Abstract Abstract
The activity of the pine wood nematode Bursaphelenchusxylophilus leads to extremely serious economic, ecological and social losses in East Asia. The nematode causes pine wilt disease, which is currently regarded as the most important forest disease in China. The pathogenic nematode feeds on dendrocola fungi to complete its cycle of infection. As the vector of the nematode, the Japanese pine sawyer (Monochamusalternatus) also carries dendrocola fungi. Pine woods, infected by B.xylophilus and tunnelled by M.alternatus, are also inhabited by ophiostomatoid fungi. These fungi are well known for their association with many bark and ambrosia beetles. They can cause sapstain and other serious tree diseases. The aims of our study were to investigate and identify the ophiostomatoid communities associated with the epidemic pine wood nematode and the pine sawyer in Pinusmassoniana and P.thunbergii forests, which are the main hosts of the pine wood nematode in China. Two hundred and forty strains of ophiostomatoid fungi were isolated from nematode and sawyer–infected trees in the coastal Shandong and Zhejiang Provinces, representing newly and historically infected areas, respectively. Six ophiostomatoid species were identified on the basis of morphological, physiological and molecular data. For the latter, DNA sequences of the internal transcribed spacer (ITS1–5.8S–ITS2) region and partial b-tubulin gene were examined. The ophiostomatoid species included one known species, Ophiostomaips, three novel species, viz. Ophiostomaalbumsp. nov., Ophiostomamassonianasp. nov. and Sporothrixzhejiangensissp. nov. and two species whose identities are still uncertain, Ophiostomacf.deltoideosporum and Graphilbumcf.rectangulosporium, due to the paucity of the materials obtained. The ophiostomatoid community was dominated by O.ips. This study revealed that a relatively high species diversity of ophiostomatoid fungi are associated with pine infected by B.xylophilus and M.alternatus in China.
Keywords: Ophiostoma , taxonomy, Sporothrix , Ophiostomaminus complex, Ophiostomaips complex
Introduction
The pathogenic pine wood nematode (PWN) Bursaphelenchusxylophilus (Steiner & Buhrer) Nickle (Aphelenchida, Parasitaphelenchidae), presumably native to North America (Steiner and Buhrer 1934, Robbins 1982, Ryss et al. 2005, Zhao et al. 2014), is a mild threat to pine trees in its native area. Nevertheless, this species and the concomitant systematic wilt symptom are responsible for pine tree deaths affecting many trees in eastern Asia, notably in Japan and China (Evans et al. 1996, Mota and Vieira 2008, Mamiya and Shoji 2009, Jung 2010, Futai 2013). Since the first report in China, in Nanjing City in 1982, the disease has spread through more than 300 counties in the provinces of Jiangsu, Zhejiang, Shandong and others, which are currently listed as PWN epidemic areas (State Forestry Administration of the People’s Republic of China 2018). The wilt disease has caused enormous losses not only to the economy and ecology, but also to society, becoming one of the most serious ecological devastation events in Chinese forests.
Bursaphelenchusxylophilus infects many species of coniferous trees, mainly from the genus Pinus (Yan et al. 2003). Pinusarmandii, P.kesiyavar.langbianensis, P.koraiensis, P.massoniana, P.tabuliformis, P.taiwanensis, P.thunbergii and P.yunnanensis are naturally infected by PWN in China (Zhao and Sun 2017). During the infection cycle, the nematode needs vector beetles for dispersal and inoculation into new hosts. The Japanese pine sawyer, Monochamusalternatus Hope (Coleoptera, Cerambycidae), is considered to be the primary PWN vector indigenous to Asia. At the initial stage of infection, PWN feeds on epithelial cells of the host pine (Mota and Vieira 2008, Zhao et al. 2008, Futai 2013). Upon tree death, it feeds on the dendrocola fungi to maintain its population and propagate (Suh et al. 2013, Zhao et al. 2013, 2014).
The ophiostomatoid fungi are one of the most common fungal groups inhabiting wood infected by B.xylophilus. Further, many ophiostomatoid reproduction structures are detected in the tunnels of M.alternatus, suggesting a relationship between the fungi and the occurrence and development of the disease. For instance, O.ips has been found in the PWN vector beetles in North America, China and Korea (Wingfield 1987, Suh et al. 2013, Zhao et al. 2014). There is some evidence that the fungi adhere to the body surface of adult M.alternatus and thus are transmitted to the twigs of healthy trees (Suh et al. 2013).
The association of PWN with ophiostomatoid fungi and bacteria likely contributes to the nematode’s pathogenicity (Zhao et al. 2013, Zhao and Sun 2017). Ophiostomaminus and Sporothrix sp. can stimulate the reproduction of PWN and, consequently, the numbers of PWN carried by the emerging beetles (Maehara and Futai 1997, Zhao et al. 2013, Zhao and Sun 2017). Moreover, the fragrant diacetone alcohol released from wood infected by Sporothrix sp. 1 can induce B.xylophilus to produce greater number of offspring and promotes beetle growth and survival (Zhao et al. 2013).
Thus far, the association with PWN and Monochamus spp. has been documented for only five species of ophiostomatoid fungi worldwide (Wingfield 1987, Maehara and Futai 1997, Hyun et al. 2007, Suh et al. 2013, Zhao et al. 2013, Zhao and Sun 2017). Determination of the identities of these species is mainly based on morphology and sequence comparisons of a single DNA locus. Given the diversity of ophiostomatoid fungi associated with other beetles, the serious impact of the nematode and sawyers on wood and the potential importance of these fungi in the disease infection cycle, studies of the diversity and occurrence of the ophiostomatoid fungi involved in the pine wilt disease should be intensified. Such studies will enable understanding of the interaction between the disease system and the fungi, ultimately helping to redress the current situation of the ceaseless outbreaks and rapid expansion of the disease.
The aims of the current study were to investigate and identify the ophiostomatoid mycobiota associated with the nematode and sawyer in the epidemic forests of Shandong and Zhejiang Provinces in eastern China to facilitate the understanding of pine wilt disease infection and prevalence mechanisms. The two coastal provinces, Shandong and Zhejiang, represent new and historic epidemic areas, with P.thunbergii and P.massoniana as hosts, respectively.
Materials and methods
Collection of samples and fungus isolations
Fungi were isolated from 98 samples of M.alternatus galleries or pupal chambers in P.massoniana and P.thunbergii in the Zhejiang and Shandong Provinces (Table 1), in November 2012. All host trees used for sample collection in this study were exhibiting weak or dying symptoms, blue stain and 4–5 instar larvae residing inside after dissecting the stems. The nematodes were also isolated from these galleries and pupal chambers by Behrman funnel. The fungi were isolated on the surface of 2% (w/v) water agar (20 g agar powder in 1000 ml of deionised water) in 9 cm wide Petri dishes and incubated at 25 °C (Seifert et al. 1993, Zhao et al. 2013, Chang et al. 2017). Subsequently, all strains were purified by hyphal tip isolation, using the procedure described by Jacobs and Wingfield (2001) and routinely grown on 2% (w/v) malt extract agar (MEA; 20 g malt extract powder and 20 g agar powder in 1000 ml of deionised water). Representative cultures were deposited in the China Forestry Culture Collection Center (CFCC), culture collection of the Chinese Academy of Forestry (CXY) and part of the Belgian Coordinated Collections of Microorganisms (MUCL), culture collection at Université Catholique de Louvain, Belgium.
Table 1.
Strains of ophiostomatoid fungi isolated from pines infested by Monochamusalternatus and pine wood nematode in the current study.
| Group | Species | Strain No. | Host | Origin (Latitude, Longitude) | Genbank No. | Collector | |
|---|---|---|---|---|---|---|---|
| ITS | β-tubulin | ||||||
| A | Sporothrixzhejiangensis sp. nov. | MUCL 55181 (CFCC52167, CXY1612) | Pinus massoniana | Yuyao, Zhejiang (29°58'10.2"N, 121°05'57.1"E) | KY094069 | MH397728 | Q. Lu, YY Lun |
| MUCL 55182 (CFCC52164, CXY1613) | P. massoniana | Yuyao, Zhejiang (29°58'10.2"N, 121°05'57.1"E) | KY094070 | MH397729 | |||
| MUCL 55183 (CFCC52165, CXY1614) | P. massoniana | Yuyao, Zhejiang (29°58'10.2"N, 121°05'57.1"E) | KY094071 | MH397730 | |||
| MUCL 55184 (CFCC52166, CXY1615) | P. massoniana | Yuyao, Zhejiang (29°58'10.2"N, 121°05'57.1"E) | KY094072 | MH397731 | |||
| B | Ophiostomaalbum sp. nov. | MUCL 55189 (CFCC52168, CXY1622) | P. massoniana | Yuyao, Zhejiang (29°58'10.2"N, 121°05'57.1"E) | KY094073 | MH360979 | |
| MUCL 55190 (CFCC52169, CXY1642) | P. massoniana | Yuyao, Zhejiang (29°58'10.2"N, 121°05'57.1"E) | KY094074 | MH360980 | |||
| CFCC52170 (CXY1643) | P. massoniana | Yuyao, Zhejiang (29°58'10.2"N, 121°05'57.1"E) | KY094075 | MH360981 | |||
| C | Ophiostoma ips | CXY1628 | P. thunbergii | Changdao, Shandong (37°59'13.5"N, 120°42'18.1"E) | KY593324 | MH324804 | |
| CXY1631 | P. thunbergii | Zhoushan, Zhejiang (29°52'51.33"N, 122°24'14.13"E) | MH324811 | MH324805 | |||
| CXY1635 | P. massoniana | Yuyao, Zhejiang (29°58'10.2"N, 121°05'57.1"E) | MH324812 | MH324808 | |||
| CXY1638 | P. thunbergii | Fuyang, Zhejiang (30°05'15.1"N, 119°58'55.1"E) | MH324813 | MH324809 | |||
| CXY1639 | P. massoniana | Weihai, Shandong (37°23'23.6"N, 122°32'33.1"E) | MH324814 | MH324810 | |||
| D | Ophiostomamassoniana sp. nov. | MUCL 55179 (CFCC51648, CXY1610) | P. massoniana | Fuyang, Zhejiang (30°05'15.1"N, 119°58'55.1"E) | KY094067 | MH370810 | |
| MUCL 55180 (CFCC51649, CXY1611) | P. massoniana | Yuyao, Zhejiang (29°59'36.87"N, 121°09'09.90"E) | KY094068 | MH370811 | |||
| E | Graphilbum cf. rectangulosporium | CXY1623 | P. massoniana | Yuyao, Zhejiang (29°59'36.87"N, 121°09'09.90"E) | MH324816 | – | |
| F | Ophiostoma cf. deltoideosporum | MUCL 55191 (CXY1640) | P. thunbergii | Weihai, Shandong (37°23'23.6"N, 122°32'33.1"E) | MH324815 | – | |
MUCL: part of the Belgian Coordinated Collections of Microorganisms; CFCC: China Forestry Culture Collection Center; Beijing, China; CXY (Culture Xingyao): culture collection of the Research Institute of Forest Ecology, Environment, and Protection, Chinese Academy of Forestry.
Sequences missing data are indicated by [–].
Culture and morphological studies
The ophiostomatoid fungal strains were incubated on 2% MEA and 2% potato dextrose agar (PDA; 200 g potato and 20 g dextrose, 20 g agar powder in 1000 ml of deionised water: the dextrose was obtained from American Amresco) in the dark at 25 °C in an incubator. Fungal growth on MEA plates was monitored daily. Hyphal tips of emerging colonies were transferred to fresh MEA plates to purify the fungi. Slides were made to observe the sexual/asexual state structures; these were mounted in lactic acid cotton blue on glass slides and examined under a BX51 OLYMPUS microscope. Fifty measurements were made of each microscopic taxonomically informative structure. The measurements are presented in the form: (minimum–) mean minus standard deviation–mean plus standard deviation (–maximum).
A 5-mm mycelium disc was cut from an actively growing fungal colony using a sterile cork borer and placed at the centre of MEA plates, with the aerial mycelium side in contact with the medium. Three replicate plates were prepared for each strain and were incubated at temperatures ranging from 5–40 °C at five-degree intervals. The colony diameters on each Petri dish were determined along two perpendicular axes every day until the entire dish was covered. The colour descriptions were provided according to Rayner (1970).
DNA extraction, PCR and sequencing reactions
DNA was extracted from freshly collected mycelia grown in liquid malt medium (20g malt extract in 1000 ml of deionised water) at 25 °C in the dark for 7 d using an Invisorb Spin Plant mini kit (Invitek, Berlin, Germany), following the manufacturer’s instructions. The internal transcribed spacer (ITS) regions and partial β–tubulin (tub2) genes were amplified using primer pairs ITS1/ITS4 (White et al. 1990) and Bt2a/Bt2b (Glass and Donaldson 1995), respectively.
PCR reactions were performed in 25 ml volumes (2.5 mM MgCl2, 1X PCR buffer, 0.2 mM dNTP, 0.2 mM of each primer and 2.5 U of Taq polymerase). The conditions for ITS and tub2 PCR amplifications were as described earlier (White et al. 1990, Glass and Donaldson 1995). PCR products were purified using an MSB Spin PCRapace kit (250) (Invitek), following the manufacturer’s instructions.
Sequencing reactions were performed using CEQ DTCS Quick Start KitH (Beckman Coulter, American), following the manufacturer’s instructions, with the same PCR primers as above. Nucleotide sequences were determined using a CEQ 2000 XL capillary automated sequencer (Beckman Coulter).
Phylogenetic analyses
Contigs were subjected to BLAST searches of the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/); published sequences of closely related species were retrieved. Alignments of the related genes (most up-to-date sequence regions deposited in the GenBank) were conducted online using MAFFT v 7.0 (https://mafft.cbrc.jp/alignment/server/index.html) (Katoh and Standley 2013) and the E-INS-i strategy. Subsequently, the datasets were checked manually by using MEGA v 5.2 (Tamura et al. 2011). Gaps were treated as a fifth base. Phylogenetic analyses were performed using maximum parsimony (MP), as implemented in PAUP* v 4.0b10 (Swofford 2003); Bayesian Inference (BI), as implemented in MrBayes v 3.1.2 (Huelsenbeck and Ronquist 2001); and Maximum Likelihood (ML), using PhyML v 3.0 (Guidon and Gascuel 2003).
The most parsimonious trees generated by MP analyses were identified by heuristic searches with a random addition sequence (1000); max trees were set to 200 and further evaluated by bootstrap analysis, retaining clades compatible with the 50% majority rule in the bootstrap consensus tree. The analysis was based on tree bisection reconnection branch swapping (TBR). The tree length (TL), consistency index (CI), retention index (RI), homoplasy index (HI) and rescaled consistency index (RC) were recorded for each dataset after tree generation.
The general-time-reversible (GTR) model for ML analyses was selected using the Akaike Information Criterion (AIC) in ModelTest v 3.7 (Posada and Crandall 1998). ML runs performed using the CIPRES cluster at the San Diego Supercomputing Center (USA). Node support was estimated from 1000 bootstrap replicates.
For BI analyses, the most appropriate substitution models were also selected using the general-time-reversible model (GRT) with AIC in ModelTest v 3.7. BI was carried out with MrBayes using the Markov Chain Monte Carlo (MCMC) approach with 5,000,000 generations, to estimate posterior probabilities.
Results
Fungal isolation and sequence comparison
In total, 240 strains belonging to Ophiostomatales were obtained from PWN-infected galleries and pupal chambers of M.alternatus. The strains were sorted into six morphological groups (groups A–F in Table 1), tentatively identified as Sporothrix, Ophiostoma and Graphilbum. After preliminary ITS sequence comparisons of all these strains, 11 strains were clearly disparate to any known species and the remaining 229 strains possessed > 99% similarity with type strain of O.ips (GenBank no. AY546704).
Phylogenetic analyses
ITS and tub2 sequences were generated for 16 strains and deposited in GenBank (Table 1). The ITS alignment matrix contained 110 sequences (Tables 1 and 2) and 651 characters, including gaps, following the preliminary determination of strain affinities using the BLAST search engine (GenBank). Due to the presence or absence in intron in the tub2 sequence in the Sporothrix and Ophiostoma lineage species (Zipfel et al. 2006, de Beer et al. 2016), three separate datasets were built for the tub2 sequences. These were Sporothrix, Ophiostomaminus complex and Ophiostomatenellum complex datasets (Linnakoski et al. 2010, de Beer et al. 2013, 2016). The Sporothrix dataset contained 8 species, 17 sequences and 403 characters, including gaps. The O.minus dataset contained 5 species, 17 sequences and 447 characters, including gaps. The O.tenellum dataset contained 8 species, 14 sequences and 280 characters, including gaps.
Table 2.
The information of references sequences used for phylogenetic analyses in this study.
| Species | Strain No. | Host/insect | Country | Genbank No. | Reference | |
|---|---|---|---|---|---|---|
| ITS | β-tubulin | |||||
| Sporothrix abietina | CBS125.89 | Abies vejari | Mexico | AF484453 | KX590755 | de Beer et al. 2003 |
| S. aurorae | CMW19362 | Pinus eliottii | South Africa | DQ396796 | DQ396800 | Francois et al. 2006 |
| S. bragantina | CBS 474.91 | Soil | Brazil | FN546965 | FN547387 | Madrid et al. 2010 |
| CBS 430.92 | Soil | Brazil | FN546964 | FN547386 | Madrid et al. 2010 | |
| S. brasiliensis | Ss383 | Felis catus | Brazil | KP890194 | FN547387 | Araujo et al. 2015 |
| S. brunneoviolacea | CBS 124562 | Soil | Spain | FN546959 | FN547385 | Madrid et al. 2010 |
| CBS 124564 | Soil | Spain | FN546958 | FN547384 | Madrid et al. 2010 | |
| S. dentifunda | CMW13016 | Quercus wood | Hungary | AY495434 | AY495445 | Aghayeva et al. 2005 |
| CMW13017 | Quercus wood | Poland | AY495435 | AY495446 | Aghayeva et al. 2005 | |
| S. epigloea | CBS 573.63 | Tremella fusiformis | Argentina | KX590817 | KX590760 | de Beer et al. 2016 |
| S. eucalyptigena | CPC 24638 | Eucalyptus marginata | Western Australia | KR476721 | N/A | Crous et al. 2015 |
| S. gemella | CMW23057 | Protea caffra | South Africa | DQ821560 | DQ821554 | Roets et al. 2008 |
| S. inflata | CMW12529 | Soil | Canada | AY495428 | AY495438 | Aghayeva et al. 2005 |
| CMW12527 | wheat-field soil | Germany | AY495426 | AY495437 | Aghayeva et al. 2005 | |
| S. nebularis | CMW27319 | Orthotomicus erosus | Spain | DQ674375 | N/A | Romón et al. 1900 |
| CMW27900 | O. erosus | Spain | DQ674376 | N/A | Romón et al. 1900 | |
| S. pallida | CBS131.56 | Stemonitis fusca | Japan | EF127880 | EF139110 | de Meyer et al. 2008 |
| CBS150.87 | S. fusca | Japan | EF127879 | EF139109 | de Meyer et al. 2008 | |
| S. palmiculminata | CMW23049 | Protea repens | South Africa | DQ316191 | DQ821543 | Francois et al. 2006 |
| S. phasma | CMW20676 | P. laurifolia | South Africa | DQ316219 | DQ821541 | Francois et al. 2006 |
| S. proteara | CMW1103 | P. caffra | South Africa | DQ316203 | DQ316165 | Francois et al. 2006 |
| S. schenckii | MITS2474 | N/A | Mexico | KP132783 | N/A | Irinyi et al. 2015 |
| CBS 938.72 | Human | Franch | KP017094 | N/A | Irinyi et al. 2015 | |
| S. fusiforis | CMW9968 | Populus nigra | Azerbaijan | AY280481 | AY280461 | Aghayeva et al. 2004 |
| S. lunata | CMW10563 | Carpinus betulus | Austria | AY280485 | AY280466 | Zhou et al. 2006 |
| S. narcissi | CBS138.50 | N/A | Canada | AY194510 | KX590765 | Jacobs et al. 2003 |
| S. splendens | CMW872 | Protea repens | South Africa | DQ316215 | DQ316177 | Francois et al. 2006 |
| S. stenoceras | CMW2524 | Acacia mearnsii | South Africa | AF484459 | AY280473 | de Beer et al. 2003 |
| CBS237.32 | pine pulp | Norway | AF484462 | N/A | de Beer et al. 2003 | |
| S. thermara | CMW38930 | Euphorbia ingens | South Africa | KR051115 | KR051103 | Ja et al. 2016 |
| CMW38929 | E. ingens | South Africa | KR051114 | KR051102 | Ja et al. 2016 | |
| S. stylites | CMW14543 | Pine utility poles | Australia | EF127883 | EF139096 | de Meyer et al. 2008 |
| Ophiostoma adjuncti | CMW135 | Pinus ponderosa | USA | AY546696 | N/A | Zhou et al. 2004 |
| O. allantosporum | CBS185.86 | P. pinaster | Europe | AY934506 | N/A | Villarreal et al. 2005 |
| O. angusticollis | Zoq16 | N/A | N/A | EU109671 | N/A | de Beer et al. 2016 |
| CBS186.86 | Pinus banksiana | USA | AY924383 | KX590757 | Villarreal et al. 2005 | |
| O. bicolor | CBS492.77 | Piceaglauca/Ips sp. | USA | DQ268604 | DQ268635 | Massoumi et al. 2007 |
| O. candidum | CMW26484 | Eucalyptus cloeziana | South Africa | HM051409 | HM041874 | Nkuekam et al. 2012 |
| CMW26483 | E. cloeziana | South Africa | HM051408 | HM041873 | Nkuekam et al. 2012 | |
| O. catonianum | C1084 | Pyrus | Italy | AF198243 | N/A | Gorton et al. 2004 |
| O. coronatum | CBS 497.77 | Pinus pinaster | Iberian Peninsula | AY924385 | KX590758 | Villarreal et al. 2005 |
| O. cupulatum | C1194 | Pseudotsuga | USA | AF198230 | N/A | Uzunovic et al. 2000 |
| O. deltoideosporum | WIN(M)41 | N/A | N/A | EU879121 | N/A | Mullineux and Hausner 2009 |
| O. fasciatum | UM56 | Pseudotsuga menziesii | CanadaCanada | EU913720 | EU913759 | Plattner et al. 2009 |
| O. floccosum | C01-021 | Girdled Picearubens | Canada | AY194504 | N/A | Jacobs et al. 2003 |
| C1086 | Soil | Sweden | AF198231 | N/A | Gorton et al. 2004 | |
| O. fumeum | CMW26813 | Eucalyptus cloeziana | South Africa | HM051412 | HM041878 | Nkuekam et al. 2012 |
| CMW26818 | E. cloeziana | South Africa | HM051415 | HM041877 | Nkuekam et al. 2012 | |
| O. fuscum | CMW23196 | Picea abies | Finland | HM031504 | HM031563 | Linnakoski et al. 2010 |
| O. himai ulmi | C1183 | Ulmus | India | AF198233 | N/A | Harrington et al. 2001 |
| C1306 | Ulmus | India | AF198234 | N/A | Harrington et al. 2001 | |
| O. ips | CMW7075 | N/A | USA | AY546704 | N/A | Zhou et al. 2004 |
| CMW22843 | Orthotomicus erosus | N/A | DQ539549 | N/A | Romón et al. 2007 | |
| O. japonicum | YCC099 | N/A | N/A | GU134169 | N/A | Yamaoka et al. 2009 |
| O. kryptum | DAOM 229701 | Piceaabies/Tetropium sp. | Austria | AY304436 | AY305685 | Jacobs and Kirisits 2013 |
| DAOM 229702 | Larixdecidua/T.gabrieli | Austria | AY304434 | AY305686 | Jacobs and Kirisits 2013 | |
| K6/3/2 | Piceaabies/Tetropium sp. | Austria | AY304428 | AY305687 | Jacobs and Kirisits 2013 | |
| O. minus | PIR 18S | N/A | N/A | AY934509 | N/A | Villarreal et al. 2005 |
| CMW22802 | Dryocoetes autographus | N/A | DQ539507 | N/A | Romón et al. 2005 | |
| RJ-T144 | Tetropium sp. | Poland | AM943886 | N/A | Jankowiak and KolařÍk 2010 | |
| CMW28117 | Piceaabies/Tomicusminor | Russia | HM031497 | HM031535 | Linnakoski et al. 2010 | |
| AU58.4 | Lodgepole pine | Canada | AF234834 | N/A | Gorton et al. 2004 | |
| DAOM 212686 | N/A | Canada | AY304438 | AY305690 | Jacobs and Kirisits 2013 | |
| O. micans | CMW:38903 | Picea crassifolia | China | KU184432 | KU184303 | Yin et al. 2016 |
| O. montium | CMW13221 | Pinusponderosa/ Dendroctonusponderosae | USA | AY546711 | N/A | Zhou et al. 2004 |
| CMW13222 | P.contorta/D.ponderosae | Canada | AY546712 | N/A | Zhou et al. 2004 | |
| O. nigrocarpum | CMW 560 | Abies sp. | USA | AY280489 | AY280479 | Aghayeva et al. 2004 |
| CMW651 | Pseudotsuga menziesii | USA | AY280490 | AY280480 | Aghayeva et al. 2004 | |
| O. nitidum | CMW:38907 | Picea crassifolia | China | KU184437 | KU184308 | Yin et al. 2016 |
| O. novo ulmi | C1185 | Ulmus | Russia | AF198235 | N/A | Harrington et al. 2001 |
| C510 | Ulmus | USA | AF198236 | N/A | Harrington et al. 2001 | |
| O. olgensis | CXY1404 | Larixgmelini/Ipssubelongatus | China | KU551299 | KU882938 | Wang et al. 2016 |
| CXY1405 | L.gmelini/I.subelongatus | China | KU551300 | KU882939 | Wang et al. 2016 | |
| CXY1410 | L.gmelini/I.subelongatus | China | KU551303 | KU882942 | Wang et al. 2016 | |
| O. pallidulum | CMW23279 | Pinussylvestris/Hylastesbrunneus | Finland | HM031509 | N/A | Linnakoski et al. 2010 |
| CMW23278 | P.sylvestris/ H.brunneus | Finland | HM031510 | HM031566 | Linnakoski et al. 2010 | |
| O. piceae | C1087 | N/A | Germany | AF198226 | N/A | Uzunovic et al. 2000 |
| C1246 | Pseudotsuga | USA | AF198227 | N/A | Uzunovic et al. 2000 | |
| O. pseudotsugae | 92-634/302/6 | Pinusmenziesii/Dendroctonusfrontalis | Canada | AY542502 | AY548744 | Gorton et al. 2004 |
| D48/3 | N/A | Canada | AY542501 | AY542511 | Gorton et al. 2004 | |
| O. proteasedis | CMW 28601 | Protea caffra | Zambia | EU660449 | EU660464 | Roets et al. 2009 |
| O. pulvinisporum | CMW9022 | Pinuspseudostrobus/Dendroctonusmexicanus | Mexico | AY546714 | DQ296100 | Zhou et al. 2004 |
| O. qinghaiense | CMW:38902 | Picea crassifolia | China | KU184445 | KU184316 | Yin et al. 2016 |
| O. querci | C970 | Quercus | United Kingdom | AF198239 | N/A | Gorton et al. 2004 |
| C969 | Quercus | United Kingdom | AF198238 | N/A | Gorton et al. 2004 | |
| C1085 | Fagus | Germany | AF198237 | N/A | Gorton et al. 2004 | |
| O. rostrocoronatum | CBS434.77 | Woodpulp | USA | AY194509 | KX590771 | Jacobs et al. 2003 |
| O. saponiodorum | CMW29497 | Piceaabies/Ipstypographus | Finland | HM031507 | HM031571 | Linnakoski et al. 2010 |
| CMW28135 | P. abies | Russia | HM031508 | N/A | Linnakoski et al. 2010 | |
| O. sejunctum | Ophi 1B | N/A | N/A | AY934520 | N/A | Villarreal et al. 2005 |
| Ophi 1A | N/A | N/A | AY934519 | N/A | Villarreal et al. 2005 | |
| O. setosum | AU160-38 | Pseutotsugae menziesii | North America | AF128929 | N/A | Uzunovic et al. 2000 |
| CMW12378 | Tsuga sp. | China | FJ430485 | FJ430515 | Grobbelaar et al. 2009 | |
| O. tenellum | CBS189.86 | Pinus banksiana | USA | AY934523 | KX590772 | Villarreal et al. 2005 |
| O. tetropii | C00-027a | Tetropium fuscum | Canada | AY194482 | NA | Jacobs et al. 2003 |
| C00-003 | T. fuscum | Canada | AY194485 | AY305701 | Jacobs et al. 2003 | |
| O. ulmi | C1182 | Ulmus | Netherlands | AF198232 | N/A | Harrington et al. 2001 |
| Graphilbum crescericum | CMW 22829 | Hylastes ater | Spain | DQ539535 | N/A | Romón et al. 2007 |
| Gra. fragrans | C1224 | Pinus sylvestris | Sweden | AF198248 | N/A | Harrington et al. 2001 |
| Gra. microcarpum | YCC612 | Japanese larch logs | Japan | GU134170 | N/A | Yamaoka et al. 2009 |
| Gra. rectangulosporium | MAFF 238951 | N/A | Japan | AB242825 | N/A | Ohtaka et al. 2006 |
| Raffaelea canadensis | CBS 168.66 | N/A | N/A | GQ225699 | N/A | Kyunghee et al. 2009 |
| Leptographium lundbergii | DAOM 64746 | N/A | N/A | EU879151 | AY534943 | Mullineux and Hausner 2009 |
| L. truncatum | WIN(M)1435 | Pinus taeda | South Africa | AY935626 | N/A | Hausner et al. 2005 |
ITS = internal transcribed spacer regions 1 and 2 of the nuclear ribosomal DNA operon, including the 5.8S region; tub2 = beta-tubulin;
N/A= represents information that are not available.
CMW = Culture Collection of the Forestry and Agricultural Biotechnology Institute; CBS = The culture collection of Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; MAFF = Ministry of Agriculture, Forestry, and Fisheries, Genetic Resource Centre, Culture Collection of National Institute of Agrobiological Resources, Japan; CXY (Culture Xingyao): Culture collection of the Research Institute of Forest Ecology, Environment and Protection, Chinese Academy of Forestry.
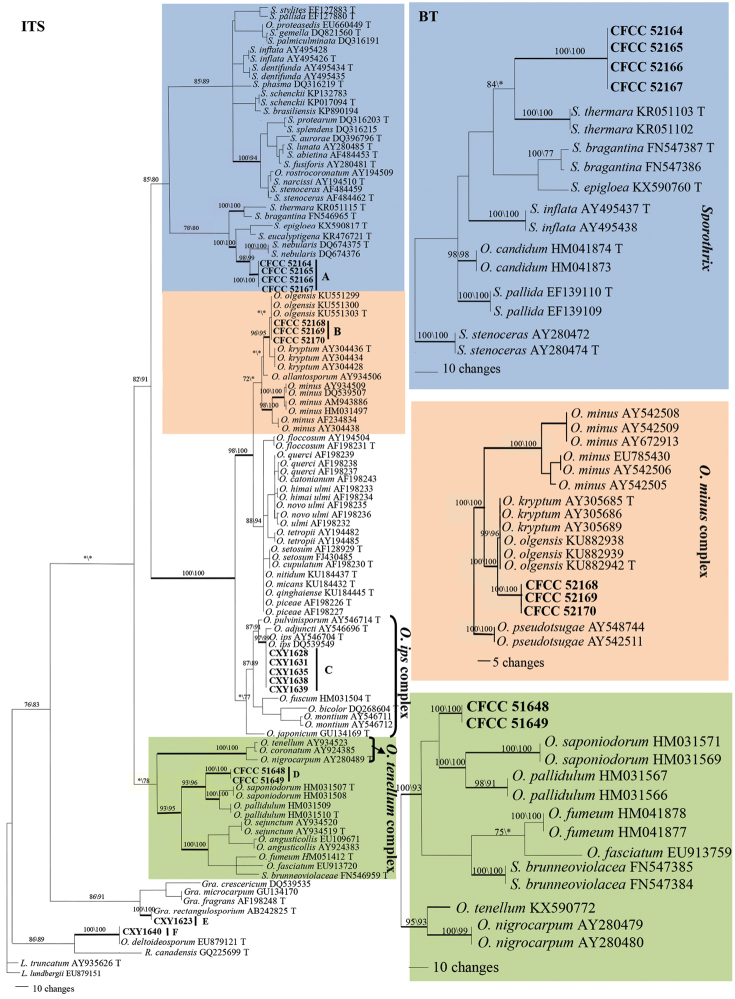
For each phylogenetic tree, MP, ML and BI analyses yielded trees with very similar topologies. Phylograms, generated by the MP analysis, are presented for all the datasets, with nodal support obtained from ML indicated at the nodes (Figure 1). In addition, posterior probabilities (above 90%), obtained from BI, are indicated by bold lines at the relevant branching points. Analyses of the ITS1–5.8S–ITS2 region revealed that the analysed strains formed six distinct clades (Figure 1).
Figure 1.
Phylograms of fungal associates of pine infected by PWN and Monochamusalternatus in China. The phylograms were generated after MP analysis of the ITS1–5.8S–ITS2 rDNA and partial tub2 sequences. Novel sequences obtained in the current study are indicated in bold type. MP bootstrap values (10,000 replicates) and ML bootstrap support values (1000 replicates) (normal type) above 70% are indicated at the nodes. Values below 70% are indicated by asterisk (*). Posterior probabilities (above 90%) obtained from BI are indicated by bold lines at the relevant branching points. Scale bar, total nucleotide differences between taxa; ML, maximum likelihood; MP, maximum parsimony; BI, Bayesian inference.
According to the ITS sequence analysis, strains of the morphological group A nested in the Sporothrix lineage, as defined by de Beer et al. (2016). They form a well-supported independent clade, closely related to S.nebularis, S.epigloea and S.eucalyptigena. Strains exhibiting morphotypes B, C and D formed three clades in the Ophiostomas. str lineage (de Beer and Wingfield 2013). Group B strains nested in the O.minus complex, with O.olgensis forming a well-supported clade, which closely related to O.kryptum (Linnakoski et al. 2010, de Beer and Wingfield 2013, Wang et al. 2016). Group C strains nested within the well-supported O.ips clade. Group D strains nested within the Ophiostoma lineage and closely related to O.saponiodorum and O.pallidulum. Finally, strains exhibiting morphotypes E and F nested in the Graphilbum and Raffaeleas. l. lineages, respectively (de Beer and Wingfield 2013) (TL=821, CI=0.5445, RI=0.8046, HI=0.4555, RC=0.4381 in the MP phylogenetic tree).
Phylogenetic inferences based on tub2 sequences revealed that clade A, B and D strains formed three well-supported independent clades within the Sporothrix and Ophiostoma lineages, respectively. Clade C strains nested within the well-supported O.ips clade (Suppl. material 1).
Considering morphological differences, strains in groups A, B and D represent three undescribed species of Sporothrix or Ophiostoma. We concluded that group C strains belong to O.ips; group E and F strains clustered together with the well-supported Graphilbumrectangulosporium and O.deltoideosporum clades, respectively. However, because of a limited number of strains, further analysis of this potential species will need to be postponed until a sufficient amount of material obtained.
Taxonomy
Based on the phylogenetic signals of the ITS and tub2 and morphological characteristics, all strains analysed in the current study were assigned to six different groups (A–F). They represent one known species, O.ips (Rumbold 1931, Upadhyay 1981, Benade et al. 1995, Rane and Tattar 1987, Suh et al. 2013, Zhao et al. 2013) and two uncertain species (Gra.cf.rectangulosporium and O.cf.deltoideosporum) and the three species are hereby described as new species.
Sporothrix zhejiangensis
Wang & Lu sp. nov.
MB825556
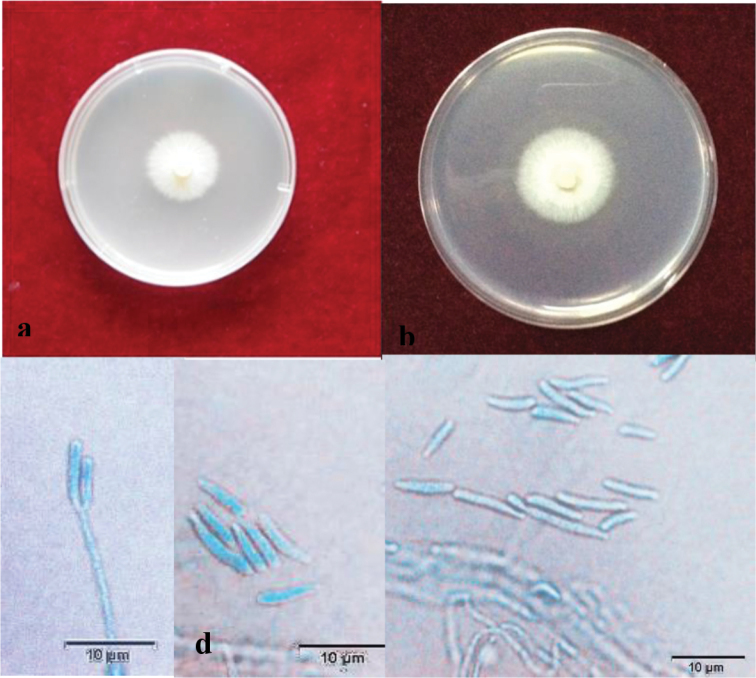
Figure 2.
Light micrographs of Sporothrixzhejiangensis. a–c Growth on 2% MEA and 2% PDA, 2 weeks after inoculation d Occasionally observed ostiolar hyphae (scale bar, 20 μm) e–f Perithecium (scale bar, 20 μm) g Pesotum-like anamorph, rhizoid, conidiophores, conidiogenous apparatus (scale bar, 20 μm), and conidia (bottom right corner) (scale bar, 10 μm) h, i Reniform ascospores without sheaths (scale bar, 10 μm) j–lSporothrix-like anamorph, conidiophores, and conidia (scale bar, 10 μm).
Etymology.
The epithet reflects Zhejiang Province in China where the species was first collected.
Type.
CHINA, Zhejiang, Yuyao City, from Monochamusalternatus gallery in Pinusmassoniana infested by numerous PWN, November 2012, collected by Q Lu and YY Lun, culture ex-holotype MUCL 55183 = CFCC52165 = CXY1614.
Description.
Sexual morph perithecial: Perithecia occasional on 2% MEA, emerging from the superficial mycelium or partly iμmersed, with a globose base, (75–)80–108(–120) μm in diameter, with some basal hyphal ornamentation, black; extending progressively into a straight, brown to black neck, (127–)156–550(–631) μm long, (26–)32–58.5(–65) μm wide at the base, (7–)7.5–10.7(–12) μm wide at the apex; ending in a crown of hyaline, (6–)9–19.5(–24) μm long ostiolar hyphae; ascospores reniform in side view, without sheath, aseptate, hyaline, (2–)2.2–3.4(–4) × (0.6–)0.74–2(–2.5) μm.
Asexual morph: pesotum-like and sporothrix-like.
Pesotum-like: Conidiophores macronematous, synnematous, abundant in 2% MEA. Synnemata occurring singly, enlarging towards both the apex and the base, dark brown at base, becoming paler toward the apex, (100–)120–260(–290) μm long including the conidiogenous apparatus, (56–)63–145(–158) μm wide at base, rhizoids present; conidiogenous cells (7–)9.5–29(–45.5) × 1–2(–1.7) μm; conidia hyaline, aseptate, single-celled, smooth, cylindrical or obovoid, (2–)2.5–4.8(–6) × (0.5–)0.8–2.1(–2.6) μm.
Sporothrix-like: Conidiophores micronematous, single on aerial mycelia, unbranched, (4.5–)9.6–31.5(–51.5) × (1.0–)1.5–2(–2.4) μm; conidia hyaline, smooth, aseptate, ellipsoid to ovoid, (2.5–)3–4.8(–5) × (0.7–)1–2.1(–2.5) μm.
Culture characteristics.
Colonies on 2% MEA medium are white, with colony edge thinning radially. Hyphae are superficial on agar. Diameter reaches 50 μm in the dark after 8 d at 25 °C, able to grow at 5 °C and 40 °C, with the optimal growth temperature of 30 °C. Growth characteristics on PDA medium are similar.
Habitat and distribution.
Galleries of Monochamusalternatus in Pinusmassoniana infested by PWN; known hitherto from Zhejiang Province, China.
Additional specimens examined.
CHINA, Zhejiang, Yuyao City, from Monochamusalternatus galleries in Pinusmassoniana infested by PWN, November 2012, collected by Q Lu and YY Lun, MUCL 55181 = CFCC 52167 = CXY1612, MUCL 55182 = CFCC 52164 = CXY1613, MUCL 55184 = CFCC 52166 = CXY1615.
Note.
Sporothrixzhejiangensis is characterised by a sexual and two asexual forms (pesotum-like and sporothrix-like). It is phylogenetically related to S.nebulare, S.eucalyptigena and S.epigloea (Figure 1). Sporothrixzhejiangensis differs from S.nebulare in both ascomatal and conidial features. The perithecial neck of S.nebulare is shorter than that of S.zhejiangensis, respectively (140–)169–293(–365) μm and (127–)156–550(–631) μm. The conidia of S.nebulare also are smaller than those of S.zhejiangensis, mostly respectively 2.9–3.7 × 1.1–1.3 μm and 3–4.8 × 1–2.1 μm (Romón et al. 1900).
Sporothrixeucalyptigena and S.epigloea produce perithecia and ascospores similar to those of S.zhejiangensis (Crous et al. 2015, Upadhyay 1981). However, S.eucalyptigena has a slightly wider neck than S.zhejiangensis (20–35 vs. 9–19.5 μm) and longer ostiolar hyphae. Furthermore, S.eucalyptigena and S.epigloea only produce a sporothrix-like asexual state and their conidia differ from those of S.zhejiangensis either in size or in shape. Sporothrixeucalyptigena has drop-shaped (lacrymoid) conidia, differing from the ellipsoid to ovoid conidia in S.zhejiangensis. Conidia of S.epigloea are larger than those of S.zhejiangensis (2.5–9 × 1–3.5 vs. 3–4.8 × 1–2.1 μm) (Crous et al. 2015). Another conspicuous difference between S.zhejiangensis and S.eucalyptigena is the growth rate; the former grows much faster than the latter (50 μm in 8 d vs. 50 μm in 30 d at 25 °C) (Upadhyay 1981).
Sporothrixzhejiangensis is also closely related to S.bragantina and S.thermara (Figure 1) (Pfenning and Oberwinkler 1993, de Beer et al. 2016). These three species display the same optimal growth temperature (30 °C) and a similar conidial shape (ellipsoid to obovoid) of their sporothrix-like morph. However, the perithecial base of S.bragantina is larger than that of S.zhejiangensis [globose base: 130–220 μm vs. (75–)80–108(–120) μm and the neck also is longer, 700–1200 μm vs. (127–)156–550(–631) μm]. The sporothrix-like conidia of S.bragantina also are larger than those of S.zhejiangensis (4–6 × 2–2.5 μm vs. 3–4.8 × 1–2.1 μm). Sporothrixthermara, hitherto, has no known sexual state. It only known by sporothrix-like state; conidia of S.thermara are larger than those of S.zhejiangensis (4–6 × 2–3 μm vs. 3–4.8 × 1–2.1 μm).
Ophiostoma album
Wang & Lu sp. nov.
MB825557
Figure 3.
Light micrographs of Ophiostomaalbum. a, b Growth on 2% MEA and 2% PDA, 2 weeks after inoculation c–eHyalorhinocladiella-like anamorph, conidiophores, and conidia (scale bar, 10 mm).
Etymology.
The epithet reflects the white colour of the colonies.
Type.
CHINA, Zhejiang, Yuyao City, from Monochamusalternatus gallery of Pinusmassoniana infested by numerous PWN, November 2012, collected by Q Lu and YY Lun, culture ex-holotype MUCL 55189 = CFCC 52168 = CXY1622.
Description.
Sexual form: Unknown. Asexual form: Hyalorhinocladiella-like. Conidiogenous cells micronematous, (4.2–)9.5–16.5(–20.5) × (0.5–)1–2(–2.5) μm; conidia hyaline, single-celled, aseptate, clavate or fusiform obovoid with pointed bases and (occasionally) rounded apices, slightly curved at the base (4–)4.2–14.5(–18) × (0.5–)1–2(–2.3) μm.
Culture characteristics.
Colonies on 2% MEA white, with the mycelium edge thinning radially; Hyphae are superficial on agar, sporulation weak. Colonies slowly growing, reaching 18.5 μm in diameter at 8 d at 25 °C, able to grow at 40 °C but not at 5 °C, with the optimal growth temperature of 35 °C. Growth characteristics on PDA culture medium are similar but the growth rate is slower than on MEA.
Habitat and distribution.
Galleries of Monochamusalternatus in Pinusmassoniana, infested by PWN, in Zhejiang Province, China.
Additional specimens examined.
CHINA, Zhejiang, Yuyao City, from Monochamusalternatus galleries of Pinusmassoniana infested by numerous PWN, November 2012, collected by Q Lu and YY Lun, MUCL 55190 = CFCC 52169 = CXY1642, CXY1643 = CFCC 52170.
Note.
Ophiostomaalbum only known in its asexual hyalorhinocladiella-like form. According to both ITS and tub2 based phylogenetic analysis, it is closely related to O.kryptum and O.olgensis in the O.minus complex (Figure 1). Ophiostomaalbum is easily distinguished from O.olgensis and O.kryptum based on their reproduction structure. Ophiostomaalbum only produces a hyalorhinocladiella-like asexual form in vitro, whereas the two other species produce both a sexual and asexual forms in vitro (Jacobs and Kirisits 2003, Wang et al. 2016). The conidial size and shape of the three species are obviously different. Ophiostomaalbum produces clavate or fusiform to obovoid and sometimes, slightly curved conidia; these are obovoid with pointed bases in both O.olgensis and O.kryptum. Furthermore, the conidia of O.album are much larger, 4.2–14.5 × 1.0–1.9 μm vs. 1.5–7 × 1.5–5 μm in the two other species.
Ophiostoma massoniana
Wang & Lu sp. nov.
MB825558
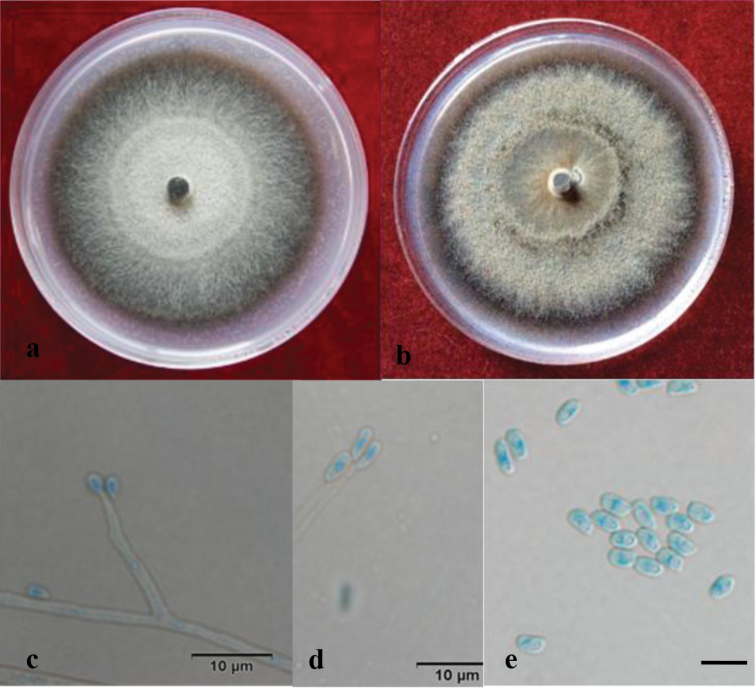
Figure 4.
Light micrographs of Ophiostomamassoniana. a, b Growth on 2% MEA and 2% PDA, 2 weeks after inoculation c–eHyalorhinocladiella-like anamorph, conidiophores, conidia (scale bar, 10 μm).
Etymology.
The epithet reflects the host tree, Pinusmassoniana.
Type.
CHINA, Zhejiang Province, Fuyang City, from Monochamusalternatus gallery in Pinusmassoniana infested by numerous PWN, November 2012, collected by Q Lu and YY Lun, culture ex-holotype, MUCL 55179 = CFCC 51648 = CXY1610.
Description.
Sexual form: Unknown. Asexual form: Hyalorhinocladiella-like. Conidiophores abundant, single, borne on aerial hyphae, (3.3–)10.5–27.5(–42.5) × (0.7–)1.3–2.0(–2.7) μm; conidia hyaline, single-celled, aseptate, obovoid or globose with pointed bases and rounded apices, (2–)2.2–3.9(–5) × (0.5–)0.7–1.7(–2) μm.
Culture characteristics.
Colonies on 2% MEA brown, the marginal hyphae sparse and radiating; some white mycelium produced early during growth that becomes black after 3–5 d. Colonies slowly growing, reaching 37.5 μm in diameter over 8 d at 25 °C, able to grow at 5 °C and 40 °C, with an optimal growth temperature of 30 °C; sporulation weak. On PDA culture medium, the colonies are dark brown; the mycelium is white, long and dense, with a daily growth of 4 μm at 25 °C.
Habitat and distribution.
Galleries of Monochamusalternatus in Pinusmassoniana infested by PWN, in Zhejiang Province, China.
Additional specimens examined.
CHINA, Zhejiang Province, Yuyao City, from Monochamusalternatus galleries in Pinusmassoniana infested by numerous PWN, November 2012, collected by Q Lu and YY Lun, MUCL 55180 = CFCC 51649 = CXY1611.
Note.
Ophiostomamassoniana, only known by its asexual, hyalorhinocladiella-like state, does not cluster in any of the 10 species complexes defined by de Beer and Wingfield (2013) in Ophiostoma s. l. According to the ITS and tub2 phylogenetic analysis, the species is related to O.saponiodorum and O.pallidulum (Figure 1). Ophiostomapallidulum also only produces asexual hyalorhinocladiella-like morphs in vitro, whereas O.saponiodorum produces a sexual and two asexual morphs (pesotum-like and hyalorhinocladiella-like). In addition, O.massoniana differs from O.saponiodorum in producing smaller conidia [(2–)2.2–3.9(–5) × (0.5–)0.7–1.7(–2) μm vs. (3–)4–6(–7) × 1–1.5(–2) μm] (Linnakoski et al. 2010). Further, the colour of O.massoniana colonies is different from that of the other two species. Namely, O.massoniana forms brown to dark brown colonies, while the other two species form pale colonies (Linnakoski et al. 2010).
Discussion
In the current study, six ophiostomatoid species were found associated with pines infected by M.alternatus and PWN in the eastern provinces of Shandong and Zhejiang in China: O.ips, the newly described S.zhejiangensis, O.album, O.massoniana and two species whose identities are uncertain; O.cf.deltoideosporum and Gra.cf.rectangulosporium. Ophiostomaips was the most frequently isolated species, accounting for over 90% of all Ophiostomatales strains.
Ophiostomaips was originally reported in association with bark beetles infecting pines in south-eastern North America (Rumbold 1931). It has been since reported in Central and South America (Mexico and Chile), Europe (Austria and Sweden), Asia (China, Japan and Korea), Africa (South Africa) and Australasia (New Zealand) (Rumbold 1931, Benade et al. 1995, Rane and Tattar 1987, Zhou et al. 2002; Lu et al. 2009, Suh et al. 2013, Zhao et al. 2013; 2014). Furthermore, O.ips is a ubiquitous sapstain fungus associated with PWN and Monochamus spp. (Zhao et al. 2014).
In China, O.ips was reportedly associated with P.massoniana infected by PWN (Zhao 1992, Zhao et al. 2006, 3013) and with P.tabuliformis infected by Dendroctonusvalens (Lu et al. 2009), two invasive pests of the local conifer ecosystems. Zhao et al. (2013) reported O.ips an isolation frequency of 37% in three ophiostomatoid fungal communities associated with PWN, much lower than that reported in the current study.
Ophiostomaips appears to have travelled long-distances in wood materials presumably originating from North America (Zhou et al. 2007). The cited study did not consider any Asian population, however. Nevertheless, the high population density of O.ips in China suggests either indigenous origin or effective adaption after the invasion to local pine forests, with a long evolution history. To verify this hypothesis, it will be necessary to analyse the dispersal routes of PWN populations in different areas globally and of the fungus–including Asian populations.
Members of Sporothrix are reportedly associated with a wide range of habitats (De Hoog 1974, Kwon-Chung and Bennet 1992, Roets et al. 2006, Zhou et al. 2006, Madrid et al. 2009), e.g. wood (Aghayeva et al. 2004), human (de Beer et al. 2016) and the soil (De Meyer et al. 2008). The genus is characterised by reniform ascospores without a mucilaginous sheath and sporothrix- and pesotum-like asexual states (Linnakoski et al. 2010, de Beer et al. 2013). Genetically, the species of the Sporothrix lineages lack the intron 4 but have intron 5 in the BT gene (Zipfel et al. 2006).
Sporothrixzhejiangensis forms an independent lineage according to both ITS and tub2 based on phylogenetic inferences. It is closely related to S.nebulare, S.eucalyptigena, S.epigloea, S.bragantina and S.thermara (Madrid et al. 2010, Romón et al. 1900, Crous et al. 2015, de Beer et al. 2016, Van der Linde et al. 2016) (Figure 1). Sporothrixnebulare was first described after isolation from Hylastesattenuatus infesting P.radiata in Spain (Romón et al. 1900). Sporothrixeucalyptigena was recently isolated from Eucalyptusmarginata (Myrtaceae) in Western Australia (Crous et al. 2015). Sporothrixepigloea was isolated from Tremellafuciformis in Argentina (Upadhyay 1981). S.bragantina was isolated from the rhizosphere soil in Brazil (Pfenning and Oberwinkler 1993) and S.thermara from Cyrtogeniusafricus galleries in diseased Euphorbiaingens trees in South Africa (Van der Linde et al. 2016). Hence, S.zhejiangensis and these five species differ with respect to their (known) hosts and geographic distributions.
Although S.zhejiangensis is unrelated to S.fusiforis, S.lunata and S.stenoceras (Figure 1), these strains exhibit a similar sexual state (Hsiau 1996, Yamaoka et al. 2000, Aghayeva et al. 2004, Zhou et al. 2004). For instance, they all develop one to two perithecial necks emerging from the globular base; occasionally, abnormal specimens of O.stenoceras develop up to five necks in vitro (Yamaoka et al. 2000).
In the current study, S.zhejiangensis was notably different from Sporothrix sp. 1 and Sporothrix sp. 2 (Zhao et al. 2013) with regard to colony characteristics (S.zhejiangensis has a white and radially thinning edge; Sporothrix sp. 1: dark, superficial mycelium; Sporothrix sp. 2: white, radially dense mycelium). Consequently, the role of S.zhejiangensis in PWN needs further research and analysis, ruling out the possibility that the species had been already discovered and its ecological role partially studied.
According to ITS phylogeny analysis, Ophiostomaalbum is related to O.olgensis (Wang et al. 2016) in a single but weakly supported clade (Figure 1). This clade nests within the O.minus complex, in which it is closely related to O.kryptum (Jacobs and Kirisits 2003). The tub2 dataset confirmed that O.album and O.olgensis formed two clades.
The O.minus complex currently includes O.minus, O.pseudotsugae, O.allantosporum, O.kryptum and O.olgensis (Jacobs and Kirisits 2003, Gorton et al. 2004, de Beer and Wingfield 2013, Wang et al. 2016). The tub2 gene of the O.minus complex members includes intron 4 but lacks intron 5 (Gorton et al. 2004). Ophiostomaalbum is phylogenetically closely related to O.olgensis and O.kryptum. Both O.olgensis and O.kryptum inhabit Larix spp. (Jacobs and Kirisits 2003; Wang et al. 2016), whereas O.album inhabits P.massoniana. Both O.olgensis and O.album occur in China, whereas O.kryptum is found in central Europe. Moreover, the three species are associated with different vectors (Jacobs and Kirisits 2003, Wang et al. 2016).
According to both ITS and tub2 phylogenetic trees, O.massoniana forms a separated well-supported clade (Figure 1). It groups with O.pallidulum and O.saponiodorum (Figure 1), which has been isolated from Pinussylvestris in Finland and Piceaabies in Russia in association with various bark beetles (Linnakoski et al. 2010). The three species produce a hyalorhinocladiella-like asexual form (Linnakoski et al. 2010; de Beer et al. 2013) and their tub2 genes lack intron 4 but contain intron 5 (Zipfel et al. 2006).
Conclusions
In the current study, a relatively large number of ophiostomatoid fungal species associated with B.xylophilus and M.alternatus in Shandong and Zhejiang Provinces in China was identified. Three novel species, O.album, O.massoniana and S.zhejiangensis were discovered and described. Fourteen additional provinces in China are currently also listed as PWN epidemic areas (State Forestry Administration of the People’s Republic of China 2018). Hence, additional ophiostomatoid fungi associated with B.xylophilus and M.alternatus should be discovered and described. Future in-depth studies of the biodiversity, biogeography and ecology of fungi associated with pine wilt disease will contribute to the understanding of disease mechanisms and provide information on effective management methods to alleviate the subsequent plant losses.
Supplementary Material
Acknowledgments
This work was supported by the National Key R&D Programme of China (2017YFD0600103) and the National Natural Science Foundation of China (Project No.: 31770682). Cony Decock gratefully acknowledges the financial support received from the Belgian State (Belgian Federal Science Policy through the BCCMTM research programme). We are grateful to Professor Yuichi Yamaoka and Hugo Madrid for their invaluable suggestions to improve the manuscript.
Citation
Wang HM, Lun YY, Lu Q, Liu HX, Decock C, Zhang XY (2018) Ophiostomatoid fungi associated with pines infected by Bursaphelenchus xylophilus and Monochamus alternatus in China, including three new species. MycoKeys 39: 1–27. https://doi.org/10.3897/mycokeys.39.27014
Contributor Information
Quan Lu, Email: luquan@caf.ac.cn.
HuiXiang Liu, Email: hxliu@sdau.edu.cn.
Supplementary materials
Figure S1. Phylogram of fungal associates of pine infected by PWN and Monochamusalternatus in China
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
HuiMin Wang, YingYing Lun, Quan Lu, HuiXiang Liu, Cony Decock, XingYao Zhang
Data type: phylogenetic data
Explanation note: The phylogram was generated after MP analysis of partial tub2 sequences. O.ips sequences obtained in the current study are designated in bold type. MP bootstrap value and BI values are indicated at the branch nodes; values below 70% are indicated by asterisk (*).
Figure S2. Phylograms of fungal associates of pine infected by PWN and Monochamusalternatus in China
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
HuiMin Wang, YingYing Lun, Quan Lu, HuiXiang Liu, Cony Decock, XingYao Zhang
Data type: phylogenetic data
Explanation note: The phylograms were generated after MP analysis of the ITS1–5.8S–ITS2 rDNA and partial tub2 sequences. Novel sequences obtained in the current study are indicated in bold type. MP bootstrap values (10,000 replicates) and ML bootstrap support values (1000 replicates) (normal type) above 70% are indicated at the nodes. Values below 70% are indicated by asterisk (*). Posterior probabilities (above 90%) obtained from BI are indicated by bold lines at the relevant branching points. Scale bar, total nucleotide differences between taxa; ML, maximum likelihood; MP, maximum parsimony; BI, Bayesian inference.
Figure S3. Three ML phylogenetic threes based on tub2 after excluding introns
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
HuiMin Wang, YingYing Lun, Quan Lu, HuiXiang Liu, Cony Decock, XingYao Zhang
Data type: phylogenetic data
References
- Aghayeva DN, Wingfield MJ, Kirisits T, Wingfield BD. (2005) Ophiostomadentifundum sp. nov. from oak in europe, characterized using molecular phylogenetic data and morphology. Mycological Research 109(Pt 10): 1127. 10.3767/003158509X468038 [DOI] [PubMed]
- Aghayeva DN, Wingfield MJ, de Beer ZW, Kirisits T. (2004) Two new Ophiostoma species with Sporothrix anamorphs from Austria and Azerbaijan. Mycologia 96(4): 866–878. 10.2307/3762119 [DOI] [PubMed] [Google Scholar]
- Araujo ML, Rodrigues AM, Fernandes GF, Camargo ZP, Hoog GS. (2015) Human Sporotrichosis beyond the epidemic front reveals classical transmission types in espírito santo, brazil. Mycoses 58(8): 485. 10.1111/myc.12346 [DOI] [PubMed]
- Benade E, Wingfield MJ, Van Wyk PS. (1995) Conidium development in the Hyalorhinocladiella anamorph of Ophiostomaips. Mycologia 87(3): 298–303. 10.2307/3760826 [DOI] [Google Scholar]
- Chang R, Duong TA, Taerum SJ, Wingfield MJ, Zhou X, De Z B. (2017) Ophiostomatoid fungi associated with conifer-infesting beetles and their phoretic mites in yunnan, China. Mycokeys 28(28): 19–64. 10.3897/mycokeys.28.21758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Guarro J, Hernández-Restrepo M, Sutton DA, Acharya K, Barber PA, Boekhout T, Dimitrov RA, Dueñas M, Dutta AK, Gené J, Gouliamova DE, Groenewald M, Lombard L, Morozova OV, Sarkar J, Smith MT, Stchigel AM, Wiederhold NP, Alexandrova AV, Antelmi I, Armengol J, Barnes I, Cano-Lira JF, Castañeda Ruiz RF, Contu M, Courtecuisse PR, da Silveira AL, Decock CA, de Goes A, Edathodu J, Ercole E, Firmino AC, Fourie A, Fournier J, Furtado EL, Geering AD, Gershenzon J, Giraldo A, Gramaje D, Hammerbacher A, He XL, Haryadi D, Khemmuk W, Kovalenko AE, Krawczynski R, Laich F, Lechat C, Lopes UP, Madrid H, Malysheva EF, Marín-Felix Y, Martín MP, Mostert L, Nigro F, Pereira OL, Picillo B, Pinho DB, Popov ES, Rodas Peláez CA, Rooney-Latham S, Sandoval-Denis M, Shivas RG, Silva V, Stoilova-Disheva MM, Telleria MT, Ullah C, Unsicker SB, van der Merwe NA, Vizzini A, Wagner HG, Wong PT, Wood AR, Groenewald JZ. (2015) Fungal Planet description sheets: 320–370. Persoonia 34: 167–266. 10.3767/003158515X688433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer ZW, Wingfield MJ. (2013) Emerging lineages in the Ophiostomatales. In: Seifert KA, de Beer ZW, Wingfield MJ. (Eds) The Ophiostomatoid Fungi: Expanding Frontiers.CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands, 21–46.
- de Beer ZW, Duong TA, Wingfield MJ. (2016) The divorce of Sporothrix and Ophiostoma: solution to a problematic relationship. Studies in Mycology 83: 165–191. 10.1016/j.simyco.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer ZW, Seifert KA, Wingfield MJ. (2013) The ophiostomatoid fungi: Their dual position in the sordariomycetes. In: Seifert KA, de Beer ZW, Wingfield MJ. (Eds) The Ophiostomatoid Fungi: Expanding Frontiers.CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands, 1–19.
- de Hoog GS. (1974) The genera Blastobotrys, Sporothrix, Calcarisporium and Calcarisporiella gen. nov. Studies in Mycology 7: 1–84. [Google Scholar]
- de Meyer EM, de Beer ZW, Summerbell RC, Moharram AM, de Hoog GS, Vismer HF, Wingfield MJ. (2008) Taxonomy and phylogeny of new wood- and soil-inhabiting species in the Ophiostomastenoceras-Sporothrixschenckii complex. Mycologia 100(4): 647–661. 10.3852/07-157R [DOI] [PubMed] [Google Scholar]
- Evans H, McNamara DG, Braasch H, Chadoeuf J, Magnusson C. (1996) Pest Risk Analysis (PRA) for the territories of the European Union (as PRA Area) on Bursaphelenchusxylophilus and its vectors in the genus Monochamus. European and Mediterranean Plant Protection Organization Bulletin 26(2): 199–249. 10.1111/j.1365-2338.1996.tb00594.x [DOI] [Google Scholar]
- Francois R, Wilhelm DBZ, Dreyer LL, Renate Z, Crous P, Wingfield MJ. (2006) Multi-gene phylogeny for Ophiostoma spp. reveals two new species from Protea infructescences. Studies in Mycology 55(1): 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai K. (2013) Pine wood nematode, Bursaphelenchusxylophilus. Annual Review of Phytopathology 51: 61–83. 10.1146/annurev-phyto-081211-172910 [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61(4): 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorton C, Kim SH, Henricot B, Webber J, Breuil C. (2004) Phylogenetic analysis of the bluestain fungus Ophiostomaminus based on partial ITS rDNA and β-tubulin gene sequences. Mycological Research 108: 759–765. 10.1017/S0953756204000012 [DOI] [PubMed] [Google Scholar]
- Grobbelaar JW, Aghayeva DN, Beer ZWD, Bloomer P, Wingfield MJ, Wingfield BD. (2009) Delimitation of Ophiostomaquercus, and its synonyms using multiple gene phylogenies. Mycological Progress 8(3): 221–236. 10.1007/s11557-009-0594-4 [DOI] [Google Scholar]
- Guidon S, Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Harrington TC, Mcnew D, Steimel J, Hofstra D, Farrell R. (2001) Phylogeny and taxonomy of the Ophiostomapiceae complex and the dutch elm disease fungi. Mycologia 93(1): 111–136. 10.2307/3761610 [DOI] [Google Scholar]
- Hsiau PTW. (1996) The taxonomy and phylogeny of the mycangial fungi from Dendroctonusbrevicomis and D.frontalis (Coleoptera: Scolytidae). PhD Thesis, Iowa State University, Ames, Iowa.
- Hausner G, Iranpour M, Kim JJ, Breuil C, Davis CN, Gibb EA, Loewen PC, Hopkin AA. (2005) Fungi vectored by the introduced bark beetle Tomicuspiniperda in Ontario, Canada, and comments on the taxonomy of Leptographiumlundbergii, Leptographiumterebrantis, Leptographiumtruncatum, and Leptographiumwingfieldii. Canadian Journal of Botany 83(10): 1222–1237. 10.1139/b05-095 [DOI] [Google Scholar]
- Hyun MW, Kim JH, Suh DY, Lee SK, Kim SH. (2007) Fungi isolated from pine wood nematode, its vector Japanese pine sawyer, and the nematode-infected Japanese black pine wood in Korea. Mycobiology 35(3): 159–161. 10.4489/MYCO.2007.35.3.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irinyi L, Serena C, Garcia-Hermoso D, Arabatzis M, Desnos-Ollivier M, Vu D, Cardinal G, Arthur I, Normand AC, Giraldo A, da Cunha KC, Sandoval-Denis M, Hendrickx M, Nishikaku AS, de Azevedo Melo AS, Merseguel KB, Khan A, Parente Rocha JA, Sampaio P, da Silva Briones MR, Carmona e Ferreira R, de Medeiros Muniz M, Castanon-Olivares LR, Estrada-Barcenas D, Cassagne C, Mary C, Duan SY, Kong F, Sun AY, Zeng X, Zhao Z, Gantois N, Botterel F, Robbertse B, Schoch C, Gams W, Ellis D, Halliday C, Chen S, Sorrell TC, Piarroux R, Colombo AL, Pais C, de Hoog S, Zancope-Oliveira RM, Taylor ML, Toriello C, de Almeida Soares CM, Delhaes L, Stubbe D, Dromer F, Ranque S, Guarro J, Cano-Lira JF, Robert V, Velegraki A, Meyer W. (2015) International society of human and animal mycology (isham)-its reference DNA barcoding database-the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Medical Mycology 53(4): 313. 10.1093/mmy/myv008 [DOI] [PubMed]
- Ja VDL, Six DL, De Beer WZ, Wingfield MJ, Roux J. (2016) Novel ophiostomatalean fungi from galleries of CyrtogeniusAfricus (Scolytinae) infesting dying EuphorbiaIngens. Antonie Van Leeuwenhoek 109(4): 589–601. 10.1007/s10482-016-0661-1 [DOI] [PubMed] [Google Scholar]
- Jacobs K, Kirisits T. (2003) Ophiostomakryptum sp. nov. from Larixdecidua and Piceaabies in Europe, similar to O.minus. Mycological Research 107(10): 1231–1242. 10.1017/S0953756203008402 [DOI] [PubMed] [Google Scholar]
- Jacobs K, Seifert KA, Harrison KJ, Kirisits T. (2003) Identity and phylogenetic relationships of ophiostomatoid fungi associated with invasive and native Tetropium species (coleoptera: cerambycidae) in Atlantic Canada. Canadian Journal of Botany 81(4): 316–329. 10.1139/b03-025 [DOI] [Google Scholar]
- Jacobs K, Wingfield MJ. (2001) Leptographium Species: tree pathogens, insect associates and agents of blue-stain. American Phytopathological Society Press, St Paul, MN.
- Jankowiak R, KolařÍk M. (2010) Diversity and pathogenicity of ophiostomatoid fungi associated with Tetropium species (coleoptera: cerambycidae) colonizing Piceaabies in Poland. Folia Microbiologica 55(2): 145–154. 10.1007/s12223-010-0022-9 [DOI] [PubMed] [Google Scholar]
- Jung J, Han H, Ryu SH, Kim W. (2010) Microsatellite variation in the pinewood nematode, Bursaphelenchusxylophilus (Steiner and Buhrer) Nickle in South Korea. Genes & Genomics 32(2): 151–158. 10.1007/s13258-009-0842-7 [DOI] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution 30(4): 772–780. 10.1093/molbev/mst010t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Bennett JE. (1992) Medical mycology. Lea & Febiger, Philadelphia, 866 pp. 10.1590/S0036-46651992000600018 [DOI]
- Kyunghee K, Youngjoon C, Sangtae S, Hyeondong S. (2009) Raffaeleaquercus-mongolicae sp. nov. associated with Platypuskoryoensis on oak in Korea. Mycotaxon 110(8): 189–197. [Google Scholar]
- Linnakoski R, de Beer ZW, Ahtiainen J, Sidorov E, Niemelä P, Pappinen A, Wingfield MJ. (2010) Ophiostoma spp. associated with pine-and spruce-infesting bark beetles in Finland and Russia. Persoonia: Molecular Phylogeny and Evolution of Fungi 25: 72. 10.3767/003158510X550845 [DOI] [PMC free article] [PubMed]
- Lu M, Zhou XD, de Beer ZW, Wingfield MJ, Sun JH. (2009) Ophiostomatoid fungi associated with the invasive pine-infesting bark beetle, Dendroctonusvalens, in China. Fungal Divers 38: 133–45. [Google Scholar]
- Madrid H, Cano J, Gene J, Bonifaz A, Toriello C. (2009) Sporothrixglobosa, a pathogenic fungus with widespread geographical distribution. Rev Iberoam Micol 26: 218–222. 10.1016/j.riam.2009.02.005 [DOI] [PubMed] [Google Scholar]
- Madrid H, Gene J, Cano J, Silvera C, Guarro J. (2010) Sporothrixbrunneoviolacea and Sporothrixdimorphospora, two new members of the Ophiostomastenoceras-Sporothrixschenckii complex. Mycologia 102(5): 1193–1203. 10.3852/09-320 [DOI] [PubMed] [Google Scholar]
- Maehara N, Futai K. (1997) Effect of fungal interactions on the numbers of the pinewood nematode, Bursaphelenchusxylophilus (Nematoda: Aphelenchoididae), carried by the Japanese pine sawyer, Monochamusalternatus (Coleoptera: Cerambycidae). Fundamental and applied Nematology 20(6): 611–618. [Google Scholar]
- Mamiya Y, Shoji T. (2009) Pathogenicity of the pinewood nematode, Bursaphelenchusxylophilus, to Japanese larch, Larixkaempferi, seedlings. J. Nematol 41: 157–162. [PMC free article] [PubMed] [Google Scholar]
- Mota M, Vieira P. (2008) Pine wilt disease: a worldwide threat to forest ecosystems. M M. Mota. Springer, Dordrecht, 428 pp. [Google Scholar]
- Massoumi AS, Kim JJ, Humble LM, Uzunovic A, Breuil C. (2007) Ophiostomatoid fungi associated with the Northern Spruce engraver, Ipsperturbatus, in Western Canada. Antonie Van Leeuwenhoek 91(1): 19–34. 10.1007/s10482-006-9092-8 [DOI] [PubMed] [Google Scholar]
- Mota MM, Vieira P. (2008) Pine Wilt Disease: A Worldwide Threat to Forest Ecosystems. Springer, Netherlands. 10.1007/978-1-4020-8455-3 [DOI]
- Mullineux T, Hausner G. (2009) Evolution of rdna its1 and ITS2 sequences and RNA secondary structures within members of the fungal genera Grosmannia and Leptographium Fungal Genetics & Biology Fg & B, 46(11): 855. 10.1016/j.fgb.2009.08.001 [DOI] [PubMed]
- Nkuekam GK, Beer ZWD, Wingfield MJ, Roux J. (2012) A diverse assemblage of Ophiostoma species, including two new taxa on Eucalypt trees in South Africa. Mycological Progress, 11(2): 515–533. 10.1007/s11557-011-0767-9 [DOI] [Google Scholar]
- Ohtaka N, Masuya H, Yamaoka Y, Kaneko S. (2006) Two new Ophiostoma species lacking conidial states isolated from bark beetles and bark beetle-infested Abies species in Japan. Revue Canadienne De Botanique, 84(84): 282–293. 10.1139/B05-164 [DOI] [Google Scholar]
- Pérez-Vera OA, Cárdenas-Soriano E, Alvarado-Rosales D, Cibrián-Tovar D, Equihua-Martínez A. (2011) Histopathology of Hartweg pine (Pinushartwegii lindl.) innoculated with three ophiostomatoid fungi. Revista Chapingo Serie Ciencias Forestales Y Del Ambiente 17(1): 91–102. 10.5154/r.rchscfa.2010.03.006 [DOI] [Google Scholar]
- Pfenning L, Oberwinkler F. (1993) Ophiostomabragantinum n. sp., a possible teleomorph of Sporothrixinflata, found in Brazil. Mycotaxon 46: 381–385. [Google Scholar]
- Plattner A, Jaejin K, Reid J, Hausner G, Youngwoon L, Yamaoka Y, Breuil C. (2009) Resolving taxonomic and phylogenetic incongruence within species Ceratocystiopsisminuta Mycologia, 101(6): 878. 10.3852/08-132 [DOI] [PubMed]
- Posada D, Crandall KA. (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rane KK, Tattar TA. (1987) Pathogenicity of blue-stain fungi associated with Dendroctonusterebrans. Plant Dis 71: 8798–8883. 10.1094/PD-71-0879 [DOI] [Google Scholar]
- Rayner RW. (1970) A mycological colour chart. Commonwealth Mycological Institute and British Mycological Society.
- Robbins K. (1982) Distribution of the pinewood nematode in the United States. In: Appleby JE, Malek RB. (Eds) Proceedings of the national pine wilt disease workshop.III. Natural History Survey, Champaign, IL, USA, 3–6.
- Roets F, Wingfield MJ, Crous PW, Dreyer LL, Savolainen V, Dreyer LL. (2009) Fungal radiation in the cape floristic region: an analysis based on Gondwanamyces and Ophiostoma. Molecular Phylogenetics & Evolution 51(1): 111–119. 10.1016/j.ympev.2008.05.041 [DOI] [PubMed] [Google Scholar]
- Roets F, de Beer ZW, Wingfield MJ, Crous PW, Dreyer LL. (2008) Ophiostomagemellus and Sporothrixvariecibatus from mites infesting Protea infructescences in South Africa. Mycologia 100: 496–510. 10.3852/07-181R [DOI] [PubMed] [Google Scholar]
- Roets F, de Beer ZW, Dreyer LL, Zipfel R, Crous PW, Wingfield MJ. (2006) Multi-gene phylogeny for Ophiostoma spp. reveals two new species from Protea infructescences. Stud Mycol 55: 199–212. 10.3114/sim.55.1.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romón P, Zhou X, Iturrondobeitia JC, Wingfield MJ, Goldarazena A. (2007) Ophiostoma species (ascomycetes: ophiostomatales) associated with bark beetles (coleoptera: Scolytinae) colonizing Pinusradiata in Northern Spain. Canadian Journal of Microbiology 53(6): 756–767. 10.1139/W07-001 [DOI] [PubMed] [Google Scholar]
- Romón P, de Beer ZW, Zhou XD, Duong TA, Wingfield BD, Wingfield MJ. (1900) Multigene phylogenies of Ophiostomataceae associated with Monterey pine bark beetles in Spain reveal three new fungal species. Mycologia 106(1): 119–32. 10.3852/13-073 [DOI] [PubMed] [Google Scholar]
- Rumbold CT. (1931) Two blue-staining fungi associated with bark beetle infestation of pines. Journal of Agricultural Research 43(10): 847–873. [Google Scholar]
- Ryss A, Vieira P, Mota M, Kulinich O. (2005) A synopsis of the genus Bursaphelenchus Fuchs, 1937 (Aphelenchida: Parasitaphelenchidae) with keys to species. Nematology 7(3): 393–458. 10.1163/156854105774355581 [DOI] [Google Scholar]
- Seifert KA, Webber JF, Wingfield MJ. (1993) Methods for studying species of Ophiostoma and Ceratocystis In: Wingfield MJ, Seifert KA, Webber JF, eds. Ceratocystis and Ophiostoma: taxonomy, ecology and pathogenicity. Minnesota, U.S.A.: The American Phytopathological Society Press, 255–259.
- State Forestry Administration of the People’s Republic of China (2018) Announcement of State Forestry Administration of the People’s Republic of China No. 1, 2018.
- Steiner G, Buhrer EM. (1934) Aphelenchoides Xylophilus N. Sp., a Nematode Associated with Blue-Stain and Other Fungi in Timber. Journal of Agricultural Research 48(10): 949–951. [Google Scholar]
- Suh DY, Hyun MW, Kim JJ, Son SY, Kim SH. (2013) 10.5941/MYCO.2013. 41.1.59 [DOI] [PMC free article] [PubMed]
- Swofford DL. (2003) PAUP*: phylogenetic analysis using parsimony, version 4.0 b10.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay HP. (1981) A monograph of Ceratocystis and Ceratocystiopsis. The University of Georgia Press, Athens, GA, USA, 95 pp. [Google Scholar]
- Uzunovic A, Seifert KA, Kim SH, Breuil C. (2000) Ophiostomasetosum, a common sapwood staining fungus from western North America, a new species of the Ophiostomapiceae, complex. Mycological Research 104(4): 486–494. 10.1017/S0953756299001446 [DOI] [Google Scholar]
- Van der Linde JA, Six DL, de Beer WZ, Wingfield MJ, Roux J. (2016) Novel ophiostomatalean fungi from galleries of Cyrtogeniusafricus (Scolytinae) infesting dying Euphorbiaingens. Antonie van Leeuwenhoek 109(4): 589–601. 10.1007/s10482-016-0661-1 [DOI] [PubMed] [Google Scholar]
- Villarreal M, Rubio V, Mtde T, Arenal F. (2005) A new Ophiostoma species isolated from Pinuspinaster in the Iberian peninsula. Mycotaxon 92(3): 259–268. [Google Scholar]
- Wang HM, Lu Q, Meng XJ, Liu XW, Decock C, Zhang XY. (2016) Ophiostomaolgensis, a new species associated with Larix spp. and Ipssubelongatus in northern China. Phytotaxa 282(4): 282–290. 10.11646/phytotaxa.282.4.5 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee SJWT, Taylor JL. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18(1): 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wingfield MJ. (1987) Fungi associated with the pinewood nematode, Bursaphelenchusxylophilus, and cerambycid beetles in Wisconsin. Mycologia 79: 325–328. 10.2307/3807667 [DOI] [Google Scholar]
- Yamaoka Y, Chung WH, Masuya H, Hizai M. (2009) Constant association of ophiostomatoid fungi with the bark beetle Ipssubelongatus, invading Japanese larch logs. Mycoscience, 50(3): 165–172. 10.1007/s10267-008-0468-7 [DOI] [Google Scholar]
- Yamaoka Y, Takahashi I, Iguchi K. (2000) Virulence of ophiostomatoid fungi associated with the spruce bark beetle Ipstypographusf.japonicus in Yezo spruce. Journal of Forest Research 5(2): 87–94. 10.1007/BF02762525 [DOI] [Google Scholar]
- Yan DH, Lu Q, Zhang XY. (2003) Pine Wilt Bursaphelenchusxylophilus (Steiner&Buhrer) Nickle. In: Zhang XY, Luo YQ. (Eds) Major Forest Diseases and Insect Pests in China.Beijing: China Forestry Publishing House, 1–29.
- Yin M, Wingfield MJ, Zhou X, Beer ZWD. (2016) Multigene phylogenies and morphological characterization of five new Ophiostoma spp. associated with Spruce-infesting bark beetles in China. Fungal Biol 120(4): 454–470. 10.1007/s11557-009-0594-4 [DOI] [PubMed] [Google Scholar]
- Zhao L, Sun J. (2017) Pinewood Nematode Bursaphelenchusxylophilus (Steiner and Buhrer) Nickle//Biological Invasions and Its Management in China. Springer, Singapore, 3–21. 10.1007/978-981-10-3427-5_1 [DOI]
- Zhao L, Mota M, Vieira P, Butcher RA, Sun J. (2014) Interspecific communication between pinewood nematode, its insect vector, and associated microbes. Trends in parasitology 30(6): 299–308. 10.1016/j.pt.2014.04.007 [DOI] [PubMed] [Google Scholar]
- Zhao L, Lu M, Niu H, Fang G, Zhang S, Sun J. (2013) A native fungal symbiont facilitates the prevalence and development of an invasive pathogen-native vector symbiosis. Ecology 94(12): 2817–2826. 10.1890/12-2229.1 [DOI] [PubMed] [Google Scholar]
- Zhao BG, Futai K, Sutherland JR, Takeuchi Y. (2008) Pine Wilt Disease. Springer, Tokyo, 144–161. 10.1007/978-4-431-75655-2 [DOI]
- Zhao GH, Chen XY, Wu YZ. (2006) Study on the Fungi from the Diseased Pine Wood Nematode. Journal of Nanjing Forestry University (Natural Sciences Edition) 30(2): 79–81. [Google Scholar]
- Zhao GH. (1992) Two new species of the genus Ceratocystis. Journal of Nanjing Forestry University 16(2): 81–86. [Google Scholar]
- Zhou XD, Burgess T, de Beer ZW, Wingfield BD, Wingfield MJ. (2002) Development of polymorphic microsatellite markers for the tree pathogen and sapstain agent, Ophiostomaips. Molecular Ecology Resources 2(3): 309–312. 10.1046/j.1471-8286.2002.00225.x-i2 [DOI] [Google Scholar]
- Zhou X, Burgess TI, de Beer ZW, Lieutier F, Yart A, Klepzig K, Carnegie A, Portales JM, Wingfield BD, Wingfield MJ. (2007) High intercontinental migration rates and population admixture in the sapstain fungus Ophiostomaips. Molecular Ecology 16(1): 89–99. 10.1111/j.1365-294X.2006.03127.x [DOI] [PubMed] [Google Scholar]
- Zhou X, de Beer ZW, Wingfield MJ. (2006) DNA sequence comparisons of Ophiostoma spp., including Ophiostomaaurorae sp. nov., associated with pine bark beetles in South Africa. Stud Mycol 55: 269–277. 10.3114/sim.55.1.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, de Beer Z, Cibrian D, Wingfield BD, Wingfield MJ. (2004) Characterisation of Ophiostoma species associated with pine bark beetles from Mexico, including O.pulvinisporum sp. nov. Mycological Research 108(Pt 6): 690–698. 10.1017/S0953756204009918 [DOI] [PubMed]
- Zipfel RD, Beer ZW de, Jacobs K, Wingfield BD, Wingfield MJ. (2006) Multigene phylogenies define Ceratocystiopsis and Grosmannia distinct from Ophiostoma. Studies in Mycology 55: 75–97. 10.3114/sim.55.1.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogram of fungal associates of pine infected by PWN and Monochamusalternatus in China
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
HuiMin Wang, YingYing Lun, Quan Lu, HuiXiang Liu, Cony Decock, XingYao Zhang
Data type: phylogenetic data
Explanation note: The phylogram was generated after MP analysis of partial tub2 sequences. O.ips sequences obtained in the current study are designated in bold type. MP bootstrap value and BI values are indicated at the branch nodes; values below 70% are indicated by asterisk (*).
Figure S2. Phylograms of fungal associates of pine infected by PWN and Monochamusalternatus in China
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
HuiMin Wang, YingYing Lun, Quan Lu, HuiXiang Liu, Cony Decock, XingYao Zhang
Data type: phylogenetic data
Explanation note: The phylograms were generated after MP analysis of the ITS1–5.8S–ITS2 rDNA and partial tub2 sequences. Novel sequences obtained in the current study are indicated in bold type. MP bootstrap values (10,000 replicates) and ML bootstrap support values (1000 replicates) (normal type) above 70% are indicated at the nodes. Values below 70% are indicated by asterisk (*). Posterior probabilities (above 90%) obtained from BI are indicated by bold lines at the relevant branching points. Scale bar, total nucleotide differences between taxa; ML, maximum likelihood; MP, maximum parsimony; BI, Bayesian inference.
Figure S3. Three ML phylogenetic threes based on tub2 after excluding introns
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
HuiMin Wang, YingYing Lun, Quan Lu, HuiXiang Liu, Cony Decock, XingYao Zhang
Data type: phylogenetic data