Abstract
Background
Microbial ecosystems that inhabit the human gut form central component of our physiology and metabolism, regulating and modulating both health and disease. Changes or disturbances in the composition and activity of this gut microbiota can result in altered immunity, inflammation, and even cancer.
Aim
To compare the composition and diversity of gut microbiota in stool samples from patient groups based on the site of neoplasm in the gastrointestinal tract (GIT) and to assess the possible contribution of the bacterial composition to tumorigenesis.
Methods
We studied gut microbiota by16S RNA gene sequencing from stool DNA of 83 patients, who were diagnosed with different GIT neoplasms, and 13 healthy individuals.
Results
As compared to healthy individuals, stools of patients with stomach neoplasms had elevated levels of Enterobacteriaceae, and those with rectal neoplasms had lower levels of Bifidobacteriaceae. Lower abundance of Lactobacillaceae was seen in patients with colon neoplasms. Abundance of Lactobacillaceae was higher in stools of GIT patients sampled after cancer treatment compared to samples collected before start of any treatment. In addition to site-specific differences, higher abundances of Ruminococcus, Subdoligranulum and lower abundances of Lachnoclostridium and Oscillibacter were observed in overall GIT neoplasms as compared to healthy controls
Conclusion
Our study demonstrates that the alterations in gut microbiota vary according to the site of GIT neoplasm. The observed lower abundance of two common families, Lactobacillaceae and Bifidobacteriaceae, and the increased abundance of Enterobacteriaceae could provide indicators of compromised gut health and potentially facilitate GIT disease monitoring.
Electronic supplementary material
The online version of this article (10.1007/s10620-018-5190-5) contains supplementary material, which is available to authorized users.
Keywords: Fecal microbiota, Gastrointestinal neoplasms, 16S rRNA gene sequencing
Introduction
Gut bacteria form a diverse and complex microbial ecosystem that plays a vital role in health and disease [1]. Bacteria that belong to the Bacteroidetes and Firmicutes phyla [2, 3] form the predominant part of the human gut microbiota, and along with Proteobacteria, Actinobacteria, Synergistetes, and Fusobacteria constitute the majority of the bacterial species found in human gastrointestinal tract (GIT) [4]. Host and factors such as age, genotype, local environment, and dietary habits exert significant effects on gut microbiota [2, 3, 5] including the development of different types of GIT tumors either through the pro-carcinogenic activities of specific pathogens, or due to the effect of microbial metabolites [6]. Short chain fatty acids and butyrate producing bacteria such as Anaerostipes species and, Butyrivibrio species have been reported to suppress inflammation and inhibit neoplastic changes while other bacterial metabolites such as secondary bile acids can induce inflammation, cause DNA damage, and enhance carcinogenesis [6, 7]. The colonization and diversity of bacteria in different regions of the GIT can vary considerably due to the variation in pH and other physiological factors along the GIT. Moreover, the pathogenicity of bacteria can be different for different regions of the GIT. Helicobacter pylori, for instance, has been associated with increased incidence of gastric cancer as well as with a reduced risk of esophageal cancer [8].
Stool specimens represent a conveniently accessible source for investigating the gut microbiota composition. Studies based on 16S rRNA gene sequence analysis in stool samples have revealed an enrichment of certain bacterial taxa in colorectal cancer (CRC) in conjunction with a depletion of others [9–11]. Bacterial species that have been reported to be linked to CRC include Streptococcus bovis, Bacteroides fragilis, Enterococcus faecalis [9], Clostridium septicum [12], Fusobacterium species [13], and Escherichia coli [14]. Higher abundances of Fusobacterium nucleatum and Bacteroides fragilis have been found to be associated with increased risk of adverse outcomes for CRC, while Faecalibacterium prausnitzii has been associated with a reduced risk [15].
Although the role of H. pylori in gastric carcinoma is well established and it is classified by the International Agency for Research on Cancer as a human carcinogen [1], the knowledge is more limited regarding the diversity of other bacterial species and their functional roles in the development and progression of stomach carcinoma. We here investigated the abundance of gut bacteria in the stool specimens of patients with different GIT malignancies in order to examine differences in taxonomic composition in stool samples based on the location of GIT tumor.
Methods
Patient Population
The study was carried out on stool samples collected from 83 GIT neoplasia patients and 13 healthy individuals (Table 1). The patients were referred to either of the three hospitals: Surgical, Meilahti, and Jorvi in Finland. Three of the authors (AK, MCH, and SK) collected the stool samples from the patients who were referred to them for surgery. All the patients and controls were of Finnish origin. Samples from 63 patients were obtained at the time of diagnosis, before start of any kind of treatment, while samples from 20 patients (13 with rectum, six with stomach, and one with small intestine neoplasms) were obtained after the patients had already received treatment, either chemotherapy and/or radiotherapy. These 20 samples from previously treated patients were categorized as a separate group (treated) regardless of tumor type/location, while the remaining 63 samples from non-treated patients were classified into five groups according to site of tumor as: stomach, small intestine, pancreas, colon, and rectum (Table 1). Patients with stomach neoplasms included seven with GIST, ten of intestinal type, 13 of diffuse type, and five others.
Table 1.
Characteristics of patients studied for stool microbiota analysis
| Tumor site | Av. age in years (range) | Gender | Total | |
|---|---|---|---|---|
| Male | Female | |||
| Controlsb | 43.8 (19–65) | 3 | 10 | 13 |
| Stomach | 69.4 (36–98) | 17 | 18 | 35 |
| Pancreas | 62.3 (57–67) | 1 | 2 | 3 |
| Small intestine | 61.5 (39–79) | 3 | 0 | 3 |
| Colon | 74.8 (64–84) | 6 | 7 | 13 |
| Rectum | 73.6 (39–85) | 7 | 2 | 9 |
| Treateda | 66.9 (53–78) | 11 | 9 | 20 |
aTreated group includes samples from those patients with gastrointestinal neoplasm, who had already undergone cancer treatment at the time of sample collection
bControls include samples from healthy individuals without any gastrointestinal disease
The study was approved by the Hospital District of Helsinki and Uusimaa (HUS) review board (ethical permission number 351/13/03/02/2014). Written informed consent was obtained from all subjects.
Stool Sample Collection
Stool samples were collected in special tubes, provided in the PSP Spin Stool DNA Plus Kit (STRATEC Biomedical AG, Germany). One spoon of stool specimen (spoon provided with the collection tubes) was transferred to the tube and mixed thoroughly to obtain a stool homogenate, followed by immediate freezing at -20 °C until DNA extraction.
Stool DNA Extraction
DNA was extracted from 1.4 ml of each stool homogenate using the PSP Spin Stool DNA Plus Kit (STRATEC Biomedical AG, Germany), according to the manufacturer’s instructions. DNA was quantified by Qubit 2.0 fluorometer (Thermo Fisher Scientific, USA) using the Qubit dsDNA BR assay kit. The extracted DNA was then stored at − 20 °C.
16S rRNA Gene Sequencing
Library Preparation
Libraries for sequencing were prepared with Ion 16S Metagenomics kit (Thermo Fisher Scientific, USA) according to supplied protocol. For each sample, two primer pools were used to amplify six hypervariable regions (primer set V2, V4, V8 and primer set V3, V6–7, and V9) of 16S rRNA gene. A volume of 1 µl of each sample DNA (3 ng/µl) was used for library preparation, and PCR was performed according to the kit’s instructions with 18-cycle PCR protocol (two reactions/sample). After PCR, the samples were purified with Agencourt AMPure XP beads (Beckman Coulter) according to kit’s protocol. Samples were end-repaired, purified with Agencourt AMPure XP beads, and the bar-coded sequencing adapters were ligated following the manufacturer’s protocol. After ligation, the libraries were purified with Agencourt AMPure XP beads and quantified in the TapeStation 4200 instrument (Agilent Technologies). Sample dilution factors were determined according to TapeStation results, and libraries were diluted in low TE (26 µM Tris, 2.6 µM EDTA) to 10 µM concentration.
Template Preparation and Sequencing
Before sequencing, the template preparation for library pools was performed either with Ion OneTouch 2 system (Thermo Fisher Scientific) using the Ion PGM™ Hi-Q™ OT2 Kit (Thermo Fisher Scientific) or Ion Chef system (Thermo Fisher Scientific) using the Ion PGM™ Hi-Q™ Chef Kit following the kit protocols. 2 µl of each 10 pM library was used for library pool, and 15–20 libraries were pooled together. 20 µl of a library pool was mixed with 5 µl of nuclease-free water and added to the amplification solution for template preparation. After the template preparation, the quality of resulting ion spheres was checked with Qubit 3.0 fluorometer (Thermo Fisher Scientific). Following quality check, the ion spheres were loaded on an Ion 318™ Chip (Thermo Fisher Scientific) and sequenced with Ion PGM system using Ion PGM Hi-Q Sequencing kit (Thermo Fisher Scientific) according to the protocol provided with the kit.
Data Analysis
Sequencing data from stool samples of 83 patients and 13 controls were used to create OTU (operational taxonomic unit) abundance tables. Between-sample normalization was done by rarifying the sequencing counts into even depth with the phyloseq R package [16] and subsequently converting the rarified read counts to relative abundances. The number of unique detected taxa included 105 families and 121 genera.
Gut microbiota community alpha diversity and observed richness were analyzed at the family and genus levels using the microbiome [17] and vegan R packages [18]. Community richness and diversity were quantified by the number of unique observed taxa and Shannon index, respectively. Significance of the group-level differences was estimated with Kruskal–Wallis test. Multiple testing correction was done separately for each group of analyses based on the Benjamini–Hochberg FDR correction [19]. The samples were grouped based on relative abundances of the taxonomic groups using hierarchical clustering (ward.D2 method in the hclust function) with Bray–Curtis distance.
Ordination with the unsupervised principal coordinates analysis (PCoA), as implemented in the phyloseq R package [16], is based on Euclidean distance between Hellinger-transformed abundance profiles [20]. Only the core genera or families that were detected in at least 20% of all samples were included in the analysis. Significance of the community-level differences between the groups was assessed with PERMANOVA for clr-transformed abundances [21] to remove compositionality bias, with the R package compositions [22]. The significance for the differences in the abundance of individual taxa was assessed with ANCOM [23], which has been recently demonstrated to reduce false discovery rate (FDR) compared to other alternatives [23, 24]. The graphs were generated with ggplot2 [25].
Results
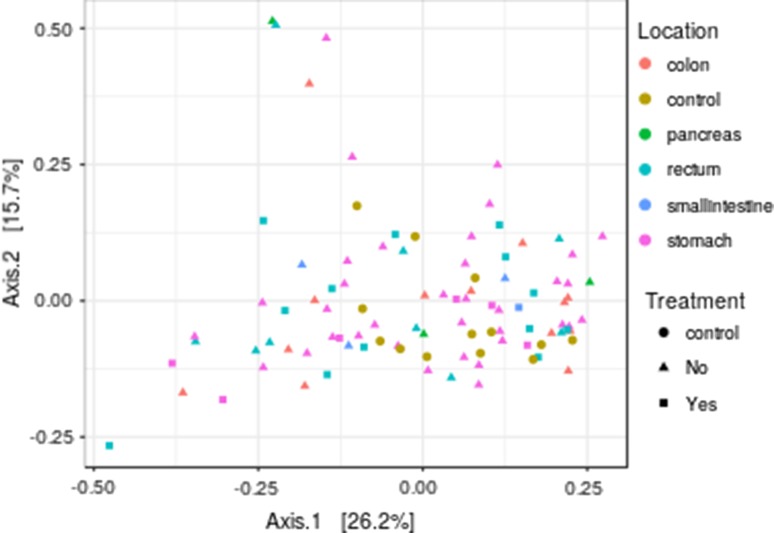
The patients were divided into five non-treated and one treated groups, and the relative abundances of the genera and families in each group were compared to those of the control group. The sample similarities (beta diversities) across the different groups based on genus-level profiles are illustrated with principal coordinates analysis in Fig. 1.
Fig. 1.
Principal coordinates analysis showing beta diversities across different gastrointestinal neoplasm groups based on genus-level bacterial profiles
Bacterial Diversity
Alpha diversity (Shannon index) and observed richness did not have significant differences between the groups (based on location of neoplasm) at the family or genus levels (adjusted Kruskal–Wallis p = 0.21 for both levels). The genus-level pairwise comparison between the groups with respect to observed richness and Shannon is shown in Fig. 2.
Fig. 2.
Observed richness and alpha diversity of bacteria at the genus level. Median relative abundance for the control group is shown by dashed line along y-axis. Adjusted p values for pairwise comparisons between gastrointestinal neoplasm groups and controls at the bacterial genus level are shown for each group
Beta diversity was not significantly different between the controls and the patients, either for the treated or the non-treated groups. Adjusted p values for pairwise comparisons at genus level were not significant in the treated/non-treated, treated/control, and non-treated/control pairwise comparisons (p > 0.2 in all comparisons).
Comparison of Each Group with the Controls
Family Level
Bacteria belonging to Enterobacteriaceae family were found to have significantly higher abundances in stools of patients with neoplasms of the stomach or the small intestine than in controls. The relative abundance of bacteria from the Lactobacillaceae family was significantly lower in the group of patients that had colon or pancreatic neoplasms, while that of Acidaminococcaceae was significantly lower than in controls only in colonic neoplasm patients (Table 2). Patients with rectal neoplasms had a significantly lower relative abundance of Bifidobacteriaceae in their stool samples than in controls. Relative abundance of bacterial families in different patient groups is shown in Supplementary Figure 1.
Table 2.
Bacteria with significant difference in relative abundance in stools of patients groups based on location of neoplasm in the gastrointestinal tract compared to healthy individuals
| Patient group | Relative abundance in patient group (%) | Relative abundance in controls (%) | Log10FC | |
|---|---|---|---|---|
| Family | ||||
| Enterobacteriaceae | Stomach | 5.1 | 0.2 | 1.5 |
| Lactobacillaceae | Pancreas | 0.0 | 1.9 | NAb |
| Enterobacteriaceae | Small intestine | 5.2 | 0.2 | 1.5 |
| Lactobacillaceae | Colon | 0.1 | 1.9 | − 1.1 |
| Acidaminococcaceae | Colon | 0.1 | 0.9 | − 1.0 |
| Bifidobacteriaceae | Rectum | 0.0 | 0.2 | NAb |
| Enterobacteriaceae | All non-treated | 5.5 | 0.2 | 1.5 |
| Bifidobacteriaceae | Treated | 0.2 | 0.2 | − 0.1 |
| Lactobacillaceae | Treated versus non-treated | 1.1 | 0.7a | 0.2 |
| Genus | ||||
| Ruminococcus | Stomach | 2.9 | 0.8 | 0.6 |
| Subdoligranulum | Stomach | 0.2 | 0.0 | 1.4 |
| Lachnoclostridium | Stomach | 0.4 | 0.6 | − 0.2 |
| Oscillibacter | Stomach | 0.1 | 0.2 | − 0.6 |
| Parabacteroides | Pancreas | 0.5 | 5.0 | − 1.0 |
| Lachnoclostridium | Small intestine | 0.10 | 0.6 | − 1.0 |
| Subdoligranulum | Colon | 0.1 | 0.0 | 1.1 |
| Lachnoclostridium | Colon | 0.2 | 0.6 | − 0.5 |
| Oscillibacter | Colon | 0.0 | 0.2 | − 1.0 |
| Bifidobacterium | Rectum | 0.0 | 0.4 | − 1.8 |
| Ruminococcus | All non-treated | 2.9 | 0.8 | 0.6 |
| Subdoligranulum | All non-treated | 0.2 | 0.0 | 1.4 |
| Lachnoclostridium | All non-treated | 0.4 | 0.6 | − 0.1 |
| Oscillibacter | All non-treated | 0.1 | 0.2 | − 0.6 |
| Ruminiclostridium | Treated | 0.0 | 0.4 | − 1.7 |
| Lachnoclostridium | Treated | 0.3 | 0.6 | − 0.4 |
| Oscillibacter | Treated | 0.0 | 0.2 | − 0.7 |
| Lactobacillus | Treated versus non-treated | 0.9 | 0.7a | 0.1 |
aRelative abundance in non-treated patients
bFold change is not shown where one of the groups has zero average abundance
Genus Level
The relative abundances of Ruminococcus and Subdoligranulum were significantly higher while those of Lachnoclostridium and Oscillibacter were significantly lower in patients with stomach neoplasms, compared to healthy individuals (Table 2). Similar to what was seen in stomach neoplasms, the relative abundance of Subdoligranulum was also significantly higher and that of Lachnoclostridium and Oscillibacter significantly lower in patients with colon neoplasms. Moreover, significantly lower relative abundance of Lachnoclostridium was also observed in patients with neoplasms of the small intestine. Samples from rectal neoplasia patients had lower relative abundance of Bifidobacterium, whereas samples from pancreatic neoplasia patients showed reduced abundance of Parabacteroides as compared to controls. Relative abundance of bacterial genera in different patient groups is shown in Supplementary Fig. 2.
Comparison of the Treated Group with the Non-treated Neoplasm Group
Lactobacillaceae at the family level and Lactobacillus at the genus level had higher relative abundance in the treated group compared to samples from the non-treated group (Table 2).
Comparison of All Neoplasms with the Controls
The Enterobacteriaceae family had a significantly higher abundance in the non-treated group than in the control group. At the genus level, Lachnoclostridium and Oscillibacter had significantly lower abundance, and Ruminococcus, and Subdoligranulum had significantly higher abundance in the non-treated group as compared to the control group. Compared to controls, the treated group (irrespective of tumor site) had significantly lower abundance of Ruminiclostridium, Lachnoclostridium and Oscillibacter at the genus level.
Discussion
We assessed and compared the stool bacterial profile of patients with GIT neoplasms grouped according to the neoplasm location, with stomach, colon, and rectum being the major groups. In addition to the altered abundances of certain bacterial taxonomic groups seen in the overall group of patients with non-treated GIT neoplasms as compared to controls, further differences were observed depending on the neoplasm location (Table 2).
At the family level, Enterobacteriaceae had significantly higher relative abundance in all non-treated patients, while based on the site of neoplasm, significantly higher abundance of Enterobacteriaceae was observed only in patients with neoplasms of the stomach or small intestine (Table 2). Lower abundances were noted for Lactobacillaceae and Acidaminococcaceae in patients with colon neoplasms, as well as for Bifidobacteriaceae in rectal neoplasms and (Table 2). The family Enterobacteriaceae includes many pathogenic bacteria, in addition to commensal ones, and gut inflammation is thought to initiate increase in Enterobacteriaceae abundance. Previous studies reviewed in Chen et al. [26] have shown increase in pathogenic bacteria together with depletion of normal healthy gut microbiota associated with colorectal cancer. In our study, the increased abundance of this family of bacteria was, however, significant only in patients with neoplasms of the stomach and small intestine.
Lactobacillaceae and Bifidobacteriaceae are the two most prominent families of probiotics that play an important role in the maintenance of GIT homeostasis. The loss of abundance of these bacteria is reported in CRC, while their administration is reported to have a protective effect on CRC development as reviewed in Zou et al. [27]. Lactobacilli are thought to prevent the development of cancer and cancer cell migration and have been reported to inhibit the growth of human colon carcinoma cells [28]. A combination of probiotic Bifidobacterium lactis and prebiotic resistant starch has been shown to prevent the development of CRC in rats and has been proposed as a chemopreventive approach for CRC [29].
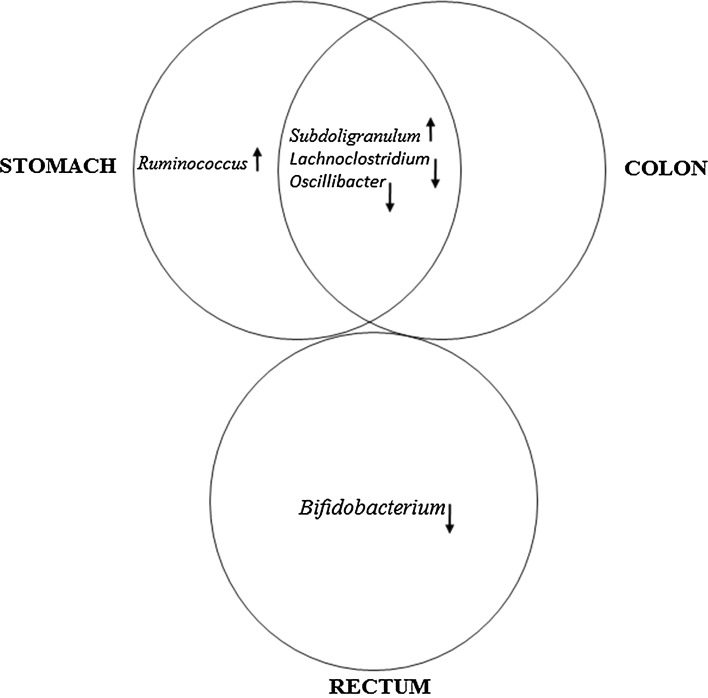
The majority of previous studies have considered colorectal cancer as a single group, and there is little information related to bacterial dysbiosis in the colon and rectal cancers separately. In the present study, we found reduced abundance of Lactobacillaceae related to neoplastic growth in colon, while reduced abundance of Bifidobacteriaceae was related to neoplasms of the rectum. Intriguingly, we also observed that at the genus level, while alterations in bacteria were similar between colon and stomach neoplasms (higher abundance of Subdoligranulum; lower abundance of Lachnoclostridium and Oscillibacter), there were no commonly altered bacteria between colon and rectal neoplasms (Fig. 3). This again suggests that there is a distinct difference in the effect of bacterial dysbiosis on neoplastic growth between colon and rectum. Although colorectal tumors are usually considered similar, there are few marked differences between the two types of tumors. At a molecular level, rectal tumors have higher rate of aneuploidy, loss of heterozygosity and TP53 mutations while colon cancers have higher microsatellite instability and more frequent CpG island methylator phenotype (reviewed in Iacopetta 2002) [30]. Bifidobacterium contributes significantly to de novo biosynthesis of folate in gut and to folate store in colon [31]. Humans cannot synthesize folate and are dependent on external sources (from food or bacterial synthesis), and the deficiency of folate can result in chromosomal instability [32, 33] and susceptibility for childhood leukemia [34]. Reduced levels of Bifidobacterium could result in decreased folate levels in the large intestine and thus increase the risk of aneuploidies associated with rectal neoplasms.
Fig. 3.
Comparison of different gastrointestinal neoplasm groups with respect to altered relative abundance of bacteria at the genus level. Elevated and reduced levels are indicated by direction of the arrows
Gut bacteria are most abundant in colon compared to other regions of the GIT, and disturbances in bacterial abundances would be expected to associate most closely with neoplastic growth located in the colorectal region. However, in our study, there were more bacteria genera altered in gastric neoplasms, which are similar to those detected in colon neoplasms, than in rectal neoplasms. Interestingly, the bacterial genera associated with gastric or colon neoplasms are related to inflammation, or metabolic diseases. Increased levels of Ruminococcus and Subdoligranulum have been reported in the stools of children with food sensitivity compared to healthy children [35]. Subdoligranulum has been found to be associated with chronic inflammation and poor metabolic control [36]. Its levels have been reported to be associated with blood markers of inflammation (C-reactive protein, CRP) and endotoxemia (lipopolysaccharide-binding protein, LBP) in Type 1 diabetes [37]. Subdoligranulum is also reported to inhibit fermentation of inulin by bifidogenic bacteria in colon, which is considered beneficial in the prevention of colon cancer [38]. Oscillibacter are known to produce anti-inflammatory metabolites and have effect on the maintenance of gut barrier integrity in mice [39]. Their abundance is reported to be affected by the presence of other gut bacteria [5]. Reduced abundance of Lachnoclostridium has been reported in stools of Hashimoto’s thyroiditis patients [40]. Increased abundance of Ruminococcus, seen exclusively in gastric neoplasms in our study, is reported in prediabetic patients and associated with impaired fasting plasma glucose, BMI, and waist circumference [41]. Moreover, their higher abundance has been reported in stools of high fat diet-induced obese rats compared to lean rats [42].
On the other hand, in patients with rectal neoplasms, Bifidobacterium had significantly lower abundance as compared to healthy individuals in our study. These bacteria have a role in maintenance of healthy gut bacterial profile by inhibiting growth of pathogens by competitive exclusion; immune function; breakdown of indigestible food component by enzyme secretion; and folate synthesis in gut (reviewed in O’Callaghan et al. 2016) [43].
In patients with neoplasms of the small intestine, higher abundance of Enterobacteriaceae and lower abundance of Lachnoclostridium were similar to those seen in stomach. Patients with pancreatic neoplasms showed, in our study, a similarly low abundance of Lactobacillaceae as seen in those with colon neoplasms and also decreased abundance of Parabacteroides. Gut Parabacteroides distasonis is reported to have anti-inflammatory and anticancer role, acting through reducing TLR4 signaling/Akt activation in mice [44], and TLRs are known to play a significant role in various pancreatic diseases, including pancreatic cancer [45]. Robust conclusions could not be drawn regarding the pancreatic and small intestinal neoplasm groups due to their small sample size.
The gut microbiota is diverse and highly individual [46]. Changes in microbiota richness have been reported in association with cancer development [47], whereas reduced alpha diversity and richness in the gut microbiota have been linked with a Western or urban lifestyle [7] and compromised health in humans [48], as well as with colon cancer in mouse models [49]. There are also reports of increased richness and diversity at the taxonomic and functional levels linked to gastrointestinal cancers [50]. We did not observe significant differences among the groups with respect to alpha diversity or taxonomic richness. The increased levels of Enterobacteriaceae and Subdoligranulum in overall GIT neoplasms could, however, indicate increase in more pathogenic bacteria in GIT neoplasms. Altered abundance of most of the bacterial genera seen in individual gastric, colon, or rectal neoplasm groups was also seen in the overall non-treated GIT neoplasm group, suggesting that a similar pattern of changes in bacterial abundance is related to development of neoplastic growth in GIT with certain taxa having more profound effect in certain specific GIT locations. Chronic inflammation, obesity, and high BMI are some of the common risk factors associated with gastric and colon neoplasms.
Whereas the heterogeneity of histological subtypes of the tumors and the small sample size of each subtype form limitations for statistical conclusions, this is, to the best of our knowledge, the first study investigating the differences in microbiota in different GIT tumor locations. The number of controls was 13, which is similar to the number of patients in each neoplasm subgroup. One limitation in our study is that the healthy control group is on average younger than the patient groups. Therefore, some of the differences in the healthy controls could be associated with their younger mean age. However, the different patient groups had relatively similar mean ages, and hence not a likely confounder in our key analysis task, which was the analysis of differences between the taxonomic compositions in fecal samples between the different patient groups.
Our comparison of bacterial profiles in the patient groups (irrespective of tumor site) collected before start of any treatment (non-treated) and those collected after treatment showed higher abundance of Lactobacillaceae at family level and Lactobacillus at genus level in the treated group compared to the whole non-treated group. Since the abundance of Lactobacillaceae was significantly lower in colon cancer patients and those with pancreatic cancer, the higher level of Lactobacillaceae in treated patients compared to non-treated patients could indicate a recovery or growth of normal beneficial bacteria after the treatment. Even though treatment types in the treated neoplasm group were heterogeneous, it is tempting to speculate that the results may indicate efficacy of the treatment regardless of type of treatment, tumor location, or histological subtype of the tumor.
In conclusion, we found significant differences in the abundances of specific bacterial taxonomic groups in stool specimens from patients with various GIT neoplasms; the differences were dependent on the location of neoplasia in the GIT. This finding could be of significance in the future as a tool for assessing neoplastic alterations in different parts of the GIT. Studies on the stool abundances of Lactobacillaceae, Bifidobacteriaceae, and Enterobacteriaceae could potentially lead to the development of a noninvasive approach to GIT disease monitoring and treatment follow-up.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1: Relative abundance of the bacterial families that were significantly different in any of the gastrointestinal neoplasm groups (marked with asterisk) as compared to controls. The number of patients in different groups is: control = 13, stomach = 35, pancreas = 3, small intestine = 3, colon = 13, rectum = 9, treated = 20. Median relative abundance for the control group is shown by dashed line along y-axis. (JPEG 68 kb)
Supplementary Figure 2: Relative abundance of the bacterial genera that were significantly different in any of the gastrointestinal neoplasm group (marked with asterisk) as compared to controls. The number of patients in different groups is: control = 13, stomach = 35, pancreas = 3, small intestine = 3, colon = 13, rectum = 9, treated = 20. Median relative abundance for the control group is shown by dashed line along y-axis. (JPEG 91 kb)
Acknowledgment
This work was supported by Sigrid Jusélius Finnish Foundation and Valtion tutkimusraha of Finland, No. VTR TYH2016244. LL was supported by Academy of Finland (Grants 307127 and 295741).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Omar Youssef, Email: omar.youssef@helsinki.fi.
Leo Lahti, Email: leo.lahti@iki.fi.
Arto Kokkola, Email: arto.kokkola@hus.fi.
Tiina Karla, Email: tiina.karla@thermofisher.com.
Milja Tikkanen, Email: milja.tikkanen@thermofisher.com.
Homa Ehsan, Email: h.ehsan3@gmail.com.
Monika Carpelan-Holmström, Email: monika.carpelan-holmstrom@hus.fi.
Selja Koskensalo, Email: selja.koskensalo@hus.fi.
Tom Böhling, Email: tom.bohling@helsinki.fi.
Hilpi Rautelin, Email: hilpi.rautelin@medsci.uu.se.
Pauli Puolakkainen, Email: pauli.puolakkainen@hus.fi.
Sakari Knuutila, Phone: +358 504482797, Email: sakari.knuutila@helsinki.fi.
Virinder Sarhadi, Email: virinder.sarhadi@helsinki.fi.
References
- 1.Gagnière J, Raisch J, Veziant J, et al. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. 2016;22:501–518. doi: 10.3748/wjg.v22.i2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falony G, Joossens M, Vieira-Silva S, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 3.Zhernakova A, Kurilshikov A, Bonder MJ, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Browne HP, Neville BA, Forster SC, Lawley TD. Transmission of the gut microbiota: spreading of health. Nat Rev Microbiol. 2017;15:531–543. doi: 10.1038/nrmicro.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker AW, Ince J, Duncan SH, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 7.O’Keefe SJD, Li JV, Lahti L, et al. Fat, fiber and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castro C, Peleteiro B, Lunet N. Modifiable factors and esophageal cancer: a systematic review of published meta-analyses. J Gastroenterol. 2018;53:37–51. doi: 10.1007/s00535-017-1375-5. [DOI] [PubMed] [Google Scholar]
- 9.Wang T, Cai G, Qiu Y, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flemer B, Lynch DB, Brown JM, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66:633–643. doi: 10.1136/gutjnl-2015-309595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirza NN, McCloud JM, Cheetham MJ. Clostridium septicum sepsis and colorectal cancer—a reminder. World J Surg Oncol. 2009;7:73. doi: 10.1186/1477-7819-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei Z, Cao S, Liu S, et al. Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients’ survival? A pilot study on relevant mechanism. Oncotarget. 2016;7:46158–46172. doi: 10.18632/oncotarget.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lahti L, Shetty S, Blake T et al. Tools for microbiome analysis in R. Microbiome Package Version 0.99.88; 2012–2017. https://github.com/microbiome/microbiome.
- 18.Oksanen J, Blanchet FG, Friendly M, et al. Vegan: community ecology package; 2017. https://cran.r-project.org/web/packages/vegan/index.html.
- 19.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 20.Legendre P, Gallagher ED. Ecologically meaningful transformations for ordination of species data. Oecologia. 2001;129:271–280. doi: 10.1007/s004420100716. [DOI] [PubMed] [Google Scholar]
- 21.Aitchison J. The statistical analysis of compositional data. In: Monographs on Statistics and Applied Probability. Chapman & Hall Ltd: London; 1986. http://dl.acm.org/citation.cfm?id=17272.
- 22.van den Boogaart KG, Tolosana R, Bren M. Compositions: compositional data analysis; 2014. https://cran.r-project.org/web/packages/compositions/index.html.
- 23.Mandal S, Van Treuren W, White RA. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis. 2015;26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss S, Xu ZZ, Peddada S, et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wickham H. ggplot2. Elegant Graphics for Data Analysis. Springer: New York; 2009. ISBN: 978-0-387-98140-6. http://ggplot2.org.
- 26.Chen J, Domingue JC, Sears CL. Microbiota dysbiosis in select human cancers: evidence of association and causality. Semin Immunol. 2017;32:25–34. doi: 10.1016/j.smim.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou S, Fang L, Lee M-H. Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol Rep. 2018;6:1–12. doi: 10.1093/gastro/gox031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen ZY, Hsieh YM, Huang CC, Tsai CC. Inhibitory effects of probiotic lactobacillus on the growth of human colonic carcinoma cell line HT-29. Molecules. 2017;22:E107. doi: 10.3390/molecules22010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Leu RK, Hu Y, Brown IL, Woodman RJ, Young GP. Synbiotic intervention of Bifidobacterium lactis and resistant starch protects against colorectal cancer development in rats. Carcinogenesis. 2010;31:246–251. doi: 10.1093/carcin/bgp197. [DOI] [PubMed] [Google Scholar]
- 30.Iacopetta B. Are there two sides to colorectal cancer? Int J Cancer. 2002;101:403–408. doi: 10.1002/ijc.10635. [DOI] [PubMed] [Google Scholar]
- 31.Rossi M, Amaretti A, Raimondi S. Folate production by probiotic bacteria. Nutrients. 2011;3:118–134. doi: 10.3390/nu3010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knuutila S, Helminen E, Vuopio P, De La Chapelle A. Increased sister chromatid exchange in megaloblastic anaemia-studies on bone marrow cells and lymphocytes. Hereditas. 1978;89:175–181. doi: 10.1111/j.1601-5223.1978.tb01273.x. [DOI] [PubMed] [Google Scholar]
- 33.Guo X, Ni J, Zhu Y, et al. Folate deficiency induces mitotic aberrations and chromosomal instability by compromising the spindle assembly checkpoint in cultured human colon cells. Mutagenesis. 2017;32:547–560. doi: 10.1093/mutage/gex030. [DOI] [PubMed] [Google Scholar]
- 34.Cantarella CD, Ragusa D, Giammanco M, Tosi S. Folate deficiency as predisposing factor for childhood leukaemia: a review of the literature. Genes Nutr. 2017;12:14. doi: 10.1186/s12263-017-0560-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen CC, Chen KJ, Kong MS, Chang HJ, Huang JL. Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr Allergy Immunol. 2016;27:254–262. doi: 10.1111/pai.12522. [DOI] [PubMed] [Google Scholar]
- 36.Chumpitazi BP, Hollister EB, Oezguen N, et al. Gut microbiota influences low fermentable substrate diet efficacy in children with irritable bowel syndrome. Gut Microbes. 2014;5:165–175. doi: 10.4161/gmic.27923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Groot PF, Belzer C, Aydin Ö, et al. Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS ONE. 2017;12:e0188475. doi: 10.1371/journal.pone.0188475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Martínez I, Walter J, Keshavarzian A, Rose DJ. In vitro characterization of the impact of selected dietary fibers on fecal microbiota composition and short chain fatty acid production. Anaerobe. 2013;23:74–81. doi: 10.1016/j.anaerobe.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 39.Lam YY, Ha CWY, Campbell CR, et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS ONE. 2012;7:e34233. doi: 10.1371/journal.pone.0034233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao F, Feng J, Li J, et al. Alterations of the gut microbiota in Hashimoto’s thyroiditis patients. Thyroid. 2018;28:175–186. doi: 10.1089/thy.2017.0395. [DOI] [PubMed] [Google Scholar]
- 41.Allin KH, Tremaroli V, Caesar R, et al. Aberrant intestinal microbiota in individuals with prediabetes. Diabetologia. 2018;61:810–820. doi: 10.1007/s00125-018-4550-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiao N, Baker SS, Nugent CA, et al. Gut microbiome may contribute to insulin resistance and systemic inflammation in obese rodents: a meta-analysis. Physiol Genomics. 2018 doi: 10.1152/physiolgenomics.00114.2017. [DOI] [PubMed] [Google Scholar]
- 43.O’Callaghan A, van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. 2016;7:925. doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koh GY, Kane A, Lee K, et al. Parabacteroides distasonis attenuates toll-like receptor 4 signaling and Akt activation and blocks colon tumor formation in high-fat diet-fed azoxymethane-treated mice. Int J Cancer. 2018 doi: 10.1002/ijc.31559. [DOI] [PubMed] [Google Scholar]
- 45.Santoni M, Andrikou K, Sotte V, et al. Toll like receptors and pancreatic diseases: from a pathogenetic mechanism to a therapeutic target. Cancer Treat Rev. 2015;41:569–576. doi: 10.1016/j.ctrv.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 46.Shetty SA, Hugenholtz F, Lahti L, Smidt H, de Vos WM. Intestinal microbiome landscaping: insight in community assemblage and implications for microbial modulation strategies. FEMS Microbiol Rev. 2017;41:182–199. doi: 10.1093/femsre/fuw045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Chatelier HL. Hawley’s Condensed Chemical Dictionary. 15. Hoboken: Wiley; 2007. [Google Scholar]
- 49.Zackular JP, Baxter NT, Iverson KD, et al. The gut microbiome modulates colon tumorigenesis. MBio. 2013;4:e00692-13. doi: 10.1128/mBio.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Relative abundance of the bacterial families that were significantly different in any of the gastrointestinal neoplasm groups (marked with asterisk) as compared to controls. The number of patients in different groups is: control = 13, stomach = 35, pancreas = 3, small intestine = 3, colon = 13, rectum = 9, treated = 20. Median relative abundance for the control group is shown by dashed line along y-axis. (JPEG 68 kb)
Supplementary Figure 2: Relative abundance of the bacterial genera that were significantly different in any of the gastrointestinal neoplasm group (marked with asterisk) as compared to controls. The number of patients in different groups is: control = 13, stomach = 35, pancreas = 3, small intestine = 3, colon = 13, rectum = 9, treated = 20. Median relative abundance for the control group is shown by dashed line along y-axis. (JPEG 91 kb)