Abstract
Background
Plectranthus edulis (Vatke) Agnew (locally known as Ethiopian dinich or Ethiopian potato) is one of the most economically important edible tuber crops indigenous to Ethiopia. Evaluating the extent of genetic diversity within and among populations is one of the first and most important steps in breeding and conservation measures. Hence, this study was aimed at evaluating the genetic diversity and population structure of this crop using collections from diverse agro-ecologies in Ethiopia.
Results
Twenty polymorphic expressed sequence tag based simple sequence repeat (EST-SSRs) markers were developed for P. edulis based on EST sequences of P. barbatus deposited in the GenBank. These markers were used for genetic diversity analyses of 287 individual plants representing 12 populations, and a total of 128 alleles were identified across the entire loci and populations. Different parameters were used to estimate the genetic diversity within populations; and gene diversity index (GD) ranged from 0.31 to 0.39 with overall mean of 0.35. Hierarchical analysis of molecular variance (AMOVA) showed significant but low population differentiation with only 3% of the total variation accounted for variation among populations. Likewise, cluster and STRUCTURE analyses did not group the populations into sharply distinct clusters, which could be attributed to historical and contemporary gene flow and the reproductive biology of the crop.
Conclusions
These newly developed EST-SSR markers are highly polymorphic within P. edulis and hence are valuable genetic tools that can be used to evaluate the extent of genetic diversity and population structure of not only P. edulis but also various other species within the Lamiaceae family. Among the 12 populations studied, populations collected from Wenbera, Awi and Wolaita showed a higher genetic diversity as compared to other populations, and hence these areas can be considered as hot spots for in-situ conservation as well as for identification of genotypes that can be used in breeding programs.
Electronic supplementary material
The online version of this article (10.1186/s12863-018-0682-z) contains supplementary material, which is available to authorized users.
Keywords: Ethiopian dinich, Expressed sequence tags, Genetic diversity, Plectranthus edulis, Population structure, Simple sequence repeats
Background
Ethiopia is one of the countries in the world that has the greatest crop genetic diversity and considered as a primary gene center for several crops [1–3] including edible roots and tubers [4]. However, the food potential of horticultural crops, particularly those of indigenous edible roots and tubers have not been fully exploited despite their significant contributions to the livelihood of subsistence farmers. Such crops are overlooked in terms of research and breeding and their production and management systems are restricted to local farmers’ varieties maintained by farmers using local knowledge [5–7].
Plectranthus edulis (Vatke) Agnew (Lamiaceae), also locally known as Ethiopian potato syno. Ethiopian dinich, is one of the most economically important indigenous tuber crops [2, 8] with wide distribution in parts of Africa, largely in wild form [9]. It produces edible stem tubers on stolon below ground. The crop can thrive and set tubers under significant environmental constraints including degraded and poor soil and can produce reasonable yield without a need for intensive management practices [10].
In Ethiopia, this edible tuber crop is commonly cultivated by smallholder farmers around homesteads, alone or mixed with cereals, fruits and pulses, largely for household consumption and rarely for marketing of its tubers. It was one of the most common Ethiopian staple crops [11] and usually called ‘hunger crop’ as it fills the food supply gap that occurs from August to November, the period before the harvest of cereal crops. It has also been widely used as a folk medicine in several parts of Africa including Ethiopia [12] and is commonly visited by honeybees for its nectar [13].
Nowadays, local gene pool of the crop is under the threat of severe genetic erosion as it is disappearing from several areas where it used to be widely cultivated and restricted to highly marginalized land and limited to few elderly farmers in some other areas. The lack of improved planting material and limited awareness about the significance of this crop among younger farmers as well as the introduction of other high yielding tuber crops like Irish potato to the area, recurrent drought and environmental degradation have contributed to the genetic erosion of this crop [5, 14, 15].
Assessing the extent of genetic diversity of crop species is a vitally important step for their effective conservation and improvement. Molecular markers, such as simple sequence repeats (SSRs) are among the most important genetic tools for such purpose [16, 17]. SSRs derived from Expressed sequence tags (ESTs) (EST-SSRs) have been widely used in many plant species including root and tuber crops, as they are relatively fast and cost-effective to develop [18], have a well-conserved flanking sequences among phylogenetically closely related species and hence are highly transferable among related taxa, and are less susceptible to null alleles [19]. Such cross-genera and cross-species transferable molecular markers are highly important genomic resources to study plant species with little or no DNA sequence information, such as P. edulis. Until recently, very few transferable EST-SSR markers have been developed within the family Lamiaceae [20, 21] and, so far, there is no report of such markers for the genus Plectranthus. Moreover, there is no publication on molecular marker based genetic diversity of P. edulis, which shows that researchers and plant breeders have not given enough attention for conservation and improvement of this crop. Consequently, its production, utilization, and improvement are highly restricted. Hence, this study was initiated with the aim of developing and validating EST-SSR markers for use in population structure and genetic diversity analyses of the crop, which eventually serve as a basis for its improvement and sustainable conservation.
Methods
Plant material
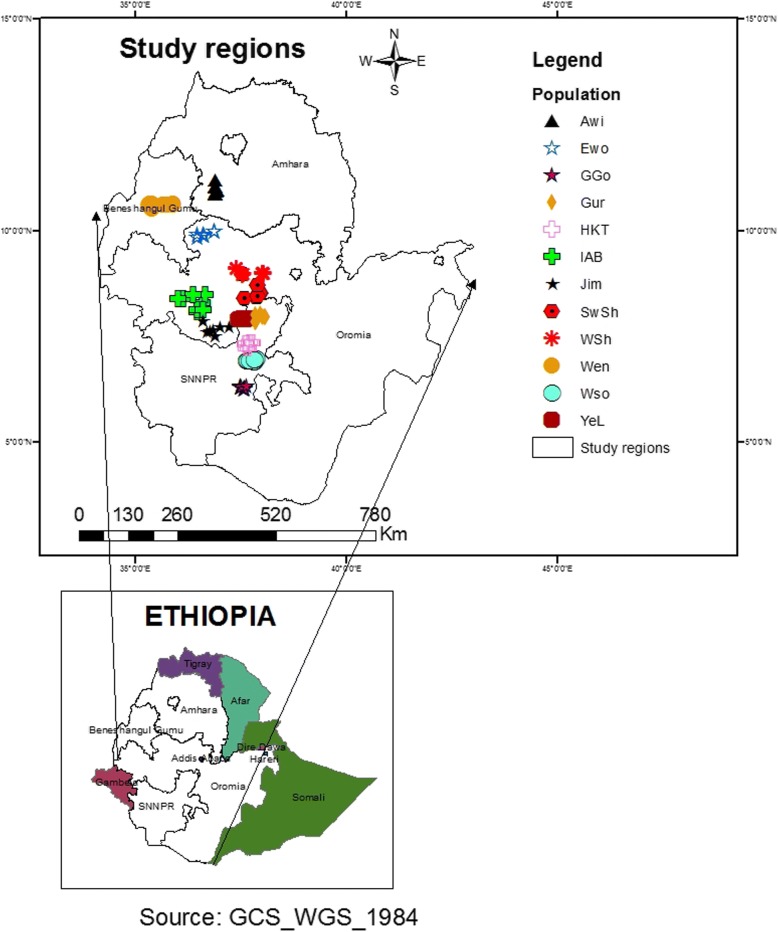
A total of 174 tuber samples of cultivated P. edulis, representing 12 populations, were randomly collected from farmers’ fields with permission from individual farmers (Fig. 1, Table 1). The identity of the samples was confirmed based on the species description provided in Flora of Ethiopia and Eritrea [22]. The collected samples were planted during the regular crop growing season (end of April) in 2016 at Holeta Agricultural Research Centre (which is a part of the Ethiopian Institute of Agricultural Research) field site located 40 Km west of Addis Ababa. This field site is located at a geographic position of 09o04’N, 38o 29′E and altitude of 2400 masl.
Fig. 1.
Map of Ethiopia showing its Federal Regions (left bottom) and tuber sample collection sites, representing the 12 studied populations, within four of the Federal Regions (left upper) (see Table 1 for full description of the populations). The map was original and constructed using geographic coordinates and elevation data gathered from each collection sites using global positioning system (GPS)
Table 1.
List of populations used in this study and their geographic positions and altitudes of collection along with their tuber and leaf sample sizes
| Population | Sample size | Altitude range (m) | Geographic position (UTM)a | ||
|---|---|---|---|---|---|
| Tuber | Leaf | Latitude range | Longitude range | ||
| SwSh | 13 | 24 | 1817–2631 | 0373275–0386714 | 0927865–0962106 |
| EW | 14 | 24 | 1991–2172 | 0221199–0266764 | 1,089,267–1,105,086 |
| Aw | 21 | 25 | 2167–2643 | 0267210–0281621 | 1,201,272–1,239,531 |
| Gur | 18 | 24 | 2069–2929 | 0371621–0397588 | 0867217–0889992 |
| HKT | 9 | 23 | 2121–2562 | 0363184–0370744 | 0813134–0832296 |
| IAB | 14 | 25 | 1650–2096 | 0133324–0784937 | 0893136–0949107 |
| Jim | 16 | 24 | 1605–2228 | 0197291–0327491 | 0829838–0875685 |
| GG | 13 | 24 | 2513–2744 | 0332363–0343464 | 0690582–0697778 |
| WS | 18 | 23 | 1979–2232 | 0349195–0372876 | 0760650–0770184 |
| Wen | 10 | 26 | 2476–2530 | 0789433–0793823 | 1,174,041–1,175,300 |
| WSh | 17 | 23 | 2249–2924 | 0322544–0394336 | 0989176–1,007,305 |
| YeL | 11 | 22 | 2466–2589 | 0337470–0339720 | 0873081–0875666 |
| Total | 174 | 287 | |||
SwSh Southwest Shewa, EW Esat Wollega, Aw Awi, Gur Gurage, HKT Hadiya, Kembata and Tembaro, IAB Illu Aba Bora, Jim Jimma, GG Gamo Gofa, WS Wollaita Sodo, Wen Wenbera, WSh West Shewa, YeL Yem Liyu; aUTM = Universal Transverse Mercator coordinate system
Leaf sample collection and DNA extraction
oung leaf tissue from 287 individual plants (one to three individual plants per tuber sample) (Table 1, Additional file 1) were separately collected in a ziplock bag, silica gel dried and transported to the Swedish University of Agricultural Sciences (SLU), Alnarp, Sweden. Genomic DNA was extracted from these samples using a modified Cetyl Trimethyl Ammonium Bromide (CTAB) protocol as described in Geleta et al. [23]. The DNA quality was assessed using 1% agarose gel electrophoresis whereas NanoDrop® ND-1000 Spectrophotometer (Saveen Warner, Sweden) was used to determine the quantity of extracted DNA.
Data mining, designing and screening EST-SSR primers
Initially, 3263 P. barbatus EST sequences were retrieved from the National Centre for Biotechnology Information (NCBI public DNA sequence database (https://www.ncbi.nlm.nih.gov/nucest/?term=Plectranthus+barbatus), and sequences containing SSRs were screened using WebSat, a web-based software for microsatellite marker development [24] (http://purl.oclc.org/NET/websat/). After excluding redundant, overlapping and short sequences, about 300 sequences containing two to six SSR motifs were identified. Of these, 40 sequences were selected for designing primers using Primer3 primer designing program [25, 26] (http://bioinfo.ut.ee/primer3-0.4.0/).
The 40 newly designed primer-pairs were tested for amplification of their target genomic regions using 36 DNA samples representing the 12 populations of P. edulis. The polymerase chain reaction (PCR) products were electrophoresed on 1.5% agarose gel containing gel-red and visualized using Saveen Werner AB UV camera equipped with SSM930CE Sony Black and white Monitor. The size of amplified products was estimated by loading GeneRuler 50 bp DNA ladder together with the samples on separate lanes. Under optimized PCR conditions, 20 of the 40 primer-pairs consistently amplified their polymorphic loci, and hence were selected for use on all samples included in the present study (Table 2).
Table 2.
Characteristics of the 20 polymorphic SSR markers developed for genetic diversity analyses of P. edulis
| Primer / locus name | GenBank ANSSa | SSR motif (5′ to 3′) | forward primer (5′ to 3′) | reverse primer (5′ to 3′) | Expected allele size (bp) | Observed allele size range (bp) | Ta (°C) |
|---|---|---|---|---|---|---|---|
| PE_01 | GB|JZ730850.1 | (TC)9 | TCACCGCAGTTTTCAGTCTCTA | ATGAGATTCGCCATAGGTTTGT | 102 | 122–146 | 59 |
| PE_02 | GB|JZ730431.1 | (AAG)6 | TGAATCTGCAAAGACATCTGCT | AACTGAGACAACTCCATTGACG | 114 | 108–126 | 57 |
| PE_03 | GB|JZ730426.1 | (TA)10 | GAGACGACGACCAATGTTGTTA | CTCTCTATCACTCCTGACGGCT | 125 | 106–142 | 56 |
| PE_04 | GB|JZ732709.1 | (ACACCC)5 | GGGGAATTAGAGATGGAAAGATAGA | TTGAGGGTGTGAAGTGGTACAG | 131 | 137–143 | 57 |
| PE_05 | GB|JZ732789.1 | (TG)10 | GCCGTATCTCCATTTGTTGATT | TCCATGCTCCTCACACATTATC | 141 | 140–168 | 57 |
| PE_06 | GB|JZ731502.1 | (AC)6 | GCTTACGCCAAGAACTGAAACT | AATAGCAATCTCTTTCCCTCCC | 164 | 162–182 | 55 |
| PE_07 | GB|JZ730947.1 | (CTC)4 | GTTGGACGACTGGGTTTTATGT | GAATGACGCTAGTTTGCTGTTG | 314 | 319–340 | 55 |
| PE_08 | GB|JZ731328.1 | (GCT)6 | CCAACAGCAATCCATATTACCA | ATTTCTCAAGTCAGTCCGAGGT | 172 | 162–189 | 57 |
| PE_09 | GB|JZ732633.1 | (CAG)6 | AACCAAATGACAGGAGCATCTT | CAATTTCTTCATACTGGGTGGC | 184 | 180–206 | 55 |
| PE_10 | GB|JZ730151.1 | (CTT)5 | GGGTAACGATTTGAATAATGCG | GTGAAGCGGGATCTACACTGA | 188 | 176–200 | 52 |
| PE_11 | GB|JZ730075.1 | (AGAGA)4 | GTCAGCCTTTCTCTCTCGTCTC | AGGGGAGTGTGTTATCAAATGG | 223 | 249–264 | 56 |
| PE_12 | GB|JZ730223.1 | (AGC)6 | GACCATCGGTAAGGAGAACTTG | GGATATGAGCTGGATAGCTGGT | 226 | 213–243 | 55 |
| PE_13 | GB|JZ731331.1 | (CAC)5 | GCAAAAGTTCTACCAGCGTTTC | GGTTTGTTGATCCCAACGTAAT | 229 | 242–251 | 57 |
| PE_14 | GB|JZ731120.1 | (GAA)10 | AATTACTTTCATGTCCAACGCC | TTTTCATCACTACCATCCCAGTC | 229 | 228–243 | 55 |
| PE_15 | GB|JZ731840.1 | (TGT)5 | AGTTGAGATTGTACTCCACGTTGT | CTACTTCAGTTCCGGCCTCTTA | 260 | 233–260 | 57 |
| PE_16 | GB|JZ732555.1 | (AC)9 | GGCGATATTCAAGAAAGCAACT | TCTTTTCACGCTTCCATCTCTT | 355 | 319–339 | 57 |
| PE_17 | GB|JZ729809.1 | (GA)8 | GGGGCTTTTCTTGTTTGAAGAT | ATTGGAGGCAACTCATCAGAAT | 361 | 370–382 | 57 |
| PE_18 | GB|JZ732589.1 | (GA)9 | ATGAAGAGTGAAGAGGCTGGAG | AGGAGCAAGCAAATAGAAATCG | 306 | 304–322 | 57 |
| PE_19 | GB|JZ732378.1 | (TGC)5 | GTGTACATGCCATAGAATGAGT | GACTTCATCATATACGGCGGTT | 167 | 180–192 | 57 |
| PE_20 | GB|JZ730154.1 | (TGC)5 | TACTTTATGGCAGAGAATGCGA | CTTGAGAGCCCTGAGACTTCAT | 178 | 200–227 | 57 |
aANSS Accession number of source sequences
Pre-amplification and capillary electrophoresis
For cost effectiveness and improved quality of amplified products, the sequences of the 20 primer-pairs were modified as follows: (1) A 18-bp universal M13 primer sequence (5′-TGTAAAACGACGGCCAGT-3′) was added as a common tail to the 5′-end of all forward primers following Oetting et al. [27]; and (2) a PIG-tail sequence of 5′-GCTTCT-3′ was added to the 5′-end of the reverse primers to prevent non-templated addition of nucleotides to amplified products as described in Brownstein et al. [28]. M13 primer 5′-end labeled with HEX™ or 6-FAM™ fluorophores was used as a third primer in each PCR amplification.
PCR was carried out using 96-well plates with 25 μl reaction volume [1× reaction buffer, 1.5 mM MgCl2, 0.3 mM dNTPs, 0.08 μM M13-tailed forward primer, 0.3 μM pig-tailed reverse primer, 0.3 μM 6-FAM or HEX labeled M13 primer, 1 U Dream Taq DNA Polymerase, and 25 ng template DNA]. A mixture of all the components except genomic DNA was included as a negative control. Amplification was performed using GeneAMP PCR 9700 thermocycler (Applied Biosystems Inc. USA) according to the following five-stage PCR protocol: (1) initial 15 min denaturation at 95 °C, (2) 35 cycles of 30 s denaturation at 94 °C, 30 s annealing at optimized annealing temperature for each primer-pair (see Table 1), and 30 s primer extension at 72 °C, (3) eight cycles of 30 s denaturation at 94 °C, 45 s annealing at 53 °C and 45 s primer extension at 72 °C, (4) additional primer extension at 72 °C for 10 min, and (5) a final 30 min primer extension at 60 °C. The PCR products were stored at 4 °C until they were electrophoresed. The PCR products were multiplexed into panels based on fragment sizes of the SSRs and the fluorescence label of the M13 primer and diluted 25× using Millipore water. Finally, 0.7 μl of multiplexed and diluted PCR products, 1.9 μl Hi-Di formamide and 0.3 μl size standard (GenScanTm600 LIZ® size standard) were mixed and ccapillary electrophoresis was conducted using Genetic Analyzer 3500 (Applied Biosystems) at SLU, Department of Plant Breeding, Alnarp, Sweden.
Allele scoring and statistical analysis
Peak identification and fragment sizing were done using GeneMarker V2.6.0 (SoftGenetics®) with default settings and 200 threshold intensities. Then, the allele size (bp) data at each locus were exported to excel for statistical analyses. Locus based diversity indices: major allele frequency (MAF), the number of alleles (NA), gene diversity (GD), and polymorphic information content (PIC) were determined using PowerMarker ver. 3.25 [29] (Table 3). The number of effective alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He) [30], Shannon’s Information Index (I), and estimate of the deviation from Hardy-Weinberg equilibrium (HWE) over the entire populations and population genetic diversity indices: Ne, percentage of polymorphic loci (PPL), Ho, He, I, Nei’s gene diversity over the entire loci were computed using GenAlEx ver. 6.501 [31] (Table 4). To determine the correlation between observed allelic diversity and sample size of populations, rarified allelic richness (Ar) and private rarified allelic richness (Ap) were estimated using HP-Rare 1.1 software [32]. To identify genetic groups, G, the number of distinct multi-locus genotypes (MLGs) present in each sample were evaluated using GenClone 2.0 [33]. To evaluate the global clonality rate of the sample, the index of clonal diversity was computed as G/N, where G is the number of MLGs and N is the total number of genotyped individuals. To analyze the distribution of genetic variation and to estimate the variance components of the populations, analysis of molecular variance (AMOVA) was computed using Arlequin ver. 3.5.2.2 [34]. Population differentiation tests: Wrights fixation index (FST) and pairwise FST were computed using GenAlEx ver. 6.501 [31], and significance was tested based on 1000 bootstraps.
Table 3.
Informativeness and levels of different diversity indices of the SSR loci across populations
| SSR loci | MAF | NA | Ne | GD | Ar | Arp | Ho | He | FST | Nm | PIC | I | PHWEa | F |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PE_01 | 0.87 | 7 | 1.35 | 0.24 | 2.91 | 0.15 | 0.22 | 0.23 | 0.08 | 2.90 | 0.23 | 0.45 | 0.530ns | 0.11 |
| PE_02 | 0.49 | 6 | 2.24 | 0.55 | 3.05 | 0.13 | 0.97 | 0.55 | 0.01 | 35.07 | 0.45 | 0.87 | 0.000* | −0.75 |
| PE_03 | 0.87 | 8 | 1.34 | 0.25 | 3.32 | 0.06 | 0.19 | 0.23 | 0.04 | 5.47 | 0.24 | 0.49 | 0.000* | 0.23 |
| PE_04 | 0.91 | 2 | 1.19 | 0.16 | 2.02 | 0.07 | 0.18 | 0.16 | 0.06 | 4.07 | 0.15 | 0.28 | 0.112ns | −0.07 |
| PE_05 | 0.79 | 10 | 1.56 | 0.36 | 3.56 | 0.18 | 0.27 | 0.35 | 0.04 | 5.52 | 0.34 | 0.66 | 0.000* | 0.27 |
| PE_06 | 0.46 | 9 | 3.17 | 0.70 | 4.97 | 0.08 | 0.86 | 0.68 | 0.03 | 8.54 | 0.66 | 1.32 | 0.000* | −0.22 |
| PE_07 | 0.97 | 9 | 1.06 | 0.05 | 1.72 | 0.46 | 0.01 | 0.05 | 0.04 | 5.67 | 0.05 | 0.12 | 0.783ns | 0.73 |
| PE_08 | 0.96 | 5 | 1.09 | 0.08 | 1.68 | 0.20 | 0.07 | 0.08 | 0.05 | 4.66 | 0.08 | 0.15 | 0.115ns | 0.18 |
| PE_09 | 0.68 | 6 | 2.04 | 0.50 | 4.28 | 0.08 | 0.38 | 0.49 | 0.03 | 7.47 | 0.47 | 0.95 | 0.000* | 0.25 |
| PE_10 | 0.72 | 5 | 1.70 | 0.42 | 2.43 | 0.11 | 0.54 | 0.40 | 0.05 | 5.23 | 0.34 | 0.62 | 0.000* | −0.29 |
| PE_11 | 0.77 | 6 | 1.66 | 0.38 | 3.11 | 0.16 | 0.39 | 0.37 | 0.03 | 7.64 | 0.35 | 0.68 | 0.335ns | −0.01 |
| PE_12 | 0.76 | 7 | 1.65 | 0.39 | 3.40 | 0.17 | 0.29 | 0.37 | 0.05 | 5.22 | 0.36 | 0.70 | 0.000* | 0.26 |
| PE_13 | 0.96 | 4 | 1.09 | 0.08 | 2.00 | 0.09 | 0.04 | 0.08 | 0.02 | 11.76 | 0.07 | 0.17 | 0.542ns | 0.53 |
| PE_14 | 0.81 | 4 | 1.44 | 0.30 | 2.24 | 0.07 | 0.30 | 0.28 | 0.09 | 2.69 | 0.26 | 0.46 | 0.965ns | 0.04 |
| PE_15 | 0.90 | 5 | 1.24 | 0.19 | 2.66 | 0.08 | 0.13 | 0.18 | 0.05 | 4.56 | 0.18 | 0.36 | 0.876ns | 0.30 |
| PE_16 | 0.70 | 12 | 1.80 | 0.46 | 4.03 | 0.30 | 0.49 | 0.43 | 0.06 | 3.70 | 0.42 | 0.81 | 0.115ns | −0.06 |
| PE_17 | 0.63 | 6 | 1.99 | 0.51 | 2.90 | 0.06 | 0.57 | 0.48 | 0.06 | 3.87 | 0.44 | 0.78 | 0.000* | − 0.10 |
| PE_18 | 0.68 | 8 | 1.96 | 0.49 | 3.40 | 0.26 | 0.43 | 0.48 | 0.03 | 8.65 | 0.44 | 0.85 | 0.128ns | 0.11 |
| PE_19 | 0.53 | 5 | 2.12 | 0.53 | 2.87 | 0.07 | 0.88 | 0.53 | 0.01 | 25.01 | 0.43 | 0.82 | 0.000* | −0.65 |
| PE_20 | 0.72 | 4 | 1.65 | 0.41 | 2.22 | 0.07 | 0.54 | 0.38 | 0.06 | 4.11 | 0.33 | 0.59 | 0.000* | −0.33 |
| Mean | 0.76 | 6.45 | 1.67 | 0.35 | 2.94 | 0.14 | 0.39 | 0.34 | 0.04 | 5.84 | 0.32 | 0.61 | −0.09 |
MAF Major allele frequency, NA Number of alleles, Ne Effective number of alleles, GD Gene diversity, Ar Allelic richness, Arp Private allelic richness, HO Observed heterozygosity, He Expected heterozygosity, Fst Inbreeding coefficient within subpopulations relative to total (genetic differentiation among subpopulations), Nm Gene flow estimated from Fst 0.25(1- Fst)/Fst, PIC Polymorphic information content, I Shannon’s Information Index, F Fixation Index, PHWEa P-value for deviation from Hardy Weinberg equilibrium, ns not significant, * = P < 0.0001 and hence highly significant
Table 4.
Summary of different population diversity indices averaged over the 20 loci
| Popa | G | N | G/N | Ar | Arp | Ne | PPL | Ho | He | GD | I |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SwSh | 8 | 24 | 0.33 | 2.78 | 0.12 | 1.60 | 95.00 | 0.43 | 0.33 | 0.34 | 0.59 |
| EW | 9 | 24 | 0.38 | 2.56 | 0.01 | 1.60 | 90.00 | 0.37 | 0.30 | 0.31 | 0.52 |
| Aw | 13 | 25 | 0.52 | 2.93 | 0.14 | 1.80 | 95.00 | 0.35 | 0.36 | 0.36 | 0.64 |
| Gur | 10 | 24 | 0.42 | 3.33 | 0.28 | 1.60 | 95.00 | 0.33 | 0.35 | 0.35 | 0.64 |
| HKT | 11 | 23 | 0.48 | 2.80 | 0.04 | 1.70 | 95.00 | 0.43 | 0.34 | 0.35 | 0.60 |
| IAB | 16 | 25 | 0.64 | 2.80 | 0.09 | 1.70 | 90.00 | 0.39 | 0.34 | 0.34 | 0.59 |
| Jim | 13 | 24 | 0.54 | 3.18 | 0.25 | 1.60 | 95.00 | 0.39 | 0.34 | 0.35 | 0.62 |
| GG | 19 | 24 | 0.79 | 2.99 | 0.17 | 1.60 | 95.00 | 0.39 | 0.34 | 0.35 | 0.61 |
| WS | 15 | 23 | 0.65 | 3.17 | 0.14 | 1.80 | 95.00 | 0.41 | 0.37 | 0.37 | 0.66 |
| Wen | 14 | 26 | 0.54 | 2.74 | 0.13 | 1.80 | 95.00 | 0.42 | 0.38 | 0.39 | 0.65 |
| WSh | 12 | 23 | 0.52 | 3.01 | 0.22 | 1.60 | 90.00 | 0.37 | 0.31 | 0.32 | 0.57 |
| YeL | 17 | 22 | 0.77 | 2.96 | 0.15 | 1.60 | 100.00 | 0.37 | 0.32 | 0.33 | 0.57 |
| Mean | 13.08 | 287c | 0.55 | 2.94 | 0.15 | 1.70 | 94.20 | 0.39 | 0.34 | 0.35 | 0.61 |
aSwSh Southwest Shewa, EW East Wollega, Aw Awi, Gur Gurage, HKT Hadiya, Kembata and Tembaro, IAB Illu Aba Bora, Jim Jimma, GG Gamo Gofa, WS Wollaita Sodo, Wen Wenbera, WSh West Shewa, YeL Yem Liyu woreda, G number of distinct multi-locus genotype (MLG), N number of samples genotyped, G/N the ratio of G to N, c total number of samples, Ar Allelic richness, Arp Private allelic richness, Ne effective number of alleles, PPL percentage of polymorphic loci, HO observed heterozygosity, He expected heterozygosity, GD gene diversity, I Shannon diversity index
Simple matching dissimilarity coefficient-based Neighbor-Joining tree and Nei’s standard genetic distance (DST, corrected) [35] based Unweighted Pair Group Method with Arithmetic Mean (UPGMA) [36] tree was constructed using DARwin var. 6.0.14 [37] and POPTREE2 [38], respectively, and significance was tested based on 1000 bootstraps [39]. The resulting trees were displayed using TreeView (Win 32) 1.6.6 program [40] and FigTree var. 1.4.3 [41]. Gene flow (Nm) among populations was estimated using the formula, Nm = 0.25(1 − Fst)/Fst [42].
A Bayesian model-based clustering algorithm in STRUCTURE ver. 2.3.4 [43, 44] was applied to infer the pattern of population structure and detection of admixture. To determine the most likely number of populations (K), a burn-in period of 50,000 was used in each run, and data were collected over 500,000 Markov Chain Monte Carlo (MCMC) replications for K = 1 to K = 12 using 20 iterations for each K. The optimum K value was predicted following the simulation method of Evanno et al. [45] using the web-based STRUCTURE HARVESTER ver. 0.6.92 [46]. Bar plot for the optimum K was determined using Clumpak beta version [47].
Results
Validation of the EST-SSRs marker and evaluation of their levels of polymorphism
The 20 newly developed EST-SSRs markers (Table 2) are predominantly trinucleotides and dinucleotides. Trinucleotide SSRs accounted for 55% of the loci with the number of repeats ranging from four to ten whereas dinucleotide SSRs accounted for 35% of the loci with the number of repeats ranging from six to ten. The remaining two loci (10%) were pentanucleotide and hexanucleotide repeats with four and five number of repeats, in that order. All the 20 loci were polymorphic and produced a total of 128 alleles (an average of 6.4 alleles per locus) (Table 3), out of which 62 (48.4%) were rare (frequency < 0.01) and 22 (17.2%) were scarce (frequency between 0.01 and 0.05). The frequency of seven alleles (5.5%) was between 0.05 and 0.1 whereas 37 alleles (28.9%) had a frequency of 0.1 or higher (Additional file 2).
The maximum number of alleles detected per locus was 12 (PE_16; Table 3). The least major allele frequency (MAF; 0.46), and largest effective number of alleles (Ne) (3.17), allelic richness (4.97), Nei’s gene diversity (GD) (0.70), polymorphic information contents (PIC) (0.66) and Shannon information index (I) (1.33) were recorded for PE_06. The highest MAF (0.97), private allelic richness (0.46), and the least Ne (1.06), I (0.12), GD (0.05) and PIC (0.05) were recorded for PE_07. In terms of the overall PIC, one SSR locus (PE_06) was found to be highly informative (PIC ≥0.5), 12 loci (60%) were moderately informative (0.5 < PIC ≥0.25), and the remaining seven were less informative (PIC < 0.25) (Table 3). The highest observed heterozygosity (Ho) (0.97), the lowest fixation index (− 0.75) and the highest value for gene flow (Nm) (35.1) were recorded for PE_02 (Table 3). Ten of the twenty loci showed a highly significant deviation from HW-equilibrium over the entire populations (Table 3).
Genetic variation within and among populations
Among the 12 populations studied, not much differences were observed in terms of a number of genetic diversity paramters, such as effective number of alleles (Ne), observed heterozygosity (Ho) and expected heterozygosity (He), gene diversity (GD) and Shannon diversity index (I). However, Wen, WS and Aw populations scored higher values in Ne, GD and I while SwSh and HKT populations scored a higher Ho as compared to the other populations. Five populations: Aw, Gur, EW, WSh, and YeL, in the order of magnitude, scored slightly less than the mean Ho whereas IAB, Jim, and GG populations had a mean Ho value. Although Ho value is the lowest for Gur population, it comes first in terms of allelic richness (Ar) including richness in private alleles (Arp) with Ar of 3.33 and Arp of 0.28 (Table 4). Jim and WSh populations ranked second and third in terms of overall Arp. The mean number of distinct multi-locus genotypes (MLGs) and index of clonality (G/N) over the entire samples (populations) was 13.08 and 0.55, respectively (Table 4). The analysis of percentage of polymorphic loci (PPL) showed that at least 90% of the loci were polymorphic in each population studied, with a mean PPL of 94.2% (Table 4).
Population genetic differentiation and gene flow
The hierarchical AMOVA was conducted without grouping the populations as well as by grouping the populations according to administrative regions (AR) and geographical regions (GR). In all cases, variation within individuals accounted for at least 97% of the total variation. The variation among populations, AR and GR accounted for only 3, 2 and 2% of the total variation, respectively. Hence, most of the within population variation is due to the heterozygosity of the individuals within each population. The overall FST value was very small (0.03) (Table 5, Additional file 3). The overall gene flow among the populations was estimated to be high (Nm = 5.84) (Table 3) whereas pairwise population differentiation test computed according to Weir [30], with exclusion of null alleles, ranged from 0.01 to 0.07.
Table 5.
Analysis of molecular variance (AMOVA) for the 12 populations without grouping and with grouping them according to their geographic regions of collections (GR) and administrative regions of collections (AR) based on data from the 20 loci
| Source of Variation | Df | SS | Variance components | %Variation | Fixation index | P value |
|---|---|---|---|---|---|---|
| Among populations | 11 | 61.94 | 0.072 Va | 2.55 | FST: 0.03 | Va and FST = 0.000 |
| Among individuals within populations | 275 | 603.72 | −0.56 Vb | −19.66 | FIS: −0.20 | Vb and FIS = 1.000 |
| Within individuals | 287 | 948.50 | 3.31 Vc | 117.11 | FIT: −0.17 | Vc and FIT = 1.000 |
| Total | 573 | 1614.17 | 2.82 | |||
| Among AR | 3 | 26.19 | 0.05 Va | 1.85 | FST: 0.02 | Va and FST = 0.000 |
| Among individuals within AR | 283 | 639.48 | −0.52 Vb | −18.44 | FIS: −0.19 | Vb and FIS = 1.000 |
| Within individuals | 287 | 948.50 | 3.31 Vc | 116.59 | FIT: −0.17 | Vc and FIT = 1.000 |
| Total | 573 | 1614.17 | 2.84 | |||
| Among GR | 3 | 24.42 | 0.04 Va | 1.53 | FST: 0.02 | Va and FST = 0.000 |
| Among individuals within GR | 283 | 641.25 | 0.52 Vb | −18.37 | FIS: −0.19 | Vb and FIS = 1.000 |
| Within individuals | 287 | 948.50 | 3.31 Vc | 116.84 | FIT: −0.17 | Vc and FIT = 1.000 |
| Total | 573 | 1614.17 | 2.83 |
AR administrative regions (Oromia, Amhara, SNNPs, and Benshangul Gumuz), GR geographic regions (Northwestern, Western, Southern, and Southwestern Ethiopia), Df Degrees of freedom, SS Sum of squares
Genetic distance between the populations
Nei’s standard genetic distance between populations ranged from 0.02 to 0.05. The highest pairwise genetic distance (0.05) was observed between four pairs of populations (Aw vs WS, Wen vs HKT, WS and YeL). The mean genetic distance of each population from the other populations ranged from 0.02 to 0.04 and, in terms of this parameter, Wen is the most distantly related population with a mean genetic distance of 0.04 (Table 6). Similarly, Weir’s estimation of population differentiation (FST) [30, 48] ranged from 0.01 to 0.07 with the highest value recorded between Wen and YeL populations.
Table 6.
Nei’s standard genetic distance between populations (below diagonal) [35], and Weir [30] estimation of population pairwise FST using the ENA correction as described in Chapuis and Estoup [48] (above diagonal) for the 12 populations studied
| Pop ID | SwSh | EW | Aw | Gur | HKT | IAB | Jim | GG | WS | Wen | WSh | YeL | Meana |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SwSh | **** | 0.02 | 0.04 | 0.03 | 0.01 | 0.01 | 0.02 | 0.04 | 0.03 | 0.05 | 0.02 | 0.03 | 0.02 |
| EW | 0.01 | **** | 0.04 | 0.02 | 0.01 | 0.01 | 0.02 | 0.03 | 0.03 | 0.05 | 0.01 | 0.02 | 0.02 |
| Aw | 0.03 | 0.03 | **** | 0.04 | 0.04 | 0.03 | 0.03 | 0.04 | 0.06 | 0.02 | 0.03 | 0.05 | 0.03 |
| Gur | 0.02 | 0.01 | 0.03 | **** | 0.03 | 0.03 | 0.02 | 0.02 | 0.01 | 0.05 | 0.01 | 0.01 | 0.02 |
| HKT | 0.01 | 0.01 | 0.03 | 0.02 | **** | 0.01 | 0.02 | 0.05 | 0.01 | 0.06 | 0.01 | 0.03 | 0.02 |
| IAB | 0.01 | 0.01 | 0.02 | 0.02 | 0.01 | **** | 0.01 | 0.04 | 0.03 | 0.05 | 0.02 | 0.02 | 0.02 |
| Jim | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.01 | **** | 0.03 | 0.03 | 0.04 | 0.01 | 0.02 | 0.02 |
| GG | 0.03 | 0.02 | 0.03 | 0.02 | 0.03 | 0.03 | 0.03 | **** | 0.04 | 0.04 | 0.01 | 0.04 | 0.03 |
| WS | 0.03 | 0.02 | 0.05 | 0.02 | 0.02 | 0.03 | 0.03 | 0.03 | **** | 0.06 | 0.02 | 0.03 | 0.03 |
| Wen | 0.04 | 0.04 | 0.02 | 0.04 | 0.05 | 0.04 | 0.04 | 0.04 | 0.05 | **** | 0.04 | 0.07 | 0.04 |
| WSh | 0.02 | 0.01 | 0.02 | 0.01 | 0.02 | 0.02 | 0.01 | 0.01 | 0.02 | 0.03 | **** | 0.02 | 0.02 |
| YeL | 0.02 | 0.02 | 0.03 | 0.01 | 0.03 | 0.02 | 0.02 | 0.03 | 0.03 | 0.05 | 0.02 | **** | 0.03 |
All pairwise FST and genetic distance values are significant at P = 0.05
ENA Exclusion of null alleles
a Mean Nei’s standard genetic distance of each population from the other populations; **** = not applicable
Cluster analysis, PCoA, and population genetic structure
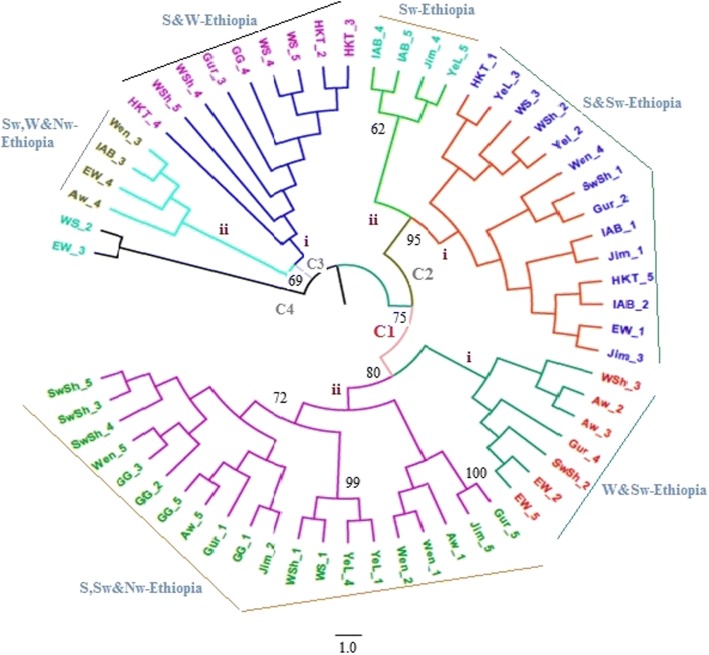
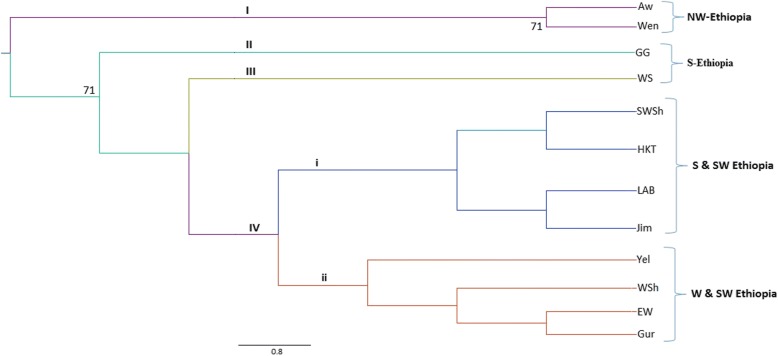
The neighbor-joining based cluster analysis of 60 individual samples randomly selected across the 12 populations (five samples per population) resulted in four major clusters (C1, C2, C3, and C4) with the first three clusters further divided into two sub-clusters. Each of the four clusters comprised individual plants from different collection zones (geographic regions). However, samples are more or less grouped according to their geographic region of origin at sub cluster levels (designated as i and ii on each major cluster) although there is considerable intermixes (Fig. 2). UPGMA [36] cluster analysis with bootstrap tests [39] has been conducted using Nei’s standard genetic distance at population level. The 12 populations formed four major clusters (I, II, III and IV) with cluster IV comprising eight populations that formed two sub-clusters (i and ii) (Fig. 3).
Fig. 2.
Neighbor-joining tree generated based on simple matching dissimilarity coefficients over 1000 replicates for 60 individual samples randomly selected from the 12 populations studied. Numbers at the roots of the branches are bootstrap values, and bootstrap values of less than 59% were not shown
Fig. 3.
Unweighted pair-group method with arithmetic mean (UPGMA) dendrogram showing genetic relationships among the 12 populations considered based on Nei’s unbiased genetic distance over 1000 replicates. Numbers at the roots of the branches are bootstrap values, and bootstrap values of less than 59% were not shown
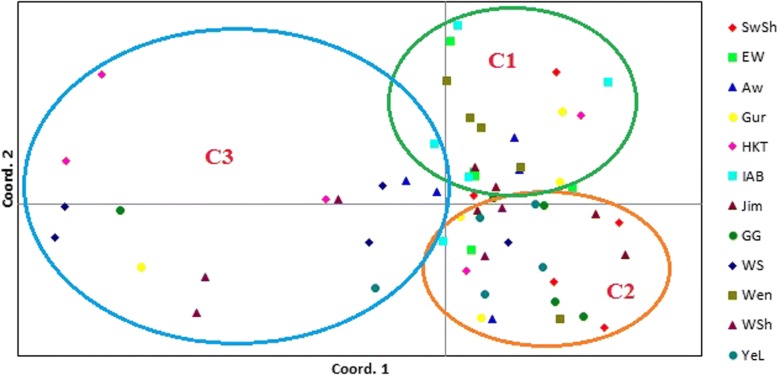
A PCoA analysis revealed that the majority of samples were placed at the center of a two-dimensional coordinate plane (Fig. 4) forming roughly three groups (C1, C2 and C3), showing poor population structure. The first three axes explained 32% of the total variation.
Fig. 4.
Principal coordinates analysis (PCoA) bi-plot showing the clustering pattern of 60 samples randomly selected from the 12 populations. Samples coded with the same symbol and color belong to the same population. Note: The percentages of variation explained by the first 3 axes (1, 2 and 3) are 14.1, 9.6, and 8.9%, respectively
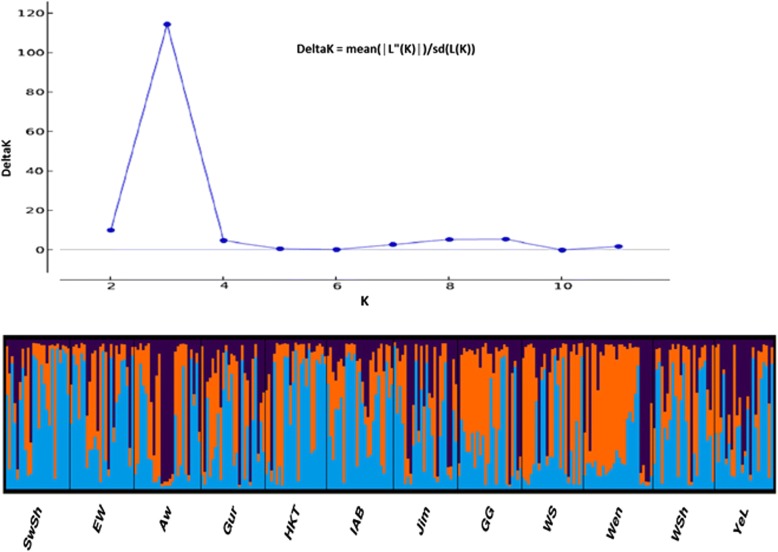
The Bayesian approach-based assignment of the 287 individual plants to different populations and determination of their population structure [45], using STRUCTURE outputs, predicted K = 3 to be the most likely number of clusters (Fig. 5a). Based on this value, Clumpak result (bar plot) showed wide admixtures and hence there was no clear geographic origin-based structuring of populations (Fig. 5b).
Fig. 5.
Delta K value estimated using Evano et al [44] method (a) and Bayesian model-based estimation of population structure (K = 3) (b) for the 287 P. edulis individual plants in twelve pre-determined populations
Discussions
EST-SSR markers validation and polymorphism evaluation
P. edulis is an indigenous tuber crop of Ethiopia that plays an indispensable role in food security of subsistence farmers in areas where it is cultivated. As the first step in the development of genomic tools and resources that can promote the conservation and breeding of this crop, we developed and validated 20 polymorphic EST-SSR markers. This work reports the transferability of SSR markers from P. barbatus to P. edulis and hence it enriches the limited reports on cross-genera [49] and cross-species [50] transferability of molecular markers, and molecular marker based genetic diversity analyses of Lamiaceae species. In the present study, the screening of 3263 P. barbatus ESTs resulted in 301 sequences (9.2%) containing SSRs. This proportion suggest that EST-SSRs are less abundant in P. barbatus than in Salvia miltiorrhiza (14.7%) [51] and Lavandula species (18.8%) [49], and slightly more abundant in P. barbatus than in Mentha piperita (8.4%) [52], which are all Lamiaceae species. The results also suggest that the abundance of the EST-SSRs in P. barbatus is higher than their abundance in cereal crops such as rice (4.7%), sorghum (3.6%), barley (3.4%) and maize (1.4%) [53]. However, other factors such as the SSR search criteria and size of the dataset might have partly contributed to the differences [54].
A maximum of two alleles are expected per individual plant at single copy microsatellite loci in diploid species. In the present study, a maximum of two alleles per plant were detected at each of the 20 loci, indicating that they are all single copy and have disomic inheritance regardless of the thus far reported basic chromosome number and ploidy level for several members of the genus Plectranthus [55]. However, the chromosome number of P. edulis has not been reported and, hence, further in-depth cytogenetic analysis is important to confirm the finding of this study.
The EST-SSR markers developed in the present study contained di-, tri- penta- and hexanucleotide repeats. Studies have shown that di, tri and tetra-nucleotide repeat SSRs are the most commonly used motifs in molecular genetic studies [56]. Tri-nucleotide repeat motifs were relatively abundant. This is attributed to the fact that they are more frequent in the EST’s coding regions, unlike in non-coding regions, in almost all taxa studied [54, 57–59] because of the positive selection for specific amino-acid stretches [60] or the prevalent selection against frameshift mutations in these regions for dinucleotides and other non-triplet repeat motifs [61]. As length and total size of perfect array of microsatellites increases, the frequency of repeats decreases and hence the informativeness increases [59, 62] owing to the higher mutation rates in longer microsatellites [63]. In agreement with this, the average number of repeats in dinucleotide SSRs is higher (8.7) than trinucleotides SSRs (5.7) among the SSRs developed in the present study. Similarly, the informativeness in terms of a number of alleles, GD and PIC was higher for di-nucleotide repeats than trinucleotides suggesting higher rate of evolution for SSRs with shorter repeat motifs than SSRs with longer repeat motifs.
The use of molecular markers for efficient selection of genotypes with desirable traits and enhancing the efficiency of breeding by allowing effective simultaneous selection of various desirable traits is a well-established approach [64, 65]. Hence, the large number of alleles detected in the present study suggests the suitability of microsatellites in general and those developed in this study in particular for genetic linkage and QTL mapping of desirable traits followed by marker assisted selection (MAS) in breeding programmes. However, most of the alleles were rare and scarce suggesting minimum selection pressure against the alleles. Otherwise, clonally propagating crops are expected to bear less proportion of such alleles as compared to seed-propagating crops. Moreover, the higher number of private alleles observed at several loci (Example: PE_07, PE_16, PE_18, PE_08) could offer a good opportunity to evaluate P. edulis genetic materials for the association of particular alleles with traits of interest and for conservation. Such alleles are useful in comparing diversity between species or populations [66] and also for measuring genetic distinctiveness of individuals in a population [67].
The average percent polymorphism per population revealed in the present study across the 20 loci (94%) is by far greater than the level reported by Kumar et al. [52] for 13 accessions of M. piperita (61%), which shares the same family with P. edulis. However, it was similar with that reported for 28 alfalfa accessions (97%) [68] and 37 Opium poppy accessions (96%) [69]. Such high percent polymorphism together with the PIC values obtained, which provides an estimate of the discriminatory power of a locus [70], and the allelic diversity suggest great potential of the markers for use in future genetic studies. However, the informativeness of a considerable number of loci is low and hence, there is a need to develop more highly informative EST-SSRs or other type of DNA markers that are suitable to characterize P. edulis genetic resources for efficient conservation and breeding.
The loci studied displayed differences between Ho and He in which half of them showed excess heterozygosity that led to a significant departure from HWE across populations. Such excess heterozygosity is expected in historically outcrossing species that maintain their heterozygosity through vegetative propagation, or if other factors such as natural and artificial selection pressure favor heterozygosity. Similar results have been reported in sweet potato [71, 72].
Population genetic diversity
Higher genetic diversity is expected in larger and older populations when compared with small and newly established ones because of higher levels of accumulated and maintained genetic variation [73] which is important in increasing fitness and therefore reduces the likelihood of local extinction [74]. However, the mean observed heterozygosity (0.39), Shannon’s information index (0.61) and Nei’s gene diversity (0.35) obtained in the present study showed a medium level of genetic variation within populations. This could be mainly due to a relatively narrow genetic basis of the populations that resulted from limited germplasm resources accessible to farmers, or due to reduction in population size both due to natural as well as human factors, such as replacing cultivation of P. edulis by other crops. In addition, farmers’ preferences for selected traits of economic importance such as tuber size, tuber skin color, tuber texture, maturity etc. and asexual mode of reproduction of the crop (clonal propagation), which was evidenced from the considerably higher global clonality index (G/N), could have contributed to limited genetic variation in P. edulis populations. There are similar reports on potato cultivars from Yunnan province, China [75].
Wen, WS, and Aw populations are genetically more diverse than the other populations as estimated by parameters such as gene diversity, heterozygosity and Shannon diversity index and hence the areas representing these populations could be considered as genetic diversity hot spots and a potential in-situ conservation sites for P. edulis. Among the populations, EW has the least genetic diversity, which might suggest current rapid genetic erosion from the area (population bottleneck) or intensive artificial selection pressure to maximize tuber yield. In terms of allelic richness, Gur, Jim, WS and WSh populations are the top four in that order, and hence are more interesting in terms of genetic and evolutionary studies on this crop because allelic richness is more informative in this regard as it is sensitive to the presence of rare alleles [76] (which is prominent in this study) and population bottlenecks when compared to other parameters such as expected heterozygosity. Moreover, these populations except WS bear a relatively high proportion of private alleles which may indicate certain level of independent evolution of their gene pools that allowed maintenance of private alleles at a population level [77].
Population genetic differentiation
AMOVA revealed that P. edulis has very low genetic differentiation among populations, which accounted only for 3% of the total genetic variation. The result is in line with previous reports on clonally propagating crops, such as Ensete (Ensete ventricosum) [78, 79], as such species tend to be more diverse within populations (but largely lower than sexually reproducing species) than among populations. Likewise, FST averaged across all loci (FST = 0.03) and pairwise FST for all pairs of populations (highest value = 0.07) was generally low to moderate on the bases of Wright [80] and Hartl and Clark [81] suggestions. Wright [80] indicated that genetic differentiation among populations can be considered high if the value of FST is greater than 0.25. This could be partly attained if gene flow, which is a powerful force to decrease differentiation among populations, is low (Nm < 1) [82]. Hence, the present study showed that P. edulis has very little population sub-structuring. The low population differentiation is supported by high gene flow (mean Nm = 18.29) owing to step-wise pollen movement across populations, germplasm exchange in the form of tubers and seeds through sharing common markets among several of the adjacent areas where different populations were collected. This study also showed the minimal effects of regions or geographic origins of populations on genetic variation in P. edulis. This could be partly explained by the extensive exchange of tubers as planting materials among farmers (gene flow), common origin of the populations, the clonally propagating nature of the crop in which only a limited number of individuals contribute tubers to the next generation, which gradually leads to recent or old population bottlenecks and hence facilitate genetic drift.
A pair-wise population genetic differentiation analysis resulted in a seven-fold variation in FST value among pairs of populations (ranging from 0.01 to 0.07). The highest population differentiation was observed between Wen and HKT, Wen and WS, Wen and YeL as well as between Aw and WS populations. Wen population showed the highest (0.05) pairwise Nei’s standard genetic distance with WS and HKT populations and is the most genetically distinct population with a mean Nei’s standard genetic distance 0.04 (Table 6). This can be partly explained by the fact that Wen and Aw populations were collected from a relatively pocket location and are separated from the other populations with a relatively longer geographic distance that probably restricted recent seed and tuber exchange. Hence, these populations may serve as potential sources of new genetic variation of important traits that can be used in breeding programs.
Population genetic relationship and structure analysis
Neighbour joining cluster analysis in which each population is represented by five individual plants revealed a weak clustering pattern confirming low genetic differentiation among the populations and suggesting that the genetic background of P. edulis populations does not always correlate with their geographical origin. Although UPGMA and PCoA analyses also showed a certain level of populations clustering according to their geographical regions, the clustering pattern is weak to support the concept of “isolation by distance” [80]. Similarly, structure analysis revealed a close relationship (weak sub-division) among the samples from the 12 collection zones, and in general, three inferred groups (K = 3) with potential admixtures and have been observed. It is interesting to indicate that all individual plants analyzed have alleles originated from the three clusters, which supports the presence of a strong gene flow that led to poor population differentiation.
Conclusions
In this study, we developed 20 EST-SSR markers for P. edulis based on EST sequences of P. barbatus deposited in the GenBank. All the markers were polymorphic in the populations studied and are valuable genetic tools to help evaluate the extent of genetic diversity and population structure of not only P. edulis but also of various species in the family Lamiaceae. These markers detected a larger number of alleles, some of which were private alleles that may be linked to important agronomic traits. The study also showed the potential of EST-SSR markers in defining how P. edulis genetic diversity is structured, and hence contribute to the development of better in-situ and ex-situ management strategies as well as selection criteria for the germplasm to be used in breeding programs for the improvement of various desirable traits in this crop. Among the 12 administrative zones and woredas considered, Wenbera, Awi and Wolaita have populations with a relatively high genetic diversity, and hence can be considered as hot spots for in-situ conservation of P. edulis as well as sources of desirable alleles for breeding values. Further studies that include germplasm from the remaining administrative zones and combine molecular characterization with agro-morphological analysis would be important to reveal additional potential sites for conservation and development of best-performing varieties. Overall, this study offered baseline information that promote further studies to exploit the high economic and endogenous values and to stop and reverse the current rapid genetic erosion of P. edulis.
Additional files
Passport data of the 174 tuber samples and 287 leaf samples representing the 12 populations used in the present study. (DOCX 36 kb)
Allele frequency distribution and overall percentage of rare alleles (f ≤ 0.01) across populations. (XLSX 9 kb)
Estimates of the overall Nei’s heterozygosity, population differentiation measures and proportion of progenies produced by selfing. (DOCX 18 kb)
Acknowledgments
This work is part of the first author’s PhD thesis. The authors would like to thank Addis Ababa University and Mada Walabu University for material and technical supports for this research, and individual farmers for allowing us to collect the tuber samples from their fields. We would also like to thank the Ethiopian Biodiversity Institute (EBI) for allowing the transfer of the study material to Sweden, and the Swedish University of Agricultural Sciences for providing laboratory facilities to conduct this research.
Funding
This work is financially supported by the Swedish Research Council (VR) through Swedish Research Link project and Addis Ababa University’s Thematic Research Project. The role of the funding bodies is limited to direct funding of the project activities in the field and laboratory that results in this manuscript.
Availability of data and materials
Full passport data of the 174 tuber samples and 287 leaf samples representing the 12 populations used in the present study is provided in Additional file 1. Allele frequency distribution and overall percentage of rare alleles (f ≤ 0.01) across populations are provided in Additional file 2. Estimates of the overall Nei’s heterozygosity, population differentiation measures and proportion of progenies produced by selfing is provided in Additional file 3.
Abbreviations
- AMOVA
Analysis of molecular variance;
- CTAB
Cetyltrimethyl ammonium bromide
- EST
Expressed sequence tags
- NCBI
National Centre for Biotechnology information
- PCoA
Principal coordinates analysis
- SNNPs
Southern Nations, Nationalities, and Peoples’ Region
- SSRs
Simple sequence repeats
- UPGMA
Unweighted pair group with arithmetic mean
Authors’ contributions
FG and MG conducted data mining, designed the SSR primers and carried out the data analysis. FG carried out the laboratory work. All co-authors participated in designing the experiment, interpreting the data, drafting and revising the manuscript; and approved the final manuscript.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fekadu Gadissa, Phone: +251-118959216, Email: fikega2000@gmail.com.
Kassahun Tesfaye, Email: kassahuntesfaye@yahoo.com.
Kifle Dagne, Email: kifledagne38@gmail.com.
Mulatu Geleta, Email: mulatu.geleta.dida@slu.se.
References
- 1.Vavilov NI. The origin, variation, immunity, and breeding of cultivated plants. Cambridge: Cambridge University Press; 1951. pp. 1–387. [Google Scholar]
- 2.Harlan J. Ethiopia: a centre of diversity. Econo Bot. 1996;23:124–132. [Google Scholar]
- 3.Westphal E. Agricultural systems in Ethiopia. Wageningen: Centre for Agricultural Publishing and Documentation; 1975. pp. 1–299. [Google Scholar]
- 4.Zohary D. Centers of diversity and centers of origin. In: Frankle OH, Bennett E, editors. Genetic resources of plants- their exploration and conservation. Oxford: Blackwell; 1970. pp. 33–42. [Google Scholar]
- 5.Mekbib Y, Deressa T. Exploration and collection of root and tuber crops in east Wollega and Ilu Ababora zones: rescuing declining genetic resources. Indian J Trad Know. 2016;15:86–92. [Google Scholar]
- 6.Asfaw Z. Survey of indigenous food plants, their preparations, and home gardens in Ethiopia. Indigenous African food crops and useful plants. Bede N. Okigbo ed. NU/INRA. Assessments Series No.: B6. ICIPE Science Press: Nairobi; 1997. p. 1–16.
- 7.Nebiyu A, Garedew W, Tofu A, Abebe W, Kifle A, and Etissa E. Crop management for other root and tuber crops (taro, cassava, and yam). In: Root and tuber crops: the untapped resources (Gebremedihin Woldegiorgis, Endale Gebre and Berga Lemaga, eds.). EIAR: Addis Ababa Ethiopia; 2008. p. 317–322.
- 8.Hedge IC. A global survey of biogeography of the Lamiaceae. In: Harley RM, Reynolds J, editors. Advances in labiate science, royal bot. Garden: Kew; 1992. pp. 7–8. [Google Scholar]
- 9.Codd LE. Plectranthus (Labiatae) and allied genera in southern Africa. Bothalia. 1975;11:371–442. doi: 10.4102/abc.v11i4.1482. [DOI] [Google Scholar]
- 10.Edward S. Crops with wild relatives found in Ethiopia. In: Engels, JMM, Hawkes, JG, Melaku W. (ed): Plant genetic resource of Ethiopia. Cambridge: Cambridge University Press; 1991. p. 42–74.
- 11.Asfaw Z, Tadesse M. Prospects for sustainable use and development of wild food plants in Ethiopia. Econ Bot. 2001;55:47–62. doi: 10.1007/BF02864545. [DOI] [Google Scholar]
- 12.Hora A. Anchote -an endemic tuber crop. Oromiya: Jimma College of Agriculture; 1995. pp. 1–47. [Google Scholar]
- 13.Reinhard F, Adi A. Honeybee flora of Ethiopia. Ethiopia: The National Herbarium, A.A.; 1994. pp. 1–510. [Google Scholar]
- 14.Demissie A. Potentially valuable crop plants in a Vavilovian center of diversity: Ethiopia. Proceedings of the international conference on crop genetic resources of Africa, Nairobi, Kenya, Attere F, Zedan H, ng, NQ & Perrino P, eds. International board for plant genetic resources, Rome; 1988.
- 15.Nebiyu A, and Awas T. Exploration and collection of root and tuber crops in Southwestern Ethiopia: Its implication for conservation research. Proceeding of the 11th Conference of the Crop Sciences Society of Ethiopia, Addis Ababa, Ethiopia; 2004.
- 16.Singh RP, Huerta-Espino J, William HM. Genetics and breeding for durable resistance to leaf and stripe rusts in wheat. Turkish J Agri Fores. 2005;29:121–127. [Google Scholar]
- 17.Leal AA, Mangolin CA, Do-Amaraljunior AT, Goncalves LSA, Scapim CA, Mott AS, Eloi IBO, Cordoves V, MFP DS. Efficiency of RAPD versus SSR markers for determining genetic diversity among popcorn lines. Genet Mol Res. 2010;9:9–18. doi: 10.4238/vol9-1gmr692. [DOI] [PubMed] [Google Scholar]
- 18.Ellis JS, Knight ME, Darvill B, Goulson D. Extremely low effective population sizes, genetic structuring, and reduced genetic diversity in a threatened umblebee species, Bombus sylvarum (Hymenoptera: Apidae) Mol Ecol. 2006;15:4375–4386. doi: 10.1111/j.1365-294X.2006.03121.x. [DOI] [PubMed] [Google Scholar]
- 19.Uchiyama K, Iwata H, Moriguchi Y, Ujino-Ihara T, Ueno S, Taguchi Y, et al. Demonstration of genome-wide association studies for identifying markers for wood property and male strobili traits in Cryptomeria japonica. PLoS One. 2013;8:798–812. doi: 10.1371/journal.pone.0079866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehme K, Ayse GI, Adnan A, Saadet TA. Cross-genera transferable e-microsatellite markers for 12 genera of the Lamiaceae family. J Sci Food Agric. 2012;93:1869–1879. doi: 10.1002/jsfa.5982. [DOI] [PubMed] [Google Scholar]
- 21.Guojie X, Chunsheng L, Luqi H, Xueyong W, Yuanyuan Z, Siqi L, Caili L, Qingjun Y, Xiaoli Z. Development of new EST-derived SSRs in Salvia miltiorrhiza (Labiatae) in China and preliminary analysis of genetic diversity and population structure. Biochem Syst Ecol. 2013;51:308–313. doi: 10.1016/j.bse.2013.09.018. [DOI] [Google Scholar]
- 22.Ryding O, Iwasson M, Morton JK, Persson E, Sebald O, Seybold S, Demissew S. Lamiaceae. In: Hedberg I, Kelbessa E, Edwards S, Demissew S, editors. Flora of Ethiopia and Eritrea. Sweden: The national herbarium, Addis Ababa University, Ethiopia and Department of Systematic Botany, Uppsala University; 2006. pp. 592–595. [Google Scholar]
- 23.Geleta M, Herrera I, Monz A, Bryngelsson T. Genetic diversity of arabica coffee (Coffea arabica L) in Nicaragua as estimated by simple sequence repeat markers. Sci World J. 2012;10:1–11. doi: 10.1100/2012/939820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martins WS, César D, Lucas S, de Souza Neves KF, Bertioli DJ. WebSat- a web software for microsatellite marker development. Bioinformation. 2009;3:282–283. doi: 10.6026/97320630003282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koressaar T, Remm M. Enhancements and modifications of primer design program, Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 26.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3 new capabilities and interfaces. Nuc Acids Res. 2012;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oetting WS, Lee HK, Flanders DJ, Wiesner GL, et al. Linkage analysis with multiplexed short tandem repeat polymorphisms using infrared fluorescence and M13 tailed primers. Genomics. 1995;30:450–458. doi: 10.1006/geno.1995.1264. [DOI] [PubMed] [Google Scholar]
- 28.Brownstein MJ, Carpten JD, Smith JR. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. BioTechniques. 1996;20:1004–1006. doi: 10.2144/96206st01. [DOI] [PubMed] [Google Scholar]
- 29.Liu KJ, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 30.Weir BS. Genetic Data Analysis II. Sunderland: Sinauer; 1996. pp. 1–445. [Google Scholar]
- 31.Peakall PR, Smouse PP. GenAlEx 6.5: genetic analysis in excel. Population genetics software for teaching and research– an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalinowski ST. HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes. 2005;5:187–189. doi: 10.1111/j.1471-8286.2004.00845.x. [DOI] [Google Scholar]
- 33.Arnaud-Haond S, Belkhir K. GENECLONE 2.0: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Notes. 2007;7:15–17. doi: 10.1111/j.1471-8286.2006.01522.x. [DOI] [Google Scholar]
- 34.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 35.Nei M. Genetic distance between populations. Am Nat. 1972;106:283–292. doi: 10.1086/282771. [DOI] [Google Scholar]
- 36.Sneath PHA, Sokal RR. Numerical taxonomy. San Francisco: WH Freeman and Company; 1973. pp. 1–573. [Google Scholar]
- 37.Perrier X, Jacquemoud-Collet JP. DARwin software. 2006. http://darwin.cirad.fr/. Accessed 6 Feb 2018.
- 38.Takezaki N, Nei M, Tamura K. POPTREE2: software for constructing population trees from allele frequency data and computing some other population statistics with windows interface. Mol Biol Evol. 2010;27:747–752. doi: 10.1093/molbev/msp312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Felsenstein J. Phylogenies and the comparative methods. Am Nat. 1985;125:1–15. doi: 10.1086/284325. [DOI] [Google Scholar]
- 40.Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp App Bioscie. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 41.Andrew R. FigTree: Tree figure drawing tool, Version 1.4.3. Institute of Evolutionary Biology. United Kingdom: University of Edinburgh; 2016. http://tree.bio.ed.ac.uk/software/figtree/.
- 42.Slatkin M, Barton NH. A comparison of three indirect methods for estimating average levels of gene flow. Evolution. 1989;43:1349–1368. doi: 10.1111/j.1558-5646.1989.tb02587.x. [DOI] [PubMed] [Google Scholar]
- 43.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multi-locus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 46.Earl DA, Von Holdt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Cons Genet Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 47.Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Res. 2015;15:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chapuis MP, Estoup A. Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol. 2007;24:621–631. doi: 10.1093/molbev/msl191. [DOI] [PubMed] [Google Scholar]
- 49.Karaca Mehmet, Ince Ayse Gul, Aydin Adnan, Ay Saadet T. Cross-genera transferable e-microsatellite markers for 12 genera of the Lamiaceae family. Journal of the Science of Food and Agriculture. 2012;93(8):1869–1879. doi: 10.1002/jsfa.5982. [DOI] [PubMed] [Google Scholar]
- 50.Adal Ayelign M., Demissie Zerihun A., Mahmoud Soheil S. Identification, validation and cross-species transferability of novel Lavandula EST-SSRs. Planta. 2014;241(4):987–1004. doi: 10.1007/s00425-014-2226-8. [DOI] [PubMed] [Google Scholar]
- 51.Xu Guojie, Liu Chunsheng, Huang Luqi, Wang Xueyong, Zhang Yuanyuan, Liu Siqi, Liao Caili, Yuan Qingjun, Zhou Xiaoli. Development of new EST-derived SSRs in Salvia miltiorrhiza (Labiatae) in China and preliminary analysis of genetic diversity and population structure. Biochemical Systematics and Ecology. 2013;51:308–313. doi: 10.1016/j.bse.2013.09.018. [DOI] [Google Scholar]
- 52.Kumar Birendra, Kumar Umesh, Yadav Hemant Kumar. Identification of EST–SSRs and molecular diversity analysis in Mentha piperita. The Crop Journal. 2015;3(4):335–342. doi: 10.1016/j.cj.2015.02.002. [DOI] [Google Scholar]
- 53.Kantety RV, La Rota M, Matthews DE, Sorrells ME. Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum, and wheat. Plant Mol Biol. 2002;48:501–10. [DOI] [PubMed]
- 54.VARSHNEY R, GRANER A, SORRELLS M. Genomics-assisted breeding for crop improvement. Trends in Plant Science. 2005;10(12):621–630. doi: 10.1016/j.tplants.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 55.de Wet JMJ. Chromosome numbers in Plectranthus and related genera. S Afri J Sci. 1958;34:153–6.
- 56.Selkoe Kimberly A., Toonen Robert J. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecology Letters. 2006;9(5):615–629. doi: 10.1111/j.1461-0248.2006.00889.x. [DOI] [PubMed] [Google Scholar]
- 57.Toth G. Microsatellites in Different Eukaryotic Genomes: Survey and Analysis. Genome Research. 2000;10(7):967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eujayl I., Sledge M. K., Wang L., May G. D., Chekhovskiy K., Zwonitzer J. C., Mian M. A. R. Medicago truncatula EST-SSRs reveal cross-species genetic markers for Medicago spp. TAG Theoretical and Applied Genetics. 2004;108(3):414–422. doi: 10.1007/s00122-003-1450-6. [DOI] [PubMed] [Google Scholar]
- 59.Wang Zhangying, Li Jun, Luo Zhongxia, Huang Lifei, Chen Xinliang, Fang Boping, Li Yujun, Chen Jingyi, Zhang Xiongjian. Characterization and development of EST-derived SSR markers in cultivated sweetpotato (Ipomoea batatas) BMC Plant Biology. 2011;11(1):139. doi: 10.1186/1471-2229-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Metzgar D, Bytof J, Wills C. Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 2000;10:72–80. [PMC free article] [PubMed]
- 61.Li You-Chun, Korol Abraham B., Fahima Tzion, Beiles Avigdor, Nevo Eviatar. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Molecular Ecology. 2002;11(12):2453–2465. doi: 10.1046/j.1365-294X.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- 62.Katti Mukund V., Ranjekar Prabhakar K., Gupta Vidya S. Differential Distribution of Simple Sequence Repeats in Eukaryotic Genome Sequences. Molecular Biology and Evolution. 2001;18(7):1161–1167. doi: 10.1093/oxfordjournals.molbev.a003903. [DOI] [PubMed] [Google Scholar]
- 63.McConnell R., Middlemist S., Scala C., Strassmann J. E., Queller D. C. An Unusually Low Microsatellite Mutation Rate in Dictyostelium discoideum, an Organism With Unusually Abundant Microsatellites. Genetics. 2007;177(3):1499–1507. doi: 10.1534/genetics.107.076067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dudley J. W. Molecular Markers in Plant Improvement: Manipulation of Genes Affecting Quantitative Traits. Crop Science. 1993;33(4):660. doi: 10.2135/cropsci1993.0011183X003300040003x. [DOI] [Google Scholar]
- 65.Edwards MD, Helentjaris T, Wright S, Stuber CW. Molecular-marker-facilitated investigations of quantitative trait loci in maize. Theor Appl Genet. 1992;83:765–74. [DOI] [PubMed]
- 66.Mahmodi Farshid, Kadir Jugah, Puteh Adam. Genetic Diversity and Pathogenic Variability ofColletotrichum truncatumCausing Anthracnose of Pepper in Malaysia. Journal of Phytopathology. 2013;162(7-8):456–465. doi: 10.1111/jph.12213. [DOI] [Google Scholar]
- 67.Kalinowski Steven T. Counting Alleles with Rarefaction: Private Alleles and Hierarchical Sampling Designs. Conservation Genetics. 2004;5(4):539–543. doi: 10.1023/B:COGE.0000041021.91777.1a. [DOI] [Google Scholar]
- 68.Wang Zan, Yan Hongwei, Fu Xinnian, Li Xuehui, Gao Hongwen. Development of simple sequence repeat markers and diversity analysis in alfalfa (Medicago sativa L.) Molecular Biology Reports. 2012;40(4):3291–3298. doi: 10.1007/s11033-012-2404-3. [DOI] [PubMed] [Google Scholar]
- 69.Şelale Hatice, Çelik Ibrahim, Gültekin Visam, Allmer Jens, Doğanlar Sami, Frary Anne. Development of EST-SSR markers for diversity and breeding studies in opium poppy. Plant Breeding. 2013;132(3):344–351. doi: 10.1111/pbr.12059. [DOI] [Google Scholar]
- 70.Marulanda M.L., López A.M., Isaza L., López P. Microsatellite isolation and characterization for Colletotrichum spp, causal agent of anthracnose in Andean blackberry. Genetics and Molecular Research. 2014;13(3):7673–7685. doi: 10.4238/2014.September.26.5. [DOI] [PubMed] [Google Scholar]
- 71.Zawedde Barbara M., Harris Craig, Alajo Agnes, Hancock Jim, Grumet Rebecca. Factors Influencing Diversity of Farmers’ Varieties of Sweet Potato in Uganda: Implications for Conservation. Economic Botany. 2014;68(3):337–349. doi: 10.1007/s12231-014-9278-3. [DOI] [Google Scholar]
- 72.Gao SJ, Han JL, Liu AP, Yan ZJ, Chang XQ. RAPD analysis on genetic divergence among populations of Oedaleus asiaticus B. Bienko and Oedaleus infernalis Saussure. Acta Agri Boreali-Sinica. 2011;26:94–100.
- 73.Rampersad Sephra N, Perez-Brito Daisy, Torres-Calzada Claudia, Tapia-Tussell Raul, Carrington Christine VF. Genetic structure and demographic history of Colletotrichum gloeosporioides sensu lato and C. truncatum isolates from Trinidad and Mexico. BMC Evolutionary Biology. 2013;13(1):130. doi: 10.1186/1471-2148-13-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Futuyma Douglas J. Specialization, Speciation, and RadiationThe Evolutionary Biology of Herbivorous Insects. 2008. Sympatric Speciation: Norm or Exception? pp. 136–148. [Google Scholar]
- 75.Liao H, Guo C. Using SSR to evaluate the genetic diversity of potato cultivars from Yunnan Province (SW China) Acta Biol Crac Ser Bot. 2014;56:16–27. [Google Scholar]
- 76.Leberg PL. Effects of bottlenecks on genetic divergence in populations of the wild Turkey. Cons Biol. 1991;5:522–530. doi: 10.1111/j.1523-1739.1991.tb00359.x. [DOI] [Google Scholar]
- 77.Slatkin M. Rare alleles as indicators of gene flow. Evolution. 1985;39:53–65. doi: 10.1111/j.1558-5646.1985.tb04079.x. [DOI] [PubMed] [Google Scholar]
- 78.Magule TO, Tesfaye B, Pagnotta MA, Enrico Pè M, Catellani M. Development of SSR markers and genetic diversity analysis in enset (Ensete ventricosum (Welw.) Cheesman), an orphan food security crop from southern Ethiopia. BMC Genet. 2015;16:98. doi: 10.1186/s12863-015-0250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tobiaw DC, Bekele E. Analysis of genetic diversity among cultivated enset (Ensete ventricosum) populations from Essera and Kefficho, southwestern part of Ethiopia using inter simple sequence repeats (ISSRs) marker. Afr J Biotechnol. 2011;70:15697–15709. [Google Scholar]
- 80.Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hartl DL, Clark AG. Principles of population genetics, third edition. Sunderland: Sinauer Associates, Inc. Publishers; Massachusetts. 1997. p. 519.
- 82.Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Passport data of the 174 tuber samples and 287 leaf samples representing the 12 populations used in the present study. (DOCX 36 kb)
Allele frequency distribution and overall percentage of rare alleles (f ≤ 0.01) across populations. (XLSX 9 kb)
Estimates of the overall Nei’s heterozygosity, population differentiation measures and proportion of progenies produced by selfing. (DOCX 18 kb)
Data Availability Statement
Full passport data of the 174 tuber samples and 287 leaf samples representing the 12 populations used in the present study is provided in Additional file 1. Allele frequency distribution and overall percentage of rare alleles (f ≤ 0.01) across populations are provided in Additional file 2. Estimates of the overall Nei’s heterozygosity, population differentiation measures and proportion of progenies produced by selfing is provided in Additional file 3.