ2A + LZ1B-T7; ZFA3-LZ2A + LZ2B-T7; ZFA3-LZ2A + LZ3B-T7; ZFA4-LZ2A + LZ1B-T7; ZFA4-LZ2A + LZ2B-T7; ZFA4-LZ2A + LZ3B-T7; ZFA5-LZ2A + LZ1B-T7; ZFA5-LZ2A + LZ2B-T7 and ZFA5-LZ2A + LZ3B-T7. The results are shown in Figure 4B (left) as bar graphs to show the error bars, as well as (right) on a 3 × 3 grid to show the two-dimensional pattern. One trend is that for every choice of LZ-T7 construct, the fold activities increase from ZFA3 to ZFA5 to ZFA4. A second trend is that the LZ2B-T7 construct gives the highest fold activities, for every choice of ZFA-LZ construct; LZ3B-T7 comes in second, and LZ1B-T7 gives the lowest fold activities. This is somewhat surprising given the previous results, where the LZ3B-T7 construct provided the highest fold activities (when paired with any of the ZFA1-LZ constructs). It is not currently clear why the best LZ pairing should vary with the choice of ZFA domain.
Growth rate curves were recorded during every experiment described above, and sample curves are presented in Supplementary Figure S11, for several ZFA-LZ variants of the bridged iiT7 system, as well as for no-plasmid cells and cells expressing comparable levels of EGFP from a fully induced ptet promoter. Some slight slowing of growth rates was observed in some cases: the no-plasmid cells had 0.83 h doubling times, while expressing EGFP from plac increased the doubling time to 0.93 h, and the ZFA{1,3,4,5}-LZ constructs had doubling times of 0.82, 0.80, 0.95 and 0.83 h respectively. This suggests that the iiT7 system does not place a substantial burden on the host cells, beyond what normally results from reporter protein expression.
CONCLUSIONS
Our results show that an orthogonal set of synthetic transcription factors (with strong on-target and low off-target activity) can be constructed in E. coli, and this represents the first programmable activation system in bacteria of which we are aware, expanding the range of options available for transcriptional activity in bacteria and offering the prospect of employing the same class of recruitment-based approaches already available in eukaryotic organisms.
In our development of the iiT7 system, we used mutation of the T7 RNA polymerase and truncation of its wild-type pt7 promoter to impair the T7 RNAP’s DNA binding capabilities, thus decoupling the T7 RNAP’s promoter recognition and transcriptional initiation functions and enabling promoter recognition to be made dependent on an auxiliary DNA binding protein. A significant enhancement in performance was obtained by replacing covalent fusion between the DNA binding domains with protein–protein interactions (through our leucine zipper pairs), which further decoupled the DNA binding process from transcriptional initiation, and likely helped to facilitate promoter escape and transcript elongation.
After testing a range of variants and optimizing its performance, the best-functioning current form of the iiT7 system (as represented by the orthogonality plot in Figure 3E) consists of the following: one protein fusion incorporating a DNA-binding domain fused to a leucine zipper (such as ZFA5-LZ2A); a second protein fusion incorporating a leucine zipper selected to form protein–protein interactions with the first construct and fused to a T7 RNAP (such as LZ2B-T7); and a promoter that incorporates both the DNA-binding target of the zinc finger array and a truncated T7 promoter sequence (such as p1zfa5-d1). Programming the system (creating new targets) involves swapping out the ZFA domains, and inserting the matching DNA binding sequences into the promoter. Tuning of the resulting levels of activation may be accomplished by varying the choice of ZFA domains, LZ domains, or both. The number of permutations available for tuning could quickly get out of control, so our advice to designers seeking to use the system (especially if working without a high-throughput screening pipeline) would be to select a LZ pair and leave that fixed while exploring the activity levels resulting from varying just the ZFA domains; changes to the LZ domains can be brought in at a later stage if needed, to tweak the activity levels.
This two component phage-based RNAP system in bacteria begins to resemble transcription in eukaryotes, where transcription factors recruit core RNAPs to minimal promoters that have little activity or specificity in isolation. Interestingly, coincident with the preparation of this manuscript, Hillen et al. showed through crystallography that the human mitochondrial RNAP (a homologue of T7 RNAP) uses a mechanism very similar to the one we have developed here, where the mitochondrial transcription factor TFAM substitutes for the ZFAs in our system (70). TFAM and the mitochondrial RNAP interact through alpha-helical domains strikingly similar to the leucine zippers we have used for the same purpose in our system (Supplementary Figure S8). It is comforting to have unintentionally converged on the same type of solution as billions of years of evolution. This convergence can also be used to inform future directions by suggesting that DNA melting can also be further decoupled from transcription in RNAP as the second mitochondrial transcription factor TFB2M assumed this role in the mitochondrial RNAP.
The iiT7 system offers a promising method for expanding the number of synthetic transcription factors available in bacterial contexts, with the large number of known ZFA and LZ components providing the basis for the modular construction of user-selected libraries of orthogonal components, each capable of high transcriptional activity upon recruitment. With further searching of the mutational space of the T7 polymerase, it may be possible to find a mutant with transcriptional activity in the absence of any element of the native T7 promoter, which would further simplify the system.
An interesting divergence in utility was discovered with the T7 RNAP HEP mutant. This mutant, while generally enhancing on-target expression from promoters based on the d1 truncation of the pt7 promoter sequence (with the exception of the ZFA1-T7(HEP) fusion) actually resulted in slightly decreased overall orthogonality due to a uniform increase in off-target activity (Supplementary Figure S9). Interestingly, the same data shows that the poor on-target activity of the ZFA1-T7(HEP) direct fusion was restored in the LZ-bridged version of the system: ZFA1-LZ2A + LZ2B-T7(HEP) had strong on-target activity, at a level nearly identical to that of its sibling ZFA2-LZ2A + LZ2B-T7(HEP). Understanding the relationship between ZFA domains and their interaction with fused and otherwise bound RNAPs requires further investigation.
Both the DNA-binding and the protein–protein interaction domains used here may be replaced with new variants: we have demonstrated a ZFA-based version of the system, but in principle any DNA-binding domain can be employed, including transcriptional-activator-like (TAL) effectors and dCas9. Separating the DNA-binding event from the recruitment through protein–protein interactions opens up the possibility of tuning or switching of the synthetic transcription factors’ activity through the growing number of known methods of external protein–protein interaction modulation, including chemical and light induction (71,72). The split iiT7 complex could be built around polynucleotide and peptide scaffolds using modality-specific binding domains, acting as a sensor (73,74) and allowing tunability for ultrasensitive responses (75). The T7 RNAP polymerase is known to operate in eukaryotes (34), opening the future possibility of creating synthetic transcription factors able to operate in either prokaryotes or eukaryotes. Though work remains to be done to fully develop the iiT7 design, this is an opportunity not only to test the universality of transcription design principles, but also to emulate the natural evolution of RNAPs from the simple single domain phage T7 to the modular and programmable polymerases of complex cells.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for biological parts received from the groups of Jeff Hasty, Keith Joung, Timothy Lu and Marcus Noyes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Sciences and Engineering Research Council (NSERC) of Canada through the Discovery Grant program (grant number 191013); Canada First Research Excellence Fund through the Medicine by Design program (grant number 170338). Funding for open access charge: NSERC Discovery Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1. Purnick P.E.M., Weiss R.. The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell Biol. 2009; 10:410–422. [DOI] [PubMed] [Google Scholar]
- 2. Lu T.K., Khalil A.S., Collins J.J.. Next-generation synthetic gene networks. Nat. Biotechnol. 2009; 27:1139–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khalil A.S., Lu T.K., Bashor C.J., Ramirez C.L., Pyenson N.C., Joung J.K., Collins J.J.. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012; 150:647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013; 154:442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maeder M.L., Linder S.J., Cascio V.M., Fu Y., Ho Q.H., Joung J.K.. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods. 2013; 10:977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Konermann S., Brigham M.D., Trevino A.E., Hsu P.D., Heidenreich M., Cong L., Platt R.J., Scott D.A., Church G.M., Zhang F.. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013; 500:472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farzadfard F., Perli S.D., Lu T.K.. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth. Biol. 2013; 2:604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Purcell O., Peccoud J., Lu T.K.. Rule-Based design of synthetic transcription factors in eukaryotes. ACS Synth. Biol. 2014; 3:737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crocker J., Stern D.L.. TALE-mediated modulation of transcriptional enhancers in vivo. Nat. Methods. 2013; 10:762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Esvelt K.M., Mali P., Braff J.L., Moosburner M., Yaung S.J., Church G.M.. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods. 2013; 10:1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Browning D.F., Busby S.J.W.. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016; 14:638–650. [DOI] [PubMed] [Google Scholar]
- 12. Shaw K. Negative transcription regulation in prokaryotes. Nat. Educ. 2008; 1:122. [Google Scholar]
- 13. Brödel A.K., Jaramillo A., Isalan M.. Intracellular directed evolution of proteins from combinatorial libraries based on conditional phage replication. Nat. Protoc. 2017; 12:1830–1843. [DOI] [PubMed] [Google Scholar]
- 14. de los Santos E.L.C., Meyerowitz J.T., Mayo S.L., Murray R.M.. Engineering transcriptional regulator effector specificity using computational design and in vitro rapid Prototyping: Developing a vanillin sensor. ACS Synth. Biol. 2016; 5:287–295. [DOI] [PubMed] [Google Scholar]
- 15. Shis D.L., Hussain F., Meinhardt S., Swint-Kruse L., Bennett M.R.. Modular, Multi-Input transcriptional logic gating with orthogonal LacI/GalR family chimeras. ACS Synth. Biol. 2014; 3:645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Taylor N.D., Garruss A.S., Moretti R., Chan S., Arbing M.A., Cascio D., Rogers J.K., Isaacs F.J., Kosuri S., Baker D. et al. Engineering an allosteric transcription factor to respond to new ligands. Nat. Methods. 2015; 13:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ellefson J.W., Ledbetter M.P., Ellington A.D.. Directed evolution of a synthetic phylogeny of programmable Trp repressors. Nat. Chem. Biol. 2018; 14:361–367. [DOI] [PubMed] [Google Scholar]
- 18. Stanton B.C., Nielsen A.A.K., Tamsir A., Clancy K., Peterson T., Voigt C.A.. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat. Chem. Biol. 2013; 10:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jayaraman P., Devarajan K., Chua T.K., Zhang H., Gunawan E., Poh C.L.. Blue light-mediated transcriptional activation and repression of gene expression in bacteria. Nucleic Acids Res. 2016; 44:6994–7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nielsen A.A.K., Der B.S., Shin J., Vaidyanathan P., Paralanov V., Strychalski E.A., Ross D., Densmore D., Voigt C.A.. Genetic circuit design automation. Science. 2016; 352:aac7341. [DOI] [PubMed] [Google Scholar]
- 21. Hubbard B.P., Badran A.H., Zuris J.A., Guilinger J.P., Davis K.M., Chen L., Tsai S.Q., Sander J.D., Joung J.K., Liu D.R.. Continuous directed evolution of DNA-binding proteins to improve TALEN specificity. Nat. Methods. 2015; 12:939–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L.A.. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013; 41:7429–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meng X., Brodsky M.H., Wolfe S.A.. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat. Biotechnol. 2005; 23:988–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee J.Y., Sung B.H., Yu B.J., Lee J.H., Lee S.H., Kim M.S., Koob M.D., Kim S.C.. Phenotypic engineering by reprogramming gene transcription using novel artificial transcription factors in Escherichia coli. Nucleic Acids Res. 2008; 36:e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Joung J.K., Ramm E.I., Pabo C.O.. A bacterial two-hybrid selection system for studying protein-DNA and protein–protein interactions. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:7382–7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Amores G.R., Guazzaroni M.-E., Silva-Rocha R.. Engineering Synthetic cis -Regulatory elements for simultaneous recognition of three transcriptional factors in bacteria. ACS Synth. Biol. 2015; 4:1287–1294. [DOI] [PubMed] [Google Scholar]
- 27. Brödel A.K., Jaramillo A., Isalan M.. Engineering orthogonal dual transcription factors for multi-input synthetic promoters. Nat. Commun. 2016; 7:13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chappell J., Takahashi M.K., Lucks J.B.. Creating small transcription activating RNAs. Nat. Chem. Biol. 2015; 11:214–220. [DOI] [PubMed] [Google Scholar]
- 29. Rhodius V.A., Segall-Shapiro T.H., Sharon B.D., Ghodasara A., Orlova E., Tabakh H., Burkhardt D.H., Clancy K., Peterson T.C., Gross C.A. et al. Design of orthogonal genetic switches based on a crosstalk map of s, anti- s, and promoters. Mol. Syst. Biol. 2014; 9:702–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bervoets I., Van Brempt M., Van Nerom K., Van Hove B., Maertens J., De Mey M., Charlier D.. A sigma factor toolbox for orthogonal gene expression in Escherichia coli. Nucleic Acids Res. 2018; 46:2133–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boulain J.-C., Dassa J., Mesta L., Savatier A., Costa N., Muller B.H., L’hostis G., Stura E.A., Troesch A., Ducancel F.. Mutants with higher stability and specific activity from a single thermosensitive variant of T7 RNA polymerase. Protein Eng. Des. Sel. 2013; 26:725–734. [DOI] [PubMed] [Google Scholar]
- 32. Wang Y., Prosen D.E., Mei L., Sullivan J.C., Finney M., Vander Horn P.B.. A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Res. 2004; 32:1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nilgiriwala K.S., Jiménez J., Rivera P.M., Del Vecchio D.. Synthetic tunable amplifying buffer circuit in E. coli. ACS Synth. Biol. 2015; 4:577–584. [DOI] [PubMed] [Google Scholar]
- 34. Pinkham J.L., Dudley A.M., Mason T.L.. T7 RNA Polymerase-Dependent expression of COXII in yeast mitochondria. Mol. Cell. Biol. 1994; 14:4643–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ghassemi F., Madadgar O., Roohvand F., Rasekhian M., Etemadzadeh M.H., Boroujeni G.R.N., Langroudi A.G., Azadmanesh K.. Translational efficiency of BVDV IRES and EMCV IRES for T7 RNA polymerase driven cytoplasmic expression in mammalian cell lines. Mol. Biol. 2017; 51:283–292. [DOI] [PubMed] [Google Scholar]
- 36. Cheetham G.M. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999; 286:2305–2309. [DOI] [PubMed] [Google Scholar]
- 37. Cheetham G.M., Jeruzalmi D., Steitz T.A.. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature. 1999; 399:80–83. [DOI] [PubMed] [Google Scholar]
- 38. Temme K., Hill R., Segall-Shapiro T.H., Moser F., Voigt C.A.. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 2012; 40:8773–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chelliserrykattil J., Cai G., Ellington A.D.. A combined in vitro/in vivo selection for polymerases with novel promoter specificities. BMC Biotechnol. 2001; 1:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Raskin C.A., Diaz G.A., McAllister W.T.. T7 RNA polymerase mutants with altered promoter specificities. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:3147–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meyer A.J., Ellefson J.W., Ellington A.D.. Directed evolution of a panel of orthogonal T7 RNA polymerase variants for in vivo or in vitro synthetic circuitry. ACS Synth. Biol. 2015; 4:1070–1076. [DOI] [PubMed] [Google Scholar]
- 42. Esvelt K.M., Carlson J.C., Liu D.R.. A system for the continuous directed evolution of biomolecules. Nature. 2011; 472:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ike K., Arasawa Y., Koizumi S., Mihashi S., Kawai-Noma S., Saito K., Umeno D.. Evolutionary design of Choline-Inducible and -Repressible T7-Based induction systems. ACS Synth. Biol. 2015; 4:1352–1360. [DOI] [PubMed] [Google Scholar]
- 44. Han T., Chen Q., Liu H.. Engineered photoactivatable genetic switches based on the bacterium phage T7 RNA polymerase. ACS Synth. Biol. 2017; 6:357–366. [DOI] [PubMed] [Google Scholar]
- 45. Schaerli Y., Gili M., Isalan M.. A split intein T7 RNA polymerase for transcriptional AND-logic. Nucleic Acids Res. 2014; 42:12322–12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Segall-Shapiro T.H., Meyer A.J., Ellington A.D., Sontag E.D., Voigt C.A.. A ‘resource allocator’ for transcription based on a highly fragmented T7 RNA polymerase. Mol. Syst. Biol. 2014; 10:742–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shis D.L., Bennett M.R.. Library of synthetic transcriptional AND gates built with split T7 RNA polymerase mutants. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:5028–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Didovyk A., Borek B., Hasty J., Tsimring L.. Orthogonal modular gene repression in escherichia coli using engineered CRISPR/Cas9. ACS Synth. Biol. 2016; 5:81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thomas F., Boyle A.L., Burton A.J., Woolfson D.N.. A set of de Novo designed parallel heterodimeric coiled coils with quantified dissociation constants in the micromolar to Sub-nanomolar regime. J. Am. Chem. Soc. 2013; 135:5161–5166. [DOI] [PubMed] [Google Scholar]
- 50. Mutalik V.K., Guimaraes J.C., Cambray G., Lam C., Christoffersen M.J., Mai Q.-A., Tran A.B., Paull M., Keasling J.D., Arkin A.P. et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. 2013; 10:354–360. [DOI] [PubMed] [Google Scholar]
- 51. Wright D.A., Thibodeau-Beganny S., Sander J.D., Winfrey R.J., Hirsh A.S., Eichtinger M., Fu F., Porteus M.H., Dobbs D., Voytas D.F. et al. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat. Protoc. 2006; 1:1637–1652. [DOI] [PubMed] [Google Scholar]
- 52. Maeder M.L., Thibodeau-Beganny S., Sander J.D., Voytas D.F., Joung J.K.. Oligomerized pool engineering (OPEN): an ‘open-source’ protocol for making customized zinc-finger arrays. Nat. Protoc. 2009; 4:1471–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sander J.D., Dahlborg E.J., Goodwin M.J., Cade L., Zhang F., Cifuentes D., Curtin S.J., Blackburn J.S., Thibodeau-Beganny S., Qi Y. et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat. Methods. 2011; 8:67–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sander J.D., Zaback P., Joung J.K., Voytas D.F., Dobbs D.. Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 2007; 35:W599–W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sander J.D., Maeder M.L., Reyon D., Voytas D.F., Joung J.K., Dobbs D.. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010; 38:W462–W468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim J.M., A. K.C.. A 2.2 anstrom resolution crystal structure of a desgined zinc finger protein bound to DNA. Nature. 1996; 3:940–945. [DOI] [PubMed] [Google Scholar]
- 57. Negi S., Imanishi M., Matsumoto M., Sugiura Y.. New redesigned zinc-finger proteins: design strategy and its application. Chemistry. 2008; 14:3236–3249. [DOI] [PubMed] [Google Scholar]
- 58. McColl D.J., Honchell C.D., Frankel A.D.. Structure-based design of an RNA-binding zinc finger. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:9521–9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kukarin A., Rong M., McAllister W.T.. Exposure of T7 RNA polymerase to the isolated binding region of the promoter allows transcription from a single-stranded template. J. Biol. Chem. 2003; 278:2419–2424. [DOI] [PubMed] [Google Scholar]
- 60. Bandwar R.P., Patel S.S.. The energetics of consensus promoter opening by T7 RNA polymerase. J. Mol. Biol. 2002; 324:63–72. [DOI] [PubMed] [Google Scholar]
- 61. Bandwar R.P., Jia Y., Stano N.M., Patel S.S.. Kinetic and thermodynamic basis of promoter strength: multiple steps of transcription initiation by T7 RNA polymerase are modulated by the promoter sequence. Biochemistry. 2002; 41:3586–3595. [DOI] [PubMed] [Google Scholar]
- 62. Imburgio D., Rong M., Ma K., McAllister W.T.. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry. 2000; 39:10419–10430. [DOI] [PubMed] [Google Scholar]
- 63. Tunitskaya V.L., Kochetkov S.N.. Structural-functional analysis of bacteriophage T7 RNA polymerase. Biochemistry (Mosc.). 2002; 67:1124–1135. [DOI] [PubMed] [Google Scholar]
- 64. Persikov A.V., Wetzel J.L., Rowland E.F., Oakes B.L., Xu D.J., Singh M., Noyes M.B.. A systematic survey of the Cys2His2 zinc finger DNA-binding landscape. Nucleic Acids Res. 2015; 43:1965–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Moll J.R., Ruvinov S.B., Pastan I., Vinson C.. Designed heterodimerizing leucine zippers with a ranger of pIs and stabilities up to 10(-15) M. Protein Sci. 2001; 10:649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Thompson K.E., Bashor C.J., Lim W.A., Keating A.E.. SYNZIP protein interaction Toolbox: in Vitro and in vivo specifications of heterospecific Coiled-Coil interaction domains. ACS Synth. Biol. 2012; 1:118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Oakley M.G., Kim P.S.. A buried polar interaction can direct the relative orientation of helices in a coiled coil. Biochemistry. 1998; 37:12603–12610. [DOI] [PubMed] [Google Scholar]
- 68. Selgrade D.F., Lohmueller J.J., Lienert F., Silver P.A.. Protein scaffold-activated protein trans-splicing in mammalian cells. J. Am. Chem. Soc. 2013; 135:7713–7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shekhawat S.S., Porter J.R., Sriprasad A., Ghosh I.. An autoinhibited Coiled-Coil design strategy for Split-Protein protease sensors. J. Am. Chem. Soc. 2009; 131:15284–15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hillen H.S., Morozov Y.I., Sarfallah A., Temiakov D., Cramer P.. Structural basis of mitochondrial transcription initiation. Cell. 2017; 171:1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. DeRose R., Miyamoto T., Inoue T.. Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch. 2013; 465:409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tucker C.L. Manipulating Cellular Processes using Optical Control of Protein–Protein Interactions. 2012; 1st ednElsevier B.V. [DOI] [PubMed] [Google Scholar]
- 73. Tang J.C.Y., Szikra T., Kozorovitskiy Y., Teixiera M., Sabatini B.L., Roska B., Cepko C.L.. A nanobody-based system using fluorescent proteins as scaffolds for cell-specific gene manipulation. Cell. 2013; 154:928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zalatan J.G., Lee M.E., Almeida R., Gilbert L.A., Whitehead E.H., La Russa M., Tsai J.C., Weissman J.S., Dueber J.E., Qi L.S. et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015; 160:339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dueber J.E., Mirsky E.A., Lim W.A.. Engineering synthetic signaling proteins with ultrasensitive input/output control. Nat. Biotechnol. 2007; 25:660–662. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.