Flagellum is the major organelle for motility in many bacterial species. While most bacteria possess external flagella, such as the multiple peritrichous flagella found in Escherichia coli and Salmonella enterica or the single polar sheathed flagellum in Vibrio spp., spirochetes uniquely assemble periplasmic flagella, which are embedded between their inner and outer membranes. Here, we show for the first time that the external flagella in Vibrio alginolyticus can be changed as periplasmic flagella by deleting two flagellar genes. The discovery here may provide new insights into the molecular basis underlying assembly, diversity, and evolution of flagella.
KEYWORDS: flagellar assembly, membrane penetration, flagellar evolution, periplasmic flagella
ABSTRACT
The bacterial flagellum has evolved as one of the most remarkable nanomachines in nature. It provides swimming and swarming motilities that are often essential for the bacterial life cycle and pathogenesis. Many bacteria such as Salmonella and Vibrio species use flagella as an external propeller to move to favorable environments, whereas spirochetes utilize internal periplasmic flagella to drive a serpentine movement of the cell bodies through tissues. Here, we use cryo-electron tomography to visualize the polar sheathed flagellum of Vibrio alginolyticus with particular focus on a Vibrio-specific feature, the H-ring. We characterized the H-ring by identifying its two components FlgT and FlgO. We found that the majority of flagella are located within the periplasmic space in the absence of the H-ring, which are different from those of external flagella in wild-type cells. Our results not only indicate the H-ring has a novel function in facilitating the penetration of the outer membrane and the assembly of the external sheathed flagella but also are consistent with the notion that the flagella have evolved to adapt highly diverse needs by receiving or removing accessary genes.
IMPORTANCE Flagellum is the major organelle for motility in many bacterial species. While most bacteria possess external flagella, such as the multiple peritrichous flagella found in Escherichia coli and Salmonella enterica or the single polar sheathed flagellum in Vibrio spp., spirochetes uniquely assemble periplasmic flagella, which are embedded between their inner and outer membranes. Here, we show for the first time that the external flagella in Vibrio alginolyticus can be changed as periplasmic flagella by deleting two flagellar genes. The discovery here may provide new insights into the molecular basis underlying assembly, diversity, and evolution of flagella.
INTRODUCTION
The flagellum is the major organelle for motility in many bacterial species. It is one of the most complex nanomachines in the bacterial kingdom. Flagella from different species share a conserved core but also adapt profound variation to accommodate different needs or functions (1, 2). While most bacteria possess external flagella, such as the multiple peritrichous flagella found in Escherichia coli and Salmonella enterica or the single polar sheathed flagellum in Vibrio spp., spirochetes uniquely assemble flagella, which are embedded in the periplasmic space between their inner and outer membranes, thus called periplasmic flagella (3). Therefore, the flagella have been a great model system for understanding the evolution and adaptation of bacterial nanomachines (4).
Peritrichous flagella have been extensively studied in E. coli and Salmonella spp. (5–9). The flagellum is composed of a long helical filament, a hook, and a motor. The motor is a complex macromolecular assembly composed of several ring structures around a rod, which functions as a drive shaft. The MS-ring consists of multiple copies of a single protein, FliF, and is embedded in the inner membrane. The C-ring is assembled in the cytoplasm and is essential for torque generation and the clockwise/counterclockwise switching of the direction of rotation. A flagellar type III export apparatus is located underneath the MS-ring and C-ring. The L-ring is located in the outer membrane. The P-ring is located in the periplasmic space and interacts with the peptidoglycan layer. The P-ring and L-ring form a bushing at the distal end of the rod. The rotation of the flagellum is driven through an interaction between the rotor and the surrounding stator complexes.
The polar sheathed flagellum from Vibrio species is quite different from the peritrichous flagella in E. coli and Salmonella spp. (9–11). The polar sheathed flagellum utilizes a sodium ion gradient as the energy resource for rotation and exhibits remarkably fast speeds of up to 1,700 Hz (12). Compared with the flagella in E. coli and Salmonella spp., the polar sheathed flagellum in Vibrio spp. possesses extra ring-like structures known as the T-ring and the H-ring (13, 14). They are essential for high-speed rotation of the Vibrio sp. flagella (15). The T-ring is located next to the P-ring and is important for incorporating the sodium-driven stator units into the basal body (13). The H-ring is known to be adjacent to the L-ring. FlgT was the first protein identified to be involved in the formation of the H-ring (15). However, the exact structure and function of the H-ring remained to be defined.
Spirochetes are a group of bacteria with distinctive morphology and motility (3). The motility of the spirochetes is driven by periplasmic flagella, which are quite different from the external flagella in E. coli and Vibrio spp. Although the highly conserved flagellar type III secretion system has been utilized to assemble the rod, the hook, and the filament in both periplasmic flagella and external flagella (3, 16, 17), one of the main differences between these two flagellar systems is whether or not the flagella penetrate the outer membrane. It is of interest to identify genes involved in outer membrane penetration.
Vibrio alginolyticus is a great model system to study polar sheathed flagella (10, 18). In particular, cryo-electron tomography (cryo-ET) has been utilized to not only visualize the sheathed flagella in V. alginolyticus and reveal several distinct features, including the membrane sheath, O-ring, T-ring, and H-ring (11), but also to provide structural evidence that proteins MotX and MotY form the T-ring adjacent to the P-ring (11, 13). Here, we attempt to understand the structure and function of the H-ring by characterizing two mutants lacking flgT or flgO, respectively. We found that periplasmic flagella assemble in both mutants. This observation suggests that the H-ring is essential for outer membrane penetration and assembly of the polar flagellum in Vibrio spp. These new findings provide a basis for the further understanding of flagellar assembly and evolution.
RESULTS
FlgO and FlgT are involved in the H-ring formation.
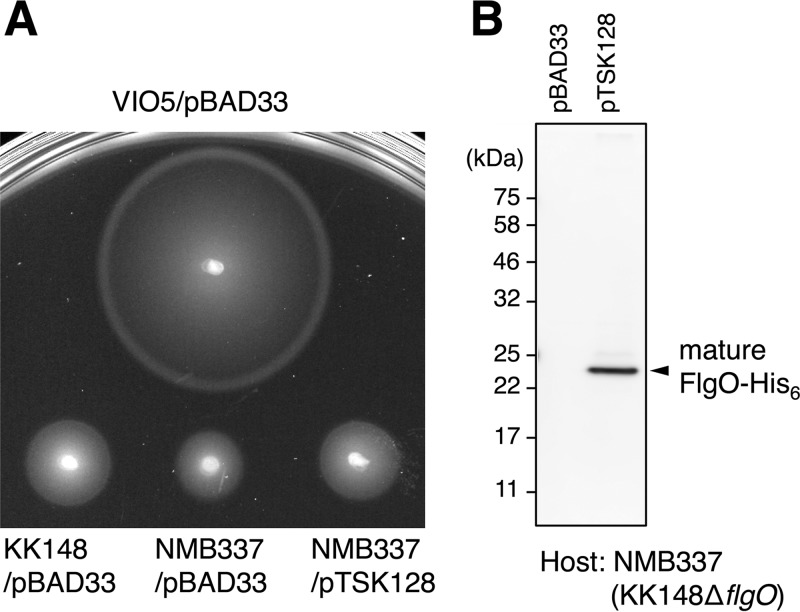
The H-ring is a Vibrio-specific flagellar feature that is important for motility. Our recent studies of the V. alginolyticus flagellar motor showed that the H-ring is a large disk underneath the outer membrane (11). FlgT is the first protein known to be involved in the H-ring formation (14, 15). Two additional outer membrane lipoproteins FlgO and FlgP were required for flagellum stability and motility of Vibrio cholerae, as the flgO and flgP mutants have reduced motility and fewer external flagella (19). The averaged motor structure of a Vibrio fischeri ΔflgP mutant showed that the PL-rings, together with the T-ring, were visible (20). Therefore, we hypothesize that both FlgO and FlgP might be involved in the formation of the H-ring complex in Vibrio spp. We constructed ΔflgO and ΔflgT mutants in the background of the multipolar flagellated strain, respectively (Table 1). The ΔflgO mutant cells were less motile than KK148 cells in a soft agar plate, whereas expression of a His-tagged flgO+ allele complemented the ΔflgO allele for motility and expression (Fig. 1A), as detected by Western blotting using anti-His tag antibody (Fig. 1B).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Reference or source |
|---|---|---|
| V. alginolyticus | ||
| VIO5 | VIK4 (Rifr Pof+ Laf−) | 42 |
| KK148 | VIO5 flhG (multi-Pof+) | 43 |
| TH7 | KK148 ΔflgT | 14 |
| NMB337 | KK148 ΔflgO | This study |
| E. coli | ||
| DH5α | Recipient for DNA manipulation | 46 |
| β3914 | Recipient for conjugational transfer of pSW7848 | 44 |
| Plasmids | ||
| pGEM-T Easy | Cloning vector, Ampr | Promega |
| pSW7848 | Suicide vector, (oriVR6Kγ oriTRP4 araC-PBAD-ccdB) Cmr | 45 |
| pBAD33 | Cmr PBAD | 47 |
| pTSK127 | pGEM-T Easy-ΔflgO | This study |
| pTSK127_2 | pSW7848-ΔflgO | This study |
| pTY57 | Cmr, PBAD with a multicloning site of pBAD24 | 48 |
| pTSK128 | pTY57-flgO::His6 | This study |
Rifr, rifampin resistant; Pof+, normal polar flagellar formation; Laf−, defective in lateral flagellar formation; multi-Pof+, multiple polar flagellar formation; Ampr, ampicillin resistant; Cmr, chloramphenicol resistant; PBAD, arabinose promoter.
FIG 1.
Lack of FlgO results in reduced motility. (A) Motility of cells in soft agar. Two-microliter aliquots of overnight cultures of each strain was spotted onto a 0.25% soft agar VPG plate containing 2.5 μg/ml chloramphenicol and 0.02% (wt/vol) l-arabinose, and the plate was incubated at 30°C for 7 h. Deletion of flgO from the strain KK148 resulted in reduced motility, and ectopic expression of FlgO fused with a hexa-histidine tag at the C terminus (FlgO-His6) from the arabinose-inducible plasmid pTSK128 restored motility (protein expression was confirmed [B]). VIO5 is the wild-type strain for polar flagellar motility; KK148 is multipolar flagellar strain and the parent of NMB337. The plasmid pBAD33 was used as the empty vector control. (B) Immunoblot analysis. Whole-cell lysates were separated by SDS-PAGE and transferred onto the PVDF membrane, and His-tagged proteins were detected by an anti-His tag antibody. The FlgO-His6 protein was detected at the size equivalent to its mature form (indicated as the filled arrow). Experiments were conducted 3 times, and the typical results are shown here.
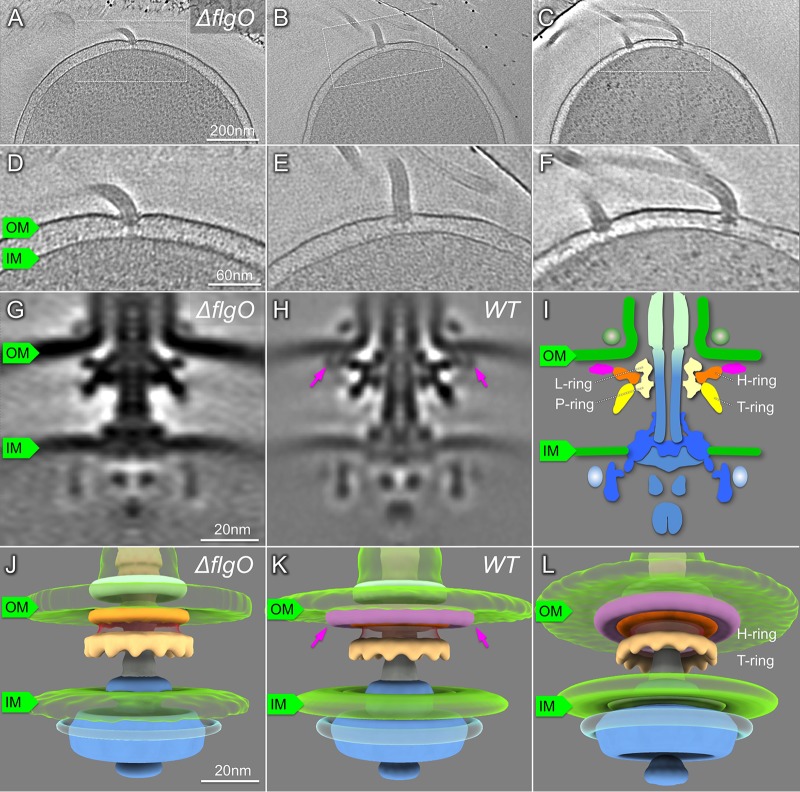
To decipher whether deletion of flgO affects the assembly of the polar sheathed flagellum and the formation of the H-ring, we examined ΔflgO mutant cells by cryo-ET. Polar sheathed flagella are clearly visible in the ΔflgO mutant (Fig. 2; Table 1). We identified flagellar motor structures from tomograms and determined the in situ motor structure from the ΔflgO strain using subtomogram averaging (Fig. 2). Compared with the motor structure from the wild type, the distal part of the H-ring density is absent in the ΔflgO motor (Fig. 2G, H, and J to L). Thus, our data suggest that FlgO is the protein component that is primarily responsible for the distal part of the H-ring. Furthermore, the distal part of the H-ring seems to anchor the whole disk onto the inner leaflet of the outer membrane, because the smaller H-ring in the ΔflgO motor appears to be less tightly associated with the outer membrane (Fig. 2G and H).
FIG 2.
Characterization of the ΔflgO flagellum in situ by cryo-ET. (A to C) A representative slice of a 3D reconstruction of the V. alginolyticus ΔflgO strain KK148 with multiple polar flagella. (D to F) Zoom-in views of the slices that are shown in panels A to C. (G) A slice of a subtomogram average of the flagellar motor. (H) A slice of a subtomogram average of the flagellar motor in KK148. The structural difference between panels G and H is indicated with arrows. (I) Schematic model of the Vibrio motor. (J) 3D surface renderings of the image from panel G. (K, L) 3D surface renderings of the image from panel H. The H-ring is labeled in orange and pink, separately; the T-ring is colored yellow. OM, outer membrane; IM, inner membrane.
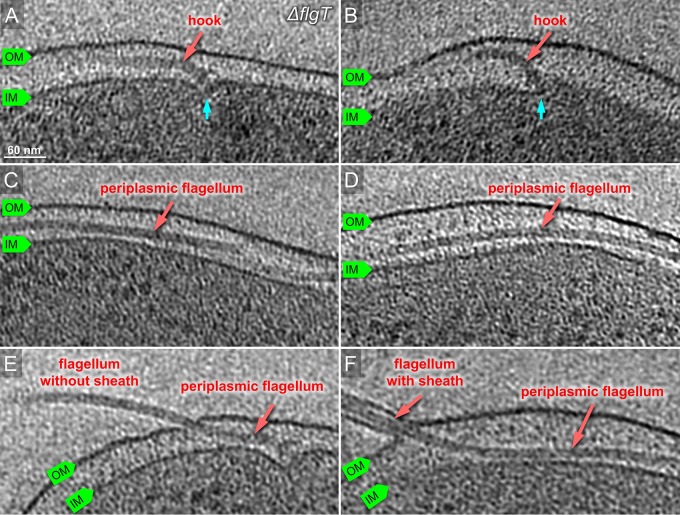
To further understand the role of the H-ring on flagellar formation, we visualized a ΔflgT mutant using cryo-ET, as FlgT is involved in the formation of the H-ring (14). Indeed, the entire H-ring density was absent in the tomograms of the flgT mutant cells, while the T-ring density remained visible (Fig. 3). This is consistent with the previous observation that the H-ring is not visible in a purified basal body of the ΔflgT mutant by negative stain electron microscopy (14). Together with the results from ΔflgO cells, we conclude that FlgO is responsible for the distal part of the H-ring and FlgT is essential for the proximal part of the H-ring. Thus, FlgT and FlgO together with FlgP contribute to the formation of the H-ring.
FIG 3.
Characterization of the ΔflgT flagellum in situ by cryo-ET. (A, B) Representative slices of tomograms from KK148 ΔflgT cells. The motor is visible beneath of outer membrane. The motor is indicated in cyan. (C, D) Representative slices of from KK148 ΔflgT cells. The flagellar filament is visible in the periplasmic space and labeled in red. (E) The flagellar filament is extended in the periplasmic space and penetrates the outer membrane without a sheath. (F) The flagellar filament penetrates from the periplasm and is sheathed.
The H-ring plays an essential role in flagellum assembly and bacterial motility.
The H-ring is tightly associated with the outer membrane (Fig. 2G and H). It has been suggested that the H-ring is important for torque generation and bacterial motility (20), although the exact role of the H-ring remains to be determined. To understand the function of the H-ring in more detail, we thoroughly screened tomograms from the ΔflgO mutant cells. Most ΔflgO mutant cells possess polar sheathed flagella, similar to those from wild-type cells. However, about 10% of the ΔflgO mutant cells displayed both polar sheathed flagella and periplasmic flagella (see Fig. S1 in the supplemental material). This observation suggested that the H-ring was likely involved in flagellar assembly, especially in the penetration of the outer membrane, to enable the formation of the external sheathed flagella.
To further understand the relationship of the H-ring and flagellar assembly, we examined more than several hundred reconstructions from ΔflgT mutant cells. We found that many hooks are severely bent beneath the PG layer and many filaments are located in the periplasmic space (Fig. S2 and Fig. 3). Because some of the filaments are much longer than the cell body, they often protrude through the PG layer and the outer membrane at the region far from the basal body (Fig. 3). Less external flagella were found on the ΔflgT cells than on wild-type cells. In total, ∼80% of the 354 flagella found in the ΔflgT cells were located in the periplasmic space. Compared to ∼10% periplasmic flagella in the ΔflgO cells and none in the wild-type cells, the lack of the H-ring had a profound influence on flagellar assembly and location. Although this result has not been visualized previously, it is consistent with the early observation that flagellated cells were rare in the flgT mutant cells (21, 22).
Whole-cell reconstructions show different flagellar assembly and location in wild-type and ΔflgT mutant cells.
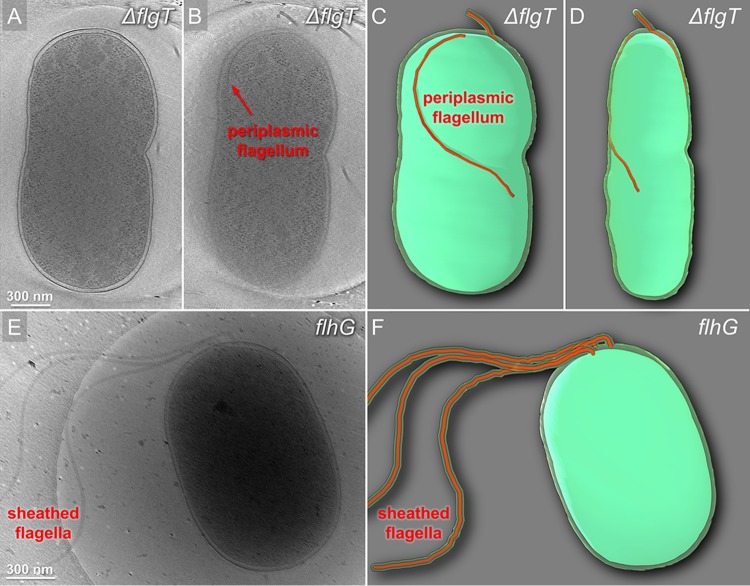
For a better understanding of the role of the H-ring, we generated the whole-cell reconstructions from the ΔflgT mutant and flhG cells. Two flagella are found in the periplasmic region of the ΔflgT cells (Fig. 4A to D). One flagellar filament extended into the cell wall and extruded through the outer membrane and was covered by the sheath at the cell pole (Fig. 4A to D). Another flagellar filament folded back toward the cell body and stayed in the periplasmic space. In contrast, no periplasmic flagella were visible in flhG (Fig. 4E and F). Three flagella directly assembled at the pole and penetrated across the outer membrane to form the long external filaments covered with the outer membrane sheath (Fig. 4E and F). Taking these results together, we conclude that the loss of the H-ring has a substantial effect on the assembly of the polar sheathed flagella.
FIG 4.
Cryo-ET reconstructions of the whole cells from KK148 and KK148 ΔflgT exhibiting dramatic differences in flagellar structures. (A, B) Tomographic slice of KK148 ΔflgT shown in different layers of the tomogram. (C, D) Images from panels A and B in a 3D segmentation to show the periplasmic flagella. (E) Representative tomogram slice of a KK148 whole cell shows multiple polar sheathed flagella. (F) 3D segmentation of the image from panel E.
DISCUSSION
The flagella have evolved as the main organelles for motility in many bacteria. Recent studies based on genome sequences and in situ structural studies by cryo-ET have demonstrated that although the flagella possess a conserved core, the overall flagellar structures appear to be strikingly diverse in different bacterial species (1). For example, a large cage-like structure surrounds the P-rings and L-rings in the Helicobacter pylori flagella (23). Vibrio sp. flagella possess the unique H-rings and T-rings essential for its motility (11, 20). To better understand the structure, function, and evolution of the flagellum, it is of particular interest to uncover unique aspects of flagella in different bacterial species. Using V. alginolyticus as the model system, we previously analyzed the Vibrio-specific T-ring, which is vital for higher torque generation and faster motility in Vibrio spp. Here, we revealed that the Vibrio-specific H-ring is required for flagellar morphogenesis and assembly.
Novel structure and function of the H-ring in Vibrio spp.
Our studies clearly indicated that FlgO forms the distal part of the H-ring and FlgT is responsible for the proximal part. Another protein component of the H-ring might be FlgP, as it is a lipoprotein localized to the outer membrane in V. cholerae (19, 24). Recent cryo-ET studies of a ΔflgP mutant from Vibrio fischeri provided evidence that most of the H-ring is absent (20), while the small density adjacent to the L-ring remains. From these results, together with our results from ΔflgO and ΔflgT, it is very likely that FlgT, FlgP, and FlgO are directly involved in the proximal, middle, and distal parts of the H-ring, respectively.
The H-ring has been suggested to be important for high torque generation (20). The H-ring is not only visible in Vibrio species but also present in Aeromonas hydrophila species (25). Flagella in both species are sodium driven, which are known to generate greater torque than flagellar motors driven by proton flow (26). The sodium-driven flagella also evolved additional accessary structures, such as the T-ring and the H-ring, to support higher torque generation (27). However, we found that reduced motility due to FlgT or FlgO dysfunction is attributed to a significant change on flagellar morphogenesis from polar flagella to periplasmic flagella in addition to an effect on torque generation. Thus, the H-ring plays important roles not only in stabilizing flagellar motors on the outer membrane but also in facilitating outer membrane penetration by external flagella.
Outer membrane penetration and implication on the evolution of flagella.
Although it has been reported previously that the conversion from external to periplasmic flagella could result from a combination of a single amino acid substitution in the flagellar rod protein FlgG and the deletion of flgIH (28), it was unexpected to find periplasmic flagella in both ΔflgT and ΔflgO mutant cells. FlgG is a conserved core protein, and the length of flagellar rod composed of multiple FlgGs is relatively constant in both spirochete periplasmic flagella and external flagella (29). On the contrary, both FlgT and FlgO are not conserved core proteins (30). In the ΔflgO mutant cells, the overall structure of the flagellar motor is similar to that in wild-type cells and only the distal portion of the H-ring structure is absent. Most flagella in the ΔflgO mutant cells are able to penetrate the outer membrane and form the external flagella covered by the sheath. However, 10% of flagella fail to penetrate the outer membrane, resulting in the formation of periplasmic flagella. In the absence of the entire H-ring, as was observed in a ΔflgT mutant strain, the majority of flagella fail to penetrate the outer membrane and form the normal, sheathed flagella. Instead, they form periplasmic flagella. Some of them are much longer than the cell body and appear to violently protrude from the cell wall without any bushing, such as the PL-rings.
The Vibrio sp. H-ring is only a part of the flagellar outer membrane complex (FOMC), which is the flagellar component associated with the outer membrane. The FOMC is often structurally variable in different Gram-negative bacterial species and is also absent in both Gram-positive bacteria and spirochetes. The fact that it is possible to generate periplasmic flagella from external flagella by altering the FOMC is particularly interesting. This allows us to speculate that periplasmic flagella might have evolved from extracellular flagella by losing genes involving in the formation of the FOMC. On the other hand, it also raises the possibility that external flagella could evolve from periplasmic flagella by receiving genes critical for the formation of the FOMC. Thus, the function of FOMC determines the bacterial flagellar morphogenesis, external or periplasmic. During flagellar assembly, the flagellar rod is also playing an important role in penetrating the outer membrane (28, 31). Thus, the change of flagellar morphogenesis would be attributed to the interactions between the flagellar rod and the FOMC. How the flagellar rod senses and coordinates with the FOMC formation awaits to be revealed.
In summary, we characterized the Vibrio-specific H-ring by using cryo-ET and genetic mutations and provided evidence that at least two proteins (FlgO and FlgT) are directly involved in the formation of the H-ring (Fig. 5). Furthermore, we discovered that the H-ring plays a novel function in facilitating the penetration of the outer membrane in Vibrio species. Thus, we conclude that the FOMC is not only working as the bushing but also functioning as an adaptor to determine the flagellar morphogenesis (Fig. 5). The discovery here may provide a new insight to understand the molecular basis underlying assembly, diversity, and evolution of flagella.
FIG 5.
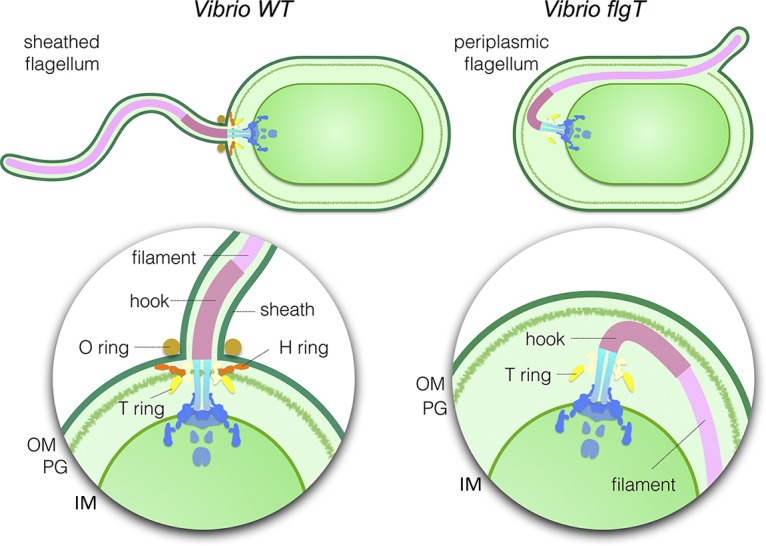
Model of polar sheathed flagellar assembly. Vibrio species have a single polar sheathed flagellum. The H-ring labeled in red is required in the assembly of the polar sheathed flagellum, in addition to flagellar stabilization. The dysfunction of FlgT causes the loss of the H-ring and, consequently, changes the polar flagellum to a periplasmic flagellum. OM, outer membrane; PG, peptidoglycan layer; IM, inner membrane.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth condition.
Bacterial strains used in this study are listed in Table 1. V. alginolyticus strains were cultured at 30°C on VC medium (0.5% [wt/vol] polypeptone, 0.5% [wt/vol] yeast extract, 3% [wt/vol] NaCl, 0.4% [wt/vol] K2HPO4, 0.2% [wt/vol] glucose) or VPG medium (1% [wt/vol] polypeptone, 3% [wt/vol] NaCl, 0.4% [wt/vol] K2HPO4, 0.5% [wt/vol] glycerol). If needed, chloramphenicol and l-arabinose were added at final concentrations of 2.5 μg/ml and 0.02% (wt/vol), respectively. E. coli was cultured at 37°C in LB medium (1% [wt/vol] Bacto tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl). If needed, chloramphenicol and ampicillin were added at final concentrations of 25 μg/ml and 100 μg/ml, respectively. The introduction of plasmids into Vibrio strains was conducted by electroporation as described previously (32).
Construction of the flgO deletion strain.
The flgO deletion strain NMB337 was generated from the multipolar flagellated strain KK148 by homologous recombination with the ΔflgO sequence (1,000 bp), which is composed of a 500-bp upstream sequence of flgO fused with a 500-bp downstream sequence of flgO, by using the method described previously (33). The ΔflgO DNA fragment was amplified by a two-step PCR; for the upstream sequence using a sense primer 1 (5′-GGGAGCTCATGGATAAATATCGACGCGAA-3′) containing a SacI site and an antisense primer 2 (5′-CATGCTTCTATCGGTTTGATTCTCCAGATAATC-3′), and for the downstream sequence using a sense primer 3 (5′-GAGAATCAAACCGATAGAAGCATGAAGAAGTT-3′) and an antisense primer 4 (5′-AAGAGCTCTGTTGCCAATCAGCCG-3′) containing a SacI site. Amplified PCR fragments for the upstream and downstream sequences were gel-purified and mixed, and then the ΔflgO DNA fragment was PCR amplified by using a sense primer 1 and an antisense primer 4. The ΔflgO fragment was cloned into the pGEM-T Easy vector using the SacI site to generate pTSK127, and then it was transferred to pSW7848 to generate pTSK127_2. By using the conjugational transfer, pTSK127_2 was introduced into KK148, and ΔflgO strains were obtained as described previously (33). The deletion was confirmed by colony PCR and DNA sequencing.
Motility assay.
Two microliters of overnight cultures of V. alginolyticus cells containing plasmids at 30°C in VC medium with chloramphenicol was spotted onto the VPG soft agar plate (VPG medium containing 0.25% [wt/vol] Bacto agar with 0.02% [wt/vol] l-arabinose and 2.5 μg/ml chloramphenicol). The plate was incubated at 30°C for 7 h.
Detection of proteins by immunoblotting.
V. alginolyticus cells of overnight cultures at 30°C in VC medium were reinoculated at a 100-fold dilution into fresh VPG medium containing 0.02% (wt/vol) l-arabinose and 2.5 μg/ml chloramphenicol. Cells were cultured at 30°C for about 3.5 h, harvested, suspended to an optical density at 660 nm equivalent to 10 in SDS loading buffer and boiled at 95°C for 5 min. These whole-cell lysate samples were separated by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane, and immunoblotting was performed using a polyclonal anti-His tag antibody (Medical and Biological Laboratories Co., Ltd., Nagoya, Japan).
Sample preparation for cryo-ET observation.
V. alginolyticus strains were cultured overnight at 30°C on VC medium and diluted 100× with fresh VC medium and cultured at 30°C at 220 rpm (BioShaker BR-23FH; Taitec). After 5 h, cells were collected and washed twice and finally diluted with TMN500 medium (50 mM Tris-HCl at pH 7.5, 5 mM glucose, 5 mM MgCl, and 500 mM NaCl). Colloidal gold solution (10-nm diameter) was added to the diluted Vibrio sp. samples to yield a 10× dilution and then deposited on a freshly glow-discharged, holey carbon grid for 1 min. The grid was blotted with filter paper and rapidly plunge-frozen in liquid ethane in a homemade plunger apparatus, as described previously (11).
Cryo-ET data collection and image processing.
The frozen-hydrated specimens of KK148 and TH7 were transferred to a Polara G2 electron microscope, and the samples of NMB337 were transferred to a Titan Krios electron microscope (FEI). Both microscopes are equipped with a 300-kV field emission gun and a direct electron detector (Gatan K2 Summit). Images collected by a Polara G2 electron microscope were observed at ×9,000 magnification and at ∼8 μm defocus, resulting in 0.42 nm/pixel. The images taken by a Titan Krios electron microscope were collected at a defocus near to 0 μm using a Volta phase plate and the energy filter with a 20-eV slit. The data were acquired automatically with SerialEM software (34). For better data collection, the phase shift is normally distributed in the range of 0.33π to 0.67π. A total dose of 50 e−/Å2 was distributed among 35 tilt images covering angles from −51° to +51° at tilt steps of 3°. For every single tilt series collection, the dose-fractionated mode was used to generate 8 to 10 frames per projection image. Collected dose-fractionated data were first subjected to the motion correction program to generate drift-corrected stack files (35, 36). The stack files were aligned using gold fiducial markers and volumes reconstructed using IMOD and Tomo3d, respectively (37, 38). In total, 137 tomograms of TH7 and 114 tomograms of NMB337 were generated.
Subtomogram analysis with i3 package.
Bacterial flagellar motors were detected manually, using the i3 program (39, 40). We selected two points on each motor, one point at the C-ring region and another near the flagellar hook. The orientation and geographic coordinates of selected particles were estimated. In total, 668 subtomograms of Vibrio sp. motors from NMB337 were used for subtomogram analysis. The i3 tomographic package was used on the basis of the “alignment by classification” method with missing wedge compensation for generating the averaged structure of the motor, as described previously (11).
Three-dimensional (3D) visualization.
Tomographic reconstructions were visualized using IMOD software (37). UCSF Chimera software was used for 3D surface rendering of subtomogram averages and molecular modeling (41).
Supplementary Material
ACKNOWLEDGMENTS
We thank Kelly Hughes for critically reading the manuscript prior to submission. We thank Didier Mazel for providing the bacterial strain and the plasmids for making gene deletion mutants in V. alginolyticus.
This work was supported by grants R01AI087946 from National Institute of Allergy and Infectious Diseases and GM107629 from the National Institute of General Medicine Sciences.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00387-18.
REFERENCES
- 1.Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Muller A, Dobro MJ, Jensen GJ. 2011. Structural diversity of bacterial flagellar motors. EMBO J 30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao X, Norris SJ, Liu J. 2014. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry 53:4323–4333. doi: 10.1021/bi500059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, Motaleb MA, Wolgemuth CW. 2012. The unique paradigm of spirochete motility and chemotaxis. Annu Rev Microbiol 66:349–370. doi: 10.1146/annurev-micro-092611-150145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pallen MJ, Penn CW, Chaudhuri RR. 2005. Bacterial flagellar diversity in the post-genomic era. Trends Microbiol 13:143–149. doi: 10.1016/j.tim.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Berg HC. 2003. The rotary motor of bacterial flagella. Annu Rev Biochem 72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 6.Minamino T, Imada K. 2015. The bacterial flagellar motor and its structural diversity. Trends Microbiol 23:267–274. doi: 10.1016/j.tim.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 9.Terashima H, Kojima S, Homma M. 2008. Flagellar motility in bacteria structure and function of flagellar motor. Int Rev Cell Mol Biol 270:39–85. doi: 10.1016/S1937-6448(08)01402-0. [DOI] [PubMed] [Google Scholar]
- 10.Zhu S, Kojima S, Homma M. 2013. Structure, gene regulation and environmental response of flagella in Vibrio. Front Microbiol 4:410. doi: 10.3389/fmicb.2013.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu S, Nishikino T, Hu B, Kojima S, Homma M, Liu J. 2017. Molecular architecture of the sheathed polar flagellum in Vibrio alginolyticus. Proc Natl Acad Sci U S A 114:10966–10971. doi: 10.1073/pnas.1712489114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magariyama Y, Sugiyama S, Muramoto K, Maekawa Y, Kawagishi I, Imae Y, Kudo S. 1994. Very fast flagellar rotation. Nature 371:752. doi: 10.1038/371752b0. [DOI] [PubMed] [Google Scholar]
- 13.Terashima H, Fukuoka H, Yakushi T, Kojima S, Homma M. 2006. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na-driven flagella and required for stator formation. Mol Microbiol 62:1170–1180. doi: 10.1111/j.1365-2958.2006.05435.x. [DOI] [PubMed] [Google Scholar]
- 14.Terashima H, Koike M, Kojima S, Homma M. 2010. The flagellar basal body-associated protein FlgT is essential for a novel ring structure in the sodium-driven Vibrio motor. J Bacteriol 192:5609–5615. doi: 10.1128/JB.00720-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terashima H, Li N, Sakuma M, Koike M, Kojima S, Homma M, Imada K. 2013. Insight into the assembly mechanism in the supramolecular rings of the sodium-driven Vibrio flagellar motor from the structure of FlgT. Proc Natl Acad Sci U S A 110:6133–6138. doi: 10.1073/pnas.1222655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao X, Zhang K, Boquoi T, Hu B, Motaleb MA, Miller KA, James ME, Charon NW, Manson MD, Norris SJ, Li C, Liu J. 2013. Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc Natl Acad Sci U S A 110:14390–14395. doi: 10.1073/pnas.1308306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macnab RM. 2004. Type III flagellar protein export and flagellar assembly. Biochim Biophys Acta 1694:207–217. doi: 10.1016/j.bbamcr.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Li N, Kojima S, Homma M. 2011. Sodium-driven motor of the polar flagellum in marine bacteria Vibrio. Genes Cells 16:985–999. doi: 10.1111/j.1365-2443.2011.01545.x. [DOI] [PubMed] [Google Scholar]
- 19.Martinez RM, Dharmasena MN, Kirn TJ, Taylor RK. 2009. Characterization of two outer membrane proteins, FlgO and FlgP, that influence Vibrio cholerae motility. J Bacteriol 191:5669–5679. doi: 10.1128/JB.00632-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beeby M, Ribardo DA, Brennan CA, Ruby EG, Jensen GJ, Hendrixson DR. 2016. Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc Natl Acad Sci U S A 113:e1917-26. doi: 10.1073/pnas.1518952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cameron DE, Urbach JM, Mekalanos JJ. 2008. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci U S A 105:8736–8741. doi: 10.1073/pnas.0803281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez RM, Jude BA, Kirn TJ, Skorupski K, Taylor RK. 2010. Role of FlgT in anchoring the flagellum of Vibrio cholerae. J Bacteriol 192:2085–2092. doi: 10.1128/JB.01562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin Z, Lin WT, Zhu S, Franco AT, Liu J. 2016. Imaging the motility and chemotaxis machineries in Helicobacter pylori by cryo-electron tomography. J Bacteriol 199:e00695-16. doi: 10.1128/JB.00695-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris DC, Peng F, Barker JR, Klose KE. 2008. Lipidation of an FlrC-dependent protein is required for enhanced intestinal colonization by Vibrio cholerae. J Bacteriol 190:231–239. doi: 10.1128/JB.00924-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merino S, Tomas JM. 2016. The FlgT protein is involved in Aeromonas hydrophila polar flagella stability and not affects anchorage of lateral flagella. Front Microbiol 7:1150. doi: 10.3389/fmicb.2016.01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lo C-J, Sowa Y, Pilizota T, Berry RM. 2013. Mechanism and kinetics of a sodium-driven bacterial flagellar motor. Proc Natl Acad Sci U S A 110:E2544–E2551. doi: 10.1073/pnas.1301664110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaban B, Coleman I, Beeby M. 2018. Evolution of higher torque in Campylobacter-type bacterial flagellar motors. Sci Rep 8:97. doi: 10.1038/s41598-017-18115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chevance FF, Takahashi N, Karlinsey JE, Gnerer J, Hirano T, Samudrala R, Aizawa S, Hughes KT. 2007. The mechanism of outer membrane penetration by the eubacterial flagellum and implications for spirochete evolution. Genes Dev 21:2326–2335. doi: 10.1101/gad.1571607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujii T, Kato T, Hiraoka KD, Miyata T, Minamino T, Chevance FFV, Hughes KT, Namba K. 2017. Identical folds used for distinct mechanical functions of the bacterial flagellar rod and hook. Nat Commun 8:14276. doi: 10.1038/ncomms14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu R, Ochman H. 2007. Stepwise formation of the bacterial flagellar system. Proc Natl Acad Sci U S A 104:7116–7121. doi: 10.1073/pnas.0700266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen EJ, Ferreira JL, Ladinsky MS, Beeby M, Hughes KT. 2017. Nanoscale-length control of the flagellar driveshaft requires hitting the tethered outer membrane. Science 356:197–200. doi: 10.1126/science.aam6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawagishi I, Imagawa M, Imae Y, McCarter L, Homma M. 1996. The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol Microbiol 20:693–699. doi: 10.1111/j.1365-2958.1996.tb02509.x. [DOI] [PubMed] [Google Scholar]
- 33.Takekawa N, Kwon S, Nishioka N, Kojima S, Homma M. 2016. HubP, a polar landmark protein, regulates flagellar number by assisting in the proper polar localization of FlhG in Vibrio alginolyticus. J Bacteriol 198:3091–3098. doi: 10.1128/JB.00462-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morado DR, Hu B, Liu J. 2016. Using tomoauto: a protocol for high-throughput automated cryo-electron tomography. J Vis Exp 30:e53608. doi: 10.3791/53608:e53608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 38.Agulleiro JI, Fernandez JJ. 2015. Tomo3D 2.0–exploitation of advanced vector extensions (AVX) for 3D reconstruction. J Struct Biol 189:147–152. doi: 10.1016/j.jsb.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 39.Winkler H. 2007. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol 157:126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 40.Winkler H, Zhu P, Liu J, Ye F, Roux KH, Taylor KA. 2009. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J Struct Biol 165:64–77. doi: 10.1016/j.jsb.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 42.Okunishi I, Kawagishi I, Homma M. 1996. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J Bacteriol 178:2409–2415. doi: 10.1128/jb.178.8.2409-2415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kusumoto A, Shinohara A, Terashima H, Kojima S, Yakushi T, Homma M. 2008. Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology 154:1390–1399. doi: 10.1099/mic.0.2007/012641-0. [DOI] [PubMed] [Google Scholar]
- 44.Le Roux F, Binesse J, Saulnier D, Mazel D. 2007. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol 73:777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Val ME, Skovgaard O, Ducos-Galand M, Bland MJ, Mazel D. 2012. Genome engineering in Vibrio cholerae: a feasible approach to address biological issues. PLoS Genet 8:e1002472. doi: 10.1371/journal.pgen.1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grant SG, Jessee J, Bloom FR, Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A 87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li N, Kojima S, Homma M. 2011. Characterization of the periplasmic region of PomB, a Na+–driven flagellar stator protein in Vibrio alginolyticus. J Bacteriol 193:3773–3784. doi: 10.1128/JB.00113-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.