Abstract
Recent studies have indicated that microRNAs (miRNAs) play an important role in hepatocellular carcinoma (HCC) progression. In this study, we showed that miR-766-3p was decreased in approximately 72% of HCC tissues and cell lines, and its low expression level was significantly correlated with tumour size, TNM stage, metastasis, and poor prognosis in HCC. Ectopic miR-766-3p expression inhibited HCC cell proliferation, colony formation, migration and invasion. In addition, we showed that miR-766-3p repressed Wnt3a expression. A luciferase reporter assay revealed that Wnt3a was a direct target of miR-766-3p, and an inverse correlation between miR-766-3p and Wnt3a expression was observed. Moreover, Wnt3a up-regulation reversed the effects of miR-766-3p on HCC progression. In addition, our study showed that miR-766-3p up-regulation decreased the nuclear β-catenin level and expression of Wnt targets (TCF1 and Survivin) and reduced the level of MAP protein regulator of cytokinesis 1 (PRC1). However, these effects of miR-766-3p were reversed by Wnt3a up-regulation. In addition, PRC1 up-regulation increased the nuclear β-catenin level and protein expression of TCF1 and Survivin. iCRT3, which disrupts the β-catenin-TCF4 interaction, repressed the TCF1, Survivin and PRC1 protein levels. Taken together, our results suggest that miR-766-3p down-regulation promotes HCC cell progression, probably by targeting the Wnt3a/PRC1 pathway, and miR-766-3p may serve as a potential therapeutic target in HCC.
Keywords: hepatocellular carcinoma, invasion, miR-766-3p, proliferation, Wnt3a
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most prevalent life-threatening human malignancies worldwide (Cancer Genome Atlas Research Network, 2017). Although many advances have been made in understanding the molecular mechanisms underlying liver carcinogenesis, HCC remains a deadly cancer whose treatment requires the use of innovative effective therapeutic options, such as gene therapy (Romano et al., 2006; Su et al., 2015).
MicroRNAs (miRNAs), one class of highly conserved small noncoding RNAs that are, on average, 22 nucleotides in length, contribute to regulation of expression of specific proteins by binding to the 3′-untranslated regions (3′UTRs) of the target genes (Leva et al., 2014; Su et al., 2015). By regulating target gene expression, miRNAs are involved in the regulation of a variety of biological processes, including cancer development (Bracken et al., 2015; Wang et al., 2018). At present, increasing experimental evidence indicates that miRNAs can function as oncogenes or tumour suppressor genes in HCC and regulate cell proliferation, apoptosis, migration and invasion (Qiu et al., 2013; Wang et al., 2018). For example, miR-21, which functions as an oncogene in HCC, is overexpressed in the disease and induces cell proliferation, invasion and chemoresistance (Qiu et al., 2013; Wagenaar et al., 2015). Conversely, miR-122, which functions as a tumour suppressor, is abnormally down-regulated in HCC (Fornari, et al., 2009; Simerzin et al., 2016). MiR-766-3p, a suspected inducer or inhibitor of tumour progression in different cancers, has recently attracted much attention because it plays pivotal roles in human cancers (Chen et al., 2017; Li et al., 2015). Importantly, however, the expression of miR-766-3p in HCC was decreased compared with its expression in noncancerous liver tissue (Wei et al., 2013), which suggested that miR-766-3p may serve as a potential tumour suppressor in HCC. Until recently, the precise contribution of miR-766-3p to HCC progression has not been reported.
Wnt3a, a member of the Wnt family, is involved in embryonic development, proliferation, differentiation, and tumourigenesis by activating the canonical Wnt signalling pathway (He et al., 2015). In cancer, Wnt3a can function as a tumour suppressor or a cancer promoter gene depending on cancer types. For instance, Wnt3a increases apoptosis of melanoma cells treated with trail (Zimmerman et al., 2013). On the other hand, Wnt3a expression is up-regulated in glioblastoma, colon cancer, and pancreatic cancer (Kaur et al., 2013; Nagano et al., 2013; Qi et al., 2014). In HCC, some reports have shown that Wnt3a is highly expressed and is associated with a poor prognosis (Pan et al., 2016). Down-regulation of Wnt3a represses HCC cell cycle, proliferation, invasion and migration (Lu et al., 2017). Although Wnt3a has been reported as an oncogene in HCC, the relationships between Wnt3a and its upstream miRNAs in HCC remain unclear.
In this study, we found that miR-766-3p suppressed cell proliferation, colony formation, migration and invasion by targeting Wnt3a and functioned as a novel tumour suppressor in HCC.
MATERIALS AND METHODS
Tissue samples
Fifty-seven pairs of human HCC and matched adjacent normal tissue samples were obtained from the Second Affiliated Hospital of Chongqing Medical University from May 2014 to February 2016. Histopathology was confirmed by two certified pathologists. All tissues were frozen and stored in liquid nitrogen. None of the patients from whom the tissues were obtained had received chemotherapy or radiotherapy. A written informed consent was obtained from all patients. The use of all human materials in this project was approved by the Ethics Committee of Chongqing Medical University and the Second Affiliated Hospital of Chongqing Medical University.
Cell culture
The SMMC7721 HCC cell line was purchased from Xiangya Central Experiment Laboratory (China), and the normal human liver cell line (LO2), as well as HCC cell lines SK-HEP-1, PLC/PRF/5, Hep3B and Huh7 were obtained from the Cell Bank Type Culture Collection of the Chinese Academy of Sciences (China). The LO2 and SMMC7721 cells were cultured in RPMI-1640 medium (Invitrogen, USA) with 10% foetal bovine serum (FBS; Life Technologies, USA). Huh7 cell line was grown in DMEM (Invitrogen, USA), supplemented with 10% FBS. SK-HEP-1, PLC/PRF/5 and Hep3B cells were cultured in MEM (Invitrogen, USA) containing 10% FBS.
RNA extraction and real-time quantitative PCR analysis
To obtain cDNA, total RNAs of frozen tissues or cells were extracted using RNeasy 96 kit (Qiagen, USA) and were reverse-transcribed. The cDNA was used to analyse miR-766-3p and Wnt3a expression. The real-time PCR was performed using a SYBR Premix Ex Taq Kit (TaKaRa, China) on a Stratagene Mx3000P Real-Time PCR System (Agilent Technologies, USA). The primers used in the reactions are listed in Table 1 and were synthesized by RiboBio (China). The expression of miR-766-3p was normalized to U6 expression. Wnt3a mRNA expression was normalized to GAPDH expression. The relative gene expression levels were calculated using the comparative threshold cycle (2−ΔΔCt) method.
Table 1.
PCR primers
| Primer name | Primer sequence |
|---|---|
| miR-766-3p RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACGCTGAGGC-3′ |
| miR-766-3p Fwd | 5′-ACTCCAGCCCCACAGC-3′ |
| U6 RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAAAATATGGAAC-3′ |
| U6 Fwd | 5′-GTGCTCGCTTCGGCAGC-3′ |
| Rev | 5′-CAGTGCAGGGTCCGAGGT-3′ |
| HOXA13 Fwd | 5′-ATCAGCCACGACGAATCTCT-3′ |
| HOXA13 Rev | 5′-GGCAAAGCAACGAGTTCTGA-3′ |
| Wnt3a Fwd | 5′-ATCGAGTTTGGTGGGATGGT-3′ |
| Wnt3a Rev | 5′-CGCTGTCGTACTTGTCCTTG-3′ |
| TSG101 Fwd | 5′-CCTCCAGTCTTCTCTCGTCC-3′ |
| TSG101 Rev | 5′-GGAGGCTGAGAAGGGTACTG-3′ |
| IGFBP2 Fwd | 5′-GAGTGTCATCTCTTCTACAATGAGC-3′ |
| IGFBP2 Rev | 5′-AATACGTGTGTCAGAACTGGAAAAT-3′ |
| GAPDH Fwd | 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ |
| GAPDH Rev | 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ |
| wt-Wnt3a 3′UTR Fwd | 5′-TAGGCGATCGCTCGAGGCACCGGCCGCGGCTCCCC-3′ |
| wt-Wnt3a 3′UTR Rev | 5′-AATTCCCGGGCTCGAGTTTCGTCTAACTCCGTTGGACAGT-3′ |
| mut-Wnt3a 3′UTR Fwd | 5′-AATGGTCCGCTTTCCTGGAGCCAATGGCCCG-3′ |
| mut-Wnt3a 3′UTR Rev | 5′-AGGAAAGCGGACCATTTCCCGCCATGAGGGGCCAGGAAGG-3′ |
Plasmid and cell transfection
The precursors of miR-766-3p (miR-766-3p) and non-targeting miRNA precursors (NC) were obtained from GeneCopoeia (China). To overexpress Wnt3a, Wnt3a CDS clone was PCR-amplified and cloned into a pcDNA3.1 vector GeneChem (China). The PRC1 ORF expression plasmid was purchased from GeneCopoeia (China).
SK-HEP-1 and PLC/PRF/5 cells were cultured in 6- or 96-well plates and grown to 70% to 80% confluence. Next, cells were transfected using Lipofectamine 2000 (Invitrogen, USA), according to the manufacturer’s instructions. The cells were then incubated for 24 h or 48 h.
Cell proliferation assay
For the cell proliferation assay, SK-HEP-1 and PLC/PRF/5 cells were cultured in a 6-well plate. Then, cells were transfected with the non-targeting miRNA precursor plasmid (NC), precursors of miR-766-3p plasmid (miR-766-3p), as well as the precursors of miR-766-3p plasmid with Wnt3a or PRC1 expression plasmid and cultured for 24 h. After transfection, the cells were cultured in 96-well plates (5 × 103 cells per well). After 1, 2, 3, or 4 days, 10 μl CCK-8 solution (Dojindo, Japan) was added into each well. The cells were incubated at 37°C for 1 h, and then the absorbance was detected at 450 nm using a microplate reader (Bio-Rad, USA).
Colony formation assay
SK-HEP-1 and PLC/PRF/5 cells after transfection were seeded into 6-well plates at a density of 200 cells/well and maintained in medium for 10 days. Then, colonies were fixed with 10% formaldehyde for 5 min and stained with 1% crystal violet for 30 s.
Transwell invasion assay
Cell invasion assays were performed using Matrigel-coated transwell cell culture inserts (Invitrogen USA). SK-HEP-1 and PLC/PRF/5 cells after transfection were seeded into the upper chamber of the insert with serum-free media. The bottom of the chamber contained complete media as a chemo-attractant. After 48 h, the cells in the upper chamber or membrane were removed, and then the cells on the lower surface of the membrane were stained with 0.1% crystal violet. The number of the cells on the lower surface was then counted with a microscope.
Wound healing assay
Cells after transfection were grown to 90% confluence, and then artificial wound tracks were created in the monolayer with a sterile micropipette tip. Cell migration ability was monitored at different time points (0 h and 48 h) under a microscope.
Xenograft tumour model
Animal experimental protocols were approved by the Ethics Committee of Chongqing Medical University and the Second Affiliated Hospital of Chongqing Medical University. SK-HEP-1 cells were transfected with non-targeting miRNA precursor plasmid (NC) and precursors of miR-766-3p plasmid (miR-766-3p) and were selected with 5 μg/ml puromycin (Thermo Scientific, USA) for three weeks. Then, MiR-766-3p- and NC-transfected SK-HEP-1 cells (3 × 106 cells) were suspended in 100 μl PBS and injected subcutaneously to the posterior flank of the BALB/c nude mice at 5 to 6 weeks of age, purchased from the Animal Center of the Chinese Academy of Science (China). Tumours were measured with a caliper every week after implantation, and the volumes of each tumour were calculated with the formula: volume = (length × width2)/2. The mice were sacrificed 5 weeks after injection. The mice were killed after 5 weeks implantation.
Western blot analysis
After transfection, SK-HEP-1 and PLC/PRF/5 cells were cultured for 72 h. Then, cells were harvested and lysed in RIPA buffer (Heart, China). The cell proteins were extracted, and their concentrations were determined by the Bradford protein assay kit (Bio-Rad, USA). NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Inc, USA) were used to isolate nuclear proteins, according to the manufacturer’s instructions. The total proteins (40 μg per well) were separated in SDS-PAGE polyacrylamide gel and then transferred to polyvinylidene difluoride membranes (Bio-Rad, USA). Next, the membranes were blocked with 5% non-fat powdered milk for 1 h and incubated overnight at 4°C with polyclonal or monoclonal antibodies. Monoclonal antibodies against human Wnt3a, β-catenin, Survivin, TCF1, PRC1 and GAPDH were purchased from Santa Cruz Biotechnology (USA). The PVDF membranes were washed 3 times with TBST and then incubated with the horseradish peroxidase-conjugated secondary antibodies for 45 min at room temperature. Finally, the membranes were washed 3 times with TBST, and the corresponding band was revealed with enhanced chemiluminescence reagents (Thermo Scientific, USA) according to the manufacturer’s instructions. GAPDH was used as an internal control.
Luciferase reporter assay
To verify the miR-766-3p-targeted 3′UTR, dual-luciferase reporter plasmids psiCHECK-2 Wnt3a 3′UTR wild-type (wt-Wnt3a) and Wnt3a 3′UTR mutant (mut-Wnt3a) were constructed. The wild-type 3′UTR of Wnt3a (1823 bp) containing the predicted miR-766-3p binding sites (1492-1498) was amplified by PCR using genomic DNA as a template and then inserted downstream of the stop codon of the firefly luciferase gene in psiCHECK-2 vector (Promega, USA). The mutation of the 3′UTR of Wnt3a (deletion of CUGGAG) was also performed by PCR. All primers used in plasmid construction were synthesized by Sangon Biotech (China) and are provided in Table 1. All constructed vectors were confirmed by sequencing.
For luciferase reporter assay, K-HEP-1 and PLC/PRF/5 cells were cultured in 24-well plates and co-transfected with Wnt3a 3′UTR wild-type/Wnt3a 3′UTR mutant plasmid and the miR-766-3p expression plasmid or the NC, according to Lipofectamine 2000 (Invitrogen, USA) reagent protocol. Cells were harvested at 48 h after transfection and lysed. The firefly luciferase activity was measured using a dual-luciferase reporter assay system (Promega, USA). Renilla luciferase activity was used for normalization.
Statistical analysis
All data were analysed using the IBM SPSS Statistics version 18.0 (IBM Corporation, USA). The results from at least 3 independent experiments were expressed as the mean ± S.D. The differences among groups were analysed using either the one-way ANOVA or the Student’s t-test. The Spearman’s correlation coefficient was used to analyse the association between miR-766-3p expression and Wnt3a expression in HCC tissues. The Pearson’s x2 test was used to analyse the relationships between miR-766-3p expression and HCC clinicopathologic characteristics. The overall survival curve in the 2 patient groups was plotted with the Kaplan–Meier method and compared by the log-rank test. Covariates with P values < 0.05 in the univariate analysis were subjected to multivariate analysis. Multivariate Cox regression models were constructed to estimate the hazard ratios (HRs) of independent factors for survival after controlling for potential confounding factors. P value < 0.05 was considered statistically significant.
RESULTS
MiR-766-3p is down-regulated in HCC tissues, which correlated with cancer progression
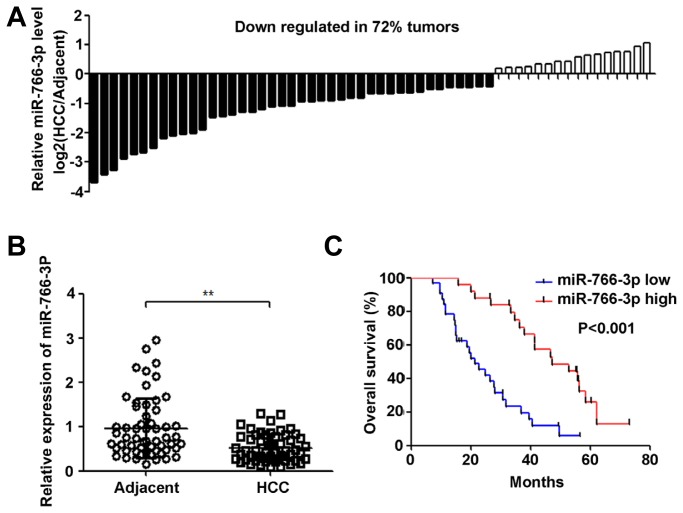
To evaluate miR-766-3p expression in HCC tissues, we analysed miR-766-3p expression in fifty-seven pairs of human HCC and matched adjacent normal tissue samples with qRT-PCR. As shown in Fig. 1A, miR-766-3p was down-regulated in approximately 72% of HCC tissues. Furthermore, miR-766-3p expression in HCC tissues was significantly lower than that in adjacent normal tissues (P < 0.01) (Fig. 1B). To analyse the correlations between the expression of miR-766-3p and patient’s clinical factors and overall survival, a Kaplan-Meier analysis and log-rank test were performed. As shown in Table 2, low miR-766-3p expression was significantly correlated with tumour size (P = 0.012), TNM stage (P = 0.003) and metastasis (P = 0.008). However, no significant correlation was observed between low miR-766-3p expression and patient age (P = 0.847), gender (P=0.494), AFP expression (P = 0.536), HBV infection (P = 0.479), or cirrhosis (P = 0.790). In addition, the results of the survival analysis shown in Fig. 1C demonstrated that HCC patients with low miR-766-3p expression had a shorter overall survival than those with high miR-766-3p expression (P < 0.001). Moreover, the results of multivariate survival analysis shown in Table 3 demonstrated that miR-766-3p was an independent prognostic indicator of the survival of patients with HCC.
Fig. 1. MiR-766-3p is down-regulated in HCC tissues and is correlated with overall survival.
(A) MiR-766-3p expression in 57 paired HCC and the matched adjacent normal tissue samples was measured by qRT-PCR. U6 expression was used as a control. (B) miR-766-3p expression in human HCC tissues was down-regulated compared with that in matched adjacent normal tissues. (C) The correlation between miR-766-3p expression and overall survival was analysed with the Kaplan–Meier method. The P value was obtained using the log-rank test. **P < 0.01.
Table 2.
Correlations between miR-766-3p expression and HCC patient clinicopathological characteristics
| miR-766-3p expression | |||
|---|---|---|---|
|
|
|||
| Clinical factor | Low expression (n = 32) | High expression (n = 25) | P value |
| Age(y) | |||
| < 60 | 20 | 15 | 0.847 |
| ≥60 | 12 | 10 | |
| Gender | |||
| Male | 17 | 11 | 0.494 |
| Female | 15 | 14 | |
| Tumour size (cm) | |||
| < 5 | 11 | 17 | 0.012* |
| ≥5 | 21 | 8 | |
| AFP (ng/ml) | |||
| < 20 | 14 | 13 | 0.536 |
| ≥20 | 18 | 12 | |
| TNM stage | |||
| I+II | 8 | 16 | 0.003** |
| III+IV | 24 | 9 | |
| HBV infection | |||
| No | 23 | 20 | 0.479 |
| Yes | 9 | 5 | |
| Cirrhosis | |||
| No | 22 | 18 | 0.790 |
| Yes | 10 | 7 | |
| Metastasis | |||
| No | 13 | 19 | 0.008** |
| Yes | 19 | 6 | |
Statistically significant (P < 0.05),
Statistically significant (P < 0.01).
Table 3.
Cox proportional hazard models for prognostic factors
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age (≥60 vs. < 60) | 0.987 (0.442–2.204) | 0.974 | ||
| Gender (female vs. male) | 1.153 (0.538–2.472) | 0.714 | ||
| Tumour size (≥5 vs. < 5) | 1.535 (0.737–3.198) | 0.252 | ||
| AFP (≥20 vs. < 20) | 1.361 (0.679–2.729) | 0.385 | ||
| TNM stage (III+IV vs. I+II) | 2.351 (1.127–4.902) | 0.023* | 2.753 (1.394–5.437) | 0.004 |
| HBV infection (Yes vs. No) | 1.014 (0.468–2.199) | 0.972 | ||
| Cirrhosis (Yes vs. No) | 3.643 (1.581–8.390) | 0.002** | 3.139 (1.540–6.399) | 0.002** |
| Metastasis (Yes vs. No) | 2.120 (1.028–4.369) | 0.042* | 2.521 (1.320–4.814) | 0.005** |
| miR-766-3p (low vs. high) | 3.699 (1.793–7.629) | < 0.001** | 3.691 (1.815–7.506) | < 0.001* |
Statistically significant (P < 0.05),
Statistically significant (P < 0.01).
MiR-766-3p overexpression inhibits HCC cell proliferation, colony formation in vitro and tumour growth in vivo
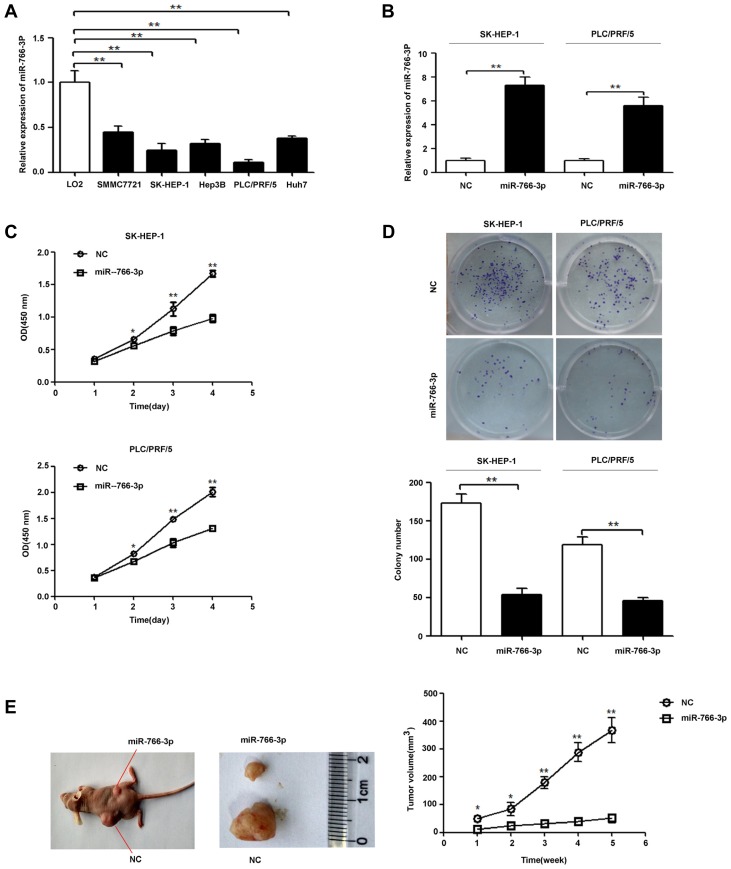
Figure 2A shows that miR-766-3p expression in HCC cell lines SMMC7721, SK-HEP-1, Hep3B, PLC/PRF/5 and Huh7 was significantly lower than that in the LO2 cell line (P < 0.01). To test the effect of miR-766-3p on HCC progression, SK-HEP-1 and PLC/PRF/5 cells were transfected with the miR-766-3p expression plasmid (miR-766-3p) or the scrambled miRNA expression plasmid (NC). As shown in Fig. 2B, cells transfected with the miR-766-3p expression plasmid displayed higher miR-766-3p expression than control cells. Furthermore, the cell proliferation assay results shown in Fig. 2C demonstrated that miR-766-3p overexpression inhibited HCC cell proliferation. Additionally, the colony formation results shown in Fig. 2D demonstrated that miR-766-3p overexpression repressed colony formation in the SK-HEP-1 and PLC/PRF/5 cell lines. To investigate the effect of miR-766-3p overexpression on HCC carcinogenesis, we inoculated nude mice with SK-HEP-1-miR-766-3p and SK-HEP-1-NC cells. The results showed that the cells transfected with the miR-766-3p expression plasmid generated smaller tumours than the NC cells transfected with the scrambled miRNA expression plasmid (Fig. 2E).
Fig. 2. MiR-766-3p up-regulation inhibits HCC cell proliferation, colony formation in vitro and tumour growth in vivo.
(A) miR-766-3p expression in HCC cell lines (SMMC7721, SK-HEP-1, Hep3B, PLC/PRF/5 and Huh7) and a normal human liver epithelial cell line (LO2) was measured by qRT-PCR. (B) SK-HEP-1 and PLC/PRF/5 cells were transfected with the miR-766-3p expression plasmid (miR-766-3p) or non-targeting miRNA precursor plasmid (NC). miR-766-3p expression in SK-HEP-1 and PLC/PRF/5 cells was detected by qRT-PCR. U6 expression was used as a control. (C) Cell proliferation was tested with CCK-8. MiR-766-3p overexpression significantly inhibited the proliferation of SK-HEP-1 and PLC/PRF/5 cells. (D) MiR-766-3p overexpression significantly reduced the colony formation of SK-HEP-1 and PLC/PRF/5 cells. (E) MiR-766-3p overexpression in SK-HEP-1 cells significantly repressed tumour growth in vivo. *P < 0.05; **P < 0.01.
MiR-766-3p overexpression repressed HCC cell invasion and migration in vitro
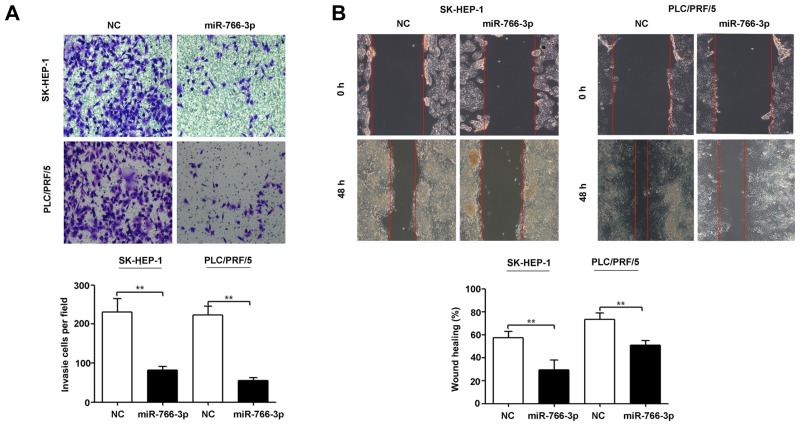
To determine the significance of miR-766-3p during invasion, we evaluated the ability of HCC cells to migrate across a layer of Matrigel deposited on a Boyden chamber when miR-766-3p was overexpressed. Our results showed that miR-766-3p up-regulation significantly suppressed SK-HEP-1 and PLC/PRF/5 cell invasion (Fig. 3A). Consistently, the same trend was observed in SK-HEP-1 and PLC/PRF/5 cell migration (Fig. 3B).
Fig. 3. MiR-766-3p up-regulation inhibits HCC cell invasion and migration in vitro.
SK-HEP-1 and PLC/PRF/5 cells were transfected with the miR-766-3p expression plasmid (miR-766-3p) or non-targeting miRNA precursor plasmid (NC). The effects of miR-766-3p overexpression on SK-HEP-1 and PLC/PRF/5 cells invasion (A) and migration (B) were measured by transwell invasion assay and wound healing assay, respectively. The experiments were independently repeated thrice. **P < 0.01.
Wnt3a is a direct target of miR-766-3p in HCC
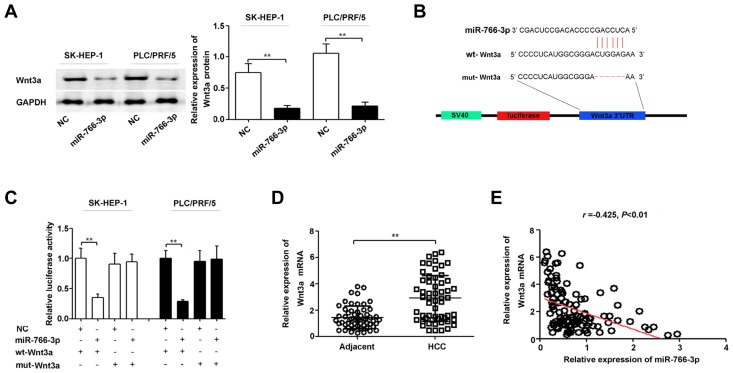
To determine the potential effectors of miR-766-3p in HCC development, we identified the target genes of miR-766-3p using a microRNA database (www.targetsan.org). We found that Wnt3a 3′UTR contains one miR-766-3p-binding site. To confirm regulation of Wnt3a by miR-766-3p, western blotting was performed for SK-HEP-1 and PLC/PRF/5 cells transfected with the miR-766-3p expression plasmid (miR-766-3p) or the scrambled miRNA expression plasmid (NC). The results showed that Wnt3a protein expression was decreased in HCC cells after miR-766-3p up-regulation (Fig. 4A). In addition, we constructed vectors containing the wild-type or mutant 3′-UTR of human Wnt3a fused downstream of the firefly luciferase gene (Fig. 4B). As shown in Fig. 4C, luciferase activity in miR-766-3p-transfected cells was significantly decreased compared with that in NC cells. In addition, miR-766-3p-mediated repression of luciferase activity was abolished by the mutations in the putative miR-766-3p binding site. The qRT-PCR results showed that Wnt3a mRNA expression in HCC tissues was significantly increased compared with that in matched adjacent normal tissues (Fig. 4D) and inversely correlated with miR-766-3p expression (r = −0.425, P < 0.01; Fig. 4E). In summary, these results indicate that Wnt3a is a direct target of miR-766-3p in HCC.
Fig. 4. Wnt3a is a direct target of miR-766-3p in HCC.
(A) SK-HEP-1 and PLC/PRF/5 cells were transfected with the miR-766-3p expression plasmid (miR-766-3p) or non-targeting miRNA precursor plasmid (NC). The effect of miR-766-3p up-regulation on Wnt3a protein expression was measured by Western blot. (B) The sequences of the putative miR-766-3p binding sites in the wild-type and mutant Wnt3a 3′UTR. (C) Luciferase reporter plasmids carrying the Wnt3a wild-type 3′UTR (wt-Wnt3a) or Wnt3a mutant 3′UTR (mut-Wnt3a) were transfected into SK-HEP-1 and PLC/PRF/5 cells with miR-766-3p or NC. MiR-766-3p up-regulation suppressed luciferase activity of the wild-type but not the mutant 3′UTR of Wnt3a. Renilla luciferase activity was used as a control. (D) Wnt3a mRNA expression levels in 57 pairs of human HCC and matched adjacent normal tissues were measured by qRT-PCR. (E) miR-766-3p expression was inversely correlated with Wnt3a miRNA expression in HCC tissues, as demonstrated by the Spearman’s correlation coefficient. **P < 0.01.
Wnt3a restoration partially reverses the suppressive effects of miR-766-3p overexpression on HCC cells in vitro
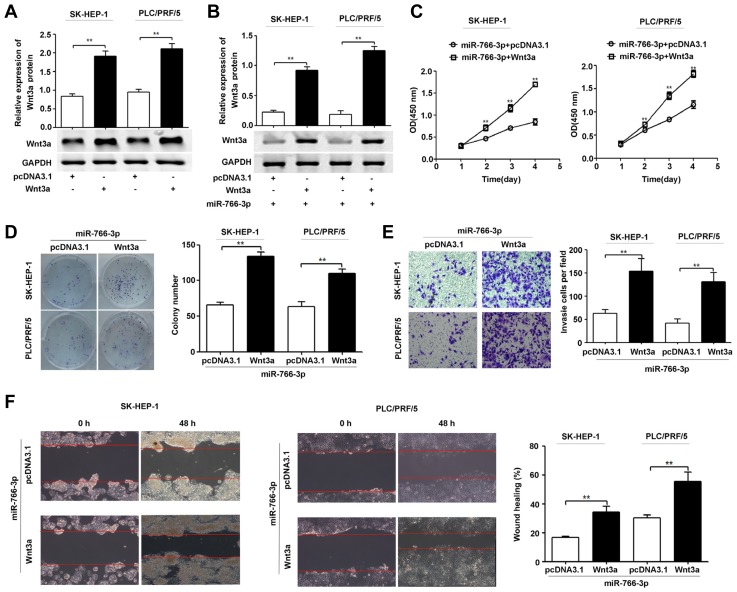
To determine the role of Wnt3a in the suppressive effects of miR-766-3p on HCC cells, we constructed a Wnt3a overexpression plasmid vector. As shown in Fig. 5A, Wnt3a plasmid-transfected cells displayed higher Wnt3a protein expression than control cells. In addition, Wnt3a overexpression restored Wnt3a protein expression in cells pretreated with the miR-766-3p expression plasmid (Fig. 5B). Moreover, Wnt3a up-regulation reversed the inhibition of cell proliferation, colony formation, invasion and migration due to miR-766-3p up-regulation (Figs. 5C–5F).
Fig. 5. Wnt3a restoration partially reverses the suppressive effects of miR-766-3p on HCC cell proliferation, colony formation, invasion and migration in vitro.
(A) SK-HEP-1 and PLC/PRF/5 cells were transfected with pcDNA3.1 or the Wnt3a expression plasmid (Wnt3a). Western blot was used to determine the Wnt3a protein expression levels in SK-HEP-1 and PLC/PRF/5 cells. (B) SK-HEP-1 and PLC/PRF/5 cells pretreated with the miR-766-3p expression plasmid were transfected with pcDNA3.1 or the Wnt3a expression plasmid. Wnt3a protein expression levels were then tested with Western blot. (C-F) SK-HEP-1 and PLC/PRF/5 cells pretreated with the miR-766-3p expression plasmid were transfected with pcDNA3.1 or the Wnt3a expression plasmid. Wnt3a protein expression levels were then tested with Western blot (B). Cell proliferation was determined by CCK-8 (C). The colony formation of SK-HEP-1 and PLC/PRF/5 cells was determined (D). The cell invasion (E) and migration (F) were measured by transwell invasion assay and wound healing assay, respectively. The experiments were repeated independently three times. GAPDH was used as a loading control. **P < 0.01.
The downstream mechanism underlying the antitumour effect of miR-766-3p/Wnt3a signalling
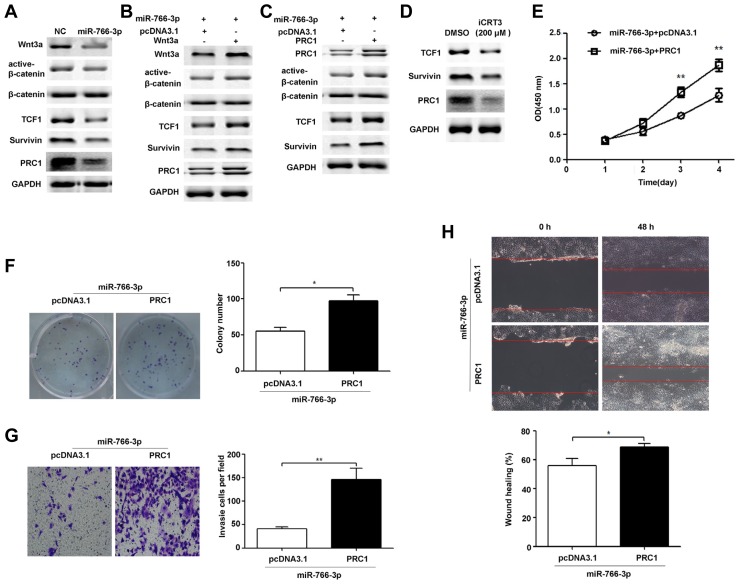
Previous reports have shown that Wnt3a triggers Wnt signalling activation and induces expression of PRC1 and other known Wnt targets (Survivin and TCF1) in HCC (Chen et al., 2016). Furthermore, PRC1 down-regulation inhibits cell proliferation, colony formation, invasion and migration. Interestingly, PRC1 down-regulation inhibits the expression of Wnt targets (Survivin and TCF1) and represses Wnt signalling activation by reducing nuclear β-catenin levels (Chen et al., 2016). Thus, we hypothesized that miR-766-3p may repress HCC progression by regulating Wnt3a/PRC1 signalling. To evaluate the effect of miR-766-3p on Wnt pathway components and PRC1 expression, we detected the protein expression of PRC1, β-catenin, TCF1 and Survivin in HCC cells transfected with the miR-766-3p expression plasmid (miR-766-3p) or the scrambled miRNA expression plasmid (NC). Our results showed that miR-766-3p up-regulation inhibited PRC1, TCF1 and Survivin protein expression and impaired the nuclear accumulation of β-catenin in HCC cells (Fig. 6A). In addition, Wnt3a up-regulation reversed the inhibition of PRC1, TCF1 and Survivin protein expression and the decrease in nuclear accumulation of β-catenin induced by miR-766-3p (Fig. 6B). To determine whether miR-766-3p could repress TCF1 and Survivin protein expression and the nuclear accumulation of β-catenin by regulating PRC1, we restored PRC1 expression in PLC/PRF/5 cells pretreated with miR-766-3p plasmid vectors. Our results showed that ectopic expression of PRC1 reversed the inhibition of TCF1 and Survivin protein expression and the decrease in the nuclear accumulation of β-catenin (Fig. 6C). Moreover, iCRT3, a potent and specific inhibitor of β-catenin-TCF4 interaction, repressed expression of Wnt targets (TCF1 and Survivin) together with the PRC1 protein level (Fig. 6D). Thus, PRC1 functions as a novel Wnt target and triggers Wnt signalling. To determine whether miR-766-3p could repress HCC progression by regulating PRC1, we restored the PRC1 expression in PLC/PRF/5 cells pretreated with miR-766-3p plasmid vectors. Our results showed that ectopic expression of PRC1 reversed the inhibition of proliferation (Fig. 6E), colony formation (Fig. 6F), invasion (Fig. 6G) and migration (Fig. 6H) induced by miR-766-3p overexpression. Taken together, miR-766-3p may control HCC cell progression by regulating Wnt3a/PRC1 signalling (Fig. 7).
Fig. 6. miR-766-3p suppresses HCC cell growth, colony formation, invasion and migration via a Wnt/PRC1 positive regulatory loop.
(A) PLC/PRF/5 cells were transfected with the miR-766-3p expression plasmid (miR-766-3p) or non-targeting miRNA precursor plasmid (NC), and then total and nuclear β-catenin levels, and protein expression levels of Wnt3a, TCF1, Survivin, and PRC1 were detected by Western blot analysis. (B) PLC/PRF/5 cells pretreated with the miR-766-3p expression plasmid were transfected with pcDNA3.1 or the Wnt3a expression plasmid, and then total and nuclear β-catenin levels, and protein expression levels of Wnt3a, TCF1, Survivin, and PRC1 were detected by Western blot analysis. (C) PLC/PRF/5 cells pretreated with the miR-766-3p expression plasmid were transfected with pcDNA3.1 or the PRC1 expression plasmid, and then total and nuclear β-catenin levels, and protein expression levels of PRC1, TCF1 and Survivin were detected by Western blot analysis. (D) PLC/PRF/5 cells were treated with DMSO or iCRT3 (200 μM) for 48 h. Then, protein expression levels of PRC1, TCF1 and Survivin were detected by Western blot analysis. (E) PLC/PRF/5 cells pretreated with the miR-766-3p expression plasmid were transfected with pcDNA3.1 or the PRC1 expression plasmid. PRC1 reversed the inhibition of proliferation (Fig. 6E), colony formation (Fig. 6F), invasion (Fig. 6G) and migration (Fig. 6H) induced by miR-766-3p overexpression. Similar results also seen in SK-HEP-1 cells are not shown in Fig. 6. * P < 0.05; **P < 0.01.
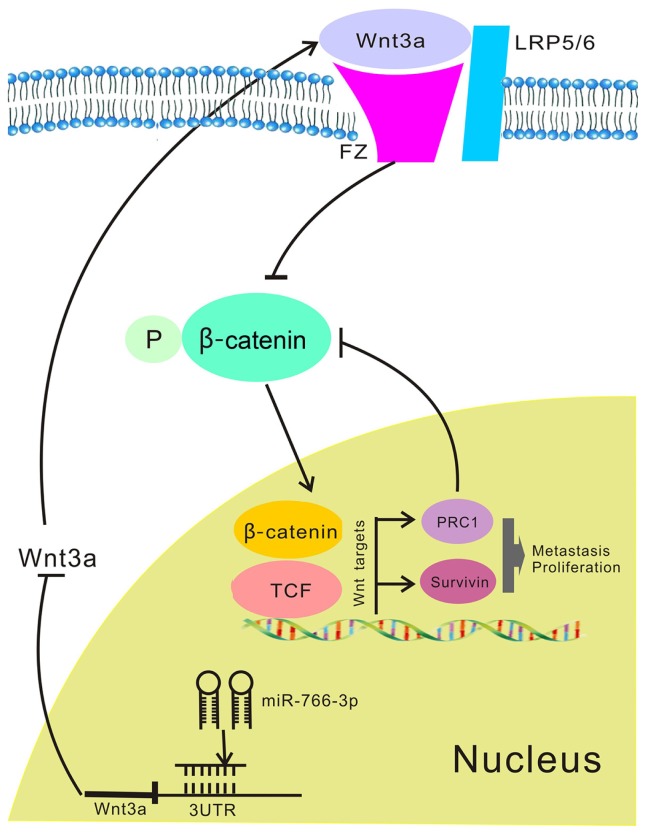
Fig. 7. The molecular mechanism underlying the tumour-suppressive effect of miR-766-3p in HCC.
Down-regulated miR-766-3p in HCC induces Wnt3a expression. Wnt3a results in activation of the canonical Wnt signalling and PRC1 expression, PRC1 as a novel Wnt target that reinforces the canonical Wnt signalling to regulate HCC cell proliferation and metastasis.
DISCUSSION
Recently, miRNAs have been shown to be involved in tumourigenesis and tumour progression and are thus a pivotal factor in a variety of cancers, including HCC (Cancer Genome Atlas Research Network, 2017; Su et al., 2015). Among the functional miRNAs is miR-766-3p, whose expression is, intriguingly, significantly deregulated in many cancers. MiR-766-3p has many prominent oncogenic and tumour-suppressive effects (Chen et al., 2017; Li et al., 2015; Wang et al., 2017). For example, miR-766-3p promotes cancer progression in colorectal cancer (Li et al., 2015) but suppresses oncogenesis in breast cancer and renal cell carcinoma (Chen et al., 2017; Wang et al., 2017). In HCC, miR-766-3p expression was decreased compared with its expression in noncancerous liver tissue (Wei et al., 2013), indicating that miR-766-3p may serve as a potential tumour suppressor in HCC. In this study, we also found that the expression levels of miR-766-3p were significantly down-regulated in HCC tissues (Figs. 1A and 1B) and cell lines (Fig. 2A). In addition, we found that the reduced miR-766-3p expression levels were significantly correlated with tumour size and advanced TNM stages, as well as metastasis and poor prognosis. Thus, the collective data provided sufficient indication that miR-766-3p might serve as a tumour suppressor gene in HCC. Furthermore, we observed that miR-766-3p up-regulation suppressed HCC cell proliferation, colony formation, cell migration and invasion in vitro (Figs. 2 and 3). In addition, our in vivo study revealed that miR-766-3p up-regulation could repress HCC xenograft tumour growth in nude mice (Fig. 2E). Taken together, these results showed that miR-766-3p functions as an inhibitor in HCC progression.
By directly regulating target genes, miRNAs can affect various pathological and physiological processes (Aushev et al., 2018; Dufresne et al., 2018). MiR-766-3p was recently reported to target many important downstream genes. For example, miR-766-3p targets SF2, which was found to promote cell proliferation in renal cell carcinoma, and SOX6, which is thought to inhibit cell proliferation in human colorectal cancer cells (Chen et al., 2017; Li et al., 2015). Since the target gene of miR-766-3p in HCC remained unclear, we attempted to determine the target genes from a group of putative candidates identified by bioinformatics tools. These candidates included HOXA13, Wnt3a, TSG101 and IGFBP2. All of these genes are up-regulated in HCC tissues and function as oncogenes in HCC progression (Hung et al., 2017; Lu et al., 2017; Pan et al., 2016; Quagliata et al., 2014; Shao et al., 2015). In addition, we found that Wnt3a mRNA expression was significantly suppressed in SK-HEP-1 cells after transfection with the miR-766-3p expression plasmid, while HOXA13, TSG101 and IGFBP2 mRNA expression did not change (Supplementary Fig. S1). We further investigated whether Wnt3a was a direct target of miR-766 in HCC. Western blot assay showed that miR-766-3p up-regulation inhibited Wnt3a protein expression in HCC cells (Fig. 4A). Moreover, the luciferase reporter assay showed that miR-766-3p significantly decreased luciferase activity by binding directly to the 3′UTR of Wnt3a (Fig. 4C). Moreover, Wnt3a was up-regulated in HCC tissues and cell lines and inversely correlated with miR-766-3p expression in HCC tissues (Figs. 4D and 4E). Furthermore, Wnt3a restoration partially rescued the effect of miR-766-3p up-regulation on HCC progression (Fig. 5). In summary, these results suggested that Wnt3a was a direct target of miR-766 in HCC.
Wnt3a has been shown to induce the nuclear accumulation of β-catenin, thereby activating the canonical Wnt signalling pathway. Activation of canonical Wnt signalling induces cell proliferation, colony formation, invasion and migration in cancer cells. For example, Wnt3a enhances cell growth and colony formation by increasing the cytosolic and nuclear levels of β-catenin in human prostate cancer cells (Verras et al., 2004). In colon cancer, Wnt3a stimulates the formation of beta-catenin/TCF complexes to enhance cell invasion and anchorage-independent growth (Kang and Min, 2010). In addition, Wnt3a induces Wnt signalling activation and increases expression of PRC1 and other known Wnt targets (Survivin and TCF1) in HCC cells (Chen et al., 2016). PRC1 has been identified as a novel Wnt target, and PRC1 down-regulation inhibits cell proliferation, colony formation, invasion and migration. Thus, we proposed that the Wnt signalling activation and the increase in PRC1 expression mediated by Wnt3a contribute to HCC progression regulated by miR-766-3p. Further investigation supported this conclusion. In this study, miR-766-3p up-regulation inhibited Wnt3a protein expression, which consequently repressed the nuclear accumulation of β-catenin as well as expression of PRC1, Survivin and TCF1 (Fig. 6A). In addition, restoration of Wnt3a rescued the effect of miR-766-3p in HCC cells (Fig. 6B). Thus, Wnt3a was indispensable for the effect of miR-766-3p on Wnt signalling activation and PRC1 expression in HCC. PRC1 has been shown to promote the nuclear accumulation of β-catenin and induce Survivin and TCF1 protein expression (Chen et al., 2016). Thus, the effect of miR-766-3p on Wnt signalling activation may be PRC1-dependent. Consistently, we found that PRC1 up-regulation rescued the effect of miR-766-3p overexpression on the nuclear accumulation of β-catenin and the protein expression of Survivin and TCF1 in HCC cells (Fig. 6C). Moreover, iCRT3, a potent and specific inhibitor of β-catenin-TCF4 interaction, repressed the protein levels of TCF1, Survivin and PRC1 (Fig. 6D). Thus, these results strongly imply that PRC1 acts as a novel Wnt target and triggers Wnt signalling. In addition, PRC1 up-regulation reversed the effect of miR-766-3p on proliferation, colony formation, invasion and migration in HCC cells. Similar results also seen in SK-HEP-1 cells were not shown in Fig. 6. Taken together, our results suggest that miR-766-3p affects HCC progression via a Wnt/PRC1 positive regulatory loop.
In conclusion, we demonstrated that miR-766-3p was down-regulated in HCC and functioned as a potential cancer inhibitor. Wnt3a was a cancer promoter and a novel target of miR-766-3p in HCC. We showed that miR-766-3p suppressed HCC cell growth and invasion via a Wnt/PRC1 positive regulatory loop. Thus, our results suggested that miR-766-3p may serve as a potential therapeutic target for HCC cancer treatment. Recently, some studies have reported that Wnt3a could induce epithelial–mesenchymal transition (EMT), thereby promoting the metastasis and progression of cancer (Qi et al., 2014; Zhang et al., 2013). Although we have shown that miR-766-3p suppressed HCC cell invasion and migration via a Wnt/PRC1 positive regulatory loop, the role and detailed mechanisms of miR-766-3p in HCC EMT require further study.
Supplementary data
ACKNOWLEDGMENTS
This project was supported by the National Natural Science Foundation of China (No. 81702357, 81470899 and 81170442).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Aushev V.N., Lee E., Zhu J., Gopalakrishnan K., Li Q., Teitelbaum S.L., Wetmur J., Degli Esposti D., Hernandez-Vargas H., Herceg Z., et al. Novel predictors of breast cancer survival derived from miRNA activity analysis. Clin Cancer Res. 2018;24:581–591. doi: 10.1158/1078-0432.CCR-17-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken C.P., Khew-Goodall Y., Goodall G.J. Network-based approaches to understand the roles of miR-200 and other microRNAs in cancer. Cancer Res. 2015;75:2594–2599. doi: 10.1158/0008-5472.CAN-15-0287. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network; Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Rajasekaran M., Xia H., Zhang X., Kong S.N., Sekar K., Seshachalam V.P., Deivasigamani A., Goh B.K., Ooi L.L., et al. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/β-catenin signalling pathway. Gut. 2016;59:1522–1534. doi: 10.1136/gutjnl-2015-310625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Xue S., Zhang J., Chen W., Gong D., Zheng J., Ma J., Xue W., Chen Y., Zhai W., et al. DNA-methylation-mediated repression of miR-766-3p promotes cell proliferation via targeting SF2 expression in renal cell carcinoma. Int J Cancer. 2017;141:1867–1878. doi: 10.1002/ijc.30853. [DOI] [PubMed] [Google Scholar]
- Dufresne S., Rebillard A., Muti P., Friedenreich C.M., Brenner D.R. A review of physical activity and circulating-mirna expression: implications in cancer risk and progression. Cancer Epidemiol Biomarkers Prev. 2018;27:11–24. doi: 10.1158/1055-9965.EPI-16-0969. [DOI] [PubMed] [Google Scholar]
- Fornari F., Gramantieri L., Giovannini C., Veronese A., Ferracin M., Sabbioni S., Calin G.A., Grazi G.L., Croce C.M., Tavolari S., et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009;69:5761–5767. doi: 10.1158/0008-5472.CAN-08-4797. [DOI] [PubMed] [Google Scholar]
- He S., Lu Y., Liu X., Huang X., Keller E.T., Qian C.N., Zhang J. Wnt3a: functions and implications in cancer. Chin J Cancer. 2015;34:554–562. doi: 10.1186/s40880-015-0052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C.S., Huang C.Y., Lee C.H., Chen W.Y., Huang M.T., Wei P.L., Chang Y.J. IGFBP2 plays an important role in heat shock protein 27-mediated cancer progression and metastasis. Oncotarget. 2017;8:54978–54992. doi: 10.18632/oncotarget.18989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D.W., Min D.S. Positive feedback regulation between phospholipase D and Wnt signaling promotes Wnt-driven anchorage-independent growth of colorectal cancer cells. PLoS One. 2010;5:e12109. doi: 10.1371/journal.pone.0012109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N., Chettiar S., Rathod S., Rath P., Muzumdar D., Shaikh M.L., Shiras A. Wnt3a mediated activation of Wnt/β-catenin signaling promotes tumor progression in glioblastoma. Mol Cell Neurosci. 2013;54:44–57. doi: 10.1016/j.mcn.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Leva G.D., Garofalo M., Croce C.M. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.C., Li C.F., Chen L.B., Li D.D., Yang L., Jin J.P., Zhang B. MicroRNA-766 targeting regulation of SOX6 expression promoted cell proliferation of human colorectal cancer. Onco Targets Ther. 2015;8:2981–2988. doi: 10.2147/OTT.S89459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., He Y., Duan J., Yang Y., Zhong C., Zhang J., Liao W., Huang X., Zhu R., Li M. Expression of Wnt3a in hepatocellular carcinoma and its effects on cell cycle and metastasis. Int J Oncol. 2017;51:1135–1145. doi: 10.3892/ijo.2017.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano H., Tomimaru Y., Eguchi H., Hama N., Wada H., Kawamoto K., Kobayashi S., Mori M., Doki Y. MicroRNA-29a induces resistance to gemcitabine through the Wnt/β-catenin signaling pathway in pancreatic cancer cells. Int J Oncol. 2013;43:1066–1072. doi: 10.3892/ijo.2013.2037. [DOI] [PubMed] [Google Scholar]
- Pan L.H., Yao M., Cai Y., Gu J.J., Yang X.L., Wang L., Yao D.F. Oncogenic Wnt3a expression as an estimable prognostic marker for hepatocellular carcinoma. World J Gastroenterol. 2016;22:3829–3836. doi: 10.3748/wjg.v22.i14.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L., Sun B., Liu Z., Cheng R., Li Y., Zhao X. Wnt3a expression is associated with epithelial-mesenchymal transition and promotes colon cancer progression. J Exp Clin Cancer Res. 2014;11:107. doi: 10.1186/s13046-014-0107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Dong S., Qiao F., Lu S., Song Y., Lao Y., Li Y., Zeng T., Hu J., Zhang L., et al. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene. 2013;32:3296–3305. doi: 10.1038/onc.2013.150. [DOI] [PubMed] [Google Scholar]
- Quagliata L., Matter M.S., Piscuoglio S., Arabi L., Ruiz C., Procino A., Kovac M., Moretti F., Makowska Z., Boldanova T., et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano P.R., McCallus D.E., Pachuk C.J. RNA interference-mediated prevention and therapy for hepatocellular carcinoma. Oncogene. 2006;25:3857–3865. doi: 10.1038/sj.onc.1209549. [DOI] [PubMed] [Google Scholar]
- Shao Z., Ji W., Liu A., Qin A., Shen L., Li G., Zhou Y., Hu X., Yu E., Jin G. TSG101 silencing suppresses hepatocellular carcinoma cell growth by inducing cell cycle arrest and autophagic cell death. Med Sci Monit. 2015;21:3371–3379. doi: 10.12659/MSM.894447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simerzin A., Zorde-Khvalevsky E., Rivkin M., Adar R., Zucman-Rossi J., Couchy G., Roskams T., Govaere O., Oren M., Giladi H., et al. The liver-specific microRNA-122*, the complementary strand of microRNA-122, acts as a tumor suppressor by modulating the p53/mouse double minute 2 homolog circuitry. Hepatology. 2016;64:1623–1636. doi: 10.1002/hep.28679. [DOI] [PubMed] [Google Scholar]
- Su X., Wang H., Ge W., Yang M., Hou J., Chen T., Li N., Cao X. An in vivo method to identify microRNA targets not predicted by computation algorithms: p21 targeting by miR-92a in cancer. Cancer Res. 2015;75:2875–2885. doi: 10.1158/0008-5472.CAN-14-2218. [DOI] [PubMed] [Google Scholar]
- Verras M., Brown J., Li X., Nusse R., Sun Z. Wnt3a growth factor induces androgen receptor-mediated transcription and enhances cell growth in human prostate cancer cells. Cancer Res. 2004;64:8860–8866. doi: 10.1158/0008-5472.CAN-04-2370. [DOI] [PubMed] [Google Scholar]
- Wagenaar T.R., Zabludoff S., Ahn S.M., Allerson C., Arlt H., Baffa R., Cao H., Davis S., Garcia-Echeverria C., Gaur R., et al. Anti-miR-21 suppresses hepatocellular carcinoma growth via broad transcriptional network deregulation. Mol Cancer Res. 2015;13:1009–10021. doi: 10.1158/1541-7786.MCR-14-0703. [DOI] [PubMed] [Google Scholar]
- Wang Q., Selth L.A., Callen D.F. MiR-766 induces p53 accumulation and G2/M arrest by directly targeting MDM4. Oncotarget. 2017;8:29914–29924. doi: 10.18632/oncotarget.15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wang Y., Liu W., van Wijnen A.J. Regulation and biological roles of the multifaceted miRNA-23b (MIR23B) Gene. 2018;642:103–109. doi: 10.1016/j.gene.2017.10.085. [DOI] [PubMed] [Google Scholar]
- Wei R., Huang G.L., Zhang M.Y., Li B.K., Zhang H.Z., Shi M., Chen X.Q., Huang L., Zhou Q.M., Jia W.H., et al. Clinical significance and prognostic value of microRNA expression signatures in hepatocellular carcinoma. Clin Cancer Res. 2013;19:4780–4791. doi: 10.1158/1078-0432.CCR-12-2728. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Bai X.L., Chen W., Ma T., Hu Q.D., Liang C., Xie S.Z., Chen C.L., Hu L.Q., Xu S.G., et al. Wnt/β-catenin signaling enhances hypoxia-induced epithelial–mesenchymal transition in hepatocellular carcinoma via crosstalk with hif-1α signaling. Carcinogenesis. 2013;34:962–973. doi: 10.1093/carcin/bgt027. [DOI] [PubMed] [Google Scholar]
- Zimmerman Z.F., Kulikauskas R.M., Bomsztyk K., Moon R.T., Chien A.J. Activation of Wnt/β-catenin signaling increases apoptosis in melanoma cells treated with trail. PLoS One. 2013;8:e69593. doi: 10.1371/journal.pone.0069593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.