MbtH-like proteins (MLPs) are essential accessory proteins for the function of many nonribosomal peptide synthetases (NRPSs). We identified 50 MLPs from diverse bacteria that are coded by genes that are not located near any NRPS-encoding biosynthetic gene clusters (BGCs). We define these as orphan MLPs because their NRPS partner(s) is unknown. Investigations into the orphan MLP from Myxococcus xanthus DK1622 determined that it interacts with NRPSs from at least seven distinct BGCs. Support for these MLP-NRPS interactions came from the use of a bacterial two-hybrid assay and copurification of the MLP with various NRPSs. The flexibility of this MLP to naturally interact with multiple NRPSs led us to hypothesize that this MLP may be used as a “universal” MLP during the construction of functional hybrid NRPSs.
KEYWORDS: MbtH-like protein, Myxococcus xanthus, natural products, nonribosomal peptide synthetase, combinatorial biosynthesis
ABSTRACT
One mechanism by which bacteria and fungi produce bioactive natural products is the use of nonribosomal peptide synthetases (NRPSs). Many NRPSs in bacteria require members of the MbtH-like protein (MLP) superfamily for their solubility or function. Although MLPs are known to interact with the adenylation domains of NRPSs, the role MLPs play in NRPS enzymology has yet to be elucidated. MLPs are nearly always encoded within the biosynthetic gene clusters (BGCs) that also code for the NRPSs that interact with the MLP. Here, we identify 50 orphan MLPs from diverse bacteria. An orphan MLP is one that is encoded by a gene that is not directly adjacent to genes predicted to be involved in nonribosomal peptide biosynthesis. We targeted the orphan MLP MXAN_3118 from Myxococcus xanthus DK1622 for characterization. The M. xanthus DK1622 genome contains 15 NRPS-encoding BGCs but only one MLP-encoding gene (MXAN_3118). We tested the hypothesis that MXAN_3118 interacts with one or more NRPS using a combination of in vivo and in vitro assays. We determined that MXAN_3118 interacts with at least seven NRPSs from distinct BGCs. We show that one of these BGCs codes for NRPS enzymology that likely produces a valine-rich natural product that inhibits the clumping of M. xanthus DK1622 in liquid culture. MXAN_3118 is the first MLP to be identified that naturally interacts with multiple NRPS systems in a single organism. The finding of an MLP that naturally interacts with multiple NRPS systems suggests it may be harnessed as a “universal” MLP for generating functional hybrid NRPSs.
IMPORTANCE MbtH-like proteins (MLPs) are essential accessory proteins for the function of many nonribosomal peptide synthetases (NRPSs). We identified 50 MLPs from diverse bacteria that are coded by genes that are not located near any NRPS-encoding biosynthetic gene clusters (BGCs). We define these as orphan MLPs because their NRPS partner(s) is unknown. Investigations into the orphan MLP from Myxococcus xanthus DK1622 determined that it interacts with NRPSs from at least seven distinct BGCs. Support for these MLP-NRPS interactions came from the use of a bacterial two-hybrid assay and copurification of the MLP with various NRPSs. The flexibility of this MLP to naturally interact with multiple NRPSs led us to hypothesize that this MLP may be used as a “universal” MLP during the construction of functional hybrid NRPSs.
INTRODUCTION
An important group of bioactive natural products produced by bacteria and fungi are the nonribosomal peptides. The biosynthesis of these structurally diverse metabolites is derived from the modular enzymology of nonribosomal peptide synthetases (NRPSs). The modularity of NRPSs resides in a set of repeating catalytic domains that work together to recognize and incorporate aryl or amino acid precursors into the final nonribosomal peptide product. Typically, a module includes an adenylation (A) domain, a thiolation (T) domain, and a condensation (C) domain cooperating in an assembly line fashion to form a nonribosomal peptide (1). A domains are essential components of NRPSs as “gatekeepers” of the assembly line by selecting the precursor introduced by each module (1). Recently, it was recognized that there are two types of A domains in bacteria. Members of the first type function independently of any additional protein factor. Members of the second class require an accessory protein from the MbtH-like protein (MLP) superfamily to be functional (2–5).
The MLP superfamily is named after MbtH, a protein encoded by the mycobactin biosynthetic gene cluster (BGC) in Mycobacterium tuberculosis (6). MLPs are small proteins of approximately 70 amino acids that are nearly always encoded within NRPS-encoding BGCs. MLPs were first identified to be essential for the production of the NRPS-assembled siderophore pyoverdine (7) and subsequently the production of clorobiocin, a calcium-dependent antibiotic, coelichelin, and enterobactin (2, 7–10). Interestingly, some of these studies also showed that MLPs encoded by one BGC are able cross talk with the NRPSs coded by other BGCs and influence metabolite production of that noncognate system (9, 11); however, this cross talk is only relevant when the gene coding for the cognate MLP is deleted.
The impact MLPs have on the production of nonribosomal peptide biosynthesis has led to biochemical and structural studies aimed at understanding the function of MLPs. The biochemical characterization of the NRPSs in the presence and absence of MLPs established that MLPs influence the solubility of overproduced NRPSs (2, 4, 5) and can also impact various aspects of NRPS enzymology, including the Km for the amino acid substrate or ATP (2–4, 12). We recently showed that incorrect MLP/NRPS pairings can result in decreased levels of amino acid thioesterification to the T domain and alterations of the overall NRPS turnover (13). These observations suggest that the long-term goal of generating hybrid NRPSs to produce “unnatural” natural products will be complicated by the impact of incorrect MLP and NRPS pairings.
In addition to biochemical studies, there have been a number of structural studies. The initial structures of isolated MLPs determined they possess a unique protein fold and do not contain a clearly identifiable catalytic site (10, 14). The structures of MLP/NRPS complexes showed that the MLPs bind to the A domains of NRPSs but at a location that suggests it does not conflict with other catalytic domains. Unfortunately, structural analyses of the NRPS EntF with and without an MLP failed to identify substantive conformational changes that would explain how an MLP imparts its activity on its partner NRPS (12, 15, 16). One original hypothesis about the function of MLPs was that they function as chaperone-like proteins to help NRPSs fold correctly (2, 4). Consistent with this hypothesis, under nondenaturing protein electrophoresis and size exclusion chromatography, an MLP/NRPS complex migrates or elutes differently than the NRPS alone, suggesting that the structural changes caused by the MLP were not detected by the previous crystallographic methods (16). In summary, the specific role of MLPs in NRPS enzymology remains unclear.
MLPs are essential components of many NRPSs, and our understanding of their role in catalysis will not only contribute to a better understanding of NRPS enzymology but also improve combinatorial biosynthesis efforts. A significant number of medically relevant nonribosomal peptides have MLP-dependent A domains, suggesting the construction of hybrid NRPSs to generate next-generation drugs will require an intimate knowledge of how MLPs impart their function on NRPSs (17). In support of this effort, our recent work investigated the effect of pairing noncognate MLPs with the enterobactin NRPS to understand how MLPs influence NRPS enzymology. We found that noncognate MLP/NRPS interactions are complex and that improper pairing is detrimental to various steps in NRPS catalysis (13).
In the present study, we identified 50 orphan MLPs from multiple bacteria that are encoded by genes which are not located near any NRPS-encoding BGCs or genes predicted to be involved in nonribosomal peptide biosynthesis. We investigated the role that one of these orphan MLPs, MXAN_3118, plays in the bacterium Myxococcus xanthus DK1622. Using a combination of assays that investigate NRPS solubility, MLP/NRPS copurification, and in vivo MLP/NRPS interactions, we determined that the only MLP encoded by M. xanthus DK1622 interacts with NRPS systems from at least seven different BGCs. This is the first example of an MLP that naturally functions with multiple NRPS systems in a single organism. As part of this investigation, we present evidence that one of the MLP-dependent NRPSs that is encoded by an orphan NRPS gene produces a valine-rich natural product that influences the clumping of M. xanthus DK1622 in minimal medium. The flexibility of MXAN_3118 to interact with multiple NRPS systems suggests that it may be exploited as a universal MLP for the combinatorial biosynthesis of functional hybrid NRPSs.
RESULTS
MXAN_3118 is an orphan MLP in M. xanthus DK1622.
The Basic Local Alignment Search Tool (BLAST) (18), with the default settings, was used to scan the predicted proteome of M. xanthus DK1622 for proteins belonging to the MLP superfamily using YbdZ from Escherichia coli (accession number YP_588441) as the search query. A single protein, MXAN_3118, was identified. Unexpectedly, the gene coding for this MLP was not near any other genes encoding NRPS enzymology or enzymology known to be associated with the production of a nonribosomal peptide, making this an orphan MLP. Analyses of fully or partially sequenced bacterial genomes using MXAN_3118 as a search query in the Integrated Microbial Genomes & Microbiome Samples from the Joint Genome Institute (19), using the default settings and analyzing the proteins encoded by the 20 kbp on either side of the gene encoding an MLP, identified 43 additional orphan MLPs in other bacteria (see Table S1 in the supplemental material). The presence of these orphan MLPs raised the question of whether they have partner NRPSs or whether they play some other role in bacterial physiology. The former proposal was considered because of our recent report that MXAN_3118 can functionally replace YbdZ, the MLP that naturally functions with the NRPS involved in enterobactin biosynthesis in E. coli (13). The latter proposal was considered since the genome of Wenzhouxiangella marina KCTC 42284 codes for an MLP but does not code for any NRPSs (20).
Interestingly, approximately half of the orphan MLPs identified were found in the Myxococcales order. Since the JGI IMG data bank did not include all available genomic information, we identified all of the MLPs harbored by Myxococcales using MXAN_3118 as a search query in BLAST using the default settings. Fifty-seven MLP homologs were identified and manually curated to identify a total of 27 orphan MLPs. A phylogenetic analysis of these 57 MLPs showed that orphan MLPs are found throughout the tree and do not cluster into a single clade (see Fig. S1). Interestingly, MLPs from Myxococcus species do form sister clades, and these are the only MLPs encoded in the genomes of these organisms. This is somewhat surprising, since these organisms are well known to be prolific producers of nonribosomal peptides (21, 22). The similarity between these MLPs may suggest they have a common role in these organisms. We focused our attention on the orphan MLP from M. xanthus DK1622, since this bacterium is well studied and can be genetically manipulated.
Identification of potential MXAN_3118 NRPS partners.
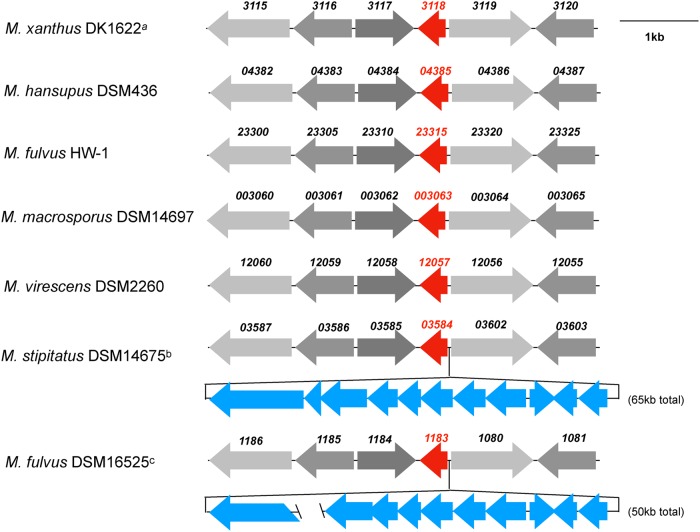
We compared the regions of the genomes surrounding the MXAN_3118 homologs in Myxococcus species. All M. xanthus strains, Myxococcus macrosporus DSM 14697, Myxococcus virescens DSM 2260, Myxococcus fulvus HW-1, and Myxococcus hansupus DSM 436 have similar genes around the gene coding the homolog of MXAN_3118 (Fig. 1). In contrast, Myxococcus stipitatus DSM 14675 and M. fulvus DSM 16525 have NRPS-encoding genes immediately upstream of the homolog of MXAN_3118. In M. stipitatus DSM 14675, the NRPS-encoding gene is associated with a putative 63-kbp BGC followed by homologs to MXAN_3119 and MXAN_3120 (Fig. 1). The genome of M. fulvus DSM 16525 was not complete at the time of our analysis, but a similar BGC is expected on the basis of one contig containing a portion of the NRPS-encoding gene immediately upstream of the MXAN_3118 homolog and a second contig containing homologs of MXAN_3119, MXAN_3120, and the majority of the genes observed in the 64-kbp BGC from M. stipitatus DSM 14675. While M. stipitatus DSM 14675 and M. fulvus DSM 16525 appear to share this BGC, it is not found in the other Myxococcus species. One hypothesis is that these other species lost the corresponding BGC but retained the gene coding for the MLP due to its requirement for one or more of the other NRPSs that are encoded by these bacteria.
FIG 1.
Schematic of the genomic regions of Myxococcus species and strains surrounding the MXAN_3118 homolog (red arrows). Similar gray shading identifies homologs of MXAN_3115 to MXAN_3117 and MXAN_3119 to MXAN_3120. The 1-kb bar indicates the scale for all genomic regions except those show in shades of blue, the sizes of which are noted in parentheses. Locus tag abbreviations: M. xanthus DK1226 (MXAN_3115 to MXAN_3120), M. virescens DSM 2260 (ga0070493_12055 to ga0070493_12060), M. fulvus HW-1 (LILAB_23300 to LILAB_23325), M. hansupus DSM 436 (A176_04382 to A176_04387), M. macrosporus DSM 14697 (MYMAC_003060 to MYMAC_003065), M. stipitatus DSM 14675 (MYSTI_03587 to MYSTI_03603), M. fulvus DSM 16525 (Ga0131203_1181 to Ga0131203_1186 and Ga0131203_1071 to Ga0131203_1081). M. xanthus DK1622a is representative of all other M. xanthus strains (DSM 16525, DZ2, and DF1). In M. stipitatus DSM 14675b, the blue genes are an NRPS-associated gene cluster covering 65 kb; the NRPS-associated gene cluster has homolog ORFs in M. fulvus DSM 16525. M. fulvus DSM 16525c genome sequence was incomplete and two contigs were identified, the first contig covers 24 kb terminating in the middle of the NRPS-encoding gene 1181 and the second contig is 26 kb starting in the middle of genes 1071 to 1079, missing the homolog for gene MYSTI_03592 in the M. fulvus DSM 16525 cluster.
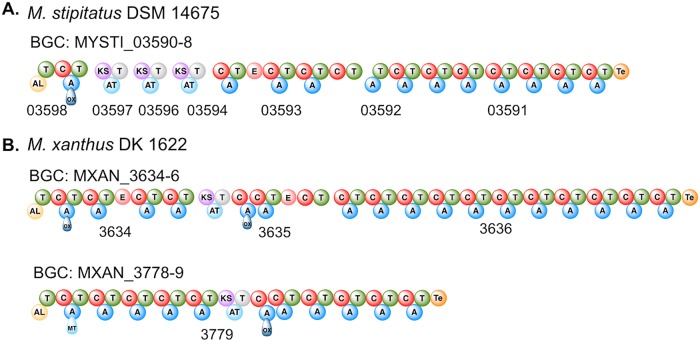
We (13) and others (12, 23) have reported that there is no signature amino acid sequence within an A domain that identifies it as an MLP-dependent or -independent megasynthase. Therefore, to identify candidate NRPSs in M. xanthus DK1622 for interactions with the MXAN_3118, we identified BGCs that code for an NRPS with significant amino acid identity/similarity (49%/64%) to MYSTI_03598 and code for enzymes that had similar characteristics to the enzymes coded by BGC in M. stipitatus DSM 14675. These characteristics were (i) initiation with an acyl coenzyme A (acyl-CoA) ligase domain, (ii) a module with an A domain disrupted by an oxidase domain, and (iii) a hybrid NRPS/polyketide synthase (PKS) system. From this analysis, we identified two potential BGCs coding for NRPS components that were candidates for MXAN_3118 interactions (Fig. 2). These clusters were MXAN_3778-3779, with MXAN-3779 coding for the hybrid NRPS/PKS that produces myxoprincomide (22), and an orphan BGC consisting of MXAN_3634 to MXAN_3636. We targeted select A domains or the associated modules from MXAN_3779 and MXAN_3634 to MXAN_3636 to evaluate them for interactions with MXAN_3118.
FIG 2.
Potential NRPS partners for the MXAN_3118 orphan MLP. (A) Schematic of the MYSTI_03590 to MYSTI_03598 NRPS/PKS hybrid megasynthase from M. stipitatus DSM 14675. (B) Schematic of the MXAN_3779 and MXAN_3634 to MXAN_3636 NRPS/PKS hybrid megasynthases from M. xanthus DK1622. Domain abbreviations: AL, acyl-CoA ligase; T, thiolation; C, condensation; ox, oxidation; KS, ketosynthase; AT, acyltransferase; E, epimerase; Te, thioesterase.
MXAN_3118 interacts with the myxoprincomide mixed NRPS/PKS megasynthase.
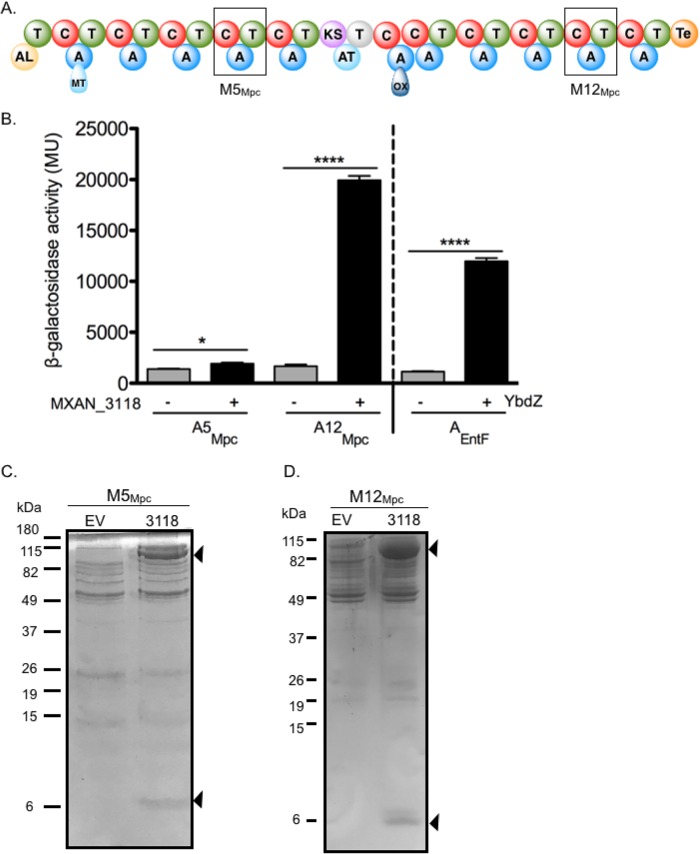
Two A domains from the myxoprincomide (Mpc) mixed NRPS/PKS were assessed to determine whether they interact with MXAN_3118. We previously established that no single assay is capable of detecting all MLP/NRPS interactions (13, 24). Thus, we used more than one assay to assess MXAN_3118 interactions with the A domains (A5Mpc and A12Mpc) of two modules (M5Mpc and M12Mpc) (Fig. 2A). One assay was an in vivo E. coli-based bacterial two-hybrid (B2H) assay to detect in vivo A domain/MXAN_3118 interactions. The other assays assessed whether the solubility of an NRPS was influenced by the presence of MXAN_3118 or whether untagged MXAN_3118 protein coeluted with a His-tagged NRPS component from a Ni-NTA column. We define an NRPS as MXAN_3118 dependent if one or more of the associated A domains or modules is positive in the B2H assay and is also positive for increased solubility in the presence of MXAN_3118 or coelutes with MXAN_3118 from a nickel-nitrilotriacetic acid (Ni-NTA) column when only the NRPS is His tagged.
A B2H system, a well-established mechanism for detecting protein-protein interactions (25), has not been previously used to assess MLP/NRPS interactions. To investigate whether this system is able to detect these interactions, the enterobactin system was used as a positive control. Briefly, the A domain of EntF was fused to the α-subunit of RNA polymerase and YbdZ, the MLP of the enterobactin system (2), was fused to λcI. Constructs expressing these fusion proteins were introduced into an E. coli ΔybdZ entF strain containing a lacZ reporter that has increased expression when there are protein-protein interactions between the two fusion proteins (25). When the YbdZ-λcI fusion protein was present in the cells along with the EntF A domain-α-subunit fusion, 10,000-fold higher LacZ activity was detected compared to that in a strain producing the EntF A domain-α-subunit fusion and the λcI protein alone (Fig. 3B), confirming this assay is able to detect MLP/NRPS interactions.
FIG 3.
Myxoprincomide (Mpc) NRPS protein interactions with MXAN_3118. (A) Schematic of the MXAN_3779 NRPS/PKS hybrid assembly line with boxes highlighting the fourth (M5Mpc) and tenth (M12Mpc) modules investigated for MXAN_3118 interactions. Domain abbreviations: AL, acyl-CoA ligase; T, thiolation; C, condensation; ox, oxidation; MT, methyltransferase; KS, ketosynthase; AT, acyltransferase; Te, thioesterase. (B) Results from the B2H assay. Quantitative β-galactosidase assay of the α-subunit-A domain fusions with MXAN_3118-λcI fusion (+) or λcI alone (−). EntF (A domain) and YbdZ (MLP) were used as controls of MLP-NRPS positive interactions. Error bars show standard deviations from three independent assays. *, P ≤ 0.05; ****, P ≤ 0.0001 by Student's t test. (C) Tris-Tricine (16.8%) polyacrylamide gels and Coomassie blue staining of His-tagged M5Mpc eluting from an Ni-NTA column after overproduction without (empty expression vector [EV]) or with (expression vector expressing MXAN_3118 [3118]) untagged MXAN_3118. Ten micrograms of protein was loaded in each lane. (D) Tris-Tricine (16.8%) polyacrylamide gels and Coomassie blue staining of His-tagged M12Mpc eluting from an Ni-NTA column after overproduction without (EV) or with (3118) untagged MXAN_3118. Seventeen micrograms of protein was loaded in each lane.
The assay was repeated with MXAN_3118 fused to the λcI protein and A5Mpc or A12Mpc fused to the α-subunit of RNA polymerase. We defined the B2H assay as testing positive for MLP/NRPS interactions when the LacZ activity detected was statistically higher than the activity detected in the negative-control strain expressing the A domain-α-subunit fusion and the λcI protein alone. Both A5Mpc and A12Mpc were positive for interactions with MXAN_3118 (Fig. 3B) when compared to the negative control. These results suggest that both A domains from the Mpc NRPS are MXAN_3118 dependent. The variability in the activity detected by the fusions is not unexpected. As we have shown (24), an MLP can have significantly different levels of interaction with each A domain from the same NRPS system.
As a second test for MXAN_3118 dependence, the two modules containing the A5Mpc and A12Mpc domains were heterologously overproduced in E. coli with N-terminal His tags, cooverproduced with and without untagged MXAN_3118, and then partially purified by Ni-NTA chromatography to determine whether the two proteins coelute. Both NRPS modules coeluted with MXAN_3118 (Fig. 3C). In control reactions, strains lacking MXAN_3118 failed to produce a module that eluted from the column or had a significantly less soluble module produced. Additionally, untagged MXAN_3118 had no affinity for the Ni-NTA resin. On the basis of these results, we conclude that at least these modules of the Mpc NRPS/PKS hybrid enzyme are MLP dependent. We did not determine whether DK1622 lacking MXAN_3118 produced Mpc, because this strain produces very low levels of Mpc. A different strain of M. xanthus had to be used to produce enough Mpc for characterization and required a 300-liter fermentation to produce enough metabolite for analysis (22).
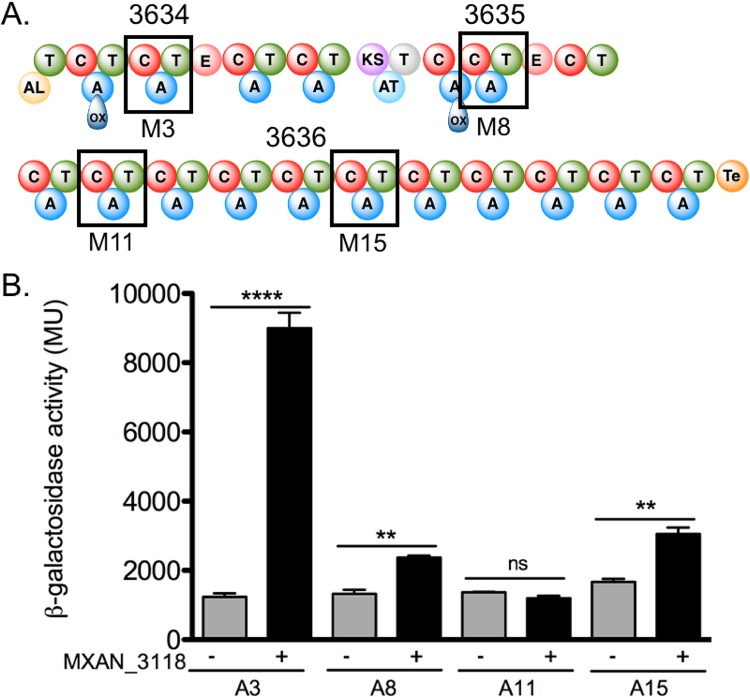
MXAN_3118 interacts with all but one of the A domains from the megasynthase encoded by the orphan BGC MXAN_3634 to MXAN_3636.
The same assays were used to assess whether components from the NRPS/PKS megasynthase coded by the BGC MXAN_3634 to MXAN_3636 interact with MXAN_3118. Initial studies focused on the representative A domains from each subunit of the megasynthase (Fig. 4A). The B2H system detected significant interactions between the A domains from modules 3, 8, and 15 but not from the A domain of module 11 (Fig. 4B). As most of these A domains were positive for interactions with MXAN_3118, we investigated all the A domains for interactions with MXAN_3118 by assessing whether the NRPS components copurified with MXAN_3118 after heterologous cooverproduction in E. coli. MXAN_3118 copurified with the A domain or the complete module from 16 of the 17 NRPS modules (see Fig. S2), including the A domain from module 11 that was negative in the B2H assay. These results support our finding that no single assay will detect all possible MLP/NRPS interactions (13). The only NRPS module that tested negative in this assay was module 7, which contains an A domain that lacks some of the signature sequences for an A domain, is disrupted by an oxidase (Ox) domain, and also lacks a T domain (Fig. 4A). As discussed in the next section, our in vitro analysis of this A domain supports the hypothesis that this A domain is inactive.
FIG 4.
MXAN_3634 to MXAN_3636 interactions with MXAN_3118. (A) Schematic of the MXAN_3634 to MXAN_3636 NRPS/PKS hybrid assembly line. Boxes highlight the third (M3), eighth (M8), eleventh (M11), and fifteenth (M15) modules investigated for MLP interactions. Domain abbreviations: AL, acyl-CoA ligase; T, thiolation; C, condensation; ox, oxidation; KS, ketosynthase; AT, acyltransferase; E, epimerase; Te, thioesterase. (B) Results from B2H assays. Quantitative β-galactosidase assay of the α-subunit-A domain fusions with MXAN_3118-λcI fusion (+) or λcI alone (−). Error bars show standard deviations from three independent cultures. **, P ≤ 0.01; ****, P ≤ 0.0001; ns, not significant by Student's t test.
Orphan BGC MXAN_3634 to MXAN_3636 codes for an MXAN_3118-dependent megasynthase that is likely to produce a valine-rich natural product that influences aggregation of M. xanthus 1622 in minimal medium.
With each A domain or module partially purified, we assayed each enzyme for amino acid specificity using standard ATP/PPi exchange assays (see Fig. S3). Each A domain or module was assayed for activation of proteinogenic amino acids. We focused our efforts on these potential substrates, since the BGC consisting of MXAN_3634 to MXAN_3636 does not code for any nonproteinogenic amino acid biosynthesis enzymes and the amino acid specificity codes of each complete A domain suggested the activation of a proteinogenic amino acid (see Table S2). Of the 17 NRPS A domains or modules, 16 of them activated an amino acid substrate. Module 7, containing an A domain we hypothesized to be inactive, failed to copurify with MXAN_3118 and also failed to activate any amino acid substrate. These data suggest that this module does not incorporate an amino acid into the mixed nonribosomal peptide-polyketide product. Nine of the sixteen remaining A domains activated the amino acid l-valine, suggesting that the final product is a valine-rich natural product. The initiating module of the megasynthase is a didomain module with a homolog of a medium-chain acyl-CoA ligase and a T domain. Repeated attempts to overproduce soluble forms of module 1 with and without MXAN_3118 in E. coli failed; thus, at this time, we do not know if this module is active.
Repeated attempts to detect a metabolite associated with MXAN_3634 to MXAN_3636 were not successful. We note that Müller and colleagues detected components of this megasynthase in proteomic studies but also failed to detect an associated metabolite (26). During our attempts to detect an associated metabolite, we observed that a strain of M. xanthus DK1622 lacking the BGC MXAN_3634 to MXAN_3636 aggregated when grown in minimal liquid medium, but the wild-type strain did not (see Fig. S4). Consistent with an essential role for MXAN_3118 in the function of the associated megasynthase, a strain lacking MXAN_3118 showed the same clumping phenotype (Fig. S4). These data support our protein-protein interaction assay results that MXAN_3118 plays a role in the function of the megasynthase consisting of MXAN_3634 to MXAN_3636.
MXAN_3118 is likely to be expressed constitutively during vegetative growth of M. xanthus DK1622.
Our observation that MXAN_3118 interacts with NRPS components coded by two distinct BGCs suggested that MXAN_3118 may play a broad role in natural product production by M. xanthus DK1622. With this in mind, we used reverse transcription-PCR (RT-PCR) to investigate when the mRNA of MXAN_3118 is produced in DK1622. We detected MXAN_3118 mRNA in both rich and minimal media (see Fig. S5), suggesting that MXAN_3118 is constitutively expressed.
MXAN_3118 interacts with at least five additional NRPS systems in M. xanthus DK1622.
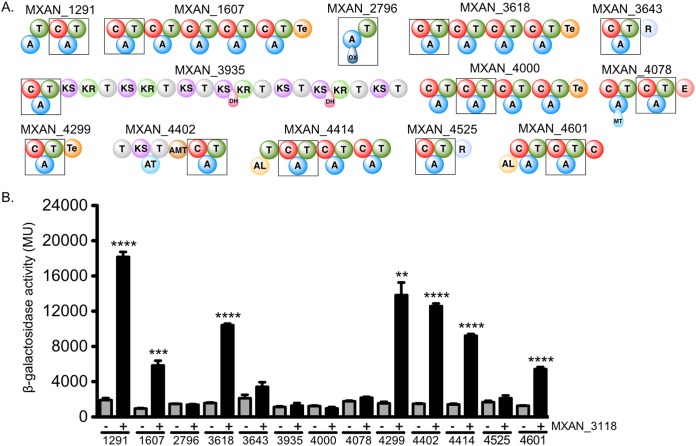
The presence of MXAN_3118 mRNA throughout the growth of M. xanthus DK1622 in both rich and minimal media suggests that the coded MLP is also present. Due to this, we investigated whether MXAN_3118 is a “universal” MLP in M. xanthus DK1622 that works with any NRPS system that requires this accessory protein. M. xanthus DK1622 has 15 BGCs that code for NRPS or NPRS/PKS megasynthases, two of which we already have provided evidence for interactions with MXAN_3118 (Fig. 3 and 4). We targeted one module from each of the remaining megasynthases for analysis by B2H and copurification assays (Fig. 5A). The B2H assay detected statistically significant interactions between MXAN_3118 and the A domains from seven NRPS systems (Fig. 5B). Of the seven B2H-positive A domains, five (MXAN_1291, MXAN_1607, MXAN_3618, MXAN_4402, and MXAN_4414) had improved solubility and copurified with MXAN_3118 when heterologously overproduced in E. coli (see Fig. S6). Interestingly, MXAN_4299 was positive in the B2H assay but did not copurify with MXAN_3118. We were unable to find an expression construct that produced soluble MXAN_4601 hence we were unable to assess whether it copurifies with MXAN_3118. In addition to the NRPS systems that were positive for in the B2H assay, we also assessed whether any of the NRPS systems that were negative in the B2H assay copurified with MXAN_3118 (Fig.S6). None of these NRPSs copurified with MXAN_3118. Thus, on the basis of our requirement for obtaining two independent lines of evidence for MXAN_3118 interactions with an NRPS, we conclude that seven NRPS systems (Mpc, MXAN_3436-3636, MXAN_1291, MXAN_1607, MXAN_3618, MXAN_4402, and MXAN_4414) in M. xanthus 1622 are MLP dependent.
FIG 5.
Protein-protein interactions of M. xanthus DK1622 NRPS and MXAN_3118 using B2H assay. (A) Schematic of the NRPS proteins from the 13 NRPS encoding gene clusters that were tested. Boxes highlight the modules containing the A domain tested. Domain abbreviations: AL, acyl-CoA ligase; T, thiolation; C, condensation; ox, oxidation; KS, ketosynthase; AT, acyltransferase; E, epimerase; Te, thioesterase; R, reductase; KR, ketoreductase; DH, dehydratase; and AMT, aminotransferase. (B) Results of a quantitative β-galactosidase assay of the α-subunit-A domain fusions with MXAN_3118-λcI fusion (+) or λcI alone (−). Error bars show standard deviations from three independent cultures. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001 by Student's t test.
DISCUSSION
Members of the MLP superfamily are nearly always encoded within NRPS-associated BGCs or are adjacent to genes involved in the biosynthesis of nonribosomal peptides. This makes it quite clear that the MLP will associate with at least one of the A domains in the associated NRPS. In contrast, here we have identified 50 orphan MLPs found across different bacterial orders that have no clear NRPS partner. In fact, we found that Wenzhouxiangella marina KCTC 42284 harbors an MLP but does not contain any NRPS-encoding genes (see Table S1 in the supplemental material). The most highly represented order coding for an orphan MLP is the Myxococcales, and we targeted a member of this order, M. xanthus DK1622, to characterize the function of an orphan MLP encoded by MXAN_3118. This is the only MLP encoded by the M. xanthus DK1622 genome, even though this bacterium has 15 BGCs that encode NRPSs (27).
Using a combination of B2H assays and heterologous protein overproduction and purification, we provide evidence that MXAN_3118 is an MLP that naturally interacts with at least seven distinct NRPS systems (Fig. 3 to 5; data summarized in Table 1). It is possible that this MLP interacts with additional NRPS systems. First, while we detected MXAN_3118 interactions with nine NRPS systems using the B2H assay, only seven of these interactions were confirmed by a solubility or copurification assay. It is possible these interactions occur, but the solubility or copurification assays were not sensitive enough to detect them. We have previously shown that some MLP/NRPS interactions are detected by in vivo phenotypic assays but fail to be detected by solubility or copurification and vice versa (13, 28). Second, we did not test all the A domains in M. xanthus DK1622 for interactions with MXAN_3118, and it is possible a different A domain from a targeted NRPS system is MLP dependent, as we have reported for the viomycin and capreomycin NRPS systems (2). Regardless, our data support the conclusion that MXAN_3118 interacts with multiple NRPS systems in M. xanthus DK1622. In the past, evidence that MLPs cross-react with more than one NRPS system was only observed when the cognate MLP-encoding gene of a BGC was deleted, opening the opportunity for the noncognate MLP to replace it for metabolite production (8, 9). Our observations with M. xanthus DK1622 differ from these prior studies, because MXAN_3118 is the only MLP in this bacterium; thus, MLP-dependent NRPS systems are dependent upon the function of this single MLP.
TABLE 1.
Summary of M. xanthus DK1622 NRPS and MXAN_3118 interactions
| NRPS locus tag (A domain)a | Result for NRPS and MXAN_3118 interaction by: |
|
|---|---|---|
| B2H | Copurification | |
| MXAN_1291 (A2) | +b | +b |
| MXAN_1607 (A1) | +b | +b |
| MXAN_2796 | − | NDc |
| MXAN_3618 (A1) | +b | +b |
| MXAN_3634 to MXAN_3636 | +b | +b |
| MXAN_3643 | − | − |
| MXAN_3935 | − | − |
| MXAN_3779 | +b | +b |
| MXAN_4000 (A2) | − | ND |
| MXAN_4078 | − | − |
| MXAN_4299 | + | − |
| MXAN_4402 (A1) | +b | +b |
| MXAN_4414 (A1) | +b | +b |
| MXAN_4525 | − | − |
| MXAN_4601 (A2) | + | ND |
A lack of parentheses indicates that all A domains of the NRPS(s) were tested.
Categorized as positive for MXAN_3118 interactions.
ND, not determined.
The ability of MXAN_3118 to interact with multiple NRPS systems strongly suggests this protein is a universal MLP in M. xanthus DK1622. Our finding that the mRNA for MXAN_3118 is detectable throughout the growth of this bacterium (Fig. S5) is consistent with this MLP playing a role in the function of multiple NRPS systems rather than a single system expressed as a specific stage of growth. With so many required interactions, it is likely that MXAN_3118 has broad NRPS interaction flexibility. This flexibility may prove invaluable for the combinatorial biosynthesis of NRPSs. The construction of hybrid NRPSs will necessitate the use of noncognate MLP/NRPS pairings. The ability of an MLP to naturally interact with multiple NRPS systems suggests that it is a candidate for a universal MLP to be included in any strain where hybrid NRPSs are being produced. We have previously shown that MXAN_3118 can functionally replace YbdZ, the natural MLP partner of the NRPS involved in enterobactin production in E. coli, even though it is evolutionarily quite distant from YbdZ (13). We propose that MXAN_3118, and potentially other members of orphan MLPs in the Myxococcales order, especially those from the Myxococcus species, may be a source of universal MLPs. These may prove to be combinatorial biosynthesis tools analogous to the broad range 4′-phosphopantetheinyl transferase Sfp that is a workhorse of combinatorial biosynthesis studies (29).
MATERIALS AND METHODS
Phylogenetic analysis.
A total of 57 Myxococcales MbtH-like protein homolog sequences were obtained from a BLAST search using MXAN_3118 as a reference sequence and aligned using MUSCLE (30). A phylogenetic analysis was performed using MrBayes software launched from the Mesquite v3.51 software package (31). The substitution model used was Felsenstein 81 plus Gamma and a Markov chain Monte Carlo setting for 10,000,000 generations. The MLP from Desulfobacula phenolica DSM 3384, accession number WP_092230082.1, was used as an outgroup. The phylogenetic tree was visualized using FigTree v1.4.3 (32).
Bacterial strains and plasmid construction.
All strains and plasmids used in this study are listed in Table S3 in the supplemental material. All overexpression vectors were constructed using a polymerase incomplete primer extension (PIPE)-based method (33). The primers used to amplify vectors and inserts used in this study are listed in Table S4. The genes coding for the NRPS or PKS components were amplified from M. xanthus DK1622 genomic DNA and cloned into the pET28b vector with an N-terminal histidine tag. The MXAN_3118 gene was cloned into pACYC Duet-1 in a manner that enabled an untagged MLP to be produced.
Cooverexpression of NRPSs with MXAN_3118 and protein purification.
Overexpression vectors containing various NRPS-encoding genes were transformed into E. coli BL21(DE3) ybdZ::acc(IV) cells containing either pACYC-Duet1-NO-MCS or pACYC-Duet1-MXAN_3118, as previously described (2, 24). Overproducing strains were grown in 3 liters of LB supplemented with kanamycin (50 μg/ml) and chloramphenicol (34 μg/ml). The cells were grown at 28°C with shaking. After reaching an optical density at 600 nm (OD600) of 0.5, the temperature was shifted to 15°C for 1 h, and then cells were induced with 100 μM isopropyl-β-d-1-thiogalactosidase (IPTG) for 15 h. The cells were harvested by centrifugation and resuspended in His tag buffer (300 mM NaCl, 20 mM HEPES [pH 7.5], 10% [vol/vol] glycerol). Protein purification using nickel affinity chromatography was performed as previously described (24). The fractions containing the protein of interest based on SDS-PAGE–Coomassie blue staining were pooled and concentrated (Millipore Centriprep YM-3K). Concentrated proteins were flash frozen in liquid nitrogen and stored at −80°C until use. NRPS/MLP coelution was evaluated using 16.8% acrylamide Tris-Tricine gels stained with Coomassie blue. All protein concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (Pierce) with bovine serum albumin as a protein standard.
ATP/PPi exchange assays.
ATP/PPi exchange assays were performed as previously described (1, 24). In summary, each assay (100 μl) contained 75 mM Tris-HCl (pH 7.5 at 25°C), 10 mM MgCl2, 5 mM dithiothreitol, 3.5 mM ATP (pH 7.0), 1 mM NaPPi, 1 mM [32P]PPi (0.9 Ci/mol; PerkinElmer),1 mM amino acid, and 2.2 to 21 μg of protein. To determine substrate specificity, enzymatic activity was first assessed using pools of amino acids. Briefly, the four pools of l-amino acids consisted of pool one (valine, methionine, cysteine, leucine, and isoleucine), pool two (proline, alanine, glycine, serine, and threonine), pool three (asparagine, aspartate, glutamate, glutamine, and phenylalanine), and pool four (tyrosine, tryptophan, histidine, lysine, and arginine). The amino acid components of the pool(s) with the highest activity were further tested to identify the amino acid substrate. All assays were performed at 25°C for 30 min. The reaction was quenched with a solution containing perchloric acid (3.5% [vol/vol]), NaPPi (100 mM), and activated charcoal (1.6% [wt/vol]). The ATP bound to charcoal was separated by centrifugation, and the pellets were washed with quench solution and water before being counted in the scintillation counter. For each amino acid/pool, the rate of the reaction (picomoles of product per minute per milligram of protein) was calculated from three independent assays performed in parallel using GraphPad Prism ver. 6.0h.
Examination of the expression of MXAN_3118 by RT-PCR.
M. xanthus DK1622 cultures were grown at 30°C in CTT (1% [wt/vol] Casitone, 8 mM MgSO4, 10 mM Tris-HCl [pH 7.6 at 25°C], 1 mM K2HPO4-KH2PO4) and A1 minimal medium [0.5% (wt/vol) potassium aspartate, 0.5% (wt/vol) sodium pyruvate, 0.5 mg/ml (NH4)2SO4, 8 mM MgSO4, 0.125 mg/ml spermidine, 0.1 mg/ml asparagine, 0.1 mg/ml isoleucine, 0.1 mg/ml phenylalanine, 0.1 mg/ml valine, 0.05 mg/ml leucine, 0.01 mg/ml methionine, 1 μg/ml cobalamin, 10 μM FeCl3, 10 μM CaCl2, 1 mM K2HPO4-KH2PO4, and 10 mM Tris-HCl (pH 7.6 at 25°C)] (34). Triplicate cultures were grown until stationary phase. The OD600 was measured every 6 to 8 h, and 500 μl of cells was harvested at different time points during vegetative growth. RNA extractions were performed according to the manufacturer's instructions (RNeasy minikit; Qiagen). All of the RNA samples were also treated with RQ1 DNase (Promega) to remove any residual genomic DNA, followed by ethanol precipitation, and were resuspended in nuclease-free water. RT-PCR was performed using the Access RT-PCR introductory system (Promega) using the primers listed in Table S4. The housekeeping gene MXAN_3078 (rpoC) was used as a positive control for each time point. The PCR products were resolved using 3% (wt/vol) agarose gels.
Bacterial two-hybrid assays.
The E. coli reporter strain FW102 F′ placOL2-62-lacZ, containing a lacZ reporter and an F′ episome (35), had both ybdZ and entF genes deleted from its genome using the temperature-sensitive plasmid pMAK705-ΔybdZ entF vector as previously described (1, 36). Plasmids encoding bacterial two-hybrid (B2H) fusion proteins (see Table S3) were constructed by cloning the portion of the NRPS-encoding gene that covered the adenylation domain into pBRα and MXAN_3118 into pACλCI between the NotI and BamHI sites in the vectors using the primers listed in Table S4. FW102 ΔybdZ entF F′ placOL2-62-lacZ was used as our reported strain and transformed with the pairs of pBRα/NRPS A domain and pAClλCI/MXAN_3118 fusion plasmids. Assays were perform as previously described (25, 37, 38), with minor modifications. Briefly, all strains were grown in 3 ml of LB containing carbenicillin (100 μg/ml), kanamycin (50 μg/ml), and chloramphenicol (34 μg/ml) (LB-Carb-Kan-Cm) at 30°C overnight in triplicates. Cultures were diluted 1:100 in LB-Carb-Kan-Cm with IPTG (100 μg/ml) and incubated at 30°C with shaking until the OD595 reached 0.5. The cultures were then incubated on ice for 20 min, and the cells were lysed with PopCulture (Millipore) and r-Lysozyme (Fisher Scientific) for 30 min. The lysed culture (30 μl) was mixed with 170 μl Z-buffer containing 0.8 mg/ml 2-nitrophenyl β-d-galactopyranoside, and the formation of o-nitrophenol was measured every minute for 60 min by following the increased absorbance at 415 nm (BioTek microplate spectrophotometer). Correction factors and Miller units were calculated as previously described (38).
M. xanthus bacterial strains and plasmids.
All strains and plasmids are listed in Table S3. The sequences of all PCR primers can be found in Table S4. M. xanthus DK1622 strains were grown at 32°C in CYE (1.0% [wt/vol] Casitone, 0.5% [wt/vol] yeast extract, 10 mM 3-[N-morpholino] propanesulfonic acid [pH 7.6 at 25°C], and 8 mM MgSO4) supplemented with 50 μg/ml kanamycin when appropriate. Deletion strains LS3135 and LS3944 were generated through the double recombination method as previously described (39) using the vector pBJ113 (40). Briefly, plasmids were created by first generating PCR fragments 1,000 bp immediately upstream of the target deletion and 1,000 bp immediately downstream. For LS3135, the DNA fragments had 5′ BamHI and 3′ NheI sites and 5′ NheI and 3′ HindIII sites, respectively. The fragments were inserted into the appropriate sites of the plasmid vector pBJ113. For LS3944, the upstream fragment had 5′ KpnI and 3′ NheI sites. The resulting plasmids were then transformed into electrocompetent M. xanthus DK1622 cells with selection for kanamycin resistance after integration by a single homologous crossover. A second recombination event was counter selected for galactose sensitivity with CYE containing 1% (wt/vol) galactose and resulted in in-frame deletions that were verified by PCR.
M. xanthus growth in minimal medium.
M. xanthus DK1622, ΔMXAN_3634-MXAN_3636 (LS3944), and ΔMXAN_3118 (LS3135) cultures were grown at 30°C in 25 ml A1 minimal medium (34). Triplicate cultures were grown for a period of 2 weeks.
Supplementary Material
ACKNOWLEDGMENTS
We thank Richard Gourse for providing the bacterial two-hybrid strains. We also thank Kurt Throckmorton, Caryn Wadler, Garret Suen, and Gaspar Bruner Montero for helpful discussions and guidance with the phylogenic analyses of the orphan MLPs.
We declare no competing financial interest.
This work was funded by the National Institutes of Health (grants GM100346 to M.G.T. and T32 GM07215 to K.J.E.-L.), the E. B. Fred Professorship (to M.G.T.), and the National Science Foundation (grant 1411891 to L.J.S.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00346-18.
REFERENCES
- 1.Felnagle EA, Jackson EE, Chan YA, Podevels AM, Berti AD, McMahon MD, Thomas MG. 2008. Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol Pharm 5:191–211. doi: 10.1021/mp700137g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felnagle EA, Barkei JJ, Park H, Podevels AM, McMahon MD, Drott DW, Thomas MG. 2010. MbtH-like proteins as integral components of bacterial nonribosomal peptide synthetases. Biochemistry 49:8815–8817. doi: 10.1021/bi1012854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imker HJ, Krahn D, Clerc J, Kaiser M, Walsh CT. 2010. N-Acylation during glidobactin biosynthesis by the tridomain nonribosomal peptide synthetase module GlbF. Chem Biol 17:1077–1083. doi: 10.1016/j.chembiol.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang W, Heemstra JR, Walsh CT, Imker HJ. 2010. Activation of the pacidamycin PacL adenylation domain by MbtH-like proteins. Biochemistry 49:9946–9947. doi: 10.1021/bi101539b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heemstra JR, Walsh CT, Sattely ES. 2009. Enzymatic tailoring of ornithine in the biosynthesis of the Rhizobium cyclic trihydroxamate siderophore vicibactin. J Am Chem Soc 131:15317–15329. doi: 10.1021/ja9056008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quadri LEN, Sello J, Keating TA, Weinreb PH, Walsh CT. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem Biol 5:631–645. doi: 10.1016/S1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- 7.Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol Microbiol 45:1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- 8.Wolpert M, Gust B, Kammerer B, Heide L. 2007. Effects of deletions of mbtH-like genes on clorobiocin biosynthesis in Streptomyces coelicolor. Microbiology 153:1413–1423. doi: 10.1099/mic.0.2006/002998-0. [DOI] [PubMed] [Google Scholar]
- 9.Lautru S, Oves-Costales D, Pernodet J-LL, Challis GL. 2007. MbtH-like protein-mediated cross-talk between non-ribosomal peptide antibiotic and siderophore biosynthetic pathways in Streptomyces coelicolor M145. Microbiology 153:1405–1412. doi: 10.1099/mic.0.2006/003145-0. [DOI] [PubMed] [Google Scholar]
- 10.Drake EJ, Cao J, Qu J, Shah MB, Straubinger RM, Gulick AM. 2007. The 1.8 A crystal structure of PA2412, an MbtH-like protein from the pyoverdine cluster of Pseudomonas aeruginosa. J Biol Chem 282:20425–20434. doi: 10.1074/jbc.M611833200. [DOI] [PubMed] [Google Scholar]
- 11.Boll B, Taubitz T, Heide L. 2011. Role of MbtH-like proteins in the adenylation of tyrosine during aminocoumarin and vancomycin biosynthesis. J Biol Chem 286:36281–36290. doi: 10.1074/jbc.M111.288092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller BR, Drake EJ, Shi C, Aldrich CC, Gulick AM. 2016. Structures of a nonribosomal peptide synthetase module bound to MbtH-like proteins support a highly dynamic domain architecture. J Biol Chem 291:22559–22571. doi: 10.1074/jbc.M116.746297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schomer RA, Thomas MG. 2017. Characterization of the functional variance in MbtH-like protein interactions with a nonribosomal peptide synthetase. Biochemistry 56:5380–5390. doi: 10.1021/acs.biochem.7b00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchko GW, Kim CY, Terwilliger TC, Myler PJ. 2010. Solution structure of Rv2377c-founding member of the MbtH-like protein family. Tuberculosis 90:245–251. doi: 10.1016/j.tube.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herbst DA, Boll B, Zocher G, Stehle T, Heide L. 2013. Structural basis of the interaction of MbtH-like proteins, putative regulators of nonribosomal peptide biosynthesis, with adenylating enzymes. J Biol Chem 288:1991–2003. doi: 10.1074/jbc.M112.420182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tarry MJ, Haque AS, Bui KH, Schmeing TM. 2017. X-ray crystallography and electron microscopy of cross- and multi-module nonribosomal peptide synthetase proteins reveal a flexible architecture. Structure 25:783.e4–793.e4. doi: 10.1016/j.str.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Baltz RH. 2011. Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. J Ind Microbiol Biotechnol 38:1747–1760. doi: 10.1007/s10295-011-1022-8. [DOI] [PubMed] [Google Scholar]
- 18.Coordinators NR. 2017. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 45:D12–D17. doi: 10.1093/nar/gkw1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SH, Choe H, Kim BK, Nasir A, Kim KM. 2015. Complete genome of the marine bacterium Wenzhouxiangella marina KCTC 42284T. Mar Genomics 24:277–280. doi: 10.1016/j.margen.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Weissman KJ, Müller R. 2010. Myxobacterial secondary metabolites: bioactivities and modes-of-action. Nat Prod Rep 27:1276–1295. doi: 10.1039/c001260m. [DOI] [PubMed] [Google Scholar]
- 22.Cortina NS, Krug D, Plaza A, Revermann O, Müller R. 2012. Myxoprincomide: a natural product from Myxococcus xanthus discovered by comprehensive analysis of the secondary metabolome. Angew Chem Int Ed Engl 51:811–816. doi: 10.1002/anie.201106305. [DOI] [PubMed] [Google Scholar]
- 23.Drake EJ, Miller BR, Shi C, Tarrasch JT, Sundlov JA, Leigh Allen C, Skiniotis G, Aldrich CC, Gulick AM. 2016. Structures of two distinct conformations of holo-non-ribosomal peptide synthetases. Nature 529:235–238. doi: 10.1038/nature16163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahon MD, Rush JS, Thomas MG. 2012. Analyses of MbtB, MbtE, and MbtF suggest revisions to the mycobactin biosynthesis pathway in Mycobacterium tuberculosis. J Bacteriol 194:2809–2818. doi: 10.1128/JB.00088-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dove SL, Joung JK, Hochschild A. 1997. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature 386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 26.Schley C, Altmeyer MO, Swart R, Mu R, Huber CG, Müller R, Huber CG. 2006. Proteome analysis of Myxococcus xanthus by off-line two-dimensional chromatographic separation using monolithic poly-(styrene-divinylbenzene) columns combined with ion-trap tandem mass spectrometry. J Proteome Res 5:2760–2768. doi: 10.1021/pr0602489. [DOI] [PubMed] [Google Scholar]
- 27.Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Eisen J, Ronning CM, Barbazuk WB, Blanchard M, Field C, Halling C, Hinkle G, Iartchuk O, Kim HS, Mackenzie C, Madupu R, Miller N, Shvartsbeyn A, Sullivan SA, Vaudin M, Wiegand R, Kaplan HB. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci U S A 103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schomer RA, Park H, Barkei JJ, Thomas MG. 2018. Alanine scanning of YbdZ, an MbtH-like protein, reveals essential residues for functional interactions with its nonribosomal peptide synthetase partner EntF. Biochemistry 57:4125–4134. doi: 10.1021/acs.biochem.8b00552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quadri LEN, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. 1998. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carder protein domains in peptide synthetases. Biochemistry 37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 30.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maddison WP, Maddison DR. 2015. Mesquite: a modular system for evolutionary analysis. Version 2.75. 2011. https://www.mesquiteproject.org/.
- 32.Rambaut A. 2016. FigTree v1.4.3. http://tree.bio.ed.ac.uk/software/figtree/.
- 33.Klock HE, Lesley SA. 2009. The polymerase incomplete primer extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol Biol 498:91–103. doi: 10.1007/978-1-59745-196-3_6. [DOI] [PubMed] [Google Scholar]
- 34.Bretscher AP, Kaiser D. 1978. Nutrition of Myxococcus xanthus, a fruiting Myxobacterium. J Bacteriol 133:763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA. 2006. Structural basis for bacterial transcription-coupled DNA repair. Cell 124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 36.Hamilton CM, Aldea M, Washburn BK, Babitzke P, Kushner SR. 1989. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol 171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banta AB, Chumanov RS, Yuan AH, Lin H, Campbell EA, Burgess RR, Gourse RL. 2013. Key features of σS required for specific recognition by Crl, a transcription factor promoting assembly of RNA polymerase holoenzyme. Proc Natl Acad Sci U S A 110:15955–15960. doi: 10.1073/pnas.1311642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thibodeau SA, Fang R, Joung JK. 2004. High-throughput β-galactosidase assay for bacterial cell-based reporter systems. Biotechniques 36:410–415. doi: 10.2144/04363BM07. [DOI] [PubMed] [Google Scholar]
- 39.Ueki T, Inouye S, Inouye M. 1996. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183:153–157. doi: 10.1016/S0378-1119(96)00546-X. [DOI] [PubMed] [Google Scholar]
- 40.Julien B, Kaiser AD, Garza A. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc Natl Acad Sci U S A 97:9098–9103. doi: 10.1073/pnas.97.16.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.