Abstract
Aims/hypothesis
The study aimed to determine whether discrete subtypes of type 1 diabetes exist, based on immunoregulatory profiles at clinical onset, as this has significant implications for disease treatment and prevention as well as the design and analysis of clinical trials.
Methods
Using a plasma-based transcriptional bioassay and a gene-ontology-based scoring algorithm, we examined local participants from the Children’s Hospital of Wisconsin and conducted an ancillary analysis of TrialNet CTLA4-Ig trial (TN-09) participants.
Results
The inflammatory/regulatory balance measured during the post-onset period was highly variable. Notably, a significant inverse relationship was identified between baseline innate inflammatory activity and stimulated C-peptide AUC measured at 3, 6, 12, 18 and 24 months post onset among placebo-treated individuals (p ≤ 0.015). Further, duration of persistent insulin secretion was negatively related to baseline inflammation (p ≤ 0.012) and positively associated with baseline abundance of circulating activated regulatory T cells (CD4+/CD45RA−/FOXP3high; p = 0.016). Based on these findings, data from participants treated with CTLA4-Ig were stratified by inflammatory activity at onset; in this way, we identified pathways and transcripts consistent with inhibition of T cell activation and enhanced immunoregulation. Variance among baseline plasma-induced signatures of TN-09 participants was further examined with weighted gene co-expression network analysis and related to clinical metrics. Four age-independent subgroups were identified that differed in terms of baseline innate inflammatory/regulatory bias, rate of C-peptide decline and response to CTLA4-Ig treatment.
Conclusions/interpretation
These data support the existence of multiple type 1 diabetes subtypes characterised by varying levels of baseline innate inflammation that are associated with the rate of C-peptide decline.
Data availability
Gene expression data files are publicly available through the National Center for Biotechnology Information Gene Expression Omnibus (accession number GSE102234).
Electronic supplementary material
The online version of this article (10.1007/s00125-018-4708-x) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Keywords: Biomarker, CTLA4-Ig, Disease heterogeneity, Honeymoon, Partial remission, Therapeutic response, Type 1 diabetes

Introduction
Type 1 diabetes arises through autoimmunity towards the pancreatic beta cells. Common features include the development of islet-antigen-specific autoantibodies and autoreactive T cells, as well as lifelong dependence on insulin-replacement therapy [1–4].
Numerous environmental factors and >40 genetic loci have been associated with susceptibility to type 1 diabetes [5, 6]. Varied combinations of these contributors likely underlie the significant disease heterogeneity observed among individuals. For example, while many individuals are diagnosed during childhood or adolescence, onset can occur at any age [7, 8]. The time between the presence of autoantibodies and clinical onset is also variable, but it is generally shorter in younger children [9]. Most individuals have residual endogenous insulin secretion at clinical diagnosis. Yet the amount of insulin secretion, measured by mixed meal tolerance test (MMTT)-stimulated C-peptide, diminishes over time and, as during the preclinical period, the rate of fall is variable, but more rapid in younger children. Trials of disease-modifying therapy aim to preserve endogenous insulin secretion after clinical diagnosis, as persistence of C-peptide, particularly MMTT-stimulated values ≥0.2 nmol/l, are associated with a decreased risk of severe hypoglycaemia and microvascular complications [10–14].
In trials of disease-modifying immunotherapy, heterogeneity in the rate of disease progression poses challenges in detecting the effect of treatment on preservation of stimulated C-peptide. As many as 17% of individuals have no fall in C-peptide during the first year after diagnosis and thus would not benefit from an intervention [15]. In contrast, 12% of individuals have levels of stimulated C-peptide that rapidly decline to less than 0.2 nmol/l during the same time frame [15]. Measures capable of predicting the post-onset disease course and/or forecasting the response to therapeutic intervention would enable individual stratification and development of individualised therapies [16].
We are able to differentiate immune states with a sensitive bioassay whereby an individual’s plasma or serum is used to induce transcription in a well-controlled reporter cell population [17–20]. After co-culture, the response is measured with a microarray then subjected to ontological analysis and follow-up studies. We have reported a disease-specific, partially IL-1-dependent, transcriptional signature associated with diabetes progression that is detectable prior to the development of autoantibodies [21]. We have also identified a familial innate inflammatory state in healthy siblings of people with diabetes that is independent of HLA, autoantibody status and disease progression [22]. Among sibling non-progressors with high-risk HLA haplotypes, this state is temporally supplanted by an IL-10/TGF-β-mediated regulatory state associated with peripheral increases in activated (CD4+/CD45RA−/forkhead box P3 [FOXP3]high) regulatory T cell (Treg) frequencies. These observations suggest that diabetes may develop because of failures in endogenous regulatory mechanisms that normally manage underlying risk.
We previously applied our bioassay to the Anti-Interleukin-1 in Diabetes Action (AIDA) and TrialNet Canakinumab (TN-14) trials [23, 24]. While these trials failed to show overall efficacy of IL-1 receptor antagonist (IL-1Ra) or canakinumab, plasma-induced transcription suggested that both therapies achieved varying degrees of immunomodulation consistent with IL-1 inhibition. At the endpoint, we quantitatively scored the signatures and identified an inverse relationship between measured inflammation and stimulated C-peptide response in treated participants.
Here, we examine post-onset diabetes progression in local patients from the Children’s Hospital of Wisconsin and participants in the TrialNet CTLA4-Ig (Abatacept) trial (TN-09) [25]. The cytotoxic T lymphocyte-associated protein 4/Fc fraction of the IgG1 fusion protein (CTLA4-Ig), also known as abatacept, inhibits CD28-mediated T cell co-stimulation by binding to CD80/86 on antigen-presenting cells. In TN-09, CTLA4-Ig slowed the decline of beta cell function in treated participants—an effect possibly mediated through modulation of central memory CD4 T cells [26]. Using plasma-induced transcription and a refined scoring algorithm, we investigated how heterogeneity at clinical diagnosis relates to persistence of insulin secretion during the post-onset period and responsiveness to CTLA4-Ig treatment.
Methods
Study participants
Local participants (n = 42) were recruited through the Children’s Hospital of Wisconsin (Milwaukee, WI, USA) and samples were collected as described previously [22]. Institutional review board approval (CHW IRB 01–15) was granted for all analyses, and informed consent/assent was obtained for all participants. In TN-09 (NCT00505375), participants were randomised in a 2:1 ratio to receive CTLA4-Ig (abatacept) (10 mg/kg, maximum 1000 mg/dose) or placebo (saline [154 mmol/l NaCl]) intravenously on days 1, 14 and 28 and monthly for a total of 27 infusions over 2 years [25]. At baseline, participants were <100 days post-diagnosis and had MMTT-stimulated C-peptide ≥0.2 nmol/l within 21 days of diagnosis. Based on availability, we procured serum collected at 0 (baseline), 3, 12 and 24 months from 74 of the 112 TN-09 participants (66.1%) for this ancillary analysis. The 54 CTLA4-Ig-treated participants analysed exhibited a higher mean baseline-adjusted 2 h AUC stimulated C-peptide response at the 24 month endpoint (0.49 ± 0.54 nmol/l) compared with the 20 placebo-treated participants (0.29 ± 0.25 nmol/l); this difference did not reach significance with the number of participants tested (p = 0.11). Relative to other metrics (electronic supplementary material [ESM] Table 1), this subset was not different compared with the complete TN-09 cohort. Compared with the local cohort, this subset exhibited an older age at onset (14.7 ± 6.8 vs 9.7 ± 3.3 years) and a shorter duration of diabetes at the baseline visit (0.24 ± 0.04 years vs 0.30 ± 0.14 years).
Preservation of insulin secretion
In the TN-09 participants, preservation of insulin secretion was defined as the duration of time that MMTT-stimulated C-peptide remained ≥0.2 nmol/l. As MMTTs were not performed in the local cohort, the duration of time that the insulin-dose-adjusted A1c (IDAA1c) remained ≤9 was used as a surrogate definition [27].
Plasma-induced transcription
Cryopreserved peripheral blood mononuclear cells (PBMCs) from a single healthy blood donor (Cellular Technology, Shaker Heights, OH, USA) were cultured in 300 μl RPMI 1640 medium with 200 μl plasma (local participants) or serum (TN-09 participants) and induced transcription was measured using Affymetrix GeneChip Human Genome U133 plus 2.0 arrays (Affymetrix, Santa Clara, CA, USA), as described previously [18, 22, 24].
Data analysis
All array data were subjected to global median normalisation with Bioconductor robust multi-array analysis [28]. The significance of differentially induced transcription was assessed through ANOVA and the rate of type I errors in multiple testing was assessed from false-discovery rates (FDRs) determined with Partek Genomics Suite 6.6 (Partek, Saint Louis, MO, USA). To optimise scoring of plasma-induced transcription, probe sets that were the best classifiers of new-onset diabetes were identified by re-analysing prior cross-sectional data [22] with random forest analysis (Salford Systems, San Diego, CA, USA), a tree-based ensemble machine learning tool. Ontological analysis used Ingenuity Pathway Analysis (IPA, QIAGEN, Redwood City, CA, USA). Hierarchical clustering was conducted with Genesis [29]. Relationships between the induced gene expression signatures and clinical phenotypes were identified with weighted gene co-expression network analysis (WGCNA) [30].
The ex vivo vs in vitro effect of CTLA4-Ig
PBMCs were pre-treated in medium with CTLA4-Ig (Bristol-Myers Squibb, New York, NY, USA) at 0 μg/ml, 25 μg/ml and 82 μg/ml for 45 min prior to the addition of plasma from a recently diagnosed local individual. Transcriptional analysis was conducted as detailed above.
Flow cytometry
PBMCs were stained with the fixable LIVE/DEAD Violet dye (Invitrogen, Carlsbad, CA, USA) for 30 min on ice, followed by blocking of Fc receptors and staining for anti-CD4 (clone RPA-T4), anti-CD25 (clone 2A3), anti-CD45RO (clone UCHL1), anti-CD45RA (clone HI100) and anti-CD127 (clone HIL-7R-M21, BD Bioscience, San Jose, CA, USA) on ice for 30 min, then intracellular staining with anti-FOXP3 (clone PCH101, Affymetrix eBioscience, San Diego, CA, USA). Analysis was conducted with an LSR II flow cytometer (BD Bioscience).
Results
Refined measurement of immunological balance
Previously, we conducted cross-sectional plasma-induced transcriptional analyses of individuals recently diagnosed with type 1 diabetes, unrelated healthy control individuals lacking family history of autoimmunity and healthy autoantibody-negative siblings of individuals with type 1 diabetes possessing high-risk (DR3 and/or DR4) or low-risk (non-DR3/non-DR4) HLA haplotypes [22]. Among these cohorts we identified 1374 differentially induced probe sets. To quantitatively measure immune activity, we developed a gene-ontology-based composite inflammatory index (I.I.com), calculated by determining the ratio of the mean intensity of the induced inflammatory genes to the mean intensity of the induced regulatory genes [22]. High scores reflect greater inflammatory bias and low scores reflect greater regulatory bias.
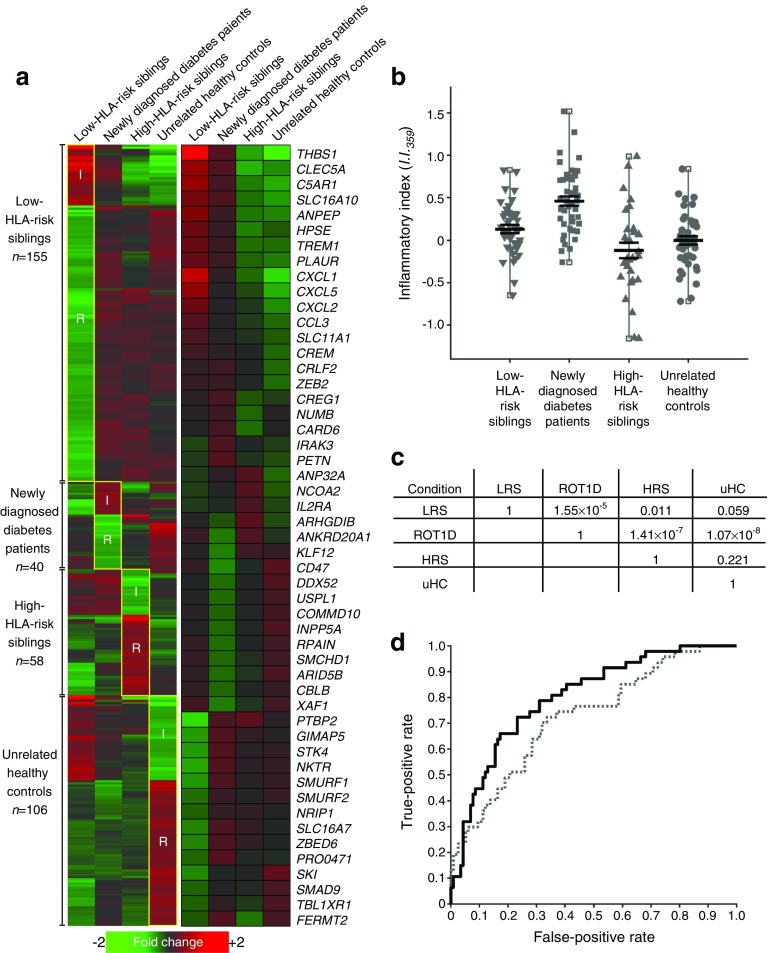
Here, in an effort to simplify the I.I.com, we used random forest analysis to identify 359 probe sets that were optimal classifiers of the diabetic cohort relative to the three control cohorts. Among these 359 probe sets, 325 (90.5%) overlapped with the 1374 probe sets identified previously [22]. Similar to the original data, this subset captured increasing IL-10/TGF-β bias and decreasing IL-1/NFκB bias across the continuum created by the siblings with low HLA risk, individuals with type 1 diabetes, siblings with high HLA risk and unrelated control individuals (Fig. 1a). This is reflected by the increased induction of chemokine transcripts by plasma of siblings with low HLA risk and individuals with diabetes, and elevated induction of regulatory transcripts (IL2RA, CBLB, SMURF1, SMURF2, SKI) by plasma of siblings with high HLA risk and unrelated control individuals. As described for I.I.com [22], an inflammatory index based on these 359 probe sets (I.I.359) was calculated by dividing the average signal intensity of the 103 probe sets generally annotated as ‘inflammatory’ by the average signal intensity of the 256 probe sets generally annotated as ‘regulatory’. Similar to I.I.com, the average I.I.359 for the individuals with diabetes was significantly higher than the other cohorts (Fig. 1b, c). The receiver operator characteristic (ROC) analysis of this subset showed improved discrimination of the diabetic and control cohorts (Fig. 1d).
Fig. 1.
Refined scoring of plasma-induced transcriptional signatures. (a) Mean expression levels of the 359 probe sets that best distinguish individuals newly diagnosed with type 1 diabetes from the low-HLA-risk sibling, high-HLA-risk sibling, and unrelated healthy control cohorts. The analysis included: individuals with diabetes (n = 47, age 10.0 ± 2.9 years, blood glucose 8.6 ± 4.1 mmol/l, HbA1c 5.8 ± 9.3 mmol/mol [7.5 ± 1.2%]; baseline samples were collected 2–7 months after diagnosis when metabolic control had been achieved); low-HLA-risk siblings (n = 42, age 8.4 ± 2.0 years, blood glucose 5.2 ± 0.9 mmol/l); high-HLA-risk siblings (n = 30, age 8.6 ± 1.9 years, blood glucose 5.2 ± 0.7 mmol/l); and unrelated healthy control individuals (n = 44, age 15.0 ± 4.1 years, blood glucose 5.1 ± 0.7 mmol/l). Random forest analysis used 12,589 probe sets identified in the six possible comparisons between the four cohorts at a fold change >1.1 and an FDR <20%. The probe sets that exhibited a random forest Gini score >3.49 when any one group was compared with the three others were retained. A second analysis identified probe sets that were regulated at log2 ratio >|0.263| (1.2-fold) and FDR <20% when one cohort was compared with any of the three others. A total of 359 transcripts met both criteria. (a) The left section shows the relative expression levels of the 359 probe sets across the four cohorts. The second analysis enabled the definition of four data subsets, the number of transcripts within each data subset is shown on the left. The transcripts generally annotated as inflammatory and regulatory within each data subset are indicated; the upregulated transcripts in the low-HLA-risk sibling and diabetic individual data subsets are generally annotated as inflammatory, while upregulated transcripts in the high-HLA-risk sibling and unrelated healthy control data subsets are generally annotated as regulatory. The annotated dataset is available from the corresponding author on request. The right section shows the mean expression levels of a subset of well-annotated transcripts. (b) Ontology-based scoring of cross-sectional samples using I.I.359 significantly discriminates the diabetic cohort from the other cohorts. The mean I.I.359 for the 47 cross-sectional diabetes participants (mean ± SE 0.46 ± 0.05) was significantly higher than that observed for the 42 siblings with low HLA risk (0.13 ± 0.05), 30 siblings with high HLA risk (−0.12 ± 0.09) and 44 unrelated healthy control participants (0.00 ± 0.05). p values are shown in panel (c); two-tailed unpaired t tests for the comparisons between each cohort. (d) ROC curve for 359 probe sets (solid line; AUC = 0.80) shows improved discrimination of the diabetic cohort from the related and unrelated control cohorts compared with the previously reported 1374 probe sets described in Chen et al [22] (dotted line; AUC = 0.72). ROT1D, recent-onset type 1 diabetes; LRS, low-HLA-risk sibling; HRS, high-HLA-risk sibling; I, inflammatory; R, regulatory; uHC, unrelated healthy control group
The relationship between baseline I.I.359 and beta cell function
Next, we examined the relationship between the baseline inflammatory/regulatory balance (I.I.359) and the decline in beta cell function among TN-09 participants. We hypothesised that baseline I.I.359 might relate to disease activity and correlate with stimulated C-peptide AUC in untreated (placebo) participants. We further hypothesised that the therapeutic effect of CTLA4-Ig would alter the natural disease course, abolishing the relationship established in the placebo arm. A significant inverse relationship was observed between baseline I.I.359 and the per cent change from baseline C-peptide AUC at 3, 6, 12, 18 and 24 months in placebo-treated participants (p ≤ 0.015); this relationship was not observed among CTLA4-Ig-treated participants, consistent with immunomodulation altering disease progression (Fig. 2a–e). Similarly, baseline I.I.359 was inversely related to the rate (slope) of C-peptide decline over the 24 month study in the placebo arm but not in the CTLA4-Ig arm (Fig. 2f).
Fig. 2.
The relationship between baseline I.I.359 and future beta cell function. A significant inverse relationship exists between baseline I.I.359 and 2 h C-peptide AUC percent change from baseline at 3, 6, 12, 18 and 24 months in 19 placebo-treated TN-09 participants (all p < 0.015) but not in 54 CTLA4-Ig-treated TN-09 participants. (a) 3 months: placebo treated, slope = −0.85, r2 = 0.48, p = 0.001; CTLA4-Ig treated, slope = 0.07, r2 = 0.002, p = 0.731. (b) 6 months: placebo treated, slope = −0.84, r2 = 0.36, p = 0.006; CTLA4-Ig treated, slope = 0.12, r2 = 0.008, p = 0.522. (c) 12 months: placebo treated, slope = −0.90; r2 = 0.29; p = 0.015; CTLA4-Ig treated, slope = −0.06; r2 = 0.002; p = 0.768. (d) 18 months: placebo treated, slope = −0.90, r2 = 0.37; p = 0.004; CTLA4-Ig treated, slope = 0.01, r2 = 0.0001, p = 0.939. (e) 24 months: placebo treated, slope = −0.91, r2 = 0.56; p = 0.0002; CTLA4-Ig treated, slope = 0.18, r2 = 0.017, p = 0.343. The data are similar if considered as baseline-normalised C-peptide AUC. (f) Baseline I.I.359 is inversely related to rate (slope) of C-peptide decline over the 24 month study period: placebo treated, slope = −0.18, r2 = 0.21; p = 0.041; CTLA4-Ig treated, slope = −0.003, r2 = 0.0003, p = 0.909. A truncated linear regression was used to estimate the slope of decrease for each individual. Larger negative slope values imply faster decline of stimulated C-peptide. The data are similar if considered as baseline-normalised C-peptide AUC. Black solid circles and solid line, placebo-treated; grey open circles and dotted line, CTLA4-Ig treated
To determine whether baseline I.I.359 is predictive of the duration of the post-onset partial remission period, we investigated the relationship between I.I.359 and the duration of MMTT-stimulated C-peptide ≥0.2 nmol/l in the TN-09 placebo arm participants (Fig. 3a) and the duration of IDAA1c ≤9 in local individuals with newly diagnosed diabetes (Fig. 3b). In both cohorts, the baseline I.I.359 was inversely related to the duration of the post-onset partial remission period (p ≤ 0.012). Further, participants with I.I.359 above the median had a significantly shorter duration of persistent insulin secretion than those below the median (Fig. 3c, d), supporting the hypothesis that individuals with higher inflammation at onset will experience accelerated decline in beta cell function. Supporting this observation was the measurement of lower abundances of peripheral activated Tregs (CD4+/CD45RA−/FOXP3high) during the immediate post-onset period in the local participants that exhibited an IDAA1c ≤9 for less than 6 months (Fig. 3e, f; p = 0.016).
Fig. 3.
Inflammation levels at onset of type 1 diabetes correlate with duration of the post-onset partial remission. (a) Among 19 TN-09 participants in the placebo arm (ages 8–35 years), Weibull (solid line, grey shaded area = 1 SE) and Cox regression (dashed line) analyses identified an inverse relationship between I.I.359 and median remission duration (p = 0.012). The partial remission period was defined as the length of time from diagnosis in which the 2 h stimulated C-peptide AUC was >0.2 nmol/l. (b) Weibull (solid line, grey shaded area = 1 SE) and Cox regression (dotted line) analyses identified a significant relationship between I.I.359 and median remission duration among 26 local individuals with diabetes (p = 6.8 × 10−10). These participants were aged 4–16 years and possessed high-risk HLA haplotypes; samples were collected 2–7 months after diagnosis (see ESM Table 1 for additional characteristics). As dynamic testing was not performed on the local cohort, the IDAA1c was determined from HbA1c and total daily insulin doses at post-onset clinic visits as described in Mortensen et al [27]. IDAA1c ≤9 is reflective of a stimulated C-peptide >0.3 nmol/l; thus, the remission duration was defined as the last quarterly clinic visit when IDAA1c was ≤9. Among TN-09 participants in the placebo arm, remission lengths determined using the IDAA1c were highly correlated with those determined through dynamic testing (r = 0.79); however, as anticipated [49], on average, the use of the IDAA1c underestimated partial remission durations relative to dynamic testing (1.2 ± 1.0 years vs 2.2 ± 1.5 years, respectively). In (a, b) participant age range is indicated by colour: green, 0–6 years; black, >6–12 years; red, >12–18 years; blue, >18 years; boxes indicate participants in remission at the last visit. (c, d) Kaplan–Meier analysis: the proportion of TN-09 placebo-arm participants (n = 19) (c) and local individuals with diabetes (n = 26) (d) in remission was compared for those with I.I.359 above the median (dashed line) and those with I.I.359 below the median (solid line). A logrank test found participants with I.I.359 above the median were significantly different in both populations (TN-09: p = 0.016; local individuals: p = 0.005). (e) Representative flow cytometry profiles showing the gating strategy for resting and activated Treg populations in cryopreserved PBMCs from local individuals with diabetes. Resting and activated CD4+ Tregs were respectively defined as CD45RA+/FOXP3low and CD45RA−/FOXP3high. Since CD45RA and CD45RO are different CD45 isoforms that are respectively expressed on naive and activated/memory T cells, the expression of CD45RO as well as CD25 confirmed the phenotype of resting (CD45RO−/CD25+) and activated (CD45RO+/CD25high) Tregs. Each analysis included fluorescence minus one controls to ensure correct gating. (f) Per cent activated Tregs among total (active + resting) Tregs in cryopreserved PBMCs collected from 23 local individuals with diabetes during the immediate post-onset period. Eight of these participants were among the 26 analysed by plasma-induced transcription. The data were classified as having shorter or longer partial remissions using the Jenks natural breaks method. The resulting groups had ages of 8.4 ± 3.3 and 11.3 ± 3.8 years, respectively (not significant, p > 0.05). The per cent activated Treg was higher in those with longer vs shorter partial remissions (p = 0.016). The data are similar if plotted as percentage of activated Treg among total CD4+ T cells
The baseline C-peptide AUC of the TN-09 participants was directly related to age at diagnosis, supporting a recognised relationship [31, 32]. However, baseline I.I.359 was independent of age in the TN-09 and local cohorts (Fig. 4), indicating that the more rapid decline of beta cell function in those individuals with higher inflammatory bias at baseline was independent of the age of clinical onset. Further, baseline I.I.359 was independent of time from clinical onset in both cohorts and, in TN-09 participants, there was no correlation between diabetes duration and the baseline stimulated C-peptide AUC.
Fig. 4.

The relationship between I.I.359 and age of clinical onset. (a) Among the 74 TN-09 participants analysed, the age at diagnosis was directly related to baseline 2 h C-peptide AUC (r2 = 0.28, p = 2.1 × 10−5). The baseline I.I.359 was independent of age in TN-09 (b) and local (c) participants (n = 26). In (a, b), black solid circles, placebo treated (n = 19); grey open circles, CTLA4-Ig treated (n = 54); dotted/dashed vertical line, 18 years of age
Stratification of individuals fosters identification of a CTLA4-Ig therapeutic response signature
Our initial strategy for defining immunomodulation achieved by CTLA4-Ig in TN-09 followed that described in our analyses of the IL-1 antagonism trials [24]. Briefly, induced transcription for each participant at 3, 12 and 24 months was normalised with that of baseline, then differentially induced transcripts between the CTLA4-Ig and placebo arms were identified. A total of 427 differentially induced probe sets (log2 ratio >|0.263|, 1.2-fold; ANOVA p < 0.05) were identified between the trial arms at ≥1 time point (ESM Fig. 1). Unacceptably, no transcript exhibited an FDR <50% at any time point. We hypothesised that the analysis could be improved by matching treated and placebo participants for baseline I.I.359 and respectively focusing on those participants with the greatest therapeutic response or the most rapid disease progression.
To identify CTLA4-Ig-treated participants with the greatest therapeutic response, we applied two criteria. The first criterion utilised the placebo arm trend line generated when baseline I.I.359 was plotted against per cent change from baseline stimulated C-peptide AUC at 3, 6, 12, 18 and 24 months (Fig. 2a–e). Participants who were >1.5 SD above this regression line at ≥3 time points were identified. The second criterion utilised the placebo arm regression line when baseline I.I.359 was plotted against the rate of C-peptide decline over the 24 month study period (Fig. 2f). Participants who were >1 SD above the trend line were identified. Eight CTLA4-Ig treated participants met both criteria. These eight participants were then compared with placebo participants (n = 7) matched for I.I.359 and residing at or below the placebo regression line depicted in Fig. 2f.
As previously reported, the decrease in beta cell function among CTLA4-Ig-treated participants paralleled that observed in the placebo arm after 6 months [25]. Therefore, we hypothesised that the maximal plasma-induced transcriptional signature representing therapeutic response would be detected at 3 months post-enrolment. In this way, we identified 1509 differentially induced probe sets between the selected eight CTLA4-Ig- and seven placebo-treated participants (mean log2 ratio >|0.263|, 1.2-fold; ANOVA p < 0.02; FDR ≤30%). These data did not significantly overlap with the cross-sectional data set. Specifically, 50/1374 (3.6%, χ2 > 0.059) and 10/359 (2.8%, χ2 = 1) transcripts overlapped those used to define I.I.com and I.I.359, respectively.
On average, the signatures of the remaining placebo- and CTLA4-Ig-treated participants were intermediate to those of the selected placebo- and CTLA4-Ig-treated participants and not distinct from one another (Fig. 5a, left). As expected, hierarchical clustering of individual selected placebo- and CTLA4-Ig-treated participants using the 1509 probe sets resulted in distinct grouping (Fig. 5a, middle). In contrast, the remaining participants exhibited imperfect clustering, suggesting that this subgroup possessed both slow-progressing and treatment-non-responder participants (Fig. 5a, right). Consistent with the resumed decline in stimulated C-peptide, at 12 months and 24 months, the plasma milieus of the selected CTLA4-Ig- and placebo-treated participants were more similar, with 236 and 0 probe sets being differentially induced to the aforementioned thresholds, respectively (Fig. 5b).
Fig. 5.
Signature of therapeutic response in TN-09. (a) Analysis of the 1509 probe sets regulated between the CTLA4-Ig responders (n = 8) and rapidly progressing placebo-treated individuals (n = 7) matched for baseline I.I.359 at the 3 month time point. The top three panels (left to right) illustrate two-way clustering of the selected and excluded participants in each arm. Left panel, mean expression of the four groups. Middle panel, participants selected for the analysis. Right panel, remaining CTLA4-Ig- and placebo-treated participants. Lower three panels, expression levels of a selected set of well-annotated transcripts. The colour bar indicates assigned treatment arm: red, placebo; blue, CTLA4-Ig. Note: samples were not available for two placebo-treated participants at the 3 month time point. The annotated dataset is available from the corresponding author on request. (b) Mean expression levels of the 1509 differentially induced probe sets at 12 and 24 months. (c) Upstream regulator analysis using IPA. A z score >2.0 is significantly activated; a z score >−2.0 is significantly inhibited. (d) CTLA4-Ig add-back experiment. The mean induced expression levels of the 1509 probe sets after plasma of five local untreated individuals with diabetes were supplemented with 0 μg/ml, 25 μg/ml or 82 μg/ml CTLA4-Ig are shown. ROT1D, recent-onset type 1 diabetes
IPA was then used to identify candidate regulators of the 1509 probe sets differentially induced between the selected CTLA4-Ig- and placebo-treated participants (Fig. 5c). Consistent with immunomodulation anticipated by CTLA4-Ig therapy, IPA identified as being significantly activated (z score >2.0): TGF-β1; histone deacetylase co-repressor 2 (HDAC2), a transcriptional repressor that governs NFκB-regulated genes [33]; aryl hydrocarbon receptor (AHR), an important modulator of adaptive responses [34]; and the transcriptional regulator sterol regulatory element-binding protein 1 (SREBF1) [35]. Further, IPA revealed significant inhibition of CD28 and CD3, molecules important in T cell activation, and lysine demethylase 5A (KDM5A), important in natural killer cell activation [36], (z score <−2.0) in CTLA4-Ig treated individuals. Transcripts annotated under these candidate upstream regulators are known to possess roles in attenuating adaptive immune responses (SKIL, SMAD2, PTPN22, AHR), mediating cell adhesion (CEACAM5, L1CAM) and regulating proliferation/apoptosis (SDC4, BAX, NDFIP2, BTG1).
The half-life of CTLA4-Ig is 14 days. At the 3 month sampling, the TN-09 dosing schedule was such that participants had last received an infusion 4 weeks prior; as such, it was considered unlikely there would be residual CTLA4-Ig in the samples. We reasoned that if the treatment arm signature was a direct consequence of carry-over CTLA4-Ig in the plasma, it would be possible to recapitulate that signature in samples of untreated individuals by spiking the cultures with CTLA4-Ig. Plasma of untreated individuals with diabetes was therefore supplemented with 0 μg/ml, 25 μg/ml (estimated steady-state trough level) or 82 μg/ml CTLA4-Ig (estimated steady-state high level). Even at the highest level, only 880 (58.3%) of the 1509 probe sets differentially induced between the selected CTLA4-Ig- and placebo-treated individuals were directionally concordant, and only 67 (4.4%) were directionally concordant and possessed an FDR <30% (Fig. 5d). This indicates that signatures of the participants in the treatment arm were largely independent of carry-over CTLA4-Ig and that signatures of the selected CTLA4-Ig-treated participants were reflective of treatment-mediated immunomodulation.
Variation in baseline signatures defines distinct subgroups that differentially respond to CTLA4-Ig
The baseline I.I.359 was significantly related to post-onset C-peptide AUC and was useful for matching participants in analyses aimed at defining the therapeutic effects of CTLA4-Ig. However, I.I.359, which is derived from cross-sectional analyses, did not completely differentiate CTLA4-Ig treatment responders from non-responders, suggesting that I.I.359 does not entirely capture the variation among newly diagnosed individuals. Therefore, we identified the 3159 most variant transcripts common to the local and TN-09 participants at baseline (Fig. 6a). This dataset was analysed with WGCNA, a software that clusters the main patterns of variation into modules of co-expressed transcripts and correlates these to phenotypes.
Fig. 6.
Analysis of highly variant transcripts in baseline signatures indicates the existence of type 1 diabetes subgroups. (a) Identification of highly variant transcripts common to TN-09 participants (n = 74) and local individuals with diabetes (n = 26). The Affymetrix U133 plus 2.0 array has probe sets for interrogation of >47,000 transcripts. The 20,000 probe sets that, on average, exhibited the lowest signal intensity were filtered from the analysis (maximum log2 intensity <4 RFU). The 7000 probe sets exhibiting the greatest median absolute deviation were retained. (b) Co-expression networks among the 3159 commonly variant transcripts were identified with WGCNA using a power of β = 12 (linear regression model fitting index R2 > 0.8). Twelve co-expression modules were identified (represented by rows, labelled by colour). The number of probe sets assigned to each module is indicated; 79/3159 (2.5%) could not be assigned to a co-expression network. Columns represent a trait or cohort subset. The correlation coefficient shown within each box is between the module eigengene and the trait or cohort; the correlation p value is given in parentheses. An eigengene is defined as the first principal component of a given module and is representative of the expression profiles of genes within that module. Red represents a positive correlation between the module eigengene and trait; green represents a negative correlation. Significant relationships were not detected with age, sex, Tanner stage, HbA1c at baseline, HLA, autoantibody status, BMI, duration of disease or complete blood cell counts. (c) Dendrogram illustrating co-expression modules. Each vertical line corresponds to a gene. The y-axis indicates the network distance (1 – topological overlap), values closer to 0 indicate greater similarity of transcript expression profiles across samples. Co-expression modules are indicated by colours in the first colour band. The additional colour bands illustrate the positively correlated (progressively red) or negatively correlated (progressively blue) transcripts for the trait/cohorts. (d) Hierarchical clustering of the baseline signatures of TN-09 participants using the 916 significantly regulated transcripts belonging to the three modules significantly correlated with 2 h AUC at baseline and the rate of C-peptide decline within the placebo-treated arm. The three modules included 85 transcripts used to calculate I.I.359 (85/359, 23.7%, χ2 < 1.0 × 10−4). Four major subgroups of individuals with new onset diabetes are indicated, none of which significantly differ by age or baseline C-peptide AUC. Colour bars indicate: fold change, baseline C-peptide AUC, rate of C-peptide decline over 24 months, selected CTLA4-Ig responders (as in Fig. 5; blue), selected rapidly progressing placebos (as in Fig. 5; ‘Selected fast placebos’; peach), age at baseline and I.I.359. The colour bar indicates assigned treatment arm: red, placebo; blue, CTLA4-Ig. The 3159 commonly variant probe sets and the modules to which they were assigned are available as an annotated dataset from the corresponding author on request
Among the 3159 transcripts, WGCNA identified 12 modules. Two modules significantly correlated with the baseline C-peptide AUC among TN-09 participants (black and yellow); a third module (blue) correlated with the rate of C-peptide decline among both placebo and CTLA4-Ig treated participants (Fig. 6b, c). These modules also correlated with I.I.359, suggesting that the inflammatory/regulatory dynamic previously identified by cross-sectional analysis is also present among newly diagnosed individuals.
To investigate whether the immune activity represented by these three modules could define subgroups among newly diagnosed individuals, the baseline signatures of TN-09 participants were subjected to hierarchical clustering. This identified four major subgroups that did not significantly differ by age or baseline C-peptide AUC (Fig. 6d). For the 916 transcripts encompassed by these modules, IPA identified numerous candidate upstream regulators, including lipopolysaccharide (1.2 × 10−31), IL-1B (1.7 × 10−18), TNF (2.4 × 10−29), IFN-γ (7.7 × 10−19), IL-10 (7.7 × 10−13) and TGF-β1 (1.8 × 10−8). The yellow and black modules included numerous proinflammatory annotations, including cytokines (IL1A, IL1B, IL6) and chemokines (CCL2, CCL3, CCL4); conversely; the blue module included many transcripts related to IL-10 and TGF-β signalling (SKI, SKIL, INPP5D, SUZ12) (Fig. 7a).
Fig. 7.
Type 1 diabetes subgroups exhibit different responses to CTLA4-Ig treatment. (a) Heatmaps illustrating expression levels of the 916 significantly regulated transcripts belonging to the three modules significantly correlated with 2 h C-peptide AUC at baseline and the placebo-treated arm of TN-09; an annotated heatmap is shown below. The four major subgroups (as shown in Fig. 6d) are indicated for each arm. Well-annotated transcripts are illustrated, and the module to which they belong is indicated by the colour bar on the left. Colour bars are provided that indicate: fold change; baseline C-peptide AUC; rate of C-peptide decline over 24 months; selected CTLA4-Ig responders (as in Fig. 5; blue); selected placebos (as in Fig. 5; ‘Selected fast placebos’; peach); age at baseline; and I.I.359. (b) TN-09 participants belonging to subgroups 1–4 exhibit different responses to CTLA4-Ig treatment. Tabulated are subgroup mean values for each metric. Statistical differences were assessed with a two-tailed unpaired Student’s t test, superscript letters denote p < 0.05 for: aCTLA4-Ig subgroup 1 vs CTLA4-Ig subgroup 2; bCTLA4-Ig subgroup 1 vs CTLA4-Ig subgroup 3; cCTLA4-Ig subgroup 1 vs CTLA4-Ig subgroup 4; dCTLA4-Ig subgroup 2 vs CTLA4-Ig subgroup 3; eCTLA4-Ig subgroup 2 vs CTLA4-Ig subgroup 4; fCTLA4-Ig subgroup 3 vs placebo subgroup 3; gplacebo subgroup 2 vs placebo subgroup 3; and hplacebo subgroup 2 vs placebo subgroup 4
The CTLA4-Ig and placebo arms of TN-09 were independently analysed from the perspective of these four subgroups. While the limited number of participants assigned to these subgroups precludes robust conclusions, significant age-independent differences were identified in the rate of C-peptide decline within each treatment arm. Among CTLA4-Ig-treated participants, those in subgroup 1 exhibited the smallest per cent change from baseline C-peptide AUC at 3, 12, 18 and 24 months and the lowest overall rate of C-peptide decline over 24 months compared with those in the other three subgroups (Fig. 7b). The CTLA4-Ig-treated participants assigned to subgroup 2 were significantly younger than their placebo-treated counterparts (Kolmogorov–Smirnov test, p < 0.04), and exhibited a faster rate of C-peptide decline and a greater per cent change from baseline C-peptide AUC at 3, 6, 12, 18 and 24 months than the other subgroups. Among the placebo-treated participants, the rate of C-peptide decline and per cent change from baseline C-peptide AUC for subgroups 3 and 4 were significantly greater than those of subgroup 2 (and greater than the single placebo-treated individual assigned to subgroup 1). Finally, within subgroup 3, CTLA4-Ig treatment significantly reduced the per cent change from baseline C-peptide AUC at 6, 18 and 24 months when compared with the placebo arm (Fig. 7b).
These data suggest that baseline plasma-induced signatures can define type 1 diabetes subgroups. Participants in subgroups 1 and 2 exhibited greater baseline regulatory bias (and lower I.I.359) and a generally slower rate of C-peptide decline. Conversely, participants in subgroups 3 and 4 exhibited greater innate inflammatory bias and a faster rate of C-peptide decline. Subgroup 3 contained most of the selected CTLA4-Ig responders and rapidly progressing individuals who received placebo. The data also suggest that participants with greater baseline inflammatory activity (subgroups 3 and 4) exhibited a better therapeutic response to CTLA4-Ig during the 24 month study period (Fig. 8).
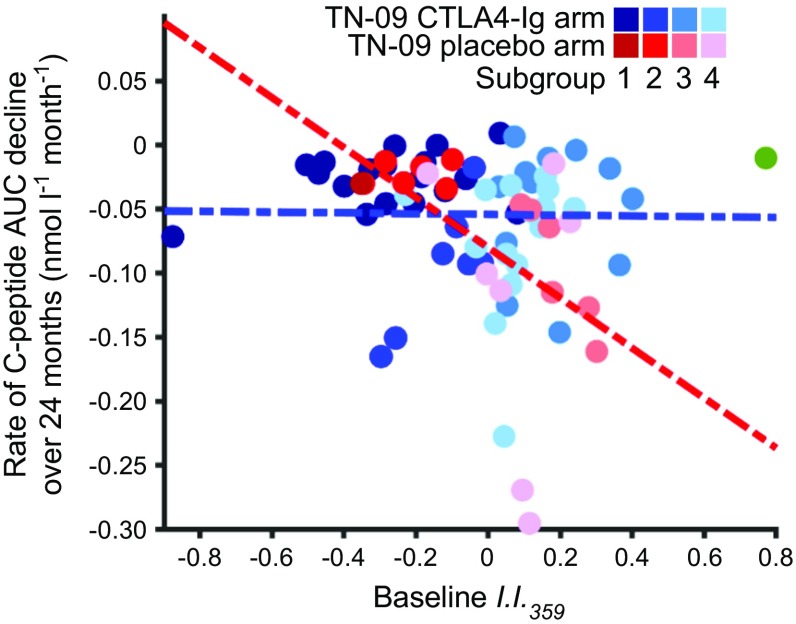
Fig. 8.
Type 1 diabetes subgroups exhibit different rates of C-peptide decline and responses to CTLA4-Ig treatment. A significant inverse relationship between baseline I.I.359 and the rate (slope) of C-peptide decline in placebo-treated TN-09 participants is described in Fig. 2f. Here, this relationship is considered from the perspective of subgroups 1–4. The green data point is the outlying CTLA4-Ig participant not assigned to a subgroup (left-most participant in Fig. 6d). Regression lines are shown for the CTLA4-Ig and placebo arms of TN-09 in blue and red, respectively
Discussion
We find that individuals with type 1 diabetes possessing lower innate inflammatory bias at the time of clinical onset, as measured through plasma-induced transcription, are more likely to have slower rates of decline in residual insulin secretion. This conclusion was supported through two independent analyses: one based on a scoring system developed from cross-sectional analyses of individuals recently diagnosed with diabetes and healthy control individuals (I.I.359); and a second that utilised WGCNA to identify clinically relevant highly variant transcripts among newly diagnosed volunteers. We also associated longer persistence of IDAA1c ≤9 with higher peripheral abundances of activated Treg, corroborating Moya et al [37]. These analyses contribute to a growing effort to predict disease progression in type 1 diabetes [26, 37–41] and are in line with studies linking elevated innate inflammation with diabetes progression [42–45]. Importantly, they support the existence of subgroups that differ in inflammatory/regulatory balance, rate of post-onset disease progression and responsiveness to therapeutic intervention with CTLA4-Ig.
These findings add to an understanding of phenotypic heterogeneity among newly diagnosed individuals. Arif et al described two distinct adaptive immune response patterns among newly diagnosed children and adolescents: (1) a partially regulated IL-10-dominated CD4 T lymphocyte response; and (2) a proinflammatory IFN-γ-dominated CD4 T lymphocyte response [46]. Similar to our analysis, and possibly paralleling subgroups 1–2 and 3–4, individuals exhibiting regulatory vs inflammatory bias were equally distributed.
Two distinct insulitic profiles have been observed in people with new-onset type 1 diabetes, further supporting the existence of subtypes [46, 47]. Individuals diagnosed below 7 years of age possessed islets with lower insulin content and greater numbers of infiltrating immunocytes, in particular CD20+ B cells. Individuals diagnosed after 13 years of age possessed islets with greater insulin content, fewer infiltrating immunocytes and low abundances of CD20+ B cells. Participants of intermediate age exhibited either one profile or the other [47]. In contrast, the subgroups defined herein did not exhibit age dependence. However, among the studied TN-09 participants, only two were diagnosed under 7 years of age; these were ascribed to subgroups 3 and 4, which exhibited inflammatory bias and a more rapid rate of C-peptide decline.
Our study of the AIDA trial [24] suggested that individuals with diabetes with the highest levels of baseline inflammation may have benefited the most from IL-1Ra treatment, as they tended to experience the greatest reduction in inflammation and retained the most beta cell function. Likewise, our findings suggest that participants with greater inflammatory bias (higher baseline I.I.359, and ascribed primarily to subgroup 3, both of which are associated with more rapid disease progression) benefited more from CTLA4-Ig treatment. One possible explanation for this observation is that participants possessing greater regulatory bias have disease activity too modest to be therapeutically altered during the trial. Whatever the biological basis, heterogeneity in disease progression among individuals is an obstacle in clinical trials, as inclusion of slow progressors impairs the ability to detect treatment-specific changes in C-peptide production.
Stratification of participants by baseline I.I.359 facilitated our analysis of the therapeutic response to CTLA4-Ig. The original analysis of TN-09 showed that CTLA4-Ig treatment delayed the decline of beta cell function [25]. Here, selected CTLA4-Ig responders exhibited signatures indicative of CD28 and CD3 inhibition, an observation consistent with co-stimulation blockade and impaired activation of auto-aggressive T cells and in line with the presumed mechanism of action of CTLA4-Ig. As demonstrated by TN-09, CTLA4-Ig treatment can suppress inflammation after disease is established, suggesting the possibility of alternative mechanisms beyond the prevention of naive T cell activation through co-stimulation blockade. In mice, CTLA4-Ig mediates regulation of activated effector T cells through a nitric oxide/Treg/TGF-β-dependent pathway [48]. Our analysis identified TGF-β1 as a potential regulator of the immunoregulatory-biased signature of the selected CTLA4-Ig- treated participants, suggesting that such alternative mechanisms may be associated with human responses.
Our findings have important implications for the stratification of individuals with diabetes. The ability to identify individuals with rapidly progressing disease would allow for more informative and targeted trials of participants most likely to benefit from therapeutic intervention. The size of this study is a limitation. Additional studies are needed to determine if the subgroups defined here are seen in other cohorts, whether they are governed by genetic and/or environmental factors and whether they differentiate progression rates prior to diabetes onset, particularly in multiple-antibody-positive ‘at-risk’ participants at earlier (preclinical) states of the disease.
Electronic supplementary material
(PDF 434 kb)
Acknowledgements
The authors thank the individuals who participated in this study as well as the physicians, nurses and staff of the Children’s Hospital of Wisconsin and the Max McGee Research Center for Juvenile Diabetes who assisted in participant recruitment and sample collection/processing.
Abbreviations
- AHR
Aryl hydrocarbon receptor
- AIDA
Anti-Interleukin-1 in Diabetes Action
- CTLA4-Ig
Cytotoxic T lymphocyte-associated protein 4/Fc fraction of the IgG1 fusion protein (abatacept)
- FDR
False-discovery rate
- HDAC2
Histone deacetylase co-repressor 2
- IDAA1c
Insulin-dose-adjusted A1c
- I.I.com
Composite inflammatory index (based on 1374 transcripts)
- I.I.359
Inflammatory index based on 359 transcripts
- IL-1Ra
IL-1 receptor antagonist
- IPA
Ingenuity Pathway Analysis
- KDM5A
Lysine demethylase 5A
- MMTT
Mixed meal tolerance test
- PBMC
Peripheral blood mononuclear cell
- ROC
Receiver operator characteristic
- SREBF1
Sterol regulatory element-binding protein 1
- TN-09
TrialNet CTLA4-Ig (Abatacept) trial
- Treg
Regulatory T cell
- WGCNA
Weighted gene co-expression network analysis
Contribution statement
SMC and Y-GC designed experiments, researched data and wrote, reviewed and edited the manuscript. SE, MK and RG recruited participants, processed samples, researched data and reviewed and edited the manuscript. SJ, PS and AS developed/tested algorithms, conducted data analysis and reviewed and revised the manuscript; CJG and CS contributed to the design of the analysis and discussed, wrote, reviewed and edited the manuscript. MJH designed experiments, analysed data and wrote, reviewed and edited the manuscript. MJH is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors approved the version of the manuscript to be published.
Funding
This work was supported by JDRF International (grants 2-SRA-2015-109-Q-R and 3-SRA-2018-478-S-B to MJH); National Institutes of Health (R56DK108802 to MJH, DP3DK098161 to MJH/CJG and 1-UL1-RR031973); the Clinical and Translational Science Institute Southeast Wisconsin; the George and Ruth Leef Family and ITU AbsorbTech; the David & Julia Uihlein Charitable Foundation; and the Children’s Hospital of Wisconsin Foundation. The funders had no role in how the studies were conducted or interpreted.
The Type 1 Diabetes TrialNet Study Group is a clinical trials network funded by the National Institutes of Health through the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases and the Eunice Kennedy Shriver National Institute of Child Health and Human Development, currently through the cooperative agreements U01 DK061010, U01 DK061034, U01 DK061042, U01 DK061058, U01 DK085465, U01 DK085461, U01 DK085466, U01 DK085476, U01 DK085499, U01 DK085509, U01 DK103180, U01 DK103153, U01 DK103266, U01 DK103282, U01 DK106984, U01 DK106994, U01 DK107013, U01 DK107014, UC4 DK106993 and JDRF. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or JDRF.
Data availability
Gene expression data files are publicly available through the National Center for Biotechnology Information Gene Expression Omnibus (accession number GSE102234).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
References
- 1.Morran MP, Vonberg A, Khadra A, Pietropaolo M. Immunogenetics of type 1 diabetes mellitus. Mol Asp Med. 2015;42:42–60. doi: 10.1016/j.mam.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kronenberg D, Knight RR, Estorninho M, et al. Circulating preproinsulin signal peptide-specific CD8 T cells restricted by the susceptibility molecule HLA-A24 are expanded at onset of type 1 diabetes and kill beta-cells. Diabetes. 2012;61:1752–1759. doi: 10.2337/db11-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arif S, Tree TI, Astill TP, et al. Autoreactive T cell responses show proinflammatory polarization in diabetes but a regulatory phenotype in health. J Clin Investig. 2004;113:451–463. doi: 10.1172/JCI19585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peakman M, Stevens EJ, Lohmann T, et al. Naturally processed and presented epitopes of the islet cell autoantigen IA-2 eluted from HLA-DR4. J Clin Investig. 1999;104:1449–1457. doi: 10.1172/JCI7936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pociot F, Lernmark A. Genetic risk factors for type 1 diabetes. Lancet. 2016;387:2331–2339. doi: 10.1016/S0140-6736(16)30582-7. [DOI] [PubMed] [Google Scholar]
- 6.Rewers M, Ludvigsson J. Environmental risk factors for type 1 diabetes. Lancet. 2016;387:2340–2348. doi: 10.1016/S0140-6736(16)30507-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thunander M, Petersson C, Jonzon K, et al. Incidence of type 1 and type 2 diabetes in adults and children in Kronoberg, Sweden. Diabetes Res Clin Pract. 2008;82:247–255. doi: 10.1016/j.diabres.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 8.Haller MJ, Atkinson MA, Schatz D. Type 1 diabetes mellitus: etiology, presentation, and management. Pediatr Clin N Am. 2005;52:1553–1578. doi: 10.1016/j.pcl.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Ziegler AG, Rewers M, Simell O, et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. Jama. 2013;309:2473–2479. doi: 10.1001/jama.2013.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Diabetes Control and Complications Trial Research Group The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med. 1993;329:977–986. doi: 10.1056/NEJM199309303291401. [DOI] [PubMed] [Google Scholar]
- 11.The Diabetes Control and Complications Trial Research Group The absence of a glycemic threshold for the development of long-term complications: the perspective of the Diabetes Control and Complications Trial. Diabetes. 1996;45:1289–1298. doi: 10.2337/diab.45.10.1289. [DOI] [PubMed] [Google Scholar]
- 12.The Diabetes Control and Complications Trial Research Group Hypoglycemia in the Diabetes Control and Complications Trial. Diabetes. 1997;46:271–286. doi: 10.2337/diab.46.2.271. [DOI] [PubMed] [Google Scholar]
- 13.The Diabetes Control and Complications Trial Research Group Effect of intensive therapy on residual beta-cell function in patients with type 1 diabetes in the Diabetes Control and Complications Trial. A randomized, controlled trial. Ann Intern Med. 1998;128:517–523. doi: 10.7326/0003-4819-128-7-199804010-00001. [DOI] [PubMed] [Google Scholar]
- 14.Steffes MW, Sibley S, Jackson M, Thomas W. Beta-cell function and the development of diabetes-related complications in the Diabetes Control and Complications Trial. Diabetes Care. 2003;26:832–836. doi: 10.2337/diacare.26.3.832. [DOI] [PubMed] [Google Scholar]
- 15.Greenbaum CJ, Beam CA, Boulware D, et al. Fall in C-peptide during first 2 years from diagnosis: evidence of at least two distinct phases from composite type 1 diabetes TrialNet data. Diabetes. 2012;61:2066–2073. doi: 10.2337/db11-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Speake C, Odegard JM. Evaluation of candidate biomarkers of type 1 diabetes via the core for assay validation. Biomark Insights. 2015;10:19–24. doi: 10.4137/BMI.S29697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cabrera SM, Chen YG, Hagopian WA, Hessner MJ. Blood-based signatures in type 1 diabetes. Diabetologia. 2016;59:414–425. doi: 10.1007/s00125-015-3843-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy H, Wang X, Kaldunski M, et al. Transcriptional signatures as a disease-specific and predictive inflammatory biomarker for type 1 diabetes. Genes Immun. 2012;13:593–604. doi: 10.1038/gene.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurram B, Salzman NH, Kaldunski ML, et al. Plasma-induced signatures reveal an extracellular milieu possessing an immunoregulatory bias in treatment-naive paediatric inflammatory bowel disease. Clin Exp Immunol. 2016;184:36–49. doi: 10.1111/cei.12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palatnik A, Ye S, Kendziorski C, et al. Identification of a serum-induced transcriptional signature associated with metastatic cervical cancer. PLoS One. 2017;12:e0181242. doi: 10.1371/journal.pone.0181242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Jia S, Geoffrey R, Alemzadeh R, Ghosh S, Hessner MJ. Identification of a molecular signature in human type 1 diabetes mellitus using serum and functional genomics. J Immunol. 2008;180:1929–1937. doi: 10.4049/jimmunol.180.3.1929. [DOI] [PubMed] [Google Scholar]
- 22.Chen YG, Cabrera SM, Jia S, et al. Molecular signatures differentiate immune states in type 1 diabetic families. Diabetes. 2014;63:3960–3973. doi: 10.2337/db14-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moran A, Bundy B, Becker DJ, et al. Interleukin-1 antagonism in type 1 diabetes of recent onset: two multicentre, randomised, double-blind, placebo-controlled trials. Lancet. 2013;381:1905–1915. doi: 10.1016/S0140-6736(13)60023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabrera SM, Wang X, Chen YG, et al. Interleukin-1 antagonism moderates the inflammatory state associated with type 1 diabetes during clinical trials conducted at disease onset. Eur J Immunol. 2016;46:1030–1046. doi: 10.1002/eji.201546005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orban T, Bundy B, Becker DJ, et al. Co-stimulation modulation with abatacept in patients with recent-onset type 1 diabetes: a randomised, double-blind, placebo-controlled trial. Lancet. 2011;378:412–419. doi: 10.1016/S0140-6736(11)60886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orban T, Beam CA, Xu P, et al. Reduction in CD4 central memory T-cell subset in costimulation modulator abatacept-treated patients with recent-onset type 1 diabetes is associated with slower C-peptide decline. Diabetes. 2014;63:3449–3457. doi: 10.2337/db14-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortensen HB, Hougaard P, Swift P, et al. New definition for the partial remission period in children and adolescents with type 1 diabetes. Diabetes Care. 2009;32:1384–1390. doi: 10.2337/dc08-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 30.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis AK, DuBose SN, Haller MJ, et al. Prevalence of detectable C-peptide according to age at diagnosis and duration of type 1 diabetes. Diabetes Care. 2015;38:476–481. doi: 10.2337/dc14-1952. [DOI] [PubMed] [Google Scholar]
- 32.The DCCT Research Group Effects of age, duration and treatment of insulin-dependent diabetes mellitus on residual beta-cell function: observations during eligibility testing for the Diabetes Control and Complications Trial (DCCT) J Clin Endocrinol Metab. 1987;65:30–36. doi: 10.1210/jcem-65-1-30. [DOI] [PubMed] [Google Scholar]
- 33.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 34.Quintana FJ, Sherr DH. Aryl hydrocarbon receptor control of adaptive immunity. Pharmacol Rev. 2013;65:1148–1161. doi: 10.1124/pr.113.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oishi Y, Spann NJ, Link VM, et al. SREBP1 contributes to resolution of pro-inflammatory TLR4 signaling by reprogramming fatty acid metabolism. Cell Metab. 2017;25:412–427. doi: 10.1016/j.cmet.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao D, Zhang Q, Liu Y, et al. H3K4me3 demethylase Kdm5a is required for NK cell activation by associating with p50 to suppress SOCS1. Cell Rep. 2016;15:288–299. doi: 10.1016/j.celrep.2016.03.035. [DOI] [PubMed] [Google Scholar]
- 37.Moya R, Robertson HK, Payne D, et al. A pilot study showing associations between frequency of CD4+ memory cell subsets at diagnosis and duration of partial remission in type 1 diabetes. Clin Immunol. 2016;166-167:72–80. doi: 10.1016/j.clim.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samandari N, Mirza AH, Nielsen LB, et al. Circulating microRNA levels predict residual beta cell function and glycaemic control in children with type 1 diabetes mellitus. Diabetologia. 2017;60:354–363. doi: 10.1007/s00125-016-4156-4. [DOI] [PubMed] [Google Scholar]
- 39.Alizadeh BZ, Hanifi-Moghaddam P, Eerligh P, et al. Association of interferon-gamma and interleukin 10 genotypes and serum levels with partial clinical remission in type 1 diabetes. Clin Exp Immunol. 2006;145:480–484. doi: 10.1111/j.1365-2249.2006.03172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanda S, Roep BO, von Herrath M. Islet antigen specific IL-10+ immune responses but not CD4+CD25+FoxP3+ cells at diagnosis predict glycemic control in type 1 diabetes. Clin Immunol. 2008;127:138–143. doi: 10.1016/j.clim.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen LB, Wang C, Sorensen K, et al. Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: evidence that miR-25 associates to residual beta-cell function and glycaemic control during disease progression. Exp Diabetes Res. 2012;2012:896362. doi: 10.1155/2012/896362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waugh K, Snell-Bergeon J, Michels A, et al. Increased inflammation is associated with islet autoimmunity and type 1 diabetes in the Diabetes Autoimmunity Study in the Young (DAISY) PLoS One. 2017;12:e0174840. doi: 10.1371/journal.pone.0174840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cabrera SM, Henschel AM, Hessner MJ. Innate inflammation in type 1 diabetes. Transl Res. 2016;167:214–227. doi: 10.1016/j.trsl.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferreira RC, Guo H, Coulson RM, et al. A type I interferon transcriptional signature precedes autoimmunity in children genetically at risk for type 1 diabetes. Diabetes. 2014;63:2538–2550. doi: 10.2337/db13-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kallionpaa H, Elo LL, Laajala E, et al. Innate immune activity is detected prior to seroconversion in children with HLA-conferred type 1 diabetes susceptibility. Diabetes. 2014;63:2402–2414. doi: 10.2337/db13-1775. [DOI] [PubMed] [Google Scholar]
- 46.Arif S, Leete P, Nguyen V, et al. Blood and islet phenotypes indicate immunological heterogeneity in type 1 diabetes. Diabetes. 2014;63:3835–3845. doi: 10.2337/db14-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leete P, Willcox A, Krogvold L, et al. Differential insulitic profiles determine the extent of beta-cell destruction and the age at onset of type 1 diabetes. Diabetes. 2016;65:1362–1369. doi: 10.2337/db15-1615. [DOI] [PubMed] [Google Scholar]
- 48.Deppong CM, Bricker TL, Rannals BD, Van Rooijen N, Hsieh CS, Green JM. CTLA4Ig inhibits effector T cells through regulatory T cells and TGF-beta. J Immunol. 2013;191:3082–3089. doi: 10.4049/jimmunol.1300830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hao W, Gitelman S, DiMeglio LA, Boulware D, Greenbaum CJ, Type 1 Diabetes TrialNet Study Group Fall in C-peptide during first 4 years from diagnosis of type 1 diabetes: variable relation to age, HbA1c, and insulin dose. Diabetes Care. 2016;39:1664–1670. doi: 10.2337/dc16-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 434 kb)
Data Availability Statement
Gene expression data files are publicly available through the National Center for Biotechnology Information Gene Expression Omnibus (accession number GSE102234).
Gene expression data files are publicly available through the National Center for Biotechnology Information Gene Expression Omnibus (accession number GSE102234).