Abstract
Background
Vacuolar invertases (VINs) have been reported to regulate plant growth and development and respond to abiotic stresses such as drought and cold. With our best knowledge, the functions of VIN genes little have been reported in tea plant (Camellia sinensis L.). Therefore, it is necessary to develop research in this field.
Results
Here, we identified a VIN gene, CsINV5, which was induced by cold acclimation and sugar treatments in the tea plant. Histochemical assays results showed that the 1154 bp 5′-flanking sequence of CsINV5 drove β-glucuronidase (GUS) gene expression in roots, stems, leaves, flowers and siliques of transgenic Arabidopsis during different developmental stages. Moreover, promoter deletion analysis results revealed that an LTRE-related motif (CCGAAA) and a WBOXHVISO1 motif (TGACT) within the promoter region of CsINV5 were the core cis-elements in response to low temperature and sugar signaling, respectively. In addition, overexpression of CsINV5 in Arabidopsis promoted taproot and lateral root elongation through glucose-mediated effects on auxin signaling. Based on physiological and RNA-seq analysis, we found that overexpression of CsINV5 improved cold tolerance in transgenic Arabidopsis mainly by increasing the contents of glucose and fructose, the corresponding ratio of hexose to sucrose, and the transcription of osmotic-stress-related genes (P5CS1, P5CS2, AtLEA3, COR413-PM1 and COR15B) to adjust its osmotic potential.
Conclusions
Comprehensive experimental results suggest that overexpression of CsINV5 may enhance the cold tolerance of plant through the modification of cellular sugar compounds contents and osmotic regulation related pathways.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1456-5) contains supplementary material, which is available to authorized users.
Keywords: CsINV5, Promoter, Cold tolerance, RNA-seq, Arabidopsis thaliana, Camellia sinensis
Background
Sugars, such as sucrose (Suc), glucose (Glc) and fructose (Fru), are reported to act not only as osmoprotectants and ROS scavengers to stabilize membranes but also as signaling molecules to regulate gene expression when plants are exposed to low temperatures [1–4]. As a source of carbohydrate and energy, Suc could be hydrolyzed into monosaccharides by two types of enzyme exist in plants: Suc synthase (Sus, E.C. 2.4.1.13) and invertase (INV, E.C. 3.2.1.26). INVs irreversibly hydrolyze Suc into Glc and Fru, playing diverse roles in growth, development and response to abiotic and biotic stresses [5–8].
Based on optimum pH, INVs are classified into acid INVs (AI) and alkaline/neutral INVs (A/N-Inv). AIs are part of glycoside hydrolase family GH32 and are further subdivided as cell wall bound INVs (CWIN) and vacuolar INVs (VIN) according to their subcellular localization. AIs are β-fructofuranosidases, N-glycosylated proteins that contain a β-fructosidase motif (NDPD/AG) and a cysteine catalytic domain (WECP/VD). CWINs generate a Suc gradient essential for phloem unloading and carbon partitioning [9] and function in the development of plant tissues such as roots, flowers [10, 11], fruits [12] and seeds [13–15]. VINs are mainly involved in regulating cell expansion [16] and sugar composition in sugar-storing sink organs [17–19]. A/N-Invs are members of glycoside hydrolase family GH100 and mainly located in the cytoplasm, mitochondria or chloroplast [20, 21]. In addition, A/N-Invs are critical players in manipulating root and reproductive development [22–24].
Apart from their critical roles in plant growth and development, many studies have demonstrated that INVs respond to biotic and abiotic stresses in plants, though the molecular mechanism remains largely unknown [5, 25–28]. One possible mechanism of VIN responses to abiotic stress is changed activity following changed transcription, thus changing the ratio of hexose (Hex) to Suc to alter osmotic pressure in response to stimuli. For example, the induction by drought stress of a VIN gene, Ivr2, in the mature leaf, leaf sheath, and primary root of rice increases VIN activity, resulting in an accumulation of Hex that may participate in a signal transduction pathway or increase osmotic pressure to improve resistance to water deficits [29]. In contrast, the expression of Ivr2 and VIN activity are reduced in young ovaries under drought stress, but the ratio of Hex to Suc still shows a close relationship with soluble INV activity, though in the ovary, sugars did not appear to contribute to osmotic acclimation [30]. In addition, Hex accumulation induced by VIN results in cold-induced sweetening in potato, whereas the suppression of VIN activity by transcription or post-translational regulation prevents this phenomenon [19, 26].
Tea plant is widely distributed in tropical and subtropical regions and represents one of the important economic crops in more than 20 countries. The growth and development of the tea plant are mainly limited by temperature, sunlight, water content, and soil fertility. Extremely low temperatures in winter and cold spells in late spring can seriously affect seasonal tea production and quality. As is well known, the freezing tolerance of plants could be induced by a period of low nonfreezing temperatures before the onset of winter, which is named as cold acclimation (CA). There are many changes occurred during CA process, involving gene expression, metabolism, and morphology [31]. In nature, CA is mainly trigged by low nonfreezing temperatures in late autumn or early winter [32], while it is induced by exposing plants to the low nonfreezing temperatures (2–6 °C) in the controlled environment chamber [33]. The freezing tolerance of acclimated plants is higher than that of nonacclimated plants. Nonacclimated Arabidopsis, for instance, had an EL50 (temperature resulted in 50% electrolyte leakage) of approximately − 4.5 °C, but the acclimated Arabidopsis had a lower EL50 values of approximately − 6 °C [34]. As previous study shown, with the average air temperature decreased to around 7 °C, the tea plants would undergo the CA process, and when the average air temperature increased to over 9 °C, tea plants would start the de-acclimation process. Moreover, a certain extent of cold acclimation can enhance the cold tolerance of tea plant [35]. Furthermore, there have many studies found the expression levels of many genes were changed in tea plants when they exposed to low nonfreezing temperatures (4 °C) or during natural CA process [36–40]. Recently, Wang et al. [41] revealed by RNA-seq analysis that the expression levels of many sugar-metabolizing genes were increased or reduced during CA, demonstrating that carbohydrate metabolism might be a major pathway in response to cold stress in the tea plant during CA. Afterwards, Yue et al. [42] further verified this hypothesis, finding that 59 sugar-related genes induced by low temperature, particularly a putative INV gene, CsINV5, which showed more than 100-fold higher expression during CA than before or after CA. The expression levels of CsINV5 was also elevated at least 3-fold in leaves and 2-fold in roots of tea plant after 5 days of low nonfreezing temperature (4 °C) treatment as compared to normal cultivated tea plant [38], indicating that CsINV5 may play an important role in cold stress response.
To better understand how CsINV5 mediate the cold tolerance of tea plant, we analyzed the functions of 5′-flanking sequence of CsINV5 drove β-glucuronidase (GUS) gene and the opening reading frame (ORF) of CsINV5 overexpressed in transgenic Arabidopsis, respectively. In the present study, we demonstrate that a low-temperature-responsive element (CCGAAA) within the promoter of CsINV5 is the core cis-element in cold stress response, and a sugar-related element (TGACT) is the main cis-element mediating sugar signaling. Furthermore, overexpression (OE) of CsINV5 in Arabidopsis not only accelerated root growth through an osmotic-independent pathway but also enhanced cold resistance, mainly through the osmotic-dependent pathway.
Results
Phylogenetic analysis of CsINV5
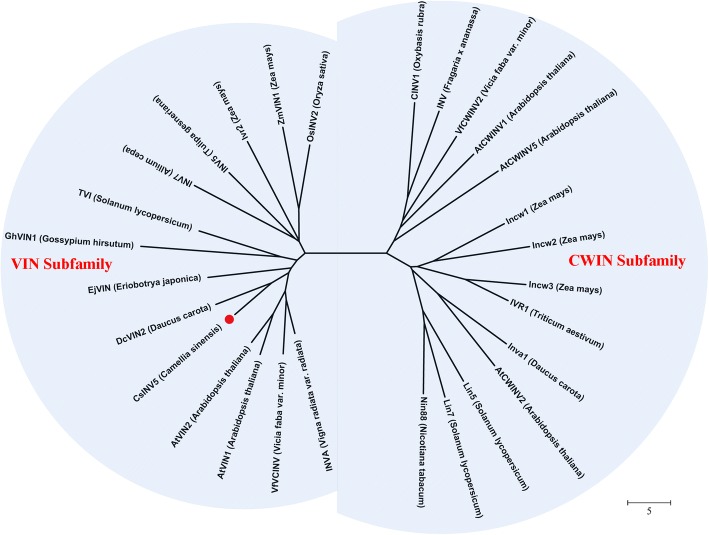
To investigate the evolutionary relationships of CsINV5, the amino acid sequence was aligned with those of known INVs, and a phylogenetic tree was constructed. As Fig. 1 shows, CsINV5 is a SAI that clusters into the VIN subfamily and has the closest relationship with DcVIN2 [43]; CsINV5 is also phylogenetically close to EjVIN1 and GhVIN1, the VINs of loquat [44] and cotton [45] respectively. In addition, the alignment result of the partial deduced protein sequences of VINs and CWINs indicated that 13 conserved regions reported from known SAIs, including four putative enzyme active site residues [46], the β-fructofuranosidase motif (NDPNG/A), and the RDP and WECP/VD motifs [47] were found in the protein sequence of CsINV5. Notably, more than 30 single amino acid differences were observed between CWINs and VINs (Additional file 1: Figure S1). In addition, we explored the genomic sequence of CsINV5 using the tea plant genome described recently by Xia et al. [48]; the genomic DNA sequence of CsINV5 contains 7 exons and 6 introns (Additional file 2: S1), and the second exon encodes a β-fructofuranosidase motif (DPN) reported as one of the smallest exons known in plants [49]. Based on the above results, we demonstrate that CsINV5 encodes a VIN protein in tea plant.
Fig. 1.
Phylogenetic analysis of CsINV5 and known SAIs. CsINV5 (KP053402, Camellia sinensis), GhVIN1 (ACQ82802, Gossypium hirsutum), INV5 (X97642, Tulipa gesneriana), INV7 (Y11230, Allium cepa), VfVCINV (Z49831, Vicia faba var. minor), DcVIN2 (Y18706, Daucus carota), EjVIN (KF938903, Eriobotrya japonica), AtVIN1 (At1g62660, Arabidopsis thaliana), AtVIN2 (At1g12240, Arabidopsis thaliana), Ivr2 (U31451, Zea mays), ZmVIN1 (U16123, Zea mays), OsINV2 (AF276703, Oryza sativa), TVI (NM_001247914, Solanum lycopersicum), INVA (D10265, Vigna radiata var. radiata), AtCWINV1 (At3G13790, Arabidopsis thaliana), AtCWINV2 (At3G52600, Arabidopsis thaliana), AtCWINV5 (At3G13784, Arabidopsis thaliana), CINV1 (X81792, Oxybasis rubra), INV (AF000520, Fragaria x ananassa), Lin7 (X91391, Solanum lycopersicum), Lin5 (X91389, Solanum lycopersicum), Nin88 (AF376773, Nicotiana tabacum), IVR1 (AF030420, Triticum aestivum), Incw1 (AF050129, Zea mays), Incw2 (AF050128, Zea mays), Incw3 (AF043346, Zea mays), Inva1 (X69321, Daucus carota), VfCWINV2 (Q43856, Vicia faba var. minor). CsINV5 is highlighted with a red dot
Expression of CsINV5 is induced by CA and sugar treatment
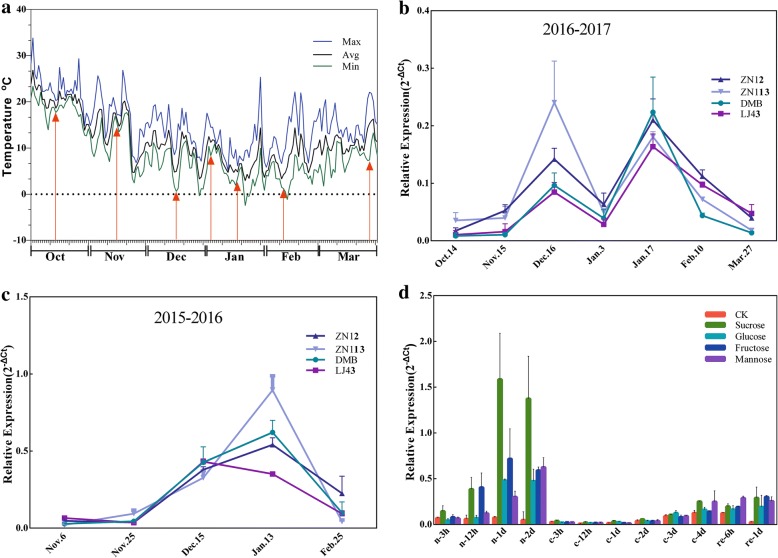
Previous study found the expression of CsINV5 was induced at a high level during CA period (from Nov.2 to Dec.19), especially in Dec.19 that the expression level of CsINV5 was elevated more than 300-fold, moreover, it was also induced at a high level during winter hardiness period (from Dec.19 to Jan.10) [42]. In the present study, we further compared the expression of CsINV5 in four-tea cultivars during CA in 2015–2016 and 2016–2017. As Fig. 2b and c show, the expression of CsINV5 was similar among different tea plant cultivars following temperature changes in winter. Briefly, CsINV5 transcripts increased during CA with decreasing temperature (from late December to early January) and decreased during de-acclimation with increasing temperature (from late January to early February). In addition, we found exogenous sugars, including Suc, Glc and Fru, enhanced the expression of CsINV5 in hydroponic seedlings of cultivar ‘LJ43’ in both normal and cold conditions (Fig. 2d), but the transcript levels of CsINV5 were higher when treated with exogenous sugars under normal conditions.
Fig. 2.
Expression analysis of CsINV5 during CA periods in 2015–2016 and 2016–2017 and under different sugar and temperature conditions. a Changes in air temperature from October 2016 to March 2017. The maximum (Max), average (Avg) and minimum (Min) daily temperatures are shown. The red arrows indicate the Avg temperature of each sampling day. b-c Expression analysis of CsINV5 in the mature leaves of four-tea cultivars during cold acclimation periods in 2015–2016 and 2016–2017 by using qRT-PCR. The expression pattern of CsINV5 in different tea cultivars was showed with different color lines. d Expression analysis of CsINV5 in the leaves of tea plant cultivar ‘LJ43’ under different sugar and temperature conditions by using qRT-PCR. ‘CK’ represents the sugar deficiency control, ‘n’ represents the normal temperature, ‘c’ represents the 4 °C treatment, ‘re’ represents the temperature recovered to normal level. All results were calculated by using the 2–ΔCt method with CsPTB as housekeeping gene. Data are shown as the means ± SE (n = 3)
Promoter analysis of CsINV5
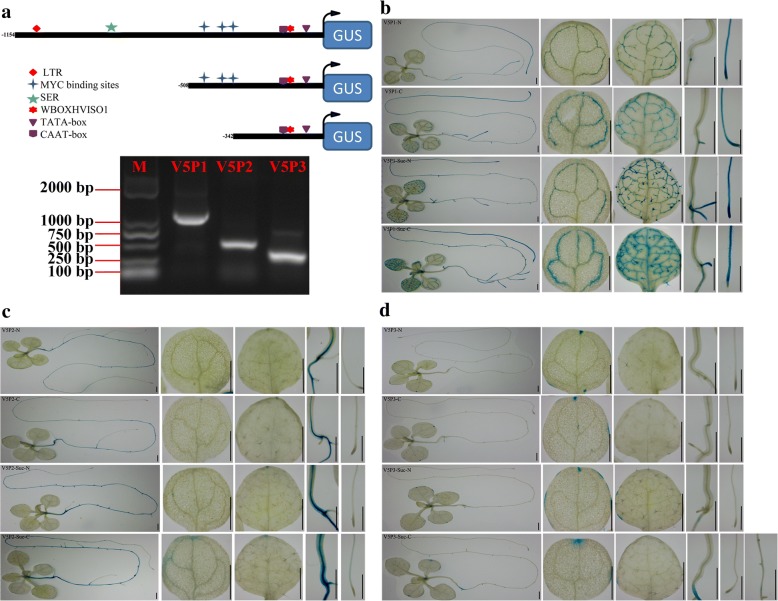
To decipher the functional mechanisms of the regulation of CsINV5 expression, the 1154 bp sequence upstream of CsINV5 was isolated from total genomic DNA of the ‘LJ43’ cultivar, and the cis-acting elements were predicted. The results showed that a TATA-box and a CAAT-box were located at − 65 bp and − 140 bp upstream of the translation start codon, respectively (Additional file 2: S1). Moreover, 18 motifs potentially involved in responding to biotic or abiotic stresses, light, hormones and sugars were found in this sequence (Table 1). Among these 18 motifs, 4 low-temperature-related cis-elements were predicted, including an LTR-related motif (CCGAAA, − 1078) and 3 MYC binding sites (CANNTG, − 453, − 369 and − 346) reported to interact with bHLH proteins at low temperatures [50]. In addition, a sugar-repressive element, SRE (TTATCCA, − 801), and a sugar-responsive element, WBOXHVISO1 (TGACT, − 124), were also found. These findings suggest that the promoter of CsINV5 contains various motifs responding to exogenous and endogenous factors.
Table 1.
Stress-, light-, hormone- and sugar-responsive elements in the 1154 bp 5′-flanking sequence of CsINV5 as predicted by the PLantCARE website
| Site name | Element | Sequence | Function | Copy | Position |
|---|---|---|---|---|---|
| Stresses | ARE | TGGTTT | Essential for the anaerobic induction | 2 | − 197, − 467 |
| HSE | AAAAAATTTC | Involved in heat stress responsiveness | 2 | −402, −551 | |
| TC-rich repeats | ATTCTCTAAC | Involved in defense and stress responsiveness | 1 | − 337 | |
| LTR | CCGAAA | Involved in low temperature responsiveness | 1 | − 1078 | |
| MYC | CAACGTG/CACATG | Low temperature-related element | 3 | − 346, − 369, −453 | |
| Light | MRE | AACCTAA | MYB binding site involved in light responsiveness | 1 | −1054 |
| I-box: | GATAAGGGT | Light responsive element | 1 | − 741 | |
| L-box | CTCACCTACCAA | Part of a light responsive element | 1 | − 491 | |
| Sp1 | CC(G/A)CCC | Light responsive element | 2 | − 170, −491 | |
| Hormones | ERE | ATTTCAAA | Ethylene-responsive element | 1 | − 399 |
| TCA-element | CAGAAAAGGA | Involved in salicylic acid responsiveness | 1 | − 142 | |
| Sugars | WBOXHVISO1 | TGACT | SUSIBA2 bind to W-box element | 1 | −124 |
| SRE | TTATCCA | Alpha-amylase; MYB proteins; gibberellin; sugar starvation; | 1 | −801 |
Tissue-specific analysis
To verify the tissue specificity of CsINV5, three independent T3 homozygous transgenic lines that contain P1154CsINV5::GUS recombinant vector (Fig. 4a), were used for tissue-specific analysis. Histochemical staining results showed that the 5′-flanking sequence of CsINV5 drove the transcription of the GUS reporter gene in various organs of transgenic Arabidopsis during different developmental stages (Fig. 3), though not in seeds. Moreover, we found that GUS activity in flowers increased with floral organ maturity, and the highest GUS activity was found in mature pollen. These results indicate that CsINV5 is commonly expressed in various organs of the tea plant during different developmental stages and may play a role in modulating floral organ maturity.
Fig. 4.
Promoter deletion analysis of CsINV5 under Suc and different temperature exposure. a Schematic representation of chimeric P1154CsINV5::GUS constructs. Different symbols indicate different motifs of interest. Numbers represent distances from the translation start codon. b-d Histochemical GUS staining of transgenic Arabidopsis with P1154CsINV5::GUS, P508CsINV5::GUS, and P342CsINV5::GUS under normal-temperature and Sucrose-deleted conditions (V5P1-N, V5P2-N, and V5P3-N), low-temperature and Sucrose-deleted conditions (V5P1-C, V5P2-C, and V5P3-C), normal-temperature and Sucrose-added conditions (V5P1-Suc-N, V5P2-Suc-N, V5P3-Suc-N), and low-temperature and Sucrose-added conditions (V5P1-Suc-C, V5P2-Suc-C, V5P3-Suc-C), respectively. Scale bars = 1 mm
Fig. 3.
Histochemical GUS staining of transgenic Arabidopsis with P1154CsINV5::GUS. a partial first rosette leaf; b partial fourth rosette leaf; c mature rosette leaf; d cauline leaf; e partial root-zone; f root tip; g main stem; h raceme; i single bud; j siliques at different development stages; k flowers; l single flower; m peduncle; n silique apex; and o seeds. Scale bars = 1 mm
Cold and sugar treatments
To explore the transcriptional regulatory mechanisms of CsINV5 under cold or sugar conditions, we evaluated the responses to cold and sugar (3% Suc) of the proximal promoter region and its deletions (Fig. 4a). Briefly, the truncated promoter P508CsINV5 (LTRE-related and SER motifs deleted) and P342CsINV5 (LTRE-related, SER and MYC motifs deleted) were fused to the GUS coding sequence, and T3 homozygous transgenic Arabidopsis plants plus the P1154CsINV5::GUS transgenic plants were used for further analysis. Both WT and the empty vector (pBI101::GUS) transgenic Arabidopsis were regarded as control.
Histochemical assays revealed that the GUS staining in the leaves and roots of P1154CsINV5::GUS transgenic plants was significantly increased by cold and/or Suc treatment (Fig. 4b). The GUS staining in P508CsINV5::GUS transgenic plants planted on Suc-deleted medium showed no difference in either normal or cold conditions. However, when planted on Suc-added medium, the GUS activity in leaves, stems and roots of the P508CsINV5::GUS transgenic plants were all increased under normal conditions. Furthermore, P508CsINV5::GUS transgenic plants on Suc-added medium showed more GUS staining in stems and roots at low temperatures than at normal temperatures (Fig. 4c). A similar result was found in P342CsINV5::GUS transgenic plants (Fig. 4d). Besides, similar results were obtained in Glc or Fru treatments (Additional file 1: Figure S2), and control results are shown in Additional file 1: Figure S3a and b. Overall, we suggest that the LTRE-related motif, not the MYC binding sites, serves as the core cis-element regulating the transcriptional activity of CsINV5 in response to cold and that the WBOXHVISO1 motif, as a sugar-responsive cis-acting motif, controls CsINV5 transcription in response to both Suc signaling and cold stress.
Overexpression analysis of CsINV5 in transgenic Arabidopsis
Overexpressed CsINV5 promotes root growth in transgenic Arabidopsis
To investigate whether CsINV5 mediates root growth, three homozygous transgenic overexpression lines (OE5–9, OE5–12 and OE5–15) with different transcript abundances were used in the following experiments (Fig. 5a). As Fig. 5b and c show, constitutively overexpressed CsINV5 in Arabidopsis promoted taproot elongation in transgenic plants compared to WT plants; the root length was significantly longer in all investigated transgenic lines than in WT. Moreover, CsINV5-OE plants showed greater development of lateral roots (Fig. 5b).
Fig. 5.
Expression of CsINV5 in WT Arabidopsis promotes root growth. a The expression of CsINV5 in the leaves of WT and CsINV5-OE plants. Data are shown as the means ± SE (n = 3). b Three CsINV5-OE lines showed a longer-root phenotype compared with WT. Scale bar = 10 mm. c The root lengths of three CsINV5-OE lines and WT Arabidopsis after 10 days of growth (each value represents the mean ± SE of 12 seedlings)
Overexpression of CsINV5 enhances cold tolerance in transgenic Arabidopsis
To explore how CsINV5 participates in low-temperature response, CsINV5-OE (OE5–9, OE5–12 and OE5–15) and WT plants were exposed to 4 °C for 6 d, the temperature was reduced to − 1 °C for 2 d, and then leaves were collected to measure the Fv/Fm. Besides, the leaves exposed to − 6 °C for 8 h were collected to measure the EL.
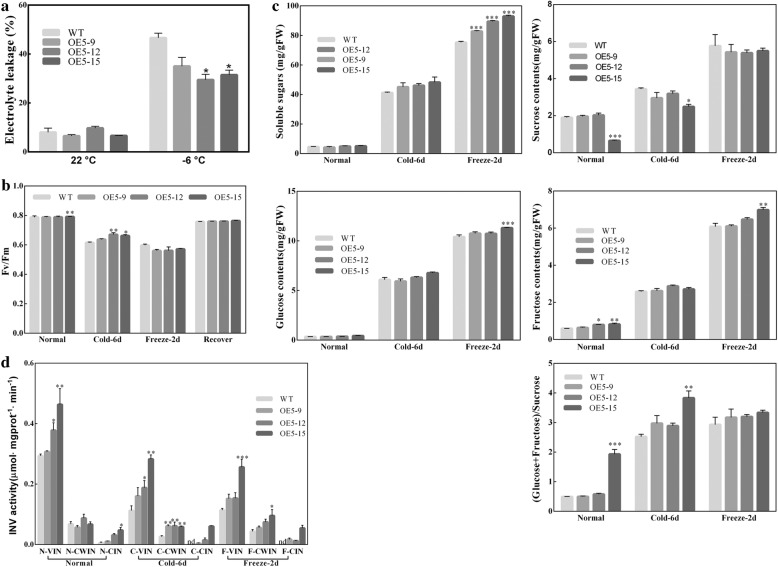
Under normal conditions, the relative EL levels showed no significant difference between CsINV5-OE and WT plants. Under cold conditions, EL in CsINV5-OE was lower than in WT, and OE5–12 and OE5–15 showed significant differences compared to WT (Fig. 6a). Fv/Fm, a key parameter for photosynthetic status [51, 52], was higher in CsINV5-OE than in WT when plants were exposed to 4 °C for 6 d and significantly different in OE5–12 and OE5–15 compared to WT. Fv/Fm values were comparable in CsINV5-OE and WT plants exposed to − 1 °C for 2 d and recovered for 1 d (Fig. 6b).
Fig. 6.
Analysis of EL, Fv/Fm, sugar contents and INV activities in CsINV5-OE plants and WT plants. a EL values in CsINV5-OE plants and WT plants under different temperature conditions. b Fv/Fm ratios in CsINV5-OE plants and WT plants under different temperature conditions. c Sugar contents, including TSS, Suc, Glc, Fru and the corresponding ratio of Hex/Suc, in CsINV5-OE and WT plants under different temperature conditions. d INV activities, including VIN, CWIN and CIN, in CsINV5-OE plants and WT plants under different conditions. Each experiment was performed with four replicates. Data are shown as the means ± SE (n = 4). Significant differences between CsINV5-OE and WT plants in the same conditions are indicated by one (P < 0.05) or two (P < 0.01) asterisks
On the other hand, all four types of sugar content (TSS, Suc, Glc and Fru) increased after cold treatment, and the contents of TSS, Glc, and Fru and the corresponding Hex/Suc ratio in CsINV5-OE were higher than in WT under chilling and freezing conditions (Fig. 6c); a significant difference existed in OE5–15 plants. In contrast, the Suc content in CsINV5-OE was slightly reduced compared to WT at low temperature. Among the INV activities, we found that VIN activity was significantly higher in CsINV5-OE than in WT under both normal and cold conditions, though the transcription of the gene (AtVIN1) corresponding to native VIN activity was reduced at low temperatures (Additional file 1: Figure S4a), which suggests that CsINV5 was successfully expressed in transgenic Arabidopsis. The activity of CWIN in CsINV5-OE was not significantly affected but slightly lower in WT plants, and CIN activity was not detected in WT plants under cold conditions. These results indicate that CsINV5-OE was associated with higher INV activity and increased the Hex/Suc ratio by hydrolyzing much more Suc into Glc and Fru. Based on these data, CsINV5 overexpression in Arabidopsis enhanced cold tolerance.
Overexpression of CsINV5 influenced the transcription of cold-related genes
To explore the molecular mechanisms underlying the CsINV5-mediated increase in cold tolerance in transgenic Arabidopsis, the leaves of WT and CsINV5-OE plants (OE5–15) under normal and cold conditions were subjected to RNA-seq analysis. After quality-based trimming, 83.59 Gb of clean data was obtained from 12 RNA-seq libraries, and the Q30 of each sample was not less than 90.65% (Additional file 3: Table S1; Submission number in NCBI: SRR6057439-SRR6057450). Twelve transcripts were selected to validate the RNA-seq data by qRT-PCR, and Additional file 1: Figure S5 shows that all had similar expression patterns between RNA-seq and qRT-PCR, suggesting that the RNA-seq data are reliable.
DEGs between WT and CsINV5-OE plants under normal or cold conditions were selected based on fold change (FC) ≥ 2 and FDR < 0.05. Under normal conditions, 1344 DEGs were obtained from WT and OE plants, while nearly three times as many DEGs (3395) were found between WT and CsINV5-OE plants under cold conditions, implying that CsINV5 played a major role in cold stress response in transgenic Arabidopsis (Fig. 7ai). Moreover, 5579 DEGs containing 893 up-regulated genes and 832 down-regulated genes were specifically differentially expressed in CsINV5-OE plants, but the other DEGs were commonly differentially expressed both in WT and CsINV5-OE plants under cold condition. However, 6297 DEGs were found in WT plants under cold conditions (Fig. 7aii), indicating that metabolic changes in WT were more extensive at low temperature.
Fig. 7.
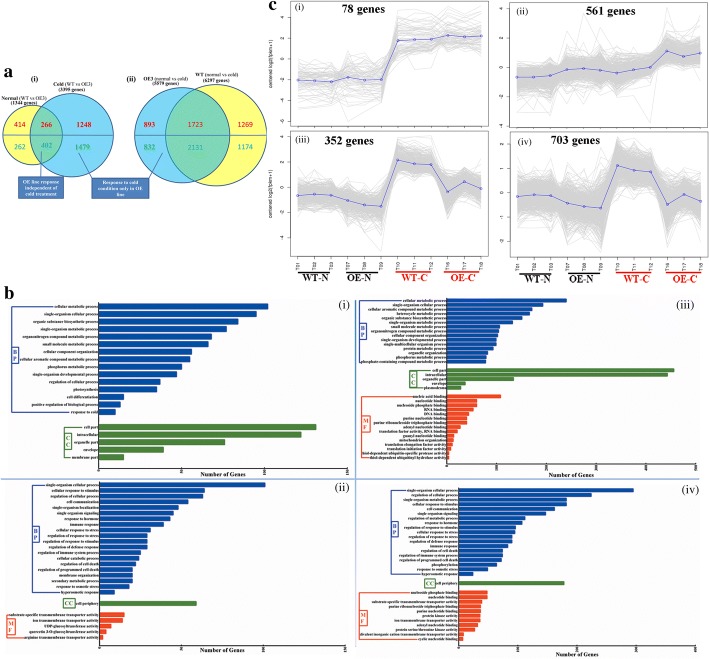
RNA-seq analysis of CsINV5-OE and WT plants. a Venn diagrams showing the numbers of DEGs (FDR < 0.05, log2FC) in CsINV5-OE plants compared to WT plants under normal and cold conditions (WT vs OE) and the DEGs (FDR < 0.05, log2FC) in CsINV5-OE plants and WT plants under normal and cold conditions. b Partial GO terms enriched in the overlap area (266 up-regulated genes (i) and 402 down-regulated genes (ii) and specifically regulated area (1248 up-regulated genes (iii) and 1479 down-regulated genes (iv) between OE and WT plants in normal and cold conditions, respectively (FDR < 0.05). c Partial co-expression patterns of DEGs between OE and WT plants in normal and cold conditions (FDR < 0.05, K-means = 10). (i-ii) DEGs up-regulated in CsINV5-OE plants compared to WT plants; (iii and iv) DEGs down-regulated in CsINV5-OE plants compared to WT plants
In CsINV5-OE plants, 668 DEGs (266 up-regulated and 402 down-regulated) were independent of temperature (Fig. 7ai). GO enrichment analysis, including biological process, cellular component and molecular function, was performed with an FDR < 0.05. For biological process, the GO terms enriched among the 266 up-regulated genes were mainly ‘photosynthesis’, ‘cell differentiation’ and ‘response to cold’, while the 402 down-regulated genes were mainly involved in responses to abiotic and biotic stimuli, defense and immunity (partial GO terms are listed in Fig. 7bi and ii and Additional file 4: Tables S2 and S3). Similarly, the 1248 up-regulated genes and 1479 down-regulated genes in CsINV5-OE plants under cold conditions showed similar GO enrichment results compared to above GO terms, respectively. For biological process, the 1248 specific up-regulated DEGs were enriched in ‘nucleic acid binding’, ‘RNA binding’, ‘DNA binding’ and ‘reactive oxygen species’, while the 1479 down-regulated DEGs were mainly involved in responses to abiotic and biotic stimuli, defense and immunity (partial GO terms are listed in Fig. 7biii and iv and Additional file 4: Tables S4 and S5).
Common DEG expression patterns were evaluated with K-means = 10 (Fig. 7c and Additional file 1: Figure S6). Six hundred and thirty-nine up-regulated DEGs clustered into two expression patterns with higher expression in CsINV5-OE than in WT under cold conditions (Fig. 7ci and ii). These genes included many cold-related genes (CPN60B, CP29, RPL23AB, etc.), osmotic-stress-related genes (ATLEA3, COR413-PM1, COR15B, P5CS1, P5CS2, etc.), and water-deprivation-related genes (CER1, CR88, etc.), which were all specifically up-regulated in CsINV5-OE (Table 2). In addition, 1055 genes that clustered as down-regulated DEGs were highly induced in WT but unchanged or slightly induced in CsINV5-OE at low temperatures (Fig. 7ciii and iv). Among these were many genes that respond to cold (CBF1, AtCDPK1, AtFER1, etc.), osmotic stress (AtMYB50, MYB108, HSP17.6II, AtSWEET15, etc.), and water deprivation (NFYA5, DREB19, AtATG18F, etc.), which were all specifically down-regulated in CsINV5-OE compared to WT (Table 2).
Table 2.
RNA-seq analysis of WT plants and CsINV5-OE plants
| ID | Gene name | Description | log2FC | |||
|---|---|---|---|---|---|---|
| WT vs OE | Normal vs Cold | |||||
| Normal | Cold | WT | OE | |||
| Specific up-regulated genes | ||||||
| Response to cold | ||||||
| AT1G20020 | ATLFNR2 | NADP (H) oxidoreductase | normal | 1.03 | normal | normal |
| AT1G55490 | CPN60B | Chaperonin 60 beta | normal | 1.94 | 1.03 | 2.01 |
| AT1G63940 | MDAR6 | Monodehydroascorbate reductase 6 | normal | 1.26 | normal | normal |
| AT1G67090 | RBCS1A | Rubisco small subunit (RBCS) multigene family | normal | 1.37 | normal | normal |
| AT2G35040 | AICARFT | IMPCHase bienzyme family protein | normal | 1.09 | normal | normal |
| AT2G37190 | RPL12A | Ribosomal protein L11 family protein | normal | 1.02 | normal | normal |
| AT2G37220 | – | Encodes a chloroplast RNA binding protein. | normal | 1.70 | normal | normal |
| AT3G08000 | – | RNA-binding (RRM/RBD/RNP motifs) family protein | normal | 1.13 | normal | normal |
| AT3G13470 | CPN60B2 | Chaperonin-60 beta2 | normal | 2.45 | normal | 2.10 |
| AT3G23700 | SRRP1 | S1 RNA-binding ribosomal protein 1 | normal | 1.59 | normal | normal |
| AT3G49910 | RPL26A | Translation protein SH3-like family protein | normal | 1.04 | normal | normal |
| AT3G53460 | CP29 | Chloroplast RNA-binding protein 29 | normal | 2.34 | normal | 1.01 |
| AT3G54050 | HCEF1 | Chloroplastic fructose 1, 6-bisphosphate phosphatase. | normal | 1.65 | normal | normal |
| AT3G55280 | RPL23AB | Ribosomal protein L23A | normal | 1.00 | normal | 1.13 |
| AT4G04330 | – | Homologue of cyanobacterial RBCX 1 | normal | 1.22 | normal | normal |
| AT4G24280 | cpHsc70–1 | Chloroplast heat shock protein 70–1 | normal | 1.47 | normal | 1.31 |
| AT5G20720 | CPN20 | Chaperonin 20 | normal | 1.71 | normal | 1.21 |
| AT5G50250 | CP31B | Chloroplast RNA-binding protein 31B | normal | 1.17 | normal | 1.04 |
| AT5G54770 | THI1 | Thiazole requiring | normal | 1.12 | normal | normal |
| Response to osmotic | ||||||
| AT1G02820 | ATLEA3 | Late embryogenesis abundant 3 (LEA3) family protein | normal | 1.40 | 4.77 | 4.95 |
| AT1G73570 | – | HCP-like superfamily protein | normal | 1.53 | normal | 1.31 |
| AT2G15970 | COR413-PM1 | Cold regulated 413 plasma memberane 1 | normal | 1.13 | 3.58 | 4.06 |
| AT2G38230 | ATPDX1.1 | Pyrodoxine biosynthesis 1.1 | normal | 1.44 | normal | normal |
| AT2G39800 | P5CS1 | Delta1-pyrroline-5-carboxylate synthase 1 | normal | 1.46 | normal | 1.83 |
| AT2G42530 | COR15B | Cold regulated 15B | normal | 2.03 | 6.50 | 7.60 |
| AT3G04770 | RPSAb | 40s ribosomal protein SA B | normal | 1.14 | normal | normal |
| AT3G14940 | ATPPC3 | Cytosolic phosphoenolpyruvate carboxylase 3 | normal | 1.64 | 1.03 | 1.16 |
| AT3G55610 | P5CS2 | Delta1-pyrroline-5-carboxylate synthase 2 | normal | 1.57 | normal | 2.47 |
| AT5G01410 | PDX1 | Aldolase-type TIM barrel family protein | normal | 1.09 | normal | 1.22 |
| AT5G44110 | ATPOP1 | Arabidopsis thaliana non-intrinsic ABC protein | normal | 1.05 | 4.91 | 5.25 |
| Response to water deprivation | ||||||
| AT1G02205 | CER1 | Fatty acid hydroxylase superfamily | normal | 1.12 | 7.08 | 4.89 |
| AT2G04030 | CR88 | Heat shock protein 88 | normal | 1.81 | normal | 1.80 |
| AT3G08000 | – | RNA-binding (RRM/RBD/RNP motifs) family protein | normal | 1.13 | normal | normal |
| Specific down-regulated genes | ||||||
| Response to cold | ||||||
| AT1G01560 | ATMPK11 | Member of MAP Kinase family | normal | −1.11 | 1.19 | normal |
| AT1G18890 | ATCDPK1 | Calcium-dependent protein kinase 1 | normal | −1.17 | 2.02 | 1.48 |
| AT1G29690 | CAD1 | Constitutively activated cell death 1 | normal | −1.82 | 2.12 | normal |
| AT1G51660 | ATMKK4 | Mitogen-activated protein kinase kinase 4 | normal | −1.12 | normal | normal |
| AT1G52890 | ANAC019 | NAC domain containing protein19 | normal | −2.73 | 1.98 | normal |
| AT1G66400 | CML23 | Calmodulin-like protein | normal | −2.24 | 2.12 | normal |
| AT2G04430 | atnudt5 | Nudix hydrolase homolog 5 | normal | −1.81 | 2.24 | normal |
| AT2G04450 | ATNUDT6 | Nucleoside diphosphates linked to some moiety X 6 | normal | −1.74 | 3.83 | 2.12 |
| AT2G17290 | CPK6 | Calcium-dependent protein kinase family protein | normal | −1.28 | 1.13 | normal |
| AT2G22300 | CAMTA3 | Calcium-binding transcription activator 3 | normal | −1.04 | normal | normal |
| AT2G30250 | WRKY25 | WRKY DNA-binding protein | normal | −1.59 | 1.17 | normal |
| AT2G38470 | WRKY33 | normal | −2.53 | 2.29 | normal | |
| AT2G40140 | CZF1 | Salt-inducible zinc finger 2 | normal | −1.38 | 2.93 | 2.30 |
| AT3G03050 | CSLD3 | Cellulose synthase like D3 | normal | −1.02 | 1.04 | normal |
| AT3G05360 | AtRLP30 | Receptor like protein 30 | normal | −1.84 | 2.77 | 1.86 |
| AT3G11820 | SYP121 | Syntaxin related protein 1 | normal | −1.42 | 1.28 | normal |
| AT3G49530 | ANAC062 | NAC domain containing protein 62 | normal | −1.70 | 2.41 | 1.03 |
| AT3G52400 | SYP122 | Syntaxin of plants 122 | normal | −2.56 | 2.19 | normal |
| AT4G02330 | ATPMEPCRB | Pectin methylesterase 41 | normal | −1.63 | 2.54 | 1.27 |
| AT4G08500 | MEKK1 | MAPK/ERK Kinase Kinase 1 | normal | −1.33 | 1.22 | normal |
| AT4G25490 | CBF1 | C-repeat/DRE binding factor 1 | normal | −1.26 | 4.98 | 3.72 |
| AT4G26120 | NPR2 | Ankyrin repeat family protein | normal | −2.03 | 1.99 | normal |
| AT5G01600 | ATFER1 | Arabidopsis thaliana ferretin 1 | normal | −1.45 | 1.84 | 1.03 |
| AT5G01820 | ATSR1 | Serine/Threonine protein Kinase 1 | normal | −1.16 | 1.47 | normal |
| AT5G02490 | MED37D | Heat shock protein 70 (Hsp 70) family protein | normal | −2.12 | 4.70 | 3.01 |
| AT5G13320 | PBS3 | AVRPPHB susceptible 3 | normal | −3.20 | 3.67 | normal |
| AT5G16910 | ATCSLD2 | Cellulose-synthase like D2 | normal | −1.40 | 1.61 | 1.08 |
| AT5G26920 | CBP60G | CAM-binding protein 60-like G | normal | −3.69 | 3.62 | normal |
| AT5G42050 | NRP | Asparagine-rich protein | normal | −1.33 | 1.58 | normal |
| AT5G57560 | TCH4 | Xyloglucan endotransglucosylase/hydrolase 22 | normal | −2.72 | 2.65 | 1.22 |
| AT5G58670 | ATPLC1 | Arabidopsis thaliana phospholipase C | normal | −1.04 | normal | normal |
| AT5G67340 | PUB2 | ARM repeat superfamily protein | normal | −2.32 | 3.82 | 2.01 |
| Response to osmotic | ||||||
| AT1G15520 | PDR12 | Arabidopsis thaliana ATP-binding cassette G40 | normal | −3.13 | 2.13 | normal |
| AT1G57560 | AtMYB50 | MYB domain protein | nd | −2.90 | 2.63 | normal |
| AT3G06490 | MYB108 | normal | −2.81 | 1.42 | normal | |
| AT3G10500 | anac053 | NAC domain containing protein 53 | normal | −1.38 | normal | normal |
| AT3G14050 | RSH2 | RELA/SPOT homolog 2 | normal | −2.10 | 2.21 | 1.04 |
| AT3G22160 | JAV1 | Jasmonate-associated VQ motif gene 1 | normal | −1.19 | 1.04 | normal |
| AT3G28210 | PMZ | Encodes a putative zinc finger protein | normal | −2.91 | 3.00 | normal |
| AT4G15120 | – | VQ motif-containing protein | normal | −2.57 | 3.07 | normal |
| AT4G34390 | XLG2 | Extra-large GTP-binding protein 2 | normal | −2.27 | 1.80 | normal |
| AT4G36990 | HSF4 | Haliana heat shock factor 4 | normal | −1.19 | 0.51 | normal |
| AT5G13170 | SAG29 | Senescense-associated gene29 | normal | −3.53 | 6.25 | 3.12 |
| AT5G39720 | AIG2L | Avirulence induced protein 2 like protein | nd | −2.90 | 2.82 | normal |
| Response to water deprivation | ||||||
| AT1G08920 | ESL1 | Early response to dehydration SIX-like 1 | normal | −1.76 | 3.42 | 2.14 |
| AT1G02930 | GSTF6 | Arabidopsis thaliana gluatione s-transferase F3 | normal | −1.54 | normal | normal |
| AT1G32870 | ANAC13 | NAC domain containing protein 13 | normal | −1.12 | 2.56 | 1.74 |
| AT1G32940 | ATSBT3.5 | Subtilase family protein | normal | −1.82 | 2.37 | 1.42 |
| AT1G32950 | – | normal | −3.30 | 4.69 | 1.15 | |
| AT1G54160 | NFYA5 | Nuclear factor Y A5 | normal | −1.29 | 1.80 | 1.20 |
| AT2G38340 | DREB19 | Dehyration response element-binding protein 19 | normal | −2.62 | 2.20 | normal |
| AT2G45570 | CYP76C2 | Cytochrome P450 | normal | −2.14 | 2.71 | 1.41 |
| AT3G56880 | – | VQ motif-containing protein | normal | −1.33 | 1.80 | normal |
| AT4G12250 | GAE5 | UDP-D-glucuronate 4-epimerase | normal | −1.26 | 1.50 | normal |
| AT5G54730 | ATATG18F | Yeast autophagy 18 F-like protein | normal | −1.04 | 1.26 | normal |
Differentially expressed genes (FDR < 0.05, log2FC) between WT plants and CsINV5-OE plants under cold conditions were annotated with the GO terms ‘response to cold’, ‘response to osmotic stress’ and ‘response to water deprivation’. The log2FC values between WT plants and CsINV5-OE plants under normal conditions, and log2FC values in CsINV5-OE plants or WT plants between normal and cold condition are also shown
“—” indicates the gene has not been named; “nd” represents that the FC value was not detected in DEGs expression analysis results. “normal” indicates the gene was not differentially expressed between these two conditions. The negative value represents the gene was down-regulated, and the positive value represents the gene was up-regulated
In summary, based on DEG expression patterns and functional annotations, we found that many cold-induced genes involved in the CDPK, MAPK, CBF/COR and ABA-dependent or -independent pathways had unchanged or slightly increased expression levels in CsINV5-OE plants exposed to cold stress. However, some genes encoding RNA-binding and chaperonin proteins and some osmotic-stress-related genes had higher expression levels in CsINV5-OE plants exposed to cold stress. Therefore, combined with the values of EL, Fv/Fm and sugar contents and the transcription levels of many osmotic-stress-related genes between WT and CsINV5-OE plants under low temperature, we demonstrate that the tolerance of CsINV5-OE to low temperature was stronger than that of WT.
Discussion
CsINV5 encodes a VIN protein involved in cold stress response
Both chilling (< 20 °C) and freezing (< 0 °C) temperatures are belong to cold stress, which detrimentally affects the growth and development of plants and seriously limits the spatial distribution of plants and crop yields [53]. Most temperate plants acquire more freezing tolerance by exposing to non-freezing low temperatures (mainly range 2 to 6 °C in the controlled environment chamber), a process known as CA. Numerous of physiological, biochemical and molecular are changed during CA, including the remodeling of cell and tissue structures, the reprogramming of metabolism and gene expression [32]. In tea plant, previous studies showed that the expression level of CsINV5 was increased 3 times in ‘LJ43’ cultivar when they exposed to a non-freezing low temperatures (4 °C) for 5 days as compared to the normal cultivated ‘LJ43’ cultivar [38]. In addition, the expression of CsINV5 was also significantly induced by natural CA process [42]. In the present study, a similar expression patterns were also found in four-tea cultivars (DMB, LJ43, ZN12, and ZN113) during CA (Fig. 2b and c), which suggest CsINV5 may function similarly in cold stress response among different tea plant cultivars, but the mechanism by which this gene participates in cold stress response remains unknown. In the present study, we found that CsINV5 encodes a soluble protein with a molecular mass of nearly 80 kDa (Additional file 1: Figure S7). The protein properties of this gene are similar to those of known VIN proteins [47, 54, 55] (Fig. 1 and Additional file 1: Figure S1), suggesting that CsINV5 encodes a functional VIN protein.
The promoter region of CsINV5 contains core motifs that respond to cold stress, sugar signaling and reproductive growth
The promoter region contains multiple cis-elements that can bind to various types of transcription factors to mediate the expression of downstream genes. By using deletion analysis, electrophoretic mobility shift analysis (EMSA) and synthetic oligonucleotides, Dunn et al. [56] identified an LTRE-related motif, CCGAAA, as the binding site of a low-mobility nuclear protein complex in the 42 bp low-temperature-responsive promoter region of blt4.9. Furthermore, they found the low-temperature responsiveness to basal levels would be decreased as this motif mutated, which suggests that CCGAAA is an alternative LTRE participating in low-temperature response. In the present study, we found that the same LTRE-related motif (CCGAAA) was proven to respond to cold stress based on our promoter deletion analysis in transgenic Arabidopsis, suggesting that the LTRE-related motif within the CsINV5 promoter may serve as the core binding site of some low-mobility nuclear protein to positively regulate the expression of CsINV5 under cold conditions.
On the other hand, many reports have shown that INV gene transcription is regulated by exogenous sugar treatment [44, 57–59]. McLaughlin and Boyer [60] reported that Suc feeding rescued the transcription of a VIN gene, ZmIVR2, which was reduced by water deficits. In addition, the results of promoter analyses have revealed the role of INV gene in response to sugar treatment [61–63]. Ou et al. [64] demonstrated that the promoter region from − 118 to − 551 bp of a VIN gene, StvacINV1, contained seven SREs (TTATCC) motifs, where they found this promoter region was required for negative response to Suc/Glc. Similarly, Rabot et al. [63] demonstrated that the − 595 to − 468 bp promoter region of RhVI1, which contains an SRE, was required for negative response to Suc and Fru in the dark. Moreover, the − 468 to − 307 bp promoter region of RhVI1, which also contains a WBOXHVISO1 (TGACT), promoted a significant induction of GUS activity by Suc and Fru in the dark, compared with mannitol treatment. SREs are recognized by three MYB transcription factors that induce the expression of α-amylase under Suc and Glc starvation in rice [65]. But the WBOXHVISO1 motif is recognized by SUSIBA2, a member of the WRKY family, which positively regulates the transcription of isoamylase in response to Suc during barley seed development [66]. In agreement with these findings, we found that the expression of CsINV5 was induced by sugar treatments at normal or low temperatures (Fig. 2d). In addition, the results of the promoter deletion analysis further confirmed this phenomenon (Fig. 4), which suggesting that the sugar-responsive element (WBOXHVISO1) but not the SRE within the promoter region of CsINV5 may act as the core motif to bind to a putative WRKY transcription factor to induce the transcription of CsINV5 in response to sugar and/or cold treatments.
In addition to response to environmental stimuli and sugar treatment, VINs also play an important role in plant reproductive growth. Direct evidence was provided by Wang and Ruan [16]; silencing a VIN gene, GhVIN1, in cotton reproductive organs reduced viable seed production due to pollination failure and impaired male and female fertility. In our study, the promoter of CsINV5 showed the highest GUS activity in mature pollen of transgenic Arabidopsis, and this result is consistent with the tissue-specific expression pattern of CsINV5 in the tea plant, where we previously found the transcription abundance of CsINV5 in flowers of ‘LJ43’ cultivar is nearly 90-fold than that in roots [38]. Thus, CsINV5 may exert strong control over reproduction from male and female fertility to floral development, but the mechanism remains to be explored.
CsINV5 overexpression in Arabidopsis promotes root elongation through an osmotic-independent pathway
Many studies have reported that VINs play an important role in regulating plant cell expansion and cell division [45, 67–69]. A similar result was obtained in our study, where the overexpression of CsINV5 enhanced both taproot and lateral root elongation in transgenic Arabidopsis, indicating that CsINV5 regulates cell expansion and cell division. However, the mechanism by which VINs regulate root elongation is not yet clear, and different mechanisms of VIN-regulated root elongation may exist in different species. As Wang et al. [45] demonstrated a VIN gene, GhVIN1, regulates the elongation of cotton fibers and the roots of transgenic Arabidopsis in osmotic-dependent and -independent manners, respectively, based on the relative contributions of sugars to sap osmolality. In Arabidopsis roots, the combined Hex and Suc concentrations are extremely low, which indicates that their osmotic contribution is negligible. Therefore, we suggest that CsINV5 increased root elongation in transgenic Arabidopsis independently of the osmotic pathway. In addition, Wang et al. [70] found that GhVIN1 may mediate Hex signaling to regulate fiber initiation, probably via regulating the transcription of several auxin signaling components and MYB transcription factors previously identified as required for fiber initiation. These results indicate that VINs may indirectly mediate root elongation by regulating the concentration of Hex to mediate the transcription of MYBs and auxin metabolism, including biosynthesis, transport and perception. In this regard, we found that many MYB genes annotated to function in cell division and many auxin-metabolism-related genes annotated to function in root development were differentially expressed in the leaves of CsINV5-OE plants under normal conditions (Additional file 5: Table S6). Whether and how these genes interacted with Hex to control cell elongation and lateral root growth remains to be tested in the future.
CsINV5 enhanced cold tolerance in CsINV5-OE plants mainly through the osmotic-dependent pathway
Based on the above, the mechanism of CsINV5 response to low temperature was evaluated by overexpression analysis. In the present study, we found the VIN activity were decreased both in WT and CsINV5-OE plants when they exposed to low temperature condition, but CsINV5-OE plants showed higher VIN activity than the WT plants both under normal temperature and cold conditions (Fig. 6). As a result, the higher VIN activity promotes the Fv/Fm values and TSS contents, especially Glc and Fru increased in CsINV5-OE plants than in WT plants under cold conditions (Fig. 6).
However, the functions of VIN genes are varied in different plant species. As Chen et al. [71] reported that the VIN activities in two evergreen woody plants were decreased as the temperature declined in winter. However, in our study, we found the expression of CsINV5 in tea plant was induced during CA with decreasing temperature (Fig. 2b and c), and a similar results was also obtained by short-term cold treatment (Additional file 1: Figure S4b), but the transcriptions of two VIN genes (AtVIN1 and AtVIN2) corresponding to native VIN activity was reduced in Arabidopsis under low temperatures condition (Additional file 1: Figure S4a). Similarly, AtVIN2 gene inhibited can significantly promote the accumulation of proline content, and then increase the cold tolerance of the transgenic Arabidopsis under cold condition [72]. In contrast, we found that overexpression of CsINV5 in transgenic Arabidopsis could improve the cold tolerance of CsINV5-OE plants, and promote the expressions of two rate-limiting enzyme genes in Pro biosynthesis, P5CS1 and P5CS2, suggesting that Pro content is higher in CsINV5-OE plants under cold condition. In a word, we suggest that the mechanisms of VIN genes in responding to low temperature are varied in different species, some genes with high expression may enhance the cold resistance of the plant, but the others may promote the cold resistance with low expressions.
Soluble sugars, especially Suc, Glc, and Fru, not only act as energy and carbon sources for plant growth and development but also function as signaling molecules, osmoprotectants and antioxidants to support abiotic stress responses [2, 73–77]. Therefore, the higher concentration of soluble sugars in CsINV5-OE plants under cold condition, may act as antioxidants to maintain ROS homeostasis, prevent membrane lipid peroxidation, and alleviate injury from low temperature. Accordingly, we found that transcription of some antioxidant biosynthesis-related genes was lower in CsINV5-OE plants than in WT plants (Additional file 1: Figure S5), indicating that CsINV5-OE plants may be less susceptible to freezing injury than WT plants due to higher sugar contents.
Suc and Glc act as signaling molecules that mediate auxin biosynthesis to regulate plant cell division and expansion [78], and this interaction also participates in responses to environmental stimuli [79, 80]. CsINV5-OE plants showed increased expression of many DEGs related to auxin biosynthesis, metabolism, signaling and transport (Additional file 5: Table S6), and most of these genes, such as the members of the SAUR and GH3 families down-regulated by Glc treatment in Arabidopsis [81], were also down-regulated in CsINV5-OE plants under cold conditions, but whether and how Glc signaling interacts with auxin signaling in CsINV5-OE plants to support cold stress response requires further study. Glc signaling requires a sensor hexokinase (HXK), which sequentially regulates plant gene expression at the transcriptional, translational, and post-translational levels [2, 82, 83]. Among the DEGs, we found a hexokinase gene, AtHXK2, up-regulated in CsINV5-OE plants under cold conditions (Additional file 5: Table S6); AtHXK2 may mediate Glc signaling to improve cold tolerance in CsINV5-OE plants.
Osmotic stress and the associated oxidative stress are common consequences induced by drought, salinity and low temperature [32, 84]. In plants, Suc hydrolyzed into Glc and Fru by VINs is thought to double its osmotic contribution, sustaining favorable cellular turgor and facilitating water influx [20, 45, 58, 69]. Higher contents of Glc and Fru, associated with higher ratios of Hex/Suc, were found in CsINV5-OE plants under both normal and cold conditions (Fig. 6) and may help sustain favorable osmotic potential, maintain normal water influx, protect plasma membrane integrity against water deprivation or oxidation, and alleviate injury caused by low temperature. Stronger evidence supporting this hypothesis was found in the RNA-seq results, where the expression of two rate-limiting enzymes in Pro biosynthesis, P5CS1 and P5CS2, were up-regulated in CsINV5-OE plants under cold conditions, suggesting that Pro content is higher in CsINV5-OE plants. Under osmotic stress, the osmoprotectant Pro is rapidly synthesized in cells to reduce water potential and prevent water deprivation [85, 86]. Moreover, GO enrichment analysis results showed that the transcription of some DEGs reported to respond to osmotic stress, such as AtLEA3, COR413-PM1 and COR15B, were up-regulated in CsINV5-OE under cold conditions, suggesting that these genes as well as P5CS1 and P5CS2 contributed to maintaining osmotic balance to prevent CsINV5-OE plants from low-temperature injury. Thus, the stable osmotic potential in CsINV5-OE plants may attenuate the perception of calcium ion signals, and then a series of cold-related genes located downstream of the calcium signaling pathway, including CDPK, MAPK, CBF/COR, and ABA signal-transduction-related genes, would maintain approximately normal transcription levels (Table 2, Additional file 5: Table S6). To summarize, the improved cold resistance of CsINV5-OE plants derives from many metabolic changes, but the osmotic-dependent pathway may be an important factor.
Conclusions
In conclusion, we found the LTRE-related and WBOXHVISO1 motifs within the promoter of CsINV5 were the core cis-elements in response to low temperature and sugar treatment; the former participated mainly in response to cold stress, and the latter positively mediated the expression of CsINV5 under both sugar and cold conditions. In addition, overexpression of CsINV5 in Arabidopsis promoted taproot and lateral root elongation through an osmotic-independent pathway and enhanced cold tolerance mainly through the osmotic-dependent pathway.
Methods
Plant materials and treatments
Four-tea cultivars (DMB, LJ43, ZN12, and ZN113) that have been grown for 15 years from the same farm (TRI, CAAS, N30°10′, E120°5′) as reported by Wang et al. [40] were used for present study. These selected cultivars were used for comparative expression analysis of CsINV5 during natural cold conditions. For natural CA analysis, the third to the fifth mature and healthy leaves from more than 10 randomly selected tea plants of each variety were collected and mixed together between 9:30 a.m. and 10:30 a.m. on each collection day. Three independent biological replicates were used for each sample.
The Arabidopsis ecotype Columbia-0 was used as wild-type (WT) and to generate the transgenic lines containing the promoter or the ORF of CsINV5. The seedlings of the selected transgenic lines and wild plants were firstly grown on 1/2MS medium (1.5% Suc, 0.8% Agar, pH 5.7) for 7 d, then transplanted to a soil mixture (3: 2: 1 peat moss: vermiculite: perlite) for 20 d in the growth chamber with photoperiod (10 h of light at 22 °C and 14 h of dark at 20 °C) and 100 μmol/m2 s as described by Wang et al. [40]. Seeds of WT and T3 homozygous transgenic lines were sterilized and vernalized, and the control vector homozygous transgenic lines were omitted as they have the same morphological as the WT as described by Wang et al. [40].
Sugar and cold treatment in tea plants
For sugar treatment, 1-year-old plants raised from cuttings of the major local cultivar ‘LJ43’ were used hydroponically. Before proceeding, the hydroponic cuttings were cultured in the greenhouse with photoperiod (14 h of light and 10 h of dark at 23 °C, 75% of humidity) and a light intensity of 12, 000 Lux as described by Qian et al. [38]; then, 3% Suc, 3% Glc, 3% Fru and 3% Man were added into the nutrient solution. All sugar treatments were carried out for 2 days at 25 °C, and the second and/or third mature leaves from the terminal bud were sampled and mixed together at 3, 12, 24, and 48 h post-treatment. Thereafter, all sugar treatments were continued for 4 days at 4 °C, and the mature leaves were collected at 3, 12, 24, 48, 72 and 96 h, respectively. Finally, all the hydroponic cuttings with sugar treatments were returned to 25 °C for 1 day, and the mature leaves sampled at 6 h and 24 h. All samples were quickly frozen in liquid nitrogen and stored at − 80 °C until used. Three independent biological replicates were performed at each sampling time point, and each replicate contained four cuttings. The nutrient solution formulation is listed in Additional file 6: Table S7.
Phylogenetic and conserved domain analysis of CsINV5
MEGA 6.0 software and the Neighbor-Joining method with 1000 bootstrap replicates were used to explore the phylogenetic relationships of CsINV5 with known VINs and CWINs, and DNAMAN software was used to investigate the characteristic amino acid sequences of CsINV5 and the known AIs. The amino acid sequences of the AI proteins used for analysis are listed in Additional file 7: Table S8.
Cloning and sequence analysis of the promoter of CsINV5
The genomic DNA used as the PCR template was extracted from the tea plant cultivar ‘LJ43’ with the cetyl trimethylammonium bromide (CTAB) method [87]. The 5′-flanking sequence of CsINV5 was amplified using the Genome Walking Kit (TaKaRa, Dalian, China) following the manufacturer’s instructions. The gene-specific primer1 (GSP1) and gene-specific primer 2 (GSP2) of CsINV5 were used for the first and the second round of PCR, respectively. Subsequently, the amplified and purified PCR product was cloned into the pMD18-T vector (TaKaRa, Dalian, China) and sequenced. Finally, the PlantCARE website was used to predict cis-acting regulatory elements. All specific primers mentioned above are listed in Additional file 6: Table S9.
Construction of vectors and plant transformation, and sugar and cold treatments in transgenic plants
To fuse the promoter of CsINV5 into the GUS-plus vector (pBI101::GUS), 5′-deletions of the CsINV5 promoter at − 1154, − 508, and − 342 were amplified by PCR using different forward primers (V5PF1, V5PF2 and V5PF3) and a single reverse primer, V5PR (Additional file 6: Table S9). The amplified and purified PCR product was cloned into the pMD18-T vector and sequenced, and then the promoter fragments were sub-cloned into the GUS-plus vector to form P1154CsINV5::GUS, P508CsINV5::GUS and P342CsINV5::GUS according to the restriction endonuclease method.
To construct the overexpression vector, the Gateway technology was used [88]. In brief, the ORF of CsINV5 without a stop codon was amplified with the primer pair V5F and V5R (Additional file 6: Table S9), and then the fragment was fused into the entry vector pENTR/D-TOPO (Invitrogen, CA, USA) according to the manufacturer’s instructions and sequenced (‘pENTR-CsINV5’). Subsequently, the CsINV5 ORF was transferred from the entry construct into the destination vector, pH7FWG2, using the LR Clonase II enzyme mix (Invitrogen, Carlsbad, CA, USA) [89].
These recombined vectors as well as the empty vector (pBI101::GUS) were introduced separately into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis via Agrobacterium-mediated transformation [90].
To explore the cold tolerance of the CsINV5-OE plants, the sterilized and vernalized seeds of both WT plants and CsINV5-OE plants (OE5–9, OE5–12 and OE5–15) with different transcript abundances of CsINV5 were grown on 1/2 MS medium for 1 week, and then transplanted to soil mixture consisting of 3: 2: 1 peat moss: vermiculite: perlite for 3 weeks. For the cold treatment, both WT plants and CsINV5-OE plants were treated at 4 °C for 6 d followed by a mild cold stress (− 1 °C) for 2 d without change the light time and intensity. Leaves were collected after 6 d of 4 °C treatment and 2 d of − 1 °C treatment to measure INV activity, and sugar contents, including total soluble sugar (TSS), Suc, Glc and Fru. Four independent biological replicates were performed, and each replicate contained four seedlings. In order to measure electrolyte leakage (EL), both WT plants and CsINV5-OE plants were treated at 4 °C for 6 d, and then exposed to − 6 °C for 8 h, as described by Rohde et al. [91]. Three independent biological replicates were performed, and each replicate contained five seedlings.
To explore the function of the promoter of CsINV5 under low temperature and sugar treatment conditions. We detected the GUS activity of the T1 positive promoter::GUS transgenic Arabidopsis by GUS staining firstly, and then three transgenic lines with relatively higher GUS activity were selected from each construct for the following experiments. The sterilized and vernalized seeds of WT plants and four independent transgenic lines of pBI101::GUS (empty vector), P1154CsINV5::GUS, P508CsINV5::GUS and P342CsINV5::GUS were sown onto a square plate with 1/2 MS medium without Suc and grown vertically for 4 d, and then the seedlings were transplanted onto a new square plate with either 1/2 MS medium without Suc or 1/2 MS medium supplemented with 3% Suc for another 7 d of growth. The seedlings in each treatment were kept at normal temperature or 4 °C for 3 d and then collected for the histochemical assay. Four independent biological replicates were performed, and each replicate contained eight seedlings.
For tissue-specific analysis, the tissues of the three independent T3 homozygous transgenic lines P1154CsINV5::GUS during different developmental periods were used to investigate GUS activity, including the leaves, roots, stems, flowers and siliques.
All fresh samples of transgenic Arabidopsis containing the promoter of CsINV5 as mentioned above were immersed in X-Gluc solution (Fermentas, Canada) for 12 h dark incubation at 37 °C, then rinsed in 70% ethanol several times. The preparations were studied with a stereomicroscope (Olympus Corporation, Japan), and the photographs were captured with an industrial digital camera (Oplenic Corporation, China).
Root development assay, and Fv/Fm, EL, sugar content and INV activity measurements
The sterilized and vernalized seeds of WT plants and CsINV5-OE plants were vertically germinated on 1/2 MS medium with 3% Suc and 1% agar for 4 d. Two seedlings of each genotype with similar growth states, representing one biological replicate, were transferred onto a new square plate and grown vertically in the same growth chamber. The root lengths were measured with a ruler at the same time every day and record the data, a total of 7 d were proceed. Finally, the phenotypes were captured with a scanner (Epson Perfection V700 Photo, Indonesia) after 7 d of development. Twelve independent biological replicates of each type of Arabidopsis were performed.
The EL was measured as described by Wang et al. [40]. To explore the maximum quantum efficiency of photosystem II (Fv/Fm), both WT plants and CsINV5-OE plants were allowed recovery growth under normal conditions for 1 day after spending 2 d at − 1 °C, and the Fv/Fm was measured under different temperature conditions. Before measurement, all plants were dark adapted for 40 min, and then the Fv/Fm was detected by a photon system instrument (Open FluorCam FC 800-O, Drasov, Czech Republic). Four independent biological replicates of each genotype were performed, and each replicate contained three seedlings.
For the sugar content assay, the collected rosette leaves from WT plants and CsINV5-OE plants were ground with liquid nitrogen. Then, 0.1 g powder was extracted with 1.0 mL distilled water and ultrasonic treatment for 30 min and subsequently shaken at 800×g for 30 min. After centrifuging at 12, 000×g for 10 min, the supernatants were used for TSS, Suc, Glc and Fru measurements with a corresponding sugar measurement kit (Suzhou Comin Biotechnology, Suzhou, China), respectively.
INV activity was assayed as described by Tomlinson et al. [92] and Wang et al. [45]. A total of 0.2 g collected leaves of each sample both WT and CsINV5-OE plants from different temperatures were used.
RNA-seq analysis
Four-week-old WT and CsINV5-OE plants (OE5–15) were grown at 4 °C for 6 d, and then the temperature was reduced to − 1 °C for 2 d before sampling the leaves. Leaves at the 22 °C were sampled as controls. All samples were snap frozen in liquid nitrogen and stored at − 80 °C until RNA isolation. Three independent biological replicates were performed at each temperature, and each replicate contained four seedlings.
For RNA-seq analysis, the total RNA of each sample was extracted as described by Wang et al. [40]. RNA concentration and integrity were measured using the NanoDrop 2000 (Thermo Scientific, USA) and RNA Nano 6000 Assay Kit on the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Thereafter, 1 μg RNA was used to construct each sequencing library with the NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. Subsequently, the index-coded samples were clustered on a cBot Cluster Generation System, and the library preparations were sequenced on an Illumina HiSeq Xten platform. After sequencing, clean data were obtained by removing reads containing adapter, reads containing poly-N and low-quality reads from the raw data. Gene function was annotated based on the following databases: Nr (NCBI non-redundant protein sequences), Nt (NCBI non-redundant nucleotide sequences), Pfam (Protein family), KOG/COG (Clusters of Orthologous Groups of proteins), Swiss-Prot (manually annotated and reviewed protein sequence database), KO (KEGG Ortholog database), and GO (Gene Ontology). Gene expression levels were estimated by fragments per kilobase of transcript per million fragments mapped (FPKM). Differential expression analysis of two conditions/groups was performed using the DESeq R package (1.10.1), and the genes with an adjusted False Discovery Rate (FDR) < 0.05 according to DESeq were considered differentially expressed. GO enrichment analysis of the differentially expressed genes (DEGs) was performed by a web database (http://omicslab.genetics.ac.cn/GOEAST/index.php). For co-expression cluster analysis, Perl, R language and the cluster package were used to construct the K-means tree.
Quantitative real-time RT-PCR (qRT-PCR) analysis
For qRT-PCR analysis in the tea plant and Arabidopsis, total RNA was extracted as described by Qian et al. [38] and Wang et al. [40], respectively. First-strand cDNA was reverse transcribed according to the user manual in the PrimeScript™ RT reagent kit with gDNA Eraser (TaKaRa, Otsu, Japan), and the qRT-PCR program was performed on a Roche 384 real-time PCR machine (Roche). The qRT-PCR reagents were the following: a total of 10 μL of reaction mixture, which included 5 μl SYBR Premix Ex Taq, 0.8 μl sense/antisense primers, 1 μl cDNA and 3.2 μl distilled water. The qRT-PCR program was performed as follows: 95 °C, 15 s; 94 °C, 5 s and 58 °C, 30 s for 40 cycles; then a melting curve. CsPTB [93] and AtEF (At5G19510) [94] were used as reference genes for quantifying the expression levels of the target genes according to the method of 2-ΔCt or 2−ΔΔCt [95]. The quantitative analysis of each RNA sample was repeated at least three times, and the representative data are expressed as the mean values ± standard error (± SE). The primers used in qRT-PCR are listed in Additional file 6: Table S10. For RNA-seq analysis, we calculated the log2 value by using the FPKM data of each sample, then the log2 value of each sample under different temperature conditions was normalized to WT samples under normal-temperature conditions; the data are shown in a bar chart with the qRT-PCR results.
Statistical analysis
Statistical differences in various physiological or molecular indicators between WT plants and CsINV5-OE plants under different conditions were tested by a one-way Analysis of Variance (ANOVA) based on Tukey’s HSD test and/or Fisher’s least significant difference (LSD) test at significance levels of p < 0.05, p < 0.01 and p < 0.001, and the WT plants and CsINV5-OE plants act as factor and the data of various physiological or molecular indicators act as dependent variables.
Additional files
Figure S1. Homologous analysis of ten SAIs protein sequences. Figure S2. Promoter deletion analysis of CsINV5 under Glc, Fru and different temperature conditions. Figure S3. Histochemical GUS staining of WT plants and transgenic Arabidopsis with the empty vector (pBI101::GUS) under Suc and different temperature exposure. Figure S4. Expression analysis of AtVIN1, AtVIN2 in Arabidopsis, and CsINV5 in tea plant under low temperature condition. Figure S5. Expression detection of selected transcripts by RNA-Seq and qRT-PCR. Figure S6. Partial co-expression patterns of DEGs between OE and WT plants in normal and cold conditions (FDR < 0.05, K-means = 10). Figure S7. Prokaryotic expression analysis of CsINV5. (DOCX 10071 kb)
S1. The genome DNA sequence of CsINV5. 1154 bp promoter sequence and 7 exons have been highlighted: (i) stress signaling (ARE, fully outlined in red; HSE, double outlined in red; TC-rich repeats, dot outlined in red; LTR, viridity highlighted; MYC, viridescence highlighted); (ii) light signaling (MRE, navy highlighted; I BOX, violet highlighted; L BOX, pink highlighted; SP1, underlined); (iii) Hormones signaling (ERE, blue highlighted; TCA-element, green highlighted); (iv) sugar repression (SRE, highlighted in deep yellow) and activation (WBOXHVISO1, highlighted in gray). The CAAT-box and TATA-box are yellow highlighted. PCR primer sequences highlighted in bold and italic. The exons were showed by red font. (DOCX 32 kb)
Table S1. RNA-seq data and quality control. (XLSX 11 kb)
Table S2-S5. Table S2. Sixty-six significant GO terms (FDR < 0.05) mapped by 266 up-regulated DEGs between CsINV5-OE and WT plants. Table S3. Fifty-five significant GO terms (FDR < 0.05) mapped by 402 down-regulated DEGs between CsINV5-OE and WT plants. Table S4. Sixty-two significant GO terms (FDR < 0.05) mapped by 1248 up-regulated DEGs between CsINV5-OE and WT plants in cold conditions condition. Table S5. Sixty-eight significant GO terms (FDR < 0.05) mapped by 1479 down-regulated DEGs between CsINV5-OE and WT plants in cold conditions condition. (XLSX 27 kb)
Table S6. Partial DEGs (FDR < 0.05, log2FC) in CsINV5-OE plants compared to WT plants in normal and cold conditions. (XLSX 21 kb)
Table S7. Nutrient solution formulation. Table S9. Primer sequences used in promoter cloning and vector construction. Table S10. Primer information used in qRT-PCR detection. (DOCX 22 kb)
Table S8. Amino acid sequences of the SAI proteins used for conserved domain analysis. (XLSX 13 kb)
Acknowledgements
We thank Dr. Lu Wang (University of Tasmania, Australia) and Prof Yongling Ruan (University of Newcastle, Australia) for providing the INV activity assay methods.
Funding
This work was supported by the National Natural Science Foundation of China (31770735, 31800588), the Earmarked Fund for China Agriculture Research System (CARS-19), the Chinese Academy of Agricultural Sciences through an Innovation Project for Agricultural Sciences and Technology (CAAS-ASTIP-2014-TRICAAS) and the Young Elite Scientist Sponsorship Program by CAST (2016QNRC001). These funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The raw data of RNA-seq experiments generated in this study have been deposited in the NCBI database under the accession number SRR6057439-SRR6057450. All the data supporting the findings of the study is contained within the manuscript and its additional information files.
Abbreviations
- CA
Cold acclimation
- CDPK
Calmodulin-dependent protein kinases
- CIN
Cytoplasm invertase
- CWIN
Cell wall binding invertase
- EL
Electrolyte leakage
- GUS
β-glucuronidase
- Hex
Hexose
- INV
Invertase
- MAPK
Mitogen-activated protein kinase
- TSS
Total soluble sugar
- VIN
Vacuolar invertase
Authors’ contributions
XW and YY designed and conducted the research; WQ, BX, LW, NL, and XH performed the experiments; CY, HC, YW, YY, and JZ sampled the materials and analyzed the data; WQ and BX wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wenjun Qian, Email: qau-WenjunQian@qau.edu.cn.
Bin Xiao, Email: 1647785720@qq.com.
Lu Wang, Email: wanglu317@tricaas.com.
Xinyuan Hao, Email: haoxy@tricaas.com.
Chuan Yue, Email: yccyyx@163.com.
Hongli Cao, Email: lili9885@126.com.
Yuchun Wang, Email: wangyuchun@tricaas.com.
Nana Li, Email: nanali@tricaas.com.
Youben Yu, Email: yyben@163.com.
Jianming Zeng, Email: zengjm@tricaas.com.
Yajun Yang, Email: yjyang@tricaas.com.
Xinchao Wang, Email: xcw75@tricaas.com.
References
- 1.Gulen H, Cansev A, Eris A. Cold hardiness of olive (Olea europaea L.) cultivars in cold-acclimated and non-acclimated stages: seasonal alteration of soluble sugars and phospholipids. J Agric Sci. 2009;147:51–69. doi: 10.1017/S0021859609008600. [DOI] [Google Scholar]
- 2.Bolouri-Moghaddam MR, Le Roy K, Xiang L, Rolland F, Van den Ende W. Sugar signalling and antioxidant network connections in plant cells. FEBS J. 2010;277:2022–2037. doi: 10.1111/j.1742-4658.2010.07633.x. [DOI] [PubMed] [Google Scholar]
- 3.Xiang L, Le Roy K, Bolouri-Moghaddam MR, Vanhaecke M, Lammens W, Rolland F, Van den Ende W. Exploring the neutral invertase-oxidative stress defence connection in Arabidopsis thaliana. J Exp Bot. 2011;62:3849–3862. doi: 10.1093/jxb/err069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van den Ende W. Multifunctional fructans and raffinose family oligosaccharides. Front Plant Sci. 2013;4:247–259. doi: 10.3389/fpls.2013.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sturm A, Chrispeels MJ. cDNA cloning of carrot extracellular beta-fructosidase and its expression in response to wounding and bacterial infection. Plant Cell. 1990;2:1107–1119. doi: 10.1105/tpc.2.11.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer JS, Mclaughlin JE. Functional reversion to identify controlling genes in multigenic responses: analysis of floral abortion. J Exp Bot. 2007;58:267–277. doi: 10.1093/jxb/erl177. [DOI] [PubMed] [Google Scholar]
- 7.Li FP, Ma CZ, Wang X, Gao CB, Zhang JF, Wang YY, Cong N, Li XH, Wen J, Yi B, Shen JX, Tu JX, Fu TD. Characterization of sucrose transporter alleles and their association with seed yield-related traits in Brassica napus L. BMC Plant Biol. 2011;11:168–182. doi: 10.1186/1471-2229-11-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulshrestha S, Tyagi P, Sindhi V, Yadavilli KS. Invertase and its applications-a brief review. J Pharm Res. 2013;7:792–797. [Google Scholar]
- 9.Sturm A. Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999;121:1–8. doi: 10.1104/pp.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zanor MI, Osorio S, Nunes-Nesi A, Carrari F, Lohse M, Usadel B, Kühn C, Bleiss W, Giavalisco P, Willmitzer L, Sulpice R, ZhouYH, Fernie AR. RNA interference of LIN5 in tomato confirms its role in controlling brix content, uncovers the influence of sugars on the levels of fruit hormones, and demonstrates the importance of sucrose cleavage for normal fruit development and fertility. Plant Physiol. 2009;150:1204–1218. doi: 10.1104/pp.109.136598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruhlmann JM, Kram BW, Carter CJ. CELL WALL INVERTASE 4 is required for nectar production in Arabidopsis. J Exp Bot. 2010;61:395–404. doi: 10.1093/jxb/erp309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes MA, Davies C, Dry IB. Isolation, functional characterization, and expression analysis of grapevine (Vitis vinifera L.) hexose transporters: differential roles in sink and source tissues. J Exp Bot. 2007;58:1985–1997. doi: 10.1093/jxb/erm061. [DOI] [PubMed] [Google Scholar]
- 13.Weber H, Borisjuk L, Wobus U. Molecular physiology of legume seed development. Annu Rev Plant Biol. 2005;56:253–279. doi: 10.1146/annurev.arplant.56.032604.144201. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Li X, Zhao Q, Jing S, Chen S, Yuan H. Identification of genes induced in response to low-temperature treatment in tea leaves. Plant Mol Biol Rep. 2008;27:257–265. doi: 10.1007/s11105-008-0079-7. [DOI] [Google Scholar]
- 15.Jin Y, Ni DA, Ruan YL. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell. 2009;21:2072–2089. doi: 10.1105/tpc.108.063719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Ruan YL. Shoot-root carbon allocation, sugar signalling and their coupling with nitrogen uptake and assimilation. Funct Plant Biol. 2016;43:105–113. doi: 10.1071/FP15249. [DOI] [PubMed] [Google Scholar]
- 17.Klann EM, Hall B, Bennett AB. Antisense acid invertase (TIV1) gene alters soluble sugar composition and size in transgenic tomato fruit. Plant Physiol. 1996;112:1321–1330. doi: 10.1104/pp.112.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greiner S, Rausch T, Sonnewald U, Herbers K. Ectopic expression of a tobacco invertase inhibitor homolog prevents cold-induced sweetening of potato tubers. Nat Biotechnol. 1999;17:708–711. doi: 10.1038/10924. [DOI] [PubMed] [Google Scholar]
- 19.Bhaskar PB, Wu L, Busse JS, Whitty BR, Hamernik AJ, Jansky SH, Buell CR, Bethke PC, Jiang J. Suppression of the vacuolar invertase gene prev ents cold-induced sweetening in potato. Plant Physiol. 2010;154:939–948. doi: 10.1104/pp.110.162545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roitsch T, González MC. Function and regulation of plant invertases: sweet sensations. Trends Plant Sci. 2004;9:606–613. doi: 10.1016/j.tplants.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Vargas W, Pontis H, Salerno GL. Differential expression of alkaline and neutral invertases in response to environmental stresses: characterization of an alkaline isoform as a stress-response enzyme in wheat leaves. Planta. 2007;226:1535–1545. doi: 10.1007/s00425-007-0590-3. [DOI] [PubMed] [Google Scholar]
- 22.Qi X, Wu Z, Li J, Mo X, Wu S, Chu J, Wu P. AtCYT-INV1, a neutral invertase, is involved in osmotic stress-induced inhibition on lateral root growth in Arabidopsis. Plant Mol Biol. 2007;64:575–587. doi: 10.1007/s11103-007-9177-4. [DOI] [PubMed] [Google Scholar]
- 23.Jia L, Zhang B, Mao C, Li J, Wu Y, Wu P, Wu ZC. OsCYT-INV1 for alkaline/neutral invertase is involved in root cell development and reproductivity in rice (Oryza sativa L.) Planta. 2008;228:51–59. doi: 10.1007/s00425-008-0718-0. [DOI] [PubMed] [Google Scholar]
- 24.Martin ML, Lechner L, Zabaleta EJ, Salerno GL. A mitochondrial alkaline/neutral invertase isoform (A/N-InvC) functions in developmental energy-demanding processes in Arabidopsis. Planta. 2013;237:813–822. doi: 10.1007/s00425-012-1794-8. [DOI] [PubMed] [Google Scholar]
- 25.Schaarschmidt S, Roitsch T, Hause B. Arbuscular mycorrhiza induces gene expression of the apoplastic invertase LIN6 in tomato (Lycopersicon esculentum) roots. J Exp Bot. 2006;57:4015–4023. doi: 10.1093/jxb/erl172. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Zhang C, Ou Y, Lin Y, Song B, Xie C, Liu J, Li XQ. Systematic analysis of potato acid invertase genes reveals that a cold-responsive member, StvacINV1, regulates cold-induced sweetening of tubers. Mol Gen Genomics. 2011;286:109–118. doi: 10.1007/s00438-011-0632-1. [DOI] [PubMed] [Google Scholar]
- 27.Kakumanu A, Ambavaram MM, Klumas C, Krishnan A, Batlang U, Myers E, Grene R, Pereira A. Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol. 2012;160:846–867. doi: 10.1104/pp.112.200444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tauzin AS, Sulzenbacher G, Lafond M, Desseaux V, Reca IB, Perrier J, Bellincampi D, Fourquet P, Lévêque C, Giardina T. Functional characterization of a vacuolar invertase from Solanum lycopersicum: post-translational regulation by N-glycosylation and a proteinaceous inhibitor. Biochimie. 2014;101:39–49. doi: 10.1016/j.biochi.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 29.Jae-Yean Kim AM. A maize vacuolar invertase, IVR2, is induced by water stress. Organ/tissue specificity and diurnal modulation of expression. Plant Physiol. 2000;124:71–84. doi: 10.1104/pp.124.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersen MN, Asch F, Wu Y, Jensen CR, Næsted H, Mogensen VO, Koch KE. Soluble invertase expression is an early target of drought stress during the critical, abortion-sensitive phase of young ovary development in Maize. Plant Physiol. 2002;130:591–604. doi: 10.1104/pp.005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xin Z, Browse J. Eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc Natl Acad Sci U S A. 1998;95:7799–7804. doi: 10.1073/pnas.95.13.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomashow MF. PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 33.Browse J, Xin ZG. Temperature sensing and cold acclimation. Curr Opin Plant Biol. 2001;4:241–246. doi: 10.1016/S1369-5266(00)00167-9. [DOI] [PubMed] [Google Scholar]
- 34.Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the arabidopsis cbf3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang YJ, Zheng LY, Wang XC. Effect of cold acclimation and ABA on cold hardiness, contents of proline of tea plants [Camellia sinensis(L.) O. Kuntze] J Tea Sci. 2004;24:177–182. [Google Scholar]
- 36.Yue C, Cao HL, Wang L, Zhou YH, Hao XY, Zeng JM, Wang XC, Yang YJ. Molecular cloning and expression analysis of tea plant aquaporin (AQP) gene family. Plant Physiol Biochem. 2014;83:65–76. doi: 10.1016/j.plaphy.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 37.Cao HL, Wang L, Yue C, Hao XY, Wang XC, Yang YJ. Isolation and expression analysis of 18 CsbZIP genes implicated in abiotic stress responses in the tea plant (Camellia sinensis) Plant Physiol Biochem. 2015;97:432–442. doi: 10.1016/j.plaphy.2015.10.030. [DOI] [PubMed] [Google Scholar]
- 38.Qian WJ, Yue C, Wang YC, Cao HL, Li NN, Wang L, Hao XY, Wang XC, Xiao B, Yang YJ. Identification of the invertase gene family (INVs) in tea plant and their expression analysis under abiotic stress. Plant Cell Rep. 2016;35:2269–2283. doi: 10.1007/s00299-016-2033-8. [DOI] [PubMed] [Google Scholar]
- 39.Li NN, Qian WJ, Wang L, Cao HL, Hao XY, Yang YJ, Wang XC. Isolation and expression features of hexose kinase genes under various abiotic stresses in the tea plant ( Camellia sinensis ) J Plant Physiol. 2017;209:95–104. doi: 10.1016/j.jplph.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Wang L, Cao HL, Qian WJ, Yao LN, Hao XY, Li NN, Yang YJ, Wang XC. Identification of a novel bZIP transcription factor in Camellia sinensis as a negative regulator of freezing tolerance in transgenic Arabidopsis. Ann Bot. 2017;119:1195–1209. doi: 10.1093/aob/mcx011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang XC, Zhao QY, Ma CL, Zhang ZH, Cao HL, Kong YM, Yue C, Hao XY, Chen L, Ma JQ, Jin JQ, Li X, Yang YJ. Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics. 2013;14:415–429. doi: 10.1186/1471-2164-14-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yue C, Cao HL, Wang L, Zhou YH, Huang YT, Hao XY, Wang YC, Wang B, Yang YJ, Wang XC. Effects of cold acclimation on sugar metabolism and sugar-related gene expression in tea plant during the winter season. Plant Mol Biol. 2015;86:591–608. doi: 10.1007/s11103-015-0345-7. [DOI] [PubMed] [Google Scholar]
- 43.Unger C, Hardegger M, Lienhard S, Sturm A. cDNA cloning of carrot (Daucus carota) soluble acid beta-fructofuranosidases and comparison with the cell wall isoenzyme. Plant Physiol. 1994;104:1351–1357. doi: 10.1104/pp.104.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Chen J, Feng J, Qin Q, Huang J. Overexpression of a loquat (Eriobotrya japonica Lindl.) vacuolar invertase affects sucrose levels and growth. Plant Cell Tiss Org. 2015;123:99–108. doi: 10.1007/s11240-015-0817-0. [DOI] [Google Scholar]
- 45.Wang L, Li XR, Lian H, Ni DA, He YK, Chen XY, Ruan YL. Evidence that high activity of vacuolar invertase is required for cotton fiber and Arabidopsis root elongation through osmotic dependent and independent pathways, respectively. Plant Physiol. 2010;154:744–756. doi: 10.1104/pp.110.162487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alberto F, Bignon C, Sulzenbacher G, Henrissat B, Czjzek M. The three-dimensional structure of invertase (beta-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases. J Biol Chem. 2004;279:18903–18910. doi: 10.1074/jbc.M313911200. [DOI] [PubMed] [Google Scholar]
- 47.Ji X, Van den Ende W, Van Laere A, Cheng S, Bennett J. Structure, evolution, and expression of the two invertase gene families of rice. J Mol Evol. 2005;60:615–634. doi: 10.1007/s00239-004-0242-1. [DOI] [PubMed] [Google Scholar]
- 48.Xia EH, Zhang HB, Sheng J, Li K, Zhang QJ, Kim C, Zhang Y, Liu Y, Zhu T, Li W, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol Plant. 2017;10:866–877. doi: 10.1016/j.molp.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 49.Bournay AS, Hedley PE, Maddison A, Waugh R, Machray GC. Exon skipping induced by cold stress in a potato invertase gene transcript. Nucleic Acids Res. 1996;24:2347–2351. doi: 10.1093/nar/24.12.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ehlert B, Hincha DK. Chlorophyll fluorescence imaging accurately quantifies freezing damage and cold acclimation responses in Arabidopsis leaves. Plant Methods. 2008;4:1–7. doi: 10.1186/1746-4811-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woo NS, Badger MR, Pogson BJ. A rapid, non-invasive procedure for quantitative assessment of drought survival using chlorophyll fluorescence. Plant Methods. 2008;4:27–41. doi: 10.1186/1746-4811-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends in Plant Sci. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 54.Kim JY, Mahe A, Guy S, Brangeon J, Roche O, Chourey PS, Prioul JL. Characterization of two members of the maize gene family, Incw3 and Incw4, encoding cell-wall invertases. Gene. 2000;245:89–102. doi: 10.1016/S0378-1119(00)00034-2. [DOI] [PubMed] [Google Scholar]
- 55.Kumari V, Das N. Vacuolar invertases in potato (Solanum tuberosum L.): molecular cloning, characterization, sequence comparison, and analysis of gene expression in the cultivars. Acta Physiol Plant. 2013;35:2055–2068. doi: 10.1007/s11738-013-1240-y. [DOI] [Google Scholar]
- 56.Dunn MA, White AJ, Vural S, Hughes MA. Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.) Plant Mol Biol. 1998;38:551–564. doi: 10.1023/A:1006098132352. [DOI] [PubMed] [Google Scholar]
- 57.Xu J, Avigne WT, McCarty DR, Koch KE. A similar dichotomy of sugar modulation and developmental expression affects both paths of sucrose metabolism: evidence from a maize invertase gene family. Plant Cell. 1996;8:1209–1220. doi: 10.1105/tpc.8.7.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruan YL, Jin Y, Yang YJ, Li GJ, Boyer JS. Sugar input, metabolism, and signaling mediated by invertase: roles in development, yield potential, and response to drought and heat. Mol Plant. 2010;3:942–955. doi: 10.1093/mp/ssq044. [DOI] [PubMed] [Google Scholar]
- 59.Huang YW, Nie YX, Wan YY, Chen SY, Sun Y, Wang XJ, Bai JG. Exogenous glucose regulates activities of antioxidant enzyme, soluble acid invertase and neutral invertase and alleviates dehydration stress of cucumber seedlings. Sci Hortic. 2013;162:20–30. doi: 10.1016/j.scienta.2013.07.031. [DOI] [Google Scholar]
- 60.McLaughlin JE, Boyer JS. Sugar-responsive gene expression, invertase activity, and senescence in aborting maize ovaries at low water potentials. Ann Bot. 2004;94:675–689. doi: 10.1093/aob/mch193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Proels RK, Roitsch T. Extracellular invertase LIN6 of tomato: a pivotal enzyme for integration of metabolic, hormonal, and stress signals is regulated by a diurnal rhythm. J Exp Bot. 2009;60:1555–1567. doi: 10.1093/jxb/erp027. [DOI] [PubMed] [Google Scholar]
- 62.Liao SC, Lin CS, Wang AY, Sung HY. Differential expression of genes encoding acid invertases in multiple shoots of bamboo in response to various phytohormones and environmental factors. J Agric Food Chem. 2013;61:4396–4405. doi: 10.1021/jf400776m. [DOI] [PubMed] [Google Scholar]
- 63.Rabot A, Portemer V, Peron T, Mortreau E, Leduc N, Hamama L, Coutos-Thévenot P, Atanassova R, Sakr S, Le Gourrierec J. Interplay of sugar, light and gibberellins in expression of Rosa hybrida vacuolar invertase 1 regulation. Plant Cell Physiol. 2014;55:1734–1748. doi: 10.1093/pcp/pcu106. [DOI] [PubMed] [Google Scholar]
- 64.Ou Y, Song B, Liu X, Xie C, Li M, Lin Y, Zhang H, Liu J. Promoter regions of potato vacuolar invertase gene in response to sugars and hormones. Plant Physiol Biochem. 2013;69:9–16. doi: 10.1016/j.plaphy.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 65.Lu CA, Ho THD, Ho SL, Yu SM. Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of α-amylase gene expression. Plant Cell. 2002;14:1963–1980. doi: 10.1105/tpc.001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun C, Palmqvist S, Olsson H, Borén M, Ahlandsberg S, Jansson C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 2003;15:2076–2092. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang GQ, Luscher M, Sturm A. Antisense repression of vacuolar and cell wall invertase in transgenic carrot alters early plant development and sucrose partitioning. Plant Cell. 1999;11:177–189. doi: 10.1105/tpc.11.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kohorn BD, Kobayashi M, Johansen S, Riese J, Huang LF, Koch K, Fu S, Dotson A, Byers N. An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant J. 2006;46:307–316. doi: 10.1111/j.1365-313X.2006.02695.x. [DOI] [PubMed] [Google Scholar]
- 69.Sergeeva LI, Keurentjes JJ, Bentsink L, Vonk J, van der Plas LH, Koornneef M, Vreugdenhil D. Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proc Natl Acad Sci U S A. 2006;103:2994–2999. doi: 10.1073/pnas.0511015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang L, Ruan YL. Unraveling mechanisms of cell expansion linking solute transport, metabolism, plasmodesmtal gating and cell wall dynamics. Plant Signal Behav. 2014;5:1561–1564. doi: 10.4161/psb.5.12.13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen T, Pei HJ, Zhang YF, Qian QL. Seasonal changes in non-structural carbohydrates and sucrose metabolism enzymes in two sabina species. Acta Physiol Plant. 2012;34:173–180. doi: 10.1007/s11738-011-0815-8. [DOI] [Google Scholar]
- 72.Klotke J, Kopka J, Gatzke N, Heyer AG. Impact of soluble sugar concentrations on the acquisition of freezing tolerance in accessio ns of Arabidopsis thaliana with contrasting cold adaptation-evidence for a role of raffinose in cold acclimation. Plant Cell Environ. 2004;27:1395–1404. doi: 10.1111/j.1365-3040.2004.01242.x. [DOI] [Google Scholar]
- 73.Couée I, Sulmon C, Gouesbet G, El Amrani A. Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J Exp Bot. 2006;57:449–459. doi: 10.1093/jxb/erj027. [DOI] [PubMed] [Google Scholar]
- 74.Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- 75.Karve A, Rauh BL, Xia X, Kandasamy M, Meagher RB, Sheen J, Moore BD. Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta. 2008;228:411–425. doi: 10.1007/s00425-008-0746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keunen E, Peshev D, Vangronsveld J, Van den Ende W, Cuypers A. Plant sugars are crucial players in the oxidative challenge during abiotic stress: extending the traditional concept. Plant Cell Environ. 2013;36:1242–1255. doi: 10.1111/pce.12061. [DOI] [PubMed] [Google Scholar]
- 77.Ruan YL. Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu Rev Plant Biol. 2014;65:33–67. doi: 10.1146/annurev-arplant-050213-040251. [DOI] [PubMed] [Google Scholar]
- 78.Wang L, Ruan YL. Regulation of cell division and expansion by sugar and auxin signaling. Front Plant Sci. 2013;4:163–172. doi: 10.3389/fpls.2013.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Min L, Li Y, Hu Q, Zhu L, Gao W, Wu Y, Ding Y, Liu S, Yang X, Zhang X. Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant Physiol. 2014;164:1293–1308. doi: 10.1104/pp.113.232314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu YH, Offler CE, Ruan YL. Cell wall invertase promotes fruit set under heat stress by suppressing ROS-independent cell death. Plant Physiol. 2016;172:163–180. doi: 10.1104/pp.16.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mishra BS, SinghM AP, Laxmi A. Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. Plos One. 2009;4:e4502. doi: 10.1371/journal.pone.0004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vargas WA, Pontis HG, Salerno GL. New insights on sucrose metabolism: evidence for an active A/N-Inv in chloroplasts uncovers a novel component of the intracellular carbon trafficking. Planta. 2008;227:795–807. doi: 10.1007/s00425-007-0657-1. [DOI] [PubMed] [Google Scholar]
- 83.Cho JI, Ryoo N, Eom JS, Lee DW, Kim HB, Jeong SW, Lee YH, Kwon YK, Cho MH, BhooSH HTR, Park YI, Hwang I, Sheen J, Jeon JS. Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol. 2009;149:745–759. doi: 10.1104/pp.108.131227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chinnusamy V, Schumaker K, Zhu JK. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot. 2004;55:225–236. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- 85.Trovato M, Mattioli R, Costantino P. Multiple roles of proline in plant stress tolerance and development. Rend Lincei. 2008;19:325–346. doi: 10.1007/s12210-008-0022-8. [DOI] [Google Scholar]
- 86.Mattioli R, Falasca G, Sabatini S, Altamura MM, Costantino P, Trovato M. The proline biosynthetic genes P5CS1 and P5CS2 play overlapping roles in Arabidopsis flower transition but not in embryo development. Plant Physiol. 2009;137:72–85. doi: 10.1111/j.1399-3054.2009.01261.x. [DOI] [PubMed] [Google Scholar]
- 87.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–941. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 89.Takahashi R, Ishimaru Y, Senoura T, Shimo H, Ishikawa S, Arao T, Nakanishi H, Nishizawa NK. The OsNRAMP1 iron transporter is involved in cd accumulation in rice. J Exp Bot. 2011;62:4843–4850. doi: 10.1093/jxb/err136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 91.Rohde P, Hincha DK, Heyer AG. Heterosis in the freezing tolerance of crosses between two Arabidopsis thaliana accessions (Columbia-0 and C24) that show differences in non-acclimated and acclimated freezing tolerance. Plant J. 2004;38:790–799. doi: 10.1111/j.1365-313X.2004.02080.x. [DOI] [PubMed] [Google Scholar]
- 92.Tomlinson KL, McHugh S, Labbe H, Grainger JL, James LE, Pomeroy KM, Mullin JW, Miller SS, Dennis DT, Miki BL. Evidence that the hexose-to-sucrose ratio does not control the switch to storage product accumulation in oilseeds: analysis of tobacco seed development and effects of overexpressing apoplastic invertase. J Exp Bot. 2004;55:2291–2303. doi: 10.1093/jxb/erh251. [DOI] [PubMed] [Google Scholar]
- 93.Hao XY, Horvath D, Chao W, Yang YJ, Wang XC, Xiao B. Identification and evaluation of reliable reference genes for quantitative real-time PCR analysis in tea plant (Camellia sinensis (L). O. Kuntze) Int J Mol Sci. 2014;15:22155–22172. doi: 10.3390/ijms151222155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008;18:385–397. doi: 10.1038/cr.2008.26. [DOI] [PubMed] [Google Scholar]
- 95.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Homologous analysis of ten SAIs protein sequences. Figure S2. Promoter deletion analysis of CsINV5 under Glc, Fru and different temperature conditions. Figure S3. Histochemical GUS staining of WT plants and transgenic Arabidopsis with the empty vector (pBI101::GUS) under Suc and different temperature exposure. Figure S4. Expression analysis of AtVIN1, AtVIN2 in Arabidopsis, and CsINV5 in tea plant under low temperature condition. Figure S5. Expression detection of selected transcripts by RNA-Seq and qRT-PCR. Figure S6. Partial co-expression patterns of DEGs between OE and WT plants in normal and cold conditions (FDR < 0.05, K-means = 10). Figure S7. Prokaryotic expression analysis of CsINV5. (DOCX 10071 kb)
S1. The genome DNA sequence of CsINV5. 1154 bp promoter sequence and 7 exons have been highlighted: (i) stress signaling (ARE, fully outlined in red; HSE, double outlined in red; TC-rich repeats, dot outlined in red; LTR, viridity highlighted; MYC, viridescence highlighted); (ii) light signaling (MRE, navy highlighted; I BOX, violet highlighted; L BOX, pink highlighted; SP1, underlined); (iii) Hormones signaling (ERE, blue highlighted; TCA-element, green highlighted); (iv) sugar repression (SRE, highlighted in deep yellow) and activation (WBOXHVISO1, highlighted in gray). The CAAT-box and TATA-box are yellow highlighted. PCR primer sequences highlighted in bold and italic. The exons were showed by red font. (DOCX 32 kb)
Table S1. RNA-seq data and quality control. (XLSX 11 kb)
Table S2-S5. Table S2. Sixty-six significant GO terms (FDR < 0.05) mapped by 266 up-regulated DEGs between CsINV5-OE and WT plants. Table S3. Fifty-five significant GO terms (FDR < 0.05) mapped by 402 down-regulated DEGs between CsINV5-OE and WT plants. Table S4. Sixty-two significant GO terms (FDR < 0.05) mapped by 1248 up-regulated DEGs between CsINV5-OE and WT plants in cold conditions condition. Table S5. Sixty-eight significant GO terms (FDR < 0.05) mapped by 1479 down-regulated DEGs between CsINV5-OE and WT plants in cold conditions condition. (XLSX 27 kb)
Table S6. Partial DEGs (FDR < 0.05, log2FC) in CsINV5-OE plants compared to WT plants in normal and cold conditions. (XLSX 21 kb)
Table S7. Nutrient solution formulation. Table S9. Primer sequences used in promoter cloning and vector construction. Table S10. Primer information used in qRT-PCR detection. (DOCX 22 kb)
Table S8. Amino acid sequences of the SAI proteins used for conserved domain analysis. (XLSX 13 kb)
Data Availability Statement
The raw data of RNA-seq experiments generated in this study have been deposited in the NCBI database under the accession number SRR6057439-SRR6057450. All the data supporting the findings of the study is contained within the manuscript and its additional information files.