Abstract
Expression of the transcription factor SOX4 is often elevated in human cancers, where it generally correlates with tumor-progression and poor-disease outcome. Reduction of SOX4 expression results in both diminished tumor-incidence and metastasis. In breast cancer, TGF-β-mediated induction of SOX4 has been shown to contribute to epithelial-to-mesenchymal transition (EMT), which controls pro-metastatic events. Here, we identify SMAD3 as a novel, functionally relevant SOX4 interaction partner. Genome-wide analysis showed that SOX4 and SMAD3 co-occupy a large number of genomic loci in a cell-type specific manner. Moreover, SOX4 expression was required for TGF-β-mediated induction of a subset of SMAD3/SOX4-co-bound genes regulating migration and extracellular matrix-associated processes, and correlating with poor-prognosis. These findings identify SOX4 as an important SMAD3 co-factor controlling transcription of pro-metastatic genes and context-dependent shaping of the cellular response to TGF-β. Targeted disruption of the interaction between these factors may have the potential to disrupt pro-oncogenic TGF-β signaling, thereby impairing tumorigenesis.
INTRODUCTION
Increased expression of SOX4 transcription factor has been observed in a large number of human cancers, suggesting a general role in tumorigenesis (1,2). In agreement with this notion, Sox4 hypomorphic mice have been found to have greatly reduced spontaneous tumor-incidence (3). Conversely, increased expression of SOX4 has been demonstrated to promote tumor progression in both hematopoietic and solid tumors (4–6).
Recently, it has become apparent that SOX4 is a key transcriptional target of the transforming growth factor-beta (TGF-β) signaling pathway in a variety of cell-types including T-cells, pituitary cells, breast epithelial cells and glioma cells (7–10). The TGF-β signaling pathway elicits pleiotropic responses in cancer, acting as both a tumor-suppressor and metastasis-promoting factor depending on the cellular context (11). In late-stage cancers, the tumor-suppressive component of the TGF-β response is lost, allowing TGF-β to enhance tumor-progression by, for example, promoting EMT (12). The metastasis promoting effects of TGF-β are regulated to a large degree by the transcriptional response mediated by the receptor associated SMAD transcription factors (SMAD2/3), which for their context-specific target gene selection require cooperative binding with distinct transcription factors and rely on a permissive epigenomic state (11,13).
SOX4 is an important component in the tumor-promoting transcriptional response induced by TGF-β in both glioma and breast cancer. In glioma, SMAD2/3 have been demonstrated to directly regulate the transcription of SOX4, which was found to be essential for the maintenance of tumor-initiating cells by promoting the SOX4-dependent regulation of SOX2 expression (14). Recently, a number of studies by ourselves and others have demonstrated that SOX4 is also involved in pro-oncogenic TGF-β responses in (tumorigenic) breast epithelial cells. The transcriptional regulation of SOX4 by TGF-β is essential for the TGF-β mediated induction of EMT, a process involving the phenotypic conversion of epithelial cells to a mesenchymal-state which confers pro-tumorigenic properties by inducing stem-cell traits and promoting metastasis and therapy resistance (5,10,15–17). Ectopic expression of SOX4 has been shown to be sufficient to induce EMT in mammary epithelial cell lines in vitro, which correlates with increased metastasis in vivo (5,10,17). The EMT-promoting effects of SOX4 have also been linked to its direct transcriptional induction of EZH2, which is part of the polycomb-repressive complex 2 (PRC2), resulting in remodeling of the epigenome (5). Recently, it has been demonstrated that SOX4 has a dichotomous role downstream of TGF-β in pancreatic ductal adenocarcinoma (PDA) where, depending on the cellular transcription factor landscape, SOX4 may mediate apoptosis or promote oncogenesis (18). These findings indicate that cooperativity between SOX4 and distinct transcription factors likely shapes the SOX4-dependent transcriptional network and cellular response.
In support of a pro-metastatic role, SOX4 has been identified in gene expression signatures mediating breast tumor metastasis to the lung and brain (19,20). Moreover, in experimental mouse models of metastasis SOX4 has been shown to be the target of microRNA-335, which is lost during tumor-progression thus promoting invasion, migration and metastasis (21). Together, these findings indicate that the expression of SOX4 is likely crucial for breast cancer progression, in part by controlling TGF-β mediated induction of EMT. Despite this important role, the mechanisms through which SOX4 mediates tumor-progression remain poorly understood.
Here, by performing an unbiased transcription factor interaction screen we identify SMAD3 as a novel interaction partner of SOX4. We demonstrate that SOX4 specifically controls a pro-oncogenic subset of SOX4/SMAD3 co-bound TGF-β target genes, associated with poor-disease outcome. Our findings thus highlight a novel role of SOX4 in the TGF-β pathway by cooperatively regulating target genes with SMAD3 in a context-dependent manner, thereby skewing TGF-β responses.
MATERIALS AND METHODS
Cell culture and co-Immunoprecipitation assays
HEK293T, MDA-MB-231, HCC1954, MCF-7 and HMLE cells were cultured as described previously (10,22,23). For co-immunoprecipitation experiments HEK293T cells were transiently transfected by overnight incubation with pre-formed PEI:DNA complexes using 10 μg of DNA and 50 μg PEI. Cells were transfected with the indicated constructs and matching empty vector controls. Cells were lysed in E1A buffer (50 mM HEPES, pH 7.0, 250 mM NaCl, 5 mM EDTA and 0.1% NP-40) and subsequently immunoprecipitation was performed using anti-HA or anti-FLAG coupled beads (Sigma Aldrich) for 2 h at 4°C. After three wash steps in E1A buffer, samples were collected and boiled for 5 min in Laemmli sample buffer. Western blot analysis was subsequently performed as described previously (23). In order to avoid background signal from the IgG heavy chain of the antibody used for immunoprecipitation, HRP-conjugated FLAG (A8592, Sigma-Aldrich) and HA (A00169-40, Genscript) antibodies were used for the detection of co-immunoprecipitated proteins. Protein expression of whole cell lysates was assessed by using the following primary antibodies: anti-FLAG (Sigma, F7425) and anti-HA (A00168-40, Genscript).
Generation of shRNA-based deletion of SOX4
To generate SOX4-depleted HMLE cells and respective control, two independent pLKO.1-puro lentiviral constructs expressing shRNA targeting SOX4 (Sigma) or expressing shRNA control were used, respectively. Lentivirus particles were produced by transfection of HEK293T cells with 3.25 ug of Pax2, 1.8 μg of pMD2.G and 5 μg of shRNA contracts. Twenty four hours after transfection, medium was replaced by MEGM:DMEM/F12 media. Viral supernatants were collected 24 h after and filtered through a 0.22 μm filter and added to the HMLE cells. Cells were selected using 1μg/ml of puromycin (invivoGen). Cells were maintained in 1 μg/ml of puromycin through the cell culture.
TF–TF interaction array
MCF7 cells were transiently transfected with FLAG-tagged Sox4 or empty vector control. Nuclear extracts were generated 48 h after transfection. Immunoprecipitation was performed using anti-FLAG coupled beads (A2220, Sigma-Aldrich). The TF–TF-screen was performed according to manufacturer's protocol (Panomics). The hybridized arrays were probed using streptavidin-HRP and were detected using ECL (GE Healthcare).
Proximity ligation assay
HMLE cells were cultured on microscope glasses (Sigma-Aldrich) and were left untreated or treated for 16 h prior to the assay. For the proximity ligation assay, cells were washed once in phosphate-buffered saline (PBS) solution and fixed using 4% paraformaldehyde in PBS for 20 min at room temperature (RT). Subsequently, cells were washed twice in PBS and permeabilized in PBS 0.25% triton for 5 min at RT. Samples were washed in PBS and blocking was performed in PBS containing 2% bovine serum albumin (BSA) for 1 h at RT. Posteriorly, cells were incubated with anti-SOX4 (Sigma-Aldrich, HPA029901) and/or anti-SMAD3 (Thermo Scientific, MA5-15663) antibodies, overnight at 4 degrees. After incubation with primary antibodies, procedure was followed according to the manufacturer's protocol (Olink Bioscience). Accordingly, cells were washed twice in PBS and subsequently incubated with the secondary mouse PLUS (DUO92001, Sigma-Aldrich) and rabbit MINUS (Duo92005, Sigma-Aldrich) probes for 1 h at 37°C in a dark humidity chamber. Cells were washed three times in PBS, followed by a ligation step for 30 min at 37°C in a dark humidity chamber (DUO92007, Sigma-Aldrich). Again, cells were washed, followed by amplification and detection for 90 min at 37°C in a dark humidity chamber (DUO92007, Sigma-Aldrich). After this, cells were washed three times in PBS and mounted in 4 ul Prolong Gold anti-fade reagent with DAPI (Invitrogen). PLA signal was assessed by confocal microscopy (Zeiss LSM 700) using the 555 nm channel.
Chromatin immunoprecipitation-sequencing (ChIP-seq)
Chromatin-immunoprecipitation was performed in HMLE, MDA-MB-231 and HCC1954 cells using five 15 cm dishes per condition, as described previously (24). HMLE cells were either treated with TGF-β (2.5 ng/ml) for 16 h or left untreated prior to the assay. Subsequently, double crosslinking was performed using di(N-succinimidyl) glutarate (DSG) for 45 min followed by a 30 min incubation with formaldehyde. The reaction was quenched using incubation with 0.1 M Glycine for 5 min after which cells were washed in PBS and nuclear extracts were generated. Sonication using covaris was subsequently performed for 8 min at maximum output, after which immunoprecipitation was performed using 15 μl of the rabbit anti-SOX4 (CS-129-100, Diagenode) or 2 μg of the rabbit anti-SMAD3 (ab28379, abcam) antibodies coupled to protein A/G sepharose beads (sc-2003, Santa Cruz Biotechnology). Sequencing libraries were generated using the TruSeq LT kit (Illumina) and sequencing was performed on NextSeq platform (Illumina). Sequencing reads were mapped to the reference genome assembly (hg19) using Bowtie2 and peak-calling was performed using MACS2 (default settings were used for Bowtie2 and MACS2). Data analysis was performed using the HOMER software package (homer.ucsd.edu/), using the functions makeTagDirectory, findMotifsGenome.pl and annotatePeaks.pl (default settings and windows as indicated in the figures. IGV was used for ChIP-seq track visualization (25). Bedtools was used to determine the overlap between peaksets (26). ChIP-seq experiments were performed using two independent experimental replicates.
ChIP-qPCR
Chromatin-immunoprecipitation was performed in HMLE cells as described above. DNA samples were amplified using SYBR green supermix (BIO-Rad), in a white 96-multiwell plate by LightCycler 96 system instrument (Roche) according to the manufacture's protocol. Expression of each gene was normalized to input and to a negative region. All the primers for the Chip-qPCR were designed based on the Chip-sequencing data: negative region: F-GAGCCAGGGTTTCTCTGATTC, R-CCTCAGTGATCAGCCCTAAATG; CDH2: F-CAGCTACTTGGGAGGCTGAG, R- GCCACTGAAGCACATTGAAA; SPOCK1 F-TCTGTGCCACGTGCTTACTC, R-GGCTGAGTCATGTCATGGTG; LBH F- GATCCCTCTGGTGCTGATGT, R-TACCTCCAGGTGGGTCAGTC; MMP10 F-CAATGGTTGCAATTCAGACG, R- TCAGTGAGCTGTTACCAGAAGC; NEDD9 F-CCCACCCCTAATTCTGAAAA, R-TCCTTTTCCTGTTCTTTCTCTTTC; KDM7A F-AAGAGGCGGTGCTCTGTAAA, R- GAGCTTTCAGTCTCGCCACTA.
RNA sequencing
HMLE cells were transduced using lentiviral transduction with scrambled control or SOX4 targeting shRNA constructs as described previously (10). The cells were treated with TGF-β for 16 h after which RNA was isolated using the RNeasy kit (Qiagen) according to the manufacturer's protocol. Purified RNA was subsequently used for RNA-seq library preparation as described previously (24), and sequenced on the SOLiD Wildfire sequencer (Applied Biosystems Life Technologies). The BWA package was used to map sequencing reads (50 bp) to the reference genome (hg19). The Cisgenome v2.0 package was used for quantification of the reads, which were additionally quantile normalized and log2 transformed after adding a small number to the RPKM to avoid log-tranformation of zero values. The RNA-sequencing was performed in duplicates for each of the conditions tested.
Transwell migration assay
Transwell assay were performed according to the manufacture's protocol (Corning). Briefly, 30 000 scrambled control or SOX4 shRNA MDA-MB-231 cells transduced with either an empty vector control construct or constitutively active TGF-β receptor construct were plated in the inner compartment of the transwell plate in 300 μl of medium, with 700 μl in the outer compartment. The plate was subsequently incubated for 24 h at 37°C and 5% CO2. Subsequently, media was removed and cells were washed, fixed and stained with DAPI to quantify the number of migrated cells using ImageJ software.
Survival analysis
The Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort includes DNA copy number, whole gene expression and breast cancer-specific survival of 1980 patients (accession number EGAS00000000083) (27). Claudin-low subtyping was performed using transcriptomic data and the predictor developed by Prat et al. (28). Signature analysis was performed using the genes that were more than 0.5 log2 fold change differentially expressed between TGF-β-treated or SOX4 knockdown cells and their respective controls. Two signatures were derived: (i) overlapping regulated genes between TGF-β and SOX4 and (ii) TGF-β without SOX4-associated genes. Samples were assigned a Spearman correlation-based score calculated between the gene expression data of each tumour and each signature. Survival was analysed using Kaplan—Meier curves and significance assessed by log-rank test. Data were analysed using R 3.2.2 and a two-sided P < 0.05 was considered significant.
Quantification of RNA expression (q-PCR)
mRNA was extracted from cells using the Rneasy Isolation Kit (Qiagen). According to the manufactures protocol for single-stranded cDNA synthesis, 500 ng of total RNA was reverse transcribed using iScript cDNA synthesis kit (BIO-Rad). cDNA samples were amplified using SYBR green supermix (BIO-Rad), in a white 96 multiwell plate by LightCycler 96 system instrument (Roche) according to the manufacture's protocol. To quantify the data, the comparative Ct method was used. Relative quantity was defined as 2-ΔΔCt and β2-Microglobulin was used as reference gene. The following primers were used: CDH2 F-AGTCACCGTGGTCAAACCAATCGA, R-TGCAGTTGACTGAGGCGGGTG; SPOCK1 F-GCTCAGATGGCCACTCCTAC, R-TCTGTGCAGGCACTCCTTTC; LBH F-TCCCCATTCACTGCCCCGAC, R-AAAGGCCATCCTTGCGGGGG; FERMT1 F- CTTGGTTCAGTGACAGCCCT, R-GGAGTCTAGCCAACCTGCAT; MMP10 F-GACAGAAGATGCATCAGGCAC, R- CATCTTGCGAAAGGCGGAAC; NEDD9 F-GCTGCCGAAATGAAGTATAAGAATC, R-GGTCAGGATGTCTCCCTTGC; KDM7A F-GTCTGAATTGGTGGAGGTCCC, R- ATCTTCTCACCCCAGAGGACA; CCDC80 F-GGATCTGTGGCGGTACTCTG, R-GTGTAATCCAATGGTGGCTCA; IL11 F- ATGAACTGTGTTTGCCGCCT, R- TCAGCTGGGAATTTGTCCCTC; STX5 F- AGTCTTTGGTCGGGTTTCGG, R- CGCATAACCTCGGACTCCC. B2M F-ATGAGTATGCCTGCCGTGTG, R-GGCATCTTCAAACCTCCATG; GAPDH F-ATGGGGAAGGTGAAGGTCG, R- GGGGTCATTGATGGCAACAATA; HRPT1 F-TGACACTGGCAAAACAATGCA, R-GGTCCTTTTCACCAGCAAGCT and ACTB F- CATGTACGTTGCTATCCAGGC, R-CTCCTTAATGTCACGCACGAT.
Signature analysis
Analysis of the SOX4-dependent TGF-β target genes was performed using the online GOBO-software (29).
Statistical analysis
Data are represented as mean ± SD of at least three independent experiments. Differences were analyzed by unpaired two-tailed t-test between two groups and by two-way ANOVA for differences between more than two groups.
RESULTS
SMAD3 is a novel SOX4 interaction partner
In order to achieve target selectivity and control of cell-type specificity, SOX transcription factors interact with distinct DNA-binding factors (30). To identify SOX4 transcription factors partners, we performed an unbiased interaction screen by utilizing a TF–TF interaction array. This technique enables the generation a global view of transcription factor interaction-networks (Supplementary Figure S1A). Since we and others have previously identified a role for SOX4 in regulating mammary epithelial homeostasis, MCF-7 cells, a well characterized breast cancer cell line, were utilized in this assay (5,10,31). Analysis of co-immunoprecipitated probes identified the specific association between Sox4 and the TGF-β pathway effector transcription factor SMAD3 (Figure 1A and Supplementary Figure S1B). As it has been previously shown that SOX4 is itself a TGF-β target gene (5,10,17), interaction between SOX4 and SMAD3 indicates that SOX4 is not only a transcriptional target of TGF-β but may also be a relevant player in directly regulating TGFβ-mediated transcriptional responses.
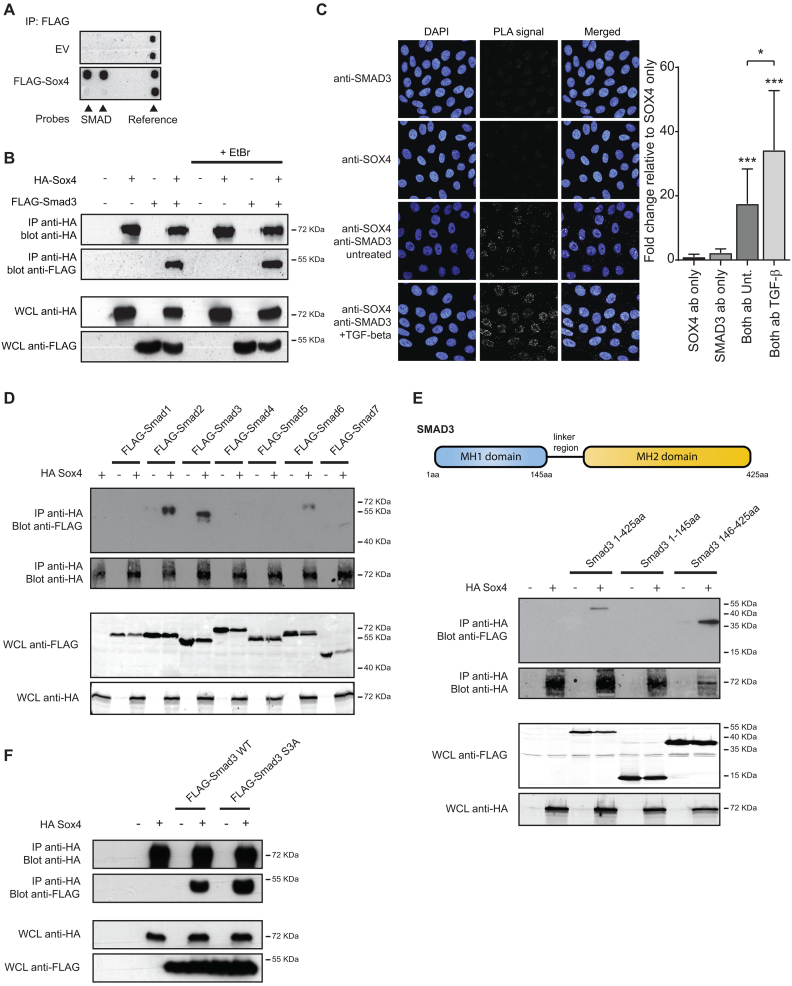
Figure 1.
SOX4 associates with the MH2-domain of SMAD3 independent of receptor-mediated phosphorylation. (A) MCF7 cells were transfected with FLAG-Sox4 or an empty vector control. Biotin-labeled transcription factor binding probes were added to the cell lysate after which Sox4 was immunoprecipitated using a FLAG- antibody. Subsequently, specifically bound probes were hybridized to the TF-TF array and visualized using streptavidin antibodies. (B) HEK293T cells were transfected with HA-Sox4 and FLAG-SMAD3. Lysates were treated with 25 μg/ml EtBr for 20 min after which Sox4 was immunoprecipitated and immunoblots were probed for HA and FLAG. Results are representative of at least three independent experiments. (C) The SOX4-SMAD3 interaction was analyzed by PLA. HMLE cells were treated with 2.5 ng/ml TGF-β overnight or left untreated. Left panel: Punctate staining indicates the specific interaction between the two proteins and DAPI was used to co-stain the nucleus. Right panel: Quantification of punctate staining relative to anti-SOX4 condition (negative control). Results are representative of three independent experiments. (D) HEK293T cells were transfected with HA-Sox4 and FLAG-SMAD1–7. Sox4 was immunoprecipitated from cell lysates and co-immunoprecipitation of SMAD proteins was assessed by immunoblot for HA and FLAG-epitope antibodies. Results are representative of at least three independent experiments. (E) Top panel: Schematic representation of SMAD3 protein. Bottom panel: HA-Sox4 was immunoprecipitated from HEK293T cells co-transfected with full-length SMAD3 (1–425aa), the N-terminal (1–145aa) or C-terminal SMAD3 regions (146-425aa). Results are representative of at least three independent experiments. (F) HEK293 cells were transfected with HA-Sox4 and Flag-SMAD3 wild-type (WT) or phosphorylation-defective SMAD3 S3A. Sox4 was immunoprecipitated from cell lysates and protein expression was assessed by immunoblot using anti-HA and Flag antibodies. Images are representative of three independent experiments.
While the TF-TF array cannot distinguish between SMAD family members, SMAD3 was initially chosen to pursue as an interaction partner since it is a TGF-β receptor SMAD, and directly binds DNA. In order to validate this interaction, co-immunoprecipitation analysis was performed utilizing HEK293T cells ectopically expressing HA-SOX4 and FLAG-SMAD3, demonstrating the association between these two proteins (Figure 1B). The association between SOX4 and SMAD3 was also observed in cell lysates treated with ethidium bromide, which disrupts DNA-mediated interactions (32), indicating that a protein-protein interaction and not DNA-mediated interaction is likely responsible for the co-immunoprecipitation of SMAD3 (Figure 1B).
To evaluate endogenous interaction between SOX4 and SMAD3, in situ proximity ligation analysis (PLA) was utilized, which selectively enables the detection of proteins that are in very close proximity. For the PLA analysis, we utilized human mammary epithelial cells (HMLEs), that we have previously demonstrated to express SOX4 and respond to TGF-β by increasing SOX4 at the transcript level (10). PLA-analysis using SOX4 and SMAD3 specific antibodies in HMLE cells demonstrated that nuclear PLA-signal could be detected only upon incubation with both antibodies, whereas only unspecific signal was observed in single antibody control conditions (Figure 1C). Increased PLA-signal was observed upon TGF-β treatment, indicating that activation of TGF-β signaling pathway is important for SOX4-SMAD3 association. Moreover, although to a lesser extent, interaction between SOX4 and SMAD3 was observed in untreated conditions, suggesting autocrine TGF-β signaling in these cells. In line with these observations, both SOX4 and SMAD3 baseline protein expression has been found decreased upon the incubation with SB431542, a specific Inhibitor of TGF-β receptor kinase (7,33). Taken together, these findings identify a novel interaction between SOX4 and SMAD3.
SOX4 interacts with SMAD3 in a phosphorylation independent manner
To further define both the specificity and functional protein domains mediating the interaction between SOX4 and the SMAD3, interaction with other members of the SMAD-family of transcription factors was evaluated. To this end, co-immunoprecipitation was performed using Sox4 and SMAD1-7. SOX4 was found to strongly associates with the receptor-SMADs of the TGF-β signaling pathway, SMAD2 and SMAD3, but not with SMAD4 (Figure 1D). In contrast to SMAD3, SMAD2 does not directly bind to DNA (34). No interaction was observed with the BMP-signaling associated receptor-SMADs, SMAD1 and SMAD5 or with the inhibitory SMAD7 and a weak interaction was found with SMAD6 (Figure 1D). These observations indicate that SOX4 is specifically associated with the effectors of TGF-β signaling pathway.
The domain structure of SMAD-proteins is well-defined, consisting of the DNA-binding Mad-homology 1 and 2 domains (MH1 and MH2), which are involved in DNA-binding and protein–protein association, respectively (34). Co-immunoprecipitation analysis demonstrated that SOX4 specifically associated with a C-terminal region of SMAD3 comprising the MH2 domain and linker region, whereas no interaction was observed with the MH1 domain (Figure 1E). Since activated SMAD3 requires phosphorylation at C-terminal serines, we next investigated whether the interaction between SOX4 and SMAD3 may be regulated by SMAD3 phosphorylation. The interaction between Sox4 and SMAD3 was also observed when these serine-residues were mutated to alanine (Figure 1F), indicating that this protein-protein interaction is independent of phosphorylation at these sites. Taken together, these findings map the interaction between SOX4 and SMAD3 and indicate that the association does not rely directly on phosphorylation of the canonical-phosphorylation sites.
Genome-wide co-occupancy of SOX4 and SMAD3 in breast cell lines is context dependent
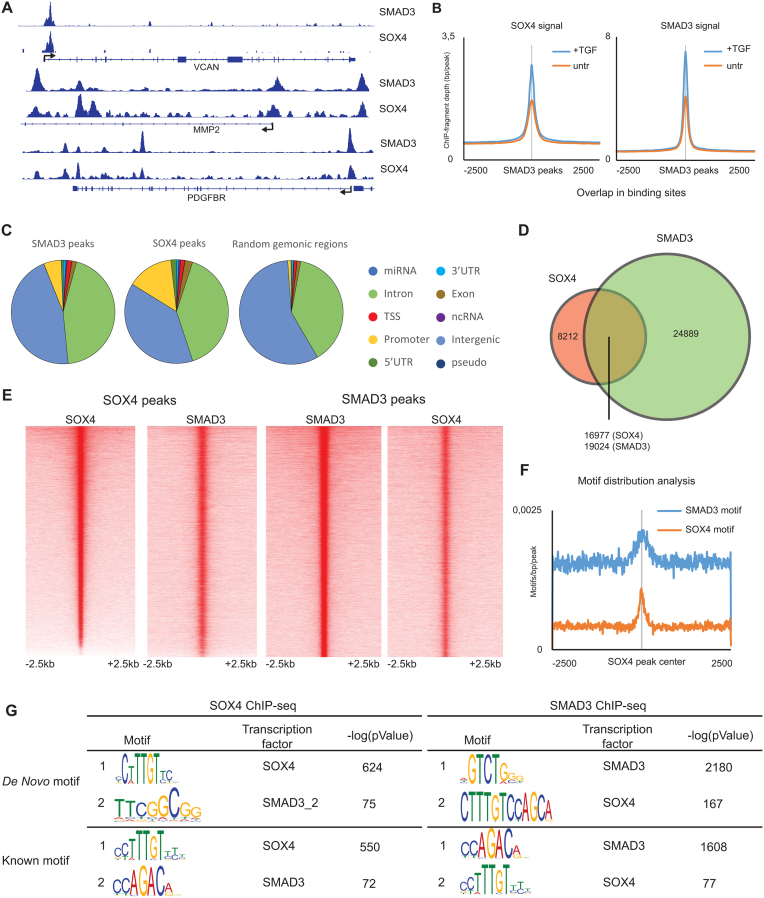
To further evaluate the association between SOX4 and SMAD3 at the DNA-level on a genome-wide scale we performed chromatin-immunoprecipitation-sequencing (ChIP-seq) for both factors in TGF-β treated HMLE cells. These cells were chosen because TGF-β has been shown to promote pro-oncogenic responses including EMT (10,35,36). We successfully identified both SOX4 and SMAD3 bound loci and binding of both factors can be observed on the selected examples for TGF-β/SOX4 target genes VCAN, MMP2 and PDGFBR, and binding of both transcription factors occurs in the same region, as indicated by overlapping peaks (Figure 2A). While SOX4 and SMAD3 co-occupancy was observed in the absence of exogenous TGF-β, it increased markedly upon TGF-β treatment (Figure 2B). These findings suggest that the transcriptional response to TGF-β may be driven by increased recruitment of both SOX4 and SMAD3, rather than by de novo recruitment or a global redistribution. The baseline binding may be the result of autocrine activation of TGF-β signaling, which has previously been demonstrated to occur in mammary epithelial cells (37). Analysis of the genomic distribution of SOX4 and SMAD3 in annotated regions demonstrated that binding of both factors is enriched at promoter regions compared to the contribution of promoter regions in the total genome (Figure 2C). To assess whether co-binding occurs on a genome-wide scale we overlapped SOX4 and SMAD3 bound loci, and found that the majority of SOX4 bound sites are co-occupied by SMAD3 (Figure 2D), The co-occupied sites, which occur at both TSS-proximal and more distal regions, are enriched for markers associated with active/open chromatin (H3K27ac, H3K4me3 and POL2) (Supplementary Figure S2A-B). This observation is consistent with the notion that SMAD3 binding is directed by the epigenome (13). Approximately, 30% of sites were exclusively bound by SOX4, and these sites were enriched at TSS-proximal regions, including 5′UTR, TSS and promoter region, and characterized by high-levels of the TSS-associated histone-mark, H3K4me3 (Figure 2D and Supplementary Figure S2A-B). Since the number of SMAD3 bound loci exceeds the number of SOX4 bound sites, a larger proportion of SMAD3 binding appears to be SOX4-independent. These SMAD3-only sites are mostly located at TSS-distal sites and are also characterized by markers of active regions in the genome (Supplementary Figure S2A and B).
Figure 2.
SOX4 and SMAD3 binding sites overlap on a genome-wide level. (A) Visualization of SOX4 and SMAD3 ChIP-seq profiles contained within the genomic region surrounding the VCAN, MMP2 and PDGFBR loci. (B) Average profile plot of SOX4 and SMAD3 signals in TGF-β treated and untreated HMLE cells. (C) Genomic-distribution of SOX4 and SMAD3 binding sites in annotated regions compared to background genomic sites. (D) Venn-diagram showing the overlap of SOX4 and SMAD3 binding sites in TGF-β treated HMLE cells (overnight 2.5 ng/ml). (E) Occupancy maps representing the intensity of SOX4 and SMAD3 binding in a 5kb regions surrounding the peak centers. (F) Motif-distribution analysis of SMAD3 and SOX4 motifs in co-bound genomic loci. (G) Motif enrichment analysis of SOX4 and SMAD3 bound sites using both de novo and known-motif discovery. miRNA: microRNA; TSS: transcription start site; 5′UTR: five prime untranslated region; 3′UTR: three prime untranslated region; ncRNA: non-coding RNA.
The large degree of overlap between SOX4 and SMAD3 is also highlighted by co-occupancy analysis of SOX4 and SMAD3 bound sites, which identified reciprocal and central enrichment of the binding of both factors (Figure 2E). Consistent with cooperative binding, motif enrichment and motif distribution analysis showed the co-occurrence of the consensus SMAD3 DNA-binding motif in SOX4 bound sites and vice versa (Figure 2F–G). When centered around the SOX4-consensus binding site within the co-occupied sites, central enrichment of the SMAD3-motif was not evident (Supplementary Figure S2C). Moreover, spaced motif analysis failed to determine a defined mode of site selection, suggesting that there is freedom in the co-binding of transcriptional complexes comprising SOX4 and SMAD3. Moreover, the central and reciprocal enrichment of these DNA-binding motifs could only be observed at co-bound sites, with exclusive enrichment of SOX4 and SMAD3 motifs at SOX4-only and SMAD3-only sites, respectively (Supplementary Figure S2D). Taken together, these findings demonstrate co-occupancy of SOX4 and SMAD3 on a genome-wide level, which is in part dependent on the co-occurrence of consensus DNA-binding motifs and relies on the activity of the TGF-β pathway.
Since the effect of TGF-β is pleiotropic and can promote dichotomous cellular responses, we assessed the potential role of the SOX4-SMAD3 interaction in two additional breast cancer lines: MDA-MB-231 and HCC1954 cells, in which TGF-β elicits contrasting responses. In HCC1954 cells, TGF-β inhibits cell proliferation and stem-like traits, whereas in MDA-MB-231 cells the response induces proliferation, breast cancer initiating cell-like characteristics and metastasis (22). ChIP-seq analysis performed in these lines demonstrated that similar to HMLE cells, SOX4 bound sites were co-occupied by SMAD3 on a genome-wide level in both MDA-MB-231 and HCC1954 cells (Supplementary Figure S3A). This co-occupancy was apparent when the consensus peaks obtained from two replicate ChIP-seq experiments were analyzed (Supplementary Figure S3A, right panel). In concordance with the HMLE data, a significant portion of the SMAD3 sites were observed to be SOX4-independent (Supplementary Figure S3A). Additionally, co-occupancy analysis demonstrated that the SOX4 signal is also centrally enriched at SMAD3 bound sites in both MDA-MB-231 and HCC1954 cells (Supplementary Figure S3B). Similarly, in these cells both SOX4 and SMAD3 binding was found to be induced by TGF-β (Supplementary Figure S3C). Interestingly, comparison of SOX4-SMAD3 co-bound sites in all cell lines demonstrated only a small degree of overlap (Supplementary Figure S3D). Analysis of H3K27Ac ChIP-seq data from HMLE, MDA-MB-231 and HCC1954 cells suggest that this context-dependent binding SOX4-SMAD3 complexes is dictated by the cell-intrinsic chromatin landscape which is defined by this active chromatin mark (Supplementary Figure S3E). Taken together, these findings demonstrate that co-occupancy between SOX4 and SMAD3 can occur in a wide variety of breast (cancer) cell lines but this has distinct context-specific roles, similar to the contextual role for TGF-β described in breast cancer.
Identification of SOX4-dependent TGF-β target genes
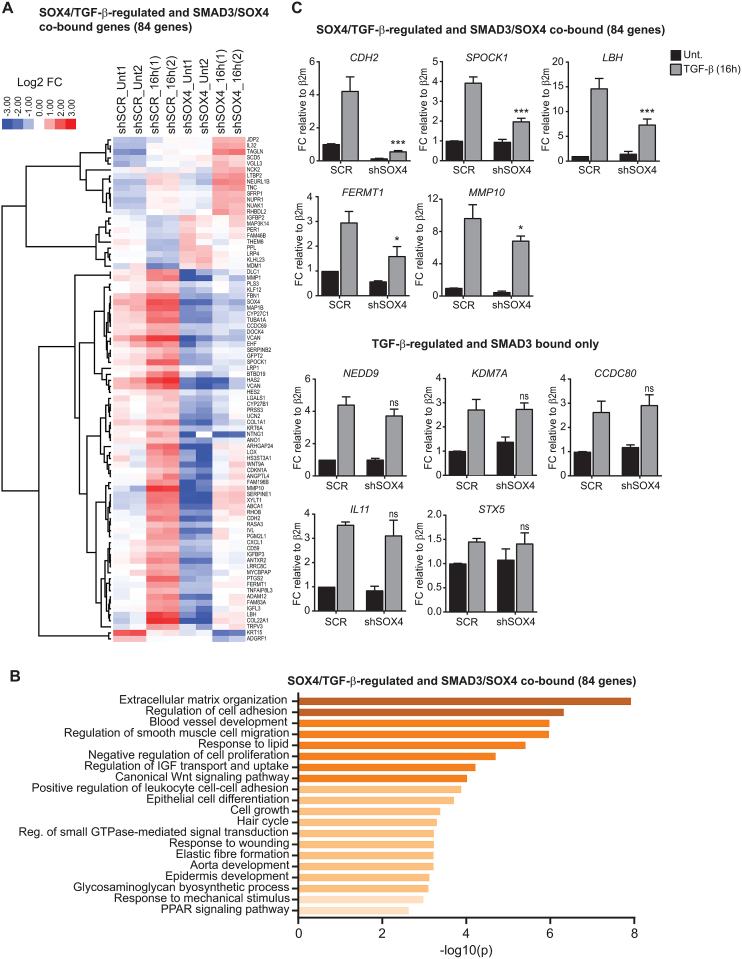
Since we observed extensive co-binding of SOX4 and SMAD3, we investigated to what degree the transcriptional response to TGF-β is SOX4-dependent. To this end, we depleted SOX4 in HMLE cells, which as discussed above, are non-tumorigenic mammary epithelial cells that upon exposure to TGF-β undergo EMT (5,10,31). Two independent shRNA constructs targeting human SOX4 were used and considering the efficiency of both shRNAs, further experiments were performed using shSOX4_1 (Supplementary Figure S4A). Subsequently, the global transcriptional response to TGF-β using RNA-sequencing (RNA-seq) was determined. Analysis of the RNA-seq data showed 167 genes differentially regulated after 16 h of TGF-β treatment in the SOX4-depleted HMLE cells compared to control cells (Supplementary Figure S4B). For the majority of the SOX4-dependent TGF-β target genes, depletion of SOX4 was found to decrease the TGFβ-mediated induction of positively regulated target genes (64%). Within the group of SOX4-dependent TGF-β target genes, 84 were found to be co-bound by SOX4 and SMAD3 in HMLE cells (Figure 3A), suggesting that these could be directly regulated through cooperative binding of both factors. Consistent with the role of SOX4 as a transcriptional activator, the majority of SOX4-SMAD3 co-bound and SOX4-dependent TGF-β target genes (84 genes) were found to be positively regulated by TGF-β in control cells and reduced upon SOX4 depletion (73%) (Figure 3A). Gene-ontology enrichment analysis of this co-bound SOX4-dependent TGF-β signature gene-set identified significant association with cellular processes connected to metastasis, including extracellular matrix remodeling, blood vessel development, cell-adhesion and cell-migration (Figure 3B). In order to validate these targets, we performed quantitative real-time PCR (qRT-PCR) on SOX4 knockdown and control (SCR) HMLE cells treated with TGF-β for 16 h (Supplementary Figure S4C). To ensure that results were not dependent on the effect of either TGF-β treatment or SOX4-knockdown on the expression of the housekeeping genes, we analyzed the expression of four different housekeeping genes in all technical and biological replicates (Supplementary Figure S5A). Results showed no significant variations within conditions for any of the four housekeeping genes, thereby we further performed our analysis using β2m. For all the selected SOX4-SMAD3 co-bound and SOX4-dependent TGF-β target genes, depletion of SOX4 resulted in the suppression of TGF-β-mediated upregulation of such targets, whereas no significant alteration in gene expression was observed of SOX4-independent TGF-β-regulated genes in SOX4-depleted HMLE cells (Figure 3C). Furthermore, the functional importance of SOX4-SMAD3 interaction was confirmed by ChIP-qPCR, where SOX4 knockdown impaired SMAD3-binding to SOX4-SMAD3 co-bound genes, whereas SMAD3 only targets were unaffected by SOX4 depletion (Supplementary Figure S5B). Taken together, our data supports a role for SOX4 in mediating TGF-β/SMADs transcriptional upregulation of genes involved in tumorigenesis.
Figure 3.
Identification of SOX4-dependent TGF-β target genes. (A) Heatmap of SOX4-SMAD3 co-bound and SOX4-dependent TGF-β target genes core identified by RNA-seq. HMLE cells transduced with scrambled control and SOX4 targeting shRNA constructs were treated with TGF-β (2.5 ng/ml) for 16 h after which RNA was isolated and analyzed by RNA-seq. Samples are displayed as biological replicates (1 and 2). (B) Gene-ontology analysis using metascape (http://metascape.org) of SOX4-SMAD3 co-bound and SOX4–dependent TGF-β target genes. Significant GO-term clusters are visualized (P < 0.05). (C) qRT-PCR results showing the effect of TGF-β (16 h) on expression of SOX4-dependent and independent TGF-β target genes. Data represented as mean ± SD, normalized for β2m (N = 3).
SOX4-dependent TGF-β target genes correlate with poor-prognosis
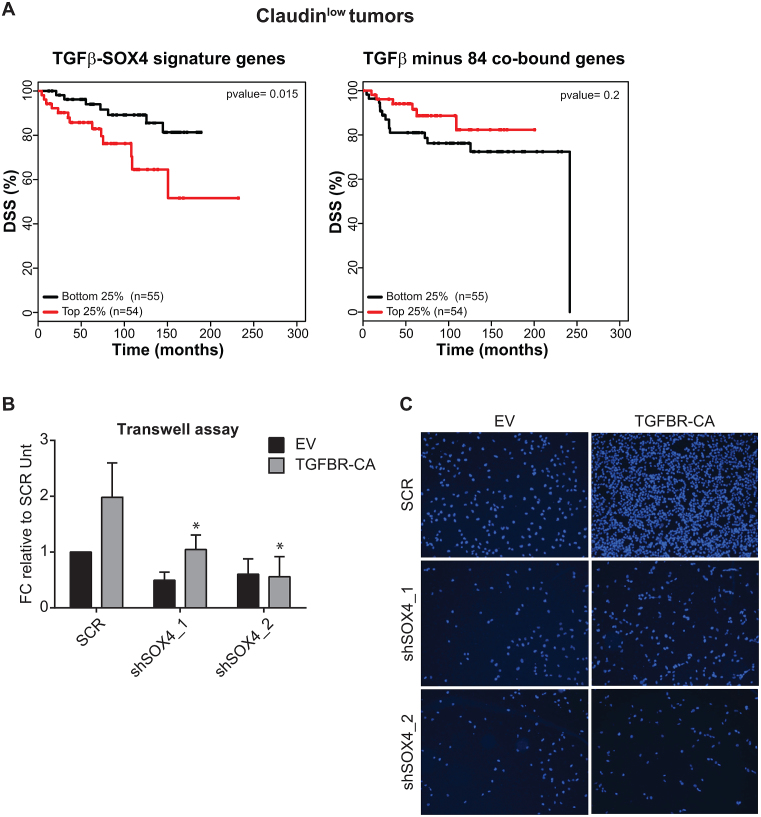
TGF-β pathway activity is associated with poor prognosis in only a subset of breast cancer patients of the claudinlow subtype. This is consistent with, TGF-β’s in vitro and in vivo pro-tumorigenic roles in claudinlow cells (22,38,39). To evaluate whether the SOX4-dependent and -independent TGF-β gene expression signatures were associated with patient survival in claudinlow tumors, gene expression data from the METABRIC cohort of breast cancer patients was stratified according to similarity to each of the signatures. Kaplan–Meier analysis of breast tumors with the 25% extreme high and low correlation values with SOX4-dependent TGF-β gene expression signature (84 genes) demonstrated a significant correlation with diminished disease-specific survival (DSS) in claudinlow tumors (Figure 4A; left panel). In contrast, analysis of the total TGF-β signature devoid of SOX4-SMAD3 co-bound and SOX4-dependent TGF-β target genes showed little-to-no association with clinical outcome (Figure 4A; right panel). This, suggests that SOX4 may redirect TGF-β cellular response toward a pro-malignant clinical outcome, where SOX4-dependent transcriptional targets play an important role in TGF-β-mediated tumor progression. In agreement with the context dependent nature of the TGF-β response, no association with survival was observed in luminal A, luminal B, HER2 and basal-like tumor subtypes for both signatures, with the exception of normal-like tumor subtype (Supplementary Figure S6A). Since SOX4 modulates a TGF-β gene transcriptional network enriched in genes associated with cell migration and that are associated with poor-prognosis in claudinlow breast cancer patients, we asked whether SOX4 is required for the TGF-β mediated induction of pro-migratory effects in the claudinlow MDA-MB-231 cell line. To this end, we performed transwell assays utilizing SOX4-depleted and control MDA-MB-231 cells (Supplementary Figure S6B) expressing a control vector or a vector containing the constitutively active TGF-β receptor. SOX4 depletion significantly reduced migration of highly metastatic MDA-MB-231 breast cancer cells ectopically expressing the constitutively active TGF-β receptor (Figure 4B and C). In addition, SOX4-knockdown also affected migration in control cells, suggesting that SOX4 affects autocrine TGF-β signaling in these cells. Taken together these findings demonstrate that the core-SOX4 dependent TGF-β signature derived from combined gene expression profiling and ChIP-seq analysis is associated with metastasis-associated processes and correlates with poor-prognosis in claudinlow breast tumors.
Figure 4.
SOX4-dependent TGF-β target genes correlate with poor-prognosis and invasiveness in breast cancer. (A) Kaplan–Meier estimates of disease-specific survival (DSS) for top (red) and low (blue) quartiles of SOX4-dependent TGF-β signature or TGF-β signature minus SOX4-SMAD3 co-bound and SOX4-dependent TGF-β target gene-set in claudinlow breast tumors subtype (B) Quantification of transwell migration of MDA-MB-231 cells transduced with empty vector (EV) or the constitutively active TGF-β receptor (TGFBR-CA) and scrambled control or SOX4 knockdown shRNA constructs. The number of cells present in the lower well was quantified after overnight incubation. Data was quantified relative to EV-scrambled control MDA-MB-231 cells and is represented as mean ± SD of three independent experiments. (C) Representative images of transwell assay stained for DAPI to visualize migrated MDA-MB-231 cells.
DISCUSSION
Although SOX4 is overexpressed in a large number of human cancers, the cellular responses elicited by this transcription factor are highly divergent, thus SOX4 can act as a tumor-suppressor or pro-oncogenic factor depending on the cell-type and context (2). In agreement with a highly context specific function, only a minor degree of overlap has been observed in SOX4 target genes between distinct cancers (10). These observations suggest that cell-type specific factors, such as the epigenome and co-factors shape the transcriptional network of SOX4. However, the contextual effects induced by SOX4 remain poorly defined. Here, we aim to identify the molecular mechanisms determining the cell context-specific transcriptional output and target gene selection to better understand SOX4-driven tumorigenesis and identify therapeutic avenues that specifically disrupt its pro-oncogenic function.
Cooperative binding is relatively common in the SOX-family of transcription factors as is exemplified by the interaction between SOX2 and OCT-4, which results in the regulation of pluripotency genes such as NANOG (30,40). Here, we identified a novel interaction between SOX4 and the TGF-β effector transcription factor SMAD3 and subsequently identified SOX4 as a mediator of the pro-oncogenic effects of the TGF-β signaling pathway. Many of the previously described SMAD3 co-factors are also transcriptional targets of the TGF-β signaling pathway thus forming a feedback loop that regulate TGF-β’s transcriptional network (11). In a similar manner, transcriptional regulation of SOX4 by TGF-β in breast cancer cells may act to promote cooperative gene regulation between SOX4 and SMAD3. In this respect is it possible that cooperative transcriptional regulation between these two factors also contributes to TGF-β responses in additional cell-types in which SOX4 is induced by TGF-β including glioma, pituitary-cells and T-cells (8,9,14).
Interestingly, other SOX-family members have also been described to interact with SMAD3 including SOX2 in lung epithelial cells and SOX9 in chondrocytes, suggesting that SMAD and SOX transcription factors have co-evolved to cooperatively regulate target genes in response to TGF-β (41,42). Depletion of SOX4 affected a specific subset of TGF-β transcriptional targets, suggesting that in HMLE cells the response to TGF-β does not solely rely on the expression of SOX4 and is likely controlled by additional (transcription) factors. This is in line with observations showing that C/EBPβ, FOXO and AP1, control distinct subsets of TGF-β target genes through cooperative binding with SMAD3 (43,44). Interestingly, we observed that SOX4-SMAD3 co-bound sites were highly cell-type specific, as indicated by the minor degree of overlap between co-bound sites in HMLE, MDA-MB-231 and HCC1954 cells, suggesting that additional cell-type specific factors in concert with the epigenome determine binding and target gene activation.
SOX4-dependent TGF-β target genes were observed to be enriched for processes such as cell migration and extracellular matrix organization. Interestingly, this subset of SOX4 dependent TGF-β target genes was also enriched for gene ontology terms related to cardiovascular system development matching phenotype connected to SOX4 depletion in knockout mice (45). This suggests that SOX4-mediated modulation of the TGF-β pathway may also contribute to the phenotypic effects observed upon depletion of SOX4 in vitro and in vivo.
In breast cancer, the SOX4-dependent TGF-β signature was found to be correlated with poor-disease outcome in the claudinlow subtype. The majority of claudinlow breast cancers are characterized by low expression levels of luminal differentiation markers and high levels of EMT markers (28). In line with these observations, SOX4 expression has been shown to contribute to TGF-β-mediated EMT in human mammary cells (5,10,31), suggesting that SOX4-mediated reduction of disease-specific survival (DSS) is achieved by regulating genes involved in the acquisition of mesenchymal traits. Differences in outcome in breast cancer have been linked to cell-type-dependent co-factors and enrichment of stem cell-like traits (22). Taken together, our results suggest a novel role for SOX4 in the TGF-β signaling pathway by acting as a SMAD3 co-factor and cooperatively regulating pro-metastatic genes contributing to poor outcome in breast cancer patients. Lack of correlation with disease outcome and SOX4-dependent TGF-β signature in other breast cancer subtypes is possibly due to differences in cell-type specific epigenomes and SOX4 co-factors.
Consistent with a pro-metastatic role in claudinlow tumors, we observed that SOX4 depletion affects MDA-MB-231 migration in the presence of TGF-β. The transcriptional response to TGF-β differs greatly between MDA-MB-231 and HCC1954 cells, resulting in contrasting effects on the TGF-β mediated induction of tumor-initiating cells (22). Accordingly, we observe that SOX4-SMAD3 co-bound sites are almost completely distinct between these two cell lines, although it remains to be determined how this affects SOX4-dependent TGF-β target gene regulation and functional effects. In line with these observations, it has been demonstrated that in pancreatic ductal adenocarcinoma (PDA), SOX4 can control either pro-apoptotic or pro-metastatic gene expression networks downstream of TGF-β, dependent on the cell-type dependent expression of SMAD4 and KLF5 (18). In our dataset, the SOX4-dependent TGF-β signature does not appear to correlate with genes associated with apoptosis and SOX4-depedent induction of pro-metastatic TGF-β target genes in breast cancer appears to be independent of KLF5 expression (Supplementary Figure S6C), suggesting that regulation of these gene-sets is dependent on distinct transcription factors and a permissive epigenome.
Taken together, our findings strongly support that SOX4 is not only a TGF-β target but also a critical effector of the TGF-β signaling pathway in breast epithelial cells. These results also highlight the importance of co-factors in determining both SOX4 and SMAD3 gene networks on a genome-wide level. Importantly, our finding that the SOX4-dependent TGF-β target genes are associated with poor-prognosis in claudinlow tumors indicates that targeting of the interaction between SOX4 and SMAD3 may have therapeutic merit in this subtype.
DATA AVAILABILITY
ChiP-seq and RNA-seq datasets have been deposited in GEO under accession number GSE104761.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Peter ten Dijke for providing the SMAD-constructs, Bernard Perreira, Jason Carroll, Michal Mokry, Ruben van Boxtel, Jorg van Loosdregt and Coffer Lab members for helpful discussions.
Author Contributions: P.J.C. supervised the study. S.V., A.R.L., A.B. and P.J.C. designed experiments. S.V., A.R.L., A.T.V., C.P. and C.F. performed experiments. S.V., A.T.V., J.L.S., J.G.C.P. and O.M.R. performed data analysis. S.V., A.R.L., C.C., A.B. and P.J.C. interpreted experiments. S.V., A.R.L., A.B. and P.J.C. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Dutch Cancer Foundation Grants [KWF UU 2013-5801, KWF UU 2015-7838]; Cancer Research UK and Fundação para a Ciência e a Tecnologia (FCT) fellowships (to J.L.S., A.R.L.). Funding for open access charge: KWF.
Conflict of interest statement. None declared.
REFERENCES
- 1. Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M.. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:9309–9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vervoort S., van Boxtel R., Coffer P.J.. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe. Oncogene. 2013; 32:3397–3409. [DOI] [PubMed] [Google Scholar]
- 3. Foronda M., Martínez P., Schoeftner S., Gómez-López G., Schneider R., Flores J.M., Pisano D.G., Blasco M.A.. Sox4 links tumor suppression to accelerated aging in mice by modulating stem cell activation. Cell Rep. 2014; 8:487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shin M.S., Fredrickson T.N., Hartley J.W., Suzuki T., Agaki K., Morse H.C.. High-throughput retroviral tagging for identification of genes involved in initiation and progression of mouse splenic marginal zone lymphomas. Cancer Res. 2004; 64:4419–4427. [DOI] [PubMed] [Google Scholar]
- 5. Tiwari N., Tiwari V.K., Waldmeier L., Balwierz P.J., Arnold P., Pachkov M., Meyer-Schaller N., Schübeler D., van Nimwegen E., Christofori G.. Sox4 is a master regulator of Epithelial-Mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013; 23:768–783. [DOI] [PubMed] [Google Scholar]
- 6. Liu P., Ramachandran S., Seyed M.A., Scharer C.D., Laycock N., Dalton W.B., Williams H., Karanam S., Datta M.W., Jaye D.L. et al. . Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006; 66:4011–4019. [DOI] [PubMed] [Google Scholar]
- 7. Ikushima H., Todo T., Ino Y., Takahashi M., Miyazawa K., Miyazono K., Aaboe M., Birkenkamp-Demtroder K., Wiuf C., Sørensen F.B. et al. . Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009; 5:504–514. [DOI] [PubMed] [Google Scholar]
- 8. Kuwahara M., Yamashita M., Shinoda K., Tofukuji S., Onodera A., Shinnakasu R., Motohashi S., Hosokawa H., Tumes D., Iwamura C. et al. . The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-β and suppresses TH2 differentiation. Nat. Immunol. 2012; 13:778–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ruebel K.H., Leontovich A.A., Tanizaki Y., Jin L., Stilling G.A., Zhang S., Coonse K., Scheithauer B.W., Lombardero M., Kovacs K. et al. . Effects of TGFbeta1 on gene expression in the HP75 human pituitary tumor cell line identified by gene expression profiling. Endocrine. 2008; 33:62–76. [DOI] [PubMed] [Google Scholar]
- 10. Vervoort S., Lourenço A.R., van Boxtel R., Coffer P.J.. SOX4 mediates TGF-β-Induced expression of mesenchymal markers during mammary cell epithelial to mesenchymal transition. PLoS One. 2013; 8:e53238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Massagué J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012; 13:616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kalluri R. EMT: When epithelial cells decide to become mesenchymal-like cells. J. Clin. Invest. 2009; 119:1417–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tufegdzic Vidakovic A., Rueda O.M., Vervoort S.J., Sati Batra A., Goldgraben M.A., Uribe-Lewis S., Greenwood W., Coffer P.J., Bruna A., Caldas C.. Context-Specific effects of TGF-β/SMAD3 in cancer are modulated by the epigenome. Cell Rep. 2015; 13:2480–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ikushima H., Todo T., Ino Y., Takahashi M., Miyazawa K., Miyazono K.. Autocrine TGF-β signaling maintains tumorigenicity of Glioma-Initiating cells through Sry-Related HMG-Box factors. Cell Stem Cell. 2009; 5:504–514. [DOI] [PubMed] [Google Scholar]
- 15. Fischer K.R., Durrans A., Lee S., Sheng J., Li F., Wong S.T.C., Choi H., El Rayes T., Ryu S., Troeger J. et al. . Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015; 527:472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kalluri R., Weinberg R.A.. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009; 119:1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang J., Liang Q., Lei Y., Yao M., Li L., Gao X., Feng J., Zhang Y., Gao H., Liu D.X. et al. . SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012; 72:4597–4608. [DOI] [PubMed] [Google Scholar]
- 18. David C.J., Huang Y.-H., Chen M., Su J., Zou Y., Bardeesy N., Iacobuzio-Donahue C.A., Massagué J.. TGF-β tumor suppression through a lethal EMT. Cell. 2016; 164:1015–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Minn A.J., Gupta G.P., Siegel P.M., Bos P.D., Shu W., Giri D.D., Viale A., Olshen A.B., Gerald W.L., Massague J.. Genes that mediate breast cancer metastasis to lung. Nature. 2005; 436:518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bos P.D., Zhang X.H.-F., Nadal C., Shu W., Gomis R.R., Nguyen D.X., Minn A.J., van de Vijver M.J., Gerald W.L., Foekens J.A. et al. . Genes that mediate breast cancer metastasis to the brain. Nature. 2009; 459:1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tavazoie S.F., Alarcón C., Oskarsson T., Padua D., Wang Q., Bos P.D., Gerald W.L., Massagué J.. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008; 451:147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bruna A., Greenwood W., Le Quesne J., Teschendorff A., Miranda-Saavedra D., Rueda O.M., Sandoval J.L., Vidakovic A.T., Saadi A., Pharoah P. et al. . TGFβ induces the formation of tumour-initiating cells in claudinlow breast cancer. Nat. Commun. 2012; 3:1055. [DOI] [PubMed] [Google Scholar]
- 23. Beekman J.M., Vervoort S.J., Dekkers F., van Vessem M.E., Vendelbosch S., Brugulat-Panès A., van Loosdregt J., Braat A.K., Coffer P.J.. Syntenin-mediated regulation of Sox4 proteasomal degradation modulates transcriptional output. Oncogene. 2012; 31:2668–2679. [DOI] [PubMed] [Google Scholar]
- 24. van Boxtel R., Gomez-Puerto C., Mokry M., Eijkelenboom A., van der Vos K.E., Nieuwenhuis E.E., Burgering B.M., Lam E.W.-F., Coffer P.J.. FOXP1 acts through a negative feedback loop to suppress FOXO-induced apoptosis. Cell Death Differ. 2013; 20:1219–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thorvaldsdóttir H., Robinson J.T., Mesirov J.P.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Curtis C., Shah S.P., Chin S.-F., Turashvili G., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y. et al. . The genomic and transcriptomic architecture of 2, 000 breast tumours reveals novel subgroups. Nature. 2012; 486:346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prat A., Parker J.S., Karginova O., Fan C., Livasy C., Herschkowitz J.I., He X., Perou C.M.. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010; 12:R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ringnér M., Fredlund E., Häkkinen J., Borg Å., Staaf J.. GOBO: Gene expression-based outcome for breast cancer online. PLoS One. 2011; 6:e17911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sarkar A., Hochedlinger K.. The Sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013; 12:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang J., Liang Q., Lei Y., Yao M., Li L., Gao X., Feng J., Zhang Y., Gao H., Liu D.-X. et al. . SOX4 induces Epithelial-Mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012; 72:4597–4608. [DOI] [PubMed] [Google Scholar]
- 32. Lai J.S., Herr W.. Ethidium bromide provides a simple tool for identifying genuine DNA- independent protein associations. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:6958–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Halder S.K., Beauchamp R.D., Datta P.K.. A specific inhibitor of TGF-beta receptor kinase, SB-431542, as a potent antitumor agent for human cancers. Neoplasia. 2005; 7:509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morikawa M., Koinuma D., Miyazono K., Heldin C.-H.. Genome-wide mechanisms of Smad binding. Oncogene. 2012; 32:1609–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mani S.A., Guo W., Liao M.-J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M. et al. . The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008; 133:704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taube J.H., Herschkowitz J.I., Komurov K., Zhou A.Y., Gupta S., Yang J., Hartwell K., Onder T.T., Gupta P.B., Evans K.W. et al. . Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:15449–15454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scheel C., Eaton E.N., Li S.H.-J., Chaffer C.L., Reinhardt F., Kah K.-J., Bell G., Guo W., Rubin J., Richardson A.L. et al. . Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011; 145:926–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lei X., Bandyopadhyay A., Le T., Sun L.. Autocrine TGFβ supports growth and survival of human breast cancer MDA-MB-231 cells. Oncogene. 2002; 21:7514–7523. [DOI] [PubMed] [Google Scholar]
- 39. Padua D., Zhang X.H.-F., Wang Q., Nadal C., Gerald W.L., Gomis R.R., Massagué J.. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008; 133:66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wilson M., Koopman P.. Matching SOX: Partner proteins and co-factors of the SOX family of transcriptional regulators. Curr. Opin. Genet. Dev. 2002; 12:441–446. [DOI] [PubMed] [Google Scholar]
- 41. Tompkins D.H., Besnard V., Lange A.W., Wert S.E., Keiser A.R., Smith A.N., Lang R., Whitsett J.A.. Sox2 is required for maintenance and differentiation of bronchiolar Clara, ciliated, and goblet cells. PLoS One. 2009; 4:e8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Furumatsu T., Tsuda M., Taniguchi N., Tajima Y., Asahara H.. Smad3 induces chondrogenesis through the activation of SOX9 via CREB-binding protein/p300 recruitment. J. Biol. Chem. 2005; 280:8343–8350. [DOI] [PubMed] [Google Scholar]
- 43. Gomis R.R., Alarcón C., Nadal C., Van Poznak C., Massagué J.. C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006; 10:203–214. [DOI] [PubMed] [Google Scholar]
- 44. Zhang Y., Feng X.H., Derynck R.. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature. 1998; 394:909–913. [DOI] [PubMed] [Google Scholar]
- 45. Schilham M.W., Oosterwegel M.A., Moerer P., Ya J., de Boer P.A.J., van de Wetering M., Verbeek S., Lamers W.H., Kruisbeek A.M., Cumano A. et al. . Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996; 380:711–714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChiP-seq and RNA-seq datasets have been deposited in GEO under accession number GSE104761.